Abstract
Among all cereals, rice is highly sensitive to cold stress, especially at the germination stage, which adversely impacts its germination ability, seed vigor, crop stand establishment, and, ultimately, grain yield. The dissection of novel quantitative trait loci (QTLs) or genes conferring a low-temperature germination (LTG) ability can significantly accelerate cold-tolerant rice breeding to ensure the wide application of rice cultivation through the direct seeding method. In this study, we identified 11 QTLs for LTG using 144 recombinant inbred lines (RILs) derived from a cross between a cold-tolerant variety, Lijiangxintuanheigu (LTH), and a cold-sensitive variety, Shennong265 (SN265). By resequencing two parents and RIL lines, a high-density bin map, including 2,828 bin markers, was constructed using 123,859 single-nucleotide polymorphisms (SNPs) between two parents. The total genetic distance corresponding to all 12 chromosome linkage maps was 2,840.12 cm. Adjacent markers were marked by an average genetic distance of 1.01 cm, corresponding to a 128.80 kb physical distance. Eight and three QTL alleles had positive effects inherited from LTH and SN265, respectively. Moreover, a pleiotropic QTL was identified for a higher number of erected panicles and a higher grain number on Chr-9 near the previously cloned DEP1 gene. Among the LTG QTLs, qLTG3 and qLTG7b were also located at relatively small genetic intervals that define two known LTG genes, qLTG3-1 and OsSAP16. Sequencing comparisons between the two parents demonstrated that LTH possesses qLTG3-1 and OsSAP16 genes, and SN-265 owns the DEP1 gene. These comparison results strengthen the accuracy and mapping resolution power of the bin map and population. Later, fine mapping was done for qLTG6 at 45.80 kb through four key homozygous recombinant lines derived from a population with 1569 segregating plants. Finally, LOC_Os06g01320 was identified as the most possible candidate gene for qLTG6, which contains a missense mutation and a 32-bp deletion/insertion at the promoter between the two parents. LTH was observed to have lower expression levels in comparison with SN265 and was commonly detected at low temperatures. In conclusion, these results strengthen our understanding of the impacts of cold temperature stress on seed vigor and germination abilities and help improve the mechanisms of rice breeding programs to breed cold-tolerant varieties.
Keywords: japonica rice, cold stress, germinability, high-density linkage map, QTLs
1. Introduction
Rice (Oryza sativa L.), which is a staple food and nutritional source for many countries, fulfills the nutritional requirements for over half of the world’s population and is cultivated across the globe, except in a few areas where icy conditions prevail during most of the year [1]. Since rice originated in tropical and sub-tropical climates, it is one of the most sensitive cereals to cold stress [2], which limits its growth, development, and yield formation, especially when cold stress prevails at the germination stage [3]. Cold stress impacts all growth stages of rice, including tillering, booting, flowering, and grain-filling, but if stress dominates at the germination stage, it proves adverse for rice development at later growth stages. Low-temperature stress during the germination stages of rice affects seedling vigor and produces poor seedling emergence and an uneven stand establishment with a lower growth rate, which delays panicle development and enhances spikelet sterility [4]. Across China’s mainland, most of the rice cultivation areas are affected by frequent cold stresses. The Chinese agricultural sector suffers from an average loss of rice of about 3–5 million tons of rice every year due to these frequent cold stresses [5]. Two kinds of cold stresses occur in Chinese rice-growing regions. The (1) “cold spring” and (2) “cold autumn wind” often cause severe yield losses in double-cropping rice regions across the Yangtze River in China. In Northeast China (NEC), commonly considered a rice region at a high latitude, and the Yunnan-Guizhou Plateau, considered a rice cultivation region at a low latitude, severe cold summer damage was observed, with an average of three to four years of cold stress. These areas are expected to encounter more severe damage in the near future due to low temperature stress [6].
Traditional genetic and molecular analyses on Arabidopsis, rice, and other model plants have revealed that C-repeat binding factors (CBFs) are mainly involved in the cold signaling pathways. Recent studies have further revealed that the protein kinases and transcription factors are also involved in cold signaling in plants [7]. Additionally, genetic research on rice has detected numerous quantitative trait loci that control cold tolerance on nearly all 12 chromosomes [8]. Among these loci, only a few quantitative trait loci (QTLs) have been thoroughly researched and cloned, while the functional mechanisms of most are still largely unknown. Among all the QTLs for low-temperature germination in rice, only qLTG3-1 and OsSAP16 were cloned. qLTG3-1 encodes a protein with glycine-rich and lipid trans-protein domain structures [9], and OsSAP16 encodes a zinc-finger protein that positively regulates germination under low temperatures [10]. The QTLs qCTS7, LTG1, COLD1, qCTS9, bZIP73, qPSR10, and HAN1 control the pathways for cold tolerance in rice. qCTS7 increases cold tolerance at the seedling stage due to its overexpression [11]. LTG1 encodes a casein kinase that plays a role in regulating the rice growth rate under cold stress [4]. A regulator of Ca2+ signaling in the plasma membrane and endoplasmic reticulum is encoded by COLD1 [12], whereas a novel protein that interacts with Brassinosteroid Insensitive-1 is encoded by qCTS9 [13]. There is a functional interaction between bZIP73 and bZIP71 that makes rice seedlings tolerant to greater cold [14]. A single-nucleotide polymorphism (SNP), SNP2G, at position 343 in qPSR10, is responsible for conferring cold tolerance during the seedling stage [15]. HAN1 encodes an oxidase that provides functional contributions to the Jasmonic acid mediated cold response in temperate japonica rice [16]. The other three QTLs, Ctb1, CTB4a, and bZIP73, control cold tolerance at the booting stage. The first encodes an F-box protein [17], the second encodes a leucine rich repeat kinase that enhances seed setting through increased ATP-synthase activity under low temperature stress [3], and the third increases the cold tolerance rate by enhancing the soluble sugar transport from anthers to pollens at the booting stage [18].
Advances in genome-wide sequencing technology have provided an effective method to detect DNA sequencing differences among closely related rice materials and to ensure the presence of sufficient markers for a genetic mapping analysis. A genotype calling method for RILs that utilizes resequencing data was developed [19], which determined the construction of resequencing bin maps and accelerated genetics-based studies for many crops, including cereals [19,20,21,22,23,24,25,26,27]. Based on the above discussions, many important advances have been achieved in the study of rice cold stress, but we still need to use high-throughput sequencing technology to mine further cold-tolerance genes from japonica rice, especially cold tolerance genes at the germination stage for breeding practice.
The current study was arranged with the following objectives: (1) constructing a high density bin map by re-sequencing a set of 144 RILs with large differences in germination abilities under cold stress; (2) identifying QTLs for LTG in RIL populations by using the built linkage map; and (3) creating an accurate map of qLTG6, with a high low-temperature germinability (LOD) score and relatively small genetic intervals.
2. Results
2.1. Phenotypic Variation among the Parent and RIL Populations
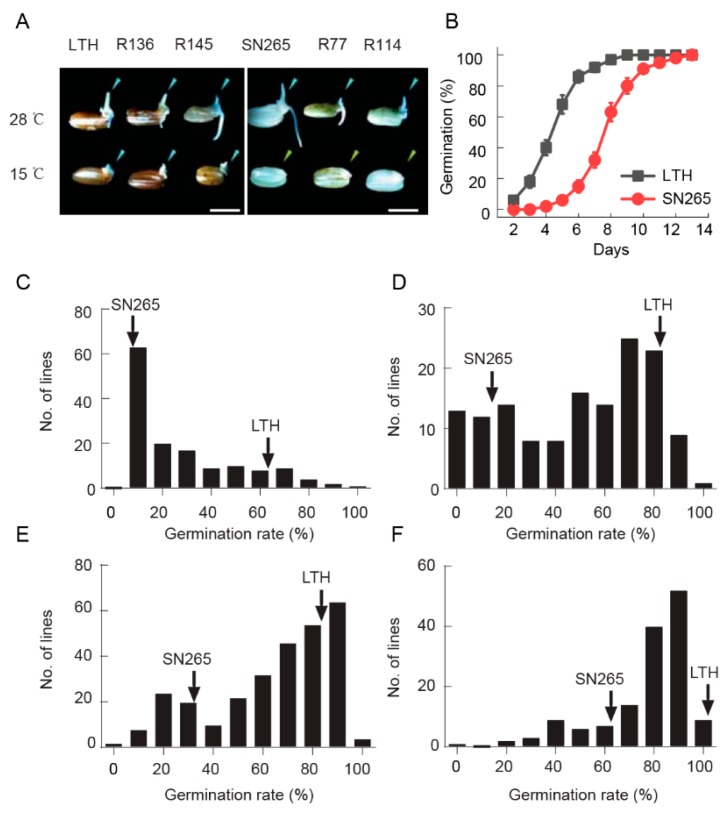
After incubating at 28 °C for three days, the germination rates of LTH and SN265 were 100%, indicating that both parents share a similar germination rate at a normal temperature, as presented in Figure 1A. However, when they were incubated at 15 °C for six days to determine their low temperature germinability, LTH and SN265 showed broad differences in their germination rates, as shown in Figure 1A,B. There was a delay of about two days in germination after incubating LTH under low temperature conditions, and, thereafter, the germination rate reached up to 90% after five days of incubation. Comparatively, for SN265, germination started after five days of incubation and, thereafter, took five more days to reach 90% of the germination of LTH, which shows a clear difference in the time taken for germination by LTH and SN265 (Figure 1B). A range of five to eight days was chosen for a proper and dynamic comparison of the low-temperature germinability among the parents and RILs (Figure 1C–F). At all four time points, the distributions of the germination percentages of the RIL populations were continuous, indicating that low-temperature germinability is controlled by QTLs. Generally, it is already known that the longer the germination time, the higher the germination percentage. Therefore, on the basis of the larger differences in germination between the parents after six days (LTH and SN265 showed 86.5% and 15.4%, respectively), those data were used for the subsequent QTL mapping.
Figure 1.
The seed germination of two parents and their recombinant inbred lines (RILs) at normal (28 °C) and low (15 °C) temperatures. (A). The germination phenotype of the two parents, Lijiangxintuanheigu (LTH) and Shennong265 (SN265), as well as the four RIL lines incubated for 3 days at 28 °C and for 6 days at 15 °C. R136 and R145 have 11 positive QTLs, whereas R77 and R144 have no positive QTLs. The bars = 5 mm; (B). The germination behavior of LTH (black) and SN265 (red) under low temperatures at 15 °C. The means are shown in triplicate; (C–F). The frequency distribution of low-temperature germinability in the RILs incubated for 5 days (C), 6 days (D), 7 days (E), and 8 days (F). Arrowheads indicate LTH or SN265.
2.2. Bin Map Construction and Comparison of the Physical Map to the Genetic Map
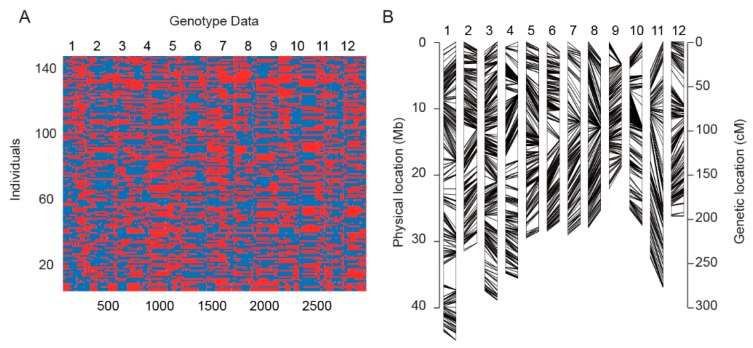
For proper identification of the SNP between the two parents as molecular markers, deep resequencing was done for LTH and SN265. The effective sequencing depths of LTH and SN265 were about 19-fold and 17-fold, respectively. After resequencing, a total of 123,859 SNPs were produced between the two parents. Construction of the genetic linkage map for the RILs was carried out by re-sequencing the 144 RIL lines, which were already derived from LTH and SN265. In this way, these 144 RIL lines produced a range of reads between about 7,448,879 and 12,118,209, with a mean value of 9,660,250. The overall effective depth coverage of these RIL lines ranged from 5.98-fold to 9.73-fold, with an average depth of 7.75-fold. About 58,738 recombination breakpoints were used to construct the fine bin map. Each RIL line was comprised of breakpoints ranging from 236 to 1007 breakpoints, with an average value of 405. The 144 RIL lines were merged into a high-density bin map comprising 2,818 recombination bins of the 12 rice chromosomes, including most recombination events (Figure 2A). The average of the physical intervals for the adjacent bins was observed between 15.00 kb and 3.60 Mb, with a mean value of 128.80 kb, where most of the bins with physical intervals less than 100.00 kb were found to be around 68.60%. Overall, only 32 bins exceeded a physical interval of 1.00 Mb, most of which were in centromeric regions. The average physical distance between the bin markers was 98.23 kb and 169.03 kb on chromosome 10 and chromosome 1, respectively. Then, we constructed 12 chromosomes with a total genetic distance of 2,840.12 cm (Figure 2B). The average genetic distance observed between the two markers was nearly 1.01 cm. Among all chromosomes under consideration, chromosome 1 was the longest, with 254 bin markers, and its genetic distance was 338.65 cm. In contrast, chromosome 9 was seen to be the shortest one, with 188 bin markers encompassing a genetic distance of 111.32 cm, as given in Table 1. Comparing the genetic distance between chromosome 9 and 11, the average values were 0.59 cm and 1.74 cm, respectively.
Figure 2.
High-resolution genotyping and comparison of the physical maps and genetic maps of RILs. (A). Aligned recombination maps of 144 RILs from a cross between LTH and SN265. The two genotypes are indicated by blue (SN265) and red (LTH); (B). Comparison of the physical maps and genetic maps. Left: physical map. Right: genetic map.
Table 1.
Characteristics of the high-density genetic map derived from a cross between LTH and SN265.
| Chr. a | No. Markers b | Genetic Distance (cm) | Physical Distance (Mb) | Avg Distance between Markers (cm/kb) | <1 Mb Gap | Min. Gap (kb) | Max. Gap (Mb) |
|---|---|---|---|---|---|---|---|
| 1 | 254 | 338.65 | 42.93 | 1.33/169.03 | 246 | 15.27 | 3.16 |
| 2 | 254 | 227.11 | 31.61 | 0.89/124.46 | 252 | 15.47 | 3.45 |
| 3 | 313 | 284.80 | 36.14 | 0.91/115.45 | 310 | 15.06 | 1.57 |
| 4 | 281 | 300.18 | 34.29 | 1.07/122.03 | 275 | 15.63 | 1.91 |
| 5 | 239 | 269.15 | 29.68 | 1.13/124.19 | 235 | 15.32 | 1.08 |
| 6 | 247 | 181.10 | 28.63 | 0.73/115.92 | 244 | 15.53 | 3.63 |
| 7 | 187 | 217.94 | 29.23 | 1.17/156.34 | 184 | 17.18 | 1.46 |
| 8 | 238 | 193.86 | 28.12 | 0.82/118.15 | 237 | 15.02 | 0.99 |
| 9 | 188 | 111.32 | 22.30 | 0.59/118.61 | 185 | 15.89 | 1.92 |
| 10 | 234 | 199.84 | 22.99 | 0.85/98.23 | 230 | 15.77 | 1.27 |
| 11 | 186 | 323.32 | 28.49 | 1.74/153.18 | 183 | 15.01 | 3.48 |
| 12 | 207 | 192.85 | 26.70 | 0.93/130.43 | 202 | 15.63 | 2.33 |
a Chr., indicates chromosome; b No. markers, the number of markers on the chromosome.
2.3. The Quality and Accuracy of the Bin Map
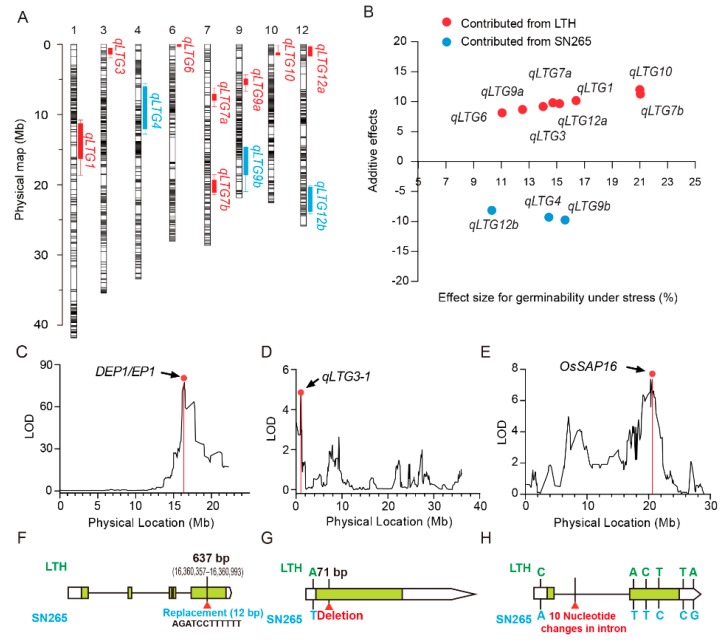
QTL mapping for the typical panicle trait named Dense and Erect Panicle1 (DEP1) was performed to estimate the accuracy and mapping resolution ability of the respective bin maps and the RILs. Careful observations were taken for the panicle curvature of the 144 RILs lines 25 days after the start of heading, whereas the erect-type panicle lines remained erect. One QTL was identified on chromosome 9, with a peak interval of 16.4 Mb, as shown in Figure 3C. The QTL peak was located on the previously characterized and cloned DEP1/EP1 gene. Sequence comparisons of the DEP1 region between LTH and SN265 illustrated the replacement of a 637-bp in the middle of exon-5 by a 12-bp sequence (Figure 3F), which caused a frame shift mutation, as described previously [28].
Figure 3.
(A). QTL scan using whole-genome sequencing and a sequence comparing the panicle curvature and low-temperature germinability QTLs near the previously identified genes. Curves indicate the chromosome locations (Mb) and the low-temperature germinability (LOD) values of the detected QTLs. Arrowheads represent the relative genetic positions of the candidate genes. (A). Genomic locations of the 11 QTLs with strong effects for low-temperature germinability (LOD > 3) identified in the RIL population; (B). Plots of the additive effect and the allele effect of the 11 QTLs. (C). A plot of the LOD values of the panicle curvature. (D). A plot of the LOD values of the low-temperature germinability on chromosome 3. (E). A plot of the LOD values of low-temperature germinability on chromosome 7. (F). Sequence comparisons of the DEP1 region between LTH and SN265. (G). Sequence comparisons of the qLTG3-1 region between LTH and SN265. (H). Sequence comparisons of the OsSAP16 region between LTH and SN265.
2.4. QTL Analysis of Low-Temperature Germinability
After combining the bin-map and phenotyping data of each RIL line, scanning of the LTG QTL was carried out. A total of 11 QTLs were identified on chromosome 1 (11.48–19.34 Mb), chromosome 3 (0.00–1.31 Mb), chromosome 4 (6.10–11.41 Mb), chromosome 6 (0.34–0.74 Mb), chromosome 7 (6.98–9.21 Mb; 18.93–21.76 Mb), chromosome 9 (5.91–6.83 Mb; 14.91–21.38 Mb), chromosome 10 (1.20–1.60 Mb), and chromosome 12 (0.86–2.35 Mb; 21.05–25.16 Mb). The confidence intervals associated with these QTLs spanned the genomic physical position from 0.07 to 7.86 Mb compared with the reference genome of rice (Table 2, Figure 3A). Five QTLs showed relatively small confidence intervals of less than 1.5 Mb, including qLTG-3 (1.31 Mb), qLTG6 (0.40 Mb), qLTG-9a (0.92 Mb), qLTG-10 (0.40 Mb), and qLTG-12a (1.49 Mb). Among these QTLs, phenotypic variation ranged from 10.30% to 21.04% (Table 2, Figure 3B). Overall, five QTLs depicted the explained phenotypic variation exceeding 15%, and two major QTLs were detected: qLTG7b, found on chromosome 7, with 21.04% phenotypic variation and 7.39 of the LOD value; and qLTG10, located on chromosome 10 and comprised of 21.00% phenotypic variation with a 7.38 LOD value. Furthermore, eight QTL alleles, qLTG1, qLTG3, qLTG6, qLTG7a, qLTG7b, qLTG9a, qLTG10, and qLTG12a, had a positive effect inherited from LTH. The other three QTL alleles, qLTG4, qLTG9b, and qLTG12b, had a positive effect inherited from SN265 (Figure 3B). Based on the high-resolution bin map, two QTLs, qLTG3 and qLTG7b, were localized to the chromosome intervals that subsumed the cloned LTG genes. The QTL named qLTG3 was located in the interval of the LTG gene described as qLTG3-1, with a 4.71 LOD value and 14.02% phenotype variance [9]. The QTL named qLTG7b had a 7.39 LOD value and 21.04% phenotypic variation and was located on OsSAP16, a rice LTG gene that has already been identified by the association study method [10]. PCR amplification and sequencing were conducted to screen out the causal polymorphisms of qLTG3-1 and OsSAP-16, which determined that LTH has two cold-tolerant genes (Figure 3E,F). Additionally, these results also indicate that the data used for scanning QTLs are entirely effective (Figure 3D,E,G,H).
Table 2.
Characteristics of the high-density genetic map derived from a cross between LTH and SN265.
| QTL | Chr. a | Peak | QTL Interval | LOD c | Var (%) d | Add. e | Positive Allele | |||
|---|---|---|---|---|---|---|---|---|---|---|
| Pos. (cm) | Pos. (Mb) b | Linkage (cm) | Physical (Mb) | Location Interval (cm/Mb) | ||||||
| qLTG1 | 1 | 116.34 | 16.90 | 103.19–128.81 | 11.48–19.34 | 25.62/7.86 | 5.59 | 16.40 | 10.16 | LTH |
| qLTG3 | 3 | 3.95 | 1.10 | 0.00–10.78 | 0.00–1.31 | 10.78/1.31 | 4.71 | 14.02 | 9.18 | LTH |
| qLTG4 | 4 | 54.58 | 6.27 | 36.64–77.67 | 6.10–11.41 | 41.03/5.31 | 4.85 | 14.43 | −9.30 | SN265 |
| qLTG6 | 6 | 0.18 | 1.34 | 0.18–1.07 | 0.34–0.74 | 0.89/0.40 | 3.64 | 11.05 | 8.13 | LTH |
| qLTG7a | 7 | 104.99 | 8.88 | 87.28–107.84 | 6.98–9.21 | 20.56/2.23 | 4.98 | 14.72 | 9.80 | LTH |
| qLTG7b | 7 | 152.12 | 20.32 | 148.13–162.80 | 18.93–21.76 | 14.67/2.84 | 7.39 | 21.04 | 11.26 | LTH |
| qLTG9a | 9 | 4.31 | 6.07 | 4.35–8.38 | 5.91–6.83 | 4.03/0.92 | 4.17 | 12.53 | 8.68 | LTH |
| qLTG9b | 9 | 79.24 | 15.27 | 71.69–102.85 | 14.91–21.38 | 31.16/6.47 | 5.29 | 15.60 | −9.76 | SN265 |
| qLTG10 | 10 | 13.93 | 1.60 | 8.55–14.06 | 1.20–1.60 | 5.51/0.40 | 7.38 | 21.00 | 12.00 | LTH |
| qLTG12a | 12 | 6.17 | 0.87 | 4.72–16.25 | 0.86–2.35 | 11.53/1.49 | 5.14 | 15.20 | 9.65 | LTH |
| qLTG12b | 12 | 131.25 | 22.79 | 111.25–145.46 | 21.05–25.16 | 34.21/4.11 | 3.41 | 10.30 | −8.12 | SN265 |
a Chr., chromosome; b Positions in the linkage map (unit: Mb); c Logarithm (base 10) of the odds for the corresponding QTL peak; d Percentage of the phenotypic variation explained by the corresponding QTL; e Additive effect of the corresponding QTL.
2.5. Fine Mapping and Candidate Gene Prediction for qLTG6
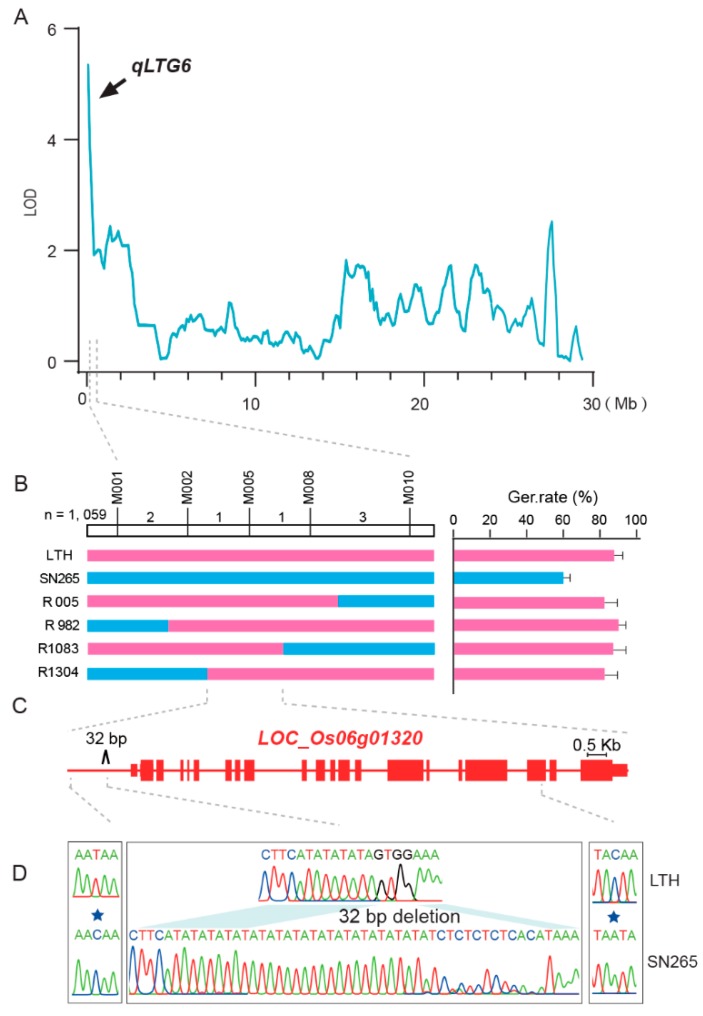
To identify the possible novel genes for low germination within the QTLs, qLTG6 was thoroughly analyzed because it has a high LOD value and relatively small genetic intervals (Figure 4A, Table 2). qLTG6 was first identified within a 400 kb interval. Then, further high-resolution mapping of qLTG6 was carried out using four key homozygous recombination lines from 1,569 segregating population plants and newly developed markers, as shown in Table 3. Finally, qLTG6 was localized to a region of a 45.8 kb physical interval between markers M002 and M008 via the progeny testing of key homozygous recombinant lines (Figure 4B). Overall, there was the prediction of about seven genes in the target region (Table 4). A re-sequencing data analysis of the delimited region only detected differences in the gene LOC_Os06g01320 (Figure 4C). To confirm and estimate these differences, PCR-based sequencing was conducted to analyze the gene body and the 3 kb promoter of LOC_Os06g01320, which revealed a 32 bp deletion and a T–C transition in the promoter. In the gene body, a C–A transition was also detected in the 18th exon, causing a substitution of Thr to Asn in LTH (Figure 4D).
Figure 4.
Identification of the candidate gene of the QTL, qLTG6. (A). Linkage analyses of the QTL, qLTG6; (B). Graphical representation of recombinants in the RILs of germinability under low temperatures, refining the location of qLTG6 in an interval defined by bin markers; (C). The identification of the candidate gene, LOC_Os06g01320, which encodes a chromodomain, helicase/ATPase, and DNA-binding domain (CHD)-related (CHR) proteins, CHR723; (D). Validation of the mutation of LOC_Os06g01320 by PCR sequencing.
Table 3.
The markers developed for the fine mapping of qLTG6 on chromosome 6.
| Molecular Marker | Primer Sequence (5′→′) |
|---|---|
| M001 | CTTCGCACTCCAGTCGCTCTCC GTTGAGGAGGTGTATGGGCTTGG |
| M002 | AGCTCACCAGGGACAACATCAAGG TTAACCAGCTCCGCCAGCATCC |
| M005 | CGCCACTGATCGATCTCCTCTCC CGAGCTGGCCTTCTTCCTTGG |
| M008 | AATTGATGCAGGTTCAGCAAGC GGAAATGTGGTTGAGAGTTGAGAGC |
| M010 | TGTTGGATTGGAATCGGAAAGC CTCTGCTGTGCTGTGCTGCTAGG |
Table 4.
Candidate genes in the 45.8 kb target region corresponding to qLTG6.
| Name | Location | Protein |
|---|---|---|
| LOC_Os06g01250 | 163205–165539 | Cytochrome P450 |
| LOC_Os06g01260 | 167364–174331 | Glutathione gamma-glutamylcysteinyltransferase 1 |
| LOC_Os06g01270 | 178580–178343 | Expressed protein |
| LOC_Os06g01280 | 180215–181423 | Retrotransposon protein |
| LOC_Os06g01290 | 182104–184623 | Expressed protein |
| LOC_Os06g01304 | 185692–191452 | Spotted leaf 11 |
| LOC_Os06g01320 | 195018–208583 | Chromodomain, helicase/ATPase, and DNA-binding domain (CHD) proteins |
2.6. Expression Analysis of LOC_Os06g01320
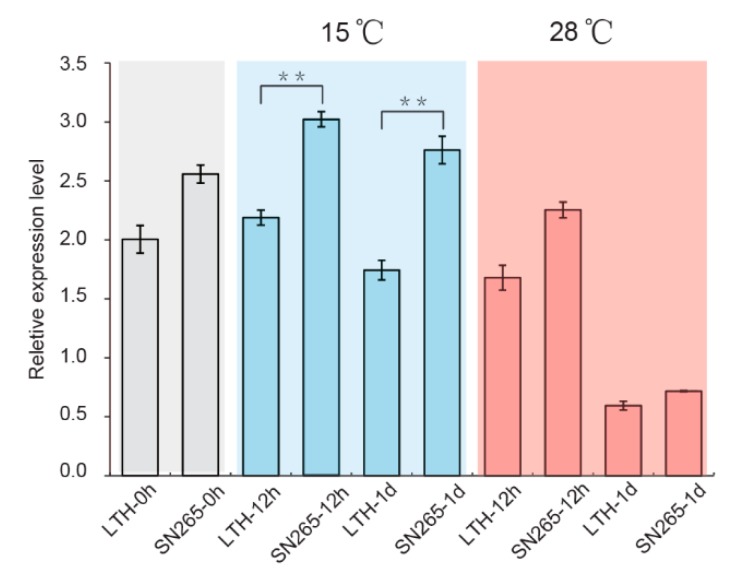
Both an SNP and deletion were detected at the promoter of LOC_Os06g01320 in LTH, which suggests that expression might vary between parents. For further investigations on these variants, qPCR analyses were executed to evaluate the expression patterns of LOC_Os06g01320 in two parents. During seed germination under optimal conditions at 28 °C, the expression levels of LOC_Os06g01320 decreased in the LTH and SN265 from 0 h to 1 d (Figure 5), which indicates that LOC_Os06g01320 is necessarily required to be repressed during seed germination. Under the conditions of low-temperature stress with a temperature range around 15 °C, the expression levels of LOC_Os06g01320 in LTH were significantly lower than SN265, both at 12 h and at 1 d. After conducting a further refinement, these results showed that LTH has lower expression levels of LOC_Os06g01320 than SN265, which could reduce the negative regulation of gene expression and promote seed germination under low-temperature stress.
Figure 5.
The expression analysis of the candidate gene of qLTG6. Relative expression of LOC_Os06g01320 at the germination stage under 15 °C and 28 °C in the LTH and SN265, respectively. ** p < 0.01, using Student’s t-test. Bar SEM, n = 3.
3. Discussion
In recent years, direct-seeded rice has received much attention in Asia because of its time and labor savings and low input demand as an alternative to conventional rice systems. However, the long-term cultivation method of seedling-transplantation has led to a loss of expressions of some low-temperature-tolerant genes, which are usually expressed at the germination stage. Poor germinability remains one of the major problems [5,21,29,30]. Therefore, screening cultivars with high germination abilities under low-temperature stress is necessary to sustain rice yield and ensure the application of direct seeding cultivation in the NEC region. In the current study, the japonica landrace variety, LTH, a locally well-adopted cultivar originating from the far Southwest province of China, was selected since it shows a high germination rate under low-temperature conditions. The germination of LTH began two days after the start of incubation at a temperature of 15 °C and took five more days to reach a germination rate of almost 90%. Screening of the low-temperature germination ability of 135 cultivars from the NEC was then done, which revealed that no cultivar was more tolerant to low-temperature stress than LTH. Therefore, the identification of tolerant genes during germination under low-temperature stress in LTH has important scientific value for improving the low-temperature germination ability of rice in the NEC.
In this study, high-throughput genotyping was employed through whole-genome resequencing, and bin map construction was carried out for QTL mapping. A total of 11 QTLs were identified on Chr.1, Chr.3, Chr.4, Chr.6, Chr.7, Chr.9, Chr.10, and Chr.12. By comparing the positions of the QTLs, 19 previously identified LTG QTLs were found to be near 10 QTLs, except for qLTG9a, as presented in Table 5. The qLTG-1 was mapped near the qCTGERM1-5 region for LTG [31]. The QTL location of qLTG-3 was very close to that of the cloned QTL qLTG3-1 [9]. The QTLs of qLTG4, qLTG6, and qLTG10 were as also found by Teng et al. [32], Ji et al. [33] and Shakiba et al. [31] to enhance low-temperature germinability. qLTG7a was mapped near qCTGERM7-1 [31], qLTG-7 [34], qGR-7, and qGI-7 [35]. Furthermore, qLTG7b was found to be mapped near qCTGERM7-4 [31], qLTG-7 [36], and OsSAP16 [10]. At the germination stage, there was a correlation between the cold tolerance of qLTG9b and the qLTG-9 region [32,33]. qLTG12a was mapped near qLTG-12a [37] and qLTG-12 [38]. qLTG12b was identified near the region of qCTGERM12-1 [31], qGR-12 [10], qLTG-12 [36], and qLTG-12 [39]. The above comparison results reflect the accuracy of this study, as well as the complexity of cold tolerance during germination. Moreover, this study also suggests that LTH’s strong low-temperature germination ability was acquired by accumulating more cold-tolerant genes. These results not only strengthen the findings of previous studies but also reflect the complexity of low-temperature germination in accelerating the breeding programs for enhanced cold-tolerance among rice cultivars.
Table 5.
Comparison of QTL positions on the rice genome.
| QTL | Chr. a | QTL interval | Prior near QTLs Location | Reference |
|---|---|---|---|---|
| Physical (Mb) | Physical (Mb) | |||
| qLTG1 | 1 | 11.48–19.34 | qCTGERM1-5 (12.71) | [31] |
| qLTG3 | 3 | 0.00–1.31 | qLTG3-1 (0.22) | [9] |
| qLTG4 | 4 | 6.10–11.41 | qLTG-4 (6.58–13.64) | [32] |
| qLTG6 | 6 | 0.34–0.74 | qLTG-6 (0.65–2.69) | [33] |
| qLTG7a | 7 | 6.98–9.21 | qCTGERM7-1 (10.46–10.65) | [31] |
| qLTG7b | 7 | 18.93–21.76 | qLTG7 (20.16–22.55) | [34] |
| qGR-7 and qGI-7(20.35–21.59) | [35] | |||
| qCTGERM7-4 (19.59–20.26) | [31] | |||
| qLTG-7 (16.88–22.52) | [36] | |||
| OsSAP16 (22.93) | [10] | |||
| qLTG9a | 9 | 5.91–6.83 | ||
| qLTG9b | 9 | 14.91–21.38 | qLTG-9 (12.29–18.90) | [32] |
| qLTG-9 (11.81–15.32) | [33] | |||
| qLTG10 | 10 | 1.20–1.60 | qCTGERM10-1 (1.40–1.53) | [31] |
| qLTG12a | 12 | 0.86–2.35 | qLTG12a (0.75) | [37] |
| qLTG-12 (2.43–3.19) | [38] | |||
| qLTG12b | 12 | 21.05–25.16 | qCTGERM12-1 (24.89–24.90) | [31] |
| qGR-12(22.78–25.15) | [10] | |||
| qLTG12 (24.52–25.08) | [36] | |||
| qLTG-12 (24.52–25.08) | [39] |
a Chr., chromosome.
Among the 11 QTLs identified in this study, qLTG-6 was ultimately narrowed down to the 45.80 kb region (Figure 4B). Currently, seven genes have been observed in the target region (Table 5) where LOC_Os06g01250 is found to encode the protein named cytochrome P450. LOC_Os06g01260 has functional activities in encoding Glutathione gamma-glutamyl cysteinyl transferase-1. Moreover, LOC_Os06g01280 encodes a retrotransposon protein. LOC_Os06g01270 and LOC_Os06g01290 encode a protein that has yet to be discovered. LOC_Os06g01304 encodes Spotted Leaf 11, whereas LOC_Os06g01320 encodes chromodomain, helicase/ATPase, and DNA-binding domain proteins. Considering the organ specificity in gene expression and molecular function information, it is difficult to ensure that a gene is a target gene. According to the sequencing data, this study identified only the C–A transition in the 18th exon of LOC_Os06g01320, which is predicted to result in the substitution of Thr to Asn in LTH. We also found a 32 bp deletion and a T–C transition in the promoter region. In addition, the expression level of LOC_Os06g01320 in LTH was found to be lower than that of SN265 under lower temperatures. This lower expression level could be associated with seed germination at low-temperature stress. The sequence and gene expression data suggest that LOC_Os06g01320 might be the most plausible prospect for qLTG-6, but the current evidence remains insufficient and will require us to carry out genetic modification complementation or gene editing verification.
4. Materials and Methods
4.1. Plant Materials
The 144 japonica rice RIL population was built via the single-seed descent method from a cross between Shennong265 (SN265) and Lijiangxintuanheigu (LTH). The SN265 is a locally well-adopted and super high yielding cultivar in Liaoning province, NEC, whereas LTH is a landrace from the Yunnan Province, in the far Southwest of China. These RILs were planted at the agricultural farm of institute of Crop Cultivation and Tillage, Heilongjiang Academy of Agricultural Sciences. The DNA of F11 RIL generations was isolated for genotyping with the specified protocols. One residual heterozygous plant was selected in the F6 population for the fine mapping of qLTG6. It had already been observed through the genotyping of 114 DNA markers that the qLTG6 region of this selected plant is heterozygous, and other chromosome regions are homozygous [40]. A total of 1,569 segregating individuals were developed from the selected heterozygous plant.
4.2. Preparation of Seeds for the Germination Test
The RILs and their parents were grown in the experimental fields in 2017, where the nursery sowing date was 15th April, and transplanting was done on 16th May, with one seedling per hill. Sixty plants of each line were planted in 4 rows, with a plant to plant distance of 13.3 × 30 cm. The planting of the segregated qLTG6 population was performed during the rice-growing seasons of 2018. The sowing and transplanting dates were 18th April and 21st May, respectively. The transplanting standards were kept consistent with those of the RILs. Field management practices were done according to the most followed agricultural practices of local farmers. The nitrogen (N), phosphorus (P), and potassium (K) fertilizers, in the form of Urea, single superphosphate, and Murate of Potash, were applied at rates of 120, 60, and 60 kg/ha, respectively. Rice harvesting was done on 30 September, and the plants were retained to dry them for three months. Immature and unfilled grains were removed to obtain high quality grain, which was stored at 5 °C to maintain the relative humidity around 10%.
4.3. Evaluation of Germinability under Cold Stress
The evaluation of the germination ratio under cold stress was performed following standard protocol. The seeds broke out of dormancy at 50 °C after 48 h. Further matured and fully-filled grains were sterilized with 0.1% mercury chloride solution for about 10 min, rinsed with tap water 3 to 4 times, and deionization was performed with deionized water 3 to 4 times. One hundred grains per line were placed on a filter paper in a petri dish, and 10 mL of distilled water was added. The petri dishes were then put in an incubator at the recommended temperature of 15 °C. To avoid any kind of reactions and contaminations, the tap water was changed every two days. The germination of the seeds was noted carefully every day. The germination rate on the 6th day was used for QTL mapping because of the large differences observed between the two parents. The germination was noted three times for each line.
4.4. DNA Extraction, Re-Sequencing, and SNP Calling
The extraction of the genomic DNA of the two parents and each RIL was performed using a modified CTAB method [41]. Biomarker Technologies were used for the re-sequencing of the parents and RILs. The procedure was performed according to Jiang et al. [41]. The short read alignment was done as described by Li and Durbin [42]. Straining of the low-quality data was performed to produce better-quality mapping. The clean data were then aligned to the Nipponbare reference genome (Os-Nipponbare-Reference-IRGSP-1.0) [43] using the BWA software [42]. The calculations of the sequencing coverage and depth were performed through Samtools [44]. Then, the Genome Analysis Toolkit (GATK) was used to detect the SNPs with default parameters [45]. The accession number raw sequence data obtained in our study have been deposited in the NCBI Short Read Archive, with accession number PRJNA587802.
4.5. Genotyping and Construction Bin Map
The estimation of genotype calling parameters, determination of the recombination breakpoint, and construction of the bin map were carried out with minor modifications via the method of Sliding Window, as described by Huang et al. [19]. The genotype of each window was determined by the SNP ratio between the two parents; if the SNP ratio of SN265 to LTH was 15:5 or higher, the genotype was considered to be a homozygous SN265 genotype, if the ratio was lower, then it was designated as a homozygous LTH genotype. Moreover, if the SNP ratio between the two parents was between 5:15 and 15:5, the genotype was recognized as heterozygous. The genotype calling parameters were performed according to the methods described by Song et al. [22]. Adjacent 15 kb segments with the same genotypes were merged as one bin marker [46]. The linkage map was built using the est.map function with the R/qtl software [47].
4.6. QTL Mapping for Low-Temperature Germinability
In this study, we used the composite interval method (CIM) in the R/qtl package [47] to detect the QTLs for LTG. The threshold level of CIM was ensured by 1000 permutations, and the QTL confidence interval was estimated by using a 1.5 LOD-drop from the peak LOD. The germination rate (%) after 6 days under controlled temperature conditions of 15 °C was used for the QTL analysis.
4.7. qRT-PCR and Expression Analysis
The extraction of the total RNA was carried out with a TRIzol reagent. The first-strand of cDNA was reverse transcribed by using a TransScript II First-Strand cDNA Synthesis SuperMix kit (Transgen). The qRT–PCR analysis was executed by using the kit named SYBR FAST qPCR (KAPA). The qPCR primer pairs sequences for LOC_Os06g01320 were 5′-CAAAAAAAAAGACAATAAGGTGGA-3′ (forward) and 5′-CAGACATTGCTTACCCTTATTTATTTT-3′ (reverse). EF-1 alpha was used as an internal control, and the sequencing was 5′-GCACGCTCTTCTTGCTTTCACTCT-3′ (forward) and 5′-AAAGGTCACCACCATACCAGGCTT-3′ (reverse).
Author Contributions
S.J., L.W., X.Y., J.W. conducted the phenotyping and genotyping of parental lines and individuals of the RIL population. C.Y. and S.J. performed the SNP calling, data analyses, and bin-map construction. X.Z., X.S., Q.X., B.L., H.L., and W.L. participated in the phenotyping and data analysis. S.J., Z.L., and W.L. designed the experiment. S.J., C.Y., Q.X. and X.S. drafted the manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by a grant from the National Key Research and Development Program of China (2016YFD0300104), the National Natural Science Foundation of China (31661143012), the Natural Science Foundation of Heilongjiang Province of China (C2016050, YQ2019C020), Heilongjiang Postdoctoral Financial Assistance (LBH-Q15133), Provincial funding for the National Key Research and Development Program in Heilongjiang Province (768001), and Heilongjiang Province Agricultural Science and Technology Innovation Project (2018CQJC002, 2019CQJC002).
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Qian Q. Genomics assisted germplasm improvement. J. Integr. Plant Biol. 2018;60:82–84. doi: 10.1111/jipb.12629. [DOI] [PubMed] [Google Scholar]
- 2.Zhang Q., Chen Q., Wang S., Hong Y., Wang Z. Rice and cold stress methods for its evaluation and summary of cold tolerance-related quantitative trait loci. Rice. 2014;7:24. doi: 10.1186/s12284-014-0024-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang Z., Li J., Pan Y., Li J., Zhou L., Shi H., Zeng Y., Guo H., Yang S., Zheng W., et al. Natural variation in CTB4a enhances rice adaptation to cold habitats. Nat. Commun. 2017;8:14788. doi: 10.1038/ncomms14788. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lu G., Wu F., Wu W., Wang H., Zheng X., Zhang Y., Chen X., Zhou K., Jin M., Cheng Z., et al. Rice LTG1 is involved in adaptive growth and fitness under low ambient temperature. Plant J. 2014;78:468–480. doi: 10.1111/tpj.12487. [DOI] [PubMed] [Google Scholar]
- 5.Liu C., Wang W., Mao B., Chu C. Cold stress tolerance in rice: Physiological changes, molecular mechanism, and future prospects. Hereditas (Beijing) 2018;40:171–185. doi: 10.16288/j.yczz.18-007. [DOI] [PubMed] [Google Scholar]
- 6.Wang Z., Wang J., Wang F., Bao Y., Wu Y., Zhang H. Genetic control of germination ability under cold stress in rice. Rice Sci. 2009;16:173–180. doi: 10.1016/S1672-6308(08)60076-1. [DOI] [Google Scholar]
- 7.Shi Y., Ding Y., Yang S. Molecular regulation of CBF signaling in cold acclimation. Trends Plant Sci. 2018;23:623–637. doi: 10.1016/j.tplants.2018.04.002. [DOI] [PubMed] [Google Scholar]
- 8.Yonemaru J., Yamamoto T., Fukuoka S., Uga Y., Hori K., Yano M. Q-TARO: QTL annotation rice online database. Rice. 2010;3:194. doi: 10.1007/s12284-010-9041-z. [DOI] [Google Scholar]
- 9.Fujino K., Sekiguchi H., Matsuda Y., Sugimoto K., Ono K., Yano M. Molecular identification of a major quantitative trait locus, qLTG3-1, controlling low-temperature germinability in rice. Proc. Natl. Acad. Sci. USA. 2008;105:12623–12628. doi: 10.1073/pnas.0805303105. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Wang X., Zou B., Shao Q., Cui Y., Lu S., Zhang Y., Huang Q., Huang J., Hua J. Natural variation reveals that OsSAP16 controls low-temperature germination in rice. J. Exp. Bot. 2018;69:413–421. doi: 10.1093/jxb/erx413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Liu F., Xu W., Song Q., Tan L., Liu J., Zhu Z., Fu Y., Su Z., Sun C. Microarray-assisted fine-mapping of quantitative trait loci for cold tolerance in rice. Mol. Plant. 2013;6:757–767. doi: 10.1093/mp/sss161. [DOI] [PubMed] [Google Scholar]
- 12.Ma Y., Dai X., Xu Y., Luo W., Zheng X., Zeng D., Pan Y., Lin X., Liu H., Zhang D., et al. COLD1 confers chilling tolerance in rice. Cell. 2015;160:1209–1221. doi: 10.1016/j.cell.2015.01.046. [DOI] [PubMed] [Google Scholar]
- 13.Zhao J., Zhang S., Dong J., Yang T., Mao X., Liu Q., Wang X., Liu B. A novel functional gene associated with cold tolerance at the seedling stage in rice. Plant Biotechnol. J. 2017;15:1141–1148. doi: 10.1111/pbi.12704. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Liu C., Ou S., Mao B., Tang J., Wang W., Wang H., Cao S., Schläppi M., Zhao B., Xiao G., et al. Early selection of bZIP73 facilitated adaptation of japonica rice to cold climates. Nat. Commun. 2018;9:3302. doi: 10.1038/s41467-018-05753-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Xiao N., Gao Y., Qian H., Gao Q., Wu Y., Zhang D., Zhang X., Yu L., Li Y., Pan C., et al. Identification of genes related to cold tolerance and a functional allele that confers cold tolerance. Plant Physiol. 2018;177:1108–1123. doi: 10.1104/pp.18.00209. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Mao D., Xin Y., Tan Y., Hu X., Bai J., Liu Z., Yu Y., Li L., Peng C., Fan T., et al. Natural variation in the HAN1 gene confers chilling tolerance in rice and allowed adaptation to a temperate climate. Proc. Natl. Acad. Sci. USA. 2019;116:3494–3501. doi: 10.1073/pnas.1819769116. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Saito K., Hayanosaito Y., Kuroki M., Sato Y. Map-based cloning of the rice cold tolerance gene ctb1. Plant Sci. 2010;179:97–102. doi: 10.1016/j.plantsci.2010.04.004. [DOI] [Google Scholar]
- 18.Liu C., Schläppi M., Mao B., Wang W., Wang A., Chu C. The bZIP73 transcription factor controls rice cold tolerance at the reproductive stage. Plant Biotechnol. J. 2019;17:1834–1849. doi: 10.1111/pbi.13104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Huang X., Feng Q., Qian Q., Zhao Q., Wang L., Wang A., Guan J., Fan D., Weng Q., Huang T., et al. High-throughput genotyping by whole-genome resequencing. Genome Res. 2009;19:1068–1076. doi: 10.1101/gr.089516.108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Chen Z., Wang B., Dong X., Liu H., Ren L., Chen J., Hauck A., Song W., Lai J. An ultra-high density bin-map for rapid QTL mapping for tassel and ear architecture in a large F2 maize population. BMC Genom. 2014;15:433. doi: 10.1186/1471-2164-15-433. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Jiang N., Shi S., Shi H., Khanzada H., Wassan G., Zhu C., Peng X., Yu Q., Chen X., He X., et al. Mapping QTL for seed germinability under low temperature using a new high-density genetic map of rice. Front. Plant Sci. 2007;8:1223. doi: 10.3389/fpls.2017.01223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Song W., Wang B., Hauck A., Dong X., Li J., Lai J. Genetic dissection of maize seedling root system architecture traits using an ultra-high density bin-map and a recombinant inbred line population. J. Integr. Plant Biol. 2016;58:266–279. doi: 10.1111/jipb.12452. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Wang B., Liu H., Liu Z., Dong X., Guo J., Li W., Chen J., Gao C., Zhu Y., Zheng X., et al. Identification of minor effect QTLs for plant architecture related traits using super high density genotyping and large recombinant inbred population in maize (Zea mays) BMC Plant Biol. 2018;18:17. doi: 10.1186/s12870-018-1233-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Wang B., Zhu Y., Zhu J., Liu Z., Liu H., Dong X., Guo J., Li W., Chen J., Gao C., et al. Identification and Fine-Mapping of a Major Maize Leaf Width QTL in a Re-sequenced Large Recombinant Inbred Lines Population. Front. Plant Sci. 2018;9:101. doi: 10.3389/fpls.2018.00101. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang L., Wang A., Huang X., Zhao Q., Dong G., Qian Q., Sang T., Han B. Mapping 49 quantitative trait loci at high resolution through sequencing-based genotyping of rice recombinant inbred lines. Theor. Appl. Genet. 2010;122:327–340. doi: 10.1007/s00122-010-1449-8. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Zhou Z., Zhang C., Zhou Y., Hao Z., Wang Z., Zeng X., Di H., Li M., Zhang D., Yong H., et al. Genetic dissection of maize plant architecture with an ultra-high density bin map based on recombinant inbred lines. BMC Genom. 2016;17:178. doi: 10.1186/s12864-016-2555-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zou G., Zhai G., Feng Q., Yan S., Wang A., Zhao Q., Shao J., Zhang Z., Zou J., Han B., et al. Identification of QTLs for eight agronomically important traits using an ultra-high-density map based on SNPs generated from high-throughput sequencing in sorghum under contrasting photoperiods. J. Exp. Bot. 2012;63:5451–5462. doi: 10.1093/jxb/ers205. [DOI] [PubMed] [Google Scholar]
- 28.Zhao M., Sun J., Xiao Z., Cheng F., Xu H., Tang L., Chen W., Xu Z., Xu Q. Variations in DENSE AND ERECT PANICLE 1 (DEP1) contribute to the diversity of the panicle trait in high-yielding japonica rice varieties in northern China. Breed. Sci. 2016;66:599–605. doi: 10.1270/jsbbs.16058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Lee J., Lee W., Kwon S. A quantitative shotgun proteomics analysis of germinated rice embryos and coleoptiles under low-temperature conditions. Proteome Sci. 2015;13:27. doi: 10.1186/s12953-015-0082-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Sasaki T., Kinoshita T., Takahashi M. Estimation of the number of genes in the germination ability at low temperature in rice genetical. J. Fac. Agric. Hokkaido Univ. 1974;57:301–312. [Google Scholar]
- 31.Shakiba E., Edwards J., Jodari F., Duke S., Baldo A., Korniliev P., McCouch S., Eizenga G. Genetic architecture of cold tolerance in rice (Oryza sativa) determined through high resolution genome-wide analysis. PLoS ONE. 2017;12:e0172133. doi: 10.1371/journal.pone.0172133. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Teng S., Zeng D., Qian Q., Yasufumi K., Huang D., Zhu L. QTL analysis of rice low temperature germinability. Chin. Sci. Bull. 2001;46:1800–1803. doi: 10.1007/BF02900554. [DOI] [Google Scholar]
- 33.Ji S., Jiang L., Wang Y., Liu S., Liu X., Zhai H., Wan J. Detection and analysis of QTL for germination rate at low temperature in rice (Oryza sativa L.) J. Nanjing Agric. Uni. 2007;30:1–6. [Google Scholar]
- 34.Iwata N., Fujino K. Genetic effects of major QTLs controlling low-temperature germinability in different genetic backgrounds in rice (Oryza sativa L.) Genome. 2010;53:763–768. doi: 10.1139/G10-060. [DOI] [PubMed] [Google Scholar]
- 35.Ji S., Jiang L., Wang Y., Zhang W., Liu X., Liu S., Chen L., Zhai H., Wan J. Quantitative trait loci mapping and stability for low temperature germination ability of rice. Plant Breed. 2009;128:387–392. doi: 10.1111/j.1439-0523.2008.01533.x. [DOI] [Google Scholar]
- 36.Hou M., Wang C., Jiang L., Wan J., Hideshi Y., Atsushi Y. Inheritance and QTL mapping of low temperature germinability in rice (Oryza sativa L.) Acta. Genet. Sin. 2004;31:701–706. [PubMed] [Google Scholar]
- 37.Fujino K., Obara M., Shimizu T., Koyanagi K.-O., Ikegaya T. Genome-wide association mapping focusing on a rice population derived from rice breeding programs in a region. Breed. Sci. 2015;65:403–410. doi: 10.1270/jsbbs.65.403. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Li L., Liu X., Xie K., Wang Y., Liu F., Lin Q., Wang W., Yang C., Lu B., Liu S., et al. qLTG-9, a stable quantitative trait locus for low-temperature germination in rice (Oryza sativa L.) Theor. Appl. Genet. 2013;126:2313–2322. doi: 10.1007/s00122-013-2137-2. [DOI] [PubMed] [Google Scholar]
- 39.Ji S., Jiang L., Wang Y., Liu S., Liu X., Zhai H., Yoshimura A., Wan J. QTL and epistasis for low temperature germinability in rice. Acta. Agron. Sin. 2008;34:551–556. [Google Scholar]
- 40.Jiang S., Zhang X., Zhang F., Xu Z., Chen W., Li Y. Identification and Fine Mapping of qCTH4, a quantitative trait loci controlling the chlorophyll content from tillering to heading in rice (Oryza sativa L.) J. Hered. 2012;103:720–726. doi: 10.1093/jhered/ess041. [DOI] [PubMed] [Google Scholar]
- 41.Jiang S., Sun S., Bai L., Ding G., Wang T., Xia T., Jiang H., Zhang X., Zhang F. Resequencing and variation identification of whole genome of the japonica rice variety “Longdao24” with high yield. PLoS ONE. 2017;12:e0181037. doi: 10.1371/journal.pone.0181037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Li H., Durbin R. Fast and accurate long-read alignment with Burrows-Wheeler transform. Bioinformatics. 2010;26:589–595. doi: 10.1093/bioinformatics/btp698. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Kawahara Y., De la Bastide M., Hamilton J., Kanamori H., McCombie W., Ouyang S., Schwartz D., Tanaka T., Wu J., Zhou S., et al. Improvement of the Oryza sativa Nipponbare reference genome using next generation sequence and optical map data. Rice. 2013;6:4. doi: 10.1186/1939-8433-6-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Li H., Handsaker B., Wysoker A., Fennell T., Ruan J., Homer N., Marth G., Abecasis G., Durbin R., Proc G. The Sequence Alignment/Map format and SAMtools. Bioinformatics. 2009;25:2078–2079. doi: 10.1093/bioinformatics/btp352. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.McKenna A., Hanna M., Banks E., Sivachenko A., Cibulskis K., Kernytsky A., Garimella K., Altshuler D., Gabriel S., Daly M., et al. The Genome Analysis Toolkit: A MapReduce framework for analyzing next-generation DNA sequencing data. Genome Res. 2010;20:1297–1303. doi: 10.1101/gr.107524.110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Han K., Jeong H., Yang H., Kang S., Kwon J., Kim S., Choi D., Kang B. An ultra-high-density bin map facilitates high-throughput QTL mapping of horticultural traits in pepper (Capsicum annuum) DNA Res. 2016;23:81–91. doi: 10.1093/dnares/dsv038. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Arends D., Prins P., Jansen R., Broman K. R/qtl: High-throughput multiple QTL mapping. Bioinformatics. 2010;26:2990–2992. doi: 10.1093/bioinformatics/btq565. [DOI] [PMC free article] [PubMed] [Google Scholar]