Significance
The first enzymes emerged ∼4 billion y ago and have subsequently become the most diverse and functionally important component of life. But what were the first enzymes doing and how did they look? We probed the properties of the first enzymes by analyzing phospho-ligand binding across all known protein evolutionary lineages. We find that phospho-ligand binding was the founding function of the most ancient enzymes. As opposed to younger evolutionary lineages, ancient enzymes preferentially use N termini of α-helices to bind phosphate moieties. The dominance of N-helix binding sites in the earliest enzymes reflects the ability of the α-helix to realize binding via short and simple sequences, including serines and threonines that interact via both the backbone and side chain.
Keywords: protein evolution, P-loop, Rossmann, phosphate, ECOD
Abstract
The ubiquity of phospho-ligands suggests that phosphate binding emerged at the earliest stage of protein evolution. To evaluate this hypothesis and unravel its details, we identified all phosphate-binding protein lineages in the Evolutionary Classification of Protein Domains database. We found at least 250 independent evolutionary lineages that bind small molecule cofactors and metabolites with phosphate moieties. For many lineages, phosphate binding emerged later as a niche functionality, but for the oldest protein lineages, phosphate binding was the founding function. Across some 4 billion y of protein evolution, side-chain binding, in which the phosphate moiety does not interact with the backbone at all, emerged most frequently. However, in the oldest lineages, and most characteristically in αβα sandwich enzyme domains, N-helix binding sites dominate, where the phosphate moiety sits atop the N terminus of an α-helix. This discrepancy is explained by the observation that N-helix binding is uniquely realized by short, contiguous sequences with reduced amino acid diversity, foremost Gly, Ser, and Thr. The latter two amino acids preferentially interact with both the backbone amide and the side-chain hydroxyl (bidentate interaction) to promote binding by short sequences. We conclude that the first αβα sandwich domains emerged from shorter and simpler polypeptides that bound phospho-ligands via N-helix sites.
Phosphate esters are the building blocks of extant life: Not only do phosphates bridge nucleotides in DNA and RNA and store energy in ATP, but phosphate is also the most common moiety in metabolites and present in nearly all key enzyme cosubstrates and cofactors (1). Accordingly, phosphate moieties are important handles for protein binding. Indeed, proteins that bind phosphate-containing ligands (“phospho-ligands”) are exceptionally common in the Protein Data Bank (PDB) (2, 3). Given the ubiquity of phosphate in biology, it is reasonable to postulate that phosphate binding was among the first protein functions to emerge. In this scenario, peptides initially served as auxiliaries to polynucleic acids and phosphonucleoside cofactors, and were later extended (via duplications and fusions) and further diverged to yield modern proteins, including enzymes that utilize phosphonucleoside cofactors (4–6). The ubiquity of phospho-ligand binding proteins is undoubtedly related to a subset of ancient and widely spread protein classes that happen to bind phospho-ligands, such as the Rossmann and P-loop NTPases. The latter, for example, can comprise up to 18% of the ORFs in a given genome (7). However, what remains unknown is whether these early emergences of phosphate binding were unique events, or if phosphate binding has emerged, independently, time and again, across ∼4 billion y of evolution. In other words, how frequently has phosphate binding emerged, and in which protein lineages?
At the structural level, multiple reports have addressed phosphate binding in terms of the mode of binding, including backbone conformations (3, 8–10). However, the hypothesis that backbone-based binding—specifically loops at the N termini of α-helices—dominates phosphate binding (11) is inevitably biased by the ubiquitous Rossmann and P-loop NTPase families. Alternative solutions to phosphate binding certainly exist, and may in fact be more common given a normalized analysis. For example, SAMHD1, a dNTP hydrolase, sculpts a phosphate-binding site entirely out of side chains, primarily Arg and Lys (12). Also, even between families with ostensibly similar binding modes, significant variations exist. While Rossmann and P-loop NTPases both realize phosphate binding at the N terminus of the first α-helix in the canonical three-layer αβα sandwich architecture, Rossmann relies on a bridging water molecule to facilitate binding (13), whereas P-loop NTPases do not. Phosphate binding in relation to catalysis is no different: While P-Loop NTPases typically recruit an Mg2+ ion to assist in phosphate binding, phosphotyrosine phosphatases, which also adopt the three-layer αβα sandwich architecture, are metal-independent. Here, a systematic evolutionary classification can map all emergences of phosphate-binding sites within independently evolved protein families, and address their structural features. For example, do N-helix binding sites, reminiscent of P-loop NTPases, preferentially emerge? Or, do binding sites formed from positively charged side chains dominate, as in the case of SAMDH1?
We surmised that evolutionary classification can also identify the trends and patterns that govern the emergence of phosphate-binding proteins. The key to an evolutionary analysis is grouping related proteins: In other words, identifying and analyzing independently emerged protein lineages. Here, we describe an “evolutionarily normalized” analysis of phosphate binding using the Evolutionary Classification of Protein Domains (ECOD) database (14). ECOD uses sequence and structure information, as well as manual curation (15), to classify protein domains, and updates every week to include new structures deposited in the PDB. By grouping related binding events together, ECOD explicitly handles redundancy in the PBD. The broadest classification level in ECOD is the X-group. X-groups correspond, in principle, to discrete events of evolutionary emergence, and no detectable sequence homology or identity of fold exists between X-groups. Conversely, F-groups, the narrowest level of classification, represent different families that diverged from a common X-group ancestor. F-groups belonging to the same X-group share the same fold, yet the sequence identity between them can be low, sometimes only at the level of short motifs.
Analyzing phosphate binding in terms of independent evolutionary lineages therefore bypasses redundancy in the PDB database, and allows key questions about phosphate binding in the protein universe to be tackled: How many independent evolutionary lineages (i.e., X-groups) bind phospho-ligands, and what fraction of all known protein lineages do these represent? For which of these lineages was phosphate binding a driver of emergence, rather than a niche functionality that was acquired at a later evolutionary stage? Which mode of phosphate binding emerged most frequently? Is the N-helix binding mode the dominant binding mode, overall, or perhaps only in certain folds? Finally and most importantly, can we deduce which phosphate-binding mode arose first, and why?
Methods
Phospho-Ligand Selection.
The 100 most-abundant small-molecule phospho-ligands in the PDB were identified using the lig_pairs.lst file downloaded from PDBsum (16). To avoid overestimation of ligand binding diversity, synonymous ligands (i.e., ligands that can bind to the same protein site, such as those related by oxidation state [e.g., NAD+ and NADH] or nonhydrolyzable analogs [e.g., ATP and its imido-derivatives]) were merged. Similarly, triphosophate, diphosphate, and monophosphate forms of the same nucleotide were merged, as these ligands frequently represent reactant-product pairs. The list of ligands used in this study, as well as the rules for ligand merging, can be found in Dataset S1.
Protein Structure Selection.
Crystal structures with a resolution threshold ≤3 Å and containing any of the 100 most-common phospho-ligands were downloaded from the PDB using BioPython (17) and XML on February 8, 2019. The PDB structures considered in the present analysis per each ligand are listed in Dataset S2.
Hydrogen Bonding Criteria.
Hydrogen bond identification followed previously established protocols (18). In principle, the structure of the phosphate anion and its various esters (hereafter referred to as phospho-ligands) consists of sp3 hybridized oxygen atoms and sp2 hybridized oxygen atoms. However, due to resonance effects in inorganic phosphate, as well as in phosphomonoesters and diesters, we treated all phosphate oxygens as being sp3 hybridized for simplicity. Depending on the substituent or ionization state, phosphate can act as either a hydrogen bond donor or acceptor (note that protonated oxygens, like substituted oxygens, are sp3 hybridized). The phosphate moiety was first treated as an acceptor, as this is by far the most common state in solution. However, for unsubstituted oxygens, if hydrogen bond angle criteria (below) were not satisfied, and the potential interaction partner can act as a hydrogen bond acceptor, angle criteria with phosphate as a hydrogen bond donor were tested. Carboxylic acids within 3.5 Å of a metal ion were assumed to interact predominantly with the metal, and were excluded. All interactions with the nonphosphate parts of the ligand were ignored. The distance and angle criteria for hydrogen bonding are as follows (see Fig. 1 for a graphical description): 1) The distance between the donor and acceptor atoms is ≤3.5 Å; 2) the acceptor angle formed by the acceptor anchor atom (e.g., P for P-O−), the acceptor atom (e.g., O for P-O−), and the donor atom (e.g., N for H-N) is between 60° and 180° for sp3 acceptors or between 90° and 180° for sp2 acceptors; and 3) the donor angle formed by the donor anchor atom (e.g., Cα for backbone amides), the donor atom (e.g., the backbone nitrogen), and the acceptor atom (e.g., O for –O-P) is between 90° and 180°, regardless of the donor hybridization state. Anchor atoms were defined as in Stickle et al. (18). Backbone amides and carbonyls were treated as sp2 donors and acceptors, respectively. Hydrogen bond-donating side chains were either sp2 (Arg [NH1, NH2, NE], His [ND1, NE2], Trp [NE1], Asn [ND2], Gln [NE2], and Tyr [OH]) or sp3 (Lys [NZ], Ser [OG], Thr [OG1], and Cys [SG]), while hydrogen bond accepting side chains were either sp2 (His [ND1, NE2], Asn [OD1], Gln [OE1], Asp [OD1, OD2], Glu [OE1, OE2], and Tyr [OH]) or sp3 (Ser [OG], Thr [OG1], Met [SD], and Cys [SG]). The analysis script for hydrogen bond detection is available upon request.
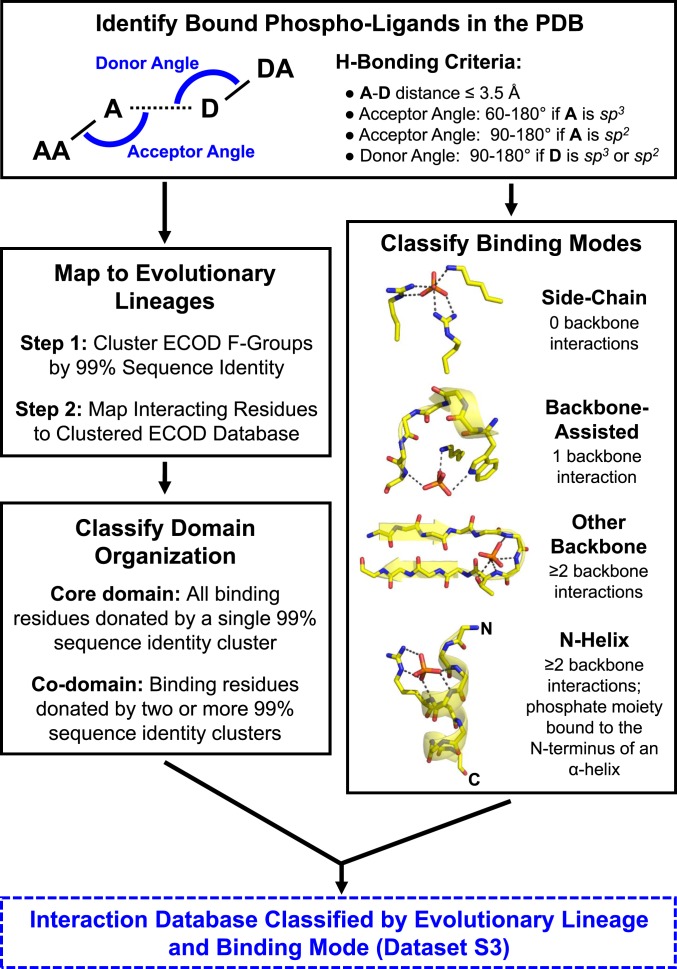
Fig. 1.
The phosphate-binding analysis pipeline (see Methods for additional details). Briefly, crystal structures in which any of the 100 most-common phospho-ligands, as well as phosphate and pyrophosphate, are bound were collected from the PDB (2). Interactions between the proteins and the phosphate moieties of these phospho-ligands were enumerated and used to classify the phosphate-binding mode. Four binding modes were defined, based on the extent that backbone amides participate in phosphate binding and whether the binding site is positioned at the N terminus of an α-helix. Interacting residues were also mapped to domains in the ECOD database. Subsequent analyses are therefore based on counting ECOD families (F-groups, which represent closely related structures) and independent evolutionary lineages (X-groups) rather than on individual PDB entries or binding events.
Classification of Interaction Type and Binding Mode.
Interactions between phosphate moieties (i.e., orthophosphate, pyrophosphate, and the phosphate moiety of a phospho-ligand) and protein residues were classified as follows: 1) A side-chain interaction occurs when only the side chain of a residue interacts with the phosphate moiety; 2) an “other” backbone interaction occurs when a backbone amide acts as a hydrogen bond donor, but the interacting residue is not at the N terminus of an α-helix; and 3) a backbone N-helix interaction occurs when the interacting amide belongs to a residue located at the N terminus of an α-helix. To assign backbone N-helix interactions, the following procedure was applied: First, all α-helices in a structure were identified using the STRIDE algorithm (19) as implemented in VMD v.1.9.4a9 (20) and the index of the first residue of each helix was retrieved. For a given phosphate moiety, if any of the phosphate oxygen atoms are within 4 Å of the amide nitrogen of the first helix residue and 6 Å of the amide nitrogen of the third helix residue (different phosphate oxygen atoms can satisfy these two criteria), the phosphate moiety was considered to be positioned above the N terminus of an α-helix. These distance criteria were validated by manual inspection, examining >10% of structures in which a phospho-ligand sits atop an α-helix. For phospho-ligands positioned above an α-helix, hydrogen bonds to the backbone amide of any of the first four helix residues were classified as a backbone N-helix interaction. In addition to α-helices, 310 helices were considered. However, these structural elements are comparatively rare, >25 times less common than α-helices, and were thus not analyzed further. Finally, we also noted the existence of bidentate interactions, when both the side chain and the backbone amide of the same amino acid interact with the phosphate moiety (or moieties) of the ligand. Bidentate interactions, for the purposes of binding mode classification, were considered as backbone interactions (i.e., other backbone interaction or backbone N-helix interaction, as appropriate). The analysis script for N-helix binding is available upon request.
Binding modes were hierarchically classified according to the types of interactions, described above, that comprise them (Fig. 1). Note that binding modes apply to the entire ligand binding site, across all phosphate moieties of the ligand. Binding events without any backbone interactions were classified as adopting a “side-chain” binding mode. Binding events with a single backbone interaction were classified as adopting a “backbone-assisted” binding mode. Binding events with at least two backbone interactions, none of which were donated by the N terminus of an α-helix, were classified as adopting an “other backbone” binding mode. Finally, binding events with at least two backbone interactions, and at least one of which donated by a residue at the N terminus of an α-helix, were classified as adopting an “N-helix” binding mode.
Evolutionary Analysis.
Interacting residues were mapped to their corresponding evolutionary lineages (X-groups, F-groups) using the ECOD database (14) version develop255, accessed on December 8, 2019 (http://prodata.swmed.edu/ecod/). By definition, structures belonging to different X-groups are nonredundant, as they relate to sequences with no detectable sequence homology (14). Related proteins from different F-groups within the same X-group have no more than 80% sequence identity. To manage redundancy within F-groups, domains were clustered by CD-HIT (21) using a 99% sequence identity cutoff. By clustering domains within an F-group, as opposed to choosing representative structures, all binding information (e.g., ligands, binding modes, domain organizations) is preserved. A database of ECOD-mapped, binding mode-classified binding events is available in Dataset S3. The domain organization of each binding event was also classified: If the phosphate moiety only interacts with a single 99% sequence identity cluster, the interacting domain is said to be a “core domain.” If multiple 99% sequence identity clusters interact with either the same phosphate moiety, or different phosphate moieties on the same ligand, all associated domains are classified as “codomains” for that binding event.
Binding Cutoffs.
To identify bona fide phospho-ligand binding sites, two criteria were considered: First, the “interaction cutoff” is the minimal number of interacting residues across all phosphate moieties of the ligand that comprise a bona fide binding event. Note that because many residues make more than one hydrogen bond, the number of hydrogen bonds between the protein and the phosphate moiety is greater than or equal to the interaction cutoff. Three residues is an appropriate threshold for defining bona fide binding events, as fewer interacting residues often (but not always) indicates nonspecific interactions. For example, only 79% of phospho-ligand binding events to the Rossmann-like X-group have three or more interacting residues (SI Appendix, Fig. S1). For all main-text figures, unless explicitly stated otherwise, an interaction cutoff of three residues was used. Analyses with alternative interaction cutoffs are provided in the SI Appendix and demonstrate the robustness of the data and our conclusions to changes in the interaction cutoff. Next, the “instance cutoff” is how many nonredundant binding events (i.e., 99% identity clusters) we observe between a phospho-ligand and a given domain (depending on context, defined as either an X-group or an F-group). Throughout the analysis, 99% identity clusters are only counted once unless they are nonredundant with respect to the property being analyzed, in which case the diversity is preserved. For example, see Fig. 3A, which displays the binding mode distribution across the PDB; if domains belonging to the same 99% sequence identity cluster bind in two different binding modes, both modes were counted. Critically, the number of X-groups identified as being phosphate binders, and the distribution of binding modes, is relatively robust to changes in either of the cutoffs (SI Appendix, Figs. S2, S3, and S5).
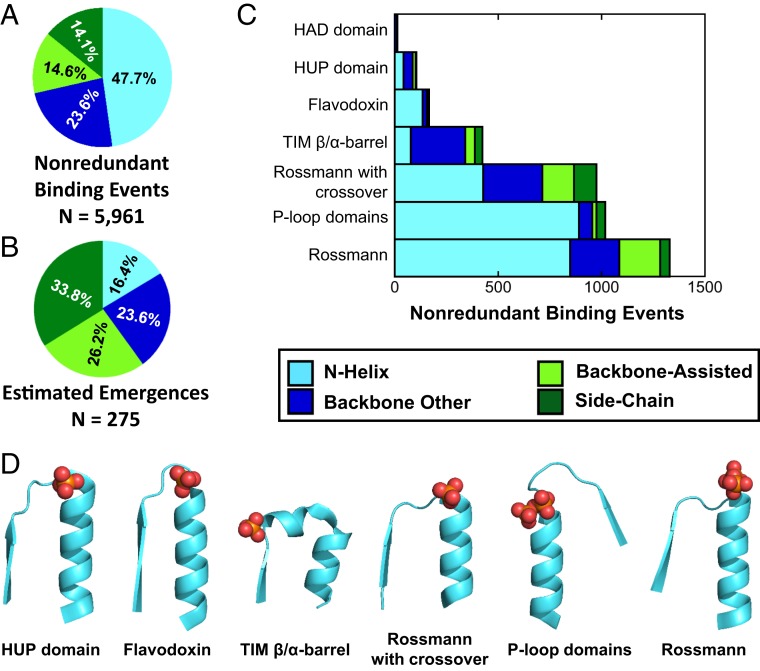
Fig. 3.
N-helix binding mode dominates the PDB but not evolutionary emergences. (A) The distribution of phospho-ligand binding modes in the PDB. About half of core phospho-ligand binding events adopt the N-helix binding mode (interaction cutoff = 3; alternative cutoffs yielded highly similar results) (SI Appendix, Fig. S5A). (B) The estimated number of emergences of each binding mode in the context of a core domain using an interaction cutoff of 3 and an instance cutoff of 1 (estimated emergences across a range of interaction cutoffs were highly similar; see SI Appendix, Fig. S5B). In contrast to the PDB dataset, along 4 billon y of evolution, the side-chain binding mode emerged most readily, approximately twice as frequently as the N-helix mode. (C) The distribution of phospho-ligand binding modes for the ancient phosphate binders indicates the dominance of the N-helix binding mode (the TIM β/α-barrel is an exception due to the unique features of its N-helix binding site; see main text for more details). The overrepresentation of the ancient binders in the PDB also explains the dominance of N-helix binding in the PDB (A). (D) Canonical N-helix binding modes for each of the ancient phosphate-binding X-groups. Most of the ancient phosphate binders use a simple strand-loop-helix motif to bind phosphate, while the TIM β/α-barrel incorporates an additional short helix oriented to interact with ligands bound in its central tunnel. PDB identifiers, from left to right: 1J21, 3OJW, 6BVE, 1N1M, 1KYI, 4WNI.
Finally, two datasets were used for analysis: One in which both phospho-ligands and inorganic phosphate (orthophosphate, or phosphate, and pyrophosphate) were considered, and another in which only phospho-ligands were considered. Note that inorganic phosphate can be the actual ligand, or may occupy a bona fide phospho-ligand binding sites even if it is not the true ligand. As such, inorganic phosphate may compensate for instances when a structure with the natural ligand is not available or for rare phospho-ligands that were not included in our ligand dataset. Thus, for estimation of the number of X-groups that bind phospho-ligands, inorganic phosphate is considered. However, because inorganic phosphate is an imperfect mimic of a phospho-ligand—which can have up to six phosphate moieties, as in inositol hexa-phosphate—inorganic phosphate was not included in analyses of binding mode and domain organization.
Estimation of Binding Mode Emergences.
The estimated number of emergences for each binding mode was calculated by counting the number of X-groups that utilize that binding mode, subject to instance and interaction cutoffs. As binding sites that use the backbone-assisted and side-chain binding modes tend to be less well-conserved between F-groups than other backbone and N-helix binding modes, this enumeration likely undercounts side-chain and backbone-assisted binding emergences.
Sulfate Analysis.
Structures with free sulfate and a resolution threshold ≤ 3 Å were downloaded from the PDB, as above. Hydrogen bond criteria and binding mode classification were applied as for phospho-ligands. Coincidental binding events were defined as having precisely three binding interactions (justification provided below).
Data Availability Statement.
All data discussed in this paper will be made available to readers upon request.
Results and Discussion
Phospho-Ligand Binding Emerged in 11 to 18% of Known Evolutionary Lineages.
How many times has phosphate binding emerged independently across ∼4 billion y of evolution? There are currently 2,344 X-groups in ECOD. Of these, by a relatively conservative estimate (i.e., interaction cutoff = 3, instance cutoff = 2) (Methods and SI Appendix, Fig. S2), 251 X-groups, or ∼11% of evolutionary lineages, bind phosphate moieties in the context of a small-molecule phospho-ligand or inorganic phosphate. Relaxing the criteria (instance cutoff = 1) yields 411 X-groups, or 18% of evolutionary lineages. Indeed, the assignment of structures as phospho-ligand binders, and the number of independent lineages, varies with the interaction and instance cutoffs, but the cutoffs used here are robust, as elaborated in SI Appendix, Fig. S2. Thus, phosphate binding is highly abundant also given an evolutionarily normalized analysis. Furthermore, enough data exist to estimate more detailed properties, such as which binding mode emerges most readily and why.
Phosphate Binding: A Founding or Niche Function?
Duplication and divergence is the mechanism that dominates the birth of new genes and families (22, 23). Hence, the prevalence of phosphate-binding proteins may reflect a more intensive divergence of these X-groups to yield large superfamilies of related proteins, rather than the existence of many independent evolutionary lineages that bind phosphate. Divergence within a lineage is reflected by the number of families, or F-groups, in a particular X-group. Indeed, phosphate-binding X-groups systematically show a higher number of F-groups compared to those X-groups where phosphate binding was not identified (Fig. 2A). However, X-groups with the greatest diversity are also more likely to display any given function, including phosphate binding. In principle, if a small fraction of F-groups bind phosphate, the progenitor likely had another function, and phosphate binding emerged later as a niche function. Conversely, if the majority of descendent F-groups show phosphate binding, then phosphate binding was present in its last common ancestor and was likely the driver of emergence of this X-group.
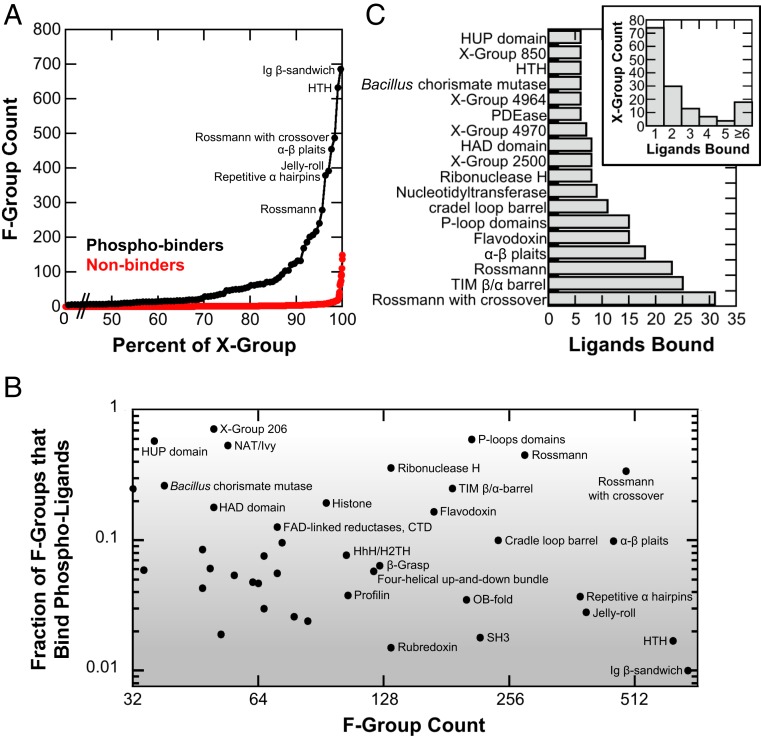
Fig. 2.
Phosphate binding: Founding or niche function? Only phospho-ligand binders were considered for this analysis (as either a core domain or a codomain); the interaction cutoff was 3. For X-groups, the instance cutoff was set to 2; for F-groups, the instance cutoff was set to 1, due to their smaller size. (A) A cumulative distribution of the F-group count for X-groups that bind phospho-ligands versus those that do not. (B) The fraction of F-groups that interact with at least one phospho-ligand (Fp). The upper right hand corner of the plot represents X-groups with a high total number of F-groups, as well as a relatively high fraction of F-groups that bind phospho-ligands. Phosphate binding is likely the founding function of those X-groups, which have also enjoyed a relatively long evolutionary history. For example, the P-loop domains-like X-group is a fundamental phosphate binder, where the “P” stands for phosphate and phospho-ligand binding is a hallmark functionality. On the other end (bottom right), the niche functionality scenario is represented by Ig-like β-sandwich, a highly diverse X-group where only about 1% of the F-groups are phospho-ligand binders. Fp is robust to the interaction cutoff (SI Appendix, Fig. S3A). (C) The number of different phospho-ligands that bind a given X-group. Instance cutoff = 2; both core domain and codomain binding events considered. While 87% of X-groups bind fewer than six phospho-ligands, most of the fundamental phosphate binder X-groups bind more than six different phospho-ligands. Ligand binding statistics are robust to the interaction cutoff (SI Appendix, Fig. S3B).
To distinguish between these two scenarios, founding or niche function, the fraction of F-groups that bind phosphate (Fp) was calculated for each X-group. A caveat of this analysis is that it cannot reveal the role of phosphate binding in X-groups with relatively few F-groups. Accordingly, plotted are only X-groups with ≥32 F-groups, which represents the top 28% of the F-group distribution for phosphate binders (Fig. 2B).
As demonstrated in Fig. 2B, niche phosphate binding is present among the most functionally diverse X-groups. Despite having hundreds of F-groups, these X-groups have only a few phosphate-binding F-groups (Fp ∼ 0.01; bottom-right corner of the plot in Fig. 2B), best exemplified by IgG-like β-sandwiches (X-group 11).
The Fundamental Phosphate Binders.
Conversely, at the top of the plot in Fig. 2B are X-groups with relatively high values of Fp, indicating that phosphate binding was likely a driver of emergence and a founding function. As expected, both P-loop domains-like (X-group 2004; Fp = 0.60) and Rossmann-like (X-group 2003; Fp = 0.45) are “fundamental phosphate binders.” Note that even for these two protein classes, for which emergence is undisputedly the result of phosphate-binding functionality (5, 7, 24), the Fp value may drop below 0.5. This reflects the ancient character (6, 25, 26) and the concomitant functional diversity of these X-groups.
Overall, 10 X-groups exhibit phosphate binding in ≥20% of F-groups (Fp ≥ 0.20; the top 24% of the Fp value distribution for X-groups with ≥32 F-groups). Among these X-groups, five are also exceptionally diverse, comprised of more than 100 F-groups, including P-loop domains-like (X-group 2004) and Rossmann-like (X-group 2003) as expected; followed by, Ribonuclease H-like (X-group 2484), other Rossmann structures with crossover (X-group 2111), and TIM β/α-barrel (X-group 2002). Indeed, these X-groups have long been associated with phospho-ligand binding (7), and phosphate binding seems to not only be the founding function, but also a driver of divergence, as reflected in the high number of F-groups. Other X-groups exhibiting high Fp values yet with fewer F-groups are NAT/Ivy (X-group 213; primarily associated with CoA utilizing enzymes) and X-group 206 [which includes ATP-grasp enzymes (27)], HUP domain-like [X-group 2005; which includes aminoacyl tRNA synthetases (28)], Bacillus chorismate mutase-like (X-group 301), and X-group 212 (which includes the second domain of ribosomal protein S5). Phosphate binding was undoubtedly the founding function of these X-groups as well.
Next in terms of divergence (0.10 < Fp < 0.20) are classes, such as Flavodoxin-like (X-group 2007) and FAD-linked reductase (X-group 244), both associated with utilizing phosphate-containing cofactors, and HAD domains-like (X-group 2006; a superfamily of enzymes, primarily phosphatases). Phosphate binding was likely present in the founding ancestor of these X-groups, although this hypothesis is not unequivocally supported by our analysis. Perhaps an interesting addition to this group are Histone-like domains (X-group 148), which serve predominantly as an accessory domain to Rossmann-like domains. In the midlow range of Fp values (∼0.01 < Fp < ∼0.10) we note the appearance of three X-groups generally associated with nucleic acid binding: OB-fold (X-group 2), HTH (X-group 101), and HhH/H2TH (X-group 102). While the OB-fold binds thiamidine diphosphate as a competitive inhibitor (29), HTH forms a dimer that binds two intercalated cyclic-di-GMP molecules in a mode highly reminiscent of nucleic acid binding. These examples suggest that phospho-ligand binding and nucleic acid binding may be readily traversable functions.
Finally, another manifestation of divergence is the diversity of phospho-ligands that an X-group binds. Approximately 87% of X-groups bind fewer than six different phospho-ligands (Fig. 2C, Inset). In contrast, many of the X-groups for which phosphate binding is the founding function bind six or more phospho-ligands. We also note that α-β plaits exhibit relatively high phospho-ligand diversity and a moderate Fp close to 0.1 (Fig. 2B). For some X-groups, like HAD domains-like, the phospho-ligands may be highly diverse, with a wide range of phospho-ester substrates (30) that are scarcely represented in the PDB and hence excluded from our analysis.
Overall, it appears that phosphate binding has emerged as both a founding and niche function. X-groups where phosphate binding was the founding function are exceptionally diverse (Fig. 2 B and C), suggesting that phosphate binding is a driver of both emergence and divergence. Phosphate is, after all, the most abundant moiety in natural metabolites (1). However, as discussed in the next section, the exceptional diversity of these fundamental phosphate-binding protein classes also relates to their ancient character, likely predating the last universal common ancestor (LUCA).
The Fundamental Phosphate Binders Are Also the Most Ancient Enzymes.
As it turns out, several of the X-groups for which phosphate binding was likely a driver of emergence are also considered to be among the most ancient protein lineages, certainly among those lineages that gave rise to enzymes [other ancient lineages include ribosomal proteins (31) and iron-sulfur proteins (4, 32)]. The most notable examples of this are P-loop domains-like, Rossmann-like, and other Rossmann structures with crossover (7, 26, 27, 33, 34). Other ancient protein lineages include the TIM β/α-barrel, HUP domains, Flavodoxin, and HAD domains X-groups. Indeed, all of these proteins are α/β proteins, which are thought to be generally older than other protein classes (35). In fact, all but one of these adopt the three-layer αβα sandwich architecture, which has been consistently identified as the oldest architecture (25, 26) or among the oldest (36). The exception, the TIM β/α-barrel, was also identified as among the oldest folds, and recent evidence suggests that the β/α-barrel and the three-layer αβα sandwich folds are evolutionarily related (37). We henceforth refer to these lineages (X-groups) as the “ancient phosphate binders” (P-loop domains-like, Rossmann-like, other Rossmann structures with crossover, Flavodoxin-like, TIM β/α-barrel, HUP-domains-like, and HAD domains-like), with an appreciation that P-loop domains-like, Rossmann-like, and other Rossmann structures with crossover likely predate the other members of the group.
Furthermore, these seven X-groups comprise domains that bind predominantly as core domains. That is, binding occurs without the involvement of additional, auxiliary domains (also known as cap domains) (SI Appendix, Fig. S4), as would be expected for ancient proteins that emerged before multidomain arrangements were feasible. Taking these data together, it seems that phospho-ligand binding likely drove the emergence of the very first αβα sandwich proteins. By analyzing the mode by which the ancient phosphate binders interact with phosphate, we may be able to deduce features of the very first phosphate-binding sites).
N-Helix Was the First Phosphate Binding Mode.
We now turn our focus to the structural details of how phospho-ligands are bound. To this end, four binding modes were defined: 1) Side-chain, in which the interactions with the phosphate are solely with side chains; 2) backbone-assisted, in which only one residue binds the phosphate via a hydrogen bond to a backbone amide, and the remaining interactions are with side chains; 3) other backbone, in which two or more backbone amides bind the phosphate; and 4) N-helix, in which two or more backbone amide hydrogen bonds are detected (as in other backbone) and the phosphate is situated at the N terminus of an α-helix (see Methods and Fig. 1 for additional details).
A simple count of nonredundant core domain phospho-ligand binding events across the entire PDB reveals that the N-helix binding mode is the most common (Fig. 3A and SI Appendix, Fig. S5A). However, the distribution of core domain phospho-ligand binding modes per independent emergence (i.e., normalized per X-group) shows a different picture; the side-chain mode dominates (Fig. 3B and SI Appendix, Fig. S5B). This discrepancy relates to the fact that the ancient phosphate binders, which are highly diverse and widely represented, exhibit a strong preference for the N-helix binding mode (Fig. 3C). Accordingly, excluding the ancient phosphate binders from consideration shifts the dominant binding mode to side-chain (31% of phospho-ligand binding events, with N-helix comprising 22%) in a simple count of nonredundant binding events in the PDB. Ligand-specific effects, however, are minor: Most ligands are bound by multiple modes and, as in the global emergence analysis, appearance of the side-chain binding mode is more common than the N-helix binding mode (SI Appendix, Fig. S6).
For four of the seven ancient phosphate binders, the N-helix binding mode is strongly preferred and only the HAD domain lacks N-helix binding events altogether. An example of the canonical N-helix binding mode is given for each of these six ancient folds (Fig. 3D). For the N-helix binders, the phosphate-binding α-helix is typically the first canonical helix in the architecture (24). For TIM β/α-barrel, the so-called standard phosphate-binding motif is traditionally described as residing between the seventh and eighth β-stands (38, 39), although earlier works have noted its helical nature (11). Manual inspection confirms participation of an α-helix in the TIM standard phosphate-binding site, as exemplified in Fig. 3D. Our automated analysis, however, systematically assigned TIM β/α-barrel binding events as adopting either the other backbone or backbone-assisted binding modes. This misassignment is because the participating helix is often short, at times distorted, and frequently only makes a single interaction with the ligand. Furthermore, the TIM β/α-barrel helix is sometimes used to bind a carboxylic acid moiety of the ligand in lieu of phosphate. Taking these data together, we reason that the N-helix binding mode of the TIM β/α-barrel is both fundamental and ancient. Finally, additional support for the observation that the N-helix mode dominates the ancient phosphate binders is the fact that these X-groups are also most frequently associated with axillary domains; consequently, emergences of codomain binding sites are skewed for N-helix relative to the core domain dataset (SI Appendix, Fig. S7).
The fact that many of the ancient phosphate binders preferentially employ an N-helix binding mode—most notably P-loop domains-like, Rossmann-like, and other Rossmann structures with crossover—suggests that this binding mode predates the others. But why should most of the ancient phosphate binders share the same binding mode at all? One possibility is that the N-helix binding mode is most likely to evolve, and hence the shared N-helix binding mode is the outcome of convergence. However, the analysis of the relative number of emergences per each binding mode (Fig. 3B) rejects this hypothesis outright. Instead, the side-chain binding mode enjoys privileged emergence, with about two times more emergences than the N-helix binding mode. Furthermore, the N-helix binding mode, which is fundamentally a backbone binding mode, has fewer emergence events than either the other backbone or backbone-assisted binding modes. The αβα sandwich itself does not appear to be limiting the space of possible solutions either, as all four types of binding modes have been realized in this architecture (Fig. 3C). In this respect, the HAD-like domain is illustrative: Despite being an ancient phosphate binder, and αβα sandwich, binding is achieved almost exclusively in the other backbone binding mode. Restriction endonuclease-like domains (X-group 2008, a αβα sandwich protein) offers another example, as the backbone-assisted binding mode is preferred in this lineage. Overall, it appears that the dominance of N-helix binding in the most ancient enzymes is unlikely to be because the N-helix binding mode is the only, or even the most preferred, mode of phosphate binding.
If dominance of the N-helix mode is not due to convergence, perhaps these six lineages, which share the αβα sandwich architecture [or relate to it, in the case of TIM β/α-barrels (37)], have diverged from a common ancestor that possessed an N-helix phosphate-binding site. Another possibility is that the oft-ignored hand of chance is to blame. While neither of these explanations can be ruled out, there is evidence in support of a third hypothesis: The N termini of α-helices are hotspots for phosphate binding, but only under restrictions related to the emergence of the earliest protein forms (i.e., convergence due to ancient constraints on protein structure).
Short, Simple Sequences Favor N-Helix Binding.
Modern proteins surely differ from their nascent precursors in at least two respects: Sequence length and amino acid composition. At the onset of protein evolution, emergence from shorter sequences was more likely (orders-of-magnitude smaller sequence space to explore) as well as necessary (4, 36, 40, 41). Second, the composition of the first proteins was likely biased for those amino acids that were freely available in the surrounding environment. The subset of amino acids that are readily produced by abiotic chemistry (reviewed in refs. 42–44) are referred to as prebiotic amino acids, and include Gly, Ala, Ser, Thr, Asp, Glu, Val, Leu, Ile, and Pro. Notably, this set of amino acids lacks Arg and Lys (45), both of which commonly mediate phosphate binding in modern proteins (as they dominate the side-chain binding mode, which is the most frequent mode of emergence) (Fig. 3B). Might the N termini of α-helices comprise hotspots for phosphate binding given short and simple sequences? How could this hypothesis be tested?
We reasoned that coincidental binding can be used to assess the above hypothesis, as coincidental, promiscuous binding represents an evolutionary potential, in this case the propensity of certain protein elements to bind phosphate. Thus, preferential coincidental binding at the N terminus of α-helices would indicate that it is a hotspot for the emergence of phosphate-binding sites. Phosphate is generally avoided in crystallization buffers (due to its poor solubility profile), and when present in a structure, it often occupies a bonafide phosphate-binding site. Sulfate, on the other hand, is widely used in crystallography as a precipitant, has negative charge and tetrahedral geometry, and may thus serve as a surrogate for phosphate-binding potential [many phosphatases, for example, are promiscuous sulfatases (46)]. We thus examined all sulfate binding events in the PDB. Sulfate interacts with fewer protein residues, on average, than either phosphate or phospho-ligands, consistent with the interpretation that in the vast majority of structures sulfate is promiscuously bound (SI Appendix, Fig. S8). Given that, only sulfate anions that interact with exactly three residues belonging to the same protein chain were considered, as fewer interacting residues may not be sufficient to indicate a potential binding site, and greater than three interacting residues is more likely to reflect an evolved phosphate-binding site.
Overall, we detected 5,232 nonredundant sulfate binding events bound by a core domain and involving three residues belonging to the same chain. Of these potential sites for emergence, 13.2% adopt an N-helix binding mode, the least-common binding mode observed (Fig. 4A), as with emergences of phospho-ligand binding sites (Fig. 3B). The overlap between these two datasets, the evolved phosphate-binding sites on the one hand (Fig. 3B) and the potential to evolve such sites as reflected by promiscuous sulfate binding on the other (Fig. 4A), shows that given a large repertoire of full-size protein domains, N-helix binding is not the most likely mode of emergence of phosphate binding.
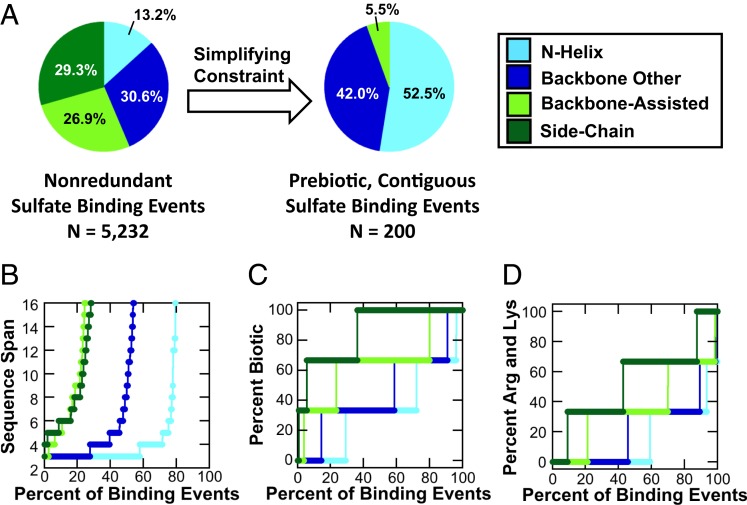
Fig. 4.
Patterns of coincidental, promiscuous sulfate binding. Overall, 5,232 core, nonredundant sulfate binding events in which three residues belonging to the same chain bind the sulfate ion were detected in the PDB. (A) When all of these sulfate binding events are considered, the N-helix binding mode is the least common (consistent with emergences across all X-groups) (Fig. 3B). However, when only short, contiguous sequences with prebiotic amino acid composition are considered, the N-helix binding mode becomes the preferred solution. (B) A cumulative distribution of all sulfate binding events that interact with a single chain indicates that N-helix binding sites are realized with shorter sequence spans compared to any other binding mode: Nearly 60% of binding events are comprised of three consecutive residues (i.e., sequence span = 3; all sulfate binding events included). (C) N-helix binding sites are preferentially realized with prebiotic sequences when all single-chain sulfate binding events are considered. The prebiotic amino acids were taken to be Gly, Ala, Ser, Thr, Asp, Glu, Val, Leu, Ile, and Pro. (D) The basic amino acids Arg and Lys are near-essential for side-chain binding (present in ∼90% of all binding events) but not for N-helix binding (present in ∼40% of all binding events).
However, once sequence length (Fig. 4B) and amino acid composition (Fig. 4 C and D) are taken into consideration, the picture changes dramatically. Short sequences strongly favor the N-helix binding mode, with ∼60% of binding events realized from a contiguous stretch of residues (span = 3). Similarly, prebiotic amino acid composition also favors the N-helix binding mode, with ∼30% of binding sites comprised entirely of prebiotic amino acids. In contrast, side-chain binding, which is overall the most preferred mode of emergence, is almost never formed from stretches of contiguous residues and is absolutely dependent on the nonprebiotic amino acids, particularly Arg and Lys (Fig. 4D), which mediate ∼90% of binding events. When only binding sites comprised of a contiguous stretch of prebiotic amino acids are considered, the N-helix binding mode is the dominant mode (52.5% of potential sites of emergence) (Fig. 4A). The overwhelming preference for the N-helix binding mode in the ancient phosphate binders (Fig. 3B and 4D) may therefore be a signature of the constraints in play at the time of their emergence.
Thr and Ser: Seeds for Phosphate Binding.
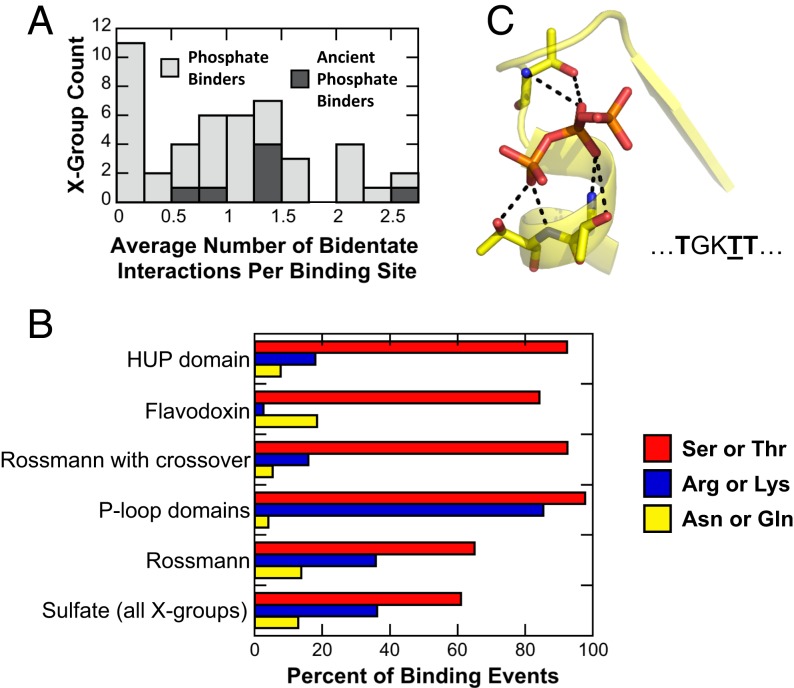
If phosphate binding initially arose from short and prebiotic sequences, devoid of basic amino acids, residues that can make multiple interactions would be strongly favored, if not critical. Specifically, bidentate interactions, in which both the side chain and the backbone amide hydrogen bond with the phospho-ligand, would be advantageous. By encoding two interactions sites within a single residue, bidentate interactions reduce the entropic cost of binding while also enabling emergence within short sequences. Indeed, we found widespread use of bidentate interactions, which is further enriched in the ancient phosphate binders (Fig. 5A); P-loop domains-like are the most distinct, making on average ∼2.5 bidentate interactions per binding event.
Fig. 5.
Bidentate interactions in phosphate binding. (A) The frequency of bidentate interactions across all X-groups. Only the N-helix mode was analyzed, as this is presumably the most ancient mode of phosphate binding. To calculate the average number of bidentate interactions per binding event for an entire X-group, the average value of each 99% sequence identity cluster within that X-group was calculated first, and then these values were averaged. The majority of X-groups make use of bidentate interactions in their binding sites. However, the ancient phosphate binders (dark shading) preferentially use bidentate interactions, with P-loop domains-like being the most extensive user (an average of ∼2.5 bidentate interactions per binding site). (B) Amino acid usage for bidentate interactions in the N-helix binding mode; only binding events with at least one bidentate interaction considered. Two prebiotic amino acids, Thr and Ser, are essential for bidentate interactions in the N-helix binding mode. In the ancient phosphate binders using the N-helix binding mode, Thr or Ser are present in 65 to 98% of nonredundant binding events that utilize at least one bidentate interaction. Basic amino acids, on the other hand, are less frequent, or even rarely employed (as in Flavodoxin and other Rossmann structures with crossover). (C) An example of the dominance of bidentate interactions in the N-helix binding mode. Shown is an enzyme belonging to the P-loop domains-like X-group, which uses three bidentate interactions per binding site [PDB ID code 1XKV (55)]. The sequence of the contiguous stretch of residues that form this ATP biding site is shown beneath the structure, with interacting Thr residues rendered in bold and the canonical Walker A Thr residue underlined.
Arg and Lys dominate the side-chain binding mode—the most common binding mode when all emergences are considered (Fig. 3B)—but do they also dominate bidentate interactions? Considering only the ancient phosphate binders, amino acid usage statistics were calculated for the binding of phospho-ligands in the N-helix mode (Fig. 5B). Thr and Ser, both prebiotic amino acids, are preferred for bidentate interactions in the ancient phosphate binders, present in 65 to 98% of nonredundant binding events that have at least one bidentate interaction and bind in the N-helix binding mode. The preference for Ser/Thr over Arg/Lys in bidentate interactions may reflect their greater overall frequency in proteins or their comparatively rigid side chains. Forming bidentate interactions, at least in the N-helix binding mode, does not enforce unfavorable rotamers, not even for the more mobile Arg and Lys side chains. The most common rotamer for bidentate interactions with Arg in all proteins binding in the N-helix mode to phospho-ligands is gauche-/trans/gauche-/trans (SI Appendix, Fig. S9), the third most common conformation [∼11%; Structural Library of Intrinsic Residue Propensities (SLIRP) database (47)]. For Ser and Thr, which have effectively two cis rotamers (trans is rare in helices, with ∼2% occurrence in SLIRP database), both rotamers can realize a bidentate interaction (SI Appendix, Fig. S9). From the standpoint of helical stability, Ser and Thr are both preferred N-terminal capping residues (48).
Concluding Remarks
Our analysis is consistent with previous work that highlighted phosphate binding as being a highly abundant (3) and uniquely ancient protein function (6, 26). For the ancient phosphate binders, the minimal structural element appears to be a β-strand followed by a phosphate-binding loop and then a flanking α-helix (β-P-loop-α) (Fig. 3D). This β-P-loop-α element may have been the ancient nucleus around which modern αβα domains condensed (5, 6, 24, 41, 49, 50).
What has been unresolved so far, however, is the question of why the first protein lineages almost exclusively employ the N terminus of an α-helix (and the preceding loop) as sites for phosphate binding. Our results show that, given intact folded domains, multiple solutions to phosphate binding exist, with the side-chain binding mode enjoying preferential emergence. Consequently, the dominance of N-helix binding among the most ancient three-layer αβα sandwich folds (and to the related TIM β/α-barrel) is even more surprising. Shared ancestry is an option to be considered; however, at present, we lack conclusive evidence for this scenario, or even a hypothesis regarding which of the seven ancient, fundamental phosphate binders noted here diverged from this putative ancestor. We propose an alternative explanation, in which the preference for α-helical binding sites in the ancient phosphate binders is a reflection of the constraints acting on the earliest proteins: Assuming short, prebiotic sequences, the N-helix is the most accessible solution to phosphate binding (this explanation may be complementary rather than contradictory to the common ancestry one). The prevalence of bidentate interactions at the N-helix underscores the importance of forming a “crown” of hydrogen bonds (8, 51), as opposed to the helix dipole (52), as previously suggested (11).
The potential for the prebiotic set of amino acids to yield functional proteins has been a matter of some debate (53). It stands to reason, however, that the first proteins must have been functional in some capacity, even with a limited amino acid alphabet, potentially lacking not just aromatic amino acids but also Arg and Lys (45). Indeed, the prebiotic set of amino acids does limit the space of possible binding solutions, as side-chain and backbone-assisted binding modes are particularly dependent on Arg and Lys (Fig. 4D). But alternative solutions do exist, and phosphate binding in the N-helix mode by short, prebiotic sequences, bolstered by bidentate interactions, appears to be a feasible evolutionary starting point.
Altogether, our analysis indicates that binding of small phospho-ligands is likely to have been the function that drove the emergence of the very first αβα sandwich proteins that, in turn, gave birth to the major enzyme superfamilies. We further conclude that these early protein forms used an N-helix binding site with a short, simple binding motif rich in Gly (41, 54), but also, as indicated by our analysis, Ser and Thr (9), because of their potential to form bidentate interactions. It also appears that N-helix binding sites bolstered by bidentate interactions were key to enabling the emergence of phospho-ligand binding even in the absence of basic amino acids.
Supplementary Material
Acknowledgments
We thank Simon Dürr for his assistance developing criteria to identify N-helix binding events in proteins; Saurav Mallik for help analyzing the sequence similarity of proteins between F-groups; and Golan Yona for programming advice. We acknowledge the reviewers for their thoughtful comments. We gratefully acknowledge funding from Israel Science Foundation Grant 980/14 (to D.S.T.), Koshland Fellowship Program Fellowship (to L.M.L.), Knut and Alice Wallenberg Foundation Grants 2013.0124 and 2018.0140 (to S.C.L.K), and Human Frontier Science Program Grant RGP0041/2017 (to S.C.L.K.). D.S.T. is the Nella and Leon Benoziyo Professor of Biochemistry.
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1911742117/-/DCSupplemental.
References
- 1.Nobeli I., Ponstingl H., Krissinel E. B., Thornton J. M., A structure-based anatomy of the E.coli metabolome. J. Mol. Biol. 334, 697–719 (2003). [DOI] [PubMed] [Google Scholar]
- 2.Berman H. M., et al. , The Protein Data Bank. Nucleic Acids Res. 28, 235–242 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hirsch A. K., Fischer F. R., Diederich F., Phosphate recognition in structural biology. Angew. Chem. Int. Ed. 46, 338–352 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Eck R. V., Dayhoff M. O., Evolution of the structure of ferredoxin based on living relics of primitive amino acid sequences. Science 152, 363–366 (1966). [DOI] [PubMed] [Google Scholar]
- 5.Romero Romero M. L., et al. , Simple yet functional phosphate-loop proteins. Proc. Natl. Acad. Sci. U.S.A. 115, E11943–E11950 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Alva V., Söding J., Lupas A. N., A vocabulary of ancient peptides at the origin of folded proteins. eLife 4, e09410 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Leipe D. D., Koonin E. V., Aravind L., Evolution and classification of P-loop kinases and related proteins. J. Mol. Biol. 333, 781–815 (2003). [DOI] [PubMed] [Google Scholar]
- 8.Watson J. D., Milner-White E. J., A novel main-chain anion-binding site in proteins: The nest. A particular combination of φ,ψ values in successive residues gives rise to anion-binding sites that occur commonly and are found often at functionally important regions. J. Mol. Biol. 315, 171–182 (2002). [DOI] [PubMed] [Google Scholar]
- 9.Copley R. R., Barton G. J., A structural analysis of phosphate and sulphate binding sites in proteins. Estimation of propensities for binding and conservation of phosphate binding sites. J. Mol. Biol. 242, 321–329 (1994). [DOI] [PubMed] [Google Scholar]
- 10.Kobayashi N., Go N., A method to search for similar protein local structures at ligand binding sites and its application to adenine recognition. Eur. Biophys. J. 26, 135–144 (1997). [DOI] [PubMed] [Google Scholar]
- 11.Hol W. G. J., van Duijnen P. T., Berendsen H. J. C., The α-helix dipole and the properties of proteins. Nature 273, 443–446 (1978). [DOI] [PubMed] [Google Scholar]
- 12.Koharudin L. M. I., et al. , Structural basis of allosteric activation of sterile α motif and histidine-aspartate domain-containing protein 1 (SAMHD1) by nucleoside triphosphates. J. Biol. Chem. 289, 32617–32627 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Bottoms C. A., Smith P. E., Tanner J. J., A structurally conserved water molecule in Rossmann dinucleotide-binding domains. Protein Sci. 11, 2125–2137 (2002). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cheng H., et al. , ECOD: An evolutionary classification of protein domains. PLOS Comput. Biol. 10, e1003926 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Cheng H., Liao Y., Schaeffer R. D., Grishin N. V., Manual classification strategies in the ECOD database. Proteins 83, 1238–1251 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Laskowski R. A., Jabłońska J., Pravda L., Vařeková R. S., Thornton J. M., PDBsum: Structural summaries of PDB entries. Protein Sci. 27, 129–134 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Cock P. J. A., et al. , Biopython: Freely available Python tools for computational molecular biology and bioinformatics. Bioinformatics 25, 1422–1423 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Stickle D. F., Presta L. G., Dill K. A., Rose G. D., Hydrogen bonding in globular proteins. J. Mol. Biol. 226, 1143–1159 (1992). [DOI] [PubMed] [Google Scholar]
- 19.Heinig M., Frishman D., STRIDE: A web server for secondary structure assignment from known atomic coordinates of proteins. Nucleic Acids Res. 32, W500-2 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Humphrey W., Dalke A., Schulten K., VMD: Visual molecular dynamics. J. Mol. Graph. 14, 33–38, 27–28 (1996). [DOI] [PubMed] [Google Scholar]
- 21.Fu L., Niu B., Zhu Z., Wu S., Li W., CD-HIT: Accelerated for clustering the next-generation sequencing data. Bioinformatics 28, 3150–3152 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohno A. J. S., Evolution by gene duplication Popul. (French Ed.) 26, 1176 (1971). [Google Scholar]
- 23.Taylor J. S., Raes J., Duplication and divergence: The evolution of new genes and old ideas. Annu. Rev. Genet. 38, 615–643 (2004). [DOI] [PubMed] [Google Scholar]
- 24.Laurino P., et al. , An ancient fingerprint indicates the common ancestry of Rossmann fold enzymes utilizing different ribose based cofactors. PLoS Biol. 14, e1002396 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Bukhari S. A., Caetano-Anollés G., Origin and evolution of protein fold designs inferred from phylogenomic analysis of CATH domain structures in proteomes. PLOS Comput. Biol. 9, e1003009 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ma B. G., et al. , Characters of very ancient proteins. Biochem. Biophys. Res. Commun. 366, 607–611 (2008). [DOI] [PubMed] [Google Scholar]
- 27.Fawaz M. V., Topper M. E., Firestine S. M., The ATP-grasp enzymes. Bioorg. Chem. 39, 185–191 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Aravind L., Anantharaman V., Koonin E. V., Monophyly of class I aminoacyl tRNA synthetase, USPA, ETFP, photolyase, and PP-ATPase nucleotide-binding domains: Implications for protein evolution in the RNA. Proteins 48, 1–14 (2002). [DOI] [PubMed] [Google Scholar]
- 29.Loll P. J., Lattman E. E., The crystal structure of the ternary complex of staphylococcal nuclease, Ca2+ and the inhibitor pdTp, refined at 1.65 Å. Proteins 5, 183–201 (1989). [DOI] [PubMed] [Google Scholar]
- 30.Huang H., et al. , Panoramic view of a superfamily of phosphatases through substrate profiling. Proc. Natl. Acad. Sci. U.S.A. 112, E1974–E1983 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Lupas A. N., Alva V., Ribosomal proteins as documents of the transition from unstructured (poly)peptides to folded proteins. J. Struct. Biol. 198, 74–81 (2017). [DOI] [PubMed] [Google Scholar]
- 32.Mutter A., et al. , De novo design of symmetric ferredoxins that shuttle electrons in vivo. Proc. Natl. Acad. Sci. U.S.A. 116, 14557–14562 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Edwards H., Abeln S., Deane C. M., Exploring fold space preferences of new-born and ancient protein superfamilies. PLOS Comput. Biol. 9, e1003325 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Sobolevsky Y., Trifonov E. N., Conserved sequences of prokaryotic proteomes and their compositional age. J. Mol. Evol. 61, 591–596 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Winstanley H. F., Abeln S., Deane C. M., How old is your fold? Bioinformatics 21 (suppl. 1), i449–i458 (2005). [DOI] [PubMed] [Google Scholar]
- 36.Lupas A. N., Ponting C. P., Russell R. B., On the evolution of protein folds: Are similar motifs in different protein folds the result of convergence, insertion, or relics of an ancient peptide world? J. Struct. Biol. 134, 191–203 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Farías-Rico J. A., Schmidt S., Höcker B., Evolutionary relationship of two ancient protein superfolds. Nat. Chem. Biol. 10, 710–715 (2014). [DOI] [PubMed] [Google Scholar]
- 38.Nagano N., Orengo C. A., Thornton J. M., One fold with many functions: The evolutionary relationships between TIM barrel families based on their sequences, structures and functions. J. Mol. Biol. 321, 741–765 (2002). [DOI] [PubMed] [Google Scholar]
- 39.Noda-Garcia L., Liebermeister W., Tawfik D. S., Metabolite-enzyme coevolution: From single enzymes to metabolic pathways and networks. Annu. Rev. Biochem. 87, 187–216 (2018). [DOI] [PubMed] [Google Scholar]
- 40.Romero Romero M. L., Rabin A., Tawfik D. S., Functional proteins from short peptides: Dayhoff’s hypothesis turns 50. Angew. Chem. Int. Ed. Engl. 55, 15966–15971 (2016). [DOI] [PubMed] [Google Scholar]
- 41.Zheng Z., Goncearenco A., Berezovsky I. N., Nucleotide binding database NBDBA collection of sequence motifs with specific protein-ligand interactions. Nucleic Acids Res. 44, D301–D307 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Longo L. M., Blaber M., Protein design at the interface of the pre-biotic and biotic worlds. Arch. Biochem. Biophys. 526, 16–21 (2012). [DOI] [PubMed] [Google Scholar]
- 43.Longo L. M., Blaber M., Prebiotic protein design supports a halophile origin of foldable proteins. Front. Microbiol. (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Trifonov E. N., Consensus temporal order of amino acids and evolution of the triplet code. Gene 261, 139–151 (2000). [DOI] [PubMed] [Google Scholar]
- 45.McDonald G. D., Storrie-Lombardi M. C., Biochemical constraints in a protobiotic earth devoid of basic amino acids: The “BAA(-) world”. Astrobiology 10, 989–1000 (2010). [DOI] [PubMed] [Google Scholar]
- 46.Lassila J. K., Herschlag D., Promiscuous sulfatase activity and thio-effects in a phosphodiesterase of the alkaline phosphatase superfamily. Biochemistry 47, 12853–12859 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Towse C. L., Rysavy S. J., Vulovic I. M., Daggett V., New dynamic rotamer libraries: Data-driven analysis of side-chain conformational propensities. Structure 24, 187–199 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Doig A. J., Baldwin R. L., N- and C-capping preferences for all 20 amino acids in α-helical peptides. Protein Sci. 4, 1325–1336 (1995). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Garboczi D. N., Shenbagamurthi P., Kirk W., Hullihen J., Pedersen P. L., Mitochondrial ATP synthase. Interaction of a synthetic 50-amino acid, β-subunit peptide with ATP. J. Biol. Chem. 263, 812–816 (1988). [PubMed] [Google Scholar]
- 50.Goncearenco A., Berezovsky I. N., Protein function from its emergence to diversity in contemporary proteins. Phys. Biol. 12, 045002 (2015). [DOI] [PubMed] [Google Scholar]
- 51.Leader D. P., Milner-White E. J., Bridging of partially negative atoms by hydrogen bonds from main-chain NH groups in proteins: The crown motif. Proteins 83, 2067–2076 (2015). [DOI] [PubMed] [Google Scholar]
- 52.Sengupta D., Behera R. N., Smith J. C., Ullmann G. M., The alpha helix dipole: Screened out? Structure 13, 849–855 (2005). [DOI] [PubMed] [Google Scholar]
- 53.Shibue R., et al. , Comprehensive reduction of amino acid set in a protein suggests the importance of prebiotic amino acids for stable proteins. Sci. Rep. 8, 1227 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Möller W., Amons R., Phosphate-binding sequences in nucleotide-binding proteins. FEBS Lett. 186, 1–7 (1985). [DOI] [PubMed] [Google Scholar]
- 55.Sugahara M., Ohshima N., Ukita Y., Sugahara M., Kunishima N., Structure of ATP-dependent phosphoenolpyruvate carboxykinase from Thermus thermophilus HB8 showing the structural basis of induced fit and thermostability. Acta Crystallogr. Biol. Crystallogr. 61, 1500–1507 (2005). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data discussed in this paper will be made available to readers upon request.