Abstract
Chronic kidney disease (CKD) is an important public health problem in the world. The aim of our research was to identify novel potential serum biomarkers of renal injury. ELISA assay showed that cytokines and chemokines IL-1β, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17, Eotaxin, FGFb, G-CSF, GM-CSF, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-1bb, RANTES, TNF-α and VEGF were significantly higher (R > 0.6, p value < 0.05) in the serum of patients with CKD compared to healthy subjects, and they were positively correlated with well-established markers (urea and creatinine). The multiple reaction monitoring (MRM) quantification method revealed that levels of HSP90B2, AAT, IGSF22, CUL5, PKCE, APOA4, APOE, APOA1, CCDC171, CCDC43, VIL1, Antigen KI-67, NKRF, APPBP2, CAPRI and most complement system proteins were increased in serum of CKD patients compared to the healthy group. Among complement system proteins, the C8G subunit was significantly decreased three-fold in patients with CKD. However, only AAT and HSP90B2 were positively correlated with well-established markers and, therefore, could be proposed as potential biomarkers for CKD.
Keywords: chronic kidney disease, inflammation, cytokines, biomarkers, 2D-DIGE, multiple reaction monitoring
1. Introduction
Chronic kidney disease (CKD) is defined by a gradual loss of kidney function for more than three months. CKD remains an increasing public health threat affecting up to 16% of the global population [1]. According to the 2016 Global Burden of Disease study, CKD caused 16.9 deaths per 100,000 in 2016 [2]. CKD is characterized by progressive kidney damage due to renal circulatory impairment, which is common for patients diagnosed with diabetes and hypertension. The treatment is not specific and aimed to improve the underlying condition as well as to manage complications.
The initial symptoms of CKD are not specific, which prevents early diagnosis. Diagnosis of CKD is based on the calculation of glomerular filtration rate (GFR), detection of albuminuria and a kidney biopsy [3,4]. These diagnostic methods are often used at the late stage of disease, when kidney damage is advanced and often irreversible. Therefore, identification of biomarkers for early diagnosis of CKD is important. Several markers of kidney inflammation were proposed for CKD diagnosis including plasma and urine interleukins and chemokines (IL-18, IL-10, IL-6, MCP-1, FGF-23) [5,6]. Also, to improve predicting CKD prognosis, the biomarkers of renal tubular injury NGAL, KIM-1, TIMP-2 and IGFBP-7 have been shown to be especially promising [7,8]. Development of novel genetic tests and identification of genomic biomarkers can also improve the management of CKD by facilitating early diagnosis and advancing patient management [9,10]. Nevertheless, investigation of novel and effective biomarkers for early CKD diagnosis remains an important aim.
Changes in blood proteins are commonly monitored to diagnose the disease, mainly because of their easy access. Also, changes in blood components can be detected in the early stage of the disease. This is especially true in CKD, where changes in kidney filtration result in significant alterations in blood chemistry. For example, increased serum creatinine and urea levels, electrolyte imbalance and increased blood coagulation are found in CKD patients. Therefore, we suggest that potential CKD biomarkers might be identified in patient serum.
Previous studies of novel, early-stage CKD biomarkers have used different proteomic approaches [11,12,13,14]. We applied a two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) approach to separate depleted serum proteins in CKD and control serum. High-abundance proteins were deleted to improve visualization of protein spots. After differentially expressed proteins were identified, these proteins were quantified with multiple reaction monitoring (MRM) in the serum samples of patients (n = 26) and control groups (n = 10).
2. Materials and Methods
2.1. Subjects
Serum samples from 26 adult patients with terminal stage CKD (18 male and 8 female; age 53.0 ± 16.2 y) and 10 control healthy subjects older than 18 y (4 male and 6 female; age 29 ± 6.7 y) were used in this study. CKD patients not treated with hemodialysis were recruited from the Department of Nephrology, Republican Clinical Hospital Ministry of Health Republic of Tatarstan. The diagnosis of CKD was confirmed by clinical tests (eGFR < 15 mL/min/1.73 m2, protein in urea > 0.033 g/L, serum creatinine > 115 mmol/L, etc.). Baseline clinical characteristics and laboratory data of the study population are summarized in Table 1. Serum samples were stored at −80 °C until they were used.
Table 1.
Characteristics of the study population.
| Parameters * | CKD Patients |
|---|---|
| Age (y) | 53.0 ± 16.2 |
| Sex (males/females) | 18/8 |
| eGFR (mL/min/1.73 m2) | 14.7 ± 3.1 |
| Diagnosis of CKD (number of patients) | 26 |
| Chronic glomerulonephritis | 18 |
| Diabetes | 3 |
| Chronic gouty nephropathy | 2 |
| Others or unknown | 3 |
| Body mass index (kg/m2) | 25.8 ± 5.6 |
| Systolic blood pressure (mmHg) | 128.7 ± 23.2 |
| Diastolic blood pressure (mmHg) | 79.2 ± 18.3 |
| Creatinine (mg/dL) | 7.25 ± 3.8 |
| Urea (mg/dL) | 135.0 ± 70.2 |
| Albumin (g/dL) | 3.8 ± 0.5 |
| Total cholesterol (mg/dL) | 177.8 ± 40.2 |
| HDL cholesterol (mg/dL) | 52.1 ± 28.4 |
| LDL cholesterol (mg/dL) | 110.6 ± 45.6 |
| Triglyceride (mg/dL) | 141.9 ± 50.1 |
* Abbreviations: CKD—chronic kidney disease; eGFR—estimated glomerular filtration rate; HDL—high-density lipoprotein; LDL—low-density lipoprotein.
2.2. Ethics Statement
The Ethics Committee of the Kazan Federal University approved this study (protocol N4/09), and informed written consent was obtained from each patient and control according to the Guidelines approved under this Protocol (article 20, Federal Law “Protection of Health Right of Citizens of Russian Federation” N323- FZ, 11.21.2011).
2.3. 2D-DIGE of Serum Proteins after Depletion
Unspecific depletion of high-abundance proteins in serum samples (200 μL) was done using a commercially available kit, ProteoMiner (Bio-Rad, Hercules, CA, USA), according to the manufacturer’s instructions with a minor modification. Briefly, 200 μL of serum was loaded into the prepared spin column with protein-binding sorbent, incubated for 2 h at room temperature, and then washed with PBS (150 mM NaCl, 10 mM NaH2PO4, pH 7.4) and water. Finally, the protein was eluted using a denaturing buffer (8 М urea, 10 mМ Tris-HCl, рН 8.5, 3% CHAPS (3-[(3-Cholamidopropyl)dimethylammonio]-1-propanesulfonate hydrate), 2% NP40 (nonylphenol ethoxylate)) and used for 2D-DIGE.
Serum proteins were separated using 2D-DIGE as previously described [15]. Isoelectric focusing (IEF) was done with commercially available 17 cm IPG (immobilized pH gradient) strips in the pH range 3–10 (Bio-Rad, Hercules, CA, USA) according to the manufacturer’s instructions. We used 150 μg of each protein sample to label with 0.4 mM cyanine dyes Cy3 or Cy5 (Lumiprobe, Moscow, Russia) before IEF. The samples were mixed, diluted in the rehydration buffer (8 М urea, 10 mМ Tris-HCl (рН 8.5), 3% CHAPS, 2% NP40, 50 mM DTT (dithiothreitol), 0.4% Pharmalyte, pH range 3–10 (GE Healthcare, Chicago, IL, USA)) and loaded into the strip for active rehydration for 12 h. Isoelectric focusing was carried out using the Protean i12 IEF cell (Bio-Rad, Hercules, CA, USA). After IEF, the strips were incubated for 10 min in the equilibration buffer (6 M urea, 0.375 M Tris-HCl, pH 8.8, 2% SDS (sodium dodecyl sulfate)), containing 2% DTT, and for 10 min in the same equilibration buffer, containing 2.5% iodoacetamide. Second-dimension electrophoresis was performed on self-cast gradient 9%–16% polyacrylamide gels. Gels were scanned with 100 μm pixel resolution using the Typhoon FLA 9500 scanner (GE Healthcare, Chicago, IL, USA). Then, the gels were stained with silver nitrate [16].
2.4. Protein Identification
Collected protein spots were destained using ferricyanide-thiosulfate as previously described [17] with some modifications. Briefly, gel pieces were washed with Milli-Q water and destained using a 30 mM potassium ferricyanide and 100 mM sodium thiosulfate solution for 5 min. Then, the gel pieces were rinsed three times with Milli-Q water, dehydrated using acetonitrile, and digested overnight (0.1% trypsin (MS grad, Promega, Madison, WI, USA) at 37 °C). Peptides were extracted using 0.2% trifluoroacetic acid (TFA) followed by incubation in 100% acetonitrile (ACN) for 30 min. Collected peptides were dried at 45 °C for 2 h, dissolved in 5 μL of 0.2% TFA and analyzed by a MALDI-TOF mass spectrometer Ultraflex (Bruker Daltonics GmbH, Bremen, Germany). The spectra were recorded in positive reflector mode from 700 to 3500 m/z. Peptide mass fingerprinting (PMF) was performed using the MASCOT software for searching matches in Swiss-Prot and NCBI databases.
2.5. Preparation of Serum Tryptic Digests
Tryptic digestion was carried out manually as previously described [18] with some modifications. Serum (8 μL) was diluted in 196.5 μL of 25 mM ammonium bicarbonate before protein denature by adding 30 μL of 10% sodium deoxycholate. Then, samples were reduced (26.1 μL of 50 mM tris(2-carboxyethyl)phosphine (TCEP) at 60 °C for 30 min) and alkylated (29.0 μL of 100 mM iodoacetamide; at 37 °C for 30 min in the dark). The remaining iodoacetamide was quenched by adding 29.0 μL of 100 mM DTT (37 °C; 30 min). Each sample was digested with 4 μL of 0.1% trypsin (MS grad, Promega, Madison, WI, USA) at 37 °C for 16 h. Digestion was quenched using formic acid at a final concentration 0.5% v/v. Samples were centrifuged (10 min at 10000× g), and supernatants were desalted and concentrated using solid phase extraction (Discovery DSC-18 (50 mg) cartridges (Supelco, Bellefonte, PA, USA)) according to the manufacturer’s protocol. Eluted peptides were dried at 45 °C for several hours and rehydrated (40 μL of 0.1% v/v formic acid in water/acetonitrile (95/5)) prior to LC-MRM/MS analysis.
2.6. LC-MRM/MS Analysis of Serum Digests
Peptides (10 μL) were separated in a reversed phase analytical column (100 × 2.1 mm i.d., Titan C18, 1.9 μm particle size (Supelco, Bellefonte, PA, USA)) with an Agilent 1290 Infinity UHPLC system coupled to a QTRAP 6500 (AB Sciex, Darmstadt, Germany) mass spectrometer. Proteins were separated using 400 μL/min flow rate and a gradient from 5%–95% mobile phase B, temperature 40 °C and a 25 min total run time. Mobile phase A consisted of 95% 0.1% v/v formic acid in water and 5% ACN, and mobile phase B consisted of 95% ACN and 5% 0.1% formic acid in water. The linear gradients were as follows (time: % B): 0.3 min: 5% B; 17 min: 40% B; 18 min: 95% B; 21.5 min: 95% B; 23 min: 5% B; 25 min: 95% B. All acquisition methods used the following parameters: 5200 V capillary voltage; source type Turbo Spray Ion Drive with temperature 500 °C; curtain gas 35 psi; declustering potential 51 V; collision energy was automatically optimized for each transition; flow rate 0.4 mL/min. Mass spectrometric data were analyzed using MultiQuant 3.0.2 software (AB Sciex, Darmstadt, Germany).
Skyline 3.6.0 [19] was used to generate precursor/fragment ion pairs, so-called MRM transitions, in silico [20]. The following options were selected: the peptide length was set to 8–25 amino acids, no post-translational modification (PTM) and one missed cleavage was allowed. In addition, two or three charge states of peptides were chosen for further MRM experiments. At least two peptides were chosen for identification of the target protein (Supplementary Table S1). The MRM method included at least three MRM transitions per peptide to select the best transition. Data analysis was done, and the areas for all the transitions were calculated using the Analyst 1.6.2 and MultiQuant 3.0.2 software (AB Sciex, Darmstadt, Germany). Used peptides with unique sequences and scheduled MRM transitions are given in Table 2.
Table 2.
Multiple reaction monitoring (MRM) quantification proteins list.
| n/n | Protein | UniProt Accession Number | Target Peptide Sequence | MRM Transition Q1 | MRM Transition Q3 | Product Ion |
|---|---|---|---|---|---|---|
| 1 | Immunoglobulin superfamily member 22 (IGSF22) | Q8N9C0 | EDSGLILLK | 494.3 | 743.5 | y7 |
| 2 | T-complex protein 1 subunit delta (CCT4) | P50991 | LVIEEAER | 479.8 | 655.4 | b6 |
| 3 | Cullin-5 (CUL5) | Q93034 | EAFQDDPR | 489.2 | 777.4 | y6 |
| 4 | Apolipoprotein A-IV (APOA4) | P06727 | LAPLAEDVR | 492.3 | 589.3 | y5 |
| 5 | Apolipoprotein E (APOE) | P02649 | LGPLVEQGR | 484.8 | 588.3 | y5 |
| 6 | Apolipoprotein A-I (APOA1) | P02647 | QGLLPVLESFK | 615.9 | 819.5 | y7 |
| 7 | Coiled-coil domain-containing protein 43 (CCDC43) | Q96MW1 | LEALGVDR | 436.7 | 446.2 | y4 |
| 8 | Coiled-coil domain-containing protein 171 (CCDC171) | Q6TFL3 | TLQEALEK | 466.3 | 830.5 | y7 |
| 9 | Putative endoplasmin-like protein (HSP90B2) | Q58FF3 | FDDSEK | 370.7 | 478.2 | y4 |
| 10 | Plasminogen (PLG) | P00747 | LSSPAVITDK | 515.8 | 769.4 | b8 |
| 11 | Phospholipase B1 (PLB1) | Q6P1J6 | TETLDLR | 424.2 | 445.2 | b4 |
| 12 | LIM and cysteine-rich domains protein 1 (LMCD1) | Q9NZU5 | YSTLTAR | 406.2 | 465.2 | b4 |
| 13 | Alpha-1-antitrypsin (AAT) | P01009 | LSITGTYDLK | 555.8 | 797.4 | y7 |
| 14 | Villin-1 (VIL1) | P09327 | AFEVPAR | 395.2 | 442.3 | y4 |
| 15 | NF-kappa-B-repressing factor (NKRF) | O15226 | EIPPADIPK | 490.3 | 736.4 | b7 |
| 16 | Amyloid protein-binding protein 2 (APPBP2) | Q92624 | VVVDVLR | 400.3 | 700.4 | y6 |
| 17 | Serine/threonine-protein phosphatase with EF-hands 2 (PPEF2) | O14830 | SLPSSPLR | 428.7 | 472.3 | y4 |
| 18 | Ras GTPase-activating protein 4 (CAPRI) | O43374 | DELDLQR | 444.7 | 531.3 | y4 |
| 19 | Cytoskeleton-associated protein 2-like (CKAP2L) | Q8IYA6 | QFVGETQSR | 526.3 | 776.4 | y7 |
| 20 | Protein kinase C epsilon type (PKCE) | Q02156 | QINQEEFK | 518.2 | 613.2 | b5 |
| 21 | Antigen KI-67 | P46013 | EDSTADDSK | 484.1 | 504.1 | b5 |
| 22 | Complement factor H (CFH) | P08603 | NGFYPATR | 463.2 | 607.3 | y5 |
| 23 | Complement C4 (C4A, C4B) |
P0C0L4 P0C0L5 |
LTSLSDR | 396.2 | 577.3 | y5 |
| 24 | Ficolin-3 (FCN3) | O75636 | VELEDFNGNR | 596.8 | 722.3 | y6 |
| 25 | C4B-binding protein alpha chain (C4BPA) | P04003 | TWYPEVPK | 510.3 | 569.3 | y5 |
| 26 | Complement C1R subcomponent (C1R) | P00736 | GGGALLGDR | 408.2 | 460.3 | y4 |
| 27 | Complement C1S subcomponent (C1S) | P09871 | LLEVPEGR | 456.8 | 686.3 | y6 |
| 28 | Complement C1q subcomponent subunit C (C1QC) | P02747 | FQSVFTVTR | 542.8 | 623.4 | y5 |
| 29 | Complement С3 (C3) | P01024 | IWDVVEK | 444.7 | 474.3 | y4 |
| 30 | Complement С5 (C5) | P01031 | GTVYNYR | 436.7 | 452.2 | y3 |
| 31 | Complement component C8 alpha chain (C8A) | P07357 | STITYR | 370.7 | 552.3 | y4 |
| 32 | Complement component C8 beta chain (C8B) | P07358 | EYESYSDFER | 662.8 | 672.3 | b5 |
| 33 | Complement component C8 gamma chain (C8G) | P07360 | QLYGDTGVLGR | 589.8 | 678.3 | b6 |
| 34 | Complement С9 (С9) | P02748 | VVEESELAR | 516.3 | 833.4 | y7 |
| 35 | Mannose-binding protein C (MBL2) | P11226 | NAAENGAIQNLIK | 678.4 | 869.4 | b9 |
| 36 | Mannan-binding lectin serine protease 2 (MASP2) | O00187 | WPEPVFGR | 494.3 | 609.3 | b5 |
| 37 | Galectin-3 (Gal-3) | P17931 | LDNNWGR | 437.7 | 671.3 | y6 |
| 38 | Galectin-3-binding protein (M2BP) | Q08380 | VEIFYR | 413.7 | 727.4 | y5 |
2.7. Analysis of Serum Levels of Cytokines
Quantitative analyses of cytokines (IL-1β, IL-2Rα, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17), Eotaxin, basic fibroblast growth factor (FGFb), granulocyte colony-stimulating factor (G-CSF), granulocyte macrophage colony-stimulating factor (GM-CSF), interferon gamma (INFγ), interferon gamma-induced protein 10 (IP-10), mast cell proteinase-1 (MCP-1), macrophage inflammatory protein 1-alpha (MIP-1α), macrophage inflammatory protein 1-beta (MIP-1 β), platelet-derived growth factor with two B subunits (PDGF-1bb), chemokine RANTES, tumor necrosis factor beta (TNF-β) and vascular endothelial growth factor (VEGF) in blood serum were performed using the multiplex analyzer Bio-Plex200 System (BioRad, Hercules, CA, USA) and “Bio-Plex Pro™ Human Cytokine 27-plex Assay” kit (BioRad, Hercules, CA, USA) according to the manufacturer‘s recommendations.
2.8. Statistical Analysis
Statistical analysis of the multiple reaction monitoring (MRM) data and ELISA data were performed in the R environment [21]. Statistically significant differences between groups of patients and healthy individuals were accepted as p < 0.05, assessed by the Wilcoxon rank sum test with Benjamini–Hochberg adjustment for MRM data and p < 0.01 for ELISA data. Correlations between the concentrations of serum proteins, cytokines, urea and creatinine were analyzed using the R Hmisc package (based on Spearman’s rank correlation coefficient).
3. Results
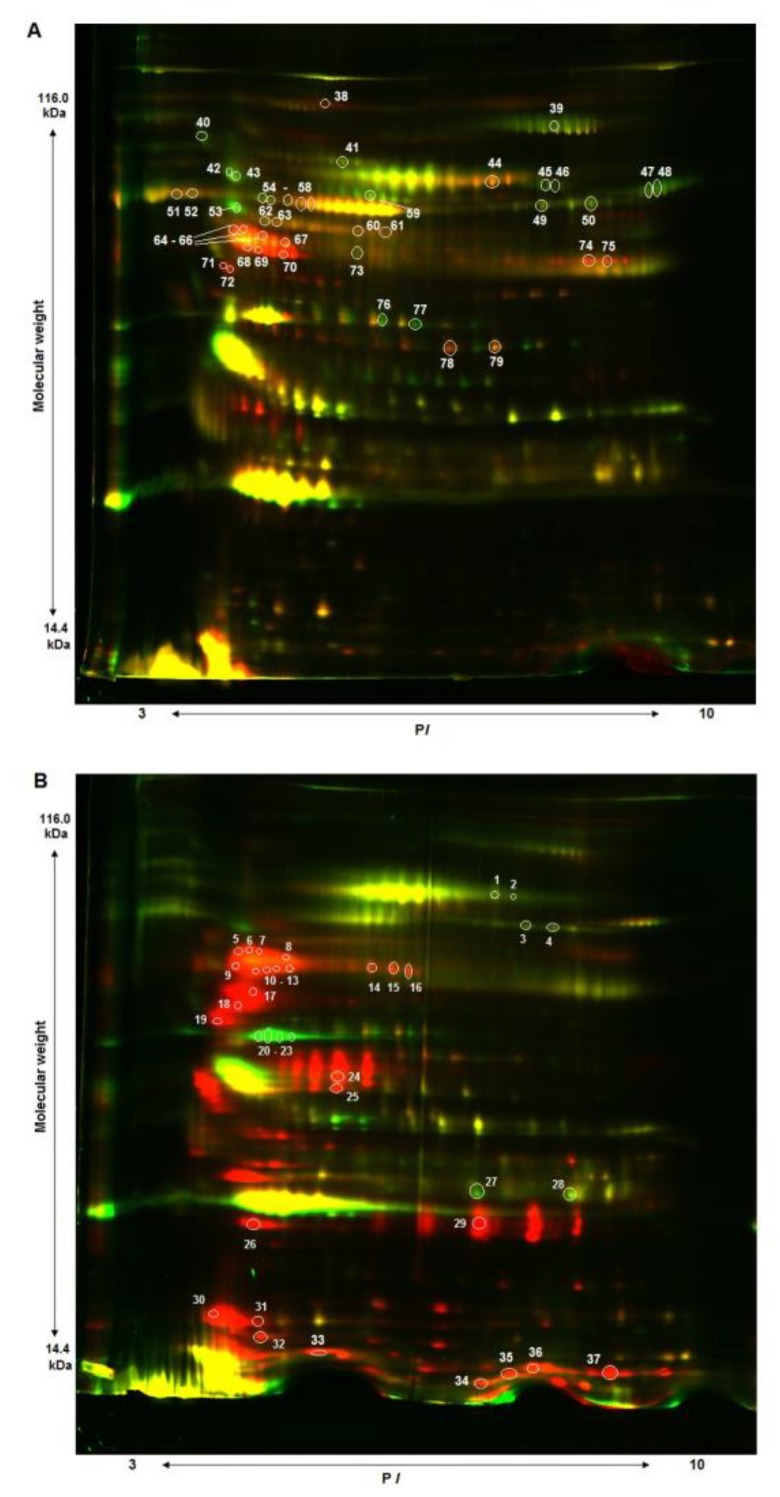
Two-dimensional fluorescence difference gel electrophoresis (2D-DIGE) is a convenient method to identify differences in protein profiles of the samples compared; however, it imposes a number of requirements on an analyte. In particular, successful separation and visualization of protein spots in gel depend upon concentrations of the proteins analyzed. The serum proteome has a dynamic range of more than ten orders of magnitude; thus, there is an excess of major proteins of albumin (more than 60% of the total amount of proteins), alpha-, beta- and gamma-globulin fractions (over 30%), with the rest being less than 10% of the total number of serum proteins [4,22,23]. Prior to 2D gel-electrophoresis, samples were depleted with the use of a commercial “ProteoMiner” kit (Bio-Rad) in order to remove major proteins. As can be seen in Figure 1, removal of most of the major fraction facilitated better blood serum protein separation. Protein spots with different expression levels between two patients and two healthy subjects were cut off and identified; these data are presented in Supplementary Table S2. Some proteins were presented by several spots on 2D gel, probably being isoforms of the same protein.
Figure 1.
2D-DIGE protein profiles of depleted serum of patients with chronic kidney disease (CKD; red fluorescent dye) and healthy individuals (green fluorescent dye). (A) serum of two women, 30 and 35 y. (B) serum of two men, 34 and 32 y.
Fifty-six unique, differentially expressed proteins between CKD patients and healthy subjects were identified with MALDI mass spectrometry, and we surmised there might be potential diagnostic markers of early CKD stages among these proteins. To test this hypothesis, 20 proteins with differential expression were quantified in the serum of CKD patients (n = 26) and healthy volunteers (n = 10) using MRM (multiple reaction monitoring). At present, MRM is an advanced method of mass spectrometry and allows simultaneous quantitation of numerous protein concentrations by the signal intensity of daughter ions, being fragments of known parent peptides [24]. For this purpose, we analyzed amino acid sequences of chosen proteins and selected relevant peptides to quantify proteins of interest using Skyline software (Table 2).
Two apolipoproteins, APOA1 and APOA4, were included in the list of proteins selected for MRM, with APOE added as a component that plays an important role in lipid metabolism and is associated with impaired hemodialysis and renal transplant functioning. Moreover, 18 complement system proteins were added into the expanded list, as they were directly involved in inflammatory reactions (Table 2).
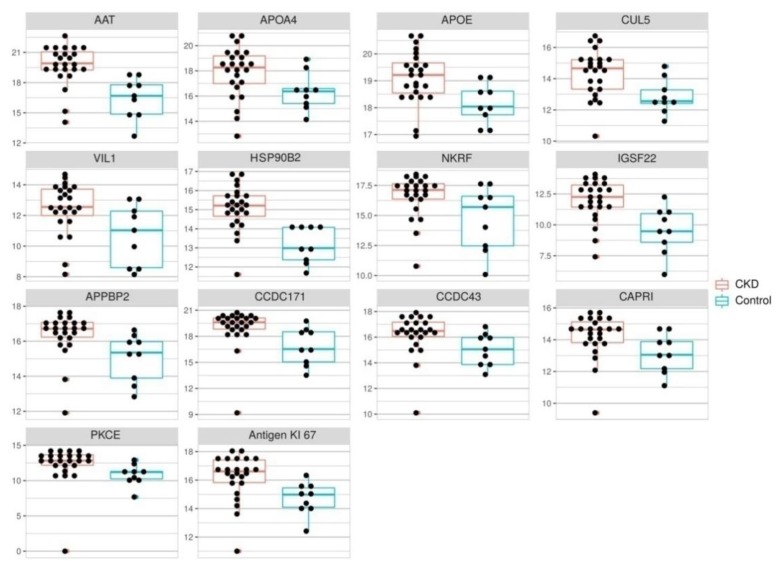
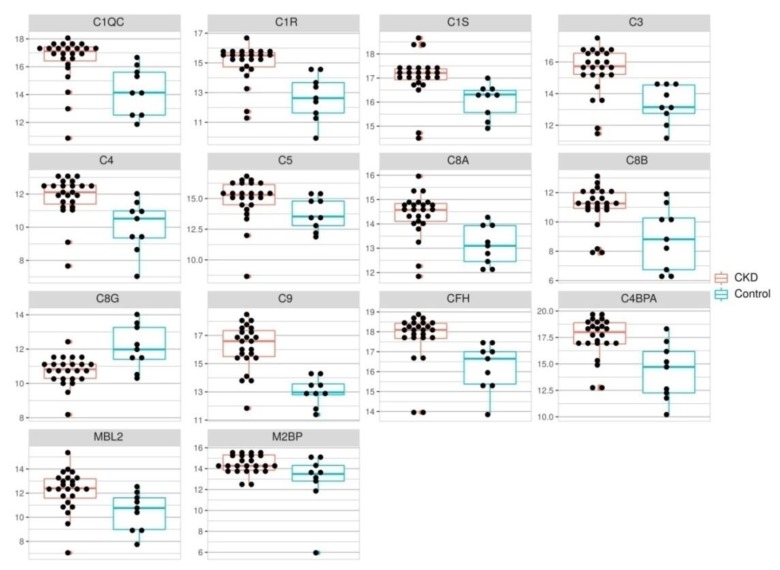
The quantification results for proteins with differential expression in 2D-DIGE and those associated with the complement system are shown in Figure 2 and Figure 3.
Figure 2.
Dot plots of complement system components levels in the serum of healthy individuals (n = 10, yellow) and patients with CKD (n = 26, blue). Levels are expressed as areas of MRM transition peaks. Wilcoxon rank sum test was performed in each case, p value < 0.05.
Figure 3.
Dot plots of differently expressed protein levels in the serum of healthy individuals (n = 10, yellow) and patients with CKD (n = 26, blue). Levels are expressed as areas of MRM transition peaks. Wilcoxon rank sum test was performed in each case, p value < 0.05.
In general, concentrations of 26 proteins enlisted in Table 2 were significantly different in patients with CKD and healthy subjects (Figure 2 and Figure 3, Supplementary Tables S3 and S4). IGSF22, HSP90B2, AAT, C4BPA, C3, C1R and C9 concentrations increased more than four times in the group of CKD patients as compared to the control. Serum CFH, CUL5, PKCE, APOA4, APOE, CCDC171, CCDC43, VIL1, antigen KI-67, NKRF, APPBP2, CAPRI, C1QC, C1S, C4, C5, C8A, C8B and MBL2 concentrations were two to three times elevated in patients with CKD. Concentrations of the remaining proteins such as CCT4, PLG, APOA1, LMCD1, CKAP2L, PLB1, PPEF2, Gal-3, MASP2, C6 and FCN3 were not significantly different between the group of CKD patients and healthy subjects. Only the concentration of C8G was significantly decreased (three-fold) in patients with CKD.
To assess the immune status serum concentrations of 27 cytokines, chemokines and growth factors such as IL-1β, IL-2Rα, IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-9, IL-10, IL-12 (p70), IL-13, IL-15, IL-17, Eotaxin, FGFb, G-CSF, GM-CSF, INFγ, IP-10, MCP-1, MIP-1α, MIP-1β, PDGF-1bb, RANTES, TNF-α and VEGF, they were quantified using Luminex xMAP technology (Figure 4). The analysis showed that serum concentrations of IL-9 and MIP-1β were elevated >500 times; those of IL-1β, IL-8, IL-12 (p70), IL-13, IL-15, IP-10, RANTES and VEGF increased >100 times; IL-6, GM-CSF, MIP-1α and PDGF-1bb levels were elevated >50 times; IL-2, IL-4, IL-7, IL-10 and IL-17 were 20–40 times higher; and those of IL-5, Eotaxin, FGFb, G-CSF, MCP-1 and TNF-α increased 6–12 times in patients with CKD (n = 19) when compared to the control group (n = 10). The INFγ concentration did not significantly differ between the groups. The blood serum concentration of soluble IL-2Rα was reduced 1.5 times in patients versus the control group.
Figure 4.
Dot plots of serum levels (pg/mL) of 26 cytokines for healthy people (n = 10, red) and patients with CKD (n = 19, blue). Wilcoxon rank sum test was performed in each case, p value < 0.01.
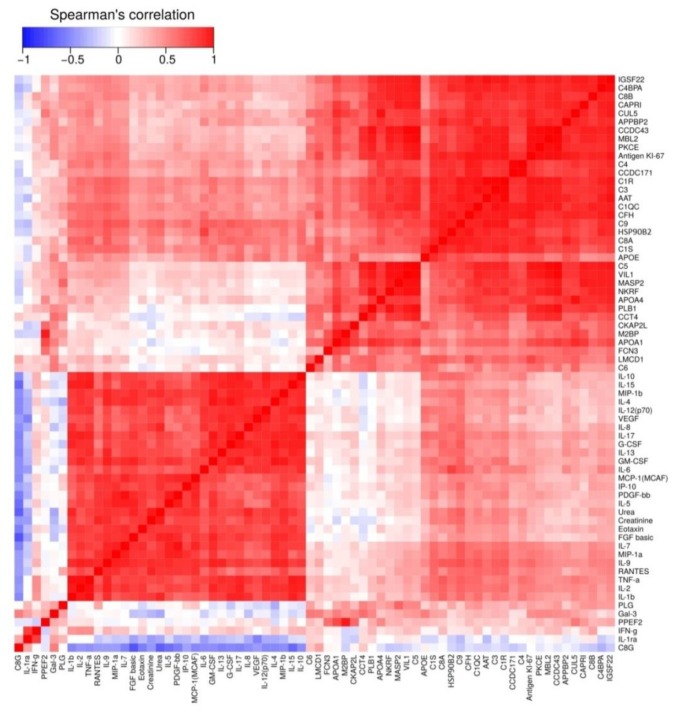
As concentrations of most proteins and cytokines analyzed were significantly different in the group of CKD patients and healthy subjects, clustering of quantitative serum protein proteomic analysis results, cytokine multiplex analysis data and creatinine and urea biochemistry results were compiled (Figure 5, Supplementary Table S5). As can be seen from the heat map data, an increase of all analyzed cytokines, except for IL-2Rα and INFγ, positively correlated with serum creatinine and urea concentrations (R > 0.6, p value < 0.05). Serum AAT concentration demonstrated a moderate, positive correlation (R = 0.44, p value < 0.05) with creatinine and some cytokines (IL-2, IL-6, IL-7, IL-9, MIP-1α, RANTES; R > 0.5, p value < 0.05). HSP90B2 positively correlated with those of creatinine and urea and most of the measured cytokines (IL-2, IL-6, IL-7, IL-8, IL-9, IL-10, IL-13, IL-17, G-CSF, GM-CSF, MIP-1α, MIP-1β, RANTES, TNF-α and VEGF; R > 0.5, p value < 0.05). Serum APOE, IGSF22, CUL5, CCDC171, CAPRI, CCDC43, Antigen KI-67, PKCE and APPBP2 levels positively correlated only with those of some cytokines such as IL-2, IL-9, MIP-1a, TNF-α and/or IL-13, IL-17, RANTES (R > 0.4, p value < 0.05). Serum APOA4, APOA1, VIL1 and NKRF concentrations had no correlations with creatinine, urea and cytokines.
Figure 5.
Heat map demonstrating correlations between urea, creatinine, cytokines and serum proteins. Hierarchical clustering of all analytes was performed by using the Euclidian distance method. The blue and red colors represent negative and positive Spearman’s rank correlation coefficients between the two analytes, respectively.
Complement system components such as CFH, C1S, C1R, C1QC, C3, C4, С8A and C9 had a positive correlation with serum urea and/or creatinine levels (R > 0.4, p value < 0.05), as well as with cytokines (IL-2, IL-7, IL-9, IL-17, MIP-1α, RANTES and TNF-α; R > 0.45, p value < 0.05). MBL2 levels mildly correlated with some cytokines (IL-2, IL-9, MIP-1a, RANTES; R < −0.45, p value < 0.05). C8G had a negative correlation with creatinine (R = −0.63, p value < 0.05) and most cytokines (IL-2, IL-4, IL-5, IL-6, IL-7, IL-8, IL-10, IL12(p70), IL-13, IL-15, IL-17, G-CSF, Eotaxin, FGFb and MIP-1β; R < −0.45, p value < 0.05). The remaining complement system proteins (i.e., C5, C6, Gal3, MASP2 and M2BP) showed no significant correlation with the well-established CKD markers and cytokines.
The raw MRM data and gel images were deposited at jPOST database [25] under the project numbed as JPST000578 and JPST000579.
4. Discussion
Modern diagnostics of CKD and monitoring of its course are based on the measurement of serum creatinine concentrations and subsequent calculation of the glomerular filtration rate [3,4]. The disadvantage of this method is its poor effectiveness in diagnosing early CKD. There are a number of studies where potential protein biomarkers of CKD were proposed and evaluated in the serum or plasma of patients [7,8,11,12,13,14]. Peptide biomarkers can also provide insight into CKD diagnosis and progression. In a recent study, 273 specific urinary peptides (CKD273 classifier) were identified as reliable predictors of CKD progression [26] and cardiovascular events in CKD [27]. In this study, we hypothesized that if some proteins showed significant quantitative changes in patient serum and correlated with well-established markers, then they would be potential biomarkers for CKD diagnosis. The first stage of our research involved the screening of serum proteins with differential expressions in patients with CKD and healthy individuals using the 2D-DIGE approach. Twenty-one proteins from the 2D dataset were selected as potential protein candidates for biomarkers of the disease (Table 2). AAT, APOA4 and VIL1 concentrations have previously been shown to increase in blood plasma as CKD progresses [28,29,30,31]. Our data confirm the increased blood serum concentrations of these proteins in patients; whereas only serum AAT values correlated with elevated levels of creatinine and cytokines. Our target proteomics results are compared with previously reported data from studies of different renal pathologies in Table 3.
Table 3.
Lists of elevated serum proteins from MRM data analysis compared to the previously reported results.
| n/n | Protein | Fold Change | SC between Protein and Creatinine | SC between Protein and Urea | Reference | Study Population | Results |
|---|---|---|---|---|---|---|---|
| 1 | APOA4 | 3.4 * ↑ | 0.07 | 0.19 | [32] | 345 CKD patients with type 2 diabetes | Increased plasma level of APOA4 |
| [33] | 177 CKD patients | Increased serum level of APOA4 were significant predictors of disease progression | |||||
| [34] | 6220 participants of general population | Increased serum level of APOA4 were significant predictors of disease progression | |||||
| 2 | APOE | 2.1 ** ↑ | 0.30 | 0.30 | [35] | 117 CKD patients | APOE was a negative predictor of eGFR reduction rate |
| [36] | 109 HD patients | APOE were significantly decreased | |||||
| [8] | 90 CKD patients | Elevated level of APOE in plasma of patients with CKD 1-2 stages | |||||
| [37] | 301 HD patients | HD patients had a significantly lower prevalence of the E4 allele and greater levels of APOE | |||||
| [38] | 7 CKD patients | Increased plasma level of APOE | |||||
| 3 | APOA1 | 1.6 * ↑ | −0.16 | −0.07 | [39] | 17,315 participants of the general population | Higher serum APOA1 was associated with lower prevalence of CKD |
| [40] | 50 patients with CKD and 198 patients on HD therapy | CKD was found to be associated with highly significant reductions in plasma APOA1 | |||||
| [8] | 90 CKD patients | No differences between plasma APOA1 level of patients with CKD 1-2 stages and healthy voluntaries | |||||
| [11] | 76 patients who received initial insertion of PD | APOA1 showed enhanced levels in PD effluents of patients with high transporter | |||||
| 4 | IGSF22 | 4.5 ** ↑ | 0.34 | 0.35 | [41] | 7 patients with clear cell carcinoma | Found in a renal cell carcinoma sample; somatic mutation |
| 5 | HSP90B2 | 4.0 ** ↑ | 0.55 ** | 0.56 ** | - | - | - |
| 6 | AAT | 8.7 ** ↑ | 0.44 * | 0.41 | [12] | 12 non-diabetic ESRD patients | HD patients had altered plasma profiles of AAT isoforms |
| [31] | 63 patients with primary membranous nephropathy | Increased urinary level of AAT | |||||
| [42] | 103 HD patients | Higher serum AAT levels select the HD patients with severe inflammation from those without | |||||
| 7 | VIL1 | 2.6 * ↑ | 0.08 | 0.18 | [43] | 3 patients with AKI after liver transplantation | VIL1 is released in plasma during AKI and shows potential as an early marker for proximal tubular injury |
| [29] | 3 renal transplant recipients | VIL1 concentrations in the urine up to 20 mg/I | |||||
| 8 | Antigen KI-67 | 3.2 * ↑ | 0.31 | 0.31 | [44] | 351 patients with clear cell carcinoma | Ki-67 are significant prognostic factors of clear cell carcinoma |
| 9 | CFH | 2.7 * ↑ | 0.43 * | 0.43 * | [45] | 63 patients with RD | Urinary CFH levels were significantly higher in patients |
| 10 | C4A | 2.8 ** ↑ | 0.42 | 0.45 * | [38] | 7 CKD patients | Increased plasma level of CA4 |
| [13] | 90 patients with CKD | Increased plasma level of CA4 | |||||
| 11 | C4BPA | 4.5 ** ↑ | 0.3 | 0.38 | - | - | - |
| 12 | C1R | 4.1 ** ↑ | 0.48 * | 0.49 * | [14] | 29 patients with CKD | Increased plasma level of C1R |
| 13 | C1S | 2.1 ** ↑ | 0.51 * | 0.51 * | [14] | 29 patients with CKD | Increased plasma level of C1S |
| 14 | C1QC | 3.7 ** ↑ | 0.46 * | 0.50 * | [46] | 62 diabetic patients | No difference |
| 15 | C3 | 4.7 ** ↑ | 0.48 * | 0.50 * | [11] | 76 patients who received initial insertion of PD | C3 showed enhanced expression in PD effluents of patients with high transporter |
| [38] | 7 CKD patients | Increased plasma level of C3 | |||||
| 16 | C5 | 2.2 * ↑ | 0.11 | 0.15 | [45] | 63 patients with RD | Increased urinary MAC (SC5b-9) |
| 17 | C8A | 2.4 ** ↑ | 0.48 ** | 0.47 * | [45] | 63 patients with RD | Increased urinary MAC (SC5b-9) |
| 18 | C8B | 2.7 ** ↑ | 0.25 | 0.38 | [45] | 63 patients with RD | Increased urinary MAC (SC5b-9) |
| 19 | C8G | 3.1 ** ↓ | −0.41 | −0.63 * | [38] | 7 CKD patients | Decreased plasma level of C8G |
| 20 | С9 | 11 ** ↑ | 0.58 ** | 0.62 ** | [45] | 63 patients with RD | Increased urinary MAC (SC5b-9) |
| [47] | 53 patients with different nephropathy | Urinary C9 was elevated in MCD, MN and FSGS groups compared with in IgA nephropathy and healthy controls | |||||
| 21 | MBL2 | 3.4 ** ↑ | 0.18 | 0.18 | [46] | 62 diabetic patients | MBL was found to increase with the progression of DN |
| 22 | CUL5 | 3.3 ** ↑ | 0.23 | 0.29 | - | - | - |
| 23 | PKCE | 3.2 ** ↑ | 0.27 | 0.28 | - | - | - |
| 24 | CCDC43 | 2.2 * ↑ | 0.18 | 0.22 | - | - | - |
| 25 | CDC171 | 3.1 ** ↑ | 0.33 | 0.38 | - | - | - |
| 26 | CAPRI | 2.1 * ↑ | 0.18 | 0.23 | - | - | - |
* p < 0.05, ** p < 0.005, ↑—increased in CKD patients, ↓—decreased in CKD patients. Abbreviations: SC—Spearman’s rank correlation coefficient, HD—hemodialysis, PD—peritoneal dialysis, ESRD—end-stage renal disease, AKI—acute kidney injury, RD—renal disease, MCD—minimal change disease, MN—membranous nephropathy, FSGS—focal segmental glomerulosclerosis, DN—diabetic nephropathy.
AAT, which is supposed to have anti-inflammatory properties, belongs to the group of serine proteinases [27]. Exogenous AAT inhibited apoptosis and inflammation in renal reperfusion [28] and reduced the urine kidney injury molecule-1 (KIM-1) concentration [30]. Several studies have demonstrated increased blood plasma AAT concentrations in hypoxia [31] and acute myocardial infarction [48,49].
Based on the MRM results, APOE concentrations were significantly higher in the group of patients with CKD, whereas serum APOA1 levels showed no changes. However, in previously reported studies, plasma APOE and APOA1 levels differed for patients with and without CKD, apparently depending on the disease stage and therapy [35,50,51]. In particular, hemodialysis and pharmacotherapy might have an impact on the levels of these proteins.
High antigen KI-67 and CUL5 expression levels in tissues are associated with cancers [44,52,53,54,55,56]. We observed increased serum concentrations of these proteins in patients with CKD in this study. Furthermore, we saw increased levels of other intracellular proteins such as NKRF, CCDC171, CAPRI, CCDC43, APPBP2, IGSF22 and PKCE, which might be associated with renal tissue necrosis and, thus, with disease progression. The literature on these proteins in CKD is limited and/or contradictory. IGSF22 is known to be similar to cytoskeletal proteins in its structure, with a one-nucleotide substitution in the IGSF22 gene associated with the development of renal carcinoma [41]. The information on PKCE is controversial; protein kinase activation in proximal nephron cells leads to impaired functioning of the mitochondria, oxidative stress, energy deficiency and cell death [57]. At the same time, there is evidence that PKCE has protective functions against ischemic injury in other tissues, particularly in the myocardium and neurons [58,59,60,61]. NKRF is a transcriptional repressor of NF-kappa-B responsive genes [62]. Increased expression of NKRF transcription factor was shown to upregulate IL-1-induced secretion of IL-8 [63]. In our study, increased serum levels of IL-1 and IL-8 were also demonstrated, suggesting that NKRF could synergize with IL-1 to induce IL-8 expression. It should be noted that serum HSP90B2 concentration positively correlated with levels of creatinine, urea and a number of cytokines. However, the data on changes of serum HSP90B2 in various pathologies are presently lacking. We have evaluated key complement system proteins known to be important components of innate immunity and to play important roles in body defense, inflammation, tissue regeneration and other physiological processes. We found that serum C1S, C1QC and C4 concentrations increased two to three times, and that of C1R was elevated four times in patients with CKD, as compared to the control subjects. Increased levels of these proteins might indicate the activation of the complement system under the classic pathway. It is noteworthy that concentrations of lectin pathway activators such as FCN3, Gal-3 and MASP2 did not show any significant difference between the groups, although the MBL2 concentration was twice as high in patients with CKD. MBL2 has been previously shown to be capable of binding with apoptotic and necrotic cells, thereby promoting the activation of phagocytosis of dying cells [64].
Serum concentrations of complement system inhibitors factor H and C4BP were elevated two and four times, respectively, in CKD patients. We found significantly elevated concentrations (two times) of late lytic cascade proteins such as C5, C8A and C8B, and both C3 and C9 were more than four times higher in CKD patients. These data are in consistent with previously reported results about higher plasma MAC (C5b-9) levels in patients with renal diseases [45,46].
It is remarkable that the serum concentration of the subunit С8G did not elevate following the increase of C8A and C8B subunits constituting a single С8 protein. Unlike C8A and C8B, the С8G subunit is a member of the lipocalin family. As Lovelace et al. have previously reported [65], С8g is not involved in the formation of a membrane attack complex, but it only enhances its activity [65]. Perhaps, the subunit С8G has a regulator function as a complement system inhibitor. Furthermore, it has been suggested that decrease in serum С8G might be specific for other chronic diseases.
Increased blood serum levels of cytokines such as IL-4, IL-5, IL-6, IL-10 and IL-13, G-CSF, eotaxin and MIP-1β might indicate the activation of Th2 cells responsible for the development of humoral immune responses [66,67]. It should be noted that increased blood levels of cytokines involved in the activation of Th1 cells as well as IL-2, GM-CSF, TNF-α, IL-12, RANTES, MIP-1α and IL-18 were previously observed in patients with CKD [68,69]. Simultaneous activation of Th1 and Th2 cells was shown in various renal pathologies [69,70,71] and in a number of other diseases [72,73,74]. Moreover, significantly increased IL-9 might suggest ongoing differentiation of Т-cells into a Th9 population [73].
5. Conclusions
Early diagnosis of renal dysfunction is essential to improve disease progress and survival of patients with CKD. In this research we quantified differentially expressed proteins and complement components using the MRM approach and measured serum concentrations of cytokines, chemokines and growth factors using the multiplex Luminex xMAP technology in patients with CKD and healthy subjects.
Our results correlate well with the data obtained by other researchers in that blood APOA4, AAT, VIL1, complement component and cytokine concentrations increased in patients with renal disorders [8,11,12,13,14,28,32,33,34,43,69,70,71]. Moreover, we found elevated serum concentrations of potential oncomarkers (CUL5, antigen KI-67) and other intracellular proteins (NKRF, CAPRI, IGSF22, APPBP2, CCD171 and CCD43) in patients with CKD. The reasons for this increase in blood serum and the role they play in renal tissue injury are not clear and require further investigations.
Patient serum levels of AAT, IGS22 and HSP90B2 had a greater than four-fold change. In addition, the data analysis showed a mild correlation between increased serum concentrations of AAT and HSP90B2 in patients with CKD and well-established markers of CKD such as creatinine and urea. Thus, we suggest that proteins AAT and HSP90B2 might be associated with kidney diseases and might be potential markers of CKD. Further investigations of these proteins as early biomarkers are needed to elucidate their clinical usefulness.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/2/257/s1, Table S1: List of peptides for MRM analysis, Table S2: Proteins differentially expressed between CKD and control groups, Table S3: Average mean, standard error and standard deviation, Table S4: Significant differences, P-value <0.05, Table S5: Spearman’s rank correlation.
Author Contributions
Conceptualization, Y.R. and S.K.; investigation Y.R., A.L. and R.K.; writing—original draft preparation, Y.R.; writing—review and editing, Y.R., S.K., A.R. and I.S.; formal analysis, A.L. and M.M.; resources, A.M. and M.H.; supervision, A.R. and I.S. All authors have read and agreed to the published version of the manuscript.
Funding
The work is performed according to the Russian Government Program of Competitive Growth of the Kazan Federal University and subsidy allocated to the Kazan Federal University for the state assignment in the sphere of scientific activities. The research was performed using the equipment of the Interdisciplinary Center for Shared Use of the Kazan Federal University supported by the Ministry of Education of Russia.
Conflicts of Interest
The authors declare no competing interests.
References
- 1.Jha V., Garcia-Garcia G., Iseki K., Li Z., Naicker S., Plattner B., Saran R., Wang A.Y., Yang C.W. Chronic kidney disease: Global dimension and perspectives. Lancet. 2013;382:260–272. doi: 10.1016/S0140-6736(13)60687-X. [DOI] [PubMed] [Google Scholar]
- 2.Naghavi M., Abajobir A.A., Abbafati C., Abbas K.M., Abd-Allah F., Abera S.F., Aboyans V., Adetokunboh O., Afshin A., Agrawal A., et al. Global, regional, and national age-sex specific mortality for 264 causes of death, 1980-2016: A systematic analysis for the Global Burden of Disease Study 2016. Lancet. 2017;390:1151–1210. doi: 10.1016/S0140-6736(17)32152-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Andrassy K.M. Comments on ‘KDIGO 2012 Clinical Practice Guideline for the Evaluation and Management of Chronic Kidney Disease’. Kidney Int. 2013;84:622–623. doi: 10.1038/ki.2013.243. [DOI] [PubMed] [Google Scholar]
- 4.Kriz W., LeHir M. Pathways to nephron loss starting from glomerular diseases-insights from animal models. Kidney Int. 2005;67:404–419. doi: 10.1111/j.1523-1755.2005.67097.x. [DOI] [PubMed] [Google Scholar]
- 5.Gentile G., Remuzzi G. Novel Biomarkers for Renal Diseases? None for the Moment (but One) J. Biomol. Screen. 2016;21:655–670. doi: 10.1177/1087057116629916. [DOI] [PubMed] [Google Scholar]
- 6.Lam C.W. 2. Inflammation, Cytokines and Chemokines in Chronic Kidney Disease. EJIFCC. 2009;20:12–20. [PMC free article] [PubMed] [Google Scholar]
- 7.Carrero J.J., Park S.H., Axelsson J., Lindholm B., Stenvinkel P. Cytokines, atherogenesis, and hypercatabolism in chronic kidney disease: A dreadful triad. Semin. Dial. 2009;22:381–386. doi: 10.1111/j.1525-139X.2009.00585.x. [DOI] [PubMed] [Google Scholar]
- 8.Luczak M., Formanowicz D., Marczak Ł., Suszyńska-Zajczyk J., Pawliczak E., Wanic-Kossowska M., Stobiecki M. iTRAQ-based proteomic analysis of plasma reveals abnormalities in lipid metabolism proteins in chronic kidney disease-related atherosclerosis. Sci. Rep. 2016;6:32511. doi: 10.1038/srep32511. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Fourdinier O., Schepers E., Metzinger-Le Meuth V., Glorieux G., Liabeuf S., Verbeke F., Vanholder R., Brigant B., Pletinck A., Diouf M., et al. Serum levels of miR-126 and miR-223 and outcomes in chronic kidney disease patients. Sci. Rep. 2019;9:4477. doi: 10.1038/s41598-019-41101-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Fujii R., Yamada H., Yamazaki M., Munetsuna E., Ando Y., Ohashi K., Ishikawa H., Shimoda H., Sakata K., Ogawa A., et al. Circulating microRNAs (miR-126, miR-197, and miR-223) are associated with chronic kidney disease among elderly survivors of the Great East Japan Earthquake. BMC Nephrol. 2019;20:474. doi: 10.1186/s12882-019-1651-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wen Q., Zhang L., Mao H.P., Tang X.Q., Rong R., Fan J.J., Yu X.Q. Proteomic analysis in peritoneal dialysis patients with different peritoneal transport characteristics. Biochem. Biophys. Res. Commun. 2013;438:473–478. doi: 10.1016/j.bbrc.2013.07.116. [DOI] [PubMed] [Google Scholar]
- 12.Lin Y.-P., Yang C.-Y., Liao C.-C., Yu W.-C., Chi C.-W., Lin C.-H. Plasma Protein Characteristics of Long-Term Hemodialysis Survivors. PLoS ONE. 2012;7:e40232. doi: 10.1371/journal.pone.0040232. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Luczak M., Formanowicz D., Marczak Ł., Pawliczak E., Wanic-Kossowska M., Figlerowicz M., Stobiecki M. Deeper insight into chronic kidney diseaserelated atherosclerosis: Comparative proteomic studies of blood plasma using 2DE and mass spectrometry. J. Transl. Med. 2015;13:20. doi: 10.1186/s12967-014-0378-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Glorieux G., Mullen W., Duranton F., Filip S., Gayrard N., Husi H., Schepers E., Neirynck N., Schanstra J.P., Jankowski J., et al. New insights in molecular mechanisms involved in chronic kidney disease using high-resolution plasma proteome analysis. Nephrol. Dial. Transplant. 2015;30:1842–1852. doi: 10.1093/ndt/gfv254. [DOI] [PubMed] [Google Scholar]
- 15.Unlu M., Morgan M.E., Minden J.S. Difference gel electrophoresis: A single gel method for detecting changes in protein extracts. Electrophoresis. 1997;18:2071–2077. doi: 10.1002/elps.1150181133. [DOI] [PubMed] [Google Scholar]
- 16.Tannu N.S., Hemby S.E. Two-dimensional fluorescence difference gel electrophoresis for comparative proteomics profiling. Nat. Protoc. 2006;1:1732–1742. doi: 10.1038/nprot.2006.256. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Wray W., Boulikas T., Wray V.P., Hancock R. Silver staining of proteins in polyacrylamide gels. Anal. Biochem. 1981;118:197–203. doi: 10.1016/0003-2697(81)90179-2. [DOI] [PubMed] [Google Scholar]
- 18.Gharahdaghi F., Weinberg C.R., Meagher D.A., Imai B.S., Mische S.M. Mass spectrometric identification of proteins from silver-stained polyacrylamide gel: A method for the removal of silver ions to enhance sensitivity. Electrophoresis. 1999;20:601–605. doi: 10.1002/(SICI)1522-2683(19990301)20:3<601::AID-ELPS601>3.0.CO;2-6. [DOI] [PubMed] [Google Scholar]
- 19.Software M.L. Skyline Targeted Mass Spec Environment. [(accessed on 6 February 2020)]; Available online: https://skyline.ms/project/home/software/Skyline/begin.view.
- 20.MacLean B., Tomazela D.M., Shulman N., Chambers M., Finney G.L., Frewen B., Kern R., Tabb D.L., Liebler D.C., MacCoss M.J. Skyline: An open source document editor for creating and analyzing targeted proteomics experiments. Bioinformatics. 2010;26:966–968. doi: 10.1093/bioinformatics/btq054. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.R-project The R Project for Statistical Computing. [(accessed on 6 February 2020)]; Available online: https://www.r-project.org/
- 22.Ihara H., Toya N., Kakinoki T., Tani A., Aoki Y., Hashizume N., Inada Y., Nanba S., Urayama T., Yoshida M. Reference ranges for serum protein fractions as determined by capillary zone electrophoresis. Jpn. J. Electroph. 2001;45:69–74. doi: 10.2198/sbk.45.69. [DOI] [Google Scholar]
- 23.Larsson A., Hansson L.O. Plasma protein fractions in healthy blood donors quantitated by an automated multicapillary electrophoresis system. J. Chromatogr. Sci. 2006;44:479–483. doi: 10.1093/chromsci/44.8.479. [DOI] [PubMed] [Google Scholar]
- 24.Domanski D., Percy A.J., Yang J., Chambers A.G., Hill J.S., Freue G.V., Borchers C.H. MRM-based multiplexed quantitation of 67 putative cardiovascular disease biomarkers in human plasma. Proteomics. 2012;12:1222–1243. doi: 10.1002/pmic.201100568. [DOI] [PubMed] [Google Scholar]
- 25.Okuda S., Watanabe Y., Moriya Y., Kawano S., Yamamoto T., Matsumoto M., Takami T., Kobayashi D., Araki N., Yoshizawa A.C., et al. jPOSTrepo: An international standard data repository for proteomes. Nucleic Acids Res. 2017;45:D1107–D1111. doi: 10.1093/nar/gkw1080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Critselis E., Lambers Heerspink H. Utility of the CKD273 peptide classifier in predicting chronic kidney disease progression. Nephrol. Dial. Transplant. 2016;31:249–254. doi: 10.1093/ndt/gfv062. [DOI] [PubMed] [Google Scholar]
- 27.Verbeke F., Siwy J., Van Biesen W., Mischak H., Pletinck A., Schepers E., Neirynck N., Magalhães P., Pejchinovski M., Pontillo C. The urinary proteomics classifier chronic kidney disease 273 predicts cardiovascular outcome in patients with chronic kidney disease. Nephrol. Dial. Transplant. 2019;gfz242:18. doi: 10.1093/ndt/gfz242. [DOI] [PubMed] [Google Scholar]
- 28.Kronenberg F., Kuen E., Ritz E., Konig P., Kraatz G., Lhotta K., Mann J.F., Muller G.A., Neyer U., Riegel W., et al. Apolipoprotein A-IV serum concentrations are elevated in patients with mild and moderate renal failure. J. Am. Soc. Nephrol. 2002;13:461–469. doi: 10.1681/ASN.V132461. [DOI] [PubMed] [Google Scholar]
- 29.Zimmerhackl L.B., Leuk B., Hoschutzky H. The cytoskeletal protein villin as a parameter for early detection of tubular damage in the human kidney. J. Chromatogr. 1991;587:81–84. doi: 10.1016/0021-9673(91)85200-Y. [DOI] [PubMed] [Google Scholar]
- 30.Salih M., Demmers J.A., Bezstarosti K., Leonhard W.N., Losekoot M., van Kooten C., Gansevoort R.T., Peters D.J., Zietse R., Hoorn E.J. Proteomics of Urinary Vesicles Links Plakins and Complement to Polycystic Kidney Disease. J. Am. Soc. Nephrol. 2016;27:3079–3092. doi: 10.1681/ASN.2015090994. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Pang L., Li Q., Li Y., Liu Y., Duan N., Li H. Urine proteomics of primary membranous nephropathy using nanoscale liquid chromatography tandem mass spectrometry analysis. Clin. Proteomics. 2018;15:5. doi: 10.1186/s12014-018-9183-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Peters K.E., Davis W.A., Ito J., Winfield K., Stoll T., Bringans S.D., Lipscombe R.J., Davis T.M.E. Identification of Novel Circulating Biomarkers Predicting Rapid Decline in Renal Function in Type 2 Diabetes: The Fremantle Diabetes Study Phase II. Diabetes Care. 2017;40:1548–1555. doi: 10.2337/dc17-0911. [DOI] [PubMed] [Google Scholar]
- 33.Boes E., Fliser D., Ritz E., König P., Lhotta K., Mann J.F., Müller G.A., Neyer U., Riegel W., Riegler P., et al. Apolipoprotein A-IV Predicts Progression of Chronic Kidney Disease: The Mild to Moderate Kidney Disease Study. Am. Soc. Nephrol. 2016;17:528–536. doi: 10.1681/ASN.2005070733. [DOI] [PubMed] [Google Scholar]
- 34.Stangl S., Kollerits B., Lamina C., Meisinger C., Huth C., Stöckl A., Dähnhardt D., Böger C.A., Krämer B.K., Peters A., et al. Association between apolipoprotein A-IV concentrations and chronic kidney disease in two large population-based cohorts: Results from the KORA studies. J. Intern. Med. 2015;278:410–423. doi: 10.1111/joim.12380. [DOI] [PubMed] [Google Scholar]
- 35.Smajic J., Hasic S., Rasic S. High-density lipoprotein cholesterol, apolipoprotein E and atherogenic index of plasma are associated with risk of chronic kidney disease. Med. Glas. (Zenica) 2018;15:115–121. doi: 10.17392/962-18. [DOI] [PubMed] [Google Scholar]
- 36.Lahrach H., Ghalim N., Taki H., Kettani A., Er-Rachdi L., Ramdani B., Saïle R. Serum paraoxonase activity, high-sensitivity C-reactive protein, and lipoprotein disturbances in end-stage renal disease patients on long-term hemodialysis. J. Clin. Lipidol. 2008;2:43–50. doi: 10.1016/j.jacl.2007.12.003. [DOI] [PubMed] [Google Scholar]
- 37.Liberopoulos E.N., Miltiadous G.A., Cariolou M., Tselepis A.D., Siamopoulos K.C., Elisaf M.S. The influence of serum apolipoprotein E concentration and polymorphism on serum lipid parameters in hemodialysis patients. Am. J. Kidney Dis. 2004;44:300–308. doi: 10.1053/j.ajkd.2004.04.035. [DOI] [PubMed] [Google Scholar]
- 38.Naseeb U., Shafqat J., Jägerbrink T., Zarina S., Alvestrand A., Jörnvall H., Axelsson J. Proteome Patterns in Uremic Plasma. Blood Purif. 2008;26:561–568. doi: 10.1159/000178773. [DOI] [PubMed] [Google Scholar]
- 39.Goek O.N., Kottgen A., Hoogeveen R.C., Ballantyne C.M., Coresh J., Astor B.C. Association of apolipoprotein A1 and B with kidney function and chronic kidney disease in two multiethnic population samples. Nephrol. Dial. Transplant. 2012;27:2839–2847. doi: 10.1093/ndt/gfr795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Calabresi L., Simonelli S., Conca P., Busnach G., Cabibbe M., Gesualdo L., Gigante M., Penco S., Veglia F., Franceschini G. Acquired lecithin:cholesterol acyltransferase deficiency as a major factor in lowering plasma HDL levels in chronic kidney disease. J. Intern. Med. 2015;277:552–561. doi: 10.1111/joim.12290. [DOI] [PubMed] [Google Scholar]
- 41.Varela I., Tarpey P., Raine K., Huang D., Ong C.K., Stephens P., Davies H., Jones D., Lin M.-L., Teague J., et al. Exome sequencing identifies frequent mutation of the SWI/SNF complex gene PBRM1 in renal carcinoma. Nature. 2011;469:539–542. doi: 10.1038/nature09639. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Borawski J., Naumnik B., Myśliwiec M. Serum alpha1-antitrypsin but not complement C3 and C4 predicts chronic inflammation in hemodialysis patients. Ren. Fail. 2003;25:589–593. doi: 10.1081/JDI-120022550. [DOI] [PubMed] [Google Scholar]
- 43.Decuypere J.P., Ceulemans L.J., Wylin T., Martinet W., Monbaliu D., Pirenne J., Jochmans I. Plasmatic Villin 1 Is a Novel In Vivo Marker of Proximal Tubular Cell Injury During Renal Ischemia-Reperfusion. Transplantation. 2017;101:e330–e336. doi: 10.1097/TP.0000000000001876. [DOI] [PubMed] [Google Scholar]
- 44.Kim S.H., Park W.S., Park E.Y., Park B., Joo J., Joung J.Y., Seo H.K., Lee K.H., Chung J. The prognostic value of BAP1, PBRM1, pS6, PTEN, TGase2, PD-L1, CA9, PSMA, and Ki-67 tissue markers in localized renal cell carcinoma: A retrospective study of tissue microarrays using immunohistochemistry. PLoS ONE. 2017;12:e0179610. doi: 10.1371/journal.pone.0179610. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Nagamachi S., Ohsawa I., Suzuki H., Sato N., Inoshita H., Hisada A., Honda D., Shimamoto M., Shimizu Y., Horikoshi S., et al. Properdin has an ascendancy over factor H regulation in complement-mediated renal tubular damage. BMC Nephrol. 2014;15:82. doi: 10.1186/1471-2369-15-82. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Zheng J.M., Ren X.G., Jiang Z.H., Chen D.J., Zhao W.J., Li L.J. Lectin-induced renal local complement activation is involved in tubular interstitial injury in diabetic nephropathy. Clin. Chim. Acta. 2018;482:65–73. doi: 10.1016/j.cca.2018.03.033. [DOI] [PubMed] [Google Scholar]
- 47.Choi Y.W., Kim Y.G., Song M., Moon J.-Y., Jeong K.-H., Lee T.-W., Ihm C.-G., Park K.-S., Lee S.-H. Potential urine proteomics biomarkers for primary nephrotic syndrome. Clin. Proteom. 2017;14:18. doi: 10.1186/s12014-017-9153-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Wenger R.H., Rolfs A., Marti H.H., Bauer C., Gassmann M. Hypoxia, a novel inducer of acute phase gene expression in a human hepatoma cell line. J. Biol. Chem. 1995;270:27865–27870. doi: 10.1074/jbc.270.46.27865. [DOI] [PubMed] [Google Scholar]
- 49.Brunetti N.D., Correale M., Pellegrino P.L., Cuculo A., Biase M.D. Acute phase proteins in patients with acute coronary syndrome: Correlations with diagnosis, clinical features, and angiographic findings. Eur. J. Intern. Med. 2007;18:109–117. doi: 10.1016/j.ejim.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 50.Gilutz H., Siegel Y., Paran E., Cristal N., Quastel M.R. Alpha 1-antitrypsin in acute myocardial infarction. Br. Heart J. 1983;49:26–29. doi: 10.1136/hrt.49.1.26. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Borges D.L., Lemes H.P., de Castro Ferreira V., Filho S.R.F. High-sensitivity C-reactive protein, apolipoproteins, and residual diuresis in chronic kidney disease patients undergoing hemodialysis. Clin. Exp. Nephrol. 2016;20:943–950. doi: 10.1007/s10157-016-1230-7. [DOI] [PubMed] [Google Scholar]
- 52.Rasmy A., Abozeed W., Elsamany S., Baiomy M.E., Nashwa A., Amrallah A., Hasaan E., Alzahrani A., Faris M., Alsaleh K., et al. Correlation of Preoperative Ki67 and Serum CA15.3 Levels with Outcome in Early Breast Cancers a Multi Institutional Study. Asian Pac. J. Cancer Prev. 2016;17:3595–3600. [PubMed] [Google Scholar]
- 53.Chauhan R., Lahiri N. Tissue- and Serum-Associated Biomarkers of Hepatocellular Carcinoma. Biomark. Cancer. 2016;8:37–55. doi: 10.4137/BIC.S34413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Okumura F., Joo-Okumura A., Nakatsukasa K., Kamura T. The role of cullin 5-containing ubiquitin ligases. Cell Div. 2016;11:1. doi: 10.1186/s13008-016-0016-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Devor E.J., Schickling B.M., Reyes H.D., Warrier A., Lindsay B., Goodheart M.J., Santillan D.A., Leslie K.K. Cullin-5, a ubiquitin ligase scaffold protein, is significantly underexpressed in endometrial adenocarcinomas and is a target of miR-182. Oncol. Rep. 2016;35:2461–2465. doi: 10.3892/or.2016.4605. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Chen P., Yao G.D. The role of cullin proteins in gastric cancer. Tumour Biol. 2016;37:29–37. doi: 10.1007/s13277-015-4154-z. [DOI] [PubMed] [Google Scholar]
- 57.Nowak G., Takacsova-Bakajsova D., Megyesi J. Deletion of protein kinase C-epsilon attenuates mitochondrial dysfunction and ameliorates ischemic renal injury. Am. J. Physiol. Renal Physiol. 2017;312:F109–F120. doi: 10.1152/ajprenal.00115.2016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Gray M.O., Karliner J.S., Mochly-Rosen D. A selective epsilon-protein kinase C antagonist inhibits protection of cardiac myocytes from hypoxia-induced cell death. J. Biol. Chem. 1997;272:30945–30951. doi: 10.1074/jbc.272.49.30945. [DOI] [PubMed] [Google Scholar]
- 59.Garlid K.D., Costa A.D., Quinlan C.L., Pierre S.V., Dos Santos P. Cardioprotective signaling to mitochondria. J. Mol. Cell. Cardiol. 2009;46:858–866. doi: 10.1016/j.yjmcc.2008.11.019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Della-Morte D., Raval A.P., Dave K.R., Lin H.W., Perez-Pinzon M.A. Post-ischemic activation of protein kinase C epsilon protects the hippocampus from cerebral ischemic injury via alterations in cerebral blood flow. Neurosci. Lett. 2011;487:158–162. doi: 10.1016/j.neulet.2010.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris-Blanco K.C., Dave K.R., Saul I., Koronowski K.B., Stradecki H.M., Perez-Pinzon M.A. Protein Kinase C Epsilon Promotes Cerebral Ischemic Tolerance Via Modulation of Mitochondrial Sirt5. Sci. Rep. 2016;6:29790. doi: 10.1038/srep29790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Nourbakhsh M., Hoffmann K., Hauser H. Interferon-b promoters contain a DNA element that acts as a position-independent silencer on the NF-kappaB site. EMBO J. 1993;12:451–459. doi: 10.1002/j.1460-2075.1993.tb05677.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Nourbakhsh M., Kalble S., Dorrie A., Hauser H., Resch K., Kracht M. The NF-kappa b repressing factor is involved in basal repression and interleukin (IL)-1-induced activation of IL-8 transcription by binding to a conserved NF-kappa b-flanking sequence element. J. Biol. Chem. 2001;276:4501–4508. doi: 10.1074/jbc.M007532200. [DOI] [PubMed] [Google Scholar]
- 64.Nauta A.J., Raaschou-Jensen N., Roos A., Daha M.R., Madsen H.O., Borrias-Essers M.C., Ryder L.P., Koch C., Garred P. Mannose-binding lectin engagement with late apoptotic and necrotic cells. Eur. J. Immunol. 2003;33:2853–2863. doi: 10.1002/eji.200323888. [DOI] [PubMed] [Google Scholar]
- 65.Lovelace L.L., Cooper C.L., Sodetz J.M., Lebioda L. Structure of human C8 protein provides mechanistic insight into membrane pore formation by complement. J. Biol. Chem. 2011;286:17585–17592. doi: 10.1074/jbc.M111.219766. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Garlisi C.G., Falcone A., Kung T.T., Stelts D., Pennline K.J., Beavis A.J., Smith S.R., Egan R.W., Umland S.P. T cells are necessary for Th2 cytokine production and eosinophil accumulation in airways of antigen-challenged allergic mice. Clin. Immunol. Immunopathol. 1995;75:75–83. doi: 10.1006/clin.1995.1055. [DOI] [PubMed] [Google Scholar]
- 67.Mosmann T.R., Coffman R.L. TH1 and TH2 cells: Different patterns of lymphokine secretion lead to different functional properties. Annu. Rev. Immunol. 1989;7:145–173. doi: 10.1146/annurev.iy.07.040189.001045. [DOI] [PubMed] [Google Scholar]
- 68.Schrum S., Probst P., Fleischer B., Zipfel P.F. Synthesis of the CC-chemokines MIP-1alpha, MIP-1beta, and RANTES is associated with a type 1 immune response. J. Immunol. 1996;157:3598–3604. [PubMed] [Google Scholar]
- 69.Romanova Y.D., Markelova M.I., Laikov A.V., Fakhrutdinova L.I., Hasanova M.I., Malanin S.Y., Chernov V.M., Salafutdinov I.I., Khaiboullina S.F. Cytokine Levels in the Serum of Patients with Chronic Kidney Insufficiency Before and After Hemodialysis. BioNanoScience. 2017;7:415–418. doi: 10.1007/s12668-016-0379-6. [DOI] [Google Scholar]
- 70.Stangou M., Spartalis M., Daikidou D.V., Kouloukourgiotou T., Sampani E., Lambropoulou I.T., Pantzaki A., Papagianni A., Efstratiadis G. Impact of Tauh1 and Tauh2 cytokines in the progression of idiopathic nephrotic syndrome due to focal segmental glomerulosclerosis and minimal change disease. J. Nephropathol. 2017;6:187–195. doi: 10.15171/jnp.2017.32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Rios D.R.A., Pinheiro M.B., de Oliveira Junior W.V., Braga Gomes K., Carvalho A.T., Martins-Filho O.A., Simoes E.S.A.C., Dusse L.M.S. Cytokine Signature in End-Stage Renal Disease Patients on Hemodialysis. Dis. Markers. 2017;2017:9678391. doi: 10.1155/2017/9678391. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Bleotu C., Chifiriuc M.C., Grigore R., Grancea C., Popescu C.R., Anton G., Cernescu C. Investigation of Th1/Th2 cytokine profiles in patients with laryngo-pharyngeal, HPV-positive cancers. Eur. Arch. Otorhinolaryngol. 2013;270:711–718. doi: 10.1007/s00405-012-2067-7. [DOI] [PubMed] [Google Scholar]
- 73.Bestetti R.B., Dellalibera-Joviliano R., Lopes G.S., Faria-Jr M., Furlan-Daniel R., Lopes K.C., Batista D.R. Determination of the Th1, Th2, Th17, and Treg cytokine profile in patients with chronic Chagas heart disease and systemic arterial hypertension. Heart Vessels. 2019;34:123–133. doi: 10.1007/s00380-018-1228-z. [DOI] [PubMed] [Google Scholar]
- 74.Khan R., Gupta S., Sharma A. Circulatory levels of T-cell cytokines (interleukin [IL]-2, IL-4, IL-17, and transforming growth factor-beta) in patients with vitiligo. J. Am. Acad. Dermatol. 2012;66:510–511. doi: 10.1016/j.jaad.2011.07.018. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.