Abstract
The genome of Propionibacterium freudenreichii ssp. freudenreichii T82, which has a chromosome containing 2,585,340 nucleotides with 67.3% GC content (guanine-cytosine content), is described in this paper. The total number of genes is 2308, of which 2260 are protein-coding genes and 48 are RNA genes. According to the genome analysis and the obtained results, the T82 strain can produce various compounds such as propionic acid, trehalose, glycogen, and B group vitamins (e.g., B6, B9, and B12). From protein-coding sequences (CDSs), genes related to stress adaptation, biosynthesis, metabolism, transport, secretion, and defense machinery were detected. In the genome of the T82 strain, sequences corresponding to the CRISPR loci (Clustered Regularly Interspaced Short Palindromic Repeats), antibiotic resistance, and restriction–modification system were found.
Keywords: genome, Propionibacterium, metabolism, resistance, stress response
1. Introduction
Bacteria of the Propionibacterium genus belong to the class Actinobacteria, the order Actinomycetales, and the family Propionibacteriaceae. Propionic acid bacteria are divided into two groups based on the environment which they inhabit: skin (acnes) and classic (dairy). The former consists of species occurring on the skin and in the mucous membranes of the oral cavity and digestive tract, these species include P. acnes, P. avidum, P. propionicum, P. granulosum, and P. lymphophilum. Classic strains include microorganisms belonging to two phylogenetic groups. The first one includes bacteria from the species P. acidipropionici, P. jensenii, and P. thoenii. The second one includes subspecies of P. freudenreichii (ssp. shermanii, ssp. freudenreichii), which differ in two features: nitrate reduction and lactose fermentation abilities [1]. The strains of P. freudenreichii ssp. freudenreichii can reduce nitrates but do not have the ability to ferment lactose. In contrast, P. freudenreichii ssp. shermanii strains metabolize lactose (they have genes encoding the enzyme ß-D-galactosidase [EC 3.2.1.23]) but do not reduce nitrates. All the classic Propionibacterium species exhibit fermentation activity and are therefore a source of useful metabolites such as propionic acid, acetic acid, trehalose, and vitamins (B12, for instance) [2,3,4,5,6,7].
Propionibacterium bacteria are applied in the cheese industry, where they are used as components of inoculants (together with lactic acid fermentation bacteria that prepare the environment for the action of Propionibacterium strains) for the production of rennet (hard) cheeses (Swiss-Emmental, Dutch-Leerdammer, and French-Comté) and Polish semi-hard cheeses (tylzyck and krolewski). Starter cultures consisting of propionic acid bacteria (PAB) and lactic acid bacteria (Lactobacillus plantarum, L. acidophilus, P. jensenii, and P. acidipropionici) are also used in the production of vegetable silage. Appropriate PAB strains are also used in feed production. They are a source of vitamin B12, increase the assimilability of iron and calcium by animals, and naturally protect the product against fungal infections (propionic acid). Some of the PAB serve as probiotics in animal nutrition.
The species P. freudenreichii regulates the intestinal microflora by stimulating the development of Bifidobacterium bacteria and, through the production of bacteriocins, protects the animal organism from potential pathogens. Moreover, PAB can neutralize mycotoxins in the digestive tract, stimulate the immune system, and are a source of trehalose and vitamins: B12, B9, and K. It has been proven that the addition of PAB to the feed increases its use and the growth of young animals [8,9,10].
Some species of PAB (including P. freudenreichii ssp. freudenreichii) have the status of GRAS (Generally Recognised As Safe) and QPS (Qualified Presumption of Safety); thus, the living cells of these microorganisms and their metabolites can be used in the production of food and feed. Additionally, because PAB can produce very important compounds such as B12 vitamin, they can be used to produce vitamin B12-enriched food for vegan diets. The biomass of PAB is rich in trehalose and other vitamins from B group. This implies a huge potential for industry. As the species P. freudenreichii can use industrial waste for fermentation, its use in everyday life can also have a beneficial effect on the environment. To date, the complete genome sequences of P. freudenreichii ssp. shermanii CIRM-BIA1 [11] and P. freudenreichii ssp. freudenreichii DSM 20271 [12] strains have been described in the literature. To fully exploit the biotechnological potential of bacteria of the genus Propionibacterium, it is necessary to know their specific molecular properties, which is greatly facilitated by the knowledge of the genome. Furthermore, screening and subsequent research of another strains are also needed. Therefore, the summary classification and set of characteristics for P. freudenreichii T82 together with the description of the genome sequence annotation are presented below.
2. Materials and Methods
2.1. Culture Conditions
The T82 strain was grown in VL medium consisting of 3.0 g meat extract, 10.0 g peptone, 5 g NaCl, 5 g yeast extract, 0.4 g L-cysteine hydrochloride, and 10 g glucose per liter and pH adjusted to 7.0. The cells were separated by centrifugation for 10 min at 10,000 rpm at 4 °C and washed once with sterile distilled water.
2.2. Genome Sequencing
Genomic DNA was isolated by CTAB/lysozyme method [13]. The quality and quantity of DNA obtained were verified by electrophoretic separation in 0.7% agarose gel and by fluorometer Qubit 2.0 (Thermo Fisher Scientific, Waltham, MA). It was mechanically fragmented with a nebulizer, and then the NGS genomic library was prepared with the KAPA Library Preparation Kit (KAPA/Roche, Basel, Switzerland). The bacterial genome library was sequenced in paired-end mode using MiSeq sequencer (Illumina, San Diego, CA) and reagents version 3 (v.3) (600 cycles). A total of 2.166.962 paired reads were obtained. Illumina sequence reads were filtered by removing poor-quality data using FastX software v.0.0.14 (http://hannonlab.cshl.edu/fastx_toolkit/). The remaining adaptor sequences were removed using Cutadapt software v.1.1 (https://github.com/marcelm/cutadapt) using default settings. The filtered data were assembled into contigs using default parameters by Newbler software v.3.0 (Roche, USA), which allowed to obtain a draft sequence of bacterial genome. Assembly metrics were generated using Quast v.5.0.2 (quast.sourceforge.net). Genome assembly resulted in generation of 58 large contigs (min. 500 bp) with a total length of 2,585,340 bp. N50 of the contigs was of 88,601 bp, the genome coverage was 211×.
2.3. Genome Annotation
Genes were identified using the RAST v.2.0 and KAAS v.2.2 (parameters for bacterial genome) tools—Predicted genes were translated and functionally described [14,15,16]. Metabolic pathway prediction (KEGG pathway mapping - Kyoto Encyclopedia of Genes and Genomes) was performed with RAST v.2.0 tool [14,15,16] and KAAS v.2.2 (BlastKoala) to assign KEGG Orthology (KO) numbers to each predicted CDS. Clusters of Orthologous Groups of Proteins (COGs) were determined using eggNOG v.4.5.1 [17]. Ribosomal RNA genes were detected using RNAmer v.1.2 [18] and tRNA genes were identified using tRNAscan-SE v.2.0 [19]. Genome mapping (visualization of the genome properties) was performed using CGView software v.1.0 [20]. Transmembrane helices and signal peptides were found with TMHMM v.2.0 [21] and SignalP v.5.0 [22], respectively. CRISPR loci (Clustered Regularly Interspaced Short Palindromic Repeats) were searched for using the CRISPRFinder server. Antibiotic resistance genes were predicted using Comprehensive Antibiotic Resistance Database (CARD). Furthermore, a comparison of nucleotide sequences was performed using the BLAST tool. Unless otherwise mentioned, default parameters were used.
2.4. Comparative Genome Analysis
The genome sequences of P. freudenreichii ssp. freudenreichii DSM 20271 (accession no. NZ_CP010341.1) and P. freudenreichii ssp. shermanii CIRM-BIA1 (accession no. NC_014215.1) were obtained from GenBank. Sequences concerning P. freudenreichii ssp. shermanii NIZO B135 were also obtained from GenBank (accession no. AY787171.1, AY787170.1, AY787169.1). Genes were identified using the RAST v.2.0 [14,15,16]. KAAS v.2.2 (BlastKoala) was performed to assign KEGG Orthology (KO) numbers to each predicted CDS. Clusters of Orthologous Groups of Proteins (COGs) were determined using eggNOG v.4.5.1 [17].
2.5. Sugars Fermentation, Trehalose, and Glycogen Concentration
The ability to ferment different carbon sources by the tested strain was detected by API 50CH test, the ability to the production of trehalose (Table 1) and glycogen (Table 1) was measured by Trehalose Megazme Assay Kit and BioVision Glycogen Assay Kit, respectively. To check the ability to produce trehalose and glycogen the T82 strain was grown in medium containing apple pomace and potato wastewater (approx. 4% of sugars) in flasks in stationary conditions (30 °C) without neutralization of pH (acid stress) through the 4 days. The cells were separated by centrifugation for 10 min at 10,000 rpm at 4 °C and washed once with sterile distilled water. Then the concentration of trehalose and glycogen was measured according to the instructions attached to the kits.
Table 1.
Classification and general features of Propionibacterium freudenreichii subspecies freudenreichii T82 according to the MIGS recommendations (Minimum Information about a Genome Sequence) [12].
| MIGS ID | Property | Term |
|---|---|---|
| Classification |
Domain Bacteria Phylum
Actinobacteria Class Actinobacteria Order Propionbacteriales Family Propionibacteriaceae Genus Propionibacterium Species Propionibacterium freudenreichii subspecies freudenreichii |
|
| GenBank accession no. | NZ_SDDY00000000.1 | |
| Bioproject (NCBI) | PRJNA224116 | |
| Biosample (NCBI) | SAMN10768998 | |
| Gram strain | Positive | |
| Cell shape | Rod | |
| Motility | No | |
| Sporulation | No | |
| Optimum temperature | 37°C | |
| Optimum pH | approx. 7.0 | |
| Trehalose | +/(50.91 mg/g d.m.) (Megazyme test) | |
| Glycogen | +/(166.07 µg/g d.m.) (BioVision Glycogen) | |
| MIGS-6 | Habitat | Unknown |
| MIGS-6.3 | Salinity | Unknown |
| MIGS-22 | Oxygen-requirement | Anaerobic/facultative anaerobic |
| MIGHS-15 | Biotic relationship | Free-living |
| MIGS-14 | Phatogenicity | Non-pathogen |
| MIGS-4 | Geographic location | Unknown |
| MIGS-5 | Sample collection | Unknown |
| MIGS-4.1 | Latitude | Unknown |
| MIGS-4.2 | Longitude | Unknown |
| MIGS-4.4 | Altitude | Unknown |
2.6. Strain Deposition and Complete Genome Sequence Data Accession Number
The sequence data for P. freudenreichii T82 genome has been deposited at GenBank under the accession number NZ_SDDY00000000.1 (Table 1).
3. Results and Discussion
3.1. General Genome Features
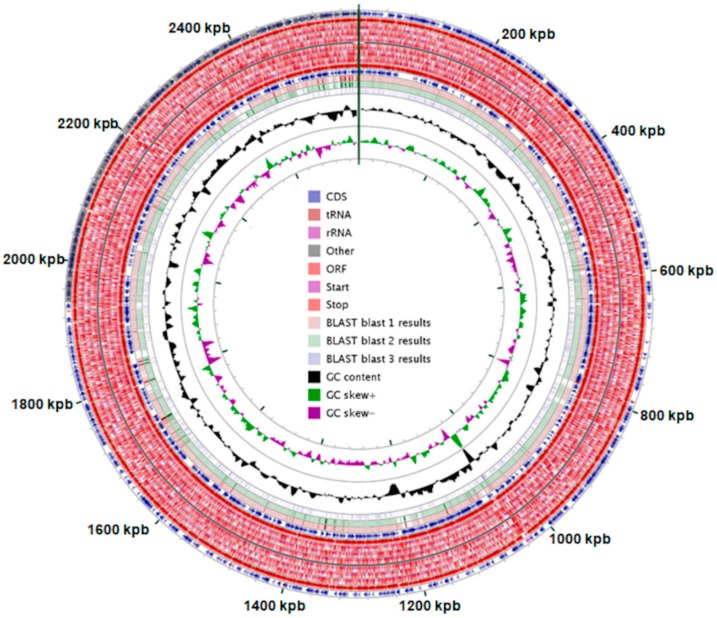
Propionibacterium freudenreichii T82 strain is a gram-positive, nonmotile, nonsporulating, mesophilic, and anaerobic to facultative anaerobic rod-shaped bacteria (Table 1). The genome of P. freudenreichii ssp. freudenreichii T82 contains 2,585,340 nucleotides with 67.3% GC content (Figure 1). The total number of genes is 2308, of which 2260 are protein-coding genes (97.9%) and 48 are RNA genes (2.08%) (Table 2). Bacteria with a small number of ribosomal operons are slow-growing organisms that can use resources efficiently and can grow under low nutrient conditions [23]. A total of 43 tRNA-encoding sequences were identified corresponding to all 20 standard amino acids (tRNAs with mismatch isotypes: (2): Ala, Gly, Pro, Thr, and Val (three sequences); Ser and Arg (4); Leu (5); Phe, Asn, Asp, His, Tyr, Trp, and Cys (1); Lys, Met, Gln, Ile, and Glu (2). The tested strain can utilize glycerol, erythritol, L-arabinose, galactose, glucose, fructose, mannose, inositol, xylitol, D-xylose, L-arabitol, potassium gluconate, esculin, and ferric citrate. The ability to ferment the last six carbon sources distinguishes the T82 strain from the DSM 20271 and CIRM-BIA1 strains (Table 3). Table 4 shows the distribution of P. freudenreichii T82 strain genes into functional categories according to COGs (Clusters of Orthologous Groups of Proteins) protein database [24], which groups proteins encoded by genomes of sequenced microorganisms into conservative families. These families are additionally divided into several superior functional groups.
Figure 1.
Genome properties of Propionibacterium freudenreichii ssp. freudenreichii T82 (in original size—Figure S11). Blast 1—Propionibacterium freudenreichii subsp. shermanii CIRM-BIA1, Blast 2—Propionibacterium freudenreichii subsp. freudenreichii DSM 20271, Blast 3—Tessaracoccus flavus. *genome mapping was performed using CGView software.
Table 2.
Genome statistics.
| Attribute | Value | % of Total |
|---|---|---|
| Genome size (bp) | 2,585,340 | 100.00 |
| DNA G + C (bp) | 1.739.933 | 67.30 |
| Total genes | 2308 | 100.00 |
| Protein coding genes | 2260 | 97.90 |
| RNA genes | 48 | 2.08 |
| tRNA | 45 | 1.95 |
| rRNA | 3 | 0.12 |
| Genes assigned to COGs | 1936 | 85.66 |
| Genes assigned to KEGG | 1222 | 49.64 |
| Gen with transmembrane helices | 665 | 28.81 |
| Genes with signal peptides | 142 | 6.15 |
Table 3.
Propionibacterium freudenreichii carbon substrate degradation.
| Carbon Source | P. freudenreichii T82 | P. freudenreichii DSM 20271 [12] | P. freudenreichii CIRM-BIA1 [11] |
|---|---|---|---|
| Glucose | + | + | + |
| Fructose | + | + | + |
| Mannose | + | - | + |
| Glycerol | + | + | + |
| Adonitol | - | + | + |
| Inositol | + | + | + |
| Erythritol | + | + | + |
| Galactose | + | + | + |
| Lactose | - | - | + |
| Lactic acid | No data | - | + |
| Gluconic acid | No data | - | - |
| Esculine hydrolisis | No data | - | + |
| L-arabinose | + | + | + |
| Ribose | - | - | - |
| Melbiose | - | - | - |
| Raffinose | - | - | - |
| Saccharose | - | - | - |
| L-arabitol | + | - | - |
| Xylitol | + | - | - |
| D-xylose | + | - | - |
| Esculin | + | - | - |
| Ferric citrate | + | - | - |
| Potassium gluconate | + | - | - |
Table 4.
Number of genes associated with general Clusters of Orthologous Groups of Proteins (COGs) functional categories.
| Code | Description | Value 1 | % | Value 2 | % | Value 3 | % |
|---|---|---|---|---|---|---|---|
| Information Storage and Processing | T82 | DSM 20271 [12] | CIRM-BIA1 [11] | ||||
| J | Translation, ribosomal structure and biogenesis | 151 | 7.79 | 153 | 7.50 | 155 | 7.65 |
| A | RNA processing and modification | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| K | Transcription | 159 | 8.21 | 159 | 7.80 | 165 | 8.14 |
| L | Replication, recombination and repair | 163 | 8.41 | 227 | 11.13 | 242 | 11.94 |
| B | Chromatin structure and dynamics | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| Cell processes and signaling | |||||||
| D | Cell cycle control, Cell division, chromosome partitioning | 30 | 1.55 | 30 | 1.47 | 32 | 1.58 |
| Y | Nuclear structure | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| V | Defense mechanisms | 39 | 2.01 | 40 | 1.96 | 42 | 2.07 |
| T | Signal transduction mechanisms | 66 | 3.41 | 69 | 3.24 | 69 | 3.41 |
| M | Cell wall/membrane biogenesis | 100 | 5.17 | 99 | 4.86 | 95 | 4.69 |
| N | Cell motility | 9 | 0.04 | 9 | 0.44 | 9 | 0.44 |
| Z | Cytoskeleton | 0 | 0.00 | 0 | 0.00 | 1 | 0.05 |
| W | Extracellular structures | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| U | Intracellular trafficking and secretion | 39 | 2.01 | 37 | 1.81 | 35 | 1.73 |
| O | Posttranslational modification, protein turnover, chaperones | 74 | 3.82 | 78 | 3.82 | 81 | 4.00 |
| Metabolism | |||||||
| C | Energy production and conversion | 136 | 7.02 | 141 | 6.91 | 140 | 6.91 |
| G | Carbohydrate transport and metabolism | 173 | 8.94 | 174 | 8.53 | 174 | 8.59 |
| E | Amino acid transport and metabolism | 210 | 10.85 | 221 | 10.85 | 214 | 10.56 |
| F | Nucleotide transport and metabolism | 65 | 3.36 | 65 | 3.19 | 64 | 3.16 |
| H | Coenzyme transport and metabolism | 113 | 5.84 | 116 | 5.70 | 112 | 5.52 |
| I | Lipid transport and metabolism | 64 | 3.31 | 65 | 3.19 | 64 | 3.16 |
| P | Inorganic ion transport and metabolism | 153 | 7.90 | 154 | 7.55 | 140 | 6.91 |
| Q | Secondary metabolites biosynthesis, transport and catabolism | 38 | 1.96 | 48 | 2.35 | 43 | 2.12 |
| Uncharacterized | |||||||
| R | General function prediction | 0 | 0.00 | 0 | 0.00 | 0 | 0.00 |
| S | Function unknown | 331 | 17.09 | 336 | 16.48 | 331 | 16.34 |
1 genes assigned to COGs—1936 (the total value is 2113, some genes were assigned to more than one class); 2 genes assigned to COGs—2039 (the total value is 2221, some genes were assigned to more than one class); 3 genes assigned to COGs—2026 (the total value is 2078, some genes were assigned to more than one class); green—the same numeric values/orange—the same percentages/white—different values.
To date, the complete sequences of P. freudenreichii ssp. shermanii CIRM-BIA1 and P. freudenreichii ssp. freudenreichii DSM 20271 strains have been described in the literature. P. freudenreichii T82 16S rRNA sequence shows 98% similarity with that of DSM 20271 strain. Comparison of genomes of the T82 and DSM 20271 strains revealed some other similarities. First, guanine and cytosine constitute 67.3% of all bases in the genomes of both microorganisms, whereas RNA constitutes only 2.08% of the genome. Moreover, the T82 strain can reduce nitrate to nitrite (respiratory nitrate reductase alpha–gamma chains [EC 1.7.99.4], nitrate and nitrite transporter) but cannot ferment lactose. Degradation of lactose is strain-dependent. In P. freudenreichii ssp. shermanii CIRM-BIA1 genome, the lactose locus contains three genes, namely PFREUD_02370, PFREUD_02360, and PFREUD_02350, which are responsible for encoding, respectively: β-galactosidase, LacZ; a galactosidase transporter, GalP; and an UDP-glucose isomerase, GalE1. In the P. freudenreichii T82 strain, sequences for only β-galactosidase were detected. All these findings indicate that the T82 strain may represent a subspecies P. freudenreichii ssp. freudenreichii.
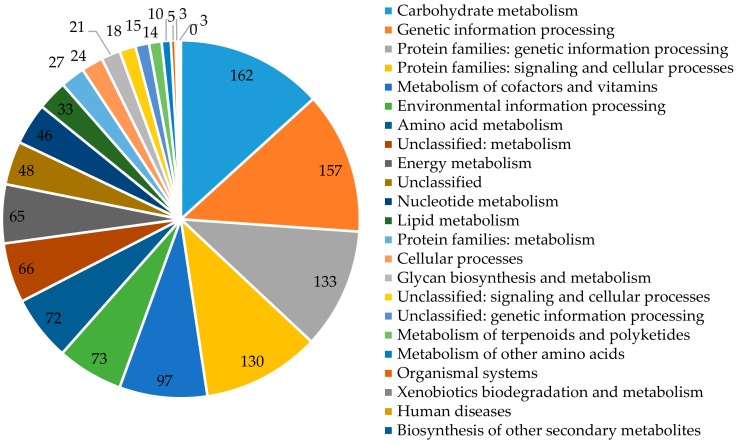
Each group is assigned a function and contains orthological proteins from at least three phylogenetic lines, which most likely evolved from a single ancestor. Functional assignment of the examined protein is based on its classification to one or more (if it is a multidomain protein) orthologous groups on the basis of sequence similarity. For P. freudenreichii T82 strain, out of 2260 identified encoding sequences, 1936 sequences were grouped in 20 COGs classes (the sum of all sequences in 20 COGs classes is 2113, because some sequences are assigned to more than one class) and 1228 in KEGG categories (Figure 2).
Figure 2.
The number of genes assigned in KEGG categories.
For COGs, coding sequences were identified, inter alia, as involved in amino acid transport and metabolism (10.85%); carbohydrate transport and metabolism (8.94%); replication, recombination and repair of nucleic acids (8.41%); translation, ribosome structure, and biogenesis (7.79%); or transcription (8.21%) (Table 4). A large number of unclassified genes (KEGG-150), with unknown function (COGs-331) and hypothetical proteins (653) show that among them may be unique genes. According to COGs, the pool of genes involved in metabolism of the T82, DSM 20271 and CIRM-BIA1 strains is 952 (49.17%), 984 (48.26%), and 951 (46.94%), respectively. Regarding KEGG-(in sequence) 50.40%, 50.00%, and 50.08% of classified genes are related to categories involving metabolism (Table S1).
3.2. Metabolism
Propionibacterium bacteria need nicotinate (B3), riboflavin (B4), pantothenate (B5), biotin (B7), cobalamin (B12), and coenzyme A (CoA) for enzymatic activity and some strains additionally require thiamine (B1) [11]. Genome analysis showed that there are complete synthesis pathways of almost all these cofactors in the T82 strain (Figures S1–S3, S5, and S7), except for pantothenate (Figure S7) and biotin (Figure S9). Results described by Koskinen et al. [12] and Falentin et al. [11] also show that PAB have to be supplemented by the addition to the medium of B5 and B7 vitamins. Moreover, according to the obtained results, it is possible that the P. freudenreichii T82 strain can produce folate (B9 from chorismite, which is also produced by the T82 strain; Figure S10) (Figure S5), thiamine (Figure S1), and pyridoxal 5′-phosphate (active form of B6) (Figure S6). According to Deptula et al. [25], the production of active therapeutic vitamin B12 by PAB requires the synthesis of 5.6-dimethylbenzimidazole (DMBI) as the initial ligand. Genomic data indicate that P. freudenreichii T82 possesses a fusion gene, bluB/cobT2 (cobalamin biosynthesis protein BluB @5.6-dimethylbenzimidazole synthase, flavin destructase family/Nicotinate-nucleotide-dimethylbenzimidazole phosphoribosyltransferase [EC 2.4.2.21]), coding for a predicted phosphoribosyltransferase/nitroreductase, which is involved in the production of DMBI and -consequently vitamin B12.
Although P. freudenreichii is usually cultured under anaerobic or relatively anaerobic conditions, the species, including the P. freudenreichii T82 strain, possess all the genes required for aerobic respiration: NADH dehydrogenase (EC 1.6.5.3, chain A-N), cytochrome complex (bd) (cytochrome d ubiquinol oxidase subunits I and II [EC 1.10.3], succinate dehydrogenase cytochrome b subunit, cytochrome c-type biogenesis protein DsbD, protein disulfide reductase [EC 1.8.1.8], putative cytochrome P450 hydroxylase) ATPase, and heme synthesis pathway (Figure S8). In anaerobic conditions, sulfates, fumarate, nitrates, and menaquinone (vitamin K2) (Figure S4) are the acceptors of electrons in P. freudenreichii [26].
Propionic acid bacteria produce propionic acid through the Wood–Werkman cycle. The species P. freudenreichii has been widely studied at the biochemical and genetic levels [11,27]. The key reaction of the Wood–Werkman cycle is the transcarboxylation reaction without free CO2. The enzyme catalyzing this reaction is methylmalonyl-CoA carboxytransferase, which transfers the carboxyl group from methylmalonyl-CoA to pyruvate with the formation of oxaloacetate and propionyl-CoA. This enzyme has been fully characterized. It is a biotin-dependent carboxytransferase (EC 2.1.3.1) that consists of three subunits (1,3S, 5S, and 12S). The methylmalonyl-CoA carboxytransferase is encoded by a polycistronic gene containing four coding sequences. Three of them encode the individual subunits of the enzyme, and one encodes the carrier protein transporting the carboxylic biotin. All of them are present in the P. freudenreichii T82 strain.
Propionibacterium freudenreichii shows many characteristics that allow them to survive in unfavorable environmental growth conditions. For example, they can store inorganic polyphosphate (polyP) as an energy reserve, while most bacteria store only ATP. The T82 strain also has this ability. Importantly, only bacteria that are particularly adapted to extreme conditions can use polyP as an energy source [28]. A key enzyme involved in the synthesis of this energy carrier is polyphosphate kinase (EC 2.7.4.1). The P. freudenreichii T82 strain also shows the presence of exopolyphosphatase (EC 3.6.1.11) and polyphosphate glucokinase (EC 2.7.1.63).
Genes potentially involved in glycogen metabolism were also identified in the genome of the T82 strain. This feature was also reported in P. freudenreichii ssp. shermanii CIRM-BIA1T strain [11]. The ability to synthesize this compound by the T82 strain depend on the sequences encoding glycogen synthase (EC 2.4.1.21), glycogen phosphorylase (EC 2.4.1.1), and enzymes branching glycogen (EC 2.4.1.18). Some of these genes were also found in P. acnes. Because P. freudenreichii and P. acnes cannot ferment extracellular glycogen, these enzymes are expected to be involved in intracellular glycogen accumulation and/or hydrolysis.
Strains of P. freudenreichii [29], including T82, can also synthesize and accumulate trehalose from glucose and pyruvate. The synthesis of trehalose by Propionibacterium occurs at the beginning of the stationary phase under oxidative, osmotic, and acidic stress conditions. Trehalose is synthesized by PAB through the synthase pathway of trehalose-6-phosphate (EC 2.4.1.15)/phosphatase (EC 3.1.3.12) (OtsA-OtsB) and catalyzed by trehalose synthase (TreS) (EC 5.4.99.16). The otsA, otsB, and treS genes were previously identified in NIZO B365, DSM 20271, and CIRM-BIA1 strains. In the P. freudenreichii T82 strain, nucleotide sequences encoding these three genes are similar to those reported by Cardoso et al. [29], Koskinen et al. [12], and Falentin et al. [11], respectively: 98.45% (NIZO B365) and 100% (DSM and CIRM-BIA1); 99.07% (NIZO B365), 99.88% (DSM), and 99.65% (CIRM-BIA1); and 99.78% (NIZO B365), 99.78% (DSM 20271), and 99.89% (CIRM-BIA1).
3.3. Resistance and Stress Response
The biosynthesis of propionic acid by Propionibacterium is inhibited mainly by a negative feedback mechanism and stress conditions. According to the RAST analysis, in theT82 strain, 57 genes are responsible for stress response and are divided into six groups: osmotic (7), oxidative (26), cold shock (2), heat shock (15), detoxification (10), and no subcategory (4) (Table S2). Therefore, as suggested by researchers, the most effective strategy to increase PAB biomass and propionic acid synthesis is improving the resistance of these bacteria to low pH and stress conditions in general. For this purpose, adaptive evolution and genome shuffling have been used [30]. It was also found that arginine deaminase (EC 3.5.3.6) and glutamate decarboxylase (EC 4.1.1.15) (the sequence encoding this gene is present in the genome of the T82 strain) play an important role in acid tolerance of P. acidipropionici [31,32]. Guan et al. [33] attempted to improve the resistance of P. jensenii ATCC 4868 strain to low pH by inducing overexpression of five genes: Arca, ARCC, gadB, GDH, and ybaS. Suwannakham et al. [34] removed the ack gene encoding acetic kinase from the genome of P. acidipropionici strain, thus increasing propionic acid production. Immobilization was also used to increase PAB resistance to stress conditions [35]. However, the current knowledge on the functioning of acid resistance in the cells of Propionibacterium remains at the microenvironmental level. Therefore, further research is needed to understand these mechanisms. System biology methods and genome analysis may be useful in this context. Technologies comparing genomics and transcriptomics can be used to induce resistance of strains to acids at the DNA and RNA level, while proteomics and metabolomics can be used to identify key proteins and metabolites as well as the pathways responsible for a particular trait. For example, Lu et al. [36] identified a previously unknown system affecting the acid resistance of Escherichia coli, namely the transformation of L-glutamine into L-glutamic acid with the simultaneous production of ammonia, alkalizing the culturing environment.
As the examples show, although it is possible to increase the yield of propionic acid by bacteria of the Propionibacterium genus, this task is quite difficult. The inhibitors of metabolic and genetic engineering of Propionibacterium are their restriction–modification (RM) systems [37,38]. These systems provide protective mechanisms against foreign DNA and bacteriophages, which, at the same time, are an obstacle to genetic manipulation. To distinguish foreign DNA from the host DNA, these systems coordinate the action of restrictive and modifying enzymes, thus protecting Propionibacterium cells from the penetration of foreign genetic material. In classical RM systems, foreign DNA is cleaved or restricted by endonucleases. Host cell DNA avoids restriction through the methylation, or modification, of certain adenine or cytosine residues in the target sequence. On the basis of subunit composition, cofactor requirements, and position of the DNA cleavage site, RM systems have been classified into four distinct groups, namely, type I, type II, type III, and type IV. In the P. freudenreichii T82 strain, the type I RM system was detected (type I restriction-modification system, DNA-methyltransferase subunits M, R, and S [EC 2.1.1.72], type I restriction-modification system, restriction subunit R [EC 3.1.21.3]) and two sequences involved in type III (type III restriction-modification system methylation subunit [EC 2.1.1.72] and type III restriction-modification enzyme helicase subunit). The type I RM system is a bifunctional, multisubunit complex containing products of the hsdR, hsdM, and hsdS genes (host specificity for DNA) [39]. HsdS interacts with the target sequence as a component of the restriction and modification complexes. HsdS consists of four domains: two variable target recognition domains (TRDs), a central-conserved domain, and a conserved C-terminus domain. The type I target sequence is asymmetric and composed of two half-sites: a 5ʹ half-site of 3–4 bp and a 3ʹ half-site of 4–5 bp, separated by a nonspecific spacer of 6–8 bp. In the HsdS subunit, each TRD recognizes one half-site, while the conserved domains are thought to interact with the HsdR and HsdM proteins in the complex.
CRISPR represent a family of DNA repeats providing acquired immunity against foreign genetic elements, for example, protection against bacteriophages [40]. They consist of short and highly conserved repeats with variable sequences called spacers. CRISPR-associated genes (Cas) are found often next to these sequences. CRISPR are found only in 40% of bacterial genomes [40]. The genome of the P. freudenreichii T82 strain is composed of three confirmed and two questionable CRISPR loci (Table S3). The detected CRISPR/CRISPR-associated (Cas) system consists of three Cas genes: Cas1, Cas2, and Cas4.
The CRISPR1 locus, located between 16 and 3173 bp (Scaffold 16—length 59,854 bp), harbors a 36 bp direct repeat (DR) sequence GCCTCAATGAAGGGCCCCTCCAGAAGGAGGGGCAAT and 43 spacer sequences. Blast results on the EMBL phage database showed that the 10 spacer show a strong similarity to the Propionibacterium phages - spacer 9 (PFR2 38/38 nt, PFR1 38/38 nt), spacer 17 (PFR2 39/39 nt, PFR1 39/39 nt), spacer 23 (PFR2 38/38 nt, PFR1 38/38 nt), 27 spacer (E6 38/38 nt, Doucette 38/38 nt), spacer 28 (E6 40/40 nt, Doucette 40/40 nt, B22 40/40 nt), spacer 29 (Doucette 35/35 nt), spacer 33 (G4 37/37 nt), spacer 38 (G4 37/37 nt, E1 37/37 nt, B22 37/37 nt, B3 37/37 nt, Anatole 37/37 nt, spacer 39 (G4 35/35 nt), spacer 43 (Doucette 37/37 nt). Two other possible CRISPR loci were identified (Table S3). The presence of CRISPR loci causes that the genome stability of a bacterial strain may be increasing, therefore, also its adaptation to the environment. What is more, it strongly suggests that P. freudenreichii T82 has had a contact with phages—they may contribute to its resistance to phage attacks [11].
RNA modification enzymes manifested as methyltransferases play a large role in antibiotic resistance in bacteria. The modifications of the ribosome almost exclusively involve methylation of various positions on the bases or at the 2’-O-ribose position. Extensive information on antibiotic resistance caused by methylation of rRNA is available. Modifications at eight 23S rRNA nucleotides (G748, A1067, C1920, A2058, G2470, U2479, A2503, and G2535) on the large ribosomal subunit have thus far been revealed as antibiotic resistance determinants [41]. In the P. freudenreichii T82 strain, the 23S rRNA with mutations at G2294A and G2295A is responsible for resistance to macrolide antibiotics. This shows that this strain had direct contact with the environment in which antibiotics were present, probably soil or cattle, which is one of the most common sources of P. freudenreichii.
Some strains of Propionibacterium are able to accumulate glycine betaine which is involved in long-term survival by acting as a chemical chaperone [11]. Genes supporting glycine betaine transport were identified in the genome of P. freudenreichii T82. On the other hand, genes responsible for encoding enzymes engaged in producing this compound were not detected—betaine aldehyde dehydrogenase (EC 1.2.1.8) and choline dehydrogenase (EC 1.1.99.1). It is possible that these genes are amongst uncharacterized coding sequences (Table S4).
Regulation of intracellular pH is crucial for survival. Analysis of the P. freudenreichii T82 strain genome shows a complete atpBEFHAGDC operon with gamma chain (atpG, PFREUD_10480) which is activated in P. freudenreichii by acid and bile salts. To keep intracellular pH homeostasis the genome of the T82 strain contains genes encoding: pyruvate-flavodoxin oxidoreductase, aspartate ammonia-lyase (EC 4.3.1.1), glutamate decarboxylase (EC 4.1.1.15), and succinate dehydrogenase (Table S4).
The genome of the P. freudenreichii T82 strain was found to encode few proteins involved in transcriptional regulation, including three genes encoding sigma factors and 59 genes responsible for transcriptional regulators. First, a relatively large number of regulatory proteins are present in the large genomes; this probably confirm that this strain has the machinery to adapt to different environmental niches. Regulatory proteins are important for the adaptation of an organism to different environments [42]. The T82 strain carries various genes encoding heat stress proteins, namely DnaJ, DnaK, GrpE, GroES, and GroEL, and cold stress proteins from the CSP family. The genes in the T82 strain genome may also be involved in oxidative stress tolerance by coding for proteins such as thioredoxin reductase, peroxidase, catalase, and superoxide dismutase. Bacteria of the P. freudenreichii genus possesses a lot of genes for disulfide-reduction and to eliminate of reactive oxygen forms (cysteine synthase, glutathione S-transferase, omega) [43]. Genes involved in the redox-dependent regulation of nucleus processes are also found. The similar situation was found for the other strains of bacteria of the Propionibacterium genus (Table S4).
Among the genes activated in response to stressors are polybibonucleotide nucleotidyltransferase and inosine dehydrogenase—it suggests the ability of the T82 strain to synthesize alarmon ppGpp during the stress conditions (Table S4).
4. Conclusions
The presence of genes involved in the metabolism of phosphates, glycogen, trehalose, and CRISPR loci and the genes responsible for stress response in the genome of P. freudenreichii T82 strain make this strain well adapted to the culturing environment (as shown by the results of this study) and capable of long-term survival under culturing conditions, especially in the stationary phase. These properties along with the ability of this strain to produce valuable metabolites indicate that this strain has a huge potential in the industry for the production of propionic acid, vitamins, and trehalose and for product enrichment with the biomass of PAB. For this purpose, further research is needed to increase cost-effectiveness and achieve high production efficiencies of specific metabolites. The first condition can be met by using industrial waste as microbiological media, which contain in its composition nutrient sources assimilable by a given strain. Production efficiency can be improved using metabolic and genetic engineering tools. Furthermore, knowledge of the genetic material of the strain used at the molecular level (matching the strain to a specific waste and improvement of the strain through genetic modification) will certainly help in the development of technological processes.
Acknowledgments
We thank Jan Gawor from the DNA Sequencing and Oligonucleotide Synthesis Laboratory, IBB Polish Academy of Science for his technical assistance during genome sequencing. Genomic DNA isolation, library construction and bacterial genome assembly were carried out at the oligo.pl—DNA Sequencing and Oligonucleotide Synthesis Laboratory IBB PAS.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/2/348/s1.
Author Contributions
K.P. collected and reviewed the literature and wrote the manuscript (drafted and critically reviewed), performed the analysis, compiled the results. E.L. reviewed the manuscript and provided supervision. E.H.-S. reviewed the manuscript and provided supervision. M.K. and A.M.K. performed the analysis, compiled the results. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the Warsaw University of Life Sciences—SGGW Internal Grant (505-10-092800-N00295-99).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Meile L., Dasen G., Miescher S., Stierli M., Teuber M. Classification of propionic acid bacteria and approaches to applied genetics. Le Lait. 1999;79:71–78. doi: 10.1051/lait:199915. [DOI] [Google Scholar]
- 2.Koussémon M., Combet-Blanc Y., Ollivier B. Glucose fermentation by Propionibacterium microaerophilum: Effect of pH on metabolism and bioenergetics. Curr. Microbiol. 2004;44:141–145. doi: 10.1007/s00284-002-3839-x. [DOI] [PubMed] [Google Scholar]
- 3.Leverrier P., Vissers J.P.C., Rouault A., Boyaval P., Jan G. Mass spectrometry proteomic analysis of stress adaptation reveals both common and distinct response pathways in Propionibacterium freudenreichii. Arch. Microbiol. 2004;181:215–230. doi: 10.1007/s00203-003-0646-0. [DOI] [PubMed] [Google Scholar]
- 4.Cardoso F.S., Gaspar P., Hugenholtz J., Ramos A., Santos H. Enhancement of trehalose production in dairy propionibacteria through manipulation of environmental conditions. Int. J Food Microbiol. 2004;91:195–204. doi: 10.1016/S0168-1605(03)00387-8. [DOI] [PubMed] [Google Scholar]
- 5.Suomalainen T.H., Sigvart-Mattila P., Mättö J., Tynkkynen S. In vitro and in vivo gastrointestinal survival, antibiotic susceptibility and genetic identification of Propionibacterium freudenreichii ssp. shermanii JS. Int. Dairy. J. 2008;18:271–278. doi: 10.1016/j.idairyj.2007.09.004. [DOI] [Google Scholar]
- 6.Deptula P., Chamlagain B., Edelman M., Sangsuwan P., Nyman T.A., Savijoki K., Piironen V., Varmanen P. Food-Like Growth Conditions Support Production of Active Vitamin B12 by Propionibacterium freudenreichii 2067 without DMBI, the Lower Ligand Base, or Cobalt Supplementation. Front. Microbiol. 2017;8:1–11. doi: 10.3389/fmicb.2017.00368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Chamlagain B., Sugito T.A., Deptula P., Edelmann M., Kariluoto S., Varmanen P., Piironen P. In situ production of active vitamin B12 in cereal matrices using Propionibacterium freudenreichii. Food Sci. Nutr. 2018;6:67–76. doi: 10.1002/fsn3.528. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Meile L., Gwenaelle L.B., Thierry A. Safety assessment of dairy microorganisms: Propionibacterium and Bifidobacterium. Int. J Food Microbiol. 2008;126:316–320. doi: 10.1016/j.ijfoodmicro.2007.08.019. [DOI] [PubMed] [Google Scholar]
- 9.Borawska B., Warmińska-Radyko I., Darewicz M. Charakterystyka i znaczenie bakterii rodzaju Propionibacterium w produkcji żywności i pasz. Med. Wet. 2010;66:534–537. [Google Scholar]
- 10.Ranadheera R.D.S.C., Baines S.K., Adams M.C. Importance of food in probiotic efficacy. Food Res. Int. 2010;43:1–7. doi: 10.1016/j.foodres.2009.09.009. [DOI] [Google Scholar]
- 11.Falentin H., Deutsch S.M., Jan G., Loux V., Thierry A., Parayre S., Maillard M.B., Dherbécourt J., Cousin F.J., Jardin J., et al. The Complete Genome of Propionibacterium freudenreichii CIRM-BIA1T, a Hardy Actinobacterium with Food and Probiotic Applications. PLoS ONE. 2010;5:1–12. doi: 10.1371/journal.pone.0011748. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Koskinen P., Deptula P., Smolander O.P., Tamene F., Kammonen J., Savijok K., Paulin L., Piironen V., Auvinen P., Varmanen P. Complete genome sequence of Propionibacterium freudenreichii DSM 20271T. Stand. Genomic Sci. 2015;10:1–6. doi: 10.1186/s40793-015-0082-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wilson K. Preparation of genomic DNA from bacteria. In: Ausubel F.M., Bent R., Kingston R.E., editors. Current Protocols in Molecular Biology. J Wiley & Sons; New York, NY, USA: 2001. [DOI] [PubMed] [Google Scholar]
- 14.Aziz R.K., Bartels D., Best A.A., DeJongh M., Disz T., Edwards R.A., Formsma K., Gerdes S., Glass E.M., Kubal M., et al. The RAST Server: Rapid Annotations using Subsystems Technology. BMC Genomics. 2008;9:1–15. doi: 10.1186/1471-2164-9-75. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Overbeek R., Olson R., Pusch G.D., Olsen G.J., Davis J.J., Disz T., Edwards R.A., Gerdes S., Parrello B., Shukla M., et al. The SEED and the Rapid Annotation of microbial genomes using Subsystems Technology (RAST) Nucleic Acids Res. 2014;42:D206–D214. doi: 10.1093/nar/gkt1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Brettin T., Davis J.J., Disz T., Edwards R.A., Gerdes S., Olsen G.J., Olson R., Overbeek R., Parrello B., Pusch G.D., et al. RASTtk: A modular and extensible implementation of the RAST algorithm for building custom annotation pipelines and annotating batches of genomes. Sci. Rep. 2015;5:1–6. doi: 10.1038/srep08365. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Huerta-Cepas J., Szklarczyk D., Forslund K., Cook H., Heller D., Walter M.C., Rattei T., Mende D.R., Sunagawa S., Kuhn M. eggNOG 4.5: A hierarchical orthology framework with improved functional annotations for eukaryotic, prokaryotic and viral sequences. Nucleic Acids Res. 2016;44:D286–D293. doi: 10.1093/nar/gkv1248. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lagesen K., Hallin P., Rødland E.A., Stærfeldt H.H., Rognes T., Ussery D.W. RNAmmer: Consistent and rapid annotation of ribosomal RNA genes. Nucleic Acids Res. 2007;35:3100–3108. doi: 10.1093/nar/gkm160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Lowe T.M., Chan P.P. tRNAscan-SE On-line: Integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res. 2016;44:54–55. doi: 10.1093/nar/gkw413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Stothard P., Wishart D.S. Circular genome visualization and exploration using CGView. Bioinformatics. 2005;21:537–539. doi: 10.1093/bioinformatics/bti054. [DOI] [PubMed] [Google Scholar]
- 21.Krogh A., Larsson B., Von Heijne G., Sonnhammer E.L. Predicting transmembrane protein topology with a hidden markov model: Application to complete genomes. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 22.Petersen T.N., Brunak S., von Heijne G., Nielsen H. SignalP 4.0: Discriminating signal peptides from transmembrane regions. Nat. Methods. 2011;8:785–786. doi: 10.1038/nmeth.1701. [DOI] [PubMed] [Google Scholar]
- 23.Kaneko T., Mori H., Iwata M., Meguro S. Growth stimulator for bifidobacteria produced by Propionibacterium freudenreichii and several intestinal bacteria. J. Dairy Sci. 1993;73:393–404. doi: 10.3168/jds.S0022-0302(94)76965-4. [DOI] [PubMed] [Google Scholar]
- 24.Tatusov R.L., Galperin M.Y., Natale D.A., Koonin E.V. The COG database: a tool for genome-scale analysis of protein functions and evolution. Nucleic Acids Res. 2008;28:33–36. doi: 10.1093/nar/28.1.33. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Deptula P., Kylli P., Chamlagain B., Holm L., Kostiainen R., Piironen V., Savijoki K., Varmanen P. BluB/CobT2 fusion enzyme activity reveals mechanisms responsible for production of active form of vitamin B12 by Propionibacterium freudenreichii. Microb. Cell Fact. 2015;14:1–12. doi: 10.1186/s12934-015-0363-9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Benz M., Schink B., Brune A. Humic acid reduction by Propionibacterium freudenreichii and other fermenting bacteria. Appl. Environ. Microbiol. 1998;65:4507–4512. doi: 10.1128/AEM.64.11.4507-4512.1998. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Beck S., Schink B. Acetate oxidation through a modified citric acid cycle in Propionibacterium freudenreichii. Arch. Microbiol. 1995;163:182–189. doi: 10.1007/BF00305351. [DOI] [Google Scholar]
- 28.Seufferheld M.J., Alvarez H.M., Farias M.E. Role of polyphosphates in microbial adaptation to extreme environments. Appl. Environ. Microbiol. 2008;74:5867–5874. doi: 10.1128/AEM.00501-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Cardoso F.S., Castro R.F., Borges N., Santos H. Biochemical and genetic characterization of the pathways for trehalose metabolism in Propionibacterium freudenreichii, and their role in stress response. Microbiology. 2007;153:270–280. doi: 10.1099/mic.0.29262-0. [DOI] [PubMed] [Google Scholar]
- 30.Guan N., Liu L., Chen R.R., Zhuge X., Xu Q., Li J., Du G., Chen J. Genome shuffling improves acid tolerance of Propionibacterium acidipropionici and propionic acid production. Adv. Chem. Res. 2012;15:143–152. [Google Scholar]
- 31.Guan N., Liu L., H-d S., Chen R.R., Zhang J., Li J., Du G., Shi Z., Chen J. Systems-level understanding how Propionibacterium acidipropionici respond to propionic acid stress at the microenvironment levels: Mechanism and application. J. Biotechnol. 2013;167:56–63. doi: 10.1016/j.jbiotec.2013.06.008. [DOI] [PubMed] [Google Scholar]
- 32.Guan N., Shin H.D., Chen R.R., Li J., Liu L., Du G., Chen J. Understanding of how Propionibacterium acidipropionici respond to propionic acid stress at the level of proteomics. Sci. Rep. 2014;6951:1–8. doi: 10.1038/srep06951. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Guan N., Li J., Shin H., Du G., Chen J., Liu L. Metabolic engineering of acid resistance elements to improve acid resistance and propionic acid production of Propionibacterium jensenii. Biotechnol. Bioeng. 2015a;113:1294–1304. doi: 10.1002/bit.25902. [DOI] [PubMed] [Google Scholar]
- 34.Suwannakham S., Huang Y., Yang S.T. Construction and characterization of ack knock-out mutants of Propionibacterium acidipropionici for enhanced propionic acid fermentation. Biotechnol. Bioeng. 2006;94:383–395. doi: 10.1002/bit.20866. [DOI] [PubMed] [Google Scholar]
- 35.Zhu L., Wei P., Cai J., Zhu X., Wang Z., Huang L., Xu Z. Improving the productivity of propionic acid with FBB-immobilized cells of an adapted acidtolerant Propionibacterium acidipropionici. Bioresour. Technol. 2012;112:248–253. doi: 10.1016/j.biortech.2012.01.055. [DOI] [PubMed] [Google Scholar]
- 36.Lu P., Ma D., Chen Y., Guo Y., Chen G.Q., Deng H., Shi Y. L-glutamine provides acid resistance for Escherichia coli through enzymatic release of ammonia. Cell Res. 2013;23:635–644. doi: 10.1038/cr.2013.13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Van Luijk N., Stierli M.P., Schwenninger S.M., Hervé C., Dasen G., Jore J.P., Pouwels P.H., Van der Werf M.J., Teuber M., Meile L. Genetics and molecular biology of propionibacteria. Le Lait. 2002;82:45–57. doi: 10.1051/lait:201004. [DOI] [Google Scholar]
- 38.Guan N., Li J., Shin H., Wu J., Du G., Shi Z., Liu L., Chen J. Comparative metabolomics analysis of the key metabolic nodes in propionic acid synthesis in Propionibacterium acidipropionici. Metabolomics. 2015b;11:1106–1116. doi: 10.1007/s11306-014-0766-3. [DOI] [Google Scholar]
- 39.Cooper L.P., Roberts G.A., White J.H., Luyten Y.A., Bower E.K.M., Morgan R.D., Roberts R.J., Lindsay J.A., Dryden D.T.F. DNA target recognition domains in the Type I restriction and modification systems of Staphylococcus aureus. Nuceleic Acid Res. 2017;45:3395–3406. doi: 10.1093/nar/gkx067. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Amitai G., Sorek R. CRISPR-Cas adaptation: Insights into the mechanism of action. Nat. Rev. Microbiol. 2016;14:67–76. doi: 10.1038/nrmicro.2015.14. [DOI] [PubMed] [Google Scholar]
- 41.Vester B., Long K.S. Resistance to linezolid caused by modifications at its binding site on the ribosome. Antimicrob. Agents Chemother. 2002;56:1–10. doi: 10.1128/AAC.05702-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Zhang W., Wang J., Zhang D., Liu H., Wang S., Wang Y., Ji H. Complete Genome Sequencing and Comparative Genome Characterization of Lactobacillus johnsonii ZLJ010, a Potential Probiotic With Health-Promoting Properties. Front. Genet. 2019;10:1–13. doi: 10.3389/fgene.2019.00812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Pophaly S.D., Singh R., Pophaly S.D., Kaushik J.K., Tomar S.K. Current status and emerging role of glutathione in food grade lactic acid bacteria. Microb. Cell Fact. 2012;11:1–14. doi: 10.1186/1475-2859-11-114. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.