Significance
For several decades, retroviral core uncoating has been thought to occur in the cytoplasm in coordination with reverse transcription, and while some recent studies have concluded that HIV-1 uncoating occurs at the nuclear envelope during nuclear import, none have concluded that uncoating occurs in the nucleus. Here, we developed methods to study HIV-1 uncoating by direct labeling and quantification of the viral capsid protein associated with infectious viral cores that produced transcriptionally active proviruses. We find that infectious viral cores in the nuclei of infected cells are largely intact and uncoat near their integration sites just before integration. These unexpected findings fundamentally change our understanding of HIV-1 postentry replication events.
Keywords: HIV-1, capsid, uncoating, integration, transcription
Abstract
HIV-1 capsid core disassembly (uncoating) must occur before integration of viral genomic DNA into the host chromosomes, yet remarkably, the timing and cellular location of uncoating is unknown. Previous studies have proposed that intact viral cores are too large to fit through nuclear pores and uncoating occurs in the cytoplasm in coordination with reverse transcription or at the nuclear envelope during nuclear import. The capsid protein (CA) content of the infectious viral cores is not well defined because methods for directly labeling and quantifying the CA in viral cores have been unavailable. In addition, it has been difficult to identify the infectious virions because only one of ∼50 virions in infected cells leads to productive infection. Here, we developed methods to analyze HIV-1 uncoating by direct labeling of CA with GFP and to identify infectious virions by tracking viral cores in living infected cells through viral DNA integration and proviral DNA transcription. Astonishingly, our results show that intact (or nearly intact) viral cores enter the nucleus through a mechanism involving interactions with host protein cleavage and polyadenylation specificity factor 6 (CPSF6), complete reverse transcription in the nucleus before uncoating, and uncoat <1.5 h before integration near (<1.5 μm) their genomic integration sites. These results fundamentally change our current understanding of HIV-1 postentry replication events including mechanisms of nuclear import, uncoating, reverse transcription, integration, and evasion of innate immunity.
The HIV-1 mature conical capsid core, composed of 250 CA hexamers and 12 pentamers (1–3), enters the cytoplasm upon fusion of the viral and host membranes and contains viral RNA and enzymes needed to complete viral replication. Determining where and when uncoating occurs is fundamental to understanding essential postentry replication events, including reverse transcription, evasion of host innate immunity, nuclear import, and integration. For the past four decades, retroviral uncoating has been thought to occur in the cytoplasm (4). Previous studies have proposed that intact viral cores with diameters of ∼61 nm (5) are too large to fit through the ∼39-nm inner diameter of nuclear pores (reviewed in ref. 6) and uncoating occurs in the cytoplasm in coordination with reverse transcription (7–12). A few recent studies have concluded that uncoating occurs at the nuclear envelope (NE) during nuclear import (12–17). Importantly, no published studies have concluded that HIV-1 viral cores remain intact or nearly intact during nuclear import and uncoat in the nucleus.
Previous studies of HIV-1 uncoating have been hampered for two major reasons. First, only one of ∼50 reverse transcription complexes/preintegration complexes (RTCs/PICs) in infected cells leads to provirus formation and productive infection (18); consequently, biochemical analyses of the population of RTCs/PICs may not reflect the properties of infectious viral complexes. Second, most previous studies of HIV-1 uncoating using imaging assays have relied on indirect CA detection methods to infer loss of CA from intracellular viral nucleoprotein complexes composed of a partial or intact viral core, viral genomic RNA or DNA, and enzymes (for simplicity, referred to as viral complexes hereafter). These indirect methods include immunofluorescence assays with anti-CA antibodies (14, 18–21) or fluorescent labeling of viral core-associated host protein cyclophilin A (CypA) (12, 15). However, immunofluorescence assays may be compromised by accessibility of the CA epitope in viral complexes, and loss of fluorescent CypA may report dissociation of the CypA from the viral core rather than viral core disassembly. Some previous studies have suggested that low levels of CA are associated with nuclear complexes (14, 15, 17, 19, 20, 22) and are thought to influence integration site selection (23, 24); in these studies, the nuclear CA amounts may have been underestimated because of reduced CA epitope accessibility (25). Although a few studies have directly labeled CA with a tetracysteine tag (26), this approach has not been widely used because of potential issues of nonspecific labeling and photobleaching.
Here, we developed methods to directly label CA with GFP and track viral cores in infected cells through viral DNA integration and proviral DNA transcription by live-cell microscopy. Astonishingly, our results show that infectious viral cores in the nucleus are intact (or nearly intact) and complete reverse transcription in the nucleus before uncoating. These observations fundamentally change our current understanding of HIV-1 postentry replication events including mechanisms of nuclear import and uncoating as well as reverse transcription, integration, and evasion of innate immunity. We also probed the mechanism of viral core nuclear import and show that intact or nearly intact viral cores gain nuclear entry through a mechanism involving interactions at the NE with host protein CPSF6.
Results
Development of a Method to Directly Label HIV-1 CA in Viral Complexes.
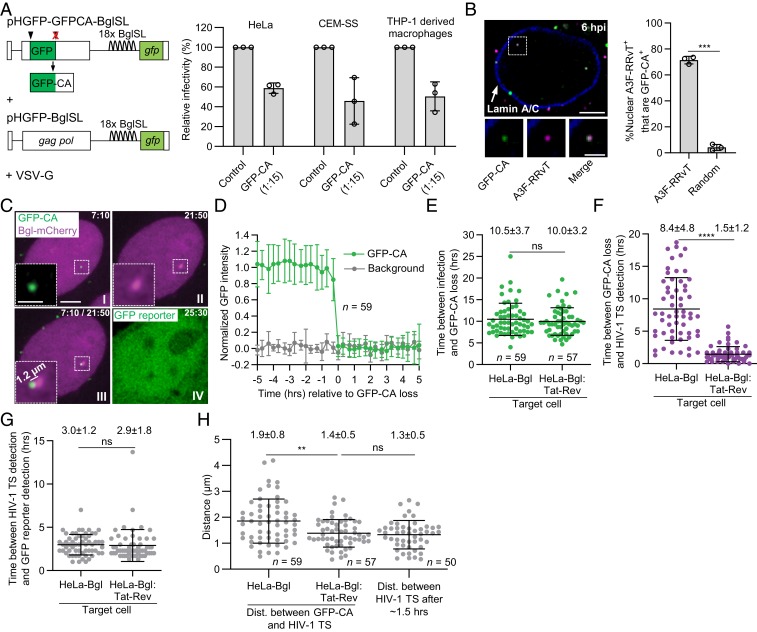
To directly label HIV-1 CA in viral complexes, we inserted GFP between matrix and CA as previously described (27, 28). In addition, mutation of the protease cleavage site between GFP and CA resulted in expression of a GFP-CA fusion protein enabling direct visualization of the viral core (Fig. 1 A, Left). This method produced virions that were labeled with a high efficiency and retained ∼50% of their infectivity in HeLa cells, CEM-SS T cells, and THP-1–derived macrophages compared to virions made in the absence of GFP-CA (Fig. 1 A, Right). Most HIV-1 virions produced by cotransfecting the GFP-CA–expressing vector and a vector expressing wild-type (WT) gag-pol at a 1:15 ratio were labeled with GFP-CA (∼96%; SI Appendix, Fig. S1 A–C). To determine whether GFP-CA remained associated with viral complexes through nuclear import, cells were infected with virions colabeled with GFP-CA and virion core-associated host restriction factor APOBEC3F (A3F) (18) fused to fluorescent protein red–red vine tomato (RRvT). The percentage of A3F-RRvT–labeled nuclear viral complexes that colocalized with detectable levels of GFP-CA was high (71%; Fig. 1B), indicating that GFP-CA remained associated with viral complexes through nuclear import at a high efficiency. The GFP-CA labeling at the 1:15 ratio did not have a significant impact on virion morphology since labeled and unlabeled virions displayed similar ratios of virions with mature and immature morphologies (SI Appendix, Fig. S1D). Sucrose-gradient fractionation of GFP-CA–labeled and unlabeled virions revealed similar proportions of intact viral cores, suggesting that GFP-CA labeling did not alter the in vitro stability of the viral cores (SI Appendix, Fig. S1 E and F).
Fig. 1.
GFP-CA–labeled viral complexes uncoat in the nucleus within ∼1.5 µm of HIV-1 transcription sites. (A) HIV-1 vectors (Left) used to produce GFP-CA–labeled virions with high infectivity in HeLa, CEM-SS, and THP-1–derived macrophages (Right) compared to unlabeled control virions (set to 100%). (B) Nucleus of a HeLa cell infected with virions colabeled with GFP-CA + A3F-RRvT and immunostained with anti-Lamin A/C antibody (Left). Most nuclear A3F-RRvT viral complexes (∼70%) have detectable GFP-CA signals 6-h postinfection (hpi) compared to random locations in the nucleus (Right). (C) Representative live-cell microscopy images of a HeLa-Bgl cell infected with GFP-CA–labeled virions. A GFP-CA–labeled nuclear viral complex uncoated and lost the GFP-CA signal 7:10 hpi (I) and HIV-1 TS appeared near the site of GFP-CA disappearance 21:50 hpi (II). The HIV-1 TS appeared 1.2 µm from the GFP-CA signal (III). GFP reporter expression detected 25:30 hpi (IV). (D) Average normalized GFP-CA intensities are stable before abrupt GFP-CA loss within a single frame (<20 min). (E) Time between infection and nuclear GFP-CA loss, (F) nuclear GFP-CA loss and HIV-1 TS appearance, and (G) HIV-1 TS appearance and gfp reporter detection for 59 and 57 infectious GFP-CA–labeled viral complexes in HeLa-Bgl cells and HeLa-Bgl:Tat-Rev cells, respectively. (H) Distance between GFP-CA signal (time point prior to GFP-CA loss) and HIV-1 TS (first time point of detection) in HeLa-Bgl cells (∼8.4 h) and HeLa-Bgl:Tat-Rev cells (∼1.5 h) compared to HIV-1 TS movements in ∼1.5 h. (Scale bars, 5 µm; Inset, 2 µm.) For A and B, data are mean ± SD from three independent experiments; P values are from paired t tests. For (E–H), lines are mean ± SD; P values are from Welch’s t tests. ****P < 0.0001; ***P < 0.001; **P < 0.01; *P < 0.05; ns, not significant (P > 0.05).
To determine the effect of GFP-CA labeling on the efficiency of NE docking and nuclear import, viral cores composed of WT CA were labeled by incorporation of integrase-YFP (14) or A3F-YFP, and their efficiency of docking at the NE and nuclear import were compared to GFP-CA–labeled viral complexes as previously described (SI Appendix, Fig. S1G) (14). Briefly, infected cells were fixed at 6 hpi and viral complexes at the NE and in the nucleus were quantified using a custom MATLAB program. Similar NE docking and nuclear import efficiency of integrase-YFP-, A3F-YFP-, and GFP-CA–labeled viral complexes indicated that the GFP-CA labeling did not significantly influence viral complex association with the nuclear pores or nuclear import.
HIV-1 Uncoating Occurs ∼1.5 μm of Integration Sites <1.5 h before Integration.
To image HIV-1 integrated proviruses, HIV-1 transcription sites (TSs) were visualized by specific recognition of RNA stem loops (29) in the HIV-1 vector RNA with mCherry-tagged bacterial protein (Bgl-mCherry; Fig. 1A). HeLa cells expressing Bgl-mCherry (HeLa-Bgl; Fig. 1C) infected with GFP-CA–labeled virions at a low multiplicity of infection (<0.1 GFP-expressing proviruses/cell; SI Appendix, Fig. S1H) were analyzed by live-cell imaging (Fig. 1C and Movie S1) to quantify nuclear GFP-CA–labeled viral complexes, HIV-1 TS, distances between GFP-CA–labeled viral complexes and HIV-1 TS, and gfp reporter expression. Live-cell imaging from ∼4–24 hpi identified intranuclear GFP-CA–labeled viral complexes that maintained a steady level of GFP-CA for several hours and abruptly lost the GFP-CA signal ∼10.5 hpi, indicating nuclear uncoating (Fig. 1 D and E; n = 59). HIV-1 TSs were detected near the sites of GFP-CA disappearance ∼8.4 h later (Fig. 1F) followed by detection of gfp reporter expression ∼3.0 h later (Fig. 1G and Movie S1).
To determine the effect of exogenous Tat and Rev expression on the time of HIV-1 TS appearance, we constructed HeLa-Bgl cells that constitutively express HIV-1 Tat and Rev proteins (HeLa-Bgl:Tat-Rev). We found that expression of Tat and Rev did not affect the kinetics of GFP-CA loss (∼10.0 hpi; Fig. 1E) or the time between HIV-1 TS detection and gfp reporter expression (∼2.9 h; Fig. 1G), but HIV-1 TSs were detected much faster (∼1.5 h) after GFP-CA loss (Fig. 1F and Movie S2). These results indicate that ∼6.9 h were needed after GFP-CA loss for Tat to reach sufficient levels of expression to produce detectable HIV-1 TSs. Treatment of cells with integrase inhibitor raltegravir (RAL) showed that most of the HIV-1 TSs detected were from integrated proviruses (SI Appendix, Fig. S1I). Interestingly, exogenous Tat-Rev expression promoted detectable transcription from unintegrated DNAs (SI Appendix, Fig. S1I), suggesting that silencing of unintegrated HIV-1 DNAs by the human silencing hub complex (30) can be suppressed or reversed by Tat expression. Comparisons with control vectors indicated that the BglSL stem loops, Vif and Vpr expression, and GFP-CA fusion protein did not influence the kinetics of nuclear uncoating or gfp reporter expression (SI Appendix, Fig. S1 J–L).
We compared the locations of HIV-1 uncoating and integration sites by adjusting for cell movements and then measuring the distances between the last frame in which GFP-CA puncta were detected and the first frame in which HIV-1 TSs were detected (Fig. 1H). The average distance in the Tat-Rev-expressing cells (∼1.4 µm) was similar to the average distance HIV-1 TS moved in ∼1.5 h (∼1.3 µm), and the previously reported constrained diffusion of genes within a 1.5-µm radius (31). The average distance in HeLa-Bgl cells was slightly higher (∼1.9 µm), perhaps due to the longer observation time (∼8.4 h vs. ∼1.5 h). These results demonstrate that viral complexes uncoat within ∼1.5 μm of the sites of integration.
Nuclear Uncoating Confers Resistance to CA-Binding Inhibitor PF-3450074 and Is Delayed by Inhibiting Reverse Transcription.
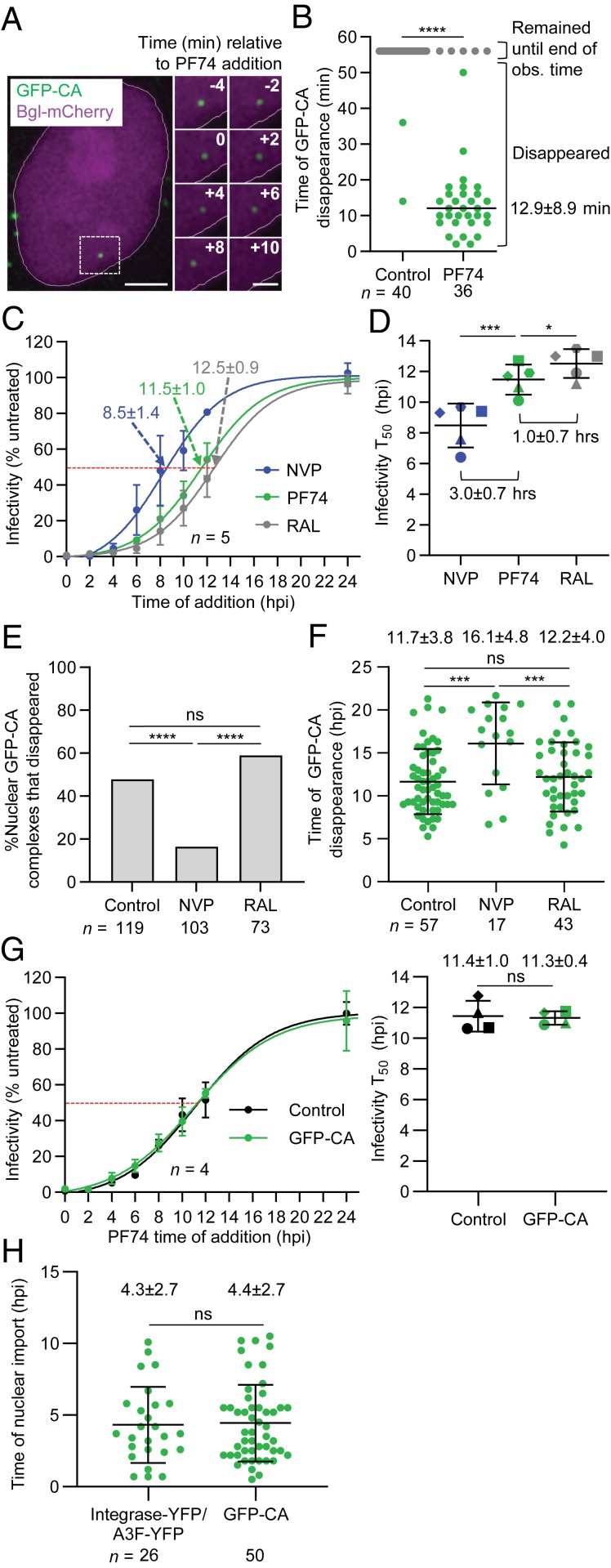
The CA-binding inhibitor PF-3450074 (PF74) binds to the N-terminal domain of one CA subunit and the C-terminal domain of an adjacent CA subunit within a hexamer and destabilizes that viral cores; interestingly, CPSF6 binds to the same site at which PF74 binds (reviewed in ref. 32). Treating infected cells after nuclear import of GFP-CA–labeled viral complexes with PF74 resulted in rapid disappearance (∼12.9 min) of 86% of the nuclear viral complexes (Fig. 2 A and B and Movie S3), indicating that nuclear viral complexes contained CA hexamers.
Fig. 2.
Determination of the sensitivity of nuclear GFP-CA–labeled viral complexes to capsid, reverse transcriptase and integrase inhibitors. (A) Representative live-cell microscopy images of a nuclear GFP-CA–labeled viral complex before and after addition of PF74 (10 µM). Numbers in white indicate time (min) relative to the time of PF74 addition. (Scale bar, 5 µm; Inset, 2 µm.) (B) Time of disappearance of GFP-CA–labeled viral complexes in untreated control cells and PF74-treated cells during ∼1-h observation time. (C) Time-of-addition assays with NVP, PF74, or RAL. The numbers indicate the time at which 50% of the viral complexes became resistant to the inhibitors (infectivity T50). (D) Comparison of average infectivity T50 for NVP, PF74, and RAL from five independent experiments. (E) Proportion of nuclear GFP-CA–labeled complexes that disappeared during the observation time (21.6 hpi). (F) Average time of GFP-CA disappearance. Lines are mean ± SD; P values are from Welch’s t tests. (G) PF74 time-of-addition assays with GFP-CA–labeled and unlabeled virions. Comparison of average time at which 50% of the viral complexes became resistant to PF74 (infectivity T50) from four independent experiments (Right). (H) Comparison of the time of nuclear import previously determined for integrase-YFP- or A3F-YFP-labeled viral complexes (14) and GFP-CA–labeled viral complexes. For B and E, P values are from Fisher’s exact tests comparing the proportion nuclear GFP-CA complexes that disappeared. For D and G, lines are mean ± SD; P values are from paired t tests. ****P < 0.0001; ***P < 0.001; *P < 0.05; ns, not significant (P > 0.05).
Next, we performed time-of-addition experiments with PF74, reverse transcriptase inhibitor nevirapine (NVP), or integrase inhibitor RAL. Addition of PF74 to cells infected with unlabeled HIV-1 virions at various times after infection showed that 50% of the viral complexes became PF74 resistant ∼11.5 hpi (Fig. 2 C and D). This average time of PF74 sensitivity loss was similar to the average time of GFP-CA loss (∼10.5 hpi; Fig. 1E), indicating that nuclear uncoating was correlated with PF74 resistance. The loss of PF74 sensitivity occurred ∼3.0 h after the loss of sensitivity to NVP (Fig. 2D), suggesting that nuclear uncoating occurred ∼3 h after completion of reverse transcription. Nevertheless, inhibition of reverse transcription with NVP was correlated with a delay or inhibition of nuclear uncoating (Fig. 2 E and F).
RAL time-of-addition experiments showed that integration was completed ∼1.0 h after uncoating (Fig. 2D) and that inhibiting integration did not affect uncoating (Fig. 2 E and F). We also determined that unlabeled- and GFP-CA–labeled viral complexes became PF74 resistant with similar kinetics (∼11.4 hpi; Fig. 2G). Since loss of sensitivity to PF74 was correlated with uncoating, the result indicated that GFP-CA labeling of viral complexes did not significantly influence their uncoating kinetics. GFP-CA–labeled and unlabeled virions exhibited the same sensitivity to NVP, PF74, and RAL with nearly identical 50% inhibitory concentrations (SI Appendix, Fig. S1M).
Finally, we determined that the average time at which GFP-CA–labeled viral complexes are imported into the nucleus was ∼4.4 hpi (Fig. 2H), which was not significantly different from our previously determined average time of import (∼4.3 hpi) for viral complexes labeled with integrase-YFP or A3F-YFP (14). Since the average time of nuclear import (∼4.4 hpi) was ∼4 h earlier than the average time of reverse transcription completion (∼8.5 hpi), we conclude that most viral complexes complete reverse transcription after nuclear import.
Nuclear Viral Complexes Retain Most of the CA Present in Intact Viral Cores.
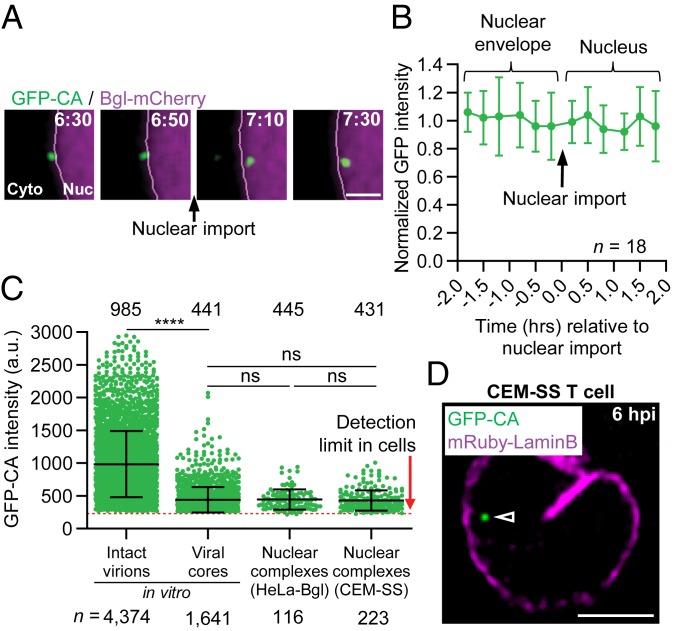
Previous studies have suggested that one uncoating step occurs at the NE during import (12–17). To determine whether some uncoating occurs during nuclear import, we compared the average GFP-CA intensities of viral complexes while they were docked at the NE for six frames before and six frames after nuclear entry and found no detectable loss of GFP-CA (Fig. 3 A and B, n = 18). Modeling 5–20% loss of GFP-CA intensities indicated that a ≥10% GFP-CA loss would have been detectable under the imaging conditions ( SI Appendix, Fig. S2A); thus, most of the CA was retained by the viral complexes during nuclear import.
Fig. 3.
Nuclear viral complexes retain most of the GFP-CA associated with in vitro viral cores. (A) Live-cell microscopy images of a GFP-CA–labeled viral complex docked at the NE and in the nucleus after import. Numbers in white indicate time postinfection. (Scale bar, 2 µm.) (B) Normalized mean GFP-CA intensities of six frames before and after nuclear import indicate no significant loss of GFP-CA. (C) Comparison of the mean GFP-CA intensities (arbitrary units; a.u.) of intact virions, in vitro viral cores, and nuclear viral complexes in infected HeLa and CEM-SS T cells. Intact virions and in vitro viral cores with GFP-CA intensities below the detection limit (<265 a.u.) in HeLa and CEM-SS cell nuclei were removed. Lines are mean ± SD; P values are from Welch’s t tests. ****P < 0.0001; ns, not significant (P > 0.05). (D) Representative image of a GFP-CA–labeled nuclear complex in an infected CEM-SS T cell expressing mRuby-LaminB 6 hpi. (Scale bar, 5 µm.)
To compare the GFP-CA intensities of nuclear viral complexes and intact viral cores, intact virions were lysed in vitro as previously reported (33), resulting in ∼55% loss of free GFP-CA that was not incorporated into viral cores (Fig. 3C and SI Appendix, Fig. S2 B–D). The mean GFP-CA intensity of 116 nuclear viral complexes from HeLa cells or 223 nuclear viral complexes from CEM-SS T cells was not significantly different from the intact viral cores (Fig. 3 C and D). Modeling 2–10% GFP-CA loss from intact viral cores indicated that ≥6% GFP-CA loss would have been detectable (SI Appendix, Fig. S2E), indicating that infectious nuclear RTCs/PICs retained ≥90% of their viral core-associated CA. In addition, the results also indicated that most reverse transcription is completed within an intact (or nearly intact) viral core since most viral complexes became NVP resistant (∼8.5 hpi) before GFP-CA loss (∼10.5 hpi).
Disruption of the CA-CPSF6 Interaction Results in Uncoating at the NE.
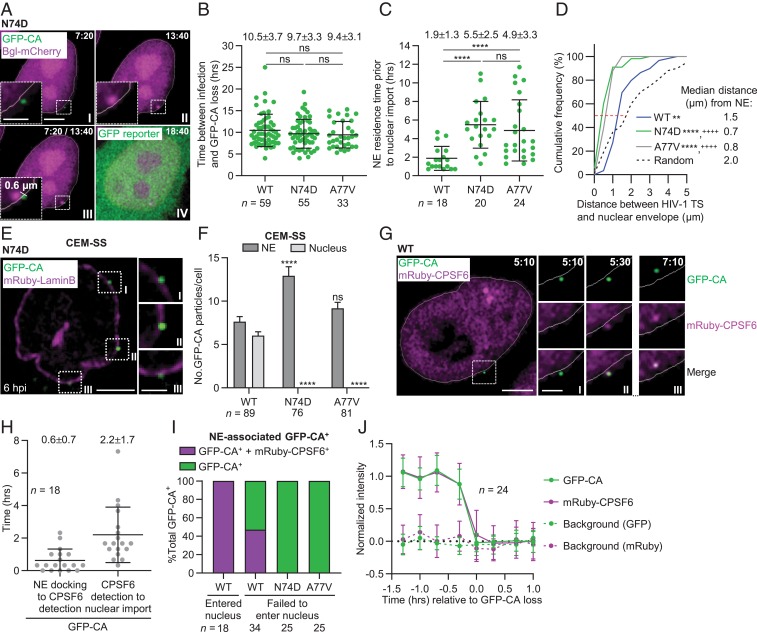
CPSF6 is a nuclear host factor that interacts with the viral core (34) and disruption of the CA-CPSF6 interaction alters the target sites of HIV-1 integration (23, 24, 35). To elucidate the role of CPSF6 in nuclear import and intranuclear trafficking, we generated GFP-CA–labeled virions containing CA mutations N74D (34) or A77V (36), which substantially reduce CPSF6 binding. As expected, the N74D and A77V mutations had minimal effects on infectivity in HeLa cells, CEM-SS cells, or THP-1–derived macrophages (SI Appendix, Fig. S3A). However, the A77V mutation has been reported to revert in humanized mice (36), indicating a lower fitness in vivo. We infected HeLa cells expressing Bgl-mCherry with the N74D and A77V mutants and observed that the kinetics of GFP-CA loss, HIV-1 TS appearance, and gfp reporter detection (Fig. 4 A and B, SI Appendix, Fig. S3 B–D, and Movie S4) were not different from the kinetics of WT virions that were described in Fig. 1 E–G.
Fig. 4.
CA-CPSF6 interaction at the NE facilitates nuclear import of viral complexes, the location of their uncoating, and the location of HIV-1 TS. (A) Representative live-cell microscopy images of a HeLa-Bgl cell infected with GFP-CA–labeled virions of CA mutant N74D. A GFP-CA–labeled viral complex uncoated at the edge of the nuclear Bgl-mCherry signal, 7:20 hpi (I) and HIV-1 TS appeared near the site of GFP-CA disappearance 13:40 hpi (II). The HIV-1 TS appeared 0.6 µm from the GFP-CA signal (III). GFP reporter expression detected 18:40 hpi (IV). (B) Time between infection and GFP-CA loss; data for WT the same as in Fig. 1E. Lines are mean ± SD; P values are from Welch’s t tests. (C) NE residence time of GFP-CA–labeled viral complexes prior to nuclear import. For N74D/A77V mutants, the time of nuclear import was assumed to occur at the time of GFP-CA loss (i.e., uncoating). (D) Cumulative frequency distribution of distances (µm) between HIV-1 TS and NE and random sites in the nuclei to NE; median distances are indicated by the red dotted line. P values are from Kolmogorov–Smirnov tests. **P < 0.01 compared to random; ****P < 0.0001 compared to random; ++++P < 0.0001 compared to WT. (E) N74D GFP-CA–labeled viral complexes localize to the NE but not in the nuclei of CEM-SS cells. (F) Quantitation of GFP-CA–labeled viral complexes at the NE and in the nucleus. Data are pooled from three independent infections (n = number of cells analyzed); P values are from paired t tests. ****P < 0.0001; ns, not significant (P > 0.05). (G) Representative live-cell microscopy images of infected HeLa:mRuby-CPSF6 cells show mRuby-CPSF6 recruitment to a GFP-CA–labeled viral complex located at or near the NE (I and II); dual-labeled GFP-CA and mRuby-CPSF6 complexes are imported into the nucleus (III). (H) Time between NE docking to CPSF6 detection and CPSF6 detection to nuclear import for 18 GFP-CA–labeled viral complexes. Lines are mean ± SD (I) Proportion of NE-associated GFP-CA+ viral complexes that are CPSF6+. The viral complexes that entered the nucleus during the observation time and those that were docked at the NE but failed to enter the nucleus were analyzed. (J) Simultaneous disappearance of intranuclear GFP-CA and mRuby-CPSF6 signals. (Scale bars, 5 µm; Inset, 2 µm.)
Next, we compared the locations of the WT, N74D, and A77V GFP-CA puncta. Bgl-mCherry protein is predominantly localized to the nucleus (SI Appendix, Fig. S3E), and the NE is localized to the periphery of the Bgl-mCherry signal. We found that, in contrast to the WT, the N74D and A77V GFP-CA puncta did not enter the nucleus but disappeared at the periphery of the Bgl-mCherry signal (SI Appendix, Fig. S3 E and F), indicating that they uncoated at the NE (Fig. 4A and SI Appendix, Fig. S3B). Interestingly, the N74D and A77V mutants exhibited GFP-CA loss at the NE at approximately the same time after infection (∼9.7 and ∼9.4 hpi, respectively) as the GFP-CA loss exhibited by WT viral complexes in the nucleus (∼10.5 hpi) (Fig. 4B). This observation suggested that the molecular events that trigger uncoating occur in the nucleus and at the NE with the same kinetics. Consistent with this finding, the NE residence time of GFP-CA–labeled viral complexes prior to nuclear import was much longer for the N74D (∼5.5 h) and A77V (∼4.9 h) mutants than for the WT complexes (∼1.9 h) (Fig. 4C). Subsequently, HIV-1 TS appeared near the site of N74D and A77V GFP-CA puncta disappearance at the NE followed by gfp reporter expression. The HIV-1 TSs were much closer to the NE (∼0.8 µm) in cells infected with N74D and A77V mutants compared to WT HIV-1 TSs (∼1.5 µm; Fig. 4D). These observations suggested that, after uncoating at the NE, the N74D/A77V PICs (without the viral cores) integrated into nearby chromatin either by entering the nucleus or by accessing the adjacent chromatin while docked at the NPC. Consistent with this hypothesis, only WT GFP-CA labeled viral complexes, but not N74D/A77V GFP-CA–labeled viral complexes, were detected in the nuclei of infected CEM-SS and HeLa cells (Fig. 4 E and F and SI Appendix, Fig. S3G). Overall, these results indicated that N74D/A77V GFP-CA–labeled viral cores did not enter the nucleus and uncoated while they were docked at the NE; furthermore, the results suggested that the CA-CPSF6 interaction is necessary for nuclear import of intact or nearly intact viral cores.
CA-CPSF6 Interaction at the NE Facilitates Nuclear Entry of Viral Cores.
To visualize CA-CPSF6 interactions, we stably knocked down endogenous CPSF6 and expressed short hairpin RNA-resistant mRuby-CPSF6, which did not significantly influence virus infectivity, efficiency of NE docking, or nuclear import efficiency (Fig. 4G and SI Appendix, Fig. S4 A–C). Live-cell imaging of 18 GFP-CA–labeled WT viral complexes that entered the nucleus showed that they all accumulated mRuby-CPSF6 ∼0.6 h after NE docking and the dual-labeled complexes translocated to the nucleus ∼2.2 h later (Fig. 4H, SI Appendix, Fig. S5 A–C, and Movie S5); similar kinetics were observed for A3F-mNeonGreen-labeled viral complexes, indicating that GFP-CA labeling of viral complexes did not influence the kinetics of accumulation of mRuby-CPSF6 or the translocation of the dual-labeled complexes into the nucleus (SI Appendix, Fig. S4D). Interestingly, a high proportion of the WT viral complexes that did not enter the nucleus (47%) were also associated with mRuby-CPSF6 (Fig. 4I), indicating that the CA-CPSF6 association at the NE is necessary but is not sufficient for nuclear import of the viral core. None of the N74D or A77V GFP-CA–labeled viral complexes at the NE colocalized with mRuby-CPSF6 (0/50), confirming that a specific CA-CPSF6 interaction is required to accumulate CPSF6 at the NE.
We sought to determine how long after nuclear import CPSF6 dissociated from the viral complexes. Analysis of 24 GFP-CA–labeled viral complexes that entered the nucleus showed that the GFP-CA and mRuby-CPSF6 signals disappeared simultaneously, indicating that CPSF6 dissociated from the viral complexes at the time of uncoating (Fig. 4J, SI Appendix, Figs. S4E and S5 D–F, and Movie S6).
Since the HeLa:mRuby-CPSF6 cells contained 62% the level of CPSF6 as the parental HeLa cells (SI Appendix, Fig. S4A), we asked whether the reduced CPSF6 levels had any effect on HIV-1 uncoating. GFP-CA–labeled viral complexes uncoated in the nuclei of HeLa:mRuby-CPSF6 cells with the same efficiency and kinetics as in HeLa-Bgl cells (SI Appendix, Fig. S4 F and G), indicating that the reduced CPSF6 levels did not significantly influence the timing or efficiency of uncoating. Finally, the mRuby-CPSF6 levels associated with GFP-CA- or integrase-superfolder GFP-labeled nuclear viral complexes were similar (SI Appendix, Fig. S4 H and I), suggesting that the nuclear viral complexes contained similar amounts of CA, which resulted in similar amounts of mRuby-CPSF6 association.
Discussion
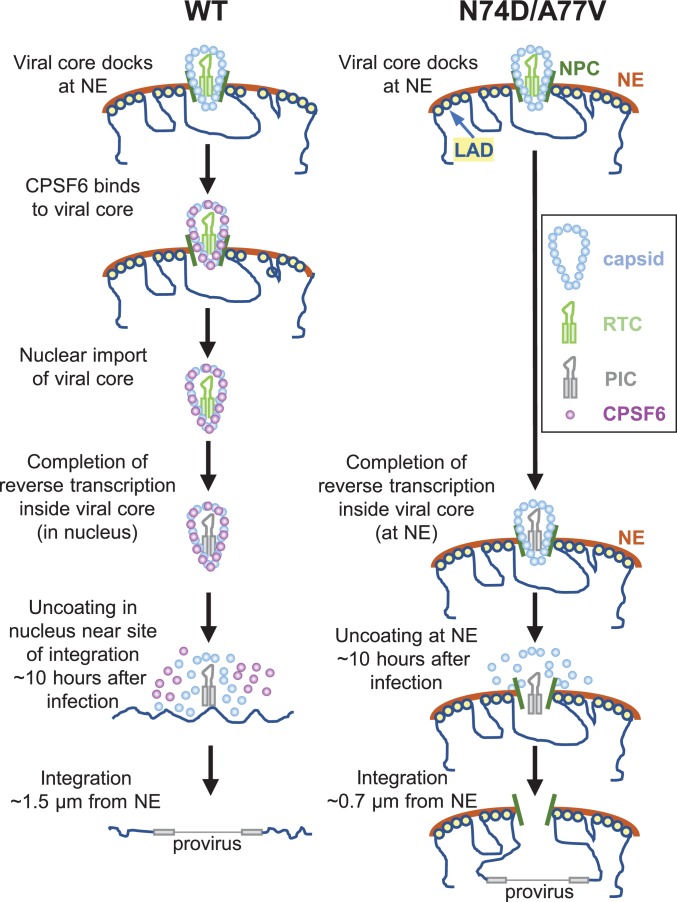
Here, we show that intact (or nearly intact) viral cores enter the nucleus and uncoat <1.5 h before integration within ∼1.5 μm of their chromosomal integration sites (model shown in Fig. 5). These results shift the current paradigm of HIV-1 postentry replication events and have important implications for the mechanisms of nuclear import and uncoating as well as reverse transcription, integration, and evasion of host innate immunity.
Fig. 5.
Model for nuclear import and uncoating of HIV-1. WT and N74D/A77V viral cores dock at the NE. CPSF6 is recruited to the WT viral cores at the NE but not to the N74D/A77V viral cores. WT viral cores are imported into the nucleus ∼1.9 h after docking at the NE; the N74D/A77V GFP-CA–labeled viral cores remain associated with the NE and are not imported into the nucleus. Reverse transcription is completed inside the intact (or nearly intact) viral core for WT (in the nucleus) and N74D/A77V mutants (at NE). The nuclear WT viral complexes and NE-associated N74D/A77V viral complexes uncoat ∼10 h after infection. WT PIC integrates into chromatin near the sites of uncoating ∼1.5 µm from the NE; the N74D/A77V PIC integrates into chromatin associated with lamina-associated domains (LADs) ∼0.7 to 0.8 µm from the NE. In addition to the localization of transcriptionally active WT and N74D/A77V proviruses to the nuclear periphery in these studies, localization of WT viral DNA (50, 51) and N74D/A77V DNA (23, 25) to the nuclear periphery was previously reported.
Our studies provide a robust method for fluorescently labeling viral cores in infected cells. GFP-CA–labeled virions were not significantly different from unlabeled virions with respect to 1) the ratio of mature and immature virions, 2) in vitro stability of viral cores during sucrose gradient fractionation, 3) proportion of viral cores that stably associated with the NE and imported into the nucleus, 4) the timing of GFP reporter expression, and 5) sensitivity to reverse transcriptase, capsid, and integrase inhibitors. Furthermore, the GFP-CA–labeling efficiency was high (96%), and virion infectivity was within twofold (∼50%) of the unlabeled virions. Most importantly, for these studies, time-of-addition assays showed that the GFP-CA–labeled and unlabeled virions became resistant to PF74 with similar kinetics, which was correlated to uncoating, indicating that GFP-CA labeling did not significantly affect the viral uncoating kinetics. Although we cannot exclude the possibility that GFP-CA labeling has some effects on HIV-1 replication that were not revealed in our experiments, we conclude that GFP-CA labeling does not substantially influence most aspects of HIV-1 replication.
Our observation that infectious viral cores uncoat in the nucleus was unexpected since most previous studies have concluded that uncoating occurs in the cytoplasm (7–12, 37), while a few recent studies have concluded that uncoating occurs at the NE during nuclear import (12–17). Previous imaging studies of HIV-1 uncoating have been hampered by the inability to fluorescently label CA directly without adversely affecting uncoating and viral replication. Although a few studies have labeled CA with a tetracysteine tag (7, 26), the method has not been widely used because of technical challenges, such as nonspecific labeling and rapid photobleaching. Immunofluorescence assays using anti-CA antibodies are widely used, and loss of the fluorescent signal is interpreted as loss of CA from viral complexes. However, reduced CA epitope accessibility by conformational changes in the viral core or association with host proteins can also result in loss of the fluorescent signal and potentially be misinterpreted as loss of CA from viral complexes. Recently, loss of fluorescently labeled CypA (CypA-dsRed) from viral complexes at the NE was interpreted as uncoating of the viral complexes at the NE during nuclear import; however, it is conceivable that CypA-dsRed dissociated from the viral complexes at the NE prior to nuclear import. Biochemical assays have also been used to study viral core uncoating in infected cells (38); however, only one in ∼50 virions leads to productive infection (18), and disassembly of a bulk population of viral cores may not reflect the behavior of the rare infectious viral cores.
Our studies provide direct evidence that CPSF6 recruitment is a critical requirement for nuclear import of intact or nearly intact viral cores. These results provide essential mechanistic insights into previous observations indicating that the CA-CPSF6 interaction is critical for integration into gene-rich euchromatin regions that are located in the interior regions of the nucleus (23, 24, 35, 39, 40). We propose the nuclear import of intact (or nearly intact) viral cores, and uncoating in the nucleus is essential for integration in gene-rich euchromatin regions since the N74D/A77V mutants, which uncoat at the NE, integrate into gene-sparse heterochromatin regions within ∼0.8 μm of the NE. The mechanism by which the N74D/A77V PICs (without the viral cores) integrate into chromatin near the NE is unclear; however, it is likely that either the mutant PICs enter the nucleus or access the adjacent chromatin while docked at the NPC. Interestingly, the N74D/A77V mutants retain their infectivity in single-cycle assays, suggesting that integration into euchromatin may be essential for maintaining viral fitness in vivo. These results indicating that HIV-1 uncoating and its regulation are linked to the selection of HIV-1 integration sites have important implications for the regulation of HIV-1 transcription and the establishment of a latent reservoir of infected cells, which are major impediments to HIV-1 eradication and cure (41).
In contrast to the current prevailing view that reverse transcription is completed in the cytoplasm after uncoating and is followed by nuclear import of the viral preintegration complex, our results indicate that viral DNA synthesis and the formation of a preintegration complex occurs within an intact (or nearly intact) core and that these steps are completed in the nucleus. Although it is generally thought that reverse transcription is completed in the cytoplasm before nuclear import, some previous studies have provided evidence indicating that viral DNA synthesis is initiated in the cytoplasm but completed in the nucleus (42–45). In support of reverse transcription taking place in an intact or nearly intact viral core, a recent study showed that CA hexamers form a positively charged channel for transport of deoxynucleotide triphosphates into viral cores, providing the substrates necessary for reverse transcription within intact viral cores (46). Our observation that uncoating occurs <1.5 h before integration implies that the viral preintegration complex is exposed to the nuclear environment for <1.5 h before viral DNA integration. We propose that the viral core may remain intact until <1.5 h before integration to ensure completion of reverse transcription, formation of a functional preintegration complex, and potential evasion of innate sensing by cytoplasmic (47) and/or nuclear DNA sensors to suppress cellular immune responses (48).
It is unclear how an intact viral core with a width of ∼61 nm (5) can be imported through a NPC with an inner diameter of ∼39 nm (49). Lack of CPSF6 recruitment to viral complexes at the NE was correlated with a failure to import viral cores, and uncoating at the NE, suggesting that the intact (or nearly intact) viral cores are, indeed, too large to be imported through nuclear pores in the absence of the CA-CPSF6 interaction at the NE. Our results suggest that CPSF6 recruitment to the viral complexes at the NE results in alteration of the NPC or the viral core structure to facilitate the nuclear import of intact (or nearly intact) viral cores in HeLa cells as well as T cells, which are the major target cells for HIV-1 infection in patients. Bejarano and colleagues recently proposed that CPSF6 binds to CA multimers and facilitates nuclear import of viral complexes in primary monocyte-derived macrophages (21) but did not determine if the nuclear viral complexes were composed of intact viral cores, partially uncoated viral cores, or CA-derived subviral structures. Although the structure of the viral complexes at the NE or after nuclear import is not known, they contain most, if not all, of the viral core-associated CA and must retain CA hexamers since they associate with CPSF6 and rapidly disassemble upon PF74 treatment.
The N74D/A77V viral complexes uncoat at the NE with the same kinetics as WT nuclear viral complexes (∼10 hpi), suggesting that the molecular trigger for uncoating is intrinsic to the viral complex and independent of its intracellular location. Inhibiting reverse transcription prevented uncoating even though it is completed ∼2 to 3 h earlier, suggesting that uncoating is not initiated upon completion of reverse transcription but requires the completion of viral DNA synthesis.
Overall, our studies help resolve a long-standing question in HIV-1 replication regarding the location and timing of uncoating and fundamentally change our understanding of HIV-1 postentry replication events. We propose that the viral core may remain intact or nearly intact until just before integration to maintain high concentrations of reverse transcriptase and integrase near the viral nucleic acid to complete DNA synthesis and ensure assembly of an integration-competent viral complex. Additionally, maintaining an intact or nearly intact viral core may facilitate evasion of innate sensing by DNA sensors to suppress cellular immune responses and ensure integration into the preferred sites of integration located in gene-rich euchromatin regions.
Materials and Methods
Experimental details and methods can be found in the SI Appendix, including sources of cell lines and procedures for their maintenance, generation of HeLa-Bgl, HeLa-Bgl:Tat-Rev, HeLa:mRuby-CPSF6, and CEM-SS-mRuby-Lamin B cell lines, construction of lentiviral vectors pHGFP-GFPCA-BglSL, pHGFP-BglSL, pHGFP(N74D)-GFPCA-BglSL, pHGFP(N74D)-BglSL, pHGFP(A77V)-GFPCA-BglSL, pHGFP(A77V)-BglSL, and procedures for virus production and infection. Details of microscopy and image processing, live-cell imaging and image analysis using custom written MATLAB programs, and fixed-cell imaging and image analysis are described in the SI Appendix. Methods for single virion analysis, in vitro analysis of intact virions and viral cores, transmission electron microscope analysis of virus pellets, fractionation of viral cores using sucrose gradients, and data analysis and statistics are also described in the SI Appendix.
Data Availability Statement.
All data generated in this study are included in the paper and SI Appendix.
Supplementary Material
Acknowledgments
We thank John Coffin, Eric Freed, and Tom Misteli for valuable discussions and suggestions during paper preparation. This work was supported, in part, by the Intramural Research Program of the NIH, National Cancer Institute, Center for Cancer Research, by Intramural AIDS Targeted Antiviral Program grant funding (to V.K.P. and to W.-S.H.) under Contract HHSN26120080001E.
Footnotes
This article is a PNAS Direct Submission.
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1920631117/-/DCSupplemental.
References
- 1.Gres A. T., et al. , STRUCTURAL VIROLOGY. X-ray crystal structures of native HIV-1 capsid protein reveal conformational variability. Science 349, 99–103 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Mattei S., Glass B., Hagen W. J., Kräusslich H. G., Briggs J. A., The structure and flexibility of conical HIV-1 capsids determined within intact virions. Science 354, 1434–1437 (2016). [DOI] [PubMed] [Google Scholar]
- 3.Zhao G., et al. , Mature HIV-1 capsid structure by cryo-electron microscopy and all-atom molecular dynamics. Nature 497, 643–646 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Aboud M., Shoor R., Salzberg S., Adsorption, penetration, and uncoating of murine leukemia virus studied by using its reverse transcriptase. J. Virol. 30, 32–37 (1979). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Briggs J. A., Wilk T., Welker R., Kräusslich H. G., Fuller S. D., Structural organization of authentic, mature HIV-1 virions and cores. EMBO J. 22, 1707–1715 (2003). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Campbell E. M., Hope T. J., HIV-1 capsid: The multifaceted key player in HIV-1 infection. Nat. Rev. Microbiol. 13, 471–483 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mamede J. I., Cianci G. C., Anderson M. R., Hope T. J., Early cytoplasmic uncoating is associated with infectivity of HIV-1. Proc. Natl. Acad. Sci. U.S.A. 114, E7169–E7178 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lukic Z., Dharan A., Fricke T., Diaz-Griffero F., Campbell E. M., HIV-1 uncoating is facilitated by dynein and kinesin 1. J. Virol. 88, 13613–13625 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Hulme A. E., Perez O., Hope T. J., Complementary assays reveal a relationship between HIV-1 uncoating and reverse transcription. Proc. Natl. Acad. Sci. U.S.A. 108, 9975–9980 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Yang Y., Fricke T., Diaz-Griffero F., Inhibition of reverse transcriptase activity increases stability of the HIV-1 core. J. Virol. 87, 683–687 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Cosnefroy O., Murray P. J., Bishop K. N., HIV-1 capsid uncoating initiates after the first strand transfer of reverse transcription. Retrovirology 13, 58 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Francis A. C., Marin M., Shi J., Aiken C., Melikyan G. B., Time-resolved imaging of single HIV-1 uncoating in vitro and in living cells. PLoS Pathog. 12, e1005709 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rasaiyaah J., et al. , HIV-1 evades innate immune recognition through specific cofactor recruitment. Nature 503, 402–405 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Burdick R. C., et al. , Dynamics and regulation of nuclear import and nuclear movements of HIV-1 complexes. PLoS Pathog. 13, e1006570 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Francis A. C., Melikyan G. B., Single HIV-1 imaging reveals progression of infection through CA-dependent steps of docking at the nuclear pore, uncoating, and nuclear transport. Cell Host Microbe 23, 536–548.e6 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fernandez J., et al. , Transportin-1 binds to the HIV-1 capsid via a nuclear localization signal and triggers uncoating. Nat. Microbiol. 4, 1840–1850 (2019). [DOI] [PubMed] [Google Scholar]
- 17.Arhel N. J., et al. , HIV-1 DNA Flap formation promotes uncoating of the pre-integration complex at the nuclear pore. EMBO J. 26, 3025–3037 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Burdick R. C., Hu W. S., Pathak V. K., Nuclear import of APOBEC3F-labeled HIV-1 preintegration complexes. Proc. Natl. Acad. Sci. U.S.A. 110, E4780–E4789 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Hulme A. E., Kelley Z., Foley D., Hope T. J., Complementary assays reveal a low level of CA associated with viral complexes in the nuclei of HIV-1-infected cells. J. Virol. 89, 5350–5361 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Peng K., et al. , Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. eLife 3, e04114 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Bejarano D. A., et al. , HIV-1 nuclear import in macrophages is regulated by CPSF6-capsid interactions at the nuclear pore complex. eLife 8, e41800 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zhou L., et al. , Transportin 3 promotes a nuclear maturation step required for efficient HIV-1 integration. PLoS Pathog. 7, e1002194 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Achuthan V., et al. , Capsid-CPSF6 interaction licenses nuclear HIV-1 trafficking to sites of viral DNA integration. Cell Host Microbe 24, 392–404.e8 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sowd G. A., et al. , A critical role for alternative polyadenylation factor CPSF6 in targeting HIV-1 integration to transcriptionally active chromatin. Proc. Natl. Acad. Sci. U.S.A. 113, E1054–E1063 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Chin C. R., et al. , Direct visualization of HIV-1 replication intermediates shows that capsid and CPSF6 modulate HIV-1 intra-nuclear invasion and integration. Cell Rep. 13, 1717–1731 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Campbell E. M., Perez O., Anderson J. L., Hope T. J., Visualization of a proteasome-independent intermediate during restriction of HIV-1 by rhesus TRIM5alpha. J. Cell Biol. 180, 549–561 (2008). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Hübner W., et al. , Sequence of human immunodeficiency virus type 1 (HIV-1) Gag localization and oligomerization monitored with live confocal imaging of a replication-competent, fluorescently tagged HIV-1. J. Virol. 81, 12596–12607 (2007). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Müller B., et al. , Construction and characterization of a fluorescently labeled infectious human immunodeficiency virus type 1 derivative. J. Virol. 78, 10803–10813 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chen J., et al. , High efficiency of HIV-1 genomic RNA packaging and heterozygote formation revealed by single virion analysis. Proc. Natl. Acad. Sci. U.S.A. 106, 13535–13540 (2009). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhu Y., Wang G. Z., Cingöz O., Goff S. P., NP220 mediates silencing of unintegrated retroviral DNA. Nature 564, 278–282 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Finn E. H., et al. , Extensive heterogeneity and intrinsic variation in spatial genome organization. Cell 176, 1502–1515.e10 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ambrose Z., Aiken C., HIV-1 uncoating: Connection to nuclear entry and regulation by host proteins. Virology 454-455, 371–379 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Márquez C. L., et al. , Kinetics of HIV-1 capsid uncoating revealed by single-molecule analysis. eLife 7, e34772 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Lee K., et al. , Flexible use of nuclear import pathways by HIV-1. Cell Host Microbe 7, 221–233 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schaller T., et al. , HIV-1 capsid-cyclophilin interactions determine nuclear import pathway, integration targeting and replication efficiency. PLoS Pathog. 7, e1002439 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Saito A., et al. , Capsid-CPSF6 interaction is dispensable for HIV-1 replication in primary cells but is selected during virus passage in vivo. J. Virol. 90, 6918–6935 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Xu H., et al. , Evidence for biphasic uncoating during HIV-1 infection from a novel imaging assay. Retrovirology 10, 70 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Yang Y., Luban J., Diaz-Griffero F., The fate of HIV-1 capsid: A biochemical assay for HIV-1 uncoating. Methods Mol. Biol. 1087, 29–36 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Ocwieja K. E., et al. , HIV integration targeting: A pathway involving transportin-3 and the nuclear pore protein RanBP2. PLoS Pathog. 7, e1001313 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhyvoloup A., et al. , Digoxin reveals a functional connection between HIV-1 integration preference and T-cell activation. PLoS Pathog. 13, e1006460 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Lusic M., Siliciano R. F., Nuclear landscape of HIV-1 infection and integration. Nat. Rev. Microbiol. 15, 69–82 (2017). [DOI] [PubMed] [Google Scholar]
- 42.Barbosa P., Charneau P., Dumey N., Clavel F., Kinetic analysis of HIV-1 early replicative steps in a coculture system. AIDS Res. Hum. Retroviruses 10, 53–59 (1994). [DOI] [PubMed] [Google Scholar]
- 43.Lee Y. M., Coffin J. M., Relationship of avian retrovirus DNA synthesis to integration in vitro. Mol. Cell. Biol. 11, 1419–1430 (1991). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Bukrinsky M. I., et al. , Association of integrase, matrix, and reverse transcriptase antigens of human immunodeficiency virus type 1 with viral nucleic acids following acute infection. Proc. Natl. Acad. Sci. U.S.A. 90, 6125–6129 (1993). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Galvis A. E., Fisher H. E., Nitta T., Fan H., Camerini D., Impairment of HIV-1 cDNA synthesis by DBR1 knockdown. J. Virol. 88, 7054–7069 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Jacques D. A., et al. , HIV-1 uses dynamic capsid pores to import nucleotides and fuel encapsidated DNA synthesis. Nature 536, 349–353 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Monroe K. M., et al. , IFI16 DNA sensor is required for death of lymphoid CD4 T cells abortively infected with HIV. Science 343, 428–432 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Diner B. A., Lum K. K., Cristea I. M., The emerging role of nuclear viral DNA sensors. J. Biol. Chem. 290, 26412–26421 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Knockenhauer K. E., Schwartz T. U., The nuclear pore complex as a flexible and dynamic gate. Cell 164, 1162–1171 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Di Primio C., et al. , Single-cell imaging of HIV-1 provirus (SCIP). Proc. Natl. Acad. Sci. U.S.A. 110, 5636–5641 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Marini B., et al. , Nuclear architecture dictates HIV-1 integration site selection. Nature 521, 227–231 (2015). [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
All data generated in this study are included in the paper and SI Appendix.