Significance
We report the discovery of functions of intrinsically disordered RNA-binding protein RBM14 in NHEJ. RBM14 and KU proteins are recruited to DSB sites and required for the generation of RNA:DNA hybrids in response to DSBs. Despite a considerable number of intrinsically disordered RNA-binding proteins being implicated in DNA damage response, little is known about their roles in DNA repair pathways. Our findings provide insight into understanding how DSBs are repaired at transcriptionally inactive genomic areas and how the RNA-binding protein RBM14 plays a role in the NHEJ process.
Keywords: DNA repair, double-strand breaks, RBM14
Abstract
Accumulating evidence suggests participation of RNA-binding proteins with intrinsically disordered domains (IDPs) in the DNA damage response (DDR). These IDPs form liquid compartments at DNA damage sites in a poly(ADP ribose) (PAR)-dependent manner. However, it is greatly unknown how the IDPs are involved in DDR. We have shown previously that one of the IDPs RBM14 is required for the canonical nonhomologous end joining (cNHEJ). Here we show that RBM14 is recruited to DNA damage sites in a PARP- and RNA polymerase II (RNAPII)-dependent manner. Both KU and RBM14 are required for RNAPII-dependent generation of RNA:DNA hybrids at DNA damage sites. In fact, RBM14 binds to RNA:DNA hybrids. Furthermore, RNA:DNA hybrids and RNAPII are detected at gene-coding as well as at intergenic areas when double-strand breaks (DSBs) are induced. We propose that the cNHEJ pathway utilizes damage-induced transcription and intrinsically disordered protein RBM14 for efficient repair of DSBs.
Recent studies showed PARP-dependent recruitment of RNA-binding proteins (RBPs) with intrinsically disordered prion-like domains (PLDs) called IDPs to microlaser-induced DNA damage sites, and that these RNA-binding proteins form liquid compartments (1, 2). These IDPs include FUS/TLS (fused in sarcoma/translocated in sarcoma), TAF15 (TATA box-binding protein-associated factor 68 kDa), EWS (Ewing sarcoma), and a number of heterogenous nuclear ribonucleoproteins (hnRNPs) (1, 2). Many point mutations within the FET genes have been associated with pathological protein aggregation in neurodegenerative diseases, such as with amyotrophic lateral sclerosis (ALS), and frontotemporal lobar degeneration (FTLD) (3–6).
Emerging evidence suggests PLDs’ physical interactions with DNA lesions and thus involvement in DNA damage response (1, 2, 7–10). RBM14 contains RNA recognition motifs (RRMs) and a PLD. We identified RBM14 by a human genomewide shRNA screen to find genes which radiosensitize GBM (glioblastoma multiforme) spheres. We have shown that RBM14 interacts with KU, and that knockdown of RBM14 inhibits NHEJ significantly. Importantly, knockdown of RBM14 radiosensitizes GBM stem-like cells and reduces tumorigenicity in vivo (11, 12). A high-level of RBM14 expression has been detected in embryonic tissues and in stem cells, and it is involved in transcription-coupled alternative splicing (13, 14).
Webs of DNA damage response pathways are organized to deal with DNA damage by regulating cell cycle and transcription and determine cell fate (14, 15). Nonhomologous end joining (NHEJ) is the prominent process to repair double-strand breaks (DSBs) in human cells. DNA-end sensor KU70/80 heterodimer together with DNA-PKcs initiates the NHEJ process, and the XRCC4/DNA-ligase IV complex ligates DSB ends (16). We previously demonstrated that RBM14 is required for efficient removal of KU proteins and recruitment/retention of XRCC4 to damaged chromatin (11).
Accumulating evidence suggests that RNA is required for the main DSB repair pathways. It was shown recently that the DNA-PK–dependent canonical NHEJ (cNHEJ) pathway utilizes nascent RNA for error-free DSB repair of transcribed genes (17). The cNHEJ is the dominant pathway for DSB repair in mammalian cells, and mammalian cNHEJ proteins form a multiprotein complex with RNA polymerase II and preferentially associate with the transcribed genes after DSB induction. Therefore, it has been hypothesized that nascent RNA can serve as a template for restoring the missing sequences, thus allowing error-free DSB repair (17). The RNA:DNA helicase Senataxin is recruited to DSB sites when they occur in transcriptionally active loci, which are prone to undergo homologous recombination (HR), and promotes Rad51 recruitment (18). DNA damage-induced small noncoding RNAs (DDRNAs) as well as long noncoding RNAs (dilncRNAs) have been detected (19–21). It was also shown in fission yeast Schizosaccharomyces pombe that RNA:DNA hybrids form as part of the HR-mediated DSB repair process, and that RNase H enzymes are essential for their degradation and efficient completion of DNA repair (22). Endogenous transcript RNA-mediated DNA recombination has been reported in budding yeast Saccharomyces cerevisiae as well (23). Genetic and biochemical studies in yeast showed that RNA transcripts facilitate homology-directed DNA repair. The HR protein RAD52 directly co-operates with RNA as a sequence-directed ribonucleoprotein complex to promote RNA–DNA repair (23–25). Rad52 is recruited to DSB sites in a RNA:DNA-dependent manner and plays pivotal roles in promoting XPG-mediated R-loop processing and initiating subsequent repair by HR (26). Most recently, single-molecule imaging study detected bidirectional transcripts with two-color labeling, revealing DSB-induced transcription initiation (27).
Despite the fact that a considerable number of RBPs have been implicated in DNA damage response (DDR), little is known about their roles in DSB repair pathways. It is proposed that the liquid-demixing state generated by the PARP-dependent recruitment of these IDPs might orchestrate dynamic formation of DNA repair pathways. To gain new insights into DSB repair mechanisms, we sought to characterize functions of RBM14 in DSB repair.
Results
RBM14 Is Recruited to DSB Sites Both at Gene-Coding and Intergenic Areas.
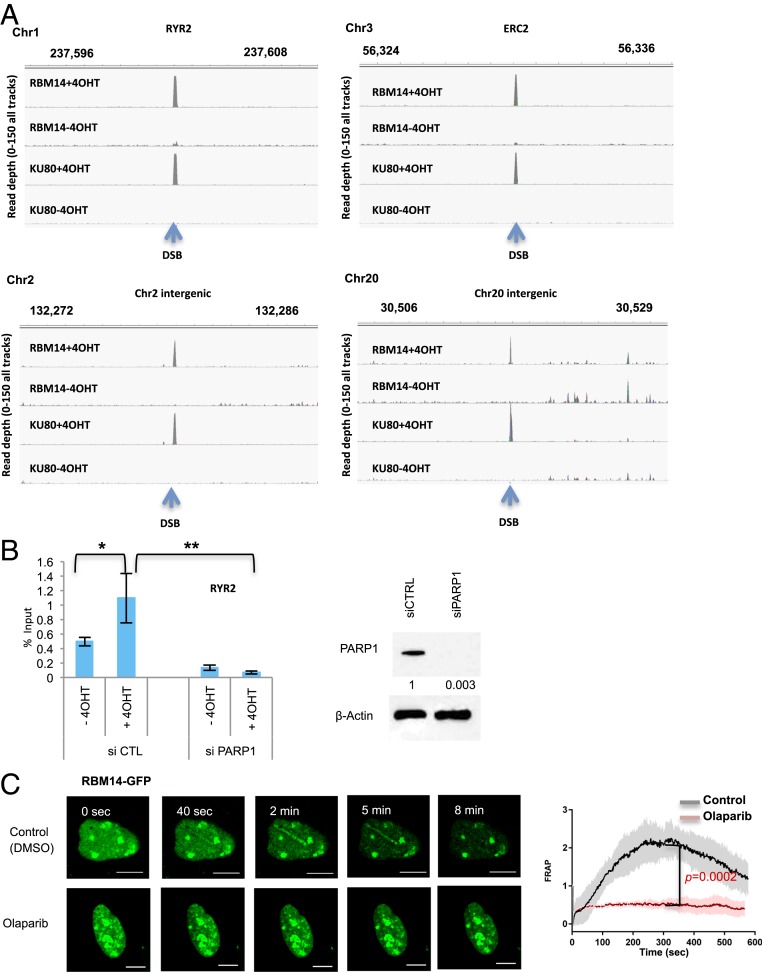
In order to investigate whether RBM14 is recruited to DSB sites, we employed the inducible DSB system (28). DSBs are induced by the eukaryotic homing endonuclease I-PpoI, which has a 15 base pair recognition sequence to cleave endogenous DNA target sites in the human genome. We identified I-PpoI target sites in the hg38 human genome that include five gene-coding sequences, four noncoding sequences, and 28S rDNA sequences (28). The addition of 4-hydroxytamoxifen (4OHT) to cells results in a time-dependent nuclear translocation of I-PpoI and induction of DSB peaks at around the 1- to 2-h time point (28). All of the chromatin immunoprecipitation sequencing (ChIP-seq), DNA–RNA immunoprecipitation sequencing (DRIP-seq), ChIP-qRT-PCR, and DRIP-qRT-PCR experiments were performed at the 2-h time point after induction of I-PpoI. Genomewide ChIP-seq analyses revealed that both RBM14 and KU80 occupy DSB sites at gene-coding as well as intergenic regions. No significant peaks were found in RBM14 and KU80 ChIP samples without I-PpoI induction (−4OHT) (Fig. 1A) (29).
Fig. 1.
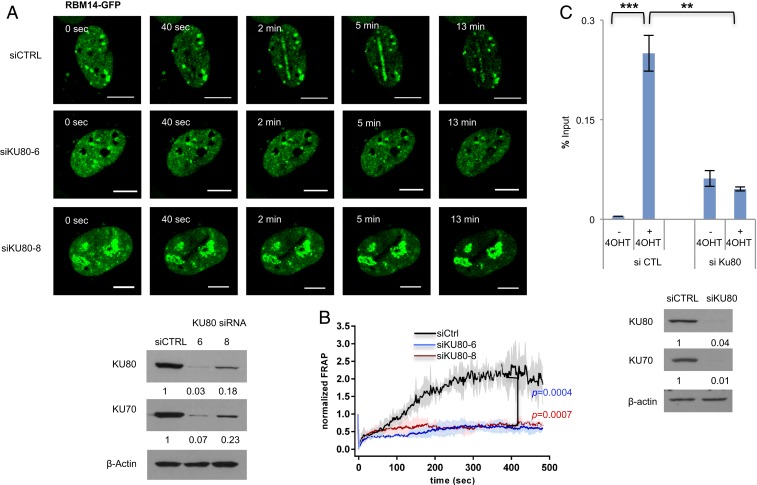
RBM14 is recruited to DNA damage sites in a PARP1-dependent manner, and RBM14 and KU are recruited to I-PpoI–induced DSB sites at both intergenic and gene-coding genomic areas. (A) Genome browser screen shots representing RBM14 ChIP-seq and KU80 ChIP-seq read depth (y axis) before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at two individual I-PpoI sites (RYR2 site, Chr20 intergenic site). The read depth of minimum 0 and maximum 150 for all tracks are shown. The I-PpoI site is indicated by an arrow. HEK293T cells treated with 4OHT (or mock) for 2 h were used for the ChIP. (B) Effects of PARP1 knockdown on recruitment of RBM14 to the RYR2 site. The bar plot shows the percentage of enrichment relative to the input of total RBM14 associated with genomic DNA detected by primers near the RYR2 site. Data shown are representative of three independent experiments. Each experiment was performed in triplicate. *P < 0.05, **P < 0.01 by two-tailed t test (Left). Knockdown level of PARP1 is shown (Right). The values indicated under the blot are the mean fold protein expression relative to control taken as 1 after normalization by β-actin (ImageJ quantification). siCTRL, control siRNA; siPARP1, PARP1 siRNA. (C) Representative live cell images of laser-irradiated U2OS cells expressing GFP-RBM14, with and without olaparib treatment. Dynamics of RBM14-GFP with and without olaparib treatment at the microlaser-damaged sites are shown (Left). Confocal images were recorded with a frame size of 512 × 512 pixels and a pixel dwell time of 2.2 μs. Live cell imaging data from microirradiation of individual cells obtained in several independent experiments performed on different days were averaged, analyzed, and displayed using ImageJ software (Right). The colored shade indicates error bars. The P values are calculated at the 300-s time point. (Scale bars, 10 μm.) FRAP, fluorescence recovery after photobleaching.
We performed RNA-seq analyses in order to examine the transcription status of the I-PpoI sites. Most of I-PpoI sites in HEK293T cells were not highly active except the Chr8 SLCO5A1 and the rDNA sites, although a low level of transcription was detected at the promoter areas (SI Appendix, Table S1). We also analyzed published HEK293T transcriptome and RNAPII ChIP-seq GEO (Gene Expression Omnibus, National Center for Biotechnology Information [NCBI]) datasets (30, 31) and found no detectable transcription and RNAPII around the intergenic I-PpoI sites. Both RBM14 and KU80 peaks were detected at four of five sites of gene-coding areas as well as three of four intergenic sites (except at the ChrX site). Interestingly, KU80 occupies the Chr8 SLCO5A1 site; however, no significant peak of RBM14 was detected (SI Appendix, Fig. S2). The INTS4 site has been shown to be inactivated due to a mutation in HEK293T cells (32).
RBM14 Is Recruited to DNA Damage Sites in a PARP-Dependent Manner.
PLD-containing RBPs, including FUS, EWS, and TAF15, seed liquid demixing and accumulate at sites of DNA damage. We have shown that RBM14 is required for DDR (12). RBM14 contains a PLD, and the purified protein forms hydrogel (33). Therefore, we tested whether RBM14 exhibits similar properties as the PLD-containing RBPs.
PARP1 knockdown significantly inhibited RBM14 occupancy at the RYR2-I-PpoI site (Fig. 1B), where transcription is not highly active (SI Appendix, Table S1). To locally induce DNA damage, we used laser microirradiation, which allowed the detection of the earliest cellular response to DNA-strand breaks in real time and with unsurpassed temporal resolution (1). All of the microlaser experiments were performed with U2OS cells, since HEK293T cells are not suited for imaging experiments due to their size. RBM14 accumulated at the sites of DNA damage quickly after microirradiation. This recruitment was inhibited by PARP inhibition with olaparib and PARP1 knockdown as in the case of the other PLD-containing RBPs (1, 2) (Fig. 1C and SI Appendix, Fig. S4). The live cell imaging of RBM14-GFP revealed that its recruitment peaks at around 5 min and disappears within 8 to 10 min with the microlaser condition that we used (10 µM BrdU, 405-nm laser, 10% power) (Fig. 1C), which is the lowest power to detect γH2AX, and GFP did not show any recruitment (SI Appendix, Fig. S3). As shown before, we also observed PARylation at the microlaser-induced DNA damage sites (34) (SI Appendix, Fig. S5). The PAR signal overlaps with γH2AX, RBM14, and KU80 (SI Appendix, Fig. S5). Indeed PARlylation at the microlaser-induced DNA damage sites was observed as early as 1 min and disappeared by 10 min after microlaser irradiation correlating with RBM14-GFP signal at the microlaser-induced DNA damage sites (Fig. 1C and SI Appendix, Fig. S5).
The Prion-Like Domain Is Required for the Recruitment of RBM14 to DNA Damage Sites.
RBM14 consists of two RRMs and an intrinsically disordered PLD (SI Appendix, Fig. S6). We investigated which domain is required for the recruitment of RBM14 to DNA damage sites by expressing truncated RBM14 proteins. GFP-RBM14-RRM that contains two RRM domains but lacks PLD failed to be recruited to microlaser-induced DNA damage sites. In contrast, GFP-RBM14-PLD that contains the C-terminal PLD but lacks RRMs was recruited to DNA damage sites as the full-length GFP-RBM14 (SI Appendix, Fig. S6). Thus, the intrinsically disordered PLD is required for the recruitment of RBM14 to DNA damage sites.
RBM14 and KU80 Are Recruited to DNA Damage Sites in an RNAPII-Dependent Manner.
Unexpected roles of transcription in DSB repair pathways have been reported recently. DDRNAs as well as dilncRNAs have been detected (19–21). In fission yeast, RNA:DNA hybrids are required for efficient DSB repair by HR, which is the dominant pathway in the organism (22). It has been shown in human cells that DSBs at transcriptionally active sites are largely unrepaired and clustered in G1 and are repaired by HR in postreplicative cells (35).
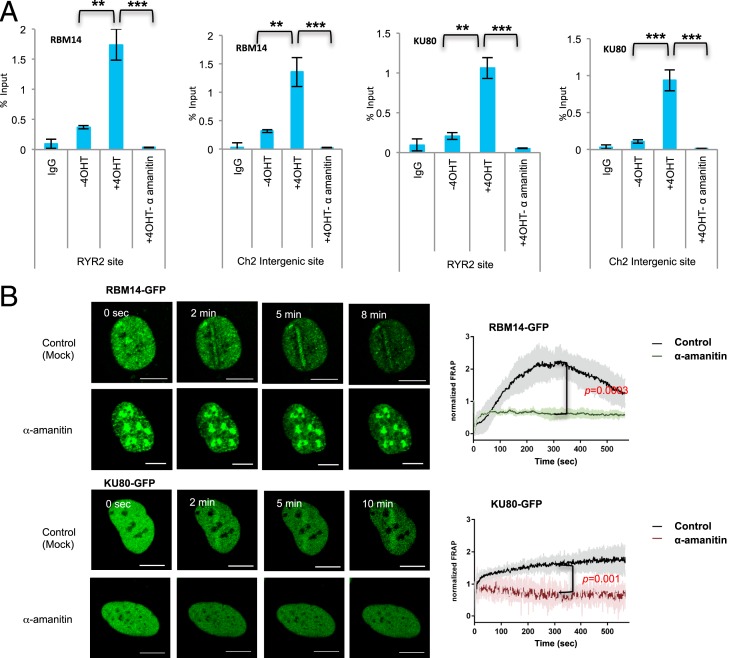
The RBM14 protein contains RRMs and has been implicated in transcription regulation (36). Thus, we investigated whether transcription is required for the recruitment of RBM14. The qRT-PCR analyses showed RNAPII-dependent association of RBM14 and KU80 protein around the I-PpoI–induced DSB sites. Importantly, the recruitment of RBM14 to both RYR2, where transcription is not highly active (SI Appendix, Table S1), and chromosome 2 intergenic sites were strongly inhibited by a transcription inhibitor, α-amanitin, suggesting that RBM14 and KU80 are recruited to near DSB sites in an RNAPII-dependent manner without ongoing transcription (Fig. 2). RNA-seq analyses of HEK293T cells published in refs. 29 and 31 as well as our RNA-seq results showed no detectable transcription at I-PpoI intergenic sites without damage induction (SI Appendix, Table S1). Transcription inhibition also strongly inhibited recruitments of both RBM14 and KU80 to microlaser-induced DNA damage sites. The other transcription inhibitor, triptolide (37), also inhibited the recruitment of RBM14 and KU80 (SI Appendix, Fig. S4). These results imply that RBM14 is involved in DNA repair that involves RNAPII-dependent transcription. The expression levels of RBM14-GFP, KU-GFP, as well as the endogenous proteins were not affected by transcription inhibition with the conditions that we used (SI Appendix, Fig. S7B). Furthermore, transcription did not affect the cell cycle state of the cells (SI Appendix, Fig. S8).
Fig. 2.
Effects of transcription inhibition on RBM14 and KU80 recruitment at DSB sites. (A) ChIP-qRT-PCR with anti-RBM14 and anti-KU80 antibodies. HEK293T cell extracts 2 h after 4OHT treatment (or mock treatment) were used for the ChIP. Recruitment of RBM14 or KU80 in either mock-treated or various transcription inhibitors-treated (α-amanitin, 50 μg/mL, 5 h) at RYR2 and Chr 2 sites. **P < 0.01, ***P < 0.001 by two-tailed t test. (B) Live cell imaging of RBM14-GFP and KU80-GFP with and without α-amanitin (50 μg/mL, 5 h) treatment. Confocal images were recorded with a frame size of 512 × 512 pixels and a pixel dwell time of 2.2 μs (Left). Data from microirradiation of individual cells obtained in several independent experiments performed on different days were averaged, analyzed, and displayed using ImageJ software (Right). The P values were calculated at the 300-s time point. The colored shade indicates error bars. (Scale bars, 10 μm.)
We also tested the recruitment of XRCC4 to the microlaser-induced DNA damage sites in the presence and absence of α-amanitin. The XRCC4 recruitment was strongly inhibited by α-amanitin (SI Appendix, Fig. S9).
RNAPII Is Recruited to I-PpoI–Induced DSB Sites in a PARP-Dependent Manner, and RNA:DNA Hybrids Are Generated at DNA Damage Sites.
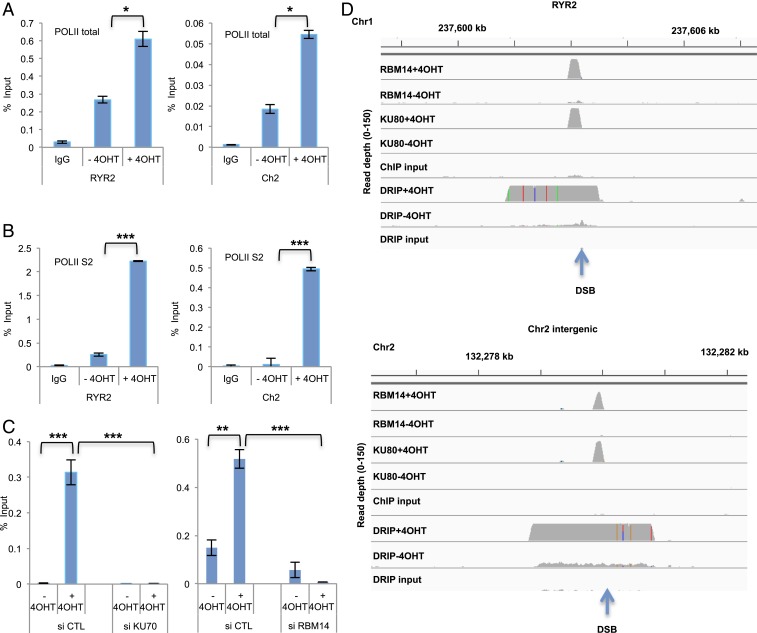
Our results indicate that transcription is induced upon DSBs. Indeed, we observed the recruitment of RNAPII to I-PpoI–induced DSB sites at RYR2 and intergenic Chr2 sites by ChIP-qRT-PCR with both anti-RNAPII-total and anti-RNAPII-S2 (active form) antibodies. Significant enrichment of RNAPII-S2 at I-PpoI sites was detected only after I-PpoI induction (+4OHT) (Fig. 3 A and B). PARP inhibition by olaparib strongly inhibited RNAPII-total and RNAPII-S2 accumulation at the RYR2 site (SI Appendix, Fig. S10A). Furthermore, knockdown of KU70 and RBM14 also induced reduction of RNAPII occupancy at the RYR2 site (Fig. 3C). We next investigated whether RNA:DNA hybrids are generated upon DSB induction. Genomewide DRIP-seq showed RNA:DNA hybrid generation upon DSB induction at the I-PpoI sites including intergenic sites (Fig. 3D) (29). PARP inhibition by olaparib strongly inhibited RNA:DNA hybrid accumulation at the RYR2 and the intergenic Chr 2 sites (SI Appendix, Fig. S10B). This assay is based on the use of the S9.6 antibody, which recognizes RNA:DNA hybrids specifically (refs. 38 and 39 and SI Appendix, Fig. S11). RNaseH1 treatment abolished S9.6 signal while RNaseA and S1 nuclease showed no effect (SI Appendix, Fig. S11A), and the DRIP-qRT-PCR with anti-S9.6 showed significant reduction of RNA:DNA hybrids by the RNaseH treatment, but not with RNaseA (SI Appendix, Fig. S11B). The DRIP-seq showed no significant peaks of RNA:DNA hybrids without I-PpoI induction except at rDNA sites, which are known to accumulate RNA:DNA hybrids (40). RNA:DNA hybrids occupied about 2 to 3 kb around DSB sites (1 to 2 kb each site), whereas both RBM14 and KU80 occupied about 100 to 150 bp at the DSB sites (about 50 bp each side) (Figs. 1A and 3D), implying that RBM14 binds to DNA ends like KU proteins.
Fig. 3.
RNAPII is recruited to DSB sites and RNA:DNA hybrids are generated around DSB sites. (A) Accumulation of total (POLII-total) and (B) active (POLII-S2) RNAPII before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at two individual I-PpoI sites, (RYR2 site, Chr2 intergenic site). The bar plot shows the percentage of enrichment relative to the input of total RNAPII and RNAPII-S2 associated with genomic DNA detected by primers near the I-PpoI sites. Data shown are representative of three independent experiments. Each experiment was performed in triplicate. *P < 0.05 by two-tailed t test. (C) Effects of KU70 (Left) and RBM14 (Right) knockdown on the recruitment of RNAPII. Accumulation of total (POLII-total) RNAPII before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at RYR2 I-PpoI site is shown. The bar plot shows the percentage of enrichment relative to the input of total RNAPII associated with genomic DNA detected by primers near the I-PpoI sites. Data shown are representative of three independent experiments. Each experiment was performed in triplicate. **P < 0.01, ***P < 0.001 by two-tailed t test. (D) Genome browser screenshot representing DRIP-seq, RBM14 ChIP-seq, and KU80 ChIP-seq before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at two individual I-PpoI sites (RYR2 site, Chr2 intergenic site).
RBM14 and KU Interact with RNA:DNA Hybrids and Are Required for RNA:DNA Hybrid Generation at DNA Damage Sites.
The RBM14 occupancy at the I-PpoI–induced DSB sites requires RNAPII (Fig. 2). Thus, we investigated whether RBM14 interacts with RNA:DNA hybrids by DRIP. We detected strong interaction between RBM14 and RNA:DNA hybrids, which is sensitive to RNaseH treatment but not to RNaseA (SI Appendix, Fig. S12A). Furthermore, we detected interaction between RBM14 and RNAPII by immunoprecipitation (SI Appendix, Fig. S12A).
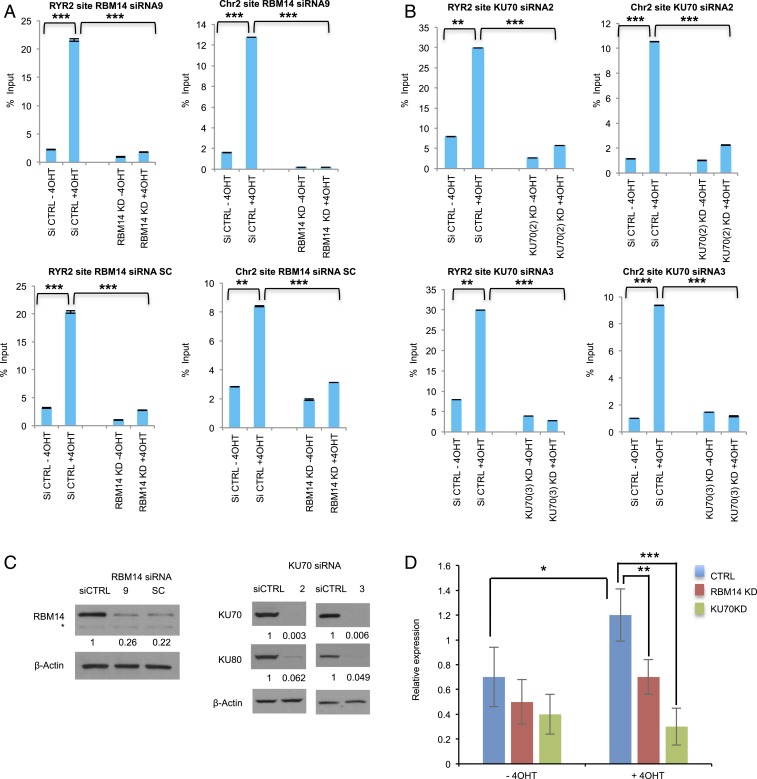
Next, we asked whether RBM14 and KU are required for generation of RNA:DNA hybrids at DNA damage sites using the S9.6 antibody. The DSB-induced RNA:DNA hybrid formation at both the RYR2 and the intergenic Chr2 sites were abolished by RBM14 knockdown as well as KU70 knockdown (Fig. 4 A and B). PARP inhibition by olaparib also significantly decreased RNA:DNA hybrid formation at the RYR2 as well as the intergenic Chr2 sites (SI Appendix, Fig. S10B), indicating that PARylation, RBM14, and KU70 are required for generation of RNA:DNA hybrids at DNA damage sites. Importantly, we detected KU70- and RBM14-dependent nascent RNA expression at the intergenic Chr2 sites (Fig. 4C).
Fig. 4.
RNA:DNA hybrids are generated at DNA damage sites in a PARP-, transcription-, RBM14-, and KU-dependent manner. DRIP-qRT-PCR before I-PpoI activation (−4OHT) and after I-PpoI activation (+4OHT) at two individual I-PpoI sites, (RYR2 site, Chr2 intergenic site) in RBM14 (A) and KU70 (B) knockdown cells. The bar plot shows the percentage of enrichment relative to the input of total RNA:DNA hybrids with genomic DNA detected by primers near the I-PpoI sites. Data shown are representative of three independent experiments. Each experiment was performed in triplicate. **P < 0.01, ***P < 0.001 by two-tailed t test. (C) Knockdown levels of RBM14 and KU70 (for the DRIP experiments) are shown. The values indicated under the blot are the mean fold protein expression relative to control taken as 1 after normalization by β-actin (ImageJ quantification). siCTRL: control siRNA. *nonspecific band. (D) qRT-PCR analyses of nascent RNA expression at the Chr 2 site. Cells were transfected with I-PpoI and RBM14 and KU70 siRNA. Cells were collected, RNA was extracted, and qRT-PCR was performed at Chr 2 site. Nascent RNA was quantified using ∆∆Ct method. Cyclophilin B was used as a reference gene to calculate the ∆Ct value for each sample then each time point was normalized to the 0-h time point using ∆∆Ct formula. To calculate the fold change in gene expression, we took the 2 to the power of negative ∆∆Ct. Data shown are representative of three independent experiments. Each experiment was performed in triplicate.
RBM14 Is Recruited to Microlaser-Induced DNA Damage Sites in a KU-Dependent Manner.
KU80-GFP was also recruited to microlaser-induced DSB sites quickly (Fig. 2B). We tested whether KU is required for RBM14 recruitment to microlaser-induced DNA damage sites. Knockdown of KU70 strongly inhibited the recruitment of RBM14 to microlaser-induced DNA damage sites (Fig. 5A), indicating that the recruitment of RBM14 requires KU. Consistent with our previous results, RBM14 is dispensable for KU recruitment (SI Appendix, Fig. S14) (11). We further performed a ChIP-qRT-PCR experiment. RBM14 occupancy at the RYR2 site was significantly reduced by KU80 knockdown (Fig. 5C).
Fig. 5.
RBM14 recruitment to DNA damage sites requires KU. (A) The effects of KU knockdown on the recruitment of RBM14 were analyzed by laser microirradiation. Representative images of cells expressing GFP-RBM14 with either negative control siRNA- or KU80 siRNAs-transfection after laser microirradiation are shown (Top). The knockdown levels of KU70/80 were confirmed by Western blot (Bottom Left). (B) Quantification of the results in A. Confocal images were recorded with a frame size of 512 × 512 pixels and a pixel dwell time of 2.2 μs. Live cell imaging data from microirradiation of individual cells obtained in several independent experiments performed on different days were averaged, analyzed, and displayed using ImageJ software. The colored shade indicates error bars. The P values were calculated at the 300-s time point. (Scale bars, 10 μm.) (C) ChIP qRT-PCR near the RYR2 site with anti-RBM14 antibody with control or KU80 siRNA (Top). Data shown are representative of three independent experiments. Each experiment was performed in triplicate. Knockdown level of the KU complex is shown (Bottom). The values indicated under the blot are the mean fold protein expression relative to control taken as 1 after normalization by β-actin (ImageJ quantification).
RBM14 Is Required for Accurate Repair of DSBs via the cNHEJ Pathway.
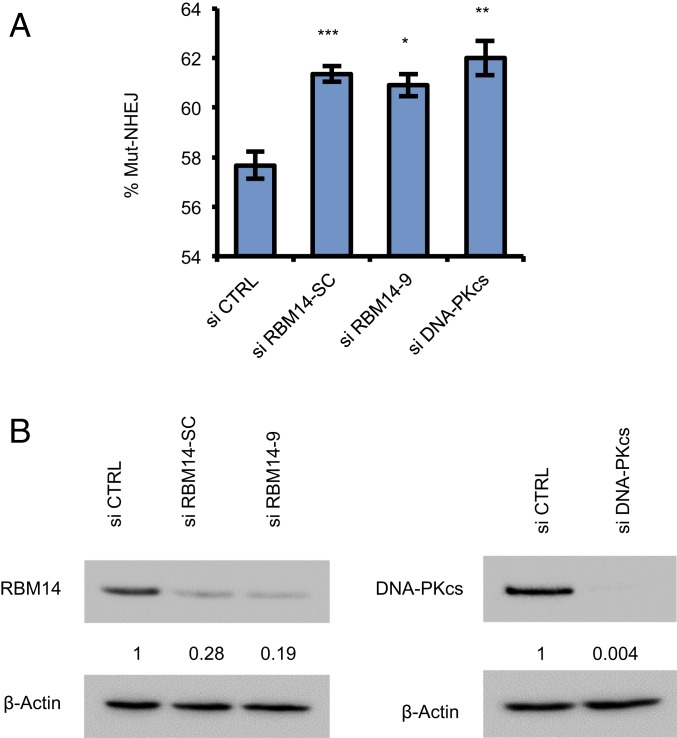
We have shown previously that knockdown of RBM14 significantly inhibits NHEJ. Furthermore, RBM14 interacts with KU, and knockdown of RBM14 reduces autophosphorylation of DNA-PKcs upon DNA damage, indicating that RBM14 is involved in DNA-PK–dependent NHEJ (11, 12). To assess the impact of RBM14 on the accuracy of NHEJ, we measured mutagenic repair in HEK293T cells using a DSB repair reporter assay for mutagenic NHEJ (Mut-NHEJ) (41). Knockdown of RBM14 significantly increased Mut-NHEJ to an extent similar to that seen following the depletion of other core components of the cNHEJ pathway, DNA-PKcs (Fig. 6), indicating that RBM14 is required for prevention of mutagenic repair. To determine the nature of the repair defects in cells depleted of RBM14, we performed DNA deep sequence analysis of the repair junctions in control cells (control small interfering RNA [si-control]) and in RBM14 knockdown cells. We extracted genomic DNA, and PCR amplified the repair junctions using primers flanking the DSB site induced by Cas9 within our integrated reporter. The analysis indicates that while the majority of the reads in control and RBM14-depleted cells contained small deletions and insertions, consistent with fluorescence-activated cell sorting (FACS) analysis, we detected a sevenfold reduction in the total reads that exhibited faithful repair (SI Appendix, Fig. S15A). Importantly, we also detected an increase (from 47 to 69.2%) in the percentage of reads that contained small deletions as well as a robust increase (from 4 to 14.7%) in the percentage of reads that contained large insertions (>10 bp) in the RBM14-depleted cells (SI Appendix, Fig. S15C). Furthermore, depletions of RBM14 resulted in a significant increase in reads that contained deletions with microhomology signature (from 60% in si-control cells to 76.2% in si-RBM14; P < 1e-15) (SI Appendix, Fig. S15B). These results show that the loss of RBM14 compromises the fidelity of DSB repair and shunts the repair of DSBs from the relatively faithful cNHEJ pathway to the error-prone alternative (alt)-NHEJ pathway which utilizes microhomologies.
Fig. 6.
Knockdown of RBM14 reduces high-fidelity cNHEJ and increases mutagenic NHEJ. (A) Knockdown of RBM14 increases error-prone, Mut-NHEJ. Knockdown of DNA-PKcs was performed in parallel. DNA DSB repair was measured in HEK293T cells containing a single copy of an integrated reporter that undergoes Cas9-mediated DNA DSBs and subsequent repair by NHEJ. Bar graphs are represented as the mean values of three independent experiments ± SD; *P < 0.05, **P < 0.01, ***P < 0.001. (B) Western blots of protein lysates from HEK293T cells containing the reporter construct and transfected with the indicated siRNAs. The values indicated under the blot are the mean fold protein expression relative to control taken as 1 after normalization by β-actin (ImageJ quantification).
Discussion
In this study, we set to understand the functions of an intrinsically disordered RNA-binding protein, RBM14 in the nonhomologous end-joining process. We found that RBM14 is recruited to microlaser-induced DNA damage sites rapidly in a PARP- and RNAPII-dependent manner and dissociates within 10 min as the other IDPs such as FUS and EWS do (1, 2). Microirradiation experiments allow us to detect immediate response of proteins; however, the damaged sites contain many different lesions, including DSBs and single-strand breaks. Therefore, we performed ChIP-seq analyses using the I-PpoI nuclease-based DSB-inducible system. We found that the recruitment of RBM14 depends on RNAPII, and that the recruitment of the NHEJ protein KU requires RNAPII at intergenic as well as gene-coding areas (Fig. 2). Both RBM14 and KU are detected at intergenic and gene-coding areas by the ChIP-seq analyses that were confirmed by the ChIP-qRT-PCR. RNA:DNA hybrids are generated upon induction of DSBs in a PARP-, RBM14-, and KU-dependent manner (Fig. 4), indicating that transcription is induced at DSB sites, and that KU- and PARP-dependent recruitment of RBM14 are prerequisite for generation of RNA:DNA hybrids at DSB sites. Indeed, the RNAPII occupancy at the DSB site required RBM14 and KU (Fig. 3C). We speculate that RBM14 and KU are required for the stable association of RNAPII to DSB sites. We have shown that RBM14 interacts with RNA:DNA hybrids (SI Appendix, Fig. S12A). Indeed, RBM14 was identified as an RNA:DNA hybrid binding protein (42). The recruitment of RBM14 to microlaser-induced DNA damage sites requires its PLD domain. The RRM domain might be required for its stable association to DNA damage sites.
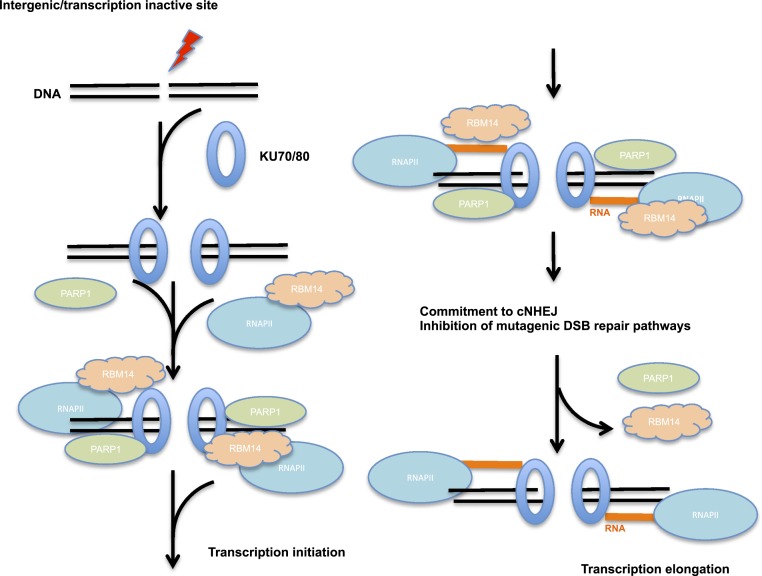
Most recently, it was shown that bidirectional transcription is induced at a DSB site by elegant single-molecule imaging systems (27). The single-molecule imaging study also resolved the paradox of DSB-induced transcription at DSB sites. Transcription silencing was detected when DSBs were induced proximal to transcription promoter sites (32, 43), which seems to be contradicting our finding shown here as well as shown by others (19, 20). The single molecule study showed that induction of a DSB near a promoter resulted in a rapid suppression of preexisting transcription from the promoter. However, DSB within the intergenic region drove formation of promoter-like nucleosome-depleted regions, and bidirectional transcription was detected (27), consistent with our results with intergenic I-PpoI–induced DSB sites. Our results indicate that RNA is transcribed at intergenic and transcriptionally silent genomic areas in response to DSB induction, and forms RNA:DNA hybrids. KU proteins are recruited quickly upon microlaser irradiation; however, the recruitment is independent of PARP (SI Appendix, Fig. S4). It has been shown that activation of PARP1 in response to bleomycin-induced DSBs depends on KU (44). Therefore, we speculate that the KU recruitment occurs prior to PARylation at DSB sites. As shown before (45), we observed that the recruitment of XRCC4, which functions at the later stage of cNHEJ, depended on PARP (SI Appendix, Fig. S9). The recruitment of XRCC4 was strongly inhibited by α-amanitin (SI Appendix, Fig. S9), indicating that XRCC4 recruitment also requires RNAPII, consistent with our previous finding that RBM14, which is needed for the RNAPII occupancy at DSB sites, is required for efficient/stable XRCC4 occupancy on damaged chromatin (11). RBM14 is recruited to microlaser-induced DNA damage sites in a PARP-, RNAPII-, and KU-dependent manner. Based on these results, we propose a model of NHEJ initiation which involves RNA:DNA hybrid formation at transcriptionally inactive genomic sites (Fig. 7): Upon induction of DSBs, the KU70/80 complex is recruited to the DSB sites. Followed by activation of PARP1, RBM14 and RNAPII are recruited to the DSB sites. RBM14 was originally discovered as a transcription coactivator activator, and was named CoAA. It was found to be involved in regulation of transcription from a set of promoters (13). Therefore, we propose that RBM14 acts as a coactivator of RNAPII for damage-induced transcription. Another IDP FUS has been shown to interact with RNAPII, and phosphorylation of RNAPII at its CTD domain induces dissociation of RNAPII from FUS releasing RNAPII for RNA synthesis (46, 47). We and others have shown interaction between RBM14 and KU proteins (12, 48). The interaction between KU and RNAPII has been shown (17). We observed weak but reproducible KU-RNAPII interaction (SI Appendix, Fig. S12B). Here, we detected DNaseI and RNase A resistant interaction between RBM14 and RNAPII by immunoprecipitation analyses (SI Appendix, Fig. S12B). Further biochemical studies with purified proteins are required to determine whether these interactions are direct or not.
Fig. 7.
The model for initiation mechanism of RNA:DNA hybrid formation. A schematic model for initiation step of RNA:DNA formation around DSB sites at transcriptionally silent genomic locations. The KU complex is recruited to DSB sites immediately followed by PARylation around the DSB sites. RNAPII and RBM14 are recruited to the DSB sites in a PARP-dependent manner. This PARP-dependent association of KU, RBM14, and RNAPII allows generation of RNA:DNA hybrids at transcriptionally inactive genomic sites when DSBs are induced. RBM14 dissociates from the DSB sites within a few minutes after induction of DNA damage when PARylation disappears at the DSB sites.
RBM14 occupies only DSB ends as the KU complex, and does not spread further where we detected RNA:DNA hybrids (about 1–2 kb around DSB sites), indicating that RBM14 does not travel with RNAPII. We speculate that RBM14 is required for activation of transcription by RNAPII at DSB sites. RBM14 accumulation peaks around 5 min after microirradiation and starts dissociating from DNA damage sites (Fig. 1C), which correlates with PARylation at DSB sites (SI Appendix, Fig. S5). This dissociation might allow RNAPII to progress RNA synthesis.
Damage-induced lncRNAs have been detected by others, and the authors hypothesized that DNA ends act as transcriptional promoters (19). Our results are consistent with the hypothesis, and further connect intrinsically disordered RNA-binding protein RBM14, PARP, and the cNHEJ protein KU to the process of transcription induction at DSB sites at transcriptionally inactive sites. DSB ends are bound by the KU complex and are PARylated immediately after induction of DSBs, and then recruit RNAPII and intrinsically disordered protein RBM14 for transcription activation. This is reminiscent of signal-induced transcription, which involves generation of topoisomerase IIβ-mediated DSB. The signal-dependent activation of gene transcription by nuclear receptors and other classes of DNA-binding transcription factors, including activating protein 1, requires DNA topoisomerase IIβ-dependent, transient, site-specific DSBs formation. Subsequent to the break, PARP1 enzymatic activity is induced, which is required for a nucleosome-specific histone H1 high-mobility group B exchange event and for local changes of chromatin architecture. The KU complex is also recruited to the sites (49). It has been shown that KU proteins are required for glucocorticoid receptor transcription activation induced by DNA topoisomerase IIβ (50). Cells induce DSBs and recruit the KU complex, PARP1, and RNAPII in order to induce signal-dependent transcription.
The importance of RNA:DNA hybrids at DSB sites has been shown. Removal of the RNA component around DSB sites results in impaired HR as well as NHEJ (22). Sequencing of RNA:DNA hybrids revealed RNA invasion around DSB sites, and removal of the RNA component from this structure results in impaired DNA repair (21). Inhibition of the RAD52-mediated RNA-dependent repair pathway leads to neuronal genomic instability and a consequent neurodegenerative phenotype such as those seen in Alzheimer’s disease (51). Further studies are needed to understand why RNA:DNA hybrids are generated upon DSBs. The chromatin architectures around DSB sites might be regulated by RNA:DNA hybrids in order to complete the NHEJ process and/or to prevent mutagenic repair processes at transcriptionally inactive sites.
There have been more reports showing damage-induced transcription. Endogenous transcript RNA-mediated DNA recombination has been reported in budding yeast S. cerevisiae (23). Generation of small RNAs which is transcribed from DSB sites has been detected in Drosophila (52). It has been shown that ∼21-nucleotide small RNAs are produced from the sequence vicinity to DSB sites in Arabidopsis and human cells (53). These small RNAs are produced by DROSHA- and DICER-like enzymes which can digest the dilncRNAs (19, 52, 53). It is not known whether these small RNAs are generated at intergenic DSB sites, and further investigation is required to answer this question.
The I-PpoI–inducible DSB system generates DSBs at five gene-coding and four intergenic genomic sites besides rDNA sites. Our RNA-seq as well as Gene Expression Omnibus (GEO) data showed that most of the gene-coding sites are not transcriptionally highly active in HEK293T cells (SI Appendix, Table S1) as well as in U2OS cells (European Molecular Biology Laboratory-European Bioinformatics Institute [EMBL-EBI] ArrayExpress, E-MTAB-6318). Interestingly, we found KU peaks at all of the I-PpoI sites and RBM14 peaks at most of the sites except at the chromosome X site and a relatively transcriptionally active SLCO5A1 site (SI Appendix, Fig. S2). Transcriptionally active sites have been shown to be repaired by HR although NHEJ proteins were also detected at the sites (35, 54). RBM14 recruitment might be inhibited at transcriptionally active DSB sites, and RBM14 might be needed for initiation of NHEJ to prevent HR. Further investigation using a different DSB system that contains more transcriptionally active sites such as the DIvA cells (35) should help to determine how the repair pathway choice between HR and NHEJ might be regulated by RBM14. Importantly, knockdown of RBM14 increased mutagenic NHEJ at DSB sites (Fig. 6), indicating that RBM14 is required to prevent mutations at DSB sites. The observed increase in Mut-NHEJ is likely a consequence of shunting from the classical cNHEJ pathway toward the mode error-prone DSB repair pathways, such as alt-NHEJ or single-strand annealing. Our results indicate that RBM14 plays a role in regulating accurate DSB repair through the cNHEJ pathway.
In conclusion, we demonstrated that transcription is induced at intergenic and transcriptionally inactive sites upon DSBs. Intrinsically disordered RNA-binding protein RBM14 is recruited to DSB sites in a PARP-dependent manner and plays a role in generating RNA:DNA hybrids for efficient NHEJ at DSB sites.
Methods
Plasmids, siRNAs, and Antibodies.
RBM14-GFP was a gift from Lan Ko, Medical College of Georgia, Augusta, GA. KU70-GFP (pEGFP-C1-FLAG-Ku70, no. 46957), KU80-GFP (pEGFP-C1-FLAG-Ku80, no. 46958), and XRCC4-GFP (pEGFP-C1-FLAG-XRCC4, no. 46959) were obtained from Addgene. Phleomycin was acquired from Sigma (P9564). The siRNAs RBM14-6 (SI02637229), RBM14-7 (SI02637236), RBM14-9 (SI03048374), KU70-3 (SI03033884), KU70-G3 (SI00423122), KU80-6 (SI02663766), KU80-8 (SI03022271), PARP1–6 (SI02662996), PARP1–12 (SI04433989), and negative control siRNA (Allstar Negative control, SI03650318) were obtained from Qiagen. siRNA RBM14-SC (SC-96838) was obtained from Santa Cruz. siRNA DNA-PKcs was obtained from Invitrogen (GAACAUGGCAGGAGAGAAUdTdT). All siRNAs were transfected with Lipofectamine RNAiMAX from Thermo Fisher Scientific (13778). Antibodies used in Western blotting were RBM14 (1:1,000, Abcam ab70636), KU70 (1:250, Abcam ab3114), KU80 (1:250, Thermo Fisher Scientific MS285P0), PARP1 (1:1,000, Abcam ab227244), DNA-PKcs (1:1,000, Abcam, ab44815), β-actin (1:1,000, Santa Cruz, sc47778), and tubulin (1:1,000, Santa Cruz).
Cell Culture.
U2OS and HEK293T cells were grown in Dulbecco’s Modified Eagle Medium (DMEM) with glutamine and 4.5 g/L glucose (Corning Cellgro 10-013-CV) supplemented with 10% fetal bovine serum (FBS) (Sigma F2442) in 5% CO2 incubator at 37 °C.
Immunofluorescence.
Following laser irradiation, cells were fixed with 4% paraformaldehyde (PFA) (wt/vol) in phosphate buffered saline (PBS) for 10 min for analysis by immunofluorescence. To preextract U2OS cells for KU staining, cells were washed with PBS buffer, and then incubated twice for 3 min at room temperature with CSK+R buffer (10 mM Pipes, pH 7.0, 100 mM NaCl, 300 mM sucrose, and 3 mM MgCl2) containing 0.7% Triton X-100 and 0.3 mg/mL RNaseA. After preextraction, cells were washed with PBS buffer and fixed with PFA. After fixation, permeabilization (0.25% Triton X-100, in PBS for 10 min) was performed. Cells were washed three times with PBS and immunostained with anti-γH2AX antibody (Millipore 05-636) or anti-PAR antibody (Enzo cat. no. ALX-804-220-R100) or anti-KU80 antibody (Abcam, ab33242) followed by blocking for 1 h with 5% BSA in PBS 0.1% Tween-20. Following three washes with PBS, the cells were incubated with a secondary antibody for 1 h. Subsequently, cells were washed with PBS three times and then mounted in vectashield mounting media (Vector Laboratories). Images were acquired using an EVOS microscope. For the 5-ethynyl uridine (EU) incorporation assay, cells were treated with dimethyl sulfoxide (DMSO) or indicated transcription inhibitors were incubated in the presence of EU (1 mM) for 1 h, followed by fixation with 4% PFA. Fixed cells were permeable with 0.1% SDS, 0.5% Triton-X in PBS for 5 min, and incubated with primary antibodies for 1 h at 37 °C. After incubation with secondary antibodies, incorporated EU was visualized by a click chemistry procedure before cells were counterstained with 4′,6-diamidino-2-phenylindole (DAPI) and mounted in the VECTASHIELD mounting medium (Vector Laboratories).
Western Blotting.
Cells were collected and lysed in RIPA buffer (50 mM Tris HCl, pH 8.0, 150 mM NaCl, 1% Nonidet P-40, 0.5% sodium deoxycholate, 0.1% SDS) containing protease and phosphatase inhibitor mixtures (Roche, Sigma) and phenylmethylsulfonyl fluoride (PMSF). The lysates were incubated for 10 min on ice, centrifuged for 15 min at 161,000 × g at 4 °C, and the supernatant was collected. Equal amounts of proteins were separated on acrylamide gels by SDS-electrophoresis and probed by antibodies.
Detailed methods are available in SI Appendix, Supplemental Methods.
Data Availability.
High throughput data have been deposited to and are publicly available at the National Center for Biotechnology Information Sequence Read Archive site, https://www.ncbi.nlm.nih.gov/sra (accession nos. SAMN11388077–SAMN11388089; BioProject ID PRJNA531840) (29).
Code Availability.
Only publicly available software was used, and no new code was developed for this study.
Supplementary Material
Acknowledgments
This work was supported by NIH grant R01 GM113253 (to M.K.). We thank Dr. Lan Ko and Dr. Archa Fox for providing the GFP-RBM14 plasmids. We thank Next Generation Sequence Core of the Sidney Kimmel Comprehensive Cancer Center at Johns Hopkins University School of Medicine for sequencing, analyses, and advice on the experiments. Work in the T.A. laboratory was supported by a Virginia Cancer Biology training grant (T32 CA009109).
Footnotes
The authors declare no competing interest.
This article is a PNAS Direct Submission.
Data deposition: High throughput data have been deposited to and are publicly available at the National Center for Biotechnology Information Sequence Read Archive site, https://www.ncbi.nlm.nih.gov/sra (accession nos. SAMN11388077–SAMN11388089; BioProject ID PRJNA531840).
This article contains supporting information online at https://www.pnas.org/lookup/suppl/doi:10.1073/pnas.1913280117/-/DCSupplemental.
References
- 1.Altmeyer M., et al. , Liquid demixing of intrinsically disordered proteins is seeded by poly(ADP-ribose). Nat. Commun. 6, 8088 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Patel A., et al. , A liquid-to-solid phase transition of the ALS protein FUS accelerated by disease mutation. Cell 162, 1066–1077 (2015). [DOI] [PubMed] [Google Scholar]
- 3.Chesi A., et al. , Exome sequencing to identify de novo mutations in sporadic ALS trios. Nat. Neurosci. 16, 851–855 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kim H. J., et al. , Mutations in prion-like domains in hnRNPA2B1 and hnRNPA1 cause multisystem proteinopathy and ALS. Nature 495, 467–473 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Li Y. R., King O. D., Shorter J., Gitler A. D., Stress granules as crucibles of ALS pathogenesis. J. Cell Biol. 201, 361–372 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nomura T., et al. , Intranuclear aggregation of mutant FUS/TLS as a molecular pathomechanism of amyotrophic lateral sclerosis. J. Biol. Chem. 289, 1192–1202 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Mastrocola A. S., Kim S. H., Trinh A. T., Rodenkirch L. A., Tibbetts R. S., The RNA-binding protein fused in sarcoma (FUS) functions downstream of poly(ADP-ribose) polymerase (PARP) in response to DNA damage. J. Biol. Chem. 288, 24731–24741 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Krietsch J., et al. , PARP activation regulates the RNA-binding protein NONO in the DNA damage response to DNA double-strand breaks. Nucleic Acids Res. 40, 10287–10301 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Britton S., et al. , DNA damage triggers SAF-A and RNA biogenesis factors exclusion from chromatin coupled to R-loops removal. Nucleic Acids Res. 42, 9047–9062 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Polo S. E., et al. , Regulation of DNA-end resection by hnRNPU-like proteins promotes DNA double-strand break signaling and repair. Mol. Cell 45, 505–516 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Simon N. E., Yuan M., Kai M., RNA-binding protein RBM14 regulates dissociation and association of non-homologous end joining proteins. Cell Cycle 16, 1175–1180 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Yuan M., Eberhart C. G., Kai M., RNA binding protein RBM14 promotes radio-resistance in glioblastoma by regulating DNA repair and cell differentiation. Oncotarget 5, 2820–2826 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Auboeuf D., et al. , CoAA, a nuclear receptor coactivator protein at the interface of transcriptional coactivation and RNA splicing. Mol. Cell. Biol. 24, 442–453 (2004). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Kai M., Roles of RNA-binding proteins in DNA damage response. Int. J. Mol. Sci. 17, 310 (2016).26927092 [Google Scholar]
- 15.Ciccia A., Elledge S. J., The DNA damage response: Making it safe to play with knives. Mol. Cell 40, 179–204 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Wang C., Lees-Miller S. P., Detection and repair of ionizing radiation-induced DNA double strand breaks: New developments in nonhomologous end joining. Int. J. Radiat. Oncol. Biol. Phys. 86, 440–449 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Chakraborty A., et al. , Classical non-homologous end-joining pathway utilizes nascent RNA for error-free double-strand break repair of transcribed genes. Nat. Commun. 7, 13049 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cohen S., et al. , Senataxin resolves RNA:DNA hybrids forming at DNA double-strand breaks to prevent translocations. Nat. Commun. 9, 533 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Michelini F., et al. , Damage-induced lncRNAs control the DNA damage response through interaction with DDRNAs at individual double-strand breaks. Nat. Cell Biol. 19, 1400–1411 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Francia S., et al. , Site-specific DICER and DROSHA RNA products control the DNA-damage response. Nature 488, 231–235 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lu W. T., et al. , Drosha drives the formation of DNA:RNA hybrids around DNA break sites to facilitate DNA repair. Nat. Commun. 9, 532 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ohle C., et al. , Transient RNA-DNA hybrids are required for efficient double-strand break repair. Cell 167, 1001–1013.e7 (2016). [DOI] [PubMed] [Google Scholar]
- 23.Keskin H., et al. , Transcript-RNA-templated DNA recombination and repair. Nature 515, 436–439 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.McDevitt S., Rusanov T., Kent T., Chandramouly G., Pomerantz R. T., How RNA transcripts coordinate DNA recombination and repair. Nat. Commun. 9, 1091 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Mazina O. M., Keskin H., Hanamshet K., Storici F., Mazin A. V., Rad52 inverse strand exchange drives RNA-templated DNA double-strand break repair. Mol. Cell 67, 19–29.e3 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Yasuhara T., et al. , Human Rad52 promotes XPG-mediated R-loop processing to initiate transcription-associated homologous recombination repair. Cell 175, 558–570.e11 (2018). [DOI] [PubMed] [Google Scholar]
- 27.Vitor A. C., et al. , Single-molecule imaging of transcription at damaged chromatin. Sci. Adv. 5, eaau1249 (2019). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Berkovich E., Monnat R. J. Jr, Kastan M. B., Roles of ATM and NBS1 in chromatin structure modulation and DNA double-strand break repair. Nat. Cell Biol. 9, 683–690 (2007). [DOI] [PubMed] [Google Scholar]
- 29.Kai M., Elseyd Z., Role of RBM14 in double-strand break repair. National Center for Biotechnology Information BioProject. http://www.ncbi.nlm.nih.gov/bioproject?term=PRJNA531840. Deposited 10 April 2019.
- 30.Richard H., et al. , Prediction of alternative isoforms from exon expression levels in RNA-seq experiments. Nucleic Acids Res. 38, e112 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Sultan M., et al. , A global view of gene activity and alternative splicing by deep sequencing of the human transcriptome. Science 321, 956–960 (2008). [DOI] [PubMed] [Google Scholar]
- 32.Pankotai T., Bonhomme C., Chen D., Soutoglou E., DNAPKcs-dependent arrest of RNA polymerase II transcription in the presence of DNA breaks. Nat. Struct. Mol. Biol. 19, 276–282 (2012). [DOI] [PubMed] [Google Scholar]
- 33.Hennig S., et al. , Prion-like domains in RNA binding proteins are essential for building subnuclear paraspeckles. J. Cell Biol. 210, 529–539 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li M., Lu L. Y., Yang C. Y., Wang S., Yu X., The FHA and BRCT domains recognize ADP-ribosylation during DNA damage response. Genes Dev. 27, 1752–1768 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Aymard F., et al. , Genome-wide mapping of long-range contacts unveils clustering of DNA double-strand breaks at damaged active genes. Nat. Struct. Mol. Biol. 24, 353–361 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Iwasaki T., Chin W. W., Ko L., Identification and characterization of RRM-containing coactivator activator (CoAA) as TRBP-interacting protein, and its splice variant as a coactivator modulator (CoAM). J. Biol. Chem. 276, 33375–33383 (2001). [DOI] [PubMed] [Google Scholar]
- 37.Titov D. V., et al. , XPB, a subunit of TFIIH, is a target of the natural product triptolide. Nat. Chem. Biol. 7, 182–188 (2011). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Boguslawski S. J., et al. , Characterization of monoclonal antibody to DNA.RNA and its application to immunodetection of hybrids. J. Immunol. Methods 89, 123–130 (1986). [DOI] [PubMed] [Google Scholar]
- 39.Nadel J., et al. , RNA:DNA hybrids in the human genome have distinctive nucleotide characteristics, chromatin composition, and transcriptional relationships. Epigenet. Chromatin 8, 46 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Wahba L., Costantino L., Tan F. J., Zimmer A., Koshland D., S1-DRIP-seq identifies high expression and polyA tracts as major contributors to R-loop formation. Genes Dev. 30, 1327–1338 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Yang C. S., et al. , Ubiquitin modification by the E3 ligase/ADP-ribosyltransferase Dtx3L/Parp9. Mol. Cell 66, 503–516.e5 (2017). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Wang I. X., et al. , Human proteins that interact with RNA/DNA hybrids. Genome Res. 28, 1405–1414 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Shanbhag N. M., Rafalska-Metcalf I. U., Balane-Bolivar C., Janicki S. M., Greenberg R. A., ATM-dependent chromatin changes silence transcription in cis to DNA double-strand breaks. Cell 141, 970–981 (2010). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Dong F., Soubeyrand S., Haché R. J., Activation of PARP-1 in response to bleomycin depends on the Ku antigen and protein phosphatase 5. Oncogene 29, 2093–2103 (2010). [DOI] [PubMed] [Google Scholar]
- 45.Luijsterburg M. S., et al. , PARP1 links CHD2-mediated chromatin expansion and H3.3 deposition to DNA repair by non-homologous end-joining. Mol. Cell 61, 547–562 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Kwon I., et al. , Phosphorylation-regulated binding of RNA polymerase II to fibrous polymers of low-complexity domains. Cell 155, 1049–1060 (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Schwartz J. C., et al. , FUS binds the CTD of RNA polymerase II and regulates its phosphorylation at Ser2. Genes Dev. 26, 2690–2695 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Ko L., Cardona G. R., Chin W. W., Thyroid hormone receptor-binding protein, an LXXLL motif-containing protein, functions as a general coactivator. Proc. Natl. Acad. Sci. U.S.A. 97, 6212–6217 (2000). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Ju B. G., et al. , A topoisomerase IIbeta-mediated dsDNA break required for regulated transcription. Science 312, 1798–1802 (2006). [DOI] [PubMed] [Google Scholar]
- 50.Trotter K. W., King H. A., Archer T. K., Glucocorticoid receptor transcriptional activation via the BRG1-dependent recruitment of TOP2β and Ku70/86. Mol. Cell. Biol. 35, 2799–2817 (2015). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Welty S., et al. , RAD52 is required for RNA-templated recombination repair in post-mitotic neurons. J. Biol. Chem. 293, 1353–1362 (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Michalik K. M., Böttcher R., Förstemann K., A small RNA response at DNA ends in Drosophila. Nucleic Acids Res. 40, 9596–9603 (2012). [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wei W., et al. , A role for small RNAs in DNA double-strand break repair. Cell 149, 101–112 (2012). [DOI] [PubMed] [Google Scholar]
- 54.Aymard F., et al. , Transcriptionally active chromatin recruits homologous recombination at DNA double-strand breaks. Nat. Struct. Mol. Biol. 21, 366–374 (2014). [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
High throughput data have been deposited to and are publicly available at the National Center for Biotechnology Information Sequence Read Archive site, https://www.ncbi.nlm.nih.gov/sra (accession nos. SAMN11388077–SAMN11388089; BioProject ID PRJNA531840) (29).