Significance
Alternative pre-mRNA splicing is a regulated process that greatly diversifies gene products by changing the exons incorporated into mRNA. This process is dysregulated in cancers. Here, we studied exon usage in aggressive prostate cancers and linked exon incorporation decisions to cancer driver genes. Through computational and experimental studies, we found that a strong cancer driver gene, Myc, was linked to exon changes in genes that themselves regulate alternative splicing. These exons often encoded premature stop codons that would decrease gene expression, suggestive of a Myc-driven autoregulatory loop to help control levels of splicing regulatory proteins.
Keywords: alternative splicing, prostate cancer, Myc, rMATS, PEGASAS
Abstract
We sought to define the landscape of alternative pre-mRNA splicing in prostate cancers and the relationship of exon choice to known cancer driver alterations. To do so, we compiled a metadataset composed of 876 RNA-sequencing (RNA-Seq) samples from five publicly available sources representing a range of prostate phenotypes from normal tissue to drug-resistant metastases. We subjected these samples to exon-level analysis with rMATS-turbo, purpose-built software designed for large-scale analyses of splicing, and identified 13,149 high-confidence cassette exon events with variable incorporation across samples. We then developed a computational framework, pathway enrichment-guided activity study of alternative splicing (PEGASAS), to correlate transcriptional signatures of 50 different cancer driver pathways with these alternative splicing events. We discovered that Myc signaling was correlated with incorporation of a set of 1,039 cassette exons enriched in genes encoding RNA binding proteins. Using a human prostate epithelial transformation assay, we confirmed the Myc regulation of 147 of these exons, many of which introduced frameshifts or encoded premature stop codons. Our results connect changes in alternative pre-mRNA splicing to oncogenic alterations common in prostate and many other cancers. We also establish a role for Myc in regulating RNA splicing by controlling the incorporation of nonsense-mediated decay-determinant exons in genes encoding RNA binding proteins.
Alternative pre-mRNA splicing is a regulated process that governs exon choice and greatly diversifies the proteome. It is an essential process that contributes to development, tissue specification, and homeostasis and is often dysregulated in disease states (1). In cancer, this includes growth signaling, epithelial-to-mesenchymal transition, resistance to apoptosis, and treatment resistance (2). In prostate cancer, our area of interest, the most notable splicing change is the emergence of the ligand-independent androgen receptor ARV7 isoform in response to hormone deprivation (3). Other examples include proangiogenic splice variants of VEGFA (4), tumorigenic variants of the transcription factors ERG and KLF6 (5, 6), and antiapoptotic splicing of BCL2L2 (7, 8). However, the intersection of upstream oncogenic signaling, pre-mRNA splicing, and the biological processes affected by those splicing events has not been defined at a global level.
Prostate cancers progress from hormone-responsive, localized disease to hormone-independent, metastatic disease accompanied by changes in gene expression and mutations that confer cell-autonomous growth and therapeutic resistance (9). The study of disease progression from primary prostate adenocarcinoma (PrAd) to metastatic, castration-resistant prostate cancer (mCRPC) and treatment-related neuroendocrine prostate cancer (NEPC) has been aided by large-scale genomic and transcriptomic studies of patient samples representing each form of the disease (10–13). Examples of driver alterations found in precursor lesions and primary tumors include TMPRSS2-ERG translocations and PTEN loss (14). Metastatic tumors are characterized by Myc and AR amplification (15, 16). NEPC includes near-universal loss of TP53 signaling by inactivating mutation as well as chromosomal loss of RB1 (17). Sequencing efforts and subsequent functional experiments have identified prostate cancer driver alterations and defined the impact of gene expression networks on prostate cancer phenotypes. These studies have led to the successful development of new therapeutics targeting AR signaling and DNA repair in advanced disease (18, 19).
Prostate cancer progression is also associated with shifts in alternative pre-mRNA splicing patterns, but this process is not well understood (20). Investigations of global changes in exon usage in prostate cancer have focused on stage- or race-specific comparisons (21–25). Comparisons of tumor-adjacent benign material and PrAd identified intron retention and exon skipping events in the biomarkers KLK3 and AMACR, respectively (22). Others studying NEPC and PrAd have shown that a network of splicing events controlled by the serine–arginine RNA-binding protein SRRM4 contributes to the neuroendocrine phenotype (26–28). Comparisons of European American and African American (AA) PrAd samples identified an AA-specific splice variant of PIK3CD that enhanced AKT/mTOR signaling (23). How these splicing alterations connect to the driver alterations described above remains to be explored.
The accumulation of RNA-sequencing (RNA-Seq) data in large databases presents a unique opportunity to conduct an analysis of alternative splicing across the full range of prostate cancer disease states. For our study, we prepared a unified dataset of large, publicly available RNA-Seq datasets representing normal tissue, tumor-adjacent benign tissue, primary adenocarcinoma, metastatic castration-resistant adenocarcinoma, and treatment-related metastatic NEPC. However, handling datasets of this size requires splicing analysis software with greater efficiency than what is currently available. To analyze these hundreds of datasets, we created an improved version of our rMATS software (dubbed rMATS-turbo) that can handle this volume of RNA-Seq data (29, 30).
We identify a high-confidence set of exons whose incorporation varies across prostate cancer disease states. By combining expression-level and exon-level analyses, we developed a pathway-guided strategy to examine the impact of oncogenic pathways on incorporation of these exons. This correlational analysis implicates Myc, mTOR, and E2F signaling in the control of exon choice in spliceosomal proteins. To further investigate the contributions of Myc signaling to exon choice, we developed unique engineered human prostate cell lines with regulated Myc expression. Functional experiments in these cell lines identify Myc-dependent exons and experimentally confirm that cassette exon choice in many splicing regulatory proteins is responsive to Myc expression level. These exons often encode frameshifts or premature termination codons (PTCs) that would result in nonsense-mediated decay (NMD). We show that an ultraconserved, NMD-determinant exon in the RNA-binding protein SRSF3 is particularly responsive to Myc signaling. Our results implicate Myc signaling as a regulator of alternative splicing-coupled NMD (AS-NMD) as part of a program of growth control.
Results
Exon-Level Analysis Defines the Landscape of Alternative Pre-mRNA Splicing Across the Prostate Cancer Disease Spectrum.
We combined RNA-Seq data from disparate published datasets representing 876 samples of normal tissue, benign tumor-adjacent material, primary adenocarcinoma, metastatic castration-resistant adenocarcinoma (mCRPC), and treatment-related NEPC (Fig. 1A) (10–13, 31, 32). Metaanalyses of RNA-Seq data with gene- or isoform-level counts are subject to confounding batch effects and rely on existing isoform annotation (33). Exon-level analysis, however, uses a ratio-based methodology to estimate exon incorporation, which may be more robust against batch effects and confounding factors in large-scale RNA-Seq datasets (34–37). In addition, exon-level analysis can detect novel exon–exon junctions and is thus independent of previous annotation.
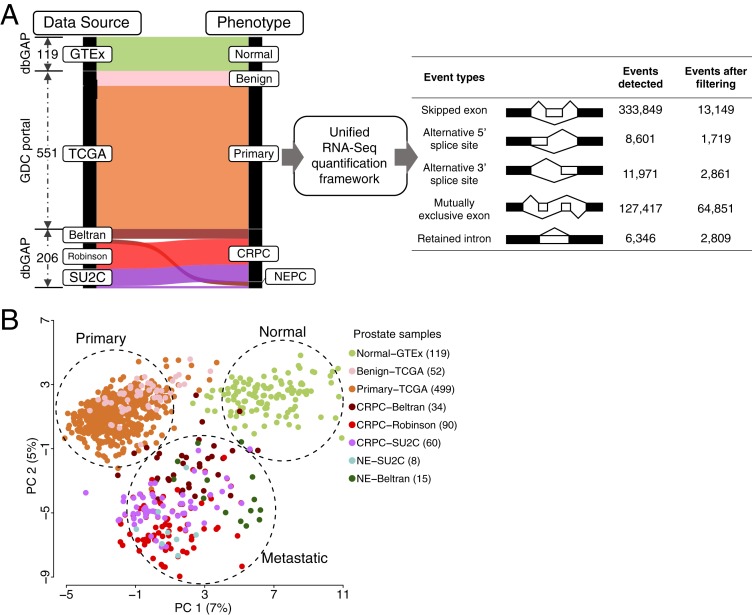
Fig. 1.
A global, exon-level analysis of alternative pre-mRNA splicing in normal prostate and prostate cancers identifies patterns of exon usage in RNA-binding proteins. (A) Schematic with alluvial plot depicting the data-processing workflow combining RNA-Seq data from various prostate tissue disease states (Left) and summary table depicting various exon events detected by rMATS-turbo before and after filtering for splice junction reads coverage, PSI range, and commonality (Right). The alluvial plot depicts the sorting of patient RNA-Seq datasets from individual studies on the Left into prostate phenotypes on the Right. (B) Scatter plot depiction of an unsupervised PCA of exon usage matrices from eight different prostate datasets representing healthy tissue, tumor-adjacent benign tissue, primary prostate cancer, metastatic castration-resistant prostate cancer (mCRPC), and treatment-associated neuroendocrine prostate cancer (NEPC).
To facilitate alternative splicing analysis in this and other large RNA-Seq datasets, we developed rMATS-turbo (also known as rMATS 4.0.2), a computational pipeline that permits the efficient capture, storage, and analysis of splicing information from very large-scale raw RNA-Seq data. This improved pipeline refactors the original ratio-based rMATS software that we developed for splicing analysis in RNA-Seq data to optimize it for very large-scale RNA-Seq datasets and is now available for public use (29, 30). It offers significant improvements in speed and data storage efficiency.
We applied rMATS-turbo to the combined RNA-Seq dataset and identified over 330,000 different cassette exons across all prostate samples. Previous estimates of the diversity of splicing events in human cells vary, but are generally of the same order of magnitude (38). We also identified tens of thousands of additional alternative splicing events (Fig. 1A), including alternative 5′ and 3′ splice sites, mutually exclusive exons, and retained introns. For this study, we focused on cassette exons, as these are the most well-defined type of alternative splicing event. We should note that although the rMATS-turbo software detected numerous mutually exclusive exons, most of these events were in fact part of more complex alternative splicing events; thus, we did not include these mutually exclusive exons in downstream analyses.
Filtering of these exons for coverage (≥10 splice junction reads per event), cross-sample variance (range of percent-spliced-in [PSI] > 5%; mean skipping or inclusion > 5%) and commonality (events detected in ≥1% of all samples) produced a set of 13,149 high-confidence exons with variable incorporation across samples (see Methods). Principal-component analysis (PCA) of this exon usage matrix grouped samples of the same disease phenotype regardless of dataset (Fig. 1B). By comparison, a similar unsupervised analysis of isoform-level count-based metric from the same metadataset grouped samples more by dataset of origin than disease phenotype (SI Appendix, Fig. S1 A and B). This result is consistent with prior observations that the exon-level splicing analysis is more robust against batch effects and other confounding factors in large-scale RNA-Seq datasets (35–37).
Combining Gene Pathway Analysis and Exon Usage Identifies Exon Correlates of Oncogenic Signaling.
Genomic studies of prostate cancer have identified driver alterations associated with disease progression (39). We sought to define how the variable cassette exons we identified and the biological processes they participate in might relate to these oncogenic signals. Instead of selecting single oncogenes for study, we developed PEGASAS (pathway enrichment-guided activity study of alternative splicing), a pathway-guided analytic strategy that uses gene signatures to estimate the activities of signaling pathways and to discover potential downstream exon changes (Fig. 2A). Gene signature-based analyses use an ensemble of features (a set of genes collectively) to estimate pathway activity and outperform single-gene measurements (40). To mitigate potential batch effects in the expression data, we utilized a rank-based metric to calculate the signature score, providing a more robust measure of pathway activity as it is in essence normalized on a per-sample basis (41).
Fig. 2.
Pathway enrichment-guided activity study of alternative splicing (PEGASAS) analysis identifies exon correlates of oncogenic signaling in prostate cancers. (A) Workflow diagram describing PEGASAS correlation of gene signature score with exon usage. Each sample is scored for a gene expression signature of interest. Gene signature scores are correlated with exon usage matrices to identify pathway-correlated exon incorporation changes. (B) Heatmap of the correlation coefficients of the exon changes correlated with gene signatures in the Molecular Signatures Database (MSigDB) hallmark gene sets as generated by PEGASAS. The 10 signatures that returned the highest number of exon correlates are shown here. Each row depicts the results of the correlation to a single hallmark signature. Each column represents a single exon. The color represents the strength and direction of the correlation (red positive, blue negative) of a single exon with each pathway. Columns are sorted by hierarchical clustering. Rows are ranked by total number of exon correlates passing statistical metrics for each pathway (# Events, bar chart). The gene ontology term with the highest enrichment for the genes containing pathway-correlated exons and the corresponding P value are also depicted. The P values correspond to the gene ontology enrichment and are not a measure of significance of pathway–exon correlation. (C) Hive plot depiction of exons correlated with selected prostate cancer-related gene signatures and the biological processes associated with genes containing those exons. All pathway-correlated exons are displayed on the left axis. Seven well-known prostate cancer driver pathways are represented as nodes on the middle axis. The area of these nodes reflects the number of exons correlated with each pathway. The right axis depicts four summary gene ontology terms. The width of the edges connecting the nodes on the middle axis to the nodes on the right axis is proportional to the enrichment of each pathway for each biological process. The size of the nodes on the right axis is proportional to the total number of exons associated with each biological process. (D) Area-proportional Venn diagram depicting the intersection of Myc-, E2F-, and MTOR-correlated exons in prostate cancer. Exons must share the same correlation direction (positive or negative) to appear in the intersection. AS, alternative splicing; K-S, Kolmogorov–Smirnov; SE, skipped exon.
We employed the hallmark gene signature sets maintained by the Molecular Signatures Database (MSigDB) (42). These 50 sets represent a diverse and well-validated array of cellular functions and signaling pathways. To assess the performance of these signatures in our combined dataset, we examined signature scores for the AR, Myc Targets V2, and MTOR gene sets across five different prostate phenotypes. Consistent with previously reported observations of pathway activation in prostate cancer progression, the androgen response gene signature scores we measured were lowest in NEPC samples (SI Appendix, Fig. S2A). Similarly, MTOR and Myc signature scores were higher in mCRPC samples than in normal tissues. The Myc and MTOR signature scores increased between normal healthy donors (Genotype-Tissue Expression [GTEx]) and tumor-adjacent normal (TCGA-PRAD), consistent with field cancerization and tumor–stromal interaction effects on gene expression reported previously by others (43).
We then scored each sample in our metadataset for all 50 pathways and correlated this score with the data matrix of over 13,000 variable cassette exons (Dataset S1). After filtering for correlation strength and false-discovery rate (FDR), each pathway returned between 11 and 1,330 exon correlates (Dataset S1). The 10 gene sets that returned the greatest number of exon correlates with a Pearson’s correlation coefficient greater than 0.3 or less than −0.3 are shown (Fig. 2B). Nine out of 10 of these gene sets had exon correlates found in genes with strong functional enrichment by gene ontology (adjusted P value < 0.05).
Cassette Exons Correlating with Myc, E2F, and MTOR Signaling Are Enriched in Splicing-Related Genes.
We next examined the biological processes specified by the genes containing the variant exons correlated with prostate cancer-relevant hallmark gene sets (Fig. 2C). We also added a signature that describes transcriptional activity due to TMPRSS signaling as this common prostate cancer alteration is not represented by a hallmark gene set (44). Here, we represent the network of data as a hive plot to show how exons (left axis) correlate with signaling pathways (middle axis) and the functional enrichment of genes containing those correlated exons (right axis) (45). Gene ontology analysis indicated that the relatively small number of exons correlated with AR or Notch were modestly enriched in cell adhesion and chromatin remodeling processes. Surprisingly, the numerous exon correlates of Myc, E2F, and MTOR were strongly enriched in genes related to the spliceosome and alternative pre-mRNA splicing. In addition, the overlap in the exon sets correlated with Myc, E2F, and MTOR was striking, with 50 to 60% of exons held in common (Fig. 2D). These pathways play central roles in growth control and are frequently codysregulated in human cancers, so a shared set of exons might be expected from a correlation analysis.
Myc-Correlated Exons Are Found in the Oncogenes SRSF3 and HRAS.
Given the centrality of Myc signaling in tumorigenesis, tumor maintenance, and tumor progression in a multitude of tissue lineages (46, 47) including the prostate, this pathway was selected for further investigation (15, 48, 49). The validity of these correlational results critically depends on the integrity of the underlying gene signature used to produce them. We therefore performed additional validation steps on the “MYC Targets V2” hallmark gene set by examining its performance in The Cancer Genome Atlas prostate adenocarcinoma RNA-Seq dataset (TCGA-PRAD) that has accompanying patient outcomes data (32). We noted that samples with genomic amplifications of Myc had higher signature scores on average, as did samples that overexpressed Myc at the mRNA level (SI Appendix, Fig. S3A). To examine whether these relatively small changes in signature score had clinical relevance, we performed Kaplan–Meier survival analyses using the “MYC Targets V2” signature, Myc genomic amplification status, or Myc single-gene overexpression status as strata. The Myc gene signature was equally predictive of overall survival as genomic amplification status and outperformed single-gene expression stratification (SI Appendix, Fig. S3B).
Convinced of the performance of the Myc signature by these additional tests, we performed further analysis of the 1,039 Myc-correlated exons we identified in the prostate metadataset (Fig. 3A and Dataset S1). Unsupervised clustering of these 1,039 exons also grouped the samples by phenotype (SI Appendix, Fig. S3C), identifying patterns in Myc-dependent exon incorporation that varied accordingly.
Fig. 3.
Exon incorporation events correlated with Myc activity are strongly enriched in RNA-binding proteins and are conserved in prostate and breast cancers. (A) Heatmap depiction of exon usage of 1,039 Myc-correlated exons across prostate cancer datasets in healthy tissue, primary adenocarcinoma, metastatic adenocarcinoma, and neuroendocrine prostate cancer (NEPC). Columns represent samples ordered by disease phenotype and sorted by Myc Targets V2 signature score within each group. The Myc score annotation is colored from white (low) to black (high) based on the rank-transformed signature score of patient samples across the datasets. Rows represent exon inclusion events ordered by hierarchical clustering. (B) Scatterplots depicting examples of cassette exons in SRSF3 and HRAS transcripts whose incorporation is negatively correlated with Myc gene signature score. (C) Sashimi plots depicting average cassette exon incorporation levels of exons in SRSF3 and HRAS in prostate cancer datasets separated by cancer phenotype. Sashimi plots depict density of exon-including and exon-skipping reads as determined by rMATS-turbo analysis. (D) Workflow diagram for performing pathway-guided alternative splicing analysis on normal and cancerous breast and lung tissues. Each sample is scored for the Myc Targets V2 signature and correlated with the exon usage matrix to identify pathway-correlated exon incorporation changes. (E) Venn diagram indicating the intersection between Myc-correlated exon sets in prostate cancers with breast and lung adenocarcinomas. Exons must share the same correlation direction (positive or negative) to appear in the intersection. (F) REVIGO chart depicting the gene ontology of genes containing the 492 Myc-correlated exons from the triple intersection described above. SE, skipped exon.
Two examples among the most strongly Myc-correlated cassette exons from our analysis are found in SRSF3 and HRAS (Fig. 3B). Incorporation of the identified alternative exon in SRSF3 is anticorrelated with the Myc signature score (Fig. 3 B, Left). When examined by cancer phenotype, incorporation of this exon decreases as prostate cancer progresses from normal tissue to primary tumor and is even lower in mCRPC samples (Fig. 3 C, Left). Incorporation of this exon in NEPC is slightly higher, consistent with the Myc signature scores in these samples (SI Appendix, Fig. S2A).
SRSF3 is a serine–arginine splicing factor that can act as a proto-oncogene and also participates in transcription termination and DNA repair (50–53). The exon in question is ultraconserved throughout evolution and contains an in-frame stop codon. Also known as a poison exon, this sequence functions as a PTC (SI Appendix, Fig. S3 D, Top). Incorporation of this PTC has been shown previously to reduce SRSF3 expression levels by inducing NMD of the transcript (54, 55). These data suggest increased Myc signaling leads to increased exon skipping, reduced NMD, and increased expression of SRSF3.
A cassette exon in HRAS was also anticorrelated with Myc activity (Fig. 3 B, Right). When examined by cancer phenotype, exon skipping increased with tumor progression (Fig. 3 C, Right). HRAS is a well-known oncogene that cooperates with Myc to induce carcinogenesis in multiple tissues (56, 57). Inclusion of the cassette exon and the stop codon it contains results in the truncated HRAS p19 product instead of the p21 form (58). HRASp19 lacks the cysteine residues in the carboxyl-terminal domain of HRASp21 required for nuclear translocation and RAS-driven transformation and may function instead as a tumor suppressor (58, 59). This exon is conserved in mammals (SI Appendix, Fig. S3 D, Bottom). Incorporation of this exon is anticorrelated with Myc activity, suggesting that Myc can drive increased expression of oncogenic HRAS by affecting its splicing.
Myc-Correlated Exons in Prostate Cancers Are Highly Conserved in Breast and Lung Adenocarcinomas.
To determine whether the observed effects of Myc activity on splicing were prostate cancer specific, we performed a similar correlation analysis on a second hormone-dependent malignancy, breast adenocarcinoma, as well as on a hormone-independent epithelial malignancy, lung adenocarcinoma. The normal tissue and cancer RNA-Seq datasets for this analysis were drawn from TCGA (TCGA-BRCA and TCGA-LUAD) datasets and the GTEx collection of normal tissue (31, 60, 61). We performed a similar correlation between Myc signature score and exon usage as described above (Fig. 3D). The Myc signature scores in breast and lung tissues behaved similarly to those in the prostate tissues, with increases in score at each step when moving from normal to tumor-adjacent normal to carcinoma (SI Appendix, Fig. S3E). We identified 2,852 Myc-correlated cassette exons in breast samples and 2,465 in lung samples using the same filtering criteria for the prostate study (SI Appendix, Fig. S3F). The exon list includes the same anticorrelated exon in SRSF3, as shown for lung samples (Fig. 3D, fourth panel). Intersecting this set with our previously defined set of Myc-responsive prostate cancer exons (Fig. 3A), we found extensive overlap and similar exon incorporation behavior in the three sets (Fig. 3E). The triple intersection was even more strongly enriched for RNA-binding proteins (Fig. 3F). Our analysis suggests the exon incorporation response to Myc overexpression is conserved across these cancers.
Creation of an Engineered Model of Advanced Prostate Cancer with Regulated Myc Expression from Benign Human Prostate Cells to Define Myc-Dependent Exon Events.
Correlation analysis strongly implicates Myc, E2F, and MTOR signaling in the control of exons related to alternative pre-mRNA splicing but cannot define the individual contribution of each pathway to the observed phenotype. We therefore sought to determine whether the Myc-correlated splicing effects we observed were indeed Myc dependent.
Numerous studies of the effect of Myc overexpression have described large numbers of Myc target genes with significant tissue heterogeneity (62, 63). The presence of complex background genetics, undefined driver alterations, and tissue culture-specific phenomena further complicate the study of Myc biology (64). We therefore constructed a model of advanced prostate cancer by the transformation of benign human prostate epithelial cells with defined oncogenes (Fig. 4A) (65). We have previously shown that the enforced expression of Myc and myristoylated (activated) AKT1 (myrAKT1) generates androgen receptor-independent adenocarcinoma (66, 67). MyrAKT1 is included to phenocopy the activation of AKT1 that follows deletion of the tumor suppressor PTEN, a common event in prostate cancer tumorigenesis. Here, we cloned the Myc cDNA into a doxycycline-inducible promoter lentiviral construct, whereas MyrAKT1 was constitutively expressed (Fig. 4B and SI Appendix, Supplementary Methods).
Fig. 4.
Enforced expression of activated AKT1 and doxycycline-regulated c-Myc initiates AR-negative PrAd in human prostate cells. (A) Workflow diagram for derivation of Myc/myrAKT1-transformed human prostate cells from benign epithelium. “B” = Trop2+/CD49fhi basal cells; “L” = Trop2+/CD49flo luminal cells. (B) Depiction of lentiviral vectors used to enforce doxycycline-regulated expression of Myc and constitutive expression of myrAKT1. Histologic sections of transduced organoids. (C) Photomicrographs and fluorescent overlay of recovered grafts and tumor outgrowth after lentiviral transduction and subcutaneous implantation in NSG mice. A, myrAKT1 transduction (RFP); C, c-Myc transduction (GFP); CA, dual transduction with c-Myc and myrAKT1 (GFP and RFP merge depicted as yellow); UT, untreated. (D) Hematoxylin and eosin (H&E) stain of histologic sections of recovered grafts and tumor outgrowths. (E) Photomicrographs of cell lines ICA-1, ICA-2, and ICA-3 derived from tumor outgrowths growing as suspended rafts in tissue culture.
After lentiviral transduction of isolated human prostate basal cells (SI Appendix, Fig. S4A), we initiated the organoid culture and subsequent subcutaneous xenograft tumor outgrowth in immunocompromised mice in the constant presence of the drug (SI Appendix, Fig. S4 B and C). As previously reported, only doubly transduced cells resulted in tumor outgrowth (Fig. 4C). The histologic appearance and marker expression patterns of the xenograft outgrowths were similar to those previously published with constitutive constructs (Fig. 4D and SI Appendix, Fig. S4D). The xenograft outgrowths were dissociated, and plated in tissue culture conditions with doxycycline to initiate autonomously growing cell lines (Fig. 4E). We repeated the entire procedure to generate three independent cell lines from the prostate epithelium of three different human specimens.
Myc Withdrawal Affects Expression of Splicing-Related Genes.
Withdrawal of doxycycline from the Myc/myrAKT1 cell lines resulted in the rapid, dose-dependent loss of Myc protein expression, consistent with its previously reported short half-life (Fig. 5A and SI Appendix, Fig. S5A) (68). The cells also rapidly slowed their growth with increased G0/G1 fraction at 24 h (SI Appendix, Fig. S5 B and C). They adopted a senescent-like phenotype after prolonged Myc withdrawal with up-regulation of P21 (Fig. 5A). A similar consequence of Myc withdrawal in oncogene-addicted transformed cells has been previously reported (69).
Fig. 5.
Myc loss in the engineered cell lines produces a senescent-like phenotype and strongly affects the expression of RNA binding proteins. (A) Western blot of lysates from ICA1 cells withdrawn from doxycycline in a time course examining Myc expression and changes in proteins related to cell cycle state. Each of the three cell lines was examined in this manner, and the data shown are representative of all three. (B) Volcano plot of gene-level expression changes after Myc withdrawal. Genes down-regulated upon Myc loss appear on the left-hand side of the plot. Gene expression changes with the Cuffdiff q-value of <0.05 appear red. (C) Selected top gene ontology terms from the gene ontology analysis of Myc-dependent gene expression changes displaying strong enrichment for RNA binding. BP, Biological Process; CC, Cellular Component; MF, Molecular Function. (D) Comparison of Myc Targets V2 signature score levels in engineered cell lines in the presence and absence of doxycycline.
We performed RNA-Seq on samples from Myc-high and Myc-low conditions to define Myc-dependent genes and exons in our model system. These samples were sequenced with high read depth (>100 M reads) to enable accurate quantification of alternative splicing in downstream analysis. Primary analysis of the RNA expression data showed that thousands of genes were highly responsive to Myc withdrawal (CuffDiff q-value < 0.05) (Fig. 5B). Gene ontology analysis identified enrichment of several growth-related biological processes among the Myc-dependent genes (Fig. 5C). Of note, genes involved in RNA processing were among the most highly enriched in this subset. This is consistent with previous reports of Myc’s broad control of the growth phenotype. The regulated Myc expression system also allowed us to independently validate the Myc signature score we used in our correlation analysis (Fig. 5D).
Experimentation Confirms Myc-Regulated Exons Are Enriched in Splicing-Related Proteins and Often Encode PTCs.
We applied rMATS-turbo to analyze Myc-regulated exon usage in our engineered cell lines. To accommodate the paired nature of the dataset (comparing Myc-high and Myc-low conditions for each), we employed the PAIRADISE statistical test to the rMATS-turbo output (70). After filtering for coverage (≥10 splice junction reads per event), effect size (|deltaPSI| > 5%), and FDR < 5%, this analysis yielded 1,970 cassette exons that significantly changed incorporation in response to Myc withdrawal (Fig. 6 A and B and Dataset S1). We note that, among the Myc-dependent exons, we again identified the alternative exons in SRSF3 and HRAS described above, experimentally demonstrating that their incorporation is dependent on Myc signaling (Fig. 6C). The relative incorporation of the poison exon in SRSF3 increased when Myc was withdrawn, which would act to decrease the amount of SRSF3 protein in response to oncogene loss. We confirmed by immunoblotting that SRSF3 protein levels decreased relative to the housekeeping protein GAPDH in this experimental setting (SI Appendix, Fig. S6A).
Fig. 6.
Exon-level splicing analysis of c-Myc/myrAKT1 transformed human prostate cells identifies Myc-dependent exon incorporation events in splicing regulatory proteins. (A) Summary table of exon incorporation changes occurring after Myc withdrawal. (B) Heatmap depicting changes in exon incorporation of 1,970 Myc-dependent cassette exons in three independent engineered cell lines. (C) Sashimi plots depicting the change in splice junction RNA-Seq reads in SRSF3 and HRAS exons in the engineered cell lines following Myc withdrawal. Sashimi plots depict density of exon-including and exon-skipping reads as determined by rMATS-turbo analysis. (D) REVIGO scatter plot depicting gene ontology terms enriched among genes containing exons whose incorporation is responsive to Myc withdrawal. Semantic distance is a measurement of relatedness between gene ontology terms calculated by REVIGO. Representative gene ontology terms have been selected to describe each cluster. The dashed line indicates adjusted P = 0.05. (E) Venn diagram depicting the overlap between Myc-dependent exons (purple) and Myc-correlated exons identified in patient tissues (green). Exons must change incorporation level with Myc in the same direction as the correlation (positive or negative) in order to appear in the intersection of the two sets. (F) Heatmap depicting the annotated outcome of exon changes in validated Myc-dependent exons. The annotation identifies exons likely to produce PTCs (orange) or frameshifts (green). SE, skipped exon.
Similar to the correlational data from the patient specimens, the Myc-dependent exons were strikingly enriched in genes affecting RNA splicing-related processes (Fig. 6D). Intersecting this set of exons with the Myc-correlated exons in patient tissue identified 147 common exons (Fig. 6E), a highly significant overlap (P = 1.03 × 10−90). The remaining exons may not be responsive to short-term withdrawal of Myc in the cell line model or may be correlated with other signaling derangements that often accompany Myc deregulation in patient cancers (e.g., E2F or MTOR).
Alternative pre-mRNA splicing can regulate transcript levels through the incorporation or skipping of NMD-determinant exons (71). We hypothesized that Myc-driven exon choice in splicing proteins could contribute to the regulation of their expression levels. To examine the functional outcome of Myc-driven splicing changes on NMD, we annotated the 147 exons in the patient data–cell line intersection for PTCs and frameshifts (Fig. 6F and Dataset S1). These 147 exons correspond to 124 genes, 30 of which were RNA-binding proteins by gene ontology designation. We annotated all these exons using the Ensembl database to identify those that contained verified PTCs. We supplemented this annotation by parsing the remaining exons to identify those predicted to produce a frameshift within the coding sequence of the parent mRNA transcript. We found that 36 of the 43 exons in RNA-binding genes encode a PTC, a frameshift, or both (SI Appendix, Table S4). These exons represent a set of Myc-responsive sequences that act to regulate transcript abundance of proteins involved in alternative pre-mRNA splicing.
Discussion
This analysis was powered by rMATS-turbo, a fast, flexible, and extensible software package that allows rigorous examination of exon usage across disparate datasets. These public datasets have moderate read depth (50 to 75 M reads) and variable read length (50 to 75 bp). Here, we have used rMATS-turbo to perform a comprehensive survey of exon usage across the entire spectrum of prostate cancer disease progression. This exon-level analysis allows the correlation of exon matrices with any continuous metadata of interest. Our PEGASAS methodology identifies putative exon targets of cancer signaling networks. Its successful application to prostate, breast, and lung cancer datasets suggests that pathway-driven analysis of alternative splicing in pancancer data will also be of interest.
The engineered human prostate cell lines we developed with regulated Myc expression represent a unique opportunity to examine the consequences of Myc withdrawal on a defined genetic background. We employed them to identify over a thousand exons that significantly altered incorporation rates in response to Myc withdrawal, again with a striking enrichment for splicing-related proteins. The effects of Myc overexpression have been shown in other cancer contexts to have deleterious effects on splicing (72, 73). In Eu-Myc lymphoma cells, a Myc-target gene, PRMT5, is essential for maintaining splicing fidelity. Similarly, a component of the core spliceosome, BUD31, was shown to be a MYC-synthetic lethal gene in a human mammary transformation model. Others have shown that Myc-driven changes in splicing are in part accomplished by the induction of the canonical serine–arginine splicing factor SRSF1 (74). Further elucidation of the events downstream from Myc overexpression that lead to splicing changes is needed.
We note that Myc dysregulates the splicing of the PTC-containing exon in the serine–arginine protein SRSF3 (54, 55). This exon is Myc-correlated in both the prostate and breast cancer datasets, Myc-regulated in our tissue culture model, and ultraconserved. SRSF3 is known to alter the splicing of a number of downstream targets, as well as to autoregulate its own splicing. In a feedback loop, high levels of SRSF3 protein bind to its pre-mRNA transcript and promote inclusion of the poison exon (55). However, in the transformed setting we examined, Myc-high states were associated with high levels of SRSF3 expression and low levels of poison exon incorporation. This suggests Myc signaling may allow escape from this autoregulatory mechanism and stabilize SRSF3 transcripts despite high SRSF3 protein levels. SRSF3 itself has been recently shown to regulate splicing of NMD-determinant exons in chromatin modifier proteins during the induction of pluripotent stem cells (75). Given the role of Myc signaling in the acquisition of stem-like phenotypes and the stem-like state of advanced cancers, the mechanism that connects Myc overexpression to splicing changes in SRSF3 deserves further exploration (76, 77).
Furthermore, the phenomenon of Myc-regulated poison exons is not limited to SRSF3. We identified a number of exons in splicing proteins from patient tissues with experimentally validated Myc dependence in vitro that also contained PTCs. Alternative splicing coupled to NMD has been widely described as a mechanism controlling levels of splicing factors and other RNA-binding proteins (78). These splicing events are often autoregulated by the encoded protein or cross-regulated by a related paralog (79). Our data on Myc regulation indicate that this system of AS-NMD is also more globally regulated as part of a program of growth control. We postulate that these exons and regulation of them by Myc may be part of an adaptive response to alter spliceosomal throughput in response to high transcriptional flux.
One limitation of our study is that RNA and protein levels of the same genes are often poorly correlated (80). The potential for premature stop codons introduced by alternative splicing to induce NMD could further skew this relationship. Further studies of the relationship between Myc levels and NMD-determinant exons in splicing-related proteins should include proteomic measurements.
Our study provides further insight into the relationship between Myc signaling and alternative splicing changes that could be used to guide the development of splicing-targeted cancer therapy (81). Future work will need to establish the specificity of these exon events for cells with oncogenic levels of Myc expression to avoid simultaneously targeting rapidly dividing normal cell types.
Methods
Descriptions of the gene ontology analysis, overlap enrichment assessment, lentiviral constructs, organotypic human prostate transformation assay, xenograft outgrowth, cell line derivation, and other tissue culture experiments are available in SI Appendix.
RNA-Seq Data Processing Framework.
A comprehensive RNA-Seq dataset was compiled from published prostate cancer and normal prostate datasets that reflect the full progression of prostate cancer. In total, 876 samples were downloaded from different sources. RNA-Seq Fastq files of normal prostate samples [GTEx Consortium (31)] and prostate cancer samples [Beltran et al. study (10), Robinson et al. study (11), and Stand-Up-To-Cancer study (12)] were downloaded from dbGAP (82, 83) via fastq-dump in SRA toolkit. RNA-Seq Fastq files from TCGA primary prostate cancer and adjacent benign samples were downloaded from GDC via gdc-client (84).
A unified RNA-Seq processing framework was constructed to perform read mapping as well as gene and isoform quantification on the collected multiphenotypic prostate RNA-Seq samples. Specifically, read mapping was done by STAR 2.5.3a (85) with a STAR 2-pass function enabled to improve the detection of splicing junctions. The STAR genome index was built with–sjdbOverhang 100 as a generic parameter to handle differences in read length of RNA-Seq samples from various sources. The genome annotation file was downloaded from GENCODE V26 (86) under human genome version hg19 (GRCh37). The subsequent gene/isoform expression quantification is performed by Cufflinks (87) with default parameters.
RNA-Seq alternative splicing quantification is conducted uniformly with a newly engineered version (version 4.0.2) of the rMATS-turbo software package (29, 30). An exon-based ratio metric, commonly defined as PSI ratio, was employed to measure the alternative splicing events. The PSI ratio is calculated as follows:
where S and I are the numbers of reads mapped to the junction supporting skipping and inclusion form, respectively. Effective length is used for normalization.
Customized scripts were applied to calculate PSI value for each individual alternative splicing event from the rMATS-turbo junction count output. To build a confident set of exon events, the splice junction of each event was required to be covered by no less than 10 splice junction reads. Additionally, each event was required to have a PSI range greater than 5% across the entire dataset (|maxPSI − minPSI| > 5%), with a mean skipping or inclusion value over 5%. Events with missing values in the majority (over 99%) of samples were removed.
Analysis and Evaluation of Alternative Splicing Profile of Prostate Cancer Metadataset.
PCA was applied to inspect the RNA-Seq–derived gene expression/alternative splicing profiles of our multiphenotypic prostate cancer dataset. First, the matrix of sample vs. fragments per kilobase of transcript per million mapped reads/PSI value was produced by customized scripts. Then, the matrix was completed and imputed by KNN method (knnImputation in DMwR package) (88) for missing values. Last, the matrix was mean centered and scaled (PSI matrix is not scaled). PCA was conducted via prcomp function in R. The top five PCs were inspected, but only the first two that describe the highest percentage of the variance are shown.
In addition, silhouette width was applied to assess the fitness of PCA clustering results derived from alternative splicing or gene/isoform expression measurements (89). Specifically, disease conditions were used as sample labels to compute the silhouette width of each cluster. Average silhouette widths were compared between PCA clustering results with different metrics (90). The R package cluster (91) was used for Silhouette calculation based on PCA results and disease phenotype labels.
PEGASAS.
In order to identify exon incorporation shifts that could correspond to oncogenic pathway alterations during tumor progression, a correlation-based analysis was developed to define signaling pathway correlated alternative splicing events. It involves two major steps.
The first step is to define signaling pathway activity and alternative splicing levels. The quantification of gene expression and alternative splicing is detailed in RNA-Seq Data Processing Framework. Signaling pathway activity can be characterized by assessing the expression level of its target genes as a set relative to other genes (42). The MSigDB (92) has compiled gene sets (42) for the use with gene set enrichment analysis (GSEA) (93) software or similar applications. Here, a group of well-defined gene sets, known as hallmarks (42), was selected to assess a wide range of pathways in prostate cancers. To measure the activity of a given signaling pathway gene set, all genes (both genes within the gene set as well as those not in the gene set) were ranked according to their gene expression values, then a weight was assigned to each gene based on the number of genes in the set (pathway or nonpathway) they belonged to. This was used to construct empirical distributions for both sets, and a two-sample Kolmogorov–Smirnov test statistic, which is the supremum of the differences between the two distributions, was computed as a measure of the activity of the signaling pathway, i.e., an “activity score.” Given the same gene set and gene annotation, the higher the score, the higher the activity of a signaling pathway in a sample. Note that the score should not be used to compare across signaling pathways as each gene set has distinct number of genes, which affects the score.
The second step is to identify pathway activity-correlated alternatively spliced exons. For each pathway, the pathway activity score defined above was correlated with all of the AS events identified by rMATS-turbo. The Pearson correlation coefficient was computed for each pathway–exon pair across samples in the dataset. A Pearson correlation coefficient with an absolute value >0.3 was considered as correlated. Data points for each pathway–exon pair were permutated 5,000 times locally to produce empirical P values to filter out faulty correlations caused by data structure or missing data points. A stringent empirical P value < 2 × 10−4 was required for this analysis. The analytical framework performs streamlined analysis of multiple gene sets (e.g., 50 hallmark gene sets). Customized scripts were implemented to generate the summary plot.
Cell Line Gene Expression and Alternative Splicing Differential Analysis.
The same RNA-Seq processing framework described above was applied to quantify gene expression and alternative splicing of Myc cell line samples. Differentially expressed genes were identified and visualized by the Cuffdiff and cummeRbund pipeline with a threshold of q-value < 0.05. Skipped exon events quantified by rMATS-turbo were analyzed by the PAIRADISE statistical model for conducting paired tests of between Myc +/− conditions (70, 87). PAIRADISE with equal.variance = TRUE was used to perform the test. The resulting events were first filtered by the coverage and deltaPSI requirements (≥10 splice junction reads per event, |deltaPSI| > 0.05). Then, an FDR 5% cutoff was applied to identify significant differential alternative splicing events between the on and off states of the engineered Myc cell line.
Code Availability.
The computational pipeline of PEGASAS is available at https://github.com/Xinglab/PEGASAS (94), and custom scripts used to perform filtering, analysis, and visualization have been deposited separately at https://github.com/Xinglab/Myc-regulated_AS_PrCa_paper (95).
Data Availability.
Raw sequencing files (fastq) from the engineered cell lines and gene expression matrices are available through Gene Expression Omnibus (accession no. GSE141633) (96). The PSI and gene expression matrices for the prostate metadataset are also available from the same source. The normal prostate expression data from GTEx used for the analyses described in the manuscript were obtained from dbGaP (https://www.ncbi.nlm.nih.gov/gap) accession no. phs000424 (accessed 1 October 2018). Data on primary prostate cancers were obtained from the TCGA Research Network and downloaded from the Genomic Data Commons (http://portal.gdc.cancer.gov/projects/TCGA-PRAD) accession no. phs000178 (accessed 1 October 2017). Additional datasets on metastatic prostate cancers are available by controlled access through dbGaP with accession nos. phs000909, phs000673, and phs000915 (accessed 1 October 2018).
Supplementary Material
Acknowledgments
This research was supported by funds from NIH/National Cancer Institute under Awards R01CA220238, U01CA233074, U24CA232979, and P50CA092131; the Parker Institute for Cancer Immunotherapy, Grant 20163828; the University of California, Los Angeles, Tumor Cell Biology Training Grant T32CA009056; and the Office of the Assistant Secretary of Defense for Health Affairs through the Prostate Cancer Research Program under Award W81XWH-16-1-0216. Opinions, interpretations, conclusions, and recommendations are those of the authors and are not necessarily endorsed by the Department of Defense. The results published here are in part based upon data generated by the TCGA Research Network (http://cancer.gov/tcga) and data generated by the Genotype-Tissue Expression (GTEx) Project (https://commonfund.nih.gov/gtex). The GTEx Project is supported by the Common Fund of the Office of the Director of the NIH, and by National Cancer Institute, National Human Genome Research Institute, National Heart, Lung, and Blood Institute, National Institute on Drug Abuse, National Institute of Mental Health, and National Institute of Neurological Disorders and Stroke.
Footnotes
Competing interest statement: O.N.W. currently has consulting, equity, and/or board relationships with Trethera Corporation, Kronos Biosciences, Sofie Biosciences, and Allogene Therapeutics. D.L.B. and Y.X. are scientific cofounders of Panorama Medicine. None of these companies contributed to or directed any of the research reported in this article.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141633 (accession no. GSE141633). The computational pipeline of PEGASAS is available at https://github.com/Xinglab/PEGASAS, and custom scripts have been deposited separately at https://github.com/Xinglab/Myc-regulated_AS_PrCa_paper.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1915975117/-/DCSupplemental.
References
- 1.Baralle F. E., Giudice J., Alternative splicing as a regulator of development and tissue identity. Nat. Rev. Mol. Cell Biol. 18, 437–451 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Liu S., Cheng C., Alternative RNA splicing and cancer. Wiley Interdiscip. Rev. RNA 4, 547–566 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ho Y., Dehm S. M., Androgen receptor rearrangement and splicing variants in resistance to endocrine therapies in prostate cancer. Endocrinology 158, 1533–1542 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Catena R., et al. , Increased expression of VEGF121/VEGF165-189 ratio results in a significant enhancement of human prostate tumor angiogenesis. Int. J. Cancer 120, 2096–2109 (2007). [DOI] [PubMed] [Google Scholar]
- 5.Narla G., et al. , KLF6-SV1 overexpression accelerates human and mouse prostate cancer progression and metastasis. J. Clin. Invest. 118, 2711–2721 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Hagen R. M., et al. , Quantitative analysis of ERG expression and its splice isoforms in formalin-fixed, paraffin-embedded prostate cancer samples: Association with seminal vesicle invasion and biochemical recurrence. Am. J. Clin. Pathol. 142, 533–540 (2014). [DOI] [PubMed] [Google Scholar]
- 7.Mercatante D. R., Bortner C. D., Cidlowski J. A., Kole R., Modification of alternative splicing of Bcl-x pre-mRNA in prostate and breast cancer cells. analysis of apoptosis and cell death. J. Biol. Chem. 276, 16411–16417 (2001). [DOI] [PubMed] [Google Scholar]
- 8.Antonopoulou E., Ladomery M., Targeting splicing in prostate cancer. Int. J. Mol. Sci. 19, E1287 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Arora K., Barbieri C. E., Molecular subtypes of prostate cancer. Curr. Oncol. Rep. 20, 58 (2018). [DOI] [PubMed] [Google Scholar]
- 10.Beltran H., et al. , Divergent clonal evolution of castration-resistant neuroendocrine prostate cancer. Nat. Med. 22, 298–305 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Robinson D., et al. , Integrative clinical genomics of advanced prostate cancer. Cell 161, 1215–1228 (2015). Correction in: Cell 162, 454 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Robinson D. R., et al. , Integrative clinical genomics of metastatic cancer. Nature 548, 297–303 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cancer Genome Atlas Research Network , The molecular taxonomy of primary prostate cancer. Cell 163, 1011–1025 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baca S. C., et al. , Punctuated evolution of prostate cancer genomes. Cell 153, 666–677 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Jenkins R. B., Qian J., Lieber M. M., Bostwick D. G., Detection of c-myc oncogene amplification and chromosomal anomalies in metastatic prostatic carcinoma by fluorescence in situ hybridization. Cancer Res. 57, 524–531 (1997). [PubMed] [Google Scholar]
- 16.Linja M. J., et al. , Amplification and overexpression of androgen receptor gene in hormone-refractory prostate cancer. Cancer Res. 61, 3550–3555 (2001). [PubMed] [Google Scholar]
- 17.Chen H., et al. , Pathogenesis of prostatic small cell carcinoma involves the inactivation of the P53 pathway. Endocr. Relat. Cancer 19, 321–331 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Tran C., et al. , Development of a second-generation antiandrogen for treatment of advanced prostate cancer. Science 324, 787–790 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Mateo J., et al. , DNA-repair defects and olaparib in metastatic prostate cancer. N. Engl. J. Med. 373, 1697–1708 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Paschalis A., et al. , Alternative splicing in prostate cancer. Nat. Rev. Clin. Oncol. 15, 663–675 (2018). [DOI] [PubMed] [Google Scholar]
- 21.Thorsen K., et al. , Alternative splicing in colon, bladder, and prostate cancer identified by exon array analysis. Mol. Cell. Proteomics 7, 1214–1224 (2008). [DOI] [PubMed] [Google Scholar]
- 22.Ren S., et al. , RNA-seq analysis of prostate cancer in the Chinese population identifies recurrent gene fusions, cancer-associated long noncoding RNAs and aberrant alternative splicings. Cell Res. 22, 806–821 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B. D., et al. , Alternative splicing promotes tumour aggressiveness and drug resistance in African American prostate cancer. Nat. Commun. 8, 15921 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Li H. R., et al. , Two-dimensional transcriptome profiling: Identification of messenger RNA isoform signatures in prostate cancer from archived paraffin-embedded cancer specimens. Cancer Res. 66, 4079–4088 (2006). [DOI] [PubMed] [Google Scholar]
- 25.Zhang C., et al. , Profiling alternatively spliced mRNA isoforms for prostate cancer classification. BMC Bioinformatics 7, 202 (2006). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Gan Y., et al. , Roles of alternative RNA splicing of the Bif-1 gene by SRRM4 during the development of treatment-induced neuroendocrine prostate cancer. EBioMedicine 31, 267–275 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Lee A. R., et al. , Alternative RNA splicing of the MEAF6 gene facilitates neuroendocrine prostate cancer progression. Oncotarget 8, 27966–27975 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Li Y., et al. , SRRM4 drives neuroendocrine transdifferentiation of prostate adenocarcinoma under androgen receptor pathway inhibition. Eur. Urol. 71, 68–78 (2017). [DOI] [PubMed] [Google Scholar]
- 29.Shen S., et al. , rMATS: Robust and flexible detection of differential alternative splicing from replicate RNA-seq data. Proc. Natl. Acad. Sci. U.S.A. 111, E5593–E5601 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Xie Z., Xing Y., rMATS-turbo. http://rnaseq-mats.sourceforge.net/. Accessed 27 January 2020.
- 31.Lonsdale J., et al. , The Genotype-Tissue Expression (GTEx) project. Nat. Genet. 45, 580–585 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Chang K., et al. , The Cancer Genome Atlas pan-cancer analysis project. Nat. Genet. 45, 1113–1120 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Leek J. T., et al. , Tackling the widespread and critical impact of batch effects in high-throughput data. Nat. Rev. Genet. 11, 733–739 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Anders S., Reyes A., Huber W., Detecting differential usage of exons from RNA-seq data. Genome Res. 22, 2008–2017 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shen S., Wang Y., Wang C., Wu Y. N., Xing Y., SURVIV for survival analysis of mRNA isoform variation. Nat. Commun. 7, 11548 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Park E., Pan Z., Zhang Z., Lin L., Xing Y., The expanding landscape of alternative splicing variation in human populations. Am. J. Hum. Genet. 102, 11–26 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Johnson N. T., Dhroso A., Hughes K. J., Korkin D., Biological classification with RNA-seq data: Can alternatively spliced transcript expression enhance machine learning classifiers? RNA 24, 1119–1132 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Djebali S., et al. , Landscape of transcription in human cells. Nature 489, 101–108 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Frank S., Nelson P., Vasioukhin V., Recent advances in prostate cancer research: Large-scale genomic analyses reveal novel driver mutations and DNA repair defects [version 1; peer review: 2 approved]. F1000Research, 7, 1173 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Mootha V. K., et al. , PGC-1alpha-responsive genes involved in oxidative phosphorylation are coordinately downregulated in human diabetes. Nat. Genet. 34, 267–273 (2003). [DOI] [PubMed] [Google Scholar]
- 41.Qiu X., Wu H., Hu R., The impact of quantile and rank normalization procedures on the testing power of gene differential expression analysis. BMC Bioinformatics 14, 124 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liberzon A., et al. , The Molecular Signatures Database (MSigDB) hallmark gene set collection. Cell Syst. 1, 417–425 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Aran D., et al. , Comprehensive analysis of normal adjacent to tumor transcriptomes. Nat. Commun. 8, 1077 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Setlur S. R., et al. , Estrogen-dependent signaling in a molecularly distinct subclass of aggressive prostate cancer. J. Natl. Cancer Inst. 100, 815–825 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Krzywinski M., Birol I., Jones S. J., Marra M. A., Hive plots—rational approach to visualizing networks. Brief. Bioinform. 13, 627–644 (2012). [DOI] [PubMed] [Google Scholar]
- 46.Zack T. I., et al. , Pan-cancer patterns of somatic copy number alteration. Nat. Genet. 45, 1134–1140 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Dang C. V., MYC on the path to cancer. Cell 149, 22–35 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Gurel B., et al. , Nuclear MYC protein overexpression is an early alteration in human prostate carcinogenesis. Mod. Pathol. 21, 1156–1167 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Koh C. M., et al. , MYC and prostate cancer. Genes Cancer 1, 617–628 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Urbanski L. M., Leclair N., Anczuków O., Alternative-splicing defects in cancer: Splicing regulators and their downstream targets, guiding the way to novel cancer therapeutics. Wiley Interdiscip. Rev. RNA 9, e1476 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Cui M., et al. , Genes involved in pre-mRNA 3′-end formation and transcription termination revealed by a lin-15 operon Muv suppressor screen. Proc. Natl. Acad. Sci. U.S.A. 105, 16665–16670 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.He X., Zhang P., Serine/arginine-rich splicing factor 3 (SRSF3) regulates homologous recombination-mediated DNA repair. Mol. Cancer 14, 158 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Jia R., Li C., McCoy J. P., Deng C. X., Zheng Z. M., SRp20 is a proto-oncogene critical for cell proliferation and tumor induction and maintenance. Int. J. Biol. Sci. 6, 806–826 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Corbo C., Orrù S., Salvatore F., SRp20: An overview of its role in human diseases. Biochem. Biophys. Res. Commun. 436, 1–5 (2013). [DOI] [PubMed] [Google Scholar]
- 55.Jumaa H., Nielsen P. J., The splicing factor SRp20 modifies splicing of its own mRNA and ASF/SF2 antagonizes this regulation. EMBO J. 16, 5077–5085 (1997). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Land H., Parada L. F., Weinberg R. A., Tumorigenic conversion of primary embryo fibroblasts requires at least two cooperating oncogenes. Nature 304, 596–602 (1983). [DOI] [PubMed] [Google Scholar]
- 57.Wang C., Lisanti M. P., Liao D. J., Reviewing once more the c-myc and ras collaboration: Converging at the cyclin D1-CDK4 complex and challenging basic concepts of cancer biology. Cell Cycle 10, 57–67 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Cohen J. B., Broz S. D., Levinson A. D., Expression of the H-ras proto-oncogene is controlled by alternative splicing. Cell 58, 461–472 (1989). [DOI] [PubMed] [Google Scholar]
- 59.Camats M., Kokolo M., Heesom K. J., Ladomery M., Bach-Elias M., P19 H-ras induces G1/S phase delay maintaining cells in a reversible quiescence state. PLoS One 4, e8513 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Pereira B., et al. , The somatic mutation profiles of 2,433 breast cancers refines their genomic and transcriptomic landscapes. Nat. Commun. 7, 11479 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Cancer Genome Atlas Research Network , Comprehensive molecular profiling of lung adenocarcinoma. Nature 511, 543–550 (2014). Correction in: Nature559, E12 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Ji H., et al. , Cell-type independent MYC target genes reveal a primordial signature involved in biomass accumulation. PLoS One 6, e26057 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zeller K. I., Jegga A. G., Aronow B. J., O’Donnell K. A., Dang C. V., An integrated database of genes responsive to the Myc oncogenic transcription factor: Identification of direct genomic targets. Genome Biol. 4, R69 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Chandriani S., et al. , A core MYC gene expression signature is prominent in basal-like breast cancer but only partially overlaps the core serum response. PLoS One 4, e6693 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Park J. W., et al. , Prostate epithelial cell of origin determines cancer differentiation state in an organoid transformation assay. Proc. Natl. Acad. Sci. U.S.A. 113, 4482–4487 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Stoyanova T., et al. , Prostate cancer originating in basal cells progresses to adenocarcinoma propagated by luminal-like cells. Proc. Natl. Acad. Sci. U.S.A. 110, 20111–20116 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Bluemn E. G., et al. , Androgen receptor pathway-independent prostate cancer is sustained through FGF signaling. Cancer Cell 32, 474–489.e6 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Dani C., et al. , Extreme instability of myc mRNA in normal and transformed human cells. Proc. Natl. Acad. Sci. U.S.A. 81, 7046–7050 (1984). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gartel A. L., et al. , Myc represses the p21(WAF1/CIP1) promoter and interacts with Sp1/Sp3. Proc. Natl. Acad. Sci. U.S.A. 98, 4510–4515 (2001). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Demirdjian L., Wu Y. N., Xing Y., PAIRADISE: Paired analysis of differential isoform expression. https://bioconductor.org/packages/release/bioc/html/PAIRADISE.html. Accessed 27 January 2020.
- 71.Lewis B. P., Green R. E., Brenner S. E., Evidence for the widespread coupling of alternative splicing and nonsense-mediated mRNA decay in humans. Proc. Natl. Acad. Sci. U.S.A. 100, 189–192 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Koh C. M., et al. , MYC regulates the core pre-mRNA splicing machinery as an essential step in lymphomagenesis. Nature 523, 96–100 (2015). [DOI] [PubMed] [Google Scholar]
- 73.Hsu T. Y., et al. , The spliceosome is a therapeutic vulnerability in MYC-driven cancer. Nature 525, 384–388 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Das S., Anczuków O., Akerman M., Krainer A. R., Oncogenic splicing factor SRSF1 is a critical transcriptional target of MYC. Cell Rep. 1, 110–117 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Ratnadiwakara M., et al. , SRSF3 promotes pluripotency through Nanog mRNA export and coordination of the pluripotency gene expression program. eLife 7, e37419 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Smith B. A., et al. , A human adult stem cell signature marks aggressive variants across epithelial cancers. Cell Rep. 24, 3353–3366.e5 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Sridharan R., et al. , Role of the murine reprogramming factors in the induction of pluripotency. Cell 136, 364–377 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Nasif S., Contu L., Mühlemann O., Beyond quality control: The role of nonsense-mediated mRNA decay (NMD) in regulating gene expression. Semin. Cell Dev. Biol. 75, 78–87 (2018). [DOI] [PubMed] [Google Scholar]
- 79.Zhou Z., Fu X. D., Regulation of splicing by SR proteins and SR protein-specific kinases. Chromosoma 122, 191–207 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Liu Y., Beyer A., Aebersold R., On the dependency of cellular protein levels on mRNA abundance. Cell 165, 535–550 (2016). [DOI] [PubMed] [Google Scholar]
- 81.Martinez-Montiel N., Rosas-Murrieta N. H., Anaya Ruiz M., Monjaraz-Guzman E., Martinez-Contreras R., Alternative splicing as a target for cancer treatment. Int. J. Mol. Sci. 19, E545 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Mailman M. D., et al. , The NCBI dbGaP database of genotypes and phenotypes. Nat. Genet. 39, 1181–1186 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Tryka K. A., et al. , NCBI’s database of genotypes and phenotypes: dbGaP. Nucleic Acids Res. 42, D975–D979 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Grossman R. L., et al. , Toward a shared vision for cancer genomic data. N. Engl. J. Med. 375, 1109–1112 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Dobin A., et al. , STAR: Ultrafast universal RNA-seq aligner. Bioinformatics 29, 15–21 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Harrow J., et al. , GENCODE: The reference human genome annotation for The ENCODE Project. Genome Res. 22, 1760–1774 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Trapnell C., et al. , Differential gene and transcript expression analysis of RNA-seq experiments with TopHat and Cufflinks. Nat. Protoc. 7, 562–578 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Torgo L., Data Mining with R: Learning with Case Studies (Chapman & Hall/CRC Data Mining and Knowledge Discovery Series, CRC Press, Taylor & Francis Group, Boca Raton, ed. 2, 2017). [Google Scholar]
- 89.Risso D., Perraudeau F., Gribkova S., Dudoit S., Vert J. P., A general and flexible method for signal extraction from single-cell RNA-seq data. Nat. Commun. 9, 284 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Rousseeuw P. J., Silhouettes: A graphical aid to the interpretation and validation of cluster analysis. J. Comput. Appl. Math. 20, 53–65 (1987). [Google Scholar]
- 91.Mächler M., Rousseeuw P., Struyf A., Hubert M., Hornik K., Cluster: Cluster analysis basics and extensions. R Package, Version 2.0.7-1 (2018).
- 92.Liberzon A., et al. , Molecular signatures database (MSigDB) 3.0. Bioinformatics 27, 1739–1740 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Subramanian A., et al. , Gene set enrichment analysis: A knowledge-based approach for interpreting genome-wide expression profiles. Proc. Natl. Acad. Sci. U.S.A. 102, 15545–15550 (2005). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Pan Y., Xing Y., Pathway Enrichment-Guided Activity Study of Alternative Splicing (PEGASAS). GitHub. https://github.com/Xinglab/PEGASAS. Deposited 11 January 2020. [Google Scholar]
- 95.Pan Y., Xing Y., Myc-regulated alternative splicing events in aggressive prostate cancers. GitHub. https://github.com/Xinglab/Myc-regulated_AS_PrCa_paper. Deposited 21 June 2019. [Google Scholar]
- 96.Phillips J. W., et al. , The landscape of alternative splicing in aggressive prostate cancers. Gene Expression Omnibus. https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE141633. Deposited 9 December 2019. [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
Raw sequencing files (fastq) from the engineered cell lines and gene expression matrices are available through Gene Expression Omnibus (accession no. GSE141633) (96). The PSI and gene expression matrices for the prostate metadataset are also available from the same source. The normal prostate expression data from GTEx used for the analyses described in the manuscript were obtained from dbGaP (https://www.ncbi.nlm.nih.gov/gap) accession no. phs000424 (accessed 1 October 2018). Data on primary prostate cancers were obtained from the TCGA Research Network and downloaded from the Genomic Data Commons (http://portal.gdc.cancer.gov/projects/TCGA-PRAD) accession no. phs000178 (accessed 1 October 2017). Additional datasets on metastatic prostate cancers are available by controlled access through dbGaP with accession nos. phs000909, phs000673, and phs000915 (accessed 1 October 2018).