Abstract
Directed breeding of horticultural crops is essential for increasing yield, nutritional content, and consumer-valued characteristics such as shape and color of the produce. However, limited genetic diversity restricts the amount of crop improvement that can be achieved through conventional breeding approaches. Natural genetic changes in cis-regulatory regions of genes play important roles in shaping phenotypic diversity by altering their expression. Utilization of CRISPR/Cas editing in crop species can accelerate crop improvement through the introduction of genetic variation in a targeted manner. The advent of CRISPR/Cas-mediated cis-regulatory region engineering (cis-engineering) provides a more refined method for modulating gene expression and creating phenotypic diversity to benefit crop improvement. Here, we focus on the current applications of CRISPR/Cas-mediated cis-engineering in horticultural crops. We describe strategies and limitations for its use in crop improvement, including de novo cis-regulatory element (CRE) discovery, precise genome editing, and transgene-free genome editing. In addition, we discuss the challenges and prospects regarding current technologies and achievements. CRISPR/Cas-mediated cis-engineering is a critical tool for generating horticultural crops that are better able to adapt to climate change and providing food for an increasing world population.
Subject terms: Molecular engineering in plants, Plant breeding, Molecular engineering in plants, Plant breeding
Introduction
Horticultural crops comprise vegetables, fruits, and ornamental flowers as well as aromatic and medicinal plants, thereby providing essential resources to society. For example, the availability and consumption of a wide variety of vegetables and fruits allow us to meet our daily dietary needs. Moreover, we enlighten our days with the abundance of floriculture products for aesthetic uses and visual enjoyment. Collectively, horticultural crops make essential contributions to humankind while also providing the economic engines that drive the success of societies all over the world1.
Despite their collective importance, the improvement of many horticultural crops has lagged behind most agronomic crops, such as rice, corn, and soybean. Yet, improvement of horticultural crops for traits such as resistance to biotic and abiotic stresses, yield, and health-related nutrients would benefit the entire sector. Genetic diversity is a critical source for crop improvement. However, this diversity is often limiting, especially for certain species2. The limited genetic diversity could result in significant obstacles for further improvement by conventional breeding approaches. Research in several crops has demonstrated that much of the genetic changes underlying traits of economic importance reside in the cis-regulatory regions of genes3,4. These changes appear to have been selected during domestication, resulting in desirable traits caused by altered gene expression3,5. The CRISPR/Cas-based platform offers a powerful tool by engineering cis-regulatory regions (cis-engineering) to introduce genetic diversity that could potentially accelerate crop improvement6–10. Despite the importance of regulatory changes in genes, the application of CRISPR/Cas-mediated cis-engineering has only been explored sporadically. The genome sequence for at least 181 horticultural species is available11 and genome editing has been used to generate primarily knockout mutations in at least 25 of them12–17. These achievements demonstrate the feasibility of applying CRISPR/Cas-mediated cis-engineering to expand the phenotypic diversity of many horticultural crops.
Natural variation in cis-regulatory regions resulting from the domestication of horticultural crops
Cis-regulatory regions are non-coding DNA sequences that control the transcription of genes18. These cis-regulatory sequences consist of combinations of CREs that affect gene expression level often in a spatiotemporal manner9,18,19. Single-nucleotide polymorphisms (SNPs), insertions, deletions, inversions, and epigenetic variations are the most common natural variation in cis-regulatory regions that are associated with domestication. Some examples from horticultural crops are discussed below.
Single-nucleotide polymorphisms
Genomic studies in horticultural crops have generated insights into the role of SNP in shaping phenotypic diversity among individuals20. During tomato (Solanum lycopersicum) domestication, selection frequently occurred for fruit size and shape, traits that show extensive variation and large increases over that of the wild relatives4. Increases in fruit weight are thought to be controlled by SNPs in the promoter of FW2.2 (SlCNR) and FW3.2 (SlKLUH)4,21,22. The lc allele contains two SNPs in a 15-bp repressor element downstream of tomato WUSCHEL (SlWUS). The SNPs are proposed to prevent the binding of the MADS-box transcription factor AGAMOUS, which is required to recruit the repressive Polycomb proteins to shut down SlWUS expression, thereby ultimately resulting in larger fruits4,23,24. In another example in tomato, two SNPs in the promoter of Slcyc-B are highly associated with high β-carotene content25.
In citrus (Citrus reticulata), a recent report found an SNP in a miniature inverted-repeat transposable element (MITE) in the promoter of carotenoid cleavage dioxygenase 4b (CCD4) to be sufficient to increase the expression of this gene, resulting in red coloration of fruit peel26. In pepper (Capsicum chinense), an SNP in the promoter of MYB31 is associated with a hyperfunctional W-box, leading to stronger binding of WRKY9. This stronger binding is associated with enhanced expression of MYB31, resulting in extremely pungent peppers27.
Insertions
Insertions are sources of genetic diversity that can alter gene expression by introducing new or disrupting existing CREs. Especially transposable elements (TEs) play important roles in creating genomic variation by altering gene regulation28,29. TE-induced variations in cis-regulatory region are also important in the shaping of domestication-related phenotypes in many horticultural crops. One example is the tomato fruit shape gene SUN. The transposition event at the sun locus mediated by the Rider retrotransposon placed a copy of SUN in addition to Rider itself in the intron of DEFL1. The ancestral copy of SUN on chromosome 10 is lowly expressed, but its derived copy on chromosome 7, where the sun locus maps, is highly expressed30. The high expression of SUN on chromosome 7 is thought to be from the promoter of DEFL1 that would now serve as an enhancer of SUN, leading to the elongated tomato fruit31. Another Rider insertion in the first intron of SEPALLATA4 (SEP4) leads to a jointless pedicel, reduced fruit dropping, which facilitates mechanical harvesting32. In grape (Vitis vinifera), the insertion of the Gret1 (Grapevine Retrotransposon 1) in the VvMYBA1 promoter leads to its inactivation, resulting in a white berry phenotype33. In blood oranges (Citrus sinensis), the insertion of a Copia-like retrotransposon controls the expression of Ruby and the cold dependency of anthocyanin production in the fruit34. In cauliflower (Brassica oleracea var botrytis), a 695-bp Harbinger DNA transposon insertion in the MYB2 promoter increases the expression of this gene, resulting in a purple phenotype35. Additionally, the differentiation of winter and spring genotypes in rapeseed (Brassica napus L.) primarily arose from a MITE transposon insertion in the upstream region of BnFLC.A1036.
Other examples of insertions that are possibly associated with TE activity are found as well. In tomato, ej2w (enhancer-of-jointless 2) is a weak loss-of-function allele, which was selected during tomato domestication and caused by a 564-bp insertion in the fifth intron of EJ2. The mutation results in unbranched inflorescences with exceptionally large sepals32. An 8-bp insertion in the promoter of SlbHLH59 significantly increased its expression in accessions producing high ascorbic acid levels37. In apple (Malus × domestica), multiple repeats of a 23-bp motif in the promoter of MYB10 generate an autoregulatory locus, which is sufficient to account for increased expression and ectopic accumulation of anthocyanins in red-fleshed apples38. Another example from apple is that a 36-bp insertion in MdSAUR37 promoter contributed to high fruit malate content39. In cucumber (Cucumis sativus L.), a 10-bp fragment was replaced by an 812-bp fragment in the promoter of CsHDZIV11/CsGL3 at the few spines 1 (fs1) locus, giving rise to higher expression of CsGL3 and fewer fruit spines40.
Deletions and inversions
Deletions are common genetic changes that provide a wealth of domesticated related phenotypic diversity. One remarkable example is a 31-kb deletion upstream of tomato OVATE Family Protein 20 (SlOFP20). The deletion is associated with reduced expression of SlOFP20 and contributes to natural fruit shape variation in the tomato germplasm41. A 3-bp deletion in the promoter of tomato Al-ACTIVATED MALATE TRANSPORTER9 (Sl-ALMT9) was selected during tomato domestication. The deletion disrupts the repression of Sl-ALMT9 by Sl-WRKY42. This results in increased Sl-ALMT9 gene expression levels, thereby conferring high fruit malate contents and aluminum tolerance in tomato42. Flowering time is an important trait for cucumber domestication. A 39.9-kb deletion and a 16.2-kb deletion located 16.5-kb upstream of cucumber FLOWERING LOCUS T (CsFT) are both associated with higher CsFT expression levels and earlier flowering43. The CsFT locus was selected during cucumber domestication and has been important in its adaptation to higher latitudes for cultivation43. Therefore, deletions can confer desirable traits through either decreased gene expression by removing enhancers and binding sites of activators or increased gene expression by removing repressors and binding sites of repressors.
Genomic inversions also play a role in plant domestication as they could have widespread cis-regulatory effects44. One of the remarkable examples of variation in locule number is controlled by a nearly 300-kb inversion of the fasciated (fas) locus in tomato. The fas locus is characterized by disruption of the promoter region of tomato CLAVATA3 (SlCLV3), leading to downregulation of the gene and larger fruit with increased number of locules24,45.
Epigenetic variations
Natural epigenetic variations contribute to heritable phenotypic diversity that is not caused by modification in the DNA sequence46–49. One of the best examples of an epiallelic variant that impacts an important agronomical trait is the Colorless Non-Ripening (Cnr) allele in tomato. The epiallele of LeSPL-CNR is responsible for colorless fruits with a substantial loss of cell-to-cell adhesion50. In Cnr mutants, hyper-methylation was found along a 286-bp CRE located ~2.4-kb upstream from the first ATG of LeSPL-CNR. This change in methylation status likely explains the reduced expression level of LeSPL-CNR and the ripening defects in Cnr fruits50. Another epigenetic mutation was found in the promoter of the tomato SlTAB2 gene. The mutation controls pigment production in tomato leaves that are affected by DNA methylation level in the promoter of the gene51. Vitamin E 3 (VTE3) is another naturally occurring epiallele controlling vitamin E accumulation in tomato fruits52. The VTE3 expression in fruits is regulated by DNA methylation in the promoter region of the gene52. Additional examples include the control of anthocyanin accumulation in apple and pear (Pyrus communis) fruit skin53–55 and sex determination in melon (Cucumis melo)56. There is also increasing evidence that promoter DNA methylation plays an important role in regulating tomato fruit ripening57,58. Notably, the tomato DML2 is critical for tomato fruit ripening by mediating DNA hypomethylation in promoters of hundreds of genes during development58.
Taken together, these studies highlight the importance of genetic and epigenetic divergence in cis-regulatory regions, including the upstream regions, introns, and downstream regions of genes. Therefore, natural genetic variants, epialleles, and functional CREs in cis-regulatory regions are excellent genome editing targets to create novel variants for the improvement of horticultural crops.
Recent progress in CRISPR/Cas-mediated cis-engineering in plants
So far, the most frequent application of CRISPR/Cas has been to target coding sequences with the goals to create null alleles59–62. Although this application greatly facilitates heritable alleles for reverse genetics studies, selection of loss-of-function mutations in coding regions may result in pleiotropic or deleterious effects45,63,64. Compared to coding sequences, modulating gene expression by cis-engineering is more likely to benefit crop improvement with less detrimental pleiotropic effects3,7,9,10,59,64.
To date, at least 15 articles described successful CRISPR/Cas-mediated cis-engineering via genome editing for 17 genes in eight plants species, including eight genes in four horticultural crops (Fig. 1a). In addition, CRISPR/Cas-mediated cis-engineering has also been achieved to edit the epigenome. However, only a handful of cases have been described in Arabidopsis that show epigenome editing by altering DNA methylation65,66 and histone acetylation67. Because of the few examples in epigenome editing, the following sections will only describe the applications of cis-engineering of DNA.
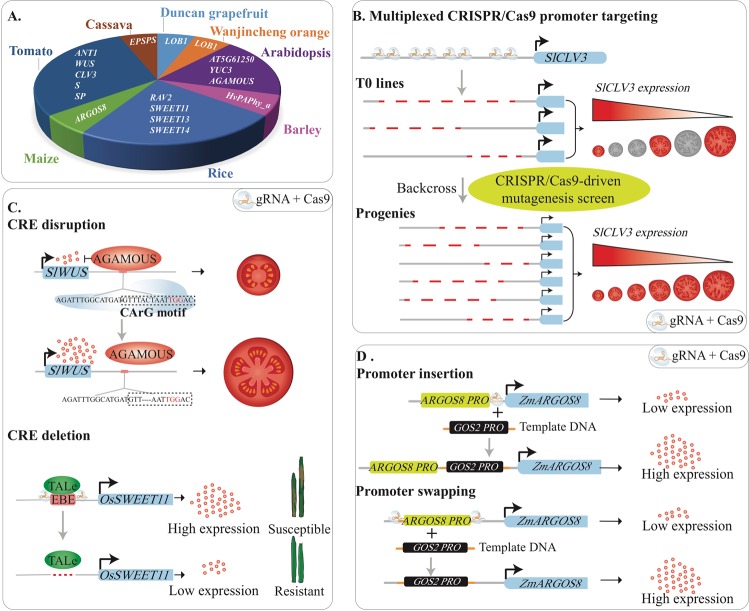
Fig. 1. Current applications of CRISPR/Cas-mediated cis-engineering.
a Summarization of current applications of CRISPR/Cas-mediated cis-engineering in plants. b A continuum of phenotypic variation can be achieved by multiplexed CRISPR/Cas9 promoter targeting and sensitized genetic screen. c Disruption of CREs with genome editing can generate gain-of-function and reduced or loss-of-function alleles. d HDR-mediated promoter insertion/swapping conferring higher gene expression resulting in desirable traits. LOB1, LATERAL ORGAN BOUNDARIES 1; YUC3, YUCCA3; ARGOS8, Auxin-Regulated Gene Involved in Organ Size 8; ANT1, Anthocyanin 1; WUS, WUSCHEL; CLV3, CLAVATA3; S, COMPOUND INFLORESCENCE; SP, SELF PRUNING; SWEET, SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERS; EPSPS, 5-enolpyruvylshikimate-3-phosphate synthase; TALe, Transcription-activator-like effector; EBE, Effector-binding element; CRE, Cis-regulatory element; PRO, promoter.
Promoter disruption
In tomato, a multiplexed CRISPR/Cas9 promoter targeting approach was used to edit the promoters of genes that control fruit size, inflorescence branching, and plant architecture7. Importantly, this approach did neither exploit nor require prior knowledge regarding the structure of promoters and other regulatory sequences. Therefore, the multiplexed CRISPR/Cas9 promoter targeting approach is generally applicable for diverse genes and traits in many crops. Notably, a CRISPR/Cas9-driven sensitized genetic screen approach can recover a collection of cis-regulatory alleles with a continuum of phenotypic effects7 (Fig. 1b), providing an avenue for expanding genetic diversity in crops.
CRE disruption/deletion
Functional CREs in cis-regulatory regions are obvious targets for expanding genetic diversity. However, only a handful of cases have been reported in plants, in which the CRE disruption/deletion was successfully applied to regulate target gene expression.
The rice RAV2 gene is transcriptionally regulated by high salinity. CRISPR/Cas-mediated cis-engineering was used to target the GT-1 element in the promoter of OsRAV2 and the results strongly indicate that the GT-1 element controls the salt response of this gene68. In barley (Hordeum vulgare), the promoter of HvPAPhy_a was targeted for three CREs, namely GCN4, Skn1, and RY69. The lines with mutations in the targeted region show a significant reduction in phytase activity, indicating the importance of these CREs for the expression of the gene. Similarly, the deletion of a 149-bp regulatory fragment containing a transcription-activator-like effector (TALe)-Binding Element (EBE) in the promoter of SUGARS WILL EVENTUALLY BE EXPORTED TRANSPORTERS 11 (SWEET11) improved rice disease resistance without affecting rice fertility64 (Fig. 1c). This result is advantageous compared to the knockout mutant of OsSWEET11 that showed a sterile phenotype, which is obviously undesirable in crop improvement. Recently, simultaneously editing of EBEs in the promoters of SWEET genes resulted in rice lines with broad-spectrum bacterial blight resistance70,71. Three recent studies in Duncan grapefruit (Citrus paradisi Macf.) and Wanjincheng orange (Citrus sinensis Osbeck) reported that canker‐resistant plants were created through CRISPR/Cas editing of the PthA4 effector binding CREs in the promoter of LATERAL ORGAN BOUNDARIES 1 (LOB1)72–74.
The CRISPR/Cas-mediated cis-engineering was also utilized to modify known CREs in introns and downstream of genes. The disruption of the CArG element, including the two causative SNPs downstream of SlWUS, is one of the remarkable examples recreating gain-of-function alleles7,75 (Fig. 1c). In Arabidopsis, a CTCTGYTY motif in the intron of YUCCA3 (YUC3) was identified by chromatin immunoprecipitation-sequencing (CHIP-seq) and is crucial for recruiting RELATIVE OF EARLY FLOWERING 6 (REF6) to its target loci76–78. The deletion of four repeats of this motif leads to diminished binding of REF6 at the mutant loci. In addition, a 450-bp CRE in the second intron of Arabidopsis AGAMOUS (AG) was deleted by CRISPR/Cas9 and verified as the activator of AG gene expression. The deletion of this CRE resulted in early flowering because of a 40% decrease in its expression79.
Promoter insertion/swapping
Promoter insertion and swapping can be achieved by homology-directed repair (HDR) with potentially great importance to crop improvement (Fig. 1d). However, HDR has been challenging due to its low efficiency in higher plants60,80. So far, only three cases have been reported, in which the promoters were accurately inserted or swapped by CRISPR/Cas9-mediated HDR81–83. A 35S promoter was inserted upstream of anthocyanin 1 (ANT1), resulting in enhanced anthocyanin accumulation and intensely purple tomato tissues81. In maize, the HDR pathway was used to insert as well as swap the native GOS2 promoter in the 5′-untranslated region of ARGOS8 (Fig. 1d). The edited plants showed increased expression of ARGOS8 and higher grain yield under drought stress conditions in field trials82. Additionally, glyphosate-tolerant cassava (Manihot esculenta) was generated by a promoter swap of the 5-enolpyruvylshikimate-3-phosphate synthase (EPSPS) gene83.
These encouraging achievements show the potential for using CRISPR/Cas-mediated cis-engineering to improve crop yield, quality, and stress resistance.
Strategies for application of CRISPR/Cas-mediated cis-engineering in horticultural crops improvement
De novo CRE discovery
Prior knowledge of CREs in cis-regulatory region is helpful to apply cis-engineering in crop improvement. Many previously described CREs, especially transcription factor-binding sites (TFBSs), in plant promoters can be identified by submitting sequences to various databases, including TRANSFAC84, PLACE85, PlantCARE86, JASPAR Core PLANTAE87, PlantTFDB88, and Plant Regulomics89. After the TFBSs have been predicted, the regions can be validated by either in vitro methods based on DNA–protein interaction, such as DNA electrophoretic mobility shift assay, DNA pull-down and yeast one-hybrid assays, or in vivo CHIP-based methods, for example, CHIP with DNA microarray (CHIP-chip) and CHIP-seq.
However, the vast majority of CREs are unknown or poorly characterized, highlighting the pressing need for de novo CRE discovery. The availability of genomic and transcriptomic data for many horticultural crops allows the identification of novel CREs using bioinformatics-based and experimental approaches11,90,91. The de novo CRE discovery is based on sequence conservation that exists among groups of genes that are co-expressed as well as gene families within a single genome, and among orthologs of multiple species91–93.
Genes that show similar expression patterns or are in the same gene family are likely to be tightly co-regulated and/or functionally related. Therefore, clustering co-expressing genes and identification of gene families are helpful to explore conserved CREs and uncover their functions for transcriptional regulation. The shared CREs can be identified by the well-established methods such as multiple EM for motif elicitation (MEME)94,95 and eXhaustive evaluation of matriX motifs (XX motif)96,97. An ensemble strategy was used for de novo soybean cyst nematode-inducible motif discovery in the upstream regulatory sequences of 18 co-expressed genes98. Another strategy to identify conserved CREs is by comparing promoter sequences of orthologous genes from different species. Phylogenetic footprinting and variations of the technique are designed for the CRE discovery approach99–103. mVISTA is a commonly used tool for comparative analysis of genomic sequences104. The comparison of the CLV3 promoters in tomato with three other Solanaceae species, S. pennellii, potato (S. tuberosum), and pepper (C. annuum) was performed using mVISTA. This resulted in the identification of three putative CREs between tomato and pepper, and four CREs between tomato and potato7. Complementary to bioinformatics-based approaches are experimental approaches, for example, deconstructive and reconstructive approaches, by which numerous inducible and tissue-specific CREs are characterized90,105.
Choice of appropriate approach for CRISPR/Cas-mediated cis-engineering
CRISPR/Cas-based technologies offer multiple strategies to engineer cis-regulatory regions according to the prior knowledge of the target region or given purpose. If no prior knowledge of the target region exists, multiplexed CRISPR/Cas promoter targeting approach can be applied to putative “negative regulators” of the desirable traits by creating a collection of reduced-function alleles (Fig. 1b). In addition, a well-defined promoter can be exchanged with the promoter of the gene of interest to increase expression level or change temporal/spatial expression pattern of the gene (Fig. 1d). For a given CRE in a target region of interest, the CRE can be disrupted or deleted on the basis of the random indel mutations introduced by non-homologous end joining (NHEJ) repair pathway7,64,69,72–75,78,79 (Fig. 1c).
CRISPR/Cas-mediated point mutations and CRE swaps are also important approaches to manipulate gene expression (Fig. 2). Apart from the above-mentioned SNPs that underlied the domestication of crops, numerous studies also documented that single-nucleotide alterations in regulatory sequences can be sufficient to produce substantial effects on gene expression106–108. For example, in soybean, nucleotide mutations in the core and flanking sequences of G-box element lead to both increases and decreases in gene expression in both native and synthetic promoters109. In apple, the presence of R6 motif, a binding site of MdMYB10, in the promoter of MdMYB10 results in auto-activation of the gene and elevated anthocyanins38. The synthetic promoters of pear MYB10 and Arabidopsis MYB75 harboring the R6 motif significantly increase the expression of these genes, leading to elevated anthocyanin levels in transgenic plants of pear and Arabidopsis110. Moreover, the insertion of the R6 motif into the promoter of the gene encoding an anthocyanin biosynthetic enzyme flavonoid 3′5′ -hydroxylase (F3′5′H) and a vitamin C biosynthesis gene GDP-L-Galactose Phosphorylase (GGP) of kiwifruit (Actinidia eriantha) altered the anthocyanin profile and increased vitamin C content in a MYB10-dependent manner, respectively110. Therefore, the R6 motif can be harnessed to generate new diversity in many horticultural species to increase anthocyanin content (Fig. 2b).
Fig. 2. Examples of the potential applications of CRISPR/Cas-mediated cis-engineering in horticultural crops.
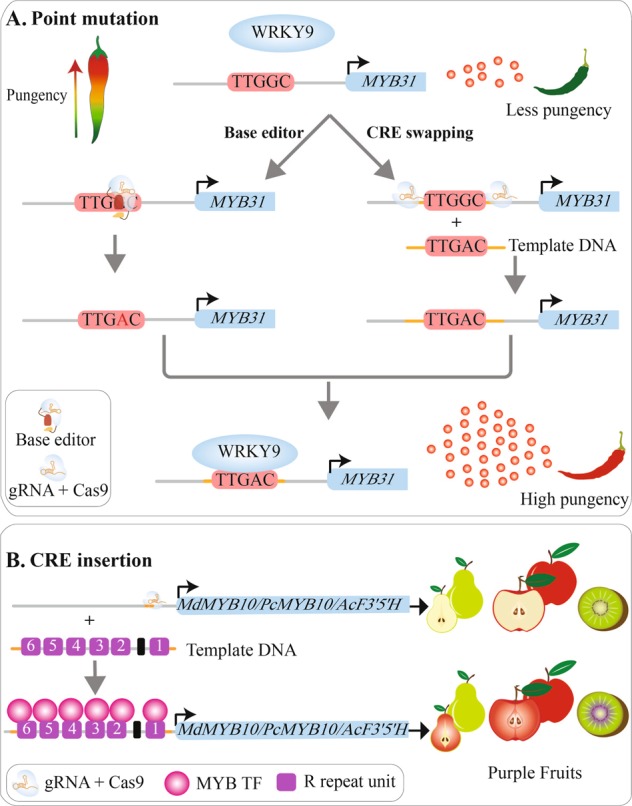
a CRISPR/Cas-mediated point mutations can be achieved by base editor or HDR-mediated CRE swapping. In some Capsicum species, a mutated W-box in the MYB31 promoter is not recognized by the activator WRKY9. Base editor and CRE swapping can change the motif TTGGC to W-box (TTGAC), which can be bound by WRKY9, resulting in increased expression of MYB31 and higher pungency level. b The R6 motif insertion mediated by HDR confers trans-regulation by flavonoid-related MYBs, which can bind the R6-containing promoters of the genes encoding enzymes of the anthocyanin biosynthetic pathway, resulting in enhanced expression of these genes and higher anthocyanin levels. CRE, cis-regulatory element; F3′5′H, flavonoid 3′5′-hydroxylase.
Transgene-free genome editing
Transgene-free genome editing is the preferred choice for the application of cis-engineering for crop improvement and commercialization of genome-edited crops. Genome editing with stable expression of CRISPR/Cas DNA involves the integration of the construct into the host genome, raising concerns about undesirable off-target changes and biosecurity60,111. Genetic segregation by selfing or crossing can be used to obtain transgene-free edited plants. Recently, several strategies have been developed to accelerate the removal of transgene components while retaining the desired mutations. These strategies include the integration of fluorescent markers112,113, herbicide-dependent isolation system114, and the programmed self-elimination system115.
An alternative approach for creating transgene-free edited plants is transient expression of CRISPR/Cas DNA as have been reported in many crops, including wheat116,117, barley118, tetraploid potato119,120, tomato121, and cotton122. Compared to stable transformation of CRISPR/Cas DNA, transient expression is especially useful in certain horticultural crops that are vegetatively propagated, self-incompatible, polyploid, and/or have long juvenile stages123.
Given that traditional breeding, including chemically and physically induced mutagenesis, and DNA-based genome editing may introduce off-target mutations, editing in a DNA-free manner via preassembled Cas9 protein-guide RNA (gRNA) ribonucleoproteins (RNPs) is an increasingly popular approach due to higher specificity, and low off-target mutations further alleviating public concerns124–127. RNPs have been adopted in the transformation of protoplasts in some horticultural crops, such as lettuce (Lactuca sativa L.)128, petunia129, apple and grape130, and potato131. However, regeneration of mature plants from the edited protoplasts is still challenging for most of the horticultural crops.
Currently available transgene-free genome editing approaches are primarily conducted through traditional transformation methods that require tissue culture, which is a labor-intensive process. Therefore, tissue culture-free methods are highly desirable and necessary for transgene-free genome editing. In planta transformation takes advantage of natural biological processes, which makes it a valuable alternative to in vitro tissue culture and regeneration132,133. Various plant cells or tissues can be the ideal transformation targets such as germline or meristematic cells116,134,135 and dormant buds136. Recently, in planta particle bombardment has been used to deliver CRISPR/Cas9 DNA into shoot apical meristems, resulting in transgene-free homozygous mutated wheat plants134. Moreover, recent efforts have been made to deliver RNPs into embryo cells in maize135 and wheat116 by particle bombardment and into zygotes by polyethylene glycol-Ca2+-mediated transfection in rice127.
Challenges and prospects
Genome complexity of horticultural crops
The genome sizes of horticultural crops are diverse, ranging from ~200-Mb in some crops, for example, peach (Prunus persica), to over 30-Gb in garlic (Allium sativum) and onion (Allium cepa)11. Many horticultural crops underwent polyploidy, posing extra challenges for genome editing using CRISPR/Cas technologies. Genome editing of polyploid crops requires increased efficiency to edit multiple alleles simultaneously. Even so, CRISPR/Cas technologies have been successfully applied in many polyploid crops due to continuous improvements, including highly active Cas nuclease, multiplex genome editing, and efficient expression systems63,137,138. In case of octoploid and highly heterozygous cultivated strawberry (Fragaria × ananassa cv. Camarosa), all five alleles of FaTM6 were successfully edited using CRISPR/Cas9-mediated dual single-guide (sg) RNA approach139. Although the genome of Fragaria × ananassa is not yet available, the diploid wild strawberry F. vesca reference genome was employed to analyze the allelic variation in the FaTM6 locus. In this regard, a workflow has been proposed for CRISPR/Cas-mediated mutagenesis for plant species that lack genome sequence information, or feature high heterozygosity or ploidy levels140. This workflow could be also applicable for many horticultural crops.
High-throughput de novo discovery of CREs in their native context
Currently, experimental validation of predicted CREs largely rely on in vitro techniques that are low accuracy and slow throughput. In recent years, new applications, such as DNase-seq (DNase I hypersensitive sites sequencing), ATAC-seq (assay for transposase-accessible chromatin using sequencing), and CHIP-seq, have significantly increased our understanding of transcriptional regulatory elements108. However, these techniques only provide circumstantial evidence and cannot assess the function of CREs in their native context108. As a complementary approach, CRISPR/Cas-based tiling screen approach was developed in mammalian cells to pinpoint functional CREs in their endogenous context141. The strategy is to densely tile gRNAs across a cis-regulatory region to map functional regulatory elements142–146. Although the CRISPR/Cas-based tiling screen approach has not been applied for pinpointing CREs at a large scale in plants, its feasibility was demonstrated in tomato by Rodríguez-Leal et al.7.
Efficient and precise genome editing
Efficient precise genome editing is required to achieve cis-engineering at the nucleotide level. Base editors, including cytidine base editors (CBEs) and adenine base editors (ABEs), are efficient tools for introducing base substitutions at target sites beyond double-strand breaks147,148. Until now, only CBEs have been optimized and applied for gene function studies in horticultural crops, including tomato121,149, potato120,121, and watermelon150. Although base editors are good alternatives to HDR-mediated point mutations, it has been challenging to achieve knock-in and replacement of desired CREs in plants. Much efforts has been made to improve the efficiency of HDR through donor design and modulating repair pathways138. Recently, a fast and precise multiplexing genome editing method was developed in moss (Physcomitrella patens)151. Co-transformation of CRISPR/Cas9 and oligonucleotide templates introduced various mutations into the moss genome with high accuracy and efficiency. It will be interesting to apply such a fast and efficient technology in horticultural crops.
Epigenome editing
The natural epimutations in plants illustrate the potential to further generating phenotypic variation46. However, only a small number of natural epialleles have been described in horticultural crops50,52–56. Fortunately, nuclease-dead Cas-mediated epigenome editing technologies hold great promise to expand phenotypic diversity in crops46,47. While some epialleles can be stably inherited over several generations, others epialleles are transient50,152–154. Thus, the stable transmission of editing induced epigenetic changes to the offspring remains unclear46,155. In addition, the expression of CRISPR components may be needed to maintain the trait in the offspring, limiting its application for crop improvement. Further development of CRISPR-based editing tools and the identification of valuable epialleles in horticultural crops will contribute to the application of epigenome editing for expanding phenotypic diversity.
Concluding remarks
We need to continuously improve horticultural commodities to meet the rising demand for food and ornamental production. The widespread applications of CRISPR/Cas technologies in horticultural crops open the possibility for accelerating new variety development12–17. Engineering cis-regulatory regions using CRISPR/Cas allows the creation of novel variants, resulting in quantitative variation, and thus holds great potential for creating phenotypic diversity. However, cis-engineering is in the beginning stages, and complex relationships between regulation of gene expression by different CREs and the resulting phenotypic changes remains to be fully elucidated7. Therefore, using these CRISPR/Cas techniques to screen for desirable traits at the phenotypic level rather than detecting gene expression differences is practical for crop improvement (Fig. 3). Although challenges remain, the application of CRISPR/Cas-mediated cis-engineering for horticultural crops improvement will further enhance breeding efforts to improve crop yield, resilience, and commercially desirable traits.
Fig. 3.
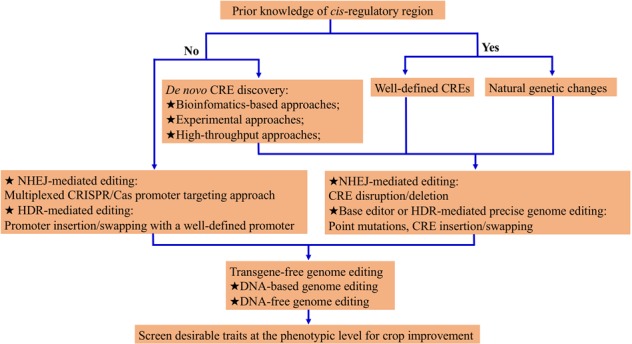
Strategies for applying CRISPR/Cas-mediated cis-engineering in horticultural crops.
Acknowledgements
Funding in the Van der Knaap laboratory is from the National Science Foundation (IOS 1564366, IOS 1732253, and USDA 2017-67013-26199).
Author contributions
Q.L. and E.v.d.K. wrote the review. M.S. prepared the figures and edited the manuscript.
Conflict of interest
The authors declare that they have no conflict of interest.
References
- 1.Van den Broeck G, Maertens M. Horticultural exports and food security in developing countries. Glob. Food Secur. 2016;10:11–20. doi: 10.1016/j.gfs.2016.07.007. [DOI] [Google Scholar]
- 2.Shi J, Lai J. Patterns of genomic changes with crop domestication and breeding. Curr. Opin. Plant Biol. 2015;24:47–53. doi: 10.1016/j.pbi.2015.01.008. [DOI] [PubMed] [Google Scholar]
- 3.Swinnen G, Goossens A, Pauwels L. Lessons from domestication: targeting cis-regulatory elements for crop improvement. Trends Plant Sci. 2016;21:506–515. doi: 10.1016/j.tplants.2016.01.014. [DOI] [PubMed] [Google Scholar]
- 4.van der Knaap E, et al. What lies beyond the eye: the molecular mechanisms regulating tomato fruit weight and shape. Front. Plant Sci. 2014;5:227. doi: 10.3389/fpls.2014.00227. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Meyer RS, Purugganan MD. Evolution of crop species: genetics of domestication and diversification. Nat. Rev. Genet. 2013;14:840–852. doi: 10.1038/nrg3605. [DOI] [PubMed] [Google Scholar]
- 6.Li X, Xie Y, Zhu Q, Liu Y-G. Targeted genome editing in genes and cis-regulatory regions improves qualitative and quantitative traits in crops. Mol. Plant. 2017;10:1368–1370. doi: 10.1016/j.molp.2017.10.009. [DOI] [PubMed] [Google Scholar]
- 7.Rodríguez-Leal D, Lemmon ZH, Man J, Bartlett ME, Lippman ZB. Engineering quantitative trait variation for crop improvement by genome editing. Cell. 2017;171:470–480. doi: 10.1016/j.cell.2017.08.030. [DOI] [PubMed] [Google Scholar]
- 8.Birchler JA. Editing the phenotype: a revolution for quantitative genetics. Cell. 2017;171:269–270. doi: 10.1016/j.cell.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 9.Wolter, F. & Puchta, H. Application of CRISPR/Cas to Understand Cis- and Trans-Regulatory Elements in Plants. In: Plant Transcription Factors. Methods in Molecular Biology (ed. Yamaguchi, N.) vol 1830. (Humana Press, New York, NY, 2018). [DOI] [PubMed]
- 10.Wolter F, Schindele P, Puchta H. Plant breeding at the speed of light: the power of CRISPR/Cas to generate directed genetic diversity at multiple sites. BMC Plant Biol. 2019;19:176. doi: 10.1186/s12870-019-1775-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Chen F, et al. Genome sequences of horticultural plants: past, present, and future. Horticult. Res. 2019;6:1–23. doi: 10.1038/s41438-019-0195-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Xu J, Hua K, Lang Z. Genome editing for horticultural crop improvement. Horticult. Res. 2019;6:1–16. doi: 10.1038/s41438-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xiong J-S, Ding J, Li Y. Genome-editing technologies and their potential application in horticultural crop breeding. Horticult. Res. 2015;2:15019. doi: 10.1038/hortres.2015.19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Karkute SG, Singh AK, Gupta OP, Singh PM, Singh B. CRISPR/Cas9 mediated genome engineering for improvement of horticultural crops. Front. Plant Sci. 2017;8:1635. doi: 10.3389/fpls.2017.01635. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Koltun A, Corte LE-D, Mertz-Henning LM, Gonçalves LS. Genetic improvement of horticultural crops mediated by CRISPR/Cas: a new horizon of possibilities. Horticult. Brasil. 2018;36:290–298. doi: 10.1590/s0102-053620180302. [DOI] [Google Scholar]
- 16.Wang T, Zhang H, Zhu H. CRISPR technology is revolutionizing the improvement of tomato and other fruit crops. Horticult. Res. 2019;6:1–13. doi: 10.1038/s41438-018-0066-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhou, J. et al. Application and future perspective of CRISPR/Cas9 genome editing in fruit crops. J. Integr. Plant Biol.10.1111/jipb.12793 (2019). [DOI] [PMC free article] [PubMed]
- 18.Wittkopp PJ, Kalay G. Cis-regulatory elements: molecular mechanisms and evolutionary processes underlying divergence. Nat. Rev. Genet. 2012;13:59. doi: 10.1038/nrg3095. [DOI] [PubMed] [Google Scholar]
- 19.Bulger M, Groudine M. Functional and mechanistic diversity of distal transcription enhancers. Cell. 2011;144:327–339. doi: 10.1016/j.cell.2011.01.024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Huq MA, et al. Identification of functional SNPs in genes and their effects on plant phenotypes. J. Plant Biotechnol. 2016;43:1–11. doi: 10.5010/JPB.2016.43.1.1. [DOI] [Google Scholar]
- 21.Frary A, et al. fw2. 2: a quantitative trait locus key to the evolution of tomato fruit size. Science. 2000;289:85–88. doi: 10.1126/science.289.5476.85. [DOI] [PubMed] [Google Scholar]
- 22.Chakrabarti M., Zhang N., Sauvage C., Munos S., Blanca J., Canizares J., Diez M. J., Schneider R., Mazourek M., McClead J., Causse M., van der Knaap E. A cytochrome P450 regulates a domestication trait in cultivated tomato. Proceedings of the National Academy of Sciences. 2013;110(42):17125–17130. doi: 10.1073/pnas.1307313110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Muños S, et al. Increase in tomato locule number is controlled by two single-nucleotide polymorphisms located near WUSCHEL. Plant Physiol. 2011;156:2244–2254. doi: 10.1104/pp.111.173997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chu YH, Jang JC, Huang Z, van der Knaap E. Tomato locule number and fruit size controlled by natural alleles of lc and fas. Plant Direct. 2019;3:e00142. doi: 10.1002/pld3.142. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Orchard, C. Naturally Occurring Variation in the Promoter of the Chromoplast-Specific Cyc-B Gene in Tomato can be used to Modulate Levels of ß-Carotene in Ripe Tomato Fruit (The Ohio State University, 2014).
- 26.Zheng X, et al. Natural variation in CCD4 promoter underpins species-specific evolution of red coloration in citrus peel. Mol. Plant. 2019;12:1294–1307. doi: 10.1016/j.molp.2019.04.014. [DOI] [PubMed] [Google Scholar]
- 27.Zhu Z, et al. Natural variations in the MYB transcription factor MYB31 determine the evolution of extremely pungent peppers. N. Phytol. 2019;223:922–938. doi: 10.1111/nph.15853. [DOI] [PubMed] [Google Scholar]
- 28.Lisch D. How important are transposons for plant evolution? Nat. Rev. Genet. 2013;14:49. doi: 10.1038/nrg3374. [DOI] [PubMed] [Google Scholar]
- 29.Olsen KM, Wendel JF. A bountiful harvest: genomic insights into crop domestication phenotypes. Annu. Rev. Plant Biol. 2013;64:47–70. doi: 10.1146/annurev-arplant-050312-120048. [DOI] [PubMed] [Google Scholar]
- 30.Xiao H, Jiang N, Schaffner E, Stockinger EJ, Van Der Knaap E. A retrotransposon-mediated gene duplication underlies morphological variation of tomato fruit. Science. 2008;319:1527–1530. doi: 10.1126/science.1153040. [DOI] [PubMed] [Google Scholar]
- 31.Jiang N, Gao D, Xiao H, Van Der Knaap E. Genome organization of the tomato sun locus and characterization of the unusual retrotransposon Rider. Plant J. 2009;60:181–193. doi: 10.1111/j.1365-313X.2009.03946.x. [DOI] [PubMed] [Google Scholar]
- 32.Soyk S, et al. Bypassing negative epistasis on yield in tomato imposed by a domestication gene. Cell. 2017;169:1142–1155. doi: 10.1016/j.cell.2017.04.032. [DOI] [PubMed] [Google Scholar]
- 33.Kobayashi S, Goto-Yamamoto N, Hirochika H. Retrotransposon-induced mutations in grape skin color. Science. 2004;304:982–982. doi: 10.1126/science.1095011. [DOI] [PubMed] [Google Scholar]
- 34.Butelli E, et al. Retrotransposons control fruit-specific, cold-dependent accumulation of anthocyanins in blood oranges. Plant Cell. 2012;24:1242–1255. doi: 10.1105/tpc.111.095232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Chiu L-W, et al. The purple cauliflower arises from activation of a MYB transcription factor. Plant Physiol. 2010;154:1470–1480. doi: 10.1104/pp.110.164160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Hou J, et al. A Tourist-like MITE insertion in the upstream region of the BnFLC. A10 gene is associated with vernalization requirement in rapeseed (Brassica napus L.) BMC Plant Biol. 2012;12:238. doi: 10.1186/1471-2229-12-238. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Ye J, et al. Genome-wide association analysis identifies a natural variation in basic helix-loop-helix transcription factor regulating ascorbate biosynthesis via d-mannose/l-galactose pathway in tomato. PLoS Genet. 2019;15:e1008149. doi: 10.1371/journal.pgen.1008149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Espley RV, et al. Multiple repeats of a promoter segment causes transcription factor autoregulation in red apples. Plant Cell. 2009;21:168–183. doi: 10.1105/tpc.108.059329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jia D, et al. Apple fruit acidity is genetically diversified by natural variations in three hierarchical epistatic genes: MdSAUR37, MdPP2CH and MdALMTII. Plant J. 2018;95:427–443. doi: 10.1111/tpj.13957. [DOI] [PubMed] [Google Scholar]
- 40.Zhang H, et al. A fragment substitution in the promoter of CsHDZIV11/CsGL3 is responsible for fruit spine density in cucumber (Cucumis sativus L.) Theor. Appl. Genet. 2016;129:1289–1301. doi: 10.1007/s00122-016-2703-5. [DOI] [PubMed] [Google Scholar]
- 41.Wu S, et al. A common genetic mechanism underlies morphological diversity in fruits and other plant organs. Nat. Commun. 2018;9:4734. doi: 10.1038/s41467-018-07216-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ye J, et al. An InDel in the promoter of Al-ACTIVATED MALATE TRANSPORTER9 selected during tomato domestication determines fruit malate contents and aluminum tolerance. Plant Cell. 2017;29:2249–2268. doi: 10.1105/tpc.17.00211. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Wang S, et al. FLOWERING LOCUS T improved cucumber adaptation to higher latitudes. Plant Physiol. 2020;182:908–918. doi: 10.1104/pp.19.01215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Said Iskander, Byrne Ashley, Serrano Victoria, Cardeno Charis, Vollmers Christopher, Corbett-Detig Russell. Linked genetic variation and not genome structure causes widespread differential expression associated with chromosomal inversions. Proceedings of the National Academy of Sciences. 2018;115(21):5492–5497. doi: 10.1073/pnas.1721275115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Xu C, et al. A cascade of arabinosyltransferases controls shoot meristem size in tomato. Nat. Genet. 2015;47:784. doi: 10.1038/ng.3309. [DOI] [PubMed] [Google Scholar]
- 46.Gallusci P, et al. Epigenetics for plant improvement: current knowledge and modeling avenues. Trends Plant Sci. 2017;22:610–623. doi: 10.1016/j.tplants.2017.04.009. [DOI] [PubMed] [Google Scholar]
- 47.Arencibia AD, D’Afonseca V, Chakravarthi M, Castiglione S. Learning from transgenics: advanced gene editing technologies should also bridge the gap with traditional genetic selection. Electron. J. Biotechnol. 2019;41:22–29. doi: 10.1016/j.ejbt.2019.06.001. [DOI] [Google Scholar]
- 48.Kumar S. Epigenomics for crop improvement: current status and future perspectives. J. Genet. Cell Biol. 2019;3:128–134. [Google Scholar]
- 49.Sun, S., Wang, X., Wang, K. & Cui, X. Dissection of complex traits of tomato in the post-genome era. Theor. Appl. Genet.10.1007/s00122-019-03478-y (2019). [DOI] [PubMed]
- 50.Manning K, et al. A naturally occurring epigenetic mutation in a gene encoding an SBP-box transcription factor inhibits tomato fruit ripening. Nat. Genet. 2006;38:948. doi: 10.1038/ng1841. [DOI] [PubMed] [Google Scholar]
- 51.Gouil Q, Novák O, Baulcombe DC. SLTAB2 is the paramutated SULFUREA locus in tomato. J. Exp. Bot. 2016;67:2655–2664. doi: 10.1093/jxb/erw096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Quadrana L, et al. Natural occurring epialleles determine vitamin E accumulation in tomato fruits. Nat. Commun. 2014;5:4027. doi: 10.1038/ncomms5027. [DOI] [PubMed] [Google Scholar]
- 53.El-Sharkawy I, Liang D, Xu K. Transcriptome analysis of an apple (Malus × domestica) yellow fruit somatic mutation identifies a gene network module highly associated with anthocyanin and epigenetic regulation. J. Exp. Bot. 2015;66:7359–7376. doi: 10.1093/jxb/erv433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Telias A, et al. Apple skin patterning is associated with differential expression of MYB10. BMC Plant Biol. 2011;11:93. doi: 10.1186/1471-2229-11-93. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Wang Z, et al. The methylation of the PcMYB10 promoter is associated with green-skinned sport in Max Red Bartlett pear. Plant Physiol. 2013;162:885–896. doi: 10.1104/pp.113.214700. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Martin A, et al. A transposon-induced epigenetic change leads to sex determination in melon. Nature. 2009;461:1135. doi: 10.1038/nature08498. [DOI] [PubMed] [Google Scholar]
- 57.Zhong S, et al. Single-base resolution methylomes of tomato fruit development reveal epigenome modifications associated with ripening. Nat. Biotechnol. 2013;31:154. doi: 10.1038/nbt.2462. [DOI] [PubMed] [Google Scholar]
- 58.Lang Zhaobo, Wang Yihai, Tang Kai, Tang Dengguo, Datsenka Tatsiana, Cheng Jingfei, Zhang Yijing, Handa Avtar K., Zhu Jian-Kang. Critical roles of DNA demethylation in the activation of ripening-induced genes and inhibition of ripening-repressed genes in tomato fruit. Proceedings of the National Academy of Sciences. 2017;114(22):E4511–E4519. doi: 10.1073/pnas.1705233114. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Pandiarajan R, Grover A. In vivo promoter engineering in plants: are we ready? Plant Sci. 2018;277:132–138. doi: 10.1016/j.plantsci.2018.10.011. [DOI] [PubMed] [Google Scholar]
- 60.Chen K, Wang Y, Zhang R, Zhang H, Gao C. CRISPR/Cas genome editing and precision plant breeding in agriculture. Annu. Rev. Plant Biol. 2019;70:667–697. doi: 10.1146/annurev-arplant-050718-100049. [DOI] [PubMed] [Google Scholar]
- 61.Korotkova A, Gerasimova S, Shumny V, Khlestkina E. Crop genes modified using the CRISPR/Cas system. Russian J. Genet. 2017;7:822–832. doi: 10.1134/S2079059717050124. [DOI] [Google Scholar]
- 62.Korotkova A, Gerasimova S, Khlestkina E. Current achievements in modifying crop genes using CRISPR/Cas system. Vavilovskii Zh. Genet. Sel. 2019;8:14422. [Google Scholar]
- 63.Morineau C, et al. Selective gene dosage by CRISPR‐Cas9 genome editing in hexaploid Camelina sativa. Plant Biotechnol. J. 2017;15:729–739. doi: 10.1111/pbi.12671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Li C, et al. A new rice breeding method: CRISPR/Cas9 system editing of the Xa13 promoter to cultivate transgene-free bacterial blight-resistant rice. Plant Biotechnol. J. 2019;18:313–315. doi: 10.1111/pbi.13217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Gallego-Bartolomé Javier, Gardiner Jason, Liu Wanlu, Papikian Ashot, Ghoshal Basudev, Kuo Hsuan Yu, Zhao Jenny Miao-Chi, Segal David J., Jacobsen Steven E. Targeted DNA demethylation of the Arabidopsis genome using the human TET1 catalytic domain. Proceedings of the National Academy of Sciences. 2018;115(9):E2125–E2134. doi: 10.1073/pnas.1716945115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Papikian A, Liu W, Gallego-Bartolomé J, Jacobsen SE. Site-specific manipulation of Arabidopsis loci using CRISPR-Cas9 SunTag systems. Nat. Commun. 2019;10:729. doi: 10.1038/s41467-019-08736-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Paixão JFR, et al. Improved drought stress tolerance in Arabidopsis by CRISPR/dCas9 fusion with a histone acetyltransferase. Sci. Rep. 2019;9:8080. doi: 10.1038/s41598-019-44571-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Duan Y-B, et al. Identification of a regulatory element responsible for salt induction of rice OsRAV2 through ex situ and in situ promoter analysis. Plant Mol. Biol. 2016;90:49–62. doi: 10.1007/s11103-015-0393-z. [DOI] [PubMed] [Google Scholar]
- 69.Holme IB, et al. Evaluation of the mature grain phytase candidate HvPAPhy_a gene in barley (Hordeum vulgare L.) using CRISPR/Cas9 and TALENs. Plant Mol. Biol. 2017;95:111–121. doi: 10.1007/s11103-017-0640-6. [DOI] [PubMed] [Google Scholar]
- 70.Oliva Ricardo, Ji Chonghui, Atienza-Grande Genelou, Huguet-Tapia José C., Perez-Quintero Alvaro, Li Ting, Eom Joon-Seob, Li Chenhao, Nguyen Hanna, Liu Bo, Auguy Florence, Sciallano Coline, Luu Van T., Dossa Gerbert S., Cunnac Sébastien, Schmidt Sarah M., Slamet-Loedin Inez H., Vera Cruz Casiana, Szurek Boris, Frommer Wolf B., White Frank F., Yang Bing. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nature Biotechnology. 2019;37(11):1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Xu Zhengyin, Xu Xiameng, Gong Qiang, Li Ziyang, Li Ying, Wang Sai, Yang Yangyang, Ma Wenxiu, Liu Longyu, Zhu Bo, Zou Lifang, Chen Gongyou. Engineering Broad-Spectrum Bacterial Blight Resistance by Simultaneously Disrupting Variable TALE-Binding Elements of Multiple Susceptibility Genes in Rice. Molecular Plant. 2019;12(11):1434–1446. doi: 10.1016/j.molp.2019.08.006. [DOI] [PubMed] [Google Scholar]
- 72.Jia H, Orbovic V, Jones JB, Wang N. Modification of the PthA4 effector binding elements in Type I Cs LOB 1 promoter using Cas9/sg RNA to produce transgenic Duncan grapefruit alleviating XccΔpthA4: dCs LOB 1.3 infection. Plant Biotechnol. J. 2016;14:1291–1301. doi: 10.1111/pbi.12495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Peng A, et al. Engineering canker‐resistant plants through CRISPR/Cas9‐targeted editing of the susceptibility gene Cs LOB 1 promoter in citrus. Plant Biotechnol. J. 2017;15:1509–1519. doi: 10.1111/pbi.12733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jia H, Orbović V, Wang N. CRISPR-LbCas12a-mediated modification of citrus. Plant Biotechnol. J. 2019;17:1928–1937. doi: 10.1111/pbi.13109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Li Tingdong, Yang Xinping, Yu Yuan, Si Xiaomin, Zhai Xiawan, Zhang Huawei, Dong Wenxia, Gao Caixia, Xu Cao. Domestication of wild tomato is accelerated by genome editing. Nature Biotechnology. 2018;36(12):1160–1163. doi: 10.1038/nbt.4273. [DOI] [PubMed] [Google Scholar]
- 76.Cui X, et al. REF6 recognizes a specific DNA sequence to demethylate H3K27me3 and regulate organ boundary formation in Arabidopsis. Nat. Genet. 2016;48:694. doi: 10.1038/ng.3556. [DOI] [PubMed] [Google Scholar]
- 77.Li C, et al. Concerted genomic targeting of H3K27 demethylase REF6 and chromatin-remodeling ATPase BRM in Arabidopsis. Nat. Genet. 2016;48:687. doi: 10.1038/ng.3555. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Li C, et al. Verification of DNA motifs in Arabidopsis using CRISPR/Cas9‐mediated mutagenesis. Plant Biotechnol. J. 2018;16:1446–1451. doi: 10.1111/pbi.12886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Yan W, Chen D, Kaufmann K. Efficient multiplex mutagenesis by RNA-guided Cas9 and its use in the characterization of regulatory elements in the AGAMOUS gene. Plant Methods. 2016;12:23. doi: 10.1186/s13007-016-0125-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Li J, et al. Gene replacements and insertions in rice by intron targeting using CRISPR–Cas9. Nat. Plants. 2016;2:16139. doi: 10.1038/nplants.2016.139. [DOI] [PubMed] [Google Scholar]
- 81.Čermák T, Baltes NJ, Čegan R, Zhang Y, Voytas DF. High-frequency, precise modification of the tomato genome. Genome Biol. 2015;16:232. doi: 10.1186/s13059-015-0796-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Shi J, et al. ARGOS 8 variants generated by CRISPR‐Cas9 improve maize grain yield under field drought stress conditions. Plant Biotechnol. J. 2017;15:207–216. doi: 10.1111/pbi.12603. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Hummel AW, et al. Allele exchange at the EPSPS locus confers glyphosate tolerance in cassava. Plant Biotechnol. J. 2018;16:1275–1282. doi: 10.1111/pbi.12868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Matys V, et al. TRANSFAC®: transcriptional regulation, from patterns to profiles. Nucleic Acids Res. 2003;31:374–378. doi: 10.1093/nar/gkg108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Higo K, Ugawa Y, Iwamoto M, Korenaga T. Plant cis-acting regulatory DNA elements (PLACE) database: 1999. Nucleic Acids Res. 1999;27:297–300. doi: 10.1093/nar/27.1.297. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Lescot M, et al. PlantCARE, a database of plant cis-acting regulatory elements and a portal to tools for in silico analysis of promoter sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Mathelier A, et al. JASPAR 2014: an extensively expanded and updated open-access database of transcription factor binding profiles. Nucleic Acids Res. 2013;42:D142–D147. doi: 10.1093/nar/gkt997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Jin J, et al. PlantTFDB 4.0: toward a central hub for transcription factors and regulatory interactions in plants. Nucleic Acids Res. 2016;45:gkw982. doi: 10.1093/nar/gkw982. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Ran X, et al. Plant Regulomics: a data‐driven interface for retrieving upstream regulators from plant multi‐omics data. Plant J. 2019;101:237–248. doi: 10.1111/tpj.14526. [DOI] [PubMed] [Google Scholar]
- 90.Mehrotra R, et al. Designer promoter: an artwork of cis engineering. Plant Mol. Biol. 2011;75:527–536. doi: 10.1007/s11103-011-9755-3. [DOI] [PubMed] [Google Scholar]
- 91.Xie D, Cai J, Chia N-Y, Ng HH, Zhong S. Cross-species de novo identification of cis-regulatory modules with GibbsModule: application to gene regulation in embryonic stem cells. Genome Res. 2008;18:1325–1335. doi: 10.1101/gr.072769.107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Gruel J, LeBorgne M, LeMeur N, Théret N. Simple shared motifs (SSM) in conserved region of promoters: a new approach to identify co-regulation patterns. BMC Bioinform. 2011;12:365. doi: 10.1186/1471-2105-12-365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Burgess D, Freeling M. The most deeply conserved noncoding sequences in plants serve similar functions to those in vertebrates despite large differences in evolutionary rates. Plant Cell. 2014;26:946–961. doi: 10.1105/tpc.113.121905. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Bailey, T. L. & Elkan, C. Fitting a mixture model by expectation maximization to discover motifs in bipolymers. Proc. Int. Conf. Intell. Syst. Mol. Biol.2, 28–36 (1994). [PubMed]
- 95.Bailey TL, et al. MEME SUITE: tools for motif discovery and searching. Nucleic Acids Res. 2009;37:W202–W208. doi: 10.1093/nar/gkp335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Hartmann H, Guthöhrlein EW, Siebert M, Luehr S, Söding J. P-value-based regulatory motif discovery using positional weight matrices. Genome Res. 2013;23:181–194. doi: 10.1101/gr.139881.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Hickman R, et al. Architecture and dynamics of the jasmonic acid gene regulatory network. Plant Cell. 2017;29:2086–2105. doi: 10.1105/tpc.16.00958. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Liu W, et al. Computational discovery of soybean promoter cis‐regulatory elements for the construction of soybean cyst nematode‐inducible synthetic promoters. Plant Biotechnol. J. 2014;12:1015–1026. doi: 10.1111/pbi.12206. [DOI] [PubMed] [Google Scholar]
- 99.Tagle DA, et al. Embryonic ε and γ globin genes of a prosimian primate (Galago crassicaudatus): nucleotide and amino acid sequences, developmental regulation and phylogenetic footprints. J. Mol. Biol. 1988;203:439–455. doi: 10.1016/0022-2836(88)90011-3. [DOI] [PubMed] [Google Scholar]
- 100.Thomas J, et al. Comparative analyses of multi-species sequences from targeted genomic regions. Nature. 2003;424:788. doi: 10.1038/nature01858. [DOI] [PubMed] [Google Scholar]
- 101.Liu B, et al. An integrative and applicable phylogenetic footprinting framework for cis-regulatory motifs identification in prokaryotic genomes. BMC Genomics. 2016;17:578. doi: 10.1186/s12864-016-2982-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Van de Velde J, Heyndrickx KS, Vandepoele K. Inference of transcriptional networks in Arabidopsis through conserved noncoding sequence analysis. Plant Cell. 2014;26:2729–2745. doi: 10.1105/tpc.114.127001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Yang J, Chen X, McDermaid A, Ma Q. DMINDA 2.0: integrated and systematic views of regulatory DNA motif identification and analyses. Bioinformatics. 2017;33:2586–2588. doi: 10.1093/bioinformatics/btx223. [DOI] [PubMed] [Google Scholar]
- 104.Frazer KA, Pachter L, Poliakov A, Rubin EM, Dubchak I. VISTA: computational tools for comparative genomics. Nucleic Acids Res. 2004;32:W273–W279. doi: 10.1093/nar/gkh458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105.Liu W, Yuan JS, Stewart CN., Jr. Advanced genetic tools for plant biotechnology. Nat. Rev. Genet. 2013;14:781–793. doi: 10.1038/nrg3583. [DOI] [PubMed] [Google Scholar]
- 106.Canver MC, et al. Characterization of genomic deletion efficiency mediated by clustered regularly interspaced palindromic repeats (CRISPR)/Cas9 nuclease system in mammalian cells. J. Biol. Chem. 2014;289:21312–21324. doi: 10.1074/jbc.M114.564625. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Mansour MR, et al. An oncogenic super-enhancer formed through somatic mutation of a noncoding intergenic element. Science. 2014;346:1373–1377. doi: 10.1126/science.1259037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.Lopes R, Korkmaz G, Agami R. Applying CRISPR–Cas9 tools to identify and characterize transcriptional enhancers. Nat. Rev. Mol. Cell. Biol. 2016;17:597–604. doi: 10.1038/nrm.2016.79. [DOI] [PubMed] [Google Scholar]
- 109.Zhang N, McHale LK, Finer JJ. Changes to the core and flanking sequences of G‐box elements lead to increases and decreases in gene expression in both native and synthetic soybean promoters. Plant Biotechnol. J. 2019;17:724–735. doi: 10.1111/pbi.13010. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110.Brendolise C, et al. Multiple copies of a simple MYB-binding site confers trans-regulation by specific flavonoid-related R2R3 MYBs in diverse species. Front. Plant Sci. 2017;8:1864. doi: 10.3389/fpls.2017.01864. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Hahn F, Nekrasov V. CRISPR/Cas precision: do we need to worry about off-targeting in plants? Plant Cell Rep. 2019;38:437–441. doi: 10.1007/s00299-018-2355-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Gao X, Chen J, Dai X, Zhang D, Zhao Y. An effective strategy for reliably isolating heritable and Cas9-free Arabidopsis mutants generated by CRISPR/Cas9-mediated genome editing. Plant Physiol. 2016;171:1794–1800. doi: 10.1104/pp.16.00663. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Aliaga-Franco N, et al. Identification of transgene-free CRISPR-edited plants of rice, tomato, and arabidopsis by monitoring DsRED fluorescence in dry seeds. Front. Plant Sci. 2019;10:1150. doi: 10.3389/fpls.2019.01150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Lu HP, et al. CRISPR-S: an active interference element for a rapid and inexpensive selection of genome-edited, transgene-free rice plants. Plant Biotechnol. J. 2017;15:1371. doi: 10.1111/pbi.12788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.He Y, et al. Programmed self-elimination of the CRISPR/Cas9 construct greatly accelerates the isolation of edited and transgene-free rice plants. Mol. Plant. 2018;11:1210–1213. doi: 10.1016/j.molp.2018.05.005. [DOI] [PubMed] [Google Scholar]
- 116.Zhang Y, et al. Efficient and transgene-free genome editing in wheat through transient expression of CRISPR/Cas9 DNA or RNA. Nat. Commun. 2016;7:12617. doi: 10.1038/ncomms12617. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Demirer GS, Zhang H, Goh NS, González-Grandío E, Landry MP. Carbon nanotube-mediated DNA delivery without transgene integration in intact plants. Nat. Protoc. 2019;14:2954–2971. doi: 10.1038/s41596-019-0208-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Gerasimova S, et al. Targeted genome modification in protoplasts of a highly regenerable Siberian barley cultivar using RNA-guided Cas9 endonuclease. Vavilovskii Zh. Genet. Sel. 2018;22:1033–1039. [Google Scholar]
- 119.Andersson M, et al. Efficient targeted multiallelic mutagenesis in tetraploid potato (Solanum tuberosum) by transient CRISPR-Cas9 expression in protoplasts. Plant Cell Rep. 2017;36:117–128. doi: 10.1007/s00299-016-2062-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120.Veillet F, et al. The Solanum tuberosum GBSSI gene: a target for assessing gene and base editing in tetraploid potato. Plant Cell Rep. 2019;38:1–16. doi: 10.1007/s00299-019-02426-w. [DOI] [PubMed] [Google Scholar]
- 121.Veillet F, et al. Transgene-free genome editing in tomato and potato plants using agrobacterium-mediated delivery of a CRISPR/Cas9 cytidine base editor. Int. J. Mol. Sci. 2019;20:402. doi: 10.3390/ijms20020402. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Demirer GS, et al. High aspect ratio nanomaterials enable delivery of functional genetic material without DNA integration in mature plants. Nat. Nanotechnol. 2019;14:456. doi: 10.1038/s41565-019-0382-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Chen L, et al. A method for the production and expedient screening of CRISPR/Cas9-mediated non-transgenic mutant plants. Horticult. Res. 2018;5:13. doi: 10.1038/s41438-018-0023-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Kouranova E, et al. CRISPRs for optimal targeting: delivery of CRISPR components as DNA, RNA, and protein into cultured cells and single-cell embryos. Hum. Gene Ther. 2016;27:464–475. doi: 10.1089/hum.2016.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Liang Z, et al. Genome editing of bread wheat using biolistic delivery of CRISPR/Cas9 in vitro transcripts or ribonucleoproteins. Nat. Protoc. 2018;13:413. doi: 10.1038/nprot.2017.145. [DOI] [PubMed] [Google Scholar]
- 126.Metje-Sprink J, Menz J, Modrzejewski D, Sprink T. DNA-free genome editing: past, present and future. Front. Plant Sci. 2018;9:1065–1080. doi: 10.3389/fpls.2018.01065. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Toda E, et al. An efficient DNA-and selectable-marker-free genome-editing system using zygotes in rice. Nat. Plants. 2019;5:363. doi: 10.1038/s41477-019-0386-z. [DOI] [PubMed] [Google Scholar]
- 128.Woo JW, et al. DNA-free genome editing in plants with preassembled CRISPR-Cas9 ribonucleoproteins. Nat. Biotechnol. 2015;33:1162. doi: 10.1038/nbt.3389. [DOI] [PubMed] [Google Scholar]
- 129.Subburaj S, et al. Site-directed mutagenesis in Petunia × hybrida protoplast system using direct delivery of purified recombinant Cas9 ribonucleoproteins. Plant Cell Rep. 2016;35:1535–1544. doi: 10.1007/s00299-016-1937-7. [DOI] [PubMed] [Google Scholar]
- 130.Malnoy M, et al. DNA-free genetically edited grapevine and apple protoplast using CRISPR/Cas9 ribonucleoproteins. Front. Plant Sci. 2016;7:1904. doi: 10.3389/fpls.2016.01904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131.Andersson M, et al. Genome editing in potato via CRISPR‐Cas9 ribonucleoprotein delivery. Physiol. Plant. 2018;164:378–384. doi: 10.1111/ppl.12731. [DOI] [PubMed] [Google Scholar]
- 132.Niazian M, Noori SS, Galuszka P, Mortazavian SMM. Tissue culture-based Agrobacterium-mediated and in planta transformation methods. Soil Water Res. 2017;53:133–143. [Google Scholar]
- 133.Nagle MF, Déjardin A, Pilate G, Strauss SH. Opportunities for innovation in genetic transformation of forest trees. Front. Plant Sci. 2018;9:1443. doi: 10.3389/fpls.2018.01443. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 134.Hamada H, et al. Biolistic-delivery-based transient CRISPR/Cas9 expression enables in planta genome editing in wheat. Sci. Rep. 2018;8:14422. doi: 10.1038/s41598-018-32714-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135.Svitashev S, Schwartz C, Lenderts B, Young JK, Cigan AM. Genome editing in maize directed by CRISPR–Cas9 ribonucleoprotein complexes. Nat. Commun. 2016;7:13274. doi: 10.1038/ncomms13274. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 136.Fujita K, Matsuoka T, Suzuki S, Takayanagi T. In planta transformation technique for grapevines (Vitis vinifera L.) using dormant buds. J. Plant Biochem. Biotechnol. 2009;18:161–167. doi: 10.1007/BF03263314. [DOI] [Google Scholar]
- 137.Zaman QU, Li C, Cheng H, Hu Q. Genome editing opens a new era of genetic improvement in polyploid crops. Crop J. 2019;7:141–150. doi: 10.1016/j.cj.2018.07.004. [DOI] [Google Scholar]
- 138.Zhang Y, Malzahn AA, Sretenovic S, Qi Y. The emerging and uncultivated potential of CRISPR technology in plant science. Nat. Plants. 2019;5:778–794. doi: 10.1038/s41477-019-0461-5. [DOI] [PubMed] [Google Scholar]
- 139.Martín-Pizarro C, Triviño JC, Posé D. Functional analysis of the TM6 MADS-box gene in the octoploid strawberry by CRISPR/Cas9-directed mutagenesis. J. Exp. Bot. 2018;70:885–895. doi: 10.1093/jxb/ery400. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140.Botella JR. Now for the hard ones: is there a limit on CRISPR genome editing in crops? J. Exp. Bot. 2019;70:734–737. doi: 10.1093/jxb/erz007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 141.Montalbano A, Canver MC, Sanjana NE. High-throughput approaches to pinpoint function within the noncoding genome. Mol. Cell. 2017;68:44–59. doi: 10.1016/j.molcel.2017.09.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142.Canver MC, et al. BCL11A enhancer dissection by Cas9-mediated in situ saturating mutagenesis. Nature. 2015;527:192. doi: 10.1038/nature15521. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143.Korkmaz G, et al. Functional genetic screens for enhancer elements in the human genome using CRISPR-Cas9. Nat. Biotechnol. 2016;34:192. doi: 10.1038/nbt.3450. [DOI] [PubMed] [Google Scholar]
- 144.Rajagopal N, et al. High-throughput mapping of regulatory DNA. Nat. Biotechnol. 2016;34:167. doi: 10.1038/nbt.3468. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 145.Sanjana NE, et al. High-resolution interrogation of functional elements in the noncoding genome. Science. 2016;353:1545–1549. doi: 10.1126/science.aaf7613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 146.Diao Y, et al. A tiling-deletion-based genetic screen for cis-regulatory element identification in mammalian cells. Nat. Methods. 2017;14:629. doi: 10.1038/nmeth.4264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147.Gaudelli NM, et al. Programmable base editing of A• T to G• C in genomic DNA without DNA cleavage. Nature. 2017;551:464. doi: 10.1038/nature24644. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 148.Komor AC, Kim YB, Packer MS, Zuris JA, Liu DR. Programmable editing of a target base in genomic DNA without double-stranded DNA cleavage. Nature. 2016;533:420. doi: 10.1038/nature17946. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149.Shimatani Z, et al. Targeted base editing in rice and tomato using a CRISPR-Cas9 cytidine deaminase fusion. Nat. Biotechnol. 2017;35:441. doi: 10.1038/nbt.3833. [DOI] [PubMed] [Google Scholar]
- 150.Tian S, et al. Engineering herbicide-resistant watermelon variety through CRISPR/Cas9-mediated base-editing. Plant Cell Rep. 2018;37:1353–1356. doi: 10.1007/s00299-018-2299-0. [DOI] [PubMed] [Google Scholar]
- 151.Yi, P. & Goshima, G. Fast, efficient, and precise gene editing in the moss Physcomitrella patens. bioRxiv10.1101/643692 (2019).
- 152.Johannes F, et al. Assessing the impact of transgenerational epigenetic variation on complex traits. PLoS Genet. 2009;5:e1000530. doi: 10.1371/journal.pgen.1000530. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153.Cortijo S, et al. Mapping the epigenetic basis of complex traits. Science. 2014;343:1145–1148. doi: 10.1126/science.1248127. [DOI] [PubMed] [Google Scholar]
- 154.Giovannoni J. Harnessing epigenome modifications for better crops. J. Exp. Bot. 2016;67:2535–2537. doi: 10.1093/jxb/erw143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 155.Hua Kai, Zhang Jinshan, Botella Jose Ramon, Ma Changle, Kong Fanjiang, Liu Baohui, Zhu Jian-Kang. Perspectives on the Application of Genome-Editing Technologies in Crop Breeding. Molecular Plant. 2019;12(8):1047–1059. doi: 10.1016/j.molp.2019.06.009. [DOI] [PubMed] [Google Scholar]