Abstract
Reports on the association of TGF-β1 polymorphisms with breast cancer (BC) have been conflicting, inconsistent, inconclusive, and controversial. PubMed, EMBASE, and Google Scholar were used to identify studies on TGF-β1 polymorphisms and BC risk. Data were extracted independently, and of the initial 3043 studies, 39 case-control studies were eligible for inclusion in the meta-analysis. Information from these studies was extracted, and the overall associations of three TGF-β1 polymorphisms (TGF-β1 29>T/C, TGF-β1-509 C/T, and TGF-β1*6A) with BC risk were analyzed using overall allele, homozygous, heterozygous, recessive, and dominant models. None of the three TGF-β1 polymorphisms studied had a significant influence on the development of BC. However, stratified analysis revealed a positive correlation between the TGF-β1 29T>C polymorphism and BC risk according to a heterozygous model of the Asian population (odds ratio (OR) = 1.115, 95% confidence interval (CI) = 1.006–1.237, p = 0.039). Interestingly, this polymorphism was associated with lower odds of BC according to a heterozygous model of the Middle Eastern population (OR = 0.602, 95% CI = 0.375–0.966, p = 0.035). Thus, our analysis of large datasets indicates that the TGF-β1 29T>C polymorphism is significantly associated with BC risk in the Asian population. In contrast, the TGF-β1*6A and TGF-β1-509 C/T polymorphisms failed to show an association with BC.
Keywords: breast cancer, polymorphism, TGFβ, meta-analysis
1. Introduction
The transcription growth factor β (TGF-β) signaling pathway has been studied extensively in several cancer types, including breast cancer (BC). The TGF-β superfamily consists of 33 structurally similar members, including bone morphogenic proteins (BMPs), TGF-β ligands, and activins, which play important roles in mammary development [1,2,3]. In humans, isoforms of TGF-β ligands that are closely related both structurally and functionally (TGF-β1, TGF-β2, and TGF-β3) have been reported to modulate cell progression, migration, and apoptosis [4]. During cancer development, TGF-β exhibits both tumorigenic and tumor-suppressive roles [5]. It has been reported that, whereas TGF-β acts as a tumor suppressor during the early stages of cancer development, it exhibits oncogenic properties, inducing cell migration and invasion, during the later stages [2,6,7].
Several polymorphisms have been identified in the TGF-β1 gene (TGFβ1), including two located in exon 1 (TGF-β1 29T>C and TGF-β1*6A) and two located in the promoter region (TGF-β1-509 C/T and TGF-β1-800 G/A) [8,9,10]. There is increasing evidence of an association between these polymorphisms and elevated cancer risk [11]. Researchers demonstrated that the TGF-β1 29T>C polymorphism increases the secretion of TGF-β levels [9]. Ziv et al. demonstrated that CC genotype of the TGF-β1 29T>C polymorphism decreases BC risk by up to 64% among Whites [12]. In contrast, Dunning et al. found that this polymorphism was associated with an increased risk of BC [9]. In a multi-ethnic cohort study, there was no association between this polymorphism and BC risk in postmenopausal Caucasian women [13]. However, a meta-analysis of case–control studies established a moderate association between the TGF-β1 29T>C polymorphism and BC susceptibility [14], and it included studies that either did not assess BC risk, lacked information regarding control samples, or did not incorporate data from more recent studies that might influence the combined ORs. Our present meta-analysis rectifies these previous mistakes by excluding studies that did not assess BC risk and/or did not have information regarding control samples.
The TGF-β1-509 C/T polymorphism, which is present in the promoter region of TGF-β1, influences its expression [15,16]. Elevated levels of TGF-β1 in plasma were associated with the T allele of TGF-β1-509 C/T [16]. Several studies have assessed the association between TGF-β1-509 C/T polymorphism and BC risk [17]. This polymorphism has also been associated with radiation-induced fibrosis; however, contrasting results have also been recently obtained [18].
The deletion of three alanine residues within the nine-alanine repeat sequence (* 9A) of the TGF-β exon 1 region results in the formation of the TGF-β1*6A variant [10]. Studies have suggested that the TGF-β1*6A variant exhibits significantly fewer tumor-suppressive properties than wild-type TGF-β1 [19]. TGF-β1*6A is a tumor-susceptibility allele that is highly correlated with the risk of various cancer types [20]. Baxter et al. found that the TGF-β1*6A variant increased BC risk by 60% [21]. However, contradictory results have also been observed [19,22,23,24,25]. Therefore, we performed the current meta-analysis to provide conclusive evidence for the associations between TGF-β1 polymorphisms and BC risk.
2. Results
2.1. Characteristics of Studies Included in the Meta-Analysis
Initially, 3043 hits were obtained from publications on PubMed, EMBASE, and Google Scholar. Among them, 373 publications were reviews, which were excluded. The remaining studies were retrieved, scrutinized, and evaluated by reading their titles and abstracts. Eighty potentially eligible studies were retrieved as full texts, 39 of which were eligible for inclusion in the meta-analysis. Precisely, 23 studies [9,12,13,26,27,28,29,30,31,32,33,34,35,36,37,38,39,40,41,42,43,44,45] covered the TGF-β1 29T>C polymorphism (also known as Pro10Leu, T869C, rs1982073, and rs1800470), whereas 13 studies [9,28,29,30,35,40,46,47,48,49,50,51,52] covered the TGF-β1-509 C/T polymorphism (also known as −1349C>T and rs1800469), and 13 studies [9,21,28,31,33,35,39,47,53,54,55,56,57] covered the TGF-β1*6A polymorphism. Genotypic and allelic frequencies from each eligible case-control study were extracted. The characteristics of all eligible studies are represented in Table 1. Studies were sub-categorized based on the ethnicities of the individuals studied (Caucasian, European, Middle Eastern, and Asian), and were analyzed separately. Supplementary Figure S1 depicts the selection of studies in a comprehensive PRISMA flow chart.
Table 1.
Characteristics of all studies included for meta-analysis.
| S.No. | First Author | Year | Ethnicity | Case | Control | Genotype Distribution | Allele Distribution (%) | Genotyping method | ||||||||
| TGFβ1 29C>T (Pro10Leu) or T869C or rs1982073 or rs1800470 | Breast cancer | Control | Breast Cancer | Control | ||||||||||||
| TT | TC | CC | TT | TC | CC | T | C | T | C | |||||||
| 1 | Ziv et al [12] | 2001 | Caucasian | 146 | 2929 | 56 | 80 | 10 | 1068 | 1413 | 448 | 192 (65.75) | 100 (34.25) | 3549 (60.58) | 2309 (39.42) | PCR-RFLP |
| 2 | Krippl et al [26] | 2003 | Caucasian | 495 | 499 | 196 | 219 | 80 | 182 | 229 | 88 | 611 (61.71) | 379 (38.29) | 593 (59.42) | 405 (40.58) | PCR |
| 3 | Dunning et al a [9] | 2003 | UK | 1,752 | 1877 | 99 | 1228 | 425 | 735 | 889 | 253 | 1426 (40.7) | 2078 (59.3) | 2359 (62.84) | 1395 (37.16) | Taqman |
| 4 | Dunning et al b [9] | 2003 | European | 415 | 574 | 116 | 220 | 79 | 202 | 274 | 98 | 452 (54.45) | 378 (45.54) | 678 (59.06) | 470 (40.94) | Taqman |
| 5 | Dunning et al c [9] | 2003 | Finnland | 481 | 451 | 255 | 191 | 35 | 232 | 191 | 28 | 701 (72.90) | 261 (27.1) | 655 (72.62) | 247 (27.38) | Taqman |
| 6 | Hishida et al [27] | 2003 | Asian | 232 | 177 | 67 | 107 | 58 | 42 | 87 | 48 | 241 (51.9) | 223 (48.1) | 171 (48.30) | 183 (51.7) | PCR-CTPP |
| 7 | Marchand et al a [13] | 2004 | Afro-American | 233 | 612 | 76 | 112 | 45 | 205 | 289 | 118 | 264 (56.65) | 202 (43.35) | 699 (57.11) | 525 (42.89) | Taqman |
| 8 | Marchand et al b [13] | 2004 | Latina | 225 | 647 | 67 | 111 | 47 | 179 | 308 | 160 | 245 (54.44) | 205 (45.56) | 666 (51.47) | 628 (48.53) | Taqman |
| 9 | Marchand et al c [13] | 2004 | White | 299 | 402 | 114 | 137 | 48 | 141 | 183 | 78 | 365 (61.0) | 233 (39) | 465 (57.84) | 339 (42.16) | Taqman |
| 10 | Marchand et al d [13] | 2004 | Japanese | 303 | 385 | 62 | 163 | 78 | 91 | 183 | 111 | 287 (47.36) | 319 (52.64) | 365 (47.40) | 405 (52.6) | Taqman |
| 11 | Marchand et al e [13] | 2004 | Hawaiian | 63 | 268 | 19 | 27 | 17 | 74 | 140 | 54 | 65 (51.6) | 61 (48.4) | 288 (53.73) | 248 (46.27) | Taqman |
| 12 | jin et al a [28] | 2004 | Finnland | 223 | 234 | 119 | 93 | 11 | 123 | 91 | 20 | 331 (74.2) | 115 (25.8) | 337 (72.0) | 131 (28.0) | PCR-RFLP, PCR-ALF |
| 13 | jin et al b [28] | 2004 | Polish | 415 | 205 | 151 | 189 | 75 | 66 | 105 | 34 | 491 (59.15) | 339 (40.85) | 237 (57.8) | 173 (42.2) | PCR-RFLP, PCR-ALF |
| 14 | jin et al c [28] | 2004 | German | 160 | 162 | 63 | 72 | 25 | 46 | 96 | 20 | 198 (61.87) | 122 (38.13) | 188 (58.02) | 136 (41.98) | PCR-RFLP, PCR-ALF |
| 15 | jin et al d [28] | 2004 | Swedish | 84 | 173 | 31 | 38 | 15 | 70 | 77 | 26 | 100 (59.5) | 68 (40.5) | 217 (62.72) | 129 (37.28) | PCR-RFLP, PCR-ALF |
| 16 | Saha et al [29] | 2004 | Asian | 23 | 84 | 10 | 13 | 0 | 47 | 33 | 4 | 33 (71.7) | 13 (28.3) | 127 (75.6) | 41 (24.4) | PCR-SSCP |
| 17 | Shin et al [30] | 2005 | Asian | 1,114 | 1189 | 258 | 554 | 302 | 255 | 615 | 319 | 1070 (48.1) | 1158 (51.9) | 1125 (47.31) | 1253 (52.69) | PCR-RFLP |
| 18 | Kaklamani et al [31] | 2005 | Caucasian | 658 | 841 | 200 | 339 | 119 | 240 | 419 | 182 | 739 (56.1) | 577 (43.9) | 899 (53.45) | 783 (46.55) | PCR-seq |
| 19 | Lee et al [32] | 2005 | Asian | 558 | 501 | 135 | 288 | 135 | 148 | 235 | 118 | 558 (50) | 558 (50) | 531 (53.0) | 471 (47.0) | PCR-CTPP |
| 20 | Feigelson et al [33] | 2006 | Caucasian | 485 | 481 | 182 | 233 | 70 | 181 | 221 | 79 | 597 (61.55) | 373 (38.45) | 583 (60.60) | 379 (39.40) | Taqman |
| 21 | Scola et al [34] | 2006 | Caucasian | 84 | 106 | 41 | 27 | 16 | 35 | 52 | 19 | 109 (64.88) | 59 (35.12) | 122 (57.55) | 90 (42.45) | PCR-RFLP |
| 22 | D Cox et al [35] | 2007 | Caucasian | 1,185 | 1651 | 469 | 548 | 168 | 613 | 797 | 241 | 1486 (62.7) | 884 (37.3) | 2023 (61.27) | 1279 (38.73) | Taqman |
| 23 | Gonullu et al [36] | 2007 | Turkey | 38 | 24 | 20 | 10 | 8 | 11 | 9 | 4 | 50 (65.8) | 26 (34.2) | 31 (64.58) | 17 (35.42) | PCR |
| 24 | A Cox et al (GESBC) [37] | 2007 | Caucasian | 556 | 713 | 169 | 284 | 103 | 255 | 338 | 120 | 622 (55.9) | 490 (44.1) | 848 (59.47) | 578 (40.53) | Taqman |
| 25 | A Cox et al (HBCS) [37] | 2007 | Caucasian | 1,073 | 1013 | 386 | 506 | 181 | 388 | 471 | 154 | 1278 (59.55) | 868 (40.45) | 1247 (61.55) | 779 (38.45) | Taqman |
| 26 | A Cox et al (IARC-Thai) [37] | 2007 | Asian | 453 | 356 | 189 | 213 | 51 | 161 | 162 | 33 | 591 (65.23) | 315 (34.77) | 484 (67.98) | 228 (32.02) | Taqman |
| 27 | A Cox et al (Kuopio) [37] | 2007 | Caucasian | 435 | 442 | 229 | 175 | 31 | 225 | 189 | 28 | 633 (72.76) | 237 (27.24) | 639 (72.29) | 245 (27.71) | Taqman |
| 28 | A Cox et al (Mayo Clinic) [37] | 2007 | Caucasian | 793 | 837 | 296 | 404 | 93 | 307 | 409 | 121 | 996 (62.8) | 590 (37.2) | 1023 (61.1) | 651 (38.89) | Taqman |
| 29 | A Cox et al (PBCS) [37] | 2007 | Caucasian | 1,841 | 2254 | 617 | 890 | 334 | 797 | 1104 | 353 | 2124 (57.67) | 1558 (42.33) | 2698 (59.85) | 1810 (40.15) | Taqman |
| 30 | A Cox et al (SEARCH) [37] | 2007 | Caucasian | 4,504 | 5689 | 1670 | 2138 | 696 | 2200 | 2716 | 773 | 5478 (60.8) | 3530 (39.2) | 7116 (62.54) | 4262 (37.46) | Taqman |
| 31 | A Cox et al (Seoul) [37] | 2007 | Asian | 643 | 529 | 162 | 327 | 154 | 155 | 253 | 121 | 651 (50.62) | 635 (49.38) | 563 (53.21) | 495 (46.79) | Taqman |
| 32 | A Cox et al (SASBAC) [37] | 2007 | Caucasian | 1,303 | 1494 | 539 | 596 | 168 | 657 | 637 | 200 | 1674 (64.2) | 932 (35.8) | 1951 (65.3) | 1037 (34.7) | Taqman |
| 33 | A Cox et al (CNIO) [37] | 2007 | Caucasian | 640 | 739 | 160 | 313 | 167 | 217 | 355 | 167 | 633 (49.45) | 647 (50.55) | 789 (53.38) | 689 (46.62) | Taqman |
| 34 | A Cox et al (USRT) [37] | 2007 | Caucasian | 705 | 1043 | 243 | 339 | 123 | 382 | 494 | 167 | 825 (58.51) | 585 (41.49) | 1258 (60.31) | 828 (39.69) | Taqman |
| 35 | Rajkumar et al [38] | 2008 | Asian | 250 | 500 | 80 | 126 | 44 | 190 | 234 | 76 | 286 (57.2) | 214 (42.8) | 614 (61.4) | 386 (38.6) | PCR-CTPP |
| 36 | Joshi et al [39] | 2011 | Asian | 203 | 384 | 67 | 104 | 32 | 114 | 181 | 89 | 238 (58.62) | 168 (41.38) | 409 (53.25) | 359 (46.75) | PCR-SSP |
| 37 | Zhang et al [40] | 2011 | Asian | 170 | 178 | 38 | 76 | 56 | 49 | 79 | 50 | 152 (44.7) | 188 (55.3) | 177 (49.72) | 179 (50.28) | PCR-RFLP |
| 38 | Pooja et al [41] | 2013 | Asian | 465 | 239 | 85 | 165 | 214 | 51 | 123 | 64 | 335 (36.1) | 593 (63.9) | 225 (47.27) | 251 (52.73) | PCR-seq |
| 39 | Eva et al [42] | 2013 | Caucasian | 274 | 252 | 99 | 131 | 44 | 87 | 117 | 48 | 329 (60.0) | 219 (40) | 291 (57.74) | 213 (42.26) | Taqman |
| 40 | Amani et al [43] | 2014 | Iranian | 100 | 104 | 45 | 28 | 27 | 33 | 41 | 30 | 118 (59) | 82 (41) | 107 (51.44) | 101 (48.56) | PCR-seq |
| 41 | Joshi et al [44] | 2014 | Asian | 172 | 229 | 60 | 81 | 31 | 61 | 114 | 54 | 201 (58.43) | 143 (41.57) | 236 (51.53) | 222 (48.47) | PCR-SSP |
| 42 | Tezerjani et al [45] | 2015 | Iranian | 60 | 60 | 18 | 29 | 13 | 16 | 32 | 12 | 65 (54.17) | 55 (45.83) | 64 (53.33) | 56 (46.67) | PCR-ARMS |
| S.No | First Author | Year | Ethnicity | Case | Control | Genotype Distribution | Allele Distribution (%) | Genotyping method | ||||||||
| TGFβ1 -509 C/T or -1349C>T or rs1800469 | Breast Cancer | Control | Breast Cancer | Control | ||||||||||||
| CC | CT | TT | CC | CT | TT | C | T | C | T | |||||||
| 1 | Dunning et al a [9] | 2003 | UK | 2439 | 2366 | 1181 | 1014 | 244 | 1194 | 977 | 195 | 3376 (69.21) | 1502 (30.79) | 3365 (71.11) | 1367 (28.89) | Taqman |
| 2 | Dunning et al b [9] | 2003 | European | 417 | 634 | 159 | 201 | 57 | 281 | 287 | 66 | 519 (62.23) | 315 (37.77) | 849 (66.96) | 419 (33.04) | Taqman |
| 3 | Dunning et al c [9] | 2003 | Finnland | 480 | 452 | 277 | 176 | 27 | 252 | 177 | 23 | 730 (76.04) | 230 (23.96) | 681 (75.33) | 223 (24.67) | Taqman |
| 4 | jin et al a [28] | 2004 | Finnland | 221 | 320 | 133 | 80 | 8 | 182 | 119 | 19 | 346 (78.28) | 96 (21.72) | 483 (75.47) | 157 (24.53) | PCR-RFLP, PCR-ALF |
| 5 | jin et al b [28] | 2004 | Polish | 170 | 188 | 71 | 81 | 18 | 74 | 95 | 19 | 223 (65.59) | 117 (34.41) | 243 (64.63) | 133 (35.37) | PCR-RFLP, PCR-ALF |
| 6 | Saha et al [29] | 2004 | Asian | 26 | 97 | 3 | 22 | 1 | 35 | 53 | 9 | 28 (53.85) | 24 (46.15) | 123 (63.4) | 71 (36.6) | PCR-SSCP |
| 7 | Shin et al [30] | 2005 | Asian | 1118 | 1206 | 260 | 559 | 299 | 260 | 628 | 318 | 1049 (48.26) | 1157 (51.74) | 1148 (47.6) | 1264 (52.4) | PCR-RFLP |
| 8 | D Cox et al [35] | 2007 | Caucasian | 1195 | 1663 | 600 | 506 | 89 | 786 | 723 | 154 | 1706 (71.38) | 684 (28.62) | 2295 (69) | 1031 (31) | Taqman |
| 9 | Jakubowska et al [46] | 2009 | Caucasian | 1011 | 1068 | 454 | 451 | 106 | 465 | 476 | 127 | 1359 (67.21) | 663 (32.79) | 1406 (65.82) | 730 (34.18) | PCR-RFLP |
| 10 | Jakubowska et al [47] | 2010 | Caucasian | 319 | 290 | 127 | 144 | 48 | 115 | 145 | 30 | 398 (62.38) | 240 (37.62) | 375 (64.66) | 205 (35.34) | PCR-RFLP |
| 11 | MARIE-GENICA [48] | 2010 | Caucasian | 3146 | 5485 | 1529 | 1315 | 302 | 2616 | 2313 | 556 | 4373 (69.5) | 1919 (30.5) | 7545 (68.78) | 3425 (31.22) | MALDI-TOF Msa and PCR-RFLP |
| 12 | Babyshkina et al [49] | 2011 | Russian | 218 | 290 | 89 | 108 | 21 | 103 | 133 | 54 | 286 (65.6) | 150 (34.4) | 339 (58.45) | 241 (41.55) | PCR-RFLP |
| 13 | Zhang et al [40] | 2011 | Asian | 170 | 178 | 28 | 93 | 49 | 41 | 84 | 53 | 149 (43.82) | 191 (56.18) | 166 (46.63) | 190 (53.37) | PCR-RFLP |
| 14 | Quan et al a [50] | 2013 | Afro-American | 454 | 405 | 268 | 164 | 22 | 215 | 165 | 25 | 700 (77.09) | 208 (22.91) | 595 (73.46) | 215 (26.54) | MassArray IPLEX Gold Assay |
| 15 | Quan et al b [50] | 2013 | European | 327 | 312 | 124 | 147 | 56 | 134 | 135 | 43 | 395 (60.4) | 259 (39.6) | 403 (64.58) | 221 (35.42) | MassArray IPLEX Gold Assay |
| 16 | Vinod et al [51] | 2013 | Asian | 153 | 128 | 64 | 66 | 23 | 70 | 36 | 22 | 194 (63.4) | 112 (36.6) | 176 (68.75) | 80 (31.25) | PCR-ARMS |
| 17 | Parvizi et al [52] | 2016 | Iranian | 100 | 100 | 31 | 50 | 19 | 21 | 48 | 31 | 112 (56) | 88 (44) | 90 (45) | 110 (55) | PCR-RFLP |
| S.No | First Author | Year | Ethinic group | Case | Control | Genotype Distribution | Allele Distribution (%) | Genotyping method | ||||||||
| TGFβ1* 6A or rs11466445 | Case | Control | Case | Control | ||||||||||||
| 9A/9A | 9A/6A | 6A/6A | 9A/9A | 9A/6A | 6A/6A | 9A | 6A | 9A | 6A | |||||||
| 1 | Pasche et al [19] | 1999 | USA | 152 | 732 | 128 | 24 | 0 | 654 | 78 | 0 | 280 (92.11) | 24 (7.89) | 1386 (94.67) | 78 (5.33) | PCR-RFLP |
| 2 | Baxter et al [21] | 2002 | United Kingdom | 355 | 248 | 268 | 83 | 4 | 207 | 39 | 2 | 619 (87.18) | 91 (12.82) | 453 (91.33) | 43 (8.67) | PCR-SSCP |
| 3 | Reiss et al [53] | 2004 | USA | 98 | 91 | 87 | 11 | 0 | 77 | 14 | 0 | 185 (94.39) | 11 (5.61) | 168 (92.31) | 14 (7.69) | PCR-RFLP |
| 4 | Caldes et al [53] | 2004 | USA | 506 | 292 | 397 | 106 | 3 | 250 | 42 | 0 | 900 (88.93) | 112 (11.07) | 542 (92.81) | 42 (7.19) | PCR-RFLP |
| 5 | Offit et al [53] | 2004 | USA | 462 | 330 | 391 | 67 | 4 | 291 | 38 | 1 | 849 (91.88) | 75 (8.12) | 620 (93.94) | 40 (6.06) | PCR-RFLP |
| 6 | jin et al a [28] | 2004 | Finnish | 221 | 234 | 177 | 38 | 6 | 171 | 60 | 3 | 392 (88.69) | 50 (11.31) | 402 (85.9) | 66 (14.1) | PCR-RFLP |
| 7 | jin et al b [28] | 2004 | Polish | 170 | 202 | 140 | 28 | 2 | 176 | 26 | 0 | 308 (90.59) | 32 (9.41) | 378 (93.56) | 26 (6.44) | PCR-RFLP |
| 8 | Kaklamani et al [31] | 2005 | USA | 611 | 690 | 515 | 92 | 4 | 612 | 77 | 1 | 1122 (91.82) | 100 (8.18) | 1301 (94.28) | 79 (5.72) | PCR-seq |
| 9 | Chen et al [54] | 2006 | USA | 115 | 130 | 92 | 23 | 0 | 111 | 18 | 1 | 207 (90) | 23 (10) | 240 (92.31) | 20 (7.69) | PCR-SSCP |
| 10 | Figelson et al [33] | 2006 | USA | 387 | 384 | 387 | 384 | PCR | ||||||||
| 11 | D Cox et al [35] | 2007 | USA | 1,187 | 1673 | 968 | 207 | 12 | 1352 | 302 | 19 | 2143 (90.27) | 231 (9.73) | 3006 (89.84) | 340 (10.16) | Taqman |
| 12 | Song et al [55] | 2007 | Sweden | 763 | 852 | 598 | 152 | 13 | 682 | 160 | 10 | 1348 (88.34) | 178 (11.66) | 1524 (89.44) | 180 (10.56) | PCR |
| 13 | Colleran et al [56] | 2010 | Ireland | 960 | 958 | 796 | 154 | 10 | 785 | 160 | 13 | 1746 (90.94) | 174 (9.06) | 1730 (90.29) | 186 (9.71) | PCR |
| 14 | Jakubowska et al [47] | 2010 | Poland | 282 | 252 | 282 | 252 | PCR-RFLP | ||||||||
| 15 | Joshi et al [39] | 2011 | Asian | 209 | 391 | 196 | 12 | 1 | 361 | 28 | 2 | 404 (96.65) | 14 (3.35) | 750 (95.91) | 782 (4.09) | PCR-SSP |
| 16 | kamali et al [57] | 2015 | Iranian | 280 | 280 | 251 | 25 | 4 | 241 | 27 | 12 | 525 (94.09) | 33 (5.91) | 509 (90.89) | 560 (9.11) | PCR |
Dunning et al. UK population designated as (a), European ethnic group designated as (b), and Finland population designated as (c). Marchand et al. African-American ethnic group designated as (a), Latinos designated as (b), Whites designated as (c), Japanese designated as (d), and Hawaiian designated as (e). Jin et al. Finland population designated as (a), Polish designated as (b), German designated as (c), and Swedish designated as (d). Quin et al. African-American ethnic group designated as (a) and European designated as (b).
2.2. Assessment of Heterogeneity among the Studies and Publication Bias
Heterogeneity was observed in the allelic models of all three TGF-β1 polymorphisms, as well as all genetic models of the TGF-β1 29T>C polymorphism and the homozygous model of the TGF-β1-509 C/T polymorphism. In contrast, none of the genetic models of the TGF-β1*6A polymorphism were heterogeneous. Heterogeneity among the studies of all three polymorphisms in all ethnic groups is represented in Supplementary Table S1. Information regarding the presence of potential publication bias in ethnic studies is illustrated in Supplementary Figure S2.
2.3. TGF-β1 29T>C, or T869C, Polymorphism
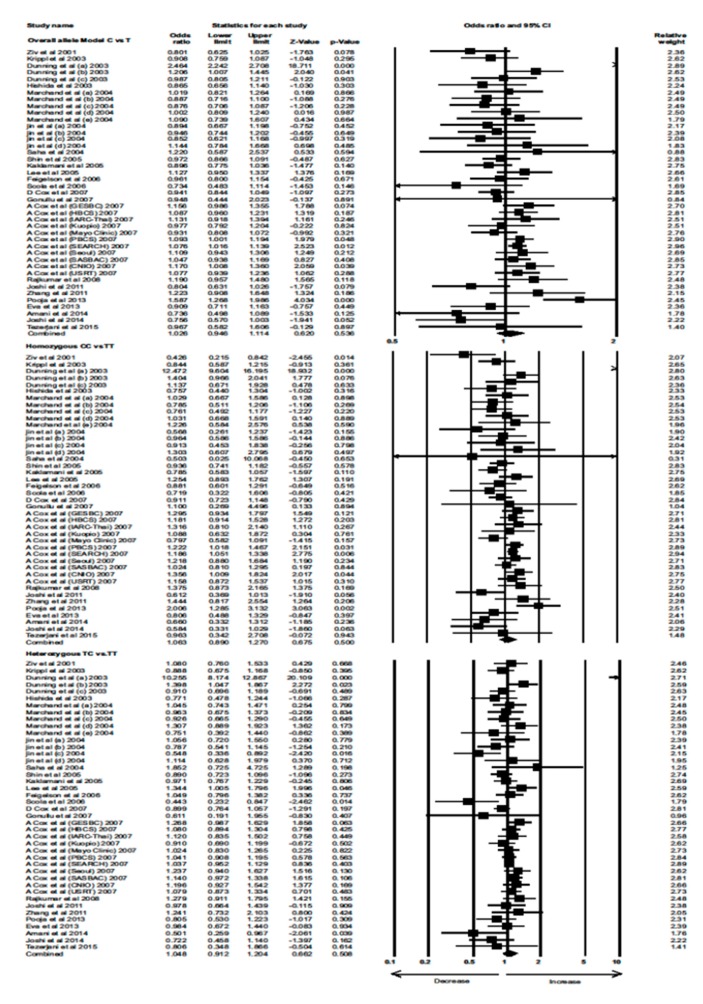
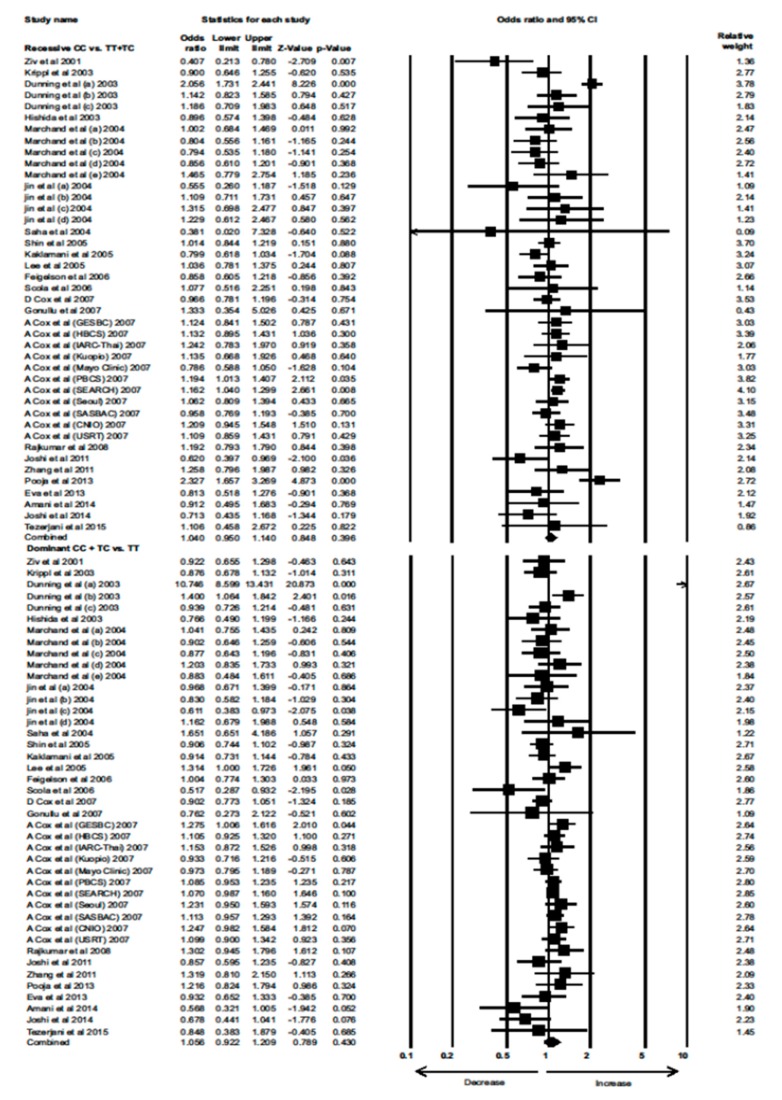
Data from 23 eligible case–control studies were extracted to evaluate the association between the TGF-β1 29T>C polymorphism and BC risk. Interestingly, neither the overall allele model nor any of the genetic models showed a significant association between the TGF-β1 29T>C polymorphism with increased risk of BC (Combined analysis: overall allele (OR) = 1.026, 95% confidence interval (CI) = 0.946–1.114, p = 0.536; homozygous: OR = 1.063, 95% CI = 0.890–1.270, p = 0.500; heterozygous: OR = 1.048, 95% CI = 0.912–1.204, p = 0.508; recessive: OR = 1.040, 95% CI = 0.950–1.140, p = 0.396; dominant: OR = 1.056, 95% CI = 0.922–1.209, p = 0.430) (Figure 1 and Figure 2).
Figure 1.
Forest plot of overall allele, homozygous, and heterozygous genotypic analyses of the TGF-β1 29T>C polymorphism and assessment of its association with BC risk using combined odds ratios (ORs) and 95% confidence intervals (CIs) of the ORs. Black squares represent the ORs of the individual studies, and horizontal lines indicate the 95% CIs of the ORs.
Figure 2.
Forest plot of recessive and dominant genotypic analyses of the TGF-β1 29T>C polymorphism and assessment of its association with BC risk using combined odds ratios (ORs) and 95% confidence intervals (CIs) of the ORs. Black squares represent the ORs of the individual studies, and horizontal lines indicate the 95% CIs of the ORs.
2.4. TGF-β1-509 C/T, or -1349C>T, Polymorphism
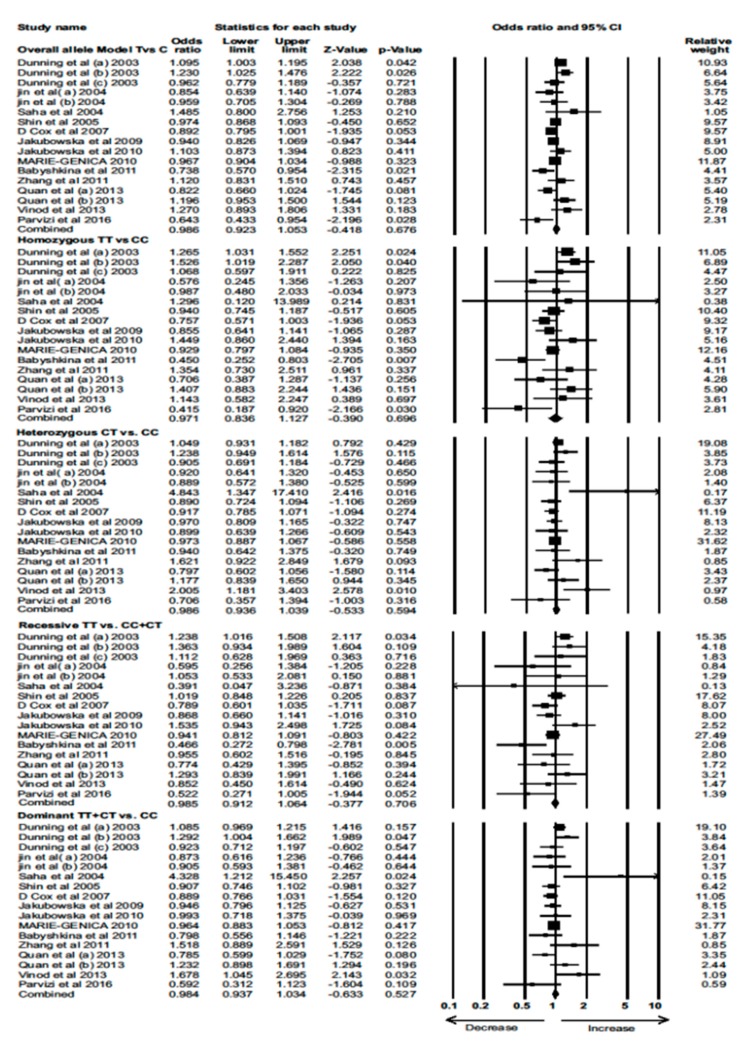
Data from 13 eligible case–control studies were extracted to evaluate the association between the TGF-β1-509 C/T polymorphism and BC risk. None of the models showed a significant association between the TGF-β1-509 C/T polymorphism and BC risk (Combined analysis: overall allele (OR) = 0.986, 95% CI = 0.923–1.053, p = 0.676; homozygous: OR = 0.971, 95% CI = 0.836–1.127, p = 0.696; heterozygous: OR = 0.986, 95% CI = 0.936–1.039, p = 0.594; recessive: OR = 0.985, 95% CI = 0.912–1.064, p = 0.706; dominant: OR = 0.984, 95% CI = 0.937–1.034, p = 0.527) (Figure 3).
Figure 3.
Forest plot of overall allele and genotypic analysis of the TGF-β1-509 C/T polymorphism and assessment of its association with BC risk using combined odds ratios (ORs) and 95% confidence intervals (CIs) of the ORs. Black squares represent the ORs of the individual studies, and horizontal lines indicate the 95% CIs of the ORs.
2.5. TGF-β1*6A polymorphism
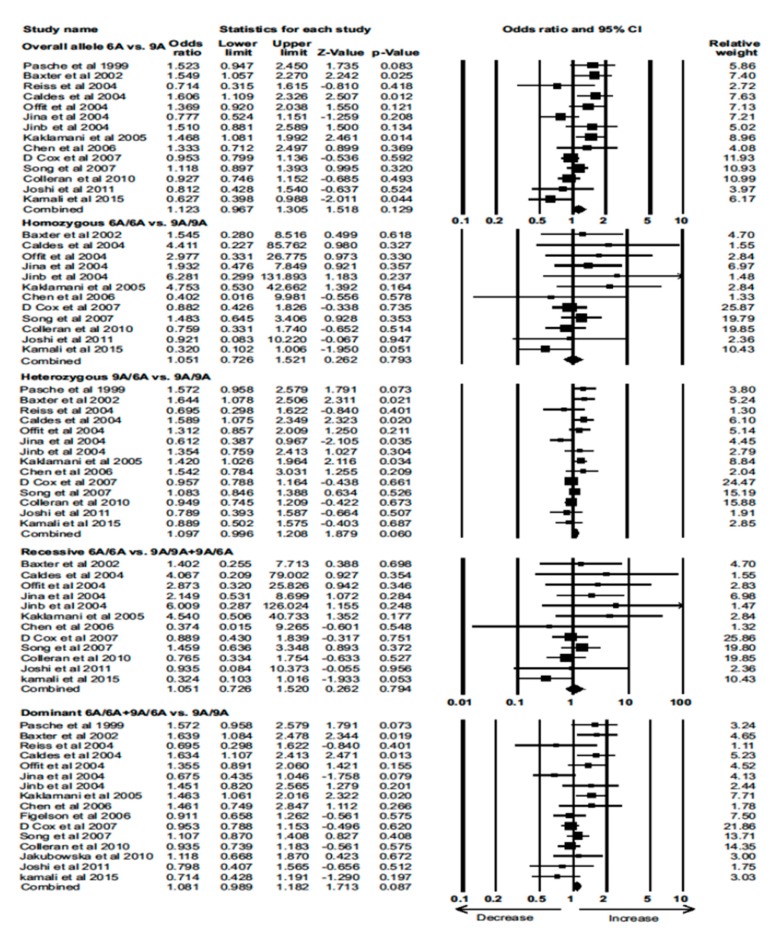
Data from 13 eligible case–control studies were extracted to evaluate the association between the TGF-β1*6A polymorphism and BC risk. Again, none of the models showed a significant association between the TGF-β1*6A polymorphism and BC (Combined analysis: overall allele (OR) = 1.123, 95% CI = 0.967–1.305, p = 0.129; homozygous: OR = 1.051, 95% CI = 0.726–1.521, p = 0.793; heterozygous: OR = 1.097, 95% CI = 0.996–1.208, p = 0.060; recessive: OR = 1.051, 95% CI = 0.726–1.520, p = 0.794; dominant: OR = 1.081, 95% CI = 0.989–1.182, p = 0.087) (Figure 4). These results for each polymorphism did not differ by genotyping method (Supplementary Figures S3 and S4).
Figure 4.
Forest plot of overall allele and genotypic analyses of the TGF-β1*6A polymorphism and assessment of its association with BC risk using pooled combined odds ratios (ORs) and 95% confidence intervals (CIs) of the ORs. Black squares represent the ORs of the individual studies, and horizontal lines indicate the 95% CIs of the ORs.
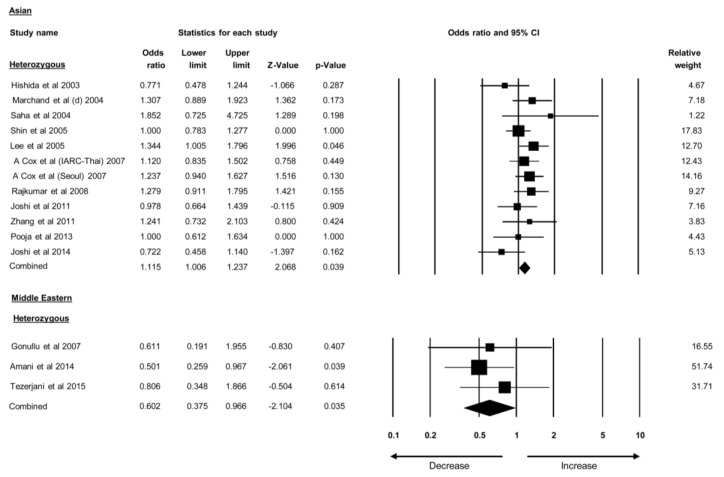
2.6. Stratified Analysis
Studies were categorized based on the ethnicities of the individuals studied. Studies covering the TGF-β1 29T>C polymorphisms were divided into four groups: Caucasian, European, Asian, and Middle Eastern. The TGF-β1 29T>C polymorphism was found to be associated with BC risk in the heterozygous model of the Asian population (OR = 1.115, 95% CI = 1.006–1.237, p = 0.039). In contrast, the same polymorphism was associated with lower odds of BC in the heterozygous model of the Middle Eastern population (OR = 0.602, 95% CI = 0.375–0.966, p = 0.035) (Figure 5). No correlation was found between the TGF-β1 29T>C polymorphism and BC risk in the Caucasian and European populations. Similarly, results of other variants of TGF-β1 polymorphism have shown no association with BC risk in ethnic groups. These findings suggested that the TGF-β1 29T>C polymorphism alone is associated with BC risk in the Asian population. The ORs and 95% CIs of all stratified ethnic group analyses are provided in Table 2.
Figure 5.
Forest plot of stratified analysis of TGF-β1 polymorphisms that show association with either BC risk or lower odds in various ethnic groups. Black squares represent the odds ratios (ORs) of the individual studies, and horizontal lines indicate the 95% confidence intervals (CIs) of the ORs.
Table 2.
Stratified analysis of TGFβ polymorphisms and their association with BC risk.
| Model | Odds Ratio (OR) | 95% CI | p-Value | |
|---|---|---|---|---|
| TGF-β1 29C>T (Pro10Leu) or T869C or rs1982073 or rs1800470 | ||||
| Caucasian | ||||
| 1 | Overall allele C vs. T | 1.008 | 0.961–1.057 | 0.749 |
| 2 | Homozygous CC vs. TT | 1.007 | 0.907–1.120 | 0.891 |
| 3 | Heterozygous TC vs. TT | 1.038 | 0.990–1.088 | 0.123 |
| 4 | Recessive CC vs. TT + TC | 0.997 | 0.910–1.093 | 0.947 |
| 5 | Dominant CC + TC vs. TT | 1.039 | 0.994–1.086 | 0.093 |
| Asian | ||||
| 6 | Overall allele C vs. T | 1.058 | 0.952–1.176 | 0.298 |
| 7 | Homozygous CC vs. TT | 1.084 | 0.894–1.314 | 0.413 |
| 8 | Heterozygous TC vs. TT | 1.115 | 1.006–1.237 | 0.039 |
| 9 | Recessive CC vs. TT + TC | 1.055 | 0.875–1.271 | 0.576 |
| 10 | Dominant CC + TC vs. TT | 1.077 | 0.980–1.182 | 0.123 |
| European | ||||
| 11 | Overall allele C vs. T | 1.142 | 0.761–1.715 | 0.521 |
| 12 | Homozygous CC vs. TT | 1.466 | 0.516–4.116 | 0.473 |
| 13 | Heterozygous TC vs. TT | 1.319 | 0.536–3.247 | 0.547 |
| 14 | Recessive CC vs. TT + TC | 1.227 | 0.882–1.707 | 0.224 |
| 15 | Dominant CC + TC vs. TT | 1.355 | 0.552–3.312 | 0.505 |
| Middle Eastern | ||||
| 16 | Overall allele C vs. T | 0.833 | 0.625–1.110 | 0.212 |
| 17 | Homozygous CC vs. TT | 0.784 | 0.461–1.332 | 0.367 |
| 18 | Heterozygous TC vs. TT | 0.602 | 0.375–0.966 | 0.035 |
| 19 | Recessive CC vs. TT + TC | 1.011 | 0.632–1.618 | 0.964 |
| 20 | Dominant CC + TC vs. TT | 0.669 | 0.438–1.020 | 0.062 |
| TGFβ-1-509 C/T or -1349C>T or rs1800469 | ||||
| Caucasian | ||||
| 21 | Overall allele T vs. C | 0.953 | 0.905–1.004 | 0.068 |
| 22 | Homozygous TT vs. CC | 0.904 | 0.803–1.018 | 0.096 |
| 23 | Heterozygous CT vs. CC | 0.957 | 0.891–1.028 | 0.228 |
| 24 | Recessive TT vs. CC + CT | 0.924 | 0.825–1.036 | 0.175 |
| 25 | Dominant TT + CT vs. CC | 0.947 | 0.885–1.031 | 0.112 |
| European | ||||
| 26 | Overall allele T vs. C | 1.016 | 0.900–1.148 | 0.795 |
| 27 | Homozygous TT vs. CC | 1.039 | 0.770–1.401 | 0.804 |
| 28 | Heterozygous CT vs. CC | 1.038 | 0.950–1.135 | 0.405 |
| 29 | Recessive TT vs. CC + CT | 1.026 | 0.782–1.346 | 0.853 |
| 30 | Dominant TT + CT vs. CC | 1.058 | 0.972–1.151 | 0.190 |
| Asian | ||||
| 31 | Overall allele T vs. C | 1.023 | 0.925–1.132 | 0.657 |
| 32 | Homozygous TT vs. CC | 1.000 | 0.813–1.230 | 0.990 |
| 33 | Heterozygous CT vs. CC | 1.633 | 0.894–2.986 | 0.111 |
| 34 | Recessive TT vs. CC + CT | 0.993 | 0.842–1.171 | 0.936 |
| 35 | Dominant TT + CT vs. CC | 1.460 | 0.886–2.408 | 0.138 |
| TGF-β *6A or rs11466445 | ||||
| European | ||||
| 36 | Overall allele 6A vs. 9A | 1.095 | 0.878–1.364 | 0.421 |
| 37 | Homozygous 6A/6A vs. 9A/9A | 1.247 | 0.749–2.075 | 0.396 |
| 38 | Heterozygous 6A/9A vs. 9A/9A | 1.056 | 0.809–1.378 | 0.690 |
| 39 | Recessive 6A/6A vs. 9A/9A + 9A/6A | 1.249 | 0.751–2.077 | 0.391 |
| 40 | Dominant 6A/6A + 9A/6A vs. 9A/9A | 1.049 | 0.839–1.311 | 0.675 |
2.7. Sensitivity Analysis
One-way sensitivity analysis was performed to analyze the effect of each study on the combined OR. During the sensitivity analysis for each polymorphism, each relevant study was deleted iteratively from the dataset prior to the analysis. No single study had a significant influence on any of the combined ORs, suggesting that our meta-analysis is relatively robust and credible (Supplementary Figures S5–S7).
2.8. Trial Sequential Analysis (TSA)
TSA was used to investigate whether the number of samples included in the present study was sufficient for detecting a possible role of each TGF-β1 polymorphism in BC. The Z curve touched the trial monitoring boundaries or reached the required information line, indicating that a sufficient number of studies were included in the meta-analysis for overall and ethnic group investigations (Supplementary Figure S8).
3. Discussion
The TGF-β1 signaling pathway has been studied extensively in several cancers, including BC. TGF-β1 plays an important role in cell differentiation, migration, invasion, and tumor growth [4]. Both tumorigenic and tumor-suppressive roles of TGF-β1 have been well established [5]. Elevated plasma levels of TGF-β1 have been associated with cancer development, and TGF-β1 polymorphisms have been found to cause high transcription and expression of TGF-β1 [9,11]. The prominent role of TGF-β1 in cancer progression underscores the importance of studying the association between TGF-β1 polymorphisms and BC risk.
The most common TGF-β1 polymorphism is a substitution of cytosine with thymine at the 29th nucleotide (TGF-β1 29 T>C), which results in the substitution of the proline at codon 10 in exon 1 with leucine [8,9,10,11]. TGF-β1 29T>C has been associated with elevated levels of TGF-β1 in plasma [9]. Studies have also suggested that TGF-β1 29T>C is associated with BC risk; however, contrasting results were reported. Dunning et al. suggested that the TGF-β1 29T>C polymorphism is associated with BC risk; they also found that this polymorphism is associated with elevated levels of the TGF-β1 protein [9]. In contrast, Ziv et al. reported no such associations [12]. Marchand et al. reported similar results in their multi-ethnic study, which suggested no association between the TGF-β1 29T>C polymorphism and BC [13]. Interestingly, Lee et al. demonstrated that the TGF-β1 29T>C polymorphism was associated with an increased risk of BC in postmenopausal women [32,33]. However, Hishida et al. observed a negative correlation between the TGF-β1 29T>C polymorphism and BC risk in premenopausal women [27]. Interestingly, Shin et al. demonstrated that no association was found in the initial stages of BC, whereas a significant correlation was observed in later stages [30].
A systematic meta-analysis was performed to provide conclusive evidence for the association of TGF-β polymorphism with breast cancer risk. Interestingly, neither the overall allele nor genotypic models showed an association between the TGF-β1 29T>C polymorphism and BC risk. However, the TGF-β1 29T>C polymorphism was found to be associated with an increased risk of BC in the heterozygous model of the Asian population. In contrast, the TGF-β129T>C polymorphism favors lower odds of BC in the heterozygous model of the Middle Eastern population.
The TGF-β1-509 C/T polymorphism is located in the promoter region of TGF-β1. Recent studies demonstrated the association between the TGF-β1-509 C/T polymorphism and elevated levels of the TGF-β protein. It has also been well established that the TGF-β1-509 C/T polymorphism is highly associated with BC risk. Both Dunning et al. [9] and Parvizi et al. [52] suggested that the polymorphism is associated with increased risk of invasive BC, whereas Cox et al. showed no such association [35]. Vinod et al. demonstrated that heterozygosity of the TGF-β1-509 C/T polymorphism was associated with BC susceptibility in an Indian population [51]. However, Babyshkina et al. reported that homozygosity of the T allele of the TGF-β1-509 C/T polymorphism was not associated with BC in a Russian population [49]. These inconclusive and controversial results suggest the necessity of a cumulative and robust analysis to arrive at a definitive conclusion regarding the association between this polymorphism and cancer risk in multiple ethnic groups. Six meta-analyses were performed to address these issues; three investigated the association between the TGF-β1-509 C/T polymorphism and multiple cancer subtypes. Huang et al., Qi et al., and Woo et al. performed meta-analyses examining the association between the TGF-β1-509 C/T polymorphism and BC specifically [14,17,58]. However, these studies were performed using a limited number of studies, and Huang et al. included a study that was not related to BC risk [58]. These drawbacks can influence the analysis and affect the results. In order to definitively determine the association between the TGF-β1-509 C/T polymorphism and BC, we performed a meta-analysis including additional studies and excluding studies not related to BC risk. Our robust analysis demonstrated that the TGF-β1-509 C/T polymorphism is not associated with BC in the overall population or within specific ethnic groups.
Researchers have identified a polyalanine variant in exon 5 of TGF-β1 and analyzed its association with BC risk. Recent studies have also suggested that the TGF-β1*6A polymorphism is associated with BC risk. Specifically, Kaklamani et al., Pasche et al., and Song et al. reported that the polymorphism is associated with an increased incidence of BC [19,31,55]. Pasche et al. also found that the TGF-β1*6A polymorphism is associated with risk for other cancers, including colorectal and ovarian cancer [19]. However, Chen et al., Colleran et al., and Cox et al. suggested that the TGF-β1*6A variant is not associated with BC risk. Meta-analyses have been performed; however, they investigated the association between the TGF-β1*6A polymorphism and all cancer types; no meta-analyses have yet correlated this polymorphism with BC specifically [20,59]. Thus, in the present study, we analyzed for the first time the association between the TGF-β1*6A polymorphism and BC risk. Our cumulative analysis suggests that the TGF-β1*6A variant is not associated with BC risk. In our analysis of the European ethnic group, the TGF-β1*6A variant also showed no association with BC risk.
Although various meta-analyses have already been reported in the literature, the present study has several advantages over them. A recent investigation by Alqumber et al. included reports published until the year 2013; in contrast, we screened for studies published up to May 2019, resulting in the inclusion of three additional studies. Furthermore, we assessed the possible association of three TGF-β1 polymorphisms (TGF-β1 29>T/C, TGF-β1-509 C/T, and TGF-β1*6A) with the risk of BC [60]. However, multiple comparisons failed to show a significant correlation between these polymorphisms with BC risk: the present study warrants the non-importance of TGF-β1 polymorphisms on the pathogenesis of BC. Finally, TSA revealed that our meta-analysis included a sufficient number of studies, performed worldwide, to dissect the possible association of TGF-β1 variants with BC risk, and further investigation is not required.
4. Materials and Methods
4.1. Literature Search Strategy and Selection of Relevant Studies
Three researchers independently conducted a comprehensive systematic electronic literature search using the online databases Pubmed, EMBASE, and Google Scholar. The following terms were used either alone or in combination: “Transforming Growth Factor β1 polymorphism”; “breast cancer”; for the TGF-β1 29T>C polymorphism: “TGF-β1 29T>C”, “TGF-β1Pro10Leu”, “TGF-β1T869C”, “rs1982073”, “rs180047029”; for the TGF-β1-509 C/T polymorphism: “TGF-β1-509 C/T”, “TGF-β1 1349C>T”, “rs1800469”; and for the TGF-β1*6A polymorphism: “TGF-β1*6A”, “rs11466445”. We also examined the cross-references in the retrieved studies for publications that were missed in the above search.
4.2. Criteria for the Inclusion and Exclusion of Studies
The present meta-analysis included studies that evaluated the association between TGF-β1 polymorphisms and BC risk, were published in English, presented original data, and provided the genotypic frequency of both case and control samples or had odds ratios (ORs) with 95% confidence interval (CI) values. The analysis excluded reviews, abstracts, duplicate or overlapping studies, studies not published in English, studies that correlated TGF-β1 polymorphisms with non-breast cancers, and studies that did not provide genotypic or allele frequencies for both case and control samples.
4.3. Data Extraction
Each publication was assessed thoroughly, and data were extracted independently by three researchers, following the same pattern and extracting the same data essential for meta-analyses that have been previously described [22,23]. The researchers conducted group discussions to resolve any discrepancies in extraction.
4.4. Statistical Analysis
Statistical analysis was performed using Comprehensive Meta-Analysis Software (CMA version 3) (Biostat, 14 North Dean Street, Englewood, NJ 07631 USA). In order to appraise the associations between TGF-β1 polymorphisms and BC risk, the combined OR, 95% CIs, and their respective p-values were calculated by comparing the cancer patient samples vs. respective healthy controls. Heterogeneity among the studies and publication bias were calculated using Begg’s funnel plots and chi-squared-based Cochran’s Q tests, respectively. Egger’s regression tests were performed to analyze and measure the symmetry of funnel plots, as described previously [22]. Asymmetrical funnel plots were made symmetrical using the “Trim and Fill” method. Random models were used to calculate the combined ORs for studies showing heterogeneity [24]. In contrast, fixed models were used to calculate the combined ORs for studies with homogeneity [25].
4.5. Trial Sequential Analysis (TSA)
An appropriate meta-analysis must include all eligible studies published to date to draw definitive conclusions. The number of studies available in the literature in the studied areas should be sufficient for decisive concluding remarks. TSA is a statistical tool developed by the Copenhagen Trial Unit, Centre for Clinical Intervention Research, Denmark, to estimate the sample size required to reach significance with definitive power. In TSA, a Z curve analysis is performed to check whether a sufficient number of samples is included in the study. If the Z curve intersects the TSA monitoring boundary before reaching the required information size or if the total number of samples exceeds the required information line, then the total number of studies included in the investigation is sufficient, and additional trials are not required. On the other hand, if the Z curve fails to touch the TSA monitoring boundaries or cross the required information line, the number of studies is limited and more are required to draw any definitive conclusions. TSA software version 0.9 (http://www.ctu.dk/tsa) was used for this analysis.
5. Conclusions
TGF-β1 plays an important role in BC progression, metastasis, stemness, and chemo-resistance. TGF-β1 has both pro-oncogenic and tumor-suppressive roles during cancer development. High levels of TGF-β1, which are influenced by TGF-β1 polymorphisms, have been associated with cancer risk. TGF-β1polymorphisms have been associated with BC risk, but conflicting results have also been reported recently. Previous meta-analyses either included studies on TGF-β1 polymorphisms that did not evaluate associations with BC or did not include new studies with huge datasets that could greatly influence their results. To address these shortcomings, a stringent and highly robust analysis was conducted to provide conclusive evidence for the association between TGF-β1 polymorphisms and BC risk. The current meta-analysis included large datasets from all recent eligible studies and excluded studies that did not evaluate the associations between these polymorphisms and BC. This study suggests that TGF-β1 polymorphisms are not associated with BC risk in the overall population. Both TGF-β1-509 C/T and TGF-β1*6A also showed no association with BC risk in stratified analyses. However, the TGF-β1 29T>C polymorphism was found to be associated with BC risk in the Asian ethnic group.
Acknowledgments
We acknowledge Beckman research institute of City of Hope for support and fund. We sincerely acknowledge our institutional scientific writer Ms. Kerin Higa for editing the manuscript.
Supplementary Materials
The following are available online at https://www.mdpi.com/2072-6694/12/2/471/s1, Figure S1: PRISMA flowchart: Search, screening and selection of eligible studies included in meta-analysis for evaluating the association of TGF-β polymorphisms with breast cancer risk. Figure S2: Funnel plot: Assessment of publication bias using Begg’s funnel plot and Egger’s regression test. Asymmetric Begg’s funnel plot was made symmetric using “Trim and Fill” method. Figure S3: Forest plot: Analyzing the influence of detection methods on association of TGF-β1 29T>C polymorphism with breast cancer risk. Studies were separated based on the methods used for the detection of polymorphism [(A) PCR-RFLP and (B) Taqman] and were analyzed individually. Figure S4: Forest plot: Studies were separated based on the methods used for the detection of TGF-β1 -509 C/T polymorphism [(A) PCR-RFLP and (B) Taqman] and were analyzed individually to assess its association with increased risk of breast cancer. Figure S5: Sensitivity analysis: One individual study of TGF-β1 29T>C was excluded each time and was analyzed to assess the effect of individual study on combined OR. Figure S6: Sensitivity analysis: Each study was assessed for the potential influence on combined OR by deleting single study each time and performing the analysis for TGF-β1 -509 C/T polymorphism. Figure S7: Sensitivity analysis of each study representing TGF-β *6A polymorphism in the meta-analysis by one study removal method. Figure S8: Trial sequence analysis of all the included studies. TSA figure for TGFβ-1 29T>C overall (A), Caucasian (B), Asian (C) showed enough number of samples enrolled for the analysis except for the American (D). TSA analysis for other two SNPs TGF-β1 -509 C/T (E) and TGF-β*6A (F) also revealed similar results. Table S1: Statistics for heterogeneity test.
Author Contributions
S.S.S.: conceptualized and obtained funding for the project; B.M.K., S.J., and A.K.P.: prepared original draft of the manuscript; D.H., R.S., S.A., and S.S.S.: reviewed and edited the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
The present work was supported in part by grants from the United States Department of Defense (W81XWH-16-1-0641) and the National Cancer Institute of the National Institutes of Health (P30CA33572). Funding from the Beckman Research Institute of City of Hope is also acknowledged.
Conflicts of Interest
Authors declare no conflict of interest.
References
- 1.Morikawa M., Derynck R., Miyazono K. TGF-beta and the TGF-beta Family: Context-Dependent Roles in Cell and Tissue Physiology. Cold Spring Harb. Perspect. Biol. 2016;8:a021873. doi: 10.1101/cshperspect.a021873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Massague J. TGFbeta signalling in context. Nat. Rev. Mol. Cell Biol. 2012;13:616–630. doi: 10.1038/nrm3434. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Moses H., Barcellos-Hoff M.H. TGF-beta biology in mammary development and breast cancer. Cold Spring Harb. Perspect. Biol. 2011;3:a003277. doi: 10.1101/cshperspect.a003277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Syed V. TGF-beta Signaling in Cancer. J. Cell. Biochem. 2016;117:1279–1287. doi: 10.1002/jcb.25496. [DOI] [PubMed] [Google Scholar]
- 5.Massague J. TGFbeta in Cancer. Cell. 2008;134:215–230. doi: 10.1016/j.cell.2008.07.001. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Heldin C.H., Vanlandewijck M., Moustakas A. Regulation of EMT by TGFbeta in cancer. FEBS Lett. 2012;586:1959–1970. doi: 10.1016/j.febslet.2012.02.037. [DOI] [PubMed] [Google Scholar]
- 7.Katsuno Y., Lamouille S., Derynck R. TGF-beta signaling and epithelial-mesenchymal transition in cancer progression. Curr. Opin. Oncol. 2013;25:76–84. doi: 10.1097/CCO.0b013e32835b6371. [DOI] [PubMed] [Google Scholar]
- 8.Syrris P., Carter N.D., Metcalfe J.C., Kemp P.R., Grainger D.J., Kaski J.C., Crossman D.C., Francis S.E., Gunn J., Jeffery S., et al. Transforming growth factor-beta1 gene polymorphisms and coronary artery disease. Clin. Sci. 1998;95:659–667. doi: 10.1042/cs0950659. [DOI] [PubMed] [Google Scholar]
- 9.Dunning A.M., Ellis P.D., McBride S., Kirschenlohr H.L., Healey C.S., Kemp P.R., Luben R.N., Chang-Claude J., Mannermaa A., Kataja V., et al. A transforming growth factorbeta1 signal peptide variant increases secretion in vitro and is associated with increased incidence of invasive breast cancer. Cancer Res. 2003;63:2610–2615. [PubMed] [Google Scholar]
- 10.Pasche B., Luo Y., Rao P.H., Nimer S.D., Dmitrovsky E., Caron P., Luzzatto L., Offit K., Cordon-Cardo C., Renault B., et al. Type I transforming growth factor beta receptor maps to 9q22 and exhibits a polymorphism and a rare variant within a polyalanine tract. Cancer Res. 1998;58:2727–2732. [PubMed] [Google Scholar]
- 11.Martelossi Cebinelli G.C., Paiva Trugilo K., Badaro Garcia S., Brajao de Oliveira K. TGF-beta1 functional polymorphisms: A review. Eur. Cytokine Netw. 2016;27:81–89. doi: 10.1684/ecn.2016.0382. [DOI] [PubMed] [Google Scholar]
- 12.Ziv E., Cauley J., Morin P.A., Saiz R., Browner W.S. Association between the T29→C polymorphism in the transforming growth factor beta1 gene and breast cancer among elderly white women: The Study of Osteoporotic Fractures. JAMA. 2001;285:2859–2863. doi: 10.1001/jama.285.22.2859. [DOI] [PubMed] [Google Scholar]
- 13.Le Marchand L., Haiman C.A., van den Berg D., Wilkens L.R., Kolonel L.N., Henderson B.E. T29C polymorphism in the transforming growth factor beta1 gene and postmenopausal breast cancer risk: The Multiethnic Cohort Study. Cancer Epidemiol. Biomarkers Prev. 2004;13:412–415. [PubMed] [Google Scholar]
- 14.Qi X., Zhang F., Yang X., Fan L., Zhang Y., Chen L., Zhou Y., Chen X., Zhong L., Jiang J. Transforming growth factor-beta1 polymorphisms and breast cancer risk: A meta-analysis based on 27 case-control studies. Breast Cancer Res. Treat. 2010;122:273–279. doi: 10.1007/s10549-010-0847-6. [DOI] [PubMed] [Google Scholar]
- 15.Guo W., Dong Z., Guo Y., Chen Z., Yang Z., Kuang G., Shan B. Polymorphisms of transforming growth factor-beta1 associated with increased risk of gastric cardia adenocarcinoma in north China. Int. J. Immunogenet. 2011;38:215–224. doi: 10.1111/j.1744-313X.2010.00991.x. [DOI] [PubMed] [Google Scholar]
- 16.Cao H., Zhou Q., Lan R., Roe O.D., Chen X., Chen Y., Wang D. A functional polymorphism C-509T in TGFbeta-1 promoter contributes to susceptibility and prognosis of lone atrial fibrillation in Chinese population. PLoS ONE. 2014;9:e112912. doi: 10.1371/journal.pone.0112912. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Woo S.U., Park K.H., Woo O.H., Yang D.S., Kim A.R., Lee E.S., Lee J.B., Kim Y.H., Kim J.S., Seo J.H. Association of a TGF-beta1 gene -509 C/T polymorphism with breast cancer risk: A meta-analysis. Breast Cancer Res. Treat. 2010;124:481–485. doi: 10.1007/s10549-010-0871-6. [DOI] [PubMed] [Google Scholar]
- 18.Grossberg A.J., Lei X., Xu T., Shaitelman S.F., Hoffman K.E., Bloom E.S., Stauder M.C., Tereffe W., Schlembach P.J., Woodward W.A., et al. Association of Transforming Growth Factor beta Polymorphism C-509T With Radiation-Induced Fibrosis Among Patients With Early-Stage Breast Cancer: A Secondary Analysis of a Randomized Clinical Trial. JAMA Oncol. 2018;4:1751–1757. doi: 10.1001/jamaoncol.2018.2583. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Pasche B., Kolachana P., Nafa K., Satagopan J., Chen Y.G., Lo R.S., Brener D., Yang D., Kirstein L., Oddoux C., et al. TbetaR-I(6A) is a candidate tumor susceptibility allele. Cancer Res. 1999;59:5678–5682. [PubMed] [Google Scholar]
- 20.Wang Y.Q., Qi X.W., Wang F., Jiang J., Guo Q.N. Association between TGFBR1 polymorphisms and cancer risk: A meta-analysis of 35 case-control studies. PLoS ONE. 2012;7:e42899. doi: 10.1371/journal.pone.0042899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Baxter S.W., Choong D.Y., Eccles D.M., Campbell I.G. Transforming growth factor beta receptor 1 polyalanine polymorphism and exon 5 mutation analysis in breast and ovarian cancer. Cancer Epidemiol. Biomarkers Prev. 2002;11:211–214. [PubMed] [Google Scholar]
- 22.Krishna B.M., Chaudhary S., Panda A.K., Mishra D.R., Mishra S.K. Her2 [1]655(Val) polymorphism and its association with breast cancer risk: An updated meta-analysis of case-control studies. Sci. Rep. 2018;8:7427. doi: 10.1038/s41598-018-25769-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Chaudhary S., Panda A.K., Mishra D.R., Mishra S.K. Association of +331G/A PgR polymorphism with susceptibility to female reproductive cancer: Evidence from a meta-analysis. PLoS ONE. 2013;8:e53308. doi: 10.1371/journal.pone.0053308. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Mantel N., Haenszel W. Statistical aspects of the analysis of data from retrospective studies of disease. J. Natl. Cancer Inst. 1959;22:719–748. [PubMed] [Google Scholar]
- 25.DerSimonian R., Laird N. Meta-analysis in clinical trials. Control. Clin. Trials. 1986;7:177–188. doi: 10.1016/0197-2456(86)90046-2. [DOI] [PubMed] [Google Scholar]
- 26.Krippl P., Langsenlehner U., Renner W., Yazdani-Biuki B., Wolf G., Wascher T.C., Paulweber B., Bahadori B., Samonigg H. The L10P polymorphism of the transforming growth factor-beta 1 gene is not associated with breast cancer risk. Cancer Lett. 2003;201:181–184. doi: 10.1016/S0304-3835(03)00468-3. [DOI] [PubMed] [Google Scholar]
- 27.Hishida A., Iwata H., Hamajima N., Matsuo K., Mizutani M., Iwase T., Miura S., Emi N., Hirose K., Tajima K. Transforming growth factor B1 T29C polymorphism and breast cancer risk in Japanese women. Breast Cancer. 2003;10:63–69. doi: 10.1007/BF02967627. [DOI] [PubMed] [Google Scholar]
- 28.Jin Q., Hemminki K., Grzybowska E., Klaes R., Soderberg M., Zientek H., Rogozinska-Szczepka J., Utracka-Hutka B., Pamula J., Pekala W., et al. Polymorphisms and haplotype structures in genes for transforming growth factor beta1 and its receptors in familial and unselected breast cancers. Int. J. Cancer. 2004;112:94–99. doi: 10.1002/ijc.20370. [DOI] [PubMed] [Google Scholar]
- 29.Saha A., Gupta V., Bairwa N.K., Malhotra D., Bamezai R. Transforming growth factor-beta1 genotype in sporadic breast cancer patients from India: Status of enhancer, promoter, 5′-untranslated-region and exon-1 polymorphisms. Eur. J. Immunogenet. 2004;31:37–42. doi: 10.1111/j.1365-2370.2004.00442.x. [DOI] [PubMed] [Google Scholar]
- 30.Shin A., Shu X.O., Cai Q., Gao Y.T., Zheng W. Genetic polymorphisms of the transforming growth factor-beta1 gene and breast cancer risk: A possible dual role at different cancer stages. Cancer Epidemiol. Biomarkers Prev. 2005;14:1567–1570. doi: 10.1158/1055-9965.EPI-05-0078. [DOI] [PubMed] [Google Scholar]
- 31.Kaklamani V.G., Baddi L., Liu J., Rosman D., Phukan S., Bradley C., Hegarty C., McDaniel B., Rademaker A., Oddoux C., et al. Combined genetic assessment of transforming growth factor-beta signaling pathway variants may predict breast cancer risk. Cancer Res. 2005;65:3454–3461. doi: 10.1158/0008-5472.CAN-04-2961. [DOI] [PubMed] [Google Scholar]
- 32.Lee K.M., Park S.K., Hamajima N., Tajima K., Yoo K.Y., Shin A., Noh D.Y., Ahn S.H., Hirvonen A., Kang D. Genetic polymorphisms of TGF-beta1 & TNF-beta and breast cancer risk. Breast Cancer Res. Treat. 2005;90:149–155. doi: 10.1007/s10549-004-3859-2. [DOI] [PubMed] [Google Scholar]
- 33.Feigelson H.S., Patel A.V., Diver W.R., Stevens V.L., Thun M.J., Calle E.E. Transforming growth factor beta receptor type I and transforming growth factor beta1 polymorphisms are not associated with postmenopausal breast cancer. Cancer Epidemiol. Biomarkers Prev. 2006;15:1236–1237. doi: 10.1158/1055-9965.EPI-06-0163. [DOI] [PubMed] [Google Scholar]
- 34.Scola L., Vaglica M., Crivello A., Palmeri L., Forte G.I., Macaluso M.C., Giacalone A., Di Noto L., Bongiovanni A., Raimondi C., et al. Cytokine gene polymorphisms and breast cancer susceptibility. Ann. N. Y. Acad. Sci. 2006;1089:104–109. doi: 10.1196/annals.1386.017. [DOI] [PubMed] [Google Scholar]
- 35.Cox D.G., Penney K., Guo Q., Hankinson S.E., Hunter D.J. TGFB1 and TGFBR1 polymorphisms and breast cancer risk in the Nurses’ Health Study. BMC Cancer. 2007;7:175. doi: 10.1186/1471-2407-7-175. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Gonullu G., Basturk B., Evrensel T., Oral B., Gozkaman A., Manavoglu O. Association of breast cancer and cytokine gene polymorphism in Turkish women. Saudi Med. J. 2007;28:1728–1733. [PubMed] [Google Scholar]
- 37.Cox A., Dunning A.M., Garcia-Closas M., Balasubramanian S., Reed M.W., Pooley K.A., Scollen S., Baynes C., Ponder B.A., Chanock S., et al. A common coding variant in CASP8 is associated with breast cancer risk. Nat. Genet. 2007;39:352–358. doi: 10.1038/ng1981. [DOI] [PubMed] [Google Scholar]
- 38.Rajkumar T., Samson M., Rama R., Sridevi V., Mahji U., Swaminathan R., Nancy N.K. TGFbeta1 (Leu10Pro), p53 (Arg72Pro) can predict for increased risk for breast cancer in south Indian women and TGFbeta1 Pro (Leu10Pro) allele predicts response to neo-adjuvant chemo-radiotherapy. Breast Cancer Res. Treat. 2008;112:81–87. doi: 10.1007/s10549-007-9821-3. [DOI] [PubMed] [Google Scholar]
- 39.Joshi N.N., Kale M.D., Hake S.S., Kannan S. Transforming growth factor beta signaling pathway associated gene polymorphisms may explain lower breast cancer risk in western Indian women. PLoS ONE. 2011;6:e21866. doi: 10.1371/journal.pone.0021866. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Zhang M., Guo L.L., Cheng Z., Liu R.Y., Lu Y., Qian Q., Lei Z., Zhang H.T. A functional polymorphism of TGFBR2 is associated with risk of breast cancer with ER(+), PR(+), ER(+)PR(+) and HER2(-) expression in women. Oncol. Lett. 2011;2:653–658. doi: 10.3892/ol.2011.312. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Pooja S., Francis A., Rajender S., Tamang R., Rajkumar R., Saini K.S., Megu K., Goel M.M., Surekha D., Rao D.R., et al. Strong impact of TGF-beta1 gene polymorphisms on breast cancer risk in Indian women: A case-control and population-based study. PLoS ONE. 2013;8:e75979. doi: 10.1371/journal.pone.0075979. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Taubenschuss E., Marton E., Mogg M., Frech B., Ehart L., Muin D., Schreiber M. The L10P polymorphism and serum levels of transforming growth factor beta1 in human breast cancer. Int. J. Mol. Sci. 2013;14:15376–15385. doi: 10.3390/ijms140815376. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Amani D., Khalilnezhad A., Ghaderi A., Niikawa N., Yoshiura K. Transforming growth factor beta1 (TGFbeta1) polymorphisms and breast cancer risk. Tumour Biol. 2014;35:4757–4764. doi: 10.1007/s13277-014-1621-x. [DOI] [PubMed] [Google Scholar]
- 44.Joshi N.N., Bhat S., Hake S., Kale M., Kannan S. Opposing effects of pro- and anti-inflammatory cytokine gene polymorphisms on the risk for breast cancer in western Indian women: A pilot study. Int. J. Immunogenet. 2014;41:242–249. doi: 10.1111/iji.12098. [DOI] [PubMed] [Google Scholar]
- 45.Dehghan Tezerjani M., Beik M., Kalantar S.M., Rasti A., Kalantar S.M., Shiryazdi S.M. Transforming growth factor Beta leucine10 proline variation and breast cancer risk in Iranian women. Iran. J. Public Health. 2015;44:427–429. [PMC free article] [PubMed] [Google Scholar]
- 46.Jakubowska A., Jaworska K., Cybulski C., Janicka A., Szymanska-Pasternak J., Lener M., Narod S.A., Lubinski J., IHCC-Breast Cancer Study Group Do BRCA1 modifiers also affect the risk of breast cancer in non-carriers? Eur. J. Cancer. 2009;45:837–842. doi: 10.1016/j.ejca.2008.10.021. [DOI] [PubMed] [Google Scholar]
- 47.Jakubowska A., Gronwald J., Menkiszak J., Gorski B., Huzarski T., Byrski T., Toloczko-Grabarek A., Gilbert M., Edler L., Zapatka M., et al. BRCA1-associated breast and ovarian cancer risks in Poland: No association with commonly studied polymorphisms. Breast Cancer Res. Treat. 2010;119:201–211. doi: 10.1007/s10549-009-0390-5. [DOI] [PubMed] [Google Scholar]
- 48.MARIE-GENICA Consortium on Genetic Susceptibility for Menopausal Hormone Therapy Related Breast Cancer Risk Genetic polymorphisms in phase I and phase II enzymes and breast cancer risk associated with menopausal hormone therapy in postmenopausal women. Breast Cancer Res. Treat. 2010;119:463–474. doi: 10.1007/s10549-009-0407-0. [DOI] [PubMed] [Google Scholar]
- 49.Babyshkina N., Malinovskaya E., Stakheyeva M., Volkomorov V., Slonimskaya E., Maximov V., Cherdyntseva N. Association of functional -509C>T polymorphism in the TGF-beta1 gene with infiltrating ductal breast carcinoma risk in a Russian Western Siberian population. Cancer Epidemiol. 2011;35:560–563. doi: 10.1016/j.canep.2011.02.002. [DOI] [PubMed] [Google Scholar]
- 50.Quan L., Gong Z., Yao S., Bandera E.V., Zirpoli G., Hwang H., Roberts M., Ciupak G., Davis W., Sucheston L., et al. Cytokine and cytokine receptor genes of the adaptive immune response are differentially associated with breast cancer risk in American women of African and European ancestry. Int. J. Cancer. 2014;134:1408–1421. doi: 10.1002/ijc.28458. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Vinod C., Jyothy A., Vijay Kumar M., Raghu Raman R., Nallari P., Venkateshwari A. Heterozygosity for TGF beta1 -509C/T Polymorphism is associated with risk for breast cancer in South Indian population. Tumour Biol. 2013;34:99–105. doi: 10.1007/s13277-012-0516-y. [DOI] [PubMed] [Google Scholar]
- 52.Parvizi S., Mohammadzadeh G., Karimi M., Noorbehbahani M., Jafary A. Effects of Two Common Promoter Polymorphisms of Transforming Growth Factor-beta1 on Breast Cancer Risks in Ahvaz, West South of Iran. Iran. J. Cancer Prev. 2016;9:e5266. doi: 10.17795/ijcp-5266. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Pasche B., Kaklamani V., Hou N., Young T., Rademaker A., Peterlongo P., Ellis N., Offit K., Caldes T., Reiss M., et al. TGFBR1*6A and cancer: A meta-analysis of 12 case-control studies. J. Clin. Oncol. 2004;22:756–758. doi: 10.1200/JCO.2004.99.271. [DOI] [PubMed] [Google Scholar]
- 54.Chen T., Jackson C.R., Link A., Markey M.P., Colligan B.M., Douglass L.E., Pemberton J.O., Deddens J.A., Graff J.R., Carter J.H. Int7G24A variant of transforming growth factor-beta receptor type I is associated with invasive breast cancer. Clin. Cancer Res. 2006;12:392–397. doi: 10.1158/1078-0432.CCR-05-1518. [DOI] [PubMed] [Google Scholar]
- 55.Song B., Margolin S., Skoglund J., Zhou X., Rantala J., Picelli S., Werelius B., Lindblom A. TGFBR1(*)6A and Int7G24A variants of transforming growth factor-beta receptor 1 in Swedish familial and sporadic breast cancer. Br. J. Cancer. 2007;97:1175–1179. doi: 10.1038/sj.bjc.6603961. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Colleran G., McInerney N., Rowan A., Barclay E., Jones A.M., Curran C., Miller N., Kerin M., Tomlinson I., Sawyer E. The TGFBR1*6A/9A polymorphism is not associated with differential risk of breast cancer. Breast Cancer Res. Treat. 2010;119:437–442. doi: 10.1007/s10549-009-0395-0. [DOI] [PubMed] [Google Scholar]
- 57.Kamali E., Hemmati S., Safari F., Tavassoli M. TGFBR1 polymorphism and risk of breast cancer in Iranian women. Int. J. Biol. Markers. 2015;30:e414–e417. doi: 10.5301/jbm.5000102. [DOI] [PubMed] [Google Scholar]
- 58.Huang Y., Hao Y., Li B., Xie J., Qian J., Chao C., Yu L. Lack of significant association between TGF-beta1-590C/T polymorphism and breast cancer risk: A meta-analysis. Med. Oncol. 2011;28:424–428. doi: 10.1007/s12032-010-9491-6. [DOI] [PubMed] [Google Scholar]
- 59.Liao R.Y., Mao C., Qiu L.X., Ding H., Chen Q., Pan H.F. TGFBR1*6A/9A polymorphism and cancer risk: A meta-analysis of 13,662 cases and 14,147 controls. Mol. Biol. Rep. 2010;37:3227–3232. doi: 10.1007/s11033-009-9906-7. [DOI] [PubMed] [Google Scholar]
- 60.Alqumber M., Sajad A.D., Haque S., Wahid M., Singh R., Akhter N. No Association of the TGF-β129T/C polymorphism with breast cancer risk in caucasian and asian populations: Evidence from a Meta-Analysis Involving 55, 841 Subjects. Asian Pac. J Cancer Prev. 2014;15:8725–8734. doi: 10.7314/APJCP.2014.15.20.8725. [DOI] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.