Significance
Chimeric antigen receptors (CARs) have established themselves as a powerful means to redirect the natural properties of T cells against tumors. However, most clinical trials fail due to limited CAR-T cell expansion. Among several factors that can influence CAR-T cell expansion, the precise location of CAR-T vector into the patient’s genome can play an essential role in the treatment outcome. An outstanding question is whether CAR-T integration at certain genomic regions can rewire the regulatory landscape of cells. We report the development of an assay for the joint profiling of the epigenome and lentiviral integration site analysis at population and single-cell resolutions. We anticipate that our method should enable discovering cellular fates associated with durable CAR-T treatment.
Keywords: epigenetics, CAR-T cell, single-cell genomics, lentiviral integration site
Abstract
Chimeric antigen receptor (CAR)-T immunotherapy has yielded impressive results in several B cell malignancies, establishing itself as a powerful means to redirect the natural properties of T lymphocytes. In this strategy, the T cell genome is modified by the integration of lentiviral vectors encoding CAR that direct tumor cell killing. However, this therapeutic approach is often limited by the extent of CAR-T cell expansion in vivo. A major outstanding question is whether or not CAR-T integration itself enhances the proliferative competence of individual T cells by rewiring their regulatory landscape. To address this question, it is critical to define the identity of an individual CAR-T cell and simultaneously chart where the CAR-T vector integrates into the genome. Here, we report the development of a method called EpiVIA (https://github.com/VahediLab/epiVIA) for the joint profiling of the chromatin accessibility and lentiviral integration site analysis at the population and single-cell levels. We validate our technique in clonal cells with previously defined integration sites and further demonstrate the ability to measure lentiviral integration sites and chromatin accessibility of host and viral genomes at the single-cell resolution in CAR-T cells. We anticipate that EpiVIA will enable the single-cell deconstruction of gene regulation during CAR-T therapy, leading to the discovery of cellular factors associated with durable treatment.
Cancer immunotherapy is emerging as an effective and dependable approach to induce durable responses and survival benefit in several cancers. Based on the success of targeting the CD19 protein in B cell malignancies, chimeric antigen receptors (CARs) have established themselves as a powerful means to redirect and enhance the natural properties of both CD8+ and CD4+ T cells against tumors. In the widely used version of this strategy, the T cell genome is modified by the integration of lentiviral or retroviral vectors encoding a CAR transgene that directs tumor cell killing. However, most clinical trials with insufficient overall efficacy have reported poor T cell persistence, suggesting that this therapeutic approach is limited by the extent of CAR-T cell expansion in vivo.
Among several factors that can influence the persistence and clonal expansion of CAR-T cells, including ex vivo culture conditions and preconditioning regimens, the precise location of CAR-T vector integration into the patient’s genome can play an essential role in the treatment outcome. A major outstanding question is whether CAR-T integration at certain genomic regions can rewire the regulatory landscape of individual cells, thereby enhancing the proliferative competence of CAR-T cells in vivo, or if passive CAR-T integration in cells with intrinsic proliferative advantage can lead to clonal expansion and successful tumor killing. To determine the extent to which these two scenarios occur in vivo, it is essential to simultaneously determine T cell fate and map where CAR-T vectors integrate into the genome.
The most widely used methods to perform lentiviral integration site analysis are ligation-mediated (LM) and linear amplification-mediated PCR (1–3). Both techniques involve ligation of a linker DNA cassette to fragmented genomic DNA, which enables PCR amplification between known sequences in the viral long-terminal repeat (LTR) and the linker DNA. High-throughput sequencing is then used to sequence the host integration site DNA between the LTR and the flanking host sequence (4). Although PCR-based techniques have been suggested to report integration events at the single-cell level (5), it is currently impossible to define the identity of an individual cell and its lentiviral integration site at the same time.
Here, we report the development of an assay called EpiVIA for the joint profiling of the epigenome and lentiviral integration site analysis at population and single-cell resolutions. We reasoned that the hyperactive Tn5 transposase, used in the assay for transposase-accessible chromatin using sequencing (ATAC-seq) (6), can also transpose proviral DNA, the genetic material of a lentivirus incorporated into the host genome. We postulated that Tn5 can insert sequencing adapters into host-viral fragments and that the precise alignment of these chimeric fragments to the host genome can pinpoint the lentiviral integration sites. We first establish the utility of our experimental and computational workflow in clonal CAR-T cells in addition to clonal HEK293T cell lines whose integration sites were measured by LM-PCR, demonstrating that integration sites measured by EpiVIA are of high accuracy. We then demonstrate the ability to measure lentiviral integration sites and chromatin accessibility at the single-cell resolution by performing single-cell (sc) ATAC-seq (7) in thousands of CAR-T cells. The application of EpiVIA across individual CAR-T cells revealed that the lentiviral integration favors introns and Alu repeats, in agreement with earlier studies reporting the HIV-1 integration preferences at such genomic regions (2, 5, 8). Strikingly, a significant number of CAR-T integration events occurred at genomic regions that were inaccessible across the population of cells. In addition to charting the chromatin accessibility state of host genome, EpiVIA was also able to detect the accessibility state of the viral genome at the single-cell resolution. Because the standard analysis of bulk and scATAC-seq datasets reveals several layers of cell identity, including the unbiased identification of regulatory elements (9), inference of transcription factor binding sites (10, 11), and nucleosome positions (12), we anticipate that EpiVIA’s addition of retroviral integration site analysis to this multifaceted assay should enable discovering cellular fates associated with durable CAR-T treatment.
Results
Reconstructing Lentiviral Integration Sites from Chromatin Accessibility Measurements Using EpiVIA.
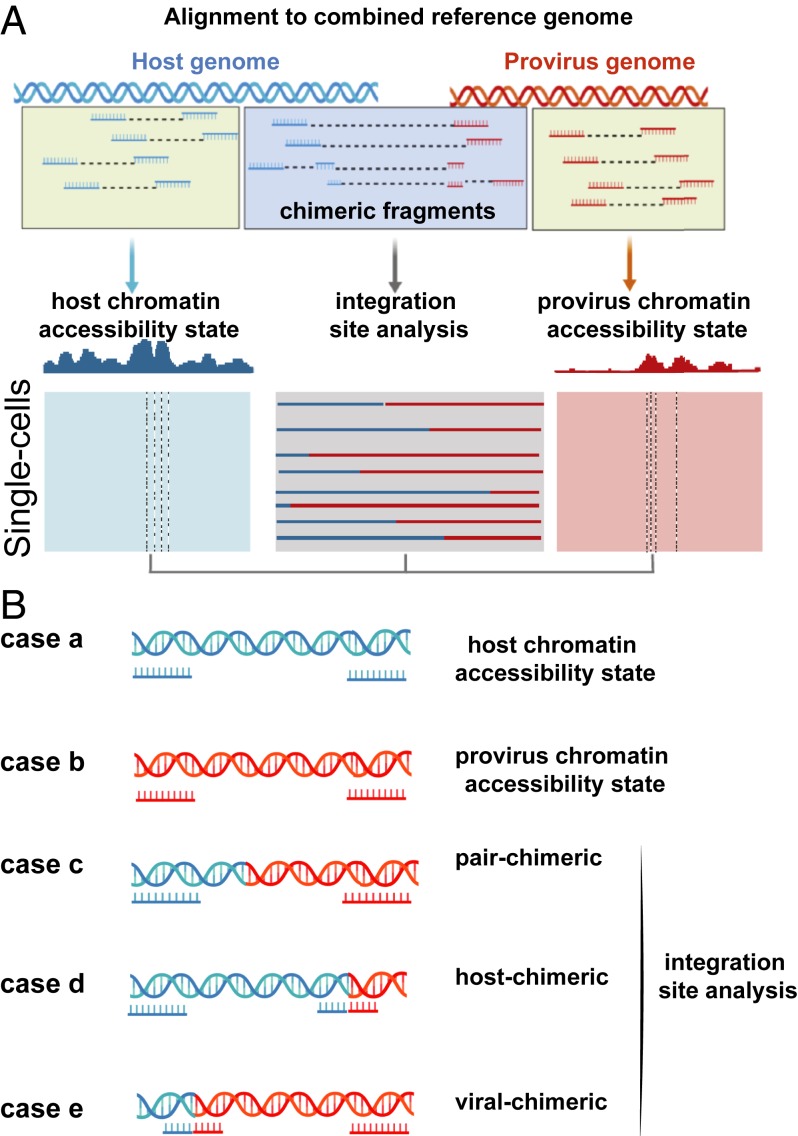
We postulated that the transposase used in the ATAC-seq protocol can also fragment the proviral genome, and that the paired-end sequencing of such fragments followed by aligning the reads to host and viral genomes can delineate the precise location of lentiviral integration events (Fig. 1A). The first step of the EpiVIA workflow is to create a combined reference genome with the provirus sequence as an extra chromosome appended to the human genome (SI Appendix, Fig. S1). We exploited bwa (13), a Burrows–Wheeler aligner, which is capable of mapping paired-end reads to two distinct chromosomes. In a combined host-viral genome, five possibilities exist for mapping the two ends of a fragment: (case A) Mapping of both ends to the host genome, (case B) mapping of both ends to the viral genome, (case C) mapping of one end to the viral genome and the other end to the host genome (referred to as “pair-chimeric”), (case D) mapping of one end to the host genome and the other end to the host and viral genomes (referred to as “host-chimeric”), and (case E) mapping of one end to the viral genome and the other end to both host and viral genomes (referred to as “viral-chimeric”) (Fig. 1B). Since cellular state information is propagated by gain and loss in the accessibility of regulatory elements, the fragments that only align to host (case A) chart the chromatin accessibility state of the host genome, forming the basis for defining cell identity. The fragments that only align to provirus (case B) can chart the chromatin accessibility state of the viral genome, assessing potential silencing of CAR transgene during treatment. On the other hand, the host sequence in chimeric host-viral fragments (cases C to E) can pinpoint CAR-T integration sites in the host genome. Because chromatin accessibility can be mapped at the single-cell level (7, 14), our approach can further enable us to simultaneously perform CAR-T integration site analysis and profile chromatin accessibility at the single-cell resolution.
Fig. 1.
Workflow of EpiVIA. (A) Schematic illustration of EpiVIA. By mapping the ATAC-seq data to the combined reference, EpiVIA is able to identify integration sites and provirus accessibility, in addition to host chromatin state. (B) Five different categories of ATAC-seq fragments. EpiVIA is able to identify five different categories of fragments based on the results of alignment to the combined reference genome.
Validation of EpiVIA’s Integration Site Analysis in Clonal Cells with Predefined Integration Sites.
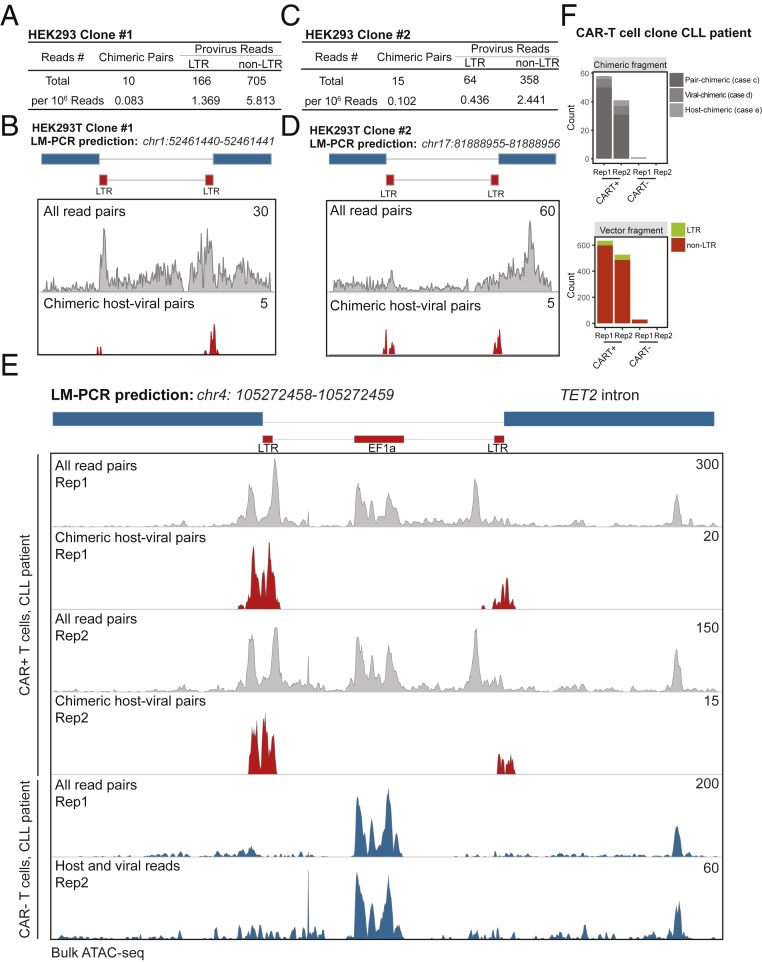
We examined if EpiVIA can reliably detect lentiviral integration events by comparing its predictions with integration site analysis performed by the LM-PCR technique in clonal contexts. First, we generated maps of chromatin accessibility using bulk ATAC-seq in two clones, each consisting of purified HEK293T cell lines with a single lentiviral integration site, which has been previously determined by LM-PCR (clones 1 and 2 used in ref. 15). We analyzed ATAC-seq data in these clonal cells by EpiVIA and found that chimeric host-viral fragments (the pair-chimeric case) mapped to the exact same lentiviral integration sites which were previously reported by LM-PCR (Fig. 2 A–D).
Fig. 2.
Validation of EpiVIA with clonal cells. (A) Chimeric and provirus fragments in HEK293 clone #1 identified by EpiVIA. (B) Genome browser view of reads and identified chimeric reads from HEK293 clone #1 mapped to the context of predefined integration site measured by LM-PCR. (C) Chimeric and provirus fragments in HEK293 clone #2 identified by EpiVIA. (D) Genome browser view of reads and identified chimeric reads from HEK293 clone #2 mapped to the context of predefined integration site measured by LM-PCR. (E) Genome browser view of reads and identified chimeric reads from two replicates of CAR+ and CAR− CD8+ cells from a CLL patient at the predefined integration site measured by LM-PCR. (F) Chimeric and provirus fragments in CAR+ and CAR− CD8+ T cells from a CLL patient identified by EpiVIA.
Second, we applied our pipeline to ATAC-seq data in T cells from a patient with chronic lymphocytic leukemia (CLL) treated with CAR-T cells (16) (Fig. 2 E and F). In this patient, following the infusion of CAR-T cells, antitumor activity was evident in the peripheral blood, lymph nodes, and bone marrow (16). At the peak of the response, 94% of CAR-T cells originated from a single T cell clone in which lentiviral vector-mediated insertion of the CAR transgene was detected by LM-PCR at an intron of the methylcytosine dioxygenase gene, TET2. It was suggested that the CAR integration on one allele together with a hypomorphic mutation in this patient’s second TET2 allele disrupted the function of the TET2 gene. Ultimately, this patient went into remission because of the clonal expansion of a single CAR-T cell and has remained cancer free in the 6 y since, with CAR-T cells derived from this single clone still circulating in his peripheral blood (16).
We examined the ability of EpiVIA to determine CAR-T integration sites in CAR+ CD8+ T cells sorted from this patient and used his CAR− CD8+ T cells as a negative control (16). We found the selective enrichment of host-viral chimeric reads at the LTRs of the lentiviral genome in CAR+ but not CAR− T cells, corroborating the integration of the provirus in CAR+ T cells (Fig. 2 E and F). Strikingly, all of the chimeric host-viral reads aligned to one region on chromosome 4, which is the intron 9 of TET2, precisely the same genomic region reported by LM-PCR (16) (Fig. 2E). ATAC-seq fragments aligned to the host’s genome further revealed the accessibility of this particular regulatory element in CAR+ T cells while this region was inaccessible in CAR− T cells (gray panel compared with blue panel in Fig. 2E). Of note, some paired-end ATAC-seq reads (Fig. 1B, case B) aligned to the lentiviral vector’s genome in both CAR+ and CAR− T cells due to sequence similarity between the CAR promoter EF1α and the host genome (Fig. 2E). Thus, EpiVIA exploited the host sequences within viral-host chimeric fragments to pinpoint the integration of the CAR vector at the TET2 gene in addition to demarcating chromatin accessibility at the site of integration. Altogether, using multiple clonal contexts with predefined integration sites, we demonstrated that EpiVIA can reliably detect lentiviral integration events in addition to mapping the chromatin accessibility state of the entire genome at the population level.
EpiVIA Can Detect CAR-T Integration Sites at the Single-Cell Level.
To investigate whether EpiVIA can link cell identity and CAR-T integration sites at the single-cell resolution, we mapped chromatin accessibility using scATAC-seq in droplets exploiting the commercially available Chromium platform (10X Genomics) for ∼5,000 human CD8+ T cells. Bulk human T cells from a healthy donor were isolated and activated in vitro with CD3/CD28 Dynabeads and high-dose IL-2 for 24 h, followed by transduction with the CAR lentivirus. The cells were then expanded over the course of 9 days. To ensure the expression of CAR and CD8 proteins, cells were further purified using magnetic beads. Sequencing of scATAC-seq libraries generated 744,921,436 read pairs and the key summary metrics from the Cell Ranger pipeline suggested high quality of single-cell chromatin accessibility data in CAR+ CD8+ T cells (SI Appendix, Fig. S2A). Importantly, the number of fragments per cell and transcriptional start site (TSS) enrichment scores were on par with existing high-quality scATAC-seq datasets (7) (SI Appendix, Fig. S2 B and C).
We first assessed if chimeric host-viral read pairs were detectable across single CAR-T cells. Remarkably, EpiVIA was able to detect 193 CAR-T integration sites in 193 individual cells where the majority of these integration events were unique to each cell (SI Appendix, Fig. S2D). While the chimeric-pair case (case C in Fig. 1B) occurred at 84% of the integration events, chimeric-host and chimeric-viral cases (cases D and E in Fig. 1B) also contributed to detected integration events across individual cells (SI Appendix, Fig. S2E). We next compared the number of cells possessing CAR-T integration sites with the number of cells with chromatin accessibility at the TSS of CD8 T cell marker genes, including CD8A, CD3D, CD3E, and CD3G, since all T cells used in our analysis express the CD8 protein. Surprisingly, the number of cells with at least one integration event was comparable to the number of cells with at least one fragment mapping to the TSS of CD8A or CD3 genes (SI Appendix, Fig. S2E), suggesting that the low detection rate of integration sites by EpiVIA is an inherent limitation of droplet-based single-cell protocols. Considering the low detection of chimeric fragments in the library, we reasoned that sequencing depth might be an important factor limiting the identification of integration sites. Therefore, we subsampled the scATAC-seq data in silico to assess the technical performance of EpiVIA with different sequencing depths (SI Appendix, Fig. S2F). Our analysis indicated that doubling the sequencing coverage (350 million reads to 700 million reads) doubles the number of detected integration events (∼100 to ∼200) and the number of cells with proviral DNA, suggesting that detecting these events largely depends on the sequencing depths (SI Appendix, Fig. S2F). The number of detected integration sites from 10 to 100% of all sequenced reads increased steadily, indicating that the detection rate at current sequencing depth is far from the upper bound. Moreover, cells with detectable integration sites had a larger number of fragments measured by scATAC-seq protocol (SI Appendix, Fig. S2G).
To comprehensively assess the sensitivity of detecting integration sites and proviral reads, we measured the chromatin accessibility of ∼1,000 FACS-sorted CAR+ T cells from the same donor using scATAC-seq, followed by sequencing of ∼407 million read pairs. The primary analysis with the Cell Ranger pipeline suggests high quality of the scATAC-seq data (SI Appendix, Fig. S3 A–C). The application of EpiVIA to these high-coverage scATAC-seq data led to the identification of 188 integration sites from 172 CAR-T cells (SI Appendix, Fig. S3D). Further down-sampling of these scATAC-seq data sets suggests that higher coverage sequencing can improve the sensitity of EpiVIA (SI Appendix, Fig. S3E). Considering the number of fragments from each cell differs a lot in scATAC-seq experiments (7), we further evaluated EpiVIA’s sensitivity based on the number of detectable fragments in each cell. We partitioned the cells in different groups by 20-quantiles according to the number of scATAC-seq fragments and calculated the lentiviral integration sites detected in each group. We found that the sensitivity of both integration sites and proviral reads were positively correlated with the number of fragments across individual cells (SI Appendix, Fig. S3F). In individual cells with more than 72,499 unique fragments, the detection rate of integration sites and proviral reads reached to 35% and 96%, respectively (SI Appendix, Fig. S3F). These results corroborate the influence of sequencing coverage in detecting lentiviral integration sites.
Genomic Features of CAR-T Integration Sites at the Single-Cell Level.
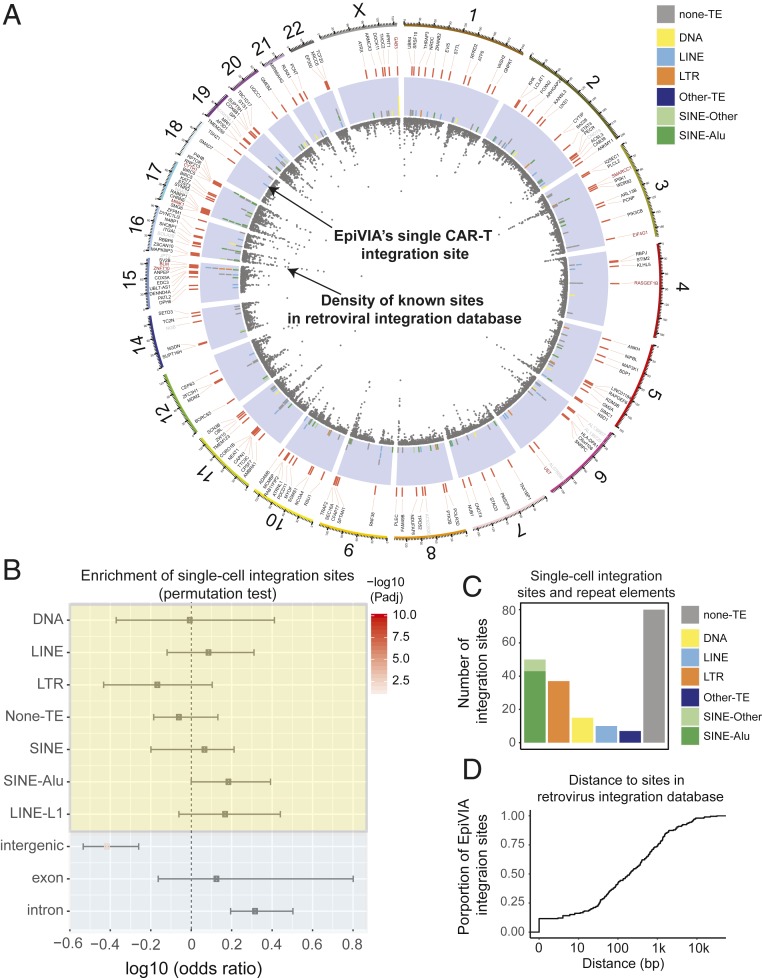
To examine the features of integration sites identified by EpiVIA, we visualized each individual integration event by a solid bar on a Circos plot (Fig. 3A, purple circle). Additional annotations, such as genomic locations of genes, classes of transposable elements (TE) harboring integration sites, and known HIV-1 integration sites distribution were also embedded in the plot. Remarkably, EpiVIA reported that 60% of single-cell integration events occurred at various classes of TEs with a bias toward Alu repeats (Fig. 3 A–C). Furthermore, single-cell integration sites were enriched at intronic regions (Fig. 3B). Taken together, our data are in agreement with the previously reported HIV integration preferences (2, 5).
Fig. 3.
Genomic features of EpiVIA identified integration sites in single cells. (A) Circos plot visualization of the integration sites across the genome and local genomic features from inner to outer circle: 1) Gray dots: the density of HIV-1 integration sites in RID; 2) purple circle: the distribution of integration sites, with the color indicating different classes of transposable elements of the host sequence; 3) genes that harbor these integration sites (gene location red bar), color of the gene names suggest the frequency of integration into these genes: black indicates the gene is a RIG; red indicates the RIG were integrated more than once in our study; gray indicates the integration into this gene only present once in our study. (B) Plot demonstrates the odds-ratio of EpiVIA’s single integration sites falling in various classes of TEs, exons, introns, and intergenic regions using permutation tests. The comparison was done between fragments used to identify integration sites in CAR-T cells and equal number of randomly selected fragments with similar GC content from the scATAC-seq data. The test was repeated for 100 times and P values were calculated by Fisher’s exact test, followed by FDR correction. (C) Bar plot demonstrates the number of integration sites located in different classes of TEs. (D) Accumulative distribution demonstrates distance of EpiVIA identified integration sites to HIV-1 integration sites reported in RID (17).
We next assessed if the integration sites measured by EpiVIA across individual cells have been previously reported by the PCR-based techniques for lentiviral integration sites in HIV-1. Enumerating HIV-1 integration events from the Retrovirus Integration Database (RID) (17), which includes in vitro HIV-1 integration sites in CD4+ T cells and HEK293T cells in addition to in vivo HIV-1 integration sites from patients collected in multiple studies (18–26) and one National Center for Biotechnology Information (NCBI) dataset (SRP065157), revealed the proximity of EpiVIA’s predictions to the previously reported HIV-1 integration sites (Fig. 3A, gray dots in inner circle). Indeed, nearly half of EpiVIA’s detected CAR-T integration sites (84) were located within 100 bp of previously measured integration sites (Fig. 3D), and 93% (148 of 159) of the genes (Fig. 3A, gene symbol in black and red) with integration sites have reported HIV-1 integration in RID, which are thus termed recurrent integration genes (RIGs) (2). Moreover, eight of these RIGs (Fig. 3A, gene symbol in red) harbor more than one independent CAR-T integration site in our analysis. The proximity to known lentiviral integration sites and identification of RIGs is in concert with the existence of integration hotspots (2), displaying the high-quality of EpiVIA’s single-cell resolution analysis.
Single-Cell Mapping of the Accessibility State of Proviral DNA.
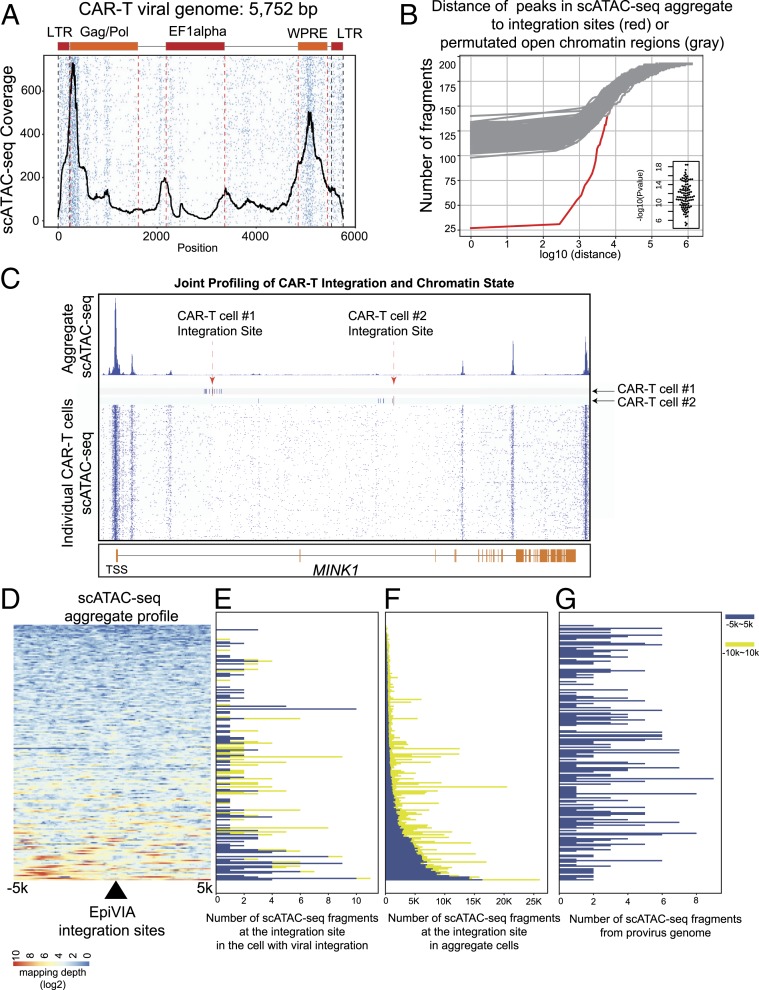
Histone proteins can be loaded onto retroviral DNA soon after nuclear entry (27). We postulated that EpiVIA’s results can be used to examine the activity of regulatory elements in the viral genome by measuring the accessibility state of viral DNA. The majority (95%) of fragments mapping to the viral vector sequence (case B, Fig. 1B) aligned to non-LTR segments since LTR sequences are mostly host-viral chimeric (SI Appendix, Fig. S4A) (case C to E, Fig. 1B). In particular, the woodchuck-hepatitis posttranscriptional response element (WPRE) required for enhancing the transgene expression, the transgene’s promoter EF1α, and the packaging signal element in Gag/Pol demonstrated accessibility across the majority of cells (Fig. 4A). Moreover, the periodicity of fragments aligning to the viral genome across individual cells is a reflection of nucleosome positioning within the viral vector, which can be utilized for examining the in vivo silencing of the transgene during CAR-T therapy (Fig. 4A).
Fig. 4.
Chromatin state of provirus sequence and host genome at integration sites. (A) Heatmap demonstrates the accessibility of different regulatory elements in the provirus genome at single-cell level. The average profile at the viral genome is depicted in black. (B) Accumulative distribution demonstrates distance between identified chimeric fragments to the peaks in aggregated scATAC-seq (red) in comparison with the distance between permuted open chromatin fragments to peaks in the aggregated scATAC-seq (gray), with the P value calculated with Wilcoxon rank-sum test. (C) Host chromatin accessibility of a gene with two independent integration events in two individual CAR-T cells. Genome browser view depicts MINK1 locus with CAR-T integration sites identified by EpiVIA in two single CAR-T cells together with scATAC-seq data across all single cells. Red arrows indicate the CAR-T integration sites and navy blue bars represent genomic locations of scATAC-seq fragments in two single CAR-T cells. Heatmap depicts the scATAC-seq fragments across all CAR-T cells. (D) Heatmap demonstrates mapping depth of aggregated scATAC-seq data in the 10-kb host genome centered on EpiVIA’s single-cell integration sites. The integration sites are sorted by the average mapping depth in this region. (E) Bar plot demonstrates the number of host fragments within 5 and 10 kb of integration sites in the cell that harbor the integration site. The integration sites are sorted in the same order as in D. (F) Bar plot demonstrates the total number of host fragments within 5 and 10 kb of integration sites across all single cells. The integration sites are sorted in the same order as in D. (G) Bar plot demonstrates the number of scATAC-seq fragments in the proviral genome from the cell that have the integration site. The integration sites are sorted in the same order as in D.
Mapping the Accessibility State of Host Chromatin at Lentiviral Integration Sites with Single-Cell Resolution.
We next examined the accessibility state of host chromatin at integration sites of individual cells. We found that ∼10% (19 of 193) of integration events measured by EpiVIA colocalized within an open chromatin region in the aggregate scATAC-seq or bulk ATAC-seq profiles, while the majority of detected integration events did not seem to occur at constitutively open chromatin regions (Fig. 4 B–G and SI Appendix, Fig. S4B). A representative example of CAR-T integration at inaccessible chromatin is MINK1 (Fig. 4C), which is a member of MAP kinase family proteins (MAP4Ks) involved in T cell development and activation (28). The genome-browser view of the 70-Kbp MINK1 locus depicts chromatin accessibility across individual CAR-T cells together with EpiVIA’s detection of two high-quality integration sites landing at introns of this gene in two CAR-T cells (Fig. 4C). Although the MINK1 promoter is constitutively accessible in majority of CAR-T cells, suggesting that the gene is transcriptionally poised or active, the two independent lentiviral integration sites (vertical red lines) occurred at intronic regions that are constitutively inaccessible across the majority of cells. Moreover, multiple open chromatin fragments in the two T cells proximal to the sites of integration suggest alterations of the chromatin landscape after CAR-T integration (vertical blue lines in Fig. 4C). We further examined this notion on all of the integration sites where proximal chromatin fragments in the individual cell carrying the lentiviral integration demonstrated local chromatin accessibility in that cell but not in the majority of other cells (Fig. 4 D–F). These results suggest that the lentiviral integration may occur at inaccessible chromatin of an individual cell and lead to gains in chromatin accessibility at the site of integration in that cell. Although it is not feasible to exclude the possibility that the CAR-T integration site in that one cell alone had been accessible before CAR-T integrated itself into the genome, it is clear that integration events do not frequently occur at sites which are constitutively open across the majority of cells. Such an interpretation is consistent with the accessibility of chromatin at the intron of TET2 gene in CAR+ but not CAR− T cells in the CLL patient treated with CAR-T therapy (Fig. 2E).
To further investigate the relationship between the chromatin accessibility state of integration sites and the accessiblity state of the provirual genome, we counted the number of viral fragments in cells with detected integration sites. Although the accessibility of the local chromatin at integration site varies a lot, we were not able to detect a correlation between number of viral fragments and the number of fragments in that one cell or in all CAR-T cells (Fig. 4G and SI Appendix, Fig. S4 D–G). Taken together, our data corroborate the accessibility of proviral genome even in cells whose integration sites demonstrate low level of accessibility.
We next aimed to evaluate histone modifications of generic CD8 T cells at CAR-T integration sites. It is evident that the inaccessibility of local chromatin at integration sites does not indicate functional repression (29), since nucleosomes with distinct histone modifications can occupy introns of actively transcribed genes (30, 31). It is well-established that HIV-1 preferentially integrates in active genes (2) and super enhancers (32). We next quantitated the levels of histone modifications in primary CD8 T cells using chromatin immunoprecipitation sequencing (ChIP-seq) features from the Roadmap Epigenomics Project (33) at EpiVIA’s CAR-T integration sites. We found that the chromatin state at the majority of integration sites was depleted of repressed histone modifications (H3K9me3), while there were higher levels of H3K27ac, H3K4me3, and H3K36me3 at CAR-T integration sites (SI Appendix, Fig. S4H), in agreement with the reported preference of HIV-1 integration. Furthermore, the level of H3K36me3 modification, which is indicative of active transcription at our integration sites, was comparable to the level of this modification across gene bodies of RIGs in a recent study (32).
Mapping the Accessible Chromatin Landscape of Single CAR-T Cells.
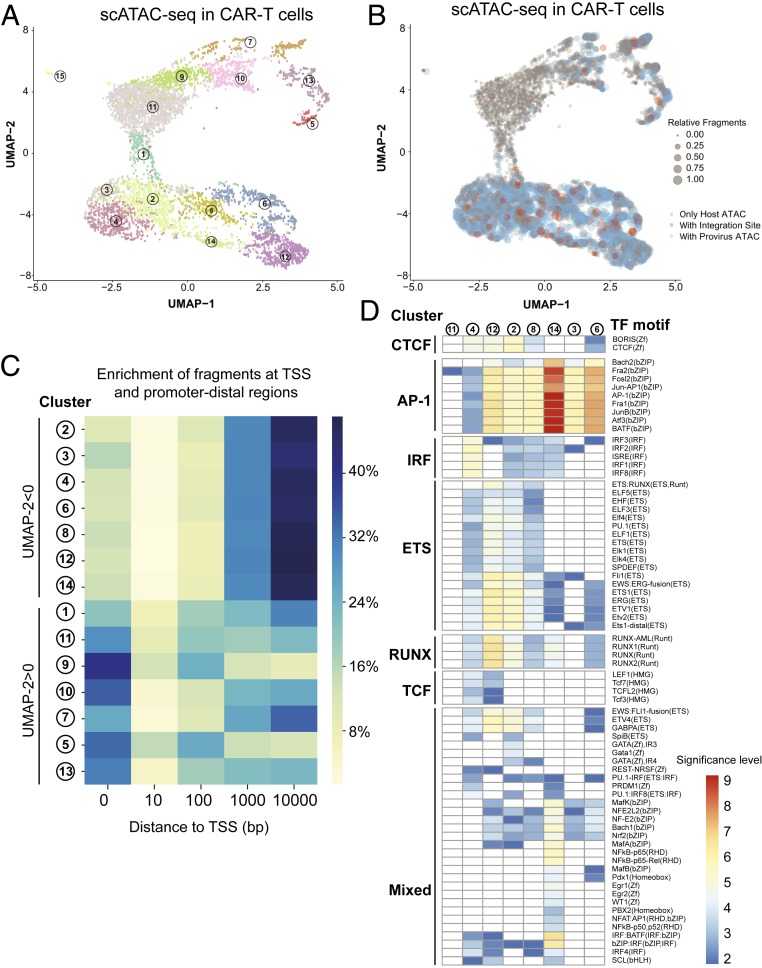
We next investigated the heterogeneity of chromatin accessibility across single CAR-T cells. Visualizing cells with uniform manifold approximation and projection (UMAP) (34), a nonlinear dimensionality-reduction technique that preserves local and global intercluster relationships, revealed cells falling into two major classes with positive or negative values for dimension 2 of UMAP (UMAP-2) (Fig. 5A). Clustering scATAC-seq profiles using SnapATAC (35) refined these two classes, identifying 15 groups of cells. Marking individual cells with detectable CAR-T integration site or the proviral genome revealed a relatively uniform distribution of CAR-T sequence across most clusters, with the exception of clusters 11 and 9, which were depleted of the viral genome (Fig. 5B and SI Appendix, Fig. S5 A and B). We examined what might have contributed to low detection of integration sites in clusters 11 and 9 and found that cells in these two clusters had the lowest average fragment numbers, suggesting that low sequencing depth in these cells contributed to limited detection of integration sites or the proviral genome (SI Appendix, Fig. S5 C and D). Hence, EpiVIA displays high detection rate in high-quality single cells.
Fig. 5.
Single-cell chromatin state of the host genome. (A) UMAP projection of scATAC-seq profiles of CAR+ CD8 T cells. Dots represent individual cells and the colors indicate the cluster of the cell identified by Snap-ATAC (35). (B) UMAP projection of scATAC-seq profiles of CAR+ CD8 T cells. The colors indicate integration site or provirus fragments present in individual cells, and the size of the dots suggests the number of fragments sequenced in the cell. (C) Heatmap visualization of distance of peaks in each cluster to the closest TSS. (D) Heatmap representation of enrichment level of transcription factor (TF) motifs in the unique peaks of each cluster. CTCF, CCCTC-binding factor; ETS, ETS proto-oncogene 1, transcription factor; IRF, interferon regulatory factor; RUNX, runt-related transcription factor (RUNX); and TCF, T cell factor.
Unlike cells in clusters 9 and 11, cells in clusters 5, 7, 10, and 13 had similar number of fragments and sequence coverage compared to other cells with negative values for UMAP-2 (SI Appendix, Fig. S5C). We next investigated what other factors contributed to the separation of cells with respect toUMAP-2. We found cells with positive values for UMAP-2 to have a smaller percentage of fragments mapped to noncoding genomic regions compared with cells with negative values for UMAP-2 (Fig. 5C). One possibility is that the CAR-T cells in clusters 5, 7, 10, and 13 represent different stages of the cell cycle in comparison with cells in clusters 2, 3, 4, 6, 8, 12, and 14. Evidence supporting this notion is the recent report on the depletion of ATAC-seq fragments at noncoding regions in mitotic cells (36). Although we cannot exclude the possibility that chromatin accessibility of cells in these clusters might reflect the nonuniformity of Tn5 treatment across individual cells, our scATAC-sq data suggest that the chromatin landscape of in vitro activated CAR-T cells is heterogeneous.
We further classified clusters of CAR-T cells based on the enrichment of transcription factor recognition motifs at differentially accessible genomic regions. While unique peaks in clusters 2, 3, 6, 8, 12, and 14 were enriched for binding sites of AP-1 family proteins, such as BATF and JUN that have key roles in effector T cell identity (37), unique peaks in cluster 4 were highly associated with HMG proteins, such as TCF-1 and LEF1, which are mostly associated with the naive or memory T cell fate (38). Moreover, the ontology analysis for genes proximal to differentially accessible peaks in aggregate scATAC-seq profiles of clusters 4 and 6 suggested gene signatures of “naive/memory” and “effector” T cells in clusters 4 and 6, respectively (SI Appendix, Fig. S6). Of note, an individual T cell with an integration site in the MINK1 locus (Fig. 4C) is grouped in cluster 6, representing the chromatin accessibility profile of effector T cells. Together, measuring the chromatin accessibility landscape of in vitro activated CAR-T cells using scATAC-seq and analyzing it by EpiVIA enables us to simultaneously 1) detect lentiviral integration sites, 2) define CD8+ T cell fate, and 3) detect the accessibility state of the viral genome at the single-cell resolution.
Discussion
Here, we report the development of EpiVIA, an approach for the joint profiling of the epigenome and lentiviral integration site analysis. To our knowledge, this tool for simultaneously detecting lentiviral integration sites, defining cell fate, and measuring the accessibility state of viral genome at bulk and single-cell levels is unique. We demonstrated EpiVIA’s accuracy using two systems where lentiviral integration sites have been previously identified using LM-PCR: T cells from a patient with CLL treated with CAR-T cells and two HEK293T clones. The adoption of single-cell chromatin accessibility profiling on thousands of cells has just become possible by a commercial system using the droplet technology (7). The scATAC-seq libraries generated by this technology capture a large scale of cells with the sacrifice of limited genomic coverage. In our measurements, the limited coverage is reflected in the percentage of cells with chromatin accessibility at the TSS of cell surface marker genes, such as CD8A, which is comparable to the percentage of cells with CAR-T integration sites. Despite these inherent limitations of scATAC-seq assay, our approach, which is also applicable at the population level, enables the simultaneous measurement of four features in a single assay: 1) Extensive profiling of T cell subsets in circulation before and after CAR-T therapy; 2) analysis of the chromatin landscape in CAR-T cells; 3) detection of CAR-T integration sites; and 4) analysis of active regulatory DNA elements in the CAR-T vector.
Numerous studies over the last few decades suggested that the retroviral integration complex displays a marked tendency to target bent DNA regions, and in particular those wrapped around nucleosomes rather than naked DNA (39–44). However, more recent studies revealed that different families of retroviruses demonstrate distinct integration preferences. While lentiviruses largely prefer the introns of transcriptionally active genes (2), particularly those distributed in the outer shell of the nucleus (45), γ-retroviruses can stably integrate into the host cells with a substantial preference for accessible chromatin regions (46–48). These studies often used methods, such as LM-PCR, to measure integration sites in a population of infected cells and relied on large-scale genomic data mining from populations of uninfected cells, assuming a generic chromatin state for distinct cell types. Despite the lessons learned from these studies highlighting that integration is not a random process and some parts of the genome are favored (49), there is a critical need to develop high-resolution techniques allowing the simultaneous mapping of retroviral integration sites and the chromatin state at the same time. EpiVIA overcomes this limitation, generating a high-resolution map of integration sites at bulk and single-cell levels. Our findings that the microenvironment of integration sites can be inaccessible chromatin are in agreement with a biophysical model for retroviral integration, suggesting the preference of integration at nucleosomal DNA (50). The application of EpiVIA across a large number of cells can generate a high-resolution atlas of retroviral integration sites combined with the chromatin states of cells carrying the viral genome.
A major application of EpiVIA is to understand the underlying mechanisms of clonal expansion. Considering that integration into genes with key roles in cell proliferation may lead to clonal expansion of the target cells (16, 21, 24), the application of EpiVIA across multiple time points after CAR-T infusion, and a comparison between responders and nonresponders to CAR-T therapy can advance our understanding of the underlying mechanisms of successful clonal expansions. Although we aimed to establish the utility of our technique in CAR-T cells, determining factors contributing to clonal expansion of a cell has broad implications in other gene therapies and also during HIV-1 infection. The ability to detect the sequence and chromatin features of the viral genome can also distinguish the replication-competent and replication-incompetent HIV-1, which could greatly advance efforts to identify the latent HIV-1 reservoirs. Although EpiVIA enables us to measure where CAR transgenes integrate in the genome and in which specific cell type, two major factors limiting the sensitivity of lentiviral integration detection are the sequencing depth and sparsity of scATAC-seq measurements. As the cost of sequencing is dropping and the coverage of scATAC-seq is increasing, we believe that repurposing scATAC-seq for detecting lentiviral integration sites and cell identity should have broad applications in many areas including gene therapy and HIV.
Materials and Methods
Workflow of EpiVIA.
EpiVIA aligns ATAC-seq fragments to the combined reference genome and can work at both bulk and single-cell levels. When analyzing scATAC-seq data, alignment results from the Cell Ranger ATAC pipeline are preferred, but bam files from snaptools are also compatible as both have barcode information for each read. EpiVIA parses each read and identifies the chimeric fragments, which are further classified into three different categories based on how the read pair is aligned to the combined reference genome: 1) Pair-chimeric are the fragments with one read mapped to the host genome and the other mapped to the provirus (or vector) genome; 2) host-chimeric are the fragments that both ends mapped to the host genome, with a small soft-clipped fragment at one end which can exactly match the start or end of LTR sequence of provirus; 3) viral-chimeric are the fragments that are properly pair-mapped to either end of the provirus genome, with a soft-clipped fragment at the end that can be mapped to the host genome with a nonzero mapping quality. Since the real chimeric fragment will not be soft-clipped at 3′ end of 5′ LTR and 5′ end of 3′ LTR, while these cases can present in the alignment results, we corrected the aligned position for these fragments. Reading the alignment result of the whole dataset is the most time-consuming step; therefore, EpiVIA has options of “–candidate_bam” and “–chimera_bam” for saving the reads used for chimeric classification and the identified chimeric reads, which can save time while redoing the analysis with different parameters.
We then applied different strategies to identify the integration site of the provirus sequence. Soft-clipping was carefully considered in identifying a potential integration site. Hence, we intended to identify the integration site with the soft-clipped read. If the read mapped to host genome is a soft-clipped alignment, we can determine the precise integration site if the clipped oligonucleotide exactly match either end of the LTR of provirus. In the other case, if the viral read that is aligned to the end of LTR is soft-clipped, we try to search the clipped fragment in the context of up or down 200 bp, where the host read aligned and an exact match can determine the precise integration site. These integration sites are further annotated with genes, transposable elements and enhancer annotation from the University of California, Santa Cruz (UCSC) genome browser database.
EpiVIA reports the results of integration events across individual integration sites at a single-cell level. In this case, we are able to identify cells that carry the same integration site which could possibly result from clonal expansions, and cells that have multiple integration sites, which might be caused by doublets in the scATAC experiment. Specifically, we found there are multiple chimeric reads aligned to the poly(G) region in “LINC00486,” which additionally has many chimeric reads with other chromosomes of the host genome, in both bulk and scATAC-seq data. Therefore, the identified integration sites in this region were excluded from the results.
We also calculated the coverage of the provirus sequence in each cell in EpiVIA. Specifically, for the fragments that are properly mapped to the LTR region of the provirus genome, which can be alternatively aligned to the other LTR, we shifted the aligned position to the 3′ LTR if the pair is reported to aligned to the 5′ LTR in the bam file. Therefore, we are able to remove the PCR duplicates in LTR region. While calculating the coverage of the LTR region, we divided the coverage to both ends.
ATAC-Seq of Clonal Cells.
ATAC-seq was performed as previously described with minor modifications (6). Fifty-thousand cells were pelleted at 550 × g and washed with 1 mL 1× PBS, followed by treatment with 50 mL lysis buffer (10 mM Tris⋅HCl [pH 7.4], 10 mM NaCl, 3 mM MgCl2, 0.1% IGEPAL CA-630). After pelleting nuclei, the pellets were resuspended in 50-mL transposition reaction with 2.5 mL Tn5 transposase (FC-121-1030; Illumina) to tag and fragment accessible chromatin. The reaction was incubated in a 37 °C water bath for 45 min. Tagmented DNA was purified using a MinElute Reaction Cleanup Kit (Qiagen) and amplified with 12 cycles of PCR. Libraries were purified using a QIAQuick PCR Purification Kit (Qiagen). Libraries were paired-end sequenced (38 bp + 37 bp) on a NextSeq 550 (Illumina).
Identification of Integration Site in Clonal Cells.
We applied EpiVIA to the ATAC-seq of HEK293T cell lines with known integration sites and the effective CAR-T clonal cells from a recently reported CLL patient (16). For the analysis of ATAC-seq data from HEK293T cell lines, we built a combined reference genome of the human reference genome GRCh38 (hg38) and p746Vector sequence with Burrows–Wheeler alignment (bwa index). Afterward, we aligned the paired end reads to the combined reference genome using bwa mem, with default parameters. The integration site was identified with our EpiVIA pipeline. Similarly, we did the same analysis with ATAC-seq data of CAR+ and CAR− cells downloaded from the NCBI (GSE112494), replacing the combined reference genome with hg38 and the vector sequence used to build the CAR-T cells.
We used Gviz (51), an R package in Bioconductor, to visualize the ATAC-seq data mapped to provirus sequence, at the integration site and nearby host genome. To visualize the data mapped to these different regions, we selected the reads mapped to the vector genome, upstream and downstream 5 kb of the integration site on host genome from alignment results with samtools, followed by converting the coordinates to a unified coordinate system. We showed EpiVIA identified chimeric reads in a separate panel.
Production of CAR-T Cells.
Bulk human T cells were activated with anti-CD3 and anti-CD28 monoclonal antibody-coated polystyrene beads for 24 h and subsequently transduced with a lentiviral vector encoding a CD19-specific CAR with 4-1BB/CD3ζ domains. T cell expansion was carried out for 9 days, as previously described, followed by cryopreservation. T cells were thawed and checked for purity based on cell-surface expression of CD3, CD4, CD8, and CAR via flow cytometry. For scATAC-seq experiments, the first experiment on ∼5,000 CD8+CAR+ T cells were isolated using bead-based positive selection, according to the manufacturer’s instructions (Miltenyi Biotech), while in the second experiment on ∼1,000 cells, they were FACS sorted with CD8 and CAR.
scATAC-Seq of CAR-T Cells.
scATAC-seq was carried out using the Chromium platform and following the standard protocols provided by 10X Genomics: 1) “Chromium Single Cell ATAC Reagent Kits” and 2) “Nuclei Isolation for Single Cell ATAC Sequencing.” Libraries were paired-end sequenced (50 bp + 50 bp) on a NextSeq 550 (Illumina).
Data Processing with 10X Cell Ranger ATAC Pipeline.
After sequencing, we used Cell Ranger ATAC pipeline (v1.1.0) to generate fastq files and to align these data to the custom reference built with its mkref module. In this pipeline, Cell Ranger generated the fastq files for the pair end ATAC-seq data, cell barcodes, and sample index. Pair-end reads were aligned to the combined reference genome with default parameters after trimming the adapter sequence for each read, and the cell barcodes sequenced were compared with the real barcode whitelist, which were further corrected for the sequencing errors. Most of the reads can be assigned to a real cell barcode, which was recorded in the “CB” tag in the final bam file. We used the real cell barcode to track the chimeric fragments and integration sites if a CB tag is assigned. Otherwise, the sequenced cell barcode read will be used, which is in the CR tag. Fragments with unique start and end positions were stored in the “fragments.tsv.gz” file (generated in Cell Ranger ATAC pipeline) with shifted coordinate accounting for the 9-bp overhang introduced by Tn5, which was used in downstream scATAC-seq analysis.
Integration Site Analysis of CAR-T scATAC-Seq Data.
Alignment results from the Cell Ranger ATAC pipeline were used as input for EpiVIA to identify chimeric fragments and integration sites at a single-cell level. As described in the workflow, we first classified the fragments into different categories and identified the integration site in each cell based on these fragments. Genomic features including gene (exon/intron), TEs, and enhancer were assigned to the integration sites in each cell based on the genome annotation file in UCSC genome browser. The number of integration sites with different genomic features were calculated and plotted with ggplot2. To test whether the integration events preferentially happen in the context of different genomic features, we used a permutation strategy to randomly select equal numbers of fragments from the fragments.tsv.gz file generated in Cell Ranger ATAC pipeline, and used Fisher’s exact test to calculate the odds ratio and P value of the enrichment of every genomic feature at integration site, followed by false-discovery rate (FDR) correction. We plotted the enrichment test results with R showing the min, max and median of odds ratio, with the median adjusted P value shown in the colored point.
To verify the integration site identified with scATAC-seq data, we downloaded previously reported HIV-1 integration sites from RID and converted the hg19 based coordinates to hg38 with liftOver and the hg19 to hg38 chain file (http://hgdownload.soe.ucsc.edu/goldenPath/hg19/liftOver/hg19ToHg38.over.chain.gz) from UCSC. Then we calculated the distance of our identified integration site to its nearest one in the database. The frequency of the integration site at different distances were calculated and plotted with R.
For the Circos plot visualization of integration sites, we calculated the density of integration sites in each 100-kb window across the genome, and plotted the density of integration sites using the scatter feature of Circos (circos.ca). The integration sites were visualized with the tile feature of Circos, which will plot the bar in different layers in the high-density regions. We used different colors to indicate the TE information at the integration site. Finally, we plotted the location and symbol of genes at the integration site with both tile and text features in Circos.
Local Chromatin State of Integration Sites.
We examined the distribution of integration sites across specific epigenetic features to assess the general local chromatin state, and compared that data with randomly selected fragments from the fragments file. We first called the peaks of CAR−, CAR+, and the aggregated scATAC-seq data with macs2 (-B -q 0.01), followed by calculating the distance of the integration sites and randomly selected fragments to the closest peaks. Then the distance of the integration sites were compared with the permuted fragments with the Wilcoxon rank-sum test. The cumulative curve was plotted with the matplotlib in python and P values were plotted with the beeswarm package in R.
For the genome browser view of host scATAC-seq fragments, we used samtools to extract the aligned reads from bam file produced by Cell Ranger ATAC pipeline at MINK1 gene region extending 2 kb at both ends. These alignment results were further processed by bamCoverage and converted to a bedgraph file. We extracted the fragments overlapped with this region with tabix (v1.9) from the fragments.tsv.gz file generated by the 10X pipeline. Fragments in each cell was identified with custom python script and the start sites of the fragments were used to record the positions. We generated a binary matrix in this region based on the presence of fragment at each locus. Specifically, we extracted the fragments from the two cells that have integration sites in this region. MINK1 gene annotation was extracted from UCSC “knownGene29.bb” file, followed by merging overlapping exons from different transcripts to generate a MINK1 exon bed file. All of this information was plotted with the ggplot2 package in R.
In the analysis of scATAC-seq fragments at the integration site, we used tabix (v1.9) to extract the fragments within certain distances (5 kb, 10 kb) from the integration sites. Then we calculated the number of fragments both in the cell that have the integration and in all of the cells, and further calculate the mapping depth in each 50-bp bin across that region. We also counted the number of proviral in the cell that have the integration, and calculated its correlation with accessibility of that cell and all of the cells using stats package from python. The aggregated mapping depth of the 5-kb region at both sides of the integration sites were visualized with pheatmap package in R. The number of fragments in each 5- and 10-kb region at both sides of the integration sites in the single cell, total and average number of fragments in other cells, were plotted with bar plot in matplotlib in python.
We downloaded the fastq files of primary CD8 T cell histone modification data from Roadmap Epigenomics Project on NCBI, including the SRR ids: SRR787510, SRR787519, SRR787520, SRR787521, SRR787545, SRR787546, SRR787547, SRR787548, SRR787549, SRR787550, SRR980433, SRR980434, SRR980435. These data were analyzed with Chip-seq pipeline from nf-core (doi: 10.1101/610741). We plotted the heatmap with deepTools (PMID: 27079975).
ATAC Profile of Provirus Genome.
The mapping depth of each position in the provirus genome is calculated with the EpiVIA pipeline. We visualized the aggregated mapping depth of the provirus genome by summing up the mapping depth of each cell at every position and plotted the information with R. LTR, Gag/Pol, EF1α promoter, and WPRE were shown according to the annotation file of the provirus genome. The ATAC-seq profile of each cell were plotted with matplotlib in python.
scATAC-Seq Analysis of CAR-T Cells.
Only the scATAC-seq data from the first experiment on ∼5,000 CAR-T cells was used for this analysis. We used SnapATAC (35) for the downstream analysis of scATAC-seq data from these CAR-T cells to investigate the host genome chromatin state of each cell. We first generated a snap file according to the tutorial on SnapATAC github page (https://github.com/r3fang/SnapATAC/wiki/FAQs#cellranger_output). To combine this with our integration site analysis, we didn’t do the filtering based on fragments in each cell. In the clustering of cells based on the presence of fragments in bins across the genome, we chose a bin size of 5 kb and used Louvain algorithm to identify the cell clusters based on the first 15 PCs. The peaks of each cluster were called with macs2 (“–nomodel–shift 37–ext 73–qvalue 1e-2 -B–SPMR–call-summits”), which were further used to build cell by peak count matrix. UMAP was used to visualize the cells in a two-dimensional (2D) plot.
We analyzed the distribution of the peaks in each cluster by calculating their distance to the closest transcription start site and visualized with heatmap from seaborn package in python. We then identified the unique peaks in each cluster by subtracting the overlapped peaks called with combined fragments in all other clusters. The motifs of these unique peaks in each cluster were searched with HOMER2 using default parameters, and the enrichment P values were visualized with pheatmap in R.
We also identified the differential accessible region with the built-in function in SnapATAC between some selected clusters. The differential accessible regions were retrieved from SnapATAC and further used to identify the motifs with HOMER2. Closest gene TSSs to each differential peak were also identified and those within 10 kb to the peaks were used to do gene enrichment analysis with metascape (http://metascape.org).
Combined Analysis of Host Chromatin State, Integration Site, and Provirus Accessibility.
We extracted the cluster and dimension reduction results from SnapATAC and combined the information of each barcode with the results from EpiVIA pipeline. Thus, we are able to do the combined analysis with the information from host chromatin state, provirus coverage, and integration site at single-cell level. We used ggplot2 to visualize the distribution of cells have integration site and provirus sequence based on the dimension reduction result with host chromatin state and calculated their distribution across different clusters. We also calculated the aggregated provirus coverage of each cluster and plotted with ggplot2 in R.
Data Availability.
All data are available on the NCBI Gene Expression Omnibus (GSE143647). The code for EpiVIA is freely available at https://github.com/VahediLab/epiVIA.
Supplementary Material
Acknowledgments
We thank Xingzhao Wen for the constructive suggestions on figures in this manuscript and EpiVIA’s code; Dr. Frederic Bushman for constructive discussion and also sharing HEK293T clones with predefined integration sites; and members of the G.V., Pear, Wherry, and Faryabi laboratories for critical discussions. This study is funded by the Penn Epigenetics pilot award, the Sloan Foundation, and NIH Grant R01HL145754 (to G.V.); the Emerging Cancer Informatics Center of Excellence funding from the Penn Institute for Biomedical Informatics and Abramson Cancer Center (G.V. and J.A.F.); NIH Grant T32 A1055428 (to N.G.); and P01CA214278, U54CA24711, U01 AG066100, and R01 CA241762, Alliance for Cancer Gene Therapy Investigator’s Award (to J.A.F.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE143647).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1919259117/-/DCSupplemental.
References
- 1.Niederer H. A., Bangham C. R., Integration site and clonal expansion in human chronic retroviral infection and gene therapy. Viruses 6, 4140–4164 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Schröder A. R., et al. , HIV-1 integration in the human genome favors active genes and local hotspots. Cell 110, 521–529 (2002). [DOI] [PubMed] [Google Scholar]
- 3.Schmidt M., et al. , High-resolution insertion-site analysis by linear amplification-mediated PCR (LAM-PCR). Nat. Methods 4, 1051–1057 (2007). [DOI] [PubMed] [Google Scholar]
- 4.Mueller P. R., Wold B., In vivo footprinting of a muscle specific enhancer by ligation mediated PCR. Science 246, 780–786 (1989). [DOI] [PubMed] [Google Scholar]
- 5.Cohn L. B., et al. , HIV-1 integration landscape during latent and active infection. Cell 160, 420–432 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buenrostro J. D., Giresi P. G., Zaba L. C., Chang H. Y., Greenleaf W. J., Transposition of native chromatin for fast and sensitive epigenomic profiling of open chromatin, DNA-binding proteins and nucleosome position. Nat. Methods 10, 1213–1218 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Satpathy A. T., et al. , Massively parallel single-cell chromatin landscapes of human immune cell development and intratumoral T cell exhaustion. Nat. Biotechnol. 37, 925–936 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lusic M., Siliciano R. F., Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 15, 69–82 (2017). [DOI] [PubMed] [Google Scholar]
- 9.Pliner H. A., et al. , Cicero predicts cis-regulatory DNA interactions from single-cell chromatin accessibility data. Mol. Cell 71, 858–871.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Schep A. N., Wu B., Buenrostro J. D., Greenleaf W. J., chromVAR: Inferring transcription-factor-associated accessibility from single-cell epigenomic data. Nat. Methods 14, 975–978 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cai S., Georgakilas G. K., Johnson J. L., Vahedi G., A cosine similarity-based method to infer variability of chromatin accessibility at the single-cell level. Front. Genet. 9, 319 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Schep A. N., et al. , Structured nucleosome fingerprints enable high-resolution mapping of chromatin architecture within regulatory regions. Genome Res. 25, 1757–1770 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Li H., Durbin R., Fast and accurate short read alignment with Burrows-Wheeler transform. Bioinformatics 25, 1754–1760 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cusanovich D. A., et al. , A single-cell atlas of in vivo mammalian chromatin accessibility. Cell 174, 1309–1324.e18 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Sherman E., et al. , INSPIIRED: A pipeline for quantitative analysis of sites of new DNA integration in cellular genomes. Mol. Ther. Methods Clin. Dev. 4, 39–49 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fraietta J. A., et al. , Disruption of TET2 promotes the therapeutic efficacy of CD19-targeted T cells. Nature 558, 307–312 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Shao W., et al. , Retrovirus Integration Database (RID): A public database for retroviral insertion sites into host genomes. Retrovirology 13, 47 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Han Y., et al. , Resting CD4+ T cells from human immunodeficiency virus type 1 (HIV-1)-infected individuals carry integrated HIV-1 genomes within actively transcribed host genes. J. Virol. 78, 6122–6133 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ikeda T., Shibata J., Yoshimura K., Koito A., Matsushita S., Recurrent HIV-1 integration at the BACH2 locus in resting CD4+ T cell populations during effective highly active antiretroviral therapy. J. Infect. Dis. 195, 716–725 (2007). [DOI] [PubMed] [Google Scholar]
- 20.Mack K. D., et al. , HIV insertions within and proximal to host cell genes are a common finding in tissues containing high levels of HIV DNA and macrophage-associated p24 antigen expression. J. Acquir. Immune Defic. Syndr. 33, 308–320 (2003). [DOI] [PubMed] [Google Scholar]
- 21.Maldarelli F., et al. , HIV latency. Specific HIV integration sites are linked to clonal expansion and persistence of infected cells. Science 345, 179–183 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sherrill-Mix S., et al. , HIV latency and integration site placement in five cell-based models. Retrovirology 10, 90 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sunshine S., et al. , HIV integration site analysis of cellular models of HIV latency with a probe-enriched next-generation sequencing assay. J. Virol. 90, 4511–4519 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wagner T. A., et al. , HIV latency. Proliferation of cells with HIV integrated into cancer genes contributes to persistent infection. Science 345, 570–573 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Singh P. K., et al. , LEDGF/p75 interacts with mRNA splicing factors and targets HIV-1 integration to highly spliced genes. Genes Dev. 29, 2287–2297 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Sharaf R., et al. , HIV-1 proviral landscapes distinguish posttreatment controllers from noncontrollers. J. Clin. Invest. 128, 4074–4085 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang G. Z., Wang Y., Goff S. P., Histones are rapidly loaded onto unintegrated retroviral DNAs soon after nuclear entry. Cell Host Microbe 20, 798–809 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Chuang H. C., Wang X., Tan T. H., MAP4K family kinases in immunity and inflammation. Adv. Immunol. 129, 277–314 (2016). [DOI] [PubMed] [Google Scholar]
- 29.Klemm S. L., Shipony Z., Greenleaf W. J., Chromatin accessibility and the regulatory epigenome. Nat. Rev. Genet. 20, 207–220 (2019). [DOI] [PubMed] [Google Scholar]
- 30.Clark D. J., Felsenfeld G., A nucleosome core is transferred out of the path of a transcribing polymerase. Cell 71, 11–22 (1992). [DOI] [PubMed] [Google Scholar]
- 31.Mieczkowski J., et al. , MNase titration reveals differences between nucleosome occupancy and chromatin accessibility. Nat. Commun. 7, 11485 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Lucic B., et al. , Spatially clustered loci with multiple enhancers are frequent targets of HIV-1 integration. Nat. Commun. 10, 4059 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kundaje A., et al. ; Roadmap Epigenomics Consortium , Integrative analysis of 111 reference human epigenomes. Nature 518, 317–330 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Becht E., et al. , Dimensionality reduction for visualizing single-cell data using UMAP. Nat. Biotechnol., 37, 38–44 (2019). [DOI] [PubMed] [Google Scholar]
- 35.Fang R., et al. ,, Fast and accurate clustering of single cell epigenomes reveals cis-regulatory elements in rare cell types. bioRxiv:10.1101/615179 (13 May 2019).
- 36.Oomen M. E., Hansen A. S., Liu Y., Darzacq X., Dekker J., CTCF sites display cell cycle-dependent dynamics in factor binding and nucleosome positioning. Genome Res. 29, 236–249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Kurachi M., et al. , The transcription factor BATF operates as an essential differentiation checkpoint in early effector CD8+ T cells. Nat. Immunol. 15, 373–383 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Johnson J. L., et al. , Lineage-determining transcription factor TCF-1 initiates the epigenetic identity of T cells. Immunity 48, 243–257.e10 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Pruss D., Reeves R., Bushman F. D., Wolffe A. P., The influence of DNA and nucleosome structure on integration events directed by HIV integrase. J. Biol. Chem. 269, 25031–25041 (1994). [PubMed] [Google Scholar]
- 40.Pruss D., Bushman F. D., Wolffe A. P., Human immunodeficiency virus integrase directs integration to sites of severe DNA distortion within the nucleosome core. Proc. Natl. Acad. Sci. U.S.A. 91, 5913–5917 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Müller H. P., Varmus H. E., DNA bending creates favored sites for retroviral integration: An explanation for preferred insertion sites in nucleosomes. EMBO J. 13, 4704–4714 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Matysiak J., et al. , Modulation of chromatin structure by the FACT histone chaperone complex regulates HIV-1 integration. Retrovirology 14, 39 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maskell D. P., et al. , Structural basis for retroviral integration into nucleosomes. Nature 523, 366–369 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Coffin J. M., Hughes S. H., Varmus H. E., Eds., Retroviruses, (Cold Spring Harbor Laboratory Press, 1997). [PubMed] [Google Scholar]
- 45.Marini B., et al. , Nuclear architecture dictates HIV-1 integration site selection. Nature 521, 227–231 (2015). [DOI] [PubMed] [Google Scholar]
- 46.LaFave M. C., et al. , MLV integration site selection is driven by strong enhancers and active promoters. Nucleic Acids Res. 42, 4257–4269 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Poletti V., Mavilio F., Interactions between retroviruses and the host cell genome. Mol. Ther. Methods Clin. Dev. 8, 31–41 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wunsche P., et al. , Mapping active gene-regulatory regions in human repopulating long-term HSCs. Cell Stem Cell 23, 132–146.e9 (2018). [DOI] [PubMed] [Google Scholar]
- 49.Hughes S. H., Coffin J. M., What integration sites tell us about HIV persistence. Cell Host Microbe 19, 588–598 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Michieletto D., Lusic M., Marenduzzo D., Orlandini E., Physical principles of retroviral integration in the human genome. Nat. Commun. 10, 575 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Hahne F., Ivanek R., Visualizing genomic data using Gviz and bioconductor. Methods Mol. Biol. 1418, 335–351 (2016). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data are available on the NCBI Gene Expression Omnibus (GSE143647). The code for EpiVIA is freely available at https://github.com/VahediLab/epiVIA.