SUMMARY
The gammaherpesviruses, including Epstein-Barr virus (EBV), Kaposi’s sarcoma-associated herpesvirus (KSHV), and murine gammaherpesvirus 68 (MHV68, MuHV-4, γHV68), are etiologic agents of a wide range of lymphomas and non-hematological malignancies. These viruses possess large and highly dense dsDNA genomes that feature >80 bidirectionally positioned open reading frames (ORFs). The abundance of overlapping transcripts and extensive splicing throughout these genomes have until now prohibited high throughput-based resolution of transcript structures. Here, we integrate the capabilities of long-read sequencing with the accuracy of short-read platforms to globally resolve MHV68 transcript structures using the transcript resolution through integration of multi-platform data (TRIMD) pipeline. This approach reveals highly complex features, including: (1) pervasive overlapping transcript structures; (2) transcripts containing intra-gene or trans-gene splices that yield chimeric ORFs; (3) antisense and intergenic transcripts containing ORFs; and (4) noncoding transcripts. This work sheds light on the underappreciated complexity of gammaherpesvirus transcription and provides an extensively revised annotation of the MHV68 transcriptome.
Graphical Abstract
In Brief
The highly dense dsDNA genomes of herpesviruses feature an abundance of overlapping transcripts and extensive splicing, greatly hindering the global identification of individual transcripts. O’Grady et al. use integrated multi-platform genomics to globally resolve transcript structures from murine gammaherpesvirus 68, providing an extensively revised genome annotation.
INTRODUCTION
Epstein-Barr virus (EBV) and Kaposi’s sarcoma-associated herpesvirus (KSHV) are ubiquitous human pathogens that establish lifelong infections and are associated with a wide range of malignancies, including B cell lymphoproliferative diseases, B cell lymphomas, nasopharyngeal carcinoma, gastric carcinoma, and Kaposi’s sarcoma. Although substantial efforts have unraveled the contribution of many EBV and KSHV genes to gammaherpesvirus infection using tissue culture models, the specific in vivo functions of many of these genetic elements remain poorly understood. In particular, the strict species specificity of gammaherpesviruses is an immense obstacle for defining the role of individual genomic features in the context of virus infection in a natural host. Murine gammaherpesvirus 68 (MHV68, MuHV-4, γHV68) is genetically related to EBV and KSHV, establishes lifelong latent infection in B cells, and is associated with the development of B cell lymphoproliferative disease and B cell lymphoma. Moreover, numerous studies have demonstrated that EBV, KSHV, and MHV68 display conservation of many critical genes and pathogenic strategies (Barton et al., 2011). Thus, MHV68 infection of mice provides an excellent system for comparative studies to elucidate the functions of the human gammaherpesvirus genes in vivo.
Gammaherpesviruses possess large double-stranded DNA (dsDNA) genomes that display features prototypical to herpesviruses, including genome sizes >110 kb, the presence of >80 open reading frames (ORFs), bidirectional gene expression, genetically and positionally conserved lytic replication genes, and GC-rich repeat sequences. The annotation of gammaherpesvirus genomes has traditionally relied on ORF-based analyses using canonical translational start and stop sequences to demarcate putative protein-coding genes (Virgin et al., 1997). Subsequent studies using labor-intensive methods such as 5′ and 3′ rapid amplification of cDNA ends, ribonuclease protection assays, and northern blotting have subsequently defined full-length transcripts for some loci. Despite progress using these targeted efforts, tiled microarray and next-generation sequencing studies have demonstrated pervasive inter-ORF transcription from gammaherpesvirus genomes. For example, using RNA-sequencing (RNA-seq) and ribosome profiling to refine the annotation of KSHV transcription start sites and initiation and termination codons, Arias et al. (2014) noted abundant transcription through numerous unexpected regions. Similar findings were reported for EBV (O’Grady et al., 2014). Likewise, Johnson et al. (2010) identified 31 inter-ORF regions of the MHV68 genome that displayed unexpected transcriptional activity (Cheng et al., 2012).
Complicating the annotation of transcript structures within these regions, empirical evidence from numerous studies has demonstrated the prevalence of multiple overlapping transcripts and extensive splicing throughout gammaherpesvirus genomes, with splicing products generated over distances as long as 100 kb (Bodescot and Perricaudet, 1986; Speck and Strominger, 1985). Thus, while short-read RNA-seq approaches have helped in the discovery of new regions of transcriptional activity within these highly compact genomes, globally resolving specific transcripts from these regions has until very recently been impossible. The advent of single-molecule real-time (SMRT) sequencing has provided a remarkable advance in sequencing longer transcript structures; however, transcript coverage is typically low, and not all sequencing products represent bona fide full-length transcripts due to 5′ sequencing truncations and potential sequencing errors (O’Grady et al., 2016). To overcome these obstacles, we recently developed the pipeline transcript resolution through integration of multi-platform data (TRIMD) to integrate parallel datasets from multiple sequencing platforms, including Pacific Biosciences (PacBio) SMRT Iso-Seq long-read sequencing, Illumina short-read RNA-Seq, and 5′ cap analysis of gene expression (deepCAGE) and applied this method to globally resolve transcript structures for EBV (O’Grady et al., 2016).
While MHV68 infection of mice is an important system to study virus and host determinants of in vivo gammaherpesvirus infection, the MHV68 genome remains poorly annotated. Although a few studies have resolved individual transcript structures for select regions of the genome (e.g., ORF50/Rta [Gray et al., 2009; Liu et al., 2000; Wakeman et al., 2014] and ORF73/mLANA [Allen et al., 2006; Cheng et al., 2012; Coleman et al., 2005]), transcriptional start and stop sequences have not been identified for the vast majority of genes, transcript structures in most inter-ORF regions have not been identified, and the original ORF-based demarcations of the MHV68 genome remain the annotation of record. Thus, we have applied multi-platform transcriptomics and TRIMD to globally resolve the MHV68 transcriptome during lytic infection. Application of this methodology has revealed highly complex transcriptional features, including multiple overlapping transcripts for many loci; trans-gene splicing yielding chimeric ORF transcripts; transcripts containing unknown ORFs; and extensive readthrough transcription. In all, we define 258 lytic MHV68 transcripts. We report transcriptional start and stop sites for 53 of the ORFs annotated in the original GenBank report (GenBank: U97553.2) (Virgin et al., 1997) and define new transcripts for 55 truncated or unknown ORFs. This work further reveals the underappreciated complexity of gammaherpesvirus transcription and provides an extensively revised annotation of the MHV68 transcriptome.
RESULTS
Sequencing and Transcript Validation
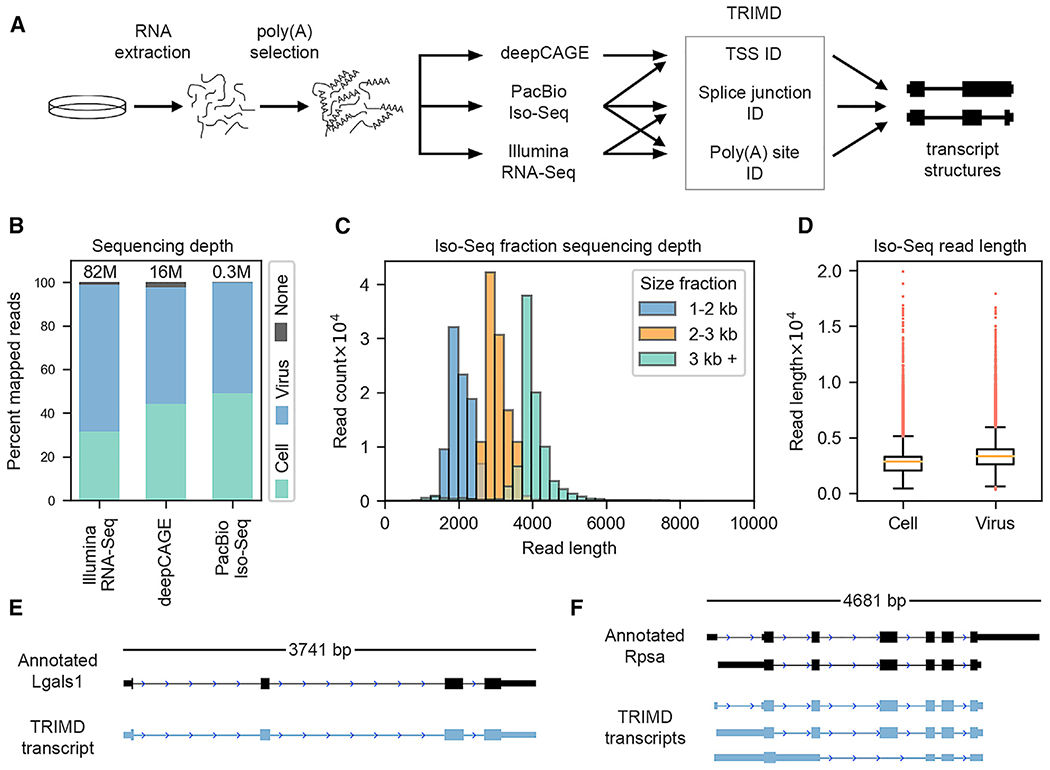
SMRT sequencing using the PacBio Iso-Seq protocol provides long reads that frequently span entire transcript isoforms. In the context of Iso-Seq, any SMRT reads that are inferred to represent full-length transcripts are informatically collapsed along with identical counterparts and reported as consensus full-length (CFL) isoforms. However, because not all CFLs represent true full-length transcripts (O’Grady et al., 2016), we incorporated the use of complementary sequencing data types from short-read platforms to corroborate Iso-Seq CFL calls. To resolve MHV68 transcript structures during lytic replication, murine NIH 3T12 fibroblast cells were infected with MHV68 at MOI 5 for 18 h. Following RNA harvest, parallel poly(A)-selected RNA samples were subjected to PacBio SMRT sequencing, Illumina RNA-Seq, and deepCAGE sequencing. These complementary datasets were then integrated using TRIMD (O’Grady et al., 2016) to validate the structural features of CFLs and thereby globally annotate the lytic MHV68 transcriptome (Figure 1A). Sequence reads were aligned and mapped using genome indexes containing the mouse mm10 and MHV68 genomes using genomic mapping and alignment program (GMAP; Iso-Seq) and spliced transcripts alignment to a reference (STAR; Illumina short-read and deepCAGE). Greater than 80 × 106, 16 × 106, and 0.3 × 106 mapped reads were obtained for Illumina RNA-Seq, deepCAGE, and Iso-Seq sequencing, respectively (Figure 1B), which was comparable to or higher than the read depths used for our previous analysis of the EBV transcriptome (O’Grady et al., 2016). Furthermore, the percentage of reads mapping to the virus was especially robust, with >50% of all reads being found to be of viral origin (Figure 1B). These substantial numbers of virus-mapped reads obtained from each platform facilitated a detailed resolution of the MHV68 lytic transcriptome.
Figure 1. Sequencing Data and Transcript Validation.
(A) Infection, sequencing, and TRIMD data integration strategy.
(B) Number of Illumina RNA-Seq, deepCAGE, and PacBio Iso-Seq reads mapped to the cellular and viral genomes.
(C) Number of PacBio Iso-Seq reads per library size fraction.
(D) Length of PacBio Iso-Seq reads mapped to the cellular and viral genomes.
(E) Annotation and TRIMD transcript of cellular gene Lgals1.
(F) Annotation and TRIMD transcripts of cellular gene Rpsa.
Although long-read sequencing can resolve transcripts >10 kb, sequencing output is typically biased for shorter transcripts. To lessen the influence of this bias, our PacBio SMRT sequencing data were generated from RNAs separated into 1–2 kb, 2–3 kb, and >3 kb size fractions. This approach led to an excellent read-length distribution, with both short and long transcripts well represented (Figure 1C) and a wide range of read lengths for both cellular (0.4–19.9 kb) and viral (0.31–7.9 kb) transcripts (Figure 1D). The depth and broad distribution of Iso-Seq CFLs, the robust Illumina RNA-Seq, and deepCAGE datasets and the recovery of complex cellular transcripts (Figures 1E and 1F), provided confidence that transcript structures could be resolved and validated across the complex and extensive MHV68 transcriptome.
A Global View of the MHV68 Transcriptome during Lytic Replication
TRIMD integrates datasets from three transcriptomic platforms to resolve and validate full-length transcript isoforms using conservative calls based on strong supporting evidence. For example, in this study, 5′ transcriptional start sites in viral Iso-Seq CFLs were considered to be valid only if they were supported by at least 135 deepCAGE tags. Similarly, to be considered a valid full-length transcript, splice junctions must also have been present in Illumina RNA-Seq reads, and CFL 3′ ends must have been represented in Illumina RNA-Seq reads and/or have been within 40 bases downstream of a canonical polyadenylation signal sequence.
To further validate TRIMD-identified transcript features and to gain insight into the kinetics of transcript expression, additional RNA-Seq and deepCAGE runs were performed on samples from (1) a full lytic infection time course (6, 12, and 18 h) in NIH 3T12 fibroblasts infected with a wild-type MHV68 isolate (MHV68) or with a bacterial artificial chromosome (BAC)-derived MHV68 carrying a β-lactamase marker (MHV68.bla) (Nealy et al., 2010); (2) latently infected HE2.1 B cells (Forrest and Speck, 2008); and (3) HE2.1 B cells reactivated from latency. TRIMD-identified transcript features that did not appear in any of these validation datasets were discarded.
Using these conservative thresholds, TRIMD-based analysis of the MHV68 lytic transcriptome identified 147 unique transcription start sites, 71 splice junctions, and 64 unique 3′ ends (Figure 2B). Of these, 89% of 5′ ends, 61% of splice junctions, and 92% of 3′ ends were detected in fully validated transcripts. As detailed below (see Figures 3 and 4), the majority of starts, ends, and splice junctions were shared by multiple overlapping transcripts. In total, from 150,634 SMRT reads that mapped to the MHV68 genome, TRIMD identified 258 unique full-length transcripts in which 5′ starts, splice junctions, and 3′ end sequences were fully validated (Figure 2A; Figure S1; Data S1, S2, S3, S4, S5, and S6).
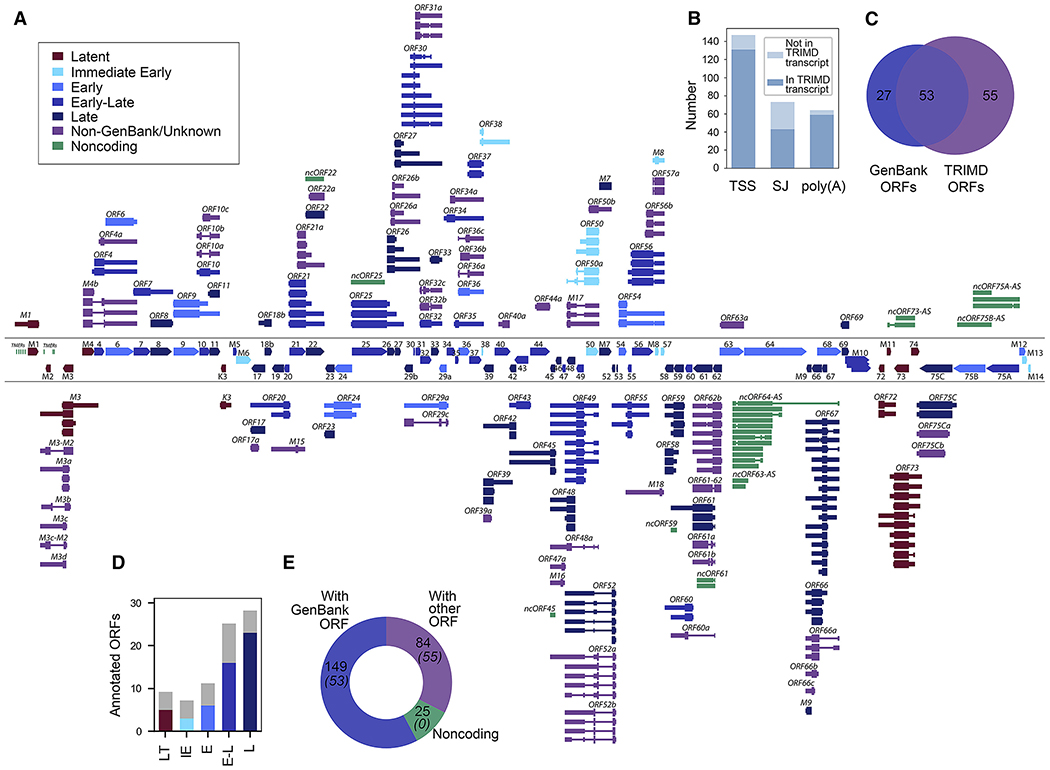
Figure 2. MHV68 Transcripts Identified by TRIMD.
(A) Overview of viral transcripts validated by TRIMD. For reference, GenBank ORFs are displayed between divider lines, with rightward ORFs at top and leftward ORFs at bottom. TRIMD transcripts are displayed outside divider lines, with rightward TRIMD transcripts displayed above divider lines and leftward TRIMD transcripts displayed below divider lines.
(B) Number of TRIMD-identified 5′ transcription start sites (TSSs), splice junctions, and polyadenylation sites.
(C) Venn diagram of GenBank-annotated and TRIMD-identified ORFs.
(D) Number of GenBank ORFs within each class identified by TRIMD.
(E) Number of TRIMD transcripts that correspond to GenBank ORFs or unknown ORFs or that are likely noncoding. The number of total ORFs as per (C) is indicated in parentheses. The analysis is based on 3 sequencing methods of 1 biological replicate.
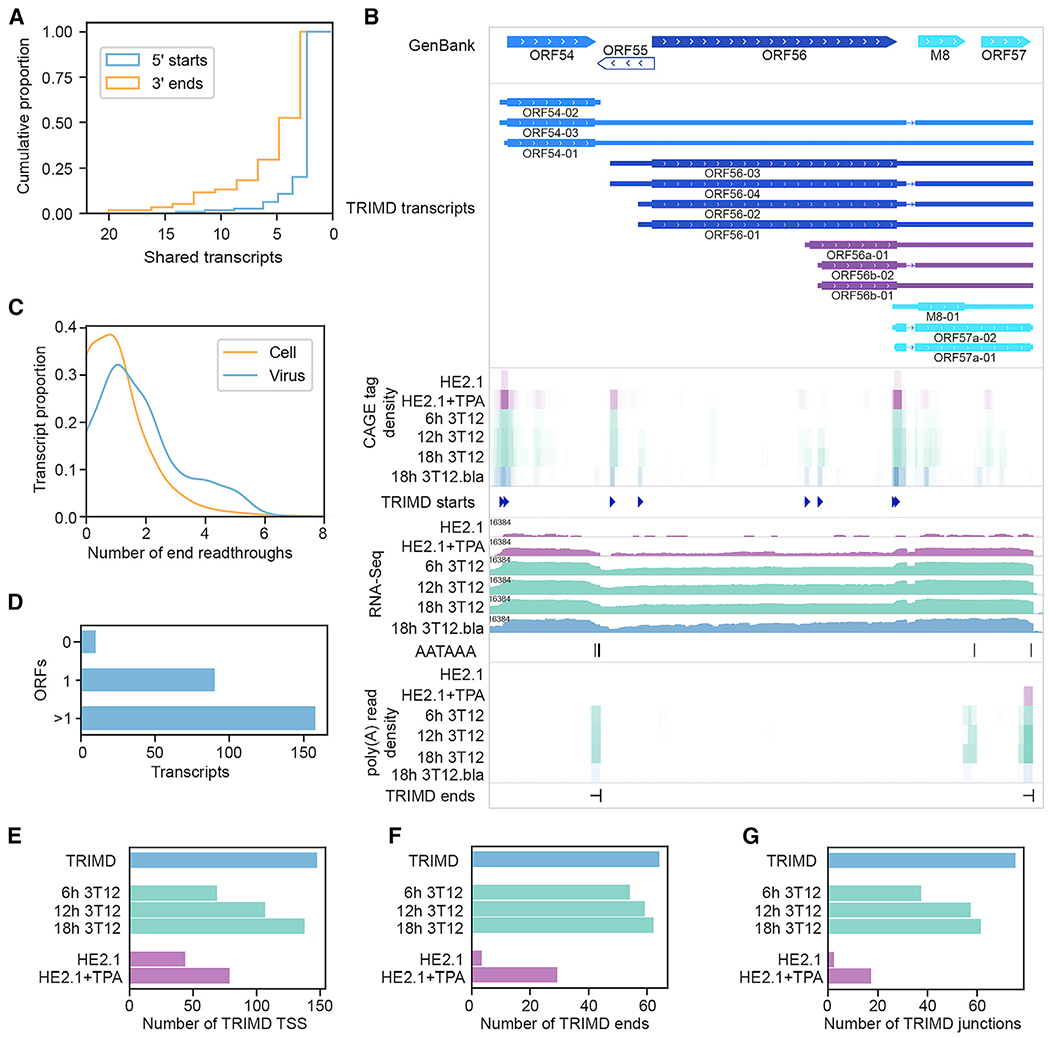
Figure 3. MHV68 Transcript Diversity.
(A) Cumulative proportion of viral 5′ TSSs and polyadenylation sites shared by ≥1 transcripts. One biological replicate, 3 sequencing methods.
(B) TRIMD-identified transcripts and supporting evidence at the ORF54-57 locus. The GenBank panel indicates reference GenBank-annotated ORFs in this locus. The TRIMD-identified transcripts are indicated (immediate early, bright blue; early, light blue; early-late, royal blue; late, dark blue; unknown ORF, purple), along with corresponding tracks for TRIMD-validated starts, poly(A) signal sequences, and ends. CAGE tag density, RNA-Seq, and poly(A) read density tracks are shown for TRIMD dataset (MHV68 marker virus infection of NIH 3T12 fibroblasts [3T12.bla]) plus validation sample sets (wild-type MHV68 infection of NIH 3T12 fibroblasts [3T12], latently infected HE2.1 B cells [HE2.1], and HE2.1 cells reactivated from latency [HE2.1 + TPA]). RNA-Seq read depth (log2 scale) is indicated. The M8/ORF57 splice variants ORF57a-01 and ORF57a-02 are not annotated in GenBank U97553, but they have been previously identified as variants of the immediate-early gene ORF57. Sequencing data tracks illustrate n = 1.
(C) Proportion of TRIMD-identified end readthroughs per transcript for cellular and viral transcripts. One biological replicate, 3 sequencing methods.
(D) Number of viral transcripts containing sequences corresponding to 0, 1, or >1 ORFs of ≥75 aa. One biological replicate, 3 sequencing methods.
(E) Number of TRIMD-identified viral TSSs detected at 6, 12, and 18 h following wild-type MHV68 infection of NIH 3T12 fibroblasts, in latently infected HE2.1 B cells, and 18h after the induction of HE2.1 B cell reactivation. The number of TRIMD-identified viral TSSs detected in TRIMD sample set (18h 3T12.bla) is shown for reference.
(F) Number of TRIMD-identified polyadenylation sites detected in validating datasets and TRIMD sample set indicated in (E).
(G) Number of TRIMD-identified splice junctions detected in validating datasets and TRIMD sample set indicated in (E).For (E)-(G) time course, and HE2.1 bars represent n = 1.
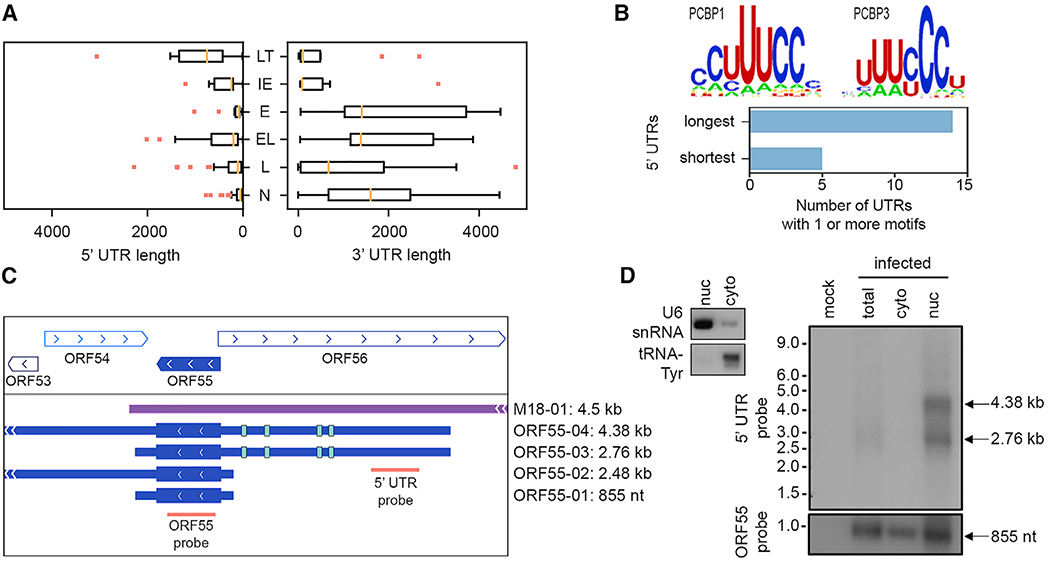
Figure 4. MHV68 Transcript 5′ and 3′ UTR Diversity.
(A) Lengths of 5′ and 3′ UTRs by class. LT, latent; IE, immediate early; E, early; EL, early-late; L, late; N, non-GenBank. Analysis based on 3 sequencing methods of 1 biological replicate.
(B) PCBP1 and PCBP3 binding motifs and enrichment in PCBP binding motifs in longest 5′ UTRs relative to shortest 5′ UTRs.
(C) ORF55 transcripts with long and short UTRs. The green bands indicate the locations of the PCBP motifs.
(D) Northern blot of total, cytoplasmic, and nuclear lysates using probes targeting transcripts containing the ORF55 ORF sequence or the ORF55 long 5′ UTR. Control probes are for nuclear (U6 small nuclear RNA [snRNA]) and cytoplasmic (tRNA-Tyr) RNA. Representative of 2 biological replicates.
To assess the coding capacity of the 258 TRIMD-validated transcripts, contiguous or spliced ORFs of at least 75 amino acids (aa) were identified using TransDecoder. Although some transcripts harbored >1 potential ORF (likely due to stop sequence readthrough; see Figure 3), downstream ORFs in polycistronic transcripts are generally thought to remain untranslated due to their lack of proximity to 5′ caps (Merrick, 2004). Thus, for nomenclature and analysis purposes, we assumed that the most upstream ORF is the primary translated reading frame. These ORFs were then compared to the original ORF-based GenBank annotation of the MHV68 genome (GenBank: U97553.2). In all, 149 of the TRIMD-validated transcripts coded for GenBank ORFs, cumulatively accounting for coverage of 53 of the 80 annotated MHV68 ORFs (Figure 2C). As expected, TRIMD transcripts identified during this robust lytic infection yielded a broad representation of all viral gene classes (Figure 2D).
In addition, TRIMD identified 84 transcripts encoding 55 primary ORFs that did not match the GenBank record (Figure 2C), although a few of these ORFs have been previously identified elsewhere (Mackett et al., 1997; Nash et al., 2001). These 84 transcripts were further evaluated using the Coding Potential Assessment Tool (CPAT) (Wang et al., 2013). Some of these transcripts did not harbor an ORF of at least 75 aa and/or were scored as noncoding transcripts by CPAT, and thus were considered herein to be noncoding RNAs (Figure 2E). Thus, TRIMD identified 258 unique MHV68 transcripts, including 233 that code for 108 different (but in some cases overlapping) proteins and 25 that we classified as noncoding.
Initiation and Termination of Transcription across the Genome
Based on visualization of the identified transcripts, a notable feature of the MHV68 lytic transcriptome is the presence of numerous shared 5′ transcriptional start sites and 3′ polyadenylation sites (Figure 3A). As previously observed for gene clusters within other herpesviruses (Cohen, 1999; Majerciak et al., 2013), the degree of 3′ end sharing is particularly prevalent, with 72% of polyadenylation sites shared by at least 2 transcripts and 14% shared by at least 8 transcripts. In contrast, 5′ start sequences are more commonly unique (Figure 3A), as exemplified by the rightward ORF56 locus (Figure 3B). The use of shared polyadenylation sites for multiple transcripts appears to reflect extensive readthrough of upstream polyadenylation signal sequences (Figure 3C), with >60% of validated transcripts (158 of 258) harboring >1 potential ORF (Figures 3B and 3D).
Transcription start and stop sites appeared with varying depths at different time points following infection. As expected, transcription start and end locations and expression patterns matched closely between the MHV68 and MHV68.bla viruses (Figures 3B and S2). Consistent with increased viral transcription over the course of infection, more transcription start sites (TSSs), polyadenylated ends, and splice junctions were detected at later time points than at earlier time points (Figures 3E–3G). Few of the transcript features identified by TRIMD in MHV68-infected fibroblasts were detected in latently infected B cells, but many were detected upon reactivation, indicating some degree of shared transcriptional mechanics between de novo MHV68 infection of fibroblasts and MHV68 reactivation in B cells (Figures 3E–3G). The limited extent of viral reactivation in HE2.1 B cells impairs the sensitivity of transcript detection, however, and the occurrence of low-level spontaneous reactivation in HE2.1 B cells complicates the comparison of latent and lytic transcriptional features.
Structural and Functional Variation in MHV68 UTRs
While previous iterations of MHV68 genome annotations define genes by reading frames only, here, we annotate MHV68 genes to include 5′ and 3′ UTR sequences. Across all 258 validated transcripts, UTR lengths varied widely, with median 5′ and 3′ UTR lengths of 114 and 1,232 nt, respectively (Figure S3). Notably, 5′ and 3′ UTR lengths varied significantly among gene classes (Figure 4A), perhaps reflecting susceptibility to or evasion of different regulatory controls. For example, while early genes universally retained very short 5′ UTRs (median, 84 nt) and long 3′ UTRs on average (median, 1,410 nt), genes associated with latency displayed the longest mean 5′ UTR lengths (median, 748 nt) and shorter 3′ UTR lengths (median, 103 nt).
The large number of 5′ transcriptional start sites and polyadenylation signal readthroughs, along with splicing (see Figure 5), resulted in a high degree of transcript diversity, with most full-length ORFs encoded by several overlapping transcripts (Figure S4) with differing 5′ and/or 3′ UTR sequences (Figure S5). For example, ORF54 is encoded by 3 transcripts with 2 different 5′ UTRs and 3 different 3′ UTRs, while ORF56 is encoded by 4 transcripts with 2 different 5′ UTRs and 2 different 3′ UTRs (Figure 3B).
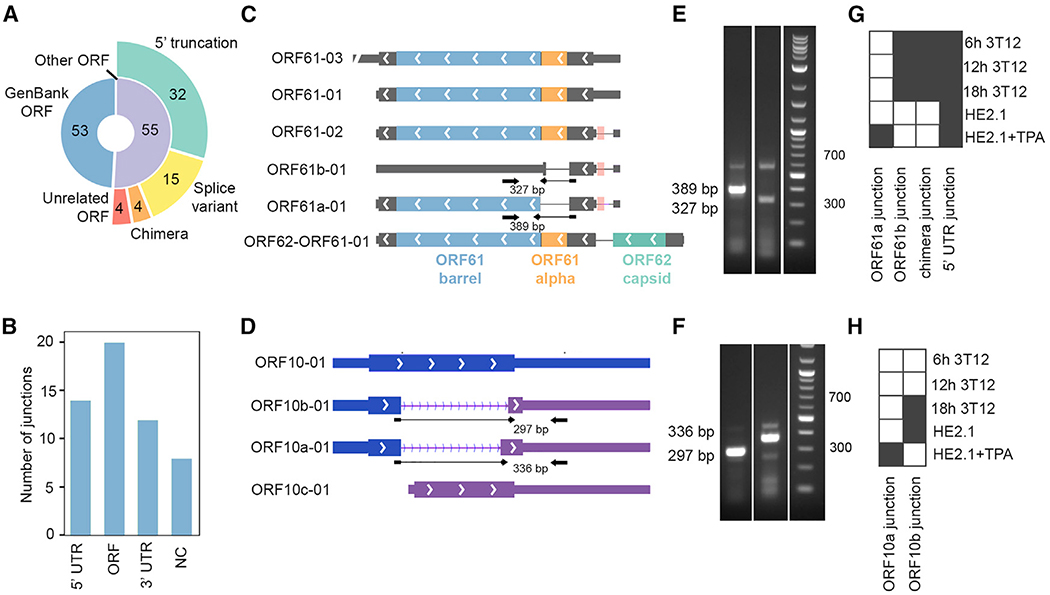
Figure 5. MHV68 ORF Diversity.
(A) Types of primary ORFs encoded by TRIMD transcripts.
(B) Locations of splice junctions within TRIMD transcripts.
(C) Transcripts containing ORF61 and its variants. The pink boxes in the splice junctions of ORF61-02, ORF61b-01, and ORF61a-01 represent a spliced-out upstream ORF. The overlapping transcripts with other primary ORFs are not displayed.
(D) Transcripts containing ORF10 and its variants. The overlapping transcripts with other primary ORFs are not displayed.
(E) RT-PCR using primers spanning ORF61 variant splice junctions. Representative of 3 biological replicates.
(F) RT-PCR using primers spanning ORF10 variant splice junctions. Representative of 3 biological replicates.
(G) Occurrence of ORF61 variant splice junctions in validation sample sets.
(H) Occurrence of ORF10 variant splice junctions in validation sample sets.
As a means to assess the relative abundance of transcripts arising from different start sites, we plotted the location of the transcription start site of every transcript, overlaid with the number of deepCAGE reads per transcript start site (Figure S6). While many ORFs showed a relatively even distribution among start sites (e.g., ORF24, ORF49), other ORFs demonstrated a clear preference for 1 start site (e.g., M3, ORF59). The functional utility of generating overlapping transcripts that originate from different start sites remains an open question. It may simply reflect the indiscriminate use of multiple active endogenous promoters during lytic infection. Alternatively, it may be a mechanism to confer transcripts with different functions and/or cellular localizations. For example, ORFs with long 5′ UTRs were significantly enriched for poly(C) binding protein (PCBP1, PCBP3) motifs (Figure 4B), raising the prospect that in some cases transcripts with long 5′ UTRs may be preferentially retained in the nucleus. Consistent with this possibility, ORF55 isoforms (Figure 4C) with long 5′ UTRs (ORF55-04, ORF55-03) were exclusively present in the nuclear fraction, whereas the isoform with the shortest 5′ UTR (ORF55-01) was evenly distributed between cytoplasmic and nuclear fractions (Figure 4D).
Detection of Known and Unknown ORFs
As described above, TRIMD identified transcripts with primary ORFs that fully spanned 53 of the ORFs recorded in GenBank: U97553.2. TRIMD also identified transcripts harboring 55 ORFs that (1) encode truncated isoforms or splice variants of known ORFs, (2) form new ORFs via spliced chimeras of GenBank ORFs, or (3) were previously unappreciated ORFs (Figure 5A). We were surprised to find that 5′ truncations represent the largest proportion, accounting for 32 of the 55 (58%) new ORFs. Although some 5′ truncated transcripts may simply represent incidental priming from minor alternate start sites downstream of the major start site, the prominence of some 5′ truncation variants relative to the full-length version suggests that proteins encoded by these transcripts may hold some mechanistic relevance. For example, while variants such as the 5′ truncated transcripts overlapping ORF75C appear to represent minor alternative isoforms of full-length transcripts (Iso-Seq read depths of 8,260 for ORF75C-01 versus read depths of 5 and 4 for ORF75a-01 and ORF75b-01), 5′ truncated transcripts in other loci such as ORF66 were as prominent as full-length ORFs (Iso-Seq read depths of 107, 111, and 167 for ORF66-01, ORF66-02, and ORF66-03, respectively, versus a read depth of 119 for ORF66a-01). Our findings are corroborated by previous work that identified some of these N-terminal truncated proteins, including ORF17a (ORF17.5), ORF47a, and ORF62a (Nash et al., 2001). Likewise, Bencun et al. (2018) have used ribosomal profiling to identify truncating downstream translation initiation sites in EBV, and N-terminal truncated proteins have also been observed in other herpesviruses (Tombácz et al., 2016, 2017).
Transcripts carrying splice junctions were a major source of both transcript and ORF diversity: in all, 43 unique splice junctions, with introns ranging from 90 to 7,702 nt, were identified in the 258 TRIMD-validated MHV68 transcripts. Of these, 26 were located in either the 5’ or the 3’ UTR and 8 were located in putative noncoding RNAs (Figure 5B). The 20 splice junctions detected in GenBank-annotated MHV68 ORFs were a major source of potential protein diversity, accounting for 27% (15 of 55) of all newly identified ORFs (Figure 5A). Like transcription start sites and poly(A) ends, splice junctions appeared with varying depths at different time points after infection, and concordance between the MHV68 and MHV68.bla viruses was high (Figure S7). Few splice junctions were detected in latently infected B cells (Figure 3G). Among these was the previously identified latency splice junction in the M2 ORF (DeZalia and Speck, 2008). The remainder of the splice junctions detected in latently infected B cells were also detected during the lytic infection of fibroblasts; thus, these likely represent lytic transcript expression in the small percentage of cells spontaneously reactivating from latency. Consistent with this possibility, multiple splice junctions that had been identified by TRIMD in lytically infected fibroblasts were also detected in B cells reactivating from latency, although sensitivity was limited by the modest induction of reactivation (Figure S7).
Some of the identified splice junctions are supported by previously published molecular identification of alternate transcripts, including ORF50a-01 and ORF50b-01 (see below) and ORF57a-01 (Mackett et al., 1997). The impact of splicing on the putative protein varied with the transcript: 15 of the transcripts featured intra-ORF splicing, in some cases removing the majority of the ORF (e.g., ORF36a-01) and in other cases removing smaller segments. In some scenarios, the removal of individual Pfam-cataloged domains (El-Gebali et al., 2019) was apparent, such as the ribonucleotide reductase all-alpha domain deleted from 2 ORF61 isoforms (ORF61a-01, ORF61b-01; Figure 5C). In 4 transcripts, splicing generated sequences encoding in-frame chimeras of partial or full-length GenBank-annotated ORFs (e.g., M3-M2-01). Of particular interest were those spliced transcripts that included partial known ORF sequences, but that were spliced into a different frame of the same ORF region (e.g., ORF10a-01, ORF10b-01; Figure 5D), potentially creating entirely new protein domains. TRIMD also identified 4 intergenic ORFs that have not previously been reported and appear to be unique to MHV68: M15, M16, M17, and M18. Although the biological functions of these putative proteins remain unknown, the presence of the spliced ORF10 and ORF61 transcripts in strand-specific PCR and validation datasets (Figures 5E–5H) lends credence to their possible relevance during MHV68 infection.
Long Noncoding Transcripts
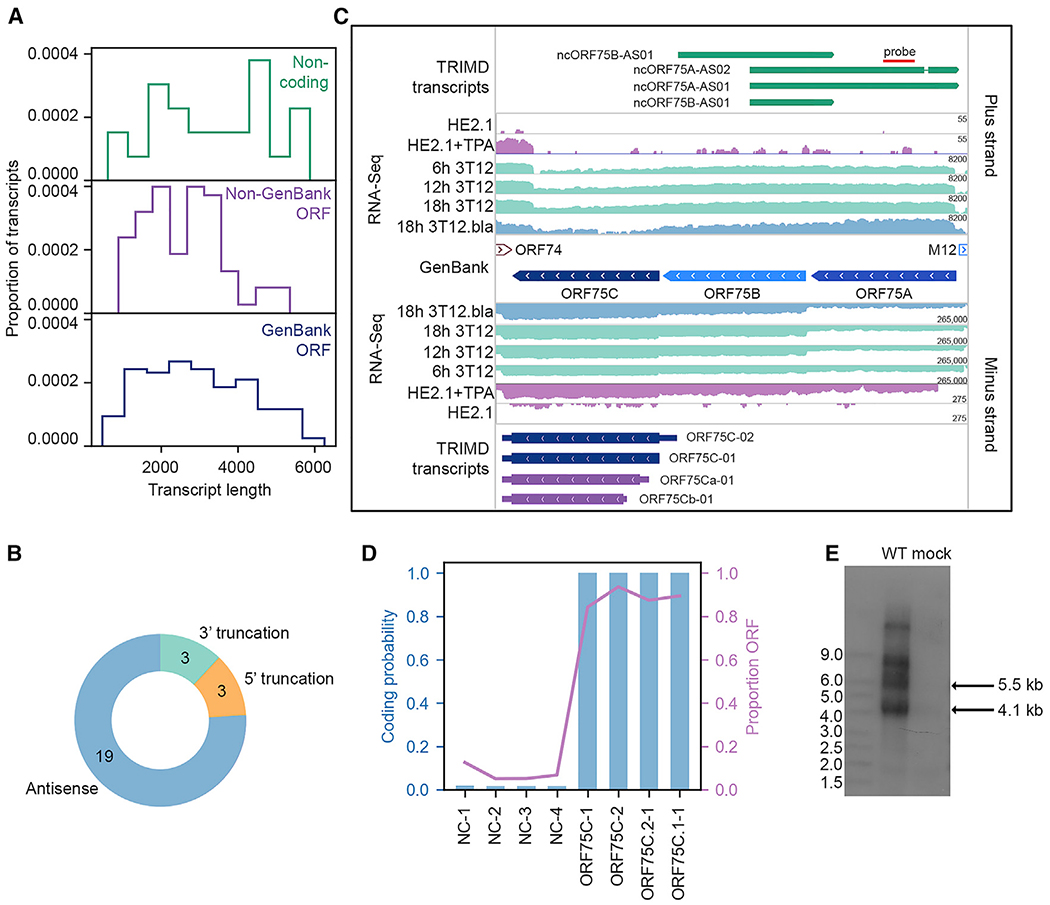
As noted above, TRIMD coupled with CPAT analysis identified 25 transcripts that displayed characteristics that are consistent with those of noncoding RNAs. These putative noncoding RNAs were not simply short transcripts: their sizes ranged from 610 to 5,884 nt (median, 3,564), generally longer than both GenBank (median, 2,864) and non-GenBank (median, 2,465) coding transcripts (Figure 6A). Notably, while the MHV68 putative noncoding RNAs were scattered throughout the genome, 13 of these transcripts were overlapping and clustered in a region directly antisense to ORF63 and ORF64 (Figure 2). Accordingly, the majority (19 of 25) of the putative noncoding transcripts lie antisense to GenBank-annotated ORFs (Figure 6B). TRIMD also identified 4 transcripts that lie antisense to the MHV68 ORF75 locus (Figure 6C), which encodes the related tegument proteins ORF75A, ORF75B, and ORF75C. Like their homologous counterparts in EBV (BNRF1) and KSHV (ORF75), the MHV68 ORF75s display homology to the cellular purine metabolism enzyme formyl-glycinamide-phosphoribosyl-amidotransferase (FGARAT) and are crucial for lytic infection (Tsai et al., 2015). Unlike the TRIMD-validated transcripts coding for full-length or N-terminal truncated versions of ORF75C, which are >85% ORF by proportion, the antisense transcripts each harbor potential ORFs in <15% of their sequence and display a CPAT coding probability of nearly 0 (Figure 6D). Consistent with TRIMD identification of antisense transcripts in this region, strand-specific northern blot detected stable expression of the 5.6-kb transcript ncORF75A-AS02 and the 4.1-kb transcript ncORF75B-AS01 (Figure 6E). This probe also detected 2 large transcripts that were not present in SMRT sequencing results, likely due to their greater length. Thus, the region of the strand antisense to the ORF75 coding genes is highly transcriptionally active and encodes numerous overlapping transcripts.
Figure 6. MHV68 Noncoding Transcripts.
(A) Proportion of noncoding transcripts, transcripts containing non-GenBank ORFs, and transcripts containing GenBank-annotated ORFs at specified transcript lengths.
(B) Type of noncoding transcript relative to overlapping ORFs.
(C) TRIMD-identified transcripts and supporting evidence at the ORF75A/B/C locus. The GenBank section indicates reference GenBank-annotated ORFs in this locus. TRIMD-identified transcripts are indicated (noncoding, dark green; early, light blue; early-late, royal blue; late, dark blue; unknown ORF, purple) for both plus and minus strands, along with corresponding RNA-seq tracks for the TRIMD dataset (18h 3T12.bla) and validation sample sets. RNA-seq read depth (log2 scale) is indicated. N = 1 for each sample. The red line indicates the probe site used in the northern blot in (E).
(D) CPAT-determined coding probability and ORF proportion for transcripts in the ORF75A/B/C locus.
(E) Northern blot for TRIMD-identified noncoding transcripts in theORF75 locus. NIH 3T12 fibroblasts were mock infected or were infected with wild-type MHV68 for 18 hours post-infection (hpi). The northern blot probe binding site is indicated in (C). Representative of 4 biological replicates.
Bi-directional Transcriptional Complexity
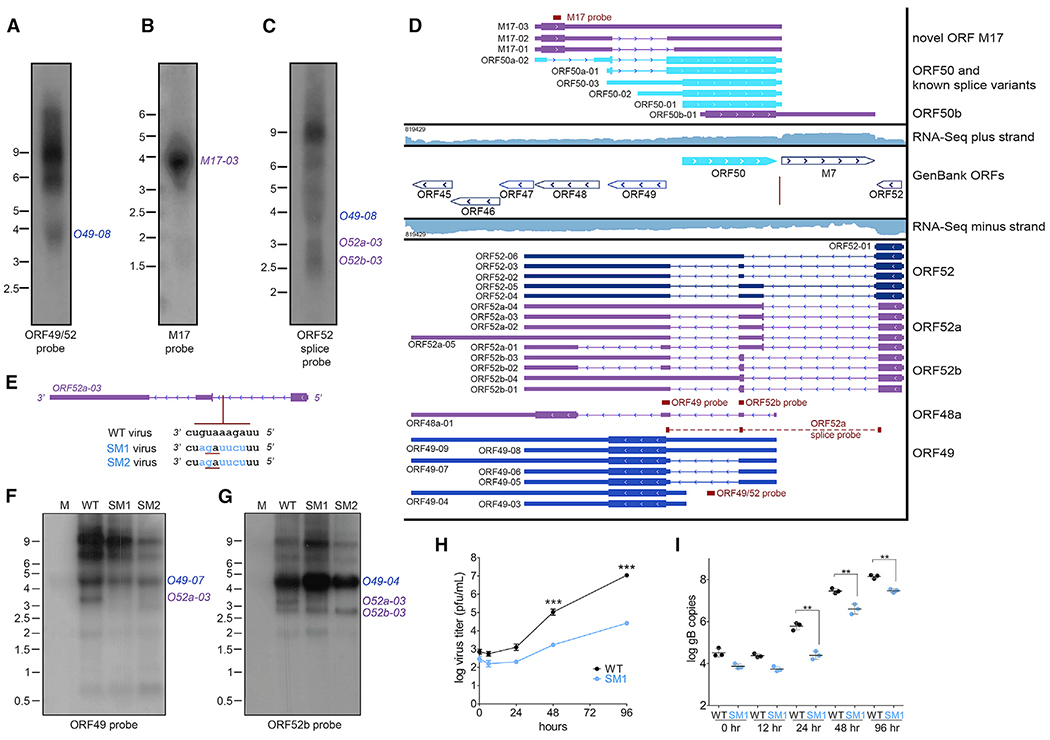
Some regions of the genome displayed particularly complex bidirectional transcriptional activity. Nowhere was this scenario more prevalent than the ORF50 locus (Figure 7), which encodes the critical latent to lytic switch protein Rta. Among gammaherpesviruses, Rta is a highly conserved protein that plays a central role in replication and reactivation (Damania et al., 2004; Wu et al., 2000). Previous work has identified multiple MHV68 ORF50 transcripts emanating from distinct upstream promoters (Gray et al., 2009; Liu et al., 2000; Wakeman et al., 2014). Our approach validated the presence of 3 of these isoforms (Figure 7D). In addition, TRIMD identified numerous other transcripts emanating from this locus, including (1) 3 sense strand readthrough transcripts that appear to encode an unknown upstream ORF (M17-01 to -03); (2) 1 sense strand transcript encoding an N-terminal truncated ORF50 (ORF50b-01); (3) 5 antisense strand ORF52 transcript isoforms with 3′ UTRs that are spliced across ORF50 (ORF52-01 to -05); and (4) 9 antisense strand transcripts encoding chimeric ORFs comprising 3′ truncated ORF52 spliced to 1 of 2 small ORFs directly antisense to ORF50 (ORF52a-01 to −05; ORF52b-01 to -04). Confirming the density of transcripts in this locus, long ORF49 transcripts (Figure 7A), M17 transcripts (Figure 7B), and multiple spliced ORF52a transcripts (Figure 7C) were detectable by northern blot.
Figure 7. Complex Transcription at the ORF50 Locus.
(A–C) Northern blot validation of TRIMD transcripts identified within the ORF50 locus using probes complementary to leftward ORF49 and ORF52 transcripts (A), rightward M17 transcripts (B), and leftward ORF52 spliced transcripts (C). The NIH 3T12 fibroblasts were infected with wild-type MHV68 for 18 h. The northern blot probe binding sites are indicated in (D). Representative of 4 biological replicates.
(D) TRIMD-identified transcripts at the ORF50 locus. The GenBank section indicates reference GenBank-annotated ORFs. The TRIMD-identified transcripts are indicated (immediate early, bright blue; early, light blue; early-late, royal blue; late, dark blue; unknown ORF, purple) for both plus (top) and minus (bottom) strands, along with corresponding RNA-seq tracks (log scale) used for the TRIMD dataset (18 h 3T12.bla). The RNA-seq read depth is indicated. The horizontal red lines indicate the probe sites used in northern blots, which are shown in (A)–(C), (F), and (G). The vertical red line indicates the location of the SM1 and SM2 virus mutations.
(E) Location and sequence of SM1 and SM2 virus mutations relative to the ORF52a-03 transcript. The altered nucleotide sequences are shown in blue. The red underline indicates the location of the new splice acceptor.
(F and G) Northern blot analysis of transcripts expressed in NIH 3T12 fibroblasts that were mock infected (“M”), infected with wild-type MHV68 (“WT”), or infected with SM1 or SM2 mutant viruses for 18 h. Northern blots were performed using probes complementary to transcripts within ORF49 (F) and with the second exon of ORF52 spliced transcripts (G). The northern blot probe binding sites are indicated in (D). Representative of 3 biological replicates.
(H) Titers of wild-type MHV68 versus SM1 mutant virus during multi-step growth in NIH 3T12 fibroblasts as determined by plaque assay. Means ± SDs for 2 biological replicates.
(I) Genome copies of wild-type MHV68 versus SM1 mutant virus during multi-step growth in NIH 3T12 fibroblasts, as determined by qPCR using primers specific for the glycoprotein B (gB) gene. Means ± SDs for 2 biological replicates.
To determine whether previously unknown transcripts within this locus contribute to viral fitness, we sought to examine the biological phenotype of mutant viruses that lacked the expression of specific transcript isoforms. In screening recombinant viruses that contained mutations within this region, we identified 1 mutant, SM1, which displayed an unexpected and apparently specific loss of the transcript ORF52a-03. The mutation within this virus incorporated a 6-nt change at a site that lies within the intronic sequence of transcripts that initiate at the ORF52 transcription start site (Figure 7E), including transcripts containing coding sequences for ORF52, ORF52a, or ORF52b. However, northern blot analyses revealed that while the SM1 mutation resulted in the complete loss of the 3.1-kb ORF52a-03 transcript, the expression of other transcripts emanating from this region were not significantly affected (Figures 7F and 7G). A second independently generated version of this mutant virus, SM2, displayed an identical loss of ORF52a-03 expression in northern blot analyses (Figures 7F and 7G). To determine whether the loss of ORF52a-03 expression altered viral fitness, we quantified SM1 virus replication in fibroblasts during multi-step growth. Notably, SM1 replication was significantly attenuated as compared to parental wild-type MHV68, in which replication was quantified using plaque assay for viral titer (Figure 7H) or qPCR for viral DNA (Figure 7I). These findings implicate the TRIMD-identified transcript ORF52a-03 as a determinant of MHV68 fitness.
DISCUSSION
Herpesviruses have complex lifestyles that require distinct transcriptional programs that operate during discrete phases of infection. Thus, by necessity, these viruses must encode a repertoire of genes frequently numbering ≥100. In the face of evolutionary pressures impeding genome size expansion, the acquisition of diversity through time results in highly dense genomes that display large numbers of genes with overlapping transcripts, alternate promoters, alternate transcript isoforms, extensive splicing, and shared transcriptional stop sites. From a research perspective, these characteristics immensely complicate the transcriptional annotation of herpesviruses. Accordingly, for many herpesvirus genomes, annotation has relied upon canonical translational start and stop sequences to define viral genes, with laborious follow-up molecular work used in a piecemeal fashion to reveal transcriptional start and stop sites, 5′ and 3′ UTRs, splice junctions, and overlapping transcripts for individual genes of interest. Such is the case for MHV68, whose full sequence was reported in 1997, along with a simple ORF-based annotation. Despite the difficult work put forth by numerous investigators on individual genes, the genome has remained poorly annotated. In work described here, we have integrated datasets from 3 complementary transcriptomics platforms to globally resolve the complex lytic transcriptome of MHV68. This annotation includes newly defined 5′ UTRs, 3′ UTRs, and splice junctions of transcripts across 53 previously reported MHV68 ORFs, as well as full-length transcripts that encode 55 ORFs that represent truncated versions of known ORFs or ORFs that have not been previously identified. In all, 258 unique MHV68 transcripts were identified, including 25 that are predicted to be noncoding.
Transcript Diversity
The map of TRIMD-validated MHV68 transcripts (Figure 2A; Figure S1) illustrates the vast extent of overlap among transcripts at individual gene loci. Although the use of PacBio and deepCAGE sequencing does not explicitly determine the relative abundance of specific transcripts, the Iso-Seq scores (which are strongly affected by transcript length) and deepCAGE tag depths (Figure S2) can provide some information about the relative abundance of overlapping transcripts, particularly when those transcripts are of similar length. The extensive overlap of transcripts is for the most part due to the use of alternate promoters, to splice variants, and to transcriptional stop read-throughs. Alternate promoter usage is a major source of transcript diversity, with upstream start sites resulting in increased 5′ UTR lengths for full-length ORFs and downstream start sites resulting in 5′ truncated ORFs. Thus, despite the large amount of overlap among transcripts, the vast majority of 5′ start sites are shared by only 2 transcripts each (Figure 3A). It is also notable from the analysis of deepCAGE tags for individual genes that while some genes clearly use a dominant promoter, others use ≥2 promoters equivalently (Figure S8). Although the high degree of 3′ stop sequence readthrough results in an enormous number of shared 3′ ends among transcripts, some genetic loci are covered by multiple transcript isoforms that use different 3′ ends.
The usage of alternate start and stop sites for overlapping isoforms varies with gene class (Figure 4A). Most notable is the difference between early lytic replication genes and latency genes. These findings may reflect different levels of regulatory controls for individual classes of genes, with early genes (short 5′ UTRs and long 3′ UTRs) benefiting from simple promoters but subject to tighter control by microRNAs (miRNAs) or 3′ UTR binding proteins, and latency genes (long 5′ UTRs and short 3′ UTRs) regulated by complex promoter elements but able to evade host miRNA and 3′ UTR binding protein control.
The benefits to the virus of multiple overlapping transcripts covering individual ORFs remains unclear. It is plausible that this strategy simply allows for differential control by regulatory elements at different stages of infection. However, this tactic may also facilitate the generation of transcripts with unique functions. For example, transcripts with long UTRs display enrichment of nuclear retention motifs and PCBP motifs (Figure 4B), suggesting that transcript isoforms with long UTRs may demonstrate a propensity for localization to the nucleus. Thus, it is possible that isoforms harboring short UTRs may be more efficiently translated (e.g., McClelland et al., 2009; Sedman et al., 1990; Singh et al., 2005), while long UTRs may impart noncoding RNA functions to some isoforms. Alternatively, it is conceivable that these overlapping transcripts may reflect promiscuous transcription due to the lack of normal regulatory controls at the late stages of infection. However, the significant difference between the number of stop sequence readthroughs of viral versus cellular transcripts (Figure 3C) strongly suggests that this diversification of viral transcripts is more likely the result of evolutionary pressure rather than transcriptional chaos.
ORF Diversity
The application of TRIMD to transcriptomic datasets from MHV68 lytic infection has revealed as many as 55 new or previously unappreciated potential ORFs. Previous efforts to identify MHV68 coding regions have by necessity been confined to computational analyses of maximized stretches of continuous genome that are bounded by canonical translation start and stop sites. Although TRIMD also requires the use of start and stop sequence constraints, the use of multiple complementary datasets allows the empirical identification and validation of ORFs that are otherwise unlikely to be predicted. Thus, this approach has revealed potential proteins that initiate from downstream start sites (N-terminal truncations), omit specific regions of an ORF (splice variants), splice from one ORF to portions of another ORF (chimeras), or derive from regions antisense to or between GenBank-annotated ORFs (unknown). While a great amount of future work will be required to determine which of these potential coding regions are translated and biologically significant, these findings demonstrate that MHV68 may encode many more proteins than has been previously appreciated.
The isoforms identified here likely represent a large majority of all potential MHV68 transcripts. In addition, for at least 8 of the GenBank-annotated ORFs for which we did not observe transcripts corresponding to the full-length ORF, we defined alternate transcripts that correspond to spliced or 5′ truncated versions of the GenBank-annotated ORF. While these have been appropriately designated as “unknown” ORFs here, it is very likely that at least some of these are the primary (or only) protein-coding transcript generated over that specific ORF. However, despite the robust number of ORFs identified by TRIMD, the cohort presented here certainly does not comprise every single protein-coding transcript generated by the virus. Although it is unlikely that every single ORF in the virus is expressed, that we did not detect all of the 80 original GenBank-annotated ORFs suggests that further detailed molecular analyses may reveal additional transcript isoforms. One arena that deserves extensive consideration is that of very long transcripts. PacBio long-read sequencing, while robust for the detection of transcripts of ≤5 kb, remains limited for the reliable sequencing of longer transcripts (O’Grady et al., 2016). Consistent with this possibility, in northern blot experiments for antisense ORF75 transcripts (Figure 6E) and for ORF52 transcripts (Figure 7), we have reproducibly detected 9-kb transcripts that are not present in pre-processing PacBio Iso-Seq datasets. Furthermore, in our previous EBV transcriptome analysis (O’Grady et al., 2016), we identified an abundant 14-kb transcript derived from the BZLF2 locus that was not detected by our TRIMD analysis. Thus, the likelihood that long transcripts run throughout the genome presents an additional layer of transcriptional complexity that must be considered in future experiments.
It is also important to note that due to readthrough of transcriptional stops, numerous isoforms carried >1 potential ORF. It is likely that the furthest upstream ORF is most frequently translated; accordingly, TRIMD annotates only the most 5′ proximal ORF. However, it remains possible that some ORFs from multi-cistronic transcripts are nevertheless translated inefficiently or through alternative translational strategies that favor the downstream ORF, as is the case for MHV68 K3 (Coleman et al., 2003). Some GenBank-annotated ORFs were fully contained within TRIMD-validated transcripts, but they were not annotated here as the primary ORF in any transcript. Thus, it is plausible that at least some of the “missing” GenBank ORFs may be translated from downstream translational start sites in these longer transcripts. Likewise, it is important to note that while non-GenBank ORFs <100 aa are below our transcript analysis thresholds, such proteins have been previously identified (May et al., 2005) and may play important roles in viral infection.
Noncoding RNAs
Using TRIMD, we have now defined 25 MHV68 transcript structures that display low coding probability (Figures 2 and 6) and thus are likely to function as noncoding RNAs. Of these transcripts, 19 lie within regions previously identified to be transcriptionally active but not covered by GenBank-annotated ORFs (Cheng et al., 2012; Johnson et al., 2010), while 6 of the transcripts overlap known coding genes. Several of the transcripts are located in clusters antisense to ORF63/64 or ORF75A/B, suggesting that individual transcripts within each set may have overlapping functionality. It is plausible that such antisense noncoding RNAs may play specific roles in the regulation of sense strand genes, as has been shown for numerous host genes (Pelechano and Steinmetz, 2013). For example, MHV68 antisense noncoding transcripts may regulate the expression of sense strand ORF75 genes, which encode an essential tegument protein that is conserved among gammaherpesviruses (Tsai et al., 2015). MHV68 expresses 3 partially homologous genes from this locus, which appear to play important but distinct roles in different stages of MHV68 infection (Cheng et al., 2012; Van Skike et al., 2018), and it is plausible that antisense noncoding transcripts may regulate their differential expression.
As is the case for the MHV68 coding transcripts, it remains likely that other noncoding transcripts have yet to be discovered. For example, northern blots to detect bona fide transcripts in 2 EGRs revealed the presence of several stable transcripts in the ORF66/67/68/69 locus that localize to the nucleus, suggesting that they may function as noncoding RNAs (Canny et al., 2014). However, those transcripts were not detected by TRIMD. As described above, transcript isoforms carrying long UTRs may also have a higher propensity for nuclear retention and therefore hold the potential for noncoding RNA activity. Thus, it is likely that additional MHV68 noncoding RNAs remain to be revealed.
Finally, it should be noted that although CPAT assessment is a reliable indicator of coding potential, it remains possible that at least some of these putative noncoding transcripts may be translated. For example, 3 of these transcripts comprise either 5′ or 3′ truncations of longer coding transcripts that retain some portion of an ORF and therefore may encode small proteins, possibly originating from a non-canonical translational start site. However, this would not exclude the possibility that these transcripts could also function as noncoding RNAs, as has been demonstrated for some putative host and viral noncoding RNAs (Nelson et al., 2016; Stern-Ginossar et al., 2012).
Transcriptional Complexity versus Biological Relevance
The MHV68 transcriptome map presented here (Figure 2) reveals the degree to which MHV68 transcript isoforms overlap during lytic infection. With a total of 258 transcripts covering 108 ORFs, many ORFs were covered by ≥2 transcripts. The magnitude of such transcriptional complexity was especially prevalent within the critical ORF50 locus, a finding that could not have been predicted a priori. Thus, this region is illustrative of the need to more carefully define herpesvirus transcripts using new cutting-edge transcriptomics approaches. Such findings are not only important for understanding the regulation of gammaherpesvirus gene expression but are also essential for designing rational virus mutations to determine the biological relevance of individual viral genes.
Does this transcriptional complexity yield biologically relevant transcript isoforms? This question remains open with regard to many of these transcripts. However, our demonstration of a fitness advantage for viruses expressing the transcript ORF52a-03 strongly suggests that some of these isoforms play functional roles in virus biology. ORF52 from MHV68, KSHV, and rhesus rhadinovirus has been previously demonstrated to be a tegument protein that carries out multiple functions, including virion tegumentation, envelopment, microtubule rearrangements, and immune evasion (Benach et al., 2007; Bortz et al., 2007; Wang et al., 2012; Wu et al., 2015; Loftus et al., 2017). This multitude of activities is carried out through the use of discreet domains. It is notable then that the unique classes of transcripts described here (ORF52a and ORF52b) are each predicted to encode ORF52 proteins that carry unique C-terminal domains.
Although the specific reason that the SM1 mutation results in the loss of ORF52a-03 expression is not yet fully clear, the altered sequence in the SM1 and SM2 mutants used here results in the introduction of a new predicted splice acceptor site. Notably, this new acceptor site lies upstream of the weak exon 1 to exon 2 splice acceptor site normally used by ORF52a-03 (while ORF52b transcripts use a canonical splice acceptor sequence). Thus, we speculate that the loss of ORF52a-03 may result from premature splicing at the new acceptor sequence, followed by nonsense-mediated decay (reviewed in Hug et al., 2016).
While previous efforts to generate mutant viruses have focused primarily on generating translational stops that would alter all of the isoforms encoding the ORF of interest, the degree of transcript overlap observed here reveals the need for more subtle mutational approaches to study individual transcript isoforms, such as approaches to regulate alternative splicing events. Follow-up studies to examine overlapping transcripts throughout the genome will require the utilization of similar approaches to alter the expression or stability of individual transcript isoforms or sets of isoforms. Nevertheless, our findings demonstrating a functional correlation between ORF52a-03 expression and enhanced viral replication strongly argue for further interrogation of many of the isoforms identified here, as well as a deeper examination of the complexity of gammaherpesvirus transcriptomes during different stages of infection.
Final Thoughts
The findings presented here provide a clear starting point for the difficult work that will be required to understand the transcriptional complexity of dense gammaherpesvirus genomes. Our application of TRIMD to MHV68 infection has resolved transcript structures across the entire MHV68 genome, revealed previously unknown potential coding sequences and noncoding RNAs, and provided insight into the transcriptional complexity of this gammaherpesvirus during lytic replication. However, it is clear that this analysis has uncovered only a portion of the potential transcripts. In particular, much further work needs to be done to resolve long transcript structures, which should become increasingly possible as long-read sequencing technology advances. Likewise, it will be important to examine transcripts that are non-polyadenylated. Most important, it will be crucial to define the biological relevance of individual genes and specific transcripts through extensive molecular and functional experiments. The results presented here yield a substantial foundation for such detailed mechanistic studies.
Finally, given the interwoven evolutionary relation of the herpesviruses to mammalian cells, the insights and patterns found here are likely paralleled in host cells. Thus, these findings also provide a guidepost for the analysis of similar transcriptional complexity in host cells, particularly in different tissues, in the presence or absence of environmental stressors such as virus infection and during malignant transformation. This and related studies will be critical to begin to refine rules for transcript variants and to eventually allow for accurate predictions of transcript isoform selection.
STAR★METHODS
CONTACT FOR REAGENT AND RESOURCE SHARING
Further information and requests for resources and reagents should be directed to and will be fulfilled by the Lead Contact, Scott A. Tibbetts (stibbe@ufl.edu).
EXPERIMENTAL MODEL AND SUBJECT DETAILS
Cell culture
NIH 3T12 fibroblasts (derived from a male BALB/c mouse) were maintained at 37°C in Dulbecco’s Modified Eagle’s Medium (DMEM) with L-glutamine, 4.5g/L glucose and sodium pyruvate (Corning, NY, USA) supplemented with 10% fetal bovine serum (Omega Scientific, CA, USA) and 1% penicillin-streptomycin (GIBCO/Thermofisher Scientific, MA, USA). Cells were passaged 1:15 every three days. Latently infected HE2.1 B cells, which are an MHV68+ cell line generated from the A20 parental B cell line (derived from a female BALB/cAnN mouse), have been previously described (Forrest and Speck, 2008). Cells were maintained at 37°C and were passaged 1:15 every three days. Reactivation of HE2.1 B cells was induced using 12-O-tetradecanoylphorbol-13-acetate (TPA) treatment (20 ng/ml) for 18 hours prior to RNA extraction.
METHOD DETAILS
Infections
NIH 3T12 fibroblasts were plated at a density of 2 × 105 cells/well in a flat-bottom 6-well TC-treated cell culture plate. Twenty four hours later, cells were infected at MOI 5 with a wild-type MHV68 (WUMS strain) or a floxed, BAC-derived wild-type MHV68 (MHV68.bla) which carries a β-lactamase marker as a C-terminal fusion to ORF73 (Nealy et al., 2010). Parallel samples were harvested for RNA extraction at 6, 12, and 18 hours.
RNA extraction
All laboratory procedures were carried out using water treated with 1% Diethyl pyrocarbonate (DEPC) (MP Biomedicals/Thermofisher), and surfaces were cleaned with RNaseZap (Ambion/Thermofisher). Cells were washed with ice cold Dulbecco’s phosphate-buffered saline (HyClone, GE Healthcare, UK). Total RNA was extracted using TRIzol reagent (Invitrogen/Thermofisher) according to the manufacturer’s protocol. Pellets were reconstituted using RNase-free water, and total RNA was quantified. RNA purity was assessed using the Thermo Scientific NanoDrop 2000 (ThermoFisher).
Sequencing
For sequencing, polyadenylated RNA was enriched using a Poly(A)Purist MAG kit (Thermofisher). For long-read sequencing, Iso-Seq library preparation and SMRT sequencing were performed by the Johns Hopkins Deep Sequencing and Microarray Core Facility (Baltimore, MD, USA) according to Pacific Biosciences protocols. To improve sequencing rates for longer transcripts, Iso-Seq libraries were separated into 1-2 kb, 2-3 kb and > 3 kb size fractions and sequenced separately in 2 SMRT cells for the 1-2 kb fraction and 3 SMRT cells each for the 2-3 kb and > 3 kb fractions. For RNA-Seq, libraries were prepared using the TruSeq stranded protocol (Illumina, CA, USA). Single-end, 101-bp sequencing was performed using an Illumina HiSeq 2500 instrument. For RNA-Seq of samples used for validation, libraries were prepared using the BGI-Seq stranded protocol and paired-end, 75-bp sequencing was performed using a BGISEQ 500 instrument. All RNA-Seq was performed by Beijing Genomics Institute (Beijing, China). For deepCAGE sequencing, nAnT-iCAGE libraries (Murata et al., 2014) were prepared and single-end, 50 bp sequencing was performed using an Illumina HiSeq 2500 instrument by DNAform (Yokohama, Japan).
Sequence alignments
All data were aligned and mapped to the mouse (mm10 assembly) and MHV68 genomes. The MHV68 genome was based on the WUMS strain genome (GenBank U97553.2)(Virgin et al., 1997) with the bla insert (Nealy et al., 2010) and LoxP scar added (Adler et al., 2000). Iso-Seq data was compiled into SMRT consensus “full-length” isoforms (CFLs) using SMRT Portal v. 1 (https://www.pacb.com/) without the Quiver algorithm. Iso-Seq CFLs were mapped with GMAP (Wu and Watanabe, 2005) release 21 July 2014. Illumina RNA-Seq and deepCAGE reads were mapped using STAR aligner (Dobin et al., 2013) with default settings.
Integrated validation of transcript structures
Transcript structures were determined using an updated version of the Transcriptome Resolution through Integration of Multi-platform Data (TRIMD) method (O’Grady et al., 2016). Briefly: clusters of non-softclipped Iso-Seq CFL 5′ ends mapping within 8 bp of each other were identified, and the consensus start site calculated as the weighted average of CFL start coordinates within the cluster. Start site clusters were identified in deepCAGE data using Paraclu (Frith et al., 2008) and requiring (i) a minimum of 135 tags/cluster (virus) or 15 tags/cluster (cell), (ii) (maximum density/baseline density) ≥ 2 and (iii) cluster length of 1-20 bp. Iso-Seq consensus start sites mapping within 2 bases of deepCAGE consensus start sites were considered validated. The higher read depth stringency for the virus (minimum 135 tags/cluster versus 15 tags/cluster) reflects the higher relative depth of sequencing reads mapping to the viral chromosome (see Figure 1A). Splice junctions identified in both CFLs (by GMAP) and Illumina reads (by STAR) were considered validated. For polyadenylation sites, clusters of non-softclipped Iso-Seq CFL 3′ ends mapping within 8 bp of each other were identified and consensus polyadenylation sites determined by calculating a weighted average within the cluster. Illumina RNA-Seq reads containing putative poly(A) tails were identified in the STAR-aligned SAM file as reads that end with a run of at least 5 As (plus strand) or 5 Ts (minus strand), at least 2 of which were softclipped. Poly(A)-tail reads aligning within 8 bp of each other were considered to represent a single polyadenylation site, and a consensus polyadenylation site was determined by calculating a weighted average of the cluster. Iso-Seq CFL consensus polyadenylation sites supported by at least 5 SMRT reads (virus) or 1 SMRT read (cell) that were located within 10 bp upstream or 4 bp downstream of an Illumina consensus polyadenylation site or within 40 bp downstream of a genomic AATAAA polyadenylation signal sequence were considered validated. Finally, CFLs whose start site, splice junctions (if any) and end site matched validated features were considered to be validated transcripts, and matching sets of CFLs were collapsed into single validated transcripts.
Validation of transcript features in other samples
TRIMD-identified transcription start sites, splice junctions and polyadenylation sites were compared to transcript features identified in deepCAGE and RNA-Seq datasets from 8 additional samples: 3T12 cells infected with unmodified wild-type MHV68 and harvested at 6 hours, 12 hours and 18 hours post-infection; 3T12 cells infected with MHV68.blaand harvested at 6 hours, 12 hours and 18 hours post-infection; HE2.1 cells and HE2.1 cells reactivated with 12-O-tetradecanoylphorbol-13-acetate (TPA). To validate TRIMD transcription start sites, deepCAGE consensus start sites were identified with Paraclu. For MHV68 and MHV68.bla infections, tag depth threshold was normalized by the ratio of the number of viral deepCAGE tags obtained from the 18hpi MHV68.bla sample used for TRIMD and the number of viral deepCAGE tags obtained from the MHV68.bla sample used for validation, and the same threshold (92 tags/cluster) applied to all infection time points for both virus strains. To compensate for the relatively low reactivation level of HE2.1 cells with TPA treatment and correspondingly low number of viral deepCAGE tags, a threshold of 3 tags/cluster was used. Other Paraclu parameters ([maximum density/baseline density] ≥ 2 and cluster length of 1-20 bp) were kept the same for the validation datasets as for the TRIMD dataset. TRIMD TSSs were considered detected if a Paraclu cluster was within 5 nt of a TRIMD Iso-Seq CFL 5′ cluster. To validate TRIMD splice junctions an exact match of splice donor and acceptor sites was required. To validate TRIMD polyadenylation sites RNA-Seq reads containing putative polyA tails were identified and clustered as above. Polyadenylation sites were considered detected in a validation dataset if the Iso-Seq 3′ end cluster was within 10 bp upstream or 4 bp downstream of a poly(A)-tail read cluster. TRIMD-identified transcript features that were not detected in any of the 8 validation datasets were discarded.
Calculation of coding potential and ORF determination
All complete viral ORFs of at least 225 nt (75 aa) in TRIMD-validated transcripts were identified with TransDecoder v. 4.1.0 (https://github.com/TransDecoder/TransDecoder/wiki). For transcripts containing multiple ORFs, the most upstream ORF was considered to be the primary ORF. ORFs in TRIMD-validated transcripts were compared to ORFs annotated in the genome of the WUMS strain of MHV68 (GenBank U97553.2)(Virgin et al., 1997). Transcripts whose primary ORF was annotated in GenBank U97553.2 were considered to encode that ORF. For transcripts whose primary ORF was not included in this GenBank record, CPAT (http://rna-cpat.sourceforge.net/)(Wang et al., 2013) was used to evaluate coding potential. If CPAT indicated the transcript was likely coding, that transcript was considered to encode a non-GenBank ORF. If CPAT indicated that the transcript was likely noncoding, it was considered noncoding. Likewise, non-GenBank transcripts without complete ORFs of at least 300 nt were considered noncoding. Two transcripts (ORF40a-01, ORF63a-01) that held consensus polyadenylation sites which fell one nt short of containing a complete ORF, but for which at least one CFL with the full ORF was identified, were adjusted manually at the 3′ end coordinate to contain the full ORF. One noncoding transcript (ncblac-AS01) was found to largely consist of sequence from the beta-lactamase insert. This transcript was omitted from most analyses but remains listed in Data S1, S2, S3, S4, S5, and S6. Transcripts containing ORFs annotated in GenBank U97553.2 were assigned to expression classes (latent, immediate early, early, early-late or late) based on previous studies (Ahn et al., 2002; Cheng et al., 2012; Ebrahimi et al., 2003; Virgin et al., 1997; Wu et al., 2009).
Transcript naming scheme
Transcripts were named using a scheme that incorporates existing ORF names, and is based upon the original sequence report and related GenBank annotations (Virgin et al., 1997). This naming scheme minimizes renaming of previously described ORFs and transcripts, and allows for the intuitive addition of transcripts discovered in the future. Transcripts encoding an ORF named in GenBank U97553.2 (as described above in “Calculation of coding potential and ORF determination”) were named for that ORF (eg ORF10). ORFs that are splice variants or truncation variants of GenBank ORFs were given that ORF name with a letter suffix (eg ORF10a, ORF10b). Chimeric ORFs were named for both contributing ORFs, with the most upstream ORF listed first (eg M3-M2). Blastp and tblastn were used to determine whether unknown ORFs were present in KSHV, EBV or Herpesvirus saimiri. Unknown ORFs that did not display homology to any ORFs in these viruses were considered unique and thus were named with an “M” and sequentially increasing numbers. All transcripts were also named with a two-digit suffix to differentiate multiple transcripts encoding the same ORF (eg ORF9-01, ORF9-02, ORF10a-01, ORF10b-01, M3-M2-01, M15-01). Noncoding transcripts were named according to the ORF with which they show the most extensive overlap, with the prefix “nc” to indicate their likely noncoding status. Where appropriate, “AS” was added to the suffix when the noncoding transcript was antisense to the ORF for which it was named (eg ncORF59-01, ncORF75A-AS01, ncORF75A-AS02).
Motif analysis of alternate 5′ UTRs
For ORFs with multiple transcripts which encode different 5′ UTRs, the sequences of the longest and the shortest UTR were extracted. The AME (Analysis of Motif Enrichment) tool (http://meme-suite.org/doc/ame.html) (McLeay and Bailey, 2010) was used to identify possible enrichment in RNA-binding protein motifs described in Ray et al., 2013 using the Wilcoxon rank-sum test with an adjusted p value threshold of 0.05 (Ray et al., 2013).
Molecular validation of transcript expression
Northern blots.
RNA extraction and Northern blot protocols were based on the protocol for longer RNA molecules (> 200) (McClure et al., 2011). For Northern blots, 8-10 μg total RNA from MHV68-infected or mock-infected murine fibroblasts was combined with a 3X volume of NorthernMax formaldehyde load dye (Ambion, ThermoFisher), and 10 μg/ml final concentration of ethidium bromide. Samples were heated at 65°C for 15 minutes, immediately placed on ice, and then loaded onto a 6% formaldehyde-containing 1% agarose gel in parallel with Millenium RNA markers (ThermoFisher). Samples were run at 105 V for 3 hr in 0.45 μM filter-sterilized 1X MOPS buffer [20mM 3-(N-morpholino) propanesulphonic acid, 5 mM sodium acetate, 1mM EDTA, pH 7.0]. RNA integrity was assessed visually using a Bio-Rad ChemiDoc XRS+ (Bio-Rad Laboratories, CA, USA). Samples were then transferred overnight onto a Whatman Nytran SuPerCharge nylon blotting membrane (Sigma-Aldrich, MO, USA) using the Whatman Turboblotter kit in 20X saline-sodium citrate (SSC) buffer. Following a 2X SSC wash, the membrane was UV-crosslinked, then stained with 0.02% methylene blue for visualization of 18 s and 28 s bands and RNA markers. The crosslinked membrane was prehybridized at 68°C for 2 hours in ULTRAhyb buffer (Ambion, Thermofisher). RNA probes complimentary to the target transcript (Table S1) were synthesized from plasmid constructs using the Maxiscript T7/Sp6 kit (ThermoFisher Scientific) as directed by the manufacturer using 10 μCi 32P α-CTP (PerkinElmer, CT, USA). The membrane was hybridized at 68-72°C overnight with labeled probe in ULTRAhyb buffer, then washes were carried out according to manufacturer’s protocol.
Nuclear/cytoplasmic RNA fractionation.
Nuclear and cytoplasmic RNA fractions were obtained as previously described (Weil et al., 2000). Briefly, following infection, cells were washed in ice cold PBS, harvested, then centrifuged (4°C, 3 min, 3000 rpm) and washed twice in cold PBS. The cell pellet was resuspended in lysis buffer (10mM Tris pH 7.8, 140mM NaCl, 1.5mM MgCl2, 10 mM EDTA, 1% NP40) supplemented with RNasin (Promega, WI, USA), incubated on ice for 5 min, then centrifuged (4°C, 5 min, 3000 rpm). The resulting supernatant fluid (containing the cytoplasmic fraction) was transferred to a new tube and subjected to high speed centrifugation (4°C, 14,000 rpm, 1 min), and the cleared supernatant fluid was removed and added to TRIzol for cytoplasmic RNA extraction. In parallel, the pellet from the cellular lysis (containing the nuclear fraction) was washed once in lysis buffer, centrifuged (4°C, 3 min, 3000 rpm), then resuspended in lysis buffer. Subsequently, TRIzol reagent was added and then the mixture was repeatedly passed through a 20 gauge needle prior to nuclear RNA extraction. Probes against U6 snRNA (nuclear) and tRNA-Try (cytoplasm) were used to confirm fractionation by Northern blot.
Gene-specific, single-stranded RT-PCR.
Total RNA was reverse transcribed using ProtoScript II Reverse Transcriptase (New England Biolabs, MA, USA). Gene-specific reverse transcription primers TV_004 and TV_007 were used for ORF10 and ORF61 transcripts, respectively. Traditional PCR was used to confirm the existence of ORF10 and ORF61 transcripts using the following primers: ORF10_c3244 forward TV_017, reverse TV_004; ORF10_c2410 forward TV_018, reverse TV_004; ORF61_c8714 forward TV_025, reverse TV_010; ORF61_c20295 forward TV_026, reverse TV_010. Each PCR reaction mixture contained 12.5 μL Q5® High-Fidelity 2X Master Mix (NEB), 10 μM each primer, 1 μL cDNA, and nuclease-free water to a final volume of 25 μl. PCR was performed with pre-denaturation at 98°C for 30 s, amplification with 35 cycles of denaturation at 98°C for 10 s, annealing at 56°C for 30 s, and extension at 72°C for 15 s, followed by a final extension at 72°C for 2 min. PCR products were loaded onto 1.5% agarose gels and the expected bands were excised and purified according to the NucleoSpin Gel and PCR Clean-Up kit instructions (Clontech/Takara, Kyoto, Japan). The purified PCR products were cloned into pCR-Blunt II-TOPO Vector (Invitrogen/Thermofisher). Subsequently, DNA minipreps were prepared from ten colonies per culture and Sanger sequenced by Genewiz (NJ, USA).
Generation of mutant viruses
The MHV68 SM1 and SM2 recombinant viruses were generated as follows: recombination competent pGS1783 cells were transformed with γHV68 BAC DNA. Targeting constructs were PCR amplified using primers containing sequence homology to MHV68, the desired point mutations flanking the Kanamycin (Kan) resistance gene, and a I-SceI cut site. pGS1783-HV68 BAC-containing cells were electroporated with the PCR product cassette and screened by restriction digest analysis. Kan resistant colonies were then induced for I-SceI expression and a second round of recombination to produce a scarless mutation. The MHV68 WUMS wild-type sequence (nt 69393 to 69405) is agacatttctaaa, the SM1 and SM2 mutant viruses sequence (altered by 6 nts, as indicated in capital letters) is agaTCtAAGAaaa. Recombinant BACs were screened via restriction digest analysis and verified by sequence confirmation. The SM1 mutant virus was constructed and validated on the wild-type MHV68 BAC backbone. The SM2 mutant virus was constructed and validated on the MHV68.bla marker virus BAC backbone, providing two independently generated viruses.
QUANTIFICATION AND STATISTICAL ANALYSIS
Plaque assay analysis of multi-step replication
NIH 3T12 fibroblasts were infected at MOI 0.05 with wild-type MHV68 or SM1 mutant virus. Both cells and supernatant were harvested at 0, 6, 12, 24, 48, and 96 hpi. Samples were freeze-thawed for 3 cycles prior to plaque assay on NIH 3T12 fibroblasts.
qPCR analysis of multi-step replication
Harvested samples were pelleted for 10min at 300xg, 4C. The supernatant was removed and DNA isolated (QIAGEN DNeasy Blood & Tissue Kit). Total DNA was normalized to 10ng/uL, and 50ng of DNA was loaded into the qPCR reaction. qPCR was carried out using Roche LightCycler 480 Probe Master-Mix kit with gB primers and probe. Standard curve was determined using gB plasmid serially diluted 1:10 from 110 copies/5uL to 10 copies/5uL.
Statistical analyses
Analyses for virus quantitation experiments were performed using GraphPad Prism software. For quantitation, mean was used for definition of center, and error bars indicate SD. Significance was determined using Student’s t- test, with significance defined as p ≥ 0.05. Asterisks within figures indicate degree of significance, as indicated in figure legends. All validation and quantitation experiments were performed using two, three, or four biological replicates. Specific sample numbers are indicated in the figure legends.
DATA AND SOFTWARE AVAILABILITY
The updated version (v17March2018) of the TRIMD software and detailed instructions for use are available at https://github.com/flemingtonlab/TRIMD. The accession number for the Iso-Seq, deepCAGE and RNA-Seq data reported in this paper is GEO: GSE117651. The accession number for the MHV68.bla genome and annotation reported in this paper is GenBank: MH636806.
Supplementary Material
KEY RESOURCES TABLE
| REAGENT or RESOURCE | SOURCE | IDENTIFIER |
|---|---|---|
| Bacterial and Virus Strains | ||
| MHV68.bla | Nealy et al., 2010 | MH636806 |
| MHV68 (WUMS strain) | Virgin et al., 1997 | GenBank U97553.2 |
| MHV68 SM1 | This paper | N/A |
| MHV68 SM2 | This paper | N/A |
| Chemicals, Peptides, and Recombinant Proteins | ||
| Poly(A)Purist MAG kit | ThermoFisher Scientific | Cat #AM1922 |
| 12-O-tetradecanoylphorbol-13-acetate (TPA) | SigmaAldrich | Cat #P1585 |
| Maxiscript T7/Sp6 transcription kit | ThermoFisher Scientific | Cat #AM1320 |
| 32P α-CTP | PerkinElmer Health Sciences Inc | Cat #BLU508X250UC |
| Critical Commercial Assays | ||
| Pacific Biosciences Iso-Seq | Pacific Biosciences | N/A |
| Illumina RNA-Seq | Illumina | N/A |
| BGI-Seq | BGI | N/A |
| Deposited Data | ||
| MHV68 WUMS strain genome | Virgin et al., 1997 | GenBank U97553.2 |
| Mus musculus genome (mm10) | Genome Reference Consortium | https://www.ncbi.nlm.nih.gov/assembly/GCF_000001635.20/ |
| Raw and analyzed data | This paper | GEO: GSE117651 |
| MHV68.bla genome and annotation | Nealy et al., 2010; This paper | GEO: MH636806 |
| Experimental Models: Cell Lines | ||
| NIH3T12 fibroblasts | ATCC | ATCC® CCL-164; RRID: CVCL_0637 |
| HE2.1 B cells | Forrest and Speck, 2008 | N/A |
| Oligonucleotides | ||
| Primers: see Table S1 | This paper | N/A |
| Software and Algorithms | ||
| TRIMD | O’Grady et al., 2016 | https://github.com/fiemingtonlab/TRIMD |
| SMRT Portal v. 1 | Pacific Biosciences | https://www.pacb.com |
| GMAP release 21 July 2014 | Wu and Watanabe, 2005 | http://research-pub.gene.com/gmap |
| STAR | Dobin et al., 2013 | https://github.com/alexdobin/STAR |
| Paraclu | Frith et al., 2008 | http://cbrc3.cbrc.jp/~martin/paraclu/ |
| CPAT | Wang et al., 2013 | http://rna-cpat.sourceforge.net |
| TransDecoder v. 4.1.0 | Open software | https://github.com/TransDecoder/TransDecoder/wiki |
| AME | McLeay and Bailey, 2010 | http://meme-suite.org/doc/ame.html |
Highlights.
Global resolution of transcript isoforms during lytic gammaherpesvirus infection
Genome-wide alternate isoform usage and readthrough transcription
Resolution of 258 transcript structures, including overlapping isoforms
Identification of long noncoding RNAs and unexpected ORFs
ACKNOWLEDGMENTS
We have sought to devise a naming scheme that is both non-biased and amenable to the addition of new transcripts identified in the future. For that reason, we used the original report of GenBank: U97553.2 as a logical foundation for building transcript names. Thus, by necessity, some protein names that are in current use were not able to be incorporated here. We have attempted to be as inclusive as possible, but we sincerely apologize for any nomenclature differences that we may have overwritten, as well as any citations that we may have overlooked.
The authors thank BGI for Illumina sequencing, the Johns Hopkins Deep Sequencing and Microarray Core Facility for Iso-Seq SMRT sequencing, DNAform for deepCAGE, and the Tulane Cancer Center Next Generation Sequence Analysis core for services and utilization of core infrastructure. We thank Dr. Rolf Renne and the Renne lab for helpful discussions, and Dr. Katrina Grau for the collation of gene class data. S.A.T. was supported by NIH R01AI108407 and P01CA214091. E.K.F. was supported by NIH P01CA214091 and R01AI106676 and DOD W81XWH-16-1-0318. L.F.v.D. and E.M.M. were supported by NIH R01AI121300. T.O.G. was supported by the Belgian National Fund for Scientific Research (FNRS) and Action de Recherches Concertée (ARC) from Uliège.
Footnotes
SUPPLEMENTAL INFORMATION
Supplemental Information can be found online at https://doi.org/10.1016/j.celrep.2019.05.086.
DECLARATION OF INTERESTS
The authors declare no competing interests.
REFERENCES
- Adler H, Messerle M, Wagner M, and Koszinowski UH (2000). Cloning and mutagenesis of the murine gammaherpesvirus 68 genome as an infectious bacterial artificial chromosome. J. Virol 74, 6964–6974. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahn JW, Powell KL, Kellam P, and Alber DG (2002). Gammaherpesvirus lytic gene expression as characterized by DNA array. J. Virol 76, 6244–6256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Allen RD 3rd, Dickerson S, and Speck SH (2006). Identification of spliced gammaherpesvirus 68 LANA and v-cyclin transcripts and analysis of their expression in vivo during latent infection. J. Virol 80, 2055–2062. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arias C, Weisburd B, Stern-Ginossar N, Mercier A, Madrid AS, Bellare P, Holdorf M, Weissman JS, and Ganem D (2014). KSHV 2.0: a comprehensive annotation of the Kaposi’s sarcoma-associated herpesvirus genome using next-generation sequencing reveals novel genomic and functional features. PLoS Pathog. 10, e1003847. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Barton E, Mandal P, and Speck SH (2011). Pathogenesis and host control of gammaherpesviruses: lessons from the mouse. Annu. Rev. Immunol 20, 351–397. [DOI] [PubMed] [Google Scholar]
- Benach J, Wang L, Chen Y, Ho CK, Lee S, Seetharaman J, Xiao R, Acton TB, Montelione GT, Deng H, et al. (2007). Structural and functional studies of the abundant tegument protein ORF52 from murine gammaherpesvirus 68. J. Biol. Chem 282, 31534–31541. [DOI] [PubMed] [Google Scholar]
- Bencun M, Klinke O, Hotz-Wagenblatt A, Klaus S, Tsai M-H, Poirey R, and Delecluse H-J (2018). Translational profiling of B cells infected with the Epstein-Barr virus reveals 5′ leader ribosome recruitment through upstream open reading frames. Nucleic Acids Res. 46, 2802–2819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bodescot M, and Perricaudet M (1986). Epstein-Barr virus mRNAs produced by alternative splicing. Nucleic Acids Res. 14, 7103–7114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bortz E, Wang L, Jia Q, Wu T-T, Whitelegge JP, Deng H, Zhou ZH, and Sun R (2007). Murine gammaherpesvirus 68 ORF52 encodes a tegument protein required for virion morphogenesis in the cytoplasm. J. Virol 81,10137–10150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Canny SP, Reese TA, Johnson LS, Zhang X, Kambal A, Duan E, Liu CY, and Virgin HW (2014). Pervasive transcription of a herpesvirus genome generates functionally important RNAs. MBio 5, e01033–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cheng BYH, Zhi J, Santana A, Khan S, Salinas E, Forrest JC, Zheng Y, Jaggi S, Leatherwood J, and Krug LT (2012). Tiled microarray identification of novel viral transcript structures and distinct transcriptional profiles during two modes of productive murine gammaherpesvirus 68 infection. J. Virol 86, 4340–4357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cohen JI (1999). Genomic structure and organization of varicella-zoster virus. Contrib. Microbiol 3, 10–20. [DOI] [PubMed] [Google Scholar]
- Coleman HM, Brierley I, and Stevenson PG (2003). An internal ribosome entry site directs translation of the murine gammaherpesvirus 68 MK3 open reading frame. J. Virol 77, 13093–13105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Coleman HM, Efstathiou S, and Stevenson PG (2005). Transcription of the murine gammaherpesvirus 68 ORF73 from promoters in the viral terminal repeats. J. Gen. Virol 86, 561–574. [DOI] [PubMed] [Google Scholar]
- Damania B, Jeong JH, Bowser BS, DeWire SM, Staudt MR, and Dittmer DP (2004). Comparison of the Rta/Orf50 transactivator proteins of gamma-2-herpesviruses. J. Virol 78, 5491–5499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- DeZalia M, and Speck SH (2008). Identification of closely spaced but distinct transcription initiation sites for the murine gammaherpesvirus 68 latency-associated M2 gene. J. Virol 82, 7411–7421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dobin A, Davis CA, Schlesinger F, Drenkow J, Zaleski C, Jha S, Batut P, Chaisson M, and Gingeras TR (2013). STAR: ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ebrahimi B, Dutia BM, Roberts KL, Garcia-Ramirez JJ, Dickinson P, Stewart JP, Ghazal P, Roy DJ, and Nash AA (2003). Transcriptome profile of murine gammaherpesvirus-68 lytic infection. J. Gen. Virol 84, 99–109. [DOI] [PubMed] [Google Scholar]
- El-Gebali S, Mistry J, Bateman A, Eddy SR, Luciani A, Potter SC, Qureshi M, Richardson LJ, Salazar GA, Smart A, et al. (2019). The Pfam protein families database in 2019. Nucleic Acids Res. 47 (D1), D427–D432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Forrest JC, and Speck SH (2008). Establishment of B-cell lines latently infected with reactivation-competent murine gammaherpesvirus 68 provides evidence for viral alteration of a DNA damage-signaling cascade. J. Virol 82, 7688–7699. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frith MC, Valen E, Krogh A, Hayashizaki Y, Carninci P, and Sandelin A (2008). A code for transcription initiation in mammalian genomes. Genome Res. 18, 1–12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gray KS, Allen RD 3rd, Farrell ML, Forrest JC, and Speck SH (2009). Alternatively initiated gene 50/RTA transcripts expressed during murine and human gammaherpesvirus reactivation from latency. J. Virol 83, 314–328. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hug N, Longman D, and Cáceres JF (2016). Mechanism and regulation of the nonsense-mediated decay pathway. Nucleic Acids Res. 44, 1483–1495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson LS, Willert EK, and Virgin HW (2010). Redefining the genetics of murine gammaherpesvirus 68 via transcriptome-based annotation. Cell Host Microbe 7, 516–526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu S, Pavlova IV, Virgin HW 4th, and Speck SH (2000). Characterization of gammaherpesvirus 68 gene 50 transcription. J. Virol 74, 2029–2037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Loftus MS, Verville N, and Kedes DH (2017). A Conserved Leucine Zipper Motif in Gammaherpesvirus ORF52 Is Critical for Distinct Microtubule Rearrangements. J. Virol 91, e00304–17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mackett M, Stewart JP, de V Pepper S, Chee M, Efstathiou S, Nash AA, and Arrand JR (1997). Genetic content and preliminary transcriptional analysis of a representative region of murine gammaherpesvirus 68. J. Gen. Virol 78, 1425–1433. [DOI] [PubMed] [Google Scholar]
- Majerciak V, Ni T, Yang W, Meng B, Zhu J, and Zheng Z-M (2013). A viral genome landscape of RNA polyadenylation from KSHV latent to lytic infection. PLoS Pathog. 9, e1003749. [DOI] [PMC free article] [PubMed] [Google Scholar]
- May JS, Coleman HM, Boname JM, and Stevenson PG (2005). Murine gammaherpesvirus-68 ORF28 encodes a non-essential virion glycoprotein. J. Gen. Virol 86,919–928. [DOI] [PubMed] [Google Scholar]
- McClelland S, Shrivastava R, and Medh JD (2009). Regulation of Translational Efficiency by Disparate-UTRs of PPAR Splice Variants. PPAR Res. 2009, 193413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- McClure LV, Lin Y-T, and Sullivan CS (2011). Detection of viral microRNAs by Northern blot analysis. Methods Mol. Biol 721, 153–171. [DOI] [PubMed] [Google Scholar]
- McLeay RC, and Bailey TL (2010). Motif Enrichment Analysis: a unified framework and an evaluation on ChIP data. BMC Bioinformatics 11, 165. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merrick WC (2004). Cap-dependent and cap-independent translation in eukaryotic systems. Gene 332, 1–11. [DOI] [PubMed] [Google Scholar]
- Murata M, Nishiyori-Sueki H, Kojima-Ishiyama M, Carninci P, Hayashizaki Y, and Itoh M (2014). Detecting expressed genes using CAGE. Methods Mol. Biol 1164, 67–85. [DOI] [PubMed] [Google Scholar]
- Nash AA, Dutia BM, Stewart JP, and Davison AJ (2001). Natural history of murine gammaherpesvirus infection. Philos. Trans. R. Soc. Lond. B Biol. Sci 356, 569–579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nealy MS, Coleman CB, Li H, and Tibbetts SA (2010). Use of a virus-encoded enzymatic marker reveals that a stable fraction of memory B cells expresses latency-associated nuclear antigen throughout chronic gammaherpesvirus infection. J. Virol 84, 7523–7534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nelson BR, Makarewich CA, Anderson DM, Winders BR, Troupes CD, Wu F, Reese AL, McAnally JR, Chen X, Kavalali ET, et al. (2016). A peptide encoded by a transcript annotated as long noncoding RNA enhances SERCA activity in muscle. Science 351, 271–275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady T, Cao S, Strong MJ, Concha M, Wang X, Splinter Bondurant S, Adams M, Baddoo M, Srivastav SK, Lin Z, et al. (2014). Global bidirectional transcription of the Epstein-Barr virus genome during reactivation. J. Virol 88, 1604–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- O’Grady T, Wang X, Höner Zu Bentrup K, Baddoo M, Concha M, and Flemington EK (2016). Global transcript structure resolution of high gene density genomes through multi-platform data integration. Nucleic Acids Res. 44, e145. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelechano V, and Steinmetz LM (2013). Gene regulation by antisense transcription. Nat. Rev. Genet 14, 880–893. [DOI] [PubMed] [Google Scholar]
- Ray D, Kazan H, Cook KB, Weirauch MT, Najafabadi HS, Li X, Gueroussov S, Albu M, Zheng H, Yang A, et al. (2013). A compendium of RNA-binding motifs for decoding gene regulation. Nature 499, 172–177. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sedman SA, Gelembiuk GW, and Mertz JE (1990). Translation initiation at a downstream AUG occurs with increased efficiency when the upstream AUG is located very close to the 5′ cap. J. Virol 64, 453–457. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh S, Bevan SC, Patil K, Newton DC, and Marsden PA (2005). Extensive variation in the 5′-UTR of Dicer mRNAs influences translational efficiency. Biochem. Biophys. Res. Commun 335, 643–650. [DOI] [PubMed] [Google Scholar]
- Speck SH, and Strominger JL (1985). Analysis of the transcript encoding the latent Epstein-Barr virus nuclear antigen I: a potentially polycistronic message generated by long-range splicing of several exons. Proc. Natl. Acad. Sci. USA 82, 8305–8309. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stern-Ginossar N, Weisburd B, Michalski A, Le VTK, Hein MY, Huang S-X, Ma M, Shen B, Qian S-B, Hengel H, et al. (2012). Decoding human cytomegalovirus. Science 338, 1088–1093. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D, Csabai Z, Oláh P, Balázs Z, Likó I, Zsigmond L, Sharon D, Snyder M, and Boldogkói Z (2016). Full-Length Isoform Sequencing Reveals Novel Transcripts and Substantial Transcriptional Overlaps in a Herpesvirus. PLoS One 11,e0162868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tombácz D, Csabai Z, Szúcs A, Balázs Z, Moldován N, Sharon D, Snyder M, and Boldogkói Z (2017). Long-Read Isoform Sequencing Reveals a Hidden Complexity of the Transcriptional Landscape of Herpes Simplex Virus Type 1. Front. Microbiol 8, 1079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tsai K, Messick TE, and Lieberman PM (2015). Disruption of host antiviral resistances by gammaherpesvirus tegument proteins with homology to the FGARAT purine biosynthesis enzyme. Curr. Opin. Virol 14, 30–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Van Skike ND, Minkah NK, Hogan CH, Wu G, Benziger PT, Oldenburg DG, Kara M, Kim-Holzapfel DM, White DW, Tibbetts SA, et al. (2018). Viral FGARAT ORF75A promotes early events in lytic infection and gammaherpesvirus pathogenesis in mice. PLoS Pathog. 14, e1006843. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Virgin HW 4th, Latreille P, Wamsley P, Hallsworth K, Weck KE, Dal Canto AJ, and Speck SH (1997). Complete sequence and genomic analysis of murine gammaherpesvirus 68. J. Virol 71, 5894–5904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wakeman BS, Johnson LS, Paden CR, Gray KS, Virgin HW, and Speck SH (2014). Identification of alternative transcripts encoding the essential murine gammaherpesvirus lytic transactivator RTA. J. Virol. 88, 5474–5490. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Guo H, Reyes N, Lee S, Bortz E, Guo F, Sun R, Tong L, and Deng H (2012). Distinct domains in ORF52 tegument protein mediate essential functions in murine gammaherpesvirus 68 virion tegumentation and secondary envelopment. J. Virol 86, 1348–1357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang L, Park HJ, Dasari S, Wang S, Kocher J-P, and Li W (2013). CPAT: Coding-Potential Assessment Tool using an alignment-free logistic regression model. Nucleic Acids Res. 41, e74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weil D, Boutain S, Audibert A, and Dautry F (2000). Mature mRNAs accumulated in the nucleus are neither the molecules in transit to the cytoplasm nor constitute a stockpile for gene expression. RNA 6, 962–975. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu TD, and Watanabe CK (2005). GMAP: a genomic mapping and alignment program for mRNA and EST sequences. Bioinformatics 21, 1859–1875. [DOI] [PubMed] [Google Scholar]
- Wu T-T, Usherwood EJ, Stewart JP, Nash AA, and Sun R (2000). Rta of murine gammaherpesvirus 68 reactivates the complete lytic cycle from latency. J. Virol 74, 3659–3667. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu T-T, Park T, Kim H,Tran T,Tong L, Martinez-Guzman D, Reyes N, Deng H, and Sun R (2009). ORF30 and ORF34 are essential for expression of late genes in murine gammaherpesvirus 68. J. Virol 83, 2265–2273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu JJ, Li W, Shao Y, Avey D, Fu B, Gillen J, Hand T, Ma S, Liu X, Miley W, et al. (2015). Inhibition of cGAS DNA Sensing by a Herpesvirus Virion Protein. Cell Host Microbe 18, 333–344. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The updated version (v17March2018) of the TRIMD software and detailed instructions for use are available at https://github.com/flemingtonlab/TRIMD. The accession number for the Iso-Seq, deepCAGE and RNA-Seq data reported in this paper is GEO: GSE117651. The accession number for the MHV68.bla genome and annotation reported in this paper is GenBank: MH636806.