Significance
Mitochondrial respiration is an ancient characteristic of eukaryotes. However, it was lost independently in multiple eukaryotic lineages as part of adaptations to an anaerobic lifestyle. We show that a similar adaptation occurred in a member of the Myxozoa, a large group of microscopic parasitic animals that are closely related to jellyfish and hydroids. Using deep sequencing approaches supported by microscopic observations, we present evidence that an animal has lost its mitochondrial genome. The myxozoan cells retain structures deemed mitochondrion-related organelles, but have lost genes related to aerobic respiration and mitochondrial genome replication. Our discovery shows that aerobic respiration, one of the most important metabolic pathways, is not ubiquitous among animals.
Keywords: Cnidaria, mitochondrial evolution, mitochondria-related organelle, MRO, cristae
Abstract
Although aerobic respiration is a hallmark of eukaryotes, a few unicellular lineages, growing in hypoxic environments, have secondarily lost this ability. In the absence of oxygen, the mitochondria of these organisms have lost all or parts of their genomes and evolved into mitochondria-related organelles (MROs). There has been debate regarding the presence of MROs in animals. Using deep sequencing approaches, we discovered that a member of the Cnidaria, the myxozoan Henneguya salminicola, has no mitochondrial genome, and thus has lost the ability to perform aerobic cellular respiration. This indicates that these core eukaryotic features are not ubiquitous among animals. Our analyses suggest that H. salminicola lost not only its mitochondrial genome but also nearly all nuclear genes involved in transcription and replication of the mitochondrial genome. In contrast, we identified many genes that encode proteins involved in other mitochondrial pathways and determined that genes involved in aerobic respiration or mitochondrial DNA replication were either absent or present only as pseudogenes. As a control, we used the same sequencing and annotation methods to show that a closely related myxozoan, Myxobolus squamalis, has a mitochondrial genome. The molecular results are supported by fluorescence micrographs, which show the presence of mitochondrial DNA in M. squamalis, but not in H. salminicola. Our discovery confirms that adaptation to an anaerobic environment is not unique to single-celled eukaryotes, but has also evolved in a multicellular, parasitic animal. Hence, H. salminicola provides an opportunity for understanding the evolutionary transition from an aerobic to an exclusive anaerobic metabolism.
The acquisition of the mitochondrion was a fundamental event in the evolution of eukaryotes, and most extant eukaryotes cannot survive without oxygen. Interestingly, the loss of aerobic respiration has occurred independently in several eukaryotic lineages that adapted to low-oxygen environments and replaced the standard mitochondrial (mt) oxidative phosphorylation pathway with novel anaerobic metabolic mechanisms (Fig. 1) (1, 2). Such anaerobic metabolism occurs within mitochondria-related organelles (MROs), which have often lost their cristae, and include hydrogenosomes and mitosomes (1, 2). There is debate regarding the existence of exclusively anaerobic animals and accompanying MROs (3). Although it was reported that some loriciferans found in anoxic conditions possess hydrogenosomes (4, 5), genomic data are not yet available for these organisms, and alternative explanations have been proposed (3). Here, we show that a myxozoan parasite (Cnidaria) has lost both its mt genome and aerobic metabolic pathways, and has a novel type of anaerobic MRO. Myxozoans are a large group of enigmatic, parasitic, cnidarians with complex life cycles that require two hosts, usually a fish and an annelid (6). They have a substantial negative economic impact on fisheries and aquaculture (7). Myxozoan mitochondria have highly divergent genome structures, with large multipartite circular mt chromosomes and unusually high evolutionary rates (8, 9). To gain further insight into the evolution of the myxozoan mt genome, we studied two closely related freshwater species, Henneguya salminicola and Myxobolus squamalis (SI Appendix, Fig. S1), both of which are parasites of salmonid fish (10–12).
Fig. 1.
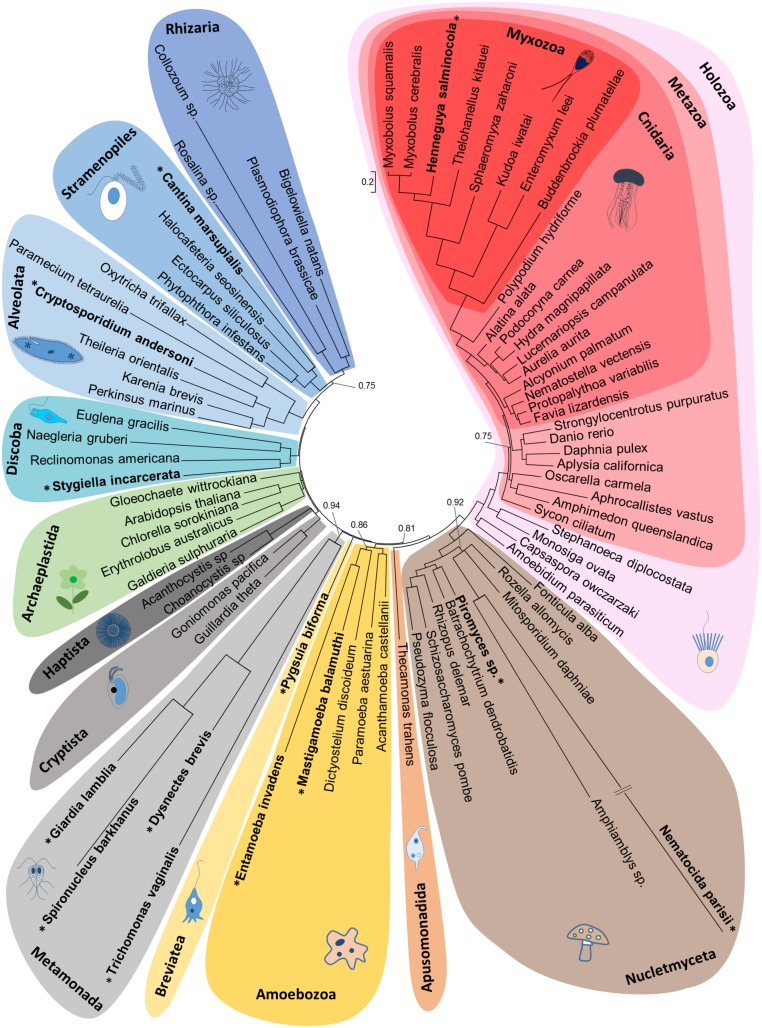
Eukaryote phylogenetic relationships inferred from a supermatrix of 9490 amino acid positions for 78 species. Bayesian majority-rule consensus tree reconstructed using the CAT + Γ model from two independent Markov-chain Monte Carlo chains. Branches with low node support (posterior probabilities PP < 0.7) were collapsed. Most nodes were highly supported (PP > 0.98), and PP are only indicated for nodes with PP < 0.98. The eukaryote classification is based on Adl et al. (47). Species known to have lost their mt genome are indicated in bold with an asterisk. Myxozoan species form a well-supported group (PP = 1.0) and our reconstructions agree with previous studies (14), which show monophyly of the fresh-water/oligochaete host lineage (10).
Results
We assembled transcriptomes and genomes from both species using identical protocols and computational pipelines. Our phylogenetic analyses based on 78 nuclear ribosomal protein-encoding genes from taxa representative of eukaryotic diversity confirmed that the organisms we sequenced are closely related myxozoans, and not contaminants (Fig. 1 and SI Appendix, Fig. S2). The genome assembly statistics revealed that H. salminicola has a more complete assembly with higher coverage and more predicted protein sequences than M. squamalis (Table 1 and SI Appendix, Figs. S3 and S4). Targeted searches in the genomes identified 75/78 nuclear ribosomal protein genes, which suggested that the completeness is >90% for both species. However, estimates of genome completeness using the Core Eukaryotic Genes Mapping Approach (CEGMA) (13) recovered only 53.6% of core eukaryotic genes for H. salminicola and 37.5% for M. squamalis. We hypothesize that the fast evolutionary rates of myxozoans (14) reduced our ability to detect many common eukaryotic genes, a challenge also known with other fast-evolving eukaryotic lineages (15). This view is supported by calculations using only the most conserved CEGMA genes, which have higher recovery in both H. salminicola and M. squamalis (76.9% and 56.9%, respectively).
Table 1.
Assembly statistics, presence of mt genome, and number of nuclear-encoded mt genes identified for myxozoan genomes (gen.) and transcriptomes (trans.)
| H. salminicola | M. squamalis | K. iwatai (14) | T. kitauei (18) | M. cerebralis (14) | |
| Gen./Trans. | Gen./Trans. | Gen./Trans. | Gen. | Trans. | |
| Genome size, Mb | 60.0/— | 53.1/— | 22.5/— | 188.5 | — |
| Coverage | 311/— | 86.1/— | 1,000/— | 37 | — |
| DNA assembly size, Mb | 61.4/— | 43.7/— | 23.7/— | 150.7 | — |
| No. of contigs | 18,330/31,825 | 37,919/11,236 | 22,174/6,528 | 5,610 | 52,821 |
| N50 | 7,570/600 | 1,286/714 | 40,195/1,662 | — | 11,965 |
| CEGs (complete) | 53.6%/26.6% | 37.5%/23.8% | 73.0%/76.6% | 46.8% | 39.1% |
| CEGs (complete group 4) | 76.9%/33.9% | 56.9%/27.7% | 96.9%/95.4% | 66.2% | 55.4% |
| CEGs (partial group 4) | 87.7%/75.4% | 76.9%/70.8% | 96.9%/96.9% | 73.9% | 84.6% |
| %GC | 29/— | 27/— | 23.6/— | 37.5 | — |
| No. of predicted proteins | 8,188/— | 5,725/— | 5,533/— | 16,638 | — |
| Presence/absence of mt genome | Absent | Present | Present (8, 9) | Present (8) | Unknown |
| No. of nuclear genes involved in mtDNA replication and translation | 6 | 58 | 52 | 41 | 49 |
| No. of nuclear genes involved in electron-transport chains | 7 | 21 | 25 | 18 | 21 |
| No. of genes involved in pyruvate metabolism | 0* | 3* | 3* | 3* | 3* |
| No. of genes involved in other mt pathways | 52 | 55 | 64 | 45 | 46 |
There are two additional proteins, involved in pyruvate metabolism and present in all Myxozoa, that appear also in other metabolic pathways.
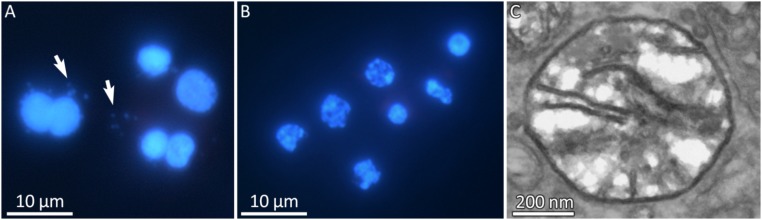
Assembly of the mt genomes revealed striking differences between the two parasites. For M. squamalis, we successfully recovered a circular mt genome composed of a single chromosome, which phylogenetic analyses confirmed was myxozoan (SI Appendix, Supplementary Results and Figs. S5 and S6). Similar to other myxozoans (8), the M. squamalis mt genome lacked tRNAs, and has a fast evolutionary rate (SI Appendix, Supplementary Results and Figs. S5 and S6). In stark contrast, we could not identify any mt sequence among the contigs of H. salminicola, despite the higher quality of that assembly compared with that of M. squamalis. To identify whether DNA was present in the myxozoan mitochondria, we stained living multicellular developing stages of M. squamalis and H. salminicola with DAPI (Fig. 2). Cells of M. squamalis showed the characteristic eukaryotic staining of both nuclei and mitochondria (as much smaller blue dots; Fig. 2A), whereas H. salminicola showed only nuclear staining (Fig. 2B). The microscopy results, together with the lack of mt contigs in the genome and transcriptome assemblies, supported our central hypothesis that this animal has lost its mt genome. Electron microscopy images, however, showed mt-like double membrane organelles with cristae in H. salminicola (Fig. 2C and SI Appendix, Fig. S7) and M. squamalis (SI Appendix, Fig. S8). Accordingly, genes involved in cristae organization were also detected in the genome of both species, in particular DNAJC11 and MTX1, which have been linked to the presence of cristae (16, 17) (Dataset S1). Together, these results confirm that an MRO without an mt genome, but with cristae, is present in this species.
Fig. 2.
Microscopic evidence for the absence of mitochondria in H. salminicola. (A and B) DAPI staining of normal 7-cell presporogonic developmental stages of two myxozoan parasites of salmonid fish. (A) M. squamalis, showing large nuclei with many smaller mitochondrial nucleosomes (arrowed). (B) H. salminicola, showing large nuclei but surprisingly no mitochondrial nucleosomes. (C) TEM image of H. salminicola mitochondrion-related organelle with few cristae. Uncropped images are available in the Figshare repository.
In animals, most of the mt proteome is encoded in the nucleus. Accordingly, we identified 51 and 57 genes involved in key mt metabolic pathways (e.g., amino acid, carbohydrate, or nucleotide metabolism) in H. salminicola and M. squamalis, respectively (Fig. 3, Table 1, and Dataset S2). This suggests that the MROs of H. salminicola still perform diverse metabolic functions, similar to the mitochondria of M. squamalis. In contrast, almost all nuclear-encoded proteins involved in mt genome replication and translation were absent from the H. salminicola genome. Using a database of 118 such nuclear-encoded genes in Drosophila, we identified 41 to 58 homologous mt genes in M. squamalis and among published myxozoan data (14, 18), but only six of these genes in H. salminicola (Table 1 and Dataset S3). In addition, we calculated that H. salminicola does not have a faster evolutionary rate than other myxozoans, which might otherwise have precluded gene discovery (Fig. 1 and SI Appendix, Fig. S2).
Fig. 3.
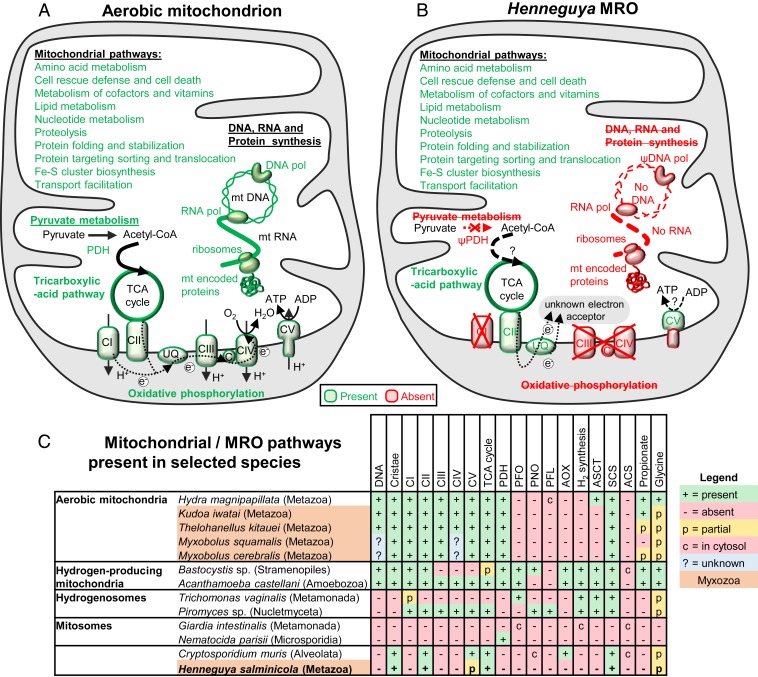
Comparison between the pathways present in (A) a typical aerobic mitochondrion and (B) the H. salminicola MRO. (C) Mitochondrial/MRO pathways present in selected species (see refs. 1 and 2). The presence and absence of organellar genomes are indicated. ACS, acetyl-CoA synthetase; acetate AOX, alternative oxidase; ASCT, acetate succinyl-CoA transferase; DNA pol, mtDNA polymerase; RNA pol, mtDNA-dependent RNA polymerase; CI-CV, respiratory complexes I-V; C, cytochrome c; PDH, pyruvate dehydrogenase; PFL, pyruvate formate lyase; PFO, pyruvate ferredoxin oxidoreductase; PNO, pyruvate NADPH oxidoreductase; SCS, succinyl-CoA synthetase; TCA cycle, tricarboxylic acid cycle; UQ, ubiquinone; e−, electrons; H+, protons; ψ indicates the presence of a pseudogene in the nuclear genome.
Interestingly, in H. salminicola, we found that the mt DNA polymerase subunit gamma-1 (19) gene is a pseudogene, as it contains three point mutations that create premature stop codons (SI Appendix, Fig. S9). Furthermore, this gene is not expressed in H. salminicola, and was absent from the H. salminicola transcriptome assembly, whereas we identified homologous contigs in all other myxozoan transcriptomes (Dataset S3). The presence of a pseudogene copy of this polymerase has several implications. First, it supports our central conclusion that H. salminicola has lost its mtDNA, as it has no mtDNA replication machinery. Second, it shows that the absence of protein homologs in this species is the result of pseudogenization, and not an assembly artifact.
The loss of the mt genome should impact aerobic respiration, since animal mt genomes code for essential proteins of the electron-transport chain (20). To verify whether the loss of the mt genome meant loss of aerobic respiration in H. salminicola, we searched for homologs of known Drosophila nuclear genes that typically encode ∼100 proteins from the mt electron-transport chain complexes (Fig. 3 and SI Appendix, Supplementary Methods). Our searches of all myxozoan genomes available revealed that nuclear genes for only seven of these mt proteins remain in H. salminicola, whereas 18 to 25 are present in other myxozoans (Fig. 3, Table 1, and Dataset S2). Specifically, all complex I, III, and IV genes that we identified in other myxozoans are absent in H. salminicola (Fig. 3B, Dataset S2, and SI Appendix, Supplementary Results and Fig. S10) or present as pseudogenes (SI Appendix, Fig. S9). Since complex IV interacts with O2 molecules, we conclude that H. salminicola might not be capable of standard cellular aerobic respiration. In concurrence with the absence of the complexes that pump protons into the mitochondrial intermembrane space (i.e., complexes I, III, and IV), most genes that encode the Fo subunit of the adenosine triphosphate (ATP) synthase complex (i.e., the proton channel of complex V) are also missing in H. salminicola (Dataset S4), while being present in Myxobolus (Dataset S4). This suggests that a proton gradient is absent across the inner organelle membrane in H. salminicola. In contrast, for complex II, which is part of the Krebs cycle, and for the F1 subunit of the ATP synthase, H. salminicola encodes a similar number of protein coding genes as other myxozoans (Dataset S4).
Discussion
Structurally, H. salminicola has lost its mt genome, but has retained an organelle that resembles a mitochondrion. However, as mitochondria are defined based on the use of oxygen as electron acceptor (21), and usually the presence of an mt genome (but see ref. 22), we conclude that H. salminicola possesses MROs rather than true mitochondria. Although MROs have evolved several times independently, some of them present striking similarities (1, 2). Not only have MROs often lost the same mt pathways (e.g., pyruvate dehydrogenase or electron transport chain enzymes) but also, in several cases, homologous enzymes, such as hydrogenases or pyruvate formate lyases, have been acquired independently by horizontal gene transfer. These enzymes allow ATP production by anaerobic pyruvate metabolism and H2 synthesis. MROs with such abilities are called hydrogen-producing mitochondria or hydrogenosomes, the latter having lost their ability to utilize oxygen (Fig. 3C) (1, 2).
As our H. salminicola assemblies did not contain any hydrogenase or other genes of prokaryotic origin (Fig. 3C and Dataset S5), we conclude that the MROs in H. salminicola are not hydrogenosomes. The presence of cristae in H. salminicola’s MRO is surprising since these membrane invaginations are usually absent in anaerobic MROs (1, 16, 17). However, we note that the MROs of H. salminicola share these characteristics with the MRO of the apicomplexan Cryptosporidium muris, which has also lost complexes I, III, and IV, but possesses an alternative oxidase and retains cristae (Fig. 3C) (23). The presence of cristae together with the identification of pseudogenes suggest that the loss of mtDNA and aerobic respiration may be a recent evolutionary event in the Henneguya lineage. Future experiments are needed to better characterize the metabolic energy pathways of H. salminicola. However, such experiments are challenging because it is currently not possible to culture H. salminicola in the laboratory.
Similar to most Myxozoa, H. salminicola likely alternates between two hosts (6). In its fish host, it undergoes proliferation and sporogenesis in pseudocysts within the white muscle (11), a tissue known to have anaerobic metabolism (24). While the obligate invertebrate host of H. salminicola is unknown, it is probably an annelid from the family Naididae, based on known life cycles of related myxozoans (25). Members of the Naididae can grow and reproduce in anoxic environments (26). As all protists that have lost their mt genomes live in anaerobic environments, we speculate that the loss of the mt genome in H. salminicola was driven by low-oxygen environments in both of its hosts.
Loss of superfluous genes likely conveys an evolutionary advantage, as it has been shown that the bioenergetic cost of a gene is higher in small genomes (27). Myxozoans have smaller genomes [22 to 180 Mb (14, 18)] than free-living Cnidaria [>250 Mb (28, 29)]. Therefore, the loss of the mt genome and associated nuclear genes involved in its replication and electron pathways may be advantageous for a myxozoan living in anaerobic environments. However, the loss of useless genes by random drift cannot be excluded. Interestingly, our results also open the way to new treatment options against this pathogen, since anaerobic protists are known to be sensitive to specific drugs (30).
Myxozoans have gone through outstanding morphological and genomic simplifications during their adaptation to parasitism from a free-living cnidarian ancestor (31). It is remarkable that these myxozoan simplifications do not appear to be ancestral, but rather the result of secondary losses (14). Here we show that at least one myxozoan species has lost a core animal feature: the genetic basis for aerobic respiration in its mitochondria. As a highly diverse group with >2,400 species, which inhabit marine, freshwater, and even terrestrial environments (32), evolutionary loss and simplification has clearly been a successful strategy for Myxozoa, which shows that less is more (33).
Materials and Methods
Samples and Sequencing.
Samples of H. salminicola and M. squamalis were identified on the basis of spore morphology (SI Appendix, Fig. S1), tissue tropism, host, and SSU rDNA sequence similarity with published sequence available at the National Center for Biotechnology Information (SI Appendix, Supplementary Methods).
DNA and RNA of H. salminicola were each extracted from a single large cyst (4 to 8 mm diameter) sampled from Chinook salmon (Oncorhynchus tschawytscha). For M. squamalis, which develops in smaller cysts (2 to 4 mm diameter), DNA and RNA were extracted from several cysts collected from a coho salmon (Oncorhynchus kisutch) and rainbow trout (Oncorhynchus mykiss), respectively. The multiisolate extract from M. squamalis may explain the differences in assembly quality between H. salminicola and M. squamalis, since polymorphism is known to complicate assembly.
DNA and RNA were extracted with the DNeasy Blood & Tissue Kit (Qiagen) and the High Pure RNA extraction kit (Roche), respectively, following manufacturers’ instructions. The samples were sent for library construction and sequencing at the Center for Genome Research and Biocomputing at Oregon State University (Corvallis, OR). Paired-end sequencing with 150-bp reads derived from fragments of average length ∼350 bp was performed on a HiSeq3000 platform.
Light Microscopy.
Myxozoan cells were prefixed with 3:1 methanol:acetic acid, and three drops of cell suspension were put on slides. DNA staining was performed with VECTASHIELD (Vectorlabs) antifade mounting medium, which contained DAPI (SI Appendix, Supplementary Methods). Cells were visualized under a Leica DMR compound fluorescence microscope at 630× and 1,000× magnification.
Electron Microscopy.
Fresh parasite pseudocysts were dissected from the tissue of a single host and then fixed in a solution comprising 1% glutaraldehyde and 2% paraformaldehyde in 0.1 M phosphate buffer. Larger pseudocysts were sliced to permit penetration of the fixative. Fixed tissue was then stained with osmium tetroxide and then uranyl acetate, before being dehydrated in a graded alcohol series and embedded in Epon resin. Ultrathin sections were mounted on copper grids and examined using a Helios 650 FEG dual-beam SEM (Thermo Scientific) in transmission mode, at the Oregon State University Electron Microscope Facility.
Filtering and Assembling the Genomic and Transcriptomic Data.
Stringent filtering, involving multiple steps, was performed to eliminate host and bacterial contamination from M. squamalis and H. salminicola data. This involved mapping reads to the corresponding host genomes and several rounds of BLAST searches against the National Center for Biotechnology Information nucleotide database (SI Appendix, Supplementary Methods). Reads from contaminant sequences were eliminated and the remaining DNA and RNA reads assembled using IDBA (version 1.1.1) (34) and Trinity (35), respectively (SI Appendix, Supplementary Methods).
Assembly of the M. squamalis mt Genome and Absence of mt Sequences in H. salminicola.
Local blastn and tblastn searches using cnidarian (including published myxozoan) mt genome and protein sequences, respectively, were performed against our myxozoan assemblies of M. squamalis and H. salminicola. After manual inspection of all sequences with E-values <1e−1, no mitochondrial sequence could be identified for H. salminicola. In contrast, a putative mitochondrial contig was identified for M. squamalis. The coverage of the mt genome was 185, about twice the nuclear coverage. To further search the H. salminicola data, Hidden Markov Model (HMM) profiles were built based on alignments of myxozoan mt proteins, using HMMer3.0 (36). These profiles were used to search protein predictions of H. salminicola by Maker2 v2.31.10 (37) (see SI Appendix, Supplementary Methods, for details regarding Maker2 annotation), but no mt proteins were identified. Similarly, HMM profiles were built based on alignments of myxozoan RNA sequences using Infernal 1.1.1 (38), following the approach of Yahalomi et al. (8), and the profiles used to search genomic and transcriptomic assemblies. Again, no mt sequences were identified.
The Perl script Novoplasty v2.6.3 (39) was used to reconstruct a first draft of the mitochondrial sequence of M. squamalis based on the mt contig identified in the BLAST search. The draft sequence was then corrected using read mapping (see SI Appendix, Supplementary Methods, for details regarding the assembly and annotation of the mt genome of M. squamalis).
Estimating Completeness of Genomic and Transcriptomic Assemblies.
CEGMA (13) was used to estimate the completeness of our assemblies. Because myxozoans show extreme evolutionary rates (14), the completeness was estimated based on the most conserved set of CEGMA genes (Group 4). We also used the program BUSCO V3 (40), but found that it performed poorly on Myxozoa, which was in concordance with other studies that show that BUSCO underestimates completeness of fast-evolving organisms (15).
Genome Size Estimation.
Genome sizes were calculated based on K-mer frequency estimation using the GenomeScope web server (41) (last accessed 2018/02). For both species, k-mer frequency histograms were generated for k = 17, using the program Jellyfish 2.2.7 (42) on the filtered reads with the following parameters: count -C -m 17 -s 1000000000 -t 10 (SI Appendix, Figs. S3 and S4).
Characterization of Myxozoan Proteins Interacting with mtDNA and mtRNA.
To create an exhaustive database of nuclear-encoded proteins that interact with mt DNA and RNA, we downloaded three protein datasets: all mt ribosomal protein sequences from FlyBase (last accessed November 2017) (43), all Drosophila melanogaster proteins with either the functional classifications “DNA and RNA” or “DNA and RNA/Protein synthesis/Others” from the MitoDrome database (last accessed January 2018) (44), and sequences of human proteins known to bind to mt RNA and described in Rackham et al. (45) from the National Center for Biotechnology Information. We then performed reciprocal blastp searches against the Drosophila proteome to identify the corresponding homologs (SI Appendix, Supplementary Methods). These sets of Drosophila proteins were used to perform reciprocal BLAST searches against the proteome of the cnidarian Hydra vulgaris. The Hydra and Drosophila sequences were then used to identify homologous sequences in the myxozoan genome and transcriptome assemblies, after they had been filtered from contaminants. Detailed information about homolog identification is provided in SI Appendix, Supplementary Methods.
Characterization of Myxozoan Mitochondrial Metabolic Pathways.
Drosophila proteins involved in the different mitochondrial metabolic pathways were downloaded from the MitoDrome database (44). Reciprocal BLAST searches were conducted using the Drosophila sequences as queries to identify homologous copies in Hydra. The Drosophila and Hydra sequences were then used as queries to identify nuclear-encoded mitochondrial proteins in the myxozoans (SI Appendix, Supplementary Methods). It is worth noting that all Kudoa proteins previously identified by Muthye and Lavrov (46) using HMM profiles were identified also in our reciprocal BLAST searches, indicating that the use of HMM profiles did not improve protein identifications in our case.
Metabolic pathway components known from protist MROs were also searched for. To do this, MRO-associated proteins from across the eukaryotes were gathered based on the supplementary data from Stairs et al. (2) and used as queries against the cnidarian assemblies (SI Appendix, Supplementary Methods).
Phylogenetic Reconstructions.
The phylogenetic analyses used a reference database of 78 ribosomal protein-coding genes, which was curated manually to avoid contamination and structural annotation errors. Two datasets were selected from this database: the first included 78 species representative of major eukaryote lineages (47), and the second included 129 species that encompassed animal diversity and their closest outgroup (choanoflagellates, ichthyosporeans, and ministeriids). In both datasets, sequences were concatenated with SCaFoS (48). After removal of any ambiguously aligned positions using Gblocks Version 0.91b (49), with default parameter values, these Eukaryota and Metazoa datasets included 9,490 and 11,352 amino acid positions, respectively. Phylogenetic reconstructions were performed using the site-heterogeneous CAT model (50), which reduces the impact of long branch attraction (51), as implemented in Phylobayes MPI Version 1.5 (52). For both datasets, two independent chains were run for 10,000 cycles. The first 5,000 trees from each chain were discarded as burn-in. Chain convergence was assessed using the bpcomp and tracecomp scripts, which are part of Phylobayes. Specifically, for both analyses, the bpcomp maxdiff values were <0.3 and the tracecomp effsize values were >70 (except for eukaryotes, where the tree length value was 21), indicating a proper convergence.
Supplementary Material
Acknowledgments
We thank Mark Dasenko and the Center for Genome Research and Biocomputing at Oregon State University (OSU), and Teresa Sawyer of the OSU Electron Microscope Facility for their assistance. This work was supported by the Binational Science Foundation (Grant No. 2015010 to D.H. and P.C.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: Voucher paratype material was deposited at the US National Parasite Collection, Smithsonian Institution, Washington, DC (https://collections.nmnh.si.edu/search/iz/) under the following accession numbers: Henneguya salminicola myxospores: USNM1611578 (from genome sample) and USNM1611579 (from transcriptome sample); Myxobolus squamalis myxospores: USNM1611580 (from genome sample); M. squamalis myxospores and developmental stages: USNM1611581 (from transcriptome sample). All sequence data have been deposited in the National Center for Biotechnology Information (NCBI) database (https://www.ncbi.nlm.nih.gov/). The H. salminicola data are available under the BioProject accession number PRJNA485580. The SSU rDNA sequence was deposited under MK480607. The raw transcriptome and genome reads are available under accession numbers SRR7754566 and SRR7754567, respectively. The transcriptome and genome shotgun assembly projects were deposited at under the accession GHBP00000000 and SGJC00000000, respectively. The M. squamalis data are available under the BioProject accession number PRJNA485581. The SSU rDNA sequence was deposited under MK480606. The raw transcriptome and genome reads are available under the accession numbers SRR7760054 and SRR7760053, respectively. The transcriptome and genome shotgun assembly projects were deposited under accession numbers GHBR00000000 and QWKW00000000, respectively. The mt genome of M. squamalis was deposited under the accession number MK087050. These accession numbers are provided in Dataset S6. The Bayesian trees and all alignments were deposited in the Dryad Digital Repository (https://doi.org/10.5061/dryad.v15dv41sm). Finally, the uncropped images underlying Fig. 2 and SI Appendix, Figs. S7 and S8 were deposited in the Figshare repository (https://doi.org/10.6084/m9.figshare.8300003, https://doi.org/10.6084/m9.figshare.9897284, https://doi.org/10.6084/m9.figshare.9897320).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1909907117/-/DCSupplemental.
References
- 1.Roger A. J., Muñoz-Gómez S. A., Kamikawa R., The origin and diversification of mitochondria. Curr. Biol. 27, R1177–R1192 (2017). [DOI] [PubMed] [Google Scholar]
- 2.Stairs C. W., Leger M. M., Roger A. J., Diversity and origins of anaerobic metabolism in mitochondria and related organelles. Philos. Trans. R. Soc. Lond. B Biol. Sci. 370, 20140326 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Bernhard J. M., et al. , Metazoans of redoxcline sediments in Mediterranean deep-sea hypersaline anoxic basins. BMC Biol. 13, 105 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Danovaro R., et al. , The first metazoa living in permanently anoxic conditions. BMC Biol. 8, 30 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Danovaro R., et al. , The challenge of proving the existence of metazoan life in permanently anoxic deep-sea sediments. BMC Biol. 14, 43 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Okamura B., Gruhl A., Bartholomew J. L., “An introduction to myxozoan evolution, ecology and development” in Myxozoan Evolution Ecology and Development, Okamura B., Gruhl A., Bartholomew J. L., Eds. (Springer International Publishing, 2015), pp. 1–20. [Google Scholar]
- 7.Fontes I., Hallett S. L., Mo T. A., “Comparative epidemiology of myxozoan diseases” in Myxozoan Evolution Ecology and Development, Okamura B., Gruhl A., Bartholomew J. L., Eds. (Springer International Publishing, 2015), pp. 317–341. [Google Scholar]
- 8.Yahalomi D., et al. , The multipartite mitochondrial genome of Enteromyxum leei (Myxozoa): Eight fast-evolving megacircles. Mol. Biol. Evol. 34, 1551–1556 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Takeuchi F., et al. , The mitochondrial genomes of a myxozoan genus Kudoa are extremely divergent in Metazoa. PLoS One 10, e0132030 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fiala I., Bartošová-Sojková P., Whipps C. M., “Classification and phylogenetics of Myxozoa” in Myxozoan Evolution Ecology and Development, Okamura B., Gruhl A., Bartholomew J. L., Eds. (Springer International Publishing, 2015), pp. 85–110. [Google Scholar]
- 11.Fish F. F., Observations on Henneguya salminicola Ward, a myxosporidian parasitic in Pacific salmon. J. Parasitol. 25, 169–172 (1939). [Google Scholar]
- 12.Polley T. M., Atkinson S. D., Jones G. R., Bartholomew J. L., Supplemental description of Myxobolus squamalis (Myxozoa). J. Parasitol. 99, 725–728 (2013). [DOI] [PubMed] [Google Scholar]
- 13.Parra G., Bradnam K., Korf I., CEGMA: A pipeline to accurately annotate core genes in eukaryotic genomes. Bioinformatics 23, 1061–1067 (2007). [DOI] [PubMed] [Google Scholar]
- 14.Chang E. S., et al. , Genomic insights into the evolutionary origin of Myxozoa within Cnidaria. Proc. Natl. Acad. Sci. U.S.A. 112, 14912–14917 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Karnkowska A., et al. , A eukaryote without a mitochondrial organelle. Curr. Biol. 26, 1274–1284 (2016). [DOI] [PubMed] [Google Scholar]
- 16.Huynen M. A., Mühlmeister M., Gotthardt K., Guerrero-Castillo S., Brandt U., Evolution and structural organization of the mitochondrial contact site (MICOS) complex and the mitochondrial intermembrane space bridging (MIB) complex. Biochim. Biophys. Acta 1863, 91–101 (2016). [DOI] [PubMed] [Google Scholar]
- 17.Muñoz-Gómez S. A., et al. , Ancient homology of the mitochondrial contact site and cristae organizing system points to an endosymbiotic origin of mitochondrial cristae. Curr. Biol. 25, 1489–1495 (2015). [DOI] [PubMed] [Google Scholar]
- 18.Yang Y., et al. , The genome of the myxosporean Thelohanellus kitauei shows adaptations to nutrient acquisition within its fish host. Genome Biol. Evol. 6, 3182–3198 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Graziewicz M. A., Longley M. J., Copeland W. C., DNA polymerase γ in mitochondrial DNA replication and repair. Chem. Rev. 106, 383–405 (2006). [DOI] [PubMed] [Google Scholar]
- 20.Lavrov D. V., Pett W., Animal mitochondrial DNA as we do not know it: Mt-genome organization and evolution in nonbilaterian lineages. Genome Biol. Evol. 8, 2896–2913 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Müller M., et al. , Biochemistry and evolution of anaerobic energy metabolism in eukaryotes. Microbiol. Mol. Biol. Rev. 76, 444–495 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.John U., et al. , An aerobic eukaryotic parasite with functional mitochondria that likely lacks a mitochondrial genome. Sci. Adv. 5, eaav1110 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Makiuchi T., Nozaki T., Highly divergent mitochondrion-related organelles in anaerobic parasitic protozoa. Biochimie 100, 3–17 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Johnston I. A., Studies on the swimming musculature of the rainbow trout. J. Fish Biol. 7, 459–467 (1975). [Google Scholar]
- 25.Alexander J. D., Kerans B. L., El-Matbouli M., Hallett S. L., Stevens L., “Annelid-myxosporean interactions” in Myxozoan Evolution Ecology and Development, Okamura B., Gruhl A., Bartholomew J. L., Eds. (Springer International Publishing, 2015), pp. 217–234. [Google Scholar]
- 26.Famme P., Knudsen J., Anoxic survival, growth and reproduction by the freshwater annelid, Tubifex sp., demonstrated using a new simple anoxic chemostat. Comp. Biochem. Physiol. A Comp. Physiol. 81, 251–253 (1985). [Google Scholar]
- 27.Lynch M., Marinov G. K., The bioenergetic costs of a gene. Proc. Natl. Acad. Sci. U.S.A. 112, 15690–15695 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chapman J. A., et al. , The dynamic genome of Hydra. Nature 464, 592–596 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Putnam N. H., et al. , Sea anemone genome reveals ancestral eumetazoan gene repertoire and genomic organization. Science 317, 86–94 (2007). [DOI] [PubMed] [Google Scholar]
- 30.Löfmark S., Edlund C., Nord C. E., Metronidazole is still the drug of choice for treatment of anaerobic infections. Clin. Infect. Dis. 50 (suppl. 1), S16–S23 (2010). [DOI] [PubMed] [Google Scholar]
- 31.Kayal E., et al. , Phylogenomics provides a robust topology of the major cnidarian lineages and insights on the origins of key organismal traits. BMC Evol. Biol. 18, 68 (2018). [Google Scholar]
- 32.Atkinson S. D., Bartholomew J. L., Lotan T., Myxozoans: Ancient metazoan parasites find a home in phylum Cnidaria. Zoology (Jena) 129, 66–68 (2018). [DOI] [PubMed] [Google Scholar]
- 33.Olson M. V., When less is more: Gene loss as an engine of evolutionary change. Am. J. Hum. Genet. 64, 18–23 (1999). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Peng Y., Leung H. C. M., Yiu S. M., Chin F. Y. L., “IDBA–A practical iterative de Bruijn graph de novo assembler” in Research in Computational Molecular Biology. RECOMB 2010, Berger B., Ed. (Lecture Notes Computer Science, 2010), vol. 6044, pp. 426–440. [Google Scholar]
- 35.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Mistry J., Finn R. D., Eddy S. R., Bateman A., Punta M., Challenges in homology search: HMMER3 and convergent evolution of coiled-coil regions. Nucleic Acids Res. 41, e121 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Holt C., Yandell M., MAKER2: An annotation pipeline and genome-database management tool for second-generation genome projects. BMC Bioinformatics 12, 491 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Nawrocki E. P., Eddy S. R., Infernal 1.1: 100-fold faster RNA homology searches. Bioinformatics 29, 2933–2935 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Dierckxsens N., Mardulyn P., Smits G., NOVOPlasty: De novo assembly of organelle genomes from whole genome data. Nucleic Acids Res. 45, e18 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Waterhouse R. M., et al. , BUSCO applications from quality assessments to gene prediction and phylogenomics. Mol. Biol. Evol. 35, 543–548 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Vurture G. W., et al. , GenomeScope: Fast reference-free genome profiling from short reads. Bioinformatics 33, 2202–2204 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Marçais G., Kingsford C., A fast, lock-free approach for efficient parallel counting of occurrences of k-mers. Bioinformatics 27, 764–770 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Gramates L. S., et al. ; The FlyBase Consortium , FlyBase at 25: Looking to the future. Nucleic Acids Res. 45, D663–D671 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.D’Elia D., et al. , The MitoDrome database annotates and compares the OXPHOS nuclear genes of Drosophila melanogaster, Drosophila pseudoobscura and Anopheles gambiae. Mitochondrion 6, 252–257 (2006). [DOI] [PubMed] [Google Scholar]
- 45.Rackham O., Mercer T. R., Filipovska A., The human mitochondrial transcriptome and the RNA-binding proteins that regulate its expression. Wiley Interdiscip. Rev. RNA 3, 675–695 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Muthye V., Lavrov D. V., Characterization of mitochondrial proteomes of nonbilaterian animals. IUBMB Life 70, 1289–1301 (2018). [DOI] [PubMed] [Google Scholar]
- 47.Adl S. M., et al. , Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 26, 12691 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Roure B., Rodriguez-Ezpeleta N., Philippe H., SCaFoS: A tool for selection, concatenation and fusion of sequences for phylogenomics. BMC Evol. Biol. 7 (suppl. 1), S2 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Castresana J., Selection of conserved blocks from multiple alignments for their use in phylogenetic analysis. Mol. Biol. Evol. 17, 540–552 (2000). [DOI] [PubMed] [Google Scholar]
- 50.Lartillot N., Philippe H., A Bayesian mixture model for across-site heterogeneities in the amino-acid replacement process. Mol. Biol. Evol. 21, 1095–1109 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Lartillot N., Brinkmann H., Philippe H., Suppression of long-branch attraction artefacts in the animal phylogeny using a site-heterogeneous model. BMC Evol. Biol. 7 (suppl. 1), S4 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lartillot N., Rodrigue N., Stubbs D., Richer J., PhyloBayes MPI: Phylogenetic reconstruction with infinite mixtures of profiles in a parallel environment. Syst. Biol. 62, 611–615 (2013). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.