Abstract
The chloroplast RNA splicing and ribosome maturation (CRM) domain-containing proteins regulate the expression of chloroplast or mitochondrial genes that influence plant growth and development. Although 14 CRM domain proteins have previously been identified in rice, there are few studies of these gene expression patterns in various tissues and under abiotic stress. In our study, we found that 14 CRM domain-containing proteins have a conservative motif1. Under salt stress, the expression levels of 14 CRM genes were downregulated. However, under drought and cold stress, the expression level of some CRM genes was increased. The analysis of gene expression patterns showed that 14 CRM genes were expressed in all tissues but especially highly expressed in leaves. In addition, we analyzed the functions of OsCFM2 and found that this protein influences chloroplast development by regulating the splicing of a group I and five group II introns. Our study provides information for the function analysis of CRM domain-containing proteins in rice.
Keywords: CRM domain, intron splicing, chloroplast development, rice, abiotic stress
1. Introduction
The chloroplast RNA splicing and ribosome maturation (CRM) domain proteins play an important role in plant growth and development by influencing RNA-binding activity according to the characteristics and structure of their CRM domains [1,2]. CRM domain proteins regulate chloroplast- or mitochondria-related gene expression through post-transcriptional regulation [3,4]. It is reported that CRM proteins participate in the process of intron splicing and affect rRNA processing in chloroplasts [5,6,7]. In plants, mutations in CRM genes lead to an albino seedling phenotype, developmental delay, or sensitivity to abiotic stress [8,9,10,11].
Based on their structure and the count of CRM domains they contain, the CRM family proteins can be divided into four subfamilies: The CRS1 subfamily, CAF subfamily, subfamily 3, and subfamily 4 [1,5]. Among these, CRS1 subfamily, CAF subfamily, and subfamily 3 proteins have been reported to regulate the splicing of group I and group II introns [5]. Meanwhile, subfamily 4 proteins are associated with the assemblage of the large ribosomal subunit [10]. The functions of CRM domain proteins in Arabidopsis thaliana and maize have been widely reported. The first CRM gene, ZmCRS1, has been identified and cloned in maize; the ZmCRS1 protein contains three CRM domains that can directly bind to, and regulate splicing of atpF intron in chloroplasts [11]. The CFM3 protein also has three CRM domains, which are related to splicing of trnG, rps16, ndhB, rpl16, petB, and petD introns in A. thaliana [12]. CFM2 contains four CRM domains and affects group I and group II introns’ splicing in A. thaliana and maize [8,13]. Previous studies have indicated that CAF1 and CAF2 belong to the CAF subfamily, and can interact with CRS2 to form CAF1–CRS2 and CAF2–CRS2 complexes, which regulate the splicing of nine group II introns in chloroplasts [14,15]. Additionally, CRM domain proteins are involved in the splicing of mitochondrial gene introns. In A. thaliana, CFM9 belongs to subfamily 3, contains one CRM domain, and influences nad1, nad2, nad4, nad5, nad7, rps3, and cox2 intron splicing in mitochondria [9]. Both AtmCSF1 and AtmCSF2 contain two CRM domains and are localized in the mitochondria. The AtmCSF1 affects cox2, nad1, nad2, nad5, nad7, and rps3 intron splicing in mitochondria [4]. AtCFM4, in subfamily 4, contains one CRM domain and localizes to the chloroplast while it affects the assemblage of 16S and the 4.5S process. It has been reported that atcfm4 mutants show sensitivity to abiotic stress, including cold and salt stress [10].
Previous studies have identified 14 CRM domain-containing proteins in rice [1,3]. Some of these CRM domain-containing proteins were found to be associated with intron splicing in chloroplast. Phenotypic analysis has shown that al2, oscfm3, and oscaf1 mutants exhibit seedlings’ albino phenotype [12,16,17]. OsCRS1 has been reported to participate in the splicing of group I and group II introns, including trnL, petD, ndhA, ndhB, and ycf3-1 [16]. Meanwhile, OsCFM3 participates in the splicing of group II introns involving rps16, rpl16, ndhB, and petD [12]. Our previous studies showed that OsCAF1 interacts with the C-terminal of OsCRS2 to form OsCAF1–OsCRS2 complex that can influence introns’ splicing of atpF, rpl2, rps12, ndhA, ndhB, and ycf3 [17]. Until now, the functions of most CRM domain proteins in rice have not been reported.
In the present study, we analyzed the genes and protein structures of 14 CRM domain proteins and identified the expression patterns of these genes in various tissues and under abiotic stress. We edited Os04g0464800 (OsCFM2) using a CRISPR/Cas9 gene editing system and obtained two allele mutants, oscfm2-1 and oscfm2-2, which both exhibited an albino seedling phenotype. We found that OsCFM2 influences group I and group II intron splicing in chloroplast and plays an important role in chloroplast development in rice.
2. Materials and Methods
2.1. Sequence Analysis and Phylogenetic Tree Construction
We acquired 14 CRM protein sequences and gene sequences from the rice genome annotation project database. The gene structures of 14 CRM genes’ family were analyzed by the online gene structure display server (GSDS, http://gsds.cbi.pku.edu.cn/) [18]. The conserved motifs of 14 CRM domain proteins were analyzed using online motif elicitation software (MEME, http://meme-suite.org/) [19]. The parameters were as follows: 10 motifs in maximum, motif width with 6–50 residues, and E-values < 1.00 × 10−20. A phylogenetic tree was built using the neighbor-joining method by 1000 bootstrap in MEGA-X software [20].
2.2. Plant Materials and Abiotic Stress Treatments
To study the expression pattern of CRM genes in various tissues, we obtained roots (R), culm (C), young leaves (YL), expansion leaves (EL), and young spikelet (YS) at the booting stage. Germinated seeds (Nipponbare) were transferred to a nutrient solution in an incubator and kept at 30 °C under a 16 h light/8 h dark cycle. Accordance to previous studies, we treated rice seedlings at the three-leaf stage with low temperature (15 °C), salt stress (100 mM NaCl), and drought stress (10% PEG) for 3 days [21,22,23]. After taking leaves, samples were stored at −80 °C for further analysis.
OsCFM2/oscfm2-1 and OsCFM2/oscfm2-2 heterozygous plants were obtained using a CRISPR/Cas9 gene editing system with the Nipponbare, which were used as wildtype (WT). The oscfm2-1 and oscfm2-2 mutants were acquired from the T1 generation of OsCFM2/oscfm2-1 and OsCFM2/oscfm2-2 heterozygous plants, respectively. The WT and two allele mutants were grown in a greenhouse under a 16 h light/8 h dark cycle at a constant temperature of 30 °C.
2.3. OsCFM2 Knockout with a CRISPR/Cas9 System
The rice gene editing methods were dependent on the CRISPR/Cas9 system [24]. For the first exon of OsCFM2, we designed one gRNA target sequence (GCTCCTCCTCTTCCTCCCCCA) to construct an intermediate vector, SK-gRNA-gOsCFM2-g1. Using Kpn I and Bgl II, intermediate vectors were digested, and then gRNA-gOsCFM2-g1 was assembled into the pC1300-Cas9 vector.
2.4. Measurement of Chlorophyll Content and Fv/Fm
Fromthe 18-day seedlings, we took 50-mg leaf samples, cut them into small pieces, and immersed them in 20 mL of 95% ethanol for 48 h in darkness at 4 °C. We then used a UV-1800PC spectrophotometer to measure the absorbance of the samples at 665, 649, and 470 nm. Then, we calculated the chlorophyll a, chlorophyll b, and total chlorophyll content according to the method of Lichtenthaler [25].
For Fv/Fm measurement, the WT and two allele mutants in the18-day seedlings were kept in darkness for 30 min. Then, we measured the Fv/Fm with a handheld fluorometer according to the manufacturer’s instructions.
2.5. Transmission Electron Microscopy (TEM)
Transmission electron microscopy analysis was carried out as previously described [26]. In the two-leaf stage, the leaves of WT plants and oscfm2-1 mutants were collected and cut into 0.1 cm × 0.1 cm pieces. Leaf samples were fixed in 2.5% glutaraldehyde and 1% OsO4. Then, samples were dehydrated via an ethanol series and embedded in resin. Samples were stained with uranyl acetate and alkaline lead citrate and after observed using a HitachiH-7500 transmission electron microscope.
2.6. RNA Extraction and Quantitative Real-Time PCR
Total rice RNA was extracted from samples using an RNA Extraction Kit (TaKaRa) according to the manufacturer’s instructions. First-strand cDNA was obtained using a ReverTra Ace qPCR RT Kit (TOYOBO). The quantitative real-time PCR (qRT-PCR) experimental procedure was as described in a previous study [27]. In the present study, the OsAction1 gene was used as an internal control, and the relative expression levels of genes were calculated using the 2−ΔΔCT method [28]. All qRT-PCR primers used are listed in Table S1.
2.7. Chloroplast Gene Intron Splicing Analysis
To analyze intron splicing in chloroplast genes, chloroplast genes containing at least one intron were selected, and amplified using the RT-PCR method. The RT-PCR procedure was as follows: 96 °C for 5 min, followed by 30 cycles of 96 °C for 40 s, 60 °C for 30 s, 72 °C for 45 s, and a final elongation step at 72 °C for 10 min. All RT-PCR primers used in the present study are listed in Table S1.
3. Results
3.1. Gene and Protein Structure Analysis of Rice CRM Domain Proteins
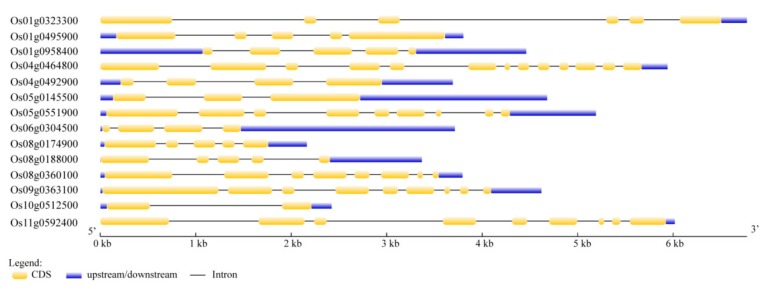
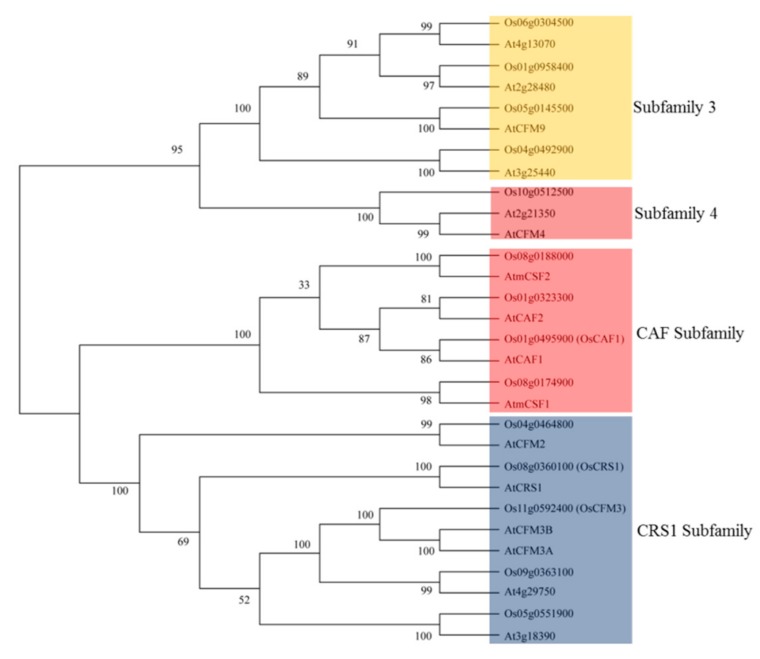
The 14 CRM genes of rice are distributed among eight chromosomes: 1, 4, 5, 6, 8, 9, 10, and 11 chromosomes. Gene structure analysis indicated that the CRM genes contained 2–11 exons and the coding sequences of these genes ranged from 759 to 3039 base pairs (bp) in length (Figure 1). The CRM domain proteins encoded by these genes contained 252–1012 amino acids and their molecular weights were in the range of 27.75–111.29 kDa (Table S2). We obtained Arabidopsis CRM domain protein sequences from the Arabidopsis information resource database to build a phylogenetic tree (Figure 2). All proteins in the CRS1 subfamily except OsCFM2 have four CRM domains, and OsCRS1, OsCFM3, Os05g0551900, and Os09g0363100 contained three CRM domains. In the CAF subfamily, all proteins contained two CRM domains while in subfamily 3 and subfamily 4, all proteins contained only one CRM domain (Table S2). Only one CRM domain protein was identified in subfamily 4. Through TargetP program prediction, it was determined that nine of these CRM domain proteins were located in the chloroplast, and five CRM domain proteins located in the mitochondrion (Table S2). Those proteins’ actual subcellular localization needed to be verified by an experiment.
Figure 1.
Structural analysis of CRM genes’ family. Yellow and blue boxes indicate coding sequences and upstream or downstream untranslated regions, respectively. Black lines indicate introns. The scale bar at the bottom shows the length of the CRM genes in Kb.
Figure 2.
Phylogenetic tree of CRM domain proteins in rice and A. thaliana. The full amino acid sequences of the CRM domain proteins were aligned by MUSCLE and the neighbor-joining tree was constructed in MEGA X.
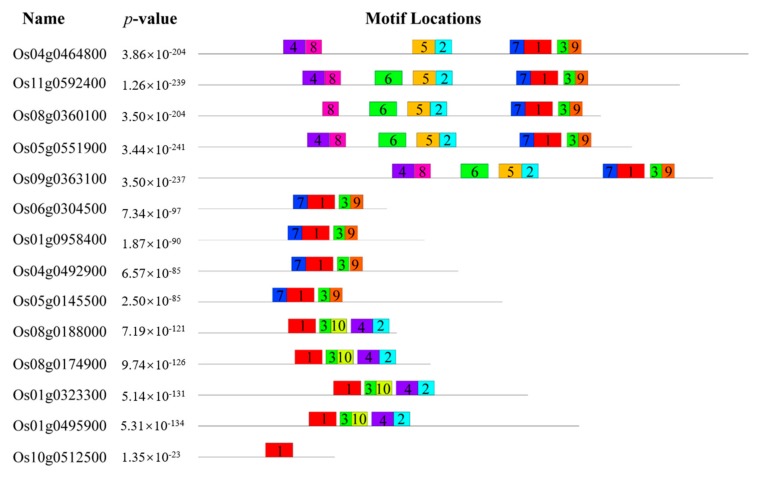
In addition, to further understand the structures of these 14 CRM domain proteins, we predicted their conserved putative motifs using the online program MEME. In total, 10 motifs were identified and named motif 1 to motif 10 (Figure S1). We found that 14 CRM domain proteins contained conserved putative motif 1. It is interesting to note that motif 1 was located in the C-terminal of the CRS1 subfamily proteins. Motif 5 and motif 8 only existed in the CRS1 subfamily in rice. Only CAF subfamily proteins contain motif10 (Figure 3).
Figure 3.
Conserved putative motif analysis of CRM domain proteins in rice. Ten putative motifs are shown in different colored boxes.
3.2. Expression Analysis of CRM Genes in Various Tissues
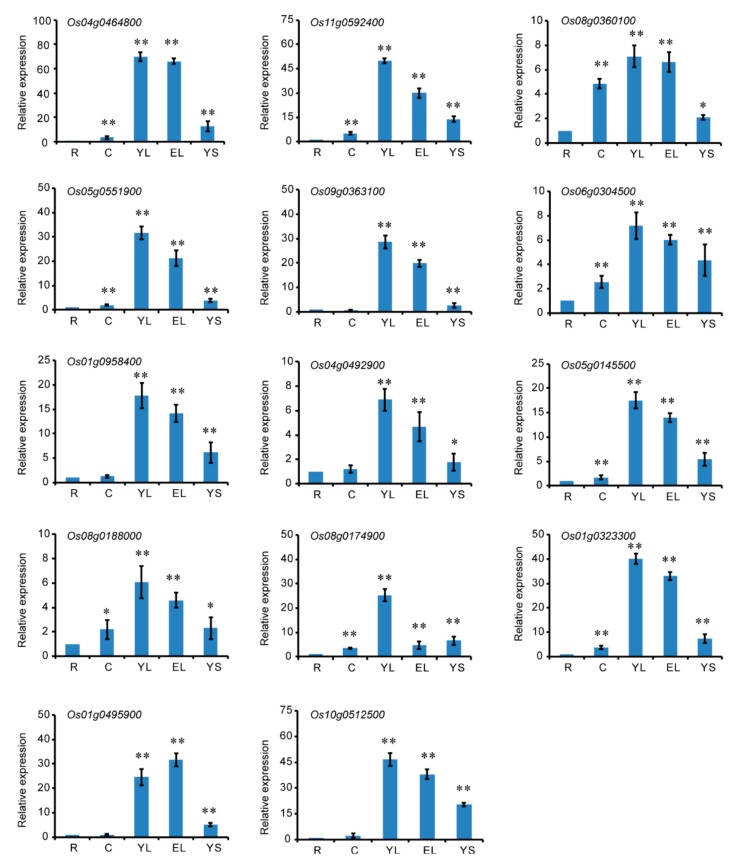
In order to study the expression patterns of CRM family genes in various tissues, we sampled roots (R), culm (C), young leaves (YL), expanded leaves (EL), and young spikelet (YS) in the booting stage for qRT-PCR analysis. The results indicated that the CRM family genes were expressed in all tissues, but particularly high expressed in green tissues, especially in YL and EL, suggesting that they might function in leaf development in rice (Figure 4). Some of the genes have previously been reported to be involved in chloroplast development, such as OsCAF1 and OsCRS1.
Figure 4.
Analysis of CRM gene expression in various rice tissues by qRT-PCR. Tissues, including root (R), culm (C), young leaf, expansion leaf (EL), and young spikelet (YS), at the booting stage. The relative expression level in each tissue was controlled by the gene expression level in the root. Data are the mean ± SD for three biological replicates. * p < 0.05, ** p < 0.01 by Student’s t-test.
3.3. Expression Analysis of CRM Genes under Abiotic Stress
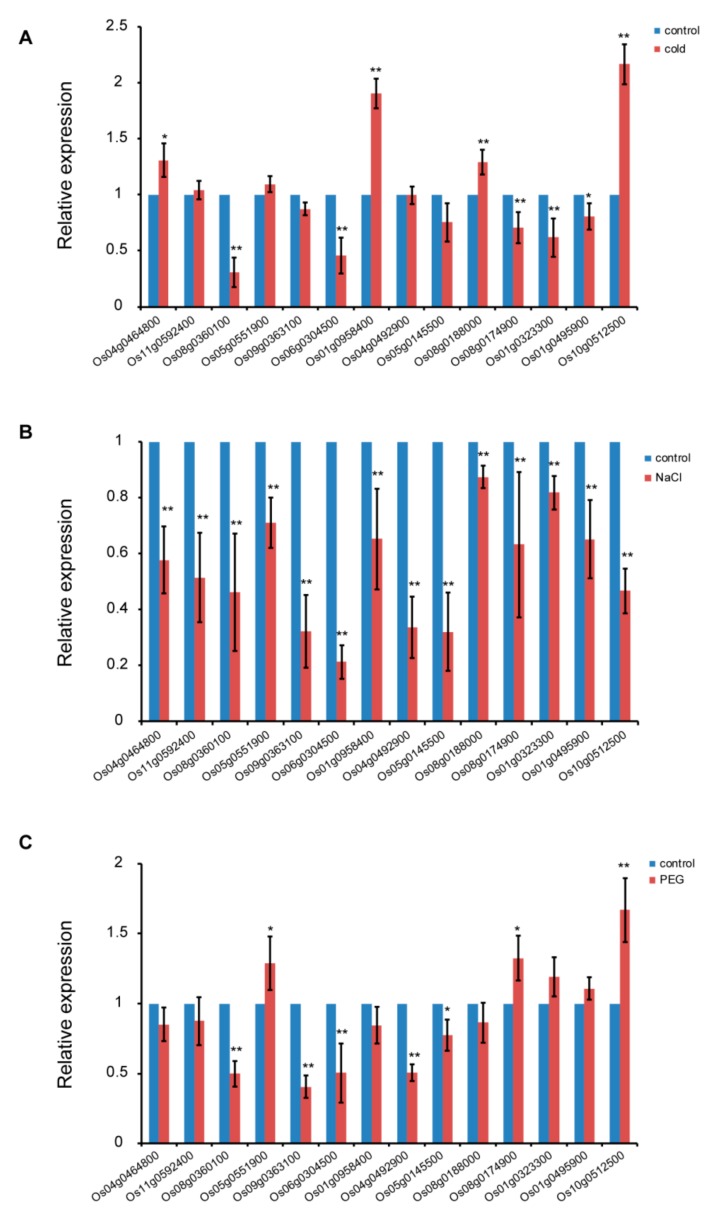
Research has shown that CRM domain proteins play an important role in plant development and growth [9,10,17]. Additionally, the CRM genes could also be involved in response to abiotic stress [29]. In A. thaliana, AtCFM4 belongs to subfamily 4, shown to be sensitive to salt and cold stresses [10]. However, the effect of abiotic stress on CRM gene expression in rice has not yet been reported. Abiotic stresses include cold temperatures, high salt, and drought, which affect rice growth. To investigate whether the expression of CRM genes was affected by abiotic stress, the expression levels of 14 CRM genes were evaluated by qRT-PCR analysis under abiotic stress. Under cold stress, the expression levels of five and four CRM genes were dramatically downregulated and upregulated, respectively, while five genes involving Os11g0592400, Os05g0551900, Os09g0363100, Os04g0492900, and Os05g0145500 expression levels showed no significant change (Figure 5A). Under salt stress, the expression levels of 14 CRM genes were significantly downregulated (Figure 5B). Under drought stress, the expression levels of five and three CRM genes were significantly downregulated and upregulated, respectively, and the expression levels of six genes, including Os04g0464800, Os11g0592400, Os01g0958400, Os08g0188000, Os01g0323300, and Os01g0495900 expression levels, showed no significant change (Figure 5C).
Figure 5.
Analysis of CRM gene expression in the leaves of three-leaf stages under (A) cold stress (15 °C), (B) salt stress (100mM NaCl), and (C) drought stress (10% PEG). Data are the mean ± SD for three biological replicates. * p < 0.05, ** p < 0.01 by Student’s t-test.
3.4. OsCFM2 (Os04g0464800) Influences Chloroplast Development in Rice
The Os04g0464800 gene belongs to the CRS1 subfamily and was homologous with AtCFM2, so we named this gene OsCFM2. The function of OsCFM2 has not been studied until now. To study the function of OsCFM2, we constructed the pC1300-Ubi::Cas9-gOsCFM2-target1 vector and transformed the vector into Nipponbare using an Agrobacterium-mediated method [30]. In the T0 generation, we obtained nine transgenic plants. The sequencing results showed that only two of these plants, OsCFM2/oscfm2-1 and OsCFM2/oscfm2-2, were heterozygotes plants. In the T1 generation, we obtained two allele mutants: oscfm2-1 and oscfm2-2, from OsCFM2/oscfm2-1 and OsCFM2/oscfm2-2 plants, respectively. Phenotypic analysis indicated that the mutants showed albinism in the seedling stage (Figure 6A). In oscfm2-1 and oscfm2-2, the deletion of 2 and 11 bp in the first exon of the OsCFM2 gene coding sequence region, respectively, led to frame-shift mutations that resulted in a premature stop codon (Figure S2). These results suggest that the mutation of OsCFM2 leads to the albino seedling phenotype.
Figure 6.
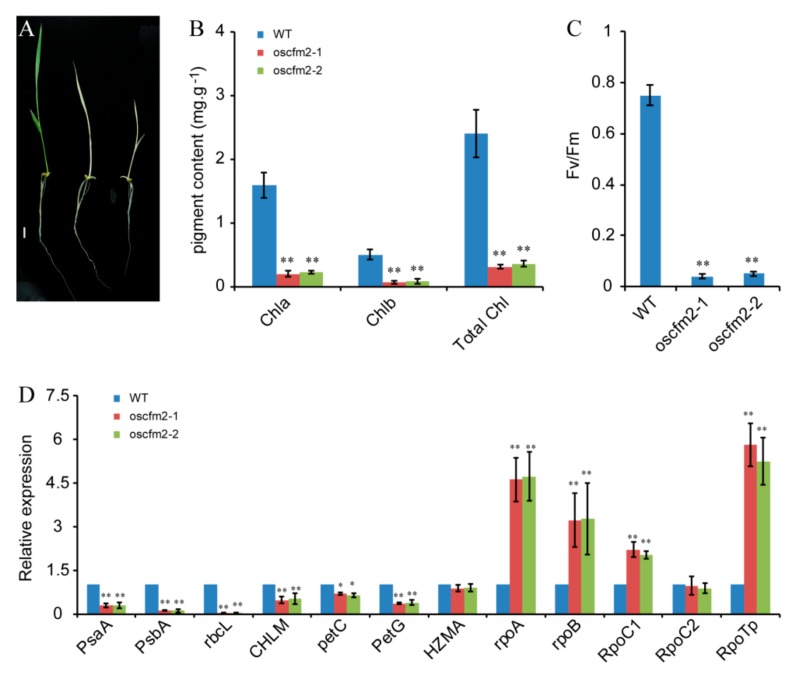
Mutation of OsCFM2 causes an albino phenotype in rice leaves. (A) Phenotypes of the WT (left) and oscfm2-1 (middle) and oscfm2-2 mutants (right) at the seedling stage. Scale bar = 0.5 cm. (B) Chlorophyll contents of WT and oscfm2-1 and oscfm2-2 mutants at the seeding stage. (C) The Fv/Fm value of the WT and oscfm2-1 and oscfm2-2 mutants at the seeding stage. (D) qRT-PCR analysis of genes related to chloroplast synthesis and photosynthesis in the WT and oscfm2-1 and oscfm2-2 mutants. Values represent the mean ± SD (n = 3). ** p < 0.01 by Student’s t-test.
The chlorophyll a, chlorophyll b, and total chlorophyll contents were markedly reduced in oscfm2-1 and oscfm2-2 mutants compared to the WT (Figure 6B). Additionally, the Fv/Fm values were significantly lower in the oscfm2-1 and oscfm2-2 mutants than in the WT (Figure 6C). We also analyzed the expression of genes related to chloroplast development and photosynthesis in oscfm2-1 and oscfm2-2 mutants and the WT. The results showed that the relative expression levels of genes associated with chloroplast development and photosynthesis were significantly changed in mutants compared to the WT. For example, the expression levels of PsaA, PsbA, rbcL, petC, PetG, and CHLM were remarkably reduced in oscfm2-1 and oscfm2-2 mutants (Figure 6D), whereas genes encoding NEP and PEP, such as rpoA, rpoB, RpoC1, and RpoTp, were significantly increased (Figure 6D).
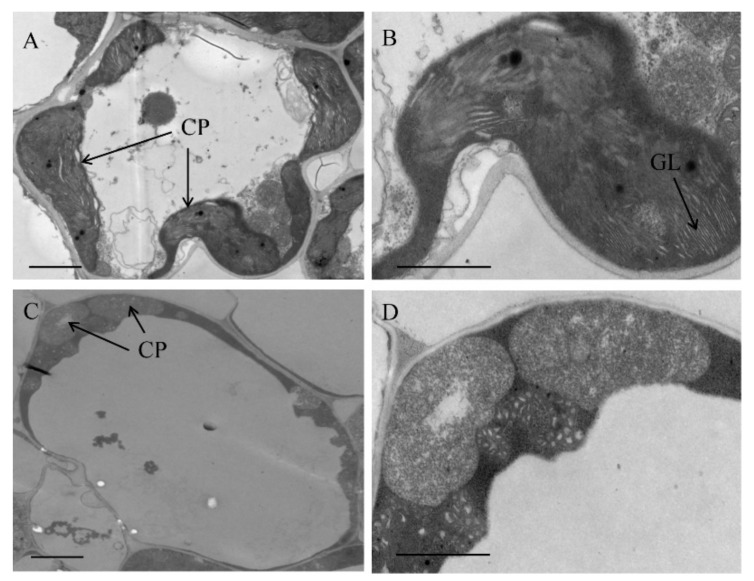
We observed the ultrastructure of chloroplasts in 18-day-old WT and oscfm2-1 mutant leaves by TEM. Normal chloroplasts and grana stacks were found in the WT (Figure 7A,B). In contrast, in oscfm2-1 mutant, the chloroplast was abnormal, and appeared to stop developing at the proplastid stage (Figure 7C,D). These observations suggest that a mutation in OsCFM2 influences chloroplast development. Taken together, our results suggest that OsCFM2’s lack of function leads to abnormal chloroplast development in rice.
Figure 7.
Chloroplast ultrastructure of WT (A, B) and oscfm2-1 mutants (C, D). cp, chloroplast, gl, grana lamella. Bars = 2.0 μm.
3.5. OsCFM2 Influences the Splicing of Group I and Group II Introns
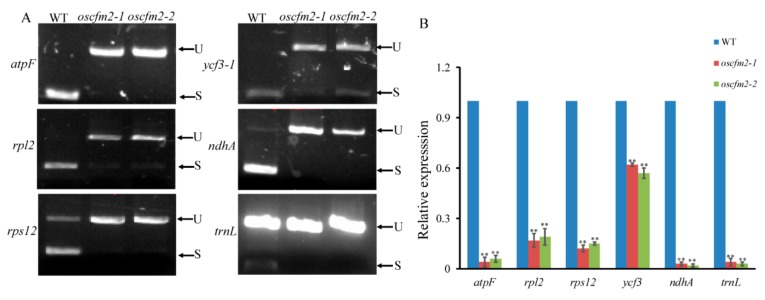
To test whether OsCFM2 affected the splicing of chloroplast gene introns, group I and group II introns were amplified from the two allele mutants and WT. The results indicated that the splicing of five group II introns, including rpl2, rps12, ycf3-1, atpF, and ndhA, were affected in mutants compared to the WT (Figure 7A). Unlike in the WT, the atpF and ndhA introns were not completely spliced in oscfm2-1 and oscfm2-2 mutants. The trnL group I intron was also not spliced in mutants (Figure 7A). The qRT-PCR analysis showed that rpl2, rps12, ycf3, atpF, ndhA, and trnL gene expression levels were decreased in oscfm2-1 and oscfm2-2 mutants compared with the WT. These results suggest that OsCFM2 may affect chloroplast development by influencing the splicing of group I and five group II introns. However, the other 10 group II introns had normal splicing in oscfm2-1 and oscfm2-2 mutants compared with the WT (Figure S3).
4. Discussion
Genomic structural analysis indicated that 14 CRM genes in rice contain introns (Figure 1). In rice, 5, 4, 4, and 1 of the 14 CRM proteins are classified as CRS1, CAF, subfamily 3, and subfamily 4 proteins, respectively (Figure 2). Protein motif analysis showed that the protein motifs in each subfamily were highly conserved (Figure 3). Proteins in the CRS1 subfamily were found to contain more motifs than those in other subfamilies. All proteins in the CRS1 subfamily except OsCRS1 contained eight motifs while the other four proteins contained nine motifs. There were five and four motifs in the CAF subfamily and subfamily 3, respectively (Figure 3). Our results indicated that the proteins in each subfamily harbored different numbers of conserved motifs, suggesting that these proteins might have different functions in rice. For example, in the CRS1 subfamily, OsCRS1 influences intron splicing in trnL and multiple group II introns [16]. Meanwhile, in the CAF subfamily, OsCAF1 regulates the splicing of some group II introns in chloroplasts [17]. The functions of the CRM domain proteins of different subfamilies may therefore be differentiated.
Expression analysis showed that the 14 CRM genes were most highly expressed in rice leaves (Figure 4); this result was consistent with previous studies. It is reported that OsCRS1 and OsCAF1 are highly expressed in rice leaves [16,17]. Phenotypic analysis showed that oscrs1 and oscaf1 mutants exhibit an albino phenotype in the seedling stage [16,17]. This result suggested that the CRM family genes might play a key role in chloroplast development in leaves. We noticed that the CRM family genes were also highly expressed in young spikelets, suggesting that they might function in spikelet development (Figure 4). It iss reported that ZmmCSF1 and AtmCSF1 were essential for seed development [4,31]. Therefore, Os08g0174900, which is homologous with AtmCSF1, may also play an important role in seed development in rice.
It has been reported that in A. thaliana, AtCFM4 and AtCFM9 influence seedling growth under cold and salt stress [9,10]. In the present study, we found that the expression levels of CRM genes were changed under abiotic stress, especially under salt stress, which might affect intron splicing in chloroplast or mitochondrial genes, leading to increased sensitivity to abiotic stress in rice (Figure 5). Further study is required to elucidate the molecular mechanism of the CRM domain protein response to abiotic stress.
The CRM domain proteins play key roles in plant growth and development [3,32]. Many intron splicing factors have been identified in different plants, involving PPR family and CRM family proteins [33,34]. For example, WSL4, SOT5, and THA8 affect chloroplast genes’ intron splicing in rice, A. thaliana, and maize, respectively [35,36,37]. There is evidence to indicate that CRM proteins have a high affinity for group II intron RNA in vitro [38]. In rice, some CRM proteins might have similar functions, influencing the splicing of group II introns. It has been reported that OsCAF1, OsCFM3, and OsCRS1 could influence the splicing of multiple group II introns in rice [12,16,17]. The molecular mechanism underlying the regulation of chloroplast gene intron splicing by CRM proteins might be different in various plants. In A. thaliana and maize, AtCRS1 and ZmCRS1 only influence atpF intron splicing while OsCRS1 influences the splicing of many introns, including atpF, petD, ndhA, ndhB, ycf3-1, and trnL [11,13,16]. In the present study, we found that OsCFM2 affected intron splicing in ycf3-1, rpl2, rps12, atpF, ndhA, and trnL, which is unlike AtCFM2 in A. thaliana (Figure 8).
Figure 8.
Intron splicing and gene expression analysis. (A) Splicing analysis of chloroplast genes introns in WT and oscfm2-1 and oscfm2-2 mutants. S indicates spliced transcripts. U indicates unspliced transcripts. (B) Expression analyzed of atpF, rpl2, rps12, ycf3, ndhA, and trnL in WT and mutants. Values represent the mean ± SD (n = 3). ** p < 0.01 by Student’s t-test.
The oscfm2-1 and oscfm2-2 mutants exhibited an albino phenotype and survived for about three weeks (Figure 6A). In mutants, chloroplast development is abnormal and chlorophyll content was significantly decreased (Figure 6B). This result suggests that disruption of the function of OsCFM2 affected chloroplast development. In the rice genome, except for OsCFM2, the CRS1 subfamily contains OsCRS1, OsCFM3, Os05g0551900, and Os09g0363100. We examined the expression levels of these four genes among WT and mutants. The results showed that their expression levels were upregulated in oscfm2-1 and oscfm2-2 mutants compared to the WT (Figure S3). This result suggests that on the one hand, there might be negative regulation between OsCFM2 and other CRS1 subfamily genes at the transcript level. On the other hand, this may be a feedback mechanism in which the cells try to increase the expression of the four CRS1 family’s genes to improve the splicing efficiency of the chloroplast genes’ intron but failed to compensate for the phenotype of the OsCFM2 mutation.
The OsCAF1 influences intron splicing in atpF, rpl2, rps12, ndhB, ycf3, and ndhA [13]. OsCRS1 not only affected the splicing of the trnL intron but also the atpF, rpl2, ndhA, ndhB, petB, and ycf3 introns in rice [17]. Therefore, OsCFM2 may have functional overlap with OsCAF1 and OsCRS1, but it is not redundant. These results indicate that OsCFM2 is functional and necessary for chloroplast development.
5. Conclusions
In the present study, we analyzed the expression patterns of 14 CRM genes’ family in different rice tissues and under abiotic stress. Moreover, we characterized the function of OsCFM2, which plays a key role in chloroplast development in rice. Our study provides information for the functional analysis of CRM domain proteins in rice.
Supplementary Materials
The following are available online at https://www.mdpi.com/2218-273X/10/2/327/s1, Figure S1: MEME analysis shows each motif of the 14 CRM protein in rice, Figure S2: Mutation types of oscfm2-1 and oscfm2-2 mutants, Figure S3: Splicing analysis of chloroplast gene introns, Figure S4: Expression analysis of CRS1 subfamily genes in WT rice plants and oscfm2-1 and oscfm2-2 mutants, Table S1: Primers for this study, Table S2 Characteristics of 14 CRM domain proteins in rice.
Author Contributions
Formal analysis, Q.Z., J.H. and L.G.; Funding acquisition, L.S., D.R., G.Z. and D.Z.; Project administration, Q.Q.; Resources, L.S., D.R. and L.Z.; Software, Z.G.; Writing—Original draft, Q.Z. and L.S.; Writing—Review and editing, Q.Z. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by the National Natural Science Foundation of China (31801333), the Central Public-interest Scientific Institution Basal Research Fund of China National Rice Research Institute (2017RG001-4), the ‘Collaborative Innovation Project’ of the Chinese Academy of Agricultural Sciences, the Zhejiang Provincial ‘Ten Thousand Talent Program’ Project (2018R52025).
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Barkan A., Klipcan L., Ostersetzer O., Kawamura T., Asakura Y., Watkins K.P. The CRM domain: An RNA binding module derived from an ancient ribosome-associated protein. RNA. 2007;13:55–64. doi: 10.1261/rna.139607. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Keren I., Klipcan L., Bezawork-Geleta A., Kolton M., Shaya F., Ostersetzer-Biran O. Characterization of the molecular basis of group II intron RNA recognition by CRS1-CRM domains. J. Biol. Chem. 2008;283:23333–23342. doi: 10.1074/jbc.M710488200. [DOI] [PubMed] [Google Scholar]
- 3.Stern D.B., Goldschmidt-Clermont M., Hanson M.R. Chloroplast RNA metabolism. Annu. Rev. Plant. Biol. 2010;61:125–155. doi: 10.1146/annurev-arplant-042809-112242. [DOI] [PubMed] [Google Scholar]
- 4.Zmudjak M., Colas des Francs-Small C., Keren I., Shaya F., Belausov E., Small I., Ostersetzer-Biran O. mCSF1, a nucleus-encoded CRM protein required for the processing of many mitochondrial introns, is involved in the biogenesis of respiratory complexes I and IV in Arabidopsis. New Phytol. 2013;199:379–394. doi: 10.1111/nph.12282. [DOI] [PubMed] [Google Scholar]
- 5.De Longevialle A.F., Small I.D., Lurin C. Nuclearly encoded splicing factors implicated in RNA splicing in higher plant organelles. Mol. Plant. 2010;3:691–705. doi: 10.1093/mp/ssq025. [DOI] [PubMed] [Google Scholar]
- 6.Barkan A. Expression of plastid genes: Organelle-specific elaborations on a prokaryotic scaffold. Plant. Physiol. 2011;155:1520–1532. doi: 10.1104/pp.110.171231. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Rochaix J.D., Ramundo S. Chloroplast signaling and quality control. Essays Biochem. 2018;62:13–20. doi: 10.1042/EBC20170048. [DOI] [PubMed] [Google Scholar]
- 8.Asakura Y., Barkan A. A CRM domain protein functions dually in group I and group II intron splicing in land plant chloroplasts. Plant. Cell. 2007;19:3864–3875. doi: 10.1105/tpc.107.055160. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lee K., Park S.J., Park Y.I., Kang H. CFM9, a mitochondrial CRM protein, is crucial for mitochondrial intron splicing, mitochondria function and Arabidopsis growth and stress responses. Plant. Cell Physiol. 2019;60:2538–2548. doi: 10.1093/pcp/pcz147. [DOI] [PubMed] [Google Scholar]
- 10.Lee K., Lee H.J., Kim D.H., Jeon Y., Pai H.S., Kang H. A nuclear-encoded chloroplast protein harboring a single CRM domain plays an important role in the Arabidopsis growth and stress response. BMC Plant. Biol. 2014;14:98. doi: 10.1186/1471-2229-14-98. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Till B., Schmitz-Linneweber C., Williams-Carrier R., Barkan A. CRS1 is a novel group II intron splicing factor that was derived from a domain of ancient origin. RNA. 2001;7:1227–1238. doi: 10.1017/S1355838201010445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Asakura Y., Bayraktar O.A., Barkan A. Two CRM protein subfamilies cooperate in the splicing of group IIB introns in chloroplasts. RNA. 2008;14:2319–2332. doi: 10.1261/rna.1223708. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Asakura Y., Barkan A. Arabidopsis orthologs of maize chloroplast splicing factors promote splicing of orthologous and species-specific group II introns. Plant. Physiol. 2006;142:1656–1663. doi: 10.1104/pp.106.088096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Ostheimer G.J., Williams-Carrier R., Belcher S., Osborne E., Gierke J., Barkan A. Group II intron splicing factors derived by diversification of an ancient RNA-binding domain. EMBO J. 2003;22:3919–3929. doi: 10.1093/emboj/cdg372. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Ostheimer G.J., Rojas M., Hadjivassiliou H., Barkan A. Formation of the CRS2-CAF2 group II intron splicing complex is mediated by a 22-amino acid motif in the COOH-terminal region of CAF2. J. Biol Chem. 2006;281:4732–4738. doi: 10.1074/jbc.M508921200. [DOI] [PubMed] [Google Scholar]
- 16.Liu C., Zhu H., Xing Y., Tan J., Chen X., Zhang J., Peng H., Xie Q., Zhang Z. Albino leaf 2 is involved in the splicing of chloroplast group I and II introns in rice. J. Exp. Bot. 2016;67:5339–5347. doi: 10.1093/jxb/erw296. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Zhang Q., Shen L., Wang Z., Hu G., Ren D., Hu J., Zhu L., Gao Z., Zhang G., Zeng D., et al. OsCAF1, a CRM domain containing protein, influences chloroplast development. Int. J. Mol. Sci. 2019;20:e4386. doi: 10.3390/ijms20184386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An upgraded gene feature visualization server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Bailey T.L., Johnson J., Grant C.E., Noble W.S. The MEME Suite. Nucleic Acids Res. 2015;43:39–49. doi: 10.1093/nar/gkv416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kumar S., Stecher G., Li M., Knyaz C., Tamura K. MEGA X: Molecular evolutionary genetics analysis across computing platforms. Mol. Biol. Evol. 2018;35:1547–1549. doi: 10.1093/molbev/msy096. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Wang Y., Cui Y., Hu G., Wang X., Chen H., Shi Q., Xiang J., Zhang Y., Zhu D., Zhang Y. Reduced bioactive gibberellin content in rice seeds under low temperature leads to decreased sugar consumption and low seed germination rates. Plant. Physiol. Biochem. 2018;133:1–10. doi: 10.1016/j.plaphy.2018.10.020. [DOI] [PubMed] [Google Scholar]
- 22.Zou J., Liu A., Chen X., Zhou X., Gao G., Wang W., Zhang X. Expression analysis of nine rice heat shock protein genes under abiotic stresses and ABA treatment. J. Plant. Physiol. 2009;166:851–861. doi: 10.1016/j.jplph.2008.11.007. [DOI] [PubMed] [Google Scholar]
- 23.Das P., Manna I., Sil P., Bandyopadhyay M., Biswas A.K. Exogenous silicon alters organic acid production and enzymatic activity of TCA cycle in two NaCl stressed indica rice cultivars. Plant. Physiol. Biochem. 2019;136:76–91. doi: 10.1016/j.plaphy.2018.12.026. [DOI] [PubMed] [Google Scholar]
- 24.Wang C., Shen L., Fu Y., Yan C., Wang K. A simple CRISPR/Cas9 system for multiplex genome editing in rice. J. Genet. Genomics. 2015;42:703–706. doi: 10.1016/j.jgg.2015.09.011. [DOI] [PubMed] [Google Scholar]
- 25.Lichtenthaler H.K. Chlorophylls and carotenoids: Pigments of photosynthetic biomembranes. Methods Enzymol. 1987;148:350–382. [Google Scholar]
- 26.Chen L., Huang L., Dai L., Gao Y., Zou W., Lu X., Wang C., Zhang G., Ren D., Hu J., et al. PALE-GREEN LEAF12 encodes a novel pentatricopeptide repeat protein required for chloroplast development and 16S rRNA processing in rice. Plant. Cell Physiol. 2019;60:587–598. doi: 10.1093/pcp/pcy229. [DOI] [PubMed] [Google Scholar]
- 27.Zhang Q., Shen L., Ren D., Hu J., Chen G., Zhu L., Gao Z., Zhang G., Guo L., Zeng D., et al. Characterization, expression, and interaction analyses of OsMORF gene family in rice. Genes. 2019;10:e694. doi: 10.3390/genes10090694. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Livak K.J., Schmittgen T.D. Analysis of relative gene expression data using real-time quantitative PCR and the 2−ΔΔCT method. Methods. 2001;25:402–408. doi: 10.1006/meth.2001.1262. [DOI] [PubMed] [Google Scholar]
- 29.Kang H., Park S.J., Kwak K.J. Plant RNA chaperones in stress response. Trends Plant. Sci. 2013;18:100–106. doi: 10.1016/j.tplants.2012.08.004. [DOI] [PubMed] [Google Scholar]
- 30.Lee K., Eggenberger A.L., Banakar R., McCaw M.E., Zhu H., Main M., Kang M., Gelvin S.B., Wang K. CRISPR/Cas9-mediated targeted T-DNA integration in rice. Plant. Mol. Biol. 2019;99:317–328. doi: 10.1007/s11103-018-00819-1. [DOI] [PubMed] [Google Scholar]
- 31.Chen Z., Wang H.C., Shen J., Sun F., Wang M., Xu C., Tan B.C. PPR-SMR1 is required for the splicing of multiple mitochondrial introns, interacts with Zm-mCSF1, and is essential for seed development in maize. J. Exp. Bot. 2019;70:5245–5258. doi: 10.1093/jxb/erz305. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Brown G.G., Colas des Francs-Small C., Ostersetzer-Biran O. Group II intron splicing factors in plant mitochondria. Front. Plant. Sci. 2014;5:35. doi: 10.3389/fpls.2014.00035. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Robles P., Quesada V. Transcriptional and post-transcriptional regulation of organellar gene expression (OGE) and its roles in plant salt tolerance. Int. J. Mol. Sci. 2019;28:20. doi: 10.3390/ijms20051056. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Rovira A.G., Smith A.G. PPR proteins—Orchestrators of organelle RNA metabolism. Physiol. Plant. 2019;166:451–459. doi: 10.1111/ppl.12950. [DOI] [PubMed] [Google Scholar]
- 35.Khrouchtchova A., Monde R.A., Barkan A. A short PPR protein required for the splicing of specific group II introns in angiosperm chloroplasts. RNA. 2012;18:1197–1209. doi: 10.1261/rna.032623.112. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Wang Y., Ren Y., Zhou K., Liu L., Wang J., Xu Y., Zhang H., Zhang L., Feng Z., Wang L., et al. White stripe leaf4 encodes a novel P-type PPR protein required for chloroplast biogenesis during early leaf development. Front. Plant. Sci. 2017;8:1116. doi: 10.3389/fpls.2017.01116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Huang W., Zhu Y., Wu W., Li X., Zhang D., Yin P., Huang J. The pentatricopeptide repeat protein SOT5/EMB2279 is required for plastid rpl2 and trnK intron splicing. Plant. Physiol. 2018;177:684–697. doi: 10.1104/pp.18.00406. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Ostersetzer O., Cooke A.M., Watkins K.P., Barkan A. CRS1, a chloroplast group II intron splicing factor, promotes intron folding through specific interactions with two intron domains. Plant. Cell. 2005;17:241–255. doi: 10.1105/tpc.104.027516. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.