Abstract
Secreted frizzled-related protein 1 (SFRP1) is a gene that belongs to the secreted glycoprotein SFRP family. SFRP1 has been classified as a tumor suppressor gene due to the loss of expression in various human cancers, which is mainly attributed by epigenetic inactivation via DNA methylation or transcriptional silencing by microRNAs. Epigenetic silencing of SFRP1 may cause dysregulation of cell proliferation, migration, and invasion, which lead to cancer cells formation, disease progression, poor prognosis, and treatment resistance. Hence, restoration of SFRP1 expression via demethylating drugs or over-expression experiments opens the possibility for new cancer therapy approach. While the role of SFRP1 as a tumor suppressor gene is well-established, some studies also reported the possible oncogenic properties of SFRP1 in cancers. In this review, we discussed in great detail the dual roles of SFRP1 in cancers—as tumor suppressor and tumor promoter. The epigenetic regulation of SFRP1 expression will also be underscored with additional emphasis on the potentials of SFRP1 in modulating responses toward chemotherapeutic and epigenetic-modifying drugs, which may encourage the development of novel drugs for cancer treatment. We also present findings from clinical trials and patents involving SFRP1 to illustrate its clinical utility, extensiveness of each research area, and progression toward commercialization. Lastly, this review provides directions for future research to advance SFRP1 as a promising cancer biomarker.
Keywords: SFRP1, DNA Methylation, cancer biomarkers, Wnt signaling pathway, gene expression, microRNA
1. Introduction
Secreted frizzled-related protein 1 (SFRP1) is a gene that belongs to the secreted glycoprotein SFRP family. This family is also composed of another four secreted glycoproteins, namely, SFRP2, SFRP3, SFRP4, and SFRP5, which have been identified in humans [1]. Among the five members of the SFRP family, SFRP1 has been extensively studied in human cancers. This gene is located within the 8p11.21 chromosome region [2] and encodes a secreted protein with 314 amino acids (35.4 kDa) [3]. The SFRP1 protein harbors two independent structural domains, namely the carboxy-terminal netrin (NTR) domain and an amino-terminal cysteine-rich domain (CRD). CRD domain is homologous to the putative Wnt-binding site of frizzled (Fz) receptors because it contains ten cysteines with a pattern of five disulfide bridges that is similar to the CRD of Fz [4]. Therefore, SFRP1 can act as a modulator of the Wnt signaling pathway.
SFRP1 has been classified as a tumor suppressor gene due to the loss of its expression in many human cancers. This may cause dysregulation of cell proliferation, migration, and invasion, which eventually lead to cancer cells’ formation. The loss of SFRP1 expression is associated with the early development of colorectal cancer (CRC) as well as prostate cancer, and is linked with disease recurrence in renal cell cancer [5].
Various mechanisms have been implicated in the loss of SFRP1 including epigenetic and genetic regulation. Endogenous SFRP1 expression increases in a dose-dependent manner after demethylating treatment, signifying DNA methylation as the main mechanism that is responsible for the silencing of SFRP1 [6]. Therefore, targeting DNA methyltransferase activity represents a promising strategy to reduce or reverse the methylation in the SFRP1 promoters. Previously, HDAC inhibitor (HDACi); romidepsin and DNA methyltransferase inhibitor (DNMTi); and 2′-deoxy-5-azacytidine (Decitabine) were used to restore SFRP1 expression in cancer cells (please refer to Section 4 below). The restoration of SFRP1 sensitized the cisplatin-resistant laryngeal carcinoma cells [7]. Moreover, reversing SFRP1 methylation using Decitabine suppressed cell proliferation, invasion, and migration of nasopharyngeal cancer [8]. Taken together, these strategies highlight the potential of using epigenetic drugs for cancer treatment. While the role of SFRP1 as a tumor suppressor gene is well-established, some studies also reported the possible oncogenic properties of SFRP1 in cancers. SFRP1 is highly expressed in the basal-like subtype [9] as well as in the triple-negative breast cancer (TNBC) [10]. Similarly, SFRP1 was also found to be over-expressed in metastatic renal cell carcinomas but not in primary tumors [11], and this was further verified in gastric cancer cells [12,13].
A comprehensive and general review on the SFRP family was published more than five years ago [14]. While SFRP2 and SFRP4 were more recently reviewed [15,16], an updated review that recapitulates the association between SFRP1 and chemoresistance is limited. A pan-cancer analysis suggested that SFRPs are strongly correlated with patient survival, but there is an inconsistency between family members and cancer types [17]. A systematic review and meta-analysis of the SFRP family also revealed that SFRP1 hypermethylation was significantly associated with cancer risk [18]. Therefore, in this review, epigenetic regulation of SFRP1 expression will be highlighted with additional emphasis on the potentials of SFRP1 in modulating responses toward chemotherapeutic and epigenetic-modifying drugs. We also provide the latest evidence of the divergent roles of SFRP1 in tumorigenesis that may encourage the development of novel drugs for cancer treatment by targeting SFRP1.
2. Epigenetic Inactivation and Genomic Alterations in SFRP1 Gene
The expression of SFRP1 mRNA is detectable in all tested human tissues and broad expression was observed in the endometrium [19], ovary [20], colon [21], prostate [22], and breast [17]. The Cancer Genome Atlas (TCGA) revealed reduced expression of SFRP1 various cancers including breast, colorectal, lung, bladder urothelial carcinoma, cervical squamous cell carcinoma, head and neck squamous cell carcinoma, glioblastoma multiforme, kidney renal clear cell carcinoma, stomach adenocarcinoma, and endometrium cancer compared to the normal tissues [23]. The downregulation of SFRP1 expression in cancer can be regulated through different mechanisms such as non-coding RNA (ncRNA), DNA methylation, allelic imbalance, or genomic alterations.
2.1. MicroRNA-27a (miR-27a)
In general, ncRNA functions to regulate gene expression at the transcriptional and post-transcriptional level. Dysregulation of ncRNAs plays a significant role in tumor initiation and progression. MicroRNAs (miRNAs) are the widely studied type of ncRNA and are closely associated with cancer. Numerous miRNAs have been demonstrated to accelerate tumorigenesis by targeting SFRP1.
SFRP1 is a prior target of miR-27a in several tumors [24,25]. miR-27a is upregulated in the gastric cancer cell line MGC803 [24], breast cancer cell lines BT-20, MCF7, T-47D, and MDA-MB-231 [26], as well as in osteosarcoma cell lines HOS, SaOS2, 143B, and MG63 [27]. This miRNA negatively regulated SFRP1 by hindering its expression. In contrast, knockdown of miR-27a in gastric cancer cell line resulted in increased SFRP1 mRNA and at the same time decreased the expression of Wnt and β-catenin. This indicates that miR-27a targets SFRP1 to activate the Wnt/β-catenin signaling pathway. The activation of this signaling pathway remarkably increased the proliferation, migration, and invasion of gastric cancer cells [24].
Moreover, a higher level of miR-27a and low level of SFRP1 were found in CRC tissues as compared to normal tissues. The authors verified the negative correlation of miR-27a with SFRP1 in colon cancer by transfecting HCT-116 cells with miR-27a mimics [25]. The low expression of SFRP1 resulted from the degradation of the mRNA through the binding of miR-27a to the SFRP1 3′-UTR and not likely from the inhibition of protein translation [25]. Mu et al. also proved that miR-27a could promote the proliferation and invasion of human osteosarcoma cells through SFRP1-dependent Wnt/β-catenin signaling pathway [28]. This study also demonstrated that knockdown of miR-27a induced the upregulation of SFRP1 and suppressed the cell proliferation and invasion in osteosarcoma cell lines [28].
Only miR-27a was studied extensively in multiple cancers to date. There are other miRNAs targeting SFRP1 being investigated to a lesser extent as displayed in Table 1.
Table 1.
Other microRNAs associated with secreted frizzled-related protein 1 (SFRP1) transcriptional silencing.
| MicroRNA | Association with SFRP1 Downregulation | Cancer | References |
|---|---|---|---|
| miR-1207 | Upregulation of miR-1207 activated Wnt signaling pathway by inhibiting SFRP1 activity and is inversely correlated with patients’ overall survival. | Ovarian | [29] |
| miR-454-3p | Over-expression of miR-454-3p reduces the SFRP1 activity by targeting 3’UTR. High expression of miR-454-3p correlates with shorter relapse-free survival of breast cancer. | Breast | [30] |
| miR-196a-1 | High expression of exosomal miR 196a-1 is associated with poor survival of gastric cancer. Ectopic miR 196a-1 expression promotes invasion of low invasive gastric cancer cells by binding to 3’UTR of SFRP1. | Gastric | [31] |
| miR-1301-3p | Significantly upregulated in prostate cancer and targets Wnt pathway inhibitors, SFRP1 and GSK3β, by directly binding to the 3’UTR. High expression of miR-1301-3p suppresses the SFRP1 expression and promotes the expansion of prostate cancer stem cells. | Prostate | [32] |
| miR-1260b | Potential candidate oncogenic miRNA in prostate cancer. Treatment with Genistein significantly downregulated miR-1260b and induces expression of SFRP1 and SMAD4 via DNA demethylation and histone modification. | Prostate | [33] |
| miR-582-3p | High expression correlates with the overall and recurrence-free survival of non-small cell lung carcinoma (NSCLC). Upregulated miR-582-3p inhibits SFRP1, AXIN2, and DKK3 expression and leads to the interaction of β-catenin to TCF-4 and subsequently activates Wnt signaling pathway. Activated Wnt pathway is associated with tumor recurrence in NSCLC. | NSCLC | [34] |
2.2. DNA Methylation Regulates SFRP1 Expression
The SFRP1 expression can be epigenetically regulated through promoter DNA methylation. Hypermethylation of the SFRP1 promoter has been recognized as a common mechanism for downregulation of this gene in cancers [35,36,37]. Methylation inactivates the SFRP1 expression by the addition of a methyl group (CH3) to the CG-rich region (CpG island), which is mainly located in the promoter regions of SFRP1 [37]. The unmethylated CpG islands enhance the accessibility of SFRP1 promoter to the transcription factor and other regulatory units such as enhancers. The presence of DNA methylation forms a heterochromatin structure and restricts the access of the transcription factor to bind to the promoter region [38]. This subsequently inhibits the initiation of gene transcription.
SFRP1 hypermethylation and subsequently reduced mRNA expression are extensively studied in CRC. Meta-analysis using TCGA data showed that hypermethylation of SFRP1 caused the downregulation of this gene and high SFRP1 methylation is associated with poorer survival among CRC patients [36]. SFRP1 methylation was also recently shown to be detectable in circulation [39], highlighting its potential as a non-invasive biomarker for CRC’s early detection. SFRP1 gene methylation was significantly associated with its reduced mRNA expression [39]. Combined with RUNX3 and carcinoembryonic antigen (CEA), this panel identified CRC with 89.41% sensitivity in tissue and 84.71% in serum [39].
In another study, the methylation of the SFRP1 gene in bladder cancer tissues occurred more frequently than its adjacent normal tissues. They discovered that SFRP1 methylation suppresses the gene expression and was associated with the pathogenesis of bladder cancer via the Wnt signaling pathway [37].
A study by Zhang et al. postulated that SFRP1 acts as a potential biomarker for NSCLC because the epigenetic silencing of this gene was associated with lymph nodes metastasis and disease progression within a year after surgery [40]. Similarly, a meta-analysis in NSCLC discovered SFRP1’s transcriptional silencing as a consequence from promoter methylation, revealing a promising application of epigenetic therapy in NSCLC [41]. In addition, by analyzing TCGA data, Liu et al. [42] identified significant correlations between SFRP1_cg15839448 promoter methylation and SFRP1 gene expression.
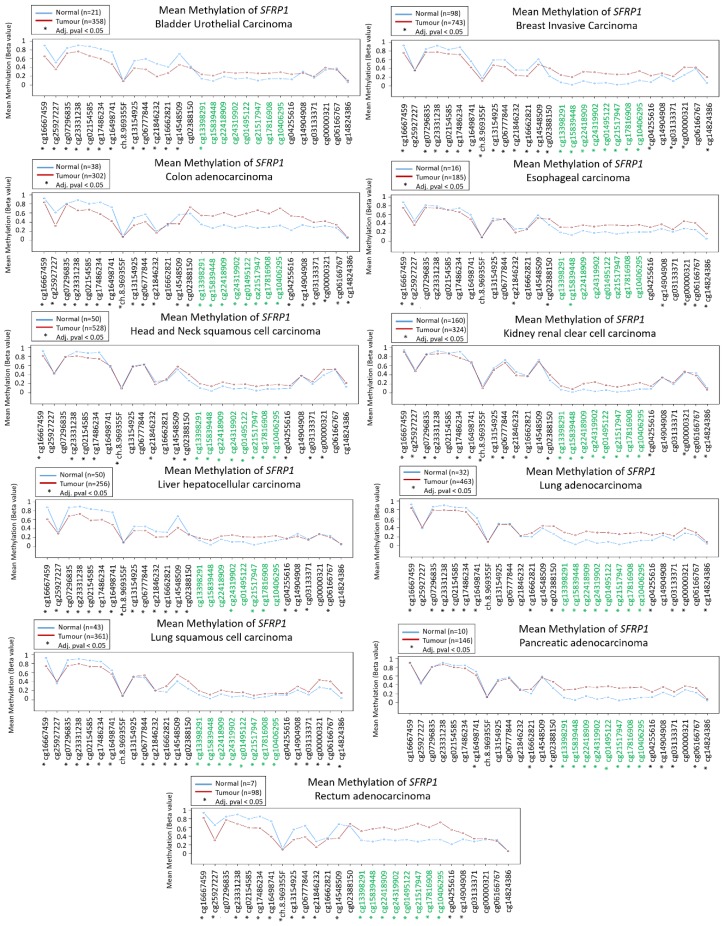
In gastric cancer, SFRP1 methylation was associated with loss of SFRP1 expression and occurred in the early event of this cancer. The epigenetic silencing of SFRP1 was also significantly correlated with tumor stage and lymph node status [43]. To illustrate the methylation signatures of SFRP1 among various cancers, the signatures from the TCGA datasets are displayed in Figure 1. Table 2 summarizes other published literature within six years pertaining to epigenetic regulation of SFRP1 in human cancers.
Figure 1.
Methylation signatures of SFRP1 in Cancer Genome Atlas (TCGA) datasets. Only selected studies with significant SFRP1 differential methylation were shown. Green indicates CpG islands and the figure was generated using Wanderer [44].
Table 2.
Regulation of SFRP1 expression by DNA methylation in cancer.
| Cancer | Description | References |
|---|---|---|
| Breast | Methylation of SFRP1 was significantly different according to the breast cancer molecular subtypes. Low methylation of SFRP1 was detected in basal-like subtype compared to luminal A, luminal B, and HER2 subtypes. SFRP1 may potentially serve as epigenetic biomarker. | [9] |
| Breast | Hypermethylation of SFRP1 was frequently found in breast cancer and caused a reduction of SFRP1 expression. Methylation of SFRP1 indicates poor prognosis in ER+/HER2. | [45] |
| Breast | Aberrant methylation of SFRP1 was observed in 96 breast cancer Chinese patients. This study also showed that methylation of SFRP1 negatively regulates the expression level. | [46] |
| Glioma | Hypermethylation of SFRP1 was associated with poor survival of patients (within 1–3 months after tumor resection). Low methylation of SFRP1 was discovered in the longer survival group. Moreover, methylated SFRP1 frequently occurred in the patients that exhibit higher-grade tumors. This study suggests the hypermethylated SFRP1 as a potential prognostic biomarker in glioma. | [47] |
| Glioma | The methylation level of SFRP1 increases with higher astrocytoma grades and is the highest in glioblastoma. SFRP1 is epigenetically silenced and involved in the progression of glioma. | [48] |
| Glioma | Hydrogen peroxide reverses the methylation of SFRP1 in U251 glioma cells. The demethylation of SFRP1 leads to the activation of this gene and was partially involved in the apoptosis process of the hydrogen-peroxide-induced U251 cells. | [49] |
| Ovarian | Loss of SFRP1 protein expression caused by promoter hypermethylation was observed in the subset of high-grade serous ovarian carcinoma. | [20] |
| Ovarian | Aberrant methylation and low expression of SFRP1 were associated with epithelial ovarian cancer. The activation of this gene inhibits the tumor growth through inactivation of the Wnt signaling pathway. This study further showed that the SFRP1 over-expression in the in vivo model could inhibit the growth of cancer cells. | [50] |
| Acute myeloid leukemia (AML) | Aberrant methylation of SFRP1 was observed in 30.2% of non-M3 AML patients, and SFRP1 expression was negatively correlated with its promoter methylation. | [51] |
| Malignant pleural mesothelioma | Long-term asbestos exposure led to hypermethylation of SFRP1 and reduced gene expression in the mesothelium. | [52] |
2.3. Allelic Imbalance of SFRP1
Besides epigenetic mechanisms, genetic events may be involved in the deregulation of SFRP1. The SFRP1 loci are commonly associated with loss of heterozygosity (LOH) in cancer, including hepatocellular carcinoma (HCC). A study by Huang and colleagues showed only 13% and 6.5% exhibit the allelic imbalance of the SFRP1 loci D8S532 and D8SAC016868, respectively. They also discovered patients that exhibit allelic imbalance of SFRP1 loci to experience a low expression of the gene [35]. This finding is supported by another study where positive LOH at the locus D8S532 is associated with downregulation of SFRP1 expression [53]. However, the allelic imbalance of SFRP1 loci in the HCC patients could not be a crucial event in suppressing SFRP1 because it only involved low frequency of LOH. In the NSCLC, SFRP1 expression is inactivated and 15 out of 40 patients exhibited allelic loss while 25 patients manifest LOH in the SFRP1 locus [54]. Fewer studies are looking into the allelic imbalance of SFRP1 since DNA methylation of the promoter SFRP1 yield more insight into how SFRP1 is downregulated.
2.4. Genomic Alterations in SFRP1
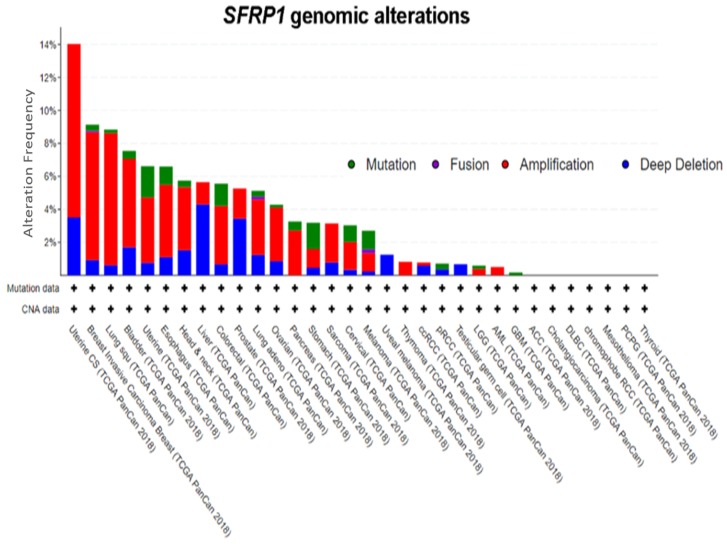
A query of 10,953 patients in 32 studies from TCGA Pan-Cancer reveals a varying degree of genomic alteration profiles (Figure 2) [55,56]. Among all of the alterations, amplification is the most frequent alteration observed, followed by deep deletion and mutation. SFRP1 is only altered in 455 (4%) among the 10,953 patients, further supporting the notion that epigenetic mechanisms play a more important role. It has also been proven that in CRC, even though cancer-associated nonsense mutation that prematurely terminates protein translation at codon 151 (N150) was identified, point mutation is not the primary cause of SFRP1 inactivation [57]. On the contrary, other evidence has shown that SFRP1 mutations found in glioblastoma and CRC promote cancer by compromising the senescence-inducing activity of SFRP1, thus failing to antagonize Wnt signaling [58]. It is known that senescence is a two-edged sword: it can promote malignant transformation by modifying the cellular microenvironment or is a potential mechanism for a cell to avoid cancer development [59]. The role of SFRP1 mutation in cancer-associated senescence warrants further research.
Figure 2.
Genomic alterations in SFRP1. Amplification is the most frequent genomic event in SFRP1. Abbreviations: ACC, adrenocortical carcinoma; BLCA, bladder urothelial carcinoma; BRCA, breast invasive carcinoma; CESC, cervical squamous cell carcinoma and endocervical adenocarcinoma; CHOL, cholangiocarcinoma; COAD, colon adenocarcinoma; COADREAD, colorectal adenocarcinoma; DLBC, lymphoid neoplasm diffuse large B-cell lymphoma; ESCA, esophageal carcinoma; GBM, glioblastoma multiforme; GBMLGG, glioblastoma multiforme Brain lower grade glioma; HNSC, head and neck squamous cell carcinoma; KICH, kidney chromophobe; KIPAN, pan-kidney cohort (KICH+KIRC+KIRP); KIRC, kidney renal clear cell carcinoma; KIRP, kidney renal papillary cell carcinoma; AML, acute myeloid leukemia; LGG, brain lower grade glioma; LIHC, liver hepatocellular carcinoma; LUAD, lung adenocarcinoma; LUSC, lung squamous cell carcinoma; MESO, mesothelioma; OV, ovarian serous cystadenocarcinoma; PAAD, pancreatic adenocarcinoma; PCPG, pheochromocytoma and Paraganglioma; PRAD, prostate adenocarcinoma; READ, rectum adenocarcinoma; SARC, sarcoma; SKCM, skin cutaneous melanoma; STAD, stomach adenocarcinoma; STES, esophagus–stomach cancers; TGCT, testicular germ cell tumors; THCA, thyroid carcinoma; THYM, thymoma; UCEC, uterine corpus endometrial carcinoma; UCS, uterine carcinosarcoma; UVM, uveal melanoma [55,56].
3. Role of SFRP1 in Cancer Pathways
SFRP1 is implicated in various cancer-related pathways. Many studies have delineated the involvement of SFRP1 as a negative modulator of the Wnt signaling pathway. Additionally, SFRP1 is also involved in Hedgehog and TGF-β signaling pathways.
3.1. Involvement of SFRP1 in the Wnt Signaling Pathway
SFRP1 modulates the Wnt signaling pathway through different modes of activation. Interestingly, NTR and CRD of SFRP1 are necessary for optimal Wnt inhibition. SFRP1 can antagonize Wnt activity by directly binding to the ligand of the Wnt protein through its NTR domain and eventually inhibits the interaction of the Wnt ligand to the Fz receptor [60]. However, one study has demonstrated that SFRP1 could also regulate the Wnt signaling pathway by circumventing the interaction with the Wnt ligand. They discovered that the binding of SFRP1 to β-catenin may inhibit the interaction of β-catenin with T-cell factor (TCF) in the nucleus, further blocking Wnt signaling activation [61].
Alternatively, SFRP1 could prevent Wnt signal transduction by interacting with the Fz receptor through their corresponding CRD motif, thereby preventing Wnt ligand interaction with the receptor [62]. In the absence of Wnt ligands, β-catenin molecules are combined with the destruction complex, which consists of scaffold proteins: axin, adenomatous polyposis coli (APC), glycogen synthase kinase (GSK-3β), and casein kinase 1 (CK1). In this state, the β-catenin molecules are phosphorylated by GSK-3β and CK1. Phosphorylation-mediated ubiquitination of β-catenin by β-TrCP and induced proteasomal degradation resulted in a low level of β-catenin in the cytoplasm. Subsequently, the cytoplasmic β-catenin could not be translocated into the nucleus. In the nucleus, the absence of β-catenin initiates a repressive complex containing T-cell factor/lymphoid enhancing factor (TCF/LEF) and transducin-like enhancer protein such as Groucho to recruit histone deacetylase (HDACs) in order to repress the Wnt target genes [60,63], thus, further inhibiting the proliferation and invasion of the cells.
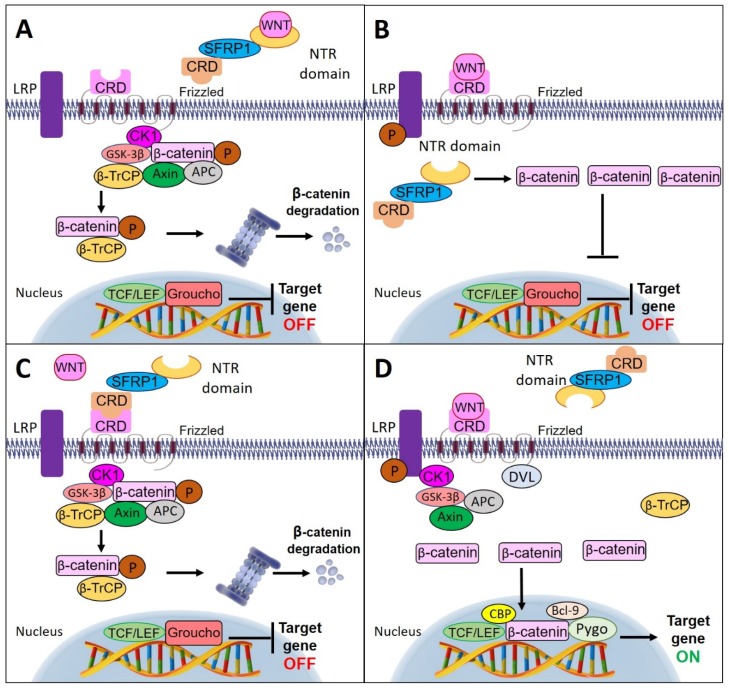
The decline of SFRP1 expression promotes the binding of Wnt ligand to the Fz receptor [61]. Upon binding, the lipoprotein receptor-related protein (LRP) is phosphorylated by CK1 and GSK-3β, which then recruits Dishevelled (Dvl) proteins to the plasma membranes where they polymerize and activate. The activated Dvl polymers inactivate the destruction complex and stabilize β-catenin by suppressing the phosphorylation process, thus, leading to the accumulation of β-catenin in the cytoplasm, and its further entry into the nucleus. Nuclear β-catenin forms an active compound with TCF/LEF proteins by replacing Groucho proteins, and thereby activating the transcription and expression of Wnt target genes [64]. Figure 3 illustrates the multiple mechanisms of SFRP1 involvement in the Wnt signaling pathway.
Figure 3.
Schematic diagram of multiple mechanisms of SFRP1 involvement in Wnt signaling pathway. (A) SFRP1 inhibits Wnt activity by directly binding to the ligand of Wnt protein through its netrin (NTR) domain. (B) SFRP1 antagonizes the Wnt signaling pathway by bypassing the interaction with the Wnt ligand and directly binding to the cytoplasmic β-catenin. (C) SFRP1 binds directly with the Fz receptor through the cycteine-rich domain (CRD) motif preventing the binding of Wnt ligands to the receptor. This causes the β-catenin molecules to combine with the destruction complex and undergo phosphorylation by glycogen synthase kinase (GSK-3β) and casein kinase (CK1). The phosphorylated β-catenin triggers ubiquitination by β-TrCP and initiates proteasomal degradation. (D) In the absence of SFRP1, the Wnt ligand binds to the Fz receptor and results in the recruitment of Dvl and destruction complex to the membrane. The destruction complex is inactivated by Dvl polymers and leads to the accumulation of β-catenin molecules in the cytoplasm. Then, the accumulated β-catenin molecules enter the nucleus and recruit histone-modifying coactivators, Pygo and Bcl-9, to activate Wnt target genes’ transcription.
3.2. Involvement of SFRP1 in the Hedgehog Signaling Pathway
The accumulation of β-catenin in the nucleus is a major indicator of activated Wnt signaling. However, some tumor cells may possess abrogated nuclear β-catenin specifically in the activated Hedgehog (Hh) signaling tumor cells [65]. The Hh signaling pathway is important for cellular growth and differentiation during embryonic development [66]. Dysregulation of Hh signaling has been implicated in several cancers including esophageal [67], gastric [68], pancreatic [69], as well as liver [70]. Previous studies showed activated Hh signaling may attenuate Wnt activity through the high expression of the Wnt inhibitor, SFRP1 [63,71,72].
Hh pathway activation is achieved by the interaction of Hh ligands to the Patched receptor (Ptch), and thus allows the accumulation of the transducer smoothened (SMO) at the cell surface. The accumulated SMO stimulates downstream components of the signaling pathway including Gli1 molecules, which then translocate into the nucleus and further activate the Hh target genes [72,73]. SFRP1 is a Hh target gene and is significantly regulated by Gli1 [71]. This targeted gene comprises of a putative Gli1 binding site, which allows the binding of Gli1 to the promoter region of SFRP1. Thus, SFRP1 expression is dependent on the Gli1 transcript [65].
SFRP1 is the hedgehog target that negatively regulates the Wnt signaling pathway. This has been confirmed by He et al. where knockdown of SFRP1 in the Gli expressing cells is able to induce cytoplasmic β-catenin expression by Wnt-1 [71]. In addition, differentiated epithelial cells exhibit activated Hh and induce the SFRP1 expression in the differentiated cells to hinder Wnt signaling activation within stem or progenitor cells [74].
3.3. Involvement of SFRP1 in TGF-β Signaling Pathway
SFRP1 over-expression restored the activity of GSK3β. Both SFRP1 and GSK3β are important to inhibit the Wnt signaling pathway [75]. However, emerging evidence showed the restoration of GSK3β in promoting the tumorigenesis is through other signaling pathways. It has been reported that GSK3β activates Rac family small GTPase-1 (Rac1) [76,77], and activation of this gene is involved in breast [78], colon [79], bladder [80], and gastric cancer [81], indicating a role of Rac1 in tumor development. Peng discovered SFRP1 over-expression activates GSK3β/Rac1 and simultaneously inhibits the pro-apoptotic effect of Smad3 in TGF-β signaling through the phosphorylation of the Smad3 linker region [82]. This study also demonstrated that the over-expression of SFRP1 activates TGF-β activity, which is correlated with cell proliferation, epithelial–mesenchymal transition (EMT), and invasion in gastric cancer cells [82].
On the contrary, SFRP1 was significantly downregulated in the TGF-β-induced EMT in lung cancer cell line A549. TGF-β suppresses SFRP1 expression and subsequently inactivates GSK3β by phosphorylating Serine 9 of GSK3β. Moreover, using an in vitro and in vivo model, ectopic expression of SFRP1 was able to inhibit TGF-β activity through suppressing Wnt pathway [83]. In breast cancer, SFRP1 knockdown activates the TGF-β signaling and further increases the expression of ZEB2, zinc finger clusters, that play a critical role in facilitating the EMT process [84]. Upon activation of TGF-β signaling, the downstream targets including Integrin β3 and PAI-1, which are responsible for cell migration and invasion, are also upregulated. Hence, this explains the migratory and invasive characteristics exhibited by SFRP1 knockdown in breast cancer cells [85]. This study also suggested the knockdown of SFRP1 can modulate TGF-β signaling not only through the Smad-dependent action but also over Smad-independent pathway through ERK1/2 phosphorylation [85].
3.4. Involvement of SFRP1 in Other Pathways
In androgen-dependent prostate cancer, loss of SFRP1 undergoes different pathways other than Wnt and Hh signaling to drive cancer cell proliferation. Kawano et al. discovered that SFRP1 represses androgen-receptor (AR) dependent transcription and subsequently inhibits cell proliferation in the androgen-dependent LNCaP cells [86]. SFRP1 negatively regulates AR through the CRD motif and binds with the Fz receptor to form SFRP1/Fz complexes. The inactivation of SFRP1 leads to the uncontrolled AR activation, which is involved in the pathogenesis of prostate cancer [87].
In addition, Bernemann and his colleagues also found that the underlying mechanism of SFRP1 in triple-negative breast cancer (TNBC) is independent of Wnt signaling pathway. They did not discover any changes in Wnt signaling activity and nuclear localization of β-catenin, besides they found the upregulation of genes that are involved in the migration process and downregulation of apoptotic genes after knockdown of SFRP1 based on their gene ontology analysis result [10]. However, this finding is in contrast with Xu et al. whereby they found that the Wnt/β-catenin signaling pathway was enriched in the TNBC [88]. The contradiction may be due to the heterogeneity of the samples, which lead to the discovery of different pathways.
One study has suggested that SFRP1 plays an additional inhibitory role in breast cancer by blocking the activity of thrombospondin-1 (TSP1), which is involved in the modulation of adhesion and migration of cancer cells. This action is through the binding of NTR-related motif of SFRP1 to the N module of TSP1. They discovered the interaction between NTR of SFRP1 and TSP1 disrupts the adhesion and migration of MDA-MB-231 breast cancer cell via α3β1 integrin [89].
4. Clinical Utility of SFRP1
4.1. SFRP1 and Chemotherapy Response
As discussed earlier, Bernemann and colleagues found that knockdown of SFRP1 is strongly correlated with TNBC subtypes. Moreover, they also found SFRP1 could be used as a potential chemotherapeutic marker to stratify patients’ response toward chemotherapy. Knockdown of SFRP1 rendered TNBC cell lines more resistance toward paclitaxel, cisplatin, and doxorubicin chemotherapy as well as radiotherapy [10].
HDAC inhibitor romidepsin and methyltransferase inhibitor decitabine are the FDA-approved epigenetic-modifying drugs for the treatment of myelodysplastic syndromes (MDS) and a subset of T cell lymphoma, respectively. In a study by Cooper and colleagues, epigenetic silencing of SFRP1 was shown to contribute to renal and breast cancer cell survival [90]. Exposure of clear cell renal cell carcinoma (ccRCC) and TNBC cells to low doses of exogenous SFRP1 resulted in dose-dependent inhibition of cancer cell through apoptosis induction [90]. Their findings also propose that SFRP1 re-expression could be used as a biomarker for romidepsin/decitabine response.
Taxanes, such as docetaxel and taxol, are microtubule-stabilizing agents used as first-line agents in the treatment of advanced lung adenocarcinoma and other solid tumors since the 1990s [91]. However, poor response to treatment remains a challenge, and a new target for the treatment of taxane-resistant patients is urgently needed. Via microarray analysis, epigenetic inactivation of SFRP1 by DNA methylation was implicated in taxane chemoresistance [92]. The authors further demonstrated that treatment with demethylating agent 5-azacytidine enhanced the sensitivity of lung cancer cell lines to taxanes, suggesting SFRP1 methylation as a clinically relevant determinant of taxanes resistance in lung cancer patients [92]. Another study revealed that downregulation of SFRP1 in laryngeal carcinoma is mediated by DNA methylation [7]. Subsequent treatment with a demethylating agent significantly increased the expression of this gene and enhanced the sensitivity of laryngeal carcinoma cells to cisplatin through inhibition of NHE1 gene.
With regards to hematological malignancy, epigenetic silencing of SFRP1 is infrequently observed in chronic myeloid leukaemia (CML). However, CML patients with methylated SFRP1 correlated with imatinib therapy resistance as well as additional second Philadelphia chromosome abnormalities [93]. Moreover, expression of antagonists SFRP1 and WIF1 was shown to sensitize chronic myeloid leukemia (CML) cells to tyrosine kinase inhibitors [94]. In in vitro experiments involving K562 cells stably expressing SFRP1, the sensitivity toward imatinib, dasatinib, and nilotinib, were 75%, 43%, and 48% more sensitive, respectively, when compared to empty vector-transfected controls [94].
In another study, the dual role of SFRP1 as a biomarker for chemoresistance was demonstrated. Upregulated SFRP1 was observed in topotecan-resistant ovarian cancer patients, whereby its mRNA level was already high before topotecan treatment [95]. While the study did not investigate the treatment effects, the authors propose that SFRP1 might act as an oncogene in patients treated with topotecan, or in a contrary manner, the cancer cells with high SFRP1 expression could be inherently more resilient to the elimination of rapidly proliferating tumor cells [95]. Collectively, these findings suggested that SFRP1 may offer therapeutic benefits and assist patient stratification in chemotherapy treatment. Evidently, more research is necessary to clarify the role of SFRP1 in modulating response to cancer treatment.
4.2. SFRP1 and Disease Prognosis
SFRP1 hypermethylation or downregulation are also associated with poor prognosis in several cancers. Kaplan–Meier analysis of nasopharyngeal cancer showed patients that exhibit low SFRP1 had significantly worse overall survival (OS), disease-free survival (DFS), and distant-metastasis-free survival (DMFS) as compared to patients that have high expression of this gene [8]. Davaadorj and his team have discovered that the loss of SFRP1 is associated with poor prognosis in hepatocellular carcinoma (HCC). They examined the expression status of SFRP1 in 63 pairs of human HCC and later they found low SFRP1 is associated with larger tumor size and vascular invasion as compared to the positive SFRP1 patients [96]. Similarly, patients with low SFRP1 expression had a poor OS in glioblastoma multiforme (GBM), relative to the positive SFRP1 patients, which seem to have favorable prognosis [97]. SFRP1 methylation is also associated with ovarian cancer recurrence and short overall survival [98].
A recent study by Kumar et al. showed promoter methylation of SFRP1 is associated with lymph node metastasis and poor mean overall survival (OS) in CRC. The majority of CRC samples in this study are methylated at SFRP1 with 72.2% methylation frequency [99]. On the contrary, Liu and colleagues postulated that the co-hypermethylation of SFRP1 and SFRP2 were suggested as independent prognostic predictors of survival advantage in postoperative CRC patients [100], whereby silencing of SFRP1 and SFRP2 by hypermethylation lead to a better prognosis. Meanwhile, in an in vivo study to correlate gut microbiota with epigenetic signature, CRC-associated microbiota induced higher numbers of hypermethylated genes in the murine colonic mucosa, among which SFRP1 was also hypermethylated [101]. Additional confirmation was obtained in 1000 patients, further demonstrating that CRC-associated dysbiosis may promote CRC development via epigenome dysregulation. The authors suggested gene methylation as a marker for CRC to predict the efficacy of prebiotic supplementation in average-risk individuals [101]. Low SFRP1 expression may also concurrently activate the Wnt pathway with WIF1 gene [102]. High expression of WIF1 was shown to be significantly correlated with big tumor diameters and deep invasion of tumor cells. However, the co-expression of high SFRP1 and WIF1 may also increase favorable OS and is associated with low TNM stage in CRC [102], postulating the role of WIF1 as an oncogene, while SFRP1 seemed to be an oncosuppressor, despite both being secreted Wnt antagonists. Nevertheless, the underlying mechanism is yet to be identified.
Hypermethylation of SFRP1 occurs in 37.5% of acute myeloid leukemia (AML), whereby patients that exhibit intermediate-risk karyotyping with concurrent methylated SFRP1 showed poor prognosis, especially in the subgroup 60 years old and younger patients [103]. Association between SFRP1 hypermethylation and poor prognosis was further supported by another study conducted among non-M3 AML patients [51].
As mentioned earlier, SFRP1 possess divergent roles depending on the context. This is shown by a study by Qu et al., which demonstrated that gastric patients with high SFRP1 exhibit poor prognosis [12]. Moreover, this study also reveals high SFRP1 was significantly associated with lymph node metastasis and worse five-year OS. Using gastric cancer cell line models, they further demonstrated that the SFRP1 upregulation activates the TGF-β signaling pathway, hence inducing cell proliferation, EMT process, and cell invasion [12]. Additionally, upregulated SFRP1 in metastatic renal cell carcinoma (RCC) was also involved in invasiveness of the metastatic cells. Knockdown of the endogenous SFRP1 in this study has been showed to reduce the invasive properties of metastatic RCC [11]. On top of that, enriched SFRP1 was also found in the metastatic osteosarcoma [104].
4.3. SFRP1 in Clinical Trials
Despite SFRP1 being a well-known gene studied in human cancer, there are only a handful of clinical trials, which involve SFRP1. Based on the hypothesis that long-term exercise may cause changes in the serum levels of SFRP1 in patients with breast cancer, a pilot trial (NCT02895178) was conducted [105]. Thirty breast cancer survivors from Wonju Severance Christian Hospital were enrolled and randomized to two different groups: 12 weeks exercise program and control group. Then, SFRP1 expression in the serum was measured for the pre- and post-treatment in both groups. This pilot study is the first study to show a decrease in serum SFRP1 levels in patients with breast cancer due to exercise training. The decrease of SFRP1 is accompanied by improving body composition such as a decrease in body fat percentage and visceral fat areas. This demonstrates that decreased serum level of SFRP1 improves the physical fitness of breast cancer survivor patients [105].
NCT01214681 is a clinical trial looking into the impact of the role of non-digestible carbohydrates (NDCs) in CRC chemoprevention [106]. A number of novel biomarkers of diet-related CRC risk measured in cancer tissue biopsies and in stool were developed, including SFRP1. The effects of supplementing healthy individuals with two NDCs, which are resistant starch (RS) and polydextrose, on fecal calprotectin concentrations and the expression of 12 Wnt-related genes including SFRP1, were investigated [107]. In addition, the trial also seeks to determine whether the effects on SFRP1 expression are regulated via the epigenetic mechanisms, which include DNA methylation and microRNA expression. Although NDC supplementation did not influence fecal calprotectin concentration, SFRP1 expression was significantly reduced by RS, which could result in increased Wnt pathway activity. However, RS and polydextrose neither affect SFRP1 methylation nor alter the expression of 10 microRNAs that were predicted to target this gene. This suggests another unknown mechanism that reduces SFRP1 expression. Nevertheless, the effects on Wnt pathway activity and downstream functional effects in the healthy colorectal mucosa warrants further investigation.
SFRP1 is among the biomarkers studied in glioblastoma clinical trial NCT00822458 [108]. The phase I clinical trial is investigating the side effects and best dose of GDC-0449, a Hedgehog signaling antagonist (Vismodegib) in treating young patients with recurrent medulloblastoma or patients who did not respond to previous treatment. Blood samples are collected periodically for pharmacokinetic studies. Archived tumor tissues were collected and analyzed for the expression of genes that activate the Hedgehog (e.g., Gli1, Gli2, SFRP1, ATOH1, and PTCH2) or Wnt (e.g., DKK2 and DKK4) cell signal pathways via in situ hybridization and real-time PCR. At the time this review was written, there is no published finding of this trial yet; therefore, the exact role of SFRP1 is undetermined.
5. SFRP1 in Patents
Despite the limited involvement of SFRP1 in cancer clinical trials, research evidence has shown the potential of SFRP1 as a diagnostic and pharmacogenetics marker, and this has led to several patents being applied or granted. For instance, Baylin and colleagues from John Hopkins University were granted a patent for methods of identifying epigenetically silenced genes that are associated with cancer in 2010 [109]. The method is demonstrated by the identification of 74 genes that are epigenetically silenced in CRC cells, including identification of methylation silencing of SFRP1 and its families such as SFRP2, SFRP4, and SFRP5. This patent covers three categories, which are C12Q1/6886 nucleic acid products used in the analysis of nucleic acids, C12Q1/6809 methods for determination or identification of nucleic acids involving differential detection, and C12Q2600/154 methylation markers.
MDxHealth SA filed for a patent of a kit involving SFRP1 in identifying and diagnosing cancer based on SFRP1 methylation status [110]. In their claim, detection of the epigenetic change of NDRG4 and at least one gene among 17 other genes including SFRP1 indicate a predisposition to gastrointestinal cancer. Granted in 2016, the kit also described the pharmacogenetic methods for determining suitable treatment regimens for cancer. Combination of NDRG4 and SFRP1 is able to predict the response of cancer treatment with a DNA damaging agent, DNA methyltransferase inhibitor, and/or an HDAC inhibitor [110]. Patients with methylated NDRG4 and SFRP1 will respond better to these therapeutic agents. The epigenetic changes of NDRG4 and SFRP1 are also indicative of the histopathological stage of gastrointestinal cancer [110]. This patent includes six categories of claims, which are C12Q1/6886 nucleic acid products used in the analysis of nucleic acids; C12Q2600/106 pharmacogenomics; C12Q2600/112 disease subtyping, staging, or classification; C12Q2600/118 prognosis of disease development; C12Q2600/154 methylation markers; C12Q2600/158 expression markers; and C12Q2600/16 primer sets for multiplex assays.
More recently, Lothe and colleagues identified CDO1, DCLK1, and SCAN18 as novel, frequently methylated genes in cholangiocarcinoma, in addition to the previously reported SFRP1 gene [111]. The combination of CDO1, DCLK1, ZSCAN18, and SFRP1 reach a sensitivity of 87% and specificity of 100% in fresh, frozen, and archival material. The group was granted a patent in 2017 under two categories, which are C12Q1/6886 nucleic acid products used in the analysis of nucleic acids and C12Q2600/154 methylation markers.
6. Conclusions
In this review, we have provided the latest evidence of epigenetic of SFRP1 and its divergent roles in carcinogenesis, highlighted its pharmacogenomics properties, and deliberated on the clinical trials and patents involving SFRP1. SFRP1 functions as a negative regulator of Wnt signaling; therefore, it has an important role in carcinogenesis. Accumulating evidence suggests that epigenetic regulation contributes to the silencing of SFRP1. Indeed, in many cancers, SFRP1 is downregulated via promoter hypermethylation. Restoration of SFRP1 expression via demethylating drugs or over-expression experiments increases sensitivity toward various chemotherapeutic agents. Given the importance of SFRP1 in cancer-related pathways, HDACi and DNMTi appear as promising epigenetic therapy to reverse SFRP1 methylation. Interestingly, a combination of chemotherapeutic and epigenetic drug is also highly synergistic at inhibiting cancer cells than single agent alone.
On the other hand, owing to its dual roles, knockdown of SFRP1 also renders certain cancer cells to be more resistant toward selected chemotherapies as well as radiotherapy. Nevertheless, the clinical utility of epigenetic drugs required extensive investigations and proper utilization, as these drugs are toxic and non-specific gene modulators. Although a lot of progress has been made regarding the regulation and function of SFRP1 in the normal and cancer cells, many questions remain unanswered. Henceforth, the investigation into the contradictory roles of SFRP1, particularly in cancer prognosis and its pharmacogenomics utilities, are indispensable.
Author Contributions
N.S.A.M. conceptualized the idea for this manuscript; R.B. and N.S.A.M. wrote the manuscript and prepared the figures; F.Y.F.T. and L.-H.L. provided critical scientific insights. All authors have read and agreed to the published version of the manuscript.
Funding
This study is supported by Geran Universiti Penyelidikan by Universiti Kebangsaan, Malaysia (GUP-2018-070) and Monash University, Malaysia.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bovolenta P., Esteve P., Ruiz J.M., Cisneros E., Lopez-Rios J. Beyond Wnt inhibition: New functions of secreted Frizzled-related proteins in development and disease. J. Cell Sci. 2008;121:737–746. doi: 10.1242/jcs.026096. [DOI] [PubMed] [Google Scholar]
- 2.Shahi M., Afzal M., Sinha S., Castresana J. Aberrant promoter methylation of SFRP1 (Secreted Frizzled-Related Protein1) gene in medulloblastoma and glioblastoma. Clin. Cancer Res. 2008;14(Suppl. 23):19. [Google Scholar]
- 3.Mao W., Wordinger R.J., Clark A.F. Focus on molecules: SFRP. Exp. Eye Res. 2010;91:552–553. doi: 10.1016/j.exer.2010.05.003. [DOI] [PubMed] [Google Scholar]
- 4.Chong J.M., Uren A., Rubin J.S., Speicher D.W. Disulfide bond assignments of secreted Frizzled-related protein-1 provide insights about Frizzled homology and netrin modules. J. Biol. Chem. 2002;277:5134–5144. doi: 10.1074/jbc.M108533200. [DOI] [PubMed] [Google Scholar]
- 5.Atschekzei F., Hennenlotter J., Jänisch S., Großhennig A., Tränkenschuh W., Waalkes S., Peters I., Dörk T., Merseburger A.S., Stenzl A., et al. SFRP1 CpG island methylation locus is associated with renal cell cancer susceptibility and disease recurrence. Epigenetics. 2012;7:447–457. doi: 10.4161/epi.19614. [DOI] [PubMed] [Google Scholar]
- 6.Hattori N., Sako M., Kimura K., Iida N., Takeshima H., Nakata Y., Kono Y., Ushijima T. Novel prodrugs of decitabine with greater metabolic stability and less toxicity. Clin. Epigenet. 2019;11:111. doi: 10.1186/s13148-019-0709-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Wu K., Li Z.H., Yi W., Wu M.H., Jiang M.J., Zhang Y., Zheng H.L., Chen W. Restoration of secreted frizzled-related protein 1 suppresses growth and increases cisplatin sensitivity in laryngeal carcinoma cells by downregulating NHE 1. Int. J. Clin. Exp. Pathol. 2017;10:8334–8343. [PMC free article] [PubMed] [Google Scholar]
- 8.Ren X.-Y., Zhou G.-Q., Jiang W., Sun Y., Xu Y.-F., Li Y.-Q., Tang X.R., Wen X., He Q.M., Yang X.J., et al. Low SFRP1 Expression Correlates with Poor Prognosis and Promotes Cell Invasion by Activating the Wnt/β-Catenin Signaling Pathway in NPC. Cancer Prev. Res. 2015;8:968–977. doi: 10.1158/1940-6207.CAPR-14-0369. [DOI] [PubMed] [Google Scholar]
- 9.Jeong Y.J., Jeong H.Y., Bong J.G., Park S.H., Oh H.K. Low methylation levels of the SFRP1 gene are associated with the basal-like subtype of breast cancer. Oncol. Rep. 2013;29:1946–1954. doi: 10.3892/or.2013.2335. [DOI] [PubMed] [Google Scholar]
- 10.Bernemann C., Hülsewig C., Ruckert C., Schäfer S., Blümel L., Hempel G., Götte M., Greve B., Barth P.J., Kiesel L., et al. Influence of secreted frizzled receptor protein 1 (SFRP1) on neoadjuvant chemotherapy in triple negative breast cancer does not rely on WNT signaling. Mol. Cancer. 2014;13:174. doi: 10.1186/1476-4598-13-174. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Saini S., Liu J., Yamamura S., Majid S., Kawakami K., Hirata H., Dahiya R. Functional Significance of Secreted Frizzled-Related Protein 1 in Metastatic Renal Cell Carcinomas. Cancer Res. 2009;69:6815–6822. doi: 10.1158/0008-5472.CAN-09-1254. [DOI] [PubMed] [Google Scholar]
- 12.Qu Y., Ray P.S., Li J., Cai Q., Bagaria S.P., Moran C., Sim M.-S., Zhang J., Turner R.R., Zhu Z., et al. High levels of secreted frizzled-related protein 1 correlate with poor prognosis and promote tumourigenesis in gastric cancer. Eur. J. Cancer. 2013;49:3718–3728. doi: 10.1016/j.ejca.2013.07.011. [DOI] [PubMed] [Google Scholar]
- 13.Lin H., Yang G., Ding B., Zhang M., Zhang M., Yan F., Qu Y., Zhang H. Secreted frizzled-related protein 1 overexpression in gastric cancer: Relationship with radiological findings of dual-energy spectral CT and PET-CT. Sci. Rep. 2017;7:1–9. doi: 10.1038/srep42020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Surana R., Sikka S., Cai W., Shin E.M., Warrier S.R., Tan H.J.G., Arfuso F., Fox S.A., Dharmarajan A.M., Kumar A.P. Secreted frizzled related proteins: Implications in cancers. Biochim. Biophys. Acta. 2014;1845:53–65. doi: 10.1016/j.bbcan.2013.11.004. [DOI] [PubMed] [Google Scholar]
- 15.Liu Y., Zhou Q., Zhou D., Huang C., Meng X., Li J. Secreted frizzled-related protein 2-mediated cancer events: Friend or foe? Pharmacol. Rep. 2017;69:403–408. doi: 10.1016/j.pharep.2017.01.001. [DOI] [PubMed] [Google Scholar]
- 16.Pawar N.M., Rao P. Secreted frizzled related protein 4 (sFRP4) update: A brief review. Cell Signal. 2018;45:63–70. doi: 10.1016/j.cellsig.2018.01.019. [DOI] [PubMed] [Google Scholar]
- 17.Vincent K.M., Postovit L.-M. A pan-cancer analysis of secreted Frizzled-related proteins: Re-examining their proposed tumour suppressive function. Sci. Rep. 2017;7:1–9. doi: 10.1038/srep42719. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yu J., Xie Y., Li M., Zhou F., Zhong Z., Liu Y., Wang F., Qi J. Association between SFRP promoter hypermethylation and different types of cancer: A systematic review and meta-analysis. Oncol. Lett. 2019;18:3481–3491. doi: 10.3892/ol.2019.10709. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Cheng C., Smith S.K., Charnock-Jones D.S. Transcript profile and localization of Wnt signaling–related molecules in human endometrium. Fertil. Steril. 2008;90:201–204. doi: 10.1016/j.fertnstert.2007.05.077. [DOI] [PubMed] [Google Scholar]
- 20.Kardum V., Karin V., Glibo M., Skrtic A., Martic T.N., Ibisevic N., Skenderi F., Vranic S., Serman L. Methylation-associated silencing of SFRP1 gene in high-grade serous ovarian carcinomas. Ann. Diagn. Pathol. 2017;31:45–49. doi: 10.1016/j.anndiagpath.2017.07.002. [DOI] [PubMed] [Google Scholar]
- 21.Wang Z., Li R., He Y., Huang S. Effects of secreted frizzled-related protein 1 on proliferation, migration, invasion, and apoptosis of colorectal cancer cells. Cancer Cell. Int. 2018;18:48. doi: 10.1186/s12935-018-0543-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.García-Tobilla P., Solórzano S.R., Salido-Guadarrama I., González-Covarrubias V., Morales-Montor G., Díaz-Otañez C.E., Rodríguez-Dorantes M. SFRP1 repression in prostate cancer is triggered by two different epigenetic mechanisms. Gene. 2016;593:292–301. doi: 10.1016/j.gene.2016.08.030. [DOI] [PubMed] [Google Scholar]
- 23.FireBrowse. [(accessed on 29 November 2019)]; Available online: http://firebrowse.org/viewGene.html?gene=SFRP.
- 24.Wu F., Li J., Guo N., Wang X.-H., Liao Y.-Q. MiRNA-27a promotes the proliferation and invasion of human gastric cancer MGC803 cells by targeting SFRP1 via Wnt/β-catenin signaling pathway. Am. J. Cancer Res. 2017;7:405–416. [PMC free article] [PubMed] [Google Scholar]
- 25.Ba S., Xuan Y., Long Z.-W., Chen H.-Y., Zheng S.-S. MicroRNA-27a Promotes the Proliferation and Invasiveness of Colon Cancer Cells by Targeting SFRP1 through the Wnt/β-Catenin Signaling Pathway. Cell. Physiol. Biochem. 2017;42:1920–1933. doi: 10.1159/000479610. [DOI] [PubMed] [Google Scholar]
- 26.Kong L.-Y., Xue M., Zhang Q.-C., Su C.-F. In vivo and in vitro effects of microRNA-27a on proliferation, migration and invasion of breast cancer cells through targeting of SFRP1 gene via Wnt/β-catenin signaling pathway. Oncotarget. 2017;8:15507–15519. doi: 10.18632/oncotarget.14662. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 27.Lin T., Ma Q., Zhang Y., Zhang H., Yan J., Gao C. MicroRNA-27a functions as an oncogene in human osteosarcoma by targeting CCNG. Oncol. Lett. 2018;15:1067–1071. doi: 10.3892/ol.2017.7389. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mu Y., Zhang L., Chen X., Chen S., Shi Y., Li J. Silencing microRNA-27a inhibits proliferation and invasion of human osteosarcoma cells through the SFRP1-dependent Wnt/β-catenin signaling pathway. Biosci. Rep. 2019;39:BSR20182366. doi: 10.1042/BSR20182366. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Wu G., Liu A., Zhu J., Lei F., Wu S., Zhang X., Ye L., Cao L., He S. MiR-1207 overexpression promotes cancer stem cell–like traits in ovarian cancer by activating the Wnt/β-catenin signaling pathway. Oncotarget. 2015;6:28882–28894. doi: 10.18632/oncotarget.4921. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Ren L., Chen H., Song J., Chen X., Lin C., Zhang X., Hou N., Pan J., Zhou Z., Wang L., et al. MiR-454-3p-Mediated Wnt/β-catenin Signaling Antagonists Suppression Promotes Breast Cancer Metastasis. Theranostics. 2019;9:449–465. doi: 10.7150/thno.29055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Feng C., She J., Chen X., Zhang Q., Zhang X., Wang Y., Ye J., Shi J., Tao J., Feng M., et al. Exosomal miR-196a-1 promotes gastric cancer cell invasion and metastasis by targeting SFRP. Nanomedicine. 2019;14:2579–2593. doi: 10.2217/nnm-2019-0053. [DOI] [PubMed] [Google Scholar]
- 32.Song X.-L., Huang B., Zhou B.-W., Wang C., Liao Z.-W., Yu Y., Zhao S.-C. miR-1301-3p promotes prostate cancer stem cell expansion by targeting SFRP1 and GSK3β. Biomed. Pharmacother. 2018;99:369–374. doi: 10.1016/j.biopha.2018.01.086. [DOI] [PubMed] [Google Scholar]
- 33.Hirata H., Hinoda Y., Shahryari V., Deng G., Tanaka Y., Tabatabai Z.L., Dahiya R. Genistein downregulates onco-miR-1260b and upregulates sFRP1 and Smad4 via demethylation and histone modification in prostate cancer cells. Br. J. Cancer. 2014;110:1645–1654. doi: 10.1038/bjc.2014.48. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Fang L., Cai J., Chen B., Wu S., Li R., Xu X., Yang Y., Guan H., Zhu X., Zhang L., et al. Aberrantly expressed miR-582-3p maintains lung cancer stem cell-like traits by activating Wnt/β-catenin signalling. Nat. Commun. 2015;6:1–15. doi: 10.1038/ncomms9640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Huang J., Zhang Y.-L., Teng X.-M., Lin Y., Zheng D.-L., Yang P.-Y., Han Z.-G. Down-regulation of SFRP1 as a putative tumor suppressor gene can contribute to human hepatocellular carcinoma. BMC Cancer. 2007;7:126. doi: 10.1186/1471-2407-7-126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Kim J., Kim S. In silico Identification of SFRP1 as a Hypermethylated Gene in Colorectal Cancers. Genomics Inform. 2014;12:171–180. doi: 10.5808/GI.2014.12.4.171. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Wang X., Wang H., Bu R., Fei X., Zhao C., Song Y. Methylation and aberrant expression of the Wnt antagonist secreted Frizzled-related protein 1 in bladder cancer. Oncol. Lett. 2012;4:334–338. doi: 10.3892/ol.2012.713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Lim D.H.K., Maher E.R. DNA methylation: A form of epigenetic control of gene expression. Obstet. Gynaecolt. 2010;12:37–42. doi: 10.1576/toag.12.1.037.27556. [DOI] [Google Scholar]
- 39.Pasha H.F., Radwan M.I., Yehia A.M., Toam M.M. Circulating methylated RUNX3 and SFRP1 genes as a noninvasive panel for early detection of colorectal cancer. Eur. J. Gastroenterol. Hepatol. 2019;31:1342. doi: 10.1097/MEG.0000000000001532. [DOI] [PubMed] [Google Scholar]
- 40.Zhang Y., Miao F., Geng J., Wang R., Chen L. Transcriptional inactivation of secreted frizzled-related protein 1 by promoter hypermethylation as a potential biomarker for non-small cell lung cancer. Neoplasma. 2010;57:228–233. doi: 10.4149/neo_2010_03_228. [DOI] [PubMed] [Google Scholar]
- 41.Taguchi Y., Iwadate M., Umeyama H. SFRP1 is a possible candidate for epigenetic therapy in non-small cell lung cancer. BMC Med. Genomics. 2016;9(Suppl. 1):28. doi: 10.1186/s12920-016-0196-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Liu S., Chen X., Chen R., Wang J., Zhu G., Jiang J., Wang H., Duan S., Huang J. Diagnostic role of Wnt pathway gene promoter methylation in non small cell lung cancer. Oncotarget. 2017;8:36354–36367. doi: 10.18632/oncotarget.16754. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao C.H., Bu X.M., Zhang N. Hypermethylation and aberrant expression of Wnt antagonist secreted frizzled-related protein 1 in gastric cancer. World J. Gastroenterol. 2007;13:2214–2217. doi: 10.3748/wjg.v13.i15.2214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Díez-Villanueva A., Mallona I., Peinado M.A. Wanderer, an interactive viewer to explore DNA methylation and gene expression data in human cancer. Epigenet. Chromatin. 2015;8:22. doi: 10.1186/s13072-015-0014-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Győrffy B., Bottai G., Fleischer T., Munkácsy G., Budczies J., Paladini L., Børresen-Dale A.-L., Kristensen V.N., Santarpia L. Aberrant DNA methylation impacts gene expression and prognosis in breast cancer subtypes. Int. J. Cancer. 2016;138:87–97. doi: 10.1002/ijc.29684. [DOI] [PubMed] [Google Scholar]
- 46.Li Z., Heng J., Yan J., Guo X., Tang L., Chen M., Peng L., Wu Y., Wang S., Xiao Z., et al. Integrated analysis of gene expression and methylation profiles of 48 candidate genes in breast cancer patients. Breast Cancer Res. Treat. 2016;160:371–383. doi: 10.1007/s10549-016-4004-8. [DOI] [PubMed] [Google Scholar]
- 47.Majchrzak-Celińska A., Słocińska M., Barciszewska A.-M., Nowak S., Baer-Dubowska W. Wnt pathway antagonists, SFRP1, SFRP2, SOX17, and PPP2R2B, are methylated in gliomas and SFRP1 methylation predicts shorter survival. J. Appl. Genet. 2016;57:189–197. doi: 10.1007/s13353-015-0312-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Kafka A., Karin-Kujundžić V., Šerman L., Bukovac A., Njirić N., Jakovčević A., Pećina-Šlaus N. Hypermethylation of Secreted Frizzled Related Protein 1 gene promoter in different astrocytoma grades. Croat. Med. J. 2018;59:213–223. doi: 10.3325/cmj.2018.59.213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Xing Z., Ni Y., Zhao J., Ma X. Hydrogen Peroxide-Induced Secreted Frizzled-Related Protein 1 Gene Demethylation Contributes to Hydrogen Peroxide-Induced Apoptosis in Human U251 Glioma Cells. DNA Cell Biol. 2017;36:347–353. doi: 10.1089/dna.2016.3594. [DOI] [PubMed] [Google Scholar]
- 50.Zhang H., Sun D., Qiu J., Yao L. SFRP1 inhibited the epithelial ovarian cancer through inhibiting Wnt/β-catenin signaling. Acta Biochim. Pol. 2019;66:393–400. doi: 10.18388/abp.2019_2757. [DOI] [PubMed] [Google Scholar]
- 51.Guo H., Zhang T., Wen X., Zhou J., Ma J., An C., Zhang W., Xu Z., Lin J., Qian J. Hypermethylation of secreted frizzled-related proteins predicts poor prognosis in non-M3 acute myeloid leukemia. Onco-Targets Ther. 2017;10:3635–3644. doi: 10.2147/OTT.S136502. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Cheng Y.Y., Mok E., Tan S., Leygo C., McLaughlin C., George A.M., Reid G. SFRP Tumour Suppressor Genes Are Potential Plasma-Based Epigenetic Biomarkers for Malignant Pleural Mesothelioma. Dis. Markers. 2017;2017:2536187. doi: 10.1155/2017/2536187. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Qiu Y., Xu L., Zhou Y.-H., Shi M., Ma Y., Li M., Li J.-C. Involvement of genetic instability in the downregulation of sFRP1 in Chinese patients with hepatocellular carcinoma. Anat. Rec. 2010;293:2020–2026. doi: 10.1002/ar.21273. [DOI] [PubMed] [Google Scholar]
- 54.Fukui T., Kondo M., Ito G., Maeda O., Sato N., Yoshioka H., Yokoi K., Ueda Y., Shimokata K., Sekido Y. Transcriptional silencing of secreted frizzled related protein 1 (SFRP1) by promoter hypermethylation in non-small-cell lung cancer. Oncogene. 2005;24:6323–6327. doi: 10.1038/sj.onc.1208777. [DOI] [PubMed] [Google Scholar]
- 55.Cerami E., Gao J., Dogrusoz U., Gross B.E., Sumer S.O., Aksoy B.A., Jacobsen A., Byrne C.J., Heuer M.L., Larsson E., et al. The cBio cancer genomics portal: An open platform for exploring multidimensional cancer genomics data. Cancer Discov. 2012;2:401–404. doi: 10.1158/2159-8290.CD-12-0095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gao J., Aksoy B.A., Dogrusoz U., Dresdner G., Gross B., Sumer S.O., Sun Y., Jacobsen A., Sinha R., Larsson E., et al. Integrative analysis of complex cancer genomics and clinical profiles using the cBioPortal. Sci. Signal. 2013;6:l1. doi: 10.1126/scisignal.2004088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Caldwell G.M., Jones C., Gensberg K., Jan S., Hardy R.G., Byrd P., Chughtai S., Wallis Y., Matthews G.M., Morton D.G. The Wnt antagonist sFRP1 in colorectal tumorigenesis. Cancer Res. 2004;64:883–888. doi: 10.1158/0008-5472.CAN-03-1346. [DOI] [PubMed] [Google Scholar]
- 58.Elzi D.J., Song M., Hakala K., Weintraub S.T., Shiio Y. Wnt Antagonist SFRP1 Functions as a Secreted Mediator of Senescence. Mol. Cell Biol. 2012;32:4388–4399. doi: 10.1128/MCB.06023-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Zeng S., Shen W.H., Liu L. Senescence and Cancer. Cancer Trans. Med. 2018;4:70–74. doi: 10.4103/ctm.ctm_22_18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Lopez-Rios J., Esteve P., Ruiz J.M., Bovolenta P. The Netrin-related domain of Sfrp1 interacts with Wnt ligands and antagonizes their activity in the anterior neural plate. Neural Dev. 2008;3:19. doi: 10.1186/1749-8104-3-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Liang C.-J., Wang Z.-W., Chang Y.-W., Lee K.-C., Lin W.-H., Lee J.-L. SFRPs Are Biphasic Modulators of Wnt-Signaling-Elicited Cancer Stem Cell Properties beyond Extracellular Control. Cell Rep. 2019;28:1511–1525. doi: 10.1016/j.celrep.2019.07.023. [DOI] [PubMed] [Google Scholar]
- 62.Agostino M., Pohl S.Ö.-G., Dharmarajan A. Structure-based prediction of Wnt binding affinities for Frizzled-type cysteine-rich domains. J. Biol. Chem. 2017;292:11218. doi: 10.1074/jbc.M117.786269. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Ding M., Wang X. Antagonism between Hedgehog and Wnt signaling pathways regulates tumorigenicity (Review) Oncol. Lett. 2017;14:6327–6333. doi: 10.3892/ol.2017.7030. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.MacDonald B.T., He X. Frizzled and LRP5/6 Receptors for Wnt/β-Catenin Signaling. Cold Spring Harb. Perspect. Biol. 2012;4:a007880. doi: 10.1101/cshperspect.a007880. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kim J.-H., Shin H.S., Lee S.H., Lee I., Lee Y.S., Park J.C., Kim Y.J., Chung J.B., Lee Y.C. Contrasting activity of Hedgehog and Wnt pathways according to gastric cancer cell differentiation: Relevance of crosstalk mechanisms. Cancer Sci. 2010;101:328–335. doi: 10.1111/j.1349-7006.2009.01395.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Jia Y., Wang Y., Xie J. The Hedgehog pathway: Role in cell differentiation, polarity and proliferation. Arch. Toxicol. 2015;89:179–191. doi: 10.1007/s00204-014-1433-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Yang L., Wang L.-S., Chen X.L., Gatalica Z., Qiu S., Liu Z., Stoner G., Zhang H., Weiss H., Xie J. Hedgehog signaling activation in the development of squamous cell carcinoma and adenocarcinoma of esophagus. Int. J. Biochem. Mol. Biol. 2012;3:46–57. [PMC free article] [PubMed] [Google Scholar]
- 68.Akyala A.I., Peppelenbosch M.P. Gastric cancer and Hedgehog signaling pathway: Emerging new paradigms. Genes Cancer. 2018;9:1–10. doi: 10.18632/genesandcancer.168. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Gu D., Schlotman K.E., Xie J. Deciphering the role of hedgehog signaling in pancreatic cancer. J. Biomed. Res. 2016;30:353–360. doi: 10.7555/JBR.30.20150107. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Machado M.V., Diehl A.M. Hedgehog Signaling in Liver Pathophysiology. J. Hepatol. 2018;68:550–562. doi: 10.1016/j.jhep.2017.10.017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.He J., Sheng T., Stelter A.A., Li C., Zhang X., Sinha M., Luxon B.A., Xie J. Suppressing Wnt Signaling by the Hedgehog Pathway through sFRP-1. J. Biol. Chem. 2006;281:35598–35602. doi: 10.1074/jbc.C600200200. [DOI] [PubMed] [Google Scholar]
- 72.Pelullo M., Zema S., Nardozza F., Checquolo S., Screpanti I., Bellavia D. Wnt, Notch, and TGF-β Pathways Impinge on Hedgehog Signaling Complexity: An Open Window on Cancer. Front. Genet. 2019;10 doi: 10.3389/fgene.2019.00711. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Yang L., Xie G., Fan Q., Xie J. Activation of the hedgehog-signaling pathway in human cancer and the clinical implications. Oncogene. 2010;29:469–481. doi: 10.1038/onc.2009.392. [DOI] [PubMed] [Google Scholar]
- 74.Katoh Y., Katoh M. WNT antagonist, SFRP1, is Hedgehog signaling target. Int. J. Mol. Med. 2006;17:171–175. doi: 10.3892/ijmm.17.1.171. [DOI] [PubMed] [Google Scholar]
- 75.Huang J., Guo X., Li W., Zhang H. Activation of Wnt/β-catenin signalling via GSK3 inhibitors direct differentiation of human adipose stem cells into functional hepatocytes. Sci. Rep. 2017;7:1–12. doi: 10.1038/srep40716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Rom S., Fan S., Reichenbach N., Dykstra H., Ramirez S.H., Persidsky Y. Glycogen Synthase Kinase 3β Inhibition Prevents Monocyte Migration across Brain Endothelial Cells via Rac1-GTPase Suppression and Down-Regulation of Active Integrin Conformation. Am. J. Pathol. 2012;181:1414–1425. doi: 10.1016/j.ajpath.2012.06.018. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Chikano Y., Domoto T., Furuta T., Sabit H., Kitano-Tamura A., Pyko I.V., Takino T., Sai Y., Hayashi Y., Sato H., et al. Glycogen Synthase Kinase 3β Sustains Invasion of Glioblastoma via the Focal Adhesion Kinase, Rac1, and c-Jun N-Terminal Kinase-Mediated Pathway. Mol. Cancer Ther. 2015;14:564–574. doi: 10.1158/1535-7163.MCT-14-0479. [DOI] [PubMed] [Google Scholar]
- 78.Rozenchan P.B., Pasini F.S., Roela R.A., Katayama M.L.H., Mundim F.G.L., Brentani H., Lyra E.C., Brentani M.M. Specific upregulation of RHOA and RAC1 in cancer-associated fibroblasts found at primary tumor and lymph node metastatic sites in breast cancer. Tumor Biol. 2015;36:9589–9597. doi: 10.1007/s13277-015-3727-1. [DOI] [PubMed] [Google Scholar]
- 79.De Toledo M., Anguille C., Roger L., Roux P., Gadea G. Cooperative Anti-Invasive Effect of Cdc42/Rac1 Activation and ROCK Inhibition in SW620 Colorectal Cancer Cells with Elevated Blebbing Activity. PLoS ONE. 2012;7:e48344. doi: 10.1371/journal.pone.0048344. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Chen X., Zhang J.-X., Luo J.-H., Wu S., Yuan G.-J., Ma N.-F., Feng Y., Cai M.-Y., Chen R.-X., Lu J., et al. CSTF2-Induced Shortening of the RAC1 3′UTR Promotes the Pathogenesis of Urothelial Carcinoma of the Bladder. Cancer Res. 2018;78:5848–5862. doi: 10.1158/0008-5472.CAN-18-0822. [DOI] [PubMed] [Google Scholar]
- 81.Ji J., Feng X., Shi M., Cai Q., Yu Y., Zhu Z., Zhang J. Rac1 is correlated with aggressiveness and a potential therapeutic target for gastric cancer. Int. J. Oncol. 2015;46:1343–1353. doi: 10.3892/ijo.2015.2836. [DOI] [PubMed] [Google Scholar]
- 82.Peng J.-X., Liang S.-Y., Li L. sFRP1 exerts effects on gastric cancer cells through GSK3β/Rac1-mediated restraint of TGFβ/Smad3 signaling. Oncol. Rep. 2019;41:224–234. doi: 10.3892/or.2018.6838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Ren J., Wang R., Huang G., Song H., Chen Y., Chen L. sFRP1 Inhibits Epithelial–Mesenchymal Transition in A549 Human Lung Adenocarcinoma Cell Line. Cancer Biother. Radiopharm. 2013;28:565–571. doi: 10.1089/cbr.2012.1453. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Garg M. Epithelial-mesenchymal transition—Activating transcription factors—Multifunctional regulators in cancer. World J. Stem Cells. 2013;5:188–195. doi: 10.4252/wjsc.v5.i4.188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Gauger K.J., Chenausky K.L., Murray M.E., Schneider S.S. SFRP1 reduction results in an increased sensitivity to TGF-β signaling. BMC Cancer. 2011;11:59. doi: 10.1186/1471-2407-11-59. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Kawano Y., Diez S., Uysal-Onganer P., Darrington R.S., Waxman J., Kypta R.M. Secreted Frizzled-related protein-1 is a negative regulator of androgen receptor activity in prostate cancer. Br. J. Cancer. 2009;100:1165–1174. doi: 10.1038/sj.bjc.6604976. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Zheng L., Sun D., Fan W., Zhang Z., Li Q., Jiang T. Diagnostic Value of SFRP1 as a Favorable Predictive and Prognostic Biomarker in Patients with Prostate Cancer. PLoS ONE. 2015;10:e0118276. doi: 10.1371/journal.pone.0118276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Xu J., Prosperi J.R., Choudhury N., Olopade O.I., Goss K.H. β-Catenin Is Required for the Tumorigenic Behavior of Triple-Negative Breast Cancer Cells. PLoS ONE. 2015;10:e0117097. doi: 10.1371/journal.pone.0117097. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Martin-Manso G., Calzada M.J., Chuman Y., Sipes J.M., Xavier C.P., Wolf V., Kuznetsova S.A., Rubin J.S., Roberts D.D. sFRP-1 binds via its netrin-related motif to the N-module of thrombospondin-1 and blocks thrombospondin-1 stimulation of MDA-MB-231 breast carcinoma cell adhesion and migration. Arch. Biochem. Biophys. 2011;509:147–156. doi: 10.1016/j.abb.2011.03.004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Cooper S.J., von Roemeling C.A., Kang K.H., Marlow L.A., Grebe S.K., Menefee M.E., Tun H.W., Colon-Otero G., Perez E.A., Copland J.A. Re-expression of tumor suppressor, sFRP1, leads to antitumor synergy of combined HDAC and methyltransferase inhibitors in chemoresistant cancers. Mol. Cancer Ther. 2012;11:2105–2115. doi: 10.1158/1535-7163.MCT-11-0873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91.Joshi M., Liu X., Belani C.P. Taxanes, past, present, and future impact on non-small cell lung cancer. Anticancer Drugs. 2014;25:571–583. doi: 10.1097/CAD.0000000000000080. [DOI] [PubMed] [Google Scholar]
- 92.Ren J., Wang R., Song H., Huang G., Chen L. Secreted Frizzled Related Protein 1 Modulates Taxane Resistance of Human Lung Adenocarcinoma. Mol. Med. 2014;20:164–178. doi: 10.2119/molmed.2013.00149. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Pehlivan M., Sercan Z., Sercan H.O. sFRP1 promoter methylation is associated with persistent Philadelphia chromosome in chronic myeloid leukemia. Leuk. Res. 2009;33:1062–1067. doi: 10.1016/j.leukres.2008.11.013. [DOI] [PubMed] [Google Scholar]
- 94.Pehlivan M., Caliskan C., Yuce Z., Sercan H.O. Forced expression of Wnt antagonists sFRP1 and WIF1 sensitizes chronic myeloid leukemia cells to tyrosine kinase inhibitors. Tumour Biol. 2017;39:1010428317701654. doi: 10.1177/1010428317701654. [DOI] [PubMed] [Google Scholar]
- 95.Menyhárt O., Fekete J.T., Győrffy B. Gene Expression Indicates Altered Immune Modulation and Signaling Pathway Activation in Ovarian Cancer Patients Resistant to Topotecan. Int. J. Mol. Sci. 2019;20:2750. doi: 10.3390/ijms20112750. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Davaadorj M., Imura S., Saito Y., Morine Y., Ikemoto T., Yamada S., Takasu C., Hiroki T., Yoshikawa M., Shimada M. Loss of SFRP1 Expression Is Associated with Poor Prognosis in Hepatocellular Carcinoma. Anticancer Res. 2016;36:659–664. [PubMed] [Google Scholar]
- 97.Chang L., Lei X., Qin Y.U., Zeng G., Zhang X., Jin H., Wang C., Wang X., Su J. Expression and prognostic value of SFRP1 and β-catenin in patients with glioblastoma. Oncol. Lett. 2016;11:69–74. doi: 10.3892/ol.2015.3873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Su H.-Y., Lai H.-C., Lin Y.-W., Chou Y.-C., Liu C.-Y., Yu M.-H. An epigenetic marker panel for screening and prognostic prediction of ovarian cancer. Int. J. Cancer. 2009;124:387–393. doi: 10.1002/ijc.23957. [DOI] [PubMed] [Google Scholar]
- 99.Kumar A., Gosipatala S.B., Pandey A., Singh P. Prognostic Relevance of SFRP1 Gene Promoter Methylation in Colorectal Carcinoma. Asian Pac. J. Cancer Prev. 2019;20:1571–1577. doi: 10.31557/APJCP.2019.20.5.1571. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.Liu X., Fu J., Bi H., Ge A., Xia T., Liu Y., Sun H., Li D., Zhao Y. DNA methylation of SFRP1, SFRP2, and WIF1 and prognosis of postoperative colorectal cancer patients. BMC Cancer. 2019;19:1212. doi: 10.1186/s12885-019-6436-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 101.Sobhani I., Bergsten E., Couffin S., Amiot A., Nebbad B., Barau C. Colorectal cancer-associated microbiota contributes to oncogenic epigenetic signatures. Proc. Natl. Acad. Sci. USA. 2019;116:24285–24295. doi: 10.1073/pnas.1912129116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Huang S., Zhong X., Gao J., Song R., Wu H., Zi S., Yang S., Du P., Cui L., Yang C., et al. Coexpression of SFRP1 and WIF1 as a prognostic predictor of favorable outcomes in patients with colorectal carcinoma. Biomed. Res. Int. 2014;2014:256723. doi: 10.1155/2014/256723. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103.Abd Elrahman M.Z., Nigm D.A., Abo Elfadle A.A. Methylated SFRP1,2 and CD25 Expression in Acute Myeloid Leukemia Play an Important Role in the Pathogenesis of the Disease and in Turn in its Treatment. J. Leuk. 2016;4:2. doi: 10.4172/2329-6917.1000219. [DOI] [Google Scholar]
- 104.Heng L., Jia Z., Bai J., Zhang K., Zhu Y., Ma J., Zhang J., Duan H. Molecular characterization of metastatic osteosarcoma: Differentially expressed genes, transcription factors and microRNAs. Mol. Med. Rep. 2017;15:2829–2836. doi: 10.3892/mmr.2017.6286. [DOI] [PubMed] [Google Scholar]
- 105.Kim T.H., Chang J.S., Park K.-S., Park J., Kim N., Lee J.I., Kong I.D. Effects of exercise training on circulating levels of Dickkpof-1 and secreted frizzled-related protein-1 in breast cancer survivors: A pilot single-blind randomized controlled trial. PLoS ONE. 2017;12:e0171771. doi: 10.1371/journal.pone.0171771. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Chemoprevention of Colorectal Cancer: The Role of Non-digestible Carbohydrates—Full Text View—ClinicalTrials.gov. [(accessed on 26 December 2019)]; Available online: https://clinicaltrials.gov/ct2/show/NCT.
- 107.Malcomson F.C., Willis N.D., McCallum I., Xie L., Ibero-Baraibar I., Leung W.C., Kelly S., Bradburn D.M., Belshaw N.J., Johnson I.T., et al. Effects of supplementation with nondigestible carbohydrates on fecal calprotectin and on epigenetic regulation of SFRP1 expression in the large-bowel mucosa of healthy individuals. Am. J. Clin. Nutr. 2017;105:400–410. doi: 10.3945/ajcn.116.135657. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108.GDC-0449 in Treating Young Patients with Medulloblastoma That Is Recurrent or Did Not Respond to Previous Treatment—Full Text View—ClinicalTrials.gov. [(accessed on 26 December 2019)]; Available online: https://clinicaltrials.gov/ct2/show/NCT.
- 109.Baylin S.B., Herman J., Suzuki H., Sidransky D. Genomic Screen for Epigenetically Silenced Genes Associated with Cancer. [(accessed on 23 December 2019)];2010 Available online: https://patents.google.com/patent/US7794929B2/en?q=sfrp1&status=GRANT.
- 110.Engeland M.V., Bruine M.A.D., Griffioen A., Louwagie J., Bierau K., Britchard G., Otto G., Penning M. Epigenetic Change in Selected Genes and Cancer. [(accessed on 23 December 2019)];2016 Available online: https://patents.google.com/patent/EP2650377B1/en?oq=sfrp1+cancer.
- 111.Lothe R.A., Lind G.E., Ahmed D., Andresen K., Skotheim R.I. Methods and Biomarkers for Detection of Gastrointestinal Cancers. [(accessed on 23 December 2019)];2017 Available online: https://patents.google.com/patent/EP2683834B1/en?q=sfrp1&status=GRANT.