Significance
New antibacterial strategies that circumvent existing mechanisms hold promise in the fight against drug-resistant bacteria. Here we present an antibacterial mechanism that safeguards cells and animals by shutting down an intracellular signal through a site-selective covalent peptide-protein reaction instead of acting on bacterium cells. The novelty of this work is multifold, including a cysteine- specific reaction at an SH3 domain of an adaptor protein in cells, selective blockage of the second SH3 domain of Nck but not the other two SH3 domains, and prevention of EPEC-induced infection in animals for at least 12 h by reactive peptide. Thus, this preventative mechanism differs significantly from the bactericidal actions of antibiotics.
Keywords: site-specific protein reaction, Nck, peptide inhibitor, EPEC, bacterial infection
Abstract
A critical problem in the fight against bacterial infection is the rising rates of resistance and the lack of new antibiotics. The discovery of new targets or new antibacterial mechanisms is a potential solution but is becoming more difficult. Here we report an antibacterial mechanism that safeguards intestine cells from enteropathogenic Escherichia coli (EPEC) by shutting down an infection-responsive signal of the host intestine cell. A key step in EPEC infection of intestinal cells involves Tir-induced actin reorganization. Nck mediates this event by binding with Tir through its SH2 domain (Nck-SH2) and with WIP through its second SH3 domain (Nck-SH3.2). Here we report the design of a synthetic peptide that reacts precisely with a unique cysteine of the Nck-SH3.2 domain, blocks the binding site of the Nck protein, and prevents EPEC infection of Caco-2 cells. Oral update of this nontoxic peptide before EPEC administration safeguards mice from EPEC infection and diarrhea. This study demonstrates domain-specific blockage of an SH3 domain of a multidomain adaptor protein inside cells and the inhibition of Tir-induced rearrangement of the host actin cytoskeleton as a previously unknown antibacterial mechanism.
The discovery of penicillin in 1929 opened a new era of the antibacterial campaign in the history of humankind (1, 2). For the first time, humans had weapons against malicious infections caused by bacteria. Since then, scientists have discovered and developed numerous medicines and treatments that can cure or prevent bacterial infection, including bactericidal agents, bacteriostatic agents, and vaccines. The bactericidal agents include β-lactam drugs, such as penicillin, and drugs that affect bacterial cell wall synthesis, including nonribosomal peptides such as polymyxins and others. Bacteriostatic agents include tetracyclines, macrolides, aminoglycosides, and chloramphenicol-type drugs, such as gentamicin and caratol, that can bind to bacterial ribosomes or nucleic acids and ultimately inhibit the synthesis of important proteins in bacteria, as well as other quinolones, such as ofloxacin, which hinder bacterial DNA replication and transcription (2, 3). Antimicrobial vaccines mainly include biological agents that enable the body to produce immune responses against microbes, including anthrax vaccine, pertussis vaccine, and others (4–6).
The use of antibiotics effectively controls life-threatening infection and reduces neonatal mortality. However, long-term use of antibiotics in large quantities has elicited a range of resistance that is endangering human health (3, 7–9). Unfortunately, accompanying the rise in global resistance is a setback in antibacterial drug discovery, including shortages of new mechanisms and new targets in recent years. Therefore, the crisis of antibacterial resistance calls for new mechanisms that are significantly different from the existing ones.
Escherichia coli diarrhea is endemic or potentially endemic to all countries and districts (10). More specifically, enteropathogenic E. coli (EPEC) causes watery diarrhea with fever and vomiting, affecting primarily children age <2 years (10). Currently, EPEC infection is treated mainly with antibiotics; however, with the emerging resistance to β-lactam antibiotics (e.g., ceftazidime), aminoglycosides, and quinolones, the control of intra-abdominal infections by multidrug-resistant Enterobacteriaceae remains an unsolved problem (11, 12).
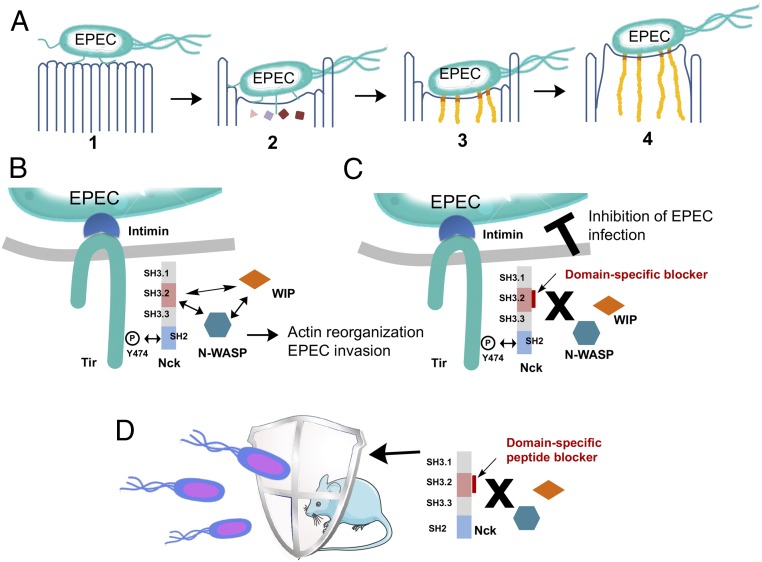
The hallmark of EPEC infection is the formation of attaching and effacing (A/E) lesions on the gut mucosa, characterized by microvilli destruction. The mechanism of EPEC infection includes three main steps: (1) a bacterium latches/adheres to the surface of an intestinal cell; (2) the bacterium injects protein Tir to the intestinal cell; and (3) an actin pedestal is then formed on the intestinal cell to form an A/E lesion, bacterium infects cells, and diarrhea commences (Fig. 1A) (13, 14). EPEC Tir protein initiates actin pedestal formation in host cells by recruiting the multidomain adaptor protein Nck via protein–protein interactions between the SH2 domain of Nck (Nck-SH2) and the phosphorylated tyrosine 474 (15). Nck serves as an adaptor between the bacterial protein Tir with the Arp2/3 complex-dependent actin polymerization pathway in the host cells. It was recently found that this Nck-mediated actin-based motility exists not only in EPEC, but also in other microbes, such as vaccinia (16, 17) and other vertebrate poxviruses (18).
Fig. 1.
Inhibiting EPEC infection by specific blockage of the Nck-SH3.2 domain. (A) The four-stage model of EPEC pathogenesis: attachment, sending in effector proteins, actin reorganization, and pedestal formation. (B) Schematic representation of Nck-mediated signaling from Tir to actin reorganization and EPEC invasion. (C) Inhibition of the Nck-SH3.2 domain as a promising strategy to block EPEC invasion to intestinal cells. (D) A new antibacterial mechanism of blocking mice diarrhea via site-selective domain-specific reaction of Nck in the intestinal cells.
Nck proteins (Nck1 and Nck2) contain four domains in tandem, including one SH2 domain and three SH3 domains. Because Nck1 (also called Nck) and Nck2 (also called Nckβ/Grb4) are 70% identical overall and their SH2, second SH3, and third SH3 domains are >90% identical, Nck1 (for simplicity, Nck hereinafter) was chosen as our target. Nck-SH2 is known to bind to the phosphorylated peptide sequence on Tir protein in EPEC and on the viral membrane protein A36 (15, 19). The second SH3 domain, Nck-SH3.2, is known to bind to WIP peptides 328NDETPRLPQRNLSLS342 and 247SNRPPLPPTPSRALD261, and Nck1-SH3.3 binds to N-WASP peptides 292PPPHSSGPPPPPARG306 and 268RQAPPPPPPSRGGPP283. Although the biochemical basis of these interactions has not been fully elucidated, Nck1-SH3.2 is known to be essential for recruiting WIP/WASP complexes, whereas the other two domains are important for stimulating actin assembly (Fig. 1B) (19). Knockdown of the Nck-SH3.2 domain was found to abolish the actin reorganization on EPEC infection (19). As Nck-SH3.2–mediated protein–protein interaction is sufficient to initiate actin assembly during EPEC infection, we envision that a peptide that blocks the protein-binding site of the Nck-SH3.2 domain will inhibit EPEC-mediated pedestal formation, which in turn will then block the entrance mechanism of EPEC and guard the cells against EPEC infection (Fig. 1C).
Domain-specific inhibition of Nck-SH3.2 domain without affecting the other two SH3 domains in the same protein remains a challenge, however. The SH3 domains bind to polyproline peptides with a promiscuous binding nature, a modest binding affinity, and a large interface area. Without the presence of “hot spot” interactions, designing a high-affinity inhibitor for a specific SH3 domain that can compete off the endogenous ligands in vivo is difficult. We recently showed that proximity-induced site-selective cysteine reactions allowed the development of domain-specific peptide-based irreversible blockers for a PDZ domain and a SH3 domain, and that the peptide inhibitors can effectively block signal transduction inside cells (20, 21). Here we describe a structure-based design of a reactive peptide that can covalently react with the Nck-SH3.2 domain at a unique cysteine residue. We report a cell-penetrating version of this reactive peptide that reacts with Nck inside cells, blocks EPEC infection to intestinal cells, and protects mice from EPEC-induced diarrhea (Fig. 1D).
Results
Designing an Nck-SH3.2–Specific Reactive Peptide.
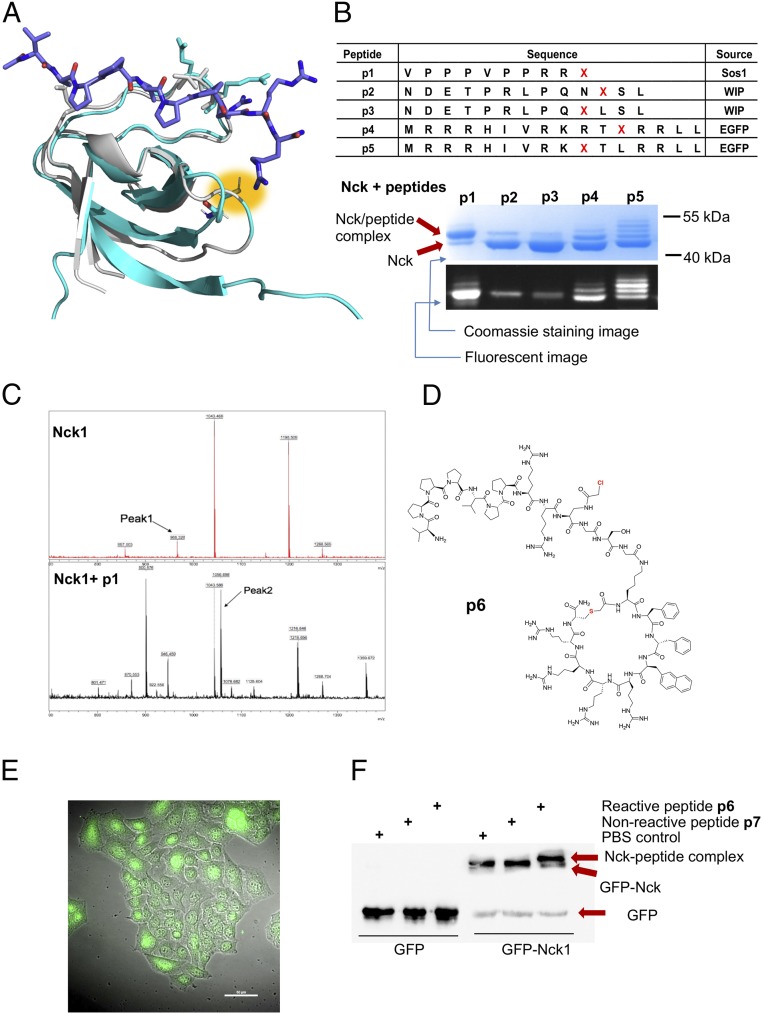
The Nck-SH3.2 domain has a unique structural feature that makes it feasible as a target of site-selective covalent peptide inhibitors. Although the complex structure of Nck-SH3.2 and peptide ligands has not yet been elucidated, a solution NMR structure of the Nck-SH3.2 protein alone without peptide ligands is available. In addition, the Nck-SH3.2 domain is highly homologous with the N-terminal SH3 domain of Grb2. Alignment of the structure of Nck-SH3.2 with Grb2-N-SH3 shows a cysteine residue, Cys-48, located at the peptide-binding site of Nck-SH3.2 (the numbering is based on that in Protein Data Bank [PDB] ID code 2CUB) (Fig. 2A) (22). The thiol group of Cys-48 points at the C terminus of the bound peptide ligand. We then envision that an Nck-binding peptide carrying a mild electrophile α-chloroacetyl group at the C terminus will bind specifically to Nck-SH3.2, position the α-chloroacetyl group in the vicinity of the thiol group of Cys-48, and spontaneously form a thioether bond between Nck-SH3.2 and the peptide (23). Such a covalent reaction will irreversibly block the peptide-binding site of SH3.2 domain while having only a minimal influence on other SH3 domains of Nck. We define peptides that spontaneously form a covalent bond with the target protein on simple incubation as “reactive peptides” (20, 21). These reactive peptides will be superior blockers, as they can outcompete endogenous ligands through covalent irreversible reactions (20, 21, 24, 25).
Fig. 2.
Nck-SH3.2–specific reactive peptides and the protein-peptide reactions in vitro and in Caco-2 cells. (A) Alignment of the solution structure of the Nck-SH3.2 domain (cyan; PDB ID code 2CUB) and crystal structure of Grb2-N-SH3 complexed with a peptide ligand (gray; PDB ID code 1GBQ). Cys-48 in Nck-SH3.2 is highlighted in yellow. (B) A list of the reactive peptides synthesized and their conjugation reactions with Nck in vitro. Fluorescently labeled synthetic peptides were mixed with recombinant Nck protein at a peptide-to-protein ratio of 5:1 at room temperature for 12 h. The reaction was thermally denatured, resolved by SDS/PAGE, imaged under a fluorescent scanner, and stained with Coomassie blue. (C) Mass spectra of the Nck-p1 complex band following in-gel tryptic digestion using Nck as a control. (D) The structure of the cell-penetrating version of p1, peptide p6. (E) Spontaneous uptake of fluorescently labeled peptide p6 [5(6)-carboxyl-fluorescein linked to the N terminus of the peptide] in Caco-2 cells. The peptide was incubated with Caco-2 cells at a final concentration of 5 µM for 2 h at 37 °C. After washing, cells were fixed with 5% paraformaldehyde for 20 min at room temperature, then observed under a fluorescent microscope. (F) Covalent reaction of p6 with GFP-Nck protein overexpressed in Caco-2 cells probed by Western blot analysis using an anti-GFP antibody.
To identify an Nck-SH3.2–binding peptide, we started with a peptide ligand of the Grb2-N-SH3 domain, which is homologous to Nck-SH3.2. Based on a polyproline peptide of Sos1 that binds with Grb2-N-SH3 with a KD of 3.5 μM (26), we synthesized a peptide library of 240 members on cellulose paper by replacing amino acids in each position with one of the 19 amino acids (except cysteine) using SPOT technology (27). Purified recombinant Nck-SH3.2 protein was then added to the cellulose for binding with the peptide on each spot. After washing, the remaining amount of protein was probed by an antibody against the His-tag at the N terminus of the Nck-SH3.2 domain and quantified (SI Appendix, Fig. S1). The results revealed that for the sequence PPPLPPRVQSLN, position P1 did not show a preference, suggesting that P1 is out of the binding pocket. Positions P2 and P3 prefer proline, although they show broad tolerance. P4 strictly binds hydrophobic amino acids. P7 exclusively binds arginine, and P8 prefers small side chains. Based on this information, we designed a reactive peptide, VPPPVPPRRX, in which X is an unnatural amino acid, a derivative of diaminopropionic acid, (2S)-2-amino-3-[(α-chloroacetyl)amino]-propionic acid, which contains an electrophile α-chloroacetyl group at its side chain (20, 21, 23).
A set of α-chloroacetyl–containing peptides, p1 to p5, designed based on the Nck-binding peptides from the WIP protein and juxtamembrane domain of the epidermal growth factor receptor (EGFR), were synthesized. When this set of reactive peptides was incubated with the purified Nck protein, covalently linked Nck-peptide complexes formed spontaneously, appearing as new protein bands with molecular weights higher than that of Nck on denatured SDS/PAGE gel (Fig. 2B). Peptide p1 showed the highest reaction efficiency, with >80% of Nck protein converted to the peptide conjugates. Furthermore, p1 formed a 1:1 adduct with the Nck protein (Fig. 2B). In contrast, multiple bands were observed in the reaction between Nck protein and peptides p4 and p5, indicating that these peptides nonselectively reacted with more than one cysteine on the Nck protein. In-gel tryptic digestion of the Nck-peptide p1 complex band identified Cys-48 in the SH3.2 domain as the primary reaction site for p1. The peptide fragment containing Cys-48 of the Nck-SH3.2 domain (C*SDGWWR; calc: 966.4, obs: 966.3, with C* denoting the cysteine residue modified by iodoacetic acid during sample preparation) disappeared after the conjugation reaction (peak 1 in the top panel). A new peak (peak 2 in the bottom panel) appeared in the Nck-p1 complex with a molecular weight matching that of the conjugated fragment C(X)SDGWWR on the matrix-assisted laser desorption/ionization time-of-flight (MALDI-TOF) MS spectrum (calc: 1,053.1, obs: 1,056.7) (Fig. 2C). These results show that based on the proximity effect, a reactive peptide p1 specifically for the Nck-SH3.2 domain was developed by selectively targeting Cys-48.
Covalent Reaction inside Caco-2 Cells.
We next sought to conduct the covalent reaction inside the cell cytosol of Caco-2 cells. To deliver peptide p1 across the membrane barrier, a cyclic cell-penetrating peptide derived from the known cell-penetrating sequence cyclo(FfΦRrRrQ) was fused with p1 to give peptide p6 (Fig. 2D and SI Appendix, Fig. S2) (28–30). A continuous line of heterogeneous human epithelial colorectal adenocarcinoma cells, Caco-2 cells, was used to mimic intestine epithelial cells (31). The fluorescent-labeled p6 peptide efficiently entered Caco-2 cells and was distributed mainly in the cytosol (Fig. 2E). After entering the cells, peptide p6 colocalized with GFP-Nck protein overexpressed in Caco-2 cells (SI Appendix, Fig. S3) and reacted with GFP-Nck protein in the cytosol of GFP-Nck–overexpressing Caco-2 cells as identified by Western blot analysis using an anti-GFP antibody (Fig. 2F). Reactions did not occur between p6 and a GFP protein or between the nonreactive peptide p7 and GFP-Nck. Due to the covalent reaction, the reactive peptide p6 also showed a higher binding signal than the nonreactive p7 in an enzyme-linked immunosorbent assay-based Nck binding assay (SI Appendix, Fig. S4). We then successfully performed a site-selective Nck reaction inside cells, despite the presence of thiol-containing molecules within the cytosol (21, 22).
A Cell-Based EPEC Infection Model.
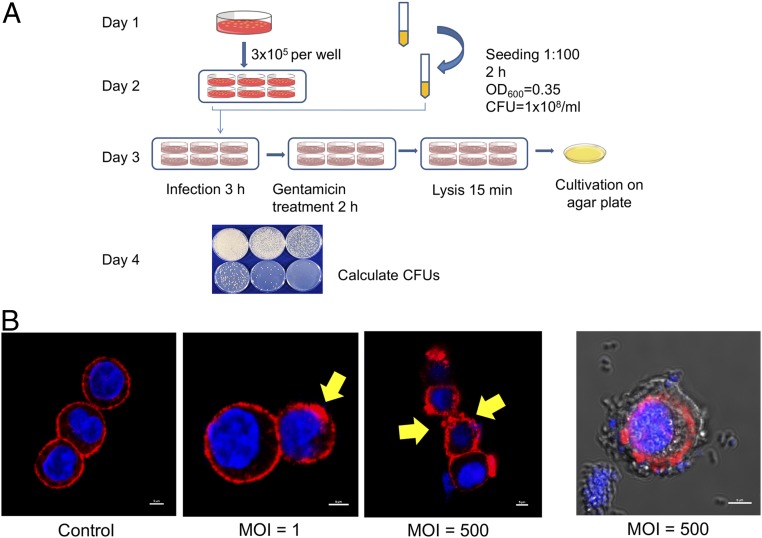
To link Nck blockage to EPEC infectivity, we next identified an infectious EPEC strain and established a cell-based infection model. Two E. coli strains were compared based on the presence of two EPEC virulence genes, bpf (on EAF plasmid) and eae (on chromosome), detected by polymerase chain reaction using selected primers (SI Appendix, Fig. S5 and Table S1) (32–35). Strain E2 was selected as the model strain, and the infectious capability was determined by an infection assay on Caco-2 cells (Fig. 3A) (36–40). The EPEC cells that associated with Caco-2 cells during infection can be divided into two subpopulations: one subpopulation that only adhere/attach to the surface of Caco-2 cells and another that enter the membrane of Caco-2 cells completely and are protected from antibiotic killing. To quantify the latter population, which has finished the infection step, a gentamicin treatment step was used to remove the EPEC population that only adhered to the surface of Caco-2 cells. Gentamicin is not permeable to the plasma membrane of Caco-2 cells, so this step will not kill EPEC cells inside Caco-2 (41, 42). At a multiplicity of infection (MOI) of 10, each agar plate developed 132 ± 22 colonies at a dilution factor of 100, giving the bacterial solution an infectivity index of 1.3 × 104 cfu/mL at an MOI of 10. At an MOI of 100, each agar plate developed 87 ± 9 colonies at a dilution factor of 1,000. This gave the same bacterial solution an infectivity index of 8.7 × 104 cfu/mL at an MOI of 100. This finding confirms that the EPEC strain E2 has a strong infection capability for Caco-2 cells. Furthermore, under a high MOI, the cell membrane formed wrinkles, inferring a drastic reorganization of the actin network (Fig. 3B). EPEC cells were clearly visible both on and inside Caco-2 cells at a high MOI.
Fig. 3.
EPEC infection of Caco-2 cells. (A) Schematic workflow showing the measurement of infectivity of EPEC. (B) EPEC infections at different MOI values show clear signs of disruption of the actin structure of Caco-2 cells. Shown is a representative image demonstrating EPEC attachment on Caco-2 cells at an MOI of 500. Blue indicates DAPI staining for cell nuclei; red, AF647 phalloidin for actin. Actin was labeled with Alexa Fluor 647 phalloidin, and nuclei were labeled with DAPI.
Reactive Peptide Defends Caco-2 Cells Against EPEC Infection.
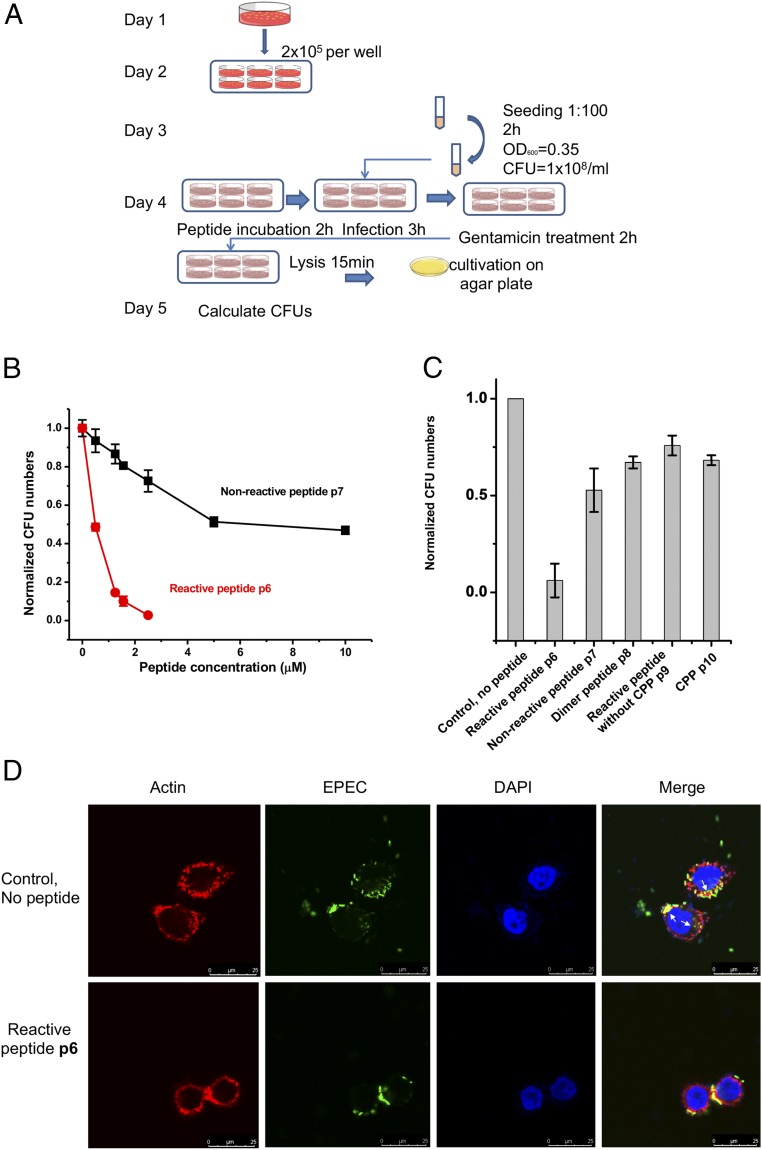
We next showed that the blockage of Nck-3.2 can defend Caco-2 cells from EPEC infection. First, incubation of peptide p6 with EPEC at various concentrations did not detectably affect the bacterial growth in bacteriological medium (SI Appendix, Fig. S6). Peptide p6 also exhibited minimal cytotoxicity to Caco-2 cells according to a 3-(4, 5-dimethylthiazol-2-yl)-2, 5-diphenyl-tetrazolium bromide (MTT) assay (SI Appendix, Fig. S7). An infection assay was designed to prove that pretreatment of Caco-2 cells with peptide p6 would prevent EPEC infection (Fig. 4A). Similarly, a gentamicin treatment step was included to quantify EPEC cells that had infected Caco-2 cells within the boundary of the Caco-2 membrane, excluding surface-attached EPEC cells. After the gentamicin treatment, cells were lysed to release EPEC cells that had been engulfed inside cells. The cell lysate was diluted until a reliable cfu count on agar plates was achieved. Peptide p6 treatment inhibited the EPEC infection to Caco-2 cells in a dose-dependent manner, and 500 nM of p6 inhibited almost 50% of the EPEC infection (Fig. 4B). Other peptides, including nonreactive peptide p7, dimeric nonreactive peptide p8, and fragments of p6 (a reactive peptide without cell-penetrating sequence p9 and the cell-penetrating sequence p10) showed significantly reduced anti-EPEC efficacy (Fig. 4C). Structures of peptides p7 to p10 are listed in SI Appendix, Fig. S2. p6-pretreated Caco-2 cells resisted EPEC infection, but not the surface attachment. EPEC cells can be found both on the surface of and inside untreated Caco-2 cells, but under the same infection conditions (without gentamicin treatment), EPEC cells were found only on the surface of p6-pretreated Caco-2 cells, with no sign of entrance into the cells (Fig. 4D). Taken together, both the reactive peptide and cell-penetrating sequence are indispensable for the efficacy of the anti-EPEC effect inside Caco-2 cells. Peptide p6 did not act on EPEC cells, nor did it cause toxicity to Caco-2 cells; instead, p6 granted Caco-2 cells an anti-EPEC effect that can prevent the infection of EPEC cells.
Fig. 4.
Reactive peptide treatment guards Caco-2 cells against EPEC infection. (A) Experimental scheme showing the procedure for measuring bacterial infectivity on peptide treatment. (B) Comparison of the inhibitory effect of reactive peptide p6 and nonreactive peptide p7 toward EPEC infection to Caco-2 cells. (C) Comparison of the anti-EPEC effect of a set of peptides. (D) Representative images of p6-treated Caco-2 cells versus untreated Caco-2 cells in response to EPEC infection. GFP-expressing EPEC can enter Caco-2 cells without p6 (white arrows) but can attach to the surface of Caco-2 cells only in the presence of p6. Cells were stained and examined using a confocal laser scanning system (Nikon C1Si) equipped with a 60× oil objective with a 20-mW 488-nm laser. Note that gentamicin treatment was not used in this experiment.
Reactive Peptide Protects Mice Against EPEC Infection.
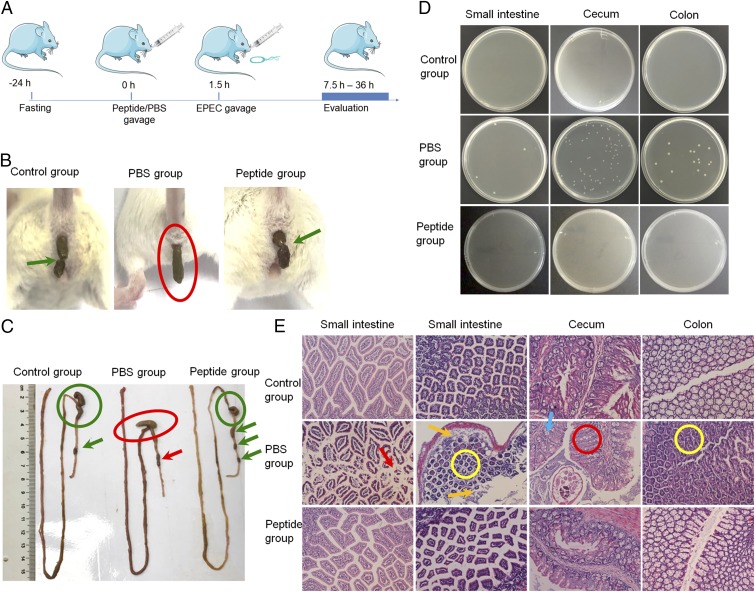
EPEC as a human enteric pathogen can also infect the mouse intestinal epithelium, induce morphological and pathophysiological changes (18, 19), and cause watery diarrhea (20). To evaluate the in vivo efficacy of the peptide treatment, we established an EPEC-induced diarrhea model (Fig. 5A) (17). In brief, mice were divided into three groups. The control group received 0.3 mL of PBS orally, followed by 0.5 mL of saline 1.5 h later. The PBS group received 0.3 mL of PBS, followed by 0.5 mL of EPEC bacterial solution (107 cfu) 1.5 h later. The peptide group was given 0.3 mL of peptide solution (3 mg of p6 dissolved in PBS), followed 1.5 h later by 0.5 mL of EPEC bacterial solution (107 cfu). The PBS group developed diarrhea based on the shape of feces, but the pretreatment group showed the same feces shape as the control (Fig. 5B). Semisolid feces were seen in the rectum of the PBS group. The feces also clumped together, while single pellets were seen in the control and peptide groups (Fig. 5C, arrow). The cecum was normally empty and contracted in the healthy controls, but the cecum of the infected group swelled, a sign of possible bacterial infiltration (Fig. 5C, circle) (43).
Fig. 5.
Peptide treatment protects mice against EPEC infection. (A) Schematic illustration of the experimental flow. (B) Peptide p6-treated mice did not develop diarrhea after EPEC gavage, whereas mice in the PBS group developed clear sign of diarrhea. Green arrows indicate healthy feces, which are grains normally ≤1/4 inch long. The red circle shows elongated feces, indicating diarrhea. (C) Morphological differences in the whole intestine among the control, PBS-treated, and peptide-treated groups. The red circle and arrow indicate signs of semisolid feces, agglomerated pellets, and swollen cecum. The green circles and arrows indicate normal morphology of intestines. (D) Tissue extracts grown on agar plates with ampicillin. Bacterial colonies were evident in the PBS group but not in the control and peptide groups. (E) Histological pictures of different parts of the whole intestine. Various signs of infection were seen in the PBS group but not in the control and peptide groups.
We also cultured extracts from different parts of the intestine on ampicillin agar plates and found bacterial colonies in the PBS group but not in the control and peptide groups (Fig. 5D). Histological analysis by hematoxylin and eosin (H&E) staining also showed clear signs of infection in the infection group, but not in the control and peptide groups. Infected stained small intestine sections showed severe apoptosis (arrows), indicating the presence of bacteria. The orange arrows in the figure point to signs of infiltration of cell necrosis. The yellow circles highlight signs of cell proliferation in the small intestines and colons of the infected group; signs of mild ulcer in the colon can be clearly seen in the yellow circle. The cecums of the infected group also showed mild inflammation of the increasing fossa hyperplasia (red circle) and goblet cell depletion (blue arrow; Fig. 5E) (44). Taken together, pretreatment by peptide p6 was shown to protect the intestine from EPEC infection.
We next asked how long this protective effect can last after one dose of peptide gavage. Mice were treated with 6 mg of peptide p6 and were then given 0.5 mL of EPEC bacterial solution (107 CFU) at 3, 6, 12, 24, and 48 h after peptide treatment (Fig. 6A). The shape of feces and intestinal morphology were then examined. We observed that a single peptide dose can protect the animals from EPEC infection for at least 12 h, because the 3 h, 6 h, and 12 h groups were completely healthy, but the 24 h and 48 h groups showed clear signs of infection and diarrhea (Fig. 6 B and C). In addition, the peptide-treated animals did not exhibit weight loss but instead showed a slight increase in body weight compared with controls without EPEC infection (SI Appendix, Fig. S8). No signs of toxicity were detected on the histological analysis (SI Appendix, Fig. S9).
Fig. 6.
One dose of peptide p6 protects mouse intestine for at least 12 h. (A) Schematic illustration of the experimental flow. (B) Diarrhea symptoms in mice at different time intervals between peptide and EPEC gavage. Red circles indicate elongated feces as a sign of diarrhea, and green arrows indicate normal feces. (C) Intestinal morphology of different groups with differing intervals between peptide and EPEC gavage. Red circles and arrows indicate semisolid feces, agglomerated pellets, and swollen cecum, whereas green circle and arrows indicate normal morphology.
To better mimic the clinical scenario, we next explored the efficacy of peptide treatment in another model in which mice received EPEC before the peptide blocker. In brief, mice in the peptide group received 0.5 mL of EPEC bacterial solution (107 CFU) followed 2 h later by 0.3 mL of peptide solution (3 mg of p6 dissolved in PBS). For comparison, mice in the PBS group received PBS only, after oral intake of EPEC solution (SI Appendix, Fig. S10A). The PBS group developed clear signs of infection, whereas the peptide-treated mice did not (SI Appendix, Fig. S10 B–D). These findings indicate that peptide treatment at 2 h after EPEC exposure can effectively block EPEC infection. A likely explanation for this is that the peptide reaction with Nck may be faster than the intestinal response to EPEC invasion, resulting in the same preventative effect. Peptide Cy5-p6 also showed a rapid accumulation in the blood at 2 h after gavage and was cleared out from the blood within 12 h and from the gastrointestinal tract within 24 h based on the quantification of Cy5 signal (SI Appendix, Fig. S11).
Conclusions
The ancient wisdom of traditional Chinese medicine puts at least equal weight on disease prevention as on treatment. Prevention of bacterial infection before symptoms emerge has been provided by vaccination, but this approach remains underrecognized in the fight against resistance (5). Here we report a nonvaccination preventive mechanism that renders the recipient cells unresponsive to the pathogenic bacteria. Focusing on the critical role of the Nck-SH3.2 domain in the pathogenesis of EPEC infection, we have developed a domain-specific site-selective reactive peptide that blocks signal transduction in cells and in animals. Blockage of EPEC infection in Caco-2 cells and in mice without toxicity has been shown. As a control, the antibiotic gentamicin did not exhibit a protective effect on EPEC infection (SI Appendix, Fig. S12). Therefore, this unique preventive mechanism that acts on the host cells rather than on the pathogen may provide insight into a previously unknown antibacterial mode of action. Whether EPEC bacterium can evolve a method to circumvent the Nck pathway is unknown. Whether this Nck-blocking antimicrobial mechanism can be applied to other human enteric pathogenic bacteria or viruses that use the same infection mechanism awaits further proof. Nevertheless, a preventive mechanism that safeguards the host cells from pathogen attack by shutting down an intracellular signal may be a promising approach to fighting drug resistance.
Methods
Materials and Instruments.
Unless noted otherwise, all reagents were used without further purification. Fmoc-protected amino acids and coupling reagents were obtained from GL Biochem. Rink amide resin was obtained from Biotage. Peptide characterization and purification were performed with reverse-phase high-performance liquid chromatography (Shimadzu; DGU-20A5). Peptide analysis was performed with an AutoFlex Speed LRF MALDI-TOF mass spectrometer (Bruker Daltonics). All gel images were captured by the Labnet ENDURO GDS Gel Documentation System or Bio-Rad ChemiDoc Image System (45). For MALDI-TOF MS analysis, 1.5 µL of digestion mixture was premixed with 1.5 µL of matrix solution (saturated α-cyano-4-hydroxycinnamic acid in TA50), and this mixture was applied onto a ground steel target that was ready for MS (45). Fluorescent images were obtained with an N-SIM superresolution microscope (Nikon). EPEC strain O127:H6 E2348/69 was a gift from Xin Deng, City University of Hong Kong.
Peptide Synthesis and Characterization.
All peptides were synthesized based on manual Fmoc-SPPS chemistry. Rink Amide-ChemMatrix resins with a loading capacity of 0.5 mmol/g were purchased from PCAS BioMatrix. For each coupling procedure, a fivefold excess of protected amino acid, HBTU, HOBt, and DIPEA (with a ratio of 1: 1: 1: 2) in dimethylformamide (DMF) was added to the resin. The deprotection reaction of Fmoc group was performed in 20% piperidine in DMF (vol/vol). The N terminus of the peptide was labeled with Cy5 dye or biotin or acetylated. After the completion of synthesis, the resin was washed and dried under vacuum. Peptides were cleaved from the resin and side chain deprotected by treatment with TFA/H2O/TIPS (95/2.5/2.5) for 2 h at room temperature, and the resin was then filtered and rinsed twice with TFA. The crude peptide was obtained by precipitation with adding cold diethyl. The peptides were purified by HPLC, lyophilized, and confirmed by MALDI-TOF MS analysis (Bruker Daltonics).
Protein-Peptide Conjugation Reactions and the Binding Assay.
Peptides (60 µM) were mixed to react with proteins (20 µM) in phosphate-citrate buffer (182 mM Na2HPO4, 9 mM citric acid, pH 7.4). The reaction solutions were then thermally denatured in the presence of loading dye to stop the reactions. After resolution by SDS/PAGE, the gel was first observed in the fluorescence channel and then stained with Coomassie brilliant blue dye. For the protein-peptide binding assay, recombinantly expressed Nck protein (2 mg) was coated in 96-well plates in carbonate/bicarbonate buffer (pH 9.6). After washing and blocking by nonfat dry milk, biotin-labeled peptides at different concentrations were added to each well, followed by a 90-min incubation at 37 °C. Diluted streptavidin-HRP solution was then added, followed by substrate solution to develop cover at 450 nm, and recorded by a plate reader.
Cell Transfection and Viability.
The GFP-Nck1 and GFP-SH3.2 expression vector was generated by cloning human Nck1 and SH3.2 into the Hind III-BamH I sites of the pEGFP-C1 expression vector. Plasmid DNA was prepared on Qiagen columns and transfected into Caco-2 cells using Lipofectamine 3000 (Invitrogen) in accordance with the manufacturer’s instructions. At 48 h after transfection, cells were analyzed in further experiments. Cell viability was measured by an MTT assay in accordance with the manufacturer’s instructions (Sigma-Aldrich). Optical density was measured at 570 nm on a microplate reader (Tecan).
Immunofluorescence Confocal Microscopy.
Caco-2 cells were grown in laser confocal culture dishes at a density of 2 × 104 cells per well for 24 h and then pretreated with peptide (5 μM) for 3 h, followed by infection with EPEC for 3 h in a 37 °C, 5% CO2 incubator. Cells were stained with DAPI (Invitrogen) and phalloidin (Alexa Fluor 594-conjugated; Invitrogen). Images were acquired with a confocal laser scanning microscope (Leica SP8). For colocalization experiments, Caco-2 cells were transfected with pEGFP-C1-Nck using Lipofectamine 3000 reagent and Opti-MEMI reduced serum medium (Gibco). DMEM with 10% FBS instead of the transfection medium was added to each well after 10 h, and cells were incubated at 37 °C with 5% CO2 for 48 h. Cells were then fixed, permeabilized, stained with DAPI dye, and analyzed by confocal microscopy (SP8; Leica).
Bacterial Counts.
A single colony of EPEC was inoculated in 2 mL of LB medium overnight. Then 1:100 expanded culture was used to achieve an OD600 of 0.35 or a EPEC density of 1 × 108 cells/mL Transfected Caco-2 cells at 5 × 106 cells/well were pretreated with peptides (5 μM) for 2 h before infection with EPEC at an MOI of 10:1 for 3 h in a 37 °C, 5% CO2 incubator. After infection, cells were washed three times with DMEM supplemented with 10% FBS to remove unattached bacteria, then treated with gentamicin (100 mg/mL) for 2 h to kill extracellular bacteria. The supernatant was discarded, and 500 μL of 1% Triton X-100 was added into cells. The cell lysates of 1,000-fold and 10,000-fold dilutions were plated on LB agar plates and incubated at 37 °C overnight, followed by colony counting.
Western Blot Analysis.
Cells were lysed using 100 μL of RIPA buffer with protease inhibitors for 15 min on ice. Lysates were resolved by SDS/PAGE using a 10% gel and then electrophoretically transferred onto a PVDF membrane. The membrane was blocked and incubated with rabbit anti-GFP antibody (Santa Cruz Biotechnology) at 1:500 dilution at 4 °C overnight, and anti-rabbit HRP-labeled secondary antibody at 1:2,000 dilution. Blots were visualized by an enhanced chemiluminescence detection system (Bio-Rad).
Animal Studies.
All procedures involving animals were approved by Soochow University’s Animal Care and Use Committee. The 6- to 8-wk-old C57BL/6J mice (Pengsheng Bio) were allowed to adapt to the new environment for 1 wk. The mice were housed in single cages and fasted for 24 h before the experiment. The treatment groups received 0.3 mL of mixture liquid, either PBS (PBS group) or a peptide solution containing 0.3 mg of peptide dissolved in a mixed solvent with DMSO and water of 2:1 ratio (peptide group), by gavage with a 4-cm-long curved needle with a steel ball at the tip. Then 0.5 mL of EPEC suspension (108 cfu/mL) was introduced into mice by gavage. After 5 h, mice were relocated to new cages to observe the feces. Mice in the normal control group received 0.5 mL of saline instead of EPEC. Over the course of infection, animals were observed daily for activity level and water intake, and weight was measured. When diarrhea was observed, animals were observed continuously for another 2 h. At the end of the experiments, the animals were killed and intestinal tissues were dissected. The contents from different parts of the intestines were plated on agar plates containing ampicillin. To measure the concentration of peptides in blood and feces, Cy5-p6 (500 μL, 3 mg/mL) was administered by gavage to mice, and at different time points, blood (10 μL) was drawn from eye sockets. In addition, mouse feces were collected, dispersed in water, sonicated, and centrifuged, and the supernatant was collected. The fluorescence signals of the blood and fecal samples were recorded with an excitation wavelength of 650 nm and emission wavelength of 670 nm and compared with a standard curve. For peptide toxicity measurements, each mouse was given a solution containing 0.3 mg of peptide dissolved in 0.3 mL of PBS. The mice were weighed at the same time each day. After 10 d, the mice were killed, and tissues (heart, liver, spleen, lung, and kidney) were sectioned for H&E staining.
Histological Analysis.
Intestines of mice were dissected, washed with PBS, fixed in 10% neutral buffered formalin, processed in a Tissue-Tek VIP 5 processor (Sakura Finetek), and embedded in paraffin. Tissue sections (5 μm thick) were cut with a microtome (Leica EM UC7) and stained with H&E. Images were acquired with a digital microscope (Olympus IX73).
Statistical Analysis.
All data are presented as mean ± SEM. Data comparisons were made using Student’s t test. Differences were considered significant at a P ≤0.05.
Data Availability Statement.
All data for the paper are contained in the main text or SI Appendix.
Supplementary Material
Acknowledgments
This work was partially funded by the University Grants Committee of Hong Kong (GRF Grants 14306317, N_CUHK422/18, 14307218, and AoE/M-09/12), the Food and Health Bureau (Grant HMRF 15140052), and the Jiangsu Key Research and Development Plan (Society Development no. BE2018639).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1914567117/-/DCSupplemental.
References
- 1.Fleming A., On the antibacterial action of cultures of a penicillium, with special reference to their use in the isolation of B. influenzae. Br. J. Exp. Pathol. 10, 226–236 (1929). [Google Scholar]
- 2.Mohr K. I., History of antibiotics research. Curr. Top. Microbiol. Immunol. 398, 237–272 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Brown E. D., Wright G. D., Antibacterial drug discovery in the resistance era. Nature 529, 336–343 (2016). [DOI] [PubMed] [Google Scholar]
- 4.Mendoza N., Ravanfar P., Satyaprakash A., Pillai S., Creed R., Existing antibacterial vaccines. Dermatol. Ther. 22, 129–142 (2009). Correction in: Dermatol. Ther.22, 269 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Jansen K. U., Knirsch C., Anderson A. S., The role of vaccines in preventing bacterial antimicrobial resistance. Nat. Med. 24, 10–19 (2018). [DOI] [PubMed] [Google Scholar]
- 6.Bloom D. E., Black S., Salisbury D., Rappuoli R., Antimicrobial resistance and the role of vaccines. Proc. Natl. Acad. Sci. U.S.A. 115, 12868–12871 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Neu H. C., The crisis in antibiotic resistance. Science 257, 1064–1073 (1992). [DOI] [PubMed] [Google Scholar]
- 8.McManus M. C., Mechanisms of bacterial resistance to antimicrobial agents. Am. J. Health Syst. Pharm. 54, 1420–1433, quiz 1444–1446 (1997). [DOI] [PubMed] [Google Scholar]
- 9.Davies J., Davies D., Origins and evolution of antibiotic resistance. Microbiol. Mol. Biol. Rev. 74, 417–433 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Biswas R., et al. , Molecular epidemiology of Escherichia coli diarrhea in children in Hong Kong. J. Clin. Microbiol. 34, 3233–3234 (1996). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Hoban D. J., Bouchillon S. K., Hawser S. P., Badal R. E., Trends in the frequency of multiple drug-resistant Enterobacteriaceae and their susceptibility to ertapenem, imipenem, and other antimicrobial agents: Data from the Study for Monitoring Antimicrobial Resistance Trends, 2002 to 2007. Diagn. Microbiol. Infect. Dis. 66, 78–86 (2010). [DOI] [PubMed] [Google Scholar]
- 12.Oteo J., Pérez-Vázquez M., Campos J., Extended-spectrum [beta]-lactamase producing Escherichia coli: Changing epidemiology and clinical impact. Curr. Opin. Infect. Dis. 23, 320–326 (2010). [DOI] [PubMed] [Google Scholar]
- 13.Kaper J. B., Nataro J. P., Mobley H. L. T., Pathogenic Escherichia coli. Nat. Rev. Microbiol. 2, 123–140 (2004). [DOI] [PubMed] [Google Scholar]
- 14.Croxen M. A., et al. , Recent advances in understanding enteric pathogenic Escherichia coli. Clin. Microbiol. Rev. 26, 822–880 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Gruenheid S., et al. , Enteropathogenic E. coli Tir binds Nck to initiate actin pedestal formation in host cells. Nat. Cell Biol. 3, 856–859 (2001). [DOI] [PubMed] [Google Scholar]
- 16.Snapper S. B., et al. , N-WASP deficiency reveals distinct pathways for cell surface projections and microbial actin-based motility. Nat. Cell Biol. 3, 897–904 (2001). [DOI] [PubMed] [Google Scholar]
- 17.Weisswange I., Newsome T. P., Schleich S., Way M., The rate of N-WASP exchange limits the extent of ARP2/3 complex-dependent actin-based motility. Nature 458, 87–91 (2009). [DOI] [PubMed] [Google Scholar]
- 18.Dodding M. P., Way M., Nck- and N-WASP-dependent actin-based motility is conserved in divergent vertebrate poxviruses. Cell Host Microbe 6, 536–550 (2009). [DOI] [PubMed] [Google Scholar]
- 19.Donnelly S. K., Weisswange I., Zettl M., Way M., WIP provides an essential link between Nck and N-WASP during Arp2/3-dependent actin polymerization. Curr. Biol. 23, 999–1006 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Yu Y., et al. , PDZ-reactive peptide activates ephrin-B reverse signaling and inhibits neuronal chemotaxis. ACS Chem. Biol. 11, 149–158 (2016). [DOI] [PubMed] [Google Scholar]
- 21.Yu Y., et al. , Targeted covalent inhibition of Grb2–Sos1 interaction through proximity-induced conjugation in breast cancer cells. Mol. Pharm. 14, 1548–1557 (2017). [DOI] [PubMed] [Google Scholar]
- 22.Wittekind M., et al. , Solution structure of the Grb2 N-terminal SH3 domain complexed with a ten-residue peptide derived from SOS: Direct refinement against NOEs, J-couplings and 1H and 13C chemical shifts. J. Mol. Biol. 267, 933–952 (1997). [DOI] [PubMed] [Google Scholar]
- 23.Lu Y., Huang F., Wang J., Xia J., Affinity-guided covalent conjugation reactions based on PDZ-peptide and SH3-peptide interactions. Bioconjug. Chem. 25, 989–999 (2014). [DOI] [PubMed] [Google Scholar]
- 24.Souroujon M. C., Mochly-Rosen D., Peptide modulators of protein-protein interactions in intracellular signaling. Nat. Biotechnol. 16, 919–924 (1998). [DOI] [PubMed] [Google Scholar]
- 25.Cardinale D., et al. , Protein-protein interface-binding peptides inhibit the cancer therapy target human thymidylate synthase. Proc. Natl. Acad. Sci. U.S.A. 108, E542–E549 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Wittekind M., et al. , Orientation of peptide fragments from Sos proteins bound to the N-terminal SH3 domain of Grb2 determined by NMR spectroscopy. Biochemistry 33, 13531–13539 (1994). [DOI] [PubMed] [Google Scholar]
- 27.Winkler D. F., Campbell W. D., The spot technique: Synthesis and screening of peptide macroarrays on cellulose membranes. Methods Mol. Biol. 494, 47–70 (2008). [DOI] [PubMed] [Google Scholar]
- 28.Qian Z., et al. , Early endosomal escape of a cyclic cell-penetrating peptide allows effective cytosolic cargo delivery. Biochemistry 53, 4034–4046 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Qian Z., et al. , Discovery and mechanism of highly efficient cyclic cell-penetrating peptides. Biochemistry 55, 2601–2612 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Pei D., Buyanova M., Overcoming endosomal entrapment in drug delivery. Bioconjug. Chem. 30, 273–283 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sambuy Y., et al. , The Caco-2 cell line as a model of the intestinal barrier: Influence of cell and culture-related factors on Caco-2 cell functional characteristics. Cell Biol. Toxicol. 21, 1–26 (2005). [DOI] [PubMed] [Google Scholar]
- 32.Aranda K. R., Fagundes-Neto U., Scaletsky I. C., Evaluation of multiplex PCRs for diagnosis of infection with diarrheagenic Escherichia coli and Shigella spp. J. Clin. Microbiol. 42, 5849–5853 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kobayashi H., et al. , Prevalence and characteristics of shiga toxin-producing Escherichia coli from healthy cattle in Japan. Appl. Environ. Microbiol. 67, 484–489 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Leomil L., Pestana de Castro A. F., Krause G., Schmidt H., Beutin L., Characterization of two major groups of diarrheagenic Escherichia coli O26 strains which are globally spread in human patients and domestic animals of different species. FEMS Microbiol. Lett. 249, 335–342 (2005). [DOI] [PubMed] [Google Scholar]
- 35.Brown M. D., Bry L., Li Z., Sacks D. B., Actin pedestal formation by enteropathogenic Escherichia coli is regulated by IQGAP1, calcium, and calmodulin. J. Biol. Chem. 283, 35212–35222 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Campellone K. G., Robbins D., Leong J. M., EspFU is a translocated EHEC effector that interacts with Tir and N-WASP and promotes Nck-independent actin assembly. Dev. Cell 7, 217–228 (2004). [DOI] [PubMed] [Google Scholar]
- 37.Campellone K. G., Leong J. M., Nck-independent actin assembly is mediated by two phosphorylated tyrosines within enteropathogenic Escherichia coli Tir. Mol. Microbiol. 56, 416–432 (2005). [DOI] [PubMed] [Google Scholar]
- 38.Brady M. J., Campellone K. G., Ghildiyal M., Leong J. M., Enterohaemorrhagic and enteropathogenic Escherichia coli Tir proteins trigger a common Nck-independent actin assembly pathway. Cell. Microbiol. 9, 2242–2253 (2007). [DOI] [PubMed] [Google Scholar]
- 39.Allen-Vercoe E., Waddell B., Toh M. C., DeVinney R., Amino acid residues within enterohemorrhagic Escherichia coli O157:H7 Tir involved in phosphorylation, alpha-actinin recruitment, and Nck-independent pedestal formation. Infect. Immun. 74, 6196–6205 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Swimm A., et al. , Enteropathogenic Escherichia coli use redundant tyrosine kinases to form actin pedestals. Mol. Biol. Cell 15, 3520–3529 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Elsinghorst E. A., Measurement of invasion by gentamicin resistance. Methods Enzymol. 236, 405–420 (1994). [DOI] [PubMed] [Google Scholar]
- 42.Weflen A. W., Alto N. M., Viswanathan V. K., Hecht G., E. coli secreted protein F promotes EPEC invasion of intestinal epithelial cells via an SNX9-dependent mechanism. Cell. Microbiol. 12, 919–929 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Savkovic S. D., Villanueva J., Turner J. R., Matkowskyj K. A., Hecht G., Mouse model of enteropathogenic Escherichia coli infection. Infect. Immun. 73, 1161–1170 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Luperchio S. A., Schauer D. B., Molecular pathogenesis of Citrobacter rodentium and transmissible murine colonic hyperplasia. Microbes Infect. 3, 333–340 (2001). [DOI] [PubMed] [Google Scholar]
- 45.Zhang Y., et al. , Site-selective lysine reactions guided by protein–peptide interaction. Biochemistry 58, 1010–1018 (2019). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data for the paper are contained in the main text or SI Appendix.