Abstract
Introduction:
Current methods for diagnosing cancer can be invasive and costly. In recent years, extensive research has been conducted on using exosomes as biomarkers for cancer detection. Exosomes are 40-150 nm-sized extracellular vesicles released by all cell types, including tumor cells. Exosomes are stable in body fluids due to their lipid bilayer member and often contain DNA, RNA, and proteins. These exosomes can be harvested from blood, plasma, serum, urine or saliva and analyzed for tumor-relevant mutations. Thus, providing an alternative to current methods of tumor detection.
Areas covered:
This review discusses the use of exosomal diagnostics in various tumor types as well as their examination in various clinical trials. The authors also discuss the limitations of exosome-based diagnostics in the clinical setting and provide examples of several studies in which the development and usage of microfluidic chips and nano sensing devices have been utilized to address these obstacles.
Expert commentary:
In recent years, exosomes and their contents have exhibited potential as novel tumor detection markers despite the labor involved in their harvest and isolation. Despite this, much work is being done to optimize exosome capture and analysis. Thus, their roles as biomarkers in the clinical setting appears promising.
Keywords: Biomarkers, cancer detection, diagnostics, exosomes, extracellular vesicles, microfluidic chips, nano-sensors, tumor detection
1. Introduction
For many tumor types, diagnosis is determined by a series of imaging scans, followed by biopsy to confirm the diagnosis [1,2]. These methods are not only invasive but can be uncomfortable for the patient as well as costly. Furthermore, in the absence of routine screening, patients are more likely to receive a late stage diagnosis. A late diagnosis has the potential drawback of decreasing the patient’s response to therapy and therefore their survivability [3–5]. This makes monitoring patient response to therapy important. Thus, it is necessary to develop new methods of diagnosing cancer that are less costly, less invasive, and have the potential for detecting tumors early and monitoring patients undergoing therapy.
In recent years, research has focused on the usefulness of exosomes in diagnosing cancer patients as well as monitoring their responses to therapy [6–13]. Exosomes are 40-150 nm extracellular vesicles comprised of a lipid bilayer membrane, complete with membrane-specific proteins [14–16]. The lipid bilayer membrane confers stability, preventing the exosomal cargo—DNA, messenger RNA (mRNA), microRNA (miRNA), and proteins— from degradation circulating throughout the body. Because of this stability, exosomes are easily harvested from a variety of accessible body fluids. This makes them attractive targets for developing new methods for detecting cancer [15,17,18].
The use of exosomes and exosomal cargo for cancer diagnostics requires identification of the most commonly deregulated genes for a specific cancer type. This is because tumors are multifactorial and exhibit what is known as tumor heterogeneity. Tumor heterogeneity describes the differences in genetic mutations and cell behaviors observed between different tumor types and their metastases as well as the same tumor type between different patients [19,20]. Tumor heterogeneity is also important in determining the proper therapy and maximizing patient survival [21,22]. It is therefore necessary to identify a panel of the most likely deregulated genes for a given tumor type or for multiple tumors. Thus, a great deal of work has incorporated bioinformatics, sequencing, and machine learning to determine deregulated genes specific to any given tumor type [7,23–25]. This review will discuss current exosomal markers that have been identified for the potential diagnosis of various tumor types, which biomarkers are useful for monitoring patients, and the limitations of current methods that must be overcome in order to bring exosomal biomarkers to the forefront of clinical applications.
1.1. Extracellular Vesicle Formation
Exosomes belong to a category of extracellular vesicles which also include apoptotic bodies, ectosomes, and microvesicles [26–31]. These vesicles were originally thought to be portions of dead cells or cellular waste. However, it is now known that exosomes exhibit important involvement in extracellular communication [32–35]. Once released, exosomes can traverse the body and localize to a specific cell. Once an exosome has localized, it will fuse with the membrane of the target cell and release its contents. These contents encompass DNA, RNA, and proteins which may proceed to carry out their functions in the new cell and contribute to tumor progression [36–39].
Exosomes are generated via a complex process. They may develop with or independent of endosomal complexes required for transport, or ESCRT [40,41]. Exosome formation which relies on ESCRT begins with the binding of ubiquitinated proteins on the endosomal membrane by ESCRT0 [42,43]. The ESCRT-0 and endosomal membrane protein complex with ESCRT-I and tumor susceptibility gene 101 (TSG101) to activate ESCRT-II. This complex contributes to the curvature of the endosome to form the intraluminal vesicles (ILVs) which make up the multivesicular body (MVB). Additionally, various cargos are deubiquitinated as they are packaged into the ILVs. Once the MVB is formed, it fuses with the cellular membrane where ESCRT-III aids in secretion of ILVs by pinching off the cellular membrane to allow their release into extracellular space as exosomes (figure 1). ESCRT-III is then disassembled by vacuolar protein sorting-associated protein 4 (VPS4) [44,45]. ESCRT-Independent exosome formation is less understood. It involves membrane folding induced by lipids and heat shock proteins to develop the ILVs while the cargo is sorted and packaged into exosomes via tetraspanins [35,45,46].
Figure 1. ESCRT-dependent sorting and development of extracellular vesicles.
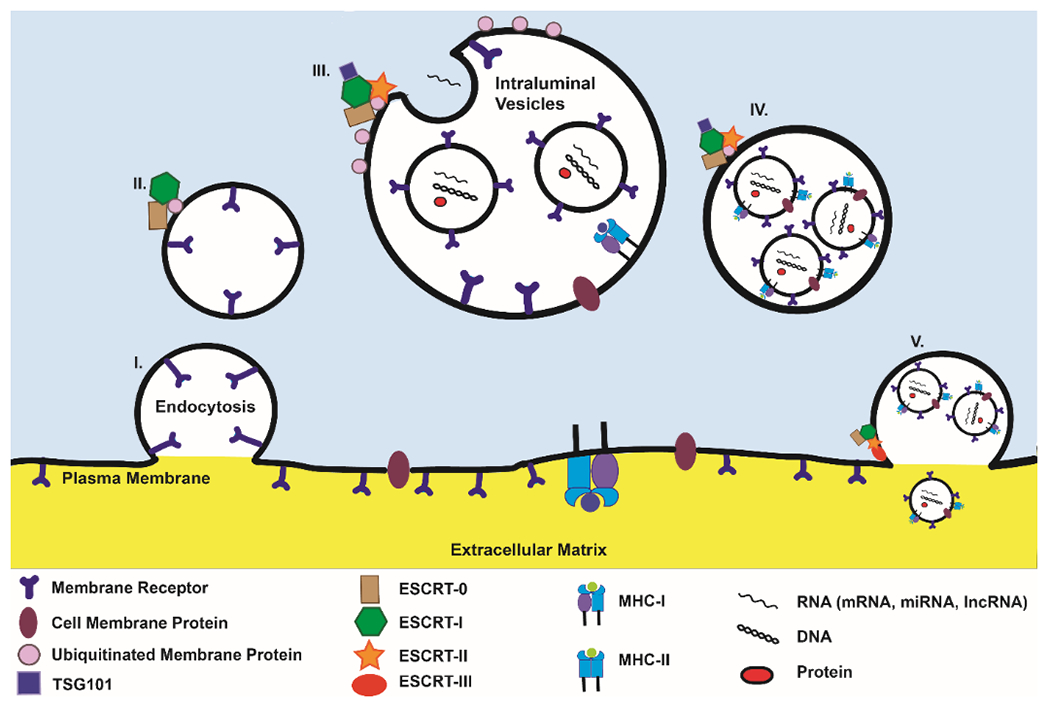
(I) ESCRT-0 initiates the extracellular vesicle formation by selecting ubiquitinated endosomal membrane proteins. (II) ESCRT-I and TSG101 complex with the ubiquitinated cargos to activate ESCRT-II. (III) ESCRT-II aids in the formation of ILVs and their consequent packaging into MVBs. (IV) The MVB contains multiple ILVs. (V) ESCRT-III deubiquitinates the MVB’s membrane and pinches the cellular membrane to aid in the release of the exosomes into extracellular space
2. Exosomal Biomarkers for Specific Cancer Diagnosis
2.1. Breast Cancer
Breast cancer is the most common type of cancer and the second leading cause of cancer-related deaths in women [47]. Breast cancer is typically diagnosed via routine mammograms or to clarify the results of a patient’s self-examination [48]. Upon a confirmation of the mass via imaging scans, the mass is analyzed via needle biopsy or surgical incision to determine malignancy. These procedures can be uncomfortable and painful to the patient. An alternative means of diagnosis could therefore benefit many patients.
Recently, circulating exosomal miRNA were analyzed as potential biomarkers for breast cancer [49]. Analysis of the media from breast cancer cell lines yielded a library of exosomal miRNA. The microRNA miR-1246 was highly expressed in two breast cancer cell lines, MCF-7 and MDA-MB-231 when compared to MCF10A, a normal mammary epithelial cell line. They then analyzed the plasma of patient-derived orthotopic xenograft mouse models to determine the expression of the miR-1246. They found that exosomal miR-1246 was significantly higher in these xenograft models than in control mice across all breast cancer subtypes, including progesterone and estrogen receptor negative subtypes, which exhibited the highest levels of miR-1246 [49]. The study concluded that exosomal miR-1246 and miR-21 in combination could offer a potential alternative to traditional methods of breast cancer diagnosis.
Another study which focused on exosomal miR-1246 confirmed the observation of increased expression levels in breast cancer. Using the MCF-7 cell line and the MCF-10a as the normal control, the laboratory was able to detect high fluorescent signaling of exosomal miR-1246 using functionalized Au nanoprobes [47]. This laboratory followed up with spiking the cell line-derived exosomes into healthy patient plasma and upon incubation with the functionalized Au probes for four hours, observed a significant increase in the fluorescent signaling of miR-1246. When they examined the plasma of healthy participants versus breast cancer patients, they found cancer patients exhibited mean signal levels of 17.73±7.73 compared to the signal levels of healthy patients with a mean of 5.27±2.61 [47].
There are currently two clinical trials focusing on the use of exosomes for breast cancer diagnosis and monitoring patient response to therapy. NCT02977468 is a phase one clinical trial aiming to assess response to Pembrolizumab (brand name Keytruda®), for treating triple negative breast cancer [50]. The study aims to monitor response to this form of therapy by analyzing the tumor samples, circulating lymphocytes, and serum exosomes. The last update occurred on September 2019 and this study was still recruiting. NCT02662621 is an interventional clinical trial focusing on the use of an exosomal detection protocol for the diagnosis of several cancer types, including breast cancer [51]. The study suggests that exosomes exhibiting the stress protein, HSP70, on the cell membrane are cancer-specific exosomes. The clinical trial aims to develop a minimally invasive diagnostic procedure that involves the isolation of these specific exosomes from blood and urine, providing an alternative to the current methods of cancer diagnoses. As of November 2019, the trial had completed but as of the writing of this work, no data has been published. Table 1 provides a summary of the current clinical trials exploring exosomes for diagnosing and monitoring cancer patients as well as their current statuses.
Table 1.
Clinical trials examining exosomes for cancer diagnosis and patient response to therapy.
| Clinical Trial | Tumor Type | Purpose | Status |
|---|---|---|---|
| NCT02977468 | Triple Negative Breast Cancer | Assess response and change in tumor to Pembrolizumab | Recruiting |
| NCT02892734 | HER2 Negative Inflammatory Breast Cancer | Monitor patient response to combination of Ipilimumab and Nivolumab | Terminated |
| NCT02662621 | Infiltrating Non-metastatic Breast Cancer Breast Cancer with First Metastasis Ovarian Cancer, Stages III and IV Metastatic NSCLC |
Quantify exosomes displaying HSP70 membrane protein in blood and urine for solid tumor diagnostic | Completed, no results posted |
| NCT02971761 | Androgen Receptor Positive and Triple Negative Breast Cancer | Monitor patient response to combination of Pembrolizumab and Enobosarm | Active, not recruiting |
| NCT03830619 | Lung Cancer | Detection of exosomal long noncoding RNA as potential lung cancer diagnosis | Recruiting |
| NCT03317080 | Lung Cancer | Monitor circulating tumor DNA in surgical patients with lung cancer | Recruiting |
| NCT02921854 | NSCLC | Monitor response to radiotherapy and chemotherapy by analyzing patient serum | Completed, no results posted |
| NCT02869685 | NSCLC | Monitoring plasma exosomal levels of PD-L1 before and after radiotherapy | Unknown |
| NCT03108677 | Primary Osterosarcoma, Lung metastases | Analyze RNA profile of circulating exosomes for early detection of metastases | Recruiting |
| NCT03228277 | T790M Positive NSCLC | Monitor response to Olmutinib in T790+ NSCLC using DNA extracted from bronchial lavage fluid | Completed, no results posted |
| NCT03432806 | Colon Cancer, Liver Cancer | Guide treatment of colon and liver cancers | Recruiting |
| NCT02439008 | Colon Cancer, Live Cancer, Kidney Neoplasms | Monitor response to radiotherapy before, during, and after administration | Terminated |
| NCT02393703 | Pancreatic Cancer | Use exosomes to monitor patients | Active, not recruiting |
| NCT03032913 | Pancreatic Cancer | Patient disease monitoring | Completed [64] |
| NCT03791073 | Pancreatic Cancer | Analyze interstitial tissue fluid for novel biomarker identification | Recruiting |
| NCT03250078 | Pancreatic Cancer | Development of blood-based screening for pancreatic cancer | Recruiting |
| NCT03334708 | Pancreatic Cancer | Early stage pancreatic cancer diagnosis | Recruiting |
| NCT03410030 | Pancreatic Cancer | Using GPC1-labeled exosomes, and additional biomarkers for monitoring patient response to combination of Ascorbic Acid, nanoparticle Paclitaxel protein bound Cisplatin and Gemcitabine | Recruiting |
2.2. Lung Cancer
Lung cancer is responsible for the most cancer deaths worldwide. There are three main types of lung cancer: small cell lung cancer (SCLC), non-small cell lung cancer (NSCLC), and lung carcinoid tumors. The two most common types, however, are NSCLC (80-85%) and SCLC (10-15%). NSCLC can be broken into further subtypes, deriving from different areas of the lungs, but all are subjected to similar treatments and have similar prognoses. There are three subtypes of lung cancer: adenocarcinomas, the most common, comprising 40% of diagnosed cases of lung cancer; squamous cell carcinomas, which contribute to 25-30% of lung tumors; and large cell carcinomas which are generally undifferentiated tumors and constitute 10-15% of lung cancers. Lung carcinoid tumors are associated with neuroendocrine cells and make up fewer than 5% of lung cancers [48].
Current lung cancer diagnostic methods involve the use of imaging scans but cannot be confirmed without examination of lung cells for presence of tumor. The least invasive procedure for detecting lung tumors involves collecting sputum from the patient. This method is effective for tumors originating in the airways, such as squamous cell carcinoma. A slightly more invasive procedure involves collection of pleural effusions, though this may not be enough to determine the type of tumor. A more invasive procedure uses needle biopsy of the tumor which could result in collapsing the lung [48]. Thus, less invasive methods for lung cancer detection are needed. Exosomes isolated from blood or serum may prove an alternative to these invasive and potentially dangerous procedures.
It is possible to differentiate between patients with lung inflammation and patients exhibiting lung adenocarcinomas by analyzing pleural effusion for exosomal RNA expression levels. A panel consisting of miR-200b, miR-200c, miR-141 and miR-375 exhibited a significant increase in expression in lung adenocarcinoma compared to benign lung inflammation. Additionally, these miRs exhibited greater than 0.95 values when analyzed using ROC curves. However, the exosomal transcript which exhibited the highest diagnostic potential was Lipocalin-2 (LCN2) with an AUC of 0.99 [52]. While this may be a promising diagnosis, the collection of pleural effusions is slightly more invasive than using blood. It would be interesting if these results were repeatable using the blood of patients with lung inflammation versus patients with lung adenocarcinoma.
One clinical trial is in progress for testing the efficacy of exosomes for monitoring patient response to lung cancer therapy. Clinical trial NCT02869685 is exploring the usefulness of Programmed Death Ligand 1 (PDL1) mRNA expression in plasma exosomes to monitor the success of radiotherapy [53]. PDL1 is a necessary receptor for activating naïve T-cells, which are important for successful destruction of tumor cells. This study was expected to conclude in June 2019; however, the status of this clinical trial remains unknown.
2.3. Gastrointestinal Cancers (Colon Cancer, Hepatocellular Carcinoma, Pancreatic Ductal Adenocarcinoma) for cancer-related deaths every year.
Diagnosing colon cancer usually involves routine screening via colonoscopy. By this method, a patient is required to consume a preparation that completely evacuates all contents of the colon while simultaneously refraining from consumption of solid foods. This process can be uncomfortable and unpleasant for the patient. One benefit to a colonoscopy is that should the physician identify polyps in the colon, they can be removed, thereby preventing the formation of a tumor. Unfortunately, only about 40% of colorectal tumors are found at this early stage. Thus, additional methods are needed to ensure a complete screening for colorectal cancer [48].
Carcinoembryonic antigen (CEA) is generally observed in fetal gastrointestinal development but resurfaces in the blood during tumorigenesis of the gastrointestinal tract. It is currently used as a marker for monitoring colon cancer patients in the hospital [54]. However, CEA is not specific to colon cancer, and thus leaves room for new, more specific markers. Lee et al. analyzed exosomes for the presence of distinguishing protein markers [54]. They used two colon cancer cell lines HT-29 and HCT-116 and one colon epithelial cell line from ATCC, CRL-1541 for analyzing the presence of distinguishing protein markers. They identified exosomal tetraspanin 1 (TSPAN1) as a potential marker for colon cancer detection. In patient plasma samples, this protein exhibited a sensitivity of 75.7%. It is possible that TSPAN1 in conjunction with CEA may provide a new diagnostic marker for colon cancer prognosis and diagnosis [54].
While the incidences of certain cancer types have exhibited an overall decline, other types, such as hepatocellular carcinoma (HCC), have increased by 3% each year since 2000 [48]. About 700,000 patients are diagnosed each year with hepatocellular carcinoma and results in 600,000 deaths per year. Additionally, HCC is often diagnosed in later stages, resulting in poor patient prognosis and outcome regardless of therapy [55].
Xue et al identified a potential biomarker of 8 exosomal miRNAs consisting of miR-17-5p, miR-29a, miR-106a, miR-122, miR-125b, miR-145, miR-192, and miR-194. Of these, they found that miR-17-5p, miR-106a, and miR-194 exhibited a correlation between expression and tumor size. Additionally, miR-106a, on its own, was correlated with a lower survival rate due to its involvement in tumorigenesis. [55]. There is therefore potential for this biomarker to be used as both a diagnostic and prognostic indicator for treatment outcome.
Pancreatic ductal adenocarcinoma (PDAC) comprises most pancreatic cancer types. It is a highly aggressive and metastatic type of tumor with a five-year survival rate of 7% [56]. Current methods of diagnosing pancreatic cancer involve imaging scans and invasive surgery to confirm the diagnosis. There is currently only one serum marker test for pancreatic cancer, sialylated Lewis (a) blood group 19-9 (CA19-9). However, CA19-9 is variable in its sensitivity and specificity, with a range of 60%−90% and 68%−91%, respectively [57]. Additionally, CA19-9 levels are increased in other gastrointestinal diseases and may not be expressed in asymptomatic pancreatic cancer patients [58]. This makes identification of new and early biomarkers for pancreatic cancer detection vital for positive patient outcome.
Glypican-1 (GPC1) is a cell-surface proteoglycan enriched on pancreatic cancer cell derived exosomes. It was originally identified as a promising target for early pancreatic cancer diagnosis [59]. A follow-up study to determine the efficacy of GPC1 as an early marker was conducted by Zhou et al [60]. They monitored PDAC patients over the course of five years and found that CA19-9 was significantly better than GPC1 in distinguishing PDAC patients from healthy patients. However, they observed that high levels of GPC1 in serum were correlated with a shorter overall survival rate. They concluded that GPC1 may be a prognostic indicator rather than an effective diagnostic.
Another study compared a panel of exosomal mRNA and small nucleolar RNA (snoRNA) to the ability of CA19-9 to distinguish between pancreatic cancer patients and healthy controls. This study found that the expressions of WAS Protein Family Member 2 (WASF2), ADP Ribosylation Factor 6 (ARF6) Small Nucleolar RNA, H/ACA Box 74A (SNORA74A), and Small Nucleolar RNA, H/ACA Box 25 (SNORA25) were able to distinguish between pancreatic cancer patients and healthy patients with a greater than 0.90 value for a ROC curve compared to CA19-9. They found that of their four-gene panel, WASF2 showed higher correlation with pancreatic cancer risk even when compared to CA19-9. Exosomal WASF2 may therefore provide a novel method for predicting a patient’s risk for developing pancreatic cancer [61].
Recently, a bioinformatics analysis identified several potential exosomal targets for pancreatic cancer diagnosis [15]. The study incorporated meta-analysis tools such as GeneCards, variant analyses, and pathway mapping to identify these putative leads. The study identified 575 protein coding genes, 26 miRNAs, one pseudogene, one long non-coding RNA, and one antisense gene directly associated with pancreatic cancer. Some of the miRNAs identified in this analysis such as miR-21, miR-34a, miR-155, miR-210, and miR-221 have previously been described as causally linked to pancreatic cancer progression [15]. Additionally, nine open reading frames were identified. Many of these targets were readily identified in exosomes found in blood, plasma, saliva, and serum, which marks them as having strong diagnostic potential. This study provides a framework with which to develop a potentially new diagnostic, and as such, further work is required to test the efficacy of the predictions laid out in this study [15].
Memorial Sloan Kettering Cancer Clinic is currently running two clinical trials. One to test the efficacy of diagnostic exosomes in colon and liver cancers, NCT03432806 [62], and pancreatic cancer, NCT03334708 [63]. Both are looking to identify biomarkers circulating in venous blood as well as analysis of respective tissues. These studies are currently recruiting. Another study, NCT03032913, was focusing on the efficacy of GPC1+ exosomes along with CA19-9 as a PDAC diagnostic. At the conclusion of this work, they found that GPC1+ exosomes could better predict PDAC in conjunction with CA19-9 than either marker on their own [64].
3. Using Exosomal Markers for Cancer Detection and Patient Monitoring
Exosomal markers are attractive targets for the cancer detection arsenal because of their increased levels in body fluids. In fact, it is well-established that tumors, and even other types of diseases, release greater numbers of exosomes compared to healthy cells, which alone may be indicative of tumor burden or presence of disease [65–67]. Though exosomes can be harvested from all body fluids, plasma is the preferred medium for detecting most solid tumors [66]. To that end, many studies analyzing plasma exosomes have observed a correlation between the number of plasma exosomes and tumor burden as well as a drop in exosome concentration post tumor treatment or removal [12,68–72]. For example, a pilot study examined CD63 and caveolin 1 (CAV1) expressing exosomes in stage IV oral squamous cell carcinoma (OSCC) patients [73]. Increase in CAV1 expression is associated with tumor progression in oral cancers, however, when oral tumors were resected, CAV1 expressing exosomes increased. On the other hand, CD63-expressing exosomes decreased within a week post-resection. In both cases, they found that lower levels of CAV1 and CD63-expressing exosomes were associated with longer term survival. This work thus supports utilizing exosomal markers for monitoring patients undergoing treatments and for determining patient prognosis.
Exosomal markers for cancer diagnosis and patient monitoring are important if the field of cancer medicine is to move forward. At present, routine screening for breast, cervical, colorectal, endometrial, lung, and prostate cancers are offered in the United States [74]. Regardless, new methods of screening that utilize exosomes may prove beneficial not just for confirming those tumor types with established routine screenings, but also for those tumors which do not yet have routine or reliable screening methods.
Exosomal-based diagnostics could provide an additional measure of verifying the presence of a concerning lesion as imaging scans are not always capable of distinguishing between what is benign or malignant [75]. For example, prostate cancer patients will often exhibit higher levels of prostate-specific antigen (PSA) in their blood, even in the presence of benign conditions [69]. However, it was observed that only patients with prostate tumors exhibited greater levels of CD81 and PSA-expressing exosomes in blood plasma [69]. A follow up study analyzed the plasma from prostate cancer patients (N=80), patients with benign prostatic hyperplasia (BPH, N=80), and healthy donors (N=80) [76]. The levels of CD81 and PSA-expressing exosomes were measured across the patient sampling. This clinical analysis was able to differentiate between prostate cancer patients and healthy donors with a sensitivity and specificity of 100%. They also found this method was able to distinguish between prostate cancer patients and patients with BPH with a sensitivity of 80% and a specificity of 98%. In both cases, PSA and CD81-expressing exosomes offer better diagnostic capability than serum PSA levels alone [77].
Likewise, another study examined exosomes as a confirmative for suspected ovarian cancer diagnosed via imaging and CA-125 expression levels [71]. They found that patients with ovarian cancer exhibited much higher levels of phosphatidylserine (PS)-positive exosomes compared to the controls [71]. A similar study used PS-labeled exosomes for detection of breast and pancreatic cancers [78]. They found an increase in tumor exosome release in early disease onset, even before clinical evidence of the tumors [78]. These studies suggest a strong potential for a pan-cancer exosome-based diagnostic.
Interestingly, studies have also found that the acidic tumor environment may contribute to the increased release of exosomes which also possess a pH lower than exosomes from healthy cells [70,79–81]. It may therefore be possible to determine the presence of cancer within a patient based off the pH of the exosomes alone. These studies thus suggest the clinical potential of exosomes for detecting tumors, confirming imaging scans, and monitoring tumor burden in patients undergoing treatment.
4. Limitations for the Use of Exosomal Biomarkers
4.1. Isolation of Exosomes
Cost effective and rapid isolation and purification of exosomal-biomarkers remains the main obstacle for their use in clinical application. There are several methods for isolation of exosomes as well as commercially available kits. Some of the more common methods include ultracentrifugation, density-gradient ultracentrifugation immunoprecipitation, ultrafiltration and size-exclusion chromatography [82,83].
Ultracentrifugation is a time-consuming method which can damage the exosomes, result in poor RNA yield, and have low reproducibility [84–86]. Density-gradient ultracentrifugation relies on developing a density gradient by using either sucrose or iodixanol and allowing the extracellular vesicles to collect at a particular density upon lengthy centrifugation. Use of iodixanol appears to be more successful at separating exosomes from viral particles than sucrose [84,87,88]. While this method may yield higher purity, it takes several days and results in sample loss of the larger exosomes, leaving only those which range in size from 50-100 nm [84,89,90]. Ultrafiltration is a method that involves the use of a series of decreasing microfilters from 0.8 to 0.1 μm [84,91]. This procedure takes just over two hours and yields pure samples at varying sizes. However, the filters can clog, and the sample is subject to protein contamination and deformation of the exosomes. Immunoprecipitation involves using antibodies specific for exosome receptors such as EpCAM, CD9, CD63, and TSG101 to name a few [83,84,92]. This process takes about four hours and yields high purity and selectivity of exosomes but is costly and may result in nonspecific binding of other molecules in the sample. Size-exclusion chromatography, also known as gel filtration, is effective for rapid processing and high yield and purity, though it does rely on ultrafiltration or ultracentrifugation to first eliminate impurities [93,94]. However, the method is costly and requires specialized equipment. Commercial kits eliminate the need for complex techniques and take about an hour to complete, with some kits requiring one overnight step. They tend to preserve the integrity of the exosomal structures but are costly and lack reliable reproducibility [84].
4.2. Conventional Methods for Detecting and Quantifying Exosomes
Once exosomes are isolated, they need to be analyzed and quantified. There are several methods for quantifying exosomes including enzyme-linked immunosorbent assay (ELISA), flow cytometry, and nanoparticle tracking analysis (NTA) [95]. Boriachek et al. has summarized these methods [95]. ELISA is a plate-based assay that utilizes specific antibody-antigen interactions for capturing specific proteins and typically produces a color change that correlates with concentration of the target. ELISAs for exosomes incorporate exosome-specific markers such as CD9, CD63, or CD81 and their antibody for capture out of patient serum or plasma [96]. For more specific capture of tumor-derived exosomes, the assay can incorporate tumor markers such as caveolin-1, GPC-1, HER2, or HSP90 [97–100]. Despite these benefits, ELISA still suffers from a fairly high degree of “biological noise” [95].
Flow cytometry is another common method for detecting and quantifying exosomes. Flow incorporates the use of fluorescent labeling and light scattering to detect and quantify the particles. A modification of traditional flow, called FACS, utilizes fluorescent labeled antibodies to sort specific exosomes. This can streamline identification and isolation of exosome species. However, this requires quite a complex setup. A major disadvantage of flow is a lack of consistent results as the sensing of the particles is dependent on the optical settings and laser setup. Another issue is that flow has difficulty in detecting the smaller exosomes. Lastly, it requires expensive equipment that is not always feasible for clinical settings or resource limited areas [95].
NTA is another method that relies on fluorescent tagging to identify and sort exosomes. It uses a laser beam to track the particles and determine size by their movement. These data points are analyzed by a computer and presented in a graph for visualization. NTA’s main advantage overflow cytometry is its ability to detect exosomes on the lower end of their accepted size range. However, NTA is incapable of determining the biochemical composition of exosomes. This coupled with a long analysis time compared to flow renders NTA disadvantageous for clinical application [95,101].
4.3. Clinical Limitations
For clinical application, exosomes must be rapidly isolated with minimal contaminants and deformities in order to ensure purity. Ultracentrifugation remains the gold standard for exosome isolation and purity though it is the most time-consuming method. Newer, more rapid, methods for isolating exosomes need to be developed if exosomes are to become appealing for clinical use.
These methods for detecting and quantifying exosomes have their advantages and disadvantages for clinical application. For example, ELISA takes time to prepare but once the assay is ready, a rapid colorimetric response can be observed for a positive result. However, ELISA is susceptible to contamination by other biomolecules in the patient fluid being analyzed. Flow cytometry can detect various exosomes and sort them by species but suffers from needing expensive equipment and production of inconsistent results. NTA also requires expensive equipment and is incapable of identifying the type of exosome it detects.
For a technique to be clinically applicable, it must be cost effective and minimize the use of expensive equipment. Perhaps more importantly, a method must be conclusive and consistent, qualities the methods discussed have difficulty achieving. Thus, a method which removes the need for costly equipment and ensures purity with minimal time would be ideal. These limitations are currently being addressed by the development of new methods of sensing and detection including electrochemical detection, microfluidics, and nano-sensors.
5. Overcoming Limitations of Exosome Diagnostic Use in the Clinic
Harvesting tumor-derived exosomes from body fluids can be laborious and costly. However, microfluidic chips and other nano-sensing technologies have shown to be useful in a variety of point-of-care scenarios encompassing cell sorting, fertility, DNA and protein sensing, viral detection, drug delivery, cancer diagnostics, and tumor modeling [102,103,112–114,104–111]. It is therefore likely that such devices will overcome the limitations that current methods exhibit for clinical application. In this section, we highlight a selection of electrochemical biosensors, nanosensors, and microfluidic chips which have been developed.
5.1. Electrochemical Biosensors
One of the more common types of biosensors are electrochemical sensors. Common sensors currently used in the clinic include blood glucose monitors and respiratory CO2 monitors. Electrochemical detection sensors utilize antibodies to interact and capture antigens. The concentration of captured antigens is determined by changes in the electric current [95,115]. This principle is easily applied to exosomes [116].
Several electrochemical devices have been developed for capture and analysis of tumor exosomes. One such device is called iMEX (integrated magnetic-electrochemical exosome) [117]. IMEX incorporates magnetic beads functionalized with the common exosome tetraspanins CD9, CD63, and CD81. The system utilizes magnets to pull these magnetic bead-captured exosomes to the electrodes for electromagnetic sensing. To determine if the device could provide clinical benefit, they used beads functionalized with ovarian cancer-specific exosome marker antibodies CD63, CA-125, EpCAM, and CD24. Through this method, iMEX was able to detect ovarian cancer exosomes from 10 μL of patient blood in about an hour’s time. Additionally, they tested the same patients post-treatment two months later and found decreased levels of EpCAM and CD24 in the patients who responded to the treatment [117].
Another method followed a similar method, using immunomagnetic sensing to capture breast cancer exosomes [118]. They used magnetic particles functionalized with antibodies for exosome tetraspanins and cancer-specific tetraspanins CD24, CD44, CD326, and CD340. The beads were isolated by a magnetic tube separator and transferred to a magnetic electrode where amperometry was used for sensing. Through this method, they identified a LOD of 105 exosomes/μL−1 in breast cancer patient serum [118].
Another electrochemical biosensor utilized CD9 antibodies functionalized to gold electrodes [119]. The CD9 labeled exosomes were captured out of MCF7 breast cancer cell culture media and then tagged with mouse IgG antibody-HRP conjugate. The electrochemical reduction of TMB by HRP was then observed. This device boasts a high degree of sensitivity with a limit of detection of 2×102 particles/μL using a minimal volume of 1.5 μL of sample [119]. As is, the device can distinguish exosomes from other extracellular vesicles. Further studies using specific breast cancer exosomal markers are needed to establish the potential of this device for clinical application. Regardless, this initial data exhibits a great deal of potential for clinical use.
Electrochemical sensors can also be applied to exosomal miRNA. Detection of exosomal miRNA is costly and requires time-consuming methods of purifying exosomes, lysing them, isolating miRNA, and then RT-qPCR analysis of the miRNA targets. In order to use exosomal miRNAs in the clinic, it is necessary to determine other methods of quantifying target miRNAs that eliminate the need for expensive equipment and time-consuming steps. Therefore, developing alternative methods of quantifying tumor-related exosomal miRNAs has been of interest. A few studies have developed a means of electrochemical detection of these miRs that eliminate much of these issues [120–122].
Two studies for electrochemical sensing of miRNAs, conducted by the same lab, involve the use of locked-nucleic acids (LNA) for capture of miR-21 from MCF-7. The first study utilizes a bipedal DNA walker and LNA to increase effectiveness of binding to the miRNA sequence [120]. Gold nanoprobes were functionalized with hairpin probes while the DNA walkers were labeled with FAM. The LNA provided a means for the walker to partially bind and upon introduction of miR-21, the walker is released. The walker moves along the first methylene blue hairpin probe, allowing it to interact with a second ferrocene labeled probe, releasing the ferrocene from probe 2 and the methylene blue (MB) from probe 1. Together, the probes guide the walker until it connects to the next miR-21. This system is capable of quantifying the miR-21 based on the electrochemical ratio of ferrocene to methylene blue [120]. Using breast cancer patients and serum from patients without breast cancer, they found their system detected 2.5 fold change higher levels of miR-21 in breast cancer patients than in the control [120].
The second LNA system utilizes two probes, one labeled with methylene blue and the other with ferrocene attached to glassy carbon electrodes [121]. Upon introduction of miR-21, the ferrocene labeled probe could functionalize with miR-21, releasing the methylene blue labeled probe. This would result in the MB probe folding into a hairpin structure and resting close to the electrode, resulting in a change of electric current. This method exhibited a close similarity to RT-qPCR values when exosomal miR-21 from MCF7 was analyzed, suggesting an alternative to RT-qPCR [121]. Further work encompassing the use of patient samples is necessary, however, to determine the efficacy of this method as a potential diagnostic.
DNA walkers have been previously described for identifying CD63 exosomes, not just exosomal miRNA, using CD63 and EpCAM aptamers for MCF7 exosome recognition [123]. This exosome capture and ratiometric system was analyzed with healthy patients and breast cancer patients. The breast cancer patients exhibited nearly 100 times more exosomes than the healthy patients [123].
A third method, called SEEmiR, uses peptide nucleic acids (PNA) for interaction with spherical nucleic acid (SNA) nanoprobes [122]. The PNA is a probe with a neutral peptide backbone that captures the target miR-21. The neutral peptide backbone decreases the charge of the gold electrode, allowing miR-21 to bind more easily to the probe. The miR-21 prehybridizes with the SNA, which is very negatively charged, providing a more sensitive binding to the PNA-functionalized electrode. When they tested SEEmiR with plasma from breast cancer patients, they found a significant difference in electrochemical responses between breast cancer patients and healthy patients [122]. In general, electrochemical biosensors may provide a more cost-effective method for detecting exosomal miRNAs.
Biosensors are ideal for clinical setting. They have the potential for highly specific and sensitive real-time detection with the added benefit of low limits of detection. This makes them ideal for monitoring patients transitioning through disease stages as well as monitoring patients as they undergo treatment [115,124]. Biosensors are also portable and cost-effective, which adds to their appeal. The disadvantage is that initial production and fabrication is complex. Special care needs to be taken when identifying the appropriate markers as well as ensuring precision in design to ensure reproducibility of the assays [95,124]. Despite this, these types of sensors have the potential to overcome the current hurdles of exosome-based diagnostics.
5.2. Mircrofluidics
Microfluidic chips also offer potential for overcoming the hurdles observed in clinical use of exosomes for cancer diagnosis and monitoring. Microfluidic chips have the potential for cost-effective and rapid application. Additionally, they require minimal amounts of sample. Microfluidic chips can have plastic (PDMA or PMMA) or silicon designs with multiple channels for isolating, washing, processing, and analyzing samples. The major drawback, however, is they typically require skilled individuals to use and may have multiple steps with limited automation [95]. Regardless, they offer a potentially rapid and cost-effective analysis of tumor exosomes.
It has been previously stated that specific surface markers distinguish exosomes from other extracellular vesicles such as CD63, MHCI, and CD83 [125–127]. The device used by Fang et al. uses CD63 and its antibody functionalized to magnetic beads. These beads are incubated overnight with either cell culture media from breast cancer and normal cell lines or plasma from breast cancer and healthy patients [125]. To analyze these captured exosomes, the solution was placed on the chip and controlled via syringe pump. On the chip, the exosomes were mixed with MHCI antibody to isolate specific tumor-associated exosomes and then subjected to immunofluorescent staining for epithelial cell adhesion molecule (EpCAM) and human epidermal growth factor 2 (HER2). This method was shown to exhibit a significant difference between healthy patients and breast cancer patients. While this particular chip requires an overnight step, the overall processing time is an improvement from other methods. Additionally, it requires a significantly smaller sample and appears to provide high yield and purity of breast cancer-specific exosomes [125].
In another study, magnetic beads functionalized with mouse T-cell immunoglobulin mucin protein 4 (Tim4) antibody were used to capture exosomes derived from hepatocellular tumors [128]. The functionalizing of the beads with Tim4-Fc antibody took 20 minutes to complete, thereby eliminating an overnight step. The chip itself incorporated Y-shaped PDMS micropillars, to form exosome capture microchambers, encased in indium tin oxide (ITO) glass electrodes. To test the ability of the chip to pull exosomes out of solution, the Tim4-antibody beads were added to the microchambers. Exosomes were isolated from cell culture media using a 0.22 μm filter to remove larger vesicles and then further processed via the commercially available ExoEasy Maxi Prep kit. The purified exosomes were then added to the chip. During the initial washing to remove non-specific exosomes, a magnet was used to hold the functionalized beads in place. To detect the exosomes, the magnet was removed, and the captured exosomes were washed into the detection area where they were immobilized once again by a magnet and tagged with Dil, a lipophilic membrane stain for fluorescent imaging. They repeated this using functionalized single-stranded DNA probes for fluorescent imaging and found just as much success in imaging the exosomes caught by the chip. Finally, when they used this device to detect liver cancer-derived exosomes from patients, they were able to process the sample within 3.5 hours using only 30 μL of patient serum with the capability of distinguishing cancer patients from healthy controls [128]. Additionally, when used for patient samples, the only pre-processing of the sample involved centrifuging to remove cells and cellular debris, thereby decreasing cost and money as the commercial kit is not necessary.
Identifying exosomes is one method for diagnosing different types of tumors, however, the contents of the exosomes may be more specific. Two chips were developed for this purpose to detect pancreatic cancer [129]. The cell line PANC1 was used and its media was collected for exosomal miRNA analysis. Upon removal of cellular debris, the cell culture media was first introduced at approximately 100 μL into the first chip. This first microfluidic device incorporated a surface acoustic wave (SAW) device to lyse the exosomes. The lysate was then collected and transferred to the second microfluidic chip device, which was functionalized with a miR-550 probe to capture the target miRNA. The second microfluidic device relies on an ion-exchange nanomembrane sensor to detect alterations in voltage current between hybridized hsa-miR-550 with its probe and the hsa-miR-550 probe on its own. This method is reported to take approximately 1.5 hours, which is a vast improvement over the current methods of exosomal RNA isolation and analysis [129].
5.3. Nanosensors
Nanosensors are a third application for overcoming the limitations of exosome-based diagnostics and monitoring in the clinic. They offer the potential for real-time monitoring and identification of multiple markers. Nanosensors can also be cost-effective while still sensitive as they usually incorporate other methods such as microfluidics, electrochemical sensing, and optics. As with the previous methods discussed, however, nanosensors suffer from reproducibility issues due to differences in fabrication from one batch to the next [130,131].
Nanosensors are diverse and can range from optical sensing to electrochemical sensors. One nanodevice for tumor exosome detection incorporates electrochemical sensing into its design. This device utilizes platinum electrodes functionalized with EpCAM antibodies for tumor-exosome capture and an additional EpCAM antibody tagged with a reporter for signal amplification [132]. The amplification occurs when the reporter interacts with streptavidin-conjugated alkaline phosphatase (ALP), causing an enzymatic reaction that acts as the first amplification signal. The second amplification signal is determined by a redox reaction of a byproduct, p-aminophenol (pAP), from the previous step as it shuttles between the anode and cathode, converting to para-quinone imine. This shuttling between electrodes and reduction produces a steady state current that is proportional to the concentration of exosomes. In this way, their device was able to detect tumor-derived exosomes as low as 10 exosomes per μL [132]. The authors do report a need for optimization of this device and streamlining of its design but do believe it has potential for point-of-care application [132]. Additionally, they used prostate cancer cell line exosomes and would thus require patient samples to determine the effectiveness of this method for point-of-care application.
Nanosensors incorporating localized surface plasmon resonance imaging (LSPRI) offer high sensitivity and portability for tumor exosome detection. This device was fabricated using gold and quartz nanopillars, each approximated to the same size as single exosomes to increase single exosome detectability [133]. The pillars were then functionalized with CD63 antibody for MCF7 breast cancer cell line exosome capture. With this device, they were able to detect single exosomes from sub-femtomolar concentrations [133]. The ability to detect single exosomes could be quite useful in distinguishing a tumor-specific exosome amidst the rest of circulating exosomes from healthy cells. This device would therefore prove invaluable in a clinical setting. However, further work is necessary to test this device using patient samples with tumor-specific antibodies such as HER2 or EpCAM.
Another device using surface plasmon resonance (SPR) to detect CD9 and CD63 exosomes with the ability to isolate clinically relevant exosome subtypes such as those expressing HER2 has been reported [134]. This group developed a SPR device that utilized CD9 and CD63 antibodies for exosome capture. Once captured, an additional antibody for HER2 was added to identify breast cancer-derived exosomes. In this way, they obtained a limit of detection of 2070 exosomes/μL. Additionally, they were able to observe a difference in spectral shift between exosomes captured from breast cancer patients (> 0.3 nm) and those from healthy patients (< 0.2 nm) [134].
A third optical nanosensor was developed that does not require light excitation to engage a response [135]. It incorporates a near-infrared semiconducting polyelectrolyte that interacts with a quencher-tagged aptamer for specific exosome populations. Upon complexing with the specific exosome, the quencher releases, producing and amplifying both a fluorescent signal and an afterglow. To overcome the issues with background fluorescence, they measured the afterglow and found it to be more reliable. They suggest the afterglow could be beneficial in measuring exosome levels with minimal purification steps [135]. This would inadvertently save time and cost when considering this method for clinical application. They tested this device with tumor aptamers for EpCAM, HER2, and MUC1 across five different cell lines (chondrocyte, HeLa, MCF7, SKOV3, and HepG2) [135]. In this combination, they found that exosomes from all cell lines contained these markers but in varying concentrations and they were thus able to distinguish between the different cell lines [135]. The sensor does require equipment to read the fluorescence and afterglow, but it’s potential for distinguishing cancer types based off concentration of specific markers suggests a potential pan-cancer diagnostic.
The issues surrounding use of exosome-based diagnostics in the clinic remain a hurdle. Exosome isolation is time-consuming and often requires costly kits or equipment. However, nanodevices may provide an alternative as they can provide a cost-effective and sensitive means of real-time detection for circulating tumor exosomes. Additionally, these devices may be useful for multi-tumor detection platforms.
6. Conclusion
In the last decade, there has been a rapid increase in interest over the use of exosomes as diagnostic tools. There are several laboratories and research groups focusing on the optimization of exosome-based diagnostics. This is because tumors release a greater number of exosomes compared to healthy cells. These tumor-specific exosomes will also exhibit tumor-specific surface markers as well as traffic tumor-specific DNA, RNA, and proteins throughout the body. Additionally, exosomes show promise in monitoring and predicting patient response to therapy [12,13,136,137]. With such beneficial traits, it’s no wonder that exosomes have become a focus for updating the current repertoire of tumor diagnostics. This is evident in the 37 clinical trials listed on the NIH clinical trials website, all of which are testing the diagnostic efficacy of tumor-derived exosomes.
However, for exosome-based diagnostics to become prevalent in the clinic, it is necessary to overcome the issues with yield, purity, and time. Current isolation techniques can be laborious, which makes them unattractive for use by physicians. Additionally, these limitations are likely to result in high cost to the patient which can limit the feasibility of such a diagnostic. Microfluidic chips and other types of nanosensing devices may provide the ability to overcome the cost and labor necessary for processing exosomes from body fluids. The biggest hurdle with using nanodevices and microfluidics, however, is their wide range of design. There is currently no standard for such devices and many struggle with consistency in fabrication. Some also require expensive equipment while others lend themselves well to portability and clinical use. Despite these disadvantages, there are many devices, each attempting to mitigate the issues in design and reproducibility while optimizing sensitivity and specificity. It is reasonable, therefore, to expect an exosome-based diagnostic to become widely used in the clinic as the field progresses and optimizes.
7. Expert Opinion
Liquid biopsies have emerged as a less invasive and alternative means of diagnosing various tumors in the last 20 years (95-97). They have great potential for diagnosing and monitoring patients, in particular those who exhibit inoperable or difficult to access tumors [141]. Liquid biopsies encompass circulating tumor cells (CTCs), cell-free DNA (cfDNA)/circulating tumor DNA (ctDNA), circulating miRNAs, and exosomes which can be harvested from various body fluids. Some of these biopsy types, such as CTCs and cf/ctDNA, may offer a more cost-effective method for identification of mutation and tumor due to advancements in next generation sequencing. However, cfDNA is usually degraded within a few hours, though in cancer patients the concentration can range up to 1000 ng/mL [141]. Meanwhile, CTCs are minimal in the blood, ranging from 1-10 CTCs per mL [142]. Exosomes, on the other hand, provide a more ample and quantifiable liquid biopsy with about 90% of cfDNA found within exosomes [143,144]. Additionally, tumor-derived exosomes possess a multitude of deregulated proteins and RNAs [145,146]. And with numbers of exosomes ranging into the hundreds of millions per mL of blood, exosomes may thus provide a more beneficial liquid biopsy diagnostic [147].
Even though tumors arising from a specific organ will harbor similar mutations within and between patients, individual polymorphisms can and often do result in differences of cellular expression [148]. Thus, as precision and personalized medicine becomes more feasible for the future, the need for tumor biomarker panels is imperative. Exosomes are known to hold several deregulated markers specific to a given tumor type and it is the opinion of the authors that such a diversity of markers is likely to prove beneficial for diagnosing a particular tumor across several patients exhibiting different polymorphisms. The issue with any new technology is, of course, the optimization of usage and its cost-effectiveness. Despite how attractive exosomes may be for diagnosing tumors, optimization of harvesting and analysis are necessary as the current methods are expensive, time consuming, and laborious. However, there are several laboratories hard at work developing such devices, which suggests a very promising future for their widespread usage in the clinic.
Despite the limitations of current methods for harvesting and analyzing exosomes, there are currently several clinical trials testing the efficacy of exosomes for monitoring patients as they progress through their cancer treatments using various chip and nano technologies as well as more traditional methods of exosome profiling. In addition, the field continues to grow as kits are produced to streamline the process of isolating and analyzing exosomes and older methods are improved upon. Thus, the future of exosomes for use in the clinic remains promising.
Acknowledgments
Funding
Florida Atlantic University
FAU Faculty Mentoring Award
Institute for Sensing and Embedded Networking Syst
Start-up support from FAU College of Engineering
U.S. Department of Health and Human Services
National Institutes of Health
NIH R15AI127214
Footnotes
Declaration of interest
The authors have no other relevant affiliations or financial involvement with any organization or entity with a financial interest in or financial conflict with the subject matter or materials discussed in the manuscript apart from those disclosed.
Reviewers Disclosure
Peer reviewers on this manuscript have no relevant financial relationships or otherwise to disclose.
References
Papers of special interest has been highlighted as (*= of importance, **= of considerable importance)
- [1].Egan TK. Monitoring Patients Undergoing Cancer Therapy. Lab. Med [Internet]. 2000. [cited 2018 Dec 1];31:666–671. Available from: https://academic.oup.com/labmed/article-lookup/doi/10.1309/R078-Y40Q-PAJP-1RPP. [Google Scholar]
- [2].Graham LJ, Shupe MP, Schneble EJ, et al. Current approaches and challenges in monitoring treatment responses in breast cancer. J. Cancer [Internet]. 2014. [cited 2019 Aug 23];5:58–68. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24396498. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Neal RD, Tharmanathan P, France B, et al. Is increased time to diagnosis and treatment in symptomatic cancer associated with poorer outcomes? Systematic review. Br. J. Cancer [Internet]. 2015. [cited 2019 Aug 2];112 Suppl:S92–107. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25734382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Car LT, Papachristou N, Urch C, et al. Preventing delayed diagnosis of cancer: clinicians’ views on main problems and solutions. J. Glob. Health [Internet]. 2016. [cited 2019 Aug 2];6:020901 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28028437. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Cassim S, Chepulis L, Keenan R, et al. Patient and carer perceived barriers to early presentation and diagnosis of lung cancer: a systematic review. BMC Cancer [Internet]. 2019. [cited 2019 Aug 2];19:25 Available from: http://www.ncbi.nlm.nih.gov/pubmed/30621616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Huang T, Deng C-X. Current Progresses of Exosomes as Cancer Diagnostic and Prognostic Biomarkers. Int. J. Biol. Sci [Internet]. 2019. [cited 2019 Aug 2];15:1–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30662342. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Soung YH, Ford S, Zhang V, et al. Exosomes in Cancer Diagnostics. Cancers (Basel). [Internet]. 2017. [cited 2019 Aug 2];9 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28085080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zöller M Exosomes in Cancer Disease. Methods Mol. Biol [Internet]. 2016. [cited 2019 Aug 2]. p. 111–149. Available from: http://www.ncbi.nlm.nih.gov/pubmed/26667458. [DOI] [PubMed] [Google Scholar]
- [9].Jalalian SH, Ramezani M, Jalalian SA, et al. Exosomes, new biomarkers in early cancer detection. Anal. Biochem [Internet]. 2019. [cited 2019 Aug 2];571:1–13. Available from: https://www.sciencedirect.com/science/article/pii/S0003269718310595. [DOI] [PubMed] [Google Scholar]
- [10].Xiong G, Feng M, Yang G, et al. The underlying mechanisms of non-coding RNAs in the chemoresistance of pancreatic cancer. Cancer Lett [Internet]. 2017/03/04 2017;397:94–102. Available from: https://www.ncbi.nlm.nih.gov/pubmed/28254409. [DOI] [PubMed] [Google Scholar]
- [11].Meng Y, Sun J, Wang X, et al. Exosomes: A Promising Avenue for the Diagnosis of Breast Cancer. Technol. Cancer Res. Treat [Internet]. 2019. [cited 2019 Aug 2];18:153303381882142 Available from: http://journals.sagepub.com/doi/10.1177/1533033818821421. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Theodoraki M-N, Yerneni S, Gooding WE, et al. Circulating exosomes measure responses to therapy in head and neck cancer patients treated with cetuximab, ipilimumab, and IMRT. Oncoimmunology [Internet]. 2019. [cited 2019 Aug 2];8:1593805 Available from: http://www.ncbi.nlm.nih.gov/pubmed/31143513. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Nardin C, Cordonnier M, Chanteloup G, et al. Circulating PD-L1-exosomes to monitor tumor response in melanoma patients. J. Clin. Oncol [Internet]. 2019. [cited 2019 Aug 2];37:9517–9517. Available from: https://ascopubs.org/doi/10.1200/JCO.2019.37.15_suppl.9517. [Google Scholar]
- [14].Kim J-H, Kim E, Lee MY. Exosomes as diagnostic biomarkers in cancer. Mol. Cell. Toxicol [Internet]. 2018. [cited 2019 Aug 19];14:113–122. Available from: http://link.springer.com/10.1007/s13273-018-0014-4. [Google Scholar]
- [15].Makler A, Narayanan R. Mining exosomal genes for pancreatic cancer targets. Cancer Genomics and Proteomics. 2017;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nedaeinia R, Manian M, Jazayeri MH, et al. Circulating exosomes and exosomal microRNAs as biomarkers in gastrointestinal cancer. Cancer Gene Ther. [Internet]. 2017. [cited 2019 Aug 19];24:48–56. Available from: http://www.nature.com/articles/cgt201677. [DOI] [PubMed] [Google Scholar]
- [17].Boukouris S, Mathivanan S. Exosomes in bodily fluids are a highly stable resource of disease biomarkers. Proteomics. Clin. Appl [Internet]. 2015. [cited 2019 Aug 2];9:358–367. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25684126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Falcon-Perez J Exosome profiling: potential in cancer diagnosis and stratification. Endocr. Abstr [Internet]. 2017. [cited 2019 Aug 3]; Available from: http://www.endocrine-abstracts.org/ea/0049/ea0049nsa2.htm. [Google Scholar]
- [19].Fisher R, Pusztai L, Swanton C. Cancer heterogeneity: implications for targeted therapeutics. Br. J. Cancer [Internet]. 2013. [cited 2019 Aug 7];108:479–485. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23299535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Jamal-Hanjani M, Quezada SA, Larkin J, et al. Translational implications of tumor heterogeneity. Clin. Cancer Res 2015;21:1258–1266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Seoane J, De Mattos-Arruda L. The challenge of intratumour heterogeneity in precision medicine. J. Intern. Med [Internet]. 2014. [cited 2019 Aug 23];276:41–51. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24661605. [DOI] [PubMed] [Google Scholar]
- [22].Stanta G, Bonin S. Overview on Clinical Relevance of Intra-Tumor Heterogeneity. Front. Med [Internet]. 2018. [cited 2019 Aug 7];5:85 Available from: http://journal.frontiersin.org/article/10.3389/fmed.2018.00085/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [23].Domenyuk V, Zhong Z, Stark A, et al. Plasma Exosome Profiling of Cancer Patients by a Next Generation Systems Biology Approach. Sci. Rep [Internet]. 2017. [cited 2019 Aug 3];7:42741 Available from: http://www.nature.com/articles/srep42741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Oliveira-Rodríguez M, López-Cobo S, Reyburn HT, et al. Development of a rapid lateral flow immunoassay test for detection of exosomes previously enriched from cell culture medium and body fluids. J. Extracell. Vesicles [Internet]. 2016. [cited 2019 May 30];5:31803 Available from: https://www.tandfonline.com/doi/full/10.3402/jev.v5.31803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [25].Zhao X, Ren Y, Cui N, et al. Identification of key microRNAs and their targets in exosomes of pancreatic cancer using bioinformatics analysis. Medicine (Baltimore). 2018;97:e12632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Cocucci E, Meldolesi J. Ectosomes and exosomes: shedding the confusion between extracellular vesicles. Trends Cell Biol. [Internet]. 2015. [cited 2018 Dec 12];25:364–372. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25683921. [DOI] [PubMed] [Google Scholar]
- [27].Raposo G, Stoorvogel W. Extracellular vesicles: Exosomes, microvesicles, and friends. J. Cell Biol [Internet]. 2013. [cited 2019 Aug 2];200:373–383. Available from: http://www.ncbi.nlm.nih.gov/pubmed/23420871. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [28].Willms E, Johansson HJ, Mäger I, et al. Cells release subpopulations of exosomes with distinct molecular and biological properties. Sci. Rep [Internet]. 2016. [cited 2019 Aug 2];6:22519 Available from: http://www.nature.com/articles/srep22519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Meldolesi J. Exosomes and Ectosomes in Intercellular Communication. Curr. Biol [Internet]. 2018. [cited 2019 Aug 2];28:R435–R444. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29689228. [DOI] [PubMed] [Google Scholar]
- [30].van Niel G, D’Angelo G, Raposo G. Shedding light on the cell biology of extracellular vesicles. Nat. Rev. Mol. Cell Biol [Internet]. 2018. [cited 2019 Aug 2];19:213–228. Available from: http://www.nature.com/articles/nrm.2017.125. [DOI] [PubMed] [Google Scholar]
- [31].Crescitelli R, Lässer C, Szabó TG, et al. Distinct RNA profiles in subpopulations of extracellular vesicles: apoptotic bodies, microvesicles and exosomes. J. Extracell. Vesicles [Internet]. 2013. [cited 2019 Aug 14];2:20677 Available from: http://www.ncbi.nlm.nih.gov/pubmed/24223256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [32].Bang C, Thum T. Exosomes: New players in cell–cell communication. Int. J. Biochem. Cell Biol [Internet]. 2012. [cited 2019 Aug 2];44:2060–2064. Available from: https://www.sciencedirect.com/science/article/pii/S1357272512002853. [DOI] [PubMed] [Google Scholar]
- [33].Su S-A, Xie Y, Fu Z, et al. Emerging role of exosome-mediated intercellular communication in vascular remodeling. Oncotarget [Internet]. 2017. [cited 2019 Aug 2];8:25700–25712. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28147325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Mathieu M, Martin-Jaular L, Lavieu G, et al. Specificities of secretion and uptake of exosomes and other extracellular vesicles for cell-to-cell communication. Nat. Cell Biol [Internet]. 2019. [cited 2019 Aug 2];21:9–17. Available from: http://www.nature.com/articles/s41556-018-0250-9. [DOI] [PubMed] [Google Scholar]
- [35].Edgar JR. Q&A: What are exosomes, exactly? 2016. [cited 2019 Apr 15];14:46 Available from: http://bmcbiol.biomedcentral.com/articles/10.1186/s12915-016-0268-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Maia J, Caja S, Strano Moraes MC, et al. Exosome-Based Cell-Cell Communication in the Tumor Microenvironment. Front. Cell Dev. Biol [Internet]. 2018. [cited 2019 Aug 2];6:18 Available from: http://journal.frontiersin.org/article/10.3389/fcell.2018.00018/full. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [37].Wortzel I, Dror S, Kenific CM, et al. Exosome-Mediated Metastasis: Communication from a Distance. Dev. Cell [Internet]. 2019. [cited 2019 Aug 2];49:347–360. Available from: http://www.ncbi.nlm.nih.gov/pubmed/31063754. [DOI] [PubMed] [Google Scholar]
- [38].Tkach M, Thé Ry C. Leading Edge Review Communication by Extracellular Vesicles: Where We Are and Where We Need to Go. 2016. [cited 2019 Aug 2]; Available from: 10.1016/j.cell.2016.01.043. [DOI]
- [39].Wan Z, Gao X, Dong Y, et al. Exosome-mediated cell-cell communication in tumor progression. Am. J. Cancer Res [Internet]. 2018. [cited 2019 Aug 2];8:1661–1673. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30323961. [PMC free article] [PubMed] [Google Scholar]
- [40].Hurley JH. ESCRT complexes and the biogenesis of multivesicular bodies. Curr. Opin. Cell Biol [Internet]. 2008. [cited 2019 Aug 2];20:4–11. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18222686. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [41].Villarroya-Beltri C, Baixauli F, Gutiérrez-Vázquez C, et al. Sorting it out: regulation of exosome loading. Semin. Cancer Biol [Internet]. 2014. [cited 2019 Aug 2];28:3–13. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24769058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tamai K, Tanaka N, Nakano T, et al. Exosome secretion of dendritic cells is regulated by Hrs, an ESCRT-0 protein. Biochem. Biophys. Res. Commun [Internet]. 2010. [cited 2019 Aug 2];399:384–390. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0006291X10014038. [DOI] [PubMed] [Google Scholar]
- [43].Colombo M, Moita C, van Niel G, et al. Analysis of ESCRT functions in exosome biogenesis, composition and secretion highlights the heterogeneity of extracellular vesicles. J. Cell Sci [Internet]. 2013. [cited 2019 Aug 2];126:5553–5565. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24105262. [DOI] [PubMed] [Google Scholar]
- [44].Adell MA, Teis D. Assembly and disassembly of the ESCRT-III membrane scission complex. FEBS Lett [Internet]. 2011/09/09 2011;585:3191–3196. Available from: https://www.ncbi.nlm.nih.gov/pubmed/21924267. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Babst M MVB vesicle formation: ESCRT-dependent, ESCRT-independent and everything in between. Curr. Opin. Cell Biol [Internet]. 2011. [cited 2019 Aug 2];23:452–457. Available from: http://www.ncbi.nlm.nih.gov/pubmed/21570275. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Zhan Y, Du L, Wang L, et al. Expression signatures of exosomal long non-coding RNAs in urine serve as novel non-invasive biomarkers for diagnosis and recurrence prediction of bladder cancer. Mol. Cancer [Internet]. 2018;17:142 Available from: https://molecular-cancer.biomedcentral.com/articles/10.1186/s12943-018-0893-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Zhai L-Y, Li M-X, Pan W-L, et al. In Situ Detection of Plasma Exosomal MicroRNA-1246 for Breast Cancer Diagnostics by a Au Nanoflare Probe. ACS Appl. Mater. Interfaces [Internet]. 2018. [cited 2019 May 7];10:39478–39486. Available from: http://pubs.acs.org/doi/10.1021/acsami.8b12725. [DOI] [PubMed] [Google Scholar]
- [48].Siegel RL, Miller KD, Jemal A. Cancer statistics, 2019. CA. Cancer J. Clin [Internet]. 2019. [cited 2019 Mar 6];69:7–34. Available from: http://www.ncbi.nlm.nih.gov/pubmed/30620402. [DOI] [PubMed] [Google Scholar]
- [49].Hannafon BN, Trigoso YD, Calloway CL, et al. Plasma exosome microRNAs are indicative of breast cancer. Breast Cancer Res. [Internet]. 2016. [cited 2019 Jan 3];18:1–14. Available from: 10.1186/s13058-016-0753-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Connolly E Effects of MK-3475 (Pembrolizumab) on the Breast Tumor Microenvironment in Triple Negative Breast Cancer (Pembro/IORT) [Internet]. United States of America: National Library of Medicine (US); 2016. Available from: https://clinicaltrials.gov/ct2/show/NCT02977468. [Google Scholar]
- [51].Isambert N Pilot Study With the Aim to Quantify a Stress Protein in the Blood and in the Urine for the Monitoring and Early Diagnosis of Malignant Solid Tumors [Internet]. Clinicaltrials.Gov. France: National Library of Medicine (US); 2018. Available from: https://clinicaltrials.gov/show/NCT02662621. [Google Scholar]
- [52].Hydbring P, De Petris L, Zhang Y, et al. Exosomal RNA-profiling of pleural effusions identifies adenocarcinoma patients through elevated miR-200 and LCN2 expression. Lung Cancer [Internet]. 2018. [cited 2019 May 7];124:45–52. Available from: https://linkinghub.elsevier.com/retrieve/pii/S0169500218304768. [DOI] [PubMed] [Google Scholar]
- [53].Sun J Clinical Research for the Consistency Analysis of PD-L1 in Lung Cancer Tissue and Plasma Exosome Before and After Radiotherapy [Internet]. China: National Library of Medicine (US); 2016. Available from: https://clinicaltrials.gov/ct2/show/NCT02869685. [Google Scholar]
- [54].Lee C-H, Im E-J, Moon P-G, et al. Discovery of a diagnostic biomarker for colon cancer through proteomic profiling of small extracellular vesicles. BMC Cancer [Internet]. 2018. [cited 2019 May 7];18:1058 Available from: https://bmccancer.biomedcentral.com/articles/10.1186/s12885-018-4952-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [55].Xue X, Zhao Y, Wang X, et al. Development and validation of serum exosomal microRNAs as diagnostic and prognostic biomarkers for hepatocellular carcinoma. J. Cell. Biochem 2019;120:135–142. [DOI] [PubMed] [Google Scholar]
- [56].Ducreux M, Cuhna AS, Caramella C, et al. Cancer of the pancreas: ESMO Clinical Practice Guidelines for diagnosis, treatment and follow-up. Ann. Oncol [Internet]. 2015. [cited 2018 Nov 26];26:v56–v68. Available from: https://academic.oup.com/annonc/article-lookup/doi/10.1093/annonc/mdv295. [DOI] [PubMed] [Google Scholar]
- [57].Kaur S, Smith LM, Patel A, et al. A Combination of MUC5AC and CA19–9 Improves the Diagnosis of Pancreatic Cancer: A Multicenter Study. Am. J. Gastroenterol 2017; [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Wu Z, Kuntz AI, Wadleigh RG. Ca 19–9 tumor marker: Is it reliable? A case report in a patient with pancreatic cancer. Clin. Adv. Hematol. Oncol 2013. [PubMed] [Google Scholar]
- [59].Melo SA, Luecke LB, Kahlert C, et al. Glypican-1 identifies cancer exosomes and detects early pancreatic cancer. Nature [Internet]. 2015/06/24 2015;523:177–182. Available from: https://www.ncbi.nlm.nih.gov/pubmed/26106858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Zhou C ya, Dong Y ping, Sun X, et al. High levels of serum glypican-1 indicate poor prognosis in pancreatic ductal adenocarcinoma. Cancer Med. [Internet]. 2018. [cited 2018 Nov 26];7:5525–5533. Available from: http://doi.wiley.com/10.1002/cam4.1833. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [61].Kitagawa T, Taniuchi K, Tsuboi M, et al. Circulating pancreatic cancer exosomal RNAs for detection of pancreatic cancer. Mol. Oncol [Internet]. 2018. [cited 2018 Nov 26]; Available from: http://www.ncbi.nlm.nih.gov/pubmed/30358104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].D’Angelica M, Wieser M. A Study of Imaging, Blood, and Tissue Samples to Guide Treatment of Colon Cancer and Related Liver Tumors [Internet]. United States of America: National Library of Medicine (US); 2018. Available from: https://clinicaltrials.gov/ct2/show/NCT03432806. [Google Scholar]
- [63].Yu K, Kelson D. A Study of Blood Based Biomarkers for Pancreas Adenocarcinoma [Internet]. United States of America: National Library of Medicine (US); 2017. Available from: https://clinicaltrials.gov/ct2/show/NCT03334708. [Google Scholar]
- [64].Buscail E, Chauvet A, Quincy P, et al. CD63-GPC1-Positive Exosomes Coupled with CA19–9 Offer Good Diagnostic Potential for Resectable Pancreatic Ductal Adenocarcinoma. Transl. Oncol 2019;12:1395–1403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Caivano A, Laurenzana I, De Luca L, et al. High serum levels of extracellular vesicles expressing malignancy-related markers are released in patients with various types of hematological neoplastic disorders. Tumor Biol. 2015;36:9739–9752. [DOI] [PubMed] [Google Scholar]
- [66].Cappello F, Logozzi M, Campanella C, et al. Exosome levels in human body fluids: A tumor marker by themselves? Eur. J. Pharm. Sci 2017;96:93–98. [DOI] [PubMed] [Google Scholar]
- [67].Shah R, Patel T, Freedman JE. Circulating Extracellular Vesicles in Human Disease. N. Engl. J. Med [Internet]. 2018. [cited 2020 Jan 7];379:958–966. Available from: http://www.nejm.org/doi/10.1056/NEJMra1704286. [DOI] [PubMed] [Google Scholar]
- [68].Osti D, Bene M Del, Rappa G, et al. Clinical significance of extracellular vesicles in plasma from glioblastoma patients. Clin. Cancer Res 2019;25:266–276. [DOI] [PubMed] [Google Scholar]
- [69].Logozzi M, Angelini DF, Iessi E, et al. Increased PSA expression on prostate cancer exosomes in in vitro condition and in cancer patients. Cancer Lett. 2017;403:318–329. [DOI] [PubMed] [Google Scholar]
- [70].Logozzi M, Spugnini E, Mizzoni D, et al. Extracellular acidity and increased exosome release as key phenotypes of malignant tumors. Cancer Metastasis Rev. Springer New York LLC; 2019. p. 93–101. [DOI] [PubMed] [Google Scholar]
- [71].Lea J, Sharma R, Yang F, et al. Detection of phosphatidylserine-positive exosomes as a diagnostic marker for ovarian malignancies: A proof of concept study. Oncotarget. 2017;8:14395–14407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Giampieri R, Piva F, Occhipinti G, et al. Clinical impact of different exosomes’ protein expression in pancreatic ductal carcinoma patients treated with standard first line palliative chemotherapy. Chalmers J, editor. PLoS One [Internet]. 2019. [cited 2020 Jan 7];14:e0215990 Available from: http://dx.plos.org/10.1371/journal.pone.0215990. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Zorrilla SR, Pérez-Sayans M, Fais S, et al. A pilot clinical study on the prognostic relevance of plasmatic exosomes levels in oral squamous cell carcinoma patients. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [74].Smith RA, Andrews KS, Brooks D, et al. Cancer screening in the United States, 2019: A review of current American Cancer Society guidelines and current issues in cancer screening. CA. Cancer J. Clin [Internet]. 2019. [cited 2020 Jan 6];69:184–210. Available from: https://onlinelibrary.wiley.com/doi/abs/10.3322/caac.21557. [DOI] [PubMed] [Google Scholar]
- [75].Schiffman JD, Fisher PG, Gibbs P. Early Detection of Cancer: Past, Present, and Future. Am. Soc. Clin. Oncol. Educ. B 2015;35:57–65. [DOI] [PubMed] [Google Scholar]
- [76].Logozzi M, Angelini DF, Giuliani A, et al. Increased plasmatic levels of psa-expressing exosomes distinguish prostate cancer patients from benign prostatic hyperplasia: A prospective study. Cancers (Basel). 2019;11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Barry MJ, Simmons LH. Prevention of Prostate Cancer Morbidity and Mortality: Primary Prevention and Early Detection. Med. Clin. North Am. W.B. Saunders; 2017. p. 787–806. [DOI] [PubMed] [Google Scholar]
- [78].Sharma R, Huang X, Brekken RA, et al. Detection of phosphatidylserine-positive exosomes for the diagnosis of early-stage malignancies. Br. J. Cancer 2017;117:545–552. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [79].Logozzi M, Mizzoni D, Capasso C, et al. Plasmatic exosomes from prostate cancer patients show increased carbonic anhydrase IX expression and activity and low pH. J. Enzyme Inhib. Med. Chem [Internet]. 2020. [cited 2020 Jan 7];35:280–288. Available from: https://www.tandfonline.com/doi/full/10.1080/14756366.2019.1697249. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [80].Logozzi M, Capasso C, Di Raimo R, et al. Prostate cancer cells and exosomes in acidic condition show increased carbonic anhydrase IX expression and activity. J. Enzyme Inhib. Med. Chem 2019;34:272–278. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Boussadia Z, Lamberti J, Mattei F, et al. Acidic microenvironment plays a key role in human melanoma progression through a sustained exosome mediated transfer of clinically relevant metastatic molecules. J. Exp. Clin. Cancer Res 2018;37. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Li P, Kaslan M, Lee SH, et al. Progress in Exosome Isolation Techniques. Theranostics [Internet]. 2017. [cited 2019 Aug 14];7:789–804. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28255367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [83].Greening DW, Xu R, Ji H, et al. A Protocol for Exosome Isolation and Characterization: Evaluation of Ultracentrifugation, Density-Gradient Separation, and Immunoaffinity Capture Methods. Methods Mol. Biol [Internet]. 2015. [cited 2019 Aug 14]. p. 179–209. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25820723. [DOI] [PubMed] [Google Scholar]
- [84].Konoshenko MY, Lekchnov EA, Vlassov A V., et al. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. Biomed Res. Int [Internet]. 2018. [cited 2018 Nov 27];2018:1–27. Available from: https://www.hindawi.com/journals/bmri/2018/8545347/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Gupta S, Rawat S, Arora V, et al. An improvised one-step sucrose cushion ultracentrifugation method for exosome isolation from culture supernatants of mesenchymal stem cells. Stem Cell Res. Ther [Internet]. 2018. [cited 2019 Aug 14];9:180 Available from: http://www.ncbi.nlm.nih.gov/pubmed/29973270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [86].Koh YQ, Almughlliq FB, Vaswani K, et al. Exosome enrichment by ultracentrifugation and size exclusion chromatography. Front. Biosci (Landmark Ed. [Internet]. 2018. [cited 2019 Aug 14];23:865–874. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28930577. [DOI] [PubMed] [Google Scholar]
- [87].Van Deun J, Mestdagh P, Sormunen R, et al. The impact of disparate isolation methods for extracellular vesicles on downstream RNA profiling. J. Extracell. Vesicles [Internet]. 2014. [cited 2019 Aug 14];3:24858 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25317274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [88].Cantin R, Diou J, Bélanger D, et al. Discrimination between exosomes and HIV-1: Purification of both vesicles from cell-free supernatants. J. Immunol. Methods [Internet]. 2008. [cited 2019 Aug 14];338:21–30. Available from: http://www.ncbi.nlm.nih.gov/pubmed/18675270. [DOI] [PubMed] [Google Scholar]
- [89].Lobb RJ, Becker M, Wen SW, et al. Optimized exosome isolation protocol for cell culture supernatant and human plasma. J. Extracell. vesicles [Internet]. 2015. [cited 2019 Aug 14];4:27031 Available from: http://www.ncbi.nlm.nih.gov/pubmed/26194179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [90].Zeringer E, Barta T, Li M, et al. Strategies for Isolation of Exosomes. Cold Spring Harb. Protoc [Internet]. 2015. [cited 2019 Aug 14];2015:pdb.top074476 Available from: http://www.ncbi.nlm.nih.gov/pubmed/25834266. [DOI] [PubMed] [Google Scholar]
- [91].Muller L, Hong C-S, Stolz DB, et al. Isolation of biologically-active exosomes from human plasma. J. Immunol. Methods [Internet]. 2014. [cited 2019 Aug 14];411:55–65. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24952243. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Oksvold MP, Neurauter A, Pedersen KW. Magnetic Bead-Based Isolation of Exosomes. Methods Mol. Biol [Internet]. 2015. [cited 2019 Aug 14]. p. 465–481. Available from: http://www.ncbi.nlm.nih.gov/pubmed/25319668. [DOI] [PubMed] [Google Scholar]
- [93].Lobb R, Möller A. Size Exclusion Chromatography: A Simple and Reliable Method for Exosome Purification. Methods Mol. Biol [Internet]. 2017. [cited 2019 Aug 14]. p. 105–110. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28828651. [DOI] [PubMed] [Google Scholar]
- [94].Vaswani K, Koh YQ, Almughlliq FB, et al. A method for the isolation and enrichment of purified bovine milk exosomes. Reprod. Biol [Internet]. 2017. [cited 2019 Aug 14];17:341–348. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29030127. [DOI] [PubMed] [Google Scholar]
- [95].Boriachek K, Islam MN, Möller A, et al. Biological Functions and Current Advances in Isolation and Detection Strategies for Exosome Nanovesicles. Small [Internet]. 2018. [cited 2020 Jan 8];14:1702153 Available from: http://doi.wiley.com/10.1002/smll.201702153. [DOI] [PubMed] [Google Scholar]
- [96].Coumans FAW, Gool EL, Nieuwland R. Bulk immunoassays for analysis of extracellular vesicles. Platelets [Internet]. 2017. [cited 2020 Jan 9];28:242–248. Available from: https://www.tandfonline.com/doi/full/10.1080/09537104.2016.1265926. [DOI] [PubMed] [Google Scholar]
- [97].Logozzi M, De Milito A, Lugini L, et al. High Levels of Exosomes Expressing CD63 and Caveolin-1 in Plasma of Melanoma Patients. Cao Y, editor. PLoS One [Internet]. 2009. [cited 2020 Jan 7];4:e5219 Available from: https://dx.plos.org/10.1371/journal.pone.0005219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Liu C, Xu X, Li B, et al. Single-Exosome-Counting Immunoassays for Cancer Diagnostics. Nano Lett. 2018;18:4226–4232. [DOI] [PubMed] [Google Scholar]
- [99].Khodashenas S, Khalili S, Forouzandeh Moghadam M. A cell ELISA based method for exosome detection in diagnostic and therapeutic applications. Biotechnol. Lett 2019;41:523–531. [DOI] [PubMed] [Google Scholar]
- [100].Zhao A, Guo L, Xu J, et al. Identification and validation of circulating exosomes‐based liquid biopsy for esophageal cancer. Cancer Med. [Internet]. 2019. [cited 2020 Jan 9];8:3566–3574. Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/cam4.2224. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Erdbrügger U, Lannigan J. Analytical challenges of extracellular vesicle detection: A comparison of different techniques. Cytom. Part A [Internet]. 2016. [cited 2020 Jan 9];89:123–134. Available from: http://doi.wiley.com/10.1002/cyto.a.22795. [DOI] [PubMed] [Google Scholar]
- [102].Asghar W, Islam M, Wadajkar AS, et al. PLGA Micro- and Nanoparticles Loaded Into Gelatin Scaffold for Controlled Drug Release. IEEE Trans. Nanotechnol [Internet]. 2012. [cited 2019 Aug 28];11:546–553. Available from: http://ieeexplore.ieee.org/document/6123209/. [Google Scholar]
- [103].Asghar W, Ilyas A, Deshmukh RR, et al. Pulsed plasma polymerization for controlling shrinkage and surface composition of nanopores. Nanotechnology [Internet]. 2011. [cited 2019 Aug 28];22:285304 Available from: http://www.ncbi.nlm.nih.gov/pubmed/21636880. [DOI] [PubMed] [Google Scholar]
- [104].Asghar W, Ilyas A, Billo JA, et al. Shrinking of Solid-state Nanopores by Direct Thermal Heating. Nanoscale Res. Lett [Internet]. 2011. [cited 2019 Aug 28];6:372 Available from: https://www.ncbi.nlm.nih.gov/pmc/articles/PMC3211463/. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Asghar W, El Assal R, Shafiee H, et al. Engineering cancer microenvironments for in vitro 3-D tumor models. Mater. Today (Kidlington) [Internet]. 2015. [cited 2019 Aug 28];18:539–553. Available from: http://www.ncbi.nlm.nih.gov/pubmed/28458612. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Asghar W, Yuksekkaya M, Shafiee H, et al. Engineering long shelf life multi-layer biologically active surfaces on microfluidic devices for point of care applications. Sci. Rep [Internet]. 2016. [cited 2019 Aug 28];6:21163 Available from: http://www.nature.com/articles/srep21163. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Asghar W, Wan Y, Ilyas A, et al. Electrical fingerprinting, 3D profiling and detection of tumor cells with solid-state micropores. Lab Chip [Internet]. 2012. [cited 2019 Aug 28];12:2345 Available from: http://www.ncbi.nlm.nih.gov/pubmed/22549275. [DOI] [PubMed] [Google Scholar]
- [108].Asghar W, Velasco V, Kingsley JL, et al. Selection of functional human sperm with higher DNA integrity and fewer reactive oxygen species. Adv. Healthc. Mater [Internet]. 2014. [cited 2019 Aug 28];3:1671–1679. Available from: http://www.ncbi.nlm.nih.gov/pubmed/24753434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Asghar W, Shafiee H, Velasco V, et al. Toxicology Study of Single-walled Carbon Nanotubes and Reduced Graphene Oxide in Human Sperm. Sci. Rep [Internet]. 2016. [cited 2019 Aug 28];6:30270 Available from: http://www.ncbi.nlm.nih.gov/pubmed/27538480. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Asghar W, Shafiee H, Chen P, et al. In Vitro Three-Dimensional Cancer Culture Models. Cancer Target. Drug Deliv [Internet]. New York, NY: Springer New York; 2013. [cited 2019 Aug 28]. p. 635–665. Available from: http://link.springer.com/10.1007/978-1-4614-7876-8_24. [Google Scholar]
- [111].Rappa K, Samargia J, Sher M, et al. Quantitative analysis of sperm rheotaxis using a microfluidic device. Microfluid. Nanofluidics [Internet]. 2018. [cited 2019 Aug 28];22:100 Available from: http://link.springer.com/10.1007/s10404-018-2117-6. [Google Scholar]
- [112].Coarsey C, Coleman B, Kabir MA, et al. Development of a flow-free magnetic actuation platform for an automated microfluidic ELISA. RSC Adv. [Internet]. 2019. [cited 2019 Aug 28];9:8159–8168. Available from: http://xlink.rsc.org/?DOI=C8RA07607C. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Sher M, Asghar W. Development of a multiplex fully automated assay for rapid quantification of CD4+ T cells from whole blood. Biosens. Bioelectron [Internet]. 2019. [cited 2019 Aug 28];142:111490 Available from: https://www.sciencedirect.com/science/article/pii/S095656631930569X. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Sharma S, Zhuang R, Long M, et al. Circulating tumor cell isolation, culture, and downstream molecular analysis. Biotechnol. Adv [Internet]. 2018. [cited 2019 Aug 28];36:1063–1078. Available from: http://www.ncbi.nlm.nih.gov/pubmed/29559380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hammond JL, Formisano N, Estrela P, et al. Electrochemical biosensors and nanobiosensors. Essays Biochem. 2016;60:69–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Fu Z, Lu Y-C, Lai JJ. Recent Advances in Biosensors for Nucleic Acid and Exosome Detection. Chonnam Med. J 2019;55:86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Jeong S, Park J, Pathania D, et al. Integrated Magneto-Electrochemical Sensor for Exosome Analysis. ACS Nano. 2016;10:1802–1809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [118].Moura SL, Martín CG, Martí M, et al. Electrochemical immunosensing of nanovesicles as biomarkers for breast cancer. Biosens. Bioelectron 2019; [DOI] [PubMed] [Google Scholar]
- [119].Doldán X, Fagúndez P, Cayota A, et al. Electrochemical Sandwich Immunosensor for Determination of Exosomes Based on Surface Marker-Mediated Signal Amplification. Anal. Chem 2016;88:10466–10473. [DOI] [PubMed] [Google Scholar]
- [120].Zhang J, Wang LL, Hou MF, et al. A ratiometric electrochemical biosensor for the exosomal microRNAs detection based on bipedal DNA walkers propelled by locked nucleic acid modified toehold mediate strand displacement reaction. Biosens. Bioelectron 2018;102:33–40. [DOI] [PubMed] [Google Scholar]
- [121].Luo L, Wang L, Zeng L, et al. A ratiometric electrochemical DNA biosensor for detection of exosomal MicroRNA. Talanta. 2020;207. [DOI] [PubMed] [Google Scholar]
- [122].Liu L, Lu H, Shi R, et al. Synergy of Peptide-Nucleic Acid and Spherical Nucleic Acid Enabled Quantitative and Specific Detection of Tumor Exosomal MicroRNA. Anal. Chem 2019; [DOI] [PubMed] [Google Scholar]
- [123].Zhao L, Sun R, He P, et al. Ultrasensitive detection of exosomes by target-triggered three-dimensional DNA walking machine and exonuclease III-assisted electrochemical ratiometric biosensing. Anal. Chem 2019; [DOI] [PubMed] [Google Scholar]
- [124].Maduraiveeran G, Sasidharan M, Ganesan V. Electrochemical sensor and biosensor platforms based on advanced nanomaterials for biological and biomedical applications. Biosens. Bioelectron. Elsevier Ltd; 2018. p. 113–129. [DOI] [PubMed] [Google Scholar]
- [125].Fang S, Tian H, Li X, et al. Clinical application of a microfluidic chip for immunocapture and quantification of circulating exosomes to assist breast cancer diagnosis and molecular classification. PLoS One [Internet]. 2017. [cited 2019 Jan 3];12:e0175050 Available from: http://www.ncbi.nlm.nih.gov/pubmed/28369094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [126].Savvateeva EN, Tikhonov AA, Butvilovskaya VI, et al. Exosomal Surface Protein Markers in Diagnosis of Colorectal Cancer. Mol. Biol [Internet]. 2017. [cited 2019 Aug 28];51:752–760. Available from: http://www.cellgs.com/Shop/Exosomes/Exosome-Purification-. [DOI] [PubMed] [Google Scholar]
- [127].Hikita T, Miyata M, Watanabe R, et al. Sensitive and rapid quantification of exosomes by fusing luciferase to exosome marker proteins. Sci. Rep [Internet]. 2018. [cited 2019 Aug 28];8:14035 Available from: http://www.nature.com/articles/s41598-018-32535-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [128].Xu H, Liao C, Zuo P, et al. Magnetic-Based Microfluidic Device for On-Chip Isolation and Detection of Tumor-Derived Exosomes. Anal. Chem 2018;90:13451–13458. [DOI] [PubMed] [Google Scholar]
- [129].Taller D, Richards K, Slouka Z, et al. On-chip surface acoustic wave lysis and ion-exchange nanomembrane detection of exosomal RNA for pancreatic cancer study and diagnosis. Lab Chip [Internet]. 2015;15:1656–1666. Available from: 10.1039/C5LC00036J. [DOI] [PubMed] [Google Scholar]
- [130].Salvati E, Stellacci F, Krol S. Nanosensors for early cancer detection and for therapeutic drug monitoring. Nanomedicine. Future Medicine Ltd; 2015. p. 3495–3512. [DOI] [PubMed] [Google Scholar]
- [131].Li W, Wang H, Zhao Z, et al. Emerging Nanotechnologies for Liquid Biopsy: The Detection of Circulating Tumor Cells and Extracellular Vesicles. Adv. Mater [Internet]. 2019. [cited 2020 Jan 14];31:1805344 Available from: https://onlinelibrary.wiley.com/doi/abs/10.1002/adma.201805344. [DOI] [PubMed] [Google Scholar]
- [132].Mathew DG, Beekman P, Lemay SG, et al. Electrochemical Detection of Tumor-Derived Extracellular Vesicles on Nanointerdigitated Electrodes. Nano Lett [Internet]. 2019. [cited 2020 Jan 14];acs.nanolett.9b02741. Available from: https://pubs.acs.org/doi/10.1021/acs.nanolett.9b02741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [133].Raghu D, Christodoulides JA, Christophersen M, et al. Nanoplasmonic pillars engineered for single exosome detection. PLoS One. 2018;13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [134].Sina AAI, Vaidyanathan R, Dey S, et al. Real time and label free profiling of clinically relevant exosomes. Sci. Rep 2016;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [135].Lyu Y, Cui D, Huang J, et al. Near-Infrared Afterglow Semiconducting Nano-Polycomplexes for the Multiplex Differentiation of Cancer Exosomes. Angew. Chemie Int. Ed [Internet]. 2019. [cited 2020 Jan 14];58:4983–4987. Available from: http://doi.wiley.com/10.1002/anie.201900092. [DOI] [PubMed] [Google Scholar]
- [136].Whiteside TL. The potential of tumor-derived exosomes for noninvasive cancer monitoring. Expert Rev. Mol. Diagn. Taylor and Francis Ltd; 2015. p. 1293–1310. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Braig Z Personalized medicine: From diagnostic to adaptive. Biomed. J. Elsevier B.V; 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [138].Bernard V, Kim DU, San Lucas FA, et al. Circulating Nucleic Acids Are Associated With Outcomes of Patients With Pancreatic Cancer. Gastroenterology [Internet]. 2019. [cited 2019 May 30];156:108–118.e4. Available from: https://www.sciencedirect.com/science/article/pii/S0016508518350066?via%3Dihub. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [139].Imamura T, Komatsu S, Ichikawa D, et al. Liquid biopsy in patients with pancreatic cancer: Circulating tumor cells and cell-free nucleic acids. World J Gastroenterol [Internet]. 2016/07/20 2016;22:5627–5641. Available from: https://www.ncbi.nlm.nih.gov/pubmed/27433079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Nuzhat Z, Kinhal V, Sharma S, et al. Tumour-derived exosomes as a signature of pancreatic cancer - liquid biopsies as indicators of tumour progression. Oncotarget [Internet]. 2017;8:17279–17291. Available from: https://www.ncbi.nlm.nih.gov/pubmed/27999198. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Sisson BA, Uvalic J, Kelly K, et al. Technical and Regulatory Considerations for Taking Liquid Biopsy to the Clinic: Validation of the JAX PlasmaMonitorTM Assay. Biomark. Insights 2019;14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [142].Alvarez Cubero MJ, Lorente JA, Robles-Fernandez I, et al. Circulating tumor cells: Markers and methodologies for enrichment and detection. Methods Mol. Biol. Humana Press Inc; 2017. p. 283–303. [DOI] [PubMed] [Google Scholar]
- [143].Fernando MR, Jiang C, Krzyzanowski GD, et al. New evidence that a large proportion of human blood plasma cell-free DNA is localized in exosomes. PLoS One. 2017;12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Thakur BK, Zhang H, Becker A, et al. Double-stranded DNA in exosomes: A novel biomarker in cancer detection. Cell Res. Nature Publishing Group; 2014. p. 766–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [145].Li A, Zhang T, Zheng M, et al. Exosomal proteins as potential markers of tumor diagnosis. J. Hematol. Oncol. BioMed Central Ltd; 2017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [146].Li Y, Zheng Q, Bao C, et al. Circular RNA is enriched and stable in exosomes: A promising biomarker for cancer diagnosis. Cell Res. Nature Publishing Group; 2015. p. 981–984. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Sauter RE Exosomes in blood and cancer. Transl. Cancer Res 2017;6:S1316–S1320. [Google Scholar]
- [148].Gupta RG, Somer RA. Intratumor Heterogeneity: Novel Approaches for Resolving Genomic Architecture and Clonal Evolution. 2017. [cited 2019 Nov 22]; Available from: www.aacrjournals.org. [DOI] [PubMed]


