Significance
We report here two previously undescribed dinoflagellates that can be models for elucidating the genome evolution associated with transforming an endosymbiotic alga into a plastid (organellogenesis). The two dinoflagellate strains possess green algal endosymbionts enclosing the plastids and relic nuclei (nucleomorphs). Our analyses indicated that DNA transfer from the nucleomorph to the host nuclear genome is in progress in both dinoflagellates, even though their endosymbiotic algae have been transformed into organelles (plastids). Moreover, the origins of the two endosymbionts were resolved at the genus level. These two features found in the two dinoflagellates are absent from well-studied nucleomorph-bearing algal lineages, namely cryptophytes and chlorarachniophytes. Consequently, the two dinoflagellates assist us in understanding endosymbiosis-driven eukaryotic genome evolution at a finer scale.
Keywords: secondary endosymbiosis, nucleomorph, endosymbiotic gene transfer, plastid, Pedinophyceae
Abstract
Nucleomorphs are relic endosymbiont nuclei so far found only in two algal groups, cryptophytes and chlorarachniophytes, which have been studied to model the evolutionary process of integrating an endosymbiont alga into a host-governed plastid (organellogenesis). However, past studies suggest that DNA transfer from the endosymbiont to host nuclei had already ceased in both cryptophytes and chlorarachniophytes, implying that the organellogenesis at the genetic level has been completed in the two systems. Moreover, we have yet to pinpoint the closest free-living relative of the endosymbiotic alga engulfed by the ancestral chlorarachniophyte or cryptophyte, making it difficult to infer how organellogenesis altered the endosymbiont genome. To counter the above issues, we need novel nucleomorph-bearing algae, in which endosymbiont-to-host DNA transfer is on-going and for which endosymbiont/plastid origins can be inferred at a fine taxonomic scale. Here, we report two previously undescribed dinoflagellates, strains MGD and TGD, with green algal endosymbionts enclosing plastids as well as relic nuclei (nucleomorphs). We provide evidence for the presence of DNA in the two nucleomorphs and the transfer of endosymbiont genes to the host (dinoflagellate) genomes. Furthermore, DNA transfer between the host and endosymbiont nuclei was found to be in progress in both the MGD and TGD systems. Phylogenetic analyses successfully resolved the origins of the endosymbionts at the genus level. With the combined evidence, we conclude that the host–endosymbiont integration in MGD/TGD is less advanced than that in cryptophytes/chrorarachniophytes, and propose the two dinoflagellates as models for elucidating organellogenesis.
The transformation of a free-living photosynthetic organism into a plastid through endosymbiosis has occurred multiple times in eukaryotic evolution. The first plastid was most likely established through “primary endosymbiosis” between a cyanobacterium and the common ancestor of red, glaucophyte, and green algae (plus descendants of green algae, i.e., land plants) (1–4). The plastids in the three lineages described above are the direct descendants of the cyanobacterial endosymbiont, and designated as “primary plastids.” After major eukaryotic lineages diverged, some heterotrophs turned into phototrophs by acquiring “secondary plastids” through algal endosymbionts bearing primary plastids (secondary endosymbioses). Secondary endosymbioses most likely occurred multiple times in eukaryotic evolution, as the host lineages bearing secondary plastids (so-called complex algae) are distantly related to one another (1–4). In addition, the origins of secondary plastids vary among complex algae; some possess red alga-derived plastids, while others possess green alga-derived plastids, strongly arguing that the two types of secondary plastids were established through separate (at least two) endosymbiotic events (1–4).
The evolutionary process of integrating an endosymbiont into the host cell (organellogenesis) has yet to be fully understood. Nevertheless, genomic data from diverse eukaryotic lineages indicated that endosymbiont genomes should have lost a massive number of genes that were dispensable for intracellular/endosymbiotic lifestyles (1–5). It is most likely that the reduction of endosymbiont genomes and integration of the endosymbiont into the host progressed simultaneously during organellogenesis (1–4). The reductive process that occurred in endosymbiont genomes seemingly has a tight correlation with genome G + C content (GC%), as reduced endosymbiont genomes are commonly poor in G and C (6–9). To interlock the host and endosymbiont metabolically and genetically, we regard, transfer of endosymbiont genes to the host nuclear genome (endosymbiotic gene transfer, EGT), coupled with the host’s invention of machineries that enable them to express the transferred genes and target the gene products to the original compartment, as critical (1–4, 10). Nevertheless, the precise process that enables organellogenesis still remains unclear.
No intracellular structures of the endosymbiotic algae were left except plastids in most of the complex algae (euglenids, chromerids, haptophytes, ochrophytes, and dinoflagellates). Only cryptophytes and chlorarachniophytes are known to retain nucleomorphs, the relic nuclei of their algal endosymbionts (1, 8, 9, 11, 12). As the two complex algae bearing nucleomorphs possess the morphological characteristics that have been lost from others, they were thought to provide clues to understand the detailed process of organellogenesis (1, 8, 9). In this regard, the genomic data of both nuclei and nucleomorphs, as well as transcriptomic and proteomic data, have been accumulated for cryptophytes and chlorarachniophytes (13–22), and genetic transformation was established for a chlorarachniophyte species (23). It has been determined that a red alga and an ulvophyte green alga are the sources of cryptophyte and chlorarachniophyte plastids, respectively (18, 24–28), but an even recent multigene phylogenetic analyses did not provide finer resolution in determining the closest living species/genus for the origins of the two plastids (18, 28–32). Such uncertainties are potential drawbacks of using cryptophytes and chlorarachniophytes as the model organisms to study the effect of the reductive process on the endosymbiont genome during secondary endosymbiosis. Without pinpointing the precise origin of the plastid (or endosymbiotic alga that gave rise to the plastid), it is difficult to reconstruct the original gene contents of the nuclear genome of the endosymbionts, and the reductive process that shaped the current nucleomorph genomes in the two algal lineages. Thus, it is ideal to find and investigate a novel nucleomorph-bearing organism, for which the endosymbiont origin is resolved at a fine evolutionary scale, and compare it with cryptophytes and chlorarachniophytes. However, no novel nucleomorph-bearing lineage has been found since the discovery of the nucleomorph in chlorarachniophytes in 1984 (12).
Dinoflagellates are a eukaryotic group belonging to Alveolata, comprising both photosynthetic and nonphotosynthetic species (33, 34). The vast majority of photosynthetic dinoflagellates possess red alga-derived plastids containing a unique carotenoid peridinin (35, 36). It is widely accepted that a peridinin-containing plastid already existed in the ancestral dinoflagellate, and photosynthetic capacity has been lost secondarily on multiple branches of the tree of dinoflagellates (37–39). In addition to multiple losses of photosynthesis, different types of noncanonical plastids lacking peridinin have been reported. So far, three types of noncanonical plastids have been found: 1) The plastids containing chlorophylls (Chls) a and c plus 19′ hexanoyloxyfucoxanthin in the family Kareniaceae (e.g., Karenia brevis); 2) those containing Chls a and b (Chls a+b) in the genus Lepidodinium (e.g., Lepidodinium chlorophorum); and 3) those containing Chls a and c plus fucoxanthin in the family Kryptoperidineaceae (e.g., Durinskia baltica) (36, 40–43). The pigment composition, together with molecular phylogenies inferred from plastid genes clearly designated the origins of the first, second, and third noncanonical plastids described above as a haptophyte, a green alga, and a diatom, respectively (44–49). In the species with the haptophyte-derived plastids, the endosymbiotic algae are regarded as being fully integrated into the dinoflagellate (host) cells, because no cellular component except the plastid remains, and gene transfer from the endosymbiont genome to the dinoflagellate genome (i.e., EGT) has been detected (47, 50–56). In Lepidodinium viride with a green alga-derived plastid, although a nucleus-like structure in the compartment corresponding to the cytoplasm of the endosymbiont alga was reported (41), it is still controversial (Discussion). On the other hand, the species with the third noncanonical plastids, derived from diatoms, are unique in maintaining major cellular components of the endosymbiont, such as the plastid, nucleus, and mitochondrion (57–59). To our knowledge, there has been no molecular evidence for the diatom endosymbionts being modified extensively during the endosymbiosis (54, 60–64).
Here, we report two undescribed dinoflagellates, strains MGD and TGD, with green alga-derived plastids containing Chls a+b. The two dinoflagellates are distinct from each other in terms of their cell morphologies, and no clear affinity between the two hosts was recovered by molecular phylogenetic analyses. In both MGD and TGD, conspicuous nucleus-like structures with DNA were identified in the periplastidal compartments (PPCs) that correspond to the endosymbiont cytoplasm. We successfully obtained evidence for the green algal endosymbionts being genetically integrated into the dinoflagellate host cells. Both green algal endosymbionts showed clear phylogenetic affinities to Pedinophyceae, a particular group of green algae. Taken together, these observations lead us to conclude that MGD and TGD are nucleomorph-bearing organisms harboring Chls a+b-containing plastids derived from endosymbiotic Pedinophyceae green algae. Finally, we propose the two dinoflagellates as models to study the genome evolution associated with secondary endosymbiosis.
Results
MGD and TGD Possess Nucleomorphs.
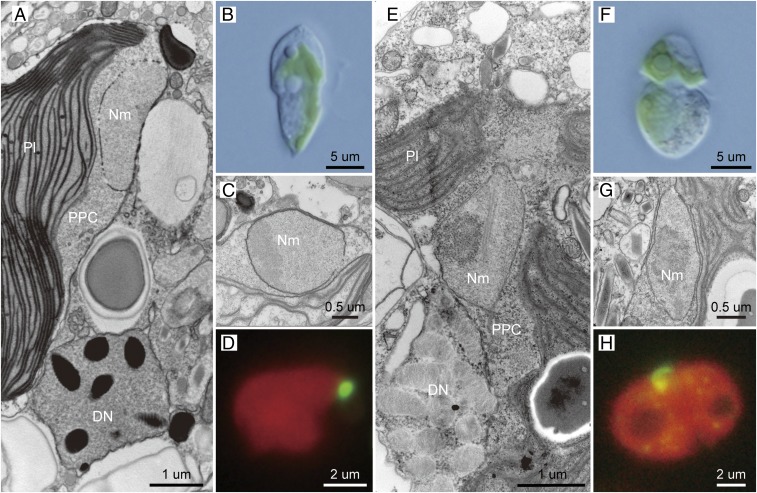
Dinoflagellate strains MGD and TGD were isolated from two distinct coastal locations in Japan and their monoclonal cultures have been maintained in the laboratory (Materials and Methods). Both strains contain permanent green-colored plastids instead of peridinin-containing plastids in major photosynthetic dinoflagellates. Their overall cell structures appeared to be distinct from each other (Fig. 1 B and F). The green-colored plastids in the two dinoflagellates are most likely of green algal origin, as pigment profiles similar to green algae were detected from MGD and TGD (SI Appendix, Fig. S1). Significantly, the cell structure and plastid shape of Lepidodinium spp. (41, 42), a previously described species bearing green alga-derived plastids, are distinctive from those of MGD and TGD (SI Appendix, Fig. S1). Under transmission electron microscopy (TEM) observations, nucleus-like structures with double-membranes were found in the spaces between the second and third plastid membranes, which corresponds to the cytoplasm of the endosymbiont algae, in MGD and TGD (Fig. 1 A, C, E, and G). Each of the dinoflagellate cells examined most likely contained a single plastid associated with a single nucleus-like structure (Fig. 1 A, D, E, and H), The nucleus-like structures in MGD and TGD are clearly distinct from characteristic dinoflagellate nuclei in the cytoplasm (Fig. 1 A and E).
Fig. 1.
Morphology of undescribed dinoflagellate strains MGD (A–D) and TGD (E–H). (A and E) Cross sections of the cell under TEM, showing the dinoflagellate nucleus (DN), nucleomorph (Nm), plastid (Pl), and PPC. (B and F) Whole-cell light micrographs. (C and G) Enlarged image of a cross section of the cell under TEM observation. (D and H) Fluorescent microscopy with SYBER green I-staining image.
We conducted fluorescent microscopic observations to examine whether the nucleus-like structure in the PPCs contains any DNA by SYBR green staining. In our preliminary observations staining whole MGD and TGD cells, a DNA signal from the endosymbiont compartment was undetectable, due to the overpowering brightness of the dinoflagellate nucleus. Thus, the plastid enclosing the nucleus-like structure was separated from the dinoflagellate nucleus prior to SYBR green staining. We confirmed that the isolated plastids were associated physically with the nucleus-like structures by TEM observation (SI Appendix, Fig. S2). The observation of the SYBR green-stained plastid samples successfully detected clear DNA signals on the surface of the plastid, a location consistent with the nucleus-like structures in the PPCs (Fig. 1 D and H). The results described above strongly suggest that the nucleus-like structures in the PPCs are derived from the genome-containing nuclei of the green algal endosymbionts in MGD and TGD.
In dinoflagellate species bearing obligate diatom endosymbionts, collectively called “dinotoms” (e.g., Kryptoperidinium foliaceum and D. baltica), the endosymbionts retain their nuclei and mitochondria as well as the plastids (44, 49). The presence of mitochondria in the endosymbiont alga-derived compartment suggests that energy production in the endosymbiont has yet to be fully brought under host control in the dinotom systems. Thus, the endosymbionts in dinotoms are much less morphologically reduced and host-governed than those in cryptophytes or chlorarachniophytes, which contain residual nuclei (nucleomorphs) and plastids, but no mitochondrion (1, 8, 9). Our TEM observations detected ribosomes but no mitochondrion in the PPCs of MGD or TGD (Fig. 1 A and E and SI Appendix, Fig. S2), suggesting that their green algal endosymbionts were ultrastructurally reduced and governed by the host to a similar degree seen in the endosymbionts of cryptophytes and chlorarachniophytes. The microscopic data described above strongly suggest that: 1) The endosymbiont compartments in both MGD and TGD are indeed endosymbiont-derived organelles, not obligate endosymbionts; and 2) the nucleus-like structures found in the PPCs of the two dinoflagellates are equivalent to cryptophyte and chlorarachniophyte nucleomorphs. Thus, we conclude that both MGD and TGD possess nucleomorphs derived from the nuclei of their green algal endosymbionts.
Recent Genetic Integration of the Green Algal Endosymbionts in MGD and TGD.
In this section, we provide the evidence for the host and endosymbiont being genetically interlocked with each other in MGD and TGD. From the transcriptomic data generated from the MGD and TGD cells cultured in the laboratory, we predicted 57,983 and 73,589 transcripts encoding putative proteins, respectively. Of the putative proteins in MGD and TGD, 534 and 961, respectively, showed high amino acid sequence similarity to nucleus-encoded proteins of free-living green algae. Hereafter, the abovementioned transcripts/proteins are referred to as “green algal transcripts/proteins.”
There are two possibilities for which genome (or genomes) the “green algal genes” encoding green algal proteins reside in, depending on how the host and endosymbiont are integrated with each other in the MGD and TGD systems. If the host–endosymbiont integration at the genetic level has yet to be established in the two systems [as proposed for the dinotom system; Hehenberger et al. (64)], green algal genes are anticipated to be found exclusively in their endosymbiont genomes. However, the endosymbiont-derived compartments of MGD and TGD are ultrastructurally more reduced than those of dinotoms, implying that the host–endosymbiont integration is more advanced in the former systems than the latter. If the host–endosymbiont integration of the MGD and TGD systems has reached the genetic level observed in cryptophytes and chlorarachniophytes (20), then some of the green algal genes were likely transferred from the endosymbiont genome to the host genome (i.e., EGT).
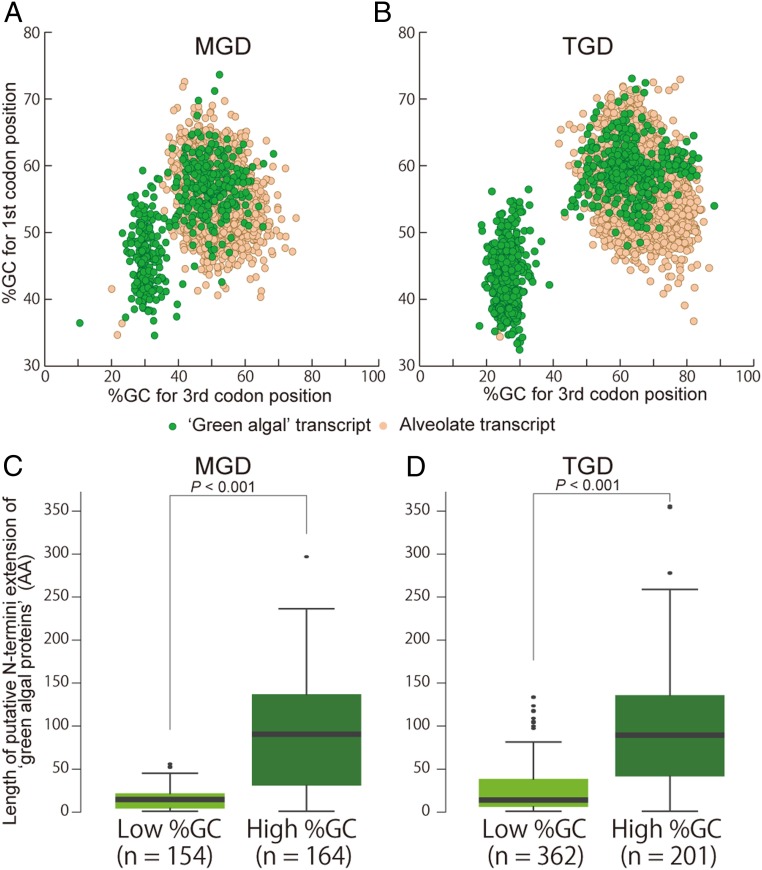
We repeated the procedure described above to retrieve the transcripts encoding the proteins conserved among alveolates (including dinoflagellates) in MGD and TGD. Such “alveolate transcripts” were most likely expressed from the host (dinoflagellate) genomes. Alveolate transcripts in MGD formed a cluster in the two dimensional plot based on the GC% of first codon positions and that of third codon positions (Fig. 2A, dots in orange). Similarly, alveolate transcripts from TGD formed a cluster in the same plot, but shifted toward higher GC% in third codon positions (Fig. 2B). In sharp contrast, green algal transcripts from both MGD and TGD were found to be split into two populations (Fig. 2 A and B, dots in green), and the population with high GC% overlapped with alveolate transcripts. Likewise, a codon usage-based analysis also split green algal transcripts into two populations and one of them overlapped with the cluster of alveolate transcripts (SI Appendix, Fig. S3).
Fig. 2.
Scatter plots showing the distribution of GC% (A and B) and box plots for putative N-terminal extension (C and D) of the transcripts found in TGD (Left) and MGD (Right). (A and B) The x and y axes show the GC% of first and third codon positions, respectively. Plots in green and orange represent the transcripts encoding the putative green algal and alveolate proteins, respectively. In both plots, green algal transcripts were divided into two populations based on GC%, and the ones with higher GC% overlapped with the masses of alveolate transcripts, which were presumably expressed from the dinoflagellate nuclear genomes. (C and D) Box plots of N-terminal extension of green algal transcripts with low and high GC%. The x axes show lengths of putative N-terminal extensions (see Material and Methods). P values displayed in the plots were calculated based on the Wilcoxon rank-sum test.
We estimated the abundance of each of the green algal/alveolate transcripts in MGD and TGD by calculating FPKM (fragments per kilo-base transcript length per million fragments mapped) (65). Green algal transcripts with high GC% and alveolate transcripts appeared to be expressed at similar levels in both dinoflagellates (SI Appendix, Fig. S4). On the other hand, the average FPKM for green algal transcripts with low GC% (334.8 and 279.6 for MGD and TGD, respectively) appeared to be higher than those for the transcripts with high GC% (5.7 and 4.8 for MGD and TGD, respectively). Similar differences in transcriptional intensity between the nuclear and nucleomorph genomes has been documented in cryptophytes and chlorarachniophytes (21, 22).
Mature mRNA molecules transcribed from dinoflagellate nuclear genomes are known to possess a particular short sequence (spliced leader or SL sequence; 5′-CCGTAGCCATTTTGGCTCAAG-3′) at their 5′ termini (66). Thus, green algal transcripts expressed from the host nuclear genome are anticipated to be preceded by the SL sequences. We examined the presence/absence of the SL sequence in six green algal transcripts identified from TGD by RT-PCR using a forward primer matching the dinoflagellate SL sequence (66) and reverse primers matching specifically to the individual transcripts. As presented in SI Appendix, Fig. S5, the 5′ termini of the three transcripts with high GC% were confirmed by PCR amplification followed by Sanger sequencing, while no amplification was observed for the three transcripts with low GC% (SI Appendix, Fig. S5, Upper). We subjected six green algal transcripts identified from MGD to the same RT-PCR experiments, and observed that the amplification of the 5′ termini occurred only for the transcripts with high GC% (SI Appendix, Fig. S5, Lower). We systematically looked for green algal transcripts bearing 5′ sequences that matched with the 3′ portion of the SL sequence (5′-TTTTGGCTCAAG-3′). We detected the partial SL sequence in 78 of the 534 green algal transcripts in MGD and the vast majority of the SL-bearing transcripts were classified into the high-GC% category, albeit a few transcripts were on the boundary between high- and low-GC% categories (SI Appendix, Fig. S6). In TGD, 9 SL-bearing transcripts, all of which are of high-GC%, were detected among the 961 green algal transcripts.
We confirmed the presence of the SL sequence in a subset of green algal transcripts (see above), and the SL-bearing transcripts are compelling evidence that the host genomes of MGD and TGD carry and transcribe genes acquired from the genomes of their green algal endosymbionts. We should be cautious not to regard the GC% of green algal transcripts as the absolute “probe” for the presence/absence of the SL sequence, but anticipate that a substantial portion of high-GC% green algal transcripts are expressed from the dinoflagellate nuclear genomes and receive the SL sequences posttranscriptionally in both MGD and TGD. Although the precise magnitude of EGT is currently uncertain in the MGD or TGD system, we here conclude that the host and endosymbiont in the two dinoflagellates have been integrated by the process of EGT to the same extent as seen in cryptophytes and chlorarachniophytes.
We additionally searched for the transcripts encoding enzymes involved in C5 pathway for the heme biosynthesis, and chlorophyll a (Chl-a) and isopentenyl diphosphate (IPP) biosynthetic pathways that are typically localized in plastids (SI Appendix, Fig. S7). Indeed, some of the transcripts were predicted to encode proteins with typical plastid-targeting signals (SI Appendix, Fig. S7). Heme and IPP pathways in both MGD and TGD appeared to be dominated by “vertically inherited (VI)-type” enzymes that share their origins with the homologs of peridinin-containing dinoflagellates. Thus, besides EGT, substantial proportions of the current plastid proteomes in MGD and TGD may have been inherited from the ancestral plastids containing peridinin. In addition to VI-type genes, “laterally acquired (LA)-type” genes from diverse organisms distantly related to dinoflagellates (host) or green algae (endosymbiont) were detected in the pathways assessed here (SI Appendix, Fig. S6). According to the shopping bag hypothesis (67), LA-type genes could have made MGD/TGD preadapted for plastid acquisition. To examine the aforementioned possibility, it is critical to examine whether each LA-type gene was acquired in the dinoflagellate genome prior to the green algal endosymbiosis by comparative studies of plastid proteomes sampled from diverse dinoflagellates, particularly those closely related to MGD and TGD.
Green algal transcripts with low GC% (Fig. 2 A and B) likely bear no SL sequence at their 5′ termini, implying the presence of a second genome with a GC% content lower than the dinoflagellate nuclear genome, in both MGD and TGD. Accumulated genomic data clearly suggest that endosymbiont and organellar genomes underwent reductive evolution (e.g., cryptophyte and chlorarachniophyte nucleomorph genomes) and tend to have low GC% (8, 9, 13–19). Considering the reduced characteristics in the endosymbiont compartments in MGD and TGD at the morphological level, we predict that the nucleomorph genomes in the two dinoflagellates bear lower GC% than those of the dinoflagellate nuclear genomes. Thus, the sources of green algal transcripts with low GC% are most likely the nucleomorph genomes. It is worth noting that housekeeping proteins, which are involved in the eukaryote-type machineries for translation, transcription, and replication, were found to be encoded almost exclusively by green algal transcripts with low GC% (i.e., putative nucleomorph transcripts) in both MGD and TGD (SI Appendix, Fig. S8 A and B). In contrast, transcripts encoding proteins involved in plastid metabolic pathways appeared to span the two populations of green algal transcripts with distinct GC% (SI Appendix, Fig. S8 C and D). Similar biased gene content in the nucleomorph genomes has been documented in the cryptophyte and chlorarachniophyte systems (8, 9, 13–19).
We identified the green algal transcripts with high GC% (i.e., putative nuclear transcripts) encoding plastid-related proteins involved in plastid function and maintenance (e.g., photosynthesis) in both MGD and TGD (SI Appendix, Fig. S8 C and D). In theory, to operate the plastids, these nucleus-encoded plastid-related proteins (many of them are presumably acquired from the green algal endosymbionts) need to be targeted to the plastids after being synthesized by the host machinery in the cytoplasm in MGD and TGD.
In photosynthetic eukaryotes with primary plastids surrounded by two membranes (e.g., green algae), many of the nucleus-encoded plastid-targeted proteins bear N-terminal extensions (so-called “transit peptides”) that work as a plastid-targeting signal. The N-terminal extensions of the nucleus-encoded proteins targeted to complex plastids, which are surrounded by three or four membranes, tend to have a bipartite structure comprising a hydrophobic “signal peptide” (SP) followed by the transit peptide-like region (68). As both MGD and TGD plastids are surrounded by four membranes (Fig. 1 A and E), the nucleus-encoded plastid-targeted proteins of the two dinoflagellates should have a bipartite plastid-targeting signal. The proteins encoded by endosymbiotically transferred green algal genes unlikely acquired their bipartite signals ab initio, and instead needed to modify their N-terminal extensions by appending the SPs to be targeted into the current MGD and TGD plastids. If the above scenario is the case, the nucleus-encoded plastid-targeted green algal proteins in MGD and TGD possess N-terminal extensions longer than those of the homologous proteins in free-living green algae with primary plastids.
It is reasonable to expect that the nucleus-encoded plastid-targeted green algal proteins are a subset of the green algal proteins encoded by the putative nuclear transcripts (i.e., green algal transcripts with high GC%). As expected, the nucleus-encoded green algal proteins tend to bear significantly longer N-terminal extensions than the nucleomorph-encoded green algal proteins (Fig. 2 C and D), implying that these N-terminal extensions have bipartite structures. Indeed, some green algal proteins in MGD and TGD were predicted to have the typical bipartite plastid-targeting signals (SI Appendix, Fig. S9). The results described above suggest that both MGD and TGD possess nucleus-encoded green algal proteins, which are localized posttranslationally in the plastid. On the other hand, for the green algal proteins encoded by green algal transcripts with low GC% (i.e., putative nucleomorph transcripts), it is unnecessary to have the N-terminal extensions with the bipartite structure.
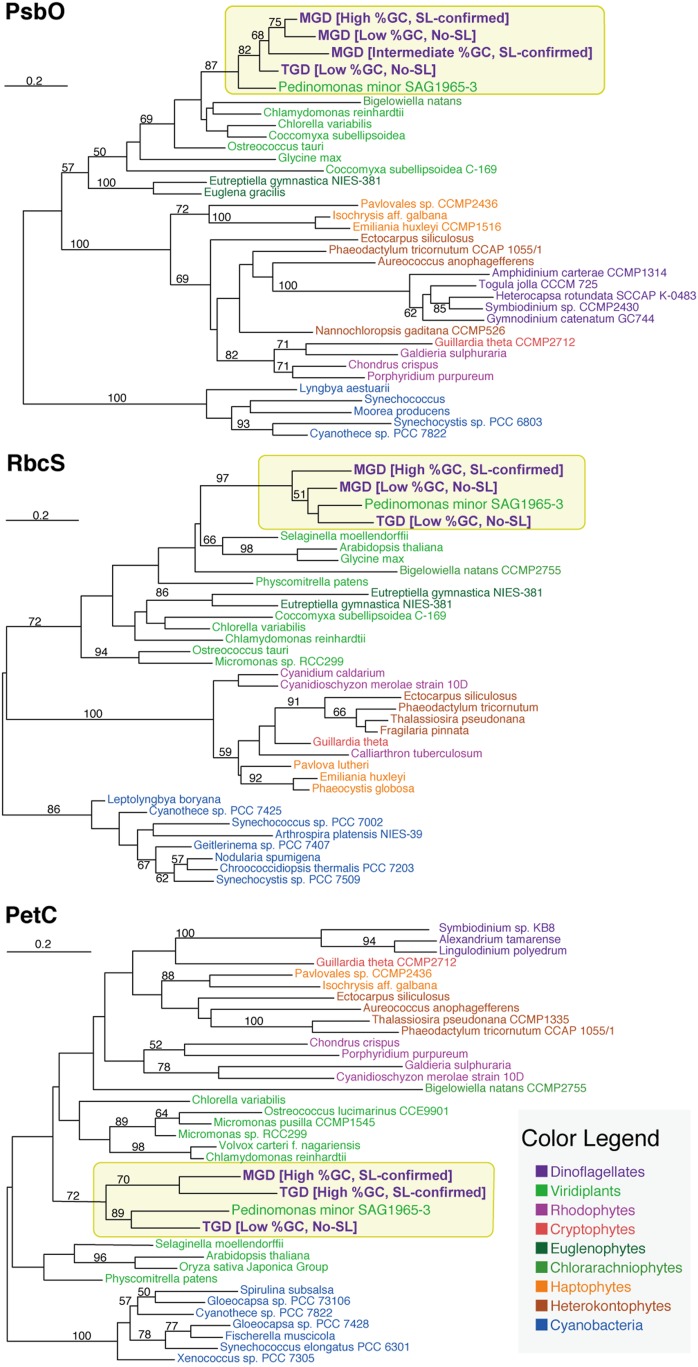
Our detailed assessment, focused on green algal transcripts, identified multiple psbO and rbcS transcripts with distinct GC% in MGD (Fig. 3). Some of these green algal transcripts showed high affinity to particular green algae, Pedinomonas spp., that were closely related to the endosymbionts that gave rise to the current plastids in MGD and TGD (see the next section for the details). We confirmed the SL sequences of the high-GC% versions of the aforementioned transcripts bioinformatically or experimentally (see above), but found no evidence for the SL sequence at the 5′ termini of the low-GC% counterparts (Fig. 3). These data suggest that MGD possesses two sets of psbO and rbcS genes; one is nucleus-encoded (high-GC% and generates the SL-bearing transcripts) and the other is nucleomorph-encoded (low-GC% and generates the SL-lacking transcripts). Similarly, TGD likely possesses both nucleus- and nucleomorph-encoded petC genes, as we found two petC transcripts; one is of high-GC% and bears the SL sequence, and the other is of low GC% and bears no SL sequence. The petC genes of TGD, as well as that of MGD, showed a phylogenetic affinity to the Pedinomonas homolog, reflecting their plastid (and endosymbiont) origins (Fig. 3).
Fig. 3.
ML trees for the green algal orthologous proteins with distinct GC%. The numbers above branches show nonparametric ML bootstrap values. Only ML bootstrap support values greater than 50% are shown on the corresponding branches.
In addition to the nuclear (high-GC%) and nucleomorph (low-GC%) versions described above, we found a third psbO transcript in MGD (Fig. 3), which has neither a high nor low GC%. The psbO transcript with an “intermediate GC%” was found to bear the SL sequence, indicating that MGD possesses a nucleus-encoded psbO gene whose GC% does not match that of other nuclear genes, including the high-GC% version of the psbO gene. We herein propose that the intermediate-GC% version of the psbO gene was transplanted in the nuclear genome before the GC% of the nucleomorph genome had been lowered to the current level. Alternatively, the intermediate-GC% psbO gene is the result of more recent EGT than the high GC% one, and the amelioration of GC% has yet to be completed in the former.
Pedinophyceae Origin of the Green Algal Endosymbionts in MGD and TGD.
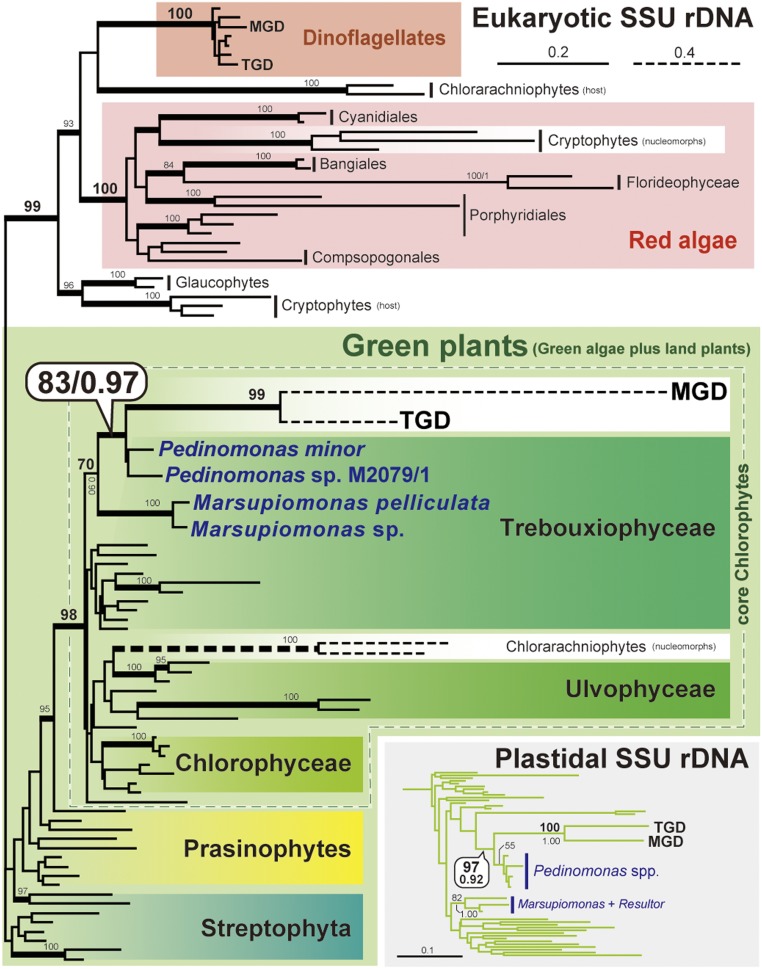
We isolated two eukaryotic small subunit rRNA (18S rRNA) genes—one with and the other without a clear phylogenetic affinity to those of dinoflagellates—from each of MGD and TGD. The 18S rRNA sequences isolated from MGD and TGD were phylogenetically analyzed along with those from red algae, green plants, including green algae and land plants, glaucophytes, cryptophytes, chlorarachniophytes, and dinoflagellates. The 18S rRNA phylogeny placed nondinoflagellate-type MGD and TGD sequences within the sequences from free-living green algae, being distant from the clade of the dinoflagellate sequences, including dinoflagellate-type MGD and TGD sequences (Fig. 4). We thus regard the positions of dinoflagellate-type and nondinoflagellate-type sequences from MGD and TGD in the 18S rRNA phylogeny as the phylogenetic positions of the host and endosymbiont in the MGD and TGD systems.
Fig. 4.
ML tree inferred from eukaryotic small subunit ribosomal RNA (18S rRNA) sequences. All of the taxon names are omitted except MGD, TGD, and Pedinophyceae green algae. Taxon labels of red and green algae per Adl et al. (89). Only ML bootstrap support values greater than 80% are shown on the corresponding branches. The branches supported by BPPs greater than 0.95 are shown as thick lines. The clade comprising MGD, TGD, and Pedinophyceae green algae inferred from plastidal small subunit rRNA (16S rRNA) sequences is shown in the box. The 16S rRNA tree including L. chlorophorum with full taxon names is provided as SI Appendix, Fig. S10.
Unlike cryptophyte or chlorarachniophyte nucleomorphs, the 18S rRNA phylogeny clarified the precise origins of the green algal endosymbionts in the MGD and TGD systems. Nondinoflagellate-type MGD and TGD sequences grouped together, and this MGD–TGD clade was placed within a clade of a small collection of green algae, Pedinophyceae, with a specific affinity to Pedinomonas spp. (Fig. 4). The monophyly of Pedinomonas plus MGD and TGD received a maximum-likelihood (ML) bootstrap value of 83% and a Bayesian posterior probability (BPP) of 0.97 (Fig. 4). The detailed origins of the endosymbionts in MGD and TGD were also assessed by phylogenetic analyses of plastidal small-subunit rRNA (16S rRNA) sequences (Fig. 4). The plastidal 16S rRNA phylogeny united MGD, TGD, and Pedinomonas spp. into a clade with an ML bootstrap value of 97% and a BPP of 0.92, excluding other Pedinophyceae considered in the analyses, Marsupiomonas spp. and Resultor mikron (Fig. 4). The two phylogenetic analyses consistently and strongly indicate that the current plastids in both MGD and TGD can be traced back to Pedinophyceae green algae closely related to Pedinomonas.
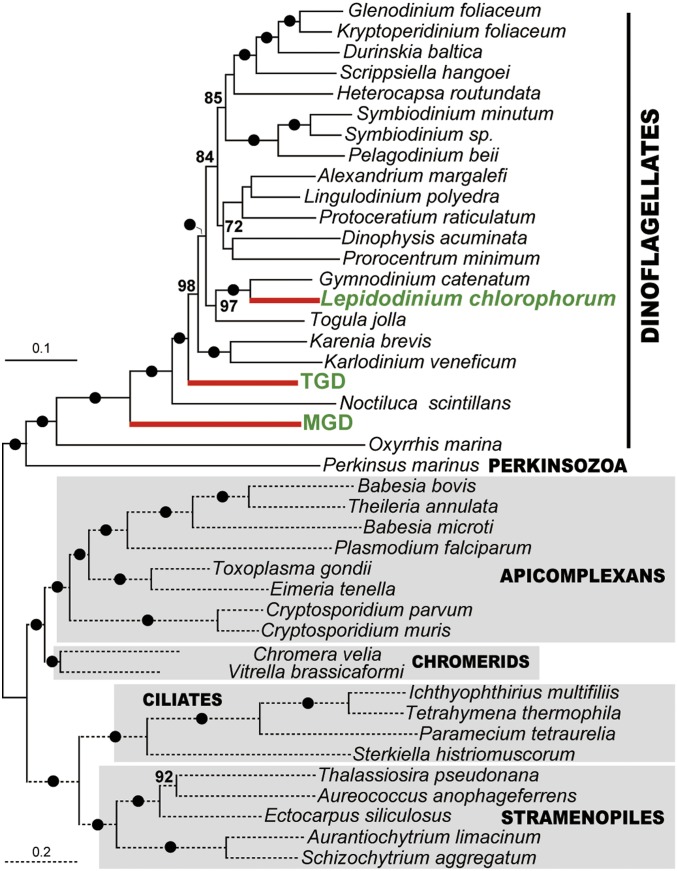
Some of us have already reported the Pedinophyceae origin of the Chls a+b-containing plastids in the dinoflagellate genus Lepidodinium (69). Indeed, in the plastidal 16S rRNA phylogeny including the L. chlorophorum sequence, L. chlorophorum robustly grouped with MGD and TGD, and this dinoflagellate clade as a whole was found to be sister to Pedinomonas (SI Appendix, Fig. S10; note that the L. chlorophorum sequence was excluded from the analyses presented in Fig. 4 due to its extremely divergent nature). The simplest interpretation of this tree topology is that L. chlorophorum, MGD, and TGD share a single ancestor with a Pedinophyceae-derived plastid. However, a taxon-rich dinoflagellate phylogeny inferred from eukaryotic large subunit rRNA (28S rRNA) sequences (SI Appendix, Fig. S11) appeared to be inconsistent with the scenario deduced from the plastid phylogeny. In the host phylogeny deduced from 28S rRNA sequences, MGD or TGD showed no particular affinity to any other dinoflagellates, while L. chlorophorum was nested within a robustly supported clade of dinoflagellates with peridinin-containing plastids (e.g., Gymnodinium and Nematodinium). The relationship among the three dinoflagellates with Pedinophyceae-derived plastids was much better resolved in a phylogenetic analysis of 75 proteins than that of the 28S rRNA sequences. In the 75-protein phylogeny, the three species were separated by robustly supported nodes (Fig. 5). We examined the relationship among L. chlorophorum, MGD, and TGD by subjecting the ML tree, wherein the three species are paraphyletic, and 15 alternative trees to an approximately unbiased test (70). In the alternative trees, all or subsets of the three species were forced to be monophyletic (SI Appendix, Fig. S12). Significantly, all of the 15 alternative trees were rejected with very small P values. Thus, we can conclude that the host lineages of the three species are highly likely to be paraphyletic.
Fig. 5.
ML tree inferred from a 75-protein alignment. The topology in question was emphasized. The 75-protein alignment contains 21,042 amino acid positions. The tree topology was generated by a ML analysis with the LG + Γ + F + C60 model. Nonparametric ML bootstrap support values (shown above the nodes) were calculated from 100 replicates with the LG + Γ + F + PMSF model. The nodes highlighted by closed dots were supported fully by nonparametric bootstrap analyses. Only bootstrap support values greater than 70% are shown.
The inconsistency between the host and plastid phylogenies demands three independent endosymbioses of Pedinophyceae green algae on the branches leading to Lepidodinium species, MGD, and TGD in dinoflagellate evolution. We currently cannot rationalize why separate dinoflagellate lineages engulfed very closely related Pedinophyceae green algae as endosymbionts. To answer the above question, we need to understand the interaction between dinoflagellates and Pedinophyceae at the genetic, physiological, and environmental levels.
Discussion
In this study, we reported previously undescribed dinoflagellates, strains MGD and TGD, both of which still retain the remnant intercellular structures of algal endosymbionts enclosing the nuclei and plastid, but no mitochondrion (Fig. 1). Multiple cases of EGT in MGD and TGD indicated that the endosymbiont-derived compartments had been integrated as organelles in the two dinoflagellates. Combining these facts, we regard MGD and TGD as nucleomorph-bearing organisms. Cryptophytes and chlorarachniophytes, have been investigated intensively as model organisms to depict organellogenesis that transformed an algal endosymbiont into a plastid (e.g., refs. 1, 8, 9, and 20). Curiously, three characteristics of the cryptophyte and chlorarachniophyte nucleomorphs were found in those of MGD and TGD (see below). Pioneering studies demonstrated that, in both cryptophytes and chlorarachniophytes: 1) the nucleomorph genomes are rich in house-keeping genes; 2) the GC% of the nucleomorph genomes are low, as seen in other reduced genomes such as plastid and mitochondrial genomes; and 3) nucleomorph genes tend to be transcribed more intensively than nuclear genes (8, 9, 13–22). Although no genome data are available for the nucleomorphs of MGD or TGD, the putative nucleomorph transcripts of the two dinoflagellates appeared to be rich in those encoding housekeeping proteins (SI Appendix, Fig. S5). We also noticed that nucleomorph genes were found to be transcribed more intensively than nuclear genes in both MGD and TGD (SI Appendix, Fig. S3). We suspect that the two characteristics in gene content and transcription shared among the nucleomorph genomes known to date might provide the critical clues to understand organellogenesis.
Besides the characteristics shared with cryptophytes and chlorarachniophytes (see above), MGD and TGD turned out to have characteristics that are not present in the two previously known nucleomorph-containing lineages. The phylogenetic analyses inferred from plastidal 16S and eukaryotic 18S rRNA sequences revealed that the plastid origins of MGD, TGD, and Lepidodinium spp. are the close relatives of a particular green algal genus, Pedinomonas (Fig. 4). In contrast, neither origin of the red alga engulfed by the ancestral cryptophyte nor that of the green alga engulfed by the ancestral chlorarachniophyte has been pinpointed to the genus level (18, 24–28). Thus, the modifications of the endosymbiont genomes during the transition from an endosymbiotic alga into the plastid can be predicted directly by comparing MGD/TGD nucleomorph genomes to those of free-living Pedinophycean green algae in the future.
The process of transferring endosymbiont genes to the host genome may have gone through a transitional state in which the particular genes co-occurred in both endosymbiont and host genomes. Nevertheless, Curtis et al. (20) proposed that both cryptophyte and chlorarachniophyte systems are in the post-EGT state, as no co-occurrence of a nucleomorph gene and its nuclear copy were detected. Nor have recent (i.e., lineage specific) EGTs been detected despite apparent examples of lineage specific nucleomorph gene loss (16, 17, 20). On top of that, no recent DNA influx from the endosymbiont to host in the cryptophyte or chlorarachniophyte system was apparent, since no nucleomorph or plastid genome copies (NUNMs or NUPTs) were found in the host genome (20). In contrast, we identified both nuclear and nucleomorph versions of psbO and rbcS genes in MGD, and those of petC in TGD (Fig. 3) (the three sets of genes showed phylogenetic affinities to the corresponding Pedinomonas homologs, suggesting that the nuclear versions were derived from the corresponding genes in the endosymbiont genome). Moreover, the MGD nuclear genome appeared to possess two psbO genes with distinct GC% (Fig. 3), which are likely the outcome of two separate EGT events. The co-occurrence of a nucleomorph gene and its nuclear copy in MGD and TGD suggest that EGT has yet to be completed in either of the two dinoflagellate systems. In this sense, MGD and TGD are excellent models to elucidate the detailed process of EGT during organellogenesis. It is also intriguing to search the evidence for DNA influx from the endosymbiont to the host in the two dinoflagellate systems. According to the limited transfer window hypothesis (20, 71, 72), NUPTs or NUNMs are unlikely to be abundant in the nuclear genomes of MGD or TGD, which have cells containing a single plastid associated with a single nucleomorph. The above question should be addressed by sequencing the nuclear, plastid, and nucleomorph genomes of both MGD and TGD in future studies.
A recent study proposed that the current plastids in Lepidodinium spp. were established more recently than the chlorarachniophyte plastids (73). Both Lepidodinium plastids and MGD/TGD plastids were derived from closely related Pedinophyceae green algae (Fig. 4), but the host phylogeny inferred from a 75-protein alignment clearly suggest that three independent endosymbioses of Pedinophyceae green algae gave rise to the current plastids in Lepidodinium spp., MGD, and TGD (Fig. 5 and SI Appendix, Fig. S12). Although a nucleus-like structure within the plastid in L. viride was reported previously (41), no clear structure such as those seen in in MGD and TGD were observed in our survey. The differences in intracellular structure between MGD/TGD and Lepidodinium spp. leads us to propose that the endosymbiosis in MGD/TGD was more recent than that in the ancestral Lepidodinium cell (or in the ancestral chlorarachniophyte cell). We suspect that 1) the clarity in plastid/nucleomorph origin and 2) traits corresponding to a transitional state of EGT (see above) found in MGD and TGD stem from their evolutionary youthfulness.
The present study reports the third and fourth nucleomorph-bearing organisms, dinoflagellate strains MGD and TGD, and unique in being discovered in the last 30 y. It is anticipated that these discoveries will shed light on the nature of endosymbiogenesis. The two dinoflagellates will be the foundation of future works that deepen our knowledge derived from the pioneering works on cryptophytes and chlorarachniophytes, which used to be the sole models to study the evolutionary process of transforming an algal endosymbiont into a plastid.
Materials and Methods
Strains and Culture Condition.
Two unique green dinoflagellate strains MGD and TGD were collected from coastal areas in Muroran, Hokkaido, Japan and Tsuruoka, Yamagata, Japan in September 2011, respectively. Single cells were isolated using a glass pipette under light microscopy. The strains were cultivated and have been maintained in half final concentration of Daigo IMK medium (Wako) with seawater at 20 °C under a light/dark cycle of 12/12 h.
Light and Fluorescent Microscopy.
Living cells were observed by Olympus IX71 inverted microscope (Olympus) with an Olympus DP71 CCD camera (Olympus). For fluorescent microscopic observation, the cells were fixed with the same volume of fixation buffer containing 2.5% glutaraldehyde, 2% paraformaldehyde, 0.2 M sucrose, and 0.1 M cacodylate at pH 7.0 for 5 min at room temperature. Fixed cells were rinsed with room temperature distilled water for 10 min three times, and were harvested by centrifugation at 2,810 × g for 10 min at 18 °C. The cells were put onto a glass coverslip coated with poly-l-Lysin (Wako), and left to stand for 30 min. DNA in the fixed cells was stained by 0.1% SYBR green I solution for 10 min at room temperature. After washing three times with distilled water for 10 min, the DNA-stained cells were mounted with ProLong Diamond Antifade Mountant (Life Technologies), and then observed using a Leica DMRD light and fluorescence microscope (Leica) with an Olympus DP73 CCD camera (Olympus).
Transmission Electron Microscopy.
The first fixation step was carried out as described above, then the cells were washed with 0.2 M cacodylate buffer. The second fixation was performed with buffer containing 1% OsO4 and 0.2 M cacodylate for 15 min at 4 °C. Cells were dehydrated in a series of baths with increasing ethanol concentrations, 10 min per bath, and were then embedded in low viscosity resin via three incubations with polypropylene oxide at room temperature over an hour. Embedded samples were polymerized for 14 h at 70 °C. Polymerized blocks were sectioned using a diamond knife and placed onto formvar-coated copper grids. Ultra-thin sections of the cells were stained with 2% uranyl acetate and lead citrate. Observations were carried out using a H7650 (Hitachi) microscope.
Plastid Isolation.
Living TGD cells were centrifuged for 10 min at 2,810 × g to make the cells burst. For MGD, the cells were put into a 25% final concentration of sucrose solution for 5 min. Then, four volumes of distilled water were added to make the cells burst by hypoosmotic shock. TGD and MGD cell pellets including the plastids freed from other cellular structures were harvested by centrifugation, and were fixed as described above. The freed plastids were sought and photos were taken under fluorescent microscopy and TEM.
Transcriptome Analyses and Protein Prediction.
Total RNAs of MGD and TGD cells harvested from monoclonal laboratory cultures were extracted using TRIzol reagent (Life Technologies). After enrichment of polyA-tailed mRNA molecules, RNA samples were reverse-transcribed and the cDNAs were ligated with adaptor primers. The cDNA libraries from MGD and TGD were then sequenced on a HiSeq 2000 instrument (Illumina); 286 and 411 million reads (paired-end) were generated from MGD and TGD libraries, respectively. Sequence reads with low sequencing quality were removed using FASTQ Trimmer and FASTQ Quality Filter programs included in the FASTX-toolkit package (http://hannonlab.cshl.edu/fastx_toolkit/). The remaining reads of MGD and TGD were then assembled into 286,124 and 393,513 transcript contigs using the TRINITY package (release 2013-02-25) (74, 75), respectively. All of the transcript contigs were subjected to blastx analysis against an in-house database of protein sequences retrieved from phylogenetically diverse organisms. ORFs encoding known proteins, identified by blastx analyses, were translated into amino acid sequences. Otherwise, the longest possible ORFs seen in individual transcripts were translated into amino acid sequences. The putative proteins predicted from the transcriptomic data were further analyzed as described below. All transcriptome data are available from the DNA Data Bank of Japan under BioProject PRJDB8237. The contig data of MGD and TGD are available from the Dryad Digital Repository (76).
Functional Annotation of the Proteins Predicted from MGD and TGD Transcriptomic Data.
The proteins predicted from MGD and TGD transcripts were roughly annotated by referring to the Kyoto Encyclopedia of Genes and Genomes (KEGG) database (77). First, a total of 193,301 proteins with KEGG orthology IDs (KO IDs) from 40 eukaryotic and 32 bacterial species were retrieved from the KEGG database. The protein sets predicted for MGD and TGD were subjected to blastp analyses against the retrieved KEGG proteins. Then the eukaryotic sequences retrieved from the KEGG database were subjected to blastp analysis in the opposite direction (i.e., blastp searches against the protein sets predicted from the MGD and TGD transcriptomes). If a particular MGD/TGD sequence and a KO ID matched in reciprocal blastp analyses, the KO ID was assigned to the sequence of interest. We assigned functional annotations to 57,983 and 73,589 of the putative proteins identified from MGD and TGD transcriptome data, respectively.
Evolutionary Origins of the Proteins Predicted from MGD and TGD Transcriptomic Data.
The phylogenetic origins of functionally annotated proteins were individually assessed as described below. Each of the MGD and TGD proteins with functional annotations was subjected to blastp analyses against a custom protein database containing genome-wide protein data from 129 phylogenetically diverse organisms (48 eukaryotic, 68 bacterial, and 13 archaeal species), and the proteins encoded in the plastid genome of a Pedinophyceae green alga, Pedinomonas minor (78), a free-living relative of the green algal endosymbionts engulfed by MGD and TGD (Discussion). We identified two sets of proteins, green algal proteins and alveolate proteins, for which the top five hits from blastp searches were occupied by sequences from members of Viridiplantae or those from alveolates, respectively. MGD and TGD proteins, whose top five blastp hits included matches to P. minor plastid genes, were designated as plastid genome-encoded proteins, and were not analyzed any further.
GC-Content and FPKM Calculation for Each Transcript.
GC-content for an entire sequence as well as for first, second, and third codon positions of each transcript was calculated using an in-house Ruby script. In this study, the green algal transcripts with >50% GC at first and >40% GC at third codon positions were designed as high-GC% green algal transcripts, and those with <60% at first and <40% at third codon positions were designed as low-GC% green algal transcripts.
To estimate expression levels for each transcript from MGD and TGD, we calculated relative abundances as FPKM. To calculate FPKM, RNA sequencing (RNA-seq) reads from MGD and TGD were separately aligned onto their respective transcriptome assembly using bowtie (79). FPKMs were then calculated using RSEM (80) based on the short-read alignments. We carried out these steps using a Perl script (align_and_estimate_abundance.pl) bundled with Trinity (v2.0.6) (74, 75, 80).
Estimation of N-Terminal Extensions in Green Algal Proteins.
Possible N-terminal extensions of the green algal proteins from MGD and TGD were determined based on blastp alignments. First, all of the green algal proteins from the two dinoflagellates were subjected to a blastp search against the custom protein database described above. For each green algal protein, we defined the mature protein region that is conserved among the top 10 hits from the blastp search. The N terminus of the putative mature protein region for each protein was set as an average of the start positions of the top 10 blast alignments (outliers detected by the Grubbs’ test were not used in the average calculation). In this way, the putative N-terminal extension of each protein corresponds to the amino acid sequence between the first methionine of the protein sequence and the estimated N terminus of the mature protein region. If no methionine was found upstream of a putative mature protein region, we concluded that that particular transcript most likely encoded an N terminus truncated protein and excluded it from the analysis described below. Finally, lengths of putative N-terminal extensions between green algal proteins encoded by high-GC% transcripts and those encoded by low-GC% transcripts (see above), were compared. Differences in the length of N-terminal extensions between the two groups of green algal proteins was tested by the Wilcoxon rank-sum test after removal of the outliers determined by the Grubbs’ test.
Confirmation of the SL Sequence at the 5′ Termini of MGD and TGD Transcripts.
Total RNA samples were extracted from the cultured MGD and TGD cells using TRIzol reagent (Life Technologies). Reverse-transcription and cDNA amplification were performed with the SMARTer PCR cDNA Synthesis Kit (Clontech Laboratories) according to the manufacturer’s instructions. DNA amplification was carried out as described below. The first PCR was performed with the cDNA sample as the template and a set of primers: An adaptor primer supplied in the kit mentioned above (5′ PCR Primer II A) and a transcript-specific primer. The thermal cycle was set as: 5 cycles consisting of 10 s at 98 °C and 1 min at 68 °C; 5 cycles consisting of 10 s at 98 °C, 20 s at 60 °C, and 1 min at 68 °C followed by 25 cycles consisting of 10 s at 98 °C, 20 s at 53 °C, 1 min at 68 °C. We conducted the second PCR with an SL sequence-specific primer (66) and another transcript-specific primer that was set in a nested position to the first primer. The amplicons of the first PCR were used as the template in the second PCR. The thermal cycle was set as: 5 cycles consisting of 10 s at 98 °C and 1 min at 68 °C; 5 cycles consisting of 10 s at 98 °C, 20 s at 60 °C, and 1 min at 68 °C followed by 25 cycles consisting of 10 s at 98 °C, 20 s at 53 °C, 1 min at 68 °C. The amplicons were sequenced by the Sanger method.
We also conducted an in silico survey of SL sequences at the 5′ termini of the transcripts reconstructed from the transcriptomic data of MGD and TGD. The 3′ portion of the SL sequence (12 nucleotides) was searched for within the first 50 nucleotides from the 5′ termini of all of the transcripts.
Phylogenetic Analyses of Ribosomal RNA Genes.
Eukaryotic small and large subunit ribosomal RNAs (rRNAs) (18S and 28S rRNA, respectively), and plastidial small-subunit ribosomal RNA (16S rRNA) sequences were amplified by standard PCR and then sequenced by the Sanger method. The determined sequences were separately aligned by the program MAFFT (81). After manual exclusion of ambiguously aligned positions, 1,658 positions and 82 taxa remained in the final 18S rRNA alignment, 732 positions and 78 taxa in the 28S rRNA alignment, and 1,232 positions and 66 taxa in the plastidal 16S rRNA alignment. ML phylogenetic analyses of the three alignments were done using RAxML v8.0.2 (82) under the GTR substitution model incorporating among-site rate variation approximated by a discrete gamma distribution with four categories. (For the 18S rRNA analysis, the proportion of invariant sites was also used to approximate among-site rate variation.) The ML trees were heuristically searched for 10 distinct maximum-parsimony trees, each of which was reconstructed by the random stepwise addition of sequences. One-thousand bootstrap replicates were generated from the 18S rRNA alignment, and analyzed with the rapid bootstrap method under the CAT model in RAxML. For the bootstrap analyses of both 28S rRNA and plastid 16S rRNA alignments, 100 replicates were generated, and individually subjected to tree searchs initiated from a single MP tree reconstructed by the random stepwise addition of sequences using RAxML.
The three alignments were also analyzed by a Bayesian method using the CAT-GTR + Γ model implemented in the program PHYLOBAYES v3.3 (83) with two independent chains. Markov chain Monte Carlo chains were run for 80,000 (18S rRNA), 80,000 (28S rRNA), and 78,000 (plastidal 16S rRNA) generations with a burn-in of 20,000 generations, respectively. We considered the two chains to have converged when the maximum difference value was less than 0.1. After “burn-in,” the consensus tree with branch lengths and BPPs were calculated from the rest of the sampled trees.
Phylogenetic Analyses of the 75-Protein Alignment.
A phylogenetic alignment comprising 75 protein sequences (SI Appendix, Table S1) was assembled by referring to a previously published multigene phylogenetic study (38). The sequences included in the 75-protein alignment were retrieved from the assembled data of MGD and TGD generated in this study, the National Center for Biotechnology Information GenBank database (https://www.ncbi.nlm.nih.gov/), and the reassembled data of the Marine Microbial Eukaryote Transcriptome Sequencing Project (https://github.com/dib-lab/dib-MMETSP) (84) by TBLASTN searches. We used the homologs in the organisms, for which high-quality genomes/transcriptomes are available, as the BLAST queries (e.g., Plasmodium falciparum, Chromera velia, and Thalassiosira pseudonana). Note than the sets of the BLAST queries included the sequences of a land plant Oyza sativa and a green alga Chlamydomonas reinhardtii to help identify and exclude the possible endosymbiont-derived (green algal) sequences in the MGD and TGD data. The retrieved sequences were aligned with MAFFT (81). After ambiguously aligned positions were removed, each single-gene alignment was subjected to an ML phylogenetic analysis to identify and remove extremely divergent sequences, as well as putative paralogous and contaminated sequences. In each of the single-gene ML analyses, the most appropriate substitution model was chosen. Finally, 75 single-gene alignments were concatenated into a single alignment with 42 taxa and 21,042 amino acid positions, and then analyzed by the ML method with the LG + Γ + F + C60 model to infer the ML tree with ultrafast bootstrap support (1,000 replicates). The statistical support for the bipartitions in the ML tree was also assessed by a nonparametric ML bootstrap analysis (100 replicates) with the LG + Γ + F + PMSF [posterior mean site frequency (85)] model. IQ-TREE (86) was used for all of the ML analyses described above. The 75-protein alignment, single-gene alignments, and a spreadsheet with calculated site coverages for the 75-protein alignment are available from Dryad Digital Repository (76).
Phylogenetic Analyses of Overlapping Green Algal Proteins.
Homologous sequences of three green algal proteins in MGD and TGD, namely PsbO, RbcS, and PetC, were retrieved from an in-house database comprising proteins sequences from phylogenetically diverse organisms by similarity searches using blastp software. Retrieved protein sequences and the homologous sequences from MGD and TGD were then aligned by MAFFT (81) with the L-INS-I method. Ambiguously aligned positions and gaps were removed from the final PsbO, RbcS, and PetC alignments, which comprise 35 sequences with 254 amino acid positions, 35 sequences with 100 amino acid positions, and 36 sequences with 178 amino acid positions, respectively. ML trees were inferred from the three alignments by IQ-TREE (86) with nonparametric bootstrapping (100 replicates) under the LG4X substitution model. The three alignments are available from the Dryad Digital Repository (76).
Phylogenetic Analyses of the Enzymes Involved in Heme, Chl-a, and IPP Biosynthetic Pathways.
We retrieved the sequences encoding the 9, 6, and 7 nucleus-encoded enzymes involved in C5 pathway for the heme biosysnthesis, and Chl-a, and IPP biosynthetic pathways, respectively, from the MGD and TGD transcriptomic data. The identified sequences were translated into amino acid sequences and added to the corresponding alignments previously generated by some of us (56). The alignments, updated by adding the MGD and TGD homologs (22 in total), were subjected to ML phylogenetic analyses to classify individual enzymes into 1) VI-type, 2) “endosymbiotically acquired (EA)-type,” or 3) LA-type (see above). We labeled the sequences, which we were unable to categorize into any of the three types with confidence as “uncertain origin” (note that this category can contain contaminated sequences). The single-gene alignments and ML tree with bootstrap support values are available from Dryad Digital Repository (76).
Approximately Unbiased Test.
To assess alternative relationships among L. chlorophorum, MGD, and TGD in the 75-protein phylogeny (Fig. 5), we pruned and regrafted one or two of the branches of interest in the ML tree to generate alternative trees with 1) the monophyly of MGD and TGD, 2) the monophyly of L. chlorophorum and MGD, 3) the monophyly of L. chlorophorum and TGD, and 4) the monophyly of the three species. The ML tree, in which the three species were paraphyletic, and the 15 alternative trees were subjected to an approximately unbiased test (70). For each test tree, site-wise log-likelihoods were calculated by IQ-TREE with the LG + Γ + F + C60 model. The resulting site-wise log-likelihood data were subsequently analyzed by Consel v0.1 with default settings (87).
Pigment Analysis.
For L. chlorophorum, we used strain NIES-1868 deposited in the National Institute for Environmental Studies (NIES) culture collection. MGD, TGD, and L. chlorophorum cells were harvested by centrifugation. After removal of a transparent viscous layer above the cell pellet, pigments were extracted with 100 µL of 100% methanol, and the pigment extract was collected into a 1.5-mL centrifuge tube after centrifugation. We repeated the extraction until the extract was no longer colored. The extracted pigments were subjected to reverse-phase HPLCy with a Waters Symmetry C8 column (150 mm × 4.6 mm; particle size 3.5 µm; pore size 100 Å). The HPLC was performed as described in Zapata et al. (88) without any modifications. The eluted pigments were detected by their absorbance at 440 nm and identified by their elution patterns compared to those reported in Zapata et al. (88).
Supplementary Material
Acknowledgments
The authors thank Dr. Bruce A. Curtis (Dalhousie University, Canada) for proofreading an early draft of this manuscript. This work was supported in part by the Japan Society for the Promotion of Sciences Grants 23117006, 16H04826, 18KK0203, and 19H03280 (to Y.I.), 25304029 and 15H04533 (to M.I.), 17H03723 and 26840123 (to G.T.) 14J05929 (to C.S.), 17K15164 (to T.N.), and 19H03274 (to R.K.); Ministry of Education, Culture, Sports, Science and Technology of Japan Grant-in-Aid for Scientific Research on Innovative Areas 3308; a research grant from The Yanmar Environmental Sustainability Support Association (to R.K.); and the “Tree of Life” research project of the University of Tsukuba. Phylogenetic analyses of the 75-protein alignment were carried out under the “Interdisciplinary Computational Science Program” in the Center for Computational Sciences, University of Tsukuba.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The transcriptome data from the dinoflagellate strains MGD and TGD are available from the DNA Data Bank Japan (BioProject PRJDB8237).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911884117/-/DCSupplemental.
References
- 1.Archibald J. M., The puzzle of plastid evolution. Curr. Biol. 19, R81–R88 (2009). [DOI] [PubMed] [Google Scholar]
- 2.Zimorski V., Ku C., Martin W. F., Gould S. B., Endosymbiotic theory for organelle origins. Curr. Opin. Microbiol. 22, 38–48 (2014). [DOI] [PubMed] [Google Scholar]
- 3.Archibald J. M., Endosymbiosis and eukaryotic cell evolution. Curr. Biol. 25, R911–R921 (2015). [DOI] [PubMed] [Google Scholar]
- 4.Nowack E. C. M., Weber A. P. M., Genomics-informed insights into endosymbiotic organelle evolution in photosynthetic eukaryotes. Annu. Rev. Plant Biol. 69, 51–84 (2018). [DOI] [PubMed] [Google Scholar]
- 5.Martin W., et al. , Evolutionary analysis of Arabidopsis, cyanobacterial, and chloroplast genomes reveals plastid phylogeny and thousands of cyanobacterial genes in the nucleus. Proc. Natl. Acad. Sci. U.S.A. 99, 12246–12251 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.McCutcheon J. P., Moran N. A., Extreme genome reduction in symbiotic bacteria. Nat. Rev. Microbiol. 10, 13–26 (2011). [DOI] [PubMed] [Google Scholar]
- 7.Smith D. R., Unparalleled GC content in the plastid DNA of Selaginella. Plant Mol. Biol. 71, 627–639 (2009). [DOI] [PubMed] [Google Scholar]
- 8.Tanifuji G., Archibald J. M., “Nucleomorph comparative genomics” in Endosymbiosis, Löffelhardt W., Ed. (Springer, 2014), pp. 197–213. [Google Scholar]
- 9.Tanifuji G., Onodera N. T., “Cryptomonads: A model organism sheds light on the evolutionary history of genome reorganization in secondary endosymbioses” in Advances in Botanical Research, Hirakawa Y., Ed. (Academic Press, 2017), pp. 263–320. [Google Scholar]
- 10.Timmis J. N., Ayliffe M. A., Huang C. Y., Martin W., Endosymbiotic gene transfer: Organelle genomes forge eukaryotic chromosomes. Nat. Rev. Genet. 5, 123–135 (2004). [DOI] [PubMed] [Google Scholar]
- 11.Greenwood A., “The Cryptophyta in relation to phylogeny and photosynthesis” in 8th International Congress of Electron Microscopy, Sanders J., Goodchild D., Eds. (Australian Academy of Sciences, Canberra, 1974), pp. 566–567. [Google Scholar]
- 12.Hibberd D. J., Norris R. E., Cytology and ultrastructure of Chlorarachnion reptans (Chlorarachniophyta Divisio Nova, Chlorarachniophyceae Classis Nova). J. Phycol. 20, 310–330 (1984). [Google Scholar]
- 13.Douglas S., et al. , The highly reduced genome of an enslaved algal nucleus. Nature 410, 1091–1096 (2001). [DOI] [PubMed] [Google Scholar]
- 14.Gilson P. R., et al. , Complete nucleotide sequence of the chlorarachniophyte nucleomorph: Nature’s smallest nucleus. Proc. Natl. Acad. Sci. U.S.A. 103, 9566–9571 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lane C. E., et al. , Nucleomorph genome of Hemiselmis andersenii reveals complete intron loss and compaction as a driver of protein structure and function. Proc. Natl. Acad. Sci. U.S.A. 104, 19908–19913 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Tanifuji G., et al. , Complete nucleomorph genome sequence of the nonphotosynthetic alga Cryptomonas paramecium reveals a core nucleomorph gene set. Genome Biol. Evol. 3, 44–54 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Moore C. E., Curtis B., Mills T., Tanifuji G., Archibald J. M., Nucleomorph genome sequence of the cryptophyte alga Chroomonas mesostigmatica CCMP1168 reveals lineage-specific gene loss and genome complexity. Genome Biol. Evol. 4, 1162–1175 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tanifuji G., et al. , Nucleomorph and plastid genome sequences of the chlorarachniophyte Lotharella oceanica: Convergent reductive evolution and frequent recombination in nucleomorph-bearing algae. BMC Genomics 15, 374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Suzuki S., Shirato S., Hirakawa Y., Ishida K., Nucleomorph genome sequences of two chlorarachniophytes, Amorphochlora amoebiformis and Lotharella vacuolata. Genome Biol. Evol. 7, 1533–1545 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Curtis B. A., et al. , Algal genomes reveal evolutionary mosaicism and the fate of nucleomorphs. Nature 492, 59–65 (2012). [DOI] [PubMed] [Google Scholar]
- 21.Tanifuji G., Onodera N. T., Moore C. E., Archibald J. M., Reduced nuclear genomes maintain high gene transcription levels. Mol. Biol. Evol. 31, 625–635 (2014). [DOI] [PubMed] [Google Scholar]
- 22.Hirakawa Y., Suzuki S., Archibald J. M., Keeling P. J., Ishida K., Overexpression of molecular chaperone genes in nucleomorph genomes. Mol. Biol. Evol. 31, 1437–1443 (2014). [DOI] [PubMed] [Google Scholar]
- 23.Hirakawa Y., Nagamune K., Ishida K., Protein targeting into secondary plastids of chlorarachniophytes. Proc. Natl. Acad. Sci. U.S.A. 106, 12820–12825 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Douglas S. E., Penny S. L., The plastid genome of the cryptophyte alga, Guillardia theta: Complete sequence and conserved synteny groups confirm its common ancestry with red algae. J. Mol. Evol. 48, 236–244 (1999). [DOI] [PubMed] [Google Scholar]
- 25.Ishida K., Cao Y., Hasegawa M., Okada N., Hara Y., The origin of chlorarachniophyte plastids, as inferred from phylogenetic comparisons of amino acid sequences of EF-Tu. J. Mol. Evol. 45, 682–687 (1997). [DOI] [PubMed] [Google Scholar]
- 26.McFadden G. I., Gilson P. R., Waller R. F., Molecular phylogeny of chlorarachniophytes based on plastid ribosomal-RNA and rbcL sequences. Arch. Protistenkd. 145, 231–239 (1995). [Google Scholar]
- 27.Rogers M. B., Gilson P. R., Su V., McFadden G. I., Keeling P. J., The complete chloroplast genome of the chlorarachniophyte Bigelowiella natans: Evidence for independent origins of chlorarachniophyte and euglenid secondary endosymbionts. Mol. Biol. Evol. 24, 54–62 (2007). [DOI] [PubMed] [Google Scholar]
- 28.Suzuki S., Hirakawa Y., Kofuji R., Sugita M., Ishida K. I., Plastid genome sequences of Gymnochlora stellata, Lotharella vacuolata, and Partenskyella glossopodia reveal remarkable structural conservation among chlorarachniophyte species. J. Plant Res. 129, 581–590 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Archibald J. M., Genomic perspectives on the birth and spread of plastids. Proc. Natl. Acad. Sci. U.S.A. 112, 10147–10153 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kim J. I., et al. , Evolutionary dynamics of Cryptophyte plastid genomes. Genome Biol. Evol. 9, 1859–1872 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Muñoz-Gómez S. A., et al. , The new red algal subphylum Proteorhodophytina comprises the largest and most divergent plastid genomes known. Curr. Biol. 27, 1677–1684.e4 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Stiller J. W., et al. , The evolution of photosynthesis in chromist algae through serial endosymbioses. Nat. Commun. 5, 5764 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Hackett J. D., Anderson D. M., Erdner D. L., Bhattacharya D., Dinoflagellates: A remarkable evolutionary experiment. Am. J. Bot. 91, 1523–1534 (2004). [DOI] [PubMed] [Google Scholar]
- 34.Taylor F. J. R., Hoppenrath M., Saldarriaga J. F., Dinoflagellate diversity and distribution. Biodivers. Conserv. 17, 407–418 (2008). [Google Scholar]
- 35.Janouškovec J., Horák A., Oborník M., Lukeš J., Keeling P. J., A common red algal origin of the apicomplexan, dinoflagellate, and heterokont plastids. Proc. Natl. Acad. Sci. U.S.A. 107, 10949–10954 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Zapata M., Fraga S., Rodríguez F., Garrido J. L., Pigment-based chloroplast types in dinoflagellates. Mar. Ecol. Prog. Ser. 465, 33–52 (2012). [Google Scholar]
- 37.Schnepf E., Elbrächter M., Dinophyte chloroplasts and phylogeny: A review. Grana 38, 81–97 (1999). [Google Scholar]
- 38.Janouškovec J., et al. , Major transitions in dinoflagellate evolution unveiled by phylotranscriptomics. Proc. Natl. Acad. Sci. U.S.A. 114, E171–E180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Saldarriaga J. F., Taylor F. J. R., Keeling P. J., Cavalier-Smith T., Dinoflagellate nuclear SSU rRNA phylogeny suggests multiple plastid losses and replacements. J. Mol. Evol. 53, 204–213 (2001). [DOI] [PubMed] [Google Scholar]
- 40.Bjørnland T., Haxo F. T., Liaaen-Jensen S., Carotenoids of the Florida red tide dinoflagellate Karenia brevis. Biochem. Syst. Ecol. 31, 1147–1162 (2003). [Google Scholar]
- 41.Watanabe M. M., et al. , A green dinoflagellate with chlorophylls a and b: Morphology, fine structure of the chloroplast and chlorophyll composition. J. Phycol. 23, 382–389 (1987). [Google Scholar]
- 42.Watanabe M. M., Suda S., Inouye I., Sawaguchi T., Chihara M., Lepidodinium viride gen. et sp. nov. (Gymnodiniales, Dinophyta), a green dinoflagellate with a chlorophyll a- and b-containing endosymbiont. J. Phycol. 26, 741–751 (1990). [Google Scholar]
- 43.Matsumoto T., Kawachi M., Miyashita H., Inagaki Y., Prasinoxanthin is absent in the green-colored dinoflagellate Lepidodinium chlorophorum strain NIES-1868: Pigment composition and 18S rRNA phylogeny. J. Plant Res. 125, 705–711 (2012). [DOI] [PubMed] [Google Scholar]
- 44.Horiguchi T., Takano Y., Serial replacement of a diatom endosymbiont in the marine dinoflagellate Peridinium quinquecorne (Peridiniales, Dinophyceae). Phycol. Res. 54, 193–200 (2006). [Google Scholar]
- 45.Matsumoto T., et al. , Green-colored plastids in the dinoflagellate genus Lepidodinium are of core chlorophyte origin. Protist 162, 268–276 (2011). [DOI] [PubMed] [Google Scholar]
- 46.Takishita K., Nakano K., Uchida A., Preliminary phylogenetic analysis of plastid-encoded genes from an anomalously pigmented dinoflagellate Gymnodinium mikimotoi (Gymnodiniales, Dinophyta). Phycol. Res. 47, 257–262 (1999). [Google Scholar]
- 47.Takishita K., et al. , Origins of plastids and glyceraldehyde-3-phosphate dehydrogenase genes in the green-colored dinoflagellate Lepidodinium chlorophorum. Gene 410, 26–36 (2008). [DOI] [PubMed] [Google Scholar]
- 48.Tengs T., et al. , Phylogenetic analyses indicate that the 19’ hexanoyloxy-fucoxanthin-containing dinoflagellates have tertiary plastids of haptophyte origin. Mol. Biol. Evol. 17, 718–729 (2000). [DOI] [PubMed] [Google Scholar]
- 49.Yamada N., Sym S. D., Horiguchi T., Identification of highly divergent diatom-derived chloroplasts in dinoflagellates, including a description of Durinskia kwazulunatalensis sp. nov. (Peridiniales, Dinophyceae). Mol. Biol. Evol. 34, 1335–1351 (2017). [DOI] [PubMed] [Google Scholar]
- 50.Takishita K., Ishida K., Maruyama T., Phylogeny of nuclear-encoded plastid-targeted GAPDH gene supports separate origins for the peridinin- and the fucoxanthin derivative-containing plastids of dinoflagellates. Protist 155, 447–458 (2004). [DOI] [PubMed] [Google Scholar]
- 51.Minge M. A., et al. , A phylogenetic mosaic plastid proteome and unusual plastid-targeting signals in the green-colored dinoflagellate Lepidodinium chlorophorum. BMC Evol. Biol. 10, 191 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Nosenko T., et al. , Chimeric plastid proteome in the Florida “red tide” dinoflagellate Karenia brevis. Mol. Biol. Evol. 23, 2026–2038 (2006). [DOI] [PubMed] [Google Scholar]
- 53.Patron N. J., Waller R. F., Keeling P. J., A tertiary plastid uses genes from two endosymbionts. J. Mol. Biol. 357, 1373–1382 (2006). [DOI] [PubMed] [Google Scholar]
- 54.Burki F., et al. , Endosymbiotic gene transfer in tertiary plastid-containing dinoflagellates. Eukaryot. Cell 13, 246–255 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bentlage B., Rogers T. S., Bachvaroff T. R., Delwiche C. F., Complex ancestries of isoprenoid synthesis in dinoflagellates. J. Eukaryot. Microbiol. 63, 123–137 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Matsuo E., Inagaki Y., Patterns in evolutionary origins of heme, chlorophyll a and isopentenyl diphosphate biosynthetic pathways suggest non-photosynthetic periods prior to plastid replacements in dinoflagellates. PeerJ 6, e5345 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Jeffrey S. W., Vesk M., Further evidence for a membrane-bound endosymbiont within the dinoflagellate Perisinium foliaceum. J. Phycol. 12, 450–455 (1976). [Google Scholar]
- 58.Tamura M., Shimada S., Horiguchi T., Galeidinium rugatum gen. et sp. nov. (Dinophyceae), a new coccoid dinoflagellate with a diatom endosymbiont. J. Phycol. 41, 658–671 (2005). [Google Scholar]
- 59.Tomas R. N., Cox E. R., Steidinger K. A., Peridinium balticum (Levander) Lemmermann, an unusual dinoflagellate with a mesokaryotic and an eucaryotic nucleus. J. Phycol. 9, 91–98 (1973). [Google Scholar]
- 60.Imanian B., Keeling P. J., The dinoflagellates Durinskia baltica and Kryptoperidinium foliaceum retain functionally overlapping mitochondria from two evolutionarily distinct lineages. BMC Evol. Biol. 7, 172 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Imanian B., Pombert J. F., Dorrell R. G., Keeling P. J., A typically unusual dinoflagellate mitochondrial genome and an unusually typical diatom mitochondrial genome constitute the mitochondrial genomes of “dinotoms”. J. Phycol. 47, S16 (2011). [Google Scholar]
- 62.Imanian B., Pombert J. F., Dorrell R. G., Burki F., Keeling P. J., Tertiary endosymbiosis in two dinotoms has generated little change in the mitochondrial genomes of their dinoflagellate hosts and diatom endosymbionts. PLoS One 7, e43763 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Hehenberger E., Imanian B., Burki F., Keeling P. J., Evidence for the retention of two evolutionary distinct plastids in dinoflagellates with diatom endosymbionts. Genome Biol. Evol. 6, 2321–2334 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Hehenberger E., Burki F., Kolisko M., Keeling P. J., Functional relationship between a dinoflagellate host and its diatom endosymbiont. Mol. Biol. Evol. 33, 2376–2390 (2016). [DOI] [PubMed] [Google Scholar]
- 65.Trapnell C., et al. , Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat. Biotechnol. 28, 511–515 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Zhang H., et al. , Spliced leader RNA trans-splicing in dinoflagellates. Proc. Natl. Acad. Sci. U.S.A. 104, 4618–4623 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Larkum A. W., Lockhart P. J., Howe C. J., Shopping for plastids. Trends Plant Sci. 12, 189–195 (2007). [DOI] [PubMed] [Google Scholar]
- 68.Bolte K., et al. , Protein targeting into secondary plastids. J. Eukaryot. Microbiol. 56, 9–15 (2009). [DOI] [PubMed] [Google Scholar]
- 69.Kamikawa R., et al. , Plastid genome-based phylogeny pinpointed the origin of the green-colored plastid in the dinoflagellate Lepidodinium chlorophorum. Genome Biol. Evol. 7, 1133–1140 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Shimodaira H., An approximately unbiased test of phylogenetic tree selection. Syst. Biol. 51, 492–508 (2002). [DOI] [PubMed] [Google Scholar]
- 71.Smith D. R., Crosby K., Lee R. W., Correlation between nuclear plastid DNA abundance and plastid number supports the limited transfer window hypothesis. Genome Biol. Evol. 3, 365–371 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Barbrook A. C., Howe C. J., Purton S., Why are plastid genomes retained in non-photosynthetic organisms? Trends Plant Sci. 11, 101–108 (2006). [DOI] [PubMed] [Google Scholar]
- 73.Jackson C., Knoll A. H., Chan C. X., Verbruggen H., Plastid phylogenomics with broad taxon sampling further elucidates the distinct evolutionary origins and timing of secondary green plastids. Sci. Rep. 8, 1523 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Grabherr M. G., et al. , Full-length transcriptome assembly from RNA-Seq data without a reference genome. Nat. Biotechnol. 29, 644–652 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Haas B. J., et al. , De novo transcript sequence reconstruction from RNA-seq using the Trinity platform for reference generation and analysis. Nat. Protoc. 8, 1494–1512 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Inagaki Y., et al. , Supplementary data for Dinoflagellates with relic endosymbiont nuclei as models for elucidating organellogenesis. Dryad, Dataset. 10.5061/dryad.k6djh9w38. Deposited 18 December 2019. [DOI] [PMC free article] [PubMed]
- 77.Kanehisa M., et al. , Data, information, knowledge and principle: Back to metabolism in KEGG. Nucleic Acids Res. 42, D199–D205 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Turmel M., Gagnon M. C., O’Kelly C. J., Otis C., Lemieux C., The chloroplast genomes of the green algae Pyramimonas, Monomastix, and Pycnococcus shed new light on the evolutionary history of prasinophytes and the origin of the secondary chloroplasts of euglenids. Mol. Biol. Evol. 26, 631–648 (2009). [DOI] [PubMed] [Google Scholar]
- 79.Langmead B., Aligning short sequencing reads with Bowtie. Curr. Protoc. Bioinformatics. Chapter 11, Unit 11.7 (2010). [DOI] [PMC free article] [PubMed]
- 80.Li B., Dewey C. N., RSEM: Accurate transcript quantification from RNA-seq data with or without a reference genome. BMC Bioinformatics 12, 323 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Katoh K., Standley D. M., MAFFT multiple sequence alignment software version 7: Improvements in performance and usability. Mol. Biol. Evol. 30, 772–780 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Stamatakis A., RAxML-VI-HPC: Maximum likelihood-based phylogenetic analyses with thousands of taxa and mixed models. Bioinformatics 22, 2688–2690 (2006). [DOI] [PubMed] [Google Scholar]
- 83.Lartillot N., Lepage T., Blanquart S., PhyloBayes 3: A Bayesian software package for phylogenetic reconstruction and molecular dating. Bioinformatics 25, 2286–2288 (2009). [DOI] [PubMed] [Google Scholar]
- 84.Johnson L. K., Alexander H., Brown C. T., Re-assembly, quality evaluation, and annotation of 678 microbial eukaryotic reference transcriptomes. Gigascience 8, giy158 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Wang H. C., Minh B. Q., Susko E., Roger A. J., Modeling site heterogeneity with posterior mean site frequency profiles accelerates accurate phylogenomic estimation. Syst. Biol. 67, 216–235 (2018). [DOI] [PubMed] [Google Scholar]
- 86.Nguyen L. T., Schmidt H. A., von Haeseler A., Minh B. Q., IQ-TREE: A fast and effective stochastic algorithm for estimating maximum-likelihood phylogenies. Mol. Biol. Evol. 32, 268–274 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Shimodaira H., Hasegawa M., CONSEL: For assessing the confidence of phylogenetic tree selection. Bioinformatics 17, 1246–1247 (2001). [DOI] [PubMed] [Google Scholar]
- 88.Zapata M., Rodríguez F., Garrido J. L., Separation of chlorophylls and carotenoids from marine phytoplankton: A new HPLC method using a reversed phase C8 column and pyridine-containing mobile phases. Mar. Ecol. Prog. Ser. 195, 29–45 (2000). [Google Scholar]
- 89.Adl S. M., et al. , Revisions to the classification, nomenclature, and diversity of eukaryotes. J. Eukaryot. Microbiol. 66, 4–119 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.