Abstract
High-grade gliomas (HGG) afflict both children and adults and respond poorly to current therapies. Epigenetic regulators have a role in gliomagenesis, but a broad, functional investigation of the impact and role of specific epigenetic targets has not been undertaken. Using a two-step, in vitro/in vivo epigenomic shRNA inhibition screen, we determine the chromatin remodeler BPTF to be a key regulator of adult HGG growth. We then demonstrate that BPTF knockdown decreases HGG growth in multiple pediatric HGG models as well. BPTF appears to regulate tumor growth through cell self-renewal maintenance, and BPTF knockdown leads these glial tumors toward more neuronal characteristics. BPTF’s impact on growth is mediated through positive effects on expression of MYC and MYC pathway targets. HDAC inhibitors synergize with BPTF knockdown against HGG growth. BPTF inhibition is a promising strategy to combat HGG through epigenetic regulation of the MYC oncogenic pathway.
Keywords: BPTF, Epigenetics, MYC, High-Grade Glioma, DIPG
Introduction
Glioblastoma (GBM) is the World Health Organization grade IV high-grade glioma (HGG) and the most common malignant primary brain tumor in adults, with over 10,000 new cases diagnosed each year in the United States.[1, 2] Even with therapy consisting of surgery, radiation therapy (RT), and chemotherapy, median survival is only 15 months.[3] HGG also occurs in children; the two most common subtypes are GBM and diffuse midline glioma (DMG), which includes most diffuse intrinsic pontine gliomas (DIPG). Overall survival of pediatric GBM, which is more common in older children, is 20 percent. Long-term survival of DMG, which is more common in younger children, is less than five percent. New treatment approaches are clearly needed for these tumors in both adults and children.
Epigenetic changes play a crucial role in cancer through activation of oncogenes, deactivation of tumor suppressor genes, and mediation of genomic stability. Multiple studies have postulated a role for epigenetic regulators in gliomagenesis.[4, 5] Although adult and pediatric HGG have diverse and somewhat divergent genetic alterations, many of the altered pathways center on epigenetic regulation.[6–8] Recently, most DIPG, and many GBM in children, were linked to mutations of histone H3, the first time histone mutations have been related to human disease,[9] leading to the new classification of DMG based on the presence of the H3K27M mutation. These findings have emphasized the likely role of epigenetic changes in driving the growth of these tumors. Epigenetic changes are also needed in normal development, however, so they may be especially important in pediatric cancer patients, representing normal development gone awry. The changes are, by definition, reversible, unlike genetic changes, and thus are realistic targets for drug design. Epigenetic-focused chemotherapeutics such as HDAC inhibitors has been used clinically in both adults and children.[10]
Initial clinical efforts aimed directly at counteracting the downstream effects of histone mutations have been disappointing. This may be because current epigenetic-targeted medications have broad effects across the epigenome, such as inhibition of DNA methylation or histone deacetylation.[11] Without specific targets, these agents have limited lasting effect and may even do harm, for example through demethylation of oncogenes as well as tumor suppressors. To this same point, genes encoding epigenetic modifiers need not be mutated to have an impact on disease, so sequencing alone is insufficient to detect relevant modifiers. A better functional understanding of the role of specific epigenetic regulators in these tumors is needed now to expand on the present descriptive studies and pave the way for translational impact through more finely targeted epigenetic modification.
Thus, given the need for new treatment approaches for both adult and pediatric HGG, as well as the potential role of epigenetic regulators in gliomagenesis, we undertook a functional screen to identify epigenetic regulators contributing to growth of these tumors.
Results
A two-step, in vitro/in vivo shRNA inhibition screen identifies BPTF as a potential regulator of HGG growth
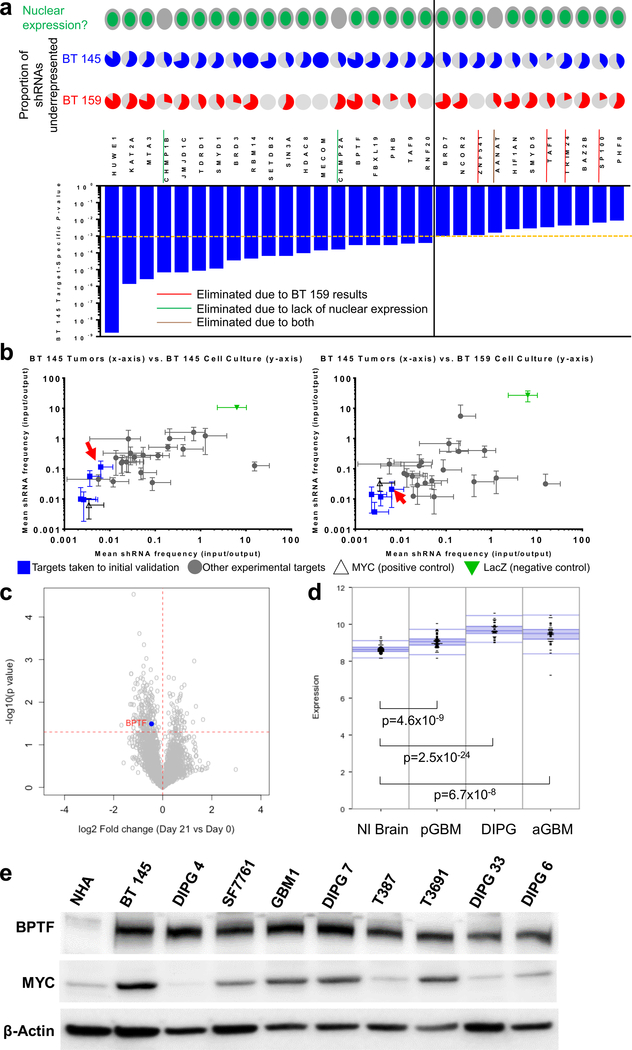
To identify the functional role of specific epigenetic regulators, we began by performing a pooled, neurosphere culture-based shRNA inhibition screen in two adult GBM cell lines, BT 145 and BT 159 (Characteristics of all cell lines are shown in Supplemental Table 1). 449 genes were selected for their known epigenetic function (methylators, deacetylators, etc.) and/or the presence of one or more epigenetic domains (bromodomain, SET domain, etc.). Five to seven shRNAs per gene were chosen and packaged in lentivirus. After infection and selection, half of the cells were reserved to allow baseline calculation of the frequency of each shRNA (input samples). The remaining cells were then allowed to grow an additional 14 population doublings to allow growth effects of gene inhibition to be fully realized (output samples). We then extracted DNA and amplified shRNAs from all samples and used Illumina sequencing to compare the frequency of each shRNA in output and input samples, since shRNAs inhibiting targets key to cell growth would decrease in frequency over the screening period. We used BT 145 as the primary cell line to determine targets for validation because of its far lower rate of shRNA absence in the output samples (19–369 shRNAs missing of 2,908 total, 0.7–12.7%) than BT 159 (1,013–2,124 shRNAs missing of 2,908 total, 34.8–73.0%), indicating less stochastic loss. 15 negative control shRNAs targeting LacZ did not change significantly in representation in either cell line. The magnitude and consistency of under-representation of the shRNAs against a given target compared to the entire population were combined to calculate gene-level p-values in BT 145. The 30 targets with the lowest p-values are shown in Figure 1a. 23 of these targets were taken forward to the secondary screen. We eliminated targets not expressed in the nucleus based on Gene Ontology cell compartment classification as unlikely to be true epigenetic regulators. We then included in the secondary screen all remaining targets with a BT 145 gene-level p-value less than 0.001. For targets with a p-value of 0.01–0.001, we also took into account proportion of shRNAs significantly under-represented in BT 159 and BT 145 output samples (shRNA-level p-value <0.05). We took forward those with ≥2 shRNAs significantly under-represented for both BT 145 and BT 159, including one shRNA in common between lines.
Figure 1:
Justification for BPTF investigation in GBM. a) Top hits from primary screen, showing gene-level p-value in BT 145 (bar graph), with proportion of shRNAs underrepresented in BT 145 and BT 159 (pie charts), and nuclear/cytoplasmic expression of protein; the dotted orange and solid black lines divide targets with BT 145 p-values <0.001 from 0.001–0.01; b) Secondary screen results showing mean underrepresentation of target shRNAs in output compared to input samples in BT 145 tumors compared to BT 145 cell culture (left) and BT 159 cell culture (right), with BPTF indicated by red arrows; c) Volcano plot showing overall fold change (x-axis) and gene-level p-value (y-axis) for a separate epigenomic shRNA screen in DIPG 4, with BPTF highlighted; d) Expression of BPTF in publicly available tumor/normal brain datasets from the R2 platform; e) Western blot of BPTF and MYC expression in normal human astrocytes (NHA), adult HGG cell lines (BT 145, T387, T3691) and pediatric HGG cell lines (DIPG 4, SF7761, GBM1, DIPG 7, DIPG 33 and DIPG 6). Error bars indicate SEM.
To validate our in vitro screen findings in multiple cell lines and models, we performed a secondary screen concurrently in neurosphere culture (BT 145 and BT 159) and in a murine orthotopic PDX model (BT 145). We again used a pooled shRNA inhibition approach. Two aliquots of cells for each line were again infected with pooled lentivirus containing 4 (BT 145) or 2 (BT 159) shRNAs per target, along with multiple negative control shRNAs targeting LacZ and 2 positive control shRNAs targeting the known glioblastoma oncogene MYC.[12, 13] The reduced number of shRNAs per target and resulting higher cell count per shRNA were used in BT 159 to mitigate the loss of library complexity observed in the primary screen. Two input samples were collected after infection as above. Two aliquots from each line were grown as above in neurosphere culture. In addition, for BT 145, 10 NSG mice underwent stereotactic injection of the entire library of shRNAs (1,000 cells per shRNA) into the right striatum of each mouse. Mice were sacrificed when they reached symptom or tumor size endpoint. DNA was then extracted, and shRNAs were amplified from all samples and sequenced using the Illumina platform. shRNA absence from output samples was 2–6 of 114 total shRNAs for the BT 145 cell culture samples (1.8–5.3%), 0–37/114 for the BT 145 PDX samples (0–32.5%), and 2–4 of 61 total shRNAs for the BT 159 cell culture samples (3.3–6.6%). Input and output counts for each shRNA were analyzed to produce gene-level relative representation and standard deviation for each target. Each line’s in vitro data were compared to the BT 145 tumor data (Figure 1b). Four targets with the most significant and consistent shRNA under-representation, including BPTF (red arrows), were consistent across datasets and are represented in blue. shRNAs targeting MYC were also significantly under-represented across datasets, while shRNAs targeting LacZ, the negative control, did not have a significant effect; although it appears that they caused over-representation in these plots, this is a result of most targets showing under-representation in the output samples. LacZ shRNAs therefore comprised a greater proportion of the population in the output samples. In a separate in vitro shRNA screen of epigenetic targets performed in DIPG 4, BPTF again emerged as a significant hit, among other targets isolated as well, with 4 out of 13 BPTF-targeted shRNAs showing decreased representation in output compared to input samples and a gene-level p-value of 0.032 (Figure 1c). There was no shRNA absence from output samples in this screen, likely because it was shorter in duration. Overall, our screening results raised BPTF as a potential regulator of adult and pediatric HGG growth in vitro and in vivo.
BPTF is overexpressed in adult and pediatric HGG
We used the R2 genomics platform to perform expression analysis on publicly-available datasets of transcriptomic data from our three tumor types of interest compared to a similar dataset from normal brain (Figure 1d). We also performed a western blot for BPTF and Myc expression in a normal human astrocyte (NHA) line and several adult and pediatric HGG cell lines (Figure 1e). BPTF was significantly overexpressed both at the gene and protein levels compared to normal brain in adult and pediatric HGG, which provided orthogonal patient sample-level support to our screening findings and encouraged further investigation of BPTF.
BPTF knockdown (KD) reduces GBM proliferation rates in vitro and in vivo
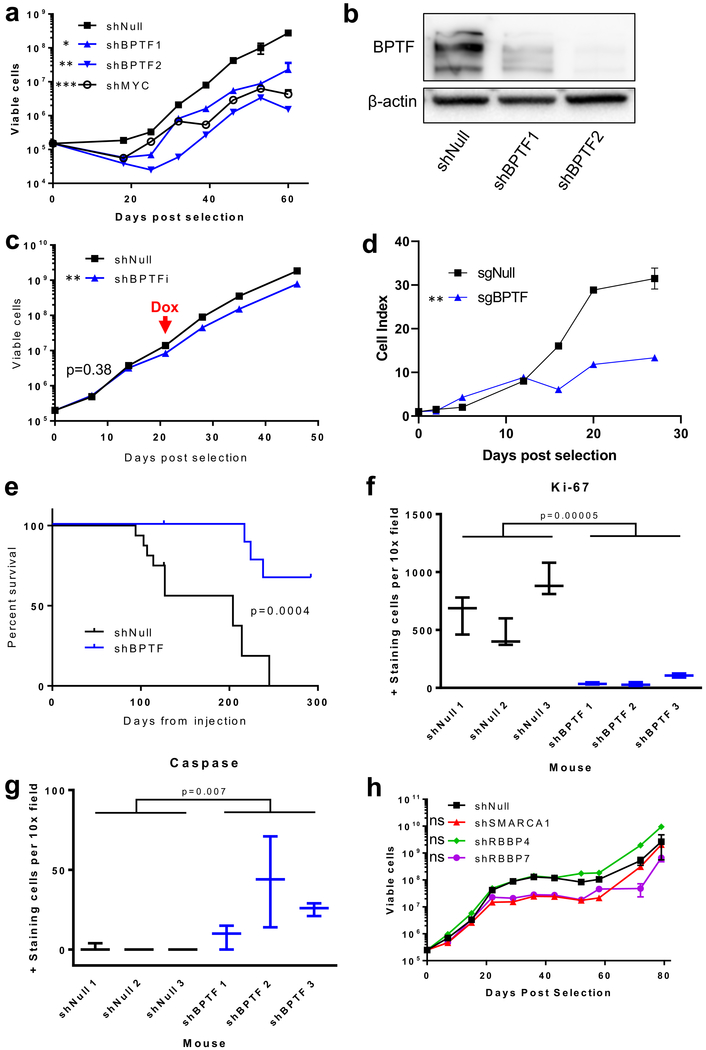
To validate BPTF’s role in the proliferation of adult GBM (aGBM), we stably transduced BT 145 cells using lentivirus to create shBPTF1, shBPTF2, positive control shMYC, and non-targeted control shNull cell lines. We measured cell proliferation in the KD compared to control cells by counting weekly for 60 days. Both shBPTF cell lines and the shMYC cell line showed decreased proliferation rates as compared to shNull (proliferation rates: shBTPF1/shNull=0.71, p<0.05; shBPTF2/shNull=0.51, p<0.01; shMYC/shNull=0.53, p<0.001, Figure 2a). By qPCR, the ratios of BPTF mRNA expression levels (ER) in BPTF KD vs. shNull were 0.35 for shBPTF1 and 0.40 for shBPTF2 (Supplemental Figure 1a). These mRNA levels correlated with decreased BPTF protein expression (BPTF protein ER in shBPTF vs shNull by immunofluorescence (IF) imaging: shBPTF1 ER=0.69, shBPTF2 ER=0.68, Supplemental Figure 1b–c). Western blot analysis confirmed that shBPTF cells had lowered BPTF protein levels versus shNull cells (Figure 2b). These findings confirmed that shRNA-mediated decreases in BPTF mRNA translate to lower levels of BPTF protein expression. We then verified BPTF’s role in BT 145 proliferation through doxycycline-inducible BPTF KD. Prior to doxycycline exposure, shBPTFi and shNull cells proliferated at a similar rate. Upon induction of BPTF shRNA through doxycycline exposure, BPTF levels dropped and the proliferation rate of shBPTFi cells decreased relative to control (Figure 2c, Supplemental Figure 1d). To confirm the results observed using shRNA-mediated knockdown, we used CRISPR-Cas9 to knock out BPTF and performed fluorescent activated cell sorting (FACS) to purify the BPTF knockout population. Proliferation rates observed between a non-targeted control versus BPTF knockout confirmed the results observed with BPTF knockdown via shRNA (Figure 2d). Levels of BPTF following the conclusion of the knockout proliferation experiment are shown in Supplemental Figure 1e. BPTF levels in the knockout line are non-zero (but still significantly less than those of the control cells), which we attribute to outgrowth of cells expressing BPTF that were not successfully eliminated during the FACS sorting.
Figure 2:
Validation of independent BPTF growth effect in BT 145. a) Growth curve showing effect of BPTF and MYC knockdown; b) Western blot showing BPTF knockdown via shRNA at the protein level; c) Growth curve showing the effect of doxycycline-inducible BPTF knockdown starting at the red arrow; d) Growth curve showing effect of BPTF knockout by CRISPR-Cas9; e) Kaplan-Meier curve showing the effect of BPTF knockdown on survival in a murine orthotopic BT 145 PDX model; f) Comparison of Ki-67 positive-staining cells by IHC for shNull compared to shBPTF PDX sections; g) Comparison of caspase positive-staining cells by IHC for shNull compared to shBPTF PDX sections; h) Growth curve showing the effect of knockdown of other NURF complex members. Error bars indicate SEM. * p<0.05; ** p<0.01; *** p<0.001; ns p>0.05
We also observed an anti-proliferative effect of shBPTF on GBM proliferation in our BT 145 murine orthotopic PDX model. Mice injected with either BT 145 shNull or shBPTF cells in the right striatum were observed weekly for clinical signs of disease. shBPTF mice showed significantly prolonged time to symptom endpoint necessitating sacrifice (IC50 204 d for shNull, not reached for shBPTF, Figure 2e). In histological sections taken at the conclusion of the experiment, IF staining demonstrated lower levels of nuclear BPTF protein in tumors originating from BPTF KD cells than those grown from control cells (ER=0.57, Supplemental Figure 1f–g). MRI images, along with hematoxylin and eosin (H&E) staining of tumor sections, show an infiltrative tumor pattern that recapitulates human GBM, as well as a decrease in tumor size and cell density with shBPTF compared to shNull (Supplemental Figure 2a–b). Immunohistochemistry (IHC) staining for the cell proliferation marker Ki-67 demonstrated this finding quantitatively, with shBPTF significantly decreasing the percentage of positive-staining cells (Figure 2f). The percentage of apoptotic cells in these tumors was significantly increased with shBPTF (Figure 2g). These findings validate the positive effect of BPTF in vitro and in vivo on GBM growth.
Knockdown of NURF complex members other than BPTF does not affect proliferation
The nucleosome remodeling factor complex (NURF), of which BPTF is the catalytically active and largest member, includes the additional subunits SMARCA1, RBBP4, and RBBP7 (Supplemental Figure 2c). To test the sufficiency of BPTF in controlling HGG cell proliferation, we stably transduced BT 145 cells with shRNAs targeting each of these NURF members or a non-targeted control (shNull). Cell growth was measured weekly for 80 days. Although a small decrease in cell proliferation rate was noted in the shSMARCA1 and shRBBP7 cell lines initially, the decrease was not statistically significant over the entire period of growth (Figure 2h). We confirmed effective targeting of the shRNAs by measuring knockdown levels via qPCR (Supplemental Figure 2d). These results suggest that BPTF may act through a non-canonical mechanism in regulating GBM growth.
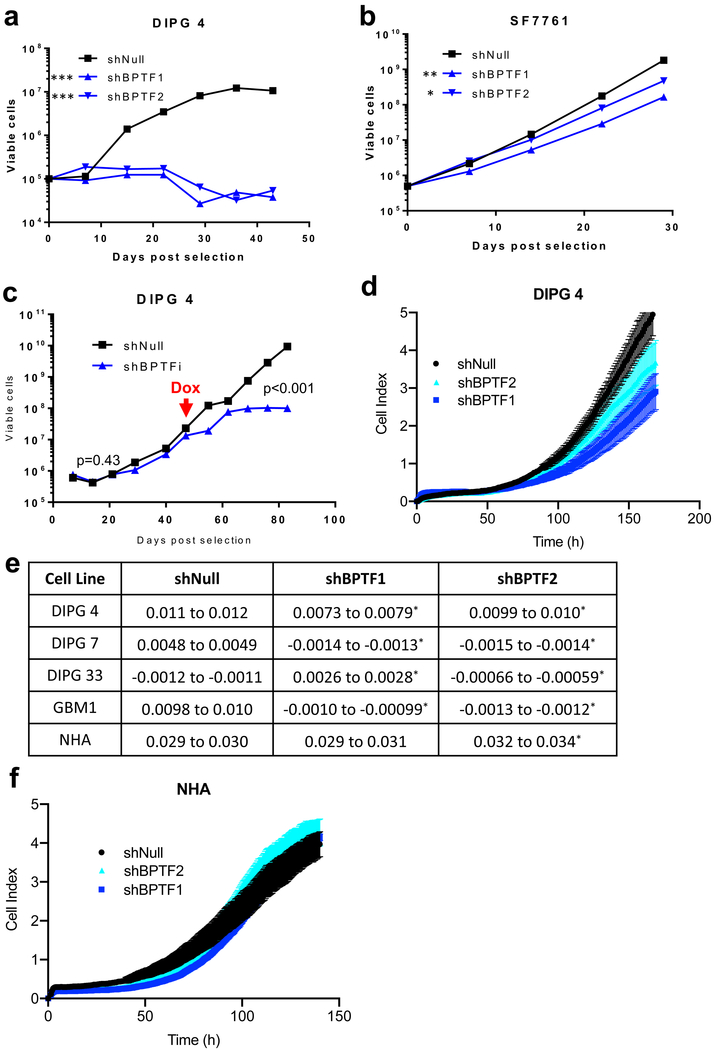
Knockdown of BPTF in pediatric HGG cell lines impairs growth
To assess the applicability of our findings on the impact of BPTF knockdown in aGBM cell lines (BT 145 and BT159) to pediatric HGG, we transduced several pediatric cell lines with shBPTF1, shBPTF2, or shNull. The pediatric HGG cell lines in which we performed the transductions were DIPG 4 (from autopsy), DIPG 7 (from autopsy), DIPG 33 (from autopsy), GBM1 (cortical pGBM from resection) and SF7761 (DIPG from biopsy). We first performed long-term proliferation assays in DIPG 4 and SF7761. Both showed a significant decrease in proliferation rate with BPTF KD, although the magnitude of impact on proliferation was greater in DIPG 4 (Figure 3a–b). We confirmed knockdown by qPCR (Supplemental Figure 3a–b). We further verified BPTF’s role in DIPG 4 proliferation in a doxycycline inducible model, in which we observed that shBPTFi and shNull cells proliferated at a similar rate prior to induction but that upon induction of BPTF shRNA, BPTF levels dropped and the proliferation rate of shBPTFi cells decreased relative to the shNull control (Figure 3c, Supplemental Figure 3c). In order to avoid potential adaptation to BPTF KD that might obscure the proliferation phenotype, we also performed short-term proliferation assays in DIPG 4, DIPG 7, DIPG 33 and GBM1 (SF7761 cells were excluded because they do not grow adherently) (Figure 3d, Supplemental Figure 3d–f). In each of these cell lines, we similarly found that the proliferation rate was significantly less in BPTF KD cells (Figure 3e, Supplemental Figure 3g). These findings suggest that BPTF’s impact on aGBM is generalizable to pediatric HGG, although potentially to varying extents in different pediatric cell lines.
Figure 3:
BPTF growth effect in other cell lines. a-b) Growth curve showing the effect of BPTF knockdown in DIPG 4 and SF7761; c) Growth curve showing the effect of doxycycline-inducible BPTF knockdown in DIPG 4 starting at the red arrow; d) Growth curve showing effect of stable BPTF knockdown in DIPG 4 cells immediately following knockdown in a short-term growth experiment; e) Effect of BPTF knockdown on proliferation in short-term assays conducted immediately after knockdown in pediatric HGG cell lines and normal human astrocyte cells (NHA); f) Growth curve showing the effect of BPTF knockdown in NHA cells. Error bars indicate SEM. * p<0.05; ** p<0.01; *** p<0.001; ns p>0.05
BPTF KD does not affect proliferation of neural stem cells
To determine whether BPTF KD affects proliferation of normal neural stem cells, we performed a proliferation assay in NHA (Figure 3f) stably transduced with shBPTF and shNull. We observed no significant difference in cell proliferation rates over the period of the experiment between shBPTF1 and shNull and a barely significant difference between shBPTF2 and shNull, with shBPTF2 proliferating at a slightly greater rate than shNull (Figure 3e). Significant BPTF mRNA knockdown was established by qPCR (Supplemental Figure 3g.)
BPTF KD affects proliferation of aGBM cell lines
To confirm whether BPTF is important to proliferation in additional preclinical models of aGBM, we utilized publicly-available data from CRISPR screens of cancer cell lines[14–16]. The screening data showed aGBM is highly dependent on BPTF for proliferation (BPTF mean dependency score (CERES): −0.61, p=1.8×10−11) (Supplemental Figure 3h). More negative scores indicate greater levels of dependence with scores less than −0.5 indicating high levels of dependence and a score of −1 indicating genes essential for cell survival. In aGBM, however, we did not find a correlation between endogenous BPTF expression level and dependence on BPTF for proliferation (CRISPR screen correlation coefficient: −0.014, 95% CI: −0.42 to 0.40). Overall, the importance of BPTF for proliferation in aGBM cell lines is similar to what we found in pediatric HGG.
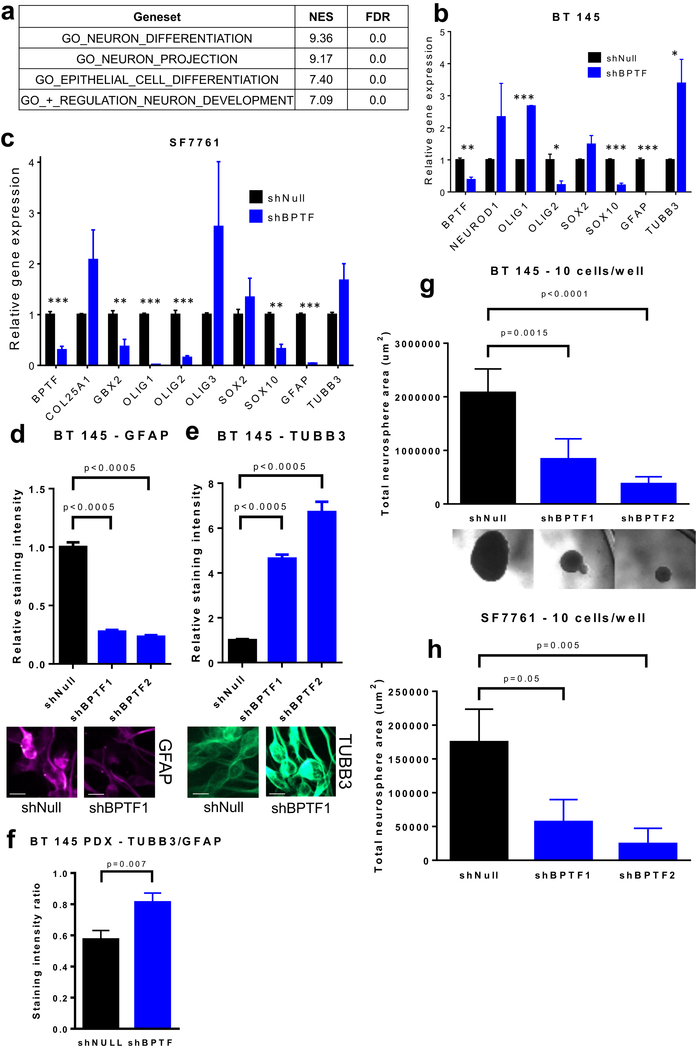
BPTF KD leads to an apparent neuronal lineage shift and decreases cell self-renewal
To test the overall effects of BPTF KD on gene expression, we performed RNA-Seq on stably transduced BT 145 shBPTF and shNull cells, followed by geneset enrichment analysis (GSEA) to investigate how BPTF KD affects the expression of genes involved in biological processes. The most overexpressed and underexpressed genesets overall can be found in Supplemental Tables 2–3. Of 3,449 gene ontology biological process genesets queried (Broad Institute MSigDB), the greatest levels of overexpression were found in genes involved in neuronal differentiation, projection, and development (Figure 4a). qPCR performed on a group of neuronal differentiation and development-related genes confirmed that BPTF KD in BT 145 cells is associated with a decrease in SOX10 and an increase in the neuronal marker TUBB3, and downregulation of the astrocytic marker GFAP (Figure 4b). We also observed upregulation of OLIG1 and NEUROD1 (although the latter did not meet criteria for statistical significance), implying differentiation away from the glial lineage and toward a neuronal phenotype. In SF7761 cells, qPCR likewise revealed a decrease in SOX10 and GFAP, along with trends toward increased expression of TUBB3 and OLIG3, similarly suggesting differentiation toward the neuronal lineage (Figure 4c). Using IF, we confirmed that BPTF KD led to significantly decreased expression of GFAP and increased expression of TUBB3 protein levels (Figure 4d–e) in BT 145 cells. TUBB3 expression was also increased significantly compared to GFAP expression in the BT 145 shBPTF PDX tumors compared to shNull (Figure 4f).
Figure 4:
Effect of BPTF knockdown on differentiation and cell self-renewal. a) GSEA statistics for gene sets related to neuronal characteristics in BT 145 cells with BPTF knockdown; b-c) Gene expression analysis via qPCR for neuronal differentiation targets in BT 145 and SF7761; d-e) Comparison of staining intensity for GFAP and TUBB3 by immunofluorescence for BPTF knockdown BT 145 cells, with representative images underneath (scale bars represent 10 μm); f) Comparison of TUBB3/GFAP staining intensity ratio for BT 145 PDX sections; g-h) Results of neurosphere dilution assays showing effect of BPTF knockdown on total neurosphere area in BT 145, with representative images overhead, and SF7761. Error bars indicate SEM. * p<0.05; ** p<0.01; *** p<0.001
We then performed a cell self-renewal assay[17, 18] in which stably transduced shBPTF or shNull BT 145 and SF7761 cells were plated at a concentration of 1 or 10 cells per well and grown in suspension. We measured neurosphere size in each well after 4–10 weeks of growth. shBPTF inhibited neurosphere formation, significantly reducing both the mean size of spheres (Figure 4g–h) and the proportion of wells in which neurospheres formed (Supplemental Figure 4a–c). We also measured the effect of MYC KD on the proportion of wells forming a neurosphere in BT 145, which showed a similar effect to BPTF KD (Supplemental Figure 4a). BPTF KD did not appear to consistently affect cell cycle in NHA, BT 145, or pHGG cell lines, nor did it cause a significant induction in apoptosis in BT 145 (Supplemental Figure 4d–e). Taken together, these results suggest that BPTF contributes to HGG proliferation most importantly through maintenance of cell self-renewal, and its loss leads these tumor cells with glial characteristics to take on the traits of differentiated neurons.
BPTF KD reduces expression of MYC and its targets
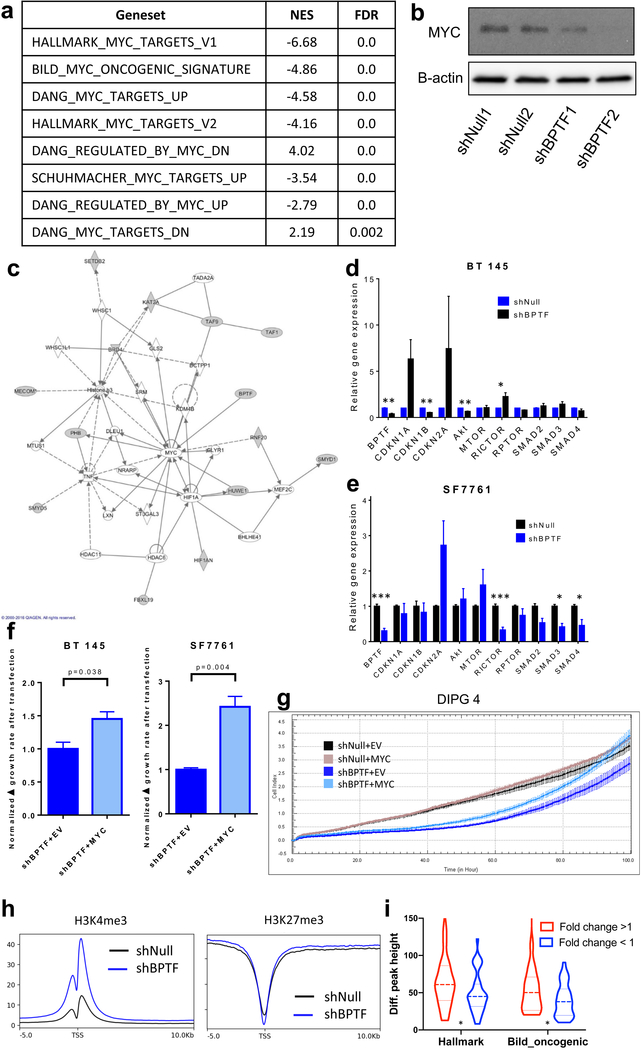
We then used our RNA-Seq data to attempt to determine a specific mechanism of action of BPTF in HGG. GSEA using the 50 hallmark gene sets in the Broad Institute MSigDB collection showed that the MYC hallmark genesets V1 and V2, respectively, were the 2nd and 12th most downregulated in BPTF KD versus control cells (Figure 5a). Other MYC genesets were also significantly downregulated, notably including a set of oncogenic targets of MYC regulation (BILD_MYC_ONCOGENIC_SIGNATURE, Figure 5a). We confirmed the decreased MYC protein levels in stably transduced shBPTF versus shNull cells by western blot (Figure 5b) and IF (Supplemental Figure 5a–b). IPA modeling of the RNA-Seq data also suggested that of the gene targets included in the secondary screen, listed in Figure 1a, only BPTF directly regulates MYC, while many of the other secondary screen targets are upstream or downstream members of a regulatory network centered on MYC (Figure 5c). In qPCR validations of the RNA-Seq data, BPTF KD correlated with suppression of CDKN1B and AKT gene expression in BT 145 cells and upregulated SMAD2 and SMAD3 in SF7761 cells (Figure 5d–e). To demonstrate that the effect of BPTF KD on the MYC pathway is an initial effect as opposed to a manifestation of long-term state change, we used short-term BPTF KD in DIPG 4 and performed qPCR for MYC targets, which recapitulated the findings from the long-term KD showing downregulation of the pathway; however, these same significant effects were absent in NHA, again suggesting a tumor-specific effect (Supplemental Figure 5c). Overall, these results show that MYC and MYC targets are broadly downregulated by BPTF KD, and that MYC is at the center of a regulatory network that comprises many of the epigenetic regulators found in our screen to be associated with HGG growth.
Figure 5:
BPTF knockdown interaction with MYC and downstream effectors. a) GSEA statistics for MYC target sets in BT 145 cells with BPTF knockdown; b) Western blot showing effect on MYC expression with BPTF knockdown; c) IPA interaction map for BPTF and other hits from the shRNA screen showing interaction with MYC; d-e) Gene expression analysis via qPCR for MYC targets in BT 145 and SF7761; f) Effect of cDNA-mediated MYC overexpression on cell growth rate compared to empty vector (EV) in BPTF knockdown BT 145 and SF7761 cells; g) Effect of cDNA-mediated MYC overexpression on cell growth compared to empty vector (EV) measured by xCELLigence in BPTF knockdown DIPG 4 cells; h) Global genome average of H3K4 and H3K27 trimethylation at transcription start sites (TSS) for BT 145 shNull and BPTF KD cells. i) H3K4me3 enrichment for upregulated and downregulated genes (shBPTF vs. shNull) in the Hallmark MYC target and Bild_Oncogenic genesets. H3K3me3 enrichment is significantly greater (p<0.05) for genes where expression increases with BPTF KD. Error bars indicate SEM. * p<0.05; ** p<0.01; *** p<0.001
To establish that the HGG growth effect of BPTF KD was mediated through the effect on MYC, we examined whether the observed reduction in proliferation rates could be reversed through increased MYC expression in shBPTF cells. Using transfection of MYC cDNA compared to an empty vector (EV), we overexpressed MYC in stably transduced shBPTF and shNull cells in BT 145, SF7761, and DIPG 4. We measured BT 145 and SF7761 cell growth in all conditions before and after transfection. MYC overexpression accelerated growth significantly in shBPTF cells compared to shBPTF-EV in both lines (Figure 5f), while MYC overexpression in shNull cells had a smaller effect (Supplemental Figure 5d). In DIPG 4, we transfected cells in these same conditions, and then 24 hours later began measuring growth on an xCELLigence plate reader. shBPTF-EV and -MYC cells initially showed decreased proliferation compared to shNull conditions, but MYC overexpression in shBPTF cells led to growth acceleration over time that reached overall growth of the shNull conditions, in which MYC overexpression had a much smaller effect, by 100 hours (Figure 5g). shBPTF-EV cells did show some growth acceleration over time in each line, which may have been due to a relatively smaller effect on growth in these slower-growing conditions from the transfection process itself compared to shNull conditions. This acceleration, however, was significantly less than with MYC overexpression. We confirmed, via qPCR, BPTF KD, as well as MYC overexpression following MYC transfection as compared to EV (Supplemental Figure 5e). We also performed a proliferation assay in the Myc-immmortalized C17.2 murine neural stem cell line, stably transduced with shBptf or shNull, and found over a longer period that there was no difference in proliferation rate as the result of Bptf KD, providing further evidence that Myc overexpression can rescue the growth effect of Bptf KD. (Supplemental Figure 6a). Significant Bptf mRNA and protein knockdown vs. shNull in the C17.2 cells was established by qPCR and IF respectively (Supplemental Figure 6b). These results suggest that the effect of BPTF knockdown on HGG growth is caused by downregulation of MYC.
We theorized that BPTF, as an epigenetic regulator, likely affected the chromatin state of MYC’s transcriptional targets, as well as possibly exerting a broader genome-wide effect. To test this idea, we performed ChIP-Seq on BPTF KD versus shNull cells, immunoprecipitating for H3K4me3 and H3K27me3. On a genome-wide average basis, knockdown of BPTF expression enriched H3K4 trimethylation and did not affect H3K27 trimethylation at transcription start sites of genes (Figure 5h). To investigate the effects of these chromatin modifications at MYC transcriptional sites, we identified the enrichment of H3K4me3 chromatin marks for genes in the HALLMARK_MYC_TARGETS_V1 and BILD_MYC_ONCOGENIC_SIGNATURE genesets and computed average enrichment for genes whose enrichment increased versus decreased as the result of BPTF KD. This analysis showed that H3K4me3 enrichment is significantly greater for MYC target genes with increased expression versus those with decreased expression as the result of BPTF KD (Figure 5i, Supplemental Table 4). Effects on these chromatin marks in representative MYC target genes CCNA2 and ODC1 are shown in Supplemental Figure 6c.
Epigenetic compound screening identifies HDAC inhibitors as synergistic with BPTF KD
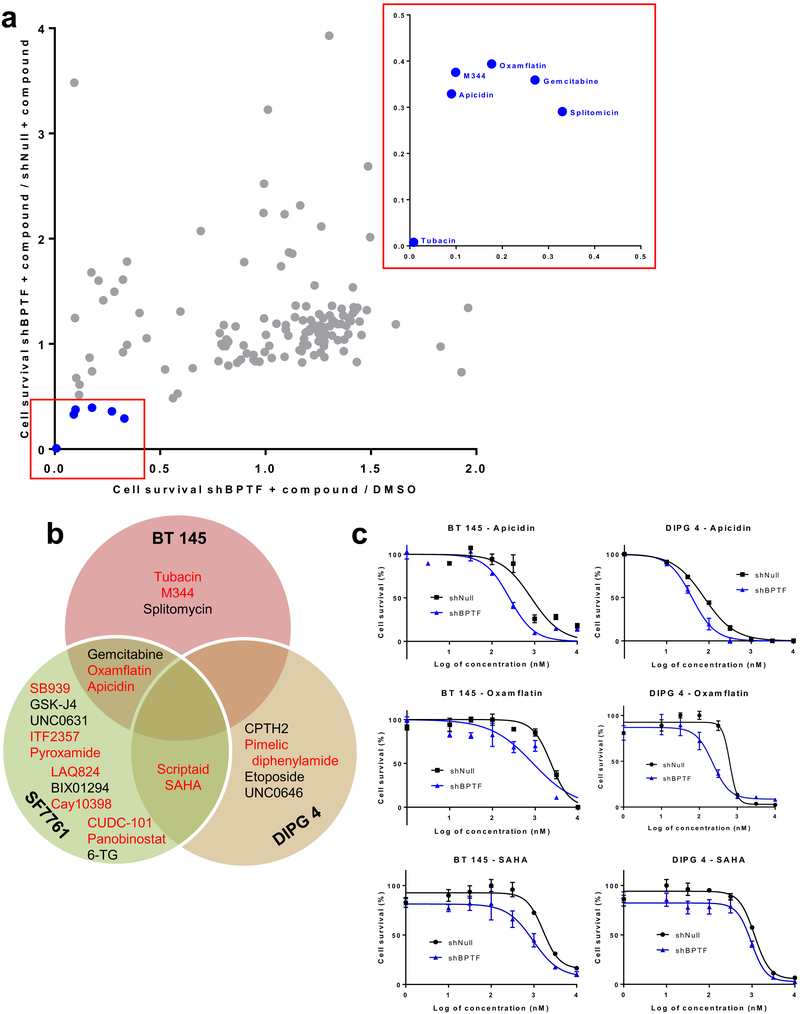
Small molecule BPTF inhibitors are currently in development,[19] but potency and specificity is still being optimized. With an eye toward eventual translational application of our results to patient treatment, in which combination therapy is crucial, we screened for potentially effective compounds targeting epigenetic regulators that act synergistically in combination with shRNA-mediated BPTF knockdown, which is an accurate model of the levels of inhibition possible pharmacologically. BT 145, SF7761, and DIPG 4 cells transduced with shBPTF or shNull were treated for 5 days with 5 μM of each compound in the panel or DMSO control. Compounds were selected as potentially synergistic with BPTF KD if they decreased viability of shBPTF cells by at least 50% compared to shNull cells in DMSO control, and also decreased viability of shBPTF cells by at least 50% compared to shNull cells exposed to that same compound.
Screening in BT 145 cells identified 6 compounds that met these criteria: gemcitabine, a nucleoside analog; splitomycin, a SIRT1 inhibitor; and four HDAC inhibitors, M344, tubacin, apicidin, and oxamflatin (Figure 6a). Apicidin, oxamflatin, and gemcitabine also met criteria in SF7761 (Figure 6b). Multiple other HDAC inhibitors met criteria in SF7761 alone, including SB939, ITF 2359, pyroxamide, LAQ824, scriptaid, CAY10398, CUDC101, SAHA, and panobinostat. Four additional compounds showed adequate effectiveness in SF7761 cells: UNC0631, a HMTase inhibitor, GSKJ4, a JMJD3/UTX inhibitor, BIX01294, a methyltransferase inhibitor, and 6-GT, an antimetabolite compound. In DIPG 4, the HDAC inhibitors scriptaid and SAHA passed screening criteria, as they did in SF7761. Other compounds effective in DIPG 4 alone included the HDAC inhibitor pimelic diphenylamide, the histone acetyltransferase inhibitor CPTH2, the topoisomerase II inhibitor etoposide, and the methyltransferase inhibitor UNC0646. HDAC inhibitors thus accounted for 14 of 23 of the hits among the three cell lines tested, including 4 of 5 shared between two lines. To validate the HDAC screening results, BT 145 and DIPG 4 cells transduced with either shBPTF or shNull were treated with three of the HDAC inhibitors for five days. Oxamflatin and apicidin both showed a significant decrease in IC50 in shBPTF cells as compared to shNull in both lines, while BPTF KD decreased the IC50 for SAHA non-significantly (Figure 6c, Supplemental Table 5). These results raise HDAC and BPTF inhibition as a potentially effective combination therapy across BPTF subtypes.
Figure 6:
Potential epigenetic-targeting compounds synergistic with BPTF knockdown. a) Scatterplot of relative cell survival of BPTF knockdown BT 145 cells treated with a panel of epigenetic-targeting compounds, with hits expanded in red box; b) Venn diagram of hits from epigenetic compound panel in BPTF knockdown BT 145, SF7761, and DIPG 4 cells; c) Dose-response curves for BPTF knockdown BT 145 and DIPG 4 cells treated with three HDAC inhibitors. Error bars indicate SEM.
Discussion
In this study, we demonstrate that BPTF is a regulator of tumor growth and mediates cell self-renewal in HGG. This target emerged from an epigenomic shRNA inhibition screen using both cell culture and orthotopic PDX models, and its overexpression appears to characterize all three of the HGG subtypes studied. BPTF is the DNA-binding member of the NURF chromatin remodeling complex and interacts with common histone modifications, giving it broad effects on transcription.[20] We took BPTF forward for further investigation from among our screening hits because it has been associated with neurologic disease[21, 22] and neurodevelopment,[23] as well as potentially implicated in multiple cancer types, including by overexpression[24, 25] or mutation,[26, 27] or by driving proliferation[28–30] or metastasis,[24, 27, 30, 31] but has not been investigated in brain cancer.
We then studied BPTF’s impact on growth and phenotype in multiple models of adult and pediatric HGG. Since we saw a significant effect of BPTF knockdown on growth in all lines except NHA, BPTF overexpression itself, which we noted in all lines compared to NHA, may be a biomarker predictive of growth response to BPTF knockdown, although the lack of correlation between endogenous BPTF expression level and proliferation dependence from publicly available cell line data calls this into question in adult HGG. We note, however, that pediatric HGG and adult HGG are biologically distinct entities. MYC overexpression was more variable in these cell lines and did not appear to correlate with the degree of effect on growth with BPTF knockdown. It is also notable that SF7761, which showed the smallest growth effect among the tumor lines, is immortalized with exogenous hTERT (See Supplemental Table 1 for cell line characteristics), while none of the other cell lines are immortalized, which may allow SF7761 to grow more successfully with BPTF knockdown. No other cell line features, including H3K27 mutation status or tumor subtype, appeared predictive of response. Identifying and validating biomarkers of BPTF dependency in each disease subtype will be a focus of ongoing work. The lack of significant effect on HGG growth via KD of other NURF members, while preliminary, raises the possibility that BPTF is either working independently of its complex to control HGG or is sufficient for this function; further inquiry into these possibilities should be the subject of ongoing investigation. While the finding of its importance to cell self-renewal of HGG fits with a recent report of a similar function for BPTF in mammary gland stem cells, the finding of increased neuronal features in these glial tumors with BPTF inhibition was unexpected and novel. Our gene expression data, especially the strong decrease in SOX10 expression with BPTF KD across multiple cell lines, suggest that BPTF expression is integral to the maintenance of stem cell potency in HGG.[32] The combined strong decrease in GFAP and increase in TUBB3 expression with BPTF KD suggest that BPTF expression may also be necessary to maintain glial lineage characteristics in HGG cells, and the trend toward overexpression of SOX2 with BPTF KD raises a potential mechanism by which the loss of BPTF may cause a direct transdifferentiation of lineage.[33–35] These findings are encouraging from a translational standpoint, since stemness is a hallmark of cancer, and brain tumors with neuronal characteristics are more responsive to chemotherapy than gliomas.
We demonstrated that BPTF appears to control HGG growth through a positive effect on MYC pathways. A recent report demonstrated that BPTF and MYC form a protein complex critical to MYC’s broad functions as a transcription factor and oncogene.[28] Our findings strongly support and add to these conclusions and are demonstrated in multiple ways. BPTF KD showed similar effects to MYC KD in our screen, and in fact, many of the epigenetic regulators implicated in contributing to HGG growth by our screen have direct or indirect effects on MYC or are affected by MYC, as shown on IPA analysis. BPTF KD downregulated MYC-dependent genesets and individual targets, which corresponds to the results of the prior report. We also found that BPTF KD decreased MYC expression, which was not demonstrated in this previous study, and that overexpression of MYC was sufficient to rescue HGG cells from the negative growth effect of BPTF KD. Decreased expression of MYC and its downstream targets through BPTF KD was a key finding of our transcriptomic studies. Our ChIP-Seq results showed that on a genome-wide average basis, BPTF KD increases global H3K4me3 enrichment at transcription start sites but had little effect on H3K27me3 levels. BPTF KD had a more nuanced effect at MYC target genes, increasing expression at some but decreasing expression at most, particularly those identified as oncogenic. This observation raises important questions about the mechanism of BPTF’s role in modifying the chromatin landscape and the reasons behind the pattern of changing H3K4me3 enrichment that we observed. For example, BPTF KD (and the accompanying decreased MYC expression) were not accompanied by corresponding changes in the expression of cell cycle regulatory genes that are typically associated with MYC-driven proliferation,[36] but instead, as noted above, by differentiation and lineage changes that would be expected to decrease proliferation. MYC has been linked to increased self-renewal of neural progenitor cells. This mechanism and its relationship to BPTF’s broader role in modiftying the chromatin landscape are the subjects of ongoing investigation.
Finally, we showed that BPTF inhibition synergizes with HDAC inhibition against HGG growth. BPTF and HDAC inhibition have not been associated in the literature, so it is intriguing that the majority of the epigenetic-targeting compounds found to synergize with BPTF KD in our screen fell into this class. A potential explanation for this finding is suggested by our IPA analysis, which identified two HDACs (6 and 11) and histone b3 in the MYC downstream network that BPTF influences. Prior work has found modulation of MYC to be an important target of HDAC inhibition.[37] Our findings will have important potential translational implications as BPTF small molecule inhibition progresses to clinical use, since multiple HDAC inhibitors, including panobinostat,[38] are under investigation for use in HGG, but combination therapy is likely to be crucial to overcoming these highly treatment-resistant tumors.
Strengths of the current study include the exclusive use of patient-derived, low-passage number cell lines throughout our work, with the goal to best represent human HGG. Including the orthotopic PDX model in our screen and validation steps allows us to bring in the influence of the tumor microenvironment and provides stronger evidence of BPTF’s influence on HGG. We included cell lines from diverse patient ages, tumor locations, treatment stages, and molecular subtypes for the major assays performed. Adult and pediatric HGG are certainly distinct in terms of specific mutations seen, but there are also similarities relevant to our study. Both adult and pediatric HGG can have dependency on MYC.[39] In addition, the effect we demonstrate of BPTF knockdown on cell self-renewal and glial characteristics would be relevant to all tumors of glial lineage. In addition, our findings on the implications of BPTF for neuronal differentiation and cell self-renewal, as well as its influence on MYC, arose from orthogonal transcriptomic and targeted assays conducted independently.
Weaknesses of the study include the absence of an available tool (either cDNA or shRNA targeting the 3’UTR) to rescue cells from BPTF knockdown and thus confirm the specificity of the effect on HGG; however, we used multiple shRNAs across the project that all had similar effects, as did CRISPR knockout. Another weakness is the lack of a small molecule BPTF inhibitor to test in parallel with shRNA-mediated KD. For now, chemical inhibitors of BPTF are still in early development. Since the heterozygous BPTF knockout model, replicating the level of BPTF knockdown likely possible pharmacologically, does not have a discernible phenotype,[40] it is an encouraging potential target; we also showed a selective growth effect of BPTF KD on HGG that spared neural stem cells. Our study demonstrates that BPTF inhibition shows strong therapeutic potential in adult and pediatric HGG, both alone and in combination with HDAC inhibitors or radiotherapy, and should encourage further small molecule development. In addition to these translational goals, future studies will also focus on developing our understanding of the specific interactions mediating BPTF’s broad influence, especially on the MYC pathway, a long-sought therapeutic target in cancer.
Supplementary Material
Acknowledgements
We would like to thank Dr. William Pomerantz for his expertise and discussion of BPTF small molecule inhibitors, and Drs. Stuart Orkin, Mark Kieran, and Jay Bradner for their advice at early stages of the project. We also acknowledge the University of Colorado Denver Research Histology Shared Resource, supported by the Cancer Center Support Grant (P30CA046934), as well as the University of Colorado Anschutz Medical Campus Advanced Light Microscopy Core Facility and Functional Genomics Core Facility, for their assistance. This work was supported by a St. Baldrick’s Scholar award, a Hyundai Hope on Wheels Young Investigator award, a Morgan Adams Foundation grant, a Pedals for Pediatrics grant, and NICHD K12HD068372 (all to ALG). The Colorado Animal Imaging Shared Resource is supported by the NCI and the University of Colorado Cancer Center (P30CA046934).
Financial Support: This work was supported by a St. Baldrick’s Scholar award, a Hyundai Hope on Wheels Young Investigator award, a Morgan Adams Foundation grant, a Pedals for Pediatrics grant, and NICHD K12HD068372 (ALG).
Footnotes
Conflict of Interest: The authors declare that they have no conflict of interest.
References
- 1.Louis DN, Ohgaki H, Wiestler OD, Cavenee WK, Burger PC, Jouvet A et al. The 2007 WHO classification of tumours of the central nervous system. Acta neuropathologica 2007; 114: 97–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Wen PY, Kesari S. Malignant gliomas in adults. The New England journal of medicine (Research Support, N.I.H., Extramural Research Support, Non-U.S. Gov’t Review) 2008; 359: 492–507. [DOI] [PubMed] [Google Scholar]
- 3.Stupp R, Mason WP, van den Bent MJ, Weller M, Fisher B, Taphoorn MJ et al. Radiotherapy plus concomitant and adjuvant temozolomide for glioblastoma. The New England journal of medicine 2005; 352: 987–996. [DOI] [PubMed] [Google Scholar]
- 4.Clarke J, Penas C, Pastori C, Komotar RJ, Bregy A, Shah AH et al. Epigenetic pathways and glioblastoma treatment. Epigenetics : official journal of the DNA Methylation Society 2013; 8: 785–795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Maleszewska M, Kaminska B. Is glioblastoma an epigenetic malignancy? Cancers 2013; 5: 1120–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Ferreira WA, Pinheiro Ddo R, Costa Junior CA, Rodrigues-Antunes S, Araujo MD, Leao Barros MB et al. An update on the epigenetics of glioblastomas. Epigenomics 2016; 8: 1289–1305. [DOI] [PubMed] [Google Scholar]
- 7.Touat M, Idbaih A, Sanson M, Ligon KL. Glioblastoma targeted therapy: updated approaches from recent biological insights. Annals of oncology : official journal of the European Society for Medical Oncology / ESMO 2017; 28: 1457–1472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lulla RR, Saratsis AM, Hashizume R. Mutations in chromatin machinery and pediatric high-grade glioma. Sci Adv 2016; 2: e1501354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Schwartzentruber J, Korshunov A, Liu XY, Jones DT, Pfaff E, Jacob K et al. Driver mutations in histone H3.3 and chromatin remodelling genes in paediatric glioblastoma. Nature 2012; 482: 226–231. [DOI] [PubMed] [Google Scholar]
- 10.Lee P, Murphy B, Miller R, Menon V, Banik NL, Giglio P et al. Mechanisms and clinical significance of histone deacetylase inhibitors: epigenetic glioblastoma therapy. Anticancer research 2015; 35: 615–625. [PMC free article] [PubMed] [Google Scholar]
- 11.Mottamal M, Zheng S, Huang TL, Wang G. Histone deacetylase inhibitors in clinical studies as templates for new anticancer agents. Molecules 2015; 20: 3898–3941. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Herms JW, von Loewenich FD, Behnke J, Markakis E, Kretzschmar HA. c-myc oncogene family expression in glioblastoma and survival. Surg Neurol 1999; 51: 536–542. [DOI] [PubMed] [Google Scholar]
- 13.Wang J, Wang H, Li Z, Wu Q, Lathia JD, McLendon RE et al. c-Myc is required for maintenance of glioma cancer stem cells. PloS one 2008; 3: e3769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Broad D DepMap Achilles 19Q1 Public, 2019. [Google Scholar]
- 15.Ghandi M, Huang FW, Jané-Valbuena J, Kryukov GV, Lo CC, McDonald ER et al. Next-generation characterization of the Cancer Cell Line Encyclopedia. Nature 2019; 569: 503–508. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Meyers RM, Bryan JG, McFarland JM, Weir BA, Sizemore AE, Xu H et al. Computational correction of copy number effect improves specificity of CRISPR-Cas9 essentiality screens in cancer cells. Nature genetics 2017; 49: 1779–1784. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Xie Q, Wu QL, Kim L, Miller TE, Liau BB, Mack SC et al. RBPJ maintains brain tumor-initiating cells through CDK9-mediated transcriptional elongation. Journal of Clinical Investigation 2016; 126: 2757–2772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Bao SD, Wu QL, McLendon RE, Hao YL, Shi Q, Hjelmeland AB et al. Glioma stem cells promote radioresistance by preferential activation of the DNA damage response. Nature 2006; 444: 756–760. [DOI] [PubMed] [Google Scholar]
- 19.Urick AK, Hawk LM, Cassel MK, Mishra NK, Liu S, Adhikari N et al. Dual Screening of BPTF and Brd4 Using Protein-Observed Fluorine NMR Uncovers New Bromodomain Probe Molecules. ACS chemical biology 2015; 10: 2246–2256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Li H, Ilin S, Wang W, Duncan EM, Wysocka J, Allis CD et al. Molecular basis for site-specific read-out of histone H3K4me3 by the BPTF PHD finger of NURF. Nature 2006; 442: 91–95. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stankiewicz P, Khan TN, Szafranski P, Slattery L, Streff H, Vetrini F et al. Haploinsufficiency of the Chromatin Remodeler BPTF Causes Syndromic Developmental and Speech Delay, Postnatal Microcephaly, and Dysmorphic Features. Am J Hum Genet 2017; 101: 503–515. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Bekpen C, Tastekin I, Siswara P, Akdis CA, Eichler EE. Primate segmental duplication creates novel promoters for the LRRC37 gene family within the 17q21.31 inversion polymorphism region. Genome research 2012; 22: 1050–1058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Ma Y, Liu X, Liu Z, Wei S, Shang H, Xue Y et al. The Chromatin Remodeling Protein Bptf Promotes Posterior Neuroectodermal Fate by Enhancing Smad2-Activated wnt8a Expression. The Journal of neuroscience : the official journal of the Society for Neuroscience 2015; 35: 8493–8506. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Gong YC, Liu DC, Li XP, Dai SP. BPTF biomarker correlates with poor survival in human NSCLC. Eur Rev Med Pharmacol Sci 2017; 21: 102–107. [PubMed] [Google Scholar]
- 25.Buganim Y, Goldstein I, Lipson D, Milyavsky M, Polak-Charcon S, Mardoukh C et al. A novel translocation breakpoint within the BPTF gene is associated with a pre-malignant phenotype. PloS one 2010; 5: e9657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Lee JH, Kim MS, Yoo NJ, Lee SH. BPTF, a chromatin remodeling-related gene, exhibits frameshift mutations in gastric and colorectal cancers. APMIS 2016; 124: 425–427. [DOI] [PubMed] [Google Scholar]
- 27.Xiao S, Liu L, Fang M, Zhou X, Peng X, Long J et al. BPTF Associated with EMT Indicates Negative Prognosis in Patients with Hepatocellular Carcinoma. Dig Dis Sci 2015; 60: 910–918. [DOI] [PubMed] [Google Scholar]
- 28.Richart L, Carrillo-de Santa Pau E, Rio-Machin A, de Andres MP, Cigudosa JC, Lobo VJ et al. BPTF is required for c-MYC transcriptional activity and in vivo tumorigenesis. Nature communications 2016; 7: 10153. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Dai M, Lu JJ, Guo W, Yu W, Wang Q, Tang R et al. BPTF promotes tumor growth and predicts poor prognosis in lung adenocarcinomas. Oncotarget 2015; 6: 33878–33892. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Dar AA, Nosrati M, Bezrookove V, de Semir D, Majid S, Thummala S et al. The role of BPTF in melanoma progression and in response to BRAF-targeted therapy. Journal of the National Cancer Institute 2015; 107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Xiao S, Liu L, Lu X, Long J, Zhou X, Fang M. The prognostic significance of bromodomain PHD-finger transcription factor in colorectal carcinoma and association with vimentin and E-cadherin. Journal of cancer research and clinical oncology 2015; 141: 1465–1474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Kim J, Lo L, Dormand E, Anderson DJ. SOX10 maintains multipotency and inhibits neuronal differentiation of neural crest stem cells. Neuron 2003; 38: 17–31. [DOI] [PubMed] [Google Scholar]
- 33.Bramanti V, Tomassoni D, Avitabile M, Amenta F, Avola R. Biomarkers of glial cell proliferation and differentiation in culture. Front Biosci (Schol Ed) 2010; 2: 558–570. [DOI] [PubMed] [Google Scholar]
- 34.Person F, Wilczak W, Hube-Magg C, Burdelski C, Moller-Koop C, Simon R et al. Prevalence of betaIII-tubulin (TUBB3) expression in human normal tissues and cancers. Tumour biology : the journal of the International Society for Oncodevelopmental Biology and Medicine 2017; 39: 1010428317712166. [DOI] [PubMed] [Google Scholar]
- 35.Su Z, Niu W, Liu ML, Zou Y, Zhang CL. In vivo conversion of astrocytes to neurons in the injured adult spinal cord. Nature communications 2014; 5: 3338. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Bouchard C, Thieke K, Maier A, Saffrich R, Hanley-Hyde J, Ansorge W et al. Direct induction of cyclin D2 by Myc contributes to cell cycle progression and sequestration of p27. EMBO J 1999; 18: 5321–5333. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Nebbioso A, Carafa V, Conte M, Tambaro FP, Abbondanza C, Martens J et al. c-Myc Modulation and Acetylation Is a Key HDAC Inhibitor Target in Cancer. Clinical cancer research : an official journal of the American Association for Cancer Research 2017; 23: 2542–2555. [DOI] [PubMed] [Google Scholar]
- 38.Grasso CS, Tang Y, Truffaux N, Berlow NE, Liu L, Debily MA et al. Functionally defined therapeutic targets in diffuse intrinsic pontine glioma. Nat Med 2015; 21: 827. [DOI] [PubMed] [Google Scholar]
- 39.Lapin DH, Tsoli M, Ziegler DS. Genomic Insights into Diffuse Intrinsic Pontine Glioma. Front Oncol 2017; 7: 57. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Landry J, Sharov AA, Piao Y, Sharova LV, Xiao H, Southon E et al. Essential role of chromatin remodeling protein Bptf in early mouse embryos and embryonic stem cells. PLoS genetics 2008; 4: e1000241. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.