Abstract
Allosteric modulators are ligands that bind to a site on the receptor that is spatially separated from the orthosteric binding site for the endogenous neurotransmitter. Allosteric modulators modulate the binding affinity, potency, and efficacy of orthosteric ligands. Muscarinic acetylcholine receptors are prototypical allosterically-modulated G-protein-coupled receptors. They are a potential therapeutic target for the treatment of psychiatric, neurologic, and internal diseases like schizophrenia, Alzheimer’s disease, Huntington disease, type 2 diabetes, or chronic pulmonary obstruction. Here, we reviewed the progress made during the last decade in our understanding of their mechanisms of binding, allosteric modulation, and in vivo actions in order to understand the translational impact of studying this important class of pharmacological agents. We overviewed newly developed allosteric modulators of muscarinic receptors as well as new spin-off ideas like bitopic ligands combining allosteric and orthosteric moieties and photo-switchable ligands based on bitopic agents.
Keywords: acetylcholine, muscarinic receptors, allosteric modulation
1. Introduction
Slow metabotropic responses to acetylcholine are mediated by muscarinic receptors. Five distinct subtypes of muscarinic acetylcholine receptors (M1–M5) have been identified in the human genome [1]. The structure of all five receptor subtypes was resolved by X-ray crystallography [2,3,4,5,6]. Muscarinic receptors are members of class A of the G-protein-coupled receptor (GPCR). M1, M3, and M5 subtypes preferentially activate phospholipase C and calcium mobilization through Gq/11, whereas M2 and M4 receptors inhibit the activity of adenylyl cyclase by activation of the α-subunit of the Gi/o family of G-proteins. The latter two receptors also modulate the conductance of ion channels (e.g., inward rectifying potassium ion channels) by βγ-dimers of the Gi/o G-proteins [7]. Muscarinic receptors mediate a wide range of physiological functions in the central and peripheral nervous system and innervated tissues. Muscarinic receptors thus represent a potential therapeutic target for the treatment of psychiatric and neurologic conditions (e.g., schizophrenia, Alzheimer’s disease, Huntington disease) [8,9] as well as internal diseases (e.g., type 2 diabetes, asthma, chronic pulmonary obstruction, incontinence) [10,11,12].
The concept of allosterism was formally introduced into the field of enzymology by Monod et al. [13] and Koshland et al. [14] in 1965 and 1966, respectively. The former model was termed concerted, the latter one sequential. Allosteric modulation of GPCR is much simpler than that of enzymes. GPCR allosteric modulators bind to a site on the receptor that is spatially distinct from that of the endogenous transmitter, acetylcholine, in the case of muscarinic receptors. Consequently, binding of an allosteric modulator and an orthosteric ligand is not mutually exclusive, i.e., both ligands may bind to the receptor simultaneously to form a ternary complex (Figure 1). Binding of allosteric modulators induces a change in the conformation of the receptor that results in changes in affinity (eventually potency and efficacy) of the orthosteric ligand [15].
Figure 1.
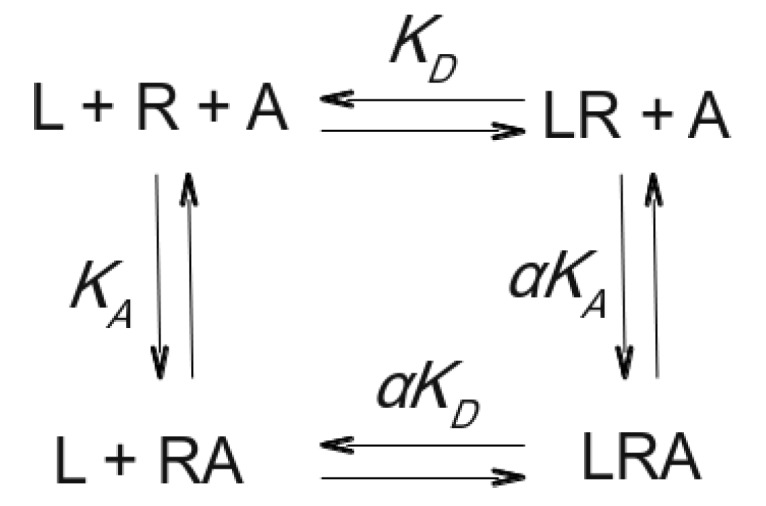
An orthosteric ligand L binds to the receptor R with equilibrium dissociation constant KD, and an allosteric modulator A binds to the receptor R with an equilibrium dissociation constant KA. The orthosteric ligand L and the allosteric modulator A can bind concurrently to the receptor R to form a ternary complex LRA. Binding of one ligand to the receptor changes the equilibrium dissociation constant of the other ligand by a factor of cooperativity α [15].
Based on the effects of an allosteric modulator on the affinity of an orthosteric ligand, allosteric modulators may be classified into three categories: 1. Positive allosteric modulators (PAM) that increase the affinity of orthosteric ligands; 2. Negative allosteric modulators (NAM) that decrease the affinity of orthosteric ligands; and 3. Neutral allosteric modulators that do not affect the affinity of the orthosteric ligand. When the intrinsic efficacy of allosteric modulator is taken into account, these three categories expand to six: 1. Pure PAMs; 2. PAM-agonists that possess intrinsic agonistic propensity in the absence of the orthosteric agonists they modulate; 3. PAM-antagonists that lower the efficacy of the agonists they modulate [16]; 4. Pure NAMs; 5. NAM-agonists that possess own agonistic propensity in the absence of the agonists and activate the receptor, while they negatively modulate endogenous agonist [17]; 6. Silent allosteric modulators (SAMs) that, although they bind to the receptor, do not affect the affinity, potency, or efficacy of the orthosteric ligand and do not have agonistic propensity on their own. In interaction with agonist, an allosteric modulator may affect both agonist affinity and efficacy. Thus, each of the six abovementioned categories has three sub-categories based on positive, negative, or neutral effects (cooperativity) of the allosteric modulator on agonist efficacy. However, six basic categories are sufficient for the general classification.
As early as in 1969, Lüllmann et al. showed in their pioneering work that alkane-bis-ammonium compounds inhibited the functional response to the conventional muscarinic agonist carbachol non-competitively [18]. Later, Clark and Mitchelson discovered that gallamine similarly inhibited the action of acetylcholine and carbachol on the function of heart atria in a non-competitive manner [19]. The concentration-response curves to the agonists were shifted to the right, but the magnitude of the progressive shifts diminished with increasing concentrations of gallamine. When the action of acetylcholine on the heart was evaluated in the combined presence of gallamine and the antagonist atropine, the inhibition of functional response to carbachol was smaller than expected for the effects of two competitive antagonists. These observations led to the conclusion that the action of gallamine takes place at an allosteric site on the receptor, resulting in negative cooperativity with the binding of both orthosteric agonists and antagonists. Since then, a wide variety of allosteric modulators has been discovered. These include inhibitors of acetylcholinesterase, ion channel blockers, various alkaloids, small peptides, etc. For review, see [20]. Thanks to early intensive research, muscarinic receptors became a useful prototype of allosterically-modulated GPCRs.
2. Advantages of Allosteric Modulators as Therapeutics
2.1. Selectivity by Targeting Less Conserved Domains on the Receptor
Muscarinic receptor subtypes share high structural homology in the transmembrane domains where the orthosteric binding site is located. On the other hand, domains out of the membrane are less conserved. Targeting allosteric domains allows achieving binding selectivity for certain receptor subtypes to the extent which is not possible with orthosteric ligands.
2.2. Conservation of Space and Time Pattern of Signaling
Theoretically, a pure PAM of acetylcholine would only induce an action when endogenous acetylcholine is released. Consequently, its action would be restricted in space and time to those synapses where signaling is currently happening. Thus, space and time patterns of signaling could be restored under diminished acetylcholine release that is typical in neurodegenerative disorders, e.g., Alzheimer’s disease. Successful action of PAMs requires functional post-synaptic receptors. Encouragingly, post-synaptic receptors are relatively intact in neurodegenerative diseases like Alzheimer’s disease or senile dementia of Lewy body type [21,22].
2.3. Absolute Selectivity
In practice, absolute selectivity due to exclusive binding to a single receptor subtype is hard to achieve. Alternatively, the absolute selectivity of an allosteric agent can be achieved by having a ligand with the desired cooperativity at a given receptor subtype and neutral (silent) cooperativity at the rest of the receptor subtypes. Selectivity may be derived from binding cooperativity as well as from effects on potency and efficacy.
2.4. Selective Blocking of Activated Receptors
PAM-antagonists that exhibit partial or absolute selectivity for a given receptor subtype may be used to selectively target receptors activated by the endogenous neurotransmitter located in specific tissues or organs [16]. This feature can be used to selectively reverse persistent excessive agonism under certain pathological conditions (e.g., bronchospasm in asthma [23]) or overstimulation of salivary and lacrimal glands after organophosphate poisoning in case of M3-selective PAM-antagonists.
3. Location of the Allosteric Binding Sites on Muscarinic Receptors
The binding site of classical allosteric modulators like gallamine, alcuronium, or alkane-bis-ammonium compounds has been located between the second (o2) and third extracellular (o3) loops (Figure 2, green) [24,25,26]. The charged EDGE motif in the o2 loop plays a critical role in the binding of these ligands [27]. In the crystal structure of the M2 receptor [4], the allosteric modulator LY2119620 (that binds to the same site between o2 and o3) does not form a hydrogen bond to E172 or E175 of the EDGE motif but makes π–π interactions to the adjacent Y177 in the o2 and Y426 in the extracellular edge of TM7.
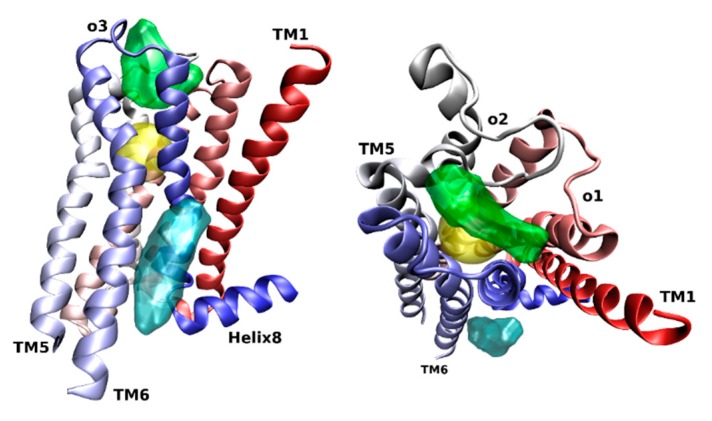
Figure 2.
Side view with TM6 front (left), and extracellular view (right) of the orthosteric (yellow), common allosteric (green), and cholesterol (cyan) binding sites at the M2 receptor (4MQT) (colored in red-white-blue gradient).
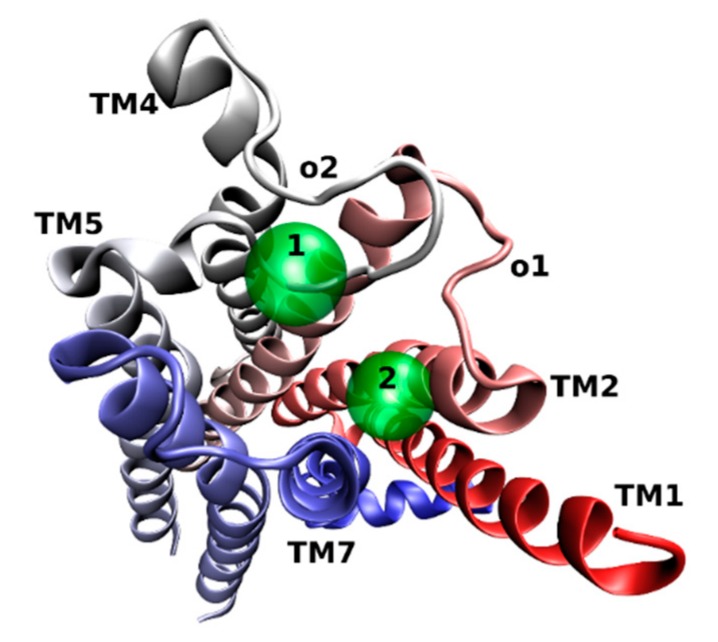
Computer modeling studies of allosteric ligand binding to M2 receptors have revealed two centers for binding of the electropositive part of allosteric ligands (Figure 3) [28]. The first center consists of Y177, N410, N419, and W422, and the second consists of Y80, Y83, T84, and T423. Further, this study showed that alkane-bis-ammonium compounds, gallamine, alcuronium, and strychnine bind to the fist center. M1-selective PAM benzyl quinolone carboxylic acid (BQCA, Figure 4) binds to Y179 and F182 in the o2 loop, and E397 and W400 in TM7 [29]. Thus, the binding site of BQCA overlaps with the common allosteric binding site. In contrast, M4-selective PAM LY2033298 (Figure 4) interacts with the o1, o2, and o3 loops [30]. Key binding residues of LY2033298 are K95 in the o1, F186 in o2, and D432 in o3 (M4 numbering). A cryptic allosteric binding pocket in the extracellular domain that is absent in existing crystal structures has been predicted by simulation of molecular dynamics and confirmed by mutagenesis experiments [31]. This cryptic pocket is dynamically formed in the vicinity of the common allosteric site center 1 by rearrangement of conserved E7.36 in TM7, Y2.64 in the o2 loop close to TM2, and conserved C45.50 in the middle of the o3 loop (general GPCR numbering [32,33]). The site is preferentially formed at the M1 receptor and has been identified as a binding site of highly M1-selective PAM BQZ12 (Figure 4) [34,35].
Figure 3.
Extracellular view of two binding centers (green) in the common allosteric binding site at the M2 receptor (4MQT) (colored in red-white-blue gradient).
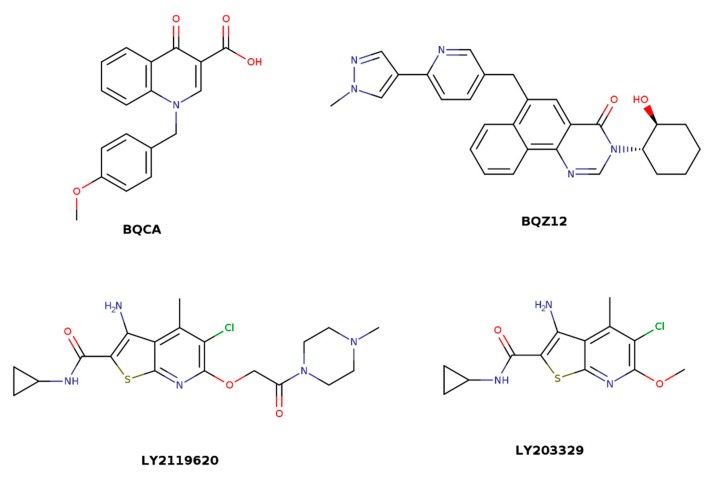
Figure 4.
Structures of M1-selective benzyl quinolone carboxylic acid (BQCA), BQZ12, M2-selective LY2119620, and M4-selective LY2033298 allosteric modulator.
While the majority of known muscarinic allosteric ligands bind to the site between the o2 and o3 loops, sterol-based WIN-compounds have been found to interact with gallamine and strychnine in a non-competitive manner [36]. Thus, the binding site for WIN-compounds is not between the o2 and o3 loops but somewhere else. However, the precise location of the WIN-compound binding site has not been determined yet. Interestingly, cholesterol also allosterically modulates the binding and function of muscarinic receptors [37,38,39,40]. Using site-directed mutagenesis, the binding site for membrane cholesterol has been located to the groove between TM6 and TM7 in the intracellular leaflet of the membrane [40]. Membrane cholesterol allosterically modulates many GPCRs [41]. Cholesterol has co-crystallized with GPCRs at various sites, both in the extracellular and intracellular leaflet of the membrane [41]. Computer modeling of the interaction between membrane cholesterol and GPCR suggests the possibility of several cholesterol binding sites per one molecule of GPCR [42]. How many cholesterol binding sites muscarinic receptors do have and whether sterol-based WIN-compounds bind to the cholesterol-binding site remains to be elucidated.
4. Molecular Mechanisms of Action of Allosteric Modulators
The maximum magnitude of the effects of an allosteric modulator at a given receptor (binding cooperativity) varies from one orthosteric ligand to another. Comparison of the binding cooperativity of a given allosteric ligand with several orthosteric ligands has suggested that cooperativity is dependent on the distance between the electropositive and electronegative part of the orthosteric ligand [43]. Thus, data has suggested that allosteric modulators change the distance between TM3 interacting with electropositive and TM6 interacting with an electronegative part of the orthosteric ligand. This hypothesis was confirmed later by crystal structures and molecular modeling [4,44]. Numerous studies have identified differential key amino acids to govern allosteric action of various allosteric modulators, suggesting the existence of multiple allosteric switches on muscarinic receptors. For example, M4-selective PAM LY2033298 gains its efficacy from interaction with K95 in the o1 loop, and its binding cooperativity results from interaction with F186 in the o2 loop (M4 numbering) [30]. In contrast, tyrosine in the o2 loop (Y179 in M1, Y177 in M2) and tryptophan in TM7 (W7.35, Ballesteros-Weinstein numbering [45]) have been identified as key residues, defining the magnitude of binding cooperativity of M1-selective PAM of acetylcholine (BQCA) as well as M2-selective PAM of iperoxo (LY2119620, Figure 4) [4,29,34]. The common feature found for all PAMs is shrinkage of the vestibule to the orthosteric binding site that is accompanied by the closure of the binding pocket [46]. Divergent mechanisms underlying how shrinkage of the vestibule is achieved probably represent the molecular basis of PAM receptor subtype selectivity.
Many GPCRs, including muscarinic receptors, activate several signaling pathways upon activation by agonists [47,48,49]. It is generally accepted that structurally different agonists induce specific changes in the conformation of GPCRs that can lead to non-uniform modulation of signaling pathways. This preferential orientation of the signaling of a GPCR towards a subset of its signal transducers is termed signaling bias [50]. In principle, the structure of the receptor in a ternary complex with agonist and the allosteric modulator is distinct from the one in a binary complex with agonist only and may result in biased signaling too. Attaining signaling bias via allosteric modulation could bring a new plethora of possibilities to modulate receptor function. It has been shown at M1 receptors that the allosteric modulator VU0029767 (Figure 5) acts as a strong PAM of acetylcholine-induced intracellular calcium mobilization while its effects on acetylcholine-induced activation of phospholipase D were marginal [51]. Based on this observation, it has been suggested that differential modulation of receptor coupling to downstream signaling pathways by allosteric modulators may result in signaling bias. However, structurally divergent M1 PAMs BQCA (Figure 4) and MIPS1674 (Figure 5) have shown only small differences in the modulation of the inositol phosphate, β-arrestin, and ERK1/2 pathways [52]. Residues near the common allosteric binding site have been implicated in mediating biased signaling even of orthosteric ligands at several GPCRs [53]. Thus, divergent mechanisms of shrinkage of the vestibule to the binding pocket may not only stand behind subtype-selective effects of allosteric modulators but also drive ligand bias that is common to the entire A class of GPCRs [54].
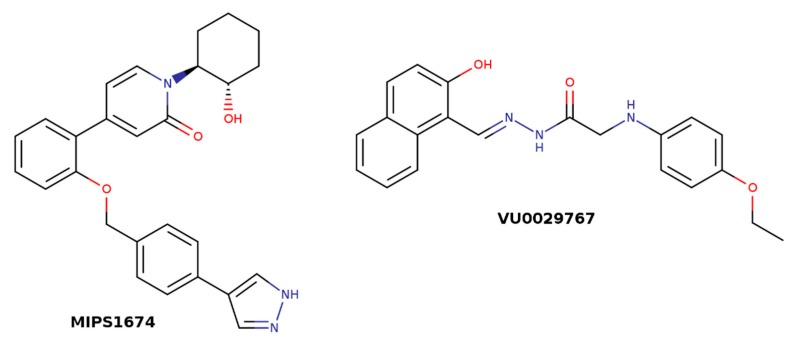
Figure 5.
Structures of M1 allosteric modulators MIPS1674 and VU0029767.
5. Role of the Common Allosteric Binding Site in the Binding of Orthosteric Ligands
The common allosteric binding site located between the o2 and o3 loops represents the vestibule to the orthosteric binding site. The model in which an orthosteric ligand binds transiently to a secondary site before moving to the orthosteric binding site (so-called tandem two-site model [55]) has been proposed to explain apparent receptor isomerization upon antagonist binding [56,57]. Interaction of orthosteric ligands with the allosteric domain between the o2 and o3 loops has been confirmed and studied in detail by computer modeling [3,58,59]. These studies showed that two-step binding is common for all orthosteric ligands studied and that orthosteric ligands interact primarily with Y177 (common allosteric center 1), N410, N419, and W422 (common allosteric center 2) (M2 numbering). Subtype variations of affinity and kinetics of orthosteric ligands may be attributed to variation in their interaction with the allosteric site [6].
The strong interaction of tiotropium with the allosteric binding site also explains the discrepancy between extremely slow binding kinetics and immediate inhibitory action of this bronchodilator [60]. Tiotropium binding to the allosteric site is fast and results in antagonism of the functional effects of the M3 receptor.
The M2-selective antagonist methoctramine competitively inhibits the binding of orthosteric ligands. High concentrations slow down the dissociation of [3H]N-methylscopolamine. Detailed analysis of methoctramine binding has revealed two modes of interaction with the receptor. Transient binding to the common allosteric site center 1 (namely E175 in the o2 loop) is followed by stable binding that occurs concurrently to both the allosteric and orthosteric sites [61]. Methoctramine is thus a bitopic dualsteric antagonist.
6. Bitopic Ligands
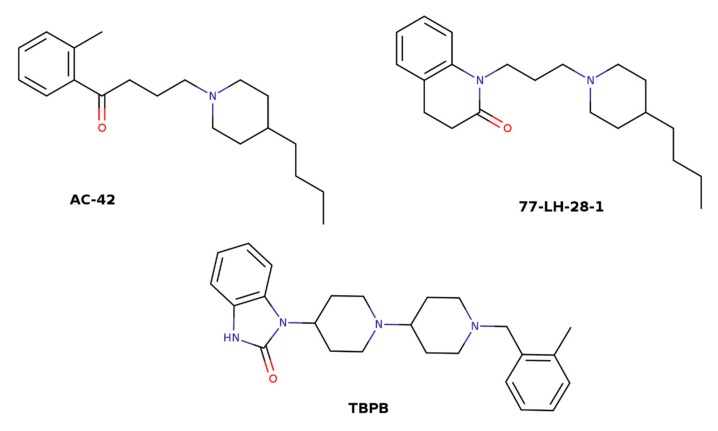
The idea of utilizing high efficacy of orthosteric ligands and subtype diversity of the allosteric binding sites has led to the development of bitopic ligands that span both sites and interact with them concurrently. In principle, a bitopic ligand consists of two molecules, one targeting the orthosteric site, and the other interacting with the allosteric site. The two parts are connected by a linker of a proper length. A combination of the nonselective muscarinic agonist iperoxo and its M1-selective PAM BQCA has led to a set of M1-selective agonists with graded efficacy that is dependent on the chemical structure of the linker [62]. The orientation of the orthosteric moiety within the orthosteric binding site is crucial for subtype selectivity [63]. Therefore, the structure of the linker affects not only efficacy but also the selectivity of bitopic ligands. Another example of a bitopic agonist that gains its selectivity from interaction with the common allosteric binding site is TBPB (Figure 6) [64,65]. Mutations of the allosteric and orthosteric binding sites have revealed that NDMC (N-desmethylclozapine) that is considered an allosteric agonist and McN-A-343 (4-(m-chlorophenyl-carbamoyloxy)-2-butynyltri-methylammonium) that is considered an orthosteric agonist are in fact bitopic ligands [66,67]. Allosteric effects of the bitopic agonists McN-A-343 and NDMC and the allosteric agonists AC-42 and 77-LH-28-1 (Figure 6) are mediated via Y177 in the o2 loop of the M2 receptor [66]. Taken together, these bitopic ligands also interact with the center 1 (Figure 3) of the common allosteric binding site (Figure 2).
Figure 6.
Structures of bitopic ligands AC-42, 77-LH-28-1, and TBPB.
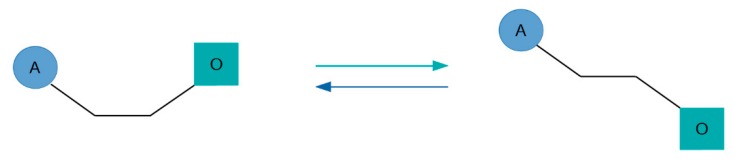
Further development of bitopic ligands has led to the discovery of the photo-switchable ligands [68]. In the case of a photo-switchable ligand, the conformation of the linker is sensitive to exposure to light of a specific wavelength, which leads to the isomerization of the linker and change in the mutual orientation of orthosteric and allosteric moieties (Figure 7). In one linker conformation, the orthosteric and allosteric moieties are in an orientation that does not allow simultaneous interaction of individual moieties with their respective binding sites that makes a ligand inactive. After linker isomerization, the orientation of individual moieties allows simultaneous interaction of individual moieties with their respective binding sites, and the ligand becomes active. Photo-switchable ligands are an invaluable tool in basic research since they allow an experimenter to apply ligand at the desired place and desired time in the blink of a laser.
Figure 7.
Scheme of isomerization of photo-switchable ligand, consisting of the orthosteric (O) and allosteric (A) moiety connected by the linker.
Besides being an interesting research tool, bitopic muscarinic ligands, consisting of the orthosteric and allosteric moiety, have also a translational potential. For example, bis(ammonio)alkane-type compounds (hybrid molecules of the orthosteric agonist iperoxo and allosteric moiety derived from naphmethonium or W84) can be used as non-opioid non-steroidal analgesics [69] or inhibitors of cell growth for glioblastoma therapy [70].
7. Novel Allosteric Modulators
Although positive allosteric modulation of acetylcholine was presented as proof of concept in the 1990s [71,72], it took more than 10 years to develop first PAMs with physiologically active properties suitable for further drug development [73,74,75,76]. However, the progress of research on muscarinic allosteric modulators has accelerated, and selective PAMs for each subtype have been discovered during the last decade.
Namely, two new classes of M1-selective PAMs have been developed. Derivatives of heterocyclic carboxamides display various degrees of intrinsic activity and various modulatory profiles of affinity and efficacy of acetylcholine [77]. Similarly, derivatives of 4-phenylpyridin-2-one (Figure 8) display a diverse range of activities, ranging from pure PAMs to pure allosteric agonists [78]. All 4-phenylpyridin-2-one derivatives have retained exquisite selectivity for the M1 receptor.
Figure 8.
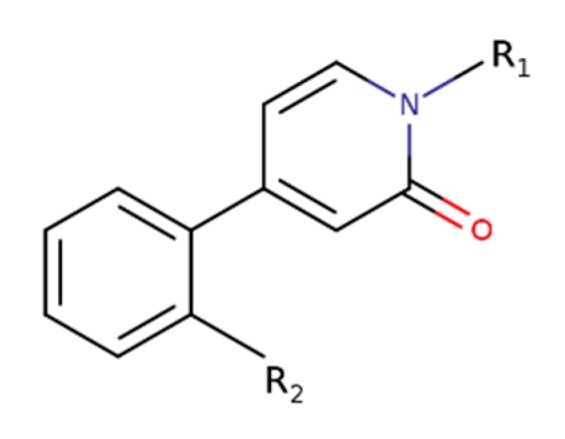
The core structure of 4-phenylpyridin-2-one derivatives.
The availability of the M2 receptor crystal structure in an inactive [2], as well as active conformation [4] and advances in computer modeling, has allowed for the acceleration of structure-based design of chemically diverse allosteric modulators of this receptor [79]. The combination of in silico simulation of molecular dynamics and virtual screening has led to the identification of allosteric modulators of the M2 receptor of chemically novel structures that have been verified as NAMs and PAMs of superagonist iperoxo in binding experiments.
Positive allosteric modulators of antagonists may turn otherwise a non-selective orthosteric antagonist to the subtype-selective antagonist with therapeutic potential. Recently, as a proof of concept, M2-selective positive modulators of orthosteric antagonists have been developed [80]. In this study, triazolo-quinazolinone analogs were identified as M2 selective positive modulators of N-methylscopolamine (NMS) by docking a large library of molecules to the allosteric binding site of the M2 receptor in an inactive conformation. Resulting allosteric modulators increased NMS affinity about 5-times and slowed NMS dissociation from the M2 receptor about 50-fold while having no such effects at the other subtypes of muscarinic receptors.
Testing of novel muscarinic allosteric modulators in animal models of human diseases has shown their good efficacy. Cholinergic neurons regulate glutamatergic neurons in the striatum that regulate selection and decision-making behavior [81]. The M1-selective PAM of acetylcholine BQCA has improved efficacy of the antipsychotics haloperidol, clozapine, and aripiprazole in the glutamatergic deficit mouse model of behavior [82]. M1 receptors play a critical role in cognitive processes [7]. Thus, positive allosteric modulation of M1 receptors appears to be the way to treat cognitive deficits in Alzheimer’s disease or schizophrenia [83]. However, cortex, hippocampus, and striatum post mortem samples from some patients with schizophrenia have shown a profound loss of muscarinic receptors and decreased responsiveness to M1-selective PAM of acetylcholine BQCA. These findings may explain why some individuals with schizophrenia may not respond to such [84].
The striatal M4 receptors attenuate dopaminergic and glutamatergic neurotransmission [85]. Thus, selective positive modulation of M4 receptors may provide a novel treatment strategy of psychotic symptoms in schizophrenia that are considered to result from dopaminergic hyperactivity [83]. A series of M4 PAMs have been found to be centrally active and efficient in an animal model of schizophrenia [86]. Huntington’s disease results from increased transmission at glutamatergic corticostriatal synapses that may be attenuated by the cholinergic system. In accordance, M4 PAMs have improved behavioral symptoms in the mouse model of Huntington disease [76].
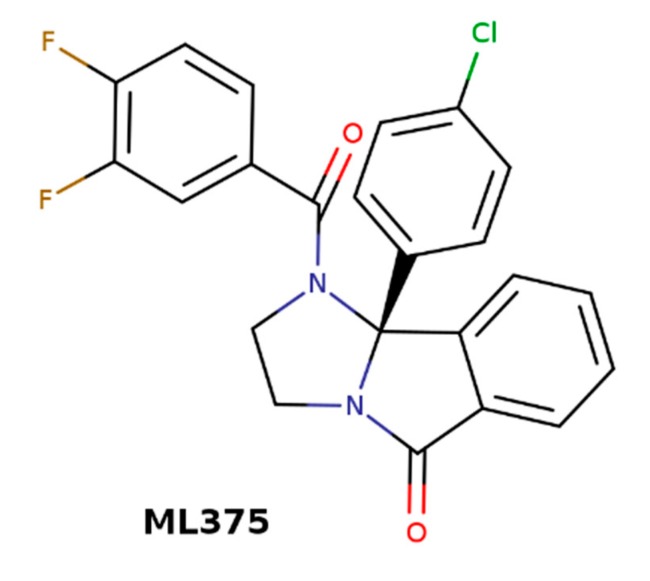
M5 muscarinic receptors are expressed solely in the substantia nigra and ventral tegmental area (VTA) [7]. Cholinergic neurons in the nucleus accumbens (NAcc) project to VTA, where they stimulate dopaminergic neurons. The VTA dopaminergic neurons project back to NAcc, where they stimulate cholinergic neurons. This positive feedback works as the reward circuit [87]. A blockade or negative allosteric modulation of M5 receptors could break the reward cycle and prevent addiction. The M5-selective NAM of acetylcholine ML375 (Figure 9) [88] has effectively prevented cocaine self-administration and the development of addiction in rats [89].
Figure 9.
Structure of M5-selective negative allosteric modulators (NAM) of acetylcholine ML375.
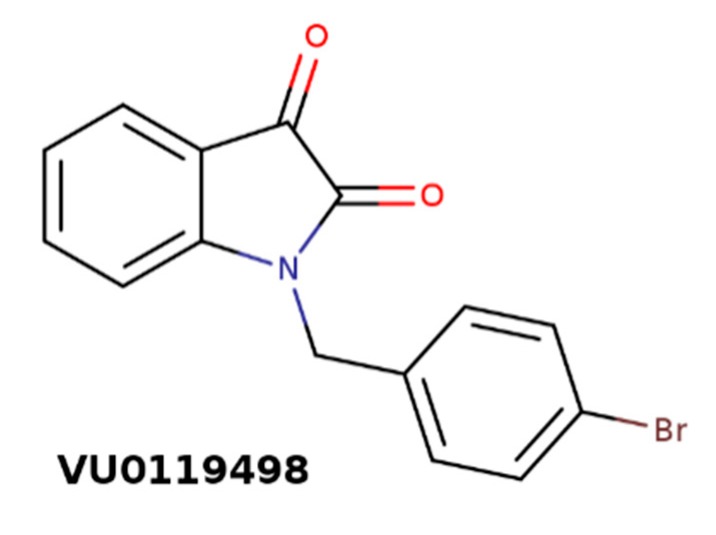
In type 2 diabetes, pancreatic β cells are unable to release sufficient amounts of insulin to maintain physiological blood glucose levels. Parasympathetic activation of pancreatic β cells by acetylcholine stimulates the release of insulin [90]. Effects of acetylcholine on insulin secretion are mediated by the M3 receptor [91]. In accordance, M3 PAM of acetylcholine VU0119498 (Figure 10) has been shown to enhance glucose-stimulated insulin secretion and greatly improve glucose tolerance in lean and obese glucose-intolerant mice [92]. These effects were absent from mice lacking M3 receptors in their β cells.
Figure 10.
Structure of M3-selective positive allosteric modulators (PAM) of acetylcholine VU0119498.
8. Perspectives
Recent findings of the diversity of actions of allosteric modulators and bitopic ligands of muscarinic receptors have greatly influenced the development of new therapies for a variety of disorders. Recent discoveries of selective positive allosteric modulators of acetylcholine with therapeutic potential in the treatment of psychiatric and neurologic disorders like Alzheimer’s or schizophrenia, as well as internal disorders like type 2 diabetes, are encouraging. Research on several muscarinic allosteric modulators has surpassed the stage of optimization of selectivity, efficacy, and bioavailability and advanced to a translational stage of medicinal research. In the field of basic research, further progress is mainly achieved by the introduction of novel concepts and techniques. Novel photo-switchable ligands may greatly increase the accuracy of timely delivery as well as allow varied permutations of experimental design. Thus, focus on the design of novel photo-switchable ligands is currently gaining momentum.
Nomenclature
| Compounds | Chemical Names |
| 77-LH-28-1 | 1-[3-(4-Butylpiperidino)propyl]-3,4-dihydroquinoline-2(1H)-one |
| AC-42 | 4-(4-Butylpiperidin-1-yl)-1-o-tolylbutan-1-one |
| Alcuronium | 2-[(1S,9Z,11S,13S,17S,25Z,27S,33S,35S,36S)-38-(2-hydroxyethylidene)-14,30-bis(prop-2-enyl)-8,24-diaza-14,30-diazoniaundecacyclo [2 5.5.2.211,14.11,26.110,17.02,7.013,17.018,23.030,33.08,35.024,36]octatriaconta-2,4,6,9,18,20,22,25-octaen-28-ylidene]ethanol |
| BQCA | 1-(4-Methoxybenzyl)-4-oxo-1,4-dihydroquinoline-3-carboxylic acid |
| BQZ12 | 3-((1S,2S)-2-hydroxycyclohexyl)-6-((6-(1-methyl-1H-pyrazol-4-yl)pyridin-3-yl)methyl)benzo [h]quinazolin-4(3H)-one |
| Gallamine | 2-[2,3-bis [2-(triethylazaniumyl)ethoxy]phenoxy]ethyl-triethylazanium |
| LY2119620 | 3-Amino-5-chloro-N-cyclopropyl-4-methyl-6-[2-(4-methylpiperazin-1-yl)-2-oxoethoxy]thieno [2,3-b]pyridine-2-carboxamide |
| LY2033298 | 3-Amino-5-Chloro-N-Cyclopropyl-6-Methoxy-4-Methylthieno [2,3-B]Pyridine-2-Carboxamide |
| McN-A-343 | 4-(m-chlorophenyl-carbamoyloxy)-2-butynyltri-methylammonium |
| Methoctramine | N,N′-bis [6-[[(2-methoxyphenyl)-methyl]hexyl]-1,8-octane]diamine |
| MIPS1674 | 1-[(1S,2S)-2-hydroxycyclohexyl]-4-[2-[[4-(1H-pyrazol-4-yl)phenyl]methoxy]phenyl]pyridin-2-one |
| ML375 | (S)-9b-(4-chlorophenyl)-1-(3,4-difluorobenzoyl)-2,3- dihydro-1H-imidazo [2,1-a]isoindol-5(9bH)-one |
| Naphmethonium | [3-(1,3-dioxobenzo [de]isoquinolin-2-yl)-2,2-dimethylpropyl]-[6-[3-(1,3-dioxoisoindol-2-yl)propyl-dimethylazaniumyl]hexyl]-dimethylazanium |
| NDMC | N-desmethylclozapine; 3-chloro-6-piperazin-1-yl-11H-benzo [b] [1,4]benzodiazepine |
| NMS | N-methylscopolamine; [(1S,2S,4R,5R)-9,9-dimethyl-3-oxa-9-azoniatricyclo [3.3.1.02,4]nonan-7-yl] (2S)-3-hydroxy-2-phenylpropanoate |
| Strychnine | (4aR,5aS,8aR,13aS,15aS,15bR)-4a,5,5a,7,8,13a,15,15a,15b,16-decahydro-2H-4,6-methanoindolo [3,2,1-ij]oxepino [2,3,4-de]pyrrolo [2,3-h]quinolin-14-one |
| TBPB | 1-(1′-(2-methylbenzyl)-1,4′-bipiperidin-4-yl)-1H-benzo [d]imidazol-2(3H)-one |
| Tiotropium | [(1S,2S,4R,5R)-9,9-dimethyl-3-oxa-9-azoniatricyclo [3.3.1.02,4]nonan-7-yl]2-hydroxy-2,2-dithiophen-2-ylacetate |
| VU0119498 | 1-(4-bromobenzyl)indole-2,3-dione |
| VU0029767 | (E)-2-(4-ethoxyphenylamino)-N′-((2-hydroxynaphthalen-1-yl)methylene)acetohydrazide |
| W84 | 6-[dimethyl-[3-(4-methyl-1,3-dioxoisoindol-2-yl)propyl]azaniumyl]hexyl-dimethyl-[3-(4-methyl-1,3-dioxoisoindol-2-yl)propyl]azanium |
Author Contributions
J.J. and E.E.E.-F. contributed to the review equally. All authors have read and agreed to the published version of the manuscript.
Funding
This research was funded by the Czech Academy of Sciences institutional support [RVO:67985823] and the Grant Agency of the Czech Republic grant [19-05318S].
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Bonner T.I., Buckley N.J., Young A.C., Brann M.R. Identification of a family of muscarinic acetylcholine receptor genes. Science. 1987;237:527–532. doi: 10.1126/science.3037705. [DOI] [PubMed] [Google Scholar]
- 2.Haga K., Kruse A.C., Asada H., Yurugi-Kobayashi T., Shiroishi M., Zhang C., Weis W.I., Okada T., Kobilka B.K., Haga T., et al. Structure of the human M2 muscarinic acetylcholine receptor bound to an antagonist. Nature. 2012;482:547–551. doi: 10.1038/nature10753. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Kruse A.C., Hu J., Pan A.C., Arlow D.H., Rosenbaum D.M., Rosemond E., Green H.F., Liu T., Chae P.S., Dror R.O., et al. Structure and dynamics of the M3 muscarinic acetylcholine receptor. Nature. 2012;482:552–556. doi: 10.1038/nature10867. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kruse A.C., Ring A.M., Manglik A., Hu J., Hu K., Eitel K., Hübner H., Pardon E., Valant C., Sexton P.M., et al. Activation and allosteric modulation of a muscarinic acetylcholine receptor. Nature. 2013;504:101–106. doi: 10.1038/nature12735. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Thal D.M., Sun B., Feng D., Nawaratne V., Leach K., Felder C.C., Bures M.G., Evans D.A., Weis W.I., Bachhawat P., et al. Crystal structures of the M1 and M4 muscarinic acetylcholine receptors. Nature. 2016;531:335–340. doi: 10.1038/nature17188. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Vuckovic Z., Gentry P.R., Berizzi A.E., Hirata K., Varghese S., Thompson G., van der Westhuizen E.T., Burger W.A.C., Rahmani R., Valant C., et al. Crystal structure of the M5 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2019;116:26001–26007. doi: 10.1073/pnas.1914446116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Eglen R.M. Handb Exp Pharmacol. Springer; Heidelberg, Germany: 2012. Overview of muscarinic receptor subtypes; pp. 3–28. [DOI] [PubMed] [Google Scholar]
- 8.Conn P.J., Jones C.K., Lindsley C.W. Subtype-selective allosteric modulators of muscarinic receptors for the treatment of CNS disorders. Trends Pharmacol Sci. 2009;30:148–155. doi: 10.1016/j.tips.2008.12.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Bock A., Schrage R., Mohr K. Allosteric modulators targeting CNS muscarinic receptors. Neuropharmacology. 2018;136:427–437. doi: 10.1016/j.neuropharm.2017.09.024. [DOI] [PubMed] [Google Scholar]
- 10.Gautam D., Han S.-J., Duttaroy A., Mears D., Hamdan F.F., Li J.H., Cui Y., Jeon J., Wess J. Role of the M3 muscarinic acetylcholine receptor in beta-cell function and glucose homeostasis. Diabetes Obes. Metab. 2007;9:158–169. doi: 10.1111/j.1463-1326.2007.00781.x. [DOI] [PubMed] [Google Scholar]
- 11.Gosens R., Zaagsma J., Meurs H., Halayko A.J. Muscarinic receptor signaling in the pathophysiology of asthma and COPD. Respir. Res. 2006;7:73. doi: 10.1186/1465-9921-7-73. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Hegde S.S. Muscarinic receptors in the bladder: From basic research to therapeutics. Br. J. Pharmacol. 2006;147:S80–S87. doi: 10.1038/sj.bjp.0706560. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Monod J., Wyman J., Changeux J.P. On the nature of allosteric transitions: A plausible model. J. Mol. Biol. 1965;12:88–118. doi: 10.1016/S0022-2836(65)80285-6. [DOI] [PubMed] [Google Scholar]
- 14.Koshland D.E., Némethy G., Filmer D. Comparison of experimental binding data and theoretical models in proteins containing subunits. Biochemistry. 1966;5:365–385. doi: 10.1021/bi00865a047. [DOI] [PubMed] [Google Scholar]
- 15.Jakubík J., El-Fakahany E.E. Allosteric Modulation of Muscarinic Receptors. In: Myslivecek J., Jakubik J., editors. Muscarinic Receptor: From Structure to Animal Models. Volume 107. Neuromethods; Humana Press; Springer Science + Business Media LLC; New York, NY, USA: 2016. pp. 95–130. [Google Scholar]
- 16.Kenakin T., Strachan R.T. PAM-Antagonists: A Better Way to Block Pathological Receptor Signaling? Trends Pharmacol. Sci. 2018;39:748–765. doi: 10.1016/j.tips.2018.05.001. [DOI] [PubMed] [Google Scholar]
- 17.Jakubík J., Bacáková L., Lisá V., El-Fakahany E.E., Tucek S. Activation of muscarinic acetylcholine receptors via their allosteric binding sites. Proc. Natl. Acad. Sci. USA. 1996;93:8705–8709. doi: 10.1073/pnas.93.16.8705. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Lüllmann H., Ohnesorge F.K., Schauwecker G.C., Wassermann O. Inhibition of the actions of carbachol and DFP on guinea pig isolated atria by alkane-bis-ammonium compounds. Eur. J. Pharmacol. 1969;6:241–247. doi: 10.1016/0014-2999(69)90181-2. [DOI] [PubMed] [Google Scholar]
- 19.Clark A.L., Mitchelson F. The inhibitory effect of gallamine on muscarinic receptors. Br. J. Pharmacol. 1976;58:323–331. doi: 10.1111/j.1476-5381.1976.tb07708.x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Jakubík J., El-Fakahany E.E. Allosteric Modulation of Muscarinic Acetylcholine Receptors. Pharmaceuticals. 2010;3:2838–2860. doi: 10.3390/ph3092838. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Mash D.C., Flynn D.D., Potter L.T. Loss of M2 muscarine receptors in the cerebral cortex in Alzheimer’s disease and experimental cholinergic denervation. Science. 1985;228:1115–1117. doi: 10.1126/science.3992249. [DOI] [PubMed] [Google Scholar]
- 22.Perry E.K., Smith C.J., Court J.A., Perry R.H. Cholinergic nicotinic and muscarinic receptors in dementia of Alzheimer, Parkinson and Lewy body types. J. Neural Transm. Park. Dis. Dement. Sect. 1990;2:149–158. doi: 10.1007/BF02257646. [DOI] [PubMed] [Google Scholar]
- 23.Buels K.S., Fryer A.D. Muscarinic receptor antagonists: Effects on pulmonary function. Handb. Exp. Pharmacol. 2012;208:317–341. doi: 10.1007/978-3-642-23274-9_14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Krejčí A., Tuček S. Changes of cooperativity between N-methylscopolamine and allosteric modulators alcuronium and gallamine induced by mutations of external loops of muscarinic M(3) receptors. Mol. Pharmacol. 2001;60:761–767. [PubMed] [Google Scholar]
- 25.Huang X.-P., Prilla S., Mohr K., Ellis J. Critical amino acid residues of the common allosteric site on the M2 muscarinic acetylcholine receptor: more similarities than differences between the structurally divergent agents gallamine and bis(ammonio)alkane-type hexamethylene-bis-[dimethyl-(3-phthalimidopropyl)ammonium]dibromide. Mol. Pharmacol. 2005;68:769–778. doi: 10.1124/mol.105.014043. [DOI] [PubMed] [Google Scholar]
- 26.Jakubík J., Krejcí A., Dolezal V. Asparagine, valine, and threonine in the third extracellular loop of muscarinic receptor have essential roles in the positive cooperativity of strychnine-like allosteric modulators. J. Pharmacol. Exp. Ther. 2005;313:688–696. doi: 10.1124/jpet.104.080358. [DOI] [PubMed] [Google Scholar]
- 27.Leppik R.A., Miller R.C., Eck M., Paquet J.L. Role of acidic amino acids in the allosteric modulation by gallamine of antagonist binding at the m2 muscarinic acetylcholine receptor. Mol. Pharmacol. 1994;45:983–990. [PubMed] [Google Scholar]
- 28.Dror R.O., Green H.F., Valant C., Borhani D.W., Valcourt J.R., Pan A.C., Arlow D.H., Canals M., Lane J.R., Rahmani R., et al. Structural basis for modulation of a G-protein-coupled receptor by allosteric drugs. Nature. 2013;503:295–299. doi: 10.1038/nature12595. [DOI] [PubMed] [Google Scholar]
- 29.Abdul-Ridha A., López L., Keov P., Thal D.M., Mistry S.N., Sexton P.M., Lane J.R., Canals M., Christopoulos A. Molecular determinants of allosteric modulation at the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 2014;289:6067–6079. doi: 10.1074/jbc.M113.539080. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Nawaratne V., Leach K., Felder C.C., Sexton P.M., Christopoulos A. Structural determinants of allosteric agonism and modulation at the M4 muscarinic acetylcholine receptor: Identification of ligand-specific and global activation mechanisms. J. Biol. Chem. 2010;285:19012–19021. doi: 10.1074/jbc.M110.125096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hollingsworth S.A., Kelly B., Valant C., Michaelis J.A., Mastromihalis O., Thompson G., Venkatakrishnan A.J., Hertig S., Scammells P.J., Sexton P.M., et al. Cryptic pocket formation underlies allosteric modulator selectivity at muscarinic GPCRs. Nat. Commun. 2019;10:3289. doi: 10.1038/s41467-019-11062-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three dimensional models and computational probing of structure function relations in G protein-coupled receptors. In: Sealfon S.C., Conn P.M., editors. Methods in Neurosciences. Volume 25. Academic Press; San Diego, CA, USA: 1995. pp. 366–428. [Google Scholar]
- 33.Isberg V., De Graaf C., Bortolato A., Cherezov V., Katritch V., Marshall F.H., Mordalski S., Pin J.-P.P., Stevens R.C., Vriend G., et al. Generic GPCR residue numbers—Aligning topology maps while minding the gaps. Trends Pharmacol. Sci. 2015;36:22–31. doi: 10.1016/j.tips.2014.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Abdul-Ridha A., Lane J.R., Mistry S.N., López L., Sexton P.M., Scammells P.J., Christopoulos A., Canals M. Mechanistic insights into allosteric structure-function relationships at the M1 muscarinic acetylcholine receptor. J. Biol. Chem. 2014;289:33701–33711. doi: 10.1074/jbc.M114.604967. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Bradley S.J., Bourgognon J.M., Sanger H.E., Verity N., Mogg A.J., White D.J., Butcher A.J., Moreno J.A., Molloy C., Macedo-Hatch T., et al. M1 muscarinic allosteric modulators slow prion neurodegeneration and restore memory loss. J. Clin. Investig. 2016;127:487–499. doi: 10.1172/JCI87526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Lazareno S., Popham A., Birdsall N.J.M. Analogs of WIN 62,577 define a second allosteric site on muscarinic receptors. Mol. Pharmacol. 2002;62:1492–1505. doi: 10.1124/mol.62.6.1492. [DOI] [PubMed] [Google Scholar]
- 37.Michal P., Rudajev V., El-Fakahany E.E., Doležal V. Membrane cholesterol content influences binding properties of muscarinic M2 receptors and differentially impacts activation of second messenger pathways. Eur. J. Pharmacol. 2009;606:50–60. doi: 10.1016/j.ejphar.2009.01.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Michal P., El-Fakahany E.E., Doležal V. Changes in Membrane Cholesterol Differentially Influence Preferential and Non-preferential Signaling of the M1 and M3 Muscarinic Acetylcholine Receptors. Neurochem. Res. 2014;40:2068–2077. doi: 10.1007/s11064-014-1325-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Janickova H., Rudajev V., Dolejsi E., Koivisto H., Jakubik J., Tanila H., El-Fakahany E.E., Dolezal V. Lipid-Based Diets Improve Muscarinic Neurotransmission in the Hippocampus of Transgenic APPswe/PS1dE9 Mice. Curr. Alzheimer Res. 2015;12:923–931. doi: 10.2174/1567205012666151027130350. [DOI] [PubMed] [Google Scholar]
- 40.Randáková A., Dolejší E., Rudajev V., Zimčík P., Doležal V., El-Fakahany E.E., Jakubík J. Role of membrane cholesterol in differential sensitivity of muscarinic receptor subtypes to persistently bound xanomeline. Neuropharmacology. 2018;133:129–144. doi: 10.1016/j.neuropharm.2018.01.027. [DOI] [PubMed] [Google Scholar]
- 41.Gimpl G. Interaction of G protein coupled receptors and cholesterol. Chem. Phys. Lipids. 2016;199:61–73. doi: 10.1016/j.chemphyslip.2016.04.006. [DOI] [PubMed] [Google Scholar]
- 42.Lee A.G. Interfacial Binding Sites for Cholesterol on G Protein-Coupled Receptors. Biophys. J. 2019;116:1586–1597. doi: 10.1016/j.bpj.2019.03.025. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zlotos D.P., Buller S., Stiefl N., Baumann K., Mohr K. Probing the pharmacophore for allosteric ligands of muscarinic M2 receptors: SAR and QSAR studies in a series of bisquaternary salts of caracurine V and related ring systems. J. Med. Chem. 2004;47:3561–3571. doi: 10.1021/jm0311341. [DOI] [PubMed] [Google Scholar]
- 44.Bermudez M., Wolber G. Structure versus function—The impact of computational methods on the discovery of specific GPCR-ligands. Bioorganic Med. Chem. 2015;23:3907–3912. doi: 10.1016/j.bmc.2015.03.026. [DOI] [PubMed] [Google Scholar]
- 45.Ballesteros J.A., Weinstein H. Integrated methods for the construction of three-dimensional models and computational probing of structure-function relations in G protein-coupled receptors. Methods Neurosci. 1995;25:366–428. [Google Scholar]
- 46.Burger W.A.C., Sexton P.M., Christopoulos A., Thal D.M. Toward an understanding of the structural basis of allostery in muscarinic acetylcholine receptors. J. Gen. Physiol. 2018;150:1360–1372. doi: 10.1085/jgp.201711979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Laugwitz K.L., Allgeier A., Offermanns S., Spicher K., Van Sande J., Dumont J.E., Schultz G. The human thyrotropin receptor: A heptahelical receptor capable of stimulating members of all four G protein families. Proc. Natl. Acad. Sci. USA. 1996;93:116–120. doi: 10.1073/pnas.93.1.116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Hermans E. Biochemical and pharmacological control of the multiplicity of coupling at G-protein-coupled receptors. Pharmacol. Ther. 2003;99:25–44. doi: 10.1016/S0163-7258(03)00051-2. [DOI] [PubMed] [Google Scholar]
- 49.Jakubik J., El-Fakahany E.E., Doležal V. Differences in kinetics of xanomeline binding and selectivity of activation of G proteins at M(1) and M(2) muscarinic acetylcholine receptors. Mol. Pharmacol. 2006;70:656–666. doi: 10.1124/mol.106.023762. [DOI] [PubMed] [Google Scholar]
- 50.Kenakin T., Christopoulos A. Signalling bias in new drug discovery: detection, quantification and therapeutic impact. Nat. Rev. Drug Discov. 2013;12:205–216. doi: 10.1038/nrd3954. [DOI] [PubMed] [Google Scholar]
- 51.Marlo J.E., Niswender C.M., Days E.L., Bridges T.M., Xiang Y., Rodriguez A.L., Shirey J.K., Brady A.E., Nalywajko T., Luo Q., et al. Discovery and characterization of novel allosteric potentiators of M1 muscarinic receptors reveals multiple modes of activity. Mol. Pharmacol. 2009;75:577–588. doi: 10.1124/mol.108.052886. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Van der Westhuizen E.T., Spathis A., Khajehali E., Jörg M., Mistry S.N., Capuano B., Tobin A.B., Sexton P.M., Scammells P.J., Valant C., et al. Assessment of the Molecular Mechanisms of Action of Novel 4-Phenylpyridine-2-One and 6-Phenylpyrimidin-4-One Allosteric Modulators at the M1 Muscarinic Acetylcholine Receptors. Mol. Pharmacol. 2018;94:770–783. doi: 10.1124/mol.118.111633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wacker D., Stevens R.C., Roth B.L. How Ligands Illuminate GPCR Molecular Pharmacology. Cell. 2017;170:414–427. doi: 10.1016/j.cell.2017.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Bermudez M., Bock A. Does Divergent Binding Pocket Closure Drive Ligand Bias for Class A GPCRs? Trends Pharmacol. Sci. 2019;40:236–239. doi: 10.1016/j.tips.2019.02.005. [DOI] [PubMed] [Google Scholar]
- 55.Jakubik J., El-Fakahany E.E., Tucek S. Evidence for a tandem two-site model of ligand binding to muscarinic acetylcholine receptors. J. Biol. Chem. 2000;275:18836–18844. doi: 10.1074/jbc.M000112200. [DOI] [PubMed] [Google Scholar]
- 56.Järv J., Hedlund B., Bartfai T. Isomerization of the muscarinic receptor. antagonist complex. J. Biol. Chem. 1979;254:5595–5598. [PubMed] [Google Scholar]
- 57.Järv J., Hedlund B., Bartfai T. Kinetic studies on muscarinic antagonist-agonist competition. J. Biol. Chem. 1980;255:2649–2651. [PubMed] [Google Scholar]
- 58.Redka D.S., Pisterzi L.F., Wells J.W. Binding of orthosteric ligands to the allosteric site of the M(2) muscarinic cholinergic receptor. Mol. Pharmacol. 2008;74:834–843. doi: 10.1124/mol.108.048074. [DOI] [PubMed] [Google Scholar]
- 59.Jakubík J., Randáková A., Zimčík P., El-Fakahany E.E., Doležal V. Binding of N-methylscopolamine to the extracellular domain of muscarinic acetylcholine receptors. Sci. Rep. 2017;7:40381. doi: 10.1038/srep40381. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Kistemaker L.E.M., Elzinga C.R.S., Tautermann C.S., Pieper M.P., Seeliger D., Alikhil S., Schmidt M., Meurs H., Gosens R. Second M3 muscarinic receptor binding site contributes to bronchoprotection by tiotropium. Br. J. Pharmacol. 2019;176:2864–2876. doi: 10.1111/bph.14707. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Jakubík J., Zimčík P., Randáková A., Fuksová K., El-Fakahany E.E., Doležal V. Molecular mechanisms of methoctramine binding and selectivity at muscarinic acetylcholine receptors. Mol. Pharmacol. 2014;86:180–192. doi: 10.1124/mol.114.093310. [DOI] [PubMed] [Google Scholar]
- 62.Bock A., Bermudez M., Krebs F., Matera C., Chirinda B., Sydow D., Dallanoce C., Holzgrabe U., De Amici M., Lohse M.J., et al. Ligand binding ensembles determine graded agonist efficacies at a G protein-coupled receptor. J. Biol. Chem. 2016;291:16375–16389. doi: 10.1074/jbc.M116.735431. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Schmitz J., Van Der Mey D., Bermudez M., Klöckner J., Schrage R., Kostenis E., Tränkle C., Wolber G., Mohr K., Holzgrabe U. Dualsteric muscarinic antagonists-orthosteric binding pose controls allosteric subtype selectivity. J. Med. Chem. 2014;57:6739–6750. doi: 10.1021/jm500790x. [DOI] [PubMed] [Google Scholar]
- 64.Avlani V.A., Langmead C.J., Guida E., Wood M.D., Tehan B.G., Herdon H.J., Watson J.M., Sexton P.M., Christopoulos A. Orthosteric and allosteric modes of interaction of novel selective agonists of the M1 muscarinic acetylcholine receptor. Mol. Pharmacol. 2010;78:94–104. doi: 10.1124/mol.110.064345. [DOI] [PubMed] [Google Scholar]
- 65.Keov P., Valant C., Devine S.M., Lane J.R., Scammells P.J., Sexton P.M., Christopoulos A. Reverse engineering of the selective agonist TBPB unveils both orthosteric and allosteric modes of action at the M₁ muscarinic acetylcholine receptor. Mol. Pharmacol. 2013;84:425–437. doi: 10.1124/mol.113.087320. [DOI] [PubMed] [Google Scholar]
- 66.Gregory K.J., Hall N.E., Tobin A.B., Sexton P.M., Christopoulos A. Identification of orthosteric and allosteric site mutations in M2 muscarinic acetylcholine receptors that contribute to ligand-selective signaling bias. J. Biol. Chem. 2010;285:7459–7474. doi: 10.1074/jbc.M109.094011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Valant C., Gregory K.J., Hall N.E., Scammells P.J., Lew M.J., Sexton P.M., Christopoulos A. A novel mechanism of G protein-coupled receptor functional selectivity. Muscarinic partial agonist McN-A-343 as a bitopic orthosteric/allosteric ligand. J. Biol Chem. 2008;283:29312–29321. doi: 10.1074/jbc.M803801200. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Agnetta L., Kauk M., Canizal M.C.A., Messerer R., Holzgrabe U., Hoffmann C., Decker M. A Photoswitchable Dualsteric Ligand Controlling Receptor Efficacy. Angew. Chem. Int. Ed. 2017;56:7282–7287. doi: 10.1002/anie.201701524. [DOI] [PubMed] [Google Scholar]
- 69.Matera C., Flammini L., Quadri M., Vivo V., Ballabeni V., Holzgrabe U., Mohr K., De Amici M., Barocelli E., Bertoni S., et al. Bis(ammonio)alkane-type agonists of muscarinic acetylcholine receptors: synthesis, in vitro functional characterization, and in vivo evaluation of their analgesic activity. Eur. J. Med. Chem. 2014;75:222–232. doi: 10.1016/j.ejmech.2014.01.032. [DOI] [PubMed] [Google Scholar]
- 70.Cristofaro I., Spinello Z., Matera C., Fiore M., Conti L., De Amici M., Dallanoce C., Tata A.M. Activation of M2 muscarinic acetylcholine receptors by a hybrid agonist enhances cytotoxic effects in GB7 glioblastoma cancer stem cells. Neurochem. Int. 2018;118:52–60. doi: 10.1016/j.neuint.2018.04.010. [DOI] [PubMed] [Google Scholar]
- 71.Jakubík J., Bacáková L., El-Fakahany E.E., Tucek S. Positive cooperativity of acetylcholine and other agonists with allosteric ligands on muscarinic acetylcholine receptors. Mol. Pharmacol. 1997;52:172–179. doi: 10.1124/mol.52.1.172. [DOI] [PubMed] [Google Scholar]
- 72.Birdsall N.J., Farries T., Gharagozloo P., Kobayashi S., Kuonen D., Lazareno S., Popham A., Sugimoto M. Selective allosteric enhancement of the binding and actions of acetylcholine at muscarinic receptor subtypes. Life Sci. 1997;60:1047–1052. doi: 10.1016/S0024-3205(97)00046-5. [DOI] [PubMed] [Google Scholar]
- 73.Bridges T.M., Marlo J.E., Niswender C.M., Jones C.K., Jadhav S.B., Gentry P.R., Plumley H.C., Weaver C.D., Conn P.J., Lindsley C.W. Discovery of the first highly M5-preferring muscarinic acetylcholine receptor ligand, an M5 positive allosteric modulator derived from a series of 5-trifluoromethoxy N-benzyl isatins. J. Med. Chem. 2009;52:3445–3448. doi: 10.1021/jm900286j. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Kuduk S.D., Chang R.K., Di Marco C.N., Pitts D.R., Greshock T.J., Ma L., Wittmann M., Seager M.A., Koeplinger K.A., Thompson C.D., et al. Discovery of a selective allosteric M1 receptor modulator with suitable development properties based on a quinolizidinone carboxylic acid scaffold. J. Med. Chem. 2011;54:4773–4780. doi: 10.1021/jm200400m. [DOI] [PubMed] [Google Scholar]
- 75.Davie B.J., Valant C., White J.M., Sexton P.M., Capuano B., Christopoulos A., Scammells P.J. Synthesis and pharmacological evaluation of analogues of benzyl quinolone carboxylic acid (BQCA) designed to bind irreversibly to an allosteric site of the M ₁ muscarinic acetylcholine receptor. J. Med. Chem. 2014;57:5405–5418. doi: 10.1021/jm500556a. [DOI] [PubMed] [Google Scholar]
- 76.Pancani T., Foster D.J., Moehle M.S., Bichell T.J., Bradley E., Bridges T.M., Klar R., Poslusney M., Rook J.M., Daniels J.S., et al. Allosteric activation of M4 muscarinic receptors improve behavioral and physiological alterations in early symptomatic YAC128 mice. Proc. Natl. Acad. Sci. USA. 2015;112:14078–14083. doi: 10.1073/pnas.1512812112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Dallagnol J.C.C., Khajehali E., Van Der Westhuizen E.T., Jörg M., Valant C., Gonçalves A.G., Capuano B., Christopoulos A., Scammells P.J. Synthesis and Pharmacological Evaluation of Heterocyclic Carboxamides: Positive Allosteric Modulators of the M1Muscarinic Acetylcholine Receptor with Weak Agonist Activity and Diverse Modulatory Profiles. J. Med. Chem. 2018;61:2875–2894. doi: 10.1021/acs.jmedchem.7b01812. [DOI] [PubMed] [Google Scholar]
- 78.Mistry S.N., Jörg M., Lim H., Vinh N.B., Sexton P.M., Capuano B., Christopoulos A., Lane J.R., Scammells P.J. 4-Phenylpyridin-2-one Derivatives: A Novel Class of Positive Allosteric Modulator of the M1 Muscarinic Acetylcholine Receptor. J. Med. Chem. 2016;59:388–409. doi: 10.1021/acs.jmedchem.5b01562. [DOI] [PubMed] [Google Scholar]
- 79.Miao Y., Goldfeld D.A., Von Moo E., Sexton P.M., Christopoulos A., McCammon J.A., Valant C. Accelerated structure-based design of chemically diverse allosteric modulators of a muscarinic G protein-coupled receptor. Proc. Natl. Acad. Sci. USA. 2016;113:E5675–E5684. doi: 10.1073/pnas.1612353113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80.Korczynska M., Clark M.J., Valant C., Xu J., Von Moo E., Albold S., Weiss D.R., Torosyan H., Huang W., Kruse A.C., et al. Structure-based discovery of selective positive allosteric modulators of antagonists for the M2 muscarinic acetylcholine receptor. Proc. Natl. Acad. Sci. USA. 2018;115:E2419–E2428. doi: 10.1073/pnas.1718037115. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Goldberg J.A., Ding J.B., Surmeier D.J. Muscarinic modulation of striatal function and circuitry. Handb. Exp. Pharmacol. 2012;208:223–241. doi: 10.1007/978-3-642-23274-9_10. [DOI] [PubMed] [Google Scholar]
- 82.Choy K.H.C., Shackleford D.M., Malone D.T., Mistry S.N., Patil R.T., Scammells P.J., Langmead C.J., Pantelis C., Sexton P.M., Lane J.R., et al. Positive Allosteric Modulation of the Muscarinic M1 Receptor Improves Efficacy of Antipsychotics in Mouse Glutamatergic Deficit Models of Behavior. J. Pharmacol. Exp. Ther. 2016;359:354–365. doi: 10.1124/jpet.116.235788. [DOI] [PubMed] [Google Scholar]
- 83.Nickols H.H., Conn J.P. Development of allosteric modulators of GPCRs for treatment of CNS disorders. Neurobiol. Dis. 2014;61:55–71. doi: 10.1016/j.nbd.2013.09.013. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Hopper S., Pavey G.M., Gogos A., Dean B. Widespread Changes in Positive Allosteric Modulation of the Muscarinic M1 Receptor in Some Participants With Schizophrenia. Int. J. Neuropsychopharmacol. 2019;22:640–650. doi: 10.1093/ijnp/pyz045. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85.Tzavara E.T., Bymaster F.P., Davis R.J., Wade M.R., Perry K.W., Wess J., McKinzie D.L., Felder C., Nomikos G.G. M4 muscarinic receptors regulate the dynamics of cholinergic and dopaminergic neurotransmission: relevance to the pathophysiology and treatment of related CNS pathologies. FASEB J. 2004;18:1410–1412. doi: 10.1096/fj.04-1575fje. [DOI] [PubMed] [Google Scholar]
- 86.Brady A.E., Jones C.K., Bridges T.M., Kennedy J.P., Thompson A.D., Heiman J.U., Breininger M.L., Gentry P.R., Yin H., Jadhav S.B., et al. Centrally active allosteric potentiators of the M4 muscarinic acetylcholine receptor reverse amphetamine-induced hyperlocomotor activity in rats. J. Pharmacol. Exp. Ther. 2008;327:941–953. doi: 10.1124/jpet.108.140350. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Yeomans J.S. Muscarinic receptors in brain stem and mesopontine cholinergic arousal functions. Handb. Exp. Pharmacol. 2012;208:243–259. doi: 10.1007/978-3-642-23274-9_11. [DOI] [PubMed] [Google Scholar]
- 88.Gentry P.R., Kokubo M., Bridges T.M., Kett N.R., Harp J.M., Cho H.P., Smith E., Chase P., Hodder P.S., Niswender C.M., et al. Discovery of the first M5-selective and CNS penetrant negative allosteric modulator (NAM) of a muscarinic acetylcholine receptor: (S)-9b-(4-chlorophenyl)-1-(3,4-difluorobenzoyl)-2,3-dihydro-1H-imidazo [2,1-a]isoindol-5(9bH)-one (ML375) J. Med. Chem. 2013;56:9351–9355. doi: 10.1021/jm4013246. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.Gunter B.W., Gould R.W., Bubser M., McGowan K.M., Lindsley C.W., Jones C.K. Selective inhibition of M5 muscarinic acetylcholine receptors attenuates cocaine self-administration in rats. Addict. Biol. 2018;23:1106–1116. doi: 10.1111/adb.12567. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90.Gilon P., Henquin J.C. Mechanisms and physiological significance of the cholinergic control of pancreatic beta-cell function. Endocr. Rev. 2001;22:565–604. doi: 10.1210/edrv.22.5.0440. [DOI] [PubMed] [Google Scholar]
- 91.Duttaroy A., Zimliki C.L., Gautam D., Cui Y., Mears D., Wess J. Muscarinic stimulation of pancreatic insulin and glucagon release is abolished in M3 muscarinic acetylcholine receptor-deficient mice. Diabetes. 2004;53:1714–1720. doi: 10.2337/diabetes.53.7.1714. [DOI] [PubMed] [Google Scholar]
- 92.Zhu L., Rossi M., Cohen A., Pham J., Zheng H., Dattaroy D., Mukaibo T., Melvin J.E., Langel J.L., Hattar S., et al. Allosteric modulation of β-cell M3 muscarinic acetylcholine receptors greatly improves glucose homeostasis in lean and obese mice. Proc. Natl. Acad. Sci. USA. 2019;116:18684–18690. doi: 10.1073/pnas.1904943116. [DOI] [PMC free article] [PubMed] [Google Scholar]