Significance
The mTORC1 signaling pathway senses nutrient signals and controls cell growth partly by regulating mRNA translation. 5′ TOP motifs are defining features of mTORC1-regulated mRNAs, but their prevalence throughout the transcriptome and the mechanism that controls them are not fully understood. Here we show that the mTORC1 effector LARP1 is the master regulator of TOP mRNAs genome-wide. We discover features of TOP sequences that tune their regulatory potential across a continuous spectrum and define a metric called a TOPscore that quantifies them. Analysis of TOPscores across the transcriptomes of 16 mammalian tissues defines both constitutive TOP mRNAs and tissue-specific variants resulting from alternative transcription initiation. These results establish global principles of a dynamic and tunable program of nutrient-regulated gene expression.
Keywords: TOP mRNA, mTORC1, translation, LARP1
Abstract
Terminal oligopyrimidine (TOP) motifs are sequences at the 5′ ends of mRNAs that link their translation to the mTOR Complex 1 (mTORC1) nutrient-sensing signaling pathway. They are commonly regarded as discrete elements that reside on ∼100 mRNAs that mostly encode translation factors. However, the full spectrum of TOP sequences and their prevalence throughout the transcriptome remain unclear, primarily because of uncertainty over the mechanism that detects them. Here, we globally analyzed translation targets of La-related protein 1 (LARP1), an RNA-binding protein and mTORC1 effector that has been shown to repress TOP mRNA translation in a few specific cases. We establish that LARP1 is the primary translation regulator of mRNAs with classical TOP motifs genome-wide, and also that these motifs are extreme instances of a broader continuum of regulatory sequences. We identify the features of TOP sequences that determine their potency and quantify these as a metric that accurately predicts mTORC1/LARP1 regulation called a TOPscore. Analysis of TOPscores across the transcriptomes of 16 mammalian tissues defines a constitutive “core” set of TOP mRNAs, but also identifies tissue-specific TOP mRNAs produced via alternative transcription initiation sites. These results establish the central role of LARP1 in TOP mRNA regulation on a transcriptome scale and show how it connects mTORC1 to a tunable and dynamic program of gene expression that is tailored to specific biological contexts.
The mTORC1 signaling pathway is a master regulator of cell growth that is essential for normal development and linked to common human diseases, including cancer, metabolic disease, and neurologic disorders (1). The pathway senses nutrient signals and orchestrates diverse growth processes that include growth-related gene expression at transcriptional and posttranscriptional levels. Acute fluctuations in mTORC1 activity, as occur upon starvation or pharmacological mTOR inhibition, primarily alter gene expression at the level of mRNA translation (2). We and others previously found that mTORC1 targets the translation of hundreds of mRNAs, about half of which encode a 5′ TOP motif (3, 4). This motif, defined as a +1 C directly adjacent to the 5′ cap structure and followed by an unbroken series of 4 to 14 pyrimidine nucleotides, renders translation of the mRNA hypersensitive to a variety of growth signals, including those transmitted by the mTORC1 pathway (5). Currently, there are 97 widely accepted TOP mRNAs, which encode mostly translation factors and nearly all ribosomal proteins (Dataset S1) (5). We refer to these as “classical” TOP mRNAs. However, large-scale analyses of mRNA 5′ sequences hint that thousands more mRNAs may encode TOP sequences (6). Additionally, many mTORC1-regulated mRNAs lack classical TOP sequences (3, 4). Whether these are nevertheless controlled through the TOP mechanism or through other mTORC1-regulated mechanisms is unknown. The full extent to which TOP sequences define mTORC1 translation targets therefore remains unclear.
A hurdle to answering this question has been persistent uncertainty over the molecular details of the TOP regulatory mechanism. Many potential regulators have been proposed, including the TIA1 and TIAR proteins, La, CNBP, and mir-10A (7). However, recent findings have hinted that the RNA-binding protein La-related protein 1 (LARP1) may have a central role (8–10). LARP1 is a large protein (150 kDa) with several RNA-binding domains. Its central region contains a La motif (LaM) domain that defines the La-related protein (LARP) superfamily, along with an adjacent RNA recognition motif-like (RRM-L) domain. Its C terminus encodes a domain known as the DM15 region. This domain is unique to LARP1 and its closely related homolog LARP1B, and is therefore also known as the LARP1 domain (11).
Several observations suggest that LARP1 directly represses TOP mRNA translation. First, it was shown using polysome profiling that LARP1 is required to repress the translation of several specific TOP mRNAs following mTOR inhibition (8, 10). Second, the DM15 region was found to directly bind a model TOP sequence and the adjacent mRNA cap structure (9). Finally, we found that this same region is necessary and sufficient to control the translation of a model TOP mRNA reporter both in cells and in vitro (10). However, polysome profiling analyses of LARP1, such as we and others have used previously (8, 10), have been criticized as potentially misleading for measuring TOP mRNA regulation (12). Other reports have argued that LARP1 instead enhances the translation of some TOP transcripts (13). It therefore remains uncertain whether LARP1 is truly a global repressor of TOP mRNAs.
In this work, we used ribosome profiling to examine the translation functions of LARP1 throughout the transcriptome. The analysis establishes that LARP1 is responsible for the global regulation of TOP mRNA translation downstream of mTORC1, and yields additional insights into the mRNA features that define LARP1 targets. We find that classical TOP motifs are instances of a broader set of 5′ regulatory sequences and show how variation in motif length and TSS heterogeneity tune an mRNA’s sensitivity to mTORC1 signaling across a continuous range. We quantify these as a metric that we call a TOPscore, and show that it accurately predicts regulation by mTORC1/LARP1. Analysis of TOPscores across the transcriptomes of 16 human tissues reveals a unique set of core “constitutive” TOP mRNAs and also instances of alternative transcription initiation that generate tissue-specific TOP transcripts with altered sensitivity to mTORC1/LARP1 regulation. Together, our findings illustrate how the 5′ sequences of mRNAs specify a nutrient-regulated program of mRNA translation that can be precisely tuned and dynamically reprogrammed across biological contexts.
Results
LARP1 Is the Primary Regulator of Known TOP mRNAs.
We first set out to determine which mRNAs are translationally regulated by mTORC1 via LARP1. To do this, we used ribosome profiling to monitor changes in translation following mTOR inhibition, mirroring the cellular response to starvation, in wild-type and LARP1-deficient HEK-293T human cells (14). LARP1-deficient cells were previously generated using CRISPR/Cas9, and are referred to here as sgLARP1 cells (10). Both cell lines treated with the mTOR inhibitor Torin 1 for 2 h were used to prepare ribosome protected fragment (RPF) and total RNA libraries (4, 15). Importantly, Torin 1 repressed global protein synthesis by ∼50% in both wild-type and sgLARP1 cells (SI Appendix, Fig. S1A). LARP1 is therefore not required for mTOR control of bulk protein synthesis.
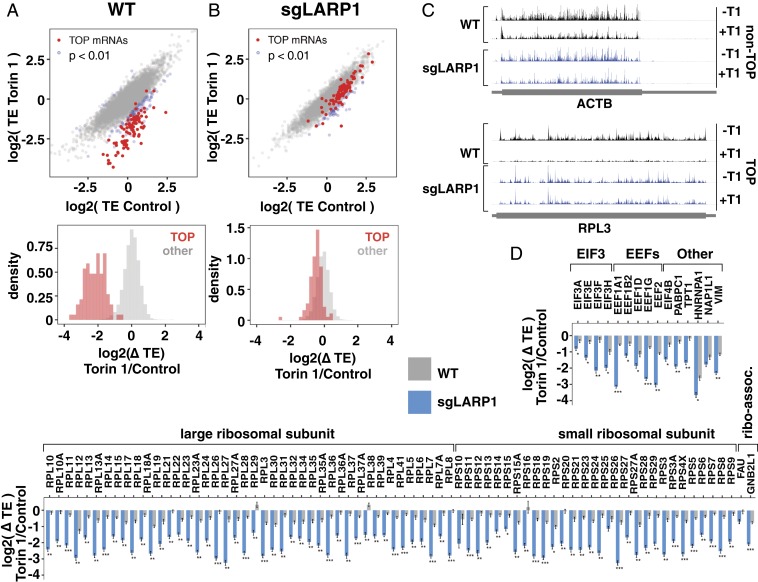
Analysis of ribosome profiling data, however, revealed significant differences at the level of individual mRNAs (Fig. 1A and Datasets S2 and S3). In wild-type cells, mTOR inhibition selectively repressed nearly all classical TOP mRNAs (90 of 92 TOP mRNAs were repressed >twofold beyond the population mean; Fig. 1A and SI Appendix, Fig. S2A) (5). We also found that mRNAs previously identified as TOP mRNAs in mouse embryonic fibroblasts (e.g., EIF4B, COX7A2L, IPO5) (4) were similarly repressed in this cell line (Dataset S3), as well as other transcripts with classical TOP motifs (PFDN5) according to experimental transcription start site (TSS) datasets (dbTSS and FANTOM5) (6, 16). In striking contrast, TOP mRNA translation was only mildly affected by mTOR inhibition in sgLARP1 cells (Fig. 1B and SI Appendix, Fig. S2 B and C). Examples are shown in Fig. 1 C and D, and contrast with similarly expressed non-TOP mRNAs (ACTB) that are unaffected by the absence of LARP1 (Fig. 1 C and D). These results argue that LARP1 is the primary repressor of mRNA with TOP motifs beyond the limited number of individual transcripts that have been previously analyzed (8, 10). Overall, LARP1-regulated transcripts accounted for ∼17.5% of all ribosome footprints in WT cells (SI Appendix, Fig. S1B).
Fig. 1.
LARP1 is the primary regulator of known TOP mRNAs. (A) mTOR controls the translation of classical TOP mRNAs. RNA-seq and ribosome profiling libraries were prepared from WT HEK-293T cells treated with 250 nM Torin 1 for 2 h (n = 2). (A, Top) Translation efficiencies (RPF/RNA) of mRNAs in control or Torin 1-treated conditions. Classical TOP mRNAs indicated in red. Differentially translated mRNAs are indicated in blue (fold change translation efficiency >2, adjusted P < 0.01). (A, Bottom) Changes in translation efficiencies between Torin 1 and control conditions for classical TOP and other mRNAs. (B) LARP1 is required for mTOR regulation of classical TOP mRNA translation. sgLARP1 cells were treated and analyzed as in A (n = 2). (C) Profiles of ribosome protected fragments (RPFs), for example, TOP (RPL3) and non-TOP (ACTB) mRNAs from A and B. (D) LARP1 regulation of classical TOP mRNAs. Changes in translation efficiency of indicated mRNAs in WT or sgLARP1 HEK-293T from A and B. Significance by Wald test (Benjamini–Hochberg adjustment; *P < 0.01, **P < 10−10, ***P < 10−50).
LARP1B Is Not a Major Regulator of TOP mRNA Translation Downstream of mTORC1.
LARP1 accounts for most TOP mRNA regulation, but we did notice some residual repression even in sgLARP1 cells (Fig. 1B). We wondered whether the LARP1 homolog, LARP1B, might account for this anomaly. LARP1B is broadly expressed and 87% identical to LARP1 within the C-terminal domain that recognizes the TOP sequence (796 to 946 of LARP1; SI Appendix, Fig. S3 A and B) (9, 10). We first generated LARP1/1B double-deficient HEK-293T (sgLARP1/1B) cells by using CRISPR/Cas9 to target LARP1B in our existing sgLARP1 cells (SI Appendix, Fig. S3C and Methods). In this context, ectopically expressed LARP1B selectively binds to synthetic capped TOP RNA sequences (SI Appendix, Fig. S3D) and controls the translation of a TOP luciferase reporter (SI Appendix, Figs. S3 E and F and S5F) similarly to LARP1. LARP1B is therefore at least capable of regulating TOP mRNAs. We next analyzed mTOR-regulated translation of endogenous mRNAs using ribosome profiling (SI Appendix, Fig. S3G). Surprisingly, TOP mRNA translation retained the same mild sensitivity to mTOR inhibition observed in sgLARP1 cells (SI Appendix, Fig. S3G and Dataset S3). LARP1B therefore appears to have a minor, if any, impact on TOP mRNA translation under these conditions. The source of the residual sensitivity of TOP mRNAs to mTOR activity in LARP1/1B-deficient cells remains unclear. We nonetheless continued our analysis using sgLARP1/1B cells to avoid the possibility that LARP1B contributes to TOP mRNA regulation under other conditions.
5′ Sequence Features Required for LARP1 Regulation.
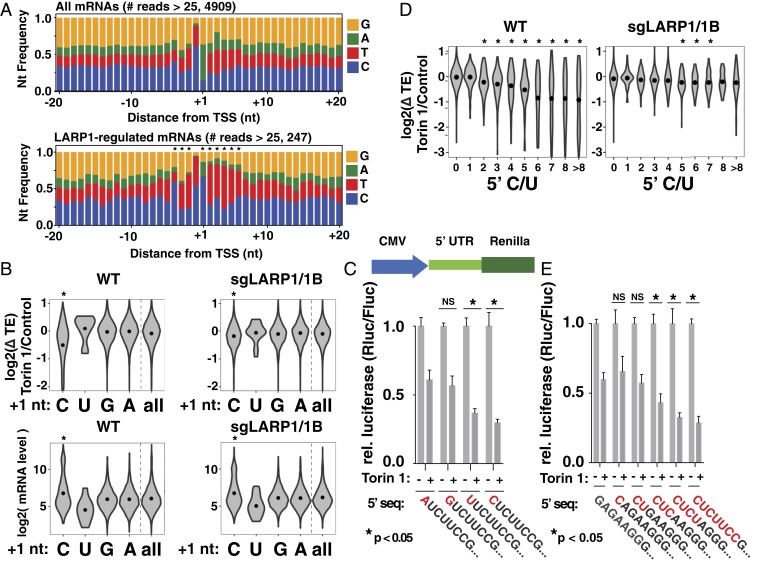
We next sought to determine the defining features of LARP1 target mRNAs. Because classical TOP motifs are anchored at the 5′ ends of mRNAs, we began by comparing the nucleotide composition around the primary TSS of all mRNAs versus the 5% of genes with the strongest LARP1-dependent repression (repression in wild-type cells minus repression in sgLARP1/1B cells). TSSs were determined by analysis of hCAGE (HeliScope cap analysis of gene expression) data for HEK-293 cells generated as part of the FANTOM5 project (Methods) (16). These results show a clear enrichment for Cs in the +1 position of LARP1-dependent mRNAs (Fig. 2A), mirroring the classical TOP definition. The next 5 positions showed an enrichment of Us, also similar to the classical TOP definition. We also observed a smaller but statistically significant enrichment for Cs or Us at the −4 to −2 upstream positions. Overall, the primary features of the most LARP1-regulated mRNAs closely resemble classical TOP motifs: a +1 C followed by a series of pyrimidines. The analysis did not provide strong evidence of a necessary downstream sequence in LARP1-dependent mRNAs (17).
Fig. 2.
Sequence features of LARP1-regulated mRNAs. (A) LARP1-regulated transcripts are enriched for 5′ nucleotides that resemble TOP motifs. Nucleotide frequencies around primary TSS (Top) and 5% most LARP1-dependent mRNAs (Lower). LARP1-dependent repression is defined as log2(ΔTEWT − ΔTEsgLARP1/1B). Significance by chi-square test (P < 10−5). (B) The +1 nucleotides of mRNAs are predictors of mTOR regulation. (Top) mTOR-regulated change in translation efficiency of mRNAs from Fig. 1 A and B in WT and sgLARP1 cells with the indicated +1 nucleotides. Analysis includes 4,910 mRNAs with >25 reads in hCAGE peak. (Bottom) Expression levels of mRNAs (average RPKM in control condition, calculated over CDS regions only) with the indicated +1 nucleotides. Dots indicate mean value. Significance by Tukey HSD test for C, U, or G versus A (*P < 10−10). (C) TOP motifs require either C or U in the +1 position. (Top) Reporter design. (Bottom) A control plasmid encoding firefly luciferase and a test plasmid encoding Renilla luciferase and the indicated 5′ sequences were transfected into WT HEK-293T cells and incubated overnight. Cells were then treated with 250 nM Torin 1 for 2 h and analyzed for luciferase levels (n = 3, error bars are SD, significance by ANOVA compared to +1 A reporter). (D) Length of TOP sequences correlate with LARP1-dependent translation repression. Violin plot shows mTOR-regulated change in translation efficiency of mRNAs from Fig. 1 A and B in WT and sgLARP1 cells binned by length of uninterrupted pyrimidines initiating at the primary TSS. Dots indicate mean value. Significance by t test comparison to 0-length group (*P < 0.01). (E) Longer C/U sequences cause greater repression. Plasmids encoding Renilla luciferase and the indicated 5′ sequences were analyzed as in C (n = 3; error bars are SD, significance by ANOVA compared to non-TOP reporter).
Cs are underrepresented in the +1 position of mRNAs, occurring on only ∼14% of mRNAs. We therefore wondered whether +1 Cs are generally associated with LARP1 regulation. To test this, we binned mRNAs according to the +1 nucleotide of the primary TSS and compared the effect of mTOR inhibition on translation efficiency (Fig. 2B). Transcripts initiating with an A or G were largely unaffected by mTOR inhibition in wild-type cells and constituted the majority (86%) of expressed mRNAs (Fig. 2B and Dataset S4). Transcripts initiating with a U also showed little sensitivity to mTOR signaling, although the number of transcripts in this category (n = 21) is too low to draw strong conclusions. In contrast, the presence of a C in the +1 position was strikingly associated with repression in wild-type cells. The impact of the +1 nucleotide was largely abolished by loss of LARP1 (Fig. 2B). Experiments using luciferase reporters confirmed the importance of the +1 C nucleotide for mTOR regulation (Fig. 2C and SI Appendix, Fig. S5A). Interestingly, these showed that TOP sequences with a +1 U are similarly regulated, even though these are almost nonexistent (∼0.4% of primary TSSs in our dataset) in the transcriptome (Fig. 2 A and C). These observations are in line with conclusions of early reporter-based studies showing the importance of the +1 C in TOP motifs (17). However, the association between +1 Cs and LARP1 regulation across the entire transcriptome is striking, and it is tempting to wonder whether the underrepresentation of these nucleotides at this position reflects a selective pressure related to LARP1 functions. Some mRNAs that are LARP1-regulated in our ribosome profiling data do appear to initiate with purines (Fig. 2A). However, these likely reflect genes with heterogenous TOP and non-TOP TSSs, mRNAs that are indirect targets of LARP1, or a small amount of error in the hCAGE dataset.
A second defining feature of classical TOP motifs is the series of 4 to 14 pyrimidines following the +1 C (or U). However, the exact number varies, and we wondered whether this might underlay a puzzle about TOP motif function: although TOP motifs are regarded as discrete “on-off” regulatory elements, many TOP mRNAs are differently sensitive to mTOR signaling. To query the relationship between motif length and regulation, we binned mRNAs according to the uninterrupted number of pyrimidines beginning at the +1 nucleotide and compared changes in translation efficiency following mTOR inhibition. Surprisingly, the translation of mRNAs with as few as two C/Us were slightly but significantly (P = 4.69 × 10−5) more sensitive to mTOR activity in wild-type cells (Fig. 2D). As the number of pyrimidines increases, so does the degree of translation repression, reaching a maximum at ∼6 nt. This relationship between the length of the polypyrimidine sequence and mTOR-regulated translation is substantially diminished in LARP1/1B-deficient cells (Fig. 2D). To directly test whether longer TOP sequences lead to stronger repression, we constructed a series of luciferase reporters with TOP motifs of increasing length. As predicted, TOP lengths were correlated with translation repression following mTOR inhibition, again reaching a maximum at 6 nt (Fig. 2E and SI Appendix, Figs. S4A and S5 B and S5C). The effect of these features was significantly diminished in sgLARP1/1B cells (SI Appendix, Figs. S4 B and C and S5 D and E). Overall, our observations argue that the “classical” TOP motif of a +1 C followed by 4 to 14 pyrimidines exists within a continuous spectrum of 5′ terminal sequences that confer a wide-range of mTOR regulation.
We also examined whether pyrimidine sequences located elsewhere in mRNA 5′ UTRs could play a role in mTOR regulation. Interestingly, we found that 5′ UTRs of the most LARP1-regulated mRNAs are slightly but significantly enriched for internal pyrimidine stretches (SI Appendix, Fig. S6). These sequences are unlikely to affect LARP1 interactions mediated through the DM15 region because of its dependence on the 5′ cap structure. However, they could conceivably contribute to mRNA binding through the LARP1 La or RRM domains.
Quantification of 5′ Sequence Regulatory Functions.
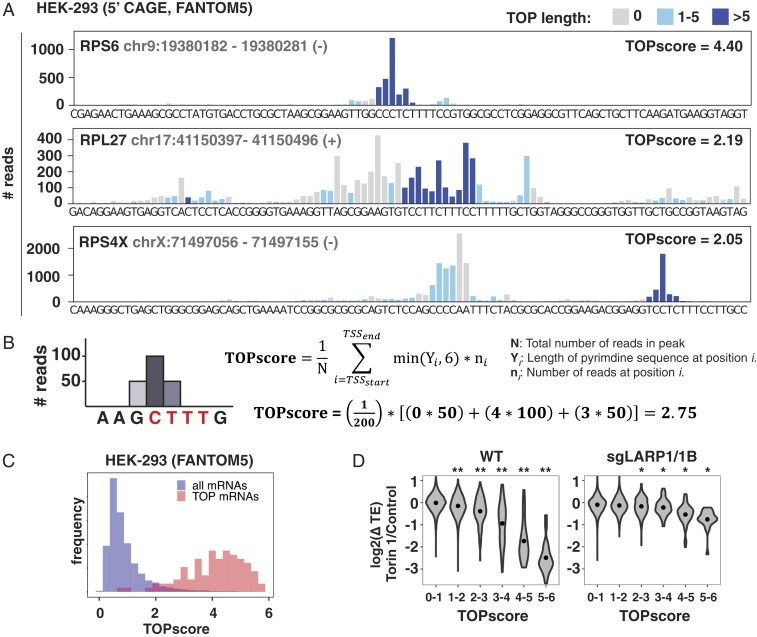
So far, our analysis considered only the sequence of the primary TSS for each gene. However, most TSSs are heterogeneous and generate mRNAs with a mixture of 5′ sequences. For example, the mRNA encoding the ribosomal protein RPL27 has many TSSs with classical TOP motifs even though its primary TSS in HEK-293 cells is actually non-TOP (Fig. 3A). We wondered whether accounting for this heterogeneity would better reflect the potential for mTORC1/LARP1 regulation on a gene level. Toward this end, we introduce here a metric called a TOPscore. This is defined as the mean length of consecutive C/U at all positions in hCAGE peaks within 1 kb of the annotated TSS, weighted by the number of hCAGE reads at each respective position (Fig. 3B). We only counted the first 6 nt at each position because TOP motifs longer than 6 nt do not increase the degree of translation repression (Fig. 2D and SI Appendix, Fig. S5A). TOPscores were calculated for 8,832 genes with greater than 100 hCAGE peak reads (Dataset S5). Of the 92 classical TOP mRNAs (5) detected, 78 had TOPscores >3 (Fig. 3C). The mean TOPscore of the 92 classical TOP mRNAs is 4.05 (SD = 1.06), compared to the mean score of 0.92 (SD = 0.70) for the 8,740 remaining mRNAs (UCSC genes/hg19). To test whether TOPscores predict LARP1 regulation, we compared them to changes in translation efficiency for WT or sgLARP1/1B cells treated with Torin 1 (Fig. 3D). Results in WT cells show a more sensitive relationship between TOPscores and mTOR regulation than when considering only the primary TSS (Fig. 3D). This relationship is severely diminished in sgLARP1/1B cells, underscoring the greater fidelity of TOPscores as a predictor of mTORC1/LARP1 regulation. In rare cases, manual inspection of TOPscores is required to eliminate artifacts of the analysis. For instance, the TSS for vimentin (VIM), a previously identified nonribosomal TOP mRNA, is 1,018 nt downstream of the annotated TSS (NM_003380), just outside the 2,000-nt window that we analyzed. The true TSS encodes a classical TOP sequence.
Fig. 3.
TOPscores predict LARP1-regulated translation. (A) hCAGE reads from HEK-293 cells mapping to the indicated genomic regions. Colors indicate 5′ TOP length on the resulting transcript. (B) Definition of the TOPscore. (C) Classical TOP mRNAs have higher TOPscores than other mRNAs. Histogram shows TOPscores of 92 classical TOP mRNAs and 8,740 remaining mRNAs calculated from hCAGE data for HEK-293 cells. (D) TOPscores are correlated with LARP1-dependent translation repression. Change in translation efficiency following mTOR inhibition from Fig. 1 A and B for mRNAs binned according to their TOPscores in WT and sgLARP1 cells as indicated. For genes with multiple transcripts, only the transcript with the highest number of hCAGE peak reads was included. Dots indicate mean value. Significance by t test comparison to group with TOPscore 0 to 1 (*P < 0.01, **P < 10−10).
Identification of Constitutive and Tissue-Specific TOP mRNAs.
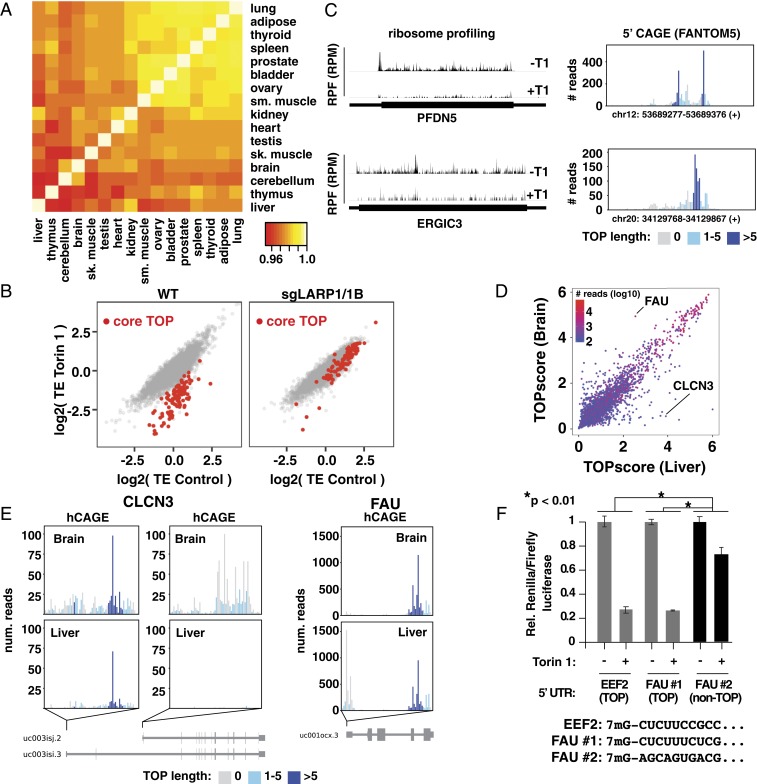
In principle, changes in TSSs might cause mRNAs to gain or lose TOP motifs in different contexts (e.g., tissues) and alter their sensitivity to mTORC1 signaling. To investigate this possibility, we calculated TOPscores from FANTOM5 hCAGE data for 16 human tissues (Dataset S6) (16). Overall, 3,241 mRNAs were detected with greater than 100 hCAGE reads in all tissues. TOPscores were tightly correlated between tissues, indicating that most TOP mRNAs retain TOP motifs across tissues (Fig. 4A). From these data, we defined a set of 105 “core” TOP mRNAs that contain TOP motifs in all tissues where they were detected (Dataset S7). Conservative criteria were used that considered for each gene only tissues in which they had more than 100 hCAGE reads and required a TOPscore greater than 3 in all tissues assessed (with a minimum of 10 tissues) and a greater than twofold decrease in translation efficiency upon mTOR inhibition in HEK-293T cells (Fig. 4B). The core TOP mRNA set includes 80 classical TOP mRNAs, but also others such as PFDN5, a member of a chaperone complex, and ERGIC3, an endoplasmic reticulum membrane-associated protein of unknown function (Fig. 4C). Given that only 3,241 mRNAs were analyzed in this dataset, and that ribosome profiling data were available for only two thirds of the genes with consistently high TOPscores, additional constitutive TOP mRNAs likely exist. Nonetheless, we conclude that there are a relatively limited number of constitutive TOP mRNAs, and that the majority of these are highly expressed genes with growth-related functions, including translation.
Fig. 4.
Variation of TOP motifs across tissues. (A) TOPscores are highly correlated across tissues. Heat map shows Pearson correlations from pairwise comparison of TOPscores for 3,241 mRNAs with >100 peak reads from indicated tissues (further details provided in Methods). (B) mTOR controls “core” TOP mRNAs via LARP1. Translation efficiencies (TEs) in control and Torin 1-treated conditions in WT and sgLARP1/1B HEK-293T cells. Core TOP mRNAs (see text) are indicated in red. (C) Ribosome footprints and hCAGE data for two “core” TOP mRNAs. (Left) Ribosome footprints from Torin 1-treated (+T1) and control (−T1) HEK293T cells for the indicated transcripts. (Right) hCAGE peaks (HEK293 cells) for the same genes. (D) Comparison of TOPscores in liver and brain. Data are colored according to the lowest number of hCAGE peak reads between these two tissues. (E) Genes producing TOP and non-TOP isoforms in brain and liver. hCAGE peaks for the indicated genes for liver and brain. (F) Alternative FAU TSSs are differently mTOR-regulated. A control plasmid encoding firefly luciferase and a test plasmid encoding Renilla luciferase and the 5′ UTR from either Eef2 (mouse), FAU TSS no. 1 (hg38, chr11:65122134), or FAU TSS no. 2 (hg38, chr11:65122188; human) with the indicated 5′ sequences were transfected into WT HEK-293T cells and incubated overnight. Cells were then treated with 250 nM Torin 1 for 2 h and analyzed for luciferase levels (n = 3; error bars are SD, significance by ANOVA).
In contrast to these constitutive TOP mRNAs, some mRNAs acquire TOP motifs in a tissue-specific manner. Overall, we also identified 60 mRNAs with TOP sequences (TOPscore >3) in at least one tissue and lacking them (TOPscore <2) in at least one other tissue. We focused our attention on two tissues that have highly divergent demands for cell growth: brain and liver. While the correlation of TOPscores between these tissues was still high (0.926), a small number of well-expressed outliers had very different TOPscores (Fig. 4D and Dataset S8). One of these is CLCN3, a poorly characterized chloride channel that is well expressed in both tissues. In the liver, it almost exclusively contains a TOP motif (Fig. 4E). In contrast, in the brain, transcription predominates at an entirely different downstream exon that lacks a TOP sequence. These alternative TSSs and their tissue-specific usage were validated by 5′ RACE and qPCR in a human liver-derived (HepG2) and brain-derived (SH-SY5Y) cell line (SI Appendix, Fig. S7 A and B). A second example is FAU, a fusion of ubiquitin with ribosomal protein S30 (Fig. 4E). FAU mRNA is transcribed with a strong TOP motif in all tissues, including the brain. In the liver, however, a second predominant TSS emerges 54 nt upstream at a position encoding a non-TOP sequence (Fig. 4E). These alternative TSSs were also validated by 5′ RACE and qPCR in a human liver-derived (HepG2) and brain-derived (SH-SY5Y) cell line (SI Appendix, Fig. S7 C and D). To test whether these alternate start sites impact mTOR regulation, we inserted both FAU 5′ UTRs into reporter vectors and compared their response to mTOR inhibition (Fig. 4F and SI Appendix, Fig. S5G). As expected, the version containing the TOP sequence was far more sensitive to mTOR inhibition than the non-TOP alternative. These results show how even relatively small changes in TSS selection can alter the posttranscriptional fate of mRNAs.
Discussion
Our results provide a comprehensive analysis of how mTORC1 uses LARP1 to control a functionally diverse, tunable, and dynamic program of gene expression. We establish LARP1 as a master repressor of TOP mRNA translation throughout the transcriptome, demonstrating the generality of a mechanism that had been previously suggested on the basis of a few examples (8–10), but also questioned (12, 13). Analysis of LARP1-target mRNAs allowed us to determine the full spectrum of mRNA 5′ sequences that define this program. The most sensitive mRNA targets of LARP1 primarily encode classical TOP motifs. These include nearly all of the 91 classical TOP mRNAs enumerated by Meyuhas and Kahan that were detected in our dataset, but also 32 other mRNAs with classical TOP sequences (+1 C, followed by 4 to 14 pyrimidines) (5). However, we also find that LARP1 recognizes a wider range of 5′ sequences. These still require a +1 C or U, but can encode as few as two additional pyrimidines (Fig. 2C). Overall, we identified 208 mRNAs (“mTOR-regulated” in Dataset S3) whose translation is repressed more than twofold by mTOR inhibition in HEK-293T cells (Padj < 0.01), about 4% of the 4,998 transcripts that passed criteria for our analysis (Dataset S2). Of these, 112 (“mTOR/LARP1-regulated” in Dataset S3) are significantly less repressed in sgLARP1/1B cells (Padj < 0.01). Many of these are directly involved in processes related to translation, like classical TOP mRNAs. Others encode proteins with distinct growth-related functions, ranging from nutrient metabolism to mitochondrial biogenesis. The majority of these are well-expressed, supporting a hypothesis whereby TOP motifs permit rapid and reversible regulation of these mRNAs without the metabolic cost of degrading and resynthesizing the transcripts.
We also find that variation in TOP sequences specifies the strength of translation regulation across a continuous range. This contrasts with the common notion that TOP motifs are functionally binary, i.e., that mRNAs are either TOP or non-TOP. First, longer 5′ sequences of pyrimidines lead to stronger repression by LARP1, reaching a plateau at six pyrimidines. This may reflect differences in affinity between the mRNA 5′ terminus and the LARP1 DM15 region, which can accommodate up to five nucleotides (9). Second, some mRNAs have heterogeneous transcription start sites that encode mixtures of TOP and non-TOP 5′ sequences. As a result, considering only an mRNA’s primary transcription start site can give a misleading impression of its sensitivity to mTORC1 regulation. The TOPscore described here incorporates information about both TOP motif length and TSS heterogeneity. Other features of mRNAs may also affect LARP1 regulation. Nonetheless, our results argue that those quantified by TOPscores are strong predictors of mTORC1/LARP1 regulation.
A final feature of TOP motifs that has been underappreciated is that their 5′ location allows their addition, modification, or removal by relatively small changes in transcription initiation. Global analyses of TSSs have found wide variation between cell types and tissues (18). We were surprised to find that many mRNAs with strong TOPscores in HEK-293 cells maintained similar TSSs across the 16 tissues we analyzed. Nonetheless, we also find clear examples of TSS switching that add or remove TOP motifs (Fig. 4 E and F). One of these is FAU, a ribosome-ubiquitin fusion protein. In the brain, FAU transcripts are almost exclusively TOP-containing, while the liver produces a mixture that is equally TOP and non-TOP. Prolonged periods of mTORC1 inhibition, such as during nutrient deprivation, could conceivably affect ribosome composition differently in these tissues in ways that modify their function. We note that our pan-tissue analysis captured only 3,241 mRNAs due to limitations in detection. We have thus likely missed other mRNAs with tissue-specific TOP sequences because of lower expression levels. Such mRNAs might be expected to encode proteins with regulatory functions, rather than the predominantly abundant and long-lived proteins encoded by classical TOP mRNAs.
An unresolved puzzle is why the translation of many TOP mRNAs remains weakly sensitive to mTORC1 activity even in the absence of LARP1. Our results rule out a role for LARP1B, although it is intriguing to wonder whether it might act downstream of other signaling pathways independent of mTORC1. One possibility is that TOP mRNAs are also intrinsically poor substrates for eIF4F, which becomes limiting when mTORC1 is inhibited. Several studies have indeed found that the cap-binding subunit of eIF4F, eIF4E, has intrinsically lower affinity for mRNAs that initiate with a C, which would include most TOP mRNAs (19, 20). It also seems likely that mTORC1 targets these mRNAs through independent mechanisms, especially given the many mTORC1-regulated but LARP1-independent transcripts in our own dataset. Deciphering the signatures and regulation of these mechanisms is still required before mTORC1 targets can be fully predicted from mRNA sequence alone.
Methods
Materials.
Reagents were obtained from the following sources: antibodies for NCBP1, eIF4E, FLAG, mTOR, and GFP from Cell Signaling Technology; DMEM and TRIzol Reagent from Life Technologies; heat-inactivated FBS from Sigma Aldrich; T4 RNA Ligase I, polynucleotide kinase, Protoscript II reverse transcriptase, Phusion DNA polymerase, Vaccinia Capping System, Oligo d(T)25 magnetic beads, streptavidin-coated magnetic beads, NEBNext Ultra II Directional RNA Library prep kit, and NEBNext rRNA Depletion kit from New England Biolabs; Ribo-Zero Gold kit from Illumina; CircLigase I from Lucigen; iTaq Universal SYBR Green Supermix and Bradford Protein Assay from Bio-Rad; RNeasy Plus Mini Kit from Qiagen; and XtremeGENE 9 transfection reagent from Roche.
Cell Culture and Preparation of Cell Extracts.
Cells were grown in high-glucose DMEM supplemented with 10% (vol/vol) heat-inactivated FBS and penicillin/streptomycin. To prepare extracts, cells rinsed once with ice-cold PBS- were lysed in ice-cold lysis buffer (40 mM Hepes [pH 7.4], 2 mM EDTA, 10 mM pyrophosphate, 10 mM glycerophosphate, 1.0% Triton X-100) in dishes. The soluble fractions of cell lysates were isolated by centrifugation at 13,000 rpm for 5 min in a microcentrifuge. Protein concentrations were normalized using the Bradford Protein Assay. Isolated proteins were denatured by the addition of sample buffer and boiling for 2 min, followed by immunoblotting.
Generation of sgLARP1/LARP1B HEK-293T Cells.
To generate sgLARP1/1B HEK-293T cells, the sgRNA sequence 5′-GACAGTGACAGCAAAGAAAAC-3′ targeting exon 4 of LARP1B was inserted into the px330 vector (21) and, along with a plasmid encoding GFP, transfected into sgLARP1 HEK-293T cells that were generated previously (10). At 48 h after transfection, cells were FACS-sorted into 96-well plates with no more than one cell per well and allowed to form colonies. Single-cell–derived colonies were screened first by monitoring LARP1B mRNA levels using qPCR. Cell lines with reduced LARP1B mRNA levels were next analyzed by cloning and sequencing the targeted region of genomic DNA. Five clones were sequenced from the sgLARP1/1B cell line used in this study, all of which contained a single nucleotide deletion resulting in a frame shift.
Synthesis of TOP and Non-TOP Short RNA Probes.
Ten-nucleotide RNA oligonucleotides with 5′ triphosphates were synthesized as described previously (22, 23). Oligonucleotides were then enzymatically capped using the Vaccinia Capping System and purified by PAGE. The capping reaction could be monitored by an obvious shift in mobility, and proceeded to near-completion. For biotinylated probes, short capped oligonucleotides were ligated to a synthetic 5′ phosphorylated and 3′ biotinylated RNA linker 5′-GTCGTCGCCGCCATCCTCGG-3′ using T4 RNA ligase I. Ligation products were purified by PAGE.
Capture of RNA Binding Proteins.
HEK-293T cells were washed once in ice cold PBS- and then lysed in low-salt lysis buffer (16 mM Hepes-KOH, pH 7.4, 10 mM KOAc, 0.5 mM Mg0Ac, 1 mM DTT, and 1% Triton X-100). Protein concentrations were then quantified using Bradford assay and normalized to equal levels. Equal volumes of extract were then incubated with 2.5 pmol of RNA probes for 1 h, followed by addition of streptavidin-coated magnetic beads. Samples were incubated for an additional 1 h and then washed six times with lysis buffer, followed by elution in lysis buffer additionally containing 1% SDS at room temperature for 30 min.
TOP mRNA Reporter Assay.
Short-lived Renilla luciferase constructs were generated by appending a tandem PEST and CL1 motif to the C terminus of the Renilla coding sequence, which we confirmed to decrease the protein half-life to ∼20 min (24). The 5′ UTRs of target genes were then amplified by PCR from genomic DNA or cDNA and inserted into the Renilla-containing vector using Gibson assembly. Eef2 mutant 5′ UTRs were constructed by PCR mutagenesis to produce reporters with the indicated 5′ terminal sequences. HEK-293T cells were transfected with 100 ng pIS0 (Addgene no. 12178, encoding firefly luciferase), 100 ng of the Renilla reporter, and 800 ng of either empty vector, GFP, LARP1, or LARP1B using XtremeGENE 9. After 24 h, cells were divided in 12-well plates at 0.3 million cells per well and incubated for an additional 24 h. Cells were then treated as indicated and analyzed using the Promega Dual-Luciferase Reporter Assay System according to the manufacturer’s instructions.
Preparation of RNA-Seq and Ribosome Profiling Libraries.
Ribosome profiling libraries were prepared similarly to previously described, with some modifications, primarily to rRNA depletion (25). Briefly, HEK-293T cells were seeded in 10-cm dishes at 5 million cells per plate, incubated overnight, and then treated with vehicle (DMSO) or 250 nM Torin 1 for 2 h. Cells were washed three times in 5 mL ice cold PBS- and then lysed in polysome lysis buffer (15 mM Hepes, pH 7.4, 150 mM NaCl, 5 mM MgCl2, 1% Triton X-100). Samples were divided and processed for either RNA-seq or ribosome profiling. For ribosome profiling, lysates were digested with RNase If for 45 min at RT and centrifuged through a 1M sucrose cushion using an MLA-150 rotor at 80,000 rpm for 4 h to pellet ribosomes. RNA was extracted from the resuspended pellet by proteinase K digest, followed by acid-phenol extraction. rRNA was removed from the sample using the Ribo-Zero Gold kit (Illumina) according to the manufacturer’s instructions, except that the final 50 °C incubation immediately prior to bead removal was omitted. Ribosome footprints were then isolated by excising and extracting 28- to 32-nt fragments from 15% polyacrylamide gels. Samples were 3′ dephosphorylated using PNK, followed by 3′ ligation using T4 RNA ligase 1 to the 3′ linker (5′-CTGTAGGCACCATCAAT-3′), and ligation products were size-selected by PAGE. RNA was then subjected to reverse transcription using SuperScript II and an RT primer (5′-AGATCGGAAGAGCGTCGTGTAGGGAAAGAGTGTAGATCTCGGTGGTCGC-spacer-TTCAGACGTGTGCTCTTCCGATCTATTGATGGTGCCTACAG-3′), after which RNA was destroyed by alkaline hydrolysis. RT products were purified by PAGE, circularized using CircLigase I, and then amplified using the Phusion DNA polymerase and primer sets compatible with the Illumina TruSeq system. (Note: RPF libraries for sgLARP1/1B cells were prepared using a slightly modified 3′ linker and RT primer based on the protocol described by McGlincy and Ingolia [26]. The 3′ linker was 5′-NNNNN-[5-nt barcode]-AGATCGGAAGAGCACACGTCTGAA-3′. RT primer was 5′-NNAGATCGGAAGAGCGTCGTGTAGGGAAAGAG-spacer-GTGACTGGAGTTCAGACGTGTGCTC.) For RNA-seq, extracts from WT HEK-293T cells were adjusted to 0.5% SDS, digested with proteinase K, and then extracted with acid-phenol. RNA was then fragmented by alkaline hydrolysis. RNA fragments of 40 to 80 nt were gel-purified and used to prepare libraries as described earlier for ribosome protected fragments. For sgLARP1 and sgLARP1/1B cells, RNA-seq libraries were prepared from extracts using the NEB Next Ultra II Directional RNA Library Prep kit along with the NEBNext rRNA Depletion kit, both according to the manufacturer’s instructions. All samples were sequenced on an Illumina HiSeq system. Two biological replicates were prepared for each library.
Bioinformatic Analysis of RNA-Seq and Ribosome Profiling Libraries.
RNA-seq and ribosome profiling datasets were first processed to remove the 3′ linker sequence using the FASTX toolkit (http://hannonlab.cshl.edu/fastx_toolkit/). Reads were then aligned to human rRNA sequences using Bowtie2 to filter out reads mapping to rRNA (27). Reads for sgLARP1/1B RPF libraries were deduplicated using the unique molecular identifier (UMI) encoded in the first five bases of the 3′ linker. The remaining reads were mapped to the human genome (hg19) using TopHat (28). Ribosome footprints and RNA levels mapping to CDS regions were counted per gene using HTSeq-count (29). Per–cell-type translation efficiencies were calculated and compared between Torin-1–treated and control conditions using DESeq2 (30), and only genes for which DESeq2 could calculate adjusted P values (Wald test, Benjamini–Hochberg adjustment) for the fold change in translation efficiency for all cell types (wild-type, LARP1 mutant, and LARP1/1B double mutant) were used for further analysis. For the same set of genes, mRNA rpkm for wild-type control conditions (Fig. 2B) were calculated by first calculating rpkm per replicate (using CDS-mapped reads and union to gene CDS lengths) and then calculating their average. Pyrimidine k-mer density per 5′UTR was calculated using UCSC annotated 5′UTR sequences (hg19 genome), summing all nonoverlapping occurrences of a pyrimidine k-mer from the +11 nucleotide to the end of the 5′UTR, using a +1 nucleotide sliding window approach, and dividing this sum by the 5′UTR length (minus 10 nt) in kb.
Analysis of FANTOM5 hCAGE Data, Calculation of TOPscores, and Identification of TSSs.
HeliScopeCAGE (hCAGE) data were downloaded from the Fantom5 project (https://fantom.gsc.riken.jp/5/). The BAM files in Table 1 were used to calculate TOPscores using custom python scripts. All hCAGE reads, including those mapping to multiple regions, were used. For each UCSC annotated transcript, hCAGE peak regions were defined in a region stretching from 1,000 nt upstream to 1,000 nt downstream from the annotated 5′ end by identifying and merging 50-nt windows that differed significantly (P < 10−9 assuming Poisson distribution) from adjacent 200-nt sequences on both sides. TOPscores were calculated using the formula in Fig. 3B on all reads in all peak regions for a given transcript. For genes with multiple transcripts, the transcript with the highest total number of reads in all peak regions was used for downstream analysis. For analyses that considered only the single primary TSS, TSSs were identified as the position with the highest number of hCAGE reads within all peak regions identified for a given transcript. Per gene, the TSS was selected according to the following criteria: (i) the highest number of reads in the peak region in which the TSS localized and (ii) the TSS with the highest number of reads if multiple peak regions had identical numbers of reads. If this still identified more than one TSS, the most upstream position was selected. Only genes with a TSS in a peak region covered by >25 reads were considered in further analyses.
Table 1.
Fantom5 data files downloaded from https://fantom.gsc.riken.jp/5/datafiles/latest/basic/
| Name | Data file |
| HEK-293 cells | human.cell_line.hCAGE/embryonic%20kidney%20cell%20line%3a%20HEK293%2fSLAM%20untreated.CNhs11046.10450–106F9.hg19.nobarcode.bam |
| Adipose | human.tissue.hCAGE/adipose%20tissue%2c%20adult%2c%20pool1.CNhs10615.10010–101C1.hg19.nobarcode.bam |
| Bladder | human.tissue.hCAGE/bladder%2c%20adult%2c%20pool1.CNhs10616.10011–101C2.hg19.nobarcode.bam |
| Brain | human.tissue.hCAGE/brain%2c%20adult%2c%20pool1.CNhs10617.10012–101C3.hg19.nobarcode.bam |
| Cerebellum | human.tissue.hCAGE/cerebellum%2c%20adult%2c%20pool1.CNhs11795.10083–102B2.hg19.nobarcode.bam |
| Heart | human.tissue.hCAGE/heart%2c%20adult%2c%20pool1.CNhs10621.10016–101C7.hg19.nobarcode.bam |
| Kidney | human.tissue.hCAGE/kidney%2c%20adult%2c%20pool1.CNhs10622.10017–101C8.hg19.nobarcode.bam |
| Liver | human.tissue.hCAGE/liver%2c%20adult%2c%20pool1.CNhs10624.10018–101C9.hg19.nobarcode.bam |
| Lung | human.tissue.hCAGE/lung%2c%20adult%2c%20pool1.CNhs10625.10019–101D1.hg19.nobarcode.bam |
| Ovary | human.tissue.hCAGE/ovary%2c%20adult%2c%20pool1.CNhs10626.10020–101D2.hg19.nobarcode.bam |
| Prostate | human.tissue.hCAGE/prostate%2c%20adult%2c%20pool1.CNhs10628.10022–101D4.hg19.nobarcode.bam |
| Skeletal muscle | human.tissue.hCAGE/skeletal%20muscle%2c%20adult%2c%20pool1.CNhs10629.10023–101D5.hg19.nobarcode.bam |
| Smooth muscle | human.tissue.hCAGE/smooth%20muscle%2c%20adult%2c%20pool1.CNhs11755.10048–101G3.hg19.nobarcode.bam |
| Spleen | human.tissue.hCAGE/spleen%2c%20adult%2c%20pool1.CNhs10631.10025–101D7.hg19.nobarcode.bam |
| Testis | human.tissue.hCAGE/testis%2c%20adult%2c%20pool1.CNhs10632.10026–101D8.hg19.nobarcode.bam |
| Thymus | human.tissue.hCAGE/thymus%2c%20adult%2c%20pool1.CNhs10633.10027–101D9.hg19.nobarcode.bam |
| Thyroid | human.tissue.hCAGE/thyroid%2c%20adult%2c%20pool1.CNhs10634.10028–101E1.hg19.nobarcode.bam |
Identification of Core TOP mRNAs.
We defined core TOP mRNAs by applying the following criteria. (i) They have a TOPscore higher than 3 in at least 10 tissues and a TOPscore <3 in 0 tissues. Only tissue TSSs with greater than 100 hCAGE peak reads are considered (215 genes meet all of these criteria). (ii) Translation data are available from our HEK-293T ribosome profiling experiment (137 genes). (iii) They are down-regulated in translation efficiency more than twofold (Torin 1/control; adjusted P value < 0.01) in wild-type cells (105 genes).
5′ Rapid Amplification of cDNA Ends (RACE).
Total RNA was extracted from HepG2 and SH-5YSY cell lines using TRIzol in combination with column purification and DNAse I treatment. FAU and CLCN3 mRNAs were reverse-transcribed with ProtoScript II using the RT primers listed in Table 2. RNA was hydrolyzed and cDNA purified using Zymo DNA purification kit and resuspended in 15 µL. A polyA tail was added to cDNA using Terminal Transferase (NEB) in a 20-µL reaction. This reaction was diluted 20× and used as a template for PCR using Phusion polymerase with reverse primer 1, reverse primer 2, and the respective forward primers (clones for CLCN3 were obtained using either of the indicated forward primers in Table 2) using the following program: 5 min at 98 °C, 5 min at 48 °C, and 40 min at 72 °C, followed by 40 cycles of 15 s at 98 °C, 30 s at 50 °C, and 1 min at 72 °C for 12 min at 72 °C. PCR product was visualized on a gel, followed by gel extraction, and digested using the Not1/Sal1 restriction enzymes. Digested products were cloned and then sequenced to determine 5′ ends.
Table 2.
Primers use for 5′ RACE
| Primer name | Sequence, 5′ to 3′ |
| FAU RT primer | CCTGTTTGGCCACCTTAGGA |
| Reverse Amp primer 1 | GACTCGAGTCGACATCGATTTTTTTTTTTTTTTTT |
| Reverse Amp primer 2 | GACTCGAGTCGACATCGA |
| FAU Forward Amp primer 1 | AATTAGCGGCCGCGTGGCCTCATCCTCCAGG |
| CLCN3 RT primer | CAGGGAAACTGCAAGAAAGG |
| CLCN3 Forward Amp primer 1 | AATTAGCGGCCGCTTTTCTCGCACCCAATCAAT |
| CLCN3 Forward Amp primer 2 | AATTAGCGGCCGCTGTTGATCCGTCTATGCCTTT |
RNA Analysis by Quantitative PCR (qPCR).
Total RNA was isolated from cells using the Qiagen RNeasy kit or TRIzol in combination with column purification. cDNA was synthesized with the ProtoScript II reverse transcriptase using oligo dT18 primers. qRT-PCR was carried out using iTaq Universal SYBR Green Supermix. Primers used are listed in Table 3.
Table 3.
Primer sets used for qPCR
| Target | Forward and reverse primers, 5′ to 3′ |
| Human LDHA | GGCTACACATCCTGGGCTAT |
| CAGCTCCTTTTGGATCCCCC | |
| Human LARP1 | CTGCCTTAATGAGCGGAAAC |
| TCGAAAAAGGCACTCCAAAC | |
| Human LARP1B | CAAAGAAAACCGGGAAACAA |
| ATCTCGCTTCCAACTTCGTG | |
| Human FAU mRNA (long 5′UTR only) | GCAGCCCACGGTCTGTACT |
| TCGAAGGTGTGTAGCTCCTG | |
| Human FAU mRNA (internal) | CTGGAGGATGAGGCCACTCT |
| TGACCTCTCACTTTTCCAGCA | |
| Human CLCN3 (TSS1) | TAGGGATCTCCAGAGCGAGA |
| GACGTTTTCTACCGCAGAGG | |
| Human CLCN3 (TSS2) | CAGCAGGGATGGAAGAA |
| AGGCAAATAAGGGTCGGAAG | |
| Human CLCN3 (internal) | TCAGCATGGGAAATGACAAA |
| TCTATTAATCCGGCCAGTGC |
Data Availability.
RNA-seq and ribosome profiling datasets have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE132703). Custom Python scripts used in this manuscript are available at GitHub, https://github.com/carsonthoreen/tss_tools.
Supplementary Material
Acknowledgments
We thank Wendy Gilbert, Chris Miller, and Susan Baserga for helpful discussions; and Michael Caplan and Biff Forbush for use of equipment and technical advice. We thank Françoise Debart and Jean-Jacques Vasseur for providing short TOP and non-TOP RNA oligonucleotides. This work was supported by the Yale Top Scholar Award (C.C.T.) and the NIH Grant (GM125955 to C.C.T.).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: The data reported in this paper have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE132703). Custom Python scripts used in this manuscript are available at GitHub, https://github.com/carsonthoreen/tss_tools.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1912864117/-/DCSupplemental.
References
- 1.Laplante M., Sabatini D. M., mTOR signaling in growth control and disease. Cell 149, 274–293 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Sonenberg N., Hinnebusch A. G., Regulation of translation initiation in eukaryotes: Mechanisms and biological targets. Cell 136, 731–745 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Hsieh A. C., et al. , The translational landscape of mTOR signalling steers cancer initiation and metastasis. Nature 485, 55–61 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thoreen C. C., et al. , A unifying model for mTORC1-mediated regulation of mRNA translation. Nature 485, 109–113 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyuhas O., Kahan T., The race to decipher the top secrets of TOP mRNAs. Biochim. Biophys. Acta 1849, 801–811 (2015). [DOI] [PubMed] [Google Scholar]
- 6.Yamashita R., et al. , Comprehensive detection of human terminal oligo-pyrimidine (TOP) genes and analysis of their characteristics. Nucleic Acids Res. 36, 3707–3715 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Patursky-Polischuk I., et al. , Reassessment of the role of TSC, mTORC1 and microRNAs in amino acids-meditated translational control of TOP mRNAs. PloS One 9, e109410 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Fonseca B. D., et al. , La-related protein 1 (LARP1) represses terminal oligopyrimidine (TOP) mRNA translation downstream of mTOR complex 1 (mTORC1). J. Biol. Chem. 290, 15996–16020 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lahr R. M., et al. , La-related protein 1 (LARP1) binds the mRNA cap, blocking eIF4F assembly on TOP mRNAs. eLife 6, e24146 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Philippe L., Vasseur J. J., Debart F., Thoreen C. C., La-related protein 1 (LARP1) repression of TOP mRNA translation is mediated through its cap-binding domain and controlled by an adjacent regulatory region. Nucleic Acids Res. 46, 1457–1469 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Bousquet-Antonelli C., Deragon J. M., A comprehensive analysis of the La-motif protein superfamily. RNA 15, 750–764 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Gentilella A., et al. , Autogenous control of 5′TOP mRNA stability by 40S ribosomes. Mol. Cell 67, 55–70.e4 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Tcherkezian J., et al. , Proteomic analysis of cap-dependent translation identifies LARP1 as a key regulator of 5'TOP mRNA translation. Genes Dev. 28, 357–371 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.van den Elzen A. M. G., Philippe L., Thoreen C. C., mTORC1 controls a dynamic program of TOP mRNA translation via LARP1. GEO Datasets: https://www.ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE132703. Deposited 13 June 2019.
- 15.Thoreen C. C., et al. , An ATP-competitive mammalian target of rapamycin inhibitor reveals rapamycin-resistant functions of mTORC1. J. Biol. Chem. 284, 8023–8032 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Lizio M., et al. ; FANTOM consortium , Gateways to the FANTOM5 promoter level mammalian expression atlas. Genome Biol. 16, 22 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Avni D., Shama S., Loreni F., Meyuhas O., Vertebrate mRNAs with a 5′-terminal pyrimidine tract are candidates for translational repression in quiescent cells: Characterization of the translational cis-regulatory element. Mol. Cell. Biol. 14, 3822–3833 (1994). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yamashita R., Wakaguri H., Sugano S., Suzuki Y., Nakai K., DBTSS provides a tissue specific dynamic view of Transcription Start Sites. Nucleic Acids Res. 38, D98–D104 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Niedzwiecka A., et al. , Biophysical studies of eIF4E cap-binding protein: Recognition of mRNA 5′ cap structure and synthetic fragments of eIF4G and 4E-BP1 proteins. J. Mol. Biol. 319, 615–635 (2002). [DOI] [PubMed] [Google Scholar]
- 20.Tamarkin-Ben-Harush A., Vasseur J. J., Debart F., Ulitsky I., Dikstein R., Cap-proximal nucleotides via differential eIF4E binding and alternative promoter usage mediate translational response to energy stress. eLife 6, e21907 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Cong L., et al. , Multiplex genome engineering using CRISPR/Cas systems. Science 339, 819–823 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lavergne T., Bertrand J. R., Vasseur J. J., Debart F., A base-labile group for 2′-OH protection of ribonucleosides: Aa major challenge for RNA synthesis. Chemistry 14, 9135–9138 (2008). [DOI] [PubMed] [Google Scholar]
- 23.Zlatev I., et al. , Efficient solid-phase chemical synthesis of 5′-triphosphates of DNA, RNA, and their analogues. Org. Lett. 12, 2190–2193 (2010). [DOI] [PubMed] [Google Scholar]
- 24.Rubio C. A., et al. , Transcriptome-wide characterization of the eIF4A signature highlights plasticity in translation regulation. Genome Biol. 15, 476 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Park Y., Reyna-Neyra A., Philippe L., Thoreen C. C., mTORC1 balances cellular amino acid supply with demand for protein synthesis through post-transcriptional control of ATF4. Cell Rep. 19, 1083–1090 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.McGlincy N. J., Ingolia N. T., Transcriptome-wide measurement of translation by ribosome profiling. Methods 126, 112–129 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Langmead B., Salzberg S. L., Fast gapped-read alignment with Bowtie 2. Nat. Methods 9, 357–359 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Trapnell C., Pachter L., Salzberg S. L., TopHat: Discovering splice junctions with RNA-seq. Bioinformatics 25, 1105–1111 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Anders S., Pyl P. T., Huber W., HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics 31, 166–169 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Love M. I., Anders S., Kim V., Huber W., RNA-seq workflow: Gene-level exploratory analysis and differential expression. F1000 Res. 4, 1070 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
RNA-seq and ribosome profiling datasets have been deposited in the Gene Expression Omnibus (GEO) database, https://www.ncbi.nlm.nih.gov/geo/ (accession no. GSE132703). Custom Python scripts used in this manuscript are available at GitHub, https://github.com/carsonthoreen/tss_tools.