Abstract
Human skin dermis contains fibroblast subpopulations in which characterization is crucial due to their roles in extracellular matrix (ECM) biology. This study investigates the properties of fibroblasts localized at the frontier of deep dermis and hypodermis, i.e., dermo-hypodermal junction fibroblasts (F-DHJ), which were compared to intermediate reticular dermis (Fr) and superficial papillary dermis (Fp) fibroblasts. F-DHJ differed from Fr and Fp cells in their wider potential for differentiation into mesodermal lineages and in their absence of contractility when integrated in a three-dimensional dermal equivalent. The transcriptomic profile of F-DHJ exhibited specificities in the expression of genes involved in ECM synthesis-processing and “tissue skeleton” organization. In accordance with transcriptome data, ECM proteins, notably Tenascin C, distributions differed between the reticular dermis and the dermo-hypodermal junction areas, which was documented in normal adult skin. Finally, genome-wide transcriptome profiling was used to evaluate the molecular proximity of F-DHJ with the two dermal fibroblast populations (Fp and Fr) and with the mesenchymal stem cells (MSCs) corresponding to five tissue origins (bone marrow, fat, amnion, chorion, and cord). This comparative analysis classified the three skin fibroblast types, including F-DHJ, as a clearly distinct group from the five MSC sample origins.
Keywords: human dermis, fibroblasts, extracellular matrix (ECM), dermo-hypodermal junction, papillary fibroblasts (Fp), reticular fibroblasts (Fr), Tenascin C (TNC), mesenchymal stem cells (MSCs)
1. Introduction
In human skin, interfollicular dermis is a heterogeneous tissue compartment, considering its fibroblast content and extracellular matrix (ECM) structure. Its segmentation into two biologically distinct territories (i.e., superficial papillary dermis and deeper reticular dermis) occurs during the embryonic development at 12 weeks of gestation in humans [1,2]. Major structural specificities of these dermal territories concern collagen reticulation and organization of the elastin network, which are dynamic characteristics in constant evolution during the intra-uterine and postnatal life (for review, see [3]).
Specificities of the different dermal territories also concern their fibroblast contents, in which characterization drives an increasing interest considering their widely expected functions in skin physiology. The existence of two dermal fibroblast populations, named papillary (Fp) and reticular (Fr) fibroblasts according to their dermal localization, was reported in human skin in the late seventies [4]. Since then, studies have been conducted to further explore their cellular properties [5] and molecular profiles [6,7]. Biological aspects that are attracting attention are the cellular and molecular changes that affect Fp and Fr cells through skin ageing [8,9].
Other fibroblast or fibroblast-like mesenchymal cell populations are present within the dermis, such as pericytes and telocytes. Pericytes appear in the fetal dermis at eight weeks of gestation in humans and acquire their mature characteristics at 21 weeks of gestation [10]. These cells contribute to the maintenance of capillary vessel integrity and may play a role in the maintenance of mesenchymal tissues in the contexts of homeostasis and/or wound healing [11]. In addition, pericytes may contribute to the niche that regulates the symmetrical versus asymmetrical division choice of epidermal keratinocyte precursors [12]. Telocytes possess an atypical fibroblast morphology characterized by long and slender moniliform cellular prolongations termed telopodes [13]. These cells serve as connecting devices, constructing homocellular junctions and connections with other cells types [14]. Telocytes are usually present at a low density (around 10 cells/mm2) [15]. These cells may participate in the stem cell niche, as shown in the intestine crypts [16]. Another described function of telocytes is the transmission of signals via atypical junctions [17] or extracellular vesicles [18], as reported in the heart. In the dermis, telocyte density augments with depth, together with the quantity of telopodes found in connection with endothelial cells, nerve endings, and hair follicle bulges [19]. Implications of telocytes in regeneration and wound healing is expected in skin but not fully demonstrated [19].
In the present study, we investigated the cellular and molecular properties of fibroblasts localized at the frontier of deep dermis and hypodermis, i.e., dermo-hypodermal junction fibroblasts (F-DHJ). Using parameters such as contractility, differentiation potential, and the supportive effect on epidermis reconstruction, we documented marked functional differences between F-DHJ and dermal (Fp and Fr) fibroblasts. At a molecular level, the study identified specific signatures in F-DHJ concerning the expression of genes involved in ECM synthesis-processing and “tissue skeleton” organization, which could explain structural properties of their tissue compartment. Finally, genome-wide transcriptome profiling was used to evaluate the molecular proximity of F-DHJ with the two dermal fibroblast populations (Fp and Fr) and the with mesenchymal stem cells (MSCs) corresponding to five tissue origins (bone marrow, fat, amnion, chorion, and cord). This approach identified skin fibroblasts and MSCs as distinct groups and will certainly contribute to the knowledge of the hierarchical clustering within the mesenchymal lineages.
2. Materials and Methods
2.1. Fibroblast Isolation and Culture
2.1.1. Human Skin Biopsy Collection
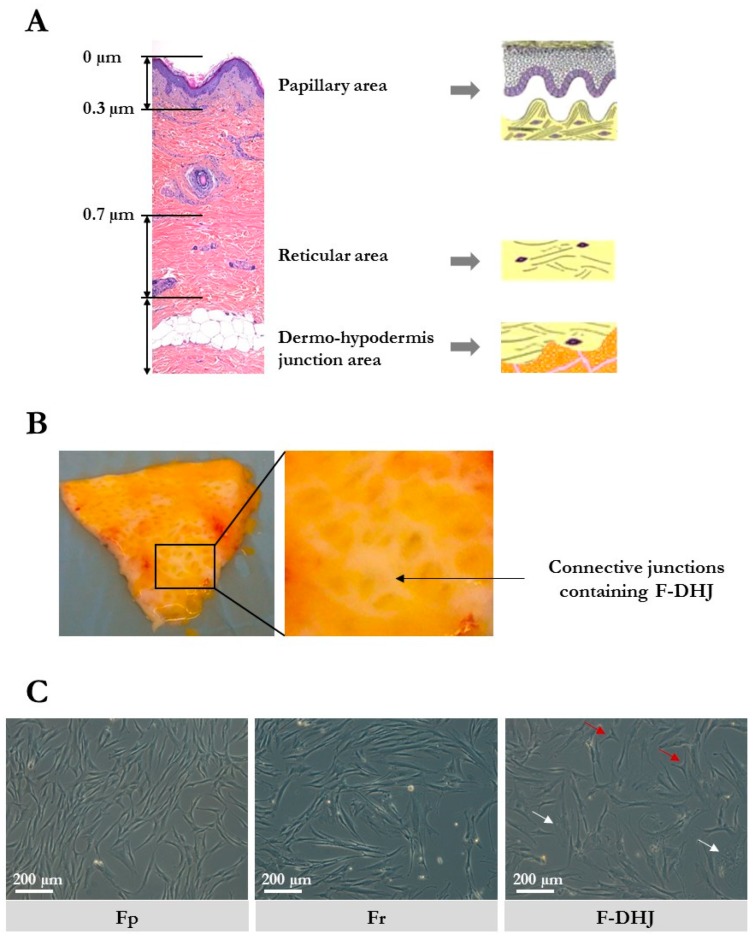
Full-thickness biopsies of human breasts and abdominal skin, collected from healthy subjects undergoing reconstructive or aesthetical surgery, were obtained from Icelltis (Toulouse, France); Alphenyx (Marseille, France); and Biopredic (Saint-Grégoire, France) under the authorizations delivered by the French Ministry of Research with the approval of the French Ethical Committee. The written informed consent was obtained from all individuals. The tissue collection used in this study included 10 biopsies of breast skin (mammoplasties) with ages ranging between 18 and 65 years and 6 biopsies of abdominal skin (abdominoplasties) with ages between 42 and 51 years. A typical skin section showing the papillary, reticular, and dermo-hypodermal dermis regions is shown in Figure 1A.
Figure 1.
Skin localization and cellular morphology of papillary dermis fibroblasts (Fp), reticular dermis fibroblasts (Fr), and dermo-hypodermal junction (DHJ) fibroblasts. (A) Representation of the papillary dermis, reticular dermis, and dermo-hypodermis junction areas. A typical full-thickness skin section is shown, as well as schemes of the three areas of interest. (B) Photographs of skin pieces taken from the below side after fat tissue removal, showing the macroscopic aspect of the conjunctival junctions that connect the dermis to the hypodermis. (C) Cellular morphology of cultured Fp, Fr, and DHJ fibroblasts. In F-DHJ cultures, red arrows point to small tricuspid cells and white arrows to large cells with a visible trabecular cytoplasmic network.
F-DHJ hypodermis was gently removed from skin biopsies by dissection using clamps and scissors to preserve the junction between hypodermis and dermis. Then, the tissue area containing the conjunctival junctions that connect the dermis to the hypodermis (Figure 1B) was harvested by dissection for extraction of fibroblasts from the demo-hypodermal junction (DHJ). Dissected pieces were checked under binocular loupe and selected according to the presence of both adipose tissue and conjunctival structures, validating their DHJ localization. F-DHJ were then extracted by tissue digestion with type II collagenase 0.2% (Gibco, France) for 2 h at 37 °C. Tissue dissociation was facilitated by 30 s of vortexing every 30 min.
2.1.2. Fp and Fr
After removing the epidermis by treatment with 2.4 U/mL dispase (Roche, Boulogne-Billancourt, France) for 16 h at 4°C and then mechanical dissection, papillary fibroblasts (Fp) were extracted by digestion of the tissue in type II collagenase 0.2% (Gibco, France) for 3 h at 37 °C. Tissue dissociation was facilitated by 30 s of vortexing every 30 min. Then, a second cut was performed on the noncut remaining part of the sample at a depth of 700 µm. This intermediate region of the dermis (depth between 300 and 700 µm) was not used for fibroblast extraction to avoid mixing papillary and reticular material. The deepest dermis slice (700 µm depth from skin surface and below) corresponded strictly to the reticular dermis and was used to extract the Fr fibroblast fraction by tissue digestion in type II collagenase 0.2% (Gibco, France) for 5 h at 37 °C. Tissue dissociation was facilitated by 30 s of vortexing every 30 min.
2.1.3. Bidimensional Mass Culture
Fp, Fr, and F-DHJ cells were cultured in similar conditions. Seeding density was 3800 cells/cm², and culture medium was composed of MEM supplemented with 10% FBS (PAN Biotech GmbH, Aidenbach, Germany); penicillin-streptomycin (20 U/mL) (Biochrom Ltd., Cambridge, UK); sodium pyruvate (Gibco, France); nonessential amino acids (Gibco, France); and glutamine (2 mM) (Invitrogen, Carlsbad, CA, USA). Cultures were incubated at 37 °C in a 90% humidified atmosphere containing 5% CO2.
2.2. Mesenchymal Stem Cell (MSC) Isolation and Culture
All human samples were collected and handled in full respect of the Declaration of Helsinki.
2.2.1. BM-MSCs
Human bone marrow MSCs (BM-MSCs) were obtained from patients undergoing routine total hip replacement surgery in Percy Hospital (Clamart, France) after written informed consent. As previously reported [20], spongious bone fragments were mixed in phosphate-buffered saline (PBS, PAN-Dominique Dutscher, Issy-les-Moulineaux, France); 1 mM EDTA (Prolabo-VWR, Paris, France); ACD-A; and 0.5% human serum-albumin (HAS, LFB). After 20 min of settling, the supernatant was collected, centrifuged at 480 g for 10 min, and filtered (70 µm). Bone marrow mononuclear cells (BM-MNCs) were counted using an automated cell analyzer (Sysmex, Villepinte, France)
2.2.2. Ad-MSCs
Human adipose tissue MSCs (Ad-MSCs) were isolated from fat obtained after liposuction surgery in Percy Hospital (Clamart, France) after written informed consent. Fat was washed by an addition of PBS supplemented with 1 µg/mL ciprofloxacin (Panpharma, Luitré, France). After centrifugation at 815 g for 2 min, the washing solution (containing blood, lipids, and adrenalin added before surgery) was discarded. This operation was repeated until washing solution was clear. Fat tissue was then enzymatically digested in 0.075% type I collagenase (75 mg/100 mL fat) for 45 min at 37 °C with agitation each 15 min. Digested fat was then centrifuged at 200 g for 5 min. The supernatant that contained lipids and adipocytes was discarded. The pellet that contained the stoma-vascular fraction was washed three times with α-MEM (Cliniscience, Nanterre, France) and filtered (70 µm). Cell numeration was performed after sample treatment with Zap Oglobin lytic reagent (Beckman Coulter, Villepinte, France).
2.2.3. Amnion, Chorion, and Umbilical Cord MSCs
Perinatal tissues were obtained from full-term deliveries after maternal written informed consent (Hôpital d’Instruction des Armées Bégin, Saint-Mandé). As previously reported [20], samples of placental membranes (amnion and chorion) and umbilical cords were incubated in an antibiotic and antifungal solution for 90 min at room temperature and then cut into pieces. Amnion and chorion 2 cm2 pieces were digested in PBS containing 0.1% type IV collagenase (Thermo-Fisher for Life Technologies, Waltham, MA, USA) and 2.4 U/mL grade II dispase (Roche, Boulogne-Billancourt, France) for 90 min at 37 °C and then in PBS containing 0.025% trypsin-EDTA (Thermo-Fisher for Life Technologies, Waltham, MA, USA) for 30 min at 37 °C. Umbilical cord 2 cm-long pieces were cut into smaller formats (around 1–2 mm3) for digestion in PBS containing 300 U/mL type I collagenase (Thermo-Fisher for Life Technologies, Waltham, MA, USA) and 1 mg/mL hyaluronidase (Calbiochem-Merck, Fontenay sous Bois, France) for 60 min at 37 °C and then in PBS containing 0.025% trypsin-EDTA (Thermo-Fisher for Life Technologies, Waltham, MA, USA) for 30 min at 37 °C. Cell samples were filtered through a 100 µm cell strainer (BD Biosciences, Le Pont de Claix, France) and then centrifuged at 200 g for 10 min. Cells were counted in a Malassez chamber using the trypan blue exclusion method.
2.2.4. Bidimensional Mass Cultures
Samples from the different tissue origins were cultured in the same conditions. Freshly-extracted cells were seeded at a density of 30000 cells/cm2 in a medium composed of α-MEM (Clinisciences, Nanterre, France) supplemented with 0.01 mg/mL ciprofloxacin; 2 U/mL heparin (Choay-Sanofi Aventis); and 5% platelet lysate (obtained from a platelet apheresis collection performed at the ‘Centre de Transfusion Sanguine des Armées’, Clamart). The medium was renewed 3 times a week. Cultures were trypsinized when reaching the stage of 80% confluence (trypsin-EDTA, Thermo-Fisher for Life Technologies, Waltham, MA, USA). Then, MSC subcultures were initiated at a density of 4,000 cells/cm2. For storage, MSC samples were frozen in α-MEM (Clinisciences, Nanterre, France) supplemented with 10% human serum-albumin and 10% DMSO (Sigma-Aldrich, St Louis, MO, USA).
2.3. Colony Assay
Cells were plated at low densities in 10 cm diameter culture-treated plastic petri dishes (400 cells/dish for Fp and 800 cells/dish for Fr and F-DHJ) and cultured during 3 weeks in a medium of similar composition to that used for mass cultures, which was renewed 3 times a week. Cultures were then fixed in 70% ethanol and stained with blue RAL. Colonies were counted manually.
2.4. Three-Dimensional Fibroblast Contractility Assay
Dermal equivalents (lattices) were produced by mixing 100000 fibroblasts in MEM containing 10% FBS and 26% (w/v) bovine type I collagen (Symatèse, Chaponost, France) in a total volume of 5 mL (3.4 mm diameter petri dishes). Spontaneous collagen polymerization occurred within a few hours of culture. Lattices were then detached from the plastic surface of petri dishes 48 h after culture initiation, enabling a contraction process that led to progressive reduction of the lattice diameter. Kinetics of contraction was characterized by measurement of the lattice diameter (millimeter scale) after 1 h, 3 h, 6 h, and 24 h. Full description of the assay principle is provided in [8].
2.5. Three-Dimensional Skin Reconstruction
Reconstructed skins were prepared as previously described [21]. Briefly, fibroblasts (1 × 106 cells per sample of reconstructed dermis) were embedded into a bovine type I collagen gel (Symatese, Chaponost, France). Thereafter, keratinocytes (50,000 cells per sample) were seeded onto the lattices and stuck to the bottom of 60 mm diameter petri dishes. The keratinocytes used in this study were frozen banked samples from a single donor amplified in a serum-containing medium in the presence of a feeder-layer of growth-arrested murine 3T3 fibroblasts [5] according to the principle described by Rheinwald and Green [22]. Developing skin reconstructs were maintained for 1 week immersed in a medium composed of MEM (Invitrogen, Carlsbad, CA, USA) supplemented with 10% FBS (Sigma, St Louis, MO, USA); epidermal growth factor (EGF) (10 ng/mL) (BD Biosciences, San Jose, CA, USA); hydrocortisone (0.4 mg/mL) (Sigma, St Louis, MO, USA); and cholera toxin (0.1 nM) (Biomol Int., Plymouth, PA, USA). Complete epidermal stratification and full differentiation was obtained 1 week after raising the system at the air-liquid interface. During the whole process of skin reconstruction, cultures were maintained at 37 °C in a fully humidified atmosphere containing 5% CO2. Reconstructed skin samples were embedded in a paraffin and used to prepare hematoxylin eosin saffron-stained sections.
2.6. Neosynthetized ECM Samples
Protocol was adapted from [23]. Fibroblasts were seeded on glass slides and cultured till postconfluence. After an additional 48 h, slides were washed twice in PBS, and cells were then lysed using a solution containing 0.5% Triton X-100 and 20 mM of NH4OH. Cell debris were washed in PBS. Slides coated with ECM components synthesized by fibroblasts were stored in PBS at 4 °C until characterization.
2.7. Mesodermal Differentiation Assays
2.7.1. Adipocyte Lineage
Fibroblasts were seeded at a density of 1400 cells/cm2 and cultured in the medium used for mass expansion and colony assay till confluency. After an additional 48 h, the fibroblast cultures medium was substituted by an adipocyte differentiation medium composed of DMEM/20% fetal calf serum (PAN Biotech GmbH, Aidenbach, Germany); 60 µM indometacin (Dr. Ehrenstorfer GmbH, Germany); 0.5 mM 3-isobutyl-1-methylxanthine (IBMX) (Sigma, St Louis, MO, USA); and 10−6 M dexamethasone (Sigma, St Louis, MO, USA). After 3 weeks of cultures in the adipocyte differentiation medium, cultures were fixed in 4% paraformaldehyde. Cells differentiated into adipocytes were visualized and quantified under microscope according to the presence of refringent lipid droplets in the cytoplasm.
2.7.2. Osteoblast Lineage
As for adipocyte differentiation, the fibroblast culture medium was substituted 48 h postconfluency by an osteoblast differentiation medium composed of MEM/10% fetal calf serum (PAN Biotech GmbH, Aidenbach, Germany); 2 mM β-glycerophosphate (Sigma, St Louis, MO, USA); and 10−7 M dexamethasone (Sigma, St Louis, MO, USA). After 3 weeks of cultures in the osteoblast differentiation medium, cultures were fixed in 4% paraformaldehyde. Cells differentiated into osteoblasts were visualized and counted after alizarin-red staining of the calcified extracellular matrix.
2.7.3. Chondrocyte Lineage
For each sample, 105 cells were centrifuged and kept as pellets for 24 h to initiate formation of spheroid structures. The fibroblast culture medium was then substituted by a chondrocyte differentiation medium composed of MEM; 0.5 µg/mL insulin (Gibco, France); 0.5 µg/mL transferrin (Sigma, St Louis, MO, USA); 0.5 ng/mL sodium selenite (Gibco, France); 6.25 µg/mL linoleic acid (Gibco, France); 6.25 µg/mL oleic acid (Gibco, France); 1.25 mg/mL bovine serum-albumin (Sigma, St Louis, MO, USA); 1 mM of sodium pyruvate (Gibco, France); 0.17 mM ascorbic acid 2-phosphate (Sigma, St Louis, MO, USA); 0.1 µM dexamethasone (Sigma, St Louis, MO, USA); 0.35 mM proline; and 0.01 µg/mL of TGF-β1 (RnD System, France). After 3 weeks of cultures in the chondrocyte differentiation medium, spheroids were included in OCT for cryosectioning. Chondrocyte differentiation was revealed by toluidine blue (Sigma, St Louis, MO, USA) and safranin O (Thermo-Fisher, France) staining and immunostaining of aggrecan (ACAN) and collagen XIα1 (ColXIα1).
2.8. Transcriptome Analysis
2.8.1. RNA Extraction
Total RNA was extracted using the RNeasy kit (QIAgen, Courtaboeuf, France), using cultured fibroblasts at the stage of 7 to 10 population doublings. To limit the impact of experimental variations on gene expression profiles, culture conditions were standardized as follows: RNA extraction was systematically performed at 80% culture confluency and 24 h after a full medium renewal. Extracted RNA samples were split into aliquots in the perspective of microarray and qRT-PCR analyses.
2.8.2. Microarray Transcriptome Profiling
Human full-genome Affymetrix GeneChip HG-U133 Plus 2.0 (PartnerChip, Evry, France) were used following the manufacturer’s recommendations. These microarrays contain 55000 probe sets (25 nucleotides per set) covering 30000 transcripts. Briefly, RNA quality and quantity were estimated using the Nanodrop (ND-1000) and BioAnalyzer 2100 systems (Agilent, Les Ulis, France). When too-high concentrations of salts or solvents were detected, RNA precipitation and washing were performed before sample processing. Quantification of array fluorescence signals was carried out using a GeneChip 3000 scanner. Then, array data were analyzed using the Affymetrix Command Console software. Quality control and statistical analyses were performed using the Affymetrix Expression Console and GeneSpring GX11 softwares.
2.8.3. qRT-PCR
RNA samples were reverse-transcribed using the random primer and Superscript II Reverse transcriptase system following the manufacturer’s instructions (Invitrogen, France). Amplifications were performed using a MyiQTM LightCycler (Biorad, Marnes-la-Coquette, France). Real-time quantitative PCR was performed using a MyiQTM LightCycler (Biorad, Marnes-la-Coquette, France) and analyzed using the iQTM 5 software. Gene expression (primers listed in Table 1) was normalized according to GAPDH and TBP transcript levels.
Table 1.
qRT-PCR primers. Primer list and references are provided.
| Gene Symbol | Supplier Reference |
|---|---|
| ACAN | QT00001365 |
| CADM1 | QT00050001 |
| COL11A1 | QT00088711 |
| DIRAS3 | QT00040558 |
| EFHD1 | QT00086163 |
| EMCN | QT00025158 |
| FGF9 | QT00000091 |
| GAPDH | QT01192646 |
| KLF9 | QT00208537 |
| LIMCH1 | QT00038794 |
| MGST1 | QT00063357 |
| NPR3 | QT00047250 |
| RHOJ | QT00092078 |
| SFRP2 | QT00073220 |
| SOST | QT00219968 |
| SOX11 | QT00221466 |
| TBP | QT00000721 |
| TGFB2 | QT02290316 |
| TOX | QT00070063 |
| UCP2 | QT00014140 |
| VCAM1 | QT00018347 |
2.9. Immunofluorescence
2.9.1. Tissue Section Staining
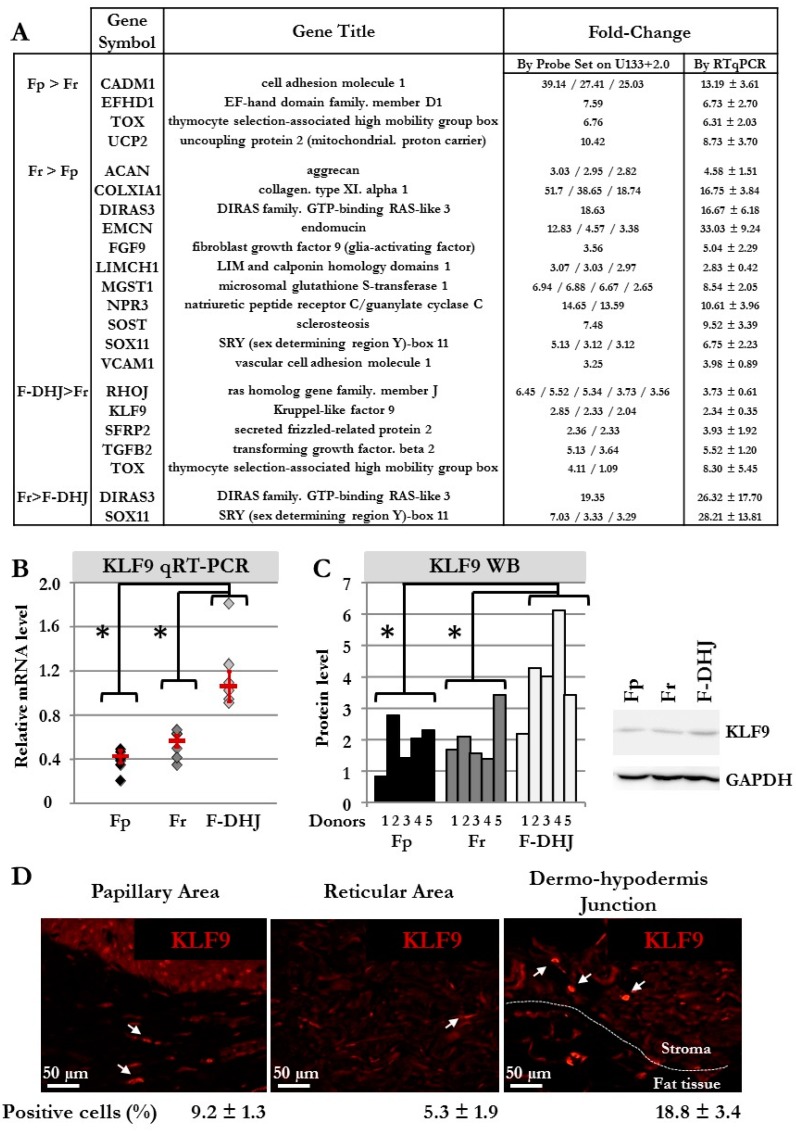
Skin samples were fixed in neutral formalin and then embedded in a paraffin. Tissue sections of 5 µm thickness were prepared. For antibody staining, sections were permeabilized in 0.1% SDS after deparaffinization and epitope retrieval in a citrate buffer (pH = 6). To limit background signals, unspecific antibody fixation sites were saturated by sample incubation in 5% BSA. Sections were incubated with primary and secondary Alexa Fluor-coupled antibodies (see Table 2 for antibody references and working dilutions). Stained skin sections were mounted in ProLong Gold supplemented with DAPI (Thermo for Molecular Probes, Waltham, MA, USA, and images were acquired using a Leica microscope coupled with a QIMAGINE RETIGA 2000R Fast 1394 camera. Signal quantification was performed using ImageJ software. Quantification of cells positive for KLF9 expression was performed by visual counting on skin samples from 4 donors. Percentages of KLF9+ cells were determined in a total of 806 cells for Fr, 289 cells for Fr, and 246 cells for F-DHJ fibroblasts.
Table 2.
Antibodies. Antibody references and working dilutions are provided.
| Protein name | Supplier | Reference | Dilution |
|---|---|---|---|
| alpha Sm actin | Sigma (Saint-Quentin Falaviers—France) | A5228 | 1/200 |
| ACAN (Aggrecan) * | Abcam (Paris—France) | ab3778 | 1/20 |
| Col XI a1 | Sigma (Saint-Quentin Falaviers—France) | SAB4500393 | 1/50 |
| Desmine (clone D33) | Dako—Agilent (France) | M0760 | 1/50 |
| GAPDH | Interchim for Meridian (France) | H86504M | 1/2000 |
| KLF9 | Abcam (Paris—France) | ab170980 | 1/100 (IHC)–1/1000 (WB) |
| Phalloïdine Rhodamin | Invitrogen (France) | R415 | 1/50 |
| TNC (Tenascin C) | Novus Biologicals (Abington—UK) | NB110-68136 | 1/50 |
| Vimentin | TEBU (Le Perray-en-Yvelines—France) | MON3005 | 1/10 |
| Goat anti-Mouse Alexa 488 | Molecular Probes Invitrogen (France) | A21121 | 1/250 |
| Goat anti-Rabbit Alexa 555 | Molecular Probes Invitrogen (France) | A21428 | 1/250 |
| Zenon Alexa 488 | Molecular Probes Invitrogen (France) | Z25002 | |
| Goat anti-Rabbit HRP | Thermo-Fisher, France | 32460 | 1/2000 |
* Pre-processing: keratanase (0.1 U/mL) + chondroitinase (0.1 U/mL)—3 h—37 °C.
2.9.2. Cell Staining
Cultured cells were fixed in 4% paraformaldehyde, permeabilized in 0.1% SDS, and incubated in 5% BSA for saturation of unspecific antibody binding sites and then with primary and secondary Alexa Fluor-coupled antibodies (see Table 2). Labeled cells were mounted in ProLong Gold supplemented with DAPI (Thermo - Molecular Probes, Waltham, MA, USA). Immunofluorescence images were acquired using a Leica microscope coupled with a QIMAGINE RETIGA 2000R Fast 1394 camera (QImageing, Canada). Signal quantification was performed using ImageJ software.
2.9.3. ECM Staining
ECM slides were incubated in 5% BSA for saturation of unspecific antibody binding sites. Incubation with Alexa Fluor-coupled antibodies (Zenon technology – Thermo – Molecular Probes, Waltham, MA, USA) was performed during 30 min at room temperature (see Table 2). Immunofluorescence images were acquired using a Leica microscope coupled with a QIMAGINE RETIGA 2000R Fast 1394 camera (QImageing, Surrey, BC, Canada). Signal quantification was performed using ImageJ software.
2.10. Western Blot Analysis
Expression of KLF9 was assessed by western blot analysis on total protein extracts from cell cultures. Protein extracts were prepared using a radioimmunoprecipitation assay (RIPA) buffer. Proteins (40 μg) were separated by 15% sodium dodecyl sulphate-polyacrylamide gel electrophoresis (PAGE) and electrotransferred onto a 0.45 µm nitrocellulose membrane. The membrane was incubated with the primary antibody, washed, and probed with the peroxidase-labeled secondary antibody. Detection was achieved by enhanced chemiluminescence (West Femto HRP substrates, ThermoFisher Scientific, France). After dehybridization, control loading was achieved by anti-glyceraldehyde-3-phosphate dehydrogenase antibodies. Densitometric analyses were performed using ImageJ.
2.11. Statistics
Error bars represent SEM. The Wilcoxon-Mann-Whitney test and the Friedman test were applied to determine p-values. Data with p < 0.05 (*) or p < 0.01 (**) were considered as statistically significant.
3. Results
3.1. Cellular Characteristics and Growth Potential Distinguish F-DHJ from Fp and Fr
The cellular morphology of the three fibroblast populations were isolated based on their skin localization, i.e., papillary dermis fibroblasts (Fp), reticular dermis fibroblasts (Fr), and dermo-hypodermal junction fibroblasts (F-DHJ) were examined in cultures and compared (Figure 1C). As previously described [4], Fp cells exhibited a thin morphology, with bi or tricuspid shapes, whereas Fr had spread morphologies and stellate shapes. F-DHJ were more heterogeneous, from small tricuspids (red arrow) to larger cells with stellate shapes (white arrow) with visible trabecular networks.
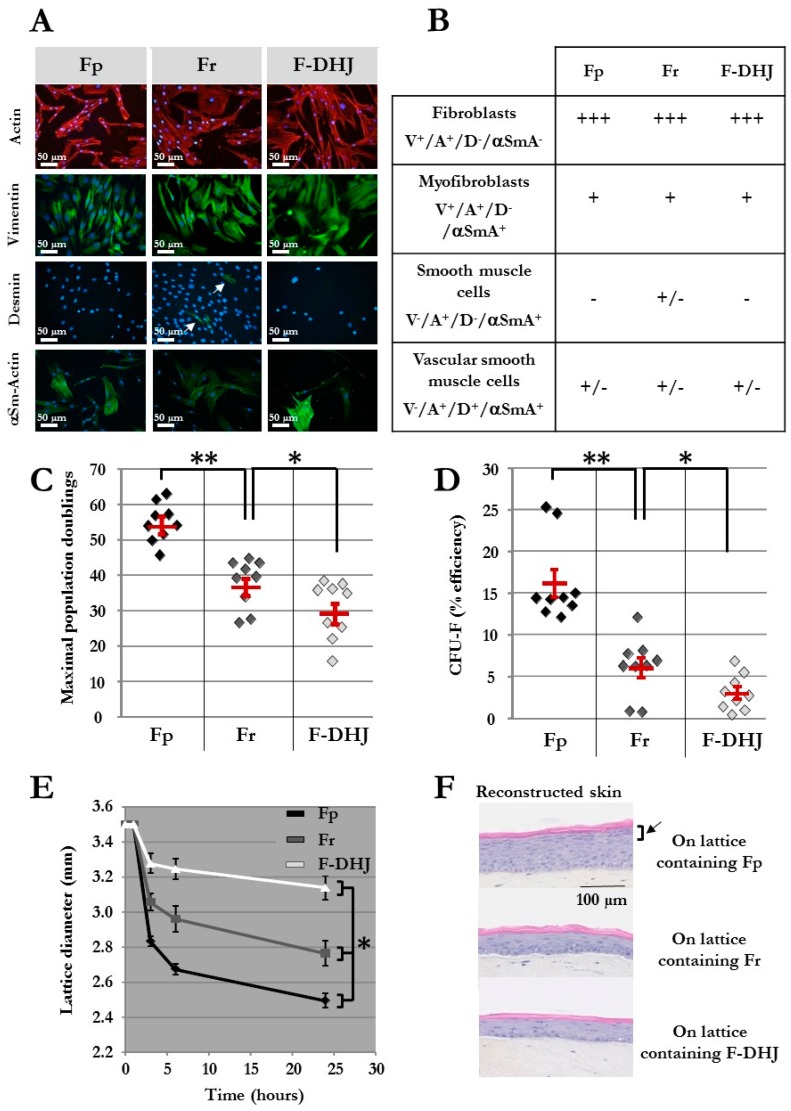
Analysis of the four markers proposed in Gabbiani’s classification [24] (Figure 2A,B) confirmed the fibroblast statuses of the Fp, Fr, and F-DHJ cellular material, as all cell types expressed almost homogenously actin (ACT) and vimentin (VIM) but expressed neither desmin (DES) nor α-smooth muscle actin (α‑SMA): ACT+/VIM+/DES−/α‑SMA− phenotype. In each population, only a minority of cells exhibited the myofibroblast ACT+/VIM+/DES−/α‑SMA+ phenotype, probably due to the cultures’ environments. Few cells corresponding to the ACT+/VIM−/DES+/α‑SMA+ vascular smooth muscle cell phenotype were also detected. In addition, the Fr population contained few ACT+/VIM−/DES−/α‑SMA+ cells, corresponding to smooth muscle cells probably originating from arrector pili muscles.
Figure 2.
Phenotypic and functional properties of Fp, Fr, and F-DHJ fibroblasts. (A) Detection of actin (ACT) and vimentin (VIM), desmin (DES), and α-smooth muscle actin (α‑SMA) by immunochemistry in cultured Fp, Fr, and F-DHJ fibroblasts in the perspective of scoring according to Gabbiani’s classification [24]. White arrow points to rare DES+ cells present within the Fr population. (B) Scoring of Fp, Fr, and F‑DHJ fibroblasts according to ACT, VIM, DES, and α‑SMA detection: (−) = not present, (+/−) = low representation, (++) = frequent representation, and (+++) = major representation. (C) Long-term growth capacity of Fp, Fr, and F‑DHJ cells. Maximal cumulative population doubling values obtained with samples from independent donors are shown. Means ± SEM are indicated (* p < 0.05, ** p < 0.01; Wilcoxon test). (D) Colony-forming unit efficiency of Fp, Fr, and F‑DHJ cells. Fibroblast colony-forming unit (CFU-F) efficiency values (% of plated cells) obtained with samples from independent donors are shown. Means ± SEM are indicated (* p < 0.05, ** p < 0.01; Wilcoxon test). (E) Contractile capacity of Fp, Fr, and F‑DHJ cells in the 3D context of collagen lattices. Kinetics of lattice diameter evolutions. Means ± SEM are indicated (values obtained with samples from 9 independent donors) (* p < 0.05, Friedman’s test). (F) Efficiency of Fp, Fr, and F‑DHJ cells in promoting epidermis organogenesis by keratinocytes in a 3D reconstructed skin model. Representative reconstructed skin sections are shown (3 independent donors, each fibroblast sample tested in triplicates). The black arrow points to the epidermal granular layer that was obtained only in the presence of Fp fibroblasts.
The proliferative capacity of Fp, Fr, and F-DHJ cells was assessed in mass long-term cultures (Figure 2C) and using a colony assay (Figure 2D) (cell samples from n = 9 individuals were studied). As previously described [4,5], the proliferative capacity of Fr was lower than that of Fp, according to both criteria. Indeed, the maximum population doublings (PD) reached by Fp was 54 ± 2 versus 37 ± 2 for Fr (p < 0.01), and colony-forming efficiency was 16.2% ± 1.7 for Fp and 6.1% ± 1.2 for Fr (p < 0.01). F-DHJ exhibited the lowest growth capacity of the three fibroblast types, with a maximum PD reaching only 29 ± 3 and colony-forming efficiency 3% ± 0.7 (p < 0.05, calculated versus Fr).
Taken together, these data show that Fp, Fr, and F-DHJ fibroblasts exhibit different cellular characteristics.
3.2. Behavior in 3D Tissue and Differentiation Potential Distinguish F-DHJ from Fp and Fr
A functional assay was designed to assess fibroblast contractile capacity in a three-dimensional environment based on a follow-up of collagen lattice contractions. Fp, Fr, and F-DHJ integrated in collagen lattices exhibited nonequivalent contraction behaviors (Figure 2E) (cell samples from n = 9 individuals were tested). Reduction of the lattice diameter was more rapid and marked with Fp than with Fr cells. In contrast, a more moderate lattice diameter reduction was observed with F-DHJ cells, indicating a lower contractile capacity for this fibroblast population. The three lattice contraction curves showed statistically significant differences (p < 0.05).
The next functional property of Fp, Fr, and F-DHJ that was investigated and compared was their efficiency in promoting epidermis organogenesis by keratinocytes in a model of in vitro three-dimensional skin reconstruction. Lattices containing either Fp, Fr, or F-DHJ cells were produced and used as dermal equivalents. On top of which, keratinocytes were then plated in order to obtain epidermis development (Figure 2F) (fibroblasts samples from n = 3 individuals were tested, in association with keratinocytes from a single donor). Fp cells were the most efficient fibroblast population for promoting the development of a correctly differentiated epidermis comprising a regular basal layer, as well as fully differentiated granular and horny layers. Epidermis reconstructs were of a lower quality with dermal lattices containing Fr fibroblasts; basal keratinocytes were of bigger sizes and less regular, and differentiation of the granular layer was incomplete. Dermal lattices populated with F-DHJ promoted poor epidermis stratification and differentiation.
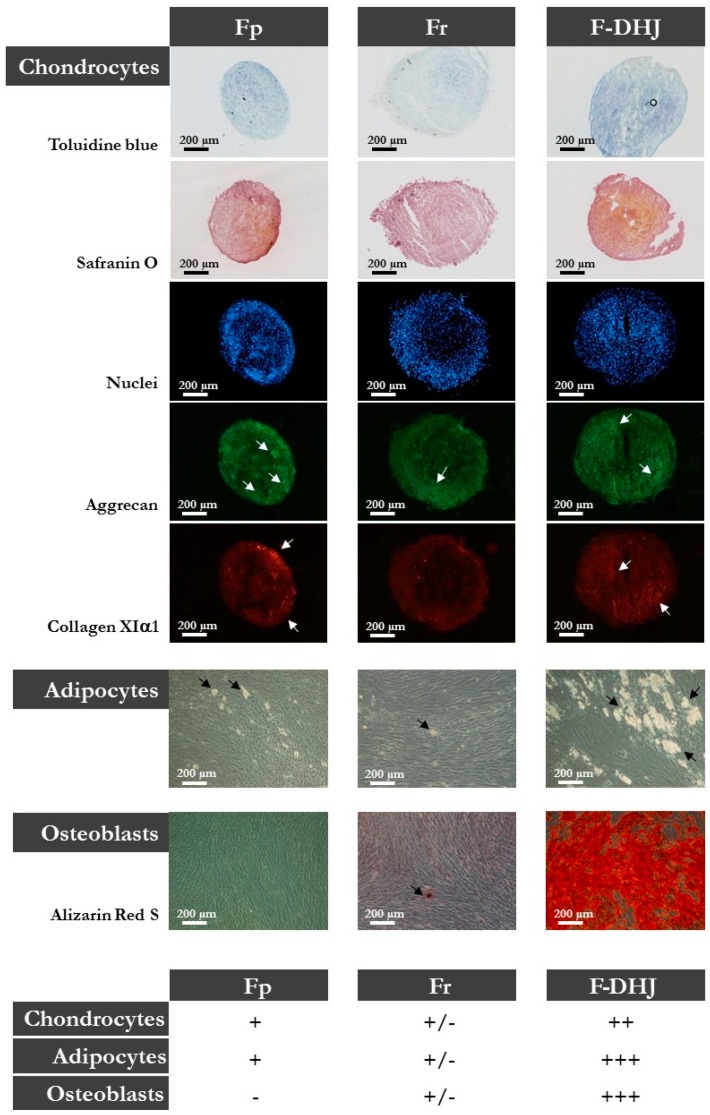
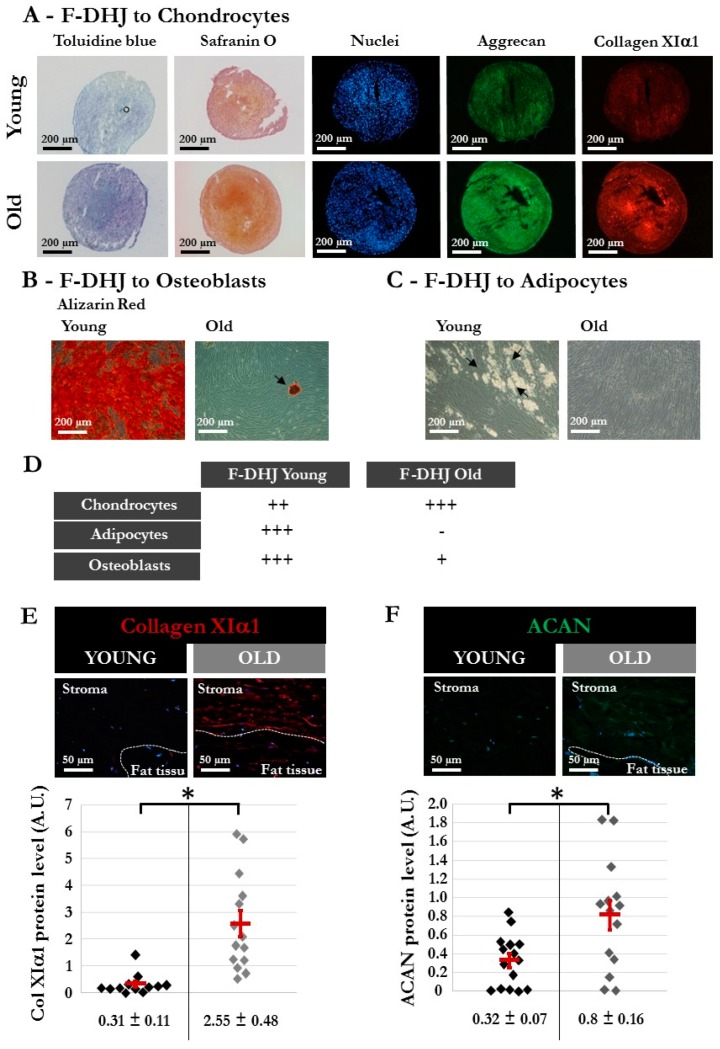
Finally, Fp, Fr, and F-DHJ were studied for their differentiation capacity into three mesodermal cell lineages: adipocytes (presence of cytoplasmic lipid droplets); osteoblasts (alizarin-red staining); and chondrocytes (toluidine blue and safranin O staining, aggrecan (ACAN) and collagen XIα1 (ColXIα1) expression). This functionality was documented using cells obtained from skin biopsies corresponding to ages ranging between 20 and 31 years (Figure 3). Interestingly, F‑DHJ exhibited a wider differentiation potential than that of Fp and Fr fibroblasts, as these cells efficiently responded to the three lineage-oriented differentiation conditions. Fp fibroblasts gave rise to fewer quantities of adipocytes and chondrocytes and did not differentiate into osteoblasts. Fr fibroblasts could give rise to differentiated cells of the three lineages but with a much lower efficiency than F-DHJ cells.
Figure 3.
Differentiation capacities of Fp, Fr, and F-DHJ fibroblasts into mesodermal lineages. Samples from 5 independent “young” donors (20, 22, 25, 28, and 31 years old) were tested for their capacity to differentiate into chondrocytes (toluidine blue and safranin O staining, aggrecan (ACAN) and collagen XIα1 (ColXIα1) expression, white arrows); adipocytes (presence of cytoplasmic lipid droplets, black arrows); and osteoblasts (alizarin-red staining). Scoring of differentiation capabilities are presented: (−) = not present, (+/−) = low representation, (++) = frequent representation, and (+++) = major representation.
Taken together, these data show that Fp, Fr, and F-DHJ fibroblasts exhibit different functional characteristics.
3.3. Molecular Profiles Distinguish the Fp, Fr, and F-DHJ Fibroblast Populations
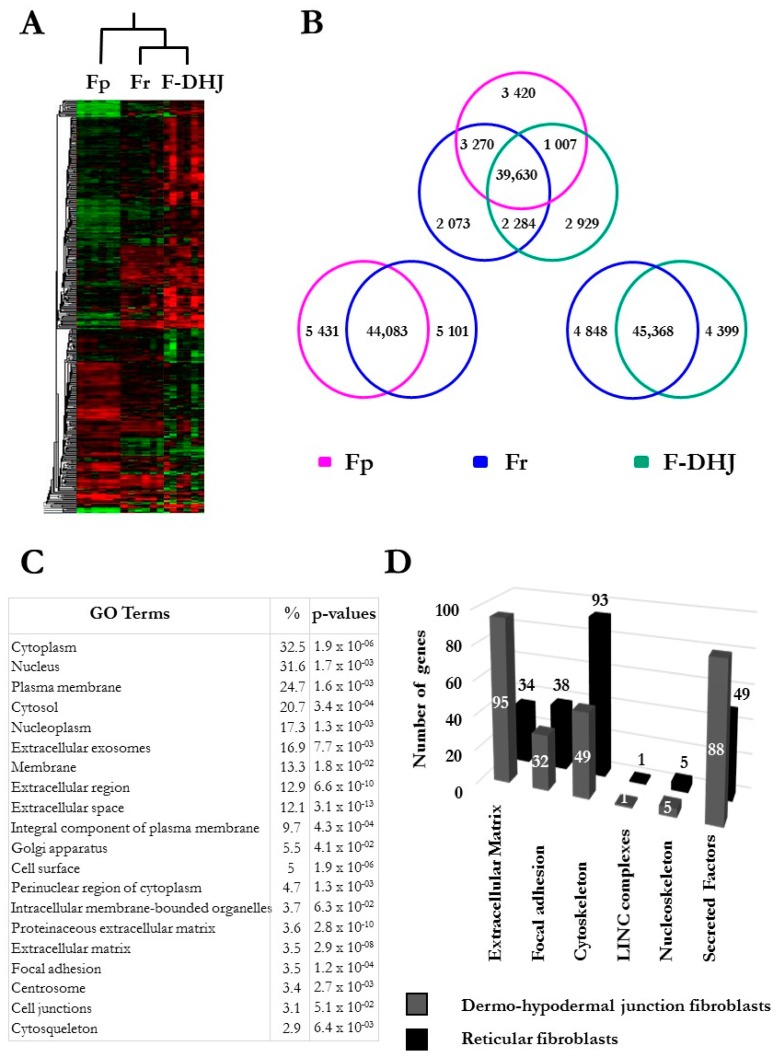
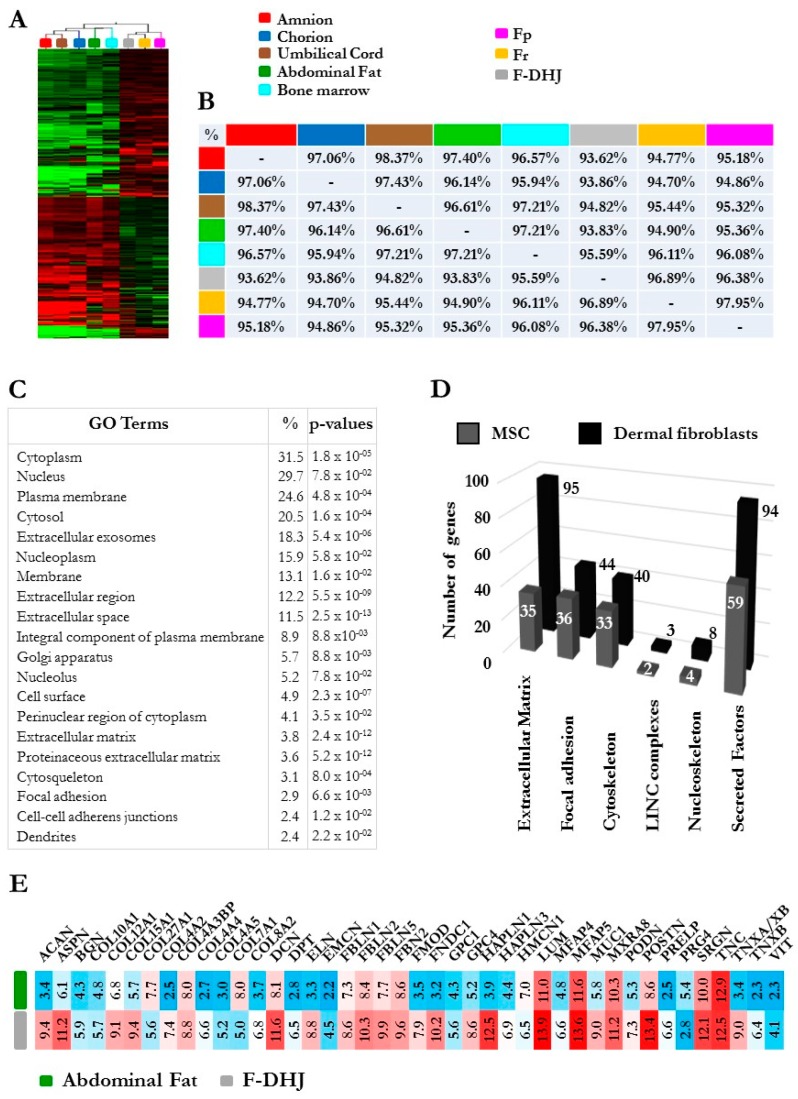
The molecular profiles of Fp, Fr, and F-DHJ cells were characterized and compared by microarray genome-wide transcriptome profiling (Figure 4 and Figure 5A). In the perspective of identifying molecular signatures distinguishing the Fp, Fr, and F-DHJ fibroblast populations whatever the donor’s age, the selected donor cohort covered both young and older ages: 22, 25, 28, 55, 61, and 65 y.o. As a first screen, a fold-change threshold value of three, together with a p-value of 0.05, were used to identify differential signals. According to these filters, a hierarchical clustering was built based on expression levels of 1078 transcripts, identifying signatures that validated at the transcriptome level of the distinct natures of Fp, Fr, and F-DHJ (Figure 4A). Next, transcriptome data were reanalyzed considering only the statistical significance threshold (p-value < 0.05) independently of fold-change values (Figure 4B). This analysis identified 3420, 2073, and 2929 probe sets, which could be used to define signatures of Fp, Fr, and F-DHJ cells, respectively. Fr cells shared the highest level of commonalities with the other fibroblast populations, probably due to their intermediate tissue localization: 3270 probe sets in common with Fp (not detected in F-DHJ) and 2284 probe sets in common with F-DHJ (not detected in Fp).
Figure 4.
Microarray analysis of the transcriptome profiles of Fp, Fr, and F-DHJ fibroblasts (donors’ ages: 22, 25, 28, 55, 61, and 65 y.o.). (A) Hierarchical clustering based on 1078 differentially expressed transcripts (fold-change cutoff at 3 and p < 0.05). (B) Venn Diagrams summarizing Fp, Fr, and F-DHJ-enriched transcriptional signatures (p < 0.05). (C) List of the 20 most significant gene ontology (GO) terms differentiating F-DHJ from Fr cells, based on 2540 probe sets (1647 transcripts) exhibiting differential signals (fold-change >1.5 and p < 0.05). (D) Signatures identifying Fr fibroblasts (black bars) and F-DHJ fibroblasts (grey bars) among transcripts related to the tissue skeleton biology (fold-change >1.5 and p < 0.05).
Figure 5.
Biomarker validations at mRNA and protein levels. (A) Selection of transcripts in which differential expression was confirmed by qRT-PCR in cell samples from 6 donors (donors’ ages: 22, 25, 28, 55, 61, and 65 y.o). (B) Detailed qRT-PCR comparative analysis of the KLF9 transcript in cells from the 6 donors (donors’ ages: 22, 25, 28, 55, 61, and 65 y.o). Means ± SEM are indicated (* p < 0.05, Wilcoxon test). (C) Western blot comparative analysis of the KLF9 protein in cells from the 5 donors (* p < 0.05, Wilcoxon test). A histogram detailing quantifications and a representative western blot gel is shown. (D) Immunofluorescence detection of the KLF9 protein in skin sections (breast). The percentage of KLF9+ cells was determined by observation under a fluorescence microscope of a total of 806 cells for Fp, 289 cells for Fr, and 246 cells for F-DHJ fibroblasts (skin sections from 4 donors were included in the analysis).
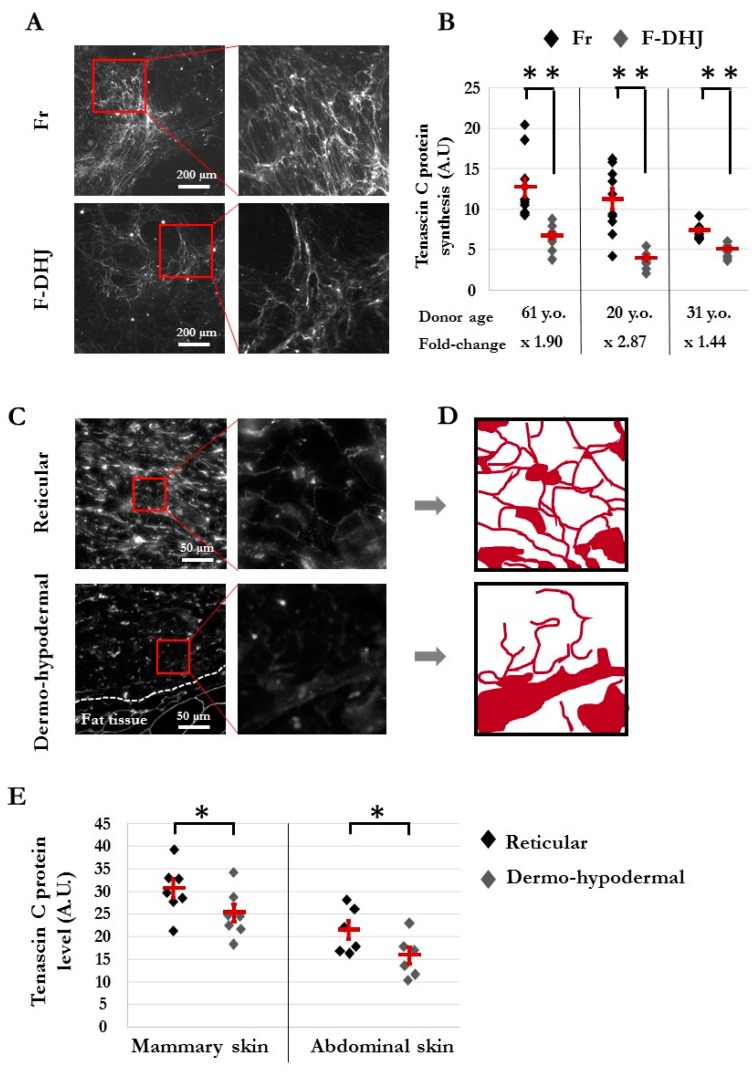
F-DHJ were then compared more specifically with Fr cells, which are in spatial proximity in the tissue. A gene ontology (GO) term analysis was performed based on 2540 probe sets (1647 genes) exhibiting differential signals between the two populations (parameters: fold-change >1.5 and p-value < 0.05) (Figure 4C). Notably, this analysis revealed marked differences between Fr and F‑DHJ concerning the expression of transcripts related to the tissue skeleton (see [9]), as 26% of the transcripts differentially expressed were linked to the structuration of this network (Figure 4D and Table 3). In particular, differentially expressed probe sets were enriched in transcripts related to ECM components, cytoskeleton, and secreted factors.
Table 3.
Transcripts related to the tissue skeleton differentially expressed in reticular dermis fibroblasts (Fr) and dermo-hypodermal junction fibroblasts (F-DHJ). This transcript list was extracted from microarray data using a fold-change >1.5 and p < 0.05 as inclusion parameters. The transcript signature with predominant expression in Fr cells concerned 297 probe sets corresponding to transcripts directly involved in the tissue skeleton structure, comprising 33 transcripts related to the extracellular matrix (ECM), 125 focal adhesion point transcripts, 60 cytoskeleton transcripts, 1 LINC complex transcript, and 8 nucleoskeleton transcripts. The transcript signature with predominant expression in F-DHJ cells concerned 359 probe sets corresponding to transcripts directly involved in the tissue skeleton structure, comprising 94 transcripts related to ECM, 76 focal adhesion point transcripts, 50 cytoskeleton transcripts, 1 LINC complex transcript, and 7 nucleoskeleton transcripts. In addition, transcripts encoding soluble factors were found in both signatures, respectively 70 and 131 for Fr and F-DHJ cells.
| UP in Fr | UP in F-DHJ | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Extracellular Matrix Genes | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205941_s_at | COL10A1 | collagen, type X, alpha 1 | 9.41 × 10−3 | 4.23 | 220518_at | ABI3BP | ABI family, member 3 (NESH) binding protein | 2.37 × 10−2 | 2.92 |
| 211343_s_at | COL13A1 | collagen, type XIII, alpha 1 | 1.44 × 10−2 | 2.26 | 1559077_at | ABI3BP | ABI family, member 3 (NESH) binding protein | 7.43 × 10−2 | 2.49 |
| 211809_x_at | COL13A1 | collagen, type XIII, alpha 1 | 2.74 × 10−2 | 1.74 | 222486_s_at | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 2.76 × 10−2 | 2.33 |
| 221900_at | COL8A2 | collagen, type VIII, alpha 2 | 2.88 × 10−1 | 2.23 | 222162_s_at | ADAMTS1 | ADAM metallopeptidase with thrombospondin type 1 motif, 1 | 4.36 × 10−2 | 1.91 |
| 226824_at | CPXM2 | carboxypeptidase X (M14 family), member 2 | 2.01 × 10−1 | 1.88 | 226997_at | ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 3.75 × 10−3 | 2.78 |
| 221541_at | CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 | 5.98 × 10−2 | 2.03 | 214913_at | ADAMTS3 | ADAM metallopeptidase with thrombospondin type 1 motif, 3 | 1.94 × 10−2 | 1.94 |
| 206595_at | CST6 | cystatin E/M | 2.34 × 10−2 | 3.12 | 237411_at | ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif, 6 | 1.36 × 10−1 | 1.61 |
| 225681_at | CTHRC1 | collagen triple helix repeat containing 1 | 1.31 × 10−2 | 2.82 | 224396_s_at | ASPN | asporin | 2.47 × 10−2 | 4.48 |
| 202450_s_at | CTSK | cathepsin K | 2.05 × 10−1 | 1.76 | 219087_at | ASPN | asporin | 1.32 × 10−2 | 3.54 |
| 213068_at | DPT | dermatopontin | 8.16 × 10−2 | 3.67 | 203477_at | COL15A1 | collagen, type XV, alpha 1 | 2.18 × 10−1 | 2.78 |
| 207977_s_at | DPT | dermatopontin | 1.24 × 10−1 | 2.85 | 209082_s_at | COL18A1 | collagen, type XVIII, alpha 1 | 6.56 × 10−4 | 2.99 |
| 222885_at | EMCN | endomucin | 6.59 × 10−2 | 2.74 | 209081_s_at | COL18A1 | collagen, type XVIII, alpha 1 | 3.92 × 10−3 | 2.83 |
| 227874_at | EMCN | endomucin | 2.70 × 10−1 | 1.81 | 208096_s_at | COL21A1 | collagen, type XXI, alpha 1 | 1.12 × 10−2 | 6.88 |
| 219436_s_at | EMCN | endomucin | 1.79 × 10−1 | 1.77 | 232458_at | COL3A1 | Collagen, type III, alpha 1 | 6.64 × 10−3 | 2.66 |
| 224374_s_at | EMILIN2 | elastin microfibril interfacer 2 | 6.75 × 10−2 | 1.68 | 211981_at | COL4A1 | collagen, type IV, alpha 1 | 2.12 × 10−3 | 1.93 |
| 203088_at | FBLN5 | fibulin 5 | 2.24 × 10−2 | 1.88 | 211980_at | COL4A1 | collagen, type IV, alpha 1 | 1.66 × 10−3 | 1.61 |
| 203638_s_at | FGFR2 | fibroblast growth factor receptor 2 | 3.21 × 10−3 | 4.52 | 222073_at | COL4A3 | collagen, type IV, alpha 3 (Goodpasture antigen) | 2.28 × 10−2 | 1.70 |
| 208228_s_at | FGFR2 | fibroblast growth factor receptor 2 | 2.04 × 10−2 | 2.52 | 229779_at | COL4A4 | collagen, type IV, alpha 4 | 2.85 × 10−8 | 5.32 |
| 210187_at | FKBP1A | FK506 binding protein 1A, 12 kDa | 8.48 × 10−2 | 1.81 | 214602_at | COL4A4 | collagen, type IV, alpha 4 | 2.30 × 10−5 | 4.16 |
| 226145_s_at | FRAS1 | Fraser syndrome 1 | 6.34 × 10−2 | 2.31 | 213110_s_at | COL4A5 | collagen, type IV, alpha 5 | 1.05 × 10−1 | 3.62 |
| 204983_s_at | GPC4 | glypican 4 | 3.14 × 10−2 | 2.07 | 52255_s_at | COL5A3 | collagen, type V, alpha 3 | 2.47 × 10−3 | 3.07 |
| 204984_at | GPC4 | glypican 4 | 1.57 × 10−2 | 2.02 | 218975_at | COL5A3 | collagen, type V, alpha 3 | 2.83 × 10−3 | 2.65 |
| 235944_at | HMCN1 | hemicentin 1 | 3.52 × 10−5 | 6.28 | 205832_at | CPA4 | carboxypeptidase A4 | 2.37 × 10−2 | 4.34 |
| 203417_at | MFAP2 | microfibrillar-associated prot 2 | 8.08 × 10−2 | 1.70 | 201116_s_at | CPE | carboxypeptidase E | 7.11 × 10−3 | 2.37 |
| 204580_at | MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 2.58 × 10−1 | 2.17 | 201117_s_at | CPE | carboxypeptidase E | 4.90 × 10−3 | 2.10 |
| 205828_at | MMP3 | matrix metallopeptidase 3 | 4.79 × 10−3 | 7.70 | 227138_at | CRTAP | cartilage associated protein | 1.31 × 10−2 | 1.63 |
| 209596_at | MXRA5 | matrix-remodelling associated 5 | 2.09 × 10−2 | 2.52 | 201360_at | CST3 | cystatin C | 6.11 × 10−2 | 1.55 |
| 236088_at | NTNG1 | netrin G1 | 2.16 × 10−2 | 2.55 | 201487_at | CTSC | cathepsin C | 6.11 × 10−5 | 2.41 |
| 222722_at | OGN | osteoglycin | 1.91 × 10−1 | 2.04 | 225646_at | CTSC | cathepsin C | 1.80 × 10−3 | 2.32 |
| 228186_s_at | RSPO3 | R-spondin 3 homolog (X. laevis) | 1.35 × 10−2 | 2.98 | 225647_s_at | CTSC | cathepsin C | 1.37 × 10−3 | 2.25 |
| 218638_s_at | SPON2 | spondin 2, extracellular matrix prot | 3.07 × 10−2 | 2.84 | 231234_at | CTSC | cathepsin C | 5.72 × 10−3 | 2.03 |
| 216005_at | TNC | Tenascin C | 1.24 × 10−2 | 2.61 | 202295_s_at | CTSH | cathepsin H | 4.00 × 10−2 | 1.85 |
| 201645_at | TNC | tenascin C | 4.34 × 10−2 | 1.59 | 209335_at | DCN | decorin | 1.02 × 10−1 | 2.05 |
| 211896_s_at | DCN | decorin | 2.16 × 10−1 | 1.69 | |||||
| 211813_x_at | DCN | decorin | 1.52 × 10−1 | 1.68 | |||||
| 201893_x_at | DCN | decorin | 1.77 × 10−1 | 1.56 | |||||
| 1568779_a_at | ECM2 | extracellular matrix protein 2 | 1.76 × 10−1 | 1.71 | |||||
| 206101_at | ECM2 | extracellular matrix protein 2 | 2.50 × 10−1 | 1.57 | |||||
| 201843_s_at | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 7.68 × 10−4 | 3.62 | |||||
| 201842_s_at | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 4.62 × 10−4 | 2.87 | |||||
| 228421_s_at | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 2.23 × 10−1 | 1.61 | |||||
| 226911_at | EGFLAM | EGF-like, fibronectin type III and laminin G domains | 3.95 × 10−3 | 4.36 | |||||
| 204834_at | FGL2 | fibrinogen-like 2 | 4.12 × 10−4 | 6.37 | |||||
| 227265_at | FGL2 | fibrinogen-like 2 | 4.28 × 10−3 | 3.56 | |||||
| 202709_at | FMOD | fibromodulin | 1.12 × 10−3 | 2.96 | |||||
| 205206_at | KAL1 | Kallmann syndrome 1 sequence | 9.65 × 10−4 | 7.84 | |||||
| 227048_at | LAMA1 | laminin, alpha 1 | 2.82 × 10−1 | 1.83 | |||||
| 216840_s_at | LAMA2 | laminin, alpha 2 | 5.17 × 10−3 | 2.80 | |||||
| 205116_at | LAMA2 | laminin, alpha 2 | 1.05 × 10−2 | 2.67 | |||||
| 213519_s_at | LAMA2 | laminin, alpha 2 | 1.10 × 10−2 | 2.57 | |||||
| 210150_s_at | LAMA5 | laminin, alpha 5 | 4.86 × 10−2 | 1.57 | |||||
| 211651_s_at | LAMB1 | laminin, beta 1 | 8.27 × 10−2 | 1.54 | |||||
| 200770_s_at | LAMC1 | laminin, gamma 1 (formerly LAMB2) | 7.61 × 10−6 | 1.91 | |||||
| 200771_at | LAMC1 | laminin, gamma 1 (formerly LAMB2) | 7.75 × 10−7 | 1.90 | |||||
| 202267_at | LAMC2 | laminin, gamma 2 | 7.41 × 10−4 | 10.0 | |||||
| 200923_at | LGALS3BP LOC100133842 | lectin, galactoside-binding, soluble, 3 binding protein similar to lectin, galactoside-binding, soluble, 3 binding protein | 1.02 × 10−2 | 2.45 | |||||
| 242767_at | LMCD1 | LIM and cysteine-rich domains 1 | 1.72 × 10−2 | 2.01 | |||||
| 202998_s_at | LOXL2 | lysyl oxidase-like 2 | 4.16 × 10−3 | 1.59 | |||||
| 227145_at | LOXL4 | lysyl oxidase-like 4 | 3.15 × 10−2 | 2.43 | |||||
| 219922_s_at | LTBP3 | latent transforming growth factor beta binding protein 3 | 7.19 × 10−2 | 1.64 | |||||
| 227308_x_at | LTBP3 | latent transforming growth factor beta binding protein 3 | 2.22 × 10−3 | 1.53 | |||||
| 213765_at | MFAP5 | microfibrillar associated prot 5 | 3.51 × 10−3 | 1.61 | |||||
| 213764_s_at | MFAP5 | microfibrillar associated prot 5 | 3.23 × 10−3 | 1.53 | |||||
| 210605_s_at | MFGE8 | milk fat globule-EGF factor 8 protein | 1.80 × 10−1 | 1.75 | |||||
| 202291_s_at | MGP | matrix Gla protein | 3.10 × 10−4 | 7.14 | |||||
| 207847_s_at | MUC1 | mucin 1, cell surface associated | 1.58 × 10−1 | 2.38 | |||||
| 213693_s_at | MUC1 | mucin 1, cell surface associated | 2.03 × 10−2 | 2.04 | |||||
| 204114_at | NID2 | nidogen 2 (osteonidogen) | 2.32 × 10−3 | 2.83 | |||||
| 223315_at | NTN4 | netrin 4 | 1.76 × 10−4 | 14.27 | |||||
| 201860_s_at | PLAT | plasminogen activator, tissue | 2.56 × 10−2 | 2.15 | |||||
| 211668_s_at | PLAU | plasminogen activator, urokinase | 2.43 × 10−1 | 1.99 | |||||
| 228224_at | PRELP | proline/arginine-rich end leucine-rich repeat protein | 2.74 × 10−2 | 3.46 | |||||
| 204223_at | PRELP | proline/arginine-rich end leucine-rich repeat protein | 3.27 × 10−2 | 3.27 | |||||
| 205923_at | RELN | reelin | 3.58 × 10−8 | 9.15 | |||||
| 202376_at | SERPINA3 | serpin peptidase inhibitor, clade A (alpha-1 antiproteinase, antitrypsin), member 3 | 6.25 × 10−2 | 2.05 | |||||
| 204614_at | SERPINB2 | serpin peptidase inhibitor, clade B (ovalbumin), member 2 | 9.41 × 10−2 | 5.27 | |||||
| 209723_at | SERPINB9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 5.41 × 10−3 | 2.25 | |||||
| 200986_at | SERPING1 | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | 2.27 × 10−1 | 1.72 | |||||
| 205352_at | SERPINI1 | serpin peptidase inhibitor, clade I (neuroserpin), member 1 | 1.54 × 10−3 | 2.71 | |||||
| 213493_at | SNED1 | sushi, nidogen and EGF-like domains 1 | 3.24 × 10−2 | 2.47 | |||||
| 213488_at | SNED1 | sushi, nidogen and EGF-like domains 1 | 2.30 × 10−1 | 1.94 | |||||
| 205236_x_at | SOD3 | superoxide dismutase 3, extracellular | 1.15 × 10−1 | 1.71 | |||||
| 202363_at | SPOCK1 | sparc/osteonectin, cwcv and kazal-like domains proteoglycan (testican) 1 | 5.31 × 10−3 | 2.13 | |||||
| 201858_s_at | SRGN | serglycin | 3.34 × 10−4 | 8.11 | |||||
| 201859_at | SRGN | serglycin | 3.81 × 10−4 | 4.87 | |||||
| 219552_at | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 1.07 × 10−1 | 1.70 | |||||
| 213247_at | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 6.04 × 10−2 | 1.70 | |||||
| 226506_at | THSD4 | thrombospondin, type I, domain containing 4 | 6.68 × 10−4 | 3.97 | |||||
| 222835_at | THSD4 | thrombospondin, type I, domain containing 4 | 5.26 × 10−5 | 3.43 | |||||
| 219058_x_at | TINAGL1 | tubulointerstitial nephritis antigen-like 1 | 8.12 × 10−3 | 2.50 | |||||
| 216333_x_at | TNXA TNXB |
tenascin XA pseudogene tenascin XB | 7.97 × 10−5 | 11.18 | |||||
| 206093_x_at | TNXA TNXB |
tenascin XA pseudogene tenascin XB | 5.32 × 10−5 | 10.76 | |||||
| 213451_x_at | TNXA TNXB |
tenascin XA pseudogene tenascin XB | 1.65 × 10−4 | 9.71 | |||||
| 208609_s_at | TNXB | tenascin XB | 7.07 × 10−5 | 8.99 | |||||
| Focal Adhesion Points | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205730_s_at | ABLIM3 | actin binding LIM protein family, member 3 | 1.06 × 10−1 | 1.53 | 200965_s_at | ABLIM1 | actin binding LIM protein 1 | 2.02 × 10−5 | 4.17 |
| 213497_at | ABTB2 | ankyrin repeat and BTB (POZ) domain containing 2 | 6.59 × 10−2 | 1.61 | 205882_x_at | ADD3 | adducin 3 (gamma) | 6.45 × 10−3 | 1.59 |
| 205268_s_at | ADD2 | adducin 2 (beta) | 8.16 × 10−4 | 7.50 | 201752_s_at | ADD3 | adducin 3 (gamma) | 9.71 × 10−3 | 1.56 |
| 205771_s_at | AKAP7 | A kinase (PRKA) anchor prot 7 | 1.03 × 10−1 | 1.58 | 227529_s_at | AKAP12 | A kinase (PRKA) anchor prot 12 | 6.81 × 10−3 | 8.03 |
| 205257_s_at | AMPH | amphiphysin | 1.45 × 10−7 | 6.02 | 227530_at | AKAP12 | A kinase (PRKA) anchor prot 12 | 4.43 × 10−3 | 6.10 |
| 1552619_a_at | ANLN | anillin, actin binding protein | 3.48 × 10−2 | 2.47 | 210517_s_at | AKAP12 | A kinase (PRKA) anchor prot 12 | 6.53 × 10−3 | 4.66 |
| 222608_s_at | ANLN | anillin, actin binding protein | 4.60 × 10−2 | 2.25 | 202920_at | ANK2 | ankyrin 2, neuronal | 8.40 × 10−3 | 2.34 |
| 203526_s_at | APC | adenomatous polyposis coli | 3.34 × 10−3 | 1.60 | 206385_s_at | ANK3 | ankyrin 3, node of Ranvier (ankyrin G) | 3.69 × 10−3 | 3.16 |
| 204492_at | ARHGAP11A | Rho GTPase activating protein 11A | 3.11 × 10−2 | 2.12 | 227337_at | ANKRD37 | ankyrin repeat domain 37 | 4.35 × 10−6 | 6.04 |
| 37577_at | ARHGAP19 | Rho GTPase activating protein 19 | 9.72 × 10−4 | 1.72 | 204671_s_at | ANKRD6 | ankyrin repeat domain 6 | 3.20 × 10−2 | 2.12 |
| 206298_at | ARHGAP22 | Rho GTPase activating protein 22 | 8.57 × 10−3 | 1.58 | 204672_s_at | ANKRD6 | ankyrin repeat domain 6 | 5.69 × 10−2 | 1.96 |
| 201288_at | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 3.85 × 10−2 | 1.54 | 228368_at | ARHGAP20 | Rho GTPase activating prot 20 | 1.15 × 10−4 | 5.72 |
| 204092_s_at | AURKA | aurora kinase A | 2.07 × 10−2 | 2.88 | 227911_at | ARHGAP28 | Rho GTPase activating prot 28 | 1.07 × 10−3 | 2.38 |
| 208079_s_at | AURKA | aurora kinase A | 3.80 × 10−2 | 2.45 | 206167_s_at | ARHGAP6 | Rho GTPase activating prot 6 | 1.09 × 10−1 | 1.65 |
| 209464_at | AURKB | aurora kinase B | 1.04 × 10−2 | 2.92 | 205109_s_at | ARHGEF4 | Rho guanine nucleotide exchange factor (GEF) 4 | 1.49 × 10−1 | 1.53 |
| 205294_at | BAIAP2 | BAI1-associated protein 2 | 2.67 × 10−3 | 1.51 | 201615_x_at | CALD1 | caldesmon 1 | 1.61 × 10−1 | 1.79 |
| 210334_x_at | BIRC5 | baculoviral IAP repeat-containing 5 | 1.01 × 10−2 | 2.69 | 201616_s_at | CALD1 | caldesmon 1 | 1.89 × 10−2 | 1.53 |
| 202094_at | BIRC5 | baculoviral IAP repeat-containing 5 | 4.10 × 10−2 | 2.63 | 236473_at | CC2D2A | coiled-coil and C2 domain containing 2A | 2.18 × 10−3 | 2.56 |
| 202095_s_at | BIRC5 | baculoviral IAP repeat-containing 5 | 1.72 × 10−2 | 2.47 | 203139_at | DAPK1 | death-associated protein kinase 1 | 2.66 × 10−2 | 3.97 |
| 220935_s_at | CDK5RAP2 | CDK5 regulatory subunit associated protein 2 | 1.91 × 10−7 | 1.78 | 229800_at | DCLK1 | Doublecortin-like kinase 1 | 1.78 × 10−1 | 1.99 |
| 204962_s_at | CENPA | centromere protein A | 3.83 × 10−2 | 2.88 | 217208_s_at | DLG1 | discs, large homolog 1 (Drosophila) | 6.19 × 10−3 | 2.32 |
| 210821_x_at | CENPA | centromere protein A | 4.21 × 10−3 | 1.98 | 202515_at | DLG1 | discs, large homolog 1 (Drosophila) | 1.59 × 10−3 | 1.72 |
| 205046_at | CENPE | centromere protein E, 312 kDa | 7.08 × 10−2 | 2.65 | 202514_at | DLG1 | discs, large homolog 1 (Drosophila) | 6.69 × 10−3 | 1.72 |
| 209172_s_at | CENPF | centromere protein F, 350/400 ka (mitosin) | 1.33 × 10−2 | 2.98 | 230229_at | DLG1 | Discs, large homolog 1 (Drosophila) | 1.49 × 10−1 | 1.63 |
| 207828_s_at | CENPF | centromere protein F, 350/400 ka (mitosin) | 2.29 × 10−2 | 2.94 | 202516_s_at | DLG1 | discs, large homolog 1 (Drosophila) | 5.92 × 10−2 | 1.60 |
| 231772_x_at | CENPH | centromere protein H | 2.74 × 10−2 | 1.82 | 203881_s_at | DMD | dystrophin | 1.06 × 10−4 | 5.02 |
| 214804_at | CENPI | centromere protein I | 5.29 × 10−2 | 1.96 | 208086_s_at | DMD | dystrophin | 2.36 × 10−2 | 1.58 |
| 207590_s_at | CENPI | centromere protein I | 1.62 × 10−2 | 1.88 | 227081_at | DNALI1 | dynein, axonemal, light intermediate chain 1 | 2.64 × 10−2 | 1.64 |
| 223513_at | CENPJ | centromere protein J | 2.83 × 10−2 | 1.69 | 226875_at | DOCK11 | dedicator of cytokinesis 11 | 4.24 × 10−4 | 1.78 |
| 222848_at | CENPK | centromere protein K | 1.00 × 10−1 | 1.94 | 1554863_s_at | DOK5 | docking protein 5 | 1.93 × 10−2 | 1.59 |
| 1554271_a_at | CENPL | centromere protein L | 1.69 × 10−1 | 1.54 | 214844_s_at | DOK5 | docking protein 5 | 2.98 × 10−3 | 1.52 |
| 218741_at | CENPM | centromere protein M | 1.86 × 10−2 | 2.36 | 220161_s_at | EPB41L4B | erythrocyte membrane protein band 4.1 like 4B | 5.64 × 10−2 | 2.33 |
| 219555_s_at | CENPN | centromere protein N | 9.25 × 10−3 | 1.89 | 209829_at | FAM65B | family with sequence similarity 65, member B | 3.97 × 10−2 | 2.58 |
| 222118_at | CENPN | centromere protein N | 1.24 × 10−1 | 1.84 | 206707_x_at | FAM65B | family with sequence similarity 65, member B | 3.02 × 10−2 | 2.20 |
| 228559_at | CENPN | centromere protein N | 8.63 × 10−2 | 1.74 | 226129_at | FAM83H | family with sequence similarity 83, member H | 2.04 × 10−2 | 1.75 |
| 226118_at | CENPO | centromere protein O | 6.22 × 10−2 | 1.78 | 227948_at | FGD4 | FYVE, RhoGEF and PH domain containing 4 | 9.89 × 10−4 | 3.94 |
| 219294_at | CENPQ | centromere protein Q | 5.50 × 10−2 | 1.56 | 230559_x_at | FGD4 | FYVE, RhoGEF and PH domain containing 4 | 3.65 × 10−3 | 2.66 |
| 205642_at | CEP110 | centrosomal protein 110 kDa | 6.59 × 10−3 | 1.87 | 225167_at | FRMD4A | FERM domain containing 4A | 1.45 × 10−2 | 2.04 |
| 239413_at | CEP152 | centrosomal protein 152 kDa | 3.34 × 10−3 | 1.71 | 225163_at | FRMD4A | FERM domain containing 4A | 8.39 × 10−3 | 1.98 |
| 218542_at | CEP55 | centrosomal protein 55 kDa | 2.63 × 10−2 | 2.44 | 225168_at | FRMD4A | FERM domain containing 4A | 2.22 × 10−2 | 1.76 |
| 206324_s_at | DAPK2 | death-associated protein kinase 2 | 7.47 × 10−2 | 1.73 | 1560031_at | FRMD4A | FERM domain containing 4A | 1.07 × 10−3 | 1.71 |
| 227666_at | DCLK2 | doublecortin-like kinase 2 | 7.38 × 10−2 | 1.52 | 208476_s_at | FRMD4A | FERM domain containing 4A | 7.85 × 10−3 | 1.69 |
| 207147_at | DLX2 | distal-less homeobox 2 | 1.94 × 10−2 | 6.12 | 1554034_a_at | FRMD4A | FERM domain containing 4A | 2.32 × 10−1 | 1.57 |
| 215116_s_at | DNM1 | dynamin 1 | 2.03 × 10−4 | 3.83 | 239290_at | FRMPD4 | FERM and PDZ domain containing 4 | 1.74 × 10−1 | 1.56 |
| 219279_at | DOCK10 | dedicator of cytokinesis 10 | 2.60 × 10−2 | 1.66 | 203037_s_at | MTSS1 | metastasis suppressor 1 | 2.31 × 10−3 | 4.32 |
| 213160_at | DOCK2 | dedicator of cytokinesis 2 | 1.12 × 10−4 | 1.76 | 212096_s_at | MTUS1 | mitochondrial tumor supp 1 | 1.02 × 10−1 | 2.47 |
| 205003_at | DOCK4 | dedicator of cytokinesis 4 | 1.44 × 10−1 | 1.54 | 212095_s_at | MTUS1 | mitochondrial tumor supp 1 | 1.08 × 10−1 | 1.74 |
| 206710_s_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 1.31 × 10−2 | 3.52 | 228098_s_at | MYLIP | myosin regulatory light chain interacting protein | 5.16 × 10−2 | 1.57 |
| 212681_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 1.38 × 10−2 | 3.22 | 220319_s_at | MYLIP | myosin regulatory light chain interacting protein | 4.55 × 10−2 | 1.50 |
| 211776_s_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 1.48 × 10−2 | 3.20 | 237206_at | MYOCD | myocardin | 8.62 × 10−3 | 3.85 |
| 218980_at | FHOD3 | formin homology 2 domain containing 3 | 7.93 × 10−3 | 3.13 | 213782_s_at | MYOZ2 | myozenin 2 | 8.68 × 10−2 | 2.16 |
| 238621_at | FMN1 | formin 1 | 6.44 × 10−3 | 2.47 | 207148_x_at | MYOZ2 | myozenin 2 | 8.91 × 10−2 | 2.07 |
| 1555471_a_at | FMN2 | formin 2 | 2.05 × 10−2 | 1.85 | 219073_s_at | OSBPL10 | oxysterol binding protein-like 10 | 3.31 × 10−2 | 2.11 |
| 223618_at | FMN2 | formin 2 | 2.05 × 10−2 | 1.82 | 209621_s_at | PDLIM3 | PDZ and LIM domain 3 | 8.38 × 10−2 | 3.31 |
| 215017_s_at | FNBP1L | formin binding protein 1-like | 3.68 × 10−3 | 1.52 | 213684_s_at | PDLIM5 | PDZ and LIM domain 5 | 6.28 × 10−3 | 1.87 |
| 230645_at | FRMD3 | FERM domain containing 3 | 2.69 × 10−1 | 1.63 | 221994_at | PDLIM5 | PDZ and LIM domain 5 | 2.46 × 10−3 | 1.81 |
| 230831_at | FRMD5 | FERM domain containing 5 | 1.02 × 10−2 | 2.87 | 203242_s_at | PDLIM5 | PDZ and LIM domain 5 | 1.86 × 10−3 | 1.68 |
| 238756_at | GAS2L3 | Growth arrest-specific 2 like 3 | 1.22 × 10−2 | 2.33 | 216804_s_at | PDLIM5 | PDZ and LIM domain 5 | 4.90 × 10−3 | 1.60 |
| 235709_at | GAS2L3 | growth arrest-specific 2 like 3 | 2.92 × 10−2 | 1.81 | 207717_s_at | PKP2 | plakophilin 2 | 2.96 × 10−2 | 3.09 |
| 226308_at | HAUS8 | HAUS augmin-like complex, subunit 8 | 3.75 × 10−2 | 1.71 | 201927_s_at | PKP4 | plakophilin 4 | 2.90 × 10−1 | 1.68 |
| 226364_at | HIP1 | Huntingtin interacting protein 1 | 1.05 × 10−3 | 2.73 | 227148_at | PLEKHH2 | pleckstrin homology domain containing, family H member 2 | 3.16 × 10−3 | 3.05 |
| 205425_at | HIP1 | huntingtin interacting protein 1 | 9.54 × 10−3 | 2.66 | 203407_at | PPL | periplakin | 7.99 × 10−3 | 3.20 |
| 218934_s_at | HSPB7 | heat shock 27 kDa protein family, member 7 (cardiovascular) | 7.51 × 10−2 | 2.16 | 226627_at | SEPT8 | septin 8 | 1.20 × 10−1 | 1.74 |
| 227750_at | KALRN | kalirin, RhoGEF kinase | 8.61 × 10−3 | 1.53 | 226438_at | SNTB1 | syntrophin, beta 1 (dystrophin-associated protein A1, 59 kDa, basic component 1) | 1.03 × 10−2 | 1.84 |
| 229125_at | KANK4 | KN motif and ankyrin repeat domains 4 | 1.76 × 10−2 | 3.51 | 214708_at | SNTB1 | syntrophin, beta 1 (dystrophin-associated protein A1, 59 kDa, basic component 1) | 3.83 × 10−2 | 1.53 |
| 204444_at | KIF11 | kinesin family member 11 | 4.00 × 10−2 | 2.43 | 227179_at | STAU2 | staufen, RNA binding protein, homolog 2 (Drosophila) | 1.58 × 10−2 | 1.89 |
| 236641_at | KIF14 | kinesin family member 14 | 1.15 × 10−2 | 3.50 | 212565_at | STK38L | serine/threonine kinase 38 like | 5.74 × 10−5 | 1.94 |
| 206364_at | KIF14 | kinesin family member 14 | 5.29 × 10−2 | 2.87 | 212572_at | STK38L | serine/threonine kinase 38 like | 5.09 × 10−3 | 1.53 |
| 219306_at | KIF15 | kinesin family member 15 | 1.87 × 10−2 | 2.65 | 202796_at | SYNPO | synaptopodin | 7.05 × 10−2 | 2.18 |
| 221258_s_at | KIF18A | kinesin family member 18A | 1.25 × 10−2 | 2.79 | 227662_at | SYNPO2 | synaptopodin 2 | 1.29 × 10−1 | 3.30 |
| 222039_at | KIF18B | kinesin family member 18B | 6.34 × 10−2 | 2.25 | 213135_at | TIAM1 | T-cell lymphoma invasion and metastasis 1 | 1.12 × 10−1 | 1.65 |
| 218755_at | KIF20A | kinesin family member 20A | 1.17 × 10−2 | 2.81 | 209904_at | TNNC1 | troponin C type 1 (slow) | 6.34 × 10−2 | 2.76 |
| 205235_s_at | KIF20B | kinesin family member 20B | 1.83 × 10−2 | 1.95 | 215389_s_at | TNNT2 | troponin T type 2 (cardiac) | 5.06 × 10−2 | 3.22 |
| 216969_s_at | KIF22 | kinesin family member 22 | 9.75 × 10−2 | 1.92 | 210276_s_at | TRIOBP | TRIO and F-actin binding prot | 6.96 × 10−2 | 1.52 |
| 202183_s_at | KIF22 | kinesin family member 22 | 6.19 × 10−3 | 1.65 | 223279_s_at | UACA | uveal autoantigen with coiled-coil domains and ankyrin repeats | 6.84 × 10−3 | 1.79 |
| 204709_s_at | KIF23 | kinesin family member 23 | 3.68 × 10−2 | 2.55 | 238868_at | UACA | uveal autoantigen with coiled-coil domains and ankyrin repeats | 1.37 × 10−1 | 1.66 |
| 244427_at | KIF23 | Kinesin family member 23 | 2.68 × 10−3 | 1.76 | |||||
| 209408_at | KIF2C | kinesin family member 2C | 3.86 × 10−2 | 2.95 | |||||
| 211519_s_at | KIF2C | kinesin family member 2C | 2.04 × 10−2 | 2.80 | |||||
| 218355_at | KIF4A | kinesin family member 4A | 3.29 × 10−2 | 2.60 | |||||
| 209680_s_at | KIFC1 | kinesin family member C1 | 1.72 × 10−2 | 2.43 | |||||
| 206316_s_at | KNTC1 | kinetochore associated 1 | 1.88 × 10−2 | 1.86 | |||||
| 224823_at | MYLK | myosin light chain kinase | 1.91 × 10−1 | 1.72 | |||||
| 236718_at | MYO10 | myosin X | 1.05 × 10−3 | 1.86 | |||||
| 244350_at | MYO10 | myosin X | 1.45 × 10−2 | 1.70 | |||||
| 241966_at | MYO5A | myosin VA (heavy chain 12, myoxin) | 2.21 × 10−2 | 1.51 | |||||
| 201774_s_at | NCAPD2 | non-SMC condensin I complex, subunit D2 | 1.30 × 10−1 | 1.57 | |||||
| 212789_at | NCAPD3 | non-SMC condensin II complex, subunit D3 | 5.55 × 10−2 | 1.56 | |||||
| 218663_at | NCAPG | non-SMC condensin I complex, subunit G | 1.03 × 10−1 | 2.23 | |||||
| 218662_s_at | NCAPG | non-SMC condensin I complex, subunit G | 8.24 × 10−2 | 2.11 | |||||
| 219588_s_at | NCAPG2 | non-SMC condensin II complex, subunit G2 | 2.03 × 10−2 | 1.86 | |||||
| 212949_at | NCAPH | non-SMC condensin I complex, subunit H | 3.01 × 10−2 | 2.58 | |||||
| 204641_at | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | 2.34 × 10−2 | 2.97 | |||||
| 211080_s_at | NEK2 | NIMA (never in mitosis gene a)-related kinase 2 | 4.89 × 10−3 | 2.64 | |||||
| 223381_at | NUF2 | NUF2, NDC80 kinetochore complex component, homolog (S. cerevisiae) | 8.44 × 10−2 | 2.67 | |||||
| 219978_s_at | NUSAP1 | nucleolar and spindle associated protein 1 | 1.39 × 10−1 | 2.48 | |||||
| 218039_at | NUSAP1 | nucleolar and spindle associated protein 1 | 2.98 × 10−2 | 2.42 | |||||
| 204972_at | OAS2 | 2’-5’-oligoadenylate synthetase 2, 69/71 kDa | 2.83 × 10−1 | 1.68 | |||||
| 209626_s_at | OSBPL3 | oxysterol binding protein-like 3 | 5.77 × 10−2 | 1.67 | |||||
| 238575_at | OSBPL6 | oxysterol binding protein-like 6 | 1.17 × 10−2 | 2.13 | |||||
| 223805_at | OSBPL6 | oxysterol binding protein-like 6 | 9.15 × 10−3 | 2.09 | |||||
| 218644_at | PLEK2 | pleckstrin 2 | 4.94 × 10−3 | 2.63 | |||||
| 218009_s_at | PRC1 | protein regulator of cytokinesis 1 | 2.03 × 10−2 | 2.37 | |||||
| 222077_s_at | RACGAP1 | Rac GTPase activating protein 1 | 1.49 × 10−2 | 1.99 | |||||
| 219263_at | RNF128 | ring finger protein 128 | 2.50 × 10−2 | 3.03 | |||||
| 230730_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 2.17 × 10−2 | 3.65 | |||||
| 213543_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 2.12 × 10−2 | 3.57 | |||||
| 228602_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 6.41 × 10−2 | 3.55 | |||||
| 214492_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 5.41 × 10−3 | 3.18 | |||||
| 210329_s_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 8.64 × 10−3 | 2.72 | |||||
| 210330_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 3.47 × 10−2 | 2.43 | |||||
| 207302_at | SGCG | sarcoglycan, gamma (35 kDa dystrophin-associated glycoprotein) | 1.42 × 10−1 | 3.09 | |||||
| 217678_at | SLC7A11 | solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | 7.43 × 10−2 | 1.53 | |||||
| 209921_at | SLC7A11 | solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | 3.18 × 10−2 | 1.53 | |||||
| 1556583_a_at | SLC8A1 | solute carrier family 8 (sodium/calcium exchanger), member 1 | 1.96 × 10−1 | 1.85 | |||||
| 241752_at | SLC8A1 | solute carrier family 8 (sodium/calcium exchanger), member 1 | 2.94 × 10−1 | 1.61 | |||||
| 200783_s_at | STMN1 | stathmin 1 | 1.16 × 10−2 | 2.14 | |||||
| 222557_at | STMN3 | stathmin-like 3 | 1.48 × 10−2 | 1.83 | |||||
| 212703_at | TLN2 | talin 2 | 5.18 × 10−4 | 1.84 | |||||
| 206117_at | TPM1 | tropomyosin 1 (alpha) | 5.66 × 10−3 | 2.26 | |||||
| 210052_s_at | TPX2 | TPX2, microtubule-associated, homolog (Xenopus laevis) | 1.78 × 10−2 | 2.56 | |||||
| 1555938_x_at | VIM | vimentin | 2.81 × 10−2 | 2.00 | |||||
| 202663_at | WIPF1 | WAS/WASL interacting protein family, member 1 | 6.39 × 10−3 | 1.64 | |||||
| 202664_at | WIPF1 | WAS/WASL interacting protein family, member 1 | 5.61 × 10−4 | 1.58 | |||||
| 202665_s_at | WIPF1 | WAS/WASL interacting protein family, member 1 | 2.97 × 10−3 | 1.51 | |||||
| Cytoskeleton | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205132_at | ACTC1 | actin, alpha, cardiac muscle 1 | 1.80 × 10−3 | 4.22 | 203563_at | AFAP1 | actin filament associated protein 1 | 2.27 × 10−3 | 1.90 |
| 230925_at | APBB1IP | amyloid beta (A4) precursor protein-binding, family B, member 1 interacting protein | 1.66 × 10−2 | 2.50 | 206488_s_at | CD36 | CD36 molecule (thrombospondin receptor) | 1.46 × 10−8 | 20.69 |
| 226292_at | CAPN5 | calpain 5 | 2.20 × 10−4 | 1.51 | 209555_s_at | CD36 | CD36 molecule (thrombospondin receptor) | 3.58 × 10−8 | 19.05 |
| 217523_at | CD44 | CD44 molecule (Indian blood group) | 1.89 × 10−2 | 1.64 | 228766_at | CD36 | CD36 molecule (thrombospondin receptor) | 2.97 × 10−6 | 11.94 |
| 220115_s_at | CDH10 | cadherin 10, type 2 (T2-cadherin) | 1.94 × 10−1 | 1.90 | 201005_at | CD9 | CD9 molecule | 7.82 × 10−4 | 2.58 |
| 207030_s_at | CSRP2 | cysteine and glycine-rich protein 2 | 1.93 × 10−2 | 1.74 | 201131_s_at | CDH1 | cadherin 1, type 1, E-cadherin (epithelial) | 4.31 × 10−2 | 2.54 |
| 211126_s_at | CSRP2 | cysteine and glycine-rich protein 2 | 2.34 × 10−2 | 1.71 | 204726_at | CDH13 | cadherin 13, H-cadherin (heart) | 3.20 × 10−2 | 2.60 |
| 214724_at | DIXDC1 | DIX domain containing 1 | 1.14 × 10−2 | 1.53 | 203256_at | CDH3 | cadherin 3, type 1, P-cadherin (placental) | 3.21 × 10−2 | 1.84 |
| 202668_at | EFNB2 | ephrin-B2 | 1.85 × 10−1 | 3.24 | 200621_at | CSRP1 | cysteine and glycine-rich protein 1 | 1.70 × 10−2 | 1.56 |
| 205031_at | EFNB3 | ephrin-B3 | 2.95 × 10−10 | 2.47 | 203716_s_at | DPP4 | dipeptidyl-peptidase 4 | 4.05 × 10−2 | 1.93 |
| 1555480_a_at | FBLIM1 | filamin binding LIM protein 1 | 1.07 × 10−2 | 1.89 | 211478_s_at | DPP4 | dipeptidyl-peptidase 4 | 2.67 × 10−1 | 1.80 |
| 1554795_a_at | FBLIM1 | filamin binding LIM protein 1 | 2.26 × 10−2 | 1.61 | 203717_at | DPP4 | dipeptidyl-peptidase 4 | 1.08 × 10−1 | 1.67 |
| 225258_at | FBLIM1 | filamin binding LIM protein 1 | 2.87 × 10−3 | 1.56 | 227955_s_at | EFNA5 | ephrin-A5 | 4.68 × 10−2 | 1.94 |
| 204379_s_at | FGFR3 | fibroblast growth factor receptor 3 | 9.77 × 10−2 | 2.01 | 214036_at | EFNA5 | ephrin-A5 | 1.32 × 10−1 | 1.53 |
| 242592_at | GPR137C | G protein-coupled receptor 137C | 1.50 × 10−2 | 2.18 | 201983_s_at | EGFR | epidermal growth factor receptor (erythroblastic leukemia viral (v-erb-b) oncogene homolog, avian) | 1.75 × 10−3 | 1.82 |
| 235961_at | GPR161 | G protein-coupled receptor 161 | 4.34 × 10−4 | 1.56 | 201809_s_at | ENG | endoglin | 2.31 × 10−3 | 2.06 |
| 230369_at | GPR161 | G protein-coupled receptor 161 | 3.44 × 10−2 | 1.53 | 201539_s_at | FHL1 | four and a half LIM domains 1 | 8.39 × 10−3 | 6.09 |
| 229055_at | GPR68 | G protein-coupled receptor 68 | 7.83 × 10−3 | 1.70 | 214505_s_at | FHL1 | four and a half LIM domains 1 | 6.48 × 10−3 | 5.76 |
| 234303_s_at | GPR85 | G protein-coupled receptor 85 | 7.25 × 10−2 | 2.04 | 210299_s_at | FHL1 | four and a half LIM domains 1 | 1.55 × 10−3 | 5.39 |
| 203632_s_at | GPRC5B | G protein-coupled receptor, family C, group 5, member B | 1.76 × 10−1 | 1.92 | 210298_x_at | FHL1 | four and a half LIM domains 1 | 6.70 × 10−3 | 5.18 |
| 222899_at | ITGA11 | integrin, alpha 11 | 1.21 × 10−2 | 1.59 | 201540_at | FHL1 | four and a half LIM domains 1 | 4.21 × 10−4 | 3.20 |
| 227314_at | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 5.68 × 10−2 | 2.53 | 222853_at | FLRT3 | fibronectin leucine rich transmembrane protein 3 | 7.40 × 10−4 | 3.36 |
| 205032_at | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 3.78 × 10−2 | 2.41 | 219250_s_at | FLRT3 | fibronectin leucine rich transmembrane protein 3 | 4.52 × 10−2 | 2.12 |
| 228080_at | LAYN | layilin | 4.84 × 10−3 | 2.59 | 212950_at | GPR116 | G protein-coupled receptor 116 | 1.98 × 10−1 | 3.52 |
| 216250_s_at | LPXN | leupaxin | 2.03 × 10−5 | 3.08 | 213094_at | GPR126 | G protein-coupled receptor 126 | 9.65 × 10−4 | 5.14 |
| 210869_s_at | MCAM | melanoma cell adhesion molecule | 1.03 × 10−6 | 7.67 | 232267_at | GPR133 | G protein-coupled receptor 133 | 1.46 × 10−2 | 2.44 |
| 209087_x_at | MCAM | melanoma cell adhesion molecule | 1.08 × 10−6 | 7.38 | 228949_at | GPR177 | G protein-coupled receptor 177 | 3.16 × 10−4 | 2.71 |
| 211340_s_at | MCAM | melanoma cell adhesion molecule | 1.07 × 10−6 | 6.81 | 228950_s_at | GPR177 | G protein-coupled receptor 177 | 3.49 × 10−3 | 2.63 |
| 209086_x_at | MCAM | melanoma cell adhesion molecule | 2.85 × 10−8 | 5.09 | 221958_s_at | GPR177 | G protein-coupled receptor 177 | 1.52 × 10−3 | 2.50 |
| 203062_s_at | MDC1 | mediator of DNA damage checkpoint 1 | 3.87 × 10−3 | 1.58 | 229105_at | GPR39 | G protein-coupled receptor 39 | 2.66 × 10−2 | 1.90 |
| 212843_at | NCAM1 | neural cell adhesion molecule 1 | 8.99 × 10−7 | 4.58 | 212070_at | GPR56 | G protein-coupled receptor 56 | 2.12 × 10−2 | 1.78 |
| 227394_at | NCAM1 | neural cell adhesion molecule 1 | 1.26 × 10−6 | 3.16 | 203108_at | GPRC5A | G protein-coupled receptor, family C, group 5, member A | 2.08 × 10−3 | 8.54 |
| 213438_at | NFASC | neurofascin homolog (chicken) | 3.12 × 10−2 | 2.57 | 202638_s_at | ICAM1 | intercellular adhesion molecule 1 | 1.31 × 10−1 | 2.21 |
| 230242_at | NFASC | neurofascin homolog (chicken) | 5.17 × 10−3 | 2.19 | 202637_s_at | ICAM1 | intercellular adhesion molecule 1 | 7.04 × 10−2 | 1.70 |
| 243645_at | NFASC | neurofascin homolog (chicken) | 2.12 × 10−3 | 2.03 | 205885_s_at | ITGA4 | integrin, alpha 4 (antigen CD49D, alpha 4 subunit of VLA-4 receptor) | 2.52 × 10−1 | 1.71 |
| 219773_at | NOX4 | NADPH oxidase 4 | 8.60 × 10−2 | 1.80 | 216331_at | ITGA7 | integrin, alpha 7 | 1.01 × 10−2 | 3.76 |
| 37966_at | PARVB | parvin, beta | 4.78 × 10−10 | 2.88 | 209663_s_at | ITGA7 | integrin, alpha 7 | 4.49 × 10−2 | 3.70 |
| 204629_at | PARVB | parvin, beta | 2.58 × 10−8 | 2.28 | 204990_s_at | ITGB4 | integrin, beta 4 | 8.14 × 10−2 | 1.67 |
| 37965_at | PARVB | parvin, beta | 7.26 × 10−5 | 1.98 | 226189_at | ITGB8 | integrin, beta 8 | 2.19 × 10−2 | 1.69 |
| 216253_s_at | PARVB | parvin, beta | 4.90 × 10−3 | 1.80 | 220765_s_at | LIMS2 | LIM and senescent cell antigen-like domains 2 | 1.80 × 10−2 | 1.80 |
| 225977_at | PCDH18 | protocadherin 18 | 4.98 × 10−3 | 2.18 | 226974_at | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like | 4.84 × 10−2 | 2.02 |
| 225975_at | PCDH18 | protocadherin 18 | 1.29 × 10−2 | 1.76 | 212448_at | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like | 9.48 × 10−2 | 1.78 |
| 207011_s_at | PTK7 | PTK7 protein tyrosine kinase 7 | 3.74 × 10−3 | 2.23 | 212445_s_at | NEDD4L | neural precursor cell expressed, developmentally down-regulated 4-like | 1.04 × 10−1 | 1.76 |
| 1555324_at | PTK7 | PTK7 protein tyrosine kinase 7 | 5.32 × 10−3 | 1.78 | 202150_s_at | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 | 3.21 × 10−3 | 1.97 |
| 207419_s_at | RAC2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 1.39 × 10−2 | 2.37 | 202149_at | NEDD9 | neural precursor cell expressed, developmentally down-regulated 9 | 3.93 × 10−4 | 1.90 |
| 213603_s_at | RAC2 | ras-related C3 botulinum toxin substrate 2 (rho family, small GTP binding protein Rac2) | 8.95 × 10−3 | 2.13 | 228635_at | PCDH10 | protocadherin 10 | 2.55 × 10−3 | 3.91 |
| 223168_at | RHOU | ras homolog gene family, member U | 1.13 × 10−5 | 3.13 | 223435_s_at | PCDHA1 PCDHA10 PCDHA11 PCDHA12 PCDHA13 PCDHA2 PCDHA3 PCDHA4 PCDHA5 PCDHA6 PCDHA7 PCDHA8 PCDHA9 PCDHAC1 PCDHAC2 |
protocadherin alpha 1 protocadherin alpha 10 protocadherin alpha 11 protocadherin alpha 12 protocadherin alpha 13 protocadherin alpha 2 protocadherin alpha 3 protocadherin alpha 4 protocadherin alpha 5 protocadherin alpha 6 protocadherin alpha 7 protocadherin alpha 8 protocadherin alpha 9 protocadherin alpha C, 1 protocadherin alpha C, 2 |
2.40 × 10−3 | 2.23 |
| 201286_at | SDC1 | syndecan 1 | 7.78 × 10−3 | 3.11 | 202565_s_at | SVIL | supervillin | 5.25 × 10−3 | 3.45 |
| 201287_s_at | SDC1 | syndecan 1 | 3.47 × 10−3 | 3.04 | 202566_s_at | SVIL | supervillin | 5.31 × 10−2 | 2.32 |
| 202898_at | SDC3 | syndecan 3 | 2.98 × 10−2 | 1.66 | 206702_at | TEK | TEK tyrosine kinase, endothelial | 4.91 × 10−4 | 3.67 |
| 218087_s_at | SORBS1 | sorbin and SH3 domain containing 1 | 1.05 × 10−2 | 4.48 | |||||
| 222513_s_at | SORBS1 | sorbin and SH3 domain containing 1 | 3.04 × 10−2 | 2.95 | |||||
| 208850_s_at | THY1 | Thy-1 cell surface antigen | 2.80 × 10−1 | 1.91 | |||||
| 213869_x_at | THY1 | Thy-1 cell surface antigen | 1.38 × 10−1 | 1.77 | |||||
| 208851_s_at | THY1 | Thy-1 cell surface antigen | 2.50 × 10−1 | 1.70 | |||||
| 217853_at | TNS3 | tensin 3 | 5.51 × 10−5 | 2.61 | |||||
| 217979_at | TSPAN13 | tetraspanin 13 | 1.08 × 10−4 | 4.67 | |||||
| 227307_at | TSPAN18 | Tetraspanin 18 | 2.78 × 10−4 | 3.95 | |||||
| 227236_at | TSPAN2 | tetraspanin 2 | 1.85 × 10−1 | 2.19 | |||||
| 214606_at | TSPAN2 | tetraspanin 2 | 1.19 × 10−1 | 1.83 | |||||
| LINC Complexes | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 203145_at | SPAG5 | sperm associated antigen 5 | 2.63 × 10−2 | 2.48 | 219888_at | SPAG4 | sperm associated antigen 4 | 8.03 × 10−2 | 1.87 |
| Nucleoskeleton | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205436_s_at | H2AFX | H2A histone family, member X | 1.88 × 10−2 | 1.71 | 215071_s_at | HIST1H2AC | histone cluster 1, H2ac | 2.13 × 10−2 | 1.70 |
| 214463_x_at | HIST1H4J | histone cluster 1, H4j | 1.24 × 10−2 | 1.53 | 214455_at | HIST1H2BC | histone cluster 1, H2bc | 1.34 × 10−2 | 1.71 |
| 201795_at | LBR | lamin B receptor | 8.68 × 10−4 | 1.88 | 236193_at | HIST1H2BC | histone cluster 1, H2bc | 1.57 × 10−2 | 1.60 |
| 203276_at | LMNB1 | lamin B1 | 8.33 × 10−2 | 2.42 | 209911_x_at | HIST1H2BD | histone cluster 1, H2bd | 1.63 × 10−2 | 1.63 |
| 209753_s_at | TMPO | thymopoietin | 5.06 × 10−3 | 1.98 | 208527_x_at | HIST1H2BE | histone cluster 1, H2be | 4.22 × 10−3 | 1.54 |
| 224944_at | TMPO | thymopoietin | 5.13 × 10−3 | 1.84 | 232035_at | HIST1H4B | Histone cluster 1, H4b | 5.60 × 10−3 | 1.96 |
| 209754_s_at | TMPO | thymopoietin | 2.51 × 10−2 | 1.83 | 208180_s_at | HIST1H4B | Histone cluster 1, H4b | 1.34 × 10−1 | 1.59 |
| 203432_at | TMPO | thymopoietin | 1.02 × 10−1 | 1.64 | |||||
| Secreted Factors | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205608_s_at | ANGPT1 | angiopoietin 1 | 1.27 × 10−4 | 3.04 | 231773_at | ANGPTL1 | angiopoietin-like 1 | 3.01 × 10−2 | 2.12 |
| 205609_at | ANGPT1 | angiopoietin 1 | 7.38 × 10−6 | 2.97 | 224339_s_at | ANGPTL1 | angiopoietin-like 1 | 7.59 × 10−2 | 1.71 |
| 213001_at | ANGPTL2 | angiopoietin-like 2 | 1.03 × 10−1 | 1.50 | 239183_at | ANGPTL1 | angiopoietin-like 1 | 8.51 × 10−2 | 1.50 |
| 220988_s_at | C1QTNF3 | C1q and tumor necrosis factor related protein 3 | 9.99 × 10−2 | 1.78 | 221009_s_at | ANGPTL4 | angiopoietin-like 4 | 6.59 × 10−3 | 2.88 |
| 1405_i_at | CCL5 | chemokine (C-C motif) ligand 5 | 1.12 × 10−1 | 1.56 | 223333_s_at | ANGPTL4 | angiopoietin-like 4 | 1.80 × 10−1 | 2.11 |
| 203666_at | CXCL12 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 6.11 × 10−3 | 2.98 | 209546_s_at | APOL1 | apolipoprotein L, 1 | 1.42 × 10−1 | 1.78 |
| 209687_at | CXCL12 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 1.99 × 10−2 | 2.24 | 221087_s_at | APOL3 | apolipoprotein L, 3 | 6.19 × 10−2 | 1.83 |
| 222484_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 5.41 × 10−4 | 7.47 | 205239_at | AREG | amphiregulin | 1.14 × 10−1 | 1.75 |
| 218002_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 1.22 × 10−3 | 6.46 | 202701_at | BMP1 | bone morphogenetic protein 1 | 5.22 × 10−2 | 1.54 |
| 204602_at | DKK1 | dickkopf homolog 1 (Xenopus laevis) | 2.42 × 10−2 | 2.04 | 205289_at | BMP2 | bone morphogenetic protein 2 | 2.65 × 10−2 | 2.54 |
| 219908_at | DKK2 | dickkopf homolog 2 (Xenopus laevis) | 7.38 × 10−4 | 4.54 | 205290_s_at | BMP2 | bone morphogenetic protein 2 | 8.44 × 10−2 | 2.25 |
| 228952_at | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 5.77 × 10−7 | 4.95 | 211518_s_at | BMP4 | bone morphogenetic protein 4 | 1.68 × 10−2 | 4.03 |
| 229088_at | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 5.38 × 10−7 | 4.83 | 206176_at | BMP6 | bone morphogenetic protein 6 | 2.52 × 10−2 | 2.33 |
| 205066_s_at | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 3.10 × 10−5 | 4.76 | 239349_at | C1QTNF7 | C1q and tumor necrosis factor related protein 7 | 2.49 × 10−1 | 1.53 |
| 205065_at | ENPP1 | ectonucleotide pyrophosphatase/phosphodiesterase 1 | 3.78 × 10−7 | 3.82 | 202357_s_at | C2 CFB |
complement component 2 complement factor B | 3.16 × 10−3 | 3.85 |
| 205110_s_at | FGF13 | fibroblast growth factor 13 | 3.58 × 10−8 | 4.73 | 217767_at | C3 | complement component 3 | 2.71 × 10−1 | 2.42 |
| 214240_at | GAL | galanin prepropeptide | 9.61 × 10−2 | 1.59 | 208451_s_at | C4A C4B |
complement component 4A (Rodgers blood group) complement component 4B (Chido blood group) | 1.98 × 10−1 | 1.83 |
| 205505_at | GCNT1 | glucosaminyl (N-acetyl) transferase 1, core 2 (beta-1,6-N-acetylglucosaminyltransferase) | 1.70 × 10−5 | 1.66 | 206407_s_at | CCL13 | chemokine (C-C motif) ligand 13 | 8.28 × 10−2 | 3.52 |
| 240509_s_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 2.66 × 10−3 | 3.23 | 216598_s_at | CCL2 | chemokine (C-C motif) ligand 2 | 5.57 × 10−4 | 8.18 |
| 235504_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 9.10 × 10−3 | 3.11 | 206508_at | CD70 | CD70 molecule | 1.14 × 10−1 | 2.44 |
| 220794_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 2.06 × 10−2 | 2.76 | 213800_at | CFH | complement factor H | 8.63 × 10−5 | 6.48 |
| 206326_at | GRP | gastrin-releasing peptide | 3.05 × 10−2 | 1.59 | 215388_s_at | CFH CFHR1 |
complement factor H complement factor H-related 1 | 4.95 × 10−6 | 10.91 |
| 203821_at | HBEGF | heparin-binding EGF-like growth factor | 1.31 × 10−1 | 1.64 | 206910_x_at | CFHR2 | complement factor H-related 2 | 8.81 × 10−3 | 1.60 |
| 203819_s_at | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | 4.18 × 10−2 | 2.51 | 209395_at | CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) | 1.10 × 10−2 | 4.08 |
| 203820_s_at | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | 6.30 × 10−2 | 2.13 | 209396_s_at | CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) | 4.55 × 10−2 | 1.94 |
| 212143_s_at | IGFBP3 | insulin-like growth factor binding protein 3 | 1.26 × 10−2 | 1.54 | 206315_at | CRLF1 | cytokine receptor-like factor 1 | 8.40 × 10−3 | 3.23 |
| 227760_at | IGFBPL1 | insulin-like growth factor binding protein-like 1 | 3.76 × 10−3 | 1.94 | 209774_x_at | CXCL2 | chemokine (C-X-C motif) ligand 2 | 1.37 × 10−1 | 3.03 |
| 204773_at | IL11RA | interleukin 11 receptor, alpha | 1.36 × 10−3 | 1.97 | 207850_at | CXCL3 | chemokine (C-X-C motif) ligand 3 | 2.50 × 10−1 | 2.07 |
| 206172_at | IL13RA2 | interleukin 13 receptor, alpha 2 | 8.39 × 10−2 | 3.56 | 219837_s_at | CYTL1 | cytokine-like 1 | 1.99 × 10−1 | 2.02 |
| 227997_at | IL17RD | interleukin 17 receptor D | 9.70 × 10−2 | 1.70 | 219501_at | ENOX1 | ecto-NOX disulfide-thiol exchanger 1 | 1.82 × 10−2 | 2.35 |
| 222062_at | IL27RA | interleukin 27 receptor, alpha | 4.50 × 10−4 | 3.73 | 226213_at | ERBB3 | v-erb-b2 erythroblastic leukemia viral oncogene homolog 3 (avian) | 1.84 × 10−2 | 2.49 |
| 205926_at | IL27RA | interleukin 27 receptor, alpha | 5.58 × 10−4 | 1.67 | 205738_s_at | FABP3 | fatty acid binding protein 3, muscle and heart (mammary-derived growth inhibitor) | 2.56 × 10−1 | 1.70 |
| 226218_at | IL7R | interleukin 7 receptor | 9.41 × 10−2 | 1.71 | 203980_at | FABP4 | fatty acid binding protein 4, adipocyte | 5.11 × 10−2 | 2.16 |
| 205798_at | IL7R | interleukin 7 receptor | 1.78 × 10−1 | 1.59 | 205117_at | FGF1 | fibroblast growth factor 1 (acidic) | 8.61 × 10−3 | 2.95 |
| 231798_at | NOG | noggin | 6.76 × 10−5 | 3.98 | 1552721_a_at | FGF1 | fibroblast growth factor 1 (acidic) | 1.47 × 10−2 | 2.71 |
| 206343_s_at | NRG1 | neuregulin 1 | 6.34 × 10−3 | 2.68 | 208240_s_at | FGF1 | fibroblast growth factor 1 (acidic) | 9.05 × 10−2 | 1.73 |
| 206237_s_at | NRG1 | neuregulin 1 | 7.90 × 10−2 | 2.15 | 231382_at | FGF18 | Fibroblast growth factor 18 | 1.71 × 10−1 | 2.09 |
| 208230_s_at | NRG1 | neuregulin 1 | 1.20 × 10−2 | 1.90 | 211029_x_at | FGF18 | fibroblast growth factor 18 | 2.54 × 10−1 | 1.64 |
| 204766_s_at | NUDT1 | nudix (nucleoside diphosphate linked moiety X)-type motif 1 | 2.22 × 10−5 | 1.59 | 221577_x_at | GDF15 | growth differentiation factor 15 | 3.41 × 10−2 | 2.03 |
| 213131_at | OLFM1 | olfactomedin 1 | 1.82 × 10−1 | 1.71 | 206614_at | GDF5 | growth differentiation factor 5 | 4.96 × 10−2 | 2.01 |
| 213125_at | OLFML2B | olfactomedin-like 2B | 2.17 × 10−1 | 1.93 | 201348_at | GPX3 | glutathione peroxidase 3 (plasma) | 2.15 × 10−2 | 3.24 |
| 218162_at | OLFML3 | olfactomedin-like 3 | 3.28 × 10−2 | 1.72 | 214091_s_at | GPX3 | glutathione peroxidase 3 (plasma) | 7.64 × 10−2 | 1.99 |
| 222719_s_at | PDGFC | platelet derived growth factor C | 2.01 × 10−3 | 1.61 | 209960_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 7.99 × 10−2 | 3.24 |
| 201578_at | PODXL | podocalyxin-like | 1.11 × 10−2 | 5.15 | 210997_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 2.26 × 10−2 | 2.96 |
| 210195_s_at | PSG1 | pregnancy specific beta-1-glycoprotein 1 | 2.14 × 10−1 | 1.82 | 210998_s_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 3.94 × 10−2 | 1.80 |
| 208134_x_at | PSG2 | pregnancy specific beta-1-glycoprotein 2 | 1.56 × 10−3 | 3.17 | 210619_s_at | HYAL1 | hyaluronoglucosaminidase 1 | 1.86 × 10−3 | 3.19 |
| 203399_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 8.06 × 10−3 | 3.33 | 209540_at | IGF1 | insulin-like growth factor 1 (somatomedin C) | 2.78 × 10−1 | 1.97 |
| 215821_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 2.08 × 10−2 | 2.47 | 209542_x_at | IGF1 | insulin-like growth factor 1 (somatomedin C) | 2.03 × 10−1 | 1.57 |
| 211741_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 1.76 × 10−2 | 2.22 | 202718_at | IGFBP2 | insulin-like growth factor binding protein 2, 36 kDa | 8.69 × 10−2 | 3.80 |
| 204830_x_at | PSG5 | pregnancy specific beta-1-glycoprotein 5 | 8.14 × 10−3 | 3.53 | 201508_at | IGFBP4 | insulin-like growth factor binding protein 4 | 1.12 × 10−1 | 1.68 |
| 209738_x_at | PSG6 | pregnancy specific beta-1-glycoprotein 6 | 7.00 × 10−3 | 3.35 | 203426_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 2.55 × 10−2 | 3.31 |
| 208106_x_at | PSG6 | pregnancy specific beta-1-glycoprotein 6 | 4.99 × 10−3 | 3.26 | 211958_at | IGFBP5 | insulin-like growth factor binding protein 5 | 2.16 × 10−1 | 2.76 |
| 209594_x_at | PSG9 | pregnancy specific beta-1-glycoprotein 9 | 5.91 × 10−3 | 3.49 | 1555997_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 1.64 × 10−1 | 2.46 |
| 207733_x_at | PSG9 | pregnancy specific beta-1-glycoprotein 9 | 1.20 × 10−2 | 3.03 | 203425_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 1.58 × 10−1 | 1.80 |
| 212187_x_at | PTGDS | prostaglandin D2 synthase 21 kDa (brain) | 5.94 × 10−3 | 1.91 | 206295_at | IL18 | interleukin 18 (interferon-gamma-inducing factor) | 3.70 × 10−2 | 3.75 |
| 211748_x_at | PTGDS | prostaglandin D2 synthase 21 kDa (brain) | 2.12 × 10−3 | 1.69 | 207526_s_at | IL1RL1 | interleukin 1 receptor-like 1 | 1.32 × 10−1 | 2.07 |
| 206631_at | PTGER2 | prostaglandin E receptor 2 (subtype EP2), 53 kDa | 4.68 × 10−2 | 1.71 | 242809_at | IL1RL1 | Interleukin 1 receptor-like 1 | 2.38 × 10−1 | 1.60 |
| 211737_x_at | PTN | pleiotrophin | 7.25 × 10−2 | 3.28 | 221111_at | IL26 | interleukin 26 | 4.68 × 10−2 | 3.44 |
| 209465_x_at | PTN | pleiotrophin | 4.83 × 10−2 | 3.24 | 209821_at | IL33 | interleukin 33 | 1.79 × 10−1 | 1.50 |
| 209466_x_at | PTN | pleiotrophin | 1.03 × 10−1 | 2.42 | 205207_at | IL6 | interleukin 6 (interferon, beta 2) | 2.42 × 10−4 | 3.20 |
| 209897_s_at | SLIT2 | slit homolog 2 (Drosophila) | 1.55 × 10−2 | 1.98 | 204863_s_at | IL6ST | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 1.97 × 10−2 | 1.99 |
| 205016_at | TGFA | transforming growth factor, alpha | 4.19 × 10−2 | 3.27 | 211000_s_at | IL6ST | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 1.21 × 10−2 | 1.88 |
| 203085_s_at | TGFB1 | transforming growth factor, beta 1 | 7.50 × 10−3 | 1.60 | 204926_at | INHBA | inhibin, beta A | 1.44 × 10−2 | 2.47 |
| 236561_at | TGFBR1 | Transforming growth factor, beta receptor 1 | 4.05 × 10−2 | 1.71 | 210511_s_at | INHBA | inhibin, beta A | 4.64 × 10−3 | 2.42 |
| 203887_s_at | THBD | thrombomodulin | 2.94 × 10−1 | 2.27 | 205266_at | LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 7.91 × 10−2 | 2.44 |
| 239336_at | THBS1 | Thrombospondin 1 | 1.84 × 10−1 | 1.58 | 219181_at | LIPG | lipase, endothelial | 3.51 × 10−2 | 2.89 |
| 227420_at | TNFAIP8L1 | tumor necrosis factor, alpha-induced protein 8-like 1 | 3.38 × 10−3 | 1.71 | 205381_at | LRRC17 | leucine rich repeat containing 17 | 2.67 × 10−4 | 18.57 |
| 219478_at | WFDC1 | WAP four-disulfide core domain 1 | 3.17 × 10−2 | 5.38 | 216320_x_at | MST1 | macrophage stimulating 1 (hepatocyte growth factor-like) | 1.05 × 10−1 | 1.53 |
| 221029_s_at | WNT5B | wingless-type MMTV integration site family, member 5B | 5.20 × 10−4 | 2.39 | 231361_at | NLGN1 | Neuroligin 1 | 4.86 × 10−2 | 3.33 |
| 223537_s_at | WNT5B | wingless-type MMTV integration site family, member 5B | 2.99 × 10−2 | 1.65 | 205893_at | NLGN1 | neuroligin 1 | 7.72 × 10−2 | 3.23 |
| 204501_at | NOV | nephroblastoma overexpressed gene | 2.01 × 10−2 | 2.95 | |||||
| 214321_at | NOV | nephroblastoma overexpressed gene | 1.69 × 10−2 | 2.60 | |||||
| 217525_at | OLFML1 | olfactomedin-like 1 | 5.70 × 10−2 | 3.66 | |||||
| 213075_at | OLFML2A | olfactomedin-like 2A | 1.64 × 10−2 | 2.12 | |||||
| 205729_at | OSMR | oncostatin M receptor | 6.53 × 10−2 | 1.71 | |||||
| 224942_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 2.20 × 10−2 | 1.88 | |||||
| 1559400_s_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 3.28 × 10−2 | 1.84 | |||||
| 201981_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 3.78 × 10−2 | 1.74 | |||||
| 224940_s_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 1.72 × 10−2 | 1.73 | |||||
| 224941_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 1.36 × 10−2 | 1.69 | |||||
| 228128_x_at | PAPPA | pregnancy-associated plasma protein A, pappalysin 1 | 2.13 × 10−2 | 1.64 | |||||
| 205560_at | PCSK5 | proprotein convertase subtilisin/kexin type 5 | 2.21 × 10−3 | 2.70 | |||||
| 213652_at | PCSK5 | Proprotein convertase subtilisin/kexin type 5 | 5.18 × 10−4 | 2.66 | |||||
| 205559_s_at | PCSK5 | proprotein convertase subtilisin/kexin type 5 | 1.23 × 10−3 | 2.52 | |||||
| 227759_at | PCSK9 | proprotein convertase subtilisin/kexin type 9 | 2.34 × 10−2 | 1.97 | |||||
| 216867_s_at | PDGFA | platelet-derived growth factor alpha polypeptide | 2.46 × 10−2 | 1.97 | |||||
| 222860_s_at | PDGFD | platelet derived growth factor D | 1.50 × 10−1 | 1.57 | |||||
| 1555778_a_at | POSTN | periostin, osteoblast specific factor | 4.41 × 10−3 | 4.30 | |||||
| 214981_at | POSTN | periostin, osteoblast specific factor | 2.20 × 10−3 | 2.96 | |||||
| 210809_s_at | POSTN | periostin, osteoblast specific factor | 8.30 × 10−3 | 2.47 | |||||
| 207808_s_at | PROS1 | protein S (alpha) | 3.88 × 10−3 | 1.51 | |||||
| 213421_x_at | PRSS3 | protease, serine, 3 | 2.41 × 10−2 | 2.68 | |||||
| 210367_s_at | PTGES | prostaglandin E synthase | 3.17 × 10−4 | 4.21 | |||||
| 207388_s_at | PTGES | prostaglandin E synthase | 4.23 × 10−2 | 2.51 | |||||
| 224950_at | PTGFRN | prostaglandin F2 receptor negative regulator | 4.86 × 10−2 | 1.78 | |||||
| 211892_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 4.91 × 10−2 | 2.06 | |||||
| 210702_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 1.13 × 10−1 | 1.70 | |||||
| 208131_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 2.43 × 10−2 | 1.67 | |||||
| 211756_at | PTHLH | parathyroid hormone-like hormone | 6.78 × 10−2 | 3.45 | |||||
| 206300_s_at | PTHLH | parathyroid hormone-like hormone | 7.08 × 10−2 | 2.86 | |||||
| 210355_at | PTHLH | parathyroid hormone-like hormone | 1.70 × 10−1 | 2.21 | |||||
| 206157_at | PTX3 | pentraxin-related gene, rapidly induced by IL-1 beta | 1.47 × 10−4 | 2.21 | |||||
| 201482_at | QSOX1 | quiescin Q6 sulfhydryl oxidase 1 | 2.19 × 10−3 | 1.68 | |||||
| 223824_at | RNLS | renalase, FAD-dependent amine oxidase | 5.18 × 10−4 | 1.74 | |||||
| 204035_at | SCG2 | secretogranin II (chromogranin C) | 2.66 × 10−2 | 2.56 | |||||
| 205475_at | SCRG1 | scrapie responsive protein 1 | 6.88 × 10−7 | 7.73 | |||||
| 213716_s_at | SECTM1 | secreted and transmembrane 1 | 3.32 × 10−2 | 3.29 | |||||
| 203071_at | SEMA3B | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3B | 6.35 × 10−2 | 2.54 | |||||
| 203788_s_at | SEMA3C | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | 6.34 × 10−4 | 2.51 | |||||
| 203789_s_at | SEMA3C | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3C | 3.13 × 10−3 | 1.64 | |||||
| 33323_r_at | SFN | stratifin | 1.80 × 10−1 | 2.42 | |||||
| 223122_s_at | SFRP2 | secreted frizzled-related prot 2 | 2.86 × 10−1 | 2.33 | |||||
| 204051_s_at | SFRP4 | secreted frizzled-related prot 4 | 4.79 × 10−2 | 4.23 | |||||
| 204052_s_at | SFRP4 | secreted frizzled-related prot 4 | 1.56 × 10−1 | 2.58 | |||||
| 210665_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 4.02 × 10−2 | 3.69 | |||||
| 210664_s_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 2.35 × 10−2 | 3.50 | |||||
| 209676_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 2.96 × 10−2 | 3.35 | |||||
| 213258_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 2.61 × 10−2 | 3.24 | |||||
| 228121_at | TGFB2 | transforming growth factor, beta 2 | 1.08 × 10−6 | 5.13 | |||||
| 209909_s_at | TGFB2 | transforming growth factor, beta 2 | 5.47 × 10−5 | 3.64 | |||||
| 204731_at | TGFBR3 | transforming growth factor, beta receptor III | 2.06 × 10−1 | 1.70 | |||||
| 203083_at | THBS2 | thrombospondin 2 | 4.84 × 10−6 | 2.83 | |||||
| 202644_s_at | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 1.61 × 10−3 | 3.88 | |||||
| 202643_s_at | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 4.35 × 10−3 | 3.52 | |||||
| 206025_s_at | TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 1.57 × 10−2 | 2.29 | |||||
| 206026_s_at | TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 3.57 × 10−2 | 2.15 | |||||
| 210260_s_at | TNFAIP8 | tumor necrosis factor, alpha-induced protein 8 | 5.99 × 10−4 | 2.01 | |||||
| 208296_x_at | TNFAIP8 | tumor necrosis factor, alpha-induced protein 8 | 1.11 × 10−3 | 1.94 | |||||
| 235737_at | TSLP | thymic stromal lymphopoietin | 1.72 × 10−1 | 2.02 | |||||
| 210513_s_at | VEGFA | vascular endothelial growth factor A | 9.18 × 10−2 | 1.57 | |||||
| 205648_at | WNT2 | wingless-type MMTV integration site family member 2 | 2.83 × 10−1 | 2.08 | |||||
| 202643_s_at | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 4.35 × 10−3 | 3.52 | |||||
| 206025_s_at | TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 1.57 × 10−2 | 2.29 | |||||
| 206026_s_at | TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 3.57 × 10−2 | 2.15 | |||||
| 210260_s_at | TNFAIP8 | tumor necrosis factor, alpha-induced protein 8 | 5.99 × 10−4 | 2.01 | |||||
| 208296_x_at | TNFAIP8 | tumor necrosis factor, alpha-induced protein 8 | 1.11 × 10−3 | 1.94 | |||||
| 235737_at | TSLP | thymic stromal lymphopoietin | 1.72 × 10−1 | 2.02 | |||||
| 210513_s_at | VEGFA | vascular endothelial growth factor A | 9.18 × 10−2 | 1.57 | |||||
| 205648_at | WNT2 | wingless-type MMTV integration site family member 2 | 2.83 × 10−1 | 2.08 | |||||
For validation of our microarray data, 19 genes were selected from the signatures that distinguished Fp, Fr, and F-DHJ identities, and transcript levels were analyzed by qRT-PCR in cell samples from the six donors (Figure 5A,B). Validation of microarray data was obtained for the 19 selected transcripts. As an attempt to identify a biomarker of F-DHJ cells, a focus was made on KLF9, which the transcript was detected as overexpressed in F-DHJ versus Fr by both technics in the six tested donors. The transcription factor KLF9 regulates the early phases of adipocyte differentiation [25], and thus, attracted attention due to the proximity of F-DHJ cells with hypodermis adipose tissue. Expression of KLF9 was analyzed at the protein level by western blot in cultured cells from six donors (Figure 5C) and by immune-fluorescence in skin biopsies from four individuals (Figure 5D). As expected from transcriptome data, the KLF9 protein was expressed at a higher level in cultured F-DHJ than in cultured Fp and Fr (p < 0.05). In skin sections, the percentage of cells expressing KLF9 was higher in F-DHJ than in Fp and Fr regions, respectively 18.8 ± 3.4% versus 9.2 ± 1.3% and 5.3 ± 1.9%.
3.4. The Dermo-Hypodermal Junction and Reticular Dermis Differ in Their Matrix Architectural Meshwork
From the lists of transcripts differentially expressed between F-DHJ and Fr, our attention was attracted by tenascin C (TNC), considering its major role in the organization of collagen fibril anchoring points. Indeed, TNC forms a typical disulfide-linked hexamer, called the hexabrachion, in which six flexible arms emanate from a central globular particle, which possibly catches and stabilizes a bifurcation of the ECM fibrils composed of FN1 and type I collagen to underlie the extracellular meshwork architecture (for review, see [26]). Our transcriptome analysis indicated a 2.61-fold lower expression of TNC in F-DHJ versus Fr cells (Table 3). To explore this property at the protein level, immunostaining of TNC was performed on samples of ECM synthesized by F-DHJ and Fr cells in vitro (Figure 6A,B) (cells from n = 3 individuals were tested). Notably, reticulation of TNC was more marked in ECM samples synthesized by Fr than in ECM secreted by F-DHJ (Figure 6A). Moreover, signal quantification indicated TNC levels lower in ECM produced by F-DHJ versus Fr (p < 0.01) (Figure 6B).
Figure 6.
Architecture of the tenascin C (TNC) meshwork produced in vitro by Fr and F-DHJ fibroblasts in skin sections. (A) Immunostaining pictures of the TNC meshwork produced by Fr and F-DHJ cells in 2D cultures. (B) Quantification of TNC secreted in 2D cultures. Cell samples from 3 donors were used. Values corresponding to 10 replicate analyses for each cell sample are shown. Means ± SEM are indicated (** p < 0.01, Wilcoxon test) A.U. for arbitrary units. (C) Photographs of TNC immunostaining in skin sections, illustrating the structural differences between reticular dermis and the dermo-hypodermal junction area (representative from 13 analyzed donors). (D) Image reconstitution of TNC meshwork architectures based on the immunostaining photographs shown in panel (C). (E) Quantification of TNC in the skin reticular and dermo-hypodermal areas. Values obtained from the analysis of skin samples from 13 donors are shown. Samples from two anatomical localizations: breast skin (7 donors of ages between 18 and 65 years) and abdominal skin (6 donors of ages between 42 and 51 years). No age-related changes in TNC synthesis/meshwork were observed. Means ± SEM are indicated (* p < 0.05, Wilcoxon test) A.U. for arbitrary units.
Architectural differences between the DHJ and reticular areas were confirmed in skin sections (Figure 6C–E). In the reticular area, TNC protein-staining revealed a thin mesh structuration around collagen bundles in agreement with the alveolar organization of this dermal territory, whereas this structuration was not present in the DHJ area (Figure 6C,D). In addition, quantification of the TNC immunostaining signals performed in sections of mammary skin (biopsies from seven individuals) and abdominal skin (six individuals) indicated a higher level of TNC in the reticular dermis area than in the HDJ area for both skin anatomical origins (p < 0.05) (Figure 6E).
3.5. F-DHJ Fibroblasts and Adipose-Derived MSCs Exhibit Distinct Transcriptome Profiles
Given the anatomical proximity between F-DHJ and MSCs derived from hypodermal adipose tissues, their molecular characteristics were explored at the level of the global transcriptome to determine whether these two cell populations have a distinct identity or not. To widen this question, the three fibroblast types (Fp, Fr, and F-DHJ) were analyzed together with MSC samples corresponding to five sources (bone marrow aspirates, adipose tissue, amnion, chorion, and umbilical cord jelly) (Figure 7). A hierarchical clustering based on 380 discriminant probe sets revealed a clear segmentation between the “fibroblast” group and the “MSC” group (Figure 7A), which confirmed the distinct identities of F-DHJ and adipose MSCs. Within the “fibroblast” group, F-DHJ appeared more similar with Fr than they were with Fp cells. Within the “MSC” group, cells from the three fetal origins (amnion, chorion, and cord) were more similar to each other than they were with the two adult origins (marrow and adipose). This clustering was confirmed when a full transcriptome analysis was considered (Pearson correlation coefficients) (Figure 7B). To document biological characteristics distinguishing the fibroblast and MSC groups, a gene ontology (GO) term analysis was performed based on 2974 probe sets (1984 genes) distinguishing the two sample groups (parameters: fold-change >2 and p-value <0.05). Among the twenty most significant GO terms, transcripts related to structuration of the tissue skeleton were largely represented, including numerous ECM, focal adhesion, cytoskeleton, LINC complexes, nucleoskeleton, and secreted factor transcripts, in which their levels distinguish fibroblasts from MSCs (Figure 7C,D and Table 4). In particular, a signature of 42 transcripts directly related to ECM structure and composition was identified (Figure 7E), constituting a pool of candidates to further explore the biological differences between F-DHJ and adipose MSCs.
Figure 7.
Comparative microarray transcriptome profiling of the three fibroblasts populations (Fp, Fr, and F-DHJ) and mesenchymal stem cell (MSC) samples corresponding to five sources (bone marrow aspirates, adipose tissue, amnion, chorion, and umbilical cord jelly). (A) Hierarchical clustering of fibroblast and MSC samples based on the 380 most discriminant probe sets showing a marked distinction between the “fibroblast” and “MSC” groups. (B) Pearson correlation coefficients evaluating sample-to-sample proximity based on comparisons of global transcriptome profiles. Notably, this analysis showed the low proximity between F-DHJ with adipose tissue MSCs (93.83% similarity) and high proximity with Fr fibroblasts (96.89% similarity). (C) List of the 20 most significant gene ontology (GO) terms differentiating the “fibroblast” and “MSC” groups based on 2974 probe sets (1984 transcripts) exhibiting differential signals (fold-change > 2 and p < 0.05). (D) Signatures identifying the “fibroblast” group (black bars) and the “MSC” group (grey bars) among transcripts related to the tissue skeleton biology (fold-change >2 and p < 0.05). (E) Focus on 42 transcripts directly involved in the structuration and composition of the ECM network and identified within the signature that distinguishes the “fibroblast” and “MSC” groups. Values were obtained by GCRMA microarray signals and corresponded to an indication of transcript levels (arbitrary units) in F-DHJ and adipose MSCs.
Table 4.
Transcripts related to the tissue skeleton, in which differential expressions distinguish the “fibroblast” and mesenchymal stem cell or “MSC” groups. This transcript list was extracted from microarray data using fold-change >2 and p < 0.05 as inclusion parameters. The transcript signature with predominant expression in the “fibroblast” group concerned 424 probe sets corresponding to transcripts directly involved in the tissue skeleton structure, comprising 145 transcripts related to ECM, 63 focal adhesion point transcripts, 68 cytoskeleton transcripts, 4 LINC complex transcripts, and 12 nucleoskeleton transcripts. The transcript signature with predominant expression in the “MSC” group concerned 241 probe sets corresponding to transcripts directly involved in the tissue skeleton structure, comprising 53 transcripts related to ECM, 63 focal adhesion point transcripts, 52 cytoskeleton transcripts, 2 LINC complex transcripts, and 7 nucleoskeleton transcripts. In addition, transcripts encoding soluble factors were found in both signatures, respectively 132 and 79 for the “fibroblast” and “MSC” groups.
| UP in Dermal Fibroblasts | UP in MSCs | ||||||||
|---|---|---|---|---|---|---|---|---|---|
| Extracellular Matrix Genes | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205679_x_at | ACAN | aggrecan | 8.39 × 10−22 | 25.41 | 209765_at | ADAM19 | ADAM metallopeptidase domain 19 (meltrin beta) | 7.41 × 10−28 | 8.60 |
| 207692_s_at | ACAN | aggrecan | 7.55 × 10−21 | 23.23 | 226997_at | ADAMTS12 | ADAM metallopeptidase with thrombospondin type 1 motif, 12 | 9.13 × 10−13 | 4.00 |
| 217161_x_at | ACAN | aggrecan | 1.14 × 10−20 | 20.79 | 214913_at | ADAMTS3 | ADAM metallopeptidase with thrombospondin type 1 motif, 3 | 1.26 × 10−10 | 4.41 |
| 232570_s_at | ADAM33 | ADAM metallopeptidase domain 33 | 2.07 × 10−9 | 4.20 | 1570351_at | ADAMTS6 | ADAM metallopeptidase with thrombospondin type 1 motif, 6 | 3.98 × 10−15 | 2.31 |
| 233868_x_at | ADAM33 | ADAM metallopeptidase domain 33 | 3.16 × 10−8 | 2.98 | 222043_at | CLU | clusterin | 5.59 × 10−4 | 2.05 |
| 214454_at | ADAMTS2 | ADAM metallopeptidase with thrombospondin type 1 motif, 2 | 4.55 × 10−14 | 2.74 | 225288_at | COL27A1 | collagen, type XXVII, alpha 1 | 3.75 × 10−9 | 2.35 |
| 229357_at | ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1, motif, 5 | 1.26 × 10−16 | 22.33 | 213110_s_at | COL4A5 | collagen, type IV, alpha 5 | 2.61 × 10−3 | 3.17 |
| 219935_at | ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 6.18 × 10−16 | 21.71 | 204136_at | COL7A1 | collagen, type VII, alpha 1 | 5.86 × 10−5 | 2.46 |
| 235368_at | ADAMTS5 | ADAM metallopeptidase with thrombospondin type 1 motif, 5 | 1.86 × 10−15 | 11.70 | 223475_at | CRISPLD1 | cysteine-rich secretory protein LCCL domain containing 1 | 2.40 × 10−5 | 2.02 |
| 219087_at | ASPN | asporin | 3.78 × 10−15 | 18.79 | 201487_at | CTSC | cathepsin C | 2.84 × 10−13 | 3.90 |
| 224396_s_at | ASPN | asporin | 6.26 × 10−6 | 3.63 | 225646_at | CTSC | cathepsin C | 1.77 × 10−7 | 2.77 |
| 201262_s_at | BGN | biglycan | 4.64 × 10−10 | 3.10 | 225647_s_at | CTSC | cathepsin C | 4.24 × 10−7 | 2.64 |
| 213905_x_at | BGN | biglycan | 5.80 × 10−7 | 2.15 | 231234_at | CTSC | cathepsin C | 7.29 × 10−8 | 2.40 |
| 201261_x_at | BGN | biglycan | 1.53 × 10−6 | 2.00 | 229115_at | DYNC1H1 | dynein, cytoplasmic 1, heavy chain 1 | 8.64 × 10−5 | 2.02 |
| 241986_at | BMPER | BMP binding endothelial regulator | 1.17 × 10−9 | 2.75 | 207379_at | EDIL3 | EGF-like repeats and discoidin I-like domains 3 | 2.94 × 10−4 | 2.09 |
| 227526_at | CDON | Cdon homolog (mouse) | 1.16 × 10−6 | 2.34 | 226911_at | EGFLAM | EGF-like, fibronectin type III and laminin G domains | 5.99 × 10−5 | 3.25 |
| 209732_at | CLEC2B | C-type lectin domain family 2, member B | 9.58 × 10−18 | 66.68 | 203184_at | FBN2 | fibrillin 2 | 5.58 × 10−5 | 6.34 |
| 1556209_at | CLEC2B | C-type lectin domain family 2, member B | 4.34 × 10−6 | 4.46 | 236028_at | IBSP | integrin-binding sialoprotein | 1.83 × 10−3 | 2.85 |
| 205200_at | CLEC3B | C-type lectin domain family 3, member B | 1.76 × 10−12 | 17.53 | 223689_at | IGF2BP1 | insulin-like growth factor 2 mRNA binding protein 1 | 1.37 × 10−17 | 5.58 |
| 217428_s_at | COL10A1 | collagen, type X, alpha 1 | 3.42 × 10−4 | 3.59 | 203819_s_at | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | 1.72 × 10−28 | 39.96 |
| 205941_s_at | COL10A1 | collagen, type X, alpha 1 | 5.32 × 10−4 | 3.30 | 203820_s_at | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | 1.52 × 10−29 | 30.51 |
| 231879_at | COL12A1 | collagen, type XII, alpha 1 | 7.44 × 10−15 | 5.04 | 216493_s_at | IGF2BP3 | insulin-like growth factor 2 mRNA binding protein 3 | 2.04 × 10−22 | 6.25 |
| 234951_s_at | COL12A1 | collagen, type XII, alpha 1 | 3.01 × 10−8 | 3.71 | 205206_at | KAL1 | Kallmann syndrome 1 sequence | 8.59 × 10−6 | 5.25 |
| 225664_at | COL12A1 | collagen, type XII, alpha 1 | 5.13 × 10−11 | 2.60 | 202728_s_at | LTBP1 | latent transforming growth factor beta binding protein 1 | 3.50 × 10−12 | 4.74 |
| 231766_s_at | COL12A1 | collagen, type XII, alpha 1 | 1.68 × 10−6 | 2.34 | 202729_s_at | LTBP1 | latent transforming growth factor beta binding protein 1 | 2.67 × 10−12 | 3.43 |
| 203477_at | COL15A1 | collagen, type XV, alpha 1 | 2.08 × 10−11 | 18.96 | 223614_at | MMP16 | matrix metallopeptidase 16 (membrane-inserted) | 7.85 × 10−11 | 3.87 |
| 211966_at | COL4A2 | collagen, type IV, alpha 2 | 2.64 × 10−4 | 2.52 | 207012_at | MMP16 | matrix metallopeptidase 16 (membrane-inserted) | 4.43 × 10−13 | 3.63 |
| 226277_at | COL4A3BP | collagen, type IV, alpha 3 (Goodpasture antigen) binding protein | 6.82 × 10−34 | 2.01 | 229346_at | NES | nestin | 6.51 × 10−18 | 6.19 |
| 229779_at | COL4A4 | collagen, type IV, alpha 4 | 2.16 × 10−6 | 3.00 | 218678_at | NES | nestin | 3.67 × 10−9 | 3.84 |
| 221900_at | COL8A2 | collagen, type VIII, alpha 2 | 1.22 × 10−7 | 4.76 | 201860_s_at | PLAT | plasminogen activator, tissue | 3.31 × 10−16 | 6.86 |
| 52651_at | COL8A2 | collagen, type VIII, alpha 2 | 3.02 × 10−8 | 3.51 | 205479_s_at | PLAU | plasminogen activator, urokinase | 4.34 × 10−28 | 22.23 |
| 205713_s_at | COMP | cartilage oligomeric matrix protein | 1.03 × 10−26 | 115.83 | 211668_s_at | PLAU | plasminogen activator, urokinase | 2.42 × 10−20 | 12.30 |
| 226824_at | CPXM2 | carboxypeptidase X (M14 family), member 2 | 1.74 × 10−8 | 5.71 | 211924_s_at | PLAUR | plasminogen activator, urokinase receptor | 7.22 × 10−17 | 2.73 |
| 208978_at | CRIP2 | cysteine-rich protein 2 | 6.66 × 10−14 | 6.14 | 210845_s_at | PLAUR | plasminogen activator, urokinase receptor | 2.18 × 10−22 | 2.69 |
| 221541_at | CRISPLD2 | cysteine-rich secretory protein LCCL domain containing 2 | 1.80 × 10−5 | 2.74 | 206007_at | PRG4 | proteoglycan 4 | 1.12 × 10−6 | 2.17 |
| 204971_at | CSTA | cystatin A (stefin A) | 9.43 × 10−5 | 3.43 | 221872_at | RARRES1 | retinoic acid receptor responder (tazarotene induced) 1 | 5.43 × 10−19 | 6.46 |
| 209101_at | CTGF | connective tissue growth factor | 5.63 × 10−10 | 2.09 | 206392_s_at | RARRES1 | retinoic acid receptor responder (tazarotene induced) 1 | 1.34 × 10−16 | 4.46 |
| 200661_at | CTSA | cathepsin A | 5.53 × 10−17 | 2.18 | 222784_at | SMOC1 | SPARC related modular calcium binding 1 | 4.56 × 10−4 | 2.10 |
| 200766_at | CTSD | cathepsin D | 5.83 × 10−18 | 2.96 | 201858_s_at | SRGN | serglycin | 1.63 × 10−6 | 6.82 |
| 203657_s_at | CTSF | cathepsin F | 1.31 × 10−37 | 7.43 | 201859_at | SRGN | serglycin | 3.48 × 10−7 | 4.90 |
| 202295_s_at | CTSH | cathepsin H | 8.10 × 10−5 | 2.03 | 209277_at | TFPI2 | tissue factor pathway inhibitor 2 | 3.21 × 10−13 | 10.83 |
| 203758_at | CTSO | cathepsin O | 5.40 × 10−11 | 2.37 | 209278_s_at | TFPI2 | tissue factor pathway inhibitor 2 | 4.82 × 10−12 | 6.13 |
| 210042_s_at | CTSZ | cathepsin Z | 4.41 × 10−13 | 2.63 | 209909_s_at | TGFB2 | transforming growth factor, beta 2 | 1.36 × 10−9 | 7.65 |
| 209335_at | DCN | decorin | 2.15 × 10−17 | 5.05 | 228121_at | TGFB2 | transforming growth factor, beta 2 | 2.09 × 10−7 | 5.04 |
| 211896_s_at | DCN | decorin | 2.64 × 10−17 | 4.08 | 220407_s_at | TGFB2 | transforming growth factor, beta 2 | 2.05 × 10−9 | 3.60 |
| 211813_x_at | DCN | decorin | 1.62 × 10−16 | 3.47 | 201042_at | TGM2 | transglutaminase 2 | 7.33 × 10−12 | 8.42 |
| 201893_x_at | DCN | decorin | 1.32 × 10−16 | 3.17 | 211573_x_at | TGM2 | transglutaminase 2 | 6.53 × 10−25 | 4.14 |
| 213068_at | DPT | dermatopontin | 2.61 × 10−22 | 47.71 | 211003_x_at | TGM2 | transglutaminase 2 | 6.51 × 10−23 | 3.00 |
| 207977_s_at | DPT | dermatopontin | 1.64 × 10−19 | 20.11 | 222835_at | THSD4 | thrombospondin, type I, domain containing 4 | 2.65 × 10−9 | 4.10 |
| 213071_at | DPT | dermatopontin | 2.41 × 10−18 | 14.09 | 226506_at | THSD4 | thrombospondin, type I, domain containing 4 | 3.76 × 10−7 | 3.35 |
| 209365_s_at | ECM1 | extracellular matrix protein 1 | 1.55 × 10−26 | 3.53 | 202643_s_at | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 1.91 × 10−9 | 4.48 |
| 206101_at | ECM2 | extracellular matrix protein 2, female organ and adipocyte specific | 6.39 × 10−13 | 10.63 | 202644_s_at | TNFAIP3 | tumor necrosis factor, alpha-induced protein 3 | 2.35 × 10−8 | 3.98 |
| 201843_s_at | EFEMP1 | EGF-containing fibulin-like extracellular matrix protein 1 | 4.72 × 10−3 | 2.04 | 206025_s_at | TNFAIP6 | tumor necrosis factor, alpha-induced protein 6 | 4.06 × 10−5 | 2.04 |
| 209356_x_at | EFEMP2 | EGF-containing fibulin-like extracellular matrix protein 2 | 2.39 × 10−21 | 3.14 | |||||
| 212670_at | ELN | elastin | 0.00 × 10+00 | 34.17 | |||||
| 222885_at | EMCN | endomucin | 6.61 × 10−8 | 4.14 | |||||
| 227874_at | EMCN | endomucin | 6.01 × 10−6 | 2.25 | |||||
| 204363_at | F3 | coagulation factor III (thromboplastin, tissue factor) | 2.04 × 10−6 | 5.30 | |||||
| 202995_s_at | FBLN1 | fibulin 1 | 4.84 × 10−8 | 2.48 | |||||
| 203886_s_at | FBLN2 | fibulin 2 | 7.69 × 10−21 | 18.44 | |||||
| 203088_at | FBLN5 | fibulin 5 | 7.88 × 10−28 | 10.14 | |||||
| 203638_s_at | FGFR2 | fibroblast growth factor receptor 2 | 1.16 × 10−11 | 6.98 | |||||
| 227265_at | FGL2 | fibrinogen-like 2 | 1.58 × 10−5 | 6.33 | |||||
| 204834_at | FGL2 | fibrinogen-like 2 | 2.20 × 10−7 | 5.59 | |||||
| 202709_at | FMOD | fibromodulin | 4.30 × 10−22 | 8.88 | |||||
| 226930_at | FNDC1 | fibronectin type III domain containing 1 | 2.46 × 10−19 | 47.43 | |||||
| 226145_s_at | FRAS1 | Fraser syndrome 1 | 0.00 × 10+00 | 76.26 | |||||
| 202755_s_at | GPC1 | glypican 1 | 3.75 × 10−15 | 2.10 | |||||
| 204984_at | GPC4 | glypican 4 | 5.19 × 10−15 | 9.94 | |||||
| 204983_s_at | GPC4 | glypican 4 | 8.43 × 10−12 | 5.40 | |||||
| 230204_at | HAPLN1 | hyaluronan and proteoglycan link protein 1 | 2.18 × 10−7 | 7.98 | |||||
| 205523_at | HAPLN1 | hyaluronan and proteoglycan link protein 1 | 5.24 × 10−7 | 7.86 | |||||
| 205524_s_at | HAPLN1 | hyaluronan and proteoglycan link protein 1 | 5.34 × 10−8 | 7.86 | |||||
| 230895_at | HAPLN1 | hyaluronan and proteoglycan link protein 1 | 6.93 × 10−7 | 7.40 | |||||
| 227262_at | HAPLN3 | hyaluronan and proteoglycan link protein 3 | 5.18 × 10−16 | 4.01 | |||||
| 235944_at | HMCN1 | hemicentin 1 | 5.33 × 10−8 | 4.24 | |||||
| 201185_at | HTRA1 | HtrA serine peptidase 1 | 3.62 × 10−10 | 3.08 | |||||
| 209541_at | IGF1 | insulin-like growth factor 1 (somatomedin C) | 4.41 × 10−6 | 2.51 | |||||
| 202718_at | IGFBP2 | insulin-like growth factor binding protein 2, 36 kDa | 1.20 × 10−2 | 2.37 | |||||
| 212143_s_at | IGFBP3 | insulin-like growth factor binding protein 3 | 1.20 × 10−23 | 28.32 | |||||
| 210095_s_at | IGFBP3 | insulin-like growth factor binding protein 3 | 5.39 × 10−19 | 9.31 | |||||
| 211959_at | IGFBP5 | insulin-like growth factor binding protein 5 | 2.35 × 10−11 | 5.19 | |||||
| 203424_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 7.05 × 10−7 | 4.65 | |||||
| 211958_at | IGFBP5 | insulin-like growth factor binding protein 5 | 1.68 × 10−7 | 3.69 | |||||
| 1555997_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 9.64 × 10−6 | 2.41 | |||||
| 203426_s_at | IGFBP5 | insulin-like growth factor binding protein 5 | 2.17 × 10−4 | 2.08 | |||||
| 203851_at | IGFBP6 | insulin-like growth factor binding protein 6 | 2.87 × 10−7 | 2.03 | |||||
| 227760_at | IGFBPL1 | insulin-like growth factor binding protein-like 1 | 6.32 × 10−20 | 2.75 | |||||
| 218574_s_at | LMCD1 | LIM and cysteine-rich domains 1 | 1.08 × 10−20 | 5.35 | |||||
| 242767_at | LMCD1 | LIM and cysteine-rich domains 1 | 2.61 × 10−5 | 2.23 | |||||
| 201744_s_at | LUM | lumican | 3.33 × 10−9 | 2.53 | |||||
| 212713_at | MFAP4 | microfibrillar-associated prot 4 | 5.47 × 10−25 | 11.79 | |||||
| 209758_s_at | MFAP5 | microfibrillar associated prot 5 | 8.86 × 10−9 | 9.84 | |||||
| 213765_at | MFAP5 | microfibrillar associated prot 5 | 1.07 × 10−7 | 9.67 | |||||
| 213764_s_at | MFAP5 | microfibrillar associated prot 5 | 8.21 × 10−8 | 8.97 | |||||
| 210605_s_at | MFGE8 | milk fat globule-EGF factor 8 protein | 2.85 × 10−8 | 3.58 | |||||
| 202291_s_at | MGP | matrix Gla protein | 5.61 × 10−5 | 5.61 | |||||
| 204475_at | MMP1 | matrix metallopeptidase 1 (interstitial collagenase) | 3.34 × 10−4 | 3.31 | |||||
| 204580_at | MMP12 | matrix metallopeptidase 12 (macrophage elastase) | 8.73 × 10−4 | 2.60 | |||||
| 205828_at | MMP3 | matrix metallopeptidase 3 (stromelysin 1, progelatinase) | 5.32 × 10−8 | 16.81 | |||||
| 213693_s_at | MUC1 | mucin 1, cell surface associated | 2.96 × 10−18 | 3.90 | |||||
| 207847_s_at | MUC1 | mucin 1, cell surface associated | 3.37 × 10−11 | 3.78 | |||||
| 209596_at | MXRA5 | matrix-remodelling associated 5 | 6.87 × 10−26 | 30.61 | |||||
| 235836_at | MXRA7 | matrix-remodelling associated 7 | 7.77 × 10−10 | 2.16 | |||||
| 213422_s_at | MXRA8 | matrix-remodelling associated 8 | 9.88 × 10−30 | 2.13 | |||||
| 214321_at | NOV | nephroblastoma overexpressed gene | 6.98 × 10−20 | 10.34 | |||||
| 204501_at | NOV | nephroblastoma overexpressed gene | 4.71 × 10−9 | 2.90 | |||||
| 1564494_s_at | P4HB | prolyl 4-hydroxylase, beta polypeptide | 7.83 × 10−23 | 3.48 | |||||
| 219295_s_at | PCOLCE2 | procollagen C-endopeptidase enhancer 2 | 3.15 × 10−6 | 7.20 | |||||
| 226522_at | PODN | podocan | 3.74 × 10−12 | 3.16 | |||||
| 1555778_a_at | POSTN | periostin, osteoblast specific factor | 1.43 × 10−3 | 2.70 | |||||
| 210809_s_at | POSTN | periostin, osteoblast specific factor | 3.60 × 10−4 | 2.31 | |||||
| 228224_at | PRELP | proline/arginine-rich end leucine-rich repeat protein | 1.49 × 10−13 | 6.34 | |||||
| 204223_at | PRELP | proline/arginine-rich end leucine-rich repeat protein | 3.05 × 10−10 | 4.16 | |||||
| 209496_at | RARRES2 | retinoic acid receptor responder (tazarotene induced) 2 | 5.58 × 10−9 | 4.86 | |||||
| 205923_at | RELN | reelin | 8.98 × 10−4 | 2.09 | |||||
| 228186_s_at | RSPO3 | R-spondin 3 homolog (Xenopus laevis) | 5.88 × 10−14 | 5.81 | |||||
| 202037_s_at | SFRP1 | secreted frizzled-related prot 1 | 1.60 × 10−4 | 2.53 | |||||
| 202035_s_at | SFRP1 | secreted frizzled-related prot 1 | 2.84 × 10−3 | 2.35 | |||||
| 202036_s_at | SFRP1 | secreted frizzled-related prot 1 | 3.39 × 10−3 | 2.18 | |||||
| 223122_s_at | SFRP2 | secreted frizzled-related prot 2 | 1.65 × 10−29 | 54.89 | |||||
| 223121_s_at | SFRP2 | secreted frizzled-related prot 2 | 5.66 × 10−18 | 13.22 | |||||
| 203813_s_at | SLIT3 | slit homolog 3 (Drosophila) | 1.22 × 10−21 | 10.53 | |||||
| 223869_at | SOST | sclerosteosis | 1.85 × 10−7 | 4.21 | |||||
| 213247_at | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 5.98 × 10−13 | 5.61 | |||||
| 219552_at | SVEP1 | sushi, von Willebrand factor type A, EGF and pentraxin domain containing 1 | 8.40 × 10−11 | 2.87 | |||||
| 205016_at | TGFA | transforming growth factor, alpha | 9.81 × 10−19 | 9.55 | |||||
| 203085_s_at | TGFB1 | transforming growth factor beta 1 |
9.57 × 10−14 | 2.17 | |||||
| 239336_at | THBS1 | Thrombospondin 1 | 7.00 × 10−26 | 7.23 | |||||
| 201107_s_at | THBS1 | thrombospondin 1 | 7.00 × 10−26 | 6.92 | |||||
| 201108_s_at | THBS1 | thrombospondin 1 | 5.67 × 10−28 | 4.10 | |||||
| 235086_at | THBS1 | Thrombospondin 1 | 1.09 × 10−16 | 3.35 | |||||
| 215775_at | THBS1 | Thrombospondin 1 | 5.62 × 10−18 | 2.96 | |||||
| 201109_s_at | THBS1 | thrombospondin 1 | 7.70 × 10−23 | 2.47 | |||||
| 201150_s_at | TIMP3 | TIMP metallopeptidase inhibitor 3 | 2.68 × 10−9 | 2.06 | |||||
| 201149_s_at | TIMP3 | TIMP metallopeptidase inhibitor 3 | 2.03 × 10−5 | 2.02 | |||||
| 201645_at | TNC | tenascin C | 6.48 × 10−7 | 2.67 | |||||
| 216005_at | TNC | Tenascin C | 1.97 × 10−3 | 2.27 | |||||
| 213451_x_at | TNXA / TNXB | tenascin XA pseudogene tenascin XB | 7.61 × 10−9 | 7.23 | |||||
| 206093_x_at | TNXA / TNXB | tenascin XA pseudogene tenascin XB | 3.25 × 10−8 | 6.27 | |||||
| 216333_x_at | TNXA / TNXB | tenascin XA pseudogene tenascin XB | 1.07 × 10−7 | 6.23 | |||||
| 208609_s_at | TNXB | tenascin XB | 2.48 × 10−5 | 3.26 | |||||
| 235616_at | TSHZ2 | teashirt zinc finger homeobox 2 |
8.68 × 10−8 | 2.28 | |||||
| 227899_at | VIT | vitrin | 6.33 × 10−10 | 3.99 | |||||
| 210102_at | VWA5A | von Willebrand factor A domain containing 5A | 9.24 × 10−6 | 2.64 | |||||
| Focal Adhesion Points | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 200965_s_at | ABLIM1 | actin binding LIM protein 1 | 5.19 × 10−5 | 2.74 | 206385_s_at | ANK3 | ankyrin 3, node of Ranvier (ankyrin G) | 4.82 × 10−8 | 3.98 |
| 205268_s_at | ADD2 | adducin 2 (beta) | 6.03 × 10−7 | 3.71 | 218950_at | ARAP3 | ArfGAP with RhoGAP domain, ankyrin repeat and PH domain 3 | 3.65 × 10−15 | 2.95 |
| 202022_at | ALDOC | aldolase C, fructose-bisphosphate | 6.50 × 10−21 | 6.09 | 227911_at | ARHGAP28 | Rho GTPase activating protein 28 | 2.26 × 10−11 | 3.59 |
| 202920_at | ANK2 | ankyrin 2, neuronal | 7.80 × 10−8 | 3.58 | 206167_s_at | ARHGAP6 | Rho GTPase activating protein 6 | 1.68 × 10−8 | 2.90 |
| 213606_s_at | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 2.12 × 10−15 | 2.88 | 1555812_a_at | ARHGDIB | Rho GDP dissociation inhibitor (GDI) beta | 4.29 × 10−5 | 2.17 |
| 201167_x_at | ARHGDIA | Rho GDP dissociation inhibitor (GDI) alpha | 2.10 × 10−14 | 2.36 | 218501_at | ARHGEF3 | Rho guanine nucleotide exchange factor (GEF) 3 | 1.16 × 10−15 | 3.82 |
| 222696_at | AXIN2 | axin 2 | 3.23 × 10−6 | 2.37 | 227372_s_at | BAIAP2L1 | BAI1-associated protein 2-like 1 | 6.04 × 10−14 | 2.08 |
| 227850_x_at | CDC42EP5 | CDC42 effector protein (Rho GTPase binding) 5 | 3.62 × 10−8 | 2.20 | 213373_s_at | CASP8 | caspase 8, apoptosis-related cysteine peptidase | 7.10 × 10−17 | 2.15 |
| 228739_at | CYS1 | cystin 1 | 4.46 × 10−26 | 5.02 | 234936_s_at | CC2D2A | coiled-coil and C2 domain containing 2A | 3.92 × 10−26 | 3.10 |
| 220559_at | EN1 | engrailed homeobox 1 | 2.00 × 10−19 | 15.83 | 203881_s_at | DMD | dystrophin | 8.99 × 10−5 | 2.84 |
| 206710_s_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 4.86 × 10−4 | 3.86 | 242283_at | DNAH14 | dynein, axonemal, heavy chain 14 | 4.14 × 10−18 | 2.72 |
| 212681_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 4.79 × 10−4 | 3.39 | 205186_at | DNALI1 | dynein, axonemal, light intermediate chain 1 | 3.02 × 10−12 | 2.71 |
| 211776_s_at | EPB41L3 | erythrocyte membrane protein band 4.1-like 3 | 5.45 × 10−4 | 3.26 | 227081_at | DNALI1 | dynein, axonemal, light intermediate chain 1 | 1.38 × 10−6 | 2.00 |
| 226129_at | FAM83H | family with sequence similarity 83, member H | 1.37 × 10−9 | 2.03 | 212838_at | DNMBP | dynamin binding protein | 2.88 × 10−16 | 2.05 |
| 212288_at | FNBP1 | formin binding protein 1 | 9.29 × 10−19 | 2.31 | 228674_s_at | EML4 | echinoderm microtubule associated protein like 4 | 1.66 × 10−19 | 2.39 |
| 230389_at | FNBP1 | formin binding protein 1 | 2.64 × 10−11 | 2.19 | 220386_s_at | EML4 | echinoderm microtubule associated protein like 4 | 1.34 × 10−16 | 2.32 |
| 230645_at | FRMD3 | FERM domain containing 3 | 8.23 × 10−12 | 3.10 | 223068_at | EML4 | echinoderm microtubule associated protein like 4 | 3.04 × 10−22 | 2.18 |
| 229893_at | FRMD3 | FERM domain containing 3 | 1.74 × 10−10 | 2.18 | 201340_s_at | ENC1 | ectodermal-neural cortex (with BTB-like domain) | 4.58 × 10−7 | 2.76 |
| 226364_at | HIP1 | Huntingtin interacting prot 1 | 2.94 × 10−7 | 2.09 | 1555137_a_at | FGD6 | FYVE, RhoGEF and PH domain containing 6 | 2.15 × 10−8 | 2.23 |
| 209558_s_at | HIP1R | huntingtin interacting prot 1 related | 3.28 × 10−15 | 2.09 | 219901_at | FGD6 | FYVE, RhoGEF and PH domain containing 6 | 1.02 × 10−8 | 2.22 |
| 226352_at | JMY | junction mediating and regulatory protein, p53 cofactor | 1.44 × 10−15 | 5.56 | 225167_at | FRMD4A | FERM domain containing 4A | 2.68 × 10−19 | 4.09 |
| 241985_at | JMY | junction mediating and regulatory protein, p53 cofactor | 1.04 × 10−16 | 2.12 | 208476_s_at | FRMD4A | FERM domain containing 4A | 3.98 × 10−15 | 3.51 |
| 226534_at | KITLG | KIT ligand | 1.06 × 10−5 | 2.21 | 225163_at | FRMD4A | FERM domain containing 4A | 3.05 × 10−17 | 3.31 |
| 213371_at | LDB3 | LIM domain binding 3 | 1.66 × 10−2 | 2.05 | 225168_at | FRMD4A | FERM domain containing 4A | 9.69 × 10−17 | 2.99 |
| 227219_x_at | MAP1LC3A | microtubule-associated protein 1 light chain 3 alpha | 1.93 × 10−18 | 2.79 | 1560031_at | FRMD4A | FERM domain containing 4A | 2.08 × 10−13 | 2.76 |
| 224378_x_at | MAP1LC3A | microtubule-associated protein 1 light chain 3 alpha | 2.64 × 10−14 | 2.72 | 230831_at | FRMD5 | FERM domain containing 5 | 1.24 × 10−19 | 6.37 |
| 232011_s_at | MAP1LC3A | microtubule-associated protein 1 light chain 3 alpha | 1.07 × 10−13 | 2.60 | 220773_s_at | GPHN | gephyrin | 9.51 × 10−19 | 2.13 |
| 208786_s_at | MAP1LC3B | microtubule-associated protein 1 light chain 3 beta | 5.66 × 10−32 | 2.26 | 223319_at | GPHN | gephyrin | 7.91 × 10−18 | 2.06 |
| 205442_at | MFAP3L | microfibrillar-associated protein 3-like | 6.52 × 10−14 | 6.61 | 202962_at | KIF13B | kinesin family member 13B | 1.09 × 10−19 | 2.74 |
| 204631_at | MYH2 | myosin, heavy chain 2, skeletal muscle, adult | 7.89 × 10−8 | 6.38 | 226003_at | KIF21A | kinesin family member 21A | 2.27 × 10−25 | 2.33 |
| 201058_s_at | MYL9 | myosin, light chain 9, regulatory | 1.46 × 10−7 | 2.00 | 231875_at | KIF21A | kinesin family member 21A | 1.40 × 10−16 | 2.21 |
| 228098_s_at | MYLIP | myosin regulatory light chain interacting protein | 5.37 × 10−27 | 7.57 | 225613_at | MAST4 | Microtubule associated serine/threonine kinase family member 4 | 2.87 × 10−16 | 3.50 |
| 223130_s_at | MYLIP | myosin regulatory light chain interacting protein | 1.59 × 10−28 | 7.13 | 225611_at | MAST4 | Microtubule associated serine/threonine kinase family member 4 | 6.91 × 10−15 | 2.95 |
| 220319_s_at | MYLIP | myosin regulatory light chain interacting protein | 7.00 × 10−16 | 2.93 | 40016_g_at | MAST4 | microtubule associated serine/threonine kinase family member 4 | 5.49 × 10−15 | 2.43 |
| 223129_x_at | MYLIP | myosin regulatory light chain interacting protein | 1.25 × 10−14 | 2.03 | 213511_s_at | MTMR1 | myotubularin related protein 1 | 2.10 × 10−20 | 2.23 |
| 202555_s_at | MYLK | myosin light chain kinase | 2.64 × 10−5 | 2.37 | 216095_x_at | MTMR1 | myotubularin related protein 1 | 4.13 × 10−21 | 2.09 |
| 224823_at | MYLK | myosin light chain kinase | 3.68 × 10−5 | 2.11 | 237206_at | MYOCD | myocardin | 7.88 × 10−3 | 2.36 |
| 212338_at | MYO1D | myosin ID | 3.13 × 10−14 | 8.06 | 219073_s_at | OSBPL10 | oxysterol binding protein-like 10 | 5.46 × 10−8 | 2.27 |
| 223464_at | OSBPL5 | oxysterol binding protein-like 5 | 1.87 × 10−17 | 2.26 | 219938_s_at | PSTPIP2 | proline-serine-threonine phosphatase interacting protein 2 | 9.14 × 10−13 | 2.08 |
| 209019_s_at | PINK1 | PTEN induced putative kinase 1 | 2.23 × 10−26 | 2.64 | 223471_at | RAB3IP | RAB3A interacting protein (rabin3) | 4.80 × 10−24 | 4.06 |
| 209018_s_at | PINK1 | PTEN induced putative kinase 1 | 8.46 × 10−26 | 2.62 | 219045_at | RHOF | ras homolog gene family, member F (in filopodia) | 3.42 × 10−18 | 3.29 |
| 226627_at | SEPT8 | septin 8 | 1.33 × 10−9 | 2.22 | 219263_at | RNF128 | ring finger protein 128 | 3.33 × 10−3 | 2.11 |
| 230730_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 3.81 × 10−13 | 10.01 | 204967_at | SHROOM2 | shroom family member 2 | 2.29 × 10−14 | 15.26 |
| 213543_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 4.91 × 10−13 | 8.70 | 213109_at | TNIK | TRAF2 and NCK interacting kinase | 8.89 × 10−6 | 2.75 |
| 228602_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 1.66 × 10−8 | 5.55 | 213107_at | TNIK | TRAF2 and NCK interacting kinase | 1.18 × 10−4 | 2.51 |
| 210330_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 1.25 × 10−12 | 4.80 | 216251_s_at | TTLL12 | tubulin tyrosine ligase-like family, member 12 | 1.63 × 10−17 | 2.07 |
| 210329_s_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 1.01 × 10−10 | 3.87 | 1552257_a_at | TTLL12 | tubulin tyrosine ligase-like family, member 12 | 1.20 × 10−18 | 2.00 |
| 214492_at | SGCD | sarcoglycan, delta (35 kDa dystrophin-associated glycoprotein) | 3.87 × 10−7 | 2.93 | 203702_s_at | TTLL4 | tubulin tyrosine ligase-like family, member 4 | 3.28 × 10−17 | 2.24 |
| 207302_at | SGCG | sarcoglycan, gamma (35 kDa dystrophin-associated glycoprotein) | 6.41 × 10−28 | 38.26 | |||||
| 228400_at | SHROOM3 | shroom family member 3 | 1.15 × 10−24 | 15.85 | |||||
| 225548_at | SHROOM3 | shroom family member 3 | 2.47 × 10−24 | 11.91 | |||||
| 217678_at | SLC7A11 | solute carrier family 7, (cationic amino acid transporter, y+ system) member 11 | 1.52 × 10−7 | 2.36 | |||||
| 203516_at | SNTA1 | syntrophin, alpha 1 (dystrophin-associated protein A1, 59 kDa, acidic component) | 1.56 × 10−19 | 2.89 | |||||
| 201061_s_at | STOM | stomatin | 2.06 × 10−23 | 2.38 | |||||
| 209306_s_at | SWAP70 | SWAP-70 protein | 3.61 × 10−37 | 2.22 | |||||
| 209307_at | SWAP70 | SWAP-70 protein | 3.18 × 10−25 | 2.07 | |||||
| 209904_at | TNNC1 | troponin C type 1 (slow) | 6.23 × 10−5 | 2.12 | |||||
| 238688_at | TPM1 | Tropomyosin 1 (alpha) | 1.82 × 10−9 | 2.70 | |||||
| 206117_at | TPM1 | tropomyosin 1 (alpha) | 6.69 × 10−11 | 2.33 | |||||
| 202479_s_at | TRIB2 | tribbles homolog 2 (Drosophila) | 1.37 × 10−11 | 2.20 | |||||
| 202478_at | TRIB2 | tribbles homolog 2 (Drosophila) | 3.53 × 10−10 | 2.09 | |||||
| 213908_at | WHAMML1 /2 | WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 /2 | 4.06 × 10−21 | 3.52 | |||||
| 1557261_at | WHAMML1 /2 | WAS protein homolog associated with actin, golgi membranes and microtubules-like 1 /2 |
2.06 × 10−15 | 3.26 | |||||
| Cytoskeleton | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 205132_at | ACTC1 | actin, alpha, cardiac muscle 1 | 1.23 × 10−34 | 78.69 | 202274_at | ACTG2 | actin, gamma 2, smooth muscle, enteric | 6.12 × 10−10 | 13.98 |
| 220115_s_at | CDH10 | cadherin 10, type 2 (T2-cadherin) | 1.12 × 10−7 | 3.11 | 210517_s_at | AKAP12 | A kinase (PRKA) anchor protein 12 | 6.41 × 10−6 | 3.58 |
| 205532_s_at | CDH6 | cadherin 6, type 2, K-cadherin (fetal kidney) | 1.02 × 10−2 | 2.36 | 227529_s_at | AKAP12 | A kinase (PRKA) anchor protein 12 | 1.17 × 10−2 | 2.42 |
| 232898_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 8.32 × 10−10 | 3.32 | 227530_at | AKAP12 | A kinase (PRKA) anchor protein 12 | 6.85 × 10−3 | 2.41 |
| 201279_s_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 1.60 × 10−18 | 2.30 | 206298_at | ARHGAP22 | Rho GTPase activating protein 22 | 4.33 × 10−30 | 4.16 |
| 201280_s_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 3.78 × 10−21 | 2.28 | 201005_at | CD9 | CD9 molecule | 7.48 × 10−6 | 2.96 |
| 201278_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 1.77 × 10−23 | 2.26 | 214297_at | CSPG4 | chondroitin sulfate proteoglycan 4 | 8.70 × 10−5 | 2.37 |
| 210757_x_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 4.14 × 10−18 | 2.17 | 220512_at | DLC1 | deleted in liver cancer 1 | 2.59 × 10−15 | 3.03 |
| 240873_x_at | DAB2 | disabled homolog 2, mitogen-responsive phosphoprotein (Drosophila) | 1.06 × 10−13 | 2.08 | 211478_s_at | DPP4 | dipeptidyl-peptidase 4 | 2.91 × 10−19 | 11.71 |
| 214724_at | DIXDC1 | DIX domain containing 1 | 1.31 × 10−9 | 2.20 | 203716_s_at | DPP4 | dipeptidyl-peptidase 4 | 2.07 × 10−21 | 10.11 |
| 224814_at | DPP7 | dipeptidyl-peptidase 7 | 1.13 × 10−21 | 2.61 | 203717_at | DPP4 | dipeptidyl-peptidase 4 | 9.35 × 10−19 | 6.79 |
| 205031_at | EFNB3 | ephrin-B3 | 1.05 × 10−9 | 2.43 | 217901_at | DSG2 | desmoglein 2 | 1.05 × 10−10 | 19.57 |
| 208228_s_at | FGFR2 | fibroblast growth factor receptor 2 | 9.30 × 10−10 | 3.71 | 1553105_s_at | DSG2 | desmoglein 2 | 4.29 × 10−9 | 7.96 |
| 204379_s_at | FGFR3 | fibroblast growth factor receptor 3 | 1.66 × 10−19 | 4.70 | 227955_s_at | EFNA5 | ephrin-A5 | 1.28 × 10−10 | 4.10 |
| 201539_s_at | FHL1 | four and a half LIM domains 1 | 2.10 × 10−4 | 2.76 | 214036_at | EFNA5 | ephrin-A5 | 1.68 × 10−7 | 2.33 |
| 214505_s_at | FHL1 | four and a half LIM domains 1 | 6.14 × 10−4 | 2.37 | 202669_s_at | EFNB2 | ephrin-B2 | 2.57 × 10−2 | 2.16 |
| 210299_s_at | FHL1 | four and a half LIM domains 1 | 2.65 × 10−4 | 2.29 | 201983_s_at | EGFR | epidermal growth factor receptor | 3.46 × 10−10 | 2.10 |
| 210298_x_at | FHL1 | four and a half LIM domains 1 | 6.34 × 10−4 | 2.27 | 218796_at | FERMT1 | fermitin family homolog 1 (Drosophila) | 2.45 × 10−12 | 4.17 |
| 208748_s_at | FLOT1 | flotillin 1 | 5.72 × 10−7 | 3.13 | 60474_at | FERMT1 | fermitin family homolog 1 (Drosophila) | 9.46 × 10−13 | 3.84 |
| 222899_at | ITGA11 | integrin, alpha 11 | 1.08 × 10−21 | 26.89 | 242422_at | G3BP1 | GTPase activating protein (SH3 domain) binding protein 1 | 3.82 × 10−5 | 2.28 |
| 215177_s_at | ITGA6 | integrin, alpha 6 | 9.34 × 10−11 | 6.34 | 206383_s_at | G3BP2 | GTPase activating protein (SH3 domain) binding protein 2 | 1.94 × 10−17 | 2.17 |
| 201656_at | ITGA6 | integrin, alpha 6 | 5.30 × 10−11 | 4.58 | 206074_s_at | HMGA1 | high mobility group AT-hook 1 | 8.66 × 10−22 | 2.89 |
| 214265_at | ITGA8 | integrin, alpha 8 | 7.06 × 10−7 | 4.79 | 208025_s_at | HMGA2 | high mobility group AT-hook 2 | 5.89 × 10−29 | 22.82 |
| 227297_at | ITGA9 | integrin, alpha 9 | 6.70 × 10−8 | 2.35 | 1567224_at | HMGA2 | high mobility group AT-hook 2 | 2.78 × 10−21 | 4.79 |
| 202803_s_at | ITGB2 | integrin, beta 2 (complement component 3 receptor 3 and 4 subunit) | 9.97 × 10−9 | 2.63 | 1558683_a_at | HMGA2 | high mobility group AT-hook 2 | 3.31 × 10−25 | 4.32 |
| 226189_at | ITGB8 | integrin, beta 8 | 8.02 × 10−14 | 6.70 | 1561633_at | HMGA2 | high mobility group AT-hook 2 | 4.45 × 10−22 | 3.95 |
| 205422_s_at | ITGBL1 | integrin, beta-like 1 (with EGF-like repeat domains) | 2.20 × 10−16 | 12.03 | 1558682_at | HMGA2 | high mobility group AT-hook 2 | 3.32 × 10−22 | 2.27 |
| 231993_at | ITGBL1 | Integrin, beta-like 1 (with EGF-like repeat domains) | 1.41 × 10−15 | 10.90 | 202638_s_at | ICAM1 | intercellular adhesion molecule 1 | 3.48 × 10−5 | 2.74 |
| 214927_at | ITGBL1 | integrin, beta-like 1 (with EGF-like repeat domains) | 2.34 × 10−13 | 8.91 | 202637_s_at | ICAM1 | intercellular adhesion molecule 1 | 6.68 × 10−8 | 2.33 |
| 1557080_s_at | ITGBL1 | integrin, beta-like 1 (with EGF-like repeat domains) | 1.48 × 10−12 | 8.22 | 213620_s_at | ICAM2 | intercellular adhesion molecule 2 | 3.73 × 10−13 | 2.64 |
| 1557079_at | ITGBL1 | Integrin, beta-like 1 (with EGF-like repeat domains) | 3.54 × 10−16 | 7.56 | 213446_s_at | IQGAP1 | IQ motif containing GTPase activating protein 1 | 1.13 × 10−6 | 2.08 |
| 228080_at | LAYN | layilin | 8.17 × 10−10 | 2.89 | 206766_at | ITGA10 | integrin, alpha 10 | 2.50 × 10−6 | 4.61 |
| 220765_s_at | LIMS2 | LIM and senescent cell antigen-like domains 2 | 1.13 × 10−10 | 2.99 | 227314_at | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 6.15 × 10−8 | 3.59 |
| 202674_s_at | LMO7 | LIM domain 7 | 4.31 × 10−14 | 2.85 | 205032_at | ITGA2 | integrin, alpha 2 (CD49B, alpha 2 subunit of VLA-2 receptor) | 4.90 × 10−7 | 2.82 |
| 242722_at | LMO7 | LIM domain 7 | 4.41 × 10−10 | 2.49 | 204627_s_at | ITGB3 | integrin, beta 3 (platelet glycoprotein IIIa, antigen CD61) | 9.77 × 10−9 | 2.61 |
| 213490_s_at | MAP2K2 | mitogen-activated protein kinase kinase 2 | 7.78 × 10−5 | 2.09 | 223800_s_at | LIMS3 | LIM and senescent cell antigen-like domains 3 | 4.73 × 10−19 | 4.89 |
| 213438_at | NFASC | neurofascin homolog (chicken) | 1.78 × 10−16 | 16.35 | 209615_s_at | PAK1 | p21 protein (Cdc42/Rac)-activated kinase 1 | 3.17 × 10−15 | 2.06 |
| 230242_at | NFASC | neurofascin homolog (chicken) | 2.51 × 10−12 | 3.55 | 228635_at | PCDH10 | protocadherin 10 | 2.29 × 10−11 | 16.71 |
| 243645_at | NFASC | neurofascin homolog (chicken) | 6.86 × 10−15 | 2.84 | 205534_at | PCDH7 | protocadherin 7 | 2.49 × 10−4 | 3.81 |
| 222455_s_at | PARVA | parvin, alpha | 5.02 × 10−28 | 2.01 | 228640_at | PCDH7 | protocadherin 7 | 7.79 × 10−3 | 2.84 |
| 37965_at | PARVB | parvin, beta | 8.71 × 10−9 | 2.05 | 205535_s_at | PCDH7 | protocadherin 7 | 6.91 × 10−5 | 2.57 |
| 225977_at | PCDH18 | protocadherin 18 | 2.01 × 10−24 | 7.05 | 219737_s_at | PCDH9 | protocadherin 9 | 2.28 × 10−3 | 2.60 |
| 225975_at | PCDH18 | protocadherin 18 | 5.95 × 10−23 | 5.41 | 238419_at | PHLDB2 | pleckstrin homology-like domain, family B, member 2 | 1.23 × 10−8 | 3.41 |
| 223854_at | PCDHB10 | protocadherin beta 10 | 2.58 × 10−11 | 2.37 | 214374_s_at | PPFIBP1 | PTPRF interacting protein, binding protein 1 (liprin beta 1) | 5.08 × 10−8 | 2.07 |
| 232099_at | PCDHB16 | protocadherin beta 16 | 7.07 × 10−15 | 3.65 | 203650_at | PROCR | protein C receptor, endothelial (EPCR) | 1.20 × 10−14 | 2.89 |
| 231725_at | PCDHB2 | protocadherin beta 2 | 1.82 × 10−30 | 10.23 | 216915_s_at | PTPN12 | protein tyrosine phosphatase, non-receptor type 12 | 2.10 × 10−8 | 2.00 |
| 212841_s_at | PPFIBP2 | PTPRF interacting protein, binding protein 2 (liprin beta 2) | 9.85 × 10−32 | 4.48 | 202565_s_at | SVIL | supervillin | 3.65 × 10−6 | 2.95 |
| 207011_s_at | PTK7 | PTK7 protein tyrosine kinase 7 | 1.50 × 10−6 | 2.06 | 206702_at | TEK | TEK tyrosine kinase, endothelial | 3.34 × 10−15 | 13.28 |
| 227557_at | SCARF2 | scavenger receptor class F, member 2 | 1.76 × 10−18 | 2.42 | 223314_at | TSPAN14 | tetraspanin 14 | 1.52 × 10−15 | 2.71 |
| 212154_at | SDC2 | syndecan 2 | 1.02 × 10−14 | 2.45 | 221002_s_at | TSPAN14 | tetraspanin 14 | 2.48 × 10−15 | 2.01 |
| 212157_at | SDC2 | syndecan 2 | 3.60 × 10−12 | 2.16 | 209890_at | TSPAN5 | tetraspanin 5 | 5.39 × 10−26 | 2.00 |
| 212158_at | SDC2 | syndecan 2 | 7.05 × 10−13 | 2.10 | 203868_s_at | VCAM1 | vascular cell adhesion molecule 1 | 2.90 × 10−4 | 4.10 |
| 202898_at | SDC3 | syndecan 3 | 2.33 × 10−9 | 2.98 | |||||
| 226438_at | SNTB1 | syntrophin, beta 1 (dystrophin-associated protein A1, 59 kDa, basic component 1) | 2.90 × 10−4 | 2.11 | |||||
| 218087_s_at | SORBS1 | sorbin and SH3 domain containing 1 | 2.49 × 10−4 | 3.11 | |||||
| 222513_s_at | SORBS1 | sorbin and SH3 domain containing 1 | 4.61 × 10−3 | 2.00 | |||||
| 225728_at | SORBS2 | sorbin and SH3 domain containing 2 | 9.66 × 10−4 | 2.52 | |||||
| 204288_s_at | SORBS2 | sorbin and SH3 domain containing 2 | 1.01 × 10−3 | 2.10 | |||||
| 202796_at | SYNPO | synaptopodin | 8.58 × 10−4 | 2.06 | |||||
| 225720_at | SYNPO2 | synaptopodin 2 | 8.30 × 10−16 | 15.40 | |||||
| 225895_at | SYNPO2 | synaptopodin 2 | 6.69 × 10−16 | 9.94 | |||||
| 225721_at | SYNPO2 | synaptopodin 2 | 2.22 × 10−16 | 9.02 | |||||
| 225894_at | SYNPO2 | synaptopodin 2 | 4.14 × 10−15 | 4.97 | |||||
| 40837_at | TLE2 | transducin-like enhancer of split 2 (E(sp1) homolog, Drosophila) | 1.54 × 10−15 | 3.71 | |||||
| 221747_at | TNS1 | tensin 1 | 4.25 × 10−10 | 2.00 | |||||
| 227307_at | TSPAN18 | Tetraspanin 18 | 2.81 × 10−9 | 4.58 | |||||
| 227236_at | TSPAN2 | tetraspanin 2 | 4.44 × 10−6 | 3.18 | |||||
| 209264_s_at | TSPAN4 | tetraspanin 4 | 3.72 × 10−11 | 2.46 | |||||
| LINC Complexes | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 209230_s_at | NUPR1 | nuclear protein 1 | 2.43 × 10−15 | 12.15 | 206550_s_at | NUP155 | nucleoporin 155 kDa | 1.03 × 10−15 | 2.04 |
| 219888_at | SPAG4 | sperm associated antigen 4 | 1.16 × 10−9 | 3.27 | 225470_at | NUP35 | nucleoporin 35 kDa | 1.38 × 10−21 | 2.06 |
| 232027_at | SYNE1 | Spectrin repeat containing, nuclear envelope 1 | 1.61 × 10−17 | 6.92 | |||||
| 209447_at | SYNE1 | spectrin repeat containing, nuclear envelope 1 | 5.46 × 10−12 | 2.25 | |||||
| Nucleoskeleton | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 215071_s_at | HIST1H2AC | histone cluster 1, H2ac | 8.12 × 10−9 | 2.61 | 227048_at | LAMA1 | laminin, alpha 1 | 7.19 × 10−11 | 4.99 |
| 209911_x_at | HIST1H2BD | histone cluster 1, H2bd | 1.44 × 10−7 | 2.08 | 211651_s_at | LAMB1 | laminin, beta 1 | 2.68 × 10−21 | 2.83 |
| 214290_s_at | HIST2H2AA3 HIST2H2AA4 | histone cluster 2, H2aa3 histone cluster 2, H2aa4 |
1.07 × 10−10 | 2.38 | 201505_at | LAMB1 | laminin, beta 1 | 6.03 × 10−20 | 2.43 |
| 218280_x_at | HIST2H2AA3 HIST2H2AA4 | histone cluster 2, H2aa3 histone cluster 2, H2aa4 |
8.30 × 10−11 | 2.22 | 242918_at | NASP | Nuclear autoantigenic sperm protein (histone-binding) | 1.21 × 10−4 | 2.15 |
| 202708_s_at | HIST2H2BE | histone cluster 2, H2be | 1.53 × 10−8 | 2.20 | 201970_s_at | NASP | nuclear autoantigenic sperm protein (histone-binding) | 2.90 × 10−16 | 2.04 |
| 221582_at | HIST3H2A | histone cluster 3, H2a | 1.51 × 10−19 | 2.39 | 209754_s_at | TMPO | thymopoietin | 7.79 × 10−15 | 2.99 |
| 205116_at | LAMA2 | laminin, alpha 2 | 1.03 × 10−10 | 4.64 | 209753_s_at | TMPO | thymopoietin | 3.32 × 10−8 | 2.02 |
| 216840_s_at | LAMA2 | laminin, alpha 2 | 4.12 × 10−10 | 4.36 | |||||
| 213519_s_at | LAMA2 | laminin, alpha 2 | 1.78 × 10−10 | 3.97 | |||||
| 202202_s_at | LAMA4 | laminin, alpha 4 | 7.35 × 10−6 | 5.39 | |||||
| 210089_s_at | LAMA4 | laminin, alpha 4 | 1.39 × 10−7 | 3.10 | |||||
| 216264_s_at | LAMB2 | laminin, beta 2 (laminin S) | 1.72 × 10−26 | 2.94 | |||||
| Secreted Factors | |||||||||
| Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC | Probe Set ID | Gene Symbol | Gene Title | adj-pval | FC |
| 229819_at | A1BG | alpha-1-B glycoprotein | 1.21 × 10−14 | 2.11 | 204694_at | AFP | alpha-fetoprotein | 2.77 × 10−12 | 2.11 |
| 202912_at | ADM | adrenomedullin | 9.27 × 10−10 | 2.59 | 221009_s_at | ANGPTL4 | angiopoietin-like 4 | 4.26 × 10−25 | 21.5 |
| 205141_at | ANG | angiogenin, ribonuclease, RNase A family, 5 | 7.82 × 10−13 | 2.35 | 223333_s_at | ANGPTL4 | angiopoietin-like 4 | 1.89 × 10−21 | 11.7 |
| 213001_at | ANGPTL2 | angiopoietin-like 2 | 5.71 × 10−23 | 7.43 | 205239_at | AREG | amphiregulin | 9.14 × 10−7 | 2.28 |
| 213004_at | ANGPTL2 | angiopoietin-like 2 | 9.64 × 10−26 | 4.96 | 211518_s_at | BMP4 | bone morphogenetic protein 4 | 6.50 × 10−3 | 2.09 |
| 219514_at | ANGPTL2 | angiopoietin-like 2 | 4.12 × 10−22 | 2.90 | 209301_at | CA2 | carbonic anhydrase II | 1.60 × 10−6 | 8.34 |
| 238987_at | B4GALT1 | UDP-Gal:betaGlcNAc beta 1,4- galactosyltransferase, polypeptide 1 | 1.32 × 10−8 | 2.15 | 216598_s_at | CCL2 | chemokine (C-C motif) ligand 2 | 7.24 × 10−8 | 6.16 |
| 206176_at | BMP6 | bone morphogenetic protein 6 | 9.19 × 10−5 | 2.75 | 205476_at | CCL20 | chemokine (C-C motif) ligand 20 | 2.08 × 10−6 | 2.24 |
| 220988_s_at | C1QTNF3 | C1q and tumor necrosis factor related protein 3 | 7.54 × 10−9 | 2.28 | 208075_s_at | CCL7 | chemokine (C-C motif) ligand 7 | 2.99 × 10−9 | 2.30 |
| 223499_at | C1QTNF5 MFRP | C1q and tumor necrosis factor related protein 5 /membrane frizzled-related protein | 2.40 × 10−25 | 8.01 | 215388_s_at | CFH CFHR1 |
complement factor H complement factor H-related 1 |
1.27 × 10−3 | 2.82 |
| 235221_at | CBLN3 | cerebellin 3 precursor | 5.03 × 10−12 | 2.22 | 209395_at | CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) | 9.49 × 10−5 | 5.50 |
| 209616_s_at | CES1 | carboxylesterase 1 (monocyte/macrophage serine esterase 1) | 1.15 × 10−4 | 2.88 | 209396_s_at | CHI3L1 | chitinase 3-like 1 (cartilage glycoprotein-39) | 3.39 × 10−4 | 3.69 |
| 205382_s_at | CFD | complement factor D (adipsin) | 2.42 × 10−18 | 5.81 | 235099_at | CMTM8 | CKLF-like MARVEL transmembrane domain containing 8 | 7.13 × 10−5 | 2.02 |
| 200884_at | CKB | creatine kinase, brain | 2.07 × 10−31 | 6.59 | 205832_at | CPA4 | carboxypeptidase A4 | 5.82 × 10−3 | 2.61 |
| 201117_s_at | CPE | carboxypeptidase E | 3.74 × 10−32 | 34.67 | 204470_at | CXCL1 | chemokine (C-X-C motif) ligand 1 | 2.10 × 10−8 | 9.57 |
| 201116_s_at | CPE | carboxypeptidase E | 9.98 × 10−32 | 26.80 | 209774_x_at | CXCL2 | chemokine (C-X-C motif) ligand 2 | 1.84 × 10−13 | 14.5 |
| 206100_at | CPM | carboxypeptidase M | 7.56 × 10−6 | 2.80 | 207850_at | CXCL3 | chemokine (C-X-C motif) ligand 3 | 3.30 × 10−17 | 15.1 |
| 201200_at | CREG1 | cellular repressor of E1A-stimulated genes 1 | 1.40 × 10−10 | 2.97 | 214974_x_at | CXCL5 | chemokine (C-X-C motif) ligand 5 | 5.88 × 10−11 | 12.1 |
| 201360_at | CST3 | cystatin C | 6.04 × 10−15 | 2.23 | 215101_s_at | CXCL5 | chemokine (C-X-C motif) ligand 5 | 5.26 × 10−10 | 8.60 |
| 206595_at | CST6 | cystatin E/M | 6.70 × 10−13 | 7.45 | 206336_at | CXCL6 | chemokine (C-X-C motif) ligand 6 (granulocyte chemotactic protein 2) | 2.59 × 10−4 | 5.46 |
| 209687_at | CXCL12 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 2.68 × 10−10 | 8.12 | 213092_x_at | DNAJC9 | DnaJ (Hsp40) homolog, subfamily C, member 9 | 5.56 × 10−13 | 2.08 |
| 203666_at | CXCL12 | chemokine (C-X-C motif) ligand 12 (stromal cell-derived factor 1) | 1.08 × 10−7 | 6.47 | 201430_s_at | DPYSL3 | dihydropyrimidinase-like 3 | 3.53 × 10−11 | 2.69 |
| 222484_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 2.55 × 10−14 | 8.48 | 201431_s_at | DPYSL3 | dihydropyrimidinase-like 3 | 1.07 × 10−11 | 2.61 |
| 218002_s_at | CXCL14 | chemokine (C-X-C motif) ligand 14 | 1.02 × 10−13 | 7.50 | 206254_at | EGF | epidermal growth factor (beta-urogastrone) | 1.25 × 10−7 | 2.37 |
| 212977_at | CXCR7 | chemokine (C-X-C motif) receptor 7 | 2.78 × 10−13 | 17.10 | 1559072_a_at | ELFN2 | extracellular leucine-rich repeat and fibronectin type III domain containing 2 | 2.43 × 10−8 | 2.17 |
| 232746_at | CXCR7 | Chemokine (C-X-C motif) receptor 7 | 4.11 × 10−6 | 2.28 | 205767_at | EREG | epiregulin | 1.96 × 10−10 | 5.08 |
| 222996_s_at | CXXC5 | CXXC finger 5 | 1.25 × 10−9 | 2.33 | 208378_x_at | FGF5 | fibroblast growth factor 5 | 2.27 × 10−12 | 2.17 |
| 233955_x_at | CXXC5 | CXXC finger 5 | 2.25 × 10−9 | 2.29 | 210310_s_at | FGF5 | fibroblast growth factor 5 | 5.16 × 10−11 | 2.13 |
| 224516_s_at | CXXC5 | CXXC finger 5 | 1.17 × 10−8 | 2.19 | 206614_at | GDF5 | growth differentiation factor 5 | 1.17 × 10−17 | 6.75 |
| 207169_x_at | DDR1 | discoidin domain receptor tyrosine kinase 1 | 2.84 × 10−10 | 2.06 | 38037_at | HBEGF | heparin-binding EGF-like growth factor | 1.51 × 10−9 | 3.17 |
| 204602_at | DKK1 | dickkopf homolog 1 (Xenopus laevis) | 9.91 × 10−7 | 2.54 | 203821_at | HBEGF | heparin-binding EGF-like growth factor | 2.98 × 10−9 | 3.15 |
| 202196_s_at | DKK3 | dickkopf homolog 3 (Xenopus laevis) | 8.90 × 10−10 | 4.17 | 209960_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 6.82 × 10−10 | 8.20 |
| 221127_s_at | DKK3 | dickkopf homolog 3 (Xenopus laevis) | 2.77 × 10−11 | 3.45 | 210997_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 2.11 × 10−6 | 4.34 |
| 214247_s_at | DKK3 | dickkopf homolog 3 (Xenopus laevis) | 1.33 × 10−8 | 2.71 | 210998_s_at | HGF | hepatocyte growth factor (hepapoietin A; scatter factor) | 1.82 × 10−4 | 2.28 |
| 230508_at | DKK3 | dickkopf homolog 3 (Xenopus laevis) | 8.73 × 10−5 | 2.07 | 206924_at | IL11 | interleukin 11 | 6.19 × 10−6 | 2.45 |
| 222802_at | EDN1 | endothelin 1 | 8.74 × 10−7 | 4.31 | 210118_s_at | IL1A | interleukin 1, alpha | 4.81 × 10−4 | 2.87 |
| 218995_s_at | EDN1 | endothelin 1 | 3.17 × 10−6 | 4.06 | 205067_at | IL1B | interleukin 1, beta | 1.25 × 10−5 | 5.09 |
| 227708_at | EEF1A1 | eukaryotic translation elongation factor 1 alpha 1 | 1.54 × 10−14 | 2.03 | 39402_at | IL1B | interleukin 1, beta | 4.60 × 10−5 | 4.09 |
| 201313_at | ENO2 | enolase 2 (gamma, neuronal) | 2.61 × 10−8 | 2.11 | 209821_at | IL33 | interleukin 33 | 5.24 × 10−5 | 2.71 |
| 210839_s_at | ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 9.19 × 10−10 | 12.01 | 204863_s_at | IL6ST | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 1.19 × 10−8 | 2.33 |
| 209392_at | ENPP2 | ectonucleotide pyrophosphatase/phosphodiesterase 2 | 2.29 × 10−10 | 10.40 | 211000_s_at | IL6ST | interleukin 6 signal transducer (gp130, oncostatin M receptor) | 3.36 × 10−8 | 2.17 |
| 205756_s_at | F8 | coagulation factor VIII, procoagulant component | 2.18 × 10−8 | 2.13 | 202859_x_at | IL8 | interleukin 8 | 4.66 × 10−8 | 8.46 |
| 226722_at | FAM20C | family with sequence similarity 20, member C | 3.87 × 10−7 | 2.58 | 211506_s_at | IL8 | interleukin 8 | 2.57 × 10−6 | 5.63 |
| 205110_s_at | FGF13 | fibroblast growth factor 13 | 7.54 × 10−9 | 3.02 | 204926_at | INHBA | inhibin, beta A | 1.82 × 10−14 | 4.83 |
| 204422_s_at | FGF2 | fibroblast growth factor 2 (basic) | 6.77 × 10−19 | 2.29 | 210511_s_at | INHBA | inhibin, beta A | 6.98 × 10−8 | 4.63 |
| 205782_at | FGF7 | fibroblast growth factor 7 (keratinocyte growth factor) | 1.37 × 10−11 | 10.94 | 205266_at | LIF | leukemia inhibitory factor (cholinergic differentiation factor) | 4.28 × 10−10 | 6.74 |
| 1554741_s_at | FGF7 KGFLP1 KGFLP2 |
fibroblast growth factor 7 (keratinocyte growth factor) keratinocyte growth factor-like protein 1 keratinocyte growth factor-like protein 2 |
6.50 × 10−11 | 7.49 | 205381_at | LRRC17 | leucine rich repeat containing 17 | 5.79 × 10−13 | 42.4 |
| 206404_at | FGF9 | fibroblast growth factor 9 (glia-activating factor) | 1.50 × 10−5 | 2.74 | 207703_at | NLGN4Y | neuroligin 4, Y-linked | 6.00 × 10−9 | 5.96 |
| 209093_s_at | GBA GBAP |
glucosidase, beta; acid (includes glucosylceramidase) glucosidase, beta; acid, pseudogene | 1.35 × 10−18 | 2.01 | 229838_at | NUCB2 | nucleobindin 2 | 4.07 × 10−22 | 2.21 |
| 205498_at | GHR | growth hormone receptor | 5.70 × 10−6 | 2.11 | 216867_s_at | PDGFA | platelet-derived growth factor alpha polypeptide | 4.78 × 10−10 | 3.93 |
| 220794_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 1.54 × 10−29 | 52.50 | 205463_s_at | PDGFA | platelet-derived growth factor alpha polypeptide | 3.82 × 10−10 | 3.83 |
| 240509_s_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 3.80 × 10−33 | 45.90 | 221898_at | PDPN | podoplanin | 2.45 × 10−8 | 3.21 |
| 235504_at | GREM2 | gremlin 2, cysteine knot superfamily, homolog (Xenopus laevis) | 2.59 × 10−33 | 37.93 | 204879_at | PDPN | podoplanin | 8.06 × 10−7 | 2.10 |
| 216041_x_at | GRN | granulin | 4.12 × 10−26 | 2.77 | 218454_at | PLBD1 | phospholipase B domain containing 1 | 7.38 × 10−7 | 2.70 |
| 200678_x_at | GRN | granulin | 8.63 × 10−27 | 2.60 | 213449_at | POP1 | processing of precursor 1, ribonuclease P/MRP subunit (S. cerevisiae) | 6.40 × 10−12 | 2.14 |
| 211284_s_at | GRN | granulin | 1.04 × 10−26 | 2.45 | 213421_x_at | PRSS3 | protease, serine, 3 | 9.97 × 10−4 | 2.31 |
| 206326_at | GRP | gastrin-releasing peptide | 1.71 × 10−5 | 2.37 | 207463_x_at | PRSS3 | protease, serine, 3 | 2.59 × 10−3 | 2.17 |
| 204773_at | IL11RA | interleukin 11 receptor, alpha | 9.87 × 10−12 | 2.32 | 206631_at | PTGER2 | prostaglandin E receptor 2 (subtype EP2), 53 kDa | 2.43 × 10−9 | 2.96 |
| 206295_at | IL18 | interleukin 18 (interferon-gamma-inducing factor) | 6.66 × 10−3 | 2.15 | 204897_at | PTGER4 | prostaglandin E receptor 4 (subtype EP4) | 1.62 × 10−5 | 4.35 |
| 202948_at | IL1R1 | interleukin 1 receptor, type I | 2.88 × 10−10 | 2.33 | 227146_at | QSOX2 | quiescin Q6 sulfhydryl oxidase 2 | 1.21 × 10−23 | 2.14 |
| 228575_at | IL20RB | interleukin 20 receptor beta | 3.21 × 10−33 | 20.03 | 204916_at | RAMP1 | receptor (G protein-coupled) activity modifying protein 1 | 5.83 × 10−5 | 2.06 |
| 221658_s_at | IL21R | interleukin 21 receptor | 5.81 × 10−10 | 2.67 | 219140_s_at | RBP4 | retinol binding protein 4, plasma | 1.87 × 10−4 | 2.28 |
| 226333_at | IL6R | interleukin 6 receptor | 1.33 × 10−9 | 2.61 | 206805_at | SEMA3A | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 1.29 × 10−5 | 2.54 |
| 206693_at | IL7 | interleukin 7 | 7.42 × 10−8 | 2.00 | 244163_at | SEMA3A | sema domain, immunoglobulin domain (Ig), short basic domain, secreted, (semaphorin) 3A | 4.25 × 10−7 | 2.02 |
| 226218_at | IL7R | interleukin 7 receptor | 4.57 × 10−14 | 12.12 | 230345_at | SEMA7A | semaphorin 7A, GPI membrane anchor (John Milton Hagen blood group) | 5.22 × 10−8 | 2.70 |
| 205798_at | IL7R | interleukin 7 receptor | 5.98 × 10−15 | 10.35 | 209723_at | SERPINB9 | serpin peptidase inhibitor, clade B (ovalbumin), member 9 | 1.23 × 10−7 | 4.73 |
| 205258_at | INHBB | inhibin, beta B | 4.04 × 10−7 | 6.33 | 205576_at | SERPIND1 | serpin peptidase inhibitor, clade D (heparin cofactor), member 1 | 1.39 × 10−6 | 2.04 |
| 205051_s_at | KIT | v-kit Hardy-Zuckerman 4 feline sarcoma viral oncogene homolog | 1.65 × 10−13 | 6.91 | 213600_at | SIPA1L3 | signal-induced proliferation-associated 1 like 3 | 4.01 × 10−23 | 2.39 |
| 207092_at | LEP | leptin | 4.09 × 10−5 | 2.83 | 37831_at | SIPA1L3 | signal-induced proliferation-associated 1 like 3 | 3.40 × 10−25 | 2.07 |
| 206584_at | LY96 | lymphocyte antigen 96 | 1.50 × 10−19 | 2.46 | 204466_s_at | SNCA | synuclein, alpha (non A4 component of amyloid precursor) | 8.83 × 10−6 | 3.50 |
| 232224_at | MASP1 | mannan-binding lectin serine peptidase 1 (C4/C2 activating component of Ra-reactive factor) | 4.44 × 10−7 | 5.65 | 201562_s_at | SORD | sorbitol dehydrogenase | 4.85 × 10−15 | 2.16 |
| 201621_at | NBL1 | neuroblastoma, suppression of tumorigenicity 1 | 2.11 × 10−9 | 2.66 | 242408_at | STYX | serine/threonine/tyrosine interacting protein | 3.00 × 10−8 | 2.02 |
| 37005_at | NBL1 | neuroblastoma, suppression of tumorigenicity 1 | 1.16 × 10−8 | 2.33 | 209676_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 4.68 × 10−5 | 2.43 |
| 205893_at | NLGN1 | neuroligin 1 | 7.98 × 10−3 | 2.09 | 213258_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 3.07 × 10−5 | 2.34 |
| 231361_at | NLGN1 | Neuroligin 1 | 4.26 × 10−3 | 2.02 | 210664_s_at | TFPI | tissue factor pathway inhibitor (lipoprotein-associated coagulation inhibitor) | 1.32 × 10−4 | 2.22 |
| 231798_at | NOG | noggin | 2.87 × 10−41 | 97.99 | 235737_at | TSLP | thymic stromal lymphopoietin | 5.21 × 10−14 | 6.10 |
| 206343_s_at | NRG1 | neuregulin 1 | 4.21 × 10−4 | 2.02 | 213425_at | WNT5A | wingless-type MMTV integration site family, member 5A | 1.06 × 10−13 | 4.23 |
| 218625_at | NRN1 | neuritin 1 | 3.68 × 10−15 | 29.54 | 205990_s_at | WNT5A | wingless-type MMTV integration site family, member 5A | 6.42 × 10−14 | 3.76 |
| 200649_at | NUCB1 | nucleobindin 1 | 7.18 × 10−12 | 2.05 | 238105_x_at | WNT7B | wingless-type MMTV integration site family, member 7B | 1.17 × 10−11 | 2.58 |
| 213131_at | OLFM1 | olfactomedin 1 | 5.16 × 10−35 | 18.89 | |||||
| 205591_at | OLFM1 | olfactomedin 1 | 0.00 × 10+00 | 15.89 | |||||
| 214620_x_at | PAM | peptidylglycine alpha-amidating monooxygenase | 8.58 × 10−23 | 2.25 | |||||
| 202336_s_at | PAM | peptidylglycine alpha-amidating monooxygenase | 1.74 × 10−22 | 2.25 | |||||
| 212958_x_at | PAM | peptidylglycine alpha-amidating monooxygenase | 1.18 × 10−21 | 2.09 | |||||
| 219304_s_at | PDGFD | platelet derived growth factor D | 1.83 × 10−15 | 9.33 | |||||
| 209652_s_at | PGF | placental growth factor | 1.69 × 10−6 | 2.11 | |||||
| 201578_at | PODXL | podocalyxin-like | 7.00 × 10−20 | 20.38 | |||||
| 207808_s_at | PROS1 | protein S (alpha) | 8.76 × 10−9 | 3.01 | |||||
| 200866_s_at | PSAP | prosaposin | 2.12 × 10−13 | 2.29 | |||||
| 208257_x_at | PSG1 | pregnancy specific beta-1-glycoprotein 1 | 7.51 × 10−7 | 6.45 | |||||
| 210195_s_at | PSG1 | pregnancy specific beta-1-glycoprotein 1 | 1.76 × 10−8 | 3.21 | |||||
| 210196_s_at | PSG1 | pregnancy specific beta-1-glycoprotein 1 | 3.24 × 10−5 | 2.18 | |||||
| 208134_x_at | PSG2 | pregnancy specific beta-1-glycoprotein 2 | 4.46 × 10−19 | 5.98 | |||||
| 211741_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 3.72 × 10−27 | 28.52 | |||||
| 203399_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 4.42 × 10−30 | 25.95 | |||||
| 215821_x_at | PSG3 | pregnancy specific beta-1-glycoprotein 3 | 1.34 × 10−22 | 7.87 | |||||
| 208191_x_at | PSG4 | pregnancy specific beta-1-glycoprotein 4 | 6.32 × 10−6 | 8.81 | |||||
| 204830_x_at | PSG5 | pregnancy specific beta-1-glycoprotein 5 | 9.42 × 10−29 | 200.65 | |||||
| 209738_x_at | PSG6 | pregnancy specific beta-1-glycoprotein 6 | 4.68 × 10−29 | 78.75 | |||||
| 208106_x_at | PSG6 | pregnancy specific beta-1-glycoprotein 6 | 2.35 × 10−28 | 51.80 | |||||
| 205602_x_at | PSG7 | pregnancy specific beta-1-glycoprotein 7 | 1.27 × 10−5 | 4.78 | |||||
| 209594_x_at | PSG9 | pregnancy specific beta-1-glycoprotein 9 | 1.83 × 10−29 | 89.31 | |||||
| 207733_x_at | PSG9 | pregnancy specific beta-1-glycoprotein 9 | 5.66 × 10−26 | 12.06 | |||||
| 212187_x_at | PTGDS | prostaglandin D2 synthase 21 kDa (brain) | 1.76 × 10−5 | 3.89 | |||||
| 211748_x_at | PTGDS | prostaglandin D2 synthase 21 kDa (brain) | 6.16 × 10−6 | 3.30 | |||||
| 211663_x_at | PTGDS | prostaglandin D2 synthase 21 kDa (brain) | 7.37 × 10−4 | 2.32 | |||||
| 213933_at | PTGER3 | prostaglandin E receptor 3 (subtype EP3) | 3.46 × 10−7 | 3.51 | |||||
| 1555097_a_at | PTGFR | prostaglandin F receptor (FP) | 5.53 × 10−8 | 2.27 | |||||
| 207177_at | PTGFR | prostaglandin F receptor (FP) | 1.00 × 10−7 | 2.21 | |||||
| 206187_at | PTGIR | prostaglandin I2 (prostacyclin) receptor (IP) | 1.11 × 10−13 | 2.31 | |||||
| 208131_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 2.20 × 10−16 | 42.66 | |||||
| 211892_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 2.07 × 10−8 | 3.10 | |||||
| 210702_s_at | PTGIS | prostaglandin I2 (prostacyclin) synthase | 7.89 × 10−9 | 2.99 | |||||
| 211756_at | PTHLH | parathyroid hormone-like hormone | 8.63 × 10−3 | 2.07 | |||||
| 215253_s_at | RCAN1 | regulator of calcineurin 1 | 1.52 × 10−6 | 2.47 | |||||
| 203498_at | RCAN2 | regulator of calcineurin 2 | 1.23 × 10−39 | 49.77 | |||||
| 226272_at | RCAN3 | RCAN family member 3 | 1.16 × 10−12 | 2.59 | |||||
| 213716_s_at | SECTM1 | secreted and transmembrane 1 | 1.48 × 10−4 | 2.14 | |||||
| 226492_at | SEMA6D | sema domain, transmembrane domain (TM), and cytoplasmic domain, (semaphorin) 6D | 1.30 × 10−4 | 2.71 | |||||
| 200986_at | SERPING1 | serpin peptidase inhibitor, clade G (C1 inhibitor), member 1 | 2.81 × 10−20 | 13.38 | |||||
| 204596_s_at | STC1 | stanniocalcin 1 | 1.03 × 10−4 | 2.89 | |||||
| 204595_s_at | STC1 | stanniocalcin 1 | 3.40 × 10−5 | 2.12 | |||||
| 203438_at | STC2 | stanniocalcin 2 | 1.40 × 10−6 | 2.84 | |||||
| 203439_s_at | STC2 | stanniocalcin 2 | 9.48 × 10−5 | 2.27 | |||||
| 212344_at | SULF1 | sulfatase 1 | 4.55 × 10−12 | 9.94 | |||||
| 212353_at | SULF1 | sulfatase 1 | 5.41 × 10−10 | 8.59 | |||||
| 212354_at | SULF1 | sulfatase 1 | 6.12 × 10−11 | 8.19 | |||||
| 224724_at | SULF2 | sulfatase 2 | 2.31 × 10−10 | 3.82 | |||||
| 207426_s_at | TNFSF4 | tumor necrosis factor (ligand) superfamily, member 4 | 1.47 × 10−2 | 2.59 | |||||
| 206907_at | TNFSF9 | tumor necrosis factor (ligand) superfamily, member 9 | 2.10 × 10−16 | 4.73 | |||||
| 219478_at | WFDC1 | WAP four-disulfide core domain 1 | 2.78 × 10−15 | 35.49 | |||||
| 205792_at | WISP2 | WNT1 inducible signaling pathway protein 2 | 3.53 × 10−4 | 2.90 | |||||
3.6. Differentiation Capacity is Reduced in F-DHJ from Aged Skin
Finally, the capacity for differentiation into the adipocyte, osteoblast, and chondrocyte lineages was compared in F-DHJ samples from “young” (between 20 and 31 years, n = five donors) and “older” (between 55 and 65 years, n = four donors) ages (Figure 8). The three-lineage mesenchymal differentiation potential of F-DHJ described in Figure 3 appeared altered in cell samples from older skin biopsies (Figure 8A–D). Although chondrocyte differentiation was maintained (Figure 8A–D), the capacity for differentiation into osteoblasts was reduced (Figure 8B,D) and differentiation into the adipocyte lineage was almost lost (Figure 8C,D). A comparative analysis of the differentiation potential of Fp, Fr, and F-DHJ from old donors indicated functional differences (data not shown). The capacity for differentiation into adipocytes persisted with a low efficiency in old Fp and Fr, although it was lost with age in F-DHJ. In contrast, differentiation into osteoblasts was not obtained with old Fp or Fr, whereas this capacity was present in old F-DHJ. Finally, we observed that the capacity for differentiation into chondrocytes was increased with age in the three cell types but remained more efficient in F-DHJ, as compared with Fp and Fr cells. In addition to these age-related changes in the F-DHJ differentiation potential, the extracellular deposition of ColXIα1 and ACAN were respectively 3.6-fold and 2.5-fold higher in skin biopsies from the older than in the young donor group (p < 0.05) (Figure 8E,F). These observations pinpoint the interest of considering F-DHJ cells in future studies on skin ageing.
Figure 8.
Different characteristics of DHJ components in skin from “young” and “old” donors. The “young” group comprised 5 donors (20, 22, 25, 28, and 31 years old) (same donors as in Figure 3), and the “old” group comprised 4 donors (55, 61, 65, and 65 years old). (A–C) Capacity of “young” and “old” F-DHJ cells to differentiate in vitro into three mesenchymal lineages: (A) chondrocytes (toluidine blue and safranin O staining, aggrecan (ACAN) and collagen XIα1 (ColXIα1) expression); (B) adipocytes (presence of cytoplasmic lipid droplets, black arrows); and (C) osteoblasts (alizarin-red staining). For panels (A–C), representative photographs are shown. (D) Summary of the differentiation capacity into chondrocytes, adipocytes, and osteoblasts of F-DHJ from “young” and “old” skin. Scoring of differentiation capabilities are presented: (−) = not present, (+) = low representation, (++) = frequent representation, and (+++) = major representation. (E,F) Immunofluorescence detection of ColXIα1 (E) and ACAN (F) in skin biopsies from “young” and “old” donors. Representative photographs are shown, in association with quantification values corresponding to a total of 12 (E) and 15 (F) regions of interest (ROI) for the 3 analyzed “young” donors (20, 22, and 28 years old) and a total of 14 (E) and 15 (F) ROI for the 3 analyzed “old” donors (57, 61, and 65 years old). Means ± SEM are indicated (* p < 0.05, Wilcoxon test).
4. Discussion
The present work investigates the properties of a fibroblast compartment localized within the conjunctival junctions that connects the dermis to the hypodermis, i.e., dermo-hypodermal junction fibroblasts (F-DHJ), which were compared to intermediate reticular dermis (Fr) and superficial papillary dermis (Fp) fibroblasts. Cellular functional assays, combined with transcriptome profiling, indicated that F-DHJ had distinct characteristics from those of Fp and Fr cells. F-DHJ had the lowest proliferation and clonogenic capacity of the three fibroblast populations in bidimensional culture conditions. Moreover, when integrated within the dermal component of an in vitro three-dimensional reconstructed skin model, F-DHJ showed a low capacity for collagen lattice contractions and had a poor capacity for promoting epidermis organogenesis by keratinocytes. Inefficient dialog with keratinocytes observed here in vitro is in agreement with F-DHJ natural deep localizations, which are not in proximity with the epidermis, unlike the superficial Fp population. The lattice contraction assay provided the opportunity to assess the contractile capacity of specific cell types in a three-dimensional matrix environment. The contraction of the lattice is proportional to the force exerted by the cells in the matrix. Parameters that impact lattice contractions include characteristics of cell matrix anchoring structures, cytoskeleton organization, and the capacity of cells to coordinate and exert unidirectional forces. These parameters are governed by components of the “tissue skeleton” network [8,9] and may participate in vivo to confer specific biophysical characteristics to the different dermal tissue compartments. Extrapolation of the in vitro observations to the specific in vivo functions of Fp, Fr, and F-DHJ will require further studies, considering the high matrix complexity of the dermis.
We observed that F-DHJ exhibited an efficient capacity for three-lineage mesenchymal differentiation (i.e., adipocyte, osteoblast, and chondrocyte lineages), which could be interpreted as an MSC-like cellular identity, considering their anatomical proximity with the hypodermis, a tissue that contains adipose MSCs. Interestingly, the hierarchical clustering built on the basis of the transcriptome profiles of the three skin fibroblast populations (Fp, Fr, and F-DHJ) and five MSC origins (bone marrow, adipose, amnion, chorion, and cord) indicated a clear “fibroblast” molecular identity of F-DHJ, which did not segregate together with the MSC group.
The molecular signature that identified F-DHJ cells comprised transcripts involved in the stabilization of monomeric proteoglycan aggregates associated with hyaluronic acid molecules, such as HAPLN1 and HAPLN3 [27], which were found overexpressed in F‑DHJ in comparison with all MSC types. Transcripts overexpressed in F-DHJ also included ACAN, which is involved in conferring tissue biomechanical resistance [28]. In addition, the overexpressed F-DHJ signature also comprised transcripts related to the collagen meshwork, such as FMOD and TNX, which are involved in collagen processing [29]; transcripts related to collagen fibril anchorage points, such as POSTN and FNDC1 [26]; and transcripts related to the elastic network, such as ELN; DCN; MFAP4 and 5; FBN2; and FBLN1, 2, and 5 [30]. On the contrary, the comparison of F-DHJ and Fr molecular profiles identified a signature of transcripts underexpressed in the F-DHJ population, which could be interpreted in accordance with the reduced ECM mesh structuration within the DHJ area, in comparison with the reticular dermis. Notably, this character was documented by lower levels of the TNC transcript in F-DHJ than in Fr cells, which is associated with a lower accumulation of the TNC protein and loss of the TNC network in the DHJ area. Thus, the molecular specificities that distinguish F-DHJ and Fr cells may contribute to the different ECM characteristics of the reticular dermis and DHJ areas.
The existence of a fibroblast population exhibiting adipocyte-like molecular characteristics within the deep reticular dermis has been reported both in mouse [31,32] and human skin [33,34]. In human skin, the capacity for adipocyte differentiation was reported to be low for FAP+/CD90- papillary fibroblasts, intermediate for FAP+/CD90+ fibroblasts from the superior reticular dermis, and high for FAP-/CD90+ deep reticular dermis fibroblasts [33]. This gradation is consistent with the data shown in the present study, showing a correlation between the capacity for adipocyte differentiation and the depth of fibroblast dermal localization. The study by Korosec et al., which used cells from skin donors of ages ranging between 26 and 61 years, did not report an age-related reduction of the adipocyte differentiation capacity [33], as documented here for F-DHJ cells, although this phenomenon has been previously described for dermal fibroblast cells [35]. In the present study, “F-DHJ” is used to name the fibroblast population that we isolated according to its junctional localization between the deep reticular dermis and the hypodermis. This terminology distinguishes the deepest dermal part from the reticular dermis compartment, which is in agreement with their particular molecular and functional characteristics that may be critical for modeling their local ECM environment.
Interestingly, data were obtained pointing to age-related changes in the DHJ region characteristics, such as augmented levels of the ECM proteins ColXIα1 and ACAN and a reduced adipocyte differentiation potential of F-DHJ in old skin. Data from the literature concerning the evolution of the dermal fibroblast capacity for differentiation into adipocytes can appear contradictory, with regard to our observation of a decreased adipogenic potential. Indeed, a study performed on mice has, on the contrary, reported the acquisition of proadipogenic traits in dermal fibroblasts from aged animals [32], in which the difference may result from physiological species-related specificities. In a recent study, a single-cell RNA-sequencing analysis of 15,000 dermal fibroblasts isolated from human skin samples from young and old donors did not detect an up-modulation of adipogenic genes associated with ageing [36]. Of note, in humans, subcutaneous fat tissue masses tend to reduce with ageing, in particular in the face (for review, see [37]).
As we performed here using collagen lattices as a dermal matrix model, human fibroblasts isolated from the deep dermis were used to populate acellular dead desepidermized dermis (DED) pieces and analyzed for their capacity to support epidermis reconstruction by keratinocytes [34]. The two studies converged to show that fibroblasts from the deep dermis do not promote the formation of a correctly differentiated multilayered epithelium, which is consistent with their distant skin localization. Interestingly, deep dermis fibroblasts spontaneously populated the deepest part of the DED [34], in which homing may be due to the recognition of specific ECM characteristics.
Fibroblast-ECM interrelations are crucial for the maintenance of dermal integrity. In a mouse model, dermal fibroblasts were studied by intravital time-lapse, which revealed active membrane dynamics characterized by protrusions that rapidly grow and shrink from a more stable cell body [38]. By this process, fibroblasts may dialog with their cellular and ECM neighbors, and thus, adapt their behaviors and fate. Accordingly, the development of membrane extensions in living cells has been proposed to compensate for the appearance of cell-free volumes due to fibroblast deaths in the dermis of aged skin [38]. These observations may be explored at a molecular level considering genes related to the network termed as “tissue skeleton” that connects the cells with their tissue environment (comprising the nucleoskeleton, the cytoskeleton, linker complexes, ECM components, and focal adhesion points), in which their expressions differ in fibroblasts according to their dermal localization and evolve with ageing (present study and [8,9]. Disruption of this multiparametric network of interactions may result in changes that affect aged dermis, including the loss of contact surfaces between fibroblasts and their surrounding ECM [39] and modification of the deposition of ECM components, such as ColXIα1 and ACAN, as shown here.
5. Patent
V.H. and D.A. are the inventors on the filed patent application numbered 1759023 (28th September 2017) entitled “Molecular signatures of aging of 3 subpopulations of dermal fibroblasts (papillary, reticular, dermo-hypodermic junction) and dermal equivalents comprising aged fibroblasts”.
V.H is the inventor on the filed patent application numbered 1855987 (June 29th 2018) entitled “Modèle de peau comprenant des fibroblastes de la jonction dermo-hypodermique pour l’identification d’actif pro-différenciant vers des lignages adipocytaire, chondroblastique et ostéoblastique”.
Author Contributions
V.H.: conceptualization, investigation, methodology, resources, formal analysis, validation, visualization, and writing—original draft preparation; V.N. and P.P.: investigation and methodology; É.B.: investigation, methodology, and resources; J.-J.L.: conceptualization, methodology, resources, validation, and writing—review and editing; D.A.: conceptualization, validation, and writing—review and editing; N.O.F.: conceptualization, methodology, formal analysis, visualization, validation, and writing—review and editing. All authors have read and agreed to the published version of the manuscript.
Funding
This experimental work was financed by L’Oréal.
Conflicts of Interest
Authors declare no conflicts of interest. V.H., V.N., P.P. and D.A. are L’Oréal employees. N.O.F. is a CEA employee and acts as the L’Oréal scientific consultant, free of charge. J.-J.L. and E.B. are external scientific collaborators from DGA.
References
- 1.Smith L.T., Holbrook K.A. Development of dermal connective tissue in human embryonic and fetal skin. Scan Electron Microsc. 1982;4:1745–1751. [PubMed] [Google Scholar]
- 2.Smith L.T., Holbrook K.A., Byers P.H. Structure of the dermal matrix during development and in the adult. J. Investig. Dermatol. 1982;79(Suppl. 1):93s–104s. doi: 10.1111/1523-1747.ep12545877. [DOI] [PubMed] [Google Scholar]
- 3.Haydont V., Bernard B.A., Fortunel N.O. Age-related evolutions of the dermis: Clinical signs, fibroblast and extracellular matrix dynamics. Mech. Ageing Dev. 2019;177:150–156. doi: 10.1016/j.mad.2018.03.006. [DOI] [PubMed] [Google Scholar]
- 4.Harper R.A., Grove G. Human skin fibroblasts derived from papillary and reticular dermis: Differences in growth potential in vitro. Science. 1979;204:526–527. doi: 10.1126/science.432659. [DOI] [PubMed] [Google Scholar]
- 5.Mine S., Fortunel N.O., Pageon H., Asselineau D. Aging alters functionally human dermal papillary fibroblasts but not reticular fibroblasts: A new view of skin morphogenesis and aging. PLoS ONE. 2008;3:e4066. doi: 10.1371/journal.pone.0004066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Janson D.G., Saintigny G., van Adrichem A., Mahé C., El Ghalbzouri A. Different gene expression patterns in human papillary and reticular fibroblasts. J. Investig. Dermatol. 2012;132:2565–2572. doi: 10.1038/jid.2012.192. [DOI] [PubMed] [Google Scholar]
- 7.Nauroy P., Barruche V., Marchand L., Nindorera-Badara S., Bordes S., Closs B., Ruggiero F. Human Dermal Fibroblast Subpopulations Display Distinct Gene Signatures Related to Cell Behaviors and Matrisome. J. Investig. Dermatol. 2017;137:1787–1789. doi: 10.1016/j.jid.2017.03.028. [DOI] [PubMed] [Google Scholar]
- 8.Haydont V., Neiveyans V., Zucchi H., Fortunel N.O., Asselineau D. Genome-wide profiling of adult human papillary and reticular fibroblasts identifies ACAN, Col XI α1, and PSG1 as general biomarkers of dermis ageing, and KANK4 as an exemplary effector of papillary fibroblast ageing, related to contractility. Mech. Ageing Dev. 2019;177:157–181. doi: 10.1016/j.mad.2018.06.003. [DOI] [PubMed] [Google Scholar]
- 9.Haydont V., Neiveyans V., Fortunel N.O., Asselineau D. Transcriptome profiling of human papillary and reticular fibroblasts from adult interfollicular dermis pinpoints the ‘tissue skeleton’ gene network as a component of skin chrono-ageing. Mech. Ageing Dev. 2019;179:60–77. doi: 10.1016/j.mad.2019.01.003. [DOI] [PubMed] [Google Scholar]
- 10.Breathnach A.S. Development and differentiation of dermal cells in man. J. Investig. Dermatol. 1978;71:2–8. doi: 10.1111/1523-1747.ep12543601. [DOI] [PubMed] [Google Scholar]
- 11.Mills S.J., Cowin A.J., Kaur P. Pericytes, mesenchymal stem cells and the wound healing process. Cells. 2013;2:621–634. doi: 10.3390/cells2030621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Zhuang L., Lawlor K.T., Schlueter H., Pieterse Z., Yu Y., Kaur P. Pericytes promote skin regeneration by inducing epidermal cell polarity and planar cell divisions. Life Sci. Alliance. 2018;1:e201700009. doi: 10.26508/lsa.201700009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Rusu M.C., Mirancea N., Mănoiu V.S., Vâlcu M., Nicolescu M.I., Păduraru D. Skin telocytes. Ann. Anat. 2012;194:359–367. doi: 10.1016/j.aanat.2011.11.007. [DOI] [PubMed] [Google Scholar]
- 14.Ceafalan L., Gherghiceanu M., Popescu L.M., Simionescu O. Telocytes in human skin--are they involved in skin regeneration? J. Cell Mol. Med. 2012;16:1405–1420. doi: 10.1111/j.1582-4934.2012.01580.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Manole C.G., Gherghiceanu M., Simionescu O. Telocyte dynamics in psoriasis. J. Cell Mol. Med. 2015;19:1504–1519. doi: 10.1111/jcmm.12601. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Shoshkes-Carmel M., Wang Y.J., Wangensteen K.J., Tóth B., Kondo A., Massasa E.E., Itzkovitz S., Kaestner K.H. Subepithelial telocytes are an important source of Wnts that supports intestinal crypts. Nature. 2018;557:242–246. doi: 10.1038/s41586-018-0084-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Gherghiceanu M., Popescu L.M. Cardiac telocytes—Their junctions and functional implications. Cell Tissue Res. 2012;348:265–279. doi: 10.1007/s00441-012-1333-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Fertig E.T., Gherghiceanu M., Popescu L.M. Extracellular vesicles release by cardiac telocytes: Electron microscopy and electron tomography. J. Cell Mol. Med. 2014;18:1938–1943. doi: 10.1111/jcmm.12436. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Manole C.G., Simionescu O. The Cutaneous Telocytes. Adv. Exp. Med. Biol. 2016;913:303–323. doi: 10.1007/978-981-10-1061-3_20. [DOI] [PubMed] [Google Scholar]
- 20.Peltzer J., Montespan F., Thepenier C., Boutin L., Uzan G., Rouas-Freiss N., Lataillade J.J. Heterogeneous functions of perinatal mesenchymal stromal cells require a preselection before their banking for clinical use. Stem Cells Dev. 2015;24:329–344. doi: 10.1089/scd.2014.0327. [DOI] [PubMed] [Google Scholar]
- 21.Asselineau D., Bernhard B., Bailly C., Darmon M. Epidermal morphogenesis and induction of the 67 kD keratin polypeptide by culture of human keratinocytes at the liquid-air interface. Exp. Cell Res. 1985;159:536–539. doi: 10.1016/S0014-4827(85)80027-6. [DOI] [PubMed] [Google Scholar]
- 22.Rheinwald J.G., Green H. Serial cultivation of strains of human epidermal keratinocytes: The formation of keratinizing colonies from single cells. Cell. 1975;6:331–343. doi: 10.1016/S0092-8674(75)80001-8. [DOI] [PubMed] [Google Scholar]
- 23.Beacham D.A., Amatangelo M.D., Cukierman E. Preparation of extracellular matrices produced by cultured and primary fibroblasts. Curr. Protoc. Cell Biol. 2007;33 doi: 10.1002/0471143030.cb1009s33. [DOI] [PubMed] [Google Scholar]
- 24.Gabbiani G. Modulation of fibroblastic cytoskeletal features during wound healing and fibrosis. Pathol. Res. Pract. 1994;190:851–853. doi: 10.1016/S0344-0338(11)80988-X. [DOI] [PubMed] [Google Scholar]
- 25.Pei H., Yao Y., Yang Y., Liao K., Wu J.R. Krüppel-like factor KLF9 regulates PPARγ transactivation at the middle stage of adipogenesis. Cell Death Differ. 2011;18:315–327. doi: 10.1038/cdd.2010.100. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Kudo A. Periostin in fibrillogenesis for tissue regeneration: Periostin actions inside and outside the cell. Cell Mol. Life Sci. 2011;68:3201–3207. doi: 10.1007/s00018-011-0784-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Spicer A.P., Joo A., Bowling R.A., Jr. A hyaluronan binding link protein gene family whose members are physically linked adjacent to chondroitin sulfate proteoglycan core protein genes: The missing links. J. Biol. Chem. 2003;278:21083–21091. doi: 10.1074/jbc.M213100200. [DOI] [PubMed] [Google Scholar]
- 28.Kiani C., Chen L., Wu Y.J., Yee A.J., Yang B.B. Structure and function of aggrecan. Cell Res. 2002;12:19–32. doi: 10.1038/sj.cr.7290106. [DOI] [PubMed] [Google Scholar]
- 29.Kadler K.E., Hill A., Canty-Laird E.G. Collagen fibrillogenesis: Fibronectin, integrins, and minor collagens as organizers and nucleators. Curr. Opin. Cell Biol. 2008;20:495–501. doi: 10.1016/j.ceb.2008.06.008. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Kielty C.M., Sherratt M.J., Shuttleworth C.A. Elastic fibres. J. Cell Sci. 2002;115:2817–2828. doi: 10.1242/jcs.115.14.2817. [DOI] [PubMed] [Google Scholar]
- 31.Driskell R.R., Lichtenberger B.M., Hoste E., Kretzschmar K., Simons B.D., Charalambous M., Ferron S.R., Herault Y., Pavlovic G., Ferguson-Smith A.C., et al. Distinct fibroblast lineages determine dermal architecture in skin development and repair. Nature. 2013;504:277–281. doi: 10.1038/nature12783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Salzer M.C., Lafzi A., Berenguer-Llergo A., Youssif C., Castellanos A., Solanas G., Peixoto F.O., Stephan-Otto Attolini C., Prats N., Aguilera M., et al. Identity Noise and Adipogenic Traits Characterize Dermal Fibroblast Aging. Cell. 2018;175:1575–1590. doi: 10.1016/j.cell.2018.10.012. [DOI] [PubMed] [Google Scholar]
- 33.Korosec A., Frech S., Gesslbauer B., Vierhapper M., Radtke C., Petzelbauer P., Lichtenberger B.M. Lineage Identity and Location within the Dermis Determine the Function of Papillary and Reticular Fibroblasts in Human Skin. J. Investig. Dermatol. 2019;139:342–351. doi: 10.1016/j.jid.2018.07.033. [DOI] [PubMed] [Google Scholar]
- 34.Philippeos C., Telerman S.B., Oulès B., Pisco A.O., Shaw T.J., Elgueta R., Lombardi G., Driskell R.R., Soldin M., Lynch M.D., et al. Spatial and Single-Cell Transcriptional Profiling Identifies Functionally Distinct Human Dermal Fibroblast Subpopulations. J. Investig. Dermatol. 2018;138:811–825. doi: 10.1016/j.jid.2018.01.016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Brun C., Ly Ka So S., Maginiot F., Bensussan A., Michel L., Larghero J., Wong H., Oddos T., Cras A. Intrinsically aged dermal fibroblasts fail to differentiate into adipogenic lineage. Exp. Dermatol. 2016;25:906–909. doi: 10.1111/exd.13080. [DOI] [PubMed] [Google Scholar]
- 36.Solé-Boldo L., Raddatz G., Schütz S., Mallm J.P., Rippe K., Lonsdorf A.S., Rodríguez-Paredes M., Lyko F. Single-cell transcriptomes of the aging human skin reveal loss of fibroblast priming. BioRxiv. 2019:633131. doi: 10.1101/633131. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wollina U., Wetzker R., Abdel-Naser M.B., Kruglikov I.L. Role of adipose tissue in facial aging. Clin. Interv. Aging. 2017;12:2069–2076. doi: 10.2147/CIA.S151599. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Marsh E., Gonzalez D.G., Lathrop E.A., Boucher J., Greco V. Positional Stability and Membrane Occupancy Define Skin Fibroblast Homeostasis In Vivo. Cell. 2018;175:1620–1633.e13. doi: 10.1016/j.cell.2018.10.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Varani J., Dame M.K., Rittie L., Fligiel S.E., Kang S., Fisher G.J., Voorhees J.J. Decreased collagen production in chronologically aged skin: Roles of age-dependent alteration in fibroblast function and defective mechanical stimulation. Am. J. Pathol. 2006;168:1861–1888. doi: 10.2353/ajpath.2006.051302. [DOI] [PMC free article] [PubMed] [Google Scholar]