While pore-forming toxins (PFTs) are a large class of molecular weapons employed by organisms across the animal kingdom (1), notably, pathogenic bacteria, many mechanistic steps of pore formation in target membranes are unknown. In PNAS, using molecular dynamics (MD) simulations, Vӧgele et al. (2) observed how central lipids corresponding to the pore lumen were expelled during the formation of Streptococcus pneumoniae pneumolysin (PLY) pores. They show that when the insertion of PLY β-hairpins was slow, central lipids escaped through the inner leaflet. When the insertion was rapid or lipid escape hindered, the membrane trapped within the lumen spontaneously formed a liposomal plug which could possibly be expelled by osmotic flows, thus unraveling additional pathways of PLY pore formation.
PLY is a β-PFT (1) whose pores have a large internal diameter of 30 to 50 nm (3), permitting the liposomal plug-formation pathway (2). Several PFT pores such as those by the α-PFT Escherichia coli cytolysin A (ClyA) (4) have smaller internal diameters lesser than 10 nm. We previously showed that oligomeric pore intermediates of ClyA opened by evacuating lipid from their interiors, forming open proteo-lipid pores partially lined with toroidal lipids (5) similar to PLY (2). However, if lipid escape were hindered during pore assembly and lipids trapped within fully oligomerized ClyA pores, how the central lipids would be evacuated from the narrow pore lumen to create open channels is unknown, since smaller diameters may prohibit liposomal plug formation (2). Therefore, in this letter, we investigate the fate of the trapped lipids within the smaller ClyA pores, whose transmembrane domain comprises α-helices in contrast to β-sheets in β-PFTs (1).
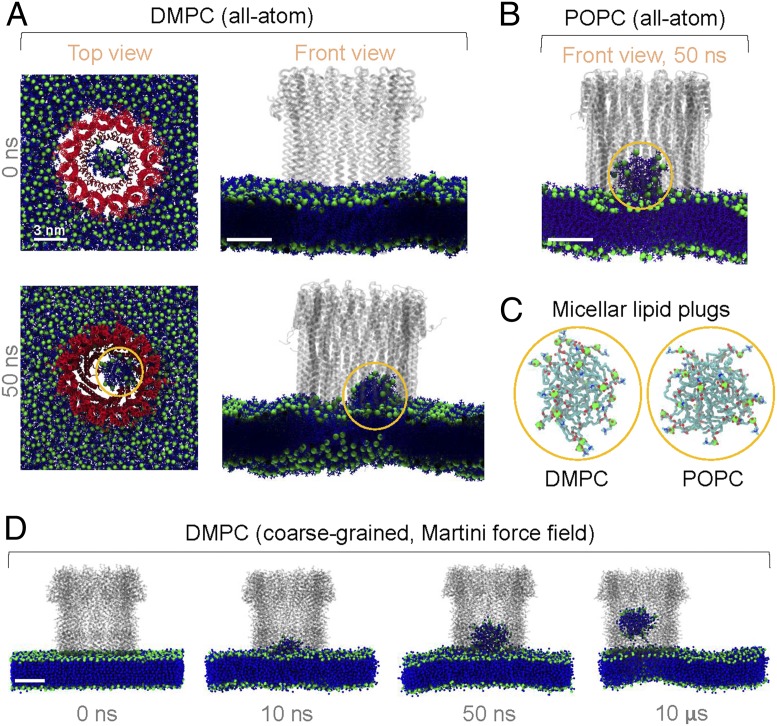
We performed all-atom MD simulations of an intact dodecameric ClyA pore inserted into two different lipid bilayers (Fig. 1 A and B), as done previously (5). Initially, the central lipids were in a lamellar configuration. Remarkably, within 50 ns, the lipids within the lumen spontaneously formed micellar lipid plugs that rose from the plane of both membranes inside the pore channel (Fig. 1 A and B). The configuration of lipids in the self-assembled micellar plugs (Fig. 1C) was similar to that in lamellar membranes, with solvated phosphatidylcholine head groups and desolvated hydrocarbon tails, thus according stability. This suggested that lipid plug formation was primarily driven by the hydrophobic effect. Compared to liposomal formation in the larger PLY pore, spatial constraints in the smaller ClyA pore led to the commensurately smaller number of central lipids favoring the micellar topology. A longer coarse-grained simulation of the ClyA pore further confirmed the spontaneous formation of a micellar plug (Fig. 1D), morphologically similar to those in all-atom simulations. The plug slowly rose through the lumen over 10 µs and could presumably be expelled by osmotic flows.
Fig. 1.
Micellar plug formation inside ClyA pores. (A and B) Snapshots of a ClyA pore in a 1,2-dimyristoyl-sn-glycero-3-phosphocholine (DMPC) membrane (A) (pore, colored red; lipids, blue; phosphorus atoms, green; solvent not shown) and a 1-palmitoyl-2-oleoyl-glycero-3-phosphocholine (POPC) membrane (B) (lipids colored violet) from all-atom MD simulations. (C) Molecular structure of the micellar plugs at 50 ns (highlighted by yellow circles in A and B). (D) Dynamics of the plug formation from Martini (6) coarse-grained simulations using gromacs 2018.6 (7, 8). We set up the initial configuration as done previously (9) and applied ElNeDyn (10) across the pore to avoid distortion of pore geometry (9).
In conclusion, Vӧgele et al. (2) show that liposomal plug formation enabled the opening of large PLY pores. Our work shows that smaller ClyA pores can expel trapped lipids and open via micellar-plug formation, suggesting that both small and large PFT nanopores, either α- or β-PFTs, exhibit generic topological features of lipid rearrangement during pore formation.
Acknowledgments
K.G.A. acknowledges funding by a grant from the Department of Science and Technology, Government of India.
Footnotes
The authors declare no competing interest.
References
- 1.Dal Peraro M., van der Goot F. G., Pore-forming toxins: Ancient, but never really out of fashion. Nat. Rev. Microbiol. 14, 77–92 (2016). [DOI] [PubMed] [Google Scholar]
- 2.Vögele M., et al. , Membrane perforation by the pore-forming toxin pneumolysin. Proc. Natl. Acad. Sci. U.S.A. 116, 13352–13357 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.van Pee K., et al. , CryoEM structures of membrane pore and prepore complex reveal cytolytic mechanism of Pneumolysin. eLife 6, e23644 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Mueller M., Grauschopf U., Maier T., Glockshuber R., Ban N., The structure of a cytolytic α-helical toxin pore reveals its assembly mechanism. Nature 459, 726–730 (2009). [DOI] [PubMed] [Google Scholar]
- 5.Desikan R., Maiti P. K., Ayappa K. G., Assessing the structure and stability of transmembrane oligomeric intermediates of an α-helical toxin. Langmuir 33, 11496–11510 (2017). [DOI] [PubMed] [Google Scholar]
- 6.Marrink S. J., Tieleman D. P., Perspective on the Martini model. Chem. Soc. Rev. 42, 6801–6822 (2013). [DOI] [PubMed] [Google Scholar]
- 7.de Jong D. H., Baoukina S., Ingólfsson H. I., Marrink S. J., Martini straight: Boosting performance using a shorter cutoff and GPUs. Comput. Phys. Commun. 199, 1–7 (2016). [Google Scholar]
- 8.Kutzner C., et al. , More bang for your buck: Improved use of GPU nodes for GROMACS 2018. J. Comput. Chem. 40, 2418–2431 (2019). [DOI] [PubMed] [Google Scholar]
- 9.Desikan R., Patra S. M., Sarthak K., Maiti P. K., Ayappa K. G., Comparison of coarse-grained (MARTINI) and atomistic molecular dynamics simulations of α and β toxin nanopores in lipid membranes. J. Chem. Sci. 129, 1017–1030 (2017). [Google Scholar]
- 10.Periole X., Cavalli M., Marrink S.-J., Ceruso M. A., Combining an elastic network with a coarse-grained molecular force field: Structure, dynamics, and intermolecular recognition. J. Chem. Theory Comput. 5, 2531–2543 (2009). [DOI] [PubMed] [Google Scholar]