Abstract
The nuclear genome drives differences in immune cell populations and differentiation potentials, in part regulated by changes in metabolism. Despite this connection, the role of mitochondrial DNA (mtDNA) polymorphisms (SNP) in this process has not been examined. Using mitochondrial nuclear exchange (MNX) mice, we and others have shown that mtDNA strongly influences varying aspects of cell biology and disease. Based upon an established connection between mitochondria and immune cell polarization, we hypothesized that mtDNA SNP alter immune cell development, trafficking, and/or differentiation. Innate and adaptive immune cell populations were isolated and characterizated from the peritoneum and spleen. While most differences between mouse strains are regulated by nuclear DNA (nDNA), there are selective changes that are mediated by mtDNA differences (e.g., macrophage (CD11c) differentiation), These findings highlight how nuclear-mitochondrial crosstalk may alter pathology and physiology via regulation of specific components of the immune system.
Keywords: mitochondria, immune, polymorphism, macrophage, T cell, differentiation
1. Introduction
Therapeutic strategies are becoming increasingly personalized, driving a need for an ever greater knowledge of the underlying biology of the individual. Our previous results using mitochondrial nuclear exchange (MNX) mice identified mitochondrial DNA single nucleotide polymorphisms (mtDNA SNP) as quantitative trait loci (QTL) that, in conjunction with the nDNA, regulate cancer progression and metastatic susceptibility [1–3]; atherosclerosis and cardiac failure [4]; selectively alter nDNA methylation, and gene expression patterns [5]. How mtDNA SNP impact the underlying biology surrounding complex disease mechanisms and homeostasis is an important question in defining future therapeutic strategies [6].
The above-referenced metastasis and tumorigenicity data were obtained by crossing transgenic mice with female MNX mice (to ensure transfer of mtDNA to progeny). However, we also recognized that any observed phenotypic changes could be a result of altered mtDNA in stromal cells. To isolate the stroma as an experimental variable, nuclear DNA-matched syngeneic tumor cells were injected into MNX mice (i.e., only the stroma had different mtDNA), and we observed profound changes in lung colonization (Brinker et al. accepted pending minor revisions). Briefly, whenever C57BL/6J mtDNA was present in stromal cells, metastasis efficiency was lower. In contrast, C3H/HeN mtDNA was correlated with increased metastasis. In this study, we therefore tested the hypothesis that mtDNA alters components of the immune system at baseline as an explanation for the observed stroma-correlated changes in cancer metastatic efficiency.
Inbred mouse strains have provided important tools for the identification of genetic influences on cancer progression and metastatic susceptibility [7]. Murine genetic diversity is reflected in overall immune development [8] and functionality [9]. Petkova et al. from Jackson laboratories observed large variability in immune profiles across seven different mouse strain lineages representing different haplogroups [10]. They observed the greatest variation in circulating lymphocytes. Haplogroup 1 (BALB/cJ and C3H/HeJ) had low lymphocyte percentages (36-78%), while Haplogroup 4 (C57BL/6J) had the highest percentages (68-80%). B cells were the predominant driver of these differences, with CD8 and CD4 T cells and NK cell variances being more subtle [10]. B cell percentages in C3H/HeJ mice (35.1% – 42.9%) and C57BL/6J (68.1%-82.0%) were at opposite ends of the spectrum, leading us to question potential mechanisms behind these differences, and the underlying genetics at play.
The potential that mitochondrial genetics contribute to immune profiles is supported by observations that metabolism is closely linked to immune differentiation and polarization [11]. T cell activation is largely dependent on the relative ratio of glycolysis and oxidative phosphorylation [12–14]. For example, upon antigen recognition by the T cell receptor, T cells undergo a metabolic switch from oxidative phosphorylation to glycolysis to facilitate the requirements necessary for expansion [15–17]. Furthermore, the ability of CD8+ memory T cells to engage fatty acid oxidation and maintain spare respiratory capacity is critical for their formation, which has important implications for tumor recurrence [18–20]. Likewise, T cell populations infiltrating infections vary by mouse strain [21, 22].
Taken together, accumulating data suggest that complex genetic mechanisms regulate baseline immune system composition as well as function. Changes in immune profiles and corresponding differential immune responses could be mediated, in part, by mt-SNP which, in turn, contribute to the phenotypic differences observed in MNX mice. This hypothesis was then translated into three corollaries: (1) mt-SNP regulate broad immune cell populations (e.g., B cells, T cells, macrophages, etc.); (2) mt-SNP alter localization of immune cell populations (e.g., spleen vs. peritoneum); and, (3) mt-SNP alter polarization/differentiation of immune (sub)populations.
The MNX mouse model affords a unique opportunity to test some of these hypotheses, isolating contibutions of the mtDNA as an experimental variable. Immune cells were harvested from two rich immune environments -- spleen and peritoneum – which differ in prevalence of innate and adaptive immune components.
2. Materials and Methods
2.1. Mitochondrial Nuclear Exchange (MNX)
Mitochondrial Nuclear Exchange (MNX) stable mouse lines were created as previously reported [4, 23]. Wild-type C57BL/6J and C3H/HeN mice were purchased from Jackson laboratories and Envigo laboratories, respectively, and WT breeding colonies were maintained. MNX counterparts were generated and breeding colonies maintained. All animal studies were approved by the Institutional Animal Care and Use Committee at the university of Kansas Medical Center. Tissues were collected from all of the strains age 1-2 months. In addition, we attempted to diminish any role for additional factors to further focus on the role of mtDNA. The first being sex. Male and female mice were collected, as sex hormones have previously been demonstrated to impact immune cell development with estrogen supporting immune enhancement, and testosterone appearing to play more of a suppressive role [24, 25]. Circadian rhythm [26–28] microbiome [11], and age [29] can greatly affect the immune system. All mice were age-matched (1-2 months), euthanized at the beginning of the light cycle (always within the first three hours after the lights have been turned on (12 h on-off cycle) (ZTO-3; 7-10 am)) to avoid changes in immune localization based on circadian rhythm, and collected and analyzed from multiple litters and multiple parents to normalize potential microbiome fluctuations as well as to mitigate inter-individual heterogeneity. Splenocytes were also collected from 6 month old mice to determine age dependent changes.
2.2. Isolation of Splenocytes
Splenocytes were harvested from wild-type and MNX mice and placed in phosphate buffered saline (Gibco) containing 3% FBS (Atlanta Biologicals). Spleens were ruptured; red blood cells lysed for 3 minutes; filtered through a 70 μm filter and then counted.
2.3. Isolation of Peritoneal exudate
For isolation of peritoneal exudate, 5 mL of ice cold PBS containing 3% FBS solution was injected into the peritoneum of a euthanized mouse. The peritoneum was then gently massaged to release cells into the buffer. Cells were then extracted using a polyethylene transfer pipet. Cells were passed through a 70 μm filter and counted.
2.4. Flow cytometry
Cells were collected for flow cytometric analysis (Spleen = 2 x 106; Peritoneum = 1 x 106 ). Cells were stained with a Live/Dead stain to exclude dead cells and Fc receptors blocked with TruStain FcX (anti-mouse CD16/32) (Biolegend). Cells were then stained with antibodies listed in Table S1, fixed with 1% paraformaldehyde overnight, and analyzed using the BD LSR II (Becton Dickinson). Data was analyzed using FlowJo (FlowJo LLC).
Antibody concentrations were titrated to reach optimal signal to noise ratio. Antibodies were then used to delineate populations from within the spleen or the peritoneum. Panels used to delineate cell populations within the spleen or peritoneum are outlined in Table S1.
2.5. Statistical analysis
Experiments included at least six mice per group in each of three independent replicates. Both male and female mice were used. Data were analyzed using SigmaPlot (Systat Software Inc.). Results were averaged +/− SEM and p-values were generated using one-way analysis of variance (ANOVA) on ranks followed by multiple comparisons analyses to determine significance between MNX and wild-type (WT) groups. A p-value < 0.05 was considered significant.
3. Results and Discussion
3.1. The nuclear genome regulates broad immune cell subclasses across mouse strain backgrounds
The immune system plays a large role in protection from diseases as well as being carefully balanced to prevent autoimmunity. Since humans have varying responses and susceptibilities to diseases [30], it is important to understand all of the contributing factors associated with differential pathogenesis. Genetic variation plays an important role in immune cell population percentages and functional activation [10, 31]}. Importantly, these differences in disease susceptibilities have also recently been associated with quantitative trait loci across individuals, which further correlates with differences in gene expression [32]. To test whether mitochondrial genetics may impact immune cell localization, function or polarization, spleens and peritoneal fluid were collected from wild-type (CC and HH) and MNX mice (CH and HC). Tissues were collected from 1-2 month old mice to identify immune population differences present in mice corresponding to the age when experimental metastasis assays were performed (Brinker et al. under review). Data is represented by white boxes (splenocytes) and grey boxes (peritoneum) to facilitate reader comparisons between figures. Using gating strategies focused on identifying primarily adaptive immunity (lymphoid) within the spleen and innate immunity (myeloid) within the peritoneum (Fig. 1A), cell numbers obtained from spleens were similar across all strains (Fig. 1B). However, a slight increase was observed in the number of cells obtained from the peritoneum with C3H nuclear DNA (HC and HH) (Fig. 1B). From these studies we identified nuclear- as well as mitochondria-driven effects (Table 2). As expected, some populations including CD4+, and CD8+ T cells remained unchanged between wild-type and MNX backgrounds (Table 2, Fig. 2 & S1), while others (see below) were different.
Figure 1. Splenocyte and peritoneal cell counts were not significantly different in MNX mice.
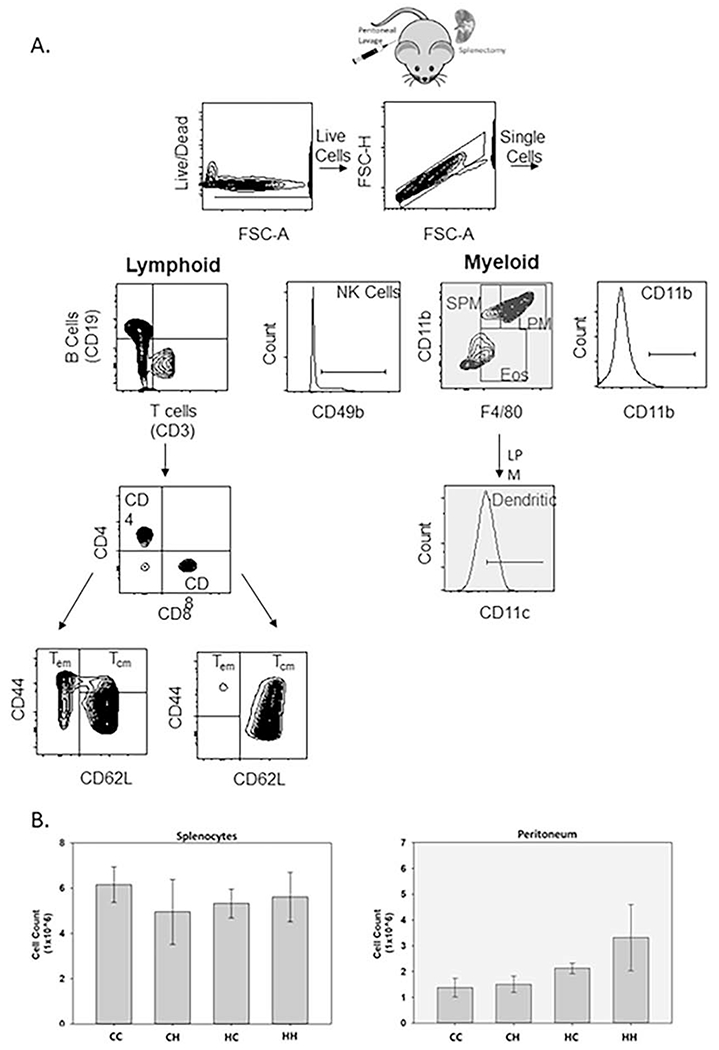
A. Spleen and peritoneal gating strategies. Dead cells were excluded using either ghost red (peritoneum) or zombie violet (spleen) viability dye in conjunction with forward scatter-area (FSC-A) to also exclude debris. Live, single cells (FSC-A/FSC-height (FSC-H)) were then gated on granulocyte and lymphocyte populations based on size and granularity (FSC-A/side scatter-area (SSC-A)). Lymphoid and myeloid populations were next analyzed for selected populations using the indicated differentiation markers. B cells (CD19+/CD3− ), T cells (CD3+/CD19−), Helper T cells (CD3+,CD4+,CD8−), Cytotoxic T cells (CD3+,CD4−,CD8+). Teff/em = T effector/effector memory cell (CD44+/CD62L−) and Tcm = T central memory cell (CD44+/CD62L+). SPM = Small Peritoneal Macrophages (CD11b+/F4/80Low), LPM = Large Peritoneal Macrophages (CD11b+/F4/80High), Eos = Eosinophils/Mast Cells (CD11bLow/F4/80Low). Throughout this manuscript myeloid populations will be signified by the grayed boxes and lymphoid populations signified by the white boxes. B. Cells were counted after red blood cell lysis and isolation into single cell suspensions. Average cell counts based on two independent experiments. Spleen (n=12) and peritoneum (n=10-13). Cells (2x106) were stained for splenocyte flow analysis, and 1x106cells were stained for peritoneum flow analysis.
Table 2:
IImmune populations change based on nuclear and mitochondrial DNA. Immune populations are organized based on regulation patterns observed amongst WT and MNX strains.
| No Difference | nDNA Mediated | mtDNA Mediated | |||
|---|---|---|---|---|---|
| Spleen | Peritoneum | Spleen | Peritoneum | Spleen | Peritoneum |
| CD3+ (T cells) | CD19+ (B cells) | CD19+ (B cells) | CD11bLow, F480Low (Eosinophils) | CD3+; CD8+; CD44+; CD62L− (CD8 Effector Memory/Effector T cells) | CD11c+ Macrophages |
| CD3+; CD8+ (Cytotoxic T cells) | CD11b+, F480low (Small Peritoneal Macrophages) | CD3+; CD4+; CD44+; CD62L− (CD4 Effector Memory) | CD3+ (T cells) | CD11b+ | |
| CD3+; CD4+ (Helper T cells) | CD11b+, F480High (Large Peritoneal Macrophages) | CD3+; CD4+; CD44+; CD62L− (CD4 Central Memory) | |||
| CD3+; CD8+; CD44+; CD62L (CD8 Central Memory) | |||||
| CD49b+ (NK Cells) | |||||
Figure 2. Nuclear haplotypes alter resident splenic and peritoneal lymphoid populations.
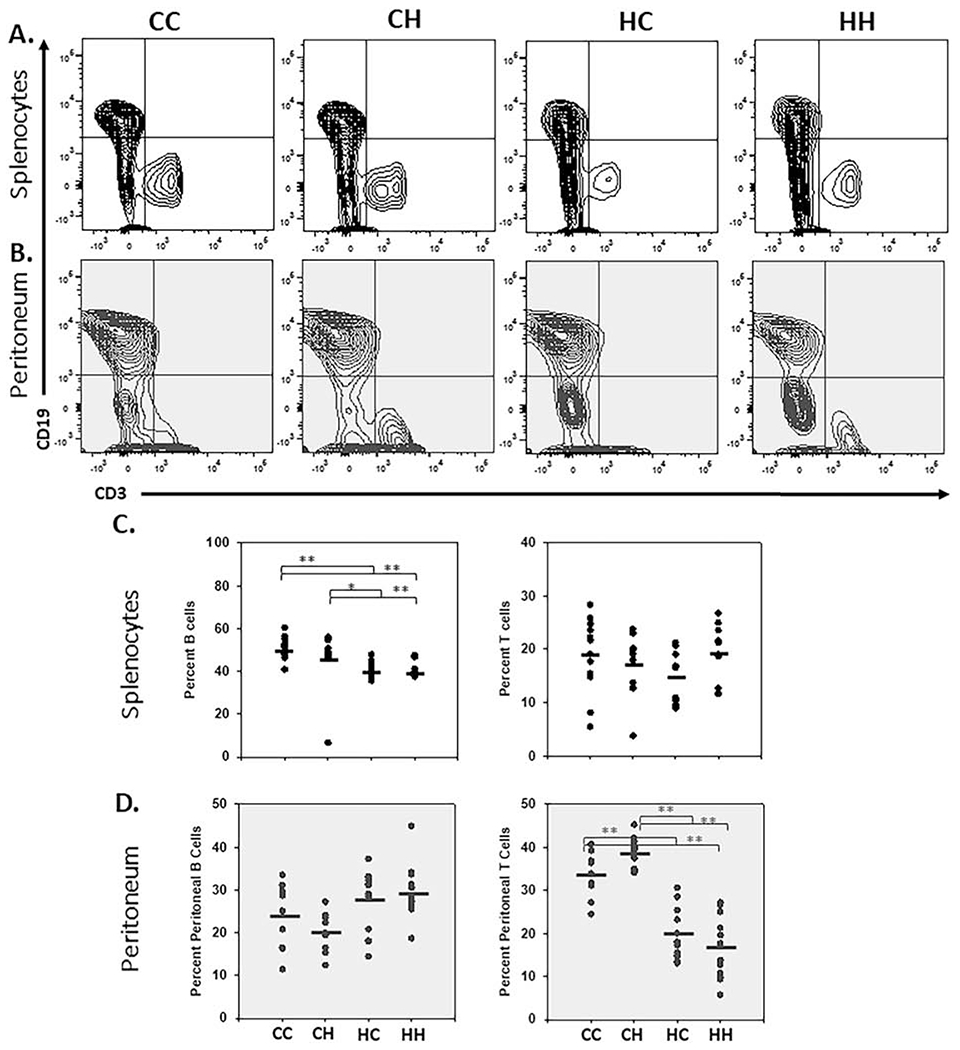
A. Cells isolated from the spleen and peritoneum (of 1-2-month-old male and female mice) were analyzed by flow cytometry. Cells gated on the lymphoid population were characterized for expression of CD19 and CD3. CD19 positive populations were gated to identify B cells and CD3 positive populations were gated to identify T cells and representative plots are depicted B. Vertical point scatter plots for T- (CD3+) and B-cell (CD19+) populations in MNX and WT mice. Horizontal lines represent the mean (spleen; n= 12, and peritoneum n=10-13). One-way ANOVA and multiple comparisons T-test (Dunn’s post-test); *, P < 0.05, **, P < 0.005)). Similar data collected at 6-months are shown in Figure S3 and S5.
To analyze the populations in more detail, we first identified the most prominent populations in the spleen and peritoneum [33, 34]. B cells within the spleen are 40-50% of splenocytes. We observed nuclear regulation with B cells accounting for 51% and 47% of CC and CH splenic lymphocytes, but only 40% and 41% for HH and HC splenocytes (p < 0.05) (Fig. 2A & 2C). In addition CD11b+/F4/80+ large peritoneal macrophages are the predominant population within the peritoneum along with CD11blow/F4/80low eosinophils, which also exhibit strong nuclear regulation (Table 2). These results suggest that major immune population differences observed across mouse strain backgrounds are likely driven by the nuclear genome. These findings are expected as many reports have highlighted a role for regulation of immune cells based on the nuclear genome [35]. In addition, based on the smaller size of the mitochondrial genome we predict that the mt-SNP act as quantitative trait loci (QTL). Quantitative traits are phenotypes that are impinged upon by multiple genes. Immune functionality is clearly polygenic. Therefore, we next hypothesized that mt-SNP may act to fine tune polarization differences amongst broader immune cell populations.
3.2. mtDNA alters basal levels of Teffector/Teffector memory populations
Mitochondrial metabolism is an important regulator of immune cell polarization. T cells require a shift between oxidative phosphorylation and glycolysis for effector and memory cell polarization [12–14]. Also, mutations that disrupt the electron transport chain result in reduced T memory cell polarization [9]. These findings led us to hypothesize that more subtle changes imparted by mt-SNP would have a greater impact on polarization potential especially in regards to populations that exhibit dynamic fluidity between polarization states [36].
CD8+ Teffector/Teffector memory (Teff/em) cells (CD44+ / CD62L−) make up a low percentage of CD8 T cells due to a lack of activating stimuli. Despite this, HH mice still exhibit a higher percentage of CD44+/CD62L− CD8+ T cells in comparison to the other backgrounds [CC 2%, CH 2%, HC 2%, HH 4%] (P < 0.001; HH vs CC) (Fig. 3A & 3C). No mitochondrial or nuclear regulation was observed for CD8+ T central memory (Tcm) cells (CD44+ / CD62L+) [CC 11%, CH 13%, HC 6%, HH 11%] (Fig. 3A & 3C).
Figure 3. Mitochondrial haplotypes alter splenic effector/effector memory CD4 and CD8 T cell populations.
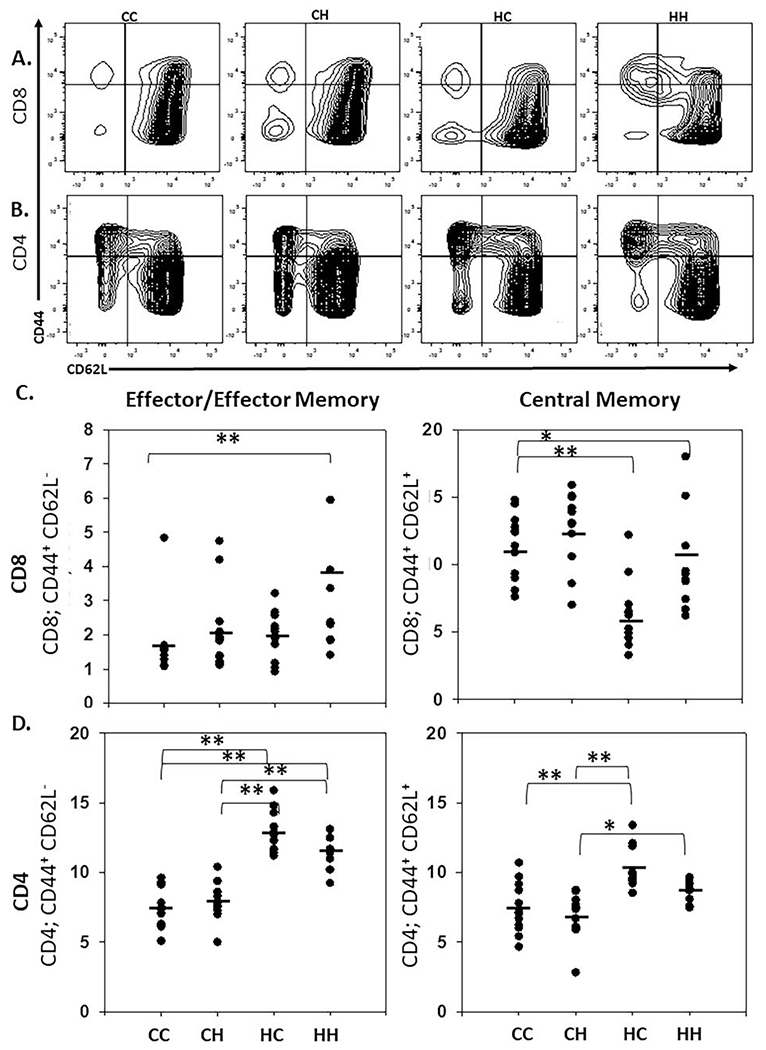
A/B. Cells isolated from the spleen (of 1-2-month-old male and female mice) were analyzed by flow cytometry. Central memory and effector/effector memory cells were then sub-gated on CD8 (A) and CD4 (B) positivity and analyzed for CD44 and CD62L. CD44+, CD62L− populations represent T effector/T effector memory cells and CD44+, CD62L+ represent T central memory cells. C/D. Vertical point scatter plots for CD8+ (C) and CD4+ (D) effector/effector-memory and central memory T cell populations in MNX and WT mice. Horizontal lines represent the mean (spleen; n= 12). One-way ANOVA and multiple comparisons T-test (Tukey and Dunn’s post-test); *, P < 0.05, **, P < 0.005)). Similar data collected at 6-months are shown in Figure S4 and S6.
CD4+ Teff/em cell populations do not appear to exhibit mtDNA-mediated differences, but rather exhibited stronger nuclear regulation [CC 7%, CH 8%, HC 13%, HH 12%] (Fig. 3B & 3D). CD4+ Tcm cells (CD44+ / CD62L+) also exhibited primarily nuclear regulation with higher percentages observed with the C3H nuclear background [CC 7%, CH 7%, HC 10%, HH 9%] (P < 0.001; CC/CH vs HC; P < 0.05; CH vs HH) (Fig. 3B & 3D).
As Tcm and Teff/em populations change and develop over time, we next analyzed splenocyte immune populations at 6 months of age. Analysis of CD19+ B cells and CD8+ T cells exhibited nuclear regulation, whereas no significant differences were observed for total or CD4+ T cells (Fig.S2). We next examined CD4 and CD8 Teff/em and Tcm differentiation. Whereas no significant difference was observed for CD4 T cell subsets, significant nuclear regulation was observed for CD8 Tcm cells ([CC 16%, CH 19%, HC 9%, HH 10%] (P < 0.001; CH vs HC, CC/CH vs HH; P < 0.05; CC vs CH) (Fig. S3), which is consistent with what we observed in 1-2 month old mice (Fig. 3C). In addition, CD8 Teff/em cells exhibited a conserved trend towards mitochondrial regulation with CH mice exhibiting higher percentages of Teff/em populations compared to HH mice ([CC 3.5%, CH 4%, HC 5%, HH 5%] (Fig. S3); however, no statistically significant difference was observed.
To further verify that these differences were not due to sex-mediated differences, we analyzed the naive immune populations (1-2 months) for both males and females. Whereas subtle variations were observed, genomic haplotypes predominated over sex-mediated effects. Both nuclear as well as mitochondrial trends remained conserved in both male and female populations when analyzed individually (Fig S4 & S5).
As T cell development varies with age [37], all analyses reported here focus on combined mice aged 4-6 weeks since that corresponds to previous reports for changes in metastatic behavior with MNX mice (Brinker et al. under review). Additionally, both male and female mice were analyzed and no sexual-dimorphisms were observed. Importantly, our observed differences in Teff/em cell differentiation were not associated with increasing age or sex (Fig S6).
An important factor for T cell differentiation is the mitochondrial spare respiratory capacity, which has previously been demonstrated to impact Tcm differentiation by providing the necessary survival capacities to become a long lived Tcm cell [15]. To further support a role for mt-SNP in regulating these differentiation potentials, our lab has previously demonstrated that the spare respiratory capacity of PyMT cancer cells harboring C57BL/6J mtDNA is reduced in comparison to cancer cells harboring FVB/NJ or BALB/cJ mtDNA [1]. However, whereas full metabolomic analysis would be required on each individual cell type (and differentiation state) to further determine the role of mt-SNP in regulating T cell metabolism, it is intriguing to speculate that mt-SNP may impact T cell metabolism through subtle changes in oxidative phosphorylation capacities. Our data, therefore, provides preliminary evidence for a role in mtDNA regulating T cell polarization and/or population skewing. Furthermore, a role for mtDNA in altered polarization towards Tcm or Teff/em populations may provide a possible alternative explanation for varying rates of immunotherapy response.
3.3. mtDNA alters CD11b expression in splenocyte populations
We next analyzed myeloid populations within the spleen. CD11b is an integrin commonly associated with innate immune activation [38] and expressed by a large proportion of myeloid populations [39]. Analysis of CD11b expression identified a correlation between CD11b expressing cells and mtDNA [CC 10%, CH 13%, HC 19%, HH 23%] (Fig. 4a). Natural Killer (NK) cells are commonly associated with cancer cell elimination within experimental metastasis models [40–42]; so, we next analyzed splenocyte CD49b expression, but found no significant baseline differences based on strain variation (Fig. 4b). Having observed a trend towards an increase in mtDNA regulation of the CD11b+ populations within the spleen, we next focused on myeloid population differentiation and recruitment in the myeloid rich environment of the peritoneum.
Figure 4. mtDNA alters CD11b expression in splenocyte populations A/B.
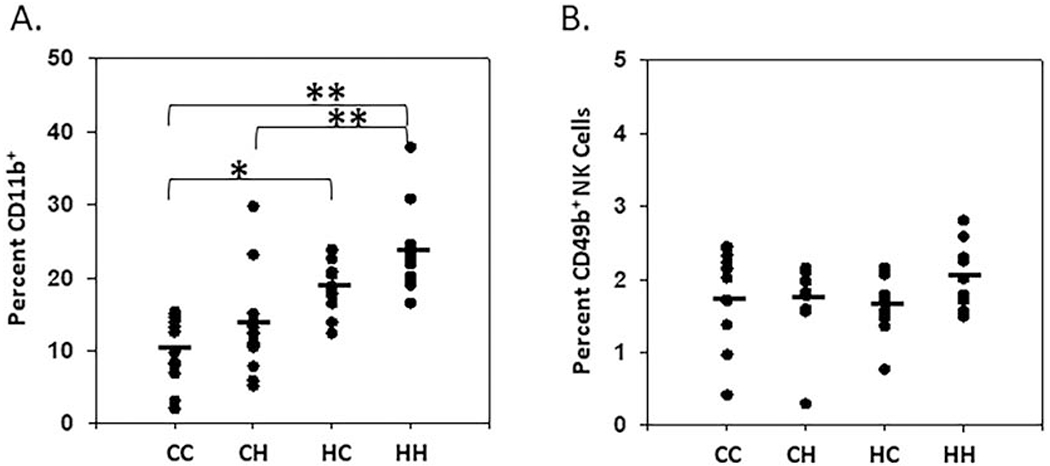
Cells isolated from the spleen (of 1-2-month-old male and female mice) were analyzed by flow cytometry. Splenocytes were analyzed for A. CD11b+ myeloid cells and B. CD49b+ Natural Killer cells (spleen; n= 12). One-way ANOVA and multiple comparisons T-test (Dunn’s post-test); *, P < 0.05, **, P < 0.005)).
3.4. mtDNA alters CD11c expression in large peritoneal macrophage populations
Similar to T cell activation, activation of other immune cell types requires a switch from a resting state of oxidative phosphorylation to an active state that is more reliant on glycolysis [43]. Macrophages exhibit a high level of plasticity [36]. Previous reports within the peritoneum have highlighted a capacity for resident macrophage differentiation into dendritic cells based on increased levels of CD11c [44]. So-called large (LPM) and small peritoneal macrophages (SPM) are the predominant macrophage populations in the peritoneum. Both traffic from different locations -- small from bone marrow and large from fetal macrophages [45]. LPM and SPM were analyzed based upon the gating protocol established by Ghosn et al., where macrophage populations were identified based on F4/80 and CD11b positivity [46]. At baseline, the majority of peritoneal macrophages are LPM based upon high levels of F4/80 (CC, 46%; CH, 46%; HC, 43%; HH, 48%) (Fig 5). There was also strong nuclear regulation of CD11blow and F4/80low cells [CC 11%; CH 15%; HC 2%; HH 3% (p = <0.001)], which have previously been demonstrated to be predominantly eosinophils [22, 47, 48] (Fig. 5A & 5C).
Figure 5. Mitochondrial haplotypes alter resident peritoneal immune cell populations.
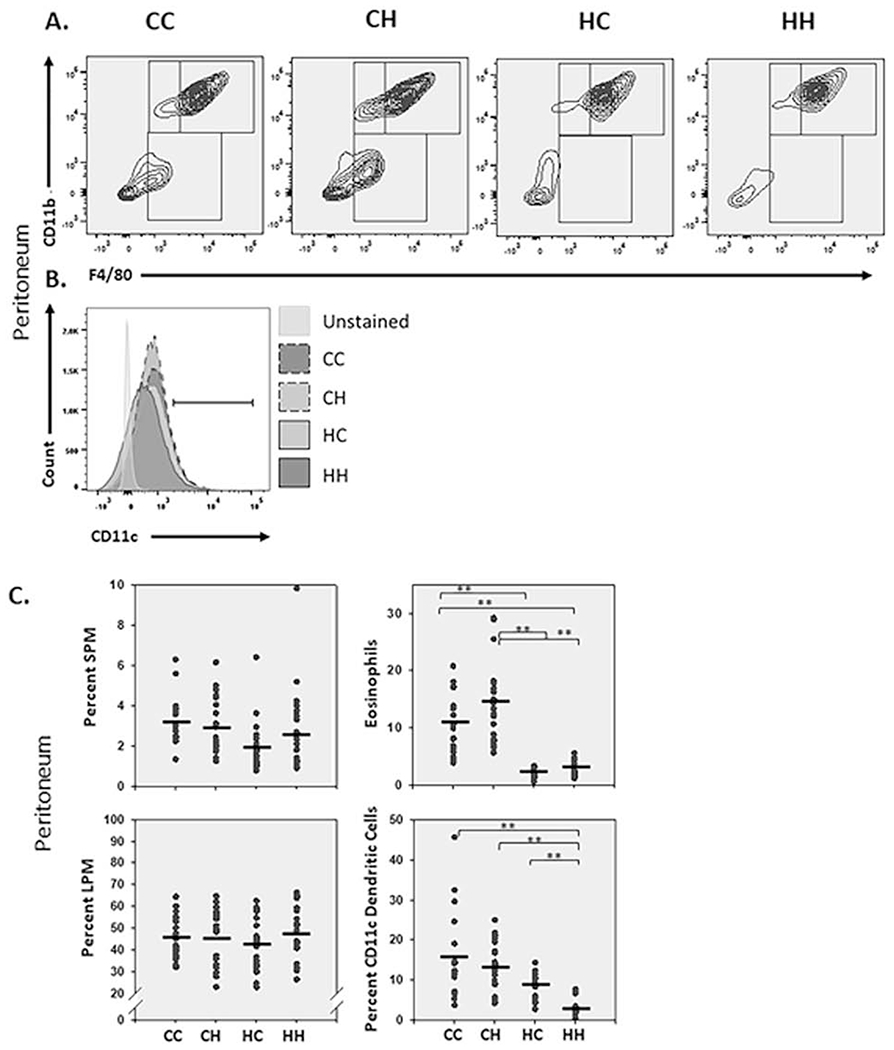
A. Cells isolated from the peritoneal cavity (in 1-2-month-old male and female mice) were analyzed by flow cytometry. Cells of the myeloid lineage were gated on CD11b and F4/80. CD11bhigh and F4/80+ cells represent large peritoneal macrophages (LPM), CD11bhigh and F4/80low represent small peritoneal macrophages (SPM), CD11blow and F4/80low represent eosinophils/mast cells. B. LPM were further analyzed for CDllc expression. C. Vertical point scatter plots for LPM, SPM and eosinophil/mast cell populations in MNX and WT mice. Black horizontal lines represent the mean (peritoneum n=18-23). One-way ANOVA and multiple comparisons T-test ((Dunn’s post-test); **, P < 0.005).
Previous work from Ados et al. demonstrated that, at baseline, CD11c+ is predominantly expressed in LPM [45], which led us to focus on CD11c dendritic cell skewing within this population. Analysis for positivity in LPM identified significant differences between wild-type HH and the three other strains CC, CH, and HC [CC 16%; CH 13%; HC 9%; HH 3% (p = <0.001)] (Fig. 5B & 5C). These data highlight a role for C57BL/6J mt-SNP in altering expression of CD11c and potential skewing of macrophage subpopulations.
Tissue resident macrophages exhibit unique epigenetic regulation based on the microenvironment [49]. Our previous work demonstrated that mt-SNP selectively alter nuclear transcription [5], suggesting that responses to microenvironmental signals may vary between strains. Consistent with this, BALB/c mice respond to infection with Leishmania major by producing a Th2 response; whereas, most other strains respond via Th1 [8]. These data further highlight how subtle genetic changes can alter how immune systems respond and/or develop.
While the mechanism(s) responsible for the differential recruitment and/or polarization are not yet fully elucidated, numerous possibilities exist. Based on given evidence CD11c expression may be regulated by differential nuclear-mitochondrial signaling which, in turn, alters cytokine/chemokine production within the peritoneal microenvironment. Together the result is population skewing and polarization. Whereas less evidence exists to suggest that the differential expression of CD11c is affecting the complement system, it is nonetheless intriguing to speculate upon this potential role. The complement system represents an ancient form of innate immunity that has evolved from a single cell origin [50]. As mitochondria are thought to have evolved from a single-celled bacterium [51], it is possible that they have retained the capacity to signal and regulate the complement system leading to the differences in CD11c expression observed.
4. Conclusion
This report is significant because it establishes, for the first time to our knowledge, that polymoprhisms in mtDNA are among the QTL for immune profiles and/or function. Mitochondrial genomes combine with nuclear genomes to affect the composition of immune cells within each tissue. Moreover, they also influence how the immune cells become activated and/or polarized.
Four polymorphisms have been reported in C57BL/6J and C3H/HeN mtDNA [6]. Briefly, NDIII has an A to C transversion at nucleotide 9461, COXIII has a G to A transition at nucleotide 9348 resulting in a isoleucine to valine substitution, and two mutations are present in tRNAARG in which C57 mitochondria have an adenine deletion at position 9821, while C3H mitochondria have an additional thymine located at 9820. At present, a definitive explanation why these polymorphic mutations would differentially affect immune profiles/function is not self-evident. COXIII mtDNA SNP status has a significant impact on mouse heart mitochondria Complex IV respiration and COX enzyme activity [4]. Interestingly, Tarasenko et al. found that cytochrome C oxidase can serve as a checkpoint following T cell activation regulating cell fate decisions [9], suggesting that COXIII SNP may impact immune activation/differentiation. Attempts to extrapolate the murine findings in this report to human mtDNA using MITOMAP [52, 53] have not yet yielded perfect synteny, which precludes definitive cause-effect conclusions to be drawn. Additionally, since >90% of mitochondrial proteins are nucleus-encoded, one must also take into account how the electron transport proteins interact with other components.
Why are these findings relevant? First, mt-SNP, along with a handful of nDNA SNP can be used to distinguish races (clades or mouse strains). Changes in immune function could be part of the explanation why there are racial disparities with regard to disease susceptibility and/or progression, especially diseases with strong immune components. Second, responses to cancer immunotherapy is heterogeneous as are side effects. Perhaps analysis of mt-QTL may provide a stronger foothold into better prediction of (non)responses or undesirable side effects. Third, mt-QTL affecting immune profiling and function may impinge upon the ability of patients to accept/reject tissue and/or bone marrow transplants. While these possibilities are speculative at this time, the size of the mitochondrial genome makes sequencing and testing of the hypotheses tractable.
Supplementary Material
Figure S1. Mitochondrial haplotypes have minimal impact on splenic CD8 and CD4 positive T cells. A. Cells isolated from the spleen (of 1-2-month-old male and female mice) were analyzed by flow cytometry. CD4 and CD8 positive cells are represented as a percentage of CD3 positive lymphocytes. B. Vertical point scatter plots for CD4+ and CD8+ T cell populations in MNX and WT mice. Horizontal lines represent the mean (spleen; n= 12). Similar data collected at 6-months are shown in Figure S3 and S5.
Figure S2. Nuclear haplotype mediated differences are maintained in 6-month old WT and MNX splenic immune cell populations. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figures 3 and S2 (spleen; n= 10-15).
Figure S3. Mitochondrial haplotypes alter splenic effector/effector memory CD8 T cell populations of 6-month old WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figure 4 (spleen; n= 10-15). Horizontal lines represent the mean (spleen; n= 12). One-way ANOVA and multiple comparisons T-test (Dunn’s posttest); **, P < 0.005)).
Figure S4. Nuclear haplotype mediated differences are maintained in both male and female WT and MNX splenic immune cell populations. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figures 3 and S2 (spleen; n= 6).
Figure S5. Mitochondrial haplotypes alter splenic effector/effector memory CD8 T cell populations in both male and female WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figure 4 (spleen; n= 6).
Figure S6. Increasing age and sex are not correlated with increasing splenic effector/effector memory CD8 and CD4T cell populations in WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and individual mice are graphed in order of increasing age from 4-7 weeks. Male mice are depicted as black bars and female mice are depicted as grey bars.
Table 1:
wild-type (WT) and MNX models utilized for this study
| Mouse Strain | nDNA | mtDNA | Abbreviation |
|---|---|---|---|
| C57BL/6J-mtMNX(C57BL/6J) | C57BL/6J | C57BL/6J | CC |
| C57BL/6J-mtMNX(C3H/HeN) | C57BL/6J | C3H/HeN | CH |
| C3H/HeN-mtMNX(C3H/HeN) | C3H/HeN | C3H/HeN | HH |
| C3H/HeN-mtMNX(C57BL/6J) | C3H/HeN | C57BL/6J | HC |
Highlights.
mtDNA SNP represent quantitative trait loci for the immune system.
Major baseline strain differences in immune cell populations appear to be driven by nuclear encoded factors.
mtDNA regulates more selective changes in immune cell populations, with the primary mtDNA-mediated effects on immune cell populations driving differentiation/polarization.
CDllc+ expression in large peritoneal macrophages appears to exhibit strong regulation by mtDNA.
Acknowledgments and Support:
DOD BCRP BC171381 (TCB); Susan G. Komen for the Cure SAC110037; National Foundation for Cancer Research NIH CA168524 (DRW); and Kansas INBRE P20 GM103418 (TCB). We are also grateful for the superb technical assistance from KU Cancer Center (CA168524) Biostatistics, Flow Cytometry (also supported by GM103326) and Transgenic and Gene Targeting Shared Resources.
Abbreviations:
- CC
C57BL/6J-mtMNX(C57BL/6J)
- CH
C57BL/6J-mtMNX(C3H/HeN)
- HC
C3H/HeN-mtMNX(C57BL/6J)
- HC
C3H/HeN-mtMNX(C3H/HeN)
- FBS
Fetal Bovine Serum
- LPM
Large Peritoneal Macrophage
- mtDNA
Mitochondrial DNA
- MNX
Mitochondrial Nuclear Exchange
- NK
Natural Killer
- nDNA
Nuclear DNA
- ANOVA
One Way Analysis of Variance
- PBS
Phosphate Buffered Saline
- SNP
Polymorphisms
- QTL
Quantitative Trait Loci
- SPM
Small Peritoneal Macrophage
- Teff
T effector
- Tcm
T central memory
- Tem
T effector memory
Footnotes
Publisher's Disclaimer: This is a PDF file of an unedited manuscript that has been accepted for publication. As a service to our customers we are providing this early version of the manuscript. The manuscript will undergo copyediting, typesetting, and review of the resulting proof before it is published in its final form. Please note that during the production process errors may be discovered which could affect the content, and all legal disclaimers that apply to the journal pertain.
References
- [1].Feeley KP, Bray AW, Westbrook DG, Johnson LW, Kesterson RA, Ballinger SW, Welch DR, Mitochondrial Genetics Regulate Breast Cancer Tumorigenicity and Metastatic Potential, Cancer Res. 75(20) (2015) 4429–36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [2].Brinker AE, Vivian CJ, Koestler DC, Tsue TT, Jensen RA, Welch DR, Mitochondrial Haplotype Alters Mammary Cancer Tumorigenicity and Metastasis in an Oncogenic Driver-Dependent Manner, Cancer Res. 77(24) (2017) 6941–6949. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [3].Beadnell TC, Scheid AD, Vivian CJ, Welch DR, Roles of the mitochondrial genetics in cancer metastasis: not to be ignored any longer, Cancer Metastasis Rev. 37(4) (2018) 615–632. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [4].Fetterman JL, Zelickson BR, Johnson LW, Moellering DR, Westbrook DG, Pompilius M, Sammy MJ, Johnson M, Dunham-Snary KJ, Cao X, Bradley WE, Zhang J, Wei CC, Chacko B, Schurr TG, Kesterson RA, Dell’italia LJ, Darley-Usmar VM, Welch DR, Ballinger SW, Mitochondrial genetic background modulates bioenergetics and susceptibility to acute cardiac volume overload, Biochem. J 455(2) (2013) 157–67. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [5].Vivian CJ, Brinker AE, Graw S, Koestler DC, Legendre C, Gooden GC, Salhia B, Welch DR, Mitochondrial Genomic Backgrounds Affect Nuclear DNA Methylation and Gene Expression, Cancer Res. 77(22) (2017) 6202–6214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [6].Scheid AD, Beadnell TC, Welch DR, The second genome: Effects of the mitochondrial genome on cancer progression, Adv. Cancer Res 142 (2019) In press. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [7].Lifsted T, Le Voyer T, Williams M, Muller W, Klein-Szanto A, Buetow KH, Hunter KW, Identification of inbred mouse strains harboring genetic modifiers of mammary tumor age of onset and metastatic progression, Int. J. Cancer 77(4) (1998) 640–4. [DOI] [PubMed] [Google Scholar]
- [8].Watanabe H, Numata K, Ito T, Takagi K, Matsukawa A, Innate immune response in Th1- and Th2-dominant mouse strains, Shock 22(5) (2004) 460–6. [DOI] [PubMed] [Google Scholar]
- [9].Tarasenko TN, Pacheco SE, Koenig MK, Gomez-Rodriguez J, Kapnick SM, Diaz F, Zerfas PM, Barca E, Sudderth J, DeBerardinis RJ, Covian R, Balaban RS, DiMauro S, McGuire PJ, Cytochrome c Oxidase Activity Is a Metabolic Checkpoint that Regulates Cell Fate Decisions During T Cell Activation and Differentiation, Cell Metab. 25(6) (2017) 1254–1268 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [10].Petkova SB, Yuan R, Tsaih SW, Schott W, Roopenian DC, Paigen B, Genetic influence on immune phenotype revealed strain-specific variations in peripheral blood lineages, Physiol. Genomics 34(3) (2008) 304–14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [11].Belkaid Y, Hand TW, Role of the microbiota in immunity and inflammation, Cell 157(1) (2014) 121–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [12].van der Windt GJ, O’Sullivan D, Everts B, Huang SC, Buck MD, Curtis JD, Chang CH, Smith AM, Ai T, Faubert B, Jones RG, Pearce EJ, Pearce EL, CD8 memory T cells have a bioenergetic advantage that underlies their rapid recall ability, Proc Natl Acad Sci U S A 110(35) (2013) 14336–41. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [13].MacIver NJ, Michalek RD, Rathmell JC, Metabolic regulation of T lymphocytes, Annu. Rev. Immunol 31 (2013) 259–83. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [14].Pearce EL, Pearce EJ, Metabolic pathways in immune cell activation and quiescence, Immunity 38(4) (2013) 633–43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [15].van der Windt GJ, Everts B, Chang CH, Curtis JD, Freitas TC, Amiel E, Pearce EJ, Pearce EL, Mitochondrial respiratory capacity is a critical regulator of CD8+ T cell memory development, Immunity 36(1) (2012) 68–78. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [16].Pearce EL, Metabolism in T cell activation and differentiation, Curr. Opin. Immunol 22(3) (2010) 314–20. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [17].Pearce EL, Poffenberger MC, Chang CH, Jones RG, Fueling immunity: insights into metabolism and lymphocyte function, Science 342(6155) (2013) 1242454. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [18].Buck MD, O’Sullivan D, Geltink R.I. Klein, Curtis JD, Chang CH, Sanin DE, Qiu J, Kretz O, Braas D, van der Windt GJ, Chen Q, Huang SC, O’Neill CM, Edelson BT, Pearce EJ, Sesaki H, Huber TB, Rambold AS, Pearce EL, Mitochondrial Dynamics Controls T Cell Fate through Metabolic Programming, Cell 166(1) (2016) 63–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [19].O’Sullivan D, van der Windt GJ, Huang SC, Curtis JD, Chang CH, Buck MD, Qiu J, Smith AM, Lam WY, DiPlato LM, Hsu FF, Birnbaum MJ, Pearce EJ, Pearce EL, Memory CD8(+) T cells use cell-intrinsic lipolysis to support the metabolic programming necessary for development, Immunity 41(1) (2014)75–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [20].Sugiura A, Rathmell JC, Metabolic Barriers to T Cell Function in Tumors, J. Immunol 200(2) (2018) 400–407. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [21].Scott P, IFN-gamma modulates the early development of Th1 and Th2 responses in a murine model of cutaneous leishmaniasis, J. Immunol 147(9) (1991) 3149–55. [PubMed] [Google Scholar]
- [22].Shimada T, Cheng L, Shi HB, Hayashi A, Motonaga C, Tang J, Enomoto K, Enomoto T, Effect of lysed Enterococcus faecalis FK-23 on allergen-induced immune responses and intestinal microflora in antibiotic-treated weaning mice, J. Investig. Allergol. Clin. Immunol 17(2) (2007) 70–6. [PubMed] [Google Scholar]
- [23].Kesterson RA, Johnson LW, Lambert LJ, Vivian JL, Welch DR, Ballinger SW, Generation of Mitochondrial-nuclear eXchange Mice via Pronuclear Transfer, Bio Protocol 6(20) (2016). [DOI] [PMC free article] [PubMed] [Google Scholar]
- [24].Cutolo M, Sulli A, Capellino S, Villaggio B, Montagna P, Seriolo B, Straub RH, Sex hormones influence on the immune system: basic and clinical aspects in autoimmunity, Lupus 13(9) (2004) 635–638. [DOI] [PubMed] [Google Scholar]
- [25].Furman D, Hejblum BP, Simon N, Jojic V, Dekker CL, Thiebaut R, Tibshirani RJ, Davis MM, Systems analysis of sex differences reveals an immunosuppressive role for testosterone in the response to influenza vaccination, Proc Natl Acad Sci U S A 111(2) (2014) 869–74. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [26].Scheiermann C, Kunisaki Y, Frenette PS, Circadian control of the immune system, Nat. Rev. Immunol 13(3) (2013) 190–8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [27].Scheiermann C, Gibbs J, Ince L, Loudon A, Clocking in to immunity, Nat. Rev. Immunol 18(7) (2018) 423–437. [DOI] [PubMed] [Google Scholar]
- [28].He W, Holtkamp S, Hergenhan SM, Kraus K, de Juan A, Weber J, Bradfield P, Grenier JMP, Pelletier J, Druzd D, Chen CS, Ince LM, Bierschenk S, Pick R, Sperandio M, Aurrand-Lions M, Scheiermann C, Circadian Expression of Migratory Factors Establishes Lineage-Specific Signatures that Guide the Homing of Leukocyte Subsets to Tissues, Immunity 49(6) (2018) 1175–1190 e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [29].Vasto S, Malavolta M, Pawelec G, Age and immunity, Immun. Ageing 3 (2006) 2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [30].Brodin P, Davis MM, Human immune system variation, Nat. Rev. Immunol 17(1) (2017) 21–29. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [31].De Jager PL, Hacohen N, Mathis D, Regev A, Stranger BE, Benoist C, ImmVar project: Insights and design considerations for future studies of “healthy” immune variation, Semin. Immunol 27(1) (2015) 51–7. [DOI] [PubMed] [Google Scholar]
- [32].Kim-Hellmuth S, Bechheim M, Putz B, Mohammadi P, Nedelec Y, Giangreco N, Becker J, Kaiser V, Fricker N, Beier E, Boor P, Castel SE, Nothen MM, Barreiro LB, Pickrell JK, Muller-Myhsok B, Lappalainen T, Schumacher J, Hornung V, Genetic regulatory effects modified by immune activation contribute to autoimmune disease associations, Nat Commun 8(1) (2017) 266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [33].Festing MF, Legg R, Eydmann T, Brammall A, Mouse strain differences in resident peritoneal cells: a flow cytometric analysis, Lab. Anim 24(1) (1990) 53–62. [DOI] [PubMed] [Google Scholar]
- [34].Chen Z, Quan L, Huang A, Zhao Q, Yuan Y, Yuan X, Shen Q, Shang J, Ben Y, Qin FX, Wu A, seq-ImmuCC: Cell-Centric View of Tissue Transcriptome Measuring Cellular Compositions of Immune Microenvironment From Mouse RNA-Seq Data, Front. Immunol 9 (2018) 1286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [35].Sellers RS, Clifford CB, Treuting PM, Brayton C, Immunological variation between inbred laboratory mouse strains: points to consider in phenotyping genetically immunomodified mice, Vet. Pathol 49(1) (2012) 32–43. [DOI] [PubMed] [Google Scholar]
- [36].Shapouri-Moghaddam A, Mohammadian S, Vazini H, Taghadosi M, Esmaeili SA, Mardani F, Seifi B, Mohammadi A, Afshari JT, Sahebkar A, Macrophage plasticity, polarization, and function in health and disease, J. Cell. Physiol 233(9) (2018) 6425–6440. [DOI] [PubMed] [Google Scholar]
- [37].Rosen JL, Tran HT, Lackey A, Viselli SM, Sex-related immune changes in young mice, Immunol. Invest 28(4) (1999) 247–56. [DOI] [PubMed] [Google Scholar]
- [38].Schmid MC, Khan SQ, Kaneda MM, Pathria P, Shepard R, Louis TL, Anand S, Woo G, Leem C, Faridi MH, Geraghty T, Rajagopalan A, Gupta S, Ahmed M, Vazquez-Padron RI, Cheresh DA, Gupta V, Varner JA, Integrin CD11b activation drives anti-tumor innate immunity, Nat Commun 9(1) (2018) 5379. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [39].Hey YY, Tan JK, O’Neill HC, Redefining Myeloid Cell Subsets in Murine Spleen, Front. Immunol 6 (2015) 652. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [40].Nieswandt B, Hafner M, Echtenacher B, Mannel DN, Lysis of tumor cells by natural killer cells in mice is impeded by platelets, Cancer Res. 59(6) (1999) 1295–1300. [PubMed] [Google Scholar]
- [41].Hanna N, The role of natural killer cells in the control of tumor growth and metastasis, Biochim. Biophys. Acta 780(3) (1985) 213–26. [DOI] [PubMed] [Google Scholar]
- [42].Hanna N, Fidler IJ, Role of natural killer cells in the destruction of circulating tumor emboli, J. Natl. Cancer Inst 65(4) (1980) 801–9. [DOI] [PubMed] [Google Scholar]
- [43].O’Neill LA, Pearce EJ, Immunometabolism governs dendritic cell and macrophage function, J. Exp. Med 213(1) (2016) 15–23. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [44].Kojima N, Kato C, Igarashi M, Ishii M, Development of peritoneal macrophage along a dendritic cell lineage in response to uptake of oligomannose-coated liposomes, Cell. Immunol 271(2) (2011) 335–41. [DOI] [PubMed] [Google Scholar]
- [45].Cassado Ados A, D’Imperio Lima MR, Bortoluci KR, Revisiting mouse peritoneal macrophages: heterogeneity, development, and function, Front. Immunol 6 (2015) 225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [46].Ghosn EE, Cassado AA, Govoni GR, Fukuhara T, Yang Y, Monack DM, Bortoluci KR, Almeida SR, Herzenberg LA, Herzenberg LA, Two physically, functionally, and developmentally distinct peritoneal macrophage subsets, Proc Natl Acad Sci U S A 107(6) (2010) 2568–73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [47].Lu Y, Sjostrand M, Malmhall C, Radinger M, Jeurink P, Lotvall J, Bossios A, New production of eosinophils and the corresponding TH1/TH2 balance in the lungs after allergen exposure in BALB/c and C57BL/6 mice, Scand. J. Immunol 71(3) (2010) 176–85. [DOI] [PubMed] [Google Scholar]
- [48].Misharin AV, Saber R, Perlman H, Eosinophil contamination of thioglycollate-elicited peritoneal macrophage cultures skews the functional readouts of in vitro assays, J. Leukoc. Biol 92(2) (2012) 325–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [49].Lavin Y, Winter D, Blecher-Gonen R, David E, Keren-Shaul H, Merad M, Jung S, Amit I, Tissue-resident macrophage enhancer landscapes are shaped by the local microenvironment, Cell 159(6) (2014) 1312–26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [50].Elvington M, Liszewski MK, Atkinson JP, Evolution of the complement system: from defense of the single cell to guardian of the intravascular space, Immunol. Rev 274(1) (2016) 9–15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [51].Gray MW, Mitochondrial evolution, Cold Spring Harb. Perspect. Biol 4(9) (2012) a011403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [52].Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC, mtDNA Variation and Analysis Using Mitomap and Mitomaster, Curr Protoc Bioinformatics 44 (2013) 1 23 1-26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- [53].Brandon MC, Lott MT, Nguyen KC, Spolim S, Navathe SB, Baldi P, Wallace DC, MITOMAP: a human mitochondrial genome database--2004 update, Nucleic Acids Res. 33(Database issue) (2005) D611–3. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Figure S1. Mitochondrial haplotypes have minimal impact on splenic CD8 and CD4 positive T cells. A. Cells isolated from the spleen (of 1-2-month-old male and female mice) were analyzed by flow cytometry. CD4 and CD8 positive cells are represented as a percentage of CD3 positive lymphocytes. B. Vertical point scatter plots for CD4+ and CD8+ T cell populations in MNX and WT mice. Horizontal lines represent the mean (spleen; n= 12). Similar data collected at 6-months are shown in Figure S3 and S5.
Figure S2. Nuclear haplotype mediated differences are maintained in 6-month old WT and MNX splenic immune cell populations. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figures 3 and S2 (spleen; n= 10-15).
Figure S3. Mitochondrial haplotypes alter splenic effector/effector memory CD8 T cell populations of 6-month old WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figure 4 (spleen; n= 10-15). Horizontal lines represent the mean (spleen; n= 12). One-way ANOVA and multiple comparisons T-test (Dunn’s posttest); **, P < 0.005)).
Figure S4. Nuclear haplotype mediated differences are maintained in both male and female WT and MNX splenic immune cell populations. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figures 3 and S2 (spleen; n= 6).
Figure S5. Mitochondrial haplotypes alter splenic effector/effector memory CD8 T cell populations in both male and female WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and vertical point scatter plots show similar trends as depicted in Figure 4 (spleen; n= 6).
Figure S6. Increasing age and sex are not correlated with increasing splenic effector/effector memory CD8 and CD4T cell populations in WT and MNX mice. Cells isolated from spleens were analyzed by flow cytometry, and individual mice are graphed in order of increasing age from 4-7 weeks. Male mice are depicted as black bars and female mice are depicted as grey bars.


