Abstract
Alcohol use disorders (AUD) are defined by several symptom criteria, which can be further dissected at the genetic level. Over the past several years, our understanding of the genetic factors influencing alcohol use and abuse has progressed tremendously; hundreds of loci have now been implicated in different aspects of alcohol use. Previously known associations with alcohol metabolizing enzymes (ADH1B, ALDH2) have been definitively replicated. Additionally, novel associations with loci containing the genes KLB, GCKR, CRHR1 and CADM2 have been reported. Downstream analyses have leveraged these genetic findings to reveal important relationships between alcohol use behaviors and both physical and mental health. AUD and aspects of alcohol misuse have been shown to overlap strongly with psychiatric disorders, whereas aspects of alcohol consumption have shown stronger links to metabolism. These results demonstrate that the genetic architecture of alcohol consumption only partially overlaps with the genetics of clinically defined AUD. We discuss the limitations of using quantitative measures of alcohol use as proxy measures for AUD, and outline how future studies will require careful phenotype harmonization to properly capture the genetic liability to AUD.
Keywords: alcoholism, alcohol consumption, AUDIT, alcohol-metabolizing genes, Genome-wide association studies, Genetics
Introduction
Alcohol abuse is a global problem, constituting the seventh leading risk factor for death and disability (1). Worldwide, over 100 million people had an alcohol use disorder (AUD) in 2016. Statistics from the National Survey on Drug Use and Health show that >85% of adults in the United States report ever having consumed alcohol, with >25% reporting binge drinking in the past month (2). The proportion of adults in the United States with an AUD is estimated to be 6.2% (2). Alcohol use behaviors are complex, and how and why people drink is partially influenced by genetic factors. However, identifying the genetic factors that increase the risk for harmful drinking has been challenging, partially because patterns of alcohol use are dynamic across the lifespan. The terms used to describe alcohol use and abuse are as diverse as the behaviors themselves. Hazardous drinking describes heavy drinking that places an individual at risk for future harm. Harmful drinking and alcohol abuse are defined as drinking that causes mental or physical damage to the individual. These descriptive terms were devised to identify individuals who would benefit from brief interventions and are assessed using screening questionnaires such as the Alcohol Use Disorders Identification Test (AUDIT). Alcohol dependence (AD) was, until recently, defined according to the DSM-IV and required the presence of 3 or more of 7 criteria in a 12-month period. The DSM-IV made a distinction between alcohol abuse and dependence that was removed under DSM-V and replaced with ‘mild’ to ‘severe’ definitions of AUD. Genetic studies encompass the wide range of alcohol use phenotypes; in this review we mirror the language used in the original studies.
AUD can be viewed as the end point of a series of transitions (Figure 1), which begin with the initiation of use, continue with the escalation to hazardous drinking and culminate in compulsive harmful use that persists despite negative consequences. Genome-wide association studies (GWAS) have been instrumental in discovering novel genetic loci associated with multiple psychiatric conditions. In the field of AUD genetics, studies have mostly focused on either levels of consumption or AUD diagnosis. Recent GWAS have now begun to identify hundreds of genome-wide significant variants, and provide evidence that the components of alcohol use behavior have a distinct genetic architecture.
Figure 1.
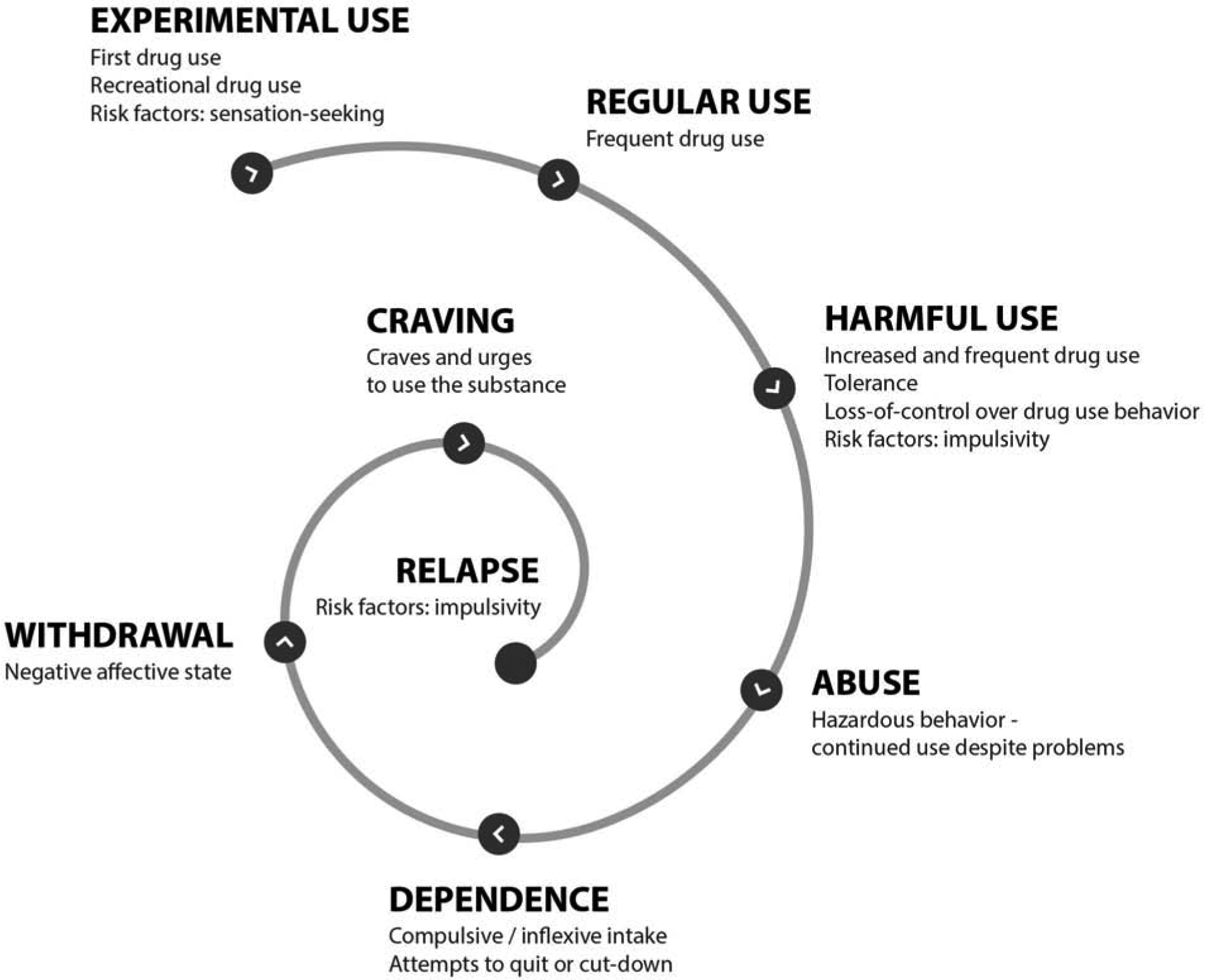
The downward spiral of alcohol use disorders. There is an initial prodromal stage during which certain individuals may be at increased risk to be exposed to alcohol. Personality traits such as sensation seeking are thought to promote alcohol experimentation, and transition to a more regular use of alcohol. As alcohol use patterns become more frequent, and tolerance develops, individuals are more likely to loss control over alcohol drinking behavior; risk factors, such as impulsivity, are considered to promote the transition to a more harmful use of alcohol. Alcohol intake may then become inflexible and compulsive, leading to hazardous or continuous alcohol use despite the negative physical and psychological consequences, and ultimately stagnating into dependence. Attempts to quit or cut-down may become apparent; these may be followed by an aversive negative affective state, or withdrawal, thereby increasing the urges to use alcohol, precipitating relapse, and thus perpetuating the spiral of alcohol use disorders.
In this review, we provide an overview of recent molecular genetic findings of alcohol use behaviors from the largest GWAS performed to date. Other reviews have elegantly summarized findings from twin and family studies of heritability, linkage, candidate gene and GWAS [e.g. (3–6)], and we extend on recent reviews of the molecular genetics of AUD (7–9) by including additional GWAS of alcohol use behaviors that identify genome-wide significant hits (P-value < 5 × 10−8). In addition, we discuss the application of polygenic methods, which provide mounting evidence that alcohol use and misuse are partially distinct. Finally, we delineate future directions to investigate the different etiologic sources that underlie the life course of alcohol use behaviors.
Design strategies for enhancing AUD genetic discovery
For decades, candidate gene studies were used to determine the contribution of specific genes that increase risk for AUD. Candidate gene studies tended to focus on genes that influenced pharmacokinetic and pharmacodynamic (e.g. dopaminergic, glutamatergic and opioid signaling systems) factors. Larger genetic studies have generally not replicated the findings from candidate gene studies (10). One exception to this are the genes encoding ethanol metabolizing enzymes, particularly alcohol dehydrogenase (ADH) and aldehyde dehydrogenase (ALDH), which have repeatedly been shown to have the largest impact on alcohol consumption and risk for AUD (7).
As study designs have evolved to incorporate GWAS, researchers have been able to scan the whole genome without any hypotheses about the underlying biology of alcohol use behaviors. Initial efforts focused on collecting clinically-defined cases of AUD, but these ascertainment strategies could not amass the large sample sizes required for GWAS (11). Accordingly, multi-ethnic and clinically-defined samples have been combined through the Psychiatric Genomic Consortium of Substance Use Disorders (PGC-SUD) working group. The efforts of the PGC-SUD have led to a trans-ancestral meta-analysis consisting of almost 15,000 AD cases and almost 38,000 controls from 28 independent cohorts (12), identifying a single locus (ADH1B), which was robustly associated with AD. More recently, using information from electronic health records to infer AUD status, a GWAS of 274,424 multi-ethnic individuals from the Million Veterans Program (MVP) cohort identified 10 loci associated with AUD (including ADH1B) (18). Kranzler et al (18) showed that alcohol consumption and AUD were genetically correlated but distinct, thus allowing them to adjust for consumption in the AUD GWAS and for AUD in the GWAS of consumption.
In parallel with these efforts, which have focused on clinical diagnoses, other GWAS have incorporated continuous measures of alcohol use. These include self-reported weekly alcohol intake or the scores from screening questionnaires such as the AUDIT (13). The AUDIT can be decomposed to provide a measure of alcohol use from the first 3 questions (AUDIT-C) and misuse from questions 4–10 (AUDIT-P). These quantitative measures are available in large population-based cohorts such as the UK Biobank (UKB), MVP and 23andMe. The GWAS meta-analysis of AUDIT identified 10 associated risk loci (14). Large consortia were also formed to collate quantitative measures of alcohol use, including AlcGen (15) and the GWAS & Sequencing Consortium of Alcohol and Nicotine Use (GSCAN). GSCAN have recently identified nearly 100 loci associated with alcohol consumption (17). The MVP study (18) also examined alcohol consumption, allowing for an explicit comparison between AUD and consumption in a single population; of the 18 loci detected in that study, 5 were common to both AUD diagnosis and alcohol consumption.
As the prior two paragraphs make clear, population based cohorts have provided larger sample sizes, which are critical for obtaining adequate power for GWAS. Their use can come at the cost of missing more severe alcohol use phenotypes. For example, the frequency of AUD in the UKB is lower than the population average [7% (19)], indicating that certain population studies may be underpowered to detect genetic effects specific to dependence (20). The frequency of AUD in the MVP, on the contrary, was much higher [20%, (18)]. Despite these limitations, population based cohorts provide a cost-effective strategy for obtaining very large samples, compared to traditional study designs that require obtaining a diagnosis from clinically trained staff.
Recent discoveries on the molecular genetics of alcohol use behaviors
Table 1 summarizes the most recent GWAS of alcohol use behaviors (N = 16); Figure 2 provides an overview of the chronology of these studies. Figure 3 shows that the list of genes identified by these studies is highly heterogeneous. These data suggest incomplete genetic overlap between measures of alcohol use behaviors (Figure 4), though ascertainment bias and limited power (see Figure 5) are likely to be additional contributing factors.
Table 1.
List of genes most commonly associated across GWAS of alcohol consumption and/or abuse. Studies are included if they demonstrate an association with the SNP denoted in the table, or with a SNP in LD (r2 > 0.6). Of note, the proximity of the listed SNPs to the nearest gene does not prove that the gene is causal.
| Chromosome | Nearest gene | SNP | Alcohol consumption | Alcohol abuse / AUD | References |
|---|---|---|---|---|---|
| 2 | GCKR | rs1260326 | Y | Y | (17, 18, 21–23) |
| 4 | KLB | rs11940694 | Y | (15, 17, 21–23) | |
| 4 | KLB | rs35538052 | Y | (18, 21) | |
| 4 | ADH1B | rs1229984 | Y | Y | (12, 17, 18, 21–23) |
| 4 | SLC39A8 | rs13107325 | Y | Y | (17, 18, 21, 22) |
| 2 | LINC01833 | rs1004787 | Y | Y | (17, 18, 21) |
| 17 | MAPT/CHRH1 | rs62062288 | Y | Y | (17, 21, 22) |
| 19 | IZUMO/FGF21 | rs281379 | Y | Y | (17, 21, 22) |
| 16 | FTO | rs35538052 | Y | (17, 18) | |
| 4 | ADH1C | rs142783062 | Y | Y | (17, 18) |
| 3 | CADM2 | rs62250685 | Y | (21, 23) |
Figure 2.
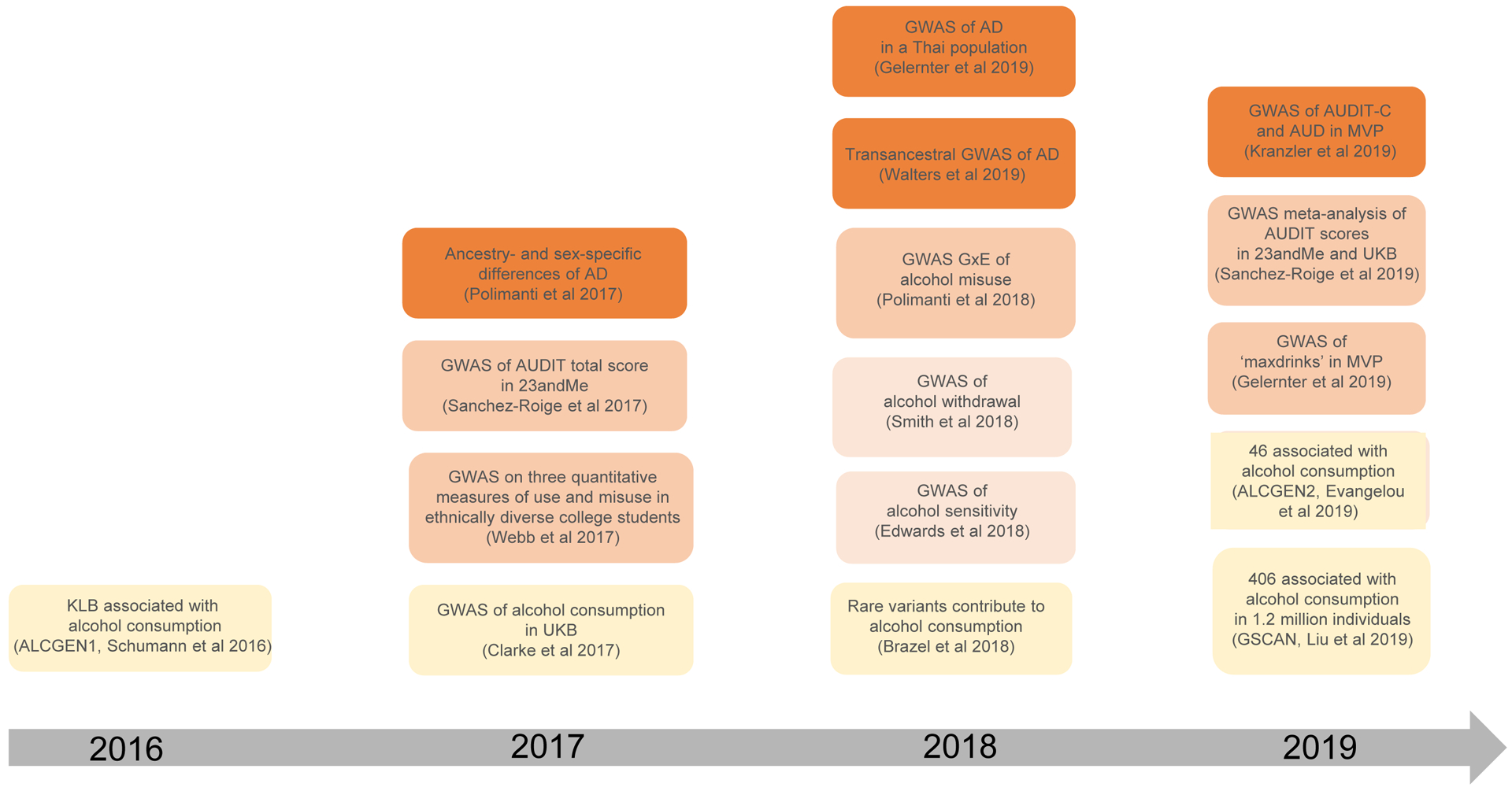
Timeline of major findings in alcohol use behaviors (alcohol use, yellow; alcohol sensitivity and withdrawal, light orange; alcohol misuse, orange; alcohol dependence and AUD, dark orange) using GWAS methods. Not all references in Table 1 are included in this figure.
Figure 3.
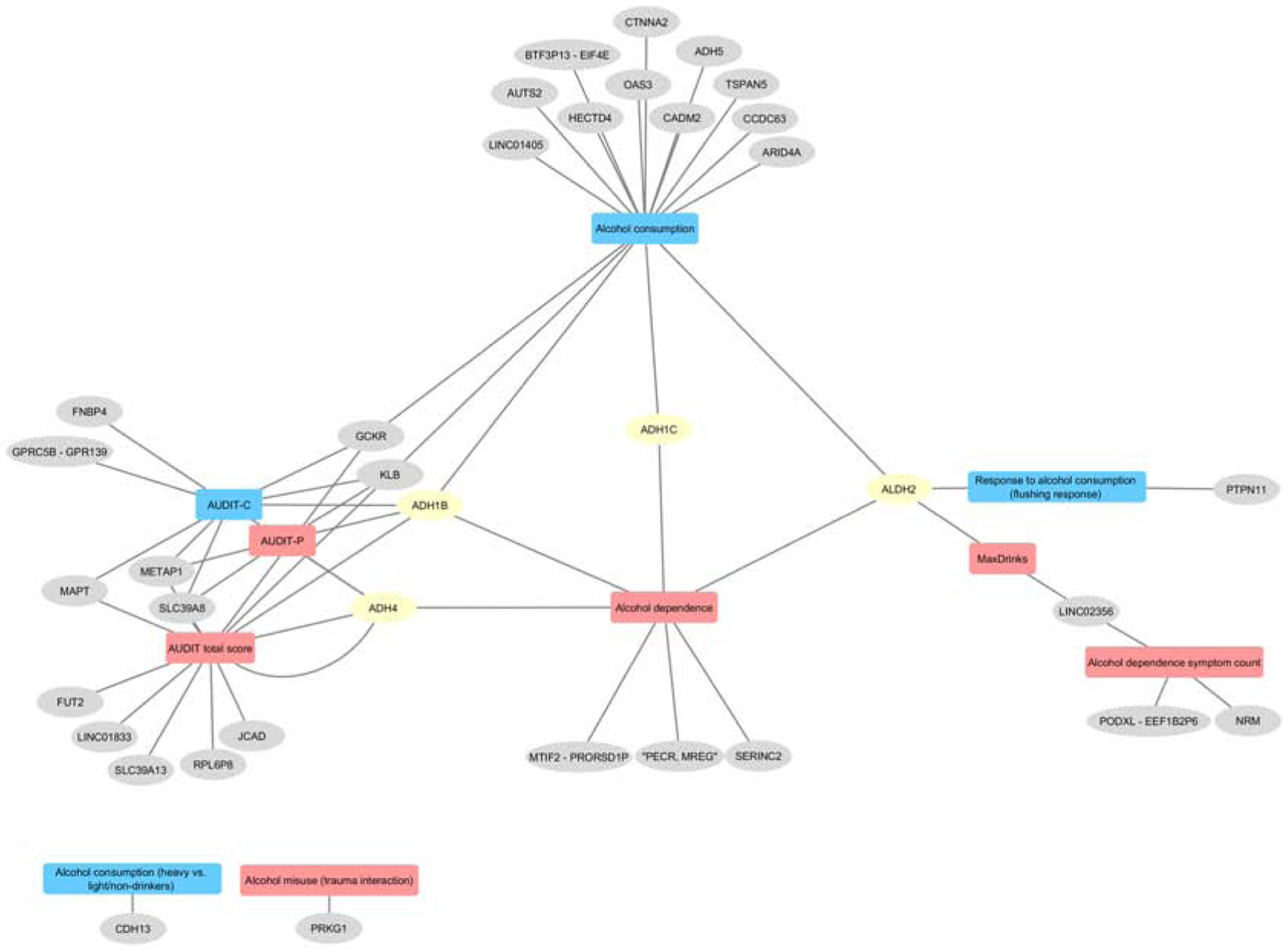
Gene-Phenotype network. Shared and specific genetic contributions at different stages or symptoms associated with alcohol use disorders, including alcohol use, indices of alcohol misuse severity (MaxDrinks, AUDIT-P), alcohol dependence, response to alcohol. Only SNPs in genes showing a significant (P < 10–8) association with multiple AUD and alcoholrelated traits, and available from the GWAS catalog at the time of this writing, are included. AUDIT, Alcohol Use Disorder Identification Test; AUDIT-C, Alcohol Use Disorder Identification Test items 1–3; AD, alcohol dependence; AUD, alcohol use disorder; UKB, UK Biobank; MVP, Million Veterans Program; AlcGen, Alcohol Genome-wide Association Consortium; GSCAN, GWAS & Sequencing Consortium of Alcohol and Nicotine use Consortium; GxE, gene by environment interaction; GWAS, genome-wide association study.
Figure 4.
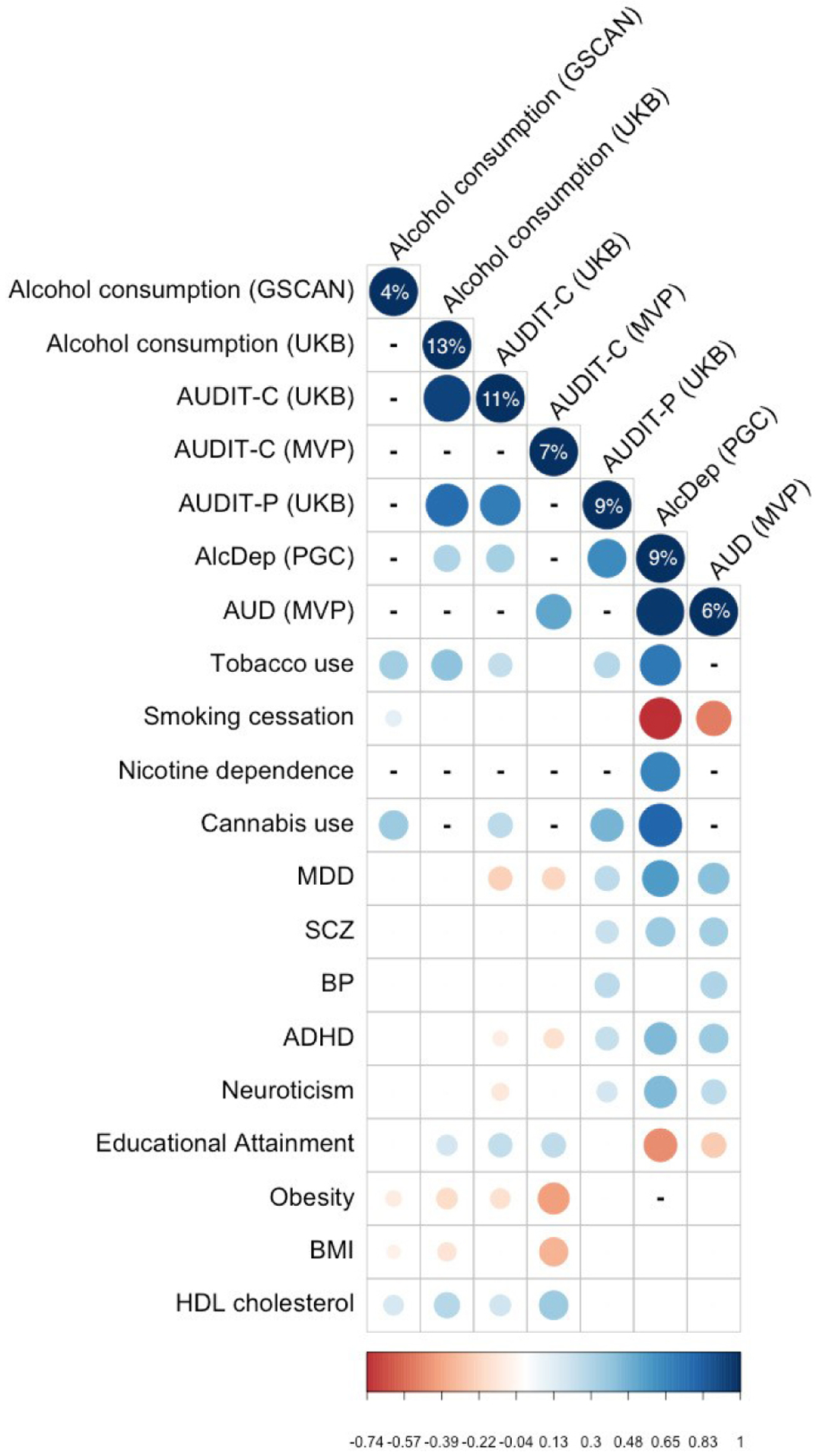
Heritability and genetic correlation estimates across alcohol use behaviors. Values (%) on the diagonal represent SNP-heritability estimates. Blank boxes represent pairs of traits that are not significantly genetically correlated; - represents a pair of traits that have not been tested.
Figure 5.
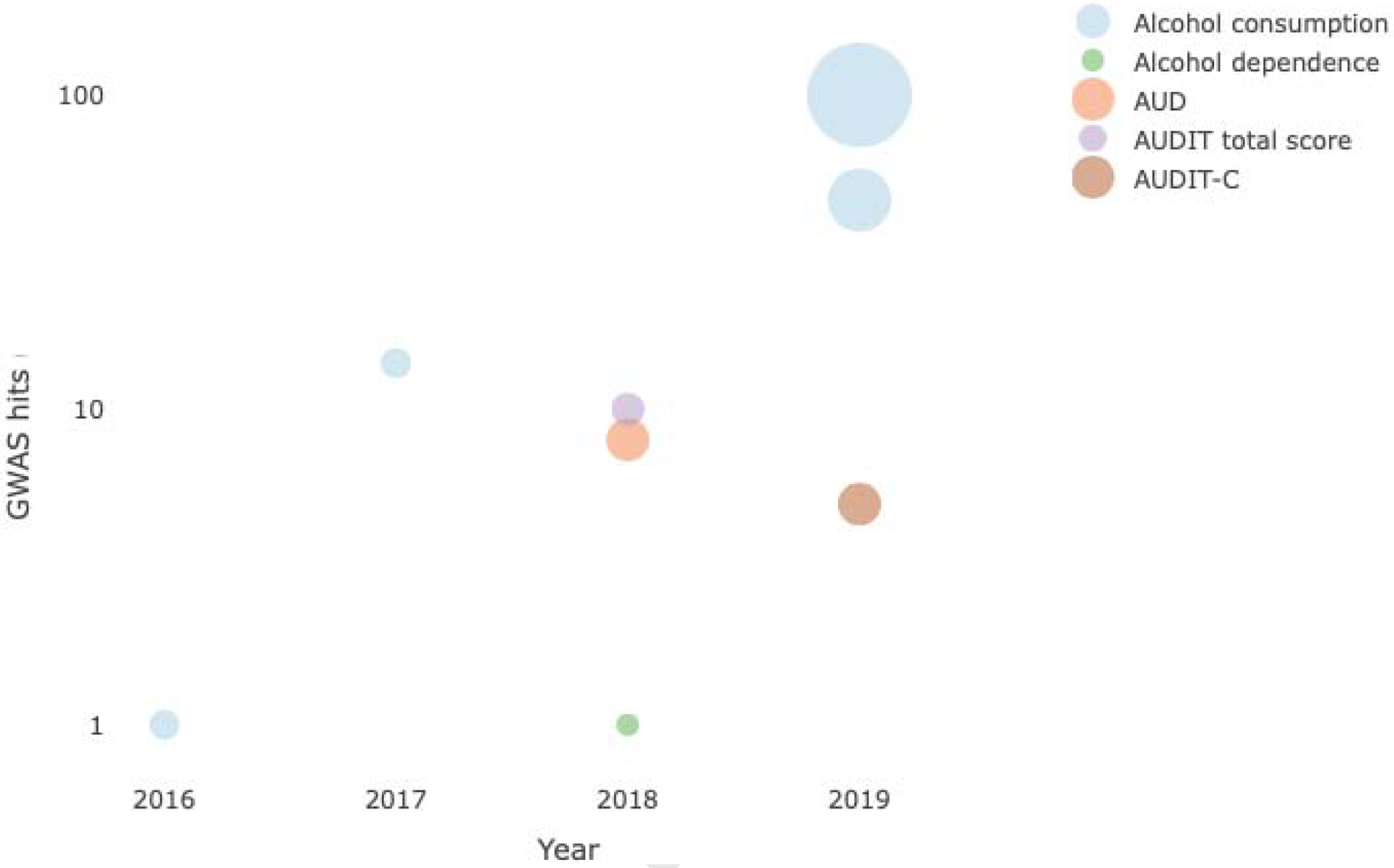
GWAS hits discovered as a function of sample size and alcohol use behaviors. AUDIT, Alcohol Use Disorder Identification Test; AUDIT-C, Alcohol Use Disorder Identification Test items 1–3. Interactive plot: http://rpubs.com/sanchezroige/475742
The 4q23 region, which contains the genes for several alcohol metabolizing enzymes, has been associated with multiple alcohol use behaviors. This association is one of the most consistently replicated findings in the field of psychiatric genetics, although the effects are clearly ancestry-specific (7). There appear to be multiple signals in this region, including ADH1C (17, 18, 21–24), ADH4 (18), ADH5 (21, 23) and the METAP1/EIF4E region (21–23). The GSCAN consortium recently showed that there are at least 13 independent signals with minor allele frequencies over 0.001 at 4q23 (21). Intriguingly, several of those loci are also strongly implicated in non-psychiatric, somatic traits (25).
Beyond the alcohol metabolizing genes, the region containing the genes beta-klotho (KLB) and the Fibroblast growth factor 21 (FGF21) has been robustly associated with alcohol consumption. The AlcGen consortium was the first to show that the A allele of rs11940694 (Figure 2), located in the intron of KLB, was associated with reduced alcohol consumption (15). This finding has since been replicated (Table 1) - the same SNP was associated with alcohol consumption (17, 18, 22, 23) and alcohol misuse (22). Beta-klotho is a transmembrane protein that acts as a cofactor for the circulating hormone fibroblast growth factor 21 (FGF21) by facilitating its binding to FGF receptors (FGFR). Interestingly the FGF21 gene, which is located on chromosome 19, was also associated with AUDIT scores at the gene-level in humans (22). Beta-klotho is primarily expressed in the liver, adipose tissue and pancreas (26), and recent studies have shown that it regulates brain specific functions related to alcohol consumption in mice. For example, mice lacking brain expressed Klb showed increased ethanol preference (15). Furthermore, FGF21 was found to suppress ethanol consumption in wild-type mice but had no effect on mice lacking Klb in the brain. Previous studies have shown that FGF21 and KLB are involved in sweet and alcohol preference in mice (27), and a recent study in humans found increased FGF21 expression in blood after binge drinking (28). These findings suggest that KLB and FGF21 act as part of a brain-liver endocrine axis that regulates alcohol consumption. Future studies could explore the effects of analogues of FGF21 on alcohol consumption, which are currently being tested in clinical trials for the treatment of type 2 diabetes and obesity (29). Although KLB and FGF21 seem to be promising avenues for translational research, it is worth noting that while SNPs in KLB are associated with alcohol consumption, they have not yet shown any association with AUD (12, 18). This implies that this system might only be relevant for the regulation of normative consumption, although studies of larger AUD populations may yet reveal a role for these loci in AUD. Furthermore, although the locus probably impacts KLB, rs11940694 was found to be an expression quantitative trait locus (eQTL) for RFC1 gene expression in the cerebellum and hemisphere (22, 23).
Another well-replicated locus associated with both alcohol consumption and AUD is the region containing the glucokinase receptor (GCKR) gene, whose product is a regulatory protein that is produced by hepatocytes and is involved in the cellular trafficking of glucokinase. A nonsynonymous SNP in GCKR, rs1260326, was robustly associated with alcohol consumption in the MVP, UKB and 23andMe samples (Table 1). Intriguingly, rs1260326 has also been previously associated with multiple metabolic traits, including diabetes, obesity and liver disease (30, 31). Given that alcohol consumption is strongly associated with both metabolic and lipid profiles (e.g. 25, 32, 33), it is not clear whether the association with rs1260326 pinpoints a pleiotropic process central to metabolic traits, or whether alcohol causally impacts glucose metabolism and lipid levels, in part via GCKR. A recent study characterized the effects of alcohol in neural cell cultures derived from induced pluripotent stem cells (iPSCs) and found that genes down-regulated upon alcohol exposure were involved in cholesterol homeostasis in the brain (34). These findings could suggest that AUD has both psychiatric and metabolic components, a theme that has also been suggested for other psychiatric disorders, such as anorexia nervosa (35). Additional evidence supporting this provocative hypothesis is the fact that several genes associated with alcohol use and dependence involve brain-endocrine-metabolic mechanisms. KLB is part of a brain-liver feedback loop, acetaldehyde modulates a number of ethanol effects in the brain, and enrichment analyses of alcohol-associated genes found glutamatergic enrichment not only in the brain but also in glucose and carbohydrate processing pathways (21). The ability to process caloric alcoholic beverages may be linked to individual differences in alcohol consumption.
In general, the ‘candidate genes’ for AUD that were examined in smaller cohorts have not been replicated by larger and better powered GWAS (10). One exception is the corticotropin releasing hormone receptor 1 (CRHR1), a candidate gene extensively studied in humans and rodents before the advent of large-scale GWAS studies (10). CRHR1 is central to the cortisol stress response as part of the hypothalamic-pituitary-axis. Extensive preclinical literature has shown that CRHR1 is associated with relapse to drug taking in mice [e.g. (36, 37)] and there is some evidence that variation in CRHR1 modulates the role of psychological stress on alcohol intake (e.g. 38, 39). Encouragingly, the genomic region surrounding CRHR1 has been associated with alcohol consumption and misuse in several recent GWAS studies (21, 22, 40). However, CRHR1 is located in an inversion polymorphism of roughly 900kb that is common in Europeans and induces extensive LD spanning many genes (41), including CRHR1 and MAPT (22). MAPT encodes the protein tau, is involved in Parkinson’s and Alzheimer’s disease. Further work is therefore required to determine which variant(s) are causal, as the inversion in this region complicates the ability of GWAS to fully address this question.
Recent GWAS have identified several regions containing a set of genes that have pleiotropic effects on many psychiatric disorders and related traits; these genes may be tagging a latent factor (“p-factor”) (42). For example, the largest GWAS of alcohol and smoking, which used over 1 million individuals, performed a multivariate GWAS approach to show that 150 loci were associated with multiple substance use phenotypes; variation at PDE4B and CUL3 were associated with both smoking (initiation, cessation, quantity) and drinks per week. Similarly, CADM2 has been recently associated with alcohol and cannabis use (21, 23, 43). CADM2 is a cell adhesion molecule (CAM) that influences brain wiring and appears to have a role in multiple neuropsychiatric disorders (44). There is now mounting evidence from independent GWAS showing an association between common genetic variants at CADM2 and risky or impulsive behaviors including risk tolerance, automobile speeding propensity, number of sexual partners (45), sensation seeking and drug experimentation (46), cannabis initiation (47), and obesity and body mass index (48–50). CADM2 has also been associated with cognitive phenotypes, including educational attainment (51, 52). We therefore hypothesize that genetic variation at CADM2 may underlie a latent personality trait or risk factor that predisposes individuals to engage in risky actions (i.e. drinking behaviors).
Despite the success of GWAS of alcohol use (Figure 4) the mechanisms by which these newly identified genetic associations exert their effects are largely unknown. More importantly, alcohol consumption and misuse (core traits associated with development of AUD) appear to have distinct genetic architectures (Table 1, Figure 3). Ever-larger studies, particularly those extending mere alcohol consumption phenotypes, are required to find the genetic variants that contribute towards the transition from normative alcohol use to misuse, and development of AUD.
Polygenic methods generate hypotheses to test across alcohol use behaviors
One successful application of GWAS has been their use for assigning polygenic risk scores (PRS), which provide estimates of an individual’s genetic risk of developing a given disorder. Reassuringly, PRS for alcohol use behaviors predict equivalent phenotypes in independent cohorts [e.g. alcohol consumption (53), AD (12), AUD symptoms (54)]. Johnson et al (2019) recently identified that, compared to PRS for alcohol consumption (AUDIT-C), PRS for alcohol misuse (AUDIT-P) were superior predictors of a range of alcohol-related phenotypes, particularly those pertaining to the domains of misuse and dependence. These findings further illustrate that alcohol consumption alone may not be a good proxy for AUD.
PRS can also be used to test specific hypotheses; for example, PRS can be used to measure how environmental, demographic, and genetic factors interact with one another. Are there developmental windows where the effects of alcohol use and misuse are more invasive? Can we identify biomarkers that would inform the transition from normative alcohol use to excessive use and dependence? For instance, the alcohol metabolizing genetic effects on alcohol use appeared to be more influential in later years of college than in earlier years (55, 56), revealing that the nature and magnitude of genetic effects vary across development.
It is worth noting important limitations of PRS analyses. First, polygenic prediction is influenced by the ancestry of the population studied. For example, PRS for AUD generated in an African American (AA) cohort explained more of the variance in AUD than PRS derived from a much larger cohort of European Americans (12). This illustrates that the prediction from one population to another does not perform well (e.g. PRS based on European Americans but used to predict in AA) (57). Second, the method of ascertainment may bias the results. As an example, PRS for DSM-IV AD derived from a population based sample predicted increased risk for AD in other population samples but did not associate with AUD symptoms in a clinically ascertained sample (54). Third, the variance explained by PRS is still low, and hence PRS have limited clinical application. For example, in the largest study of alcohol consumption (21), the alcohol consumption PRS accounted for only ~2.5% of the variance in alcohol use in two independent datasets. Recent work suggested that predictions may improve by incorporating functional genomic information. For example, McCartney et al (58) showed that, compared to conventional PRS, risk scores that took into account DNA methylation were better predictors of alcohol consumption (12.5% vs PRS 0.7%; but see (59)). Nonetheless, the way in which such methods can be used for prevention or treatments of AUD has yet to be established. Lastly, it remains to be determined the nature of these associations. Mendelian randomization analyses can serve to further understand and explore the correlations between alcohol use behaviors and comorbid traits (see Supplemental 1).
Alcohol consumption and misuse show a distinct genetic architecture
Before the era of large-scale genomic research, twin and family-based studies identified a high degree of genetic overlap between the genetic risk for AUD and psychopathology by modeling correlations among family members (e.g. (60)). With the recent development of linkage disequilibrium score regression (LDSC), it is now possible to estimate the genetic correlations between specific alcohol use behaviors (Figure 1, Figure 4) and a plethora of psychiatric, health and educational outcomes using GWAS summary statistics. Most notably, the genetic overlap between alcohol consumption and AD was positive but relatively modest (rg = 0.38–0.52, 12, 18), suggesting that, although the use of alcohol is necessary to develop AD, some of the genetic liability is specific to either levels of consumption or AD.
Another consistent finding from genetic correlation analyses has been that alcohol consumption and AUD show distinct patterns of genetic overlap with disease traits (Figure 4). Counterintuitively, alcohol consumption tends to correlate with desirable attributes including educational attainment and is negatively genetically correlated with coronary heart disease, type 2 diabetes and BMI (18, 21–23). These genetic correlations are unlike those observed when analyzing alcohol dependent individuals: AD was negatively genetically correlated with educational attainment (18) and positively genetically correlated with other psychiatric diseases, including major depressive disorder (MDD), bipolar disorder, schizophrenia and attentiondeficit/hyperactivity disorder (ADHD, 12, 18). Importantly, alcohol consumption (AUDIT-C) and misuse (AUDIT-P or AUD) measured in the same population (UKB or MVP) showed distinct patterns of genetic association with psychopathology and health outcomes (18, 22). This set of findings emphasize the importance of deep phenotyping and demonstrates that alcohol consumption and problematic drinking have distinct genetic influences.
Ascertainment bias may explain some of the paradoxical genetic correlations associated with alcohol consumption (61). Population based cohorts, such as UKB and 23andMe, are based on voluntary participation and tend to attract individuals with higher education levels and socioeconomic status than the general population and, crucially, lower levels of problem drinking. In contrast, ascertainment in the PGC and MVP cohorts (12, 18) was based on DSMIV AD diagnosis and ICD (International Classification of Diseases) codes for AUD, respectively. Collider bias (the biased estimation of the causal effect of an exposure on an outcome) has been proposed to underlie some of the genetic correlations between alcohol consumption and BMI (62); however, BMI has been consistently negatively correlated with alcohol use in several subsequent studies (18, 21, 22, 63). Furthermore, it is also possible that the genetic overlap between AD and aspects of alcohol consumption are dependent on the specific patterns of drinking. For example, Polimanti et al (64) identified a positive genetic correlation between AD and alcohol drinking quantity (rg = 0.75), but not frequency.
Limitations and future directions
Prior to the availability of large population studies and collaborative consortia efforts, few genes were reliably associated with AUD. The use of intermediate traits or endophenotypes (such as alcohol consumption as an intermediate phenotype for AUD) has become increasingly common and hundreds of new loci have now been associated with alcohol use behaviors. Using intermediate phenotypes also facilitates translational research; we can mimic aspects of human alcohol use using animal models, including alcohol consumption, novelty response, impulsivity, withdrawal and sensitivity (e.g. 65, 66). Animal models provide an opportunity to evaluate the role of newly identified genes (Table 1) at the molecular, cellular and circuit level. We may also be able to perform human genetic studies of specific components of AUD such as DSM-IV AD criterion count (67) and alcohol withdrawal (68). To date these traits have only been studied in smaller samples but this approach will be invaluable as sample sizes increase.
Another challenge for AUD genetics is that AUD is a dynamic phenotype, even more so than other psychiatric conditions, and therefore may necessitate yet larger sample sizes. Ever-larger studies, particularly those extending mere alcohol consumption phenotypes, are required to find the genetic variants that contribute towards the transition from normative alcohol use to misuse, and development of AUD. Furthermore, genetic risk unfolds across development, particularly during adolescence, when drug experimentation is more prominent and when the brain is most vulnerable to the deleterious effects of alcohol (69). The Adolescent Brain Cognitive Development (ABCD), with neuroimaging, genotyping and extensive longitudinal phenotypic information including alcohol use behaviors (70), offers new avenues for research, namely to understand how genetic risk interacts with the environment across critical developmental windows. Population biobanks aligning genotype data from thousands of individuals to electronic health records are also promising emerging platforms to accelerate AUD genetic research (71).
Despite these caveats, the GWAS described in Table 1 have already vastly expanded our understanding of the genetic architecture of alcohol use behaviors. It is evident that alcohol use behaviors, like all complex traits, are highly polygenic (11). The proportion of variance explained by genetic variants on GWAS chips (SNP-heritability) ranges from 4 to 13% (Figure 4). It is possible that a significant portion of the heritability can be explained by SNPs not tagged by GWAS chips, including rare variants (46). For instance, a recent study showed that rare variants explained 1–2% of phenotypic variance and 11–18% of total SNP heritability of substance use phenotypes (72). Nonetheless, rare variants are often not analyzed when calculating SNP heritability, which can lead to an underestimate of polygenic effects, as well as missing biologically relevant contributions for post-GWAS analyses (73). Equally important is the need to include other sources of -omics data when interpreting genetic findings, and the need to increase population diversity (see Supplemental 2). Therefore, a multifaceted approach targeting both rare and common variation, including functional data, and assembling much larger datasets for meta-analyses (particularly for alcohol misuse and clinical phenotypes) in ethnically diverse populations, is critical for identifying the key genes and pathways important in AUD.
Conclusion
AUD is a complex, heterogeneous disorder encompassing a variety of behavioral, psychological, and physiological traits with a complex longitudinal structure, thus posing an enormous challenge for genetic analysis. Instead, AUD can be fractionated into dimensions or symptoms. Several recent GWAS have used this approach, and it is now common to study quantitative measures, including alcohol consumption and aspects of disordered drinking, in large population samples. As a result, GWAS of alcohol use, misuse and AUD are now beginning to uncover genetic signals that have the potential to be further analyzed at the molecular, cellular, and circuit level in cellular and animal model systems. Findings from polygenic prediction and genetic correlation analyses, which are major trends in psychiatric genetics, have demonstrated that alcohol use behaviors share a common genetic basis with numerous psychiatric, educational and health outcomes. Unsurprisingly, even though studying alcohol consumption has shown some utility, it is apparent that this phenotype cannot be used as a proxy for AUD. We anticipate that big datasets, including those from electronic health records, will revolutionize the field in the years to come.
Supplementary Material
Acknowledgements and Disclosures
The authors report no biomedical financial interests or potential conflicts of interest.
Funding
SSR was supported by the Frontiers of Innovation Scholars Program (#3-P3029), the Interdisciplinary Research Fellowship in NeuroAIDS (MH081482), a pilot award from the NIH (DA037844) and the 2018 NARSAD Young Investigator Grant (#27676). SSR and AAP were supported by funds from the California Tobacco-Related Disease Research Program (TRDRP; #28IR-0070 and T29KT-0526). AAP was supported by NIH grants AA026281 and P50DA037844. TKC was supported by a Wellcome Trust (Wellcome Trust Strategic Award “STratifying Resilience and Depression Longitudinally” (STRADL) Reference 104036/Z/14/Z).
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- 1.Degenhardt L, Charlson F, Ferrari A, Santomauro D, Erskine H, Mantilla-Herrara A, et al. (2018): The global burden of disease attributable to alcohol and drug use in 195 countries and territories, 1990–2016: a systematic analysis for the Global Burden of Disease Study 2016. The Lancet Psychiatry. 5: 987–1012. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Substance Abuse and Mental Health Services Administration (2017): Key substance use and mental health indicators in the United States: Results from the 2016 National Survey on Drug Use and Health (HHS Publication No. SMA 17–5044, NSDUH Series H-52). Rockville, MD: Center for Behavioral Health Statistics and Quality, Substance Abuse and Mental Health Services Administration. [Google Scholar]
- 3.Tawa EA, Hall SD, Lohoff FW (2016): Overview of the Genetics of Alcohol Use Disorder. Alcohol Alcohol. 51: 507–514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Hart AB, Kranzler HR (2015): Alcohol Dependence Genetics: Lessons Learned From Genome-Wide Association Studies (GWAS) and Post-GWAS Analyses. Alcohol Clin Exp Res. 39: 1312–1327. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hancock DB, Markunas CA, Bierut LJ, Johnson EO (2018): Human Genetics of Addiction: New Insights and Future Directions. Current Psychiatry Reports. 20: 8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Agrawal A, Verweij KJH, Gillespie NA, Heath AC, Lessov-Schlaggar CN, Martin NG, et al. (2012): The genetics of addiction-a translational perspective. Transl Psychiatry. 2: e140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Edenberg HJ, McClintick JN (2018): Alcohol Dehydrogenases, Aldehyde Dehydrogenases, and Alcohol Use Disorders: A Critical Review. Alcohol Clin Exp Res. 42: 2281–2297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Edenberg HJ, Gelernter J, Agrawal A (2019): Genetics of Alcoholism. Curr Psychiatry Rep. 21: 26. [DOI] [PubMed] [Google Scholar]
- 9.Deak JD, Miller AP, Gizer IR (2018): Genetics of alcohol use disorder: a review. Curr Opin Psychol. 27: 56–61. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Olfson E, Bierut LJ (2012): Convergence of genome-wide association and candidate gene studies for alcoholism. Alcohol Clin Exp Res. 36: 2086–2094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Visscher PM, Wray NR, Zhang Q, Sklar P, McCarthy MI, Brown MA, Yang J (2017): 10 Years of GWAS Discovery: Biology, Function, and Translation. Am J Hum Genet. 101: 5–22. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Walters RK, Polimanti R, Johnson EC, McClintick JN, Adams MJ, Adkins AE, et al. (2018): Transancestral GWAS of alcohol dependence reveals common genetic underpinnings with psychiatric disorders. Nature Neuroscience. 21: 1656–1669. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Saunders JB, Aasland OG, Babor TF, de la Fuente JR, Grant M (1993): Development of the Alcohol Use Disorders Identification Test (AUDIT): WHO Collaborative Project on Early Detection of Persons with Harmful Alcohol Consumption--II. Addiction. 88: 791–804. [DOI] [PubMed] [Google Scholar]
- 14.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, Adams MJ, et al. (2019): Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry. 176: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, et al. (2016): KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA. 113: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Schumann G, Liu C, O’Reilly P, Gao H, Song P, Xu B, et al. (2016): KLB is associated with alcohol drinking, and its gene product β-Klotho is necessary for FGF21 regulation of alcohol preference. Proc Natl Acad Sci USA. 113: 14372–14377. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Evangelou E, Gao H, Chu C, Ntritsos G, Blakeley P, Butts AR, et al. (2018): Genome-wide association and functional studies identify 46 novel loci for alcohol consumption and suggest common genetic mechanisms with neuropsychiatric disorders. preprint, Genetics. doi: 10.1101/453332. [DOI] [Google Scholar]
- 18.Kranzler HR, Zhou H, Kember RL, Vickers Smith R, Justice AC, Damrauer S, et al. (2019): Genome-wide association study of alcohol consumption and use disorder in 274,424 individuals from multiple populations. Nat Commun. 10: 1499. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Grant BF, Goldstein RB, Saha TD, Chou SP, Jung J, Zhang H, et al. (2015): Epidemiology of DSM-5 Alcohol Use Disorder: Results From the National Epidemiologic Survey on Alcohol and Related Conditions III. JAMA Psychiatry. 72: 757–766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Davis KAS, Coleman JRI, Adams M, Allen N, Breen G, Cullen B, et al. (2018): Mental health in UK Biobank: development, implementation and results from an online questionnaire completed by 157 366 participants. BJPsych Open. 4: 83–90. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 21.Liu M, Jiang Y, Wedow R, Li Y, Brazel DM, Chen F, et al. (2019): Association studies of up to 1.2 million individuals yield new insights into the genetic etiology of tobacco and alcohol use. Nat Genet. 51: 237–244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Sanchez-Roige S, Palmer AA, Fontanillas P, Elson SL, 23andMe Research Team, the Substance Use Disorder Working Group of the Psychiatric Genomics Consortium, Adams MJ, et al. (2019): Genome-Wide Association Study Meta-Analysis of the Alcohol Use Disorders Identification Test (AUDIT) in Two Population-Based Cohorts. Am J Psychiatry. 176: 107–118. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Clarke T-K, Adams MJ, Davies G, Howard DM, Hall LS, Padmanabhan S, et al. (2017): Genome-wide association study of alcohol consumption and genetic overlap with other health-related traits in UK Biobank (N=112 117). Mol Psychiatry. 22: 1376–1384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Sanchez-Roige S, Fontanillas P, Elson SL, 23andMe Research Team, Gray JC, de Wit H, et al. (2019): Genome-wide association study of alcohol use disorder identification test (AUDIT) scores in 20 328 research participants of European ancestry. Addict Biol. 24: 121–131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Horwitz T, Lam K, Chen Y, Xia Y, Liu C (2019): A decade in psychiatric GWAS research. Mol Psychiatry. 24: 378–389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ito S, Kinoshita S, Shiraishi N, Nakagawa S, Sekine S, Fujimori T, Nabeshima YI (2000): Molecular cloning and expression analyses of mouse betaklotho, which encodes a novel Klotho family protein. Mech Dev. 98: 115–119. [DOI] [PubMed] [Google Scholar]
- 27.Talukdar S, Owen BM, Song P, Hernandez G, Zhang Y, Zhou Y, et al. (2016): FGF21 Regulates Sweet and Alcohol Preference. Cell Metab. 23: 344–349. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Søberg S, Andersen ES, Dalsgaard NB, Jarlhelt I, Hansen NL, Hoffmann N, et al. (2018): FGF21, a liver hormone that inhibits alcohol intake in mice, increases in human circulation after acute alcohol ingestion and sustained binge drinking at Oktoberfest. Mol Metab. 11: 96–103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Talukdar S, Zhou Y, Li D, Rossulek M, Dong J, Somayaji V, et al. (2016): A Long-Acting FGF21 Molecule, PF-05231023, Decreases Body Weight and Improves Lipid Profile in Non-human Primates and Type 2 Diabetic Subjects. Cell Metab. 23: 427–440. [DOI] [PubMed] [Google Scholar]
- 30.Raimondo A, Rees MG, Gloyn AL (2015): Glucokinase regulatory protein: complexity at the crossroads of triglyceride and glucose metabolism. Curr Opin Lipidol. 26: 88–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Rasheed H, Stamp LK, Dalbeth N, Merriman TR (2017): Interaction of the GCKR and A1CF loci with alcohol consumption to influence the risk of gout. Arthritis Res Ther. 19: 161. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ridker PM, Pare G, Parker A, Zee RYL, Danik JS, Buring JE, et al. (2008): Loci related to metabolic-syndrome pathways including LEPR,HNF1A, IL6R, and GCKR associate with plasma C-reactive protein: the Women’s Genome Health Study. Am J Hum Genet. 82: 1185–1192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Jamalpour S, Zain SM, Mosavat M, Mohamed Z, Omar SZ (2018): A case-control study and meta-analysis confirm glucokinase regulatory gene rs780094 is a risk factor for gestational diabetes mellitus. Gene. 650: 34–40. [DOI] [PubMed] [Google Scholar]
- 34.Jensen KP, Lieberman R, Kranzler HR, Gelernter J, Clinton K, Covault J (2019): Alcoholresponsive genes identified in human iPSC-derived neural cultures. Transl Psychiatry. 9: 96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Duncan L, Yilmaz Z, Gaspar H, Walters R, Goldstein J, Anttila V, et al. (2017): Significant Locus and Metabolic Genetic Correlations Revealed in Genome-Wide Association Study of Anorexia Nervosa. Am J Psychiatry. 174: 850–858. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Volkow ND, Koob GF, McLellan AT (2016): Neurobiologic Advances from the Brain Disease Model of Addiction. N Engl J Med. 374: 363–371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Koob GF (2014): Neurocircuitry of alcohol addiction: synthesis from animal models. Handb Clin Neurol. 125: 33–54. [DOI] [PubMed] [Google Scholar]
- 38.Clarke T-K, Schumann G (2009): Gene-environment interactions resulting in risk alcohol drinking behaviour are mediated by CRF and CRF1. Pharmacol Biochem Behav. 93: 230–236. [DOI] [PubMed] [Google Scholar]
- 39.Treutlein J, Kissling C, Frank J, Wiemann S, Dong L, Depner M, et al. (2006): Genetic association of the human corticotropin releasing hormone receptor 1 (CRHR1) with binge drinking and alcohol intake patterns in two independent samples. Mol Psychiatry. 11: 594–602. [DOI] [PubMed] [Google Scholar]
- 40.Gelernter J, Sun N, Polimanti R, Pietrzak R, Levey DF, Lu Q, et al. (2019): Genomewide Association Study of Maximum Habitual Alcohol Intake in >140,000 US European- and African-American Veterans Yields Novel Risk Loci. Biological Psychiatry. S0006322319311564. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Stefansson H, Helgason A, Thorleifsson G, Steinthorsdottir V, Masson G, Barnard J, et al. (2005): A common inversion under selection in Europeans. Nat Genet. 37: 129–137. [DOI] [PubMed] [Google Scholar]
- 42.Selzam S, Coleman JRI, Caspi A, Moffitt TE, Plomin R (2018): A polygenic p factor for major psychiatric disorders. Translational Psychiatry. 8: 205. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, et al. (2018): GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 21: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Sakurai T (2017): The role of cell adhesion molecules in brain wiring and neuropsychiatric disorders. Mol Cell Neurosci. 81: 4–11. [DOI] [PubMed] [Google Scholar]
- 45.Karlsson Linnér R, Biroli P, Kong E, Meddens SFW, Wedow R, Fontana MA, et al. (2019): Genome-wide association analyses of risk tolerance and risky behaviors in over 1 million individuals identify hundreds of loci and shared genetic influences. Nat Genet. 51: 245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Sanchez-Roige S, Fontanillas P, Elson SL, Gray JC, de Wit H, MacKillop J, Palmer AA (2019): Genome-Wide Association Studies of Impulsive Personality Traits (BIS-11 and UPPS-P) and Drug Experimentation in up to 22,861 Adult Research Participants Identify Loci in the CACNA1I and CADM2 genes. J Neurosci. 39: 2562–2572. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Pasman JA, Verweij KJH, Gerring Z, Stringer S, Sanchez-Roige S, Treur JL, et al. (2018): GWAS of lifetime cannabis use reveals new risk loci, genetic overlap with psychiatric traits, and a causal influence of schizophrenia. Nat Neurosci. 21: 1161–1170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Locke AE, Kahali B, Berndt SI, Justice AE, Pers TH, Day FR, et al. (2015): Genetic studies of body mass index yield new insights for obesity biology. Nature. 518: 197–206. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Akiyama M, Okada Y, Kanai M, Takahashi A, Momozawa Y, Ikeda M, et al. (2017): Genome-wide association study identifies 112 new loci for body mass index in the Japanese population. Nat Genet. 49: 1458–1467. [DOI] [PubMed] [Google Scholar]
- 50.Graff M, Scott RA, Justice AE, Young KL, Feitosa MF, Barata L, et al. (2017): Genome-wide physical activity interactions in adiposity - A meta-analysis of 200,452 adults. PLoS Genet. 13: e1006528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Okbay A, Beauchamp JP, Fontana MA, Lee JJ, Pers TH, Rietveld CA, et al. (2016): Genome-wide association study identifies 74 loci associated with educational attainment. Nature. 533: 539–542. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Davies G, Marioni RE, Liewald DC, Hill WD, Hagenaars SP, Harris SE, et al. (2016): Genome-wide association study of cognitive functions and educational attainment in UK Biobank (N=112 151). Molecular Psychiatry. 21: 758–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Marees AT, Hammerschlag AR, Bastarache L, de Kluiver H, Vorspan F, van den Brink W, et al. (2018): Exploring the role of low-frequency and rare exonic variants in alcohol and tobacco use. Drug Alcohol Depend. 188: 94–101. [DOI] [PubMed] [Google Scholar]
- 54.Savage JE, Salvatore JE, Aliev F, Edwards AC, Hickman M, Kendler KS, et al. (2018): Polygenic Risk Score Prediction of Alcohol Dependence Symptoms Across PopulationBased and Clinically Ascertained Samples. Alcohol Clin Exp Res. 42: 520–530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Thomas NS, Adkins A, Aliev F, Edwards AC, Webb BT, Tiarsmith EC, et al. (2018): Alcohol Metabolizing Polygenic Risk for Alcohol Consumption in European American College Students. J Stud Alcohol Drugs. 79: 627–634. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Olfson E, Edenberg HJ, Nurnberger J, Agrawal A, Bucholz KK, Almasy LA, et al. (2014): An ADH1B variant and peer drinking in progression to adolescent drinking milestones: evidence of a gene-by-environment interaction. Alcohol Clin Exp Res. 38: 2541–2549. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Martin AR, Gignoux CR, Walters RK, Wojcik GL, Neale BM, Gravel S, et al. (2017): Human Demographic History Impacts Genetic Risk Prediction across Diverse Populations. Am J Hum Genet. 100: 635–649. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.McCartney DL, Hillary RF, Stevenson AJ, Ritchie SJ, Walker RM, Zhang Q, et al. (2018): Epigenetic prediction of complex traits and death. Genome Biol. 19: 136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Salvatore JE, Savage JE, Barr P, Wolen AR, Aliev F, Vuoksimaa E, et al. (2018): Incorporating Functional Genomic Information to Enhance Polygenic Signal and Identify Variants Involved in Gene-by-Environment Interaction for Young Adult Alcohol Problems. Alcohol Clin Exp Res. 42: 413–423. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kendler KS, Prescott CA, Myers J, Neale MC (2003): The structure of genetic and environmental risk factors for common psychiatric and substance use disorders in men and women. Arch Gen Psychiatry. 60: 929–937. [DOI] [PubMed] [Google Scholar]
- 61.Adams M, Hill WD, Howard DM, Davis KAS, Deary IJ, Hotopf M, McIntosh AM (2018): Factors associated with sharing email information and mental health survey participation in two large population cohorts. preprint, Genetics. doi: 10.1101/471433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Holmes MV, Davey Smith G (2019): Problems in interpreting and using GWAS of conditional phenotypes illustrated by “alcohol GWAS.” Mol Psychiatry. 24: 167–168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Clarke T-K, McIntosh AM (2018): Response to “Problems in interpreting and using GWAS of conditional phenotypes illustrated by alcohol GWAS.” preprint, Genetics. doi: 10.1101/290965. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Polimanti R, Peterson RE, Ong J-S, MacGregor S, Edwards AC, Clarke T-K, et al. (2019): Evidence of causal effect of major depression on alcohol dependence: findings from the psychiatric genomics consortium. Psychol Med. 1–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Foroud T, Phillips TJ (2012): Assessing the genetic risk for alcohol use disorders. Alcohol Res. 34: 266–272. [PMC free article] [PubMed] [Google Scholar]
- 66.Foroud T, Edenberg HJ, Crabbe JC (2010): Genetic research: who is at risk for alcoholism. Alcohol Res Health. 33: 64–75. [PMC free article] [PubMed] [Google Scholar]
- 67.Lai D, Wetherill L, Bertelsen S, Carey CE, Kamarajan C, Kapoor M, et al. (2019): Genomewide association studies of alcohol dependence, DSM-IV criterion count and individual criteria. Genes Brain Behav. 18: e12579. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Smith AH, Ovesen PL, Skeldal S, Yeo S, Jensen KP, Olsen D, et al. (2018): Risk Locus Identification Ties Alcohol Withdrawal Symptoms to SORCS2. Alcohol Clin Exp Re. 42: 2337–2348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Dick DM, Barr PB, Cho SB, Cooke ME, Kuo SI-C, Lewis TJ, et al. (2018): Post-GWAS in Psychiatric Genetics: A Developmental Perspective on the “Other” Next Steps. Genes Brain Behav. 17: e12447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Volkow ND, Koob GF, Croyle RT, Bianchi DW, Gordon JA, Koroshetz WJ, et al. (2018): The conception of the ABCD study: From substance use to a broad NIH collaboration. Developmental Cognitive Neuroscience. 32: 4–7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Sanchez-Roige S, Palmer AA (2019): Electronic Health Records Are the Next Frontier for the Genetics of Substance Use Disorders. Trends Genet. 35: 317–318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Brazel DM, Jiang Y, Hughey JM, Turcot V, Zhan X, Gong J, et al. (2018): Exome Chip Metaanalysis Fine Maps Causal Variants and Elucidates the Genetic Architecture of Rare Coding Variants in Smoking and Alcohol Use. Biol Psychiatry.. doi: 10.1016/j.biopsych.2018.11.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Wainschtein P, Jain DP, Yengo L, Zheng Z, TOPMed Anthropometry Working Group, TransOmics for Precision Medicine Consortium, Cupples LA, et al. (2019): Recovery of trait heritability from whole genome sequence data. preprint, Genetics. doi: 10.1101/588020. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


