Abstract
The influence of sodium chlorate (SC), ferulic acid (FA), and essential oils (EO) was examined on the survivability of two porcine diarrhetic enterotoxigenic Escherichia coli (ETEC) strains (F18 and K88) and populations of porcine fecal bacteria. Fecal bacterial populations were examined by denaturing gradient gel electrophoresis (DGGE) and identification by 16S gene sequencing. The treatments were control (no additives), 10 mM SC, 2.5 mg FA /mL, a 1.5% vol/vol solution of an EO mixture as well as mixtures of EO + SC, EO + FA, and FA + SC at each of the aforementioned concentrations. EO were a commercial blend of oregano oil and cinnamon oil with water and citric acid. Freshly collected porcine feces in half-strength Mueller Hinton broth was inoculated with E. coli F18 (Trial 1) or E. coli K88 (Trial 2). The fecal-E. coli suspensions were transferred to crimp top tubes preloaded with the treatment compounds. Quantitative enumeration was at 0, 6, and 24 h. All treatments reduced (P < 0.05) the counts of E. coli F18 at 6 and 24 h. With the exception of similarity coefficient (%SC), all the other treatments reduced (P < 0.05) the K88 counts at 24 h. The most effective treatments to reduce the F18 and K88 CFU numbers were those containing EO. Results of DGGE revealed that Dice percentage similarity coefficients (%SC) of bacterial profiles among treatment groups varied from 81.3% to 100%SC. The results of gene sequencing showed that, except for SC at 24 h, all the other treatments reduced the counts of the family Enterobacteriaceae, while Lactobacillaceae and Ruminococcaceae increased and Clostridiaceae decreased in all treatments. In conclusion, all treatments were effective in reducing the ETEC, but EO mixture was the most effective. The porcine microbial communities may be influenced by the studied treatments.
Keywords: essential oils, ferulic acid, pathogenic microorganisms, porcine fecal, sodium chlorate
Introduction
In intensive production, confined pigs with high productive performance are under physiological and metabolic stresses and are susceptible to diseases, particularly those caused by Salmonella and Escherichia coli. To maintain pig production, antibiotics have been used as growth promoters and in the treatment of diseases. However, antibacterial resistance has limited the use of antibiotics. Additionally, consumer trends are moving toward preferences for more “organic” meat produced from animals with minimum or no antibiotic additives. In this context, there is much interest to investigate strategies to maintain efficient pig production with considerations for limited or no antibiotic use to control pathogenic bacteria.
The influence of sodium chlorate (SC), ferulic acid (FA), and natural antibiotic compounds from plants on inhibition on gut pathogenic bacteria has been explored. SC in pigs has reduced Salmonella and E. coli counts (Anderson et al., 2001a, 2001b), bacterial shedding (Burkey et al., 2004), and also has been used to control pathogens in the gastrointestinal tract of ruminants (Anderson et al., 2002, 2005; Callaway et al., 2003, 2004; Edrington et al., 2003; Taylor et al., 2012; Arzola et al., 2014; Copado et al., 2014). FA has shown antimicrobial effect against E. coli, Bacillus cereus, Listeria monocytogenes, Fusarium culmorum, Saccharomyces cerevisiae (Merkl et al., 2010), Pseudomonas aeruginosa, and Staphylococcus aureus (Borges et al., 2013). Extracts from plants containing FA and other flavonoids had inhibitory effects against S. aureus, E. coli, Salmonella Typhi, S. Typhimurium, and Vibrio cholera (Mukherjee et al., 2018). Cinnamaldehyde in cinnamon essential oils (EO) fed to pigs reduced fecal E. coli counts (Yan and Kim, 2012). Cinnamon EO also had antibacterial activities against different Salmonella serotypes (Olaimat et al., 2019). A reduction in E. coli growth using cinnamon and oregano EO and their active components cinnamaldehyde and carvacrol, respectively, was observed by Gilling et al. (2019). Carvacrol has inhibited the growth of Enterobacter spp. and Serratia spp. via membrane damage (Bnyan et al., 2014). Also, oregano oil reduced E. coli in the guts of pigs (Cheng et al., 2018) and had bactericidal effect on S. aureus, Enterococcus faecalis, E. coli, Klebsiella pneumoniae, and P. aeruginosa (Man et al., 2019).
There are few reports on the growth of pathogenic bacteria that may affect the productive performance of pigs, when treated with different EO or combinations of EO with other compounds, such as FA and SC. The objective of the present study was to test the hypothesis that SC, FA, EO, as mixtures of EO + SC, EO + FA, and FA + SC, reduce the survivability of enterotoxigenic E. coli (F18 and K88) and may influence on the porcine fecal microbial community diversity.
Material and Methods
Feces were freshly collected at Texas A&M University on the day of study from a commercial Hampshire × York × Landrace pig fed a standard grower diet without added antibiotics. Escherichia coli strains K88 and F18 had been graciously provided by Dr Nancy Cornick, of Iowa State University Ames, IA, and were made resistant to nalidixic acid (Sigma, Saint Louis, MO, USA) and novobiocin (Sigma, Saint Louis, MO, USA) via successive culture in tryptic soy broth (Difco, Becton Dickinson, Sparks, MD) as described by Levent et al. (2016). Cultures prepared as inocula for incubations of mixed populations of fecal microbes were grown overnight at 37 °C in tryptic soy broth supplemented with 25 and 20 µg novobiocin and nalidixic acid per milliliter of medium, respectively.
Microbiological study
Studies with incubations of mixed populations of porcine fecal microbes were conducted to test seven treatments: control (no additives); 10 mM SC (Fisher Scientific, Fire Lawn, NJ, USA); 2.5 mg/mL FA (Fisher Scientific); and 1.5% (vol/vol) mixture of EO or combinations to achieve as mixtures of EO + SC, EO + FA, and FA + SC at concentrations stated above. EO are a blend of oregano oil and cinnamon oil with water and citric acid. The commercial product was: activo (EW Nutrition USA, Des Moines, IA). The doses of SC, FA , and EO used here were based on doses reported earlier (Anderson et al., 2007, Borges et al., 2013; Arzola et al., 2019). Fecal populations were prepared by mixing 1.5 g of freshly collected feces with 300 mL of anaerobically prepared (100% N2 gas) one-half-strength Mueller Hinton broth (Difco) inoculated with 0.3 mL from an overnight grown culture of a novobiocin- and nalidixic acid-resistant strain of E. coli F18 (trial 1) or K88 (trial 2). This fecal suspension (10 mL) was transferred, while under a stream of N2 gas, to 18 × 150 mm crimp top tubes (three tubes/treatment) preloaded with small volumes (≤0.3 mL) of stock concentrations of the treatment compounds. All tubes were closed with rubber stoppers, crimped with aluminum seals, and incubated anaerobically at 39 °C. Fluid samples (1 mL) collected at 0, 6, and 24 h were 10-fold serially diluted in phosphate buffer (pH 6.5) and plated to MacConkey agar (Difco) supplemented with 25 and 20 µg novobiocin and nalidixic acid per milliliter of medium, respectively, for quantitative enumeration (log10 colony-forming units; CFU/g) after 24 h of aerobic incubation at 37 °C. The limit of detection value which was 20 CFU/mL incubation fluid (equivalent to 1.3 log10 CFU/mL) based on the spreading of 0.5 mL of our undiluted sample to our selective and differential MacConkey agar supplemented with novobiocin and nalidixic acid.
Fecal populations of bacteria study
Denaturing gradient gel electrophoresis
To assess microbial band patterns as influenced by treatments, genomic DNA was extracted from the culture samples collected from each treatment in the third replicate of the experiment. Extraction was accomplished according to the protocol of the NucleoSpinVR Tissue kit (Macherey-Nagel, Germany) and was quantified using an ENDURO VR Touch Gel Documentation system (Labnet, Edison NJ). Polymerase chain reaction (PCR) amplification of the V3 region of the 16S rRNA gene for denaturing gradient gel electrophoresis (DGGE) analysis of the bacterial band patterns was carried out according to the methods described by Muyzer and Smalla (1998) and Hume et al. (2003).
Changes in culture bacterial populations were determined by analysis of band patterns to determine the Dice percentage similarity coefficient (%SC) and dendrograms were constructed using unweighted pair group method using arithmetic averages options in Gel Compare II 6.6 (Applied Maths, Inc., Austin, TX, USA).
Porcine microbial 16S rRNA Illumina sequencing
Genomic DNA of anaerobic culture was extracted using the QIAmp DNA Mini Kit (QIAgen Corporation, Valencia, CA). The DNA was quantified using a Nanodrop spectrophotometer (ND-1000; Wilmington, DE) for genome concentrations (ng/uL) as well as purity (A260/280 and A260/230). Subsequently, in a two-step protocol, samples were amplified for sequencing using a forward (28F—5′-GAGTTTGATCNTGGCTCAG-3′) and reverse (388R—5′-TGCTGCCTCCCGTAGGAGT-3′) fusion primer. In the first step, the amplifications were performed with Qiagen HotStar Taq master mix (Qiagen) in a 25-µL reaction mixture containing 1 µL of each 5-µM primer, and 1 µL of template DNA. Reactions were performed on an ABI Veriti Thermal Cycler (Applied Biosystems, Carlsbad, California) under the following thermal profile: 95 °C for 5 min, then 35 cycles of 94 °C for 30 s, 54 °C for 40 s, 72 °C for 1 min, followed by one cycle of 72 °C for 10 min, and 4 °C hold. The products from the first stage amplification were added to a second PCR reaction to qualitatively determine concentrations. The primers for the second PCR were designed based on the Illumina Nextera PCR primers as follows: Forward—5′-TCGTCGGCAGCGTCAGATGTGTATAAGAGACAG-3′ and Reverse—5′-GTCTCGTGGGCTCGGAGATGTGTATAAGAGACAG-3′. The second-stage amplification was run the same as the first stage except for 10 cycles. The product contained raw sequences of 16S rRNA amplicon. The sequencing procedures were done at Research and Testing Laboratories, Lubbock, TX, USA.
Sequencing data were processed using the QIIME 1.9.1 pipeline (Caporaso et al., 2010) under default parameters. Operational taxonomic units (OTUs) were clustered using UCLUST at 97% similarity. The Greengenes database (5.13) with PyNAST was used to align cluster representative sequences (DeSantis et al., 2006; McDonald et al., 2012). After quality filtration, 2,273,780 were obtained and were subsequently standardized to the lowest sequencing depth level (60,830) then alpha, beta, and richness were calculated. UniFrac-based principle coordinate analysis (PCoA) was conducted to visualize the grouping of similar microbiome environments.
Statistical analysis
Log10 CFU of E. coli per gram incubation mixture was subjected to analysis of variance in a completely randomized design. The statistical model for the trial was as follows: Yij = μ+ Ti + Eij, where Yij is the response variable, μ is the general mean, Ti is the treatment effect, and Eij is the residual error. Means of treatments were compared using the Tukey’s test with treatment effects considered significant at P < 0.05. Data were analyzed using the GLM procedure of SAS (SAS Institute, Inc., Cary, NC)
Results
Microbiological study
In trial 1, at 6 h of incubation, greater (P < 0.05) E. coli F18 counts were in the control than in treated groups, followed by FA, sodium chlorate, FA + SC, and EO + SC (Table 1). EO and EO + FA had similar (P > 0.05) E.coli F18 CFU numbers between them and had lower (P < 0.05) counts than the other groups, except for the treatment of EO + SC which was similar (P > 0.05) to them. After 24 h of incubation, greater (P < 0.05) E. coli F18 CFU counts occurred in the control than in treated groups, followed by SC and FA (Table 1). EO, EO + SC, EO + FA, and FA + SC had similar (P > 0.05) CFU/g between them and had lower (P < 0.05) counts than the other groups. Escherichia coli F18 counts in incubations treated with FA noticeably decreased at 6 h and 24 h compared with control. Treatment with FA + SC resulted in only a moderate decrease in CFU/g incubation mixture from 6 to 24 h.
Table 1.
Influence of treatments on the ETEC F18 log10 CFU/g incubation mixture at different times post-inoculation
| Treatment | Time post-inoculation, h | ||
|---|---|---|---|
| 0 | 6 | 24 | |
| Control | 5.62 | 7.70a | 6.72a |
| EO | 5.62 | 1.30e | 1.30d |
| SC | 5.62 | 2.94c | 3.06b |
| FA | 5.62 | 5.73b | 1.82c |
| EO + SC | 5.62 | 1.63de | 1.30d |
| EO + FA | 5.62 | 1.30e | 1.30d |
| FA + SC | 5.48 | 2.30cd | 1.30d |
| P-value | 0.99 | <0.0001 | <0.0001 |
| SEM | 0.250 | 0.190 | 0.099 |
a–dMeans (n = 3) in each column with different superscript are statistically different (P < 0.05).
In trial 2, at 6 h of incubation, greater (P < 0.05) E. coli K88 counts were seen in the control than in treated groups, followed by FA, SC, and FA + SC (Table 2). Escherichia coli K88 treated with EO, EO + SC, and EO + FA had similar (P > 0.05) CFU/g incubation mixture between them and had lower (P < 0.05) counts than the other groups.
Table 2.
Influence of treatments on the ETEC K88 log10 CFU/g incubation mixture at different times post-inoculation
| Treatment | Time post-inoculation, h | ||
|---|---|---|---|
| 0 | 6 | 24 | |
| Control | 5.65 | 8.74a | 6.48a |
| EO | 5.65 | 1.30d | 1.30c |
| SC | 5.65 | 5.85b | 6.41a |
| FA | 5.65 | 5.84b | 3.48b |
| EO + SC | 5.65 | 2.15d | 1.30c |
| EO + FA | 5.65 | 1.30d | 1.30c |
| FA + SC | 5.65 | 4.63c | 2.09bc |
| P-value | 1.00 | <0.0001 | <0.0001 |
| SEM | 0.197 | 0.190 | 0.318 |
a–dMeans (n = 3) in each column with different superscripts are statistically different (P < 0.05).
At 24 h of incubation, control and SC groups had similar (P > 0.05) CFU/g incubation mixture between them and had greater (P < 0.05) counts than the other groups; the FA and FA + SC treatments had intermediate values. Lower (P < 0.05) E. coli K88 CFU/g incubation mixture was observed in treatments with EO, EO + SC, and EO + FA than in the other groups.
As observed in Table 2, E. coli K88 numbers in the control group increased at 6 h then decreased at 24 h. Cell numbers in the SC treatment did not change during the 24 h, and in the FA group, CFU counts did no change at 6 h, then decreased at 24 h, having intermediate CFU numbers when compared with other treatments. The FA + SC CFU/g decreased at 6 h and decreased again at 24 h. Escherichia coli K88 exhibited no reductions in counts when the essential oil mixture was combined with FA or SC at 24 h of incubation.
Fecal populations of bacteria study
Denaturing gradient gel electrophoresis
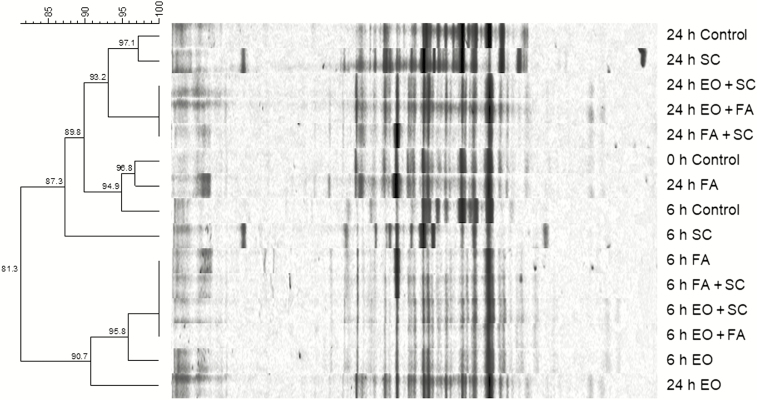
Results of bacterial 16S rRNA DGGE analysis of pig fecal populations revealed that Dice percentage similarity coefficients among the seven treatment groups from pig feces varied from 81.3% to 100%. The fecal populations within groups: FA, SC + FA, EO + SC, and EO + FA were identical at 6 h with 100%SC. Those same populations were very similar at 24 h with an 89.8%SC, remained similar at 24 h to the 6 h cultures with an 87.3%SC, and that clad included the 0-h and 6-h control groups (Figure 1). In general, there was an 81.3%SC somewhat similar banding patterns between 6-h and 24-h treatment groups. The 24-h essential oil mixture incubation had a 90.7%SC with a 6-h clad with 95.8%SC and represented very similar band patterns and possibly the same patterns, respectively. At 6 h, the control and SC treatments had 87.3% similarity, which increased by 24 h to 97.1%SC, possibly the same or very similar band patterns.
Figure 1.
Dendrogram of cultured fecal bacteria 16S rRNA band pattern comparison following denaturing gradient gel electrophoresis. Treatments designations: control, no additives; 1.5% vol/vol of EO mixture; 2.5 mg/mL of FA; 10 mM of SC, and mixtures of EO + SC, EO + FA, and FA + SC at each of the respective concentrations.
Porcine microbial 16S rRNA Illumina sequencing
The percentage relative abundance of the main phyla and classes are presented in Table 3. The percentage relative abundance of the main orders and families are presented in Table 4. The percentage relative abundance of the genus, as well as alpha diversity and richness across time and treatment, are presented in Tables 5 and 6, respectively.
Table 3.
Effect of time and treatment on the relative percentage abundance of main phyla and classes in the porcine microbial community as determined by 16SrRNA gene sequencing
| Treatment1 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | EO | FA | SC | EO + SC | FA + SC | EO + FA | |||||||||
| 0 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | |
| Main phyla | |||||||||||||||
| Firmicutes | 78.48 | 64.16 | 72.40 | 78.49 | 79.35 | 90.35 | 86.24 | 93.71 | 71.68 | 76.06 | 84.59 | 83.85 | 84.30 | 80.68 | 85.78 |
| Proteobacteria | 9.78 | 34.89 | 25.32 | 3.38 | 4.78 | 2.09 | 2.87 | 1.91 | 22.73 | 3.45 | 4.26 | 2.93 | 2.02 | 4.78 | 4.02 |
| Bacteroidetes | 8.93 | 0.40 | 1.81 | 8.20 | 7.33 | 3.28 | 3.48 | 1.12 | 4.48 | 8.34 | 7.02 | 4.82 | 4.13 | 8.61 | 6.94 |
| Euryarchaeota | 0.56 | 0.23 | 0.27 | 6.13 | 5.62 | 3.00 | 6.08 | 0.45 | 0.05 | 7.17 | 1.54 | 5.52 | 6.98 | 1.45 | 0.64 |
| Spirochaetes | 0.98 | 0.08 | 0.06 | 2.03 | 1.50 | 0.70 | 0.59 | 0.29 | 0.10 | 3.37 | 1.16 | 1.51 | 1.59 | 2.78 | 1.20 |
| Main classes | |||||||||||||||
| Clostridia | 54.81 | 55.31 | 67.54 | 55.36 | 55.07 | 24.25 | 28.75 | 8.79 | 31.49 | 55.22 | 57.00 | 45.50 | 34.59 | 56.02 | 57.47 |
| Bacilli | 23.23 | 8.76 | 4.63 | 22.54 | 23.89 | 65.80 | 57.25 | 84.78 | 39.93 | 20.24 | 27.15 | 37.81 | 49.41 | 23.83 | 27.76 |
| Gammaproteobacteria | 8.99 | 34.20 | 23.85 | 2.69 | 3.80 | 1.76 | 2.56 | 1.64 | 22.37 | 2.79 | 3.52 | 2.38 | 1.69 | 3.65 | 3.18 |
| Bacteroidia | 8.93 | 0.40 | 1.81 | 8.20 | 7.33 | 3.28 | 3.48 | 1.12 | 4.48 | 8.34 | 7.02 | 4.82 | 4.13 | 8.61 | 6.94 |
| Methanobacteria | 0.52 | 0.23 | 0.14 | 5.53 | 5.34 | 2.91 | 6.02 | 0.44 | 0.03 | 6.70 | 1.25 | 5.35 | 6.87 | 0.66 | 0.29 |
1Treatment designations: Control, no additives; 1.5% vol/vol solution of EO mixture; 2.5 mg/mL of FA; 10 Mm of SC, as well as mixtures of EO + SC, EO + FA, and FA + SC at each of the aforementioned concentrations.
Table 4.
Effect of time and treatment on the relative percentage abundance of main orders and families in the porcine microbial community as determined by 16S rRNA gene sequencing
| Treatment1 | |||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | EO | FA | SC | EO + SC | FA + SC | EO + FA | |||||||||
| 0 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | |
| Main orders | |||||||||||||||
| Clostridiales | 54.81 | 55.31 | 67.54 | 55.35 | 55.06 | 24.25 | 28.75 | 8.79 | 31.49 | 55.22 | 57.00 | 45.50 | 34.59 | 56.02 | 57.47 |
| Lactobacillales | 23.08 | 8.74 | 4.61 | 22.34 | 23.75 | 65.70 | 57.15 | 84.73 | 39.64 | 20.05 | 26.99 | 37.61 | 49.27 | 23.41 | 27.56 |
| Enterobacteriales | 8.75 | 34.19 | 23.84 | 2.37 | 3.57 | 1.67 | 2.49 | 1.62 | 22.35 | 2.42 | 3.29 | 2.23 | 1.58 | 3.36 | 2.95 |
| Bacteroidales | 8.93 | 0.40 | 1.81 | 8.20 | 7.33 | 3.28 | 3.48 | 1.12 | 4.48 | 8.34 | 7.02 | 4.82 | 4.13 | 8.61 | 6.94 |
| Methanobacteriales | 0.52 | 0.23 | 0.14 | 5.53 | 5.34 | 2.91 | 6.02 | 0.44 | 0.03 | 6.70 | 1.25 | 5.35 | 6.87 | 0.66 | 0.29 |
| Spirochaetales | 0.98 | 0.08 | 0.06 | 2.02 | 1.50 | 0.70 | 0.59 | 0.29 | 0.10 | 3.36 | 1.16 | 1.51 | 1.59 | 2.78 | 1.20 |
| Main families | |||||||||||||||
| Lactobacillaceae | 17.31 | 4.90 | 2.64 | 18.41 | 19.52 | 63.48 | 55.08 | 11.08 | 4.52 | 16.51 | 22.61 | 34.28 | 46.47 | 18.83 | 23.41 |
| Ruminococcaceae | 31.51 | 1.89 | 11.79 | 29.41 | 27.32 | 13.13 | 15.68 | 3.88 | 12.02 | 29.28 | 28.97 | 24.43 | 18.37 | 28.39 | 28.99 |
| Clostridiaceae | 4.83 | 51.32 | 47.37 | 4.30 | 4.43 | 1.87 | 2.07 | 1.90 | 9.80 | 4.32 | 4.28 | 3.49 | 2.97 | 5.15 | 5.14 |
| Streptococcaceae | 5.75 | 3.77 | 1.93 | 3.93 | 4.22 | 2.21 | 2.06 | 72.90 | 33.33 | 3.54 | 4.37 | 3.33 | 2.80 | 4.58 | 4.14 |
| Enterobacteriaceae | 8.75 | 34.19 | 23.84 | 2.37 | 3.57 | 1.67 | 2.49 | 1.62 | 22.35 | 2.42 | 3.29 | 2.23 | 1.58 | 3.36 | 2.95 |
| Lachnospiraceae | 6.75 | 0.90 | 5.81 | 10.07 | 10.74 | 4.11 | 4.82 | 1.24 | 1.63 | 9.84 | 10.98 | 8.05 | 5.96 | 11.02 | 10.89 |
| Clostridiales (order) | 9.36 | 0.55 | 0.94 | 8.67 | 9.52 | 3.91 | 4.77 | 1.27 | 0.59 | 8.87 | 9.83 | 7.35 | 5.67 | 8.74 | 9.50 |
1Treatments designations: Control, no additives; 1.5% vol/vol solution of EO mixture; 2.5 mg/mL of FA; 10 Mm of SC, as well as mixtures of EO + SC, EO + FA, and FA + SC at each of the aforementioned concentrations.
Table 5.
Effect of time and treatment on the relative percentage abundance of genera in the porcine microbial community as determined by 16S rRNA gene sequencing
| Top 20 genera | Treatment1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | EO | FA | SC | EO + SC | FA + SC | EO + FA | |||||||||
| 0 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | |
| Lactobacillus | 17.31 | 4.90 | 2.64 | 18.41 | 19.52 | 63.48 | 55.08 | 11.08 | 4.52 | 16.51 | 22.61 | 34.28 | 46.47 | 18.82 | 23.41 |
| Ruminococcaceae (family) | 28.84 | 1.71 | 9.62 | 26.64 | 24.42 | 11.89 | 14.14 | 3.51 | 10.90 | 26.43 | 25.69 | 22.26 | 16.47 | 25.86 | 25.88 |
| Streptococcus | 5.75 | 3.77 | 1.93 | 3.93 | 4.21 | 2.18 | 2.04 | 72.89 | 33.33 | 3.53 | 4.37 | 3.32 | 2.79 | 4.57 | 4.13 |
| Enterobacteriaceae (family) | 8.75 | 34.18 | 23.83 | 2.36 | 3.57 | 1.67 | 2.49 | 1.62 | 22.34 | 2.42 | 3.29 | 2.22 | 1.58 | 3.35 | 2.94 |
| Clostridiaceae (family) | 2.72 | 42.50 | 36.73 | 2.24 | 2.29 | 0.96 | 1.11 | 1.27 | 1.62 | 2.25 | 2.13 | 1.74 | 1.60 | 2.72 | 2.56 |
| Clostridiales (order) | 9.36 | 0.55 | 0.94 | 8.67 | 9.52 | 3.91 | 4.77 | 1.27 | 0.59 | 8.87 | 9.83 | 7.35 | 5.67 | 8.74 | 9.50 |
| Lachnospiraceae (family) | 3.35 | 0.45 | 2.35 | 5.06 | 5.16 | 1.85 | 2.26 | 0.58 | 1.28 | 4.96 | 4.97 | 3.66 | 2.76 | 5.61 | 4.89 |
| Methanobrevibacter | 0.52 | 0.23 | 0.14 | 5.53 | 5.34 | 2.91 | 6.02 | 0.44 | 0.03 | 6.70 | 1.25 | 5.35 | 6.87 | 0.66 | 0.29 |
| Bacteroidales (order) | 4.20 | 0.19 | 0.56 | 3.29 | 3.02 | 1.38 | 1.47 | 0.55 | 1.24 | 3.33 | 2.80 | 1.98 | 1.75 | 3.54 | 2.76 |
| Clostridium | 1.27 | 8.73 | 8.16 | 0.96 | 0.97 | 0.41 | 0.48 | 0.40 | 0.52 | 1.01 | 0.95 | 0.71 | 0.63 | 0.95 | 1.05 |
| SMB53 | 0.84 | 0.09 | 2.47 | 1.09 | 1.16 | 0.50 | 0.47 | 0.23 | 7.63 | 1.06 | 1.20 | 1.04 | 0.73 | 1.47 | 1.52 |
| S24-7 (family) | 2.15 | 0.09 | 0.13 | 2.61 | 1.94 | 1.02 | 1.06 | 0.33 | 0.09 | 2.65 | 1.82 | 1.50 | 1.14 | 2.83 | 1.94 |
| Ruminococcus | 1.95 | 0.32 | 3.27 | 2.05 | 2.09 | 0.93 | 1.00 | 0.44 | 0.46 | 2.09 | 2.22 | 1.79 | 1.37 | 2.16 | 2.24 |
| Coprococcus | 1.03 | 0.10 | 0.19 | 2.20 | 2.39 | 0.89 | 1.13 | 0.15 | 0.08 | 2.12 | 2.65 | 1.86 | 1.46 | 2.15 | 2.67 |
| Oscillospira | 1.69 | 0.11 | 1.88 | 1.71 | 1.85 | 0.80 | 1.03 | 0.21 | 0.83 | 1.75 | 2.15 | 1.32 | 1.16 | 1.55 | 2.02 |
| Treponema | 0.98 | 0.08 | 0.06 | 2.02 | 1.50 | 0.70 | 0.59 | 0.29 | 0.10 | 3.36 | 1.16 | 1.51 | 1.59 | 2.78 | 1.20 |
| Christensenellaceae family) | 1.27 | 0.12 | 0.11 | 1.26 | 1.26 | 0.58 | 0.70 | 0.20 | 0.07 | 1.29 | 1.32 | 0.96 | 0.76 | 0.95 | 1.21 |
| Dorea | 0.91 | 0.07 | 0.19 | 1.09 | 1.21 | 0.58 | 0.59 | 0.15 | 0.06 | 1.09 | 1.31 | 1.01 | 0.67 | 1.35 | 1.33 |
| CF231 | 1.21 | 0.02 | 0.11 | 1.10 | 1.18 | 0.39 | 0.49 | 0.06 | 0.02 | 1.02 | 1.21 | 0.70 | 0.64 | 1.03 | 1.08 |
1Treatments designations: Control, no additives; 1.5% vol/vol solution of EO mixture; 2.5 mg/mL of FA; 10 Mm of SC, as well as mixtures of EO + SC, EO + FA, and FA + SC at each of the aforementioned concentrations.
Table 6.
Alpha diversity of the porcine microbial community as determined by 16S rRNA gene sequencing
| α-Diveristy | Treatment1 | ||||||||||||||
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| Control | EO | FA | SC | EO + SC | FA + SC | EO + FA | |||||||||
| 0 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | 6 h | 24 h | |
| PD whole tree | 70.73 | 42.47 | 47.32 | 72.94 | 72.68 | 60.10 | 61.07 | 47.97 | 47.66 | 74.59 | 71.14 | 68.99 | 64.35 | 76.36 | 72.48 |
| Chao1 | 1,893.5 | 928.86 | 1,048.4 | 1,924.6 | 1,967.9 | 1,509.0 | 1,530.7 | 1,136.0 | 1,174.8 | 1,926.8 | 1,865.4 | 1,772.9 | 1,650.4 | 2,004.3 | 1,897.0 |
| Observed OTUs | 1,267.3 | 605.90 | 725.50 | 1,308.4 | 1,307.7 | 994.40 | 1,016.6 | 729.40 | 752.10 | 1,359.2 | 1,271.2 | 1,210.4 | 1,101.0 | 1,362.1 | 1,298.3 |
| Shannon | 6.73 | 2.82 | 3.96 | 7.03 | 6.92 | 4.70 | 4.88 | 2.76 | 4.00 | 7.11 | 6.87 | 6.26 | 5.25 | 7.02 | 6.89 |
| Simpson | 0.97 | 0.71 | 0.81 | 0.98 | 0.97 | 0.83 | 0.83 | 0.51 | 0.84 | 0.98 | 0.97 | 0.95 | 0.85 | 0.98 | 0.97 |
| ACE | 2,220.8 | 1,328.7 | 1,405.9 | 2,073.1 | 2,289.3 | 1,837.6 | 2,068.1 | 1,636.7 | 1,560.1 | 2,260.7 | 2,276.1 | 2,227.8 | 2,124.5 | 2,351.6 | 2,214.3 |
1Treatments designations: Control, no additives; 1.5% vol/vol solution of EO mixture; 2.5 mg/mL of FA; 10 Mm of SC, as well as mixtures of EO + SC, EO + FA, and FA + SC at each of the aforementioned concentrations.
Archaea represented 3.05% of the population and Bacteria 96.95%. Firmicutes was the most abundant phyla across all parameters (80.67%) followed by Proteobacteria (8.61%) and Bacteroides (5.26%). Considering 24 h of incubation, Firmicutes in treatment groups approached in number to the control; however, Proteobacteria in treatment groups had lower numerical values than control, and Bacteroides had the opposite trend.
The predominant classes were Clostridia (45.81%), Bacilli (34.46%), and Gammaproteobacteria (7.94%). At 24 h of incubation, Clostridia and Gammaproteobacteria in treatment groups had lower numerical values than control; however, Bacilli increased in numbers.
The predominant orders were Clostridiales (45.81%), Lactobacillales (34.31%), and Enterobacteriales (7.78%). At 24 h of incubation, Clostridiales and Enterobacteriales in treatment groups had lower numerical values than control; however, Lactobacillales increased in numbers.
Lactobacillaceae (23.93%) family was the most abundant, followed by Ruminococcaceae (20.34%), although both families had lower numbers in control than treated groups.
The Lactobacillus genus represented 23.94%, followed by Streptococcus with 10.16%. The Lactobacillus genus has greater numbers in control than treated groups. The Streptococcus had only moderate changes between control and treatment groups.
The number of observed OTUs for all treatments was greater than control at 24 h (725.5). Treatments EO, EO + SC, and EO + FA resulted in an increase in observed OTUs over time. All treatments resulted in a greater number of observed OTUs at time 24 h as compared with the control. This change was reflected in other diversity metrics (Shannon, Simpson, ACE [abundance-based coverage estimator], Chao1, and PD [Phylogenetic diversity] whole tree), which were all elevated at time 24 h as compared with the control.
The beta diversity across treatment and time was evaluated with PCoA. The communities of EO + SC, EO + FA, SC + FA, and EO were the most similar to the 0-h control and drifted the least with time. SC is most spatial heterogeneous treatment as compared with the 0-h control, followed by FA.
Discussion
Microbiological study
SC was effective in controlling E. coli F18; however, the effect was not maintained over 24 h with E. coli K88. In agreement, other authors have reported enteric bacteria reductions with administration of SC in pigs (Anderson et al., 2001a, 2001b; Burkey et al., 2004) and lambs (Edrington et al., 2003; Arzola et al., 2014); however, a low dose of SC in ewes had minimal effect on ruminal or fecal microbial diversity (Copado et al., 2014).
FA was effective in controlling E. coli F18 and K88. The effect was most marked at 24 h than at 6 h. In agreement, Merkl et al. (2010) also reported antimicrobial effects of FA. The antimicrobial activity of FA is mainly causing changes in the cell membrane properties (Borges et al., 2013; Lemos et al., 2014). Extracts from plants containing FA and other flavonoids also showed inhibitory activity against S. aureus, E. coli, Salmonella Typhi, S. Typhimurium, and V. cholera (Mukherjee et al., 2018).
In the current study, EO alone or in combination with FA were the most effective in reducing E. coli counts, although the effect was not summative. The EO consisted of a blend of oregano oil and cinnamon oil. The antibacterial effect of cinnamon oil was reported by Prabuseenivasan et al. (2006) and Olaimat et al. (2019). The high antibacterial effect of cinnamon EO is attributed to the compound trans-cinnamaldehyde (El Atki et al., 2019). Cinnamaldehyde in feed has reduced fecal E. coli counts in pigs (Yan and Kim, 2012), and the oral supplementation also reduced the uropathogenic E. coli colonization in the urinary bladder and urethra in mice (Narayanan et al., 2017). Similar to the findings from the present study, Gilling et al. (2019) observed a significant reduction in E. coli counts using cinnamon and oregano EO and their active components cinnamaldehyde and carvacrol, respectively.
Oregano oil has shown bactericidal effect on S. aureus, E. faecalis, E. coli, K. pneumoniae, and P. aeruginosa (Man et al., 2019) and reduced E. coli in the guts of pigs (Cheng et al., 2018). The oregano EO or their active compound carvacrol inhibit bacteria growth through cell membrane damage increasing membrane permeability and of the cell wall disruption (Bnyan et al., 2014; Alexopoulos et al., 2017).
Ribeiro-Santos et al. (2018) did not observe synergistic effect with combinations of oregano, cinnamon, and sweet fennel EO against E. coli. In the present study, EO were a blend of oregano oil and cinnamon oil. The bacterial growth inhibition was not greater when compared with EO alone vs. EO + FA or vs. EO + SC.
Fecal populations of bacteria study
Results of DGGE analysis of fecal bacteria populations revealed that the Dice percentage similarity coefficient among the seven treatment groups from pig feces varied from 81.3% to 100%. Current results showed similar fecal populations among treatments, indicating that treatments did not have overly adverse effects on the populations during their inhibition of survival of E. coli. Two main objectives of anti-pathogen treatment strategies are to reduce or eliminate the pathogen load, while minimizing non-beneficial treatment effects on microbial populations. A healthy population is essential for maintaining the overall well-being of the animal, with an added objective of production enhancement from nonantibiotic supplementation. Treatment populations’ DGGE band patterns at 6 h shifted slightly from those of the 0-h and 6-h control groups. By 24 h, band patterns of treatment and control populations were very similar, thus reflecting that potentially negative effects of treatments were limited. Essentially, the two stated objects of treatment effects were seen in reduced pathogen counts, while minimizing drastic shifts in DGGE band patterns of commensal populations.
The study of gene sequencing showed that treatments reduced the counts of the Enterobacteriaceae (family) and Clostridiaceae, while the Lactobacillaceae and Ruminococcaceae were increased. Based on earlier reports of the porcine fecal microbiome (Leser et al., 2002; Zhao et al., 2015), current taxonomic profile is normal. Whereas reductions in the relative abundance of Enterobacteriaceae in the present study are in agreement with the reduction of E. coli F18 or K88 counts observed with treatments during the microbiological study, the possible contribution of other members of this family to the decreased abundance of Enterobacteriaceae cannot be ruled out.
In the current study, a reduction of Lactobacillaceae was observed in the control group, while, with the exception of SC, an increase of Lactobacillaceae was observed in all the other tested treatments. In agreement, Chan et al. (2018) reported antibacterial properties of phenolic compounds extracted from edible plants against food-borne pathogenic bacteria but not against selected lactic acid bacteria. Similar responses were found by Si et al. (2006) using EO to reduce bacterial pathogens but with minimal effect on the total number of lactobacilli in the swine intestinal tract. Also, Namkung et al. (2004) reported that plant extracts supplementation in the diet reduced coliform bacteria but not the lactobacilli in the pig gut. Further research is warranted to elucidate the increase of Lactobacillaceae and its family members associated with pathogenic bacteria diminution in the digestive tract of the pig when EO from plants are administered.
Conclusion
Escherichia coli counts were reduced by the tested compounds: EO, SC, FA, EO + SC, EO + FA, and FA + SC. EO were a blend of oregano oil and cinnamon oil. The most effective treatment reducing E. coli growth was the essential oil blend. The DGGE profile analysis coupled with 16S sequencing demonstrated that bacterial populations were not drastically altered by treatment and population increases in potential beneficial Lactobacillaceae.
Glossary
Abbreviations
- DGGE
denaturing gradient gel electrophoresis
- EO
essential oils
- ETEC
enterotoxigenic Escherichia coli
- FA
ferulic acid
- OTUs
operational taxonomic units
- PCoA
principle coordinate analysis
- PCR
polymerase chain reaction
- SC
sodium chlorate
Conflict of interest statement
The authors declare no real or perceived conflicts of interest.
Literature Cited
- Alexopoulos A., Plessas S., Kimbaris A., Varvatou M., Mantzourani I., and Fournomiti M.. . 2017. Mode of antimicrobial action of Origanum vulgare essential oil against clinical pathogens. Curr. Res. Nutr. Food Sci. 5:109–115. doi: 10.12944/CRNFSJ.5.2.07 [DOI] [Google Scholar]
- Anderson R. C., Buckley S. A., Callaway T. R., Genovese K. J., Kubena L. F., Harvey R. B., and Nisbet D. J.. . 2001a. Effect of sodium chlorate on Salmonella typhimurium concentrations in the weaned pig gut. J. Food Prot. 64:255–258. doi: 10.4315/0362-028x-64.2.255 [DOI] [PubMed] [Google Scholar]
- Anderson R. C., Callaway T. R., Anderson T. J., Kubena L. F., Keith N. K., and Nisbet D. J.. . 2002. Bactericidal effect of sodium chlorate on Escherichia coli concentrations in bovine ruminal and fecal contents in vivo. Microb. Ecol. Health Dis. 14: 24–29. doi: 10.1080/089106002760002720 [DOI] [Google Scholar]
- Anderson R. C., Callaway T. R., Buckley S. A., Anderson T. J., Genovese K. J., Sheffield C. L., and Nisbet D. J.. . 2001b. Effect of oral sodium chlorate administration on Escherichia coli O157:H7 in the gut of experimentally infected pigs. Int. J. Food Microbiol. 71:125–130. doi: 10.1016/s0168-1605(01)00562-1 [DOI] [PubMed] [Google Scholar]
- Anderson R. C., Carr M. A., Miller R. K., King D. A., Carstens G. E., Genovese K. J., Callaway T. R., Edrington T. S., Jung Y.-S., McReynolds J. L., . et al. 2005. Effects of experimental chlorate preparations as feed and water supplements on Escherichia coli colonization and contamination of beef cattle and carcasses. Food Microbiol. 22:439–447. doi: 10.1016/j.fm.2004.09.002 [DOI] [Google Scholar]
- Anderson R. C., Jung Y. S., Oliver C. E., Horrocks S. M., Genovese K. J., Harvey R. B., Callaway T. R., Edrington T. S., and Nisbet D. J.. . 2007. Effects of nitrate or nitro supplementation, with or without added chlorate, on Salmonella enterica serovar Typhimurium and Escherichia coli in swine feces. J. Food Prot. 70:308–315. doi: 10.4315/0362-028x-70.2.308 [DOI] [PubMed] [Google Scholar]
- Arzola C., Copado R., Epps S. V., Rodriguez-Almeida F., Ruiz-Barrera O., Rodriguez-Muela C., Corral-Luna A., Castillo-Castillo Y., and Diaz-Plascencia D.. . 2014. Effects of repeated-low level sodium chlorate administration on ruminal and fecal coliforms in sheep. J. Environ. Sci. Health. B 49:966–970. doi: 10.1080/03601234.2014.951585 [DOI] [PubMed] [Google Scholar]
- Arzola C., Latham E., Anderson R., Salinas-Chavira J., Castillo Y., Ontiveros M., Felix-Portillo M., Olivas-Palacios A. L., Armendariz-Rivas R. L., and Arzola-Rubio A.. . 2019. Antibacterial activity of ferulic acid and sodium chlorate against Escherichia coli F18 and K88 during incubations with porcine fecal bacteria. J. Anim. Sci. 97(suppl. S3):296–297. doi: 10.1093/jas/skz258.599 [DOI] [Google Scholar]
- Borges A., Ferreira C., Saavedra M. J., and Simões M.. . 2013. Antibacterial activity and mode of action of ferulic and gallic acids against pathogenic bacteria. Microb. Drug Resist. 19:256–265. doi: 10.1089/mdr.2012.0244 [DOI] [PubMed] [Google Scholar]
- Bnyan I. A., Abid A. T., and Obied H. N.. . 2014. Antibacterial activity of carvacrol against different types of bacteria. J. Nat. Sci. Res. 4:13–17. Available from https://www.iiste.org/Journals/index.php/JNSR/article/view/13191/13559 [Google Scholar]
- Burkey T. E., Dritz S. S., Nietfeld J. C., Johnson B. J., and Minton J. E.. . 2004. Effect of dietary mannanoligosaccharide and sodium chlorate on the growth performance, acute-phase response, and bacterial shedding of weaned pigs challenged with Salmonella enterica serotype Typhimurium. J. Anim. Sci. 82:397–404. doi: 10.2527/2004.822397x [DOI] [PubMed] [Google Scholar]
- Callaway T. R., Anderson R. C., Edrington T. S., Bischoff K. M., Genovese K. J., Poole T. L., Byrd J. A., Harvey R. B., and Nisbet D. J.. . 2004. Effects of sodium chlorate on antibiotic resistance in Escherichia coli O157:H7. Foodborne Pathog. Dis. 1:59–63. doi: 10.1089/153531404772914473 [DOI] [PubMed] [Google Scholar]
- Callaway T. R., Edrington T. S., Anderson R. C., Genovese K. J., Poole T. L., Elder R. O., Byrd J. A., Bischoff K. M., and Nisbet D. J.. . 2003. Escherichia coli O157:H7 populations in sheep can be reduced by chlorate supplementation. J. Food Prot. 66:194–199. doi: 10.4315/0362-028x-66.2.194 [DOI] [PubMed] [Google Scholar]
- Caporaso J. G., Kuczynski J., Stombaugh J., Bittinger K., Bushman F. D., Costello E. K., Fierer N., Peña A. G., Goodrich J. K., Gordon J. I., . et al. 2010. QIIME allows analysis of high-throughput community sequencing data. Nat. Methods 7:335–336. doi: 10.1038/nmeth.f.303 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan C. L., Gan R. Y., Shah N. P., and Corke H.. . 2018. Polyphenols from selected dietary spices and medicinal herbs differentially affect common food-borne pathogenic bacteria and lactic acid bacteria. Food Control. 92:437–443. doi: 10.1016/j.foodcont.2018.05.032 [DOI] [Google Scholar]
- Cheng C., Xia M., Zhang X., Wang C., Jiang S., and Peng J.. . 2018. Supplementing oregano essential oil in a reduced-protein diet improves growth performance and nutrient digestibility by modulating intestinal bacteria, intestinal morphology, and antioxidative capacity of growing-finishing pigs. Animals 8:E159. doi: 10.3390/ani8090159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Copado R., Arzola C., Epps S. V., Rodriguez-Almeida F., Ruiz O., Rodriguez-Muela C., Castillo Y. C., Corral-Luna A., and Salinas J.. . 2014. Effect of repeated suboptimal chlorate treatment on ruminal and fecal bacterial diversity. J. Food Prot. 77:1588–1592. doi: 10.4315/0362-028X.JFP-14-140 [DOI] [PubMed] [Google Scholar]
- DeSantis T. Z., Hugenholtz P., Larsen N., Rojas M., Brodie E. L., Keller K., Huber T., Dalevi D., Hu P., and Andersen G. L.. . 2006. Greengenes, a chimera-checked 16S rRNA gene database and workbench compatible with ARB. Appl. Environ. Microbiol. 72:5069–5072. doi: 10.1128/AEM.03006-05 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Edrington T. S., Callaway T. R., Anderson R. C., Genovese K. J., Jung Y. S., McReynolds J. L., Bischoff K.M., and Nisbet D. J.. . 2003. Reduction of E. coli O157: H7 populations in sheep by supplementation of an experimental sodium chlorate product. Small Rumin. Res. 49:173–181. doi: 10.1016/S0921-4488(03)00099-3 [DOI] [Google Scholar]
- El Atki Y., Aouam I., El Kamari F., Taroq A., Nayme K., Timinouni M., Lyoussi B., and Abdellaoui A.. . 2019. Antibacterial activity of cinnamon essential oils and their synergistic potential with antibiotics. J. Adv. Pharm. Technol. Res. 10:63–67. doi: 10.4103/japtr.JAPTR_366_18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gilling D. H., Ravishankar S., and Bright K. R.. . 2019. Antimicrobial efficacy of plant essential oils and extracts against Escherichia coli. J. Environ. Sci. Health. A. Tox. Hazard. Subst. Environ. Eng. 54:608–616. doi: 10.1080/10934529.2019.1574153 [DOI] [PubMed] [Google Scholar]
- Hume M. E., Kubena L. F., Edrington T. S., Donskey C. J., Moore R. W., Ricke S. C., and Nisbet D. J.. . 2003. Poultry digestive microflora biodiversity as indicated by denaturing gradient gel electrophoresis. Poult. Sci. 82:1100–1107. doi: 10.1093/ps/82.7.1100 [DOI] [PubMed] [Google Scholar]
- Lemos M., Borges A., Teodósio J., Araújo P., Mergulhão F., Melo L., and Simões M.. . 2014. The effects of ferulic and salicylic acids on Bacillus cereus and Pseudomonas fluorescens single-and dual-species biofilms. Int. Biodeter. Biodegr. 86:42–51. doi: 10.1016/j.ibiod.2013.06.011 [DOI] [Google Scholar]
- Leser T. D., Amenuvor J. Z., Jensen T. K., Lindecrona R. H., Boye M., and Møller K.. . 2002. Culture-independent analysis of gut bacteria: the pig gastrointestinal tract microbiota revisited. Appl. Environ. Microbiol. 68:673–690. doi: 10.1128/aem.68.2.673-690.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Levent G., Harvey R. B., Ciftcioglu G., Beier R. C., Genovese K. J., He H. L., Anderson R. C., and Nisbet D. J.. . 2016. In vitro effects of Thymol-β-D-glucopyranoside on Salmonella enterica serovar Typhimurium and Escherichia coli K88. J. Food Prot. 79:299–303. doi: 10.4315/0362-028X.JFP-15-360 [DOI] [PubMed] [Google Scholar]
- Man A., Santacroce L., Jacob R., Mare A., and Man L.. . 2019. Antimicrobial activity of six essential oils against a group of human pathogens: a comparative study. Pathogens 8:E15. doi: 10.3390/pathogens8010015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McDonald D., Price M. N., Goodrich J., Nawrocki E. P., DeSantis T. Z., Probst A., Andersen G. L., Knight R., and Hugenholtz P.. . 2012. An improved Greengenes taxonomy with explicit ranks for ecological and evolutionary analyses of bacteria and archaea. ISME J. 6:610–618. doi: 10.1038/ismej.2011.139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Merkl R., HRádkoVá I., Filip V., and ŠMIdRkal J.. . 2010. Antimicrobial and antioxidant properties of phenolic acids alkyl esters. Czech. J. Food Sci. 28:275–279. doi: 10.17221/132/2010-CJFS [DOI] [Google Scholar]
- Mukherjee S., Pal S., Chakraborty R., Koley H., and Dhar P.. . 2018. Biochemical assessment of extract from Oxalis corniculata L.: its role in food preservation, antimicrobial and antioxidative paradigms using in situ and in vitro models. Indian J. Exp. Biol. 56:230–243. http://nopr.niscair.res.in/handle/123456789/44190 [Google Scholar]
- Muyzer G., and Smalla K.. . 1998. Application of denaturing gradient gel electrophoresis (DGGE) and temperature gradient gel electrophoresis (TGGE) in microbial ecology. Antonie Van Leeuwenhoek 73:127–141. doi: 10.1023/a:1000669317571 [DOI] [PubMed] [Google Scholar]
- Namkung H., Li M., Gong J., Yu H., Cottrill M., and de Lange C. F. M.. . 2004. Impact of feeding blends of organic acids and herbal extracts on growth performance, gut microbiota and digestive function in newly weaned pigs. Can. J. Anim. Sci. 84:697–704. doi: 10.4141/A04-005 [DOI] [Google Scholar]
- Narayanan A., Muyyarikkandy M. S., Mooyottu S., Venkitanarayanan K., and Amalaradjou M. A.. . 2017. Oral supplementation of trans-cinnamaldehyde reduces uropathogenic Escherichia coli colonization in a mouse model. Lett. Appl. Microbiol. 64:192–197. doi: 10.1111/lam.12713 [DOI] [PubMed] [Google Scholar]
- Olaimat A. N., Al-Holy M. A., Abu Ghoush M. H., Al-Nabulsi A. A., Osaili T. M., and Holley R. A.. . 2019. Inhibitory effects of cinnamon and thyme essential oils against Salmonella spp. in hummus (chickpea dip). J. Food Process Pres. 43:13925–13934. doi: 10.1111/jfpp.13925 [DOI] [Google Scholar]
- Prabuseenivasan S., Jayakumar M., and Ignacimuthu S.. . 2006. In vitro antibacterial activity of some plant essential oils. BMC Complement. Altern. Med. 6:39. doi: 10.1186/1472-6882-6-39 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ribeiro-Santos R., Ventura L. A. F., Santos D. C., Melo N. R., and Costa B. S.. . 2018. Effects of oregano, cinnamon, and sweet fennel essential oils and their blends on foodborne microorganisms. Int. Food Res. J. 25:540–544. Available from http://www.ifrj.upm.edu.my/25%20(02)%202018/(12).pdf [Google Scholar]
- Si W., Gong J., Tsao R., Zhou T., Yu H., Poppe C., Johnson R., and Du Z.. . 2006. Antimicrobial activity of essential oils and structurally related synthetic food additives towards selected pathogenic and beneficial gut bacteria. J. Appl. Microbiol. 100:296–305. doi: 10.1111/j.1365-2672.2005.02789.x [DOI] [PubMed] [Google Scholar]
- Taylor J. B., Dungan R. S., and Lewis G. S.. . 2012. Sodium chlorate reduces the presence of Escherichia coli in feces of lambs and ewes managed in shed-lambing systems. J. Anim. Sci. 90:381–386. doi: 10.2527/jas.2011-4270 [DOI] [PubMed] [Google Scholar]
- Yan L., and Kim I. H.. . 2012. Effect of eugenol and cinnamaldehyde on the growth performance, nutrient digestibility, blood characteristics, fecal microbial shedding and fecal noxious gas content in growing pigs. Asian-Australas. J. Anim. Sci. 25:1178–1183. doi: 10.5713/ajas.2012.12111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhao W., Wang Y., Liu S., Huang J., Zhai Z., He C., Ding J., Wang J., Wang H., Fan W., . et al. 2015. The dynamic distribution of porcine microbiota across different ages and gastrointestinal tract segments. PLoS One 10:e0117441. doi: 10.1371/journal.pone.0117441 [DOI] [PMC free article] [PubMed] [Google Scholar]