Abstract
Extracellular vesicles (EVs) have been recently identified as vital components of cell-based therapies based on the observation that conditioned media from cultured stromal cells reproduce some of the beneficial effects of intact cells. In order to obtain clinically active EVs derived from Mesenchymal Stromal Cells (MSCs) different procedures have been reported in the literature. Usually, non-confluent cells are incubated with culture medium for 48 h either with EV-depleted Fetal Bovine Serum (FBS) or without FBS. Our aim was to compare the effects of EVs isolated by ultracentrifugation from human umbilical cord MSC conditioned media obtained using these two conditions: with EV-depleted FBS (UC) or without FBS (UCw/o) on the mRNA expression levels of extracellular matrix related genes using the mouse chondrogenic cell line ATDC-5. We observed a deleterious effect on chondrogenic cells treated with UCw/o, showing higher mRNA expression levels of different metalloproteinases and decorin (Dcn) and lower collagen (Col1a1 and Col2a1) and aggrecan (Acan) mRNA levels. To elucidate whether this deleterious effect was induced by the EVs or by any proteins co-purified in the EV pellet, we used size exclusion chromatography (SEC) to further purify the EV pellet, obtaining an EV enriched fraction (EV or EVw/o) and a protein enriched fraction (Prot or Protw/o). Our results pointed that the negative effect on the chondrogenic cell line was due to the contaminant proteins coisolated with the EVs by ultracentrifugation and not from the EVs themselves. Thus, these results highlight the importance of working with well purified EV preparations to specifically achieve their therapeutic effect.
Keywords: extracellular vesicles, ultracentrifugation, size exclusion chromatography, ATDC-5 cell line, gene expression, collagen, human umbilical cord mesenchymal stromal cells, FBS, conditioned media, purity
1. Introduction
Mesenchymal Stromal Cells (MSCs) play an important role in cell therapy and in regenerative medicine due to their capability to differentiate in different cell types depending on the experimental or physiological conditions [1]. However, there are evidences that this beneficial effect cannot be attributed to cell survival and differentiation, and cells are rather thought to act in a paracrine manner [2]. This is based, on one hand, on in vivo studies showing low and transient stem cell engraftment and differentiation at injury sites [3,4,5,6,7] and, on the other hand, on the observation that some of the beneficial effects are reproduced when conditioned medium from cultured stem cells is used [8,9,10,11]. This paracrine effect exerted by stem cells depends not only on their capacity to secrete soluble factors [12], but also on the release of extracellular vesicles (EVs) [13,14,15].
EVs carry proteins, lipids and nucleic acids that have functional effects on other cells, thus, being in part responsible for the cell-to-cell communication of MSCs [16]. EV regenerative potential has been widely studied in different approaches including kidney, liver, heart, bone, cartilage or neural injuries, proving that EVs and their cargos are related to cell proliferation, cell regenerative mechanisms or prevention of apoptosis [17,18]. Moreover, EVs present the ability to cross biological barriers with the possibility to reach target cells easily, they are immunologically compatible and their membranes and cargo can be modified [19]. Hence, EVs are acquiring a potential role as drug carriers [17] and therapeutic agents [20]. In fact, currently different phase I and II clinical trials in different areas are being undertaken [21,22,23,24,25,26,27,28].
To obtain EVs from MSCs and use them in therapeutics, MSCs are harvested from culture media. However, there is no agreement on whether using cell culture media with or without Fetal Bovine Serum (FBS) to obtain EVs [29], since there exists a risk of contamination with EV derived from FBS when used as supplement in the cell culture media, even if this FBS has been previously EV-depleted as suggested by Shelke et al. [30]. The same risk exists when using human platelet lysate instead of FBS, since it also contains EVs [31]. Moreover, the market is developing media to be used for cell culture without the need of adding FBS although currently these solutions are not as widely used as conventional culture media.
Previous studies have shown that serum-free conditions lead to increased EV production with protein composition that differs from EVs isolated in serum-containing media [32] and in MISEV2018 it is recommended to avoid FBS if possible for EV production [33]. However, the therapeutic effects and safety of EVs secreted in these conditions have not been tested. In addition, cellular stress under FBS starvation conditions may increase contaminants, such as apoptotic bodies [34]. For these reasons, there are some divergences in protocols when EVs are isolated from culture media. It is not clear whether it is better to use or not EV-depleted FBS in cell culture media [35,36,37]. Additionally, there are a lot of different protocols to isolate EVs such as ultracentrifugation, which is the gold standard protocol, centrifugation with sucrose gradient, ultrafiltration, size exclusion chromatography (SEC) or precipitation with polymers [38,39]; obtaining different EV purity grade according to the used method. In fact, SEC allows to obtain enriched EVs isolates with a better purity grade compared to other approaches [40,41,42].
Thus our aim was to compare the effects of EVs isolated by ultracentrifugation from MSC conditioned media obtained using EV-depleted FBS or incubating the cells in the absence of FBS. Then, in view of our results, we explored whether purer EVs obtained by SEC may influence the EV effect.
2. Materials and Methods
2.1. Mesenchymal Stromal Cells Culture, EVs Production and Ultracentrifugation
Human Umbilical Cord Mesenchymal Stromal Cells (MSCs) were obtained from the IdISBa Biobanc, with the approval of the Ethical Committee of Balearic Islands (CEI-IB). MSCs were cultured in proliferative medium DMEM–low glucose (Biowest, Nuaillé, France) supplemented with 20% FBS Embryonic Stem Cells tested (Biowest) and 1% penicillin-streptomycin (Biowest). For EV production, MSCs were used between passages 6 and 14.
MSCs were grown in 75 cm2 flasks with proliferative medium until 60% of confluence. Then, cells were washed twice with PBS and proliferative medium was replaced by DMEM–low glucose medium supplemented with 1% penicillin-streptomycin and 20% EV-depleted FBS Embryonic Stem Cells tested to obtain conditioned medium or supplemented just with 1% penicillin-streptomycin to obtain conditioned medium without FBS. EV-depleted FBS was obtained by centrifugation at 120,000 × g (sw 32 Ti rotor, 38.5 mL polypropylene tubes, Beckman Coulter, Brea, CA, USA) for 18 h at 4 °C and supernatant filtered on 0.22 µm porous membrane (Sartorius, Goettingen, Germany) as previously described [30]. After 48 h of incubation with the cells, conditioned media with EV-depleted FBS and conditioned media without FBS were collected and centrifuged at 1500 × g for 15 min at 4 °C to remove cell debris. Then, supernatants were filtered through a 0.22 µm porous membrane and centrifuged at 10,000× g for 30 min at 4 °C to remove largest EVs. Supernatants were centrifuged at 120,000 × g for 18 h at 4 °C (as previously reported [43]) and pellets were suspended in 300 µL of PBS (Biowest) obtaining EVs from conditioned media with EV-depleted FBS (UC) and EVs from conditioned media without FBS (UCw/o). Three independent isolation experiments (using 12 different 75 cm2 flasks in each) were performed for EV isolation and then, the obtained EVs were pooled and aliquoted in order to avoid thawing-freezing cycles and stored at −80 °C until use. Pooling was needed to get enough amount in order to perform further studies as it is done with RNA samples for microarray experiments [44]. Pooling of samples can be used assuming that the measurements taken on the pool are equal to the average of the measurements taken on the individuals [45].
After collecting conditioned media, MSCs cultured with EV-depleted FBS and without FBS were checked: bright-side field images were taken and cell monolayer was used to evaluate cell surface profile (CD105-phycoeritrin (PE), CD90-fluorescein (FITC), CD73-PE, CD34-PE, CD45-FITC and HLA-DR-FITC, Thermo Fisher) [46] by standard flow cytometry as previously described [47].
2.2. Size Exclusion Chromatography
SEC of UC and UCw/o samples was performed as previously described by Böing et al. [48]. Briefly, 14 mL of Sepharose CL-2B (Sigma-Aldrich, St. Louis, MO, USA) was stacked into 10 mL syringe—1.5 cm inner diameter × 7.5 cm column length (BD biosciences, Franklin Lakes, NJ, USA)—stuffed with nylon stocking (Calzedonia, Italy) and washed with 10 mL PBS.
One milliliter of UC or UCw/o sample was loaded on SEC columns and eluted with PBS. Twenty fractions of 0.5 mL eluate were collected. For each fraction, total protein content was quantified and EV markers were identified by western blot. Fractions 6-12 enriched in EVs (EV and EVw/o) and 13–22 enriched in proteins (Prot and Protw/o) were pooled and characterized.
2.3. Total Protein Quantification
Total protein content was quantified by BCA Protein Assay kit (Thermo Fisher, Waltham, MA, USA), using a bovine serum albumin standard included in the assay or by reading absorbance at λ = 280 nm with the NanoDrop spectrophotometer (NanoDrop Technologies, Wilmington, DE, USA).
2.4. Atomic Force Microscopy (AFM)
EV samples (1 µg protein) were diluted in a final volume of 200 µL PBS and 50 µL were adsorbed to freshly cleaved Ruby Muscovite mica (NanoAndMore GmbH, Germany) discs for 10 min. The discs were rinsed with deionized water and dried gently under nitrogen stream. Atomic force microscope (Veeco, Oyster Bay, NY, USA) in tapping mode and aluminum coated silicon probe tips (Mikromasch, Lady’s Island, SC, USA) were used for imaging. Topographic height, amplitude and phase images were recorded at 512 pixels × 512 pixels at a scanning rate of 1 Hz. Images were processed using NanoScope Image software (Veeco Metrology, NY, USA).
2.5. Transmission Electron Microscopy (TEM)
EV samples (10 µg protein) were mixed 1:1 with 4% formaldehyde (Sigma-Aldrich) and 10 µL were fixed on copper Formvar-Carbon coated grids (Ted Pella, CA, USA) for 20 min. These grids were washed with PBS and incubated with 1% glutaraldehyde (Sigma-Aldrich) for 5 min. Finally, grids were washed with deionized water.
To contrast samples, grids were stained with 2% phosphotungstic acid for 1 min and then air dried. Images of EVs were taken with transmission electron microscope Hitachi H600 (Hitachi, Tokyo, Japan) at 50 kV.
2.6. Nanoparticle Tracking Analysis (NTA) and Purity Ratio
The number of particles in EV samples was analyzed using a ZetaView® Nanoparticle Tracking Analyzer device (Particle Metrix GmbH, Meerbush, Germany). Before each session, size and concentration of standard polystyrene beads (100 nm) were measured. Samples were diluted 1:2000 before analyzing 22 position tracks per video.
Once the number of particles in EV samples was set, purity ratio was calculated as described by Webber et al. [49] using the following Equation (1):
| (1) |
2.7. Western Blot
EV samples were prepared with non-reducing loading buffer (without β-mercaptoethanol) to detect tetraspanins presence and loaded with the same amount of protein (10 µg) in a 12% SDS-PAGE gels. Proteins were transferred onto nitrocellulose membrane (GE Healthcare, Pittsburgh, PA, USA) by humid transference, blocked with 10% dry skimmed milk (Central Lechera Asturiana, Asturias, Spain) in TBS containing 10% Tween-20 (Scharlab, Barcelona, Spain) and incubated overnight at 4 °C with the following primary antibodies: anti-human CD9 monoclonal antibody (clone Ts9 diluted 1:2000, Thermo Fisher) and anti-human CD63 monoclonal antibody (clone TS63, diluted 1:2000, Abcam, Cambridge, UK). Then, membranes were incubated for 1 h with HRP-coupled secondary antibody (Thermo Fisher) diluted 1:2000. For membrane exposure, membranes were incubated with Clarity Western ECL Substrate (Bio-Rad, Hercules, CA, USA) and chemiluminescence was detected with C-DiGit® Blot scanner (LI-COR Biosciences, Lincoln, NE, USA). Images were processed with Image Studio Digits Software version 4.0 (LI-COR Biosciences).
In order to confirm the correct transferring of the proteins, before blocking, membranes were then incubated with 0.2% (w/v) Ponceau S (Sigma-Aldrich) in 3% (v/v) acetic acid solution (Sigma-Aldrich) for 5 min. Then, membranes were washed with deionized water. After taking images, membranes were finally washed with TBS for 5 min.
2.8. Chondrogenic Cell Line Culture: Functional Study
ATDC-5 cells (25,000 cells/well) were seeded in 48 well plates with DMEM-F12 medium (Biowest) supplemented with 5% FBS Premium (Biowest), 1% penicillin-streptomycin, supplemented with 10 µg/mL transferrin (Sigma-Aldrich) and 5 ng/mL sodium selenite (Sigma-Aldrich). When cells reached confluence 10 µg/mL insulin (Sigma-Aldrich) were added to the medium to induce cell differentiation. Medium was refreshed every other day.
At day 15, cells were incubated with DMEM-F12 medium supplemented with 5% EV-depleted FBS Premium, 1% penicillin-streptomycin, 10 µg/mL transferrin and 5 ng/mL sodium selenite, and treated for 48 h either with one dose of UC (5 µg protein/1.51 × 107 particles), UCw/o (5 µg protein /1.37 × 109 particles), EV (1.37 × 109 particles), EVw/o (1.37 × 109 particles), Prot (5 µg protein) and Protw/o (5 µg protein). Two independent experiments were performed using 6 replicates in each study.
2.9. Lactate Dehydrogenase Activity
Lactate dehydrogenase (LDH) activity was measured from cell culture media after 48 h of treatment, following the manufacturer’s instructions (Cytotoxicity Detection kit, Roche Diagnostics, Manheim, Germany). Data were presented relative to cells treated with vehicle control (low control, 0% of cell death) and cells treated with 1% Triton X-100 (high control, 100% cell death). Cytotoxicity percentage was calculated using the following Equation (2):
| (2) |
2.10. Cell Metabolic Activity
Metabolic activity was measured using PrestoBlueTM Cell Viability Reagent (Life Technologies, Carlsbad, CA) after 48 h of treatment following the manufacturer’s instructions. Absorbance data were normalized to the control group that was set as 100% of metabolic activity with the following Equation (3):
| (3) |
2.11. Collagen Quantification
Collagen quantification by Sirius red staining was performed as previously described by Gómez-Florit et al. [50]. Briefly, after 48 h of treatment, ATDC-5 cells were washed with PBS and dried overnight at 37 °C in a humidified atmosphere. A solution of 0.1% Sirius red F3BA (Sigma-Aldrich) in saturated picric acid (Sigma-Aldrich) was used to stain collagen for 1 h. A wash with 10 mM HCl (Scharlab, Barcelona, Spain) was performed to remove unbound dye. Then, dye was solubilized with 100 mM NaOH (Scharlab) and absorbance was measured at 540 nm. Results were compared with a calfskin collagen standard curve included in the assay.
2.12. RNA Isolation and Real-Time RT-PCR Analysis
Total RNA isolation was performed using Tripure® (Roche Diagnostics) and quantified at λ = 260 nm by NanoDrop spectrophotometer. To obtain cDNA, the same amount of RNA (500 ng) was reverse transcribed at 42 °C for 60 min using High Capacity RNA-to-cDNA kit (Applied Biosystems, Foster City, CA, USA).
Real-time PCR was performed for several target genes and two reference genes (Table 1) with the Lightcycler480® (Roche Diagnostics) using SYBR green detection. For cDNA amplification, a pre-incubation cycle of 5 min at 95 °C was followed by 45 cycles of cDNA denaturation for 10 s at 95 °C, an annealing step of 10 s at 60 °C and finally an extension step for 10 s at 72 °C. After each cycle, fluorescence was measured at 72 °C. Water (Sigma-Aldrich) was used as negative control without cDNA. To allow relative quantification after PCR, standard curves were constructed from standard reactions for each target and reference genes. The crossing point readings for each sample were used to calculate the amount of either the target or the reference relative to a standard curve, using the Second Derivative Maximum Method provided by the LightCycler480® analysis software version 1.5 (Roche Diagnostics). All samples were normalized by the mean of the expression levels of reference genes and changes were related to the control groups that were set to 100%.
Table 1.
Primers of reference and target genes used in real-time PCR.
| Related Function | Gene | Primer Sequence (5′→3′) | Product Size (bp) | GeneBank Accession Number |
|---|---|---|---|---|
| ECM component | Collagen type I, alpha 1 (Col1a1) | S: AGAGCATGACCGATGGATTC A: CCTTCTTGAGGTTGCCAGTC |
177 | NM_007742.4 |
| ECM component | Collagen type II, alpha 1 (Col2a1) | S: CCTGCAGGTGCTTCTGGTAA A: TAAAGCCAGCAATGCCAGGT |
184 | NM_031163.3 |
| ECM component | Decorin (Dcn) | S: TTGATGCACCCAGCCTGAAA A: TGTGAAGGTAGACGACCTGG |
195 | NM_001190451.2 |
| ECM component | Aggrecan (Acan) | S: TGACGGACACTCTCTGCAAT A: CACGGTGCCCTTTTTACACG |
163 | NM_007424.2 |
| ECM turnover | Matrix metalloproteinase-3 (Mmp3) | S: TAAAGACAGGCACTTTTGGCG A: GGAGACCCAGGGTGTGAATG |
218 | NM_010809.2 |
| ECM turnover | Matrix metalloproteinase-13 (Mmp13) | S: GCCATTACCAGTCTCCGAGG A: GAGCCCAGAATTTTCTCCCTCT |
196 | NM_008607.2 |
| ECM turnover | Metallopeptidase inhibitor 1 (Timp1) | S: GATCGGGGCTCCTAGAGACA A: AGCCCTTATGACCAGGTCCG |
168 | NM_011593.2 |
| Reference gene | 18s ribosomal RNA (Rn18s) | S: GTAACCCGTTGAACCCCATT A: CCATCCAATCGGTAGTAGCG |
151 | NR_003278.3 |
| Reference gene | Glyceraldehyde 3-phosphate dehydrogenase (Gapdh) | S: ACCCAGAAGACTGTGGATGG A: CACATTGGGGGTAGGAACAC |
171 | NM_008084.3 |
2.13. Statistical Analysis
Two independent experiments were performed, and values represent either mean values with minima and maximum or mean ± standard deviation (SD), n = 11 or n = 12. The Kolmogorov–Smirnov test was used to assume parametric or non-parametric distribution for the normality tests. Differences between groups with parametric distribution were assessed by ANOVA followed by Bonferroni test as a post-hoc. When data were non-parametric, the Kruskal–Wallis test was assessed. Results were considered statistically significant at p < 0.05. SPSS program for Windows, version 25.0 (SPSS Inc., Chicago, IL, USA) was used.
3. Results
3.1. Characterization of MSC-Derived EVs Isolated by Ultracentrifugation
Conditioned media from MSCs cultured with EV-depleted FBS and without FBS was used to isolate EVs. On the one hand, MSCs analyzed by flow cytometry did not express CD34, CD45 and HLA-DR, whereas they were positive for CD105, CD90 and CD73, showing a typical profile of MSCs [46] (Supplementary Materials; Figures S1–S3) and morphology was not different as shown in bright-side field images (Supplementary Materials; Figure S4).
On the other hand, EV pellets (UC and UCw/o) were isolated from conditioned medium with EV-depleted FBS and conditioned medium without FBS by ultracentrifugation, which is considered a gold standard protocol [38]. EVs presence was confirmed by TEM and AFM as shown in Figure 1A. As shown in Table 2, EVs from UCw/o were larger than EVs from UC, while higher protein content was quantified for UC compared to UCw/o isolates, having this one a higher purity ratio as calculated following Webber et al. [49] parameters. For both EVs isolates, typical EV markers (CD9 and CD63) were identified by western blot (Figure 1B). Furthermore, when evaluating the negative controls, neither cytochrome C (a mitochondria marker) nor bovine serum albumin (as marker of FBS contaminant) was detected in these isolates, showing no presence of contamination due to the isolating process (Supplementary Materials; Figure S5).
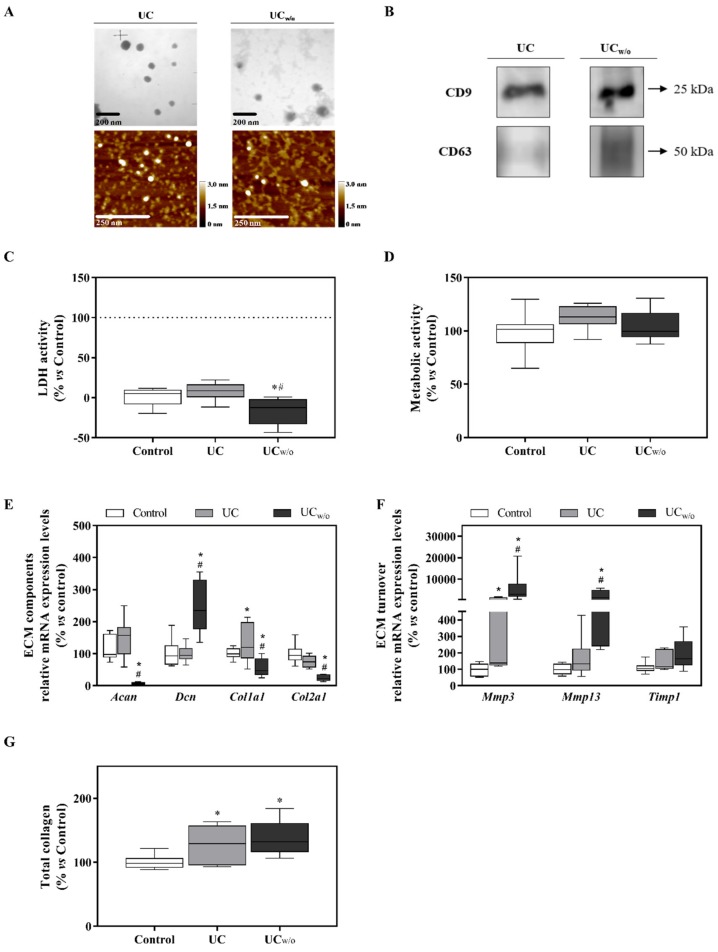
Figure 1.
UC and UCw/o isolates characterization and their effects on the ATDC-5 chondrogenic cell line. (A) Representative TEM and AFM images for each group of EVs with their respective scale bars. (B) Typical EV markers (CD9 and CD63) identified by western blot, loading the same amount of protein for both samples. (C) Lactate dehydrogenase activity was measured from cell culture media of ATDC-5 after 48 h of treatment with UC or UCw/o. Negative control (0%) was culture media from cells treated with the vehicle control. Positive control (100%) was culture media from cells treated with 1% Triton X-100. (D) Metabolic activity measured after 48 h of treatment. Data were normalized to the vehicle control group that was set to 100%. (E,F) mRNA expression levels of ECM components (Acan, Dcn, Col1a1 and Col2a1) genes or ECM turnover genes (Mmp3, Mmp13 and Timp1) of ATDC-5 after treatment with UC or UCw/o for 48 h. Data represent fold changes of target genes normalized to reference genes (Rn18s and Gapdh) and relative to group control that was set as 100%. (G) Total collagen deposition in ATDC-5 cells treated with UC or UCw/o for 48 h. Data were normalized to the vehicle control group that was set to 100%. Values presented in box and whisker plots represent the median, minima and maxima of two independent experiments (n = 11). Results were statistically compared by ANOVA and Bonferroni as post hoc for metabolic activity and Col2a1 mRNA expression levels; and by Kruskal–Wallis for Acan, Dcn, Col1a1, Mmp3 and Mmp13 mRNA expression levels and total collagen deposition: * p < 0.05 versus the control; # p < 0.05 versus UC.
Table 2.
Particles characterization in terms of quantity, size and purity.
| Sample | Number Particles (Particles/mL) | NTA Size | Protein Content (µg/µL) | Purity Ratio (Particles/µg) § |
|---|---|---|---|---|
| UC | 6.30 × 1010 | 150 ± 61.2 | 21.4 | 2.94 × 109 |
| UCw/o | 2.20 × 1011 | 154 ± 59.8 * | 0.800 * | 2.75 × 1011 |
| EV | 1.20 × 1011 | 122 ± 73.5 | 0.050 | 2.40 × 1012 |
| EVw/o | 6.40 × 1010 | 147 ± 78.4 * | 0.080 | 8.00 × 1011 |
| Prot | 7.50 × 109 | 192 ± 107.1 | 3.52 | 2.13 × 109 |
| Protw/o | 5.10 × 1010 | 187 ± 119.7 | 0.270* | 1.89 × 1011 |
§As described by Webber et al. [49]: High Purity: >3 × 1010 particles/µg; low purity: 2 × 109–2 × 1010 particles/µg; unpure: <1.5 × 109. Results were statistically compared by Student t-test: * p < 0.05 UC vs. UCw/o, EV vs. EVw/o or Prot vs. Protw/o.
3.2. Functional Study: UC and UCw/o
To evaluate the effect exerted by EVs isolated by ultracentrifugation from different conditions on cells, a functional study was performed using the ATDC-5 chondrogenic cell line. Cells were differentiated for 15 days and then treated for 48 h with the isolated EVs. As shown in Figure 1D, no differences were found as regards to metabolic activity, however, UCw/o induced lower LDH activity released to the cell culture medium compared to UC or control (Figure 1C).
Different genes related with extracellular matrix (ECM) composition and turnover were studied to evaluate the effect of treatments in their mRNA expression levels (Figure 1E,F). On one hand, UCw/o treatment had a deleterious effect on Acan, Col1a1 and Col2a1 expression levels, which decreased significantly compared to control and treatment with UC. However, Dcn expression levels increased compared to control and UC treatment. On the other hand, UCw/o treatment significantly increased mRNA expression levels for Mmp3 and Mmp13 compared to control and UC treatment, while no differences were shown for Timp1.
Moreover, higher collagen deposition was found for both treated groups compared to control (Figure 1G).
3.3. EV and Protein Fractions Characterization
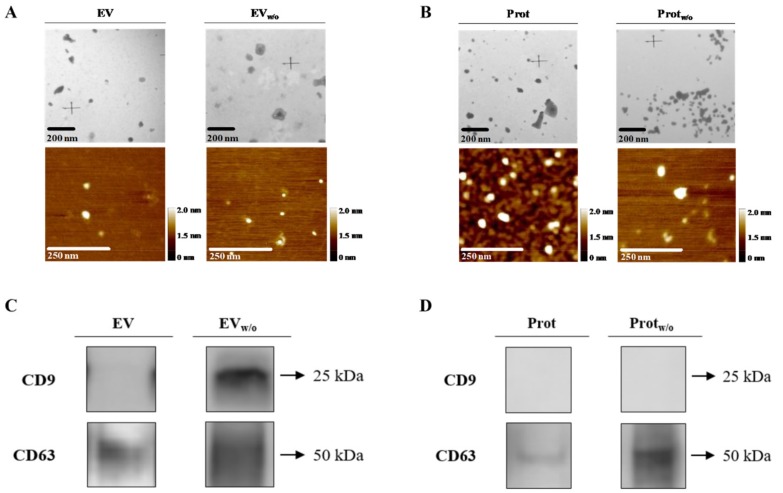
Since a deleterious effect was observed in the UCw/o treated group, and in order to elucidate whether this effect was induced by the EVs themselves or by any proteins co-purified with the EV pellet, we used SEC in order to obtain an EV enriched fraction (EV or EVw/o) pooled from fractions 6-12 and a protein enriched fraction (Prot or Protw/o) pooled from fractions 13-22 (individual characterization of each fraction is shown in supplementary data; Figures S6 and S7).
As expected, both EV fractions obtained after SEC presented a high purity ratio, even EV isolates from UC (EV), which were classified as unpure (Table 2). Following this purification step, a higher number of EVs was obtained in EVw/o, showing higher size compared to EV. Furthermore, typical EV marker tetraspanins appeared: CD9 in EVw/o and CD63 in both EV enriched fractions (Figure 2C) in contrast to protein enriched fractions (Figure 2D). As found on UC samples, neither cytochrome C nor bovine serum albumin was detected for any of the evaluated fractions (Figure S5).
Figure 2.
EV, EVw/o, Prot and Protw/o isolates characterization by TEM, AFM and western blot. (A,B) Representative TEM and AFM images for each group of EVs or pooled protein fractions with their respective scale bars. (C,D) Typical EV markers (CD9 and CD63) identified by western blot from EV, EVw/o samples or Prot and Protw/o, loading the same amount of protein for both group samples.
Prot and Protw/o were also characterized to confirm that, although, some EVs were also present as shown by TEM and AFM (Figure 2B), a lower number of particles/mL were quantified (Table 2). Protein content of Protw/o was statistically lower than Prot and purity ratio was in the high purity range for Protw/o and low purity range for Prot. Tetraspanin CD63 was identified, too, but showing a very low signal in Prot compared to Protw/o (Figure 2D).
3.4. Functional Study with the SEC Purified Fractions
Once purer EVs were obtained, a functional study was performed to clarify if the deleterious effect found for UCw/o was due to the EVs secreted by the cells when cultured without FBS or if such an effect was caused by proteins co-purified with the EVs. As shown in Figure 3A,C,I, no significant changes in LDH activity, metabolic activity or total collagen deposition were observed when chondrogenic cells were treated with EV or EVw/o; and only a slight decrease in Col1a1 mRNA levels was found in cells treated with EVw/o (Figure 3E). On the contrary, a similar response to UCw/o was found for cells treated with Protw/o; inducing a down-regulation of the ECM components Acan and Col2a1, and an up-regulation of Dcn, Mmp3 and Mmp13 and Timp1 mRNA levels (Figure 3F,H).
Figure 3.
EV, EVw/o, Prot and Protw/o isolates effects on ATDC-5 chondrogenic cell line. (A,B) Lactate dehydrogenase activity was measured from cell culture media of ATDC-5 after 48 h of treatment with EV and EVw/o or Prot and Protw/o. Negative control (0%) was the culture media from cells treated with the vehicle control. Positive control (100%) was the culture media from cells treated with 1% Triton X-100. (C,D) Metabolic activity measured after 48 h of treatment. Data were normalized to the vehicle control group that was set to 100%. (E–H) mRNA expression levels of ECM components genes (Acan, Dcn, Col1a1 and Col2a1) or ECM turnover genes (Mmp3, Mmp13 and Timp1) of ATDC-5 after treatment with EV and EVw/o or Prot and Protw/o for 48 h. Data represent fold changes of target genes, normalized to reference genes (Rn18s and Gapdh) and relative to the group control that was set as 100%. Values presented in box and whisker plots represent the median, minima and maxima of two independent experiments (n = 11) Results in subfigures A,C–E,G and I were statistically compared by ANOVA and Bonferroni as post hoc for cytotoxicity, metabolic activity, Acan mRNA expression levels and total collagen deposition; and by Kruskal–Wallis for Dcn, Col2a1, Mmp3 and Mmp13 mRNA expression levels: * p < 0.05 versus the control; # p < 0.05 versus Prot. Results in subfigures B,D,F,H and J were statistically compared by ANOVA and Bonferroni as post hoc for Acan, Col1a1, Mmp3, Mmp13 and Timp1 mRNA expression levels; and by Kruskal–Wallis for Col2a1 mRNA expression levels: * p < 0.05 versus Control; # p < 0.05 versus EV.
4. Discussion
Over the last years, the interest on MSC-derived EVs has increased. EV isolation from MSCs requires a protocol that assures that these EVs can have a therapeutic use. Hence, it is important to decide whether to use or not EV-depleted FBS when isolating MSC derived EVs. Nevertheless, our results demonstrate that the purity of EV preparations is more important than the usage or not of EV-depleted FBS in determining their final effect in chondrocytes.
When isolating EVs from cell culture conditioned media, different procedures have been reported in the literature. Usually, non-confluent cells are incubated with culture medium for 48 h either with EV-depleted FBS or without FBS, though, as far as we are concerned, a direct comparison of the effect of EVs obtained by these two protocols has not been previously reported. On one hand, the use of FBS, even when having been previously EV-depleted, entails a risk of contamination with FBS-derived EVs; or when using FBS alternatives such as human platelet lysate, since it also contains EVs [30,31,45]. On the other hand, even if MISEV2018 [33] recommends serum-free cell culture to obtain EVs, using serum-free conditions increases the risk of contaminants due to cellular stress [34]. In fact, here, we show evidences that cells cultured without FBS supplementation need to be highly purified to avoid the effect of co-purified proteins that, in some cases, might be deleterious.
We observed a higher amount of protein content in UC, yielding a sample with a poor purity ratio. This finding was not surprising, since the proteins coming from the FBS supplement are expected to be in the medium. Interestingly, although the EVs of the UCw/o group showed higher purity and lower protein contamination, this group showed a deleterious effect on the chondrocyte cell line, down-regulating mRNA expression levels of ECM related-components and up-regulating mRNA expression levels of different matrix-metalloproteinases, thus this effect would lead to collagen degradation, and thus could contribute to tissue break-down. It has previously been reported that protein presence and composition in the neuroblastoma cells-derived EVs may be different when obtained from cells cultured in medium with EV-depleted FBS or medium without FBS [32]. Furthermore, contamination with proteins generally occurs when cells are cultured in serum-containing media but also in serum-free conditions, mainly because cells produce soluble proteins that may interfere in later analysis [51].
Some studies have shown that EVs harvested under stress conditions have a stress effect on non-stressed cells due to their cargo [52,53,54]. Thus, our first thought was that the UCw/o group had EVs with a cargo that was inducing the deleterious effect on chondrocytes. However, using ultracentrifugation as EV isolation method we could not discard the fact that stress inducing contaminants were co-precipitated with our EV sample. In fact, some studies showed that centrifugations at 100,000 × g form protein aggregates that co-precipitate with the EVs, leading to low purity samples, or even when harvesting the UC pellet some remaining supernatant, extremely rich in proteins, could be taken affecting the purity of the EV preparation. The yield obtained using ultracentrifugation is also low, and EVs can be damaged after centrifugation at that speed [44,55,56]. Even if ultracentrifugation has been considered for years the gold standard protocol, currently, extensive research on EVs has shown that other protocols for EV isolation allow us to obtain purer EVs and assure their usage in therapeutics. In view of our first results, we next performed SEC as a strategy to obtain purer EVs, as it was done in previous studies [44]. This technique allows the separation of EV fractions from protein fractions, thus obtaining purer and more functional EVs without altering their characteristics [41,47]. In fact, EV and EVw/o fractions showed lower levels of protein content compared to UC or UCw/o, yielding as well to purer EV samples. In addition, what is more important, the same effect on chondrogenic cells was observed for EV and EVw/o treatments, and, at the same time, the deleterious effect pattern caused by UCw/o appeared in cells treated with Protw/o. Thus, pointing that the deleterious effect is not caused by EVw/o cargo but instead by contamination with co-precipitated elements when they are isolated by ultracentrifugation, this contamination can be removed by using a purification method such as SEC. These results were unexpected taking into account previous reports isolating EVs from conditioned media using EV-depleted FBS by ultracentrifugation, which showed that the deleterious effects were caused by EV cargo and not by the presence of stressing proteins on the EV fraction [52,53,54].
5. Conclusions
Our results show that the negative effect on the chondrogenic cell line is due to the contaminant proteins co-isolated with the EVs by ultracentrifugation and not from the EVs themselves, highlighting the importance of working with well purified EV preparations to specifically achieve their therapeutic effect.
From the results obtained in this study we show evidences that cells cultured without FBS supplementation need to be highly purified to avoid the effect of co-purified proteins that, in some cases, might be deleterious.
Acknowledgments
The authors thank SCT-UIB for access to their facilities; Aina Marina Garau Ramon for her help during her final degree project; and Lucía Castaño Castaño for English proofreading of the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4409/9/2/422/s1, Figure S1: MSC profile by cytometry after conditioning for 48 h in proliferative media with 20% FBS. Figure S2: MSC profile by cytometry after conditioning for 48 h in medium with 20% EV-depleted FBS. Figure S3: MSC profile by cytometry after conditioning for 48 h in medium without FBS. Figure S4: Representative bright-field images of MSCs in culture under all conditions. Figure S5: EV markers on UC, UCw/o, EV, EVw/o, Prot and Protw/o samples and cell lysates. Figure S6: Presence of proteins and EV marker in SEC fractions collected from UC and UCw/o samples. Figure S7: Total protein quantification of SEC fractions collected from UC and UCw/o samples.
Author Contributions
M.A.F.-G., M.A.R., M.M. and J.M.R. contributed to the conception and design. J.C. and A.G. provided hUC-MSCs. M.A.F.-G. and M.A.R. produced EV samples, acquired and analyzed data. M.A.F.-G. wrote the main manuscripts text. M.A.F.-G., M.A.R., M.M. and J.M.R. contributed to interpretation of the results and M.A.F.-G., M.A.R., J.C., A.G., M.M. and J.M.R. reviewed the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by Instituto de Salud Carlos III, co-funded by the ESF European Social Fund and the ERDF European Regional Development Fund (grant number MS16/00124; CP16/00124), Ministerio de Economía y Competividad (IEDI-2017-00941), and the Direcció General d’Investigació, Conselleria d’Investigació, Govern Balear (grant number FPI/2046/2017).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Vizoso F.J., Eiro N., Cid S., Schneider J., Perez-Fernandez R. Mesenchymal Stem Cell Secretome: Toward Cell-Free Therapeutic Strategies in Regenerative Medicine. Int. J. Mol. Sci. 2017;18:1852. doi: 10.3390/ijms18091852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Caplan A.I., Correa D. The MSC: An Injury Drugstore. Cell Stem Cell. 2011;9:11–15. doi: 10.1016/j.stem.2011.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Ma S., Xie N., Li W., Yuan B., Shi Y., Wang Y. Immunobiology of mesenchymal stem cells. Cell Death Differ. 2014;21:216–225. doi: 10.1038/cdd.2013.158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Bennet L., Tan S., Heuij L.V.D., Derrick M., Groenendaal F., Van Bel F., Juul S., Back S.A., Northington F., Robertson N.J., et al. Cell therapy for neonatal hypoxia-ischemia and cerebral palsy. Ann. Neurol. 2012;71:589–600. doi: 10.1002/ana.22670. [DOI] [PubMed] [Google Scholar]
- 5.Chen J., Li Y., Katakowski M., Chen X., Wang L., Lu D., Lü M., Gautam S.C., Chopp M. Intravenous bone marrow stromal cell therapy reduces apoptosis and promotes endogenous cell proliferation after stroke in female rat. J. Neurosci. Res. 2003;73:778–786. doi: 10.1002/jnr.10691. [DOI] [PubMed] [Google Scholar]
- 6.Chimenti I., Smith R.R., Li T.-S., Gerstenblith G., Messina E., Giacomello A., Marbán E. Relative roles of direct regeneration versus paracrine effects of human cardiosphere-derived cells transplanted into infarcted mice. Circ. Res. 2010;106:971–980. doi: 10.1161/CIRCRESAHA.109.210682. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Katsha A.M., Ohkouchi S., Xin H., Kanehira M., Sun R., Nukiwa T., Saijo Y. Paracrine factors of multipotent stromal cells ameliorate lung injury in an elastase-induced emphysema model. Mol. Ther. 2011;19:196–203. doi: 10.1038/mt.2010.192. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Lee J.W., Fang X., Gupta N., Serikov V., Matthay M.A. Allogeneic human mesenchymal stem cells for treatment of E. coli endotoxin-induced acute lung injury in the ex vivo perfused human lung. Proc. Natl. Acad. Sci. USA. 2009;106:16357–16362. doi: 10.1073/pnas.0907996106. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Timmers L., Lim S.K., Arslan F., Armstrong J.S., Hoefer I.E., Doevendans P.A., Piek J.J., El Oakley R.M., Choo A., Lee C.N., et al. Reduction of myocardial infarct size by human mesenchymal stem cell conditioned medium. Stem Cell Res. 2008;1:129–137. doi: 10.1016/j.scr.2008.02.002. [DOI] [PubMed] [Google Scholar]
- 10.Gnecchi M., He H., Noiseux N., Liang O.D., Zhang L., Morello F., Mu H., Melo L.G., Pratt R.E., Ingwall J.S., et al. Evidence supporting paracrine hypothesis for Akt-modified mesenchymal stem cell-mediated cardiac protection and functional improvement. FASEB J. 2006;20:661–669. doi: 10.1096/fj.05-5211com. [DOI] [PubMed] [Google Scholar]
- 11.Gnecchi M., He H., Liang O.D., Melo L.G., Morello F., Mu H., Noiseux N., Zhang L., Pratt R.E., Ingwall J.S., et al. Paracrine action accounts for marked protection of ischemic heart by Akt-modified mesenchymal stem cells. Nat. Med. 2005;11:367–368. doi: 10.1038/nm0405-367. [DOI] [PubMed] [Google Scholar]
- 12.Caplan A.I., Dennis J.E. Mesenchymal stem cells as trophic mediators. J. Cell. Biochem. 2006;98:1076–1084. doi: 10.1002/jcb.20886. [DOI] [PubMed] [Google Scholar]
- 13.Heldring N., Mäger I., Wood M.J., Le Blanc K., Andaloussi S.E. Therapeutic Potential of Multipotent Mesenchymal Stromal Cells and Their Extracellular Vesicles. Hum. Gene Ther. 2015;26:506–517. doi: 10.1089/hum.2015.072. [DOI] [PubMed] [Google Scholar]
- 14.Bruno S., Grange C., Deregibus M.C., Calogero R.A., Saviozzi S., Collino F., Morando L., Busca A., Falda M., Bussolati B., et al. Mesenchymal stem cell-derived microvesicles protect against acute tubular injury. J. Am. Soc. Nephrol. 2009;20:1053–1067. doi: 10.1681/ASN.2008070798. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Lai R.C., Arslan F., Lee M.M., Sze N.S.K., Choo A., Chen T.S., Salto-Tellez M., Timmers L., Lee C.N., El Oakley R.M., et al. Exosome secreted by MSC reduces myocardial ischemia/reperfusion injury. Stem Cell Res. 2010;4:214–222. doi: 10.1016/j.scr.2009.12.003. [DOI] [PubMed] [Google Scholar]
- 16.Colombo M., Raposo G., Théry C. Biogenesis, Secretion, and Intercellular Interactions of Exosomes and Other Extracellular Vesicles. Annu. Rev. Cell Dev. Boil. 2014;30:255–289. doi: 10.1146/annurev-cellbio-101512-122326. [DOI] [PubMed] [Google Scholar]
- 17.Lai R.C., Yeo R.W.Y., Tan K.H., Lim S.K. Exosomes for drug delivery—A novel application for the mesenchymal stem cell. Biotechnol. Adv. 2013;31:543–551. doi: 10.1016/j.biotechadv.2012.08.008. [DOI] [PubMed] [Google Scholar]
- 18.Codispoti B., Marrelli M., Paduano F., Tatullo M. NANOmetric BIO-Banked MSC-Derived Exosome (NANOBIOME) as a Novel Approach to Regenerative Medicine. J. Clin. Med. 2018;7:357. doi: 10.3390/jcm7100357. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Marcus M.E., Leonard J.N. FedExosomes: Engineering Therapeutic Biological Nanoparticles that Truly Deliver. Pharm. 2013;6:659–680. doi: 10.3390/ph6050659. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Lener T., Gimona M., Aigner L., Börger V., Buzas E., Camussi G., Chaput N., Chatterjee D., Court F.A., Del Portillo H.A., et al. Applying extracellular vesicles based therapeutics in clinical trials–an ISEV position paper. J. Extracell. Vesicles. 2015;4:30087. doi: 10.3402/jev.v4.30087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Näslund T.I., Gehrmann U., Qazi K.R., Karlsson M.C.I., Gabrielsson S. Dendritic Cell–Derived Exosomes Need To Activate Both T and B Cells To Induce Antitumor Immunity. J. Immunol. 2013;190:2712–2719. doi: 10.4049/jimmunol.1203082. [DOI] [PubMed] [Google Scholar]
- 22.Lugini L., Cecchetti S., Huber V., Luciani F., Macchia G., Spadaro F., Paris L., Abalsamo L., Colone M., Molinari A., et al. Immune Surveillance Properties of Human NK Cell-Derived Exosomes. J. Immunol. 2012;189:2833–2842. doi: 10.4049/jimmunol.1101988. [DOI] [PubMed] [Google Scholar]
- 23.Beauvillain C., Ruiz S., Guiton R., Bout D., Dimier-Poisson I. A vaccine based on exosomes secreted by a dendritic cell line confers protection against T. gondii infection in syngeneic and allogeneic mice. Microbes Infect. 2007;9:1614–1622. doi: 10.1016/j.micinf.2007.07.002. [DOI] [PubMed] [Google Scholar]
- 24.Beauvillain C., Juste M.O., Dion S., Pierre J., Dimier-Poisson I. Exosomes are an effective vaccine against congenital toxoplasmosis in mice. Vaccine. 2009;27:1750–1757. doi: 10.1016/j.vaccine.2009.01.022. [DOI] [PubMed] [Google Scholar]
- 25.Herrera M.B., Fonsato V., Gatti S., Deregibus M.C., Sordi A., Cantarella D., Calogero R., Bussolati B., Tetta C., Camussi G. Human liver stem cell-derived microvesicles accelerate hepatic regeneration in hepatectomized rats. J. Cell Mol. Med. 2010;14:1605–1618. doi: 10.1111/j.1582-4934.2009.00860.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Tan C.Y., Lai R.C., Wong W., Dan Y.Y., Lim S.-K., Ho H.K. Mesenchymal stem cell-derived exosomes promote hepatic regeneration in drug-induced liver injury models. Stem Cell Res. Ther. 2014;5:76. doi: 10.1186/scrt465. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Li T., Yan Y., Wang B., Qian H., Zhang X., Shen L., Wang M., Zhou Y., Zhu W., Li W., et al. Exosomes derived from human umbilical cord mesenchymal stem cells alleviate liver fibrosis. Stem Cells Dev. 2013;22:845–854. doi: 10.1089/scd.2012.0395. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Xin H., Li Y., Cui Y., Yang J.J., Zhang Z.G., Chopp M. Systemic Administration of Exosomes Released from Mesenchymal Stromal Cells Promote Functional Recovery and Neurovascular Plasticity After Stroke in Rats. Br. J. Pharmacol. 2013;33:1711–1715. doi: 10.1038/jcbfm.2013.152. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Van Der Valk J., Mellor D., Brands R., Fischer R., Gruber F., Gstraunthaler G., Hellebrekers L., Hyllner J., Jonker F.H., Prieto P., et al. The humane collection of fetal bovine serum and possibilities for serum-free cell and tissue culture. Toxicol. Vitr. 2004;18:1–12. doi: 10.1016/j.tiv.2003.08.009. [DOI] [PubMed] [Google Scholar]
- 30.Shelke G.V., Lässer C., Gho Y.S., Lötvall J. Importance of exosome depletion protocols to eliminate functional and RNA-containing extracellular vesicles from fetal bovine serum. J. Extracell. Vesicles. 2014;3:24783. doi: 10.3402/jev.v3.24783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Torreggiani E., Perut F., Roncuzzi L., Zini N., Baglìo S.R., Baldini N. Exosomes: Novel effectors of human platelet lysate activity. Eur. Cell Mater. 2014;28:137–151. doi: 10.22203/eCM.v028a11. [DOI] [PubMed] [Google Scholar]
- 32.Li J., Lee Y., Johansson H.J., Mäger I., Vader P., Nordin J.Z., Wiklander O.P.B., Lehtiö J., Wood M.J.A., El Andaloussi S. Serum-free culture alters the quantity and protein composition of neuroblastoma-derived extracellular vesicles. J. Extracell. Vesicles. 2015;4:26883. doi: 10.3402/jev.v4.26883. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Théry C., Witwer K.W., Aikawa E., Alcaraz M.J., Anderson J.D., Andriantsitohaina R., Antoniou A., Arab T., Archer F., Atkin-Smith G.K., et al. Minimal information for studies of extracellular vesicles 2018 (MISEV2018): A position statement of the International Society for Extracellular Vesicles and update of the MISEV2014 guidelines. J. Extracell. Vesicles. 2018;7:1535750. doi: 10.1080/20013078.2018.1535750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Yamashita T., Takahashi Y., Takakura Y. Possibility of Exosome-Based Therapeutics and Challenges in Production of Exosomes Eligible for Therapeutic Application. Boil. Pharm. Bull. 2018;41:835–842. doi: 10.1248/bpb.b18-00133. [DOI] [PubMed] [Google Scholar]
- 35.Witwer K.W., Buzás E.I., Bemis L.T., Bora A., Lässer C., Lötvall J., Hoen E.N.N.-T., Piper M.G., Sivaraman S., Skog J., et al. Standardization of sample collection, isolation and analysis methods in extracellular vesicle research. J. Extracell. Vesicles. 2013;2:18389. doi: 10.3402/jev.v2i0.20360. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kowal J., Arras G., Colombo M., Jouve M., Morath J.P., Primdal-Bengtson B., Dingli F., Loew D., Tkach M., Théry C. Proteomic comparison defines novel markers to characterize heterogeneous populations of extracellular vesicle subtypes. Proc. Natl. Acad. Sci. USA. 2016;113:E968–E977. doi: 10.1073/pnas.1521230113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Harting M.T., Srivastava A.K., Zhaorigetu S., Bair H., Prabhakara K.S., Toledano Furman N.E., Vykoukal J.V., Ruppert K.A., Cox C.S., Olson S.D. Inflammation-Stimulated Mesenchymal Stromal Cell-Derived Extracellular Vesicles Attenuate Inflammation. Stem Cells. 2018;36:79–90. doi: 10.1002/stem.2730. [DOI] [PubMed] [Google Scholar]
- 38.Momen-Heravi F. Isolation of Extracellular Vesicles by Ultracentrifugation. Adv. Struct. Safety Stud. 2017;1660:25–32. doi: 10.1007/978-1-4939-7253-1_3. [DOI] [PubMed] [Google Scholar]
- 39.Konoshenko M.Y., Lekchnov E.A., Vlassov A.V., Laktionov P.P. Isolation of Extracellular Vesicles: General Methodologies and Latest Trends. BioMed Res. Int. 2018;2018:8545347. doi: 10.1155/2018/8545347. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Lozano-Ramos I., Bancu I., Oliveira-Tercero A., Armengol M.P., Menezes-Neto A., Del Portillo H.A., Lauzurica-Valdemoros R., Borràs F.E. Size-exclusion chromatography-based enrichment of extracellular vesicles from urine samples. J. Extracell. Vesicles. 2015;4:27369. doi: 10.3402/jev.v4.27369. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Gámez-Valero A., Monguió-Tortajada M., Carreras-Planella L., Franquesa M., Beyer K., Borràs F.E. Size-Exclusion Chromatography-based isolation minimally alters Extracellular Vesicles’ characteristics compared to precipitating agents. Sci. Rep. 2016;6:33641. doi: 10.1038/srep33641. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Takov K., Yellon D.M., Davidson S.M. Comparison of small extracellular vesicles isolated from plasma by ultracentrifugation or size-exclusion chromatography: Yield, purity and functional potential. J. Extracell. Vesicles. 2019;8:1560809. doi: 10.1080/20013078.2018.1560809. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Freitas D., Balmaña M., Poças J., Campos D., Osório H., Konstantinidi A., Vakhrushev S.Y., Magalhães A., Reis C.A. Different isolation approaches lead to diverse glycosylated extracellular vesicle populations. J. Extracell. Vesicles. 2019;8:1621131. doi: 10.1080/20013078.2019.1621131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Rood I.M., Deegens J.K., Merchant M.L., Tamboer W.P., Wilkey D.W., Wetzels J.F., Klein J.B. Comparison of three methods for isolation of urinary microvesicles to identify biomarkers of nephrotic syndrome. Kidney Int. 2010;78:810–816. doi: 10.1038/ki.2010.262. [DOI] [PubMed] [Google Scholar]
- 45.Karp N.A., Lilley K.S. Investigating sample pooling strategies for DIGE experiments to address biological variability. Proteom. 2009;9:388–397. doi: 10.1002/pmic.200800485. [DOI] [PubMed] [Google Scholar]
- 46.Dominici M., Le Blanc K., Mueller I., Slaper-Cortenbach I., Marini F., Krause D., Deans R., Keating A., Prockop D., Horwitz E. Minimal criteria for defining multipotent mesenchymal stromal cells. The International Society for Cellular Therapy position statement. Cytotherapy. 2006;8:315–317. doi: 10.1080/14653240600855905. [DOI] [PubMed] [Google Scholar]
- 47.Forteza-Genestra M.A., Antich-Rosselló M., Monjo M., Ramis J.M. Use of Extracellular Vesicles for Cell-Free Regenerative Medicine in Osteochondral and Bone-Related Therapies. Biomed. J. Sci. Tech. Res. 2018;3:36–38. doi: 10.26717/BJSTR.2018.03.000888. [DOI] [Google Scholar]
- 48.Böing A.N., Van Der Pol E., Grootemaat A.E., Coumans F.A.W., Sturk A., Nieuwland R. Single-step isolation of extracellular vesicles by size-exclusion chromatography. J. Extracell. Vesicles. 2014;3:42. doi: 10.3402/jev.v3.23430. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Webber J., Clayton A. How pure are your vesicles? J. Extracell. Vesicles. 2013;2:1–6. doi: 10.3402/jev.v2i0.19861. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Rubert M., Ramis J.M., Vondrasek J., Gayá A., Lyngstadaas S.P., Monjo M. Synthetic Peptides Analogue to Enamel Proteins Promote Osteogenic Differentiation of MC3T3-E1 and Mesenchymal Stem Cells. J. Biomater. Tissue Eng. 2011;1:198–209. doi: 10.1166/jbt.2011.1018. [DOI] [Google Scholar]
- 51.Coumans F.A., Brisson A.R., Buzas E.I., Dignat-George F., Drees E.E., El-Andaloussi S., Emanueli C., Gasecka A., Hendrix A., Hill A.F., et al. Methodological Guidelines to Study Extracellular Vesicles. Circ. Res. 2017;120:1632–1648. doi: 10.1161/CIRCRESAHA.117.309417. [DOI] [PubMed] [Google Scholar]
- 52.Bewicke-Copley F., Mulcahy L.A., Jacobs L.A., Samuel P., Akbar N., Pink R.C., Carter D.R.F. Extracellular vesicles released following heat stress induce bystander effect in unstressed populations. J. Extracell. Vesicles. 2017;6:1340746. doi: 10.1080/20013078.2017.1340746. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.De Jong O.G., Verhaar M.C., Chen Y., Vader P., Gremmels H., Posthuma G., Schiffelers R.M., Gucek M., van Balkom B.W.M. Cellular stress conditions are reflected in the protein and RNA content of endothelial cell-derived exosomes. J. Extracell. Vesicles. 2012;1:1–12. doi: 10.3402/jev.v1i0.18396. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Collett G.P., Redman C.W., Sargent I.L., Vatish M. Endoplasmic reticulum stress stimulates the release of extracellular vesicles carrying danger-associated molecular pattern (DAMP) molecules. Oncotarget. 2018;9:6707–6717. doi: 10.18632/oncotarget.24158. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Bard M.P., Hegmans J.P., Hemmes A., Luider T.M., Willemsen R., Severijnen L.-A.A., Van Meerbeeck J.P., Burgers S.A., Hoogsteden H.C., Lambrecht B.N. Proteomic Analysis of Exosomes Isolated from Human Malignant Pleural Effusions. Am. J. Respir. Cell Mol. Boil. 2004;31:114–121. doi: 10.1165/rcmb.2003-0238OC. [DOI] [PubMed] [Google Scholar]
- 56.Mol E.A., Goumans M.-J., Doevendans P.A., Sluijter J.P., Vader P. Higher functionality of extracellular vesicles isolated using size-exclusion chromatography compared to ultracentrifugation. Nanomed. Nanotechnol. Boil. Med. 2017;13:2061–2065. doi: 10.1016/j.nano.2017.03.011. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.