Abstract
The directed movements of individual, groups, or sheets of cells at specific times in particular locations bring about form and complexity to developing organisms. Cells move by extending protrusions, such as macropinosomes, pseudopods, lamellipods, filopods, or blebs. Although many of the cytoskeletal components within these structures are known, less is known about the mechanisms that determine their location, number, and characteristics. Recent evidence suggests that control may be exerted by a signal transduction excitable network whose components and activities, including Ras, PI3K, TorC2, and phosphoinositides, self-organize on the plasma membrane and propagate in waves. The waves drive the various types of protrusions, which in turn, determine the modes of cell migration. Acute perturbations at specific points in the network produce abrupt shifts in protrusion type, including transitions from pseudopods to filopods or lamellipods. These observations have also contributed to a delineation of the signal transduction network, including candidate fast positive and delayed negative feedback loops. The network contains many oncogenes and tumor suppressors, and other molecules which have recently been implicated in developmental and metabolic abnormalities. Thus, the concept of signal transduction network excitability in cell migration can be used to understand disease states and morphological changes occurring in development.
Keywords: Excitability, Migratory modes, PTEN, PIP3, RasGAPs/GEFs, Wave patterns
1. Introduction
Throughout phylogeny, migrating cells possess an internal compass which enables them to sense and move towards or away from gradients of extracellular, soluble chemoattractants or repellants, respectively. In-depth studies over the past century have revealed that chemotaxis plays a crucial role in the development and physiology of uni- and multicellular organisms. In bacteria, archaea and protozoa, cells perform chemotaxis for the purposes of seeking nutrients in the environment, intercellular aggregation and multicellular morphogenesis, and spreading infection within the host [1–4]. Apart from chemotaxis, cells possess the ability to sense and move along gradients of other environmental stimuli, such as light, electric fields, surface stiffness, shear forces, temperature, and substrate-bound signaling molecules [5–16].
In metazoans, directed cell migration is required during embryonic development and in adults. During embryogenesis, concerted movement of epithelial sheets bring about gastrulation, while precursor cells residing in different stem cell regions, such as neural crest, brain ventricles, somites and primitive streak, leave their niche and move towards their target sites[17–22]. In adults, guided migration is critical for several processes such as trafficking of immune cells towards invading pathogens, coordinated movement of keratinocytes and fibroblasts into the wound site during its healing, motility of sperm towards the egg during fertilization, homing of endogenous stem cells to maintain tissue homeostasis [23–28].
Irregularities in directional migration gives rise to congenital abnormalities, such as neuronal migration disorders, and inflammatory diseases, such as various allergies and infections, atherosclerosis, angiogenesis and cancer metastasis[29–34]. This review aims to summarize recent models and molecular mechanisms controlling individual cell migration. The concepts outlined here also shed light on understanding collective cell migration in development and diseases.
2. Varied cellular protrusions in a vast range of physiological functions
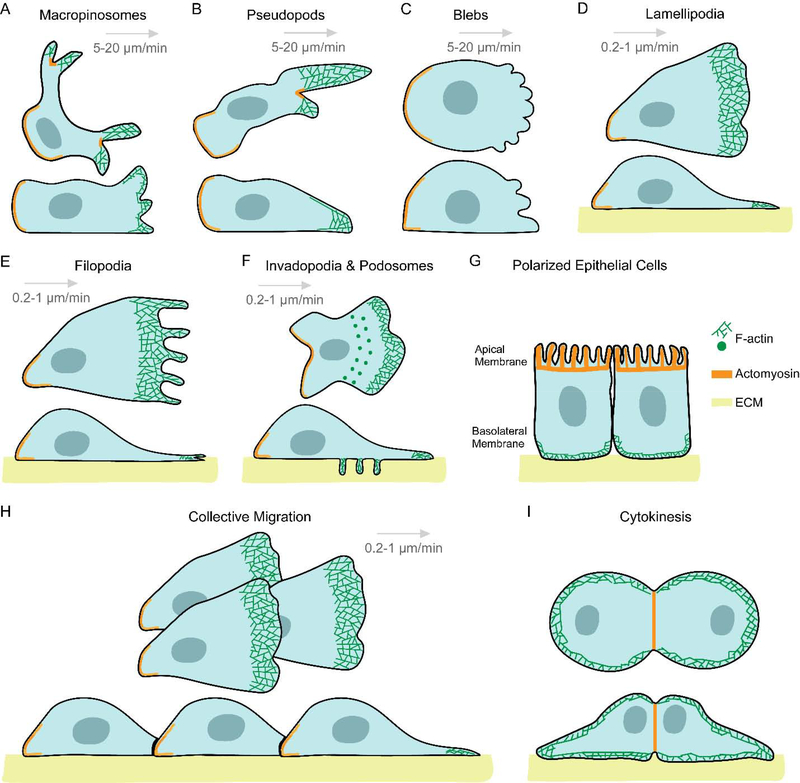
Migratory cells move with the help of a diverse array of morphological appendages, which lead to different migration modes. Bacteria, protozoa and sperm rely on flagella and cilia for propulsion and movement[1, 35]. Leukocytes and amoebae move by rhythmically extending and retracting discrete actin-rich protrusions, which may appear as wide cup-like structures at the top and sides of the cell or narrower ones situated closer to the substratum. These are traditionally referred to as macropinosomes or pseudopods, respectively (Figure 1A and B)[36–39]. In certain cases of amoeboid motion, due to contractile pressure, the plasma membrane detaches from the actomyosin cortex and causes cytoplasmic extensions or ‘blebs’ to appear (Figure 1C)[20, 40–42]. Keratocytes glide with the help of a single, broad, actin-filled, anterior protrusion and can preserve their shape and direction during motion[43, 44]. Mesenchymal cells, such as fibroblasts, move on the extracellular matrix (ECM) with the help of sheet-like lamellipodia and thin, finger-like filopodia, which form at the leading edge of the cell (Figure 1D and E)[45, 46]. The attachment to the ECM causes these cells to move at a speed of 0.2–1 μm/min, while cells displaying amoeboid or keratocyte-like migration move at a speed of 5–20 μm/min[47].
Figure 1. Snapshots of diverse cellular protrusions in a variety of physiological functions.
Throughout the illustration, all cells and their protrusions are shown in top (upper) and side (lower) views, except for polarized epithelial cells (G), which is shown only in side view. F-actin is denoted as mesh of green lines, acto-myosin as a heavy orange line, and the extracellular matrix (ECM) is shown in yellow. For migratory cells, the direction of migration is from left to right, as shown with a grey arrow. The respective migratory speeds are mentioned below each grey arrow. Wide and cup-shaped macropinosomes (A) or narrower and longer pseudopods (B) are F-actin rich structures which extend and retract rhythmically near the top and sides of the amoeboid cell, or near the substratum, respectively. (C) In some instances, cells move with the help of bulky cytoplasmic structures or ‘blebs’ which form by detachment of cell membrane from the actomyosin cortex due to contractile pressure. Cells move at a fast speed of 5–20 μm/min with the help of macropinosomes, pseudopods or blebs. (D) Fibroblasts move on the ECM with the help of a broad, thin, F-actin filled anterior protrusion called lamellipodia. The actin network of these structures sometimes protrudes further to form thin, finger-like projections called filopodia (E). Due to the focal adhesion-based attachment of these cells to the ECM, they move at a far slower speed of 0.2–1 μm/min. (F) Macrophages and some cancer cells form polymerized actin-rich ventral protrusions, referred to as podosomes or invadopodia, which also secrete metalloproteases to degrade the ECM. (G) Epithelial cells polarize along the apical-basal axis and migrate very slowly as epithelial sheets. (H) During oogenesis and embryogenesis, actin polymerization directly pushes forward the plasma membrane of Drosophila border cells and Xenopus neural crest cells in the form of a broad lamellipodia, and help the cells to migrate collectively. (I) During cytokinesis, F-actin and actomyosin localize to the poles and cleavage furrow, giving the dividing cell the appearance of two cells migrating away from each other.
Under different physiological conditions, variations of this general scheme can give rise to a diversity of protrusions and migration modes in cells. 1) Blood cells of myeloid and monocytic lineages or metastatic cancer cells form actin-rich ventral projections called podosomes or invadopodia, respectively, which degrade the ECM through secretion of matrix metalloproteinases, and help cells migrate across extracellular barriers (Figure 1F)[48]. Interestingly, in patients suffering from Wiskott–Aldrich syndrome, dendritic cells and macrophages have defective podosome formation[49]. 2) Polarized epithelial cells display acto-myosin enriched apical regions with broad, F-actin rich lamella-like protrusions at the baso-lateral surface. Their migration is limited due to intercellular interactions (Figure 1G)[50, 51]. 3) During oogenesis and embryogenesis, cells move collectively, including Drosophila border cell clusters, which detach from the follicular epithelium and migrate toward the oocyte [52, 53]; Drosophila tracheal cell clusters, which branch out to form the tracheal system[54] and zebrafish lateral line primordium, which migrates to form the lateral line sensory system (Figure 1H)[55]. 4) Though not technically cell migration, the process of cytokinesis resembles two cells migrating away from each other. Consistently, F-actin rich protrusions and myosin, which segregate to the anterior or posterior regions of migrating cells move to the poles or furrow of dividing cells during cytokinesis (Figure 1I)[56, 57]. Thus, varied actin-dependent protrusions are crucial for a vast range of cellular functions, including morphogenesis, tissue regeneration, immunity and cell division.
3. Spatiotemporal regulation of signal transduction and cytoskeletal networks
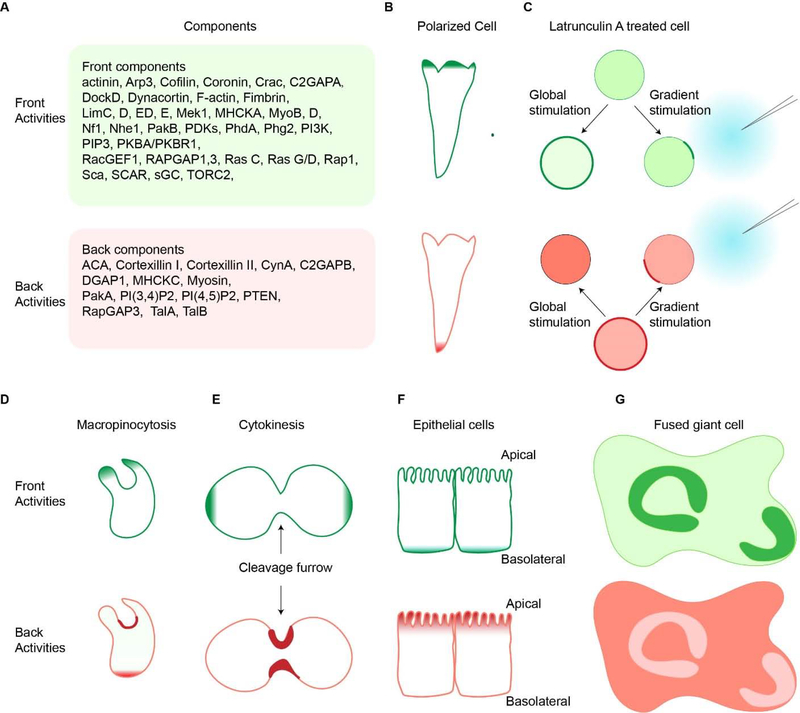
In migrating cells, many of the signaling molecules, including both lipids and proteins, that mediate polarity and directed cell migration are selectively localized or activated at the leading edge. While others, initially present on the membrane/cortex dissociate from the protrusions. These asymmetrically distributed molecules are referred to as “front” or “back”, respectively. A growing list of such spatially restricted molecules is shown in Figure 2A. For example, front events such as Ras, TORC2, PI3K activation, PKB localization or PIP3 accumulation occur at the tips of protrusions of the vegetative and polarized cells, and back events such as PTEN or Myosin II dissociation from pseudopods and localization to the tail of the cells [58–65]. We use the terms “events” or “activities” throughout the review to refer to localization/accumulation or activation/deactivation of components. These complementary distributions of molecules and activities are a crucial first step in establishing polarity and guiding cell migration and are maintained even in the absence of a chemoattractant gradient (Figure 2B). The same complementary pattern of front and back events is observed even when the actin skeleton is disrupted by Latrunculin A. As shown in Figure 2C, in Latrunculin A-treated cells, front components such as PI3K are localized in the cytosol and back components like PTEN are on the membrane. In response to uniform chemoattractant, front components redistribute evenly over the cortex or membrane, whereas back components translocate into the cytosol. When the cells are exposed to a gradient of chemoattractant, front events show an accumulation in a ‘dancing crescent’ whose orientation oscillates around the direction of the gradient. The behavior of the back events such as PI(3,4)P2 accumulation is diametrically opposed and faces away from the gradient [66, 67] (Figure 2C).
Figure 2. Complementary front and back cellular events.
A. Components of front (up) and back (bottom) are listed. (B). Front activities, such as Ras or PI3K activation, occur at the protrusions of migrating polarized cells, respectively (denoted in dark green, top row). These front activities are complemented with back activities, which localize to the trailing edge of the cells (denoted in red, top row). (C). Upon global or gradient chemoattractant stimulation, latrunculin A-treated cells also show opposite distribution of front (top) and back (bottom) activities. (D). During macropinocytosis, front activities is at the macropinocytic cup, and back activities are at the back of the cup and cell. (E). During cytokinesis, these front molecules are found at the poles of the dividing cells and the back molecules accumulate at the cleavage furrow. (F). In epithelial cells, the front events are localized to the basolateral surfaces (green, top), and the back events are at the apical side (red, bottom). (G). This complimentary pattern of front and back molecules is conserved in fused Dictyostelium cells.
There are situations which parallel migration where these complementary distributions are conserved. For example, during macropinocytosis, the extending edges of the forming cups are decorated with front components like PIP3, whereas back components like PTEN localize to the base (Figure 2D)[68]. During cytokinesis, back components move uniformly to the membrane, while front components localized to the cytosol. As the cell elongates, front proteins associate with the cell membrane at the poles while back proteins accumulate at the cleavage furrow together with the acto-myosin ring (Figure 2E) [69]. In epithelial cells, front events are localized to the basolateral surfaces, and back events are at the apical side[70] (Figure 2F). In multinucleated mutants or fused giant Dictyostelium cells, which display waves of actin polymerization, the same complementary patterns of front and back events are again observed as shown in Figure 2G[71]. Most of the basal surface is occupied by back markers, except in the region of actin polymerization where the front molecules appear.
We have observed that cytoskeletal activity alone produces only transient, narrow extensions. Upstream signal transduction networks are needed to provide coordination of cytoskeletal activities across protrusive cellular structures. When accompanied by signal transduction events, such as Ras and PI3K activation or PTEN inhibition, the protrusions are sustained, reaching out wider or longer[72]. The complementary distributions of these front and back events observed have an important role in regulating protrusions. In Dictyostelium, the coordinated activity of at least thirteen biosensors suggests that activation of the entire signal transduction network accompanies each protrusion. The molecular components involved in cell migration are remarkably conserved between the social amoeba, Dictyostelium and mammalian cells. Dictyostelium cells are easily cultivable in the laboratory, well suited for live cell imaging and are naturally migratory cells and have a haploid genome which is completely sequenced and annotated. This makes the amoeba an excellent model system for studies of eukaryotic cell migration[73–75].
Furthermore, as described below, small shifts in these upstream events can elicit a striking transition in different protrusive activities and migratory modes[58, 76, 77]. These observations are not accounted for in the classical view of cell migration. It is believed that different types of protrusions, such as filopodia and lamellipodia, are the results of activations of specific regulators of the actin-myosin cytoskeleton, such as Arp2/3 and formins [78–80], and signal transduction networks merely provide directionality to these protrusions. In the following sections, we discuss emerging insights into the role of spontaneous signal transduction events in the formation of the protrusions.
4. Excitability of signal transduction and cytoskeletal networks
Accumulating evidence points to the fact that these signal transduction and cytoskeletal activities are excitable. Excitability has served as an emerging framework to decipher numerous biological systems, from neuronal action potentials to cardiac calcium waves, from yeast glycolytic oscillations to vertebrate segmentation. Excitable systems have characteristic hallmarks such as all-or-none response, refractoriness and wave propagation. Quantitative measurement in Dictyostelium has directly demonstrated the excitable nature of the signal transduction and cytoskeletal networks. For example, Huang et al. showed that activation of Ras, in response to short and long supra-threshold stimuli is the same and are followed by a refractory period of ~50 seconds [81]. Consistently, signal transduction and cytoskeletal constituents propagate as waves on the cortex and annihilate when oppositely-directed waves collide, further supporting the existence of excitability (see Figure 2E)[82, 83]. With similar observations being described in other cell types including neutrophils [84–86], macrophages [87], and mast cells [88–90], excitability is likely a conserved feature of many molecular networks.
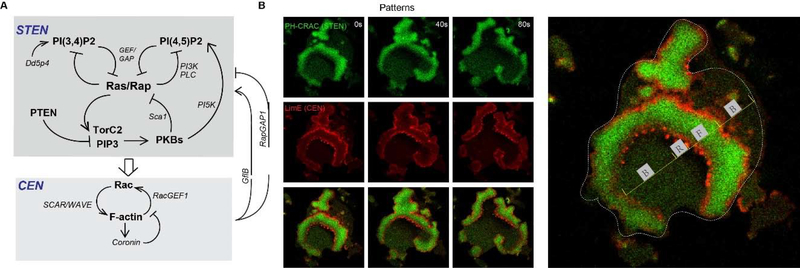
The signal transduction and cytoskeletal events appear to operate as distinct, yet closely interacting, excitable systems. In Dictyostelium, molecular events in cell migration can be broadly classified into two networks, the Signal Transduction Excitable Network (STEN) and the Cytoskeletal Excitable Network (CEN) (Figure 3A). Differences between STEN and CEN are manifested in their responding kinetics to global stimulus, as well as localization morphologies in spontaneous wave patterns. In response to cAMP addition, STEN components, including phospholipids, Ras/Rap small GTPases, and PKBs, show a level/activity increase with kinetics slower than CEN components including F-actin, Rac, coronin, and HPSC300. Consistently, in spontaneous traveling waves, STEN components show diffuse bands while CEN display sharp ‘two-peak’ morphologies and puncta associated with the trailing peak (Figure 3B). These differences in kinetics and localization highlight the intrinsic differences in excitability between STEN and CEN. On the other hand, STEN and CEN are closely coupled and entrained. Acute perturbations at different nodes suggest that STEN drives CEN to form waves. CEN also regulates the triggering of STEN, possibly through RapGAP1 and GflB [76, 77]. Future work is required to reveal more detailed biochemical interactions and integrate them on a systematic level.
Figure 3. Summary of STEN and CEN networks with positive and negative feedback loops.
(A) Positive feedback in STEN is brought about by mutual inhibition between Ras/Rap activity and PI(3,4)P2 and PI(4,5)P2 at the cell cortex and delayed negative feedback due to PKB activation by PIP3 and TorC2. (B). The corresponding B, F, and R states are depicted on basal surface of a fused Dictyostelium cell. Time-lapse Confocal images (left) showing distribution of LimE (red) and PHcrac (green) during wave propagation at the basal surface of a migrating fused Dictyostelium cell. White dashed line marks the outline of the fused cell (Right). F represents the F state, B represents the B state and R represents the R state. The B-state region is characterized by low Rap/Ras, TorC2, and PI3K activity and high PI(3,4)P2 and PI(4,5)P2 levels. As a region transitions to the F-state, Rap/Ras activity increases, PI(3,4)P2 decreases strongly, and PI(4,5)P2 decreases slightly. There is a slower rise in PKB activity due to the elevation of TorC2 and PIP3. A refractory period (R state) follows associated with lower Rap/Ras activity and higher PI(3,4)P2/PI(4,5)P2 levels and PKB activity. The PKBs feed into CEN, promoting F-actin polymerization and loss of cortical myosin. In turn, the activated cytoskeletal activity provides fast positive and slow negative feedbacks to STEN.
5. Wave patterns, cortical dynamics and cellular protrusions
The idea that wave propagation controls protrusion formation has been supported by recent evidence. It was found that fast undulations of CEN cover the cell periphery, but sustained protrusions are only formed when significant STEN activities drive CEN to support wave propagation [72, 77, 81, 91]. Furthermore, the dynamic properties of wave propagation closely mirror those of pseudopods in migrating cells. Innately, waves of STEN and CEN have limited range – they gradually decrease speed and eventually extinguish rather than propagating persistently. Similarly, pseudopods only grow to a defined size before collapsing rather than expanding continually[36, 92–94]. Thus, STEN and CEN waves, serving as the drivers of protrusions, can account for the extension and retraction dynamics of pseudopods.
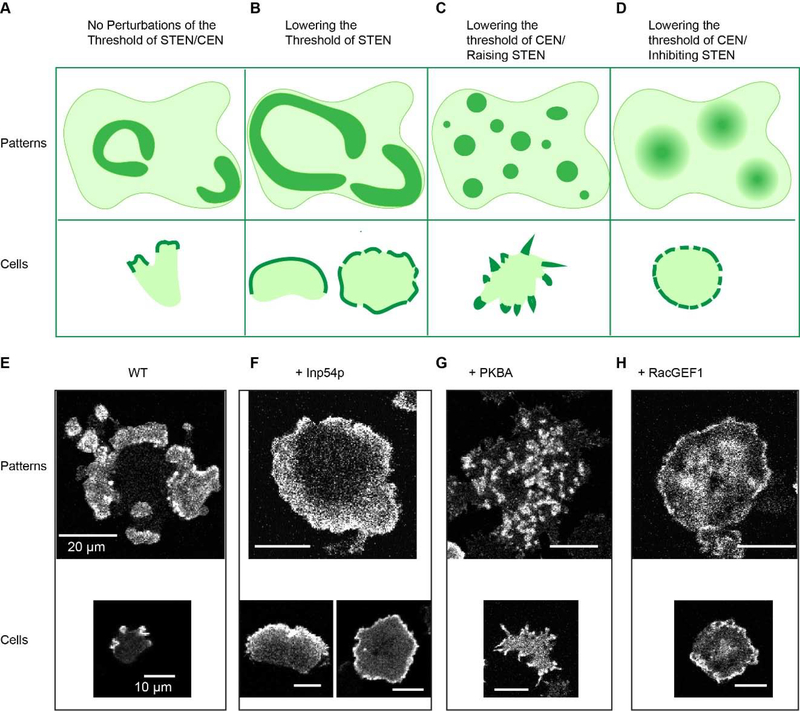
Recent experiments with acute perturbations further support the causal relationship between wave properties and protrusion determination (Figure 4). First, within minutes of lowering threshold of STEN activation, by decreasing PIP2 or increasing Ras/Rap activities using chemically induced dimerization, waves propagate with elevated speed and range. Consequently, narrower pseudopods become wider lamellipodia-like protrusions and the cell migratory mode shifts from amoeboid to keratocyte-like and oscillatory (Figures 4B and F). Second, increasing PKBs activities produces negative feedback in STEN, but also promotes coupling between STEN and CEN. This raises the threshold of STEN, and simultaneously lowers threshold of CEN, leading to slower wave propagation and more wave initiation centers (Figure 3A). This generates numerous spiky filopodia-like protrusions (Figures 4C and G). Similarly, recruiting RacGEF1 to the membrane produces positive feedback in CEN, and delayed negative feedbacks in STEN. This lowers the threshold of CEN and increases threshold of STEN, leading to disrupted wave patterns and many diffuse actin patches (Figure 3A). As a result, sustained protrusions cannot be formed due to a lack of wave propagation, and the cells generate ruffles on the cortex and lose the ability to move (Figures 4D and H)[77]. All these transitions occur in a much faster time scale than that of gene regulation. Thus, tweaking the feedback loops within and between STEN and CEN leads to a range of wave patterns, which results in a spectrum of interchangeable cellular protrusions.
Figure 4. Cartoon of Perturbations and different wave patterns & cellular protrusions.
(A-D). Cartoons depicting the cortical wave patterns corresponding to cellular morphology in fused giant cells (top) or single cells (bottom). Cells without perturbations of the threshold of STEN/CEN (A); with perturbations of lowering the threshold of STEN (B); with perturbations of lowering of the threshold for CEN/raising that of STEN (C); with perturbations of lowering of the threshold of CEN/removing STEN (D). (E-H) Confocal images corresponding to (A-D), showing LimE patterns (CEN) on the basal surface of fused Dictyostelium cells (top) and on the protrusions of single cells (bottom). Cells without perturbations of the threshold of STEN/CEN. (E); with perturbations of lowering PI(4,5)P2 by recruitment of 5-phosphatase Inp54p (lowers the threshold for STEN) (F); with perturbations of recruitment of PKBA (lowers the threshold of CEN and raises that of STEN) (G); with perturbations of recruiting RacGEF1 (lowers the threshold of CEN/extinguishes STEN) (H). Scale bar top 20μm, bottom 10μm.
Future work is needed to advance the hypothesis that set points of the STEN-CEN machinery determine various types of cellular protrusions. Sophisticated higher resolution imaging is required to directly visualize and analyze wave propagation patterns at sites of different protrusions, given the challenges posed by the highly dynamic and three-dimensional nature of the cortex of migrating cells. Moreover, the extent to which this model is conserved must be tested. It is possible that different cells types have different expression levels of the STEN-CEN components innately, which holds the networks of each cell at a specific set point. Thus, different cells display specific types of protrusions predominantly. Finally, the connection between the wave properties and specific cytoskeleton nucleators needs investigation. It is interesting to note that blebs, usually thought to be controlled by hydrostatic pressure rather than any actin regulators, are nevertheless associated with Ras activation (Figure 5), pointing to the possibility of using the STEN-CEN framework to fully comprehend bleb formation.
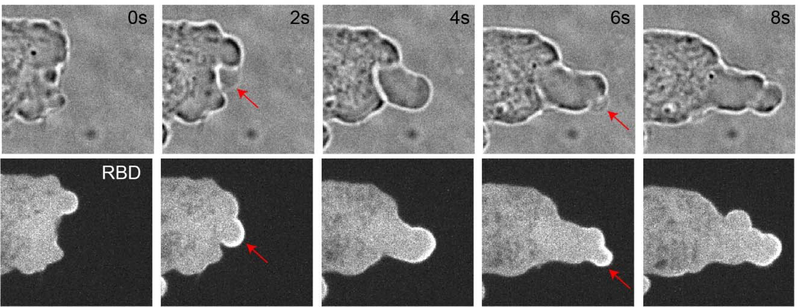
Figure 5. Blebs are associated with Ras activation.
The time-lapse confocal images of the same cell were obtained every 2 s. Confocal images showing blebs under bright field (top) and Ras activity sensor, Ras-binding domain of Raf-1 (RBD-GFP) under confinement in a Dictyostelium cell. Red arrows point to newly forming blebs.
6. Molecular mechanisms giving rise to excitability and wave patterns
Activator-inhibitor systems are typically used to explain wave propagation and excitability. In these systems, the activator is controlled by an autocatalytic loop, and generates the inhibitor, which provides negative feedback to the activator[95, 96]. Further refinement of this model proposes that local regions of the cell cortex transition between inactive, active, and refractory states, designated as B, F, and R, respectively. Mutual inhibition between the B and F states creates the positive feedback loop. The F and R states are related through a delayed negative feedback loop. In resting cells, most of the cortex is in the B state. Once initiated, waves propagate outwardly because diffusion of F state components triggers activation in adjoining B but not R region[97]. Computational analyses based on such reaction–diffusion equations are able to simulate the distributions of F, B and R during wave propagation.
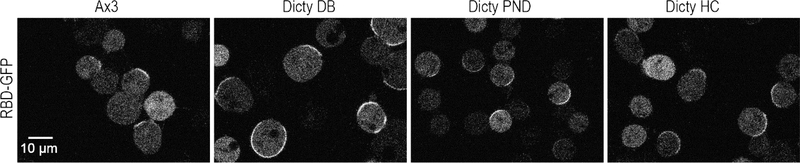
Mutually inhibitory interactions between Ras/Rap and PI(3,4)P2/ PI(4,5)P2 activities comprise a positive feedback loop, consistent with assigning these components as F and B, respectively. When cells are stimulated, Ras is activated, while PI(3,4)P2 levels drop significantly, and PI(4,5)P2 levels drop slightly. When PI(3,4)P2 levels are lowered with genetic or synthetic tools in the cells, Ras/Rap activities are elevated (Figure 3A). Purified Lowe oculocerebrorenal syndrome protein (OCRL) homolog, Dd5P4, has been shown to generate PI(3,4)P2 from PI(3,4,5)P3 in vitro, and Li and Edwards et al have reported PI(3,4)P2 level is low in Dd5P4 null cells. An example is shown in figure 6, wild-type cells display one to three small patches of Ras activity, but the Dd5P4 null cells, which have lower PI(3,4)P2 levels, display higher Ras activities. Similarly, Miao et al found that acute lowering of PI(4,5)P2 levels promotes Ras/Rap activities. These regulations of Ras/Rap activities by PI(3,4)P2/ PI(4,5)P2 may occur through GEF and GAP proteins. In fact, Li and Edwards et al found that PI(3,4)P2 binds to and regulates RasGAP and RapGAP proteins [67, 76]. These results add to the emerging role of PI(3,4)P2 in multiple cellular processes as elaborated in Box 1.
Figure 6. RBD activities in wild-type Ax3 cells and in Dd5P4- cells.
Confocal images showing RBD patches in wild-type Ax3 cells and three independently generated Dd5P4- cell lines. Dd5P4- cells from Dictybase stock center, Dd5P4- cells from Devreotes lab, Dd5P4- cells from Cai lab. Cells are treated with Latrunculin A for 20 min. Scale bar 10μm.
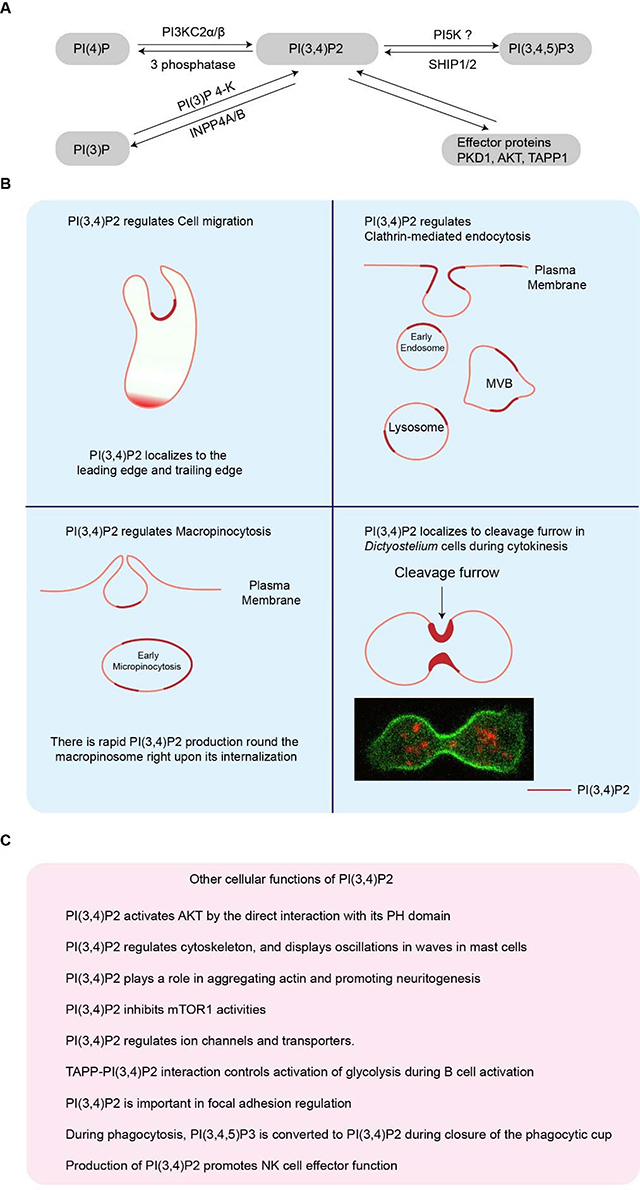
Box 1. The cellular functions of PI(3,4)P2.
(A). PI(3,4)P2 metabolism pathways. (B). Dynamics, localization, and roles of PI(3,4)P2 during cell migration, endocytosis, macropinocytosis and cytokinesis[137–139]. PI(3,4)P2 localizes to the leading edge and lagging edge during cell migration [64, 67, 140, 141]. PI(3,4)P2 localizes to the early endosome, multivesicular body (MVB), and lysosome during clathrin mediated endocytosis [142–145]. PI(3,4)P2 localizes to the back of the macropinocytic cups and early macropinosomes during micropinocytosis [144, 146–150]. PI(3,4)P2 localizes to the cleavage furrow during cytokinesis (C). The other cellular functions of PI(3,4)P2 highlighted in pink box. PI(3,4)P2 activates AKT by the direct interaction with its PH domain[151]. PI(3,4)P2 regulates cytoskeleton, and displays oscillations in waves in mast cells [87, 89, 152]. PI(3,4)P2 plays a role in aggregating actin and promoting neuritogenesis [153]. PI(3,4)P2 inhibits mTOR1 activities[154]. PI(3,4)P2 regulates ion channels and transporters [155, 156]. TAPP-PI(3,4)P2 interaction controls activation of glycolysis during B cell activation[157]. PI(3,4)P2 is important in focal adhesion regulation[158]. During phagocytosis, PI(3,4,5)P3 is converted to PI(3,4)P2 during closure of the phagocytic cup[159]. Production of PI(3,4)P2 promotes NK cell effector function[160].
As positive feedback grows, negative feedback builds in a delayed manner. Charest et al. found that Dictyostelium cells lacking PKBA and PKBR1 display persistently high RasC activity in pull-down assays, suggesting a negative feedback loop involving the phosphorylation of upstream components by downstream PKBs[98–100]. Miao et al. showed that in immobilized cells lacking PKBs, RBD patches are more frequent but the patches are rapidly quenched by recruitment of PKBA [77]. These observations suggest that activation of PKBs serves as a negative feedback loop. Possible mechanisms may include inhibition of RasGEF, Aimless, and activation of PI5K to increase PI(4,5)P2 synthesis. Interestingly, PIP3 as an activator of PKBA, plays a negative role in STEN, while it is also an important positive regulator of actin polymerization. This may occur through a series of PKBA substrates (Figure 3A).
Some other feedback loops have also been described. For example, a positive-feedback loop appears to link cytoskeletal events and PIP3 because inhibition of either reduces the spontaneous activation of the other [101, 102]. There is other evidence that such a positive feedback path involving Ras exists[103]. It has been reported that the cortex of vertebrate and invertebrate oocytes and embryos is also an excitable medium. Positive feedback is mediated by Rho autoactivation and negative feedback is mediated by F-actin-mediated Rho inhibition [104]. Taken together, these suggest that individual molecular regulators of various protrusions are feeding into the same overall molecular machinery. Global properties and feedback loops of the Ras/Rap-centered STEN and Rac/F-actin centered CEN are the true determinants of controlling the entire spectrum of protrusions observed in cell migration.
7. Dysregulations of signaling events in development and disease
We suggest that a variety of diseases involving altered cell migration or morphology may be viewed as changes in the set point of the excitable networks. Many of the signaling molecules involved in the feedback loops are often mutated or deleted in a variety of cancers (Figure 3A). Examples include PIK3CA (p110a) is mutated in a range of tumors [105], PTEN is a well-established tumor suppressor in a large proportion of human cancers[106], some 5-phosphatases such as SHIP2 and PIB5PA and the 4-phosphatase INPP4B act as tumor suppressors in various cancers[107–110], Myosin II is reported to be involved in cancers and regional activation of Myosin II in amoeboid cancer cells drives tumor progression [111–114]. Of course, Ras is a ubiquitous oncogene, with activating mutations in kRas, nRas, or hRas found in 27% of all human cancers[115]. Finally, Rho GTPases are altered in multiple different cancers as well as melanoma[116]. As we have observed in Dictyostelium and cultured mammalian cells, loss or gain of function in these oncogenes and tumor suppressors in vivo likely leads to alterations of the threshold of excitable networks, and consequently, the morphological changes typically associated with cancer cells.
These signaling molecules are also involved in other human diseases, which might also be traced to effects on excitable networks. PTEN is implicated in a subset of autism and Alzheimer’s disease[117, 118]. Mutations in SHIP2 are associated with Type II diabetes [119, 120], hypertension [121] and Alzheimer’s disease [122]. PTEN or SHIP1 deletions in neutrophils show increased motility and excessive recruitment to inflamed sites [64, 123]. Mutations in the PI 5-phosphatase, INPP5E, are causative of MORM syndrome [124]. Mutations in the genes encoding erythroid membrane skeletal proteins lead to hereditary haemolytic anaemias [125]. The mechanisms responsible for the pathophysiology of Wiskott–Aldrich syndrome (WAS) are directly linked to lack of Wiskott–Aldrich syndrome protein (WASP) and deficient actin organization in haematopoietic cells [126].
Cell migration is extensively employed during development to form the embryo. Defects in migration often result in central nervous system malformations [127–130]. Autism Spectrum Disorder (ASD) is associated with changes in the morphology of dendritic spines[131]. Moreover, several syndromes such as Waardenburg, Carpenter, Alagille and CHARGE, have been traced to impaired migration of neural crest cells, resulting in deafness, pigmentation changes, abnormalities of the bones of skull and digits, defective heart, liver and genitals, and growth retardation in adults [132–136].
Conclusions
The excitable network hypothesis for cell migration may provide a novel framework for understanding morphological changes in cells. In this review, we explained how waves of signal transduction activities drive various types of protrusions, which in turn, determine the modes of cell migration. Going forward, it will be important to determine the extent to which the same or similar excitable networks are used in migrating cells in developing embryos as have been elaborated for Dictyostelium and several mammalian cells lines.
Acknowledgements
We thank all members of the P.N.D. laboratory for helpful suggestions for this work. We thank the Johns Hopkins University (JHU) Microscope Facility for confocal microscopy. We thank Tatsat Banerjee for conceptualizing Figure 2B–D. We thank Dr. Huaqing Cai (Institute of Biophysics Chinese Academy of Sciences, China) and Jane Borleis for Dd5p4 null cells. We also thank Dr. Matthieu Piel’s lab (Institut Curie, France) for conducting confinement experiment. This work was supported by NIH grant R35 GM118177 (to P.N.D.), AFOSR MURI FA95501610052, DARPA HR0011-16-C-0139, as well as NIH Grant S10 OD016374 (to S. Kuo of the JHU Microscope Facility).
Footnotes
Disclosure statement
The authors are not aware of any affiliations, memberships, funding, or financial holdings that might be perceived as affecting the objectivity of this review.
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
Reference
- [1].Alexandre G, Chemotaxis Control of Transient Cell Aggregation, J Bacteriol 197(20) (2015) 3230–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Matilla MA, Krell T, The effect of bacterial chemotaxis on host infection and pathogenicity, FEMS Microbiol Rev 42(1) (2018). [DOI] [PubMed] [Google Scholar]
- [3].Miller LD, Russell MH, Alexandre G, Diversity in bacterial chemotactic responses and niche adaptation, Adv Appl Microbiol 66 (2009) 53–75. [DOI] [PubMed] [Google Scholar]
- [4].Swaney KF, Huang CH, Devreotes PN, Eukaryotic chemotaxis: a network of signaling pathways controls motility, directional sensing, and polarity, Annu Rev Biophys 39 (2010) 265–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Armitage JP, Hellingwerf KJ, Light-induced behavioral responses (;phototaxis’) in prokaryotes, Photosynth Res 76(1–3) (2003) 145–55. [DOI] [PubMed] [Google Scholar]
- [6].Cortese B, Palama IE, D’Amone S, Gigli G, Influence of electrotaxis on cell behaviour, Integr Biol (Camb) 6(9) (2014) 817–30. [DOI] [PubMed] [Google Scholar]
- [7].Gao RC, Zhang XD, Sun YH, Kamimura Y, Mogilner A, Devreotes PN, Zhao M, Different roles of membrane potentials in electrotaxis and chemotaxis of dictyostelium cells, Eukaryot Cell 10(9) (2011) 1251–6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [8].Zhao M, Song B, Pu J, Wada T, Reid B, Tai G, Wang F, Guo A, Walczysko P, Gu Y, Sasaki T, Suzuki A, Forrester JV, Bourne HR, Devreotes PN, McCaig CD, Penninger JM, Electrical signals control wound healing through phosphatidylinositol-3-OH kinase-gamma and PTEN, Nature 442(7101) (2006) 457–60. [DOI] [PubMed] [Google Scholar]
- [9].Harland B, Walcott S, Sun SX, Adhesion dynamics and durotaxis in migrating cells, Phys Biol 8(1) (2011) 015011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Lo CM, Wang HB, Dembo M, Wang YL, Cell movement is guided by the rigidity of the substrate, Biophys J 79(1) (2000) 144–52. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Artemenko Y, Axiotakis L Jr., Borleis J, Iglesias PA, Devreotes PN, Chemical and mechanical stimuli act on common signal transduction and cytoskeletal networks, Proc Natl Acad Sci U S A 113(47) (2016) E7500–E7509. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].Ramot D, MacInnis BL, Lee HC, Goodman MB, Thermotaxis is a robust mechanism for thermoregulation in Caenorhabditis elegans nematodes, J Neurosci 28(47) (2008) 12546–57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].Whitaker BD, Poff KL, Thermal adaptation of thermosensing and negative thermotaxis in Dictyostelium, Exp Cell Res 128(1) (1980) 87–93. [DOI] [PubMed] [Google Scholar]
- [14].Zetter BR, The cellular basis of site-specific tumor metastasis, N Engl J Med 322(9) (1990) 605–12. [DOI] [PubMed] [Google Scholar]
- [15].Taraboletti G, Roberts DD, Liotta LA, Thrombospondin-induced tumor cell migration: haptotaxis and chemotaxis are mediated by different molecular domains, J Cell Biol 105(5) (1987) 2409–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Nabeshima K, Kataoka H, Koono M, Enhanced migration of tumor cells in response to collagen degradation products and tumor cell collagenolytic activity, Invasion Metastasis 6(5) (1986) 270–86. [PubMed] [Google Scholar]
- [17].Keller R, Cell migration during gastrulation, Curr Opin Cell Biol 17(5) (2005) 533–41. [DOI] [PubMed] [Google Scholar]
- [18].Leptin M, Gastrulation movements: the logic and the nuts and bolts, Dev Cell 8(3) (2005) 305–20. [DOI] [PubMed] [Google Scholar]
- [19].Theveneau E, Mayor R, Neural crest delamination and migration: from epithelium-to-mesenchyme transition to collective cell migration, Dev Biol 366(1) (2012) 34–54. [DOI] [PubMed] [Google Scholar]
- [20].Blaser H, Reichman-Fried M, Castanon I, Dumstrei K, Marlow FL, Kawakami K, Solnica-Krezel L, Heisenberg CP, Raz E, Migration of zebrafish primordial germ cells: a role for myosin contraction and cytoplasmic flow, Dev Cell 11(5) (2006) 613–27. [DOI] [PubMed] [Google Scholar]
- [21].Richardson BE, Lehmann R, Mechanisms guiding primordial germ cell migration: strategies from different organisms, Nat Rev Mol Cell Biol 11(1) (2010) 37–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [22].Klambt C, Modes and regulation of glial migration in vertebrates and invertebrates, Nat Rev Neurosci 10(11) (2009) 769–79. [DOI] [PubMed] [Google Scholar]
- [23].Nourshargh S, Alon R, Leukocyte migration into inflamed tissues, Immunity 41(5) (2014) 694–707. [DOI] [PubMed] [Google Scholar]
- [24].Weninger W, Biro M, Jain R, Leukocyte migration in the interstitial space of non-lymphoid organs, Nat Rev Immunol 14(4) (2014) 232–46. [DOI] [PubMed] [Google Scholar]
- [25].Shaw TJ, Martin P, Wound repair at a glance, J Cell Sci 122(Pt 18) (2009) 3209–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Baumann K, Stem cells: moving out of the niche, Nat Rev Mol Cell Biol 15(2) (2014) 79. [DOI] [PubMed] [Google Scholar]
- [27].Wright DE, Wagers AJ, Gulati AP, Johnson FL, Weissman IL, Physiological migration of hematopoietic stem and progenitor cells, Science 294(5548) (2001) 1933–6. [DOI] [PubMed] [Google Scholar]
- [28].Morisawa M, Yoshida M, Activation of motility and chemotaxis in the spermatozoa: From invertebrates to humans, Reprod Med Biol 4(2) (2005) 101–114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].van Galen EJ, Ramakers GJ, Rho proteins, mental retardation and the neurobiological basis of intelligence, Prog Brain Res 147 (2005) 295–317. [DOI] [PubMed] [Google Scholar]
- [30].Vicente-Manzanares M, Sancho D, Yanez-Mo M, Sanchez-Madrid F, The leukocyte cytoskeleton in cell migration and immune interactions, Int Rev Cytol 216 (2002) 233–89. [DOI] [PubMed] [Google Scholar]
- [31].Montoya MC, Sancho D, Vicente-Manzanares M, Sanchez-Madrid F, Cell adhesion and polarity during immune interactions, Immunol Rev 186 (2002) 68–82. [DOI] [PubMed] [Google Scholar]
- [32].Friedl P, Gilmour D, Collective cell migration in morphogenesis, regeneration and cancer, Nat Rev Mol Cell Biol 10(7) (2009) 445–57. [DOI] [PubMed] [Google Scholar]
- [33].Lakshman R, Finn A, Neutrophil disorders and their management, J Clin Pathol 54(1) (2001) 7–19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [34].Friedl P, Alexander S, Cancer invasion and the microenvironment: plasticity and reciprocity, Cell 147(5) (2011) 992–1009. [DOI] [PubMed] [Google Scholar]
- [35].Mitchell DR, The evolution of eukaryotic cilia and flagella as motile and sensory organelles, Adv Exp Med Biol 607 (2007) 130–40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [36].Van Haastert PJ, Chemotaxis: insights from the extending pseudopod, J Cell Sci 123(Pt 18) (2010) 3031–7. [DOI] [PubMed] [Google Scholar]
- [37].King JS, Kay RR, The origins and evolution of macropinocytosis, Philos Trans R Soc Lond B Biol Sci 374(1765) (2019) 20180158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [38].Francis EA, Heinrich V, Extension of chemotactic pseudopods by nonadherent human neutrophils does not require or cause calcium bursts, Sci Signal 11(521) (2018). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Moreau HD, Blanch-Mercader C, Attia R, Maurin M, Alraies Z, Sanseau D, Malbec O, Delgado MG, Bousso P, Joanny JF, Voituriez R, Piel M, Lennon-Dumenil AM, Macropinocytosis Overcomes Directional Bias in Dendritic Cells Due to Hydraulic Resistance and Facilitates Space Exploration, Dev Cell 49(2) (2019) 171–188 e5. [DOI] [PubMed] [Google Scholar]
- [40].Yoshida K, Soldati T, Dissection of amoeboid movement into two mechanically distinct modes, J Cell Sci 119(Pt 18) (2006) 3833–44. [DOI] [PubMed] [Google Scholar]
- [41].Wolf K, Mazo I, Leung H, Engelke K, von Andrian UH, Deryugina EI, Strongin AY, Brocker EB, Friedl P, Compensation mechanism in tumor cell migration: mesenchymal-amoeboid transition after blocking of pericellular proteolysis, J Cell Biol 160(2) (2003) 267–77. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [42].Tyson RA, Zatulovskiy E, Kay RR, Bretschneider T, How blebs and pseudopods cooperate during chemotaxis, Proc Natl Acad Sci U S A 111(32) (2014) 11703–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [43].Barnhart EL, Allen GM, Julicher F, Theriot JA, Bipedal locomotion in crawling cells, Biophys J 98(6) (2010) 933–42. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Keren K, Pincus Z, Allen GM, Barnhart EL, Marriott G, Mogilner A, Theriot JA, Mechanism of shape determination in motile cells, Nature 453(7194) (2008) 475–80. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [45].Mejillano MR, Kojima S, Applewhite DA, Gertler FB, Svitkina TM, Borisy GG, Lamellipodial versus filopodial mode of the actin nanomachinery: pivotal role of the filament barbed end, Cell 118(3) (2004) 363–73. [DOI] [PubMed] [Google Scholar]
- [46].Sixt M, Cell migration: fibroblasts find a new way to get ahead, J Cell Biol 197(3) (2012) 347–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Hou Y, Hedberg S, Schneider IC, Differences in adhesion and protrusion properties correlate with differences in migration speed under EGF stimulation, BMC Biophys 5 (2012) 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [48].Murphy DA, Courtneidge SA, The ‘ins’ and ‘outs’ of podosomes and invadopodia: characteristics, formation and function, Nat Rev Mol Cell Biol 12(7) (2011) 413–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Linder S, Nelson D, Weiss M, Aepfelbacher M, Wiskott-Aldrich syndrome protein regulates podosomes in primary human macrophages, Proc Natl Acad Sci U S A 96(17) (1999) 9648–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Roignot J, Peng X, Mostov K, Polarity in mammalian epithelial morphogenesis, Cold Spring Harb Perspect Biol 5(2) (2013). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Martin-Belmonte F, Perez-Moreno M, Epithelial cell polarity, stem cells and cancer, Nat Rev Cancer 12(1) (2011) 23–38. [DOI] [PubMed] [Google Scholar]
- [52].Dai W, Montell DJ, Live Imaging of Border Cell Migration in Drosophila, Methods Mol Biol 1407 (2016) 153–68. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Bianco A, Poukkula M, Cliffe A, Mathieu J, Luque CM, Fulga TA, Rorth P, Two distinct modes of guidance signalling during collective migration of border cells, Nature 448(7151) (2007) 362–5. [DOI] [PubMed] [Google Scholar]
- [54].Ghabrial AS, Krasnow MA, Social interactions among epithelial cells during tracheal branching morphogenesis, Nature 441(7094) (2006) 746–9. [DOI] [PubMed] [Google Scholar]
- [55].Haas P, Gilmour D, Chemokine signaling mediates self-organizing tissue migration in the zebrafish lateral line, Dev Cell 10(5) (2006) 673–80. [DOI] [PubMed] [Google Scholar]
- [56].West-Foyle H, Robinson DN, Cytokinesis mechanics and mechanosensing, Cytoskeleton (Hoboken) 69(10) (2012) 700–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [57].Falnikar A, Baas PW, Neuronal migration re-purposes mechanisms of cytokinesis, Cell Cycle 12(23) (2013) 3577–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [58].Devreotes PN, Bhattacharya S, Edwards M, Iglesias PA, Lampert T, Miao Y, Excitable Signal Transduction Networks in Directed Cell Migration, Annu Rev Cell Dev Biol 33 (2017) 103–125. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [59].Xu X, Wen X, Veltman DM, Keizer-Gunnink I, Pots H, Kortholt A, Jin T, GPCR-controlled membrane recruitment of negative regulator C2GAP1 locally inhibits Ras signaling for adaptation and long-range chemotaxis, Proc Natl Acad Sci U S A 114(47) (2017) E10092–E10101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [60].Parent CA, Making all the right moves: chemotaxis in neutrophils and Dictyostelium, Curr Opin Cell Biol 16(1) (2004) 4–13. [DOI] [PubMed] [Google Scholar]
- [61].Kriebel PW, Barr VA, Rericha EC, Zhang G, Parent CA, Collective cell migration requires vesicular trafficking for chemoattractant delivery at the trailing edge, J Cell Biol 183(5) (2008) 949–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [62].Filic V, Marinovic M, Faix J, Weber I, The IQGAP-related protein DGAP1 mediates signaling to the actin cytoskeleton as an effector and a sequestrator of Rac1 GTPases, Cell Mol Life Sci 71(15) (2014) 2775–85. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [63].Tsai TY, Collins SR, Chan CK, Hadjitheodorou A, Lam PY, Lou SS, Yang HW, Jorgensen J, Ellett F, Irimia D, Davidson MW, Fischer RS, Huttenlocher A, Meyer T, Ferrell JE Jr., Theriot JA, Efficient Front-Rear Coupling in Neutrophil Chemotaxis by Dynamic Myosin II Localization, Dev Cell 49(2) (2019) 189–205 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [64].Lam PY, Yoo SK, Green JM, Huttenlocher A, The SH2-domain-containing inositol 5-phosphatase (SHIP) limits the motility of neutrophils and their recruitment to wounds in zebrafish, J Cell Sci 125(Pt 21) (2012) 4973–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [65].Matsuoka S, Ueda M, Mutual inhibition between PTEN and PIP3 generates bistability for polarity in motile cells, Nat Commun 9(1) (2018) 4481. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [66].Senoo H, Cai H, Wang Y, Sesaki H, Iijima M, The novel RacE-binding protein GflB sharpens Ras activity at the leading edge of migrating cells, Mol Biol Cell 27(10) (2016) 1596–605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [67].Li X, Edwards M, Swaney KF, Singh N, Bhattacharya S, Borleis J, Long Y, Iglesias PA, Chen J, Devreotes PN, Mutually inhibitory Ras-PI(3,4)P2 feedback loops mediate cell migration, Proc Natl Acad Sci U S A 115(39) (2018) E9125–E9134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [68].Veltman DM, Williams TD, Bloomfield G, Chen BC, Betzig E, Insall RH, Kay RR, A plasma membrane template for macropinocytic cups, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [69].Janetopoulos C, Borleis J, Vazquez F, Iijima M, Devreotes P, Temporal and spatial regulation of phosphoinositide signaling mediates cytokinesis, Dev Cell 8(4) (2005) 467–77. [DOI] [PubMed] [Google Scholar]
- [70].Shewan A, Eastburn DJ, Mostov K, Phosphoinositides in cell architecture, Cold Spring Harb Perspect Biol 3(8) (2011) a004796. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [71].Gerisch G, Schroth-Diez B, Muller-Taubenberger A, Ecke M, PIP3 waves and PTEN dynamics in the emergence of cell polarity, Biophys J 103(6) (2012) 1170–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [72].Huang CH, Tang M, Shi C, Iglesias PA, Devreotes PN, An excitable signal integrator couples to an idling cytoskeletal oscillator to drive cell migration, Nat Cell Biol 15(11) (2013) 1307–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [73].Muller-Taubenberger A, Kortholt A, Eichinger L, Simple system--substantial share: the use of Dictyostelium in cell biology and molecular medicine, Eur J Cell Biol 92(2) (2013) 45–53. [DOI] [PubMed] [Google Scholar]
- [74].Chisholm RL, Firtel RA, Insights into morphogenesis from a simple developmental system, Nat Rev Mol Cell Biol 5(7) (2004) 531–41. [DOI] [PubMed] [Google Scholar]
- [75].Mahadeo DC, Parent CA, Signal relay during the life cycle of Dictyostelium, Curr Top Dev Biol 73 (2006) 115–40. [DOI] [PubMed] [Google Scholar]
- [76].Miao Y, Bhattacharya S, Edwards M, Cai H, Inoue T, Iglesias PA, Devreotes PN, Altering the threshold of an excitable signal transduction network changes cell migratory modes, Nat Cell Biol 19(4) (2017) 329–340. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [77].Miao Y, Bhattacharya S, Banerjee T, Abubaker-Sharif B, Long Y, Inoue T, Iglesias PA, Devreotes PN, Wave patterns organize cellular protrusions and control cortical dynamics, Mol Syst Biol 15(3) (2019) e8585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [78].Bailly M, Ichetovkin I, Grant W, Zebda N, Machesky LM, Segall JE, Condeelis J, The F-actin side binding activity of the Arp2/3 complex is essential for actin nucleation and lamellipod extension, Curr Biol 11(8) (2001) 620–5. [DOI] [PubMed] [Google Scholar]
- [79].Blanchoin L, Amann KJ, Higgs HN, Marchand JB, Kaiser DA, Pollard TD, Direct observation of dendritic actin filament networks nucleated by Arp2/3 complex and WASP/Scar proteins, Nature 404(6781) (2000) 1007–11. [DOI] [PubMed] [Google Scholar]
- [80].Yang C, Czech L, Gerboth S, Kojima S, Scita G, Svitkina T, Novel roles of formin mDia2 in lamellipodia and filopodia formation in motile cells, PLoS Biol 5(11) (2007) e317. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [81].Tang M, Wang M, Shi C, Iglesias PA, Devreotes PN, Huang CH, Evolutionarily conserved coupling of adaptive and excitable networks mediates eukaryotic chemotaxis, Nat Commun 5 (2014) 5175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [82].Gerhardt M, Ecke M, Walz M, Stengl A, Beta C, Gerisch G, Actin and PIP3 waves in giant cells reveal the inherent length scale of an excited state, J Cell Sci 127(Pt 20) (2014) 4507–17. [DOI] [PubMed] [Google Scholar]
- [83].Vicker MG, F-actin assembly in Dictyostelium cell locomotion and shape oscillations propagates as a self-organized reaction-diffusion wave, FEBS Lett 510(1–2) (2002) 5–9. [DOI] [PubMed] [Google Scholar]
- [84].Weiner OD, Marganski WA, Wu LF, Altschuler SJ, Kirschner MW, An actin-based wave generator organizes cell motility, PLoS Biol 5(9) (2007) e221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [85].Hepper I, Schymeinsky J, Weckbach LT, Jakob SM, Frommhold D, Sixt M, Laschinger M, Sperandio M, Walzog B, The mammalian actin-binding protein 1 is critical for spreading and intraluminal crawling of neutrophils under flow conditions, J Immunol 188(9) (2012) 4590–601. [DOI] [PubMed] [Google Scholar]
- [86].Wang MJ, Artemenko Y, Cai WJ, Iglesias PA, Devreotes PN, The directional response of chemotactic cells depends on a balance between cytoskeletal architecture and the external gradient, Cell Rep 9(3) (2014) 1110–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [87].Masters TA, Sheetz MP, Gauthier NC, F-actin waves, actin cortex disassembly and focal exocytosis driven by actin-phosphoinositide positive feedback, Cytoskeleton (Hoboken) 73(4) (2016) 180–96. [DOI] [PubMed] [Google Scholar]
- [88].Wu M, Wu X, De Camilli P, Calcium oscillations-coupled conversion of actin travelling waves to standing oscillations, Proc Natl Acad Sci U S A 110(4) (2013) 1339–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [89].Xiong D, Xiao S, Guo S, Lin Q, Nakatsu F, Wu M, Frequency and amplitude control of cortical oscillations by phosphoinositide waves, Nat Chem Biol 12(3) (2016) 159–66. [DOI] [PubMed] [Google Scholar]
- [90].Colin-York H, Li D, Korobchevskaya K, Chang VT, Betzig E, Eggeling C, Fritzsche M, Cytoskeletal actin patterns shape mast cell activation, Commun Biol 2 (2019) 93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [91].Shi C, Huang CH, Devreotes PN, Iglesias PA, Interaction of motility, directional sensing, and polarity modules recreates the behaviors of chemotaxing cells, PLoS Comput Biol 9(7) (2013) e1003122. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [92].Van Haastert PJ, A stochastic model for chemotaxis based on the ordered extension of pseudopods, Biophys J 99(10) (2010) 3345–54. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [93].Insall RH, Understanding eukaryotic chemotaxis: a pseudopod-centred view, Nat Rev Mol Cell Biol 11(6) (2010) 453–8. [DOI] [PubMed] [Google Scholar]
- [94].Andrew N, Insall RH, Chemotaxis in shallow gradients is mediated independently of PtdIns 3-kinase by biased choices between random protrusions, Nat Cell Biol 9(2) (2007) 193–200. [DOI] [PubMed] [Google Scholar]
- [95].Meinhardt H, de Boer PA, Pattern formation in Escherichia coli: a model for the pole-to-pole oscillations of Min proteins and the localization of the division site, Proc Natl Acad Sci U S A 98(25) (2001) 14202–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [96].Xiong Y, Huang CH, Iglesias PA, Devreotes PN, Cells navigate with a local-excitation, global-inhibition-biased excitable network, Proc Natl Acad Sci U S A 107(40) (2010) 17079–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [97].Iglesias PA, Devreotes PN, Biased excitable networks: how cells direct motion in response to gradients, Curr Opin Cell Biol 24(2) (2012) 245–53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [98].Charest PG, Shen Z, Lakoduk A, Sasaki AT, Briggs SP, Firtel RA, A Ras signaling complex controls the RasC-TORC2 pathway and directed cell migration, Dev Cell 18(5) (2010) 737–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [99].Kamimura Y, Xiong Y, Iglesias PA, Hoeller O, Bolourani P, Devreotes PN, PIP3-independent activation of TorC2 and PKB at the cell’s leading edge mediates chemotaxis, Curr Biol 18(14) (2008) 1034–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [100].Fets L, Nichols JM, Kay RR, A PIP5 kinase essential for efficient chemotactic signaling, Curr Biol 24(4) (2014) 415–21. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [101].Inoue T, Meyer T, Synthetic activation of endogenous PI3K and Rac identifies an AND-gate switch for cell polarization and migration, PLoS One 3(8) (2008) e3068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [102].Weiner OD, Neilsen PO, Prestwich GD, Kirschner MW, Cantley LC, Bourne HR, A PtdInsP(3)- and Rho GTPase-mediated positive feedback loop regulates neutrophil polarity, Nat Cell Biol 4(7) (2002) 509–13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [103].Sasaki AT, Janetopoulos C, Lee S, Charest PG, Takeda K, Sundheimer LW, Meili R, Devreotes PN, Firtel RA, G protein-independent Ras/PI3K/F-actin circuit regulates basic cell motility, J Cell Biol 178(2) (2007) 185–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [104].Bement WM, Leda M, Moe AM, Kita AM, Larson ME, Golding AE, Pfeuti C, Su KC, Miller AL, Goryachev AB, von Dassow G, Activator-inhibitor coupling between Rho signalling and actin assembly makes the cell cortex an excitable medium, Nat Cell Biol 17(11) (2015) 1471–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [105].Samuels Y, Waldman T, Oncogenic mutations of PIK3CA in human cancers, Curr Top Microbiol Immunol 347 (2010) 21–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [106].Milella M, Falcone I, Conciatori F, Cesta Incani U, Del Curatolo A, Inzerilli N, Nuzzo CM, Vaccaro V, Vari S, Cognetti F, Ciuffreda L, PTEN: Multiple Functions in Human Malignant Tumors, Front Oncol 5 (2015) 24. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [107].Fuhler GM, Brooks R, Toms B, Iyer S, Gengo EA, Park MY, Gumbleton M, Viernes DR, Chisholm JD, Kerr WG, Therapeutic potential of SH2 domain-containing inositol-5’-phosphatase 1 (SHIP1) and SHIP2 inhibition in cancer, Mol Med 18 (2012) 65–75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [108].Fernandes S, Iyer S, Kerr WG, Role of SHIP1 in cancer and mucosal inflammation, Ann N Y Acad Sci 1280 (2013) 6–10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [109].Gewinner C, Wang ZC, Richardson A, Teruya-Feldstein J, Etemadmoghadam D, Bowtell D, Barretina J, Lin WM, Rameh L, Salmena L, Pandolfi PP, Cantley LC, Evidence that inositol polyphosphate 4-phosphatase type II is a tumor suppressor that inhibits PI3K signaling, Cancer Cell 16(2) (2009) 115–25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [110].Ivetac I, Gurung R, Hakim S, Horan KA, Sheffield DA, Binge LC, Majerus PW, Tiganis T, Mitchell CA, Regulation of PI(3)K/Akt signalling and cellular transformation by inositol polyphosphate 4-phosphatase-1, EMBO Rep 10(5) (2009) 487–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [111].Georgouli M, Herraiz C, Crosas-Molist E, Fanshawe B, Maiques O, Perdrix A, Pandya P, Rodriguez-Hernandez I, Ilieva KM, Cantelli G, Karagiannis P, Mele S, Lam H, Josephs DH, Matias-Guiu X, Marti RM, Nestle FO, Orgaz JL, Malanchi I, Fruhwirth GO, Karagiannis SN, Sanz-Moreno V, Regional Activation of Myosin II in Cancer Cells Drives Tumor Progression via a Secretory Cross-Talk with the Immune Microenvironment, Cell 176(4) (2019) 757–774 e23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [112].Aguilar-Cuenca R, Juanes-Garcia A, Vicente-Manzanares M, Myosin II in mechanotransduction: master and commander of cell migration, morphogenesis, and cancer, Cell Mol Life Sci 71(3) (2014) 479–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [113].Lee W, Lim S, Kim Y, The role of myosin II in glioma invasion: A mathematical model, PLoS One 12(2) (2017) e0171312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [114].Surcel A, Schiffhauer ES, Thomas DG, Zhu Q, DiNapoli KT, Herbig M, Otto O, West-Foyle H, Jacobi A, Krater M, Plak K, Guck J, Jaffee EM, Iglesias PA, Anders RA, Robinson DN, Targeting Mechanoresponsive Proteins in Pancreatic Cancer: 4-Hydroxyacetophenone Blocks Dissemination and Invasion by Activating MYH14, Cancer Res 79(18) (2019) 4665–4678. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [115].Hobbs GA, Der CJ, Rossman KL, RAS isoforms and mutations in cancer at a glance, J Cell Sci 129(7) (2016) 1287–92. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [116].Haga RB, Ridley AJ, Rho GTPases: Regulation and roles in cancer cell biology, Small GTPases 7(4) (2016) 207–221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [117].Zhou J, Parada LF, PTEN signaling in autism spectrum disorders, Curr Opin Neurobiol 22(5) (2012) 873–9. [DOI] [PubMed] [Google Scholar]
- [118].Griffin RJ, Moloney A, Kelliher M, Johnston JA, Ravid R, Dockery P, O’Connor R, O’Neill C, Activation of Akt/PKB, increased phosphorylation of Akt substrates and loss and altered distribution of Akt and PTEN are features of Alzheimer’s disease pathology, J Neurochem 93(1) (2005) 105–17. [DOI] [PubMed] [Google Scholar]
- [119].Kagawa S, Sasaoka T, Yaguchi S, Ishihara H, Tsuneki H, Murakami S, Fukui K, Wada T, Kobayashi S, Kimura I, Kobayashi M, Impact of SRC homology 2-containing inositol 5’-phosphatase 2 gene polymorphisms detected in a Japanese population on insulin signaling, J Clin Endocrinol Metab 90(5) (2005) 2911–9. [DOI] [PubMed] [Google Scholar]
- [120].Nicolas A, Kenna KP, Renton AE, Ticozzi N, Faghri F, Chia R, Dominov JA, Kenna BJ, Nalls MA, Keagle P, Rivera AM, van Rheenen W, Murphy NA, van Vugt J, Geiger JT, Van der Spek RA, Pliner HA, Shankaracharya B.N. Smith, Marangi G, Topp SD, Abramzon Y, Gkazi AS, Eicher JD, Kenna A, Consortium I, Mora G, Calvo A, Mazzini L, Riva N, Mandrioli J, Caponnetto C, Battistini S, Volanti P, La Bella V, Conforti FL, Borghero G, Messina S, Simone IL, Trojsi F, Salvi F, Logullo FO, D’Alfonso S, Corrado L, Capasso M, Ferrucci L, A.L.S.C.C. Genomic Translation for, Moreno CAM, Kamalakaran S, Goldstein DB, Consortium ALSS, Gitler AD, Harris T, Myers RM, Consortium NA, Phatnani H, Musunuri RL, Evani US, Abhyankar A, Zody MC, Answer ALSF, Kaye J, Finkbeiner S, Wyman SK, LeNail A, Lima L, Fraenkel E, Svendsen CN, Thompson LM, Van Eyk JE, Berry JD, Miller TM, Kolb SJ, Cudkowicz M, Baxi E, A.L.S. Clinical Research in, C. Related Disorders for Therapeutic Development, Benatar M, Taylor JP, Rampersaud E, Wu G, Wuu J, Consortium S, Lauria G, Verde F, Fogh I, Tiloca C, Comi GP, Soraru G, Cereda C, French ALSC, Corcia P, Laaksovirta H, Myllykangas L, Jansson L, Valori M, Ealing J, Hamdalla H, Rollinson S, Pickering-Brown S, Orrell RW, Sidle KC, Malaspina A, Hardy J, Singleton AB, Johnson JO, Arepalli S, Sapp PC, McKenna-Yasek D, Polak M, Asress S, Al-Sarraj S, King A, Troakes C, Vance C, de Belleroche J, Baas F, Ten Asbroek A, Munoz-Blanco JL, Hernandez DG, Ding J, Gibbs JR, Scholz SW, Floeter MK, Campbell RH, Landi F, Bowser R, Pulst SM, Ravits JM, MacGowan DJL, Kirby J, Pioro EP, Pamphlett R, Broach J, Gerhard G, Dunckley TL, Brady CB, Kowall NW, Troncoso JC, Le Ber I, Mouzat K, Lumbroso S, Heiman-Patterson TD, Kamel F, Van Den Bosch L, Baloh RH, Strom TM, Meitinger T, Shatunov A, Van Eijk KR, de Carvalho M, Kooyman M, Middelkoop B, Moisse M, McLaughlin RL, Van Es MA, Weber M, Boylan KB, Van Blitterswijk M, Rademakers R, Morrison KE, Basak AN, Mora JS, Drory VE, Shaw PJ, Turner MR, Talbot K, Hardiman O, Williams KL, Fifita JA, Nicholson GA, Blair IP, Rouleau GA, Esteban-Perez J, Garcia-Redondo A, Al-Chalabi A, Project Min EALSSC, Rogaeva E, Zinman L, Ostrow LW, Maragakis NJ, Rothstein JD, Simmons Z, Cooper-Knock J, Brice A, Goutman SA, Feldman EL, Gibson SB, Taroni F, Ratti A, Gellera C, Van Damme P, Robberecht W, Fratta P, Sabatelli M, Lunetta C, Ludolph AC, Andersen PM, Weishaupt JH, Camu W, Trojanowski JQ, Van Deerlin VM, Brown RH Jr., van den Berg LH, Veldink JH, Harms MB, Glass JD, Stone DJ, Tienari P, Silani V, Chio A, Shaw CE, Traynor BJ, Landers JE, Genome-wide Analyses Identify KIF5A as a Novel ALS Gene, Neuron 97(6) (2018) 1268–1283 e6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [121].Marion E, Kaisaki PJ, Pouillon V, Gueydan C, Levy JC, Bodson A, Krzentowski G, Daubresse JC, Mockel J, Behrends J, Servais G, Szpirer C, Kruys V, Gauguier D, Schurmans S, The gene INPPL1, encoding the lipid phosphatase SHIP2, is a candidate for type 2 diabetes in rat and man, Diabetes 51(7) (2002) 2012–7. [DOI] [PubMed] [Google Scholar]
- [122].Kam TI, Park H, Gwon Y, Song S, Kim SH, Moon SW, Jo DG, Jung YK, FcgammaRIIb-SHIP2 axis links Abeta to tau pathology by disrupting phosphoinositide metabolism in Alzheimer’s disease model, Elife 5 (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [123].Sarraj B, Massberg S, Li Y, Kasorn A, Subramanian K, Loison F, Silberstein LE, von Andrian U, Luo HR, Myeloid-specific deletion of tumor suppressor PTEN augments neutrophil transendothelial migration during inflammation, J Immunol 182(11) (2009) 7190–200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [124].Jacoby M, Cox JJ, Gayral S, Hampshire DJ, Ayub M, Blockmans M, Pernot E, Kisseleva MV, Compere P, Schiffmann SN, Gergely F, Riley JH, Perez-Morga D, Woods CG, Schurmans S, INPP5E mutations cause primary cilium signaling defects, ciliary instability and ciliopathies in human and mouse, Nat Genet 41(9) (2009) 1027–31. [DOI] [PubMed] [Google Scholar]
- [125].Birkenmeier CS, Barker JE, Hereditary haemolytic anaemias: unexpected sequelae of mutations in the genes for erythroid membrane skeletal proteins, J Pathol 204(4) (2004) 450–9. [DOI] [PubMed] [Google Scholar]
- [126].Calle Y, Chou HC, Thrasher AJ, Jones GE, Wiskott-Aldrich syndrome protein and the cytoskeletal dynamics of dendritic cells, J Pathol 204(4) (2004) 460–9. [DOI] [PubMed] [Google Scholar]
- [127].Hatten ME, Central nervous system neuronal migration, Annu Rev Neurosci 22 (1999) 511–39. [DOI] [PubMed] [Google Scholar]
- [128].McLennan R, Dyson L, Prather KW, Morrison JA, Baker RE, Maini PK, Kulesa PM, Multiscale mechanisms of cell migration during development: theory and experiment, Development 139(16) (2012) 2935–44. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [129].Fraser MM, Bayazitov IT, Zakharenko SS, Baker SJ, Phosphatase and tensin homolog, deleted on chromosome 10 deficiency in brain causes defects in synaptic structure, transmission and plasticity, and myelination abnormalities, Neuroscience 151(2) (2008) 476–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [130].Chow LM, Endersby R, Zhu X, Rankin S, Qu C, Zhang J, Broniscer A, Ellison DW, Baker SJ, Cooperativity within and among Pten, p53, and Rb pathways induces high-grade astrocytoma in adult brain, Cancer Cell 19(3) (2011) 305–16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [131].Penzes P, Cahill ME, Jones KA, VanLeeuwen JE, Woolfrey KM, Dendritic spine pathology in neuropsychiatric disorders, Nat Neurosci 14(3) (2011) 285–93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [132].Hutson MR, Kirby ML, Neural crest and cardiovascular development: a 20-year perspective, Birth Defects Res C Embryo Today 69(1) (2003) 2–13. [DOI] [PubMed] [Google Scholar]
- [133].Gitler AD, Brown CB, Kochilas L, Li J, Epstein JA, Neural crest migration and mouse models of congenital heart disease, Cold Spring Harb Symp Quant Biol 67 (2002) 57–62. [DOI] [PubMed] [Google Scholar]
- [134].Stoller JZ, Epstein JA, Cardiac neural crest, Semin Cell Dev Biol 16(6) (2005) 704–15. [DOI] [PubMed] [Google Scholar]
- [135].Heanue TA, Pachnis V, Enteric nervous system development and Hirschsprung’s disease: advances in genetic and stem cell studies, Nat Rev Neurosci 8(6) (2007) 466–79. [DOI] [PubMed] [Google Scholar]
- [136].Kurosaka S, Kashina A, Cell biology of embryonic migration, Birth Defects Res C Embryo Today 84(2) (2008) 102–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [137].Hawkins PT, Stephens LR, Emerging evidence of signalling roles for PI(3,4)P2 in Class I and II PI3K-regulated pathways, Biochem Soc Trans 44(1) (2016) 307–14. [DOI] [PubMed] [Google Scholar]
- [138].Ramos AR, Elong Edimo W, Erneux C, Phosphoinositide 5-phosphatase activities control cell motility in glioblastoma: Two phosphoinositides PI(4,5)P2 and PI(3,4)P2 are involved, Adv Biol Regul 67 (2018) 40–48. [DOI] [PubMed] [Google Scholar]
- [139].Fukumoto M, Ijuin T, Takenawa T, PI(3,4)P2 plays critical roles in the regulation of focal adhesion dynamics of MDA-MB-231 breast cancer cells, Cancer Sci 108(5) (2017) 941–951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [140].Yoo SK, Deng Q, Cavnar PJ, Wu YI, Hahn KM, Huttenlocher A, Differential regulation of protrusion and polarity by PI3K during neutrophil motility in live zebrafish, Dev Cell 18(2) (2010) 226–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [141].Li H, Wu X, Hou S, Malek M, Kielkowska A, Noh E, Makondo KJ, Du Q, Wilkins JA, Johnston JB, Gibson SB, Lin F, Marshall AJ, Phosphatidylinositol-3,4-Bisphosphate and Its Binding Protein Lamellipodin Regulate Chemotaxis of Malignant B Lymphocytes, J Immunol 196(2) (2016) 586–95. [DOI] [PubMed] [Google Scholar]
- [142].Wallroth A, Haucke V, Phosphoinositide conversion in endocytosis and the endolysosomal system, J Biol Chem 293(5) (2018) 1526–1535. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [143].Watt SA, Kimber WA, Fleming IN, Leslie NR, Downes CP, Lucocq JM, Detection of novel intracellular agonist responsive pools of phosphatidylinositol 3,4-bisphosphate using the TAPP1 pleckstrin homology domain in immunoelectron microscopy, Biochem J 377(Pt 3) (2004) 653–63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [144].Posor Y, Eichhorn-Gruenig M, Puchkov D, Schoneberg J, Ullrich A, Lampe A, Muller R, Zarbakhsh S, Gulluni F, Hirsch E, Krauss M, Schultz C, Schmoranzer J, Noe F, Haucke V, Spatiotemporal control of endocytosis by phosphatidylinositol-3,4-bisphosphate, Nature 499(7457) (2013) 233–7. [DOI] [PubMed] [Google Scholar]
- [145].Posor Y, Eichhorn-Grunig M, Haucke V, Phosphoinositides in endocytosis, Biochim Biophys Acta 1851(6) (2015) 794–804. [DOI] [PubMed] [Google Scholar]
- [146].Valenzuela-Iglesias A, Sharma VP, Beaty BT, Ding Z, Gutierrez-Millan LE, Roy P, Condeelis JS, Bravo-Cordero JJ, Profilin1 regulates invadopodium maturation in human breast cancer cells, Eur J Cell Biol 94(2) (2015) 78–89. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [147].Maekawa M, Terasaka S, Mochizuki Y, Kawai K, Ikeda Y, Araki N, Skolnik EY, Taguchi T, Arai H, Sequential breakdown of 3-phosphorylated phosphoinositides is essential for the completion of macropinocytosis, Proc Natl Acad Sci U S A 111(11) (2014) E978–87. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [148].Dormann D, Weijer G, Dowler S, Weijer CJ, In vivo analysis of 3-phosphoinositide dynamics during Dictyostelium phagocytosis and chemotaxis, J Cell Sci 117(Pt 26) (2004) 6497–509. [DOI] [PubMed] [Google Scholar]
- [149].Li H, Marshall AJ, Phosphatidylinositol (3,4) bisphosphate-specific phosphatases and effector proteins: A distinct branch of PI3K signaling, Cell Signal 27(9) (2015) 1789–98. [DOI] [PubMed] [Google Scholar]
- [150].Erneux C, Edimo WE, Deneubourg L, Pirson I, SHIP2 multiple functions: a balance between a negative control of PtdIns(3,4,5)P(3) level, a positive control of PtdIns(3,4)P(2) production, and intrinsic docking properties, J Cell Biochem 112(9) (2011) 2203–9. [DOI] [PubMed] [Google Scholar]
- [151].Franke TF, Kaplan DR, Cantley LC, Toker A, Direct regulation of the Akt proto-oncogene product by phosphatidylinositol-3,4-bisphosphate, Science 275(5300) (1997) 665–8. [DOI] [PubMed] [Google Scholar]
- [152].Yang Y, Xiong D, Pipathsouk A, Weiner OD, Wu M, Clathrin Assembly Defines the Onset and Geometry of Cortical Patterning, Dev Cell 43(4) (2017) 507–521 e4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [153].Zhang SX, Duan LH, He SJ, Zhuang GF, Yu X, Phosphatidylinositol 3,4-bisphosphate regulates neurite initiation and dendrite morphogenesis via actin aggregation, Cell Res 27(2) (2017) 253–273. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [154].Marat AL, Wallroth A, Lo WT, Muller R, Norata GD, Falasca M, Schultz C, Haucke V, mTORC1 activity repression by late endosomal phosphatidylinositol 3,4-bisphosphate, Science 356(6341) (2017) 968–972. [DOI] [PubMed] [Google Scholar]
- [155].Rohacs T, Lopes CM, Jin T, Ramdya PP, Molnar Z, Logothetis DE, Specificity of activation by phosphoinositides determines lipid regulation of Kir channels, Proc Natl Acad Sci U S A 100(2) (2003) 745–50. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [156].Yamaguchi S, Kurokawa T, Taira I, Aoki N, Sakata S, Okamura Y, Homma KJ, Potential role of voltage-sensing phosphatases in regulation of cell structure through the production of PI(3,4)P2, J Cell Physiol 229(4) (2014) 422–33. [DOI] [PubMed] [Google Scholar]
- [157].Jayachandran N, Mejia EM, Sheikholeslami K, Sher AA, Hou S, Hatch GM, Marshall AJ, TAPP Adaptors Control B Cell Metabolism by Modulating the Phosphatidylinositol 3-Kinase Signaling Pathway: A Novel Regulatory Circuit Preventing Autoimmunity, J Immunol 201(2) (2018) 406–416. [DOI] [PubMed] [Google Scholar]
- [158].Oikawa T, Takenawa T, PtdIns(3,4)P2 instigates focal adhesions to generate podosomes, Cell Adh Migr 3(2) (2009) 195–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [159].Loovers HM, Kortholt A, de Groote H, Whitty L, Nussbaum RL, van Haastert PJ, Regulation of phagocytosis in Dictyostelium by the inositol 5-phosphatase OCRL homolog Dd5P4, Traffic 8(5) (2007) 618–28. [DOI] [PubMed] [Google Scholar]
- [160].Sauer K, Park E, Siegemund S, French AR, Wahle JA, Sternberg L, Rigaud S, Jonsson AH, Yokoyama WM, Huang YH, Inositol tetrakisphosphate limits NK cell effector functions by controlling PI3K signaling, Blood 121(2) (2013) 286–97. [DOI] [PMC free article] [PubMed] [Google Scholar]