Abstract
Purpose
Pandrug-resistant K. pneumoniae is still rare in China, and the lack of studies limits our understanding of the emergence mechanism of pandrug-resistant K. pneumoniae. Here, we report the rapid emergence of a pandrug-resistant K. pneumoniae ST11 strain in an inpatient after treatment with multiple broad-spectrum antibiotics in China.
Patients and Methods
K. pneumoniae strain KP65 was isolated from a 55-year-old male patient hospitalized in the Department of Intensive Care Unit (ICU) of a teaching hospital in China. Antimicrobial susceptibility testing was conducted according to the guidelines of the Clinical and Laboratory Standards Institute (CLSI). The complete genome sequence of the strain was determined using the Illumina NovaSeq 6000 platform and long-read MinION sequencer. Genomic features and resistance mechanisms of the strain were comprehensively analysed using various bioinformatics approaches.
Results
K. pneumoniae strain KP65 was found to be resistant to all antibiotics tested, including beta-lactams, aminoglycosides, quinolones, tigecycline and colistin. Seven resistance genes were identified in the genome. The carbapenem-resistant gene blaKPC-2 and extended-spectrum β-lactamase (ESBL)-producing gene blaCTX-M-65 are located on the IncFII-type plasmid pKPC-2-KP65. No mcr genes were detected in the genome, but an IS5 insertion element was found at position 117 of the mgrB gene. Regarding the rpsJ gene, single-base substitution, G169C, leading to the amino acid substitution V57L was also identified. According to in silico MLST analysis, K. pneumoniae KP65 belongs to sequence type ST11. The closest relative of K. pneumoniae KP65 is another ST11 K. pneumoniae strain, which was isolated from a bloodstream infection in Hangzhou, differing by only 53 cgMLST loci.
Conclusion
Under the selective pressure of antibiotics, the KPC-2-producing K. pneumoniae ST11 strain can easily evolve pandrug resistance through chromosomal mutations. More attention is required to monitor the prevalence of the KPC-2-producing K. pneumoniae ST11 strain in China.
Keywords: Klebsiella pneumoniae, mgrB, rpsJ, ST11, blaKPC-2
Introduction
Klebsiella pneumoniae is an important pathogen that often causes nosocomial infections in hospitalized patients. In the past decade, the clinical isolation rate of K. pneumoniae has been rising, and in China, K. pneumoniae has become the second largest clinically isolated gram-negative bacilli after Escherichia coli.1 The emergence of carbapenem-resistant K. pneumoniae poses a serious threat to public health due to the limited therapeutic options to treat such infections.2 K. pneumoniae carbapenemase (KPC)-type carbapenemase, an Ambler class A β-lactamase, is one of the most important carbapenemases commonly found in K. pneumoniae. Acquisition of KPC-type carbapenemase has contributed to resistance to all β-lactams. Additionally, nationwide surveillance of clinical carbapenem-resistant K. pneumoniae strains has revealed that sequence type (ST) 11 is the dominant clone of KPC-producing K. pneumoniae in China.3
Most blaKPC-carrying K. pneumoniae strains are multidrug-resistant (MDR) bacteria that are often co-resistant to β-lactams, quinolones, aminoglycosides and fosfomycin.4 Tigecycline and colistin are two effective antibiotics for KPC-producing K. pneumoniae infections.5 Tigecycline was effective against most KPC-producing K. pneumoniae at the beginning of its clinical use,6,7 and colistin is currently recognized as the last resort for treating infections due to carbapenem-resistant K. pneumoniae strains.8 Carbapenem-resistant K. pneumoniae that are co-resistant to tigecycline and colistin often represent the emergence of pandrug-resistant bacteria, which are called super bacteria and make clinical treatment difficult.
Pandrug-resistant K. pneumoniae has been reported sporadically in many countries.9–13 However, pandrug-resistant K. pneumoniae in China is still rare and has only been reported by a few studies.14 In the present study, one pandrug-resistant K. pneumoniae strain, KP65, was collected from a urine specimen of an inpatient in a teaching hospital in China. The entire genome sequence of the strain was determined, and genomic features, plasmids and resistance mechanisms were comprehensively analysed.
Materials and Methods
Patient and Isolate
A 55-year-old male patient diagnosed with hypoxic-ischaemic encephalopathy and aspiration pneumonia was hospitalized in the Department of Intensive Care Unit (ICU) of a teaching hospital in Zhejiang Province, China, in March 2018. Two days after admission, he experienced fever and showed elevated inflammatory markers (including CRP, PCT, among others) suggestive of infection. He was administered piperacillin/tazobactam as empiric treatment. One week later, a carbapenem-resistant Acinetobacter baumannii strain was isolated from his sputum sample. The antibiotic treatment was changed to cefoperazone/sulbactam combined with tigecycline. This treatment lasted for twenty days until a carbapenem-resistant K. pneumoniae strain was isolated from both his sputum and urine samples. Antibiotic treatment was then changed to imipenem combined with tigecycline. Three days later, a carbapenem-resistant K. pneumoniae strain was isolated from his blood sample, and antibiotic treatment was changed to polymyxin B for one week. A pandrug-resistant K. pneumoniae strain, KP65, was collected from the urine sample of the patient six days after the end of polymyxin B treatment. The isolate was preliminarily identified using the VITEK MS system (bioMérieux, France) and was further confirmed by 16S rRNA gene sequencing.
Antimicrobial Susceptibility Test
Antimicrobial susceptibility testing was conducted using the VITEK 2 system (bioMérieux, France) with a gram-negative antimicrobial susceptibility testing card (AST-GN16) and standard broth microdilution tests following the guidelines of the Clinical and Laboratory Standards Institute (CLSI). For tigecycline MIC detection, the medium must be prepared fresh on the day of use. As there are no CLSI breakpoints for tigecycline and colistin, the EUCAST standard was adopted.15
Whole-Genome Sequencing
The whole-genome sequence of the strain was determined using the Illumina NovaSeq 6000 platform (Illumina Inc., San Diego, CA, USA) and a long-read MinION sequencer (Nanopore, Oxford, UK). Both short Illumina reads and long MinION reads were hybrid assembled using Unicycler (v0.4.7) under the conservative mode. Complete circular contigs were generated and corrected using Pilon with Illumina reads for several rounds until no change was detected. The complete genome sequence was automatically annotated by the NCBI Prokaryotic Genome Annotation Pipeline (PGAP) server.
Genomic Features and Plasmid Characterization
Multilocus sequence typing (MLST), antimicrobial resistance genes and plasmid replicons were analysed using the BacWGSTdb server.16,17 Circular comparisons based on concentric rings between the blaKPC-2-carrying plasmid and similar plasmids were conducted with a BLAST Ring Image Generator (BRIG).18
Phylogenetic Relationship Analysis
The phylogenetic relationship between KP65 and other K. pneumoniae strains was analysed using the BacWGSTdb server with core genome multilocus sequence typing (cgMLST) approaches with a 100 threshold for phylogenetic analysis.16 The database currently contains 7480 K. pneumoniae strains.
Nucleotide Sequence Accession Numbers
The complete sequences of the isolate K. pneumoniae KP65 and plasmids have been deposited in GenBank under accession numbers CP044258-CP044260. The version described in this paper is the first version.
Ethical Approval
This study was conducted in accordance with the Declaration of Helsinki and approved by the Ethics Committee of Zhejiang Provincial People’s Hospital. Written informed consent from the patient was exempted by the Ethics Committee of Zhejiang Provincial People’s Hospital because the present study focused only on bacteria. Isolation of clinical K. pneumoniae strain KP65 was part of routine hospital laboratory procedure.
Results and Discussion
K. pneumoniae strain KP65 is a pandrug-resistant strain resistant to all antibiotics tested, including beta-lactams, aminoglycosides, quinolones, carbapenems, tigecycline and colistin. The MICs of the antibiotics tested are presented in Table 1.
Table 1.
MICs of the Antibiotics Tested in K. pneumoniae KP65
| Antibiotics | MIC (mg/L) | |
|---|---|---|
| BKP19 | Interpretive Categoriesc | |
| Cefazolina | ≥64 | R |
| Ceftriaxonea | ≥64 | R |
| Cefepimea | ≥64 | R |
| Cefoxitina | ≥64 | R |
| Aztreonama | ≥64 | R |
| Piperacillin/tazobatama | ≥128 | R |
| Imipenemb | >128 | R |
| Meropenemb | >128 | R |
| Amikacina | ≥64 | R |
| Ciprofloxacina | ≥4 | R |
| Levofloxacina | ≥8 | R |
| Tigecyclineb | 32 | R |
| Colistinb | >64 | R |
Notes: aTested by AST-GN16. bTested by standard broth microdilution tests. cMIC, minimum inhibitory concentration.
Abbreviation: R, resistant.
The genome of K. pneumoniae strain KP65 consisted of 3 contigs comprising 5,592,496 bp. Among them, one contig (contig 1: 5,438,014 bp) belongs to the chromosome and the others to two plasmids (contig pKPC-2-KP65: 144,422 bp and contig 3: 10,060 bp). Three plasmid replicons were identified in the genome: one on contig3 (ColRNAI) and another two on plasmid pKPC-2-KP65 (IncFII and IncR). The PGAP server predicted a total of 5384 protein-coding sequences, 85 tRNA genes and 25 rRNA operons.
The resistance genes and plasmid replicons present in the genome of the isolate are presented in Table 2. We identified the beta-lactam resistance genes blaSHV-11, blaTEM-1B, blaCTX-M-65 and blaKPC-2; the aminoglycoside resistance gene rmtB; and the fosfomycin resistance genes fosA and fosA3. Among them, blaSHV-11 and fosA are located on the chromosome. The other resistance genes, including the carbapenem-resistance gene blaKPC-2 and extended-spectrum β-lactamase (ESBL)-producing gene blaCTX-M-65, are located on plasmid pKPC-2-KP65. The pKPC-2-KP65 plasmid contributes the most to the KP65 resistance phenotype.
Table 2.
Antimicrobial Resistance Genes and Plasmid Replicons in Isolates K. pneumoniae Strain KP65
| Antimicrobial Resistance Gene | Contig | Identity (%) | Position | Antimicrobial Resistance Category |
|---|---|---|---|---|
| blaCTX-M-65 | pKPC-2-KP65 | 100.00 | 5660..6535 | Beta-lactam |
| fosA3 | pKPC-2-KP65 | 100.00 | 12397..12813 | Fosfomycin |
| blaTEM-1B | pKPC-2-KP65 | 100.00 | 15580..16440 | Beta-lactam |
| rmtB | pKPC-2-KP65 | 100.00 | 16610..17365 | Aminoglycoside |
| blaKPC-2 | pKPC-2-KP65 | 100.00 | 92566..93447 | Beta-lactam |
| blaSHV-11 | 1 | 100.00 | 2783701..2784561 | Beta-lactam |
| fosA | 1 | 99.27 | 4752290..4752701 | Fosfomycin |
| Plasmid replicon | Contig | Identity (%) | Position | Annotation |
| IncR | pKPC-2-KP65 | 99.20 | 27677..27927 | Gram-negative |
| IncFII | pKPC-2-KP65 | 100.00 | 144166..144422 | Gram-negative |
| ColRNAI | 3 | 98.46 | 5781..5910 | Gram-negative |
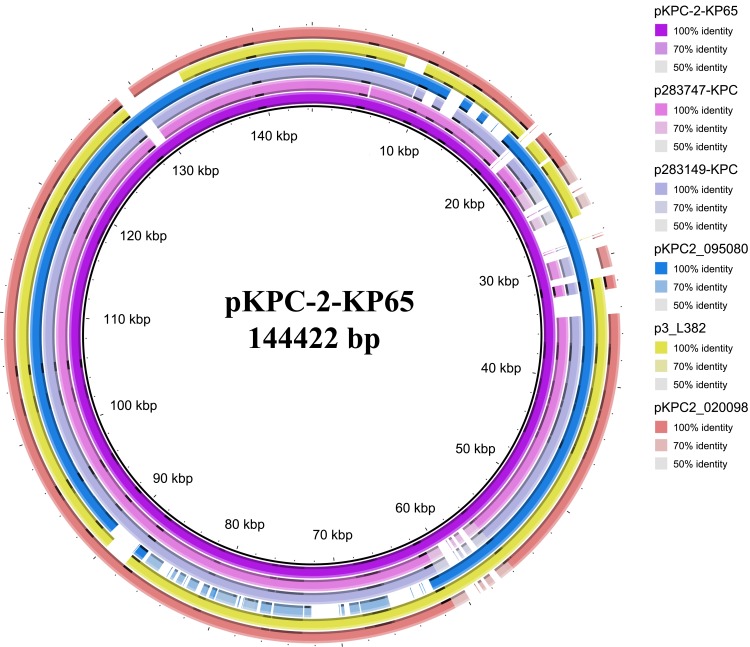
Local alignment of plasmid pKPC-2-KP65 with similar plasmids deposited in the NCBI GenBank database indicated that it is a novel plasmid that has not been reported (Figure 1). Several similar blaKPC-2-carrying plasmids were found in the database, with the most closely related plasmid showing 90% coverage and 99.98% identity to pKPC-2-KP65 (K. pneumoniae strain 283747 plasmid p283747-KPC, accession no. MF168406).19
Figure 1.
Circular comparison between the blaKPC-2-carrying plasmid pKPC-2-KP65 and similar plasmids. p283747-KPC: K pneumoniae strain 283747 plasmid p283747-KPC, accession no. MF168406, size 148,021 bp. p283149-KPC: K pneumoniae strain 283149 plasmid p283149-KPC, accession no. MK036886, size 136,186 bp. pKPC2_09508: K. pneumoniae strain WCHKP2080 plasmid pKPC2_095080, accession no. CP036362, size 120,195 bp. p3_L382: K pneumoniae strain L482 plasmid p3_L382, accession no. CP033962, size 136,159 bp. pKPC2_020098: K. pneumoniae strain WCHKP020098 plasmid pKPC2_020098, accession no. CP036306, size 149,519 bp.
No mcr genes were detected in the genome, but an IS5 insertion element was found at position 117 of the mgrB gene, a negative regulator of the PhoP/PhoQ two-component system contributing to colistin resistance. The insertion interrupts the coding sequence of mgrB and may contribute to the colistin resistance of KP65.20 Moreover, a single-base substitution, G169C, leading to the amino acid substitution V57L, was detected in the rpsJ gene. This mutation may contribute to tigecycline resistance in this isolate.21,22 Both mutations occur on the chromosome. As the strain was isolated from urine samples of the patient after a period of tigecycline and colistin use, these mutations may have occurred due to antibiotic stress from tigecycline and colistin. Our findings support the hypothesis that there is a risk of mutation-mediated resistance under antibiotic pressure in KPC-producing K. pneumoniae during tigecycline and colistin treatment, leading to treatment failure. This is also an important mechanism for the emergence of pandrug-resistant K. pneumoniae strains in hospitalized patients.
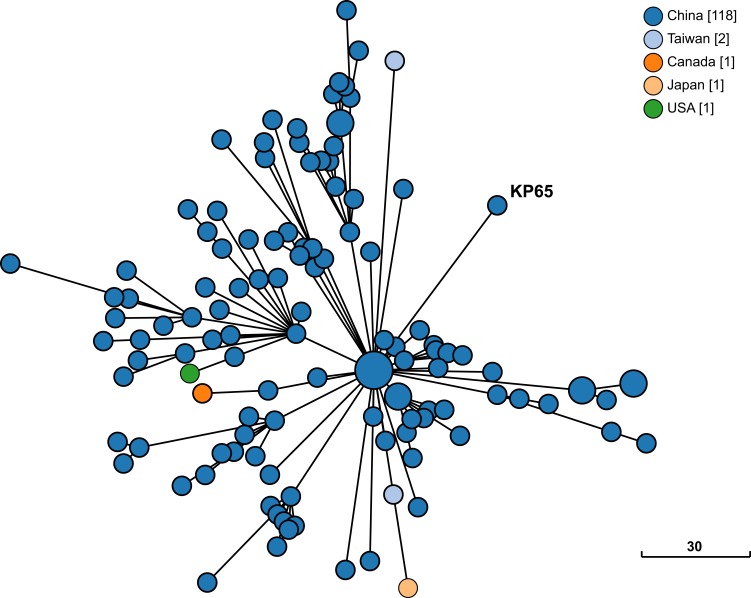
The phylogenetic relationship between K. pneumoniae KP65 and other K. pneumoniae strains currently deposited in the BacWGSTdb server was analysed by cgMLST approaches (Figure 2). A total of 122 phylogenetically related strains were identified in the database; among them, 117 strains were from China. The data suggest that the closest relative of K. pneumoniae KP65 is another ST11 K. pneumoniae strain, sklx10042 (Accession No. NXDY00000000), which was isolated from a bloodstream infection in Hangzhou in 2013; the sequences differ by only 53 cgMLST loci. Compared with K. pneumoniae KP65, no insertion, deletion or point mutation events were found in the mgrB and rpsJ genes in these classic ST11 KPC-2-producing strains, constituting a different antimicrobial resistance profile in these strains.
Figure 2.
Phylogenetic relationship between K. pneumoniae strain KP65 and closely related K. pneumoniae strains currently available in BacWGSTdb. Clonal relationships between different isolates are depicted by the line length connecting each circle. The numbers given in square brackets indicate the number of isolates from each country.
The emergence of pandrug-resistant K. pneumoniae strains makes clinical treatment difficult. Moreover, clonal dissemination of such pandrug-resistant K. pneumoniae clinical strains has led to public health concerns. Therefore, the prevalence of the KPC-2-producing K. pneumoniae ST11 strain, especially the emergence of colistin and tigecycline co-resistant strains, needs our attention. Classification management systems of antibiotics, strict medication guidelines for tigecycline and colistin, and combined use of two or three antibiotics may be an effective way to prevent the emergence of pandrug-resistant K. pneumoniae.
Conclusion
Our study reports the rapid emergence of a pandrug-resistant K. pneumoniae ST11 strain in an inpatient after treatment with multiple broad-spectrum antibiotics in China. More attention is required to monitor the prevalence of the KPC-2-producing K. pneumoniae ST11 strain in China. These strains can easily evolve pandrug resistance through chromosomal mutations under the selective pressure of antibiotics.
Acknowledgments
This study was supported by grants from the National Natural Science Foundation of China (81702042), Natural Science Foundation of Zhejiang Province (LQ19H200003) and the Zhejiang Provincial Medical and Health Science and Technology Plan (2018KY344 and 2019KY311).
Disclosure
The authors report no conflicts of interest in this work.
References
- 1.Hu F, Zhu D, Wang F, Wang M. Current status and trends of antibacterial resistance in China. Clin Infect Dis. 2018;67(suppl_2):S128–S134. doi: 10.1093/cid/ciy657 [DOI] [PubMed] [Google Scholar]
- 2.Tumbarello M, Trecarichi EM, De Rosa FG, et al. Infections caused by KPC-producing Klebsiella pneumoniae: differences in therapy and mortality in a multicentre study. J Antimicrob Chemother. 2015;70(7):2133–2143. doi: 10.1093/jac/dkv086 [DOI] [PubMed] [Google Scholar]
- 3.Qi Y, Wei Z, Ji S, Du X, Shen P, Yu Y. ST11, the dominant clone of KPC-producing Klebsiella pneumoniae in China. J Antimicrob Chemother. 2011;66(2):307–312. doi: 10.1093/jac/dkq431 [DOI] [PubMed] [Google Scholar]
- 4.Bassetti M, Giacobbe DR, Giamarellou H, et al. Management of KPC-producing Klebsiella pneumoniae infections. Clin Microbiol Infect. 2018;24(2):133–144. doi: 10.1016/j.cmi.2017.08.030 [DOI] [PubMed] [Google Scholar]
- 5.Logan LK, Weinstein RA. The epidemiology of carbapenem-resistant enterobacteriaceae: the impact and evolution of a global menace. J Infect Dis. 2017;215(suppl_1):S28–S36. doi: 10.1093/infdis/jiw282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Pankey GA. Tigecycline. J Antimicrob Chemother. 2005;56(3):470–480. doi: 10.1093/jac/dki248 [DOI] [PubMed] [Google Scholar]
- 7.He F, Fu Y, Chen Q, et al. Tigecycline susceptibility and the role of efflux pumps in tigecycline resistance in KPC-producing Klebsiella pneumoniae. PLoS One. 2015;10(3):e0119064. doi: 10.1371/journal.pone.0119064 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Porreca AM, Sullivan KV, Gallagher JC. The epidemiology, evolution, and treatment of KPC-producing organisms. Curr Infect Dis Rep. 2018;20(6):13. doi: 10.1007/s11908-018-0617-x [DOI] [PubMed] [Google Scholar]
- 9.Longo LGA, de Sousa VS, Kraychete GB, et al. Colistin resistance emerges in pandrug-resistant Klebsiella pneumoniae epidemic clones in Rio de Janeiro, Brazil. Int J Antimicrob Agents. 2019;54(5):579–586. doi: 10.1016/j.ijantimicag.2019.08.017 [DOI] [PubMed] [Google Scholar]
- 10.Sieswerda E, van den Brand M, van den Berg RB, et al. Successful rescue treatment of sepsis due to a pandrug-resistant, NDM-producing Klebsiella pneumoniae using aztreonam powder for nebulizer solution as intravenous therapy in combination with ceftazidime/avibactam. J Antimicrob Chemother. 2020;75(3):773–775. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zowawi HM, Forde BM, Alfaresi M, et al. Stepwise evolution of pandrug-resistance in Klebsiella pneumoniae. Sci Rep. 2015;5:15082. doi: 10.1038/srep15082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Avgoulea K, Di Pilato V, Zarkotou O. Characterization of extensively drug-resistant or pandrug-resistant sequence type 147 and 101 OXA-48-producing Klebsiella pneumoniae causing bloodstream infections in patients in an intensive care unit. Antimicrob Agents Chemother. 2018;62(7):e02457–17. doi: 10.1128/AAC.02457-17 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Lazaro-Perona F, Sotillo A, Troyano-Hernaez P, Gomez-Gil R, de la Vega-Bueno A, Mingorance J. Genomic path to pandrug resistance in a clinical isolate of Klebsiella pneumoniae. Int J Antimicrob Agents. 2018;52(5):713–718. doi: 10.1016/j.ijantimicag.2018.08.012 [DOI] [PubMed] [Google Scholar]
- 14.Jia H, Chen H, Ruan Z. Unravelling the genome sequence of a pandrug-resistant Klebsiella pneumoniae isolate with sequence type 11 and capsular serotype KL64 from China. J Glob Antimicrob Resist. 2019;19:40–42. doi: 10.1016/j.jgar.2019.08.013 [DOI] [PubMed] [Google Scholar]
- 15.EUCAST. Clinical breakpoint tables for interpretation of MICs and zone diameters. Version 8.0. 2018.
- 16.Ruan Z, Feng Y. BacWGSTdb, a database for genotyping and source tracking bacterial pathogens. Nucleic Acids Res. 2016;44(D1):D682–D687. doi: 10.1093/nar/gkv1004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Ruan Z, Yu Y, Feng Y. The global dissemination of bacterial infections necessitates the study of reverse genomic epidemiology. Brief Bioinform. 2019:1–10 doi: 10.1093/bib/bbz010 [DOI] [PubMed] [Google Scholar]
- 18.Alikhan NF, Petty NK, Ben Zakour NL, Beatson SA. BLAST Ring Image Generator (BRIG): simple prokaryote genome comparisons. BMC Genomics. 2011;12:402. doi: 10.1186/1471-2164-12-402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Jiang Z, Li M, Zhou D, et al. Sequencing and comparative genomics analysis of Klebsiella pneumoniae plasmid p1512-KPC. Acta Microbiol Sin. 2019;59(2):349–363. [Google Scholar]
- 20.Uz Zaman T, Albladi M, Siddique MI, Aljohani SM, Balkhy HH. Insertion element mediated mgrB disruption and presence of ISKpn28 in colistin-resistant Klebsiella pneumoniae isolates from Saudi Arabia. Infect Drug Resist. 2018;11:1183–1187. doi: 10.2147/IDR.S161146 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Beabout K, Hammerstrom TG, Perez AM, et al. The ribosomal S10 protein is a general target for decreased tigecycline susceptibility. Antimicrob Agents Chemother. 2015;59(9):5561–5566. doi: 10.1128/AAC.00547-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.He F, Shi Q, Fu Y, Xu J, Yu Y, Du X. Tigecycline resistance caused by rpsJ evolution in a 59-year-old male patient infected with KPC-producing Klebsiella pneumoniae during tigecycline treatment. Infect Genet Evol. 2018;66:188–191. doi: 10.1016/j.meegid.2018.09.025 [DOI] [PubMed] [Google Scholar]