Abstract
NCAPG is a subunit of condensin I that plays a crucial role in chromatin condensation during mitosis. NCAPG has been demonstrated to be associated with farm animal growth traits. However, its role in regulating myoblast differentiation is still unclear. We used myoblasts derived from fetal bovine tissue as an in vitro model and found that NCAPG was expressed during myogenic differentiation in the cytoplasm and nucleus. Silencing NCAPG prolonged the mitosis and impaired the differentiation due to increased myoblast apoptosis. After 1.5 days of differentiation, silencing NCAPG enhanced muscle-specific gene expression. An assay for transposase-accessible chromatin- high throughput sequencing (ATAC-seq) revealed that silencing NCAPG altered chromatin accessibility to activating protein 1 (AP-1) and its subunits. Knocking down the expression of the AP-1 subunits fos-related antigen 2 (FOSL2) or junB proto-oncogene (JUNB) enhanced part of the muscle-specific gene expression. In conclusion, our data provide valuable evidence about NCAPG’s function in myogenesis, as well as its potential role in gene expression.
Keywords: NCAPG, bovine myoblasts, myogenesis, chromatin accessibility, AP-1
1. Introduction
The growth rate is the most important trait in farm animal production. Genome variations regulating animal growth and development have been widely studied in farm animals. NCAPG is a gene well-documented to be associated with body size, average daily gain, feed intake efficiency, fetal growth, carcass trait, and muscle development in beef cattle [1,2,3,4,5,6,7]; body weight in sheep [8]; and withers height in horses [9,10]. Moreover, in clinical practice, NCAPG is a crucial gene that is correlated with height [11].
NCAPG is a subunit of the condensin I complex, which is essential for the structural maintenance of chromosomes [12]. The core subunits of condensin are SMC2 and SMC4, both of which are the members of structural maintenance of chromosome (SMC) family of chromosomal ATPases [13]. Condensin I also has unique non-SMC subunits: CAP-G, CAP-H, and CAP-D2 [12]. Previous studies have revealed that both condensin members play essential but distinct roles in mitosis. Condensin I is sequestered in the cytoplasm during the interphase and gains access to chromosomes only after the nuclear envelope break down (NEBD) in the prometaphase. Condensin II functions in the nucleus from the interphase through the prophase as an early regulator of chromosome condensation. After NEBD, condensin I engages in further chromosome compaction to facilitate final mitosis [14,15,16]. In one study, the abrogation of the CAP-D2 subunit resulted in a significant delay of prometaphase to anaphase progression compared to the control group [15]. This provided direct evidence for the relationship between NCAPG and growth traits.
Skeletal muscle development is one of the most important traits in farm animal production because skeletal muscle account for about half of an animal’s body weight. Muscle development is classified into prenatal and postnatal stages. The fetal stage is crucial for skeletal muscle development, as the number of muscle fibers barely increases after birth [17,18,19].
However, the regulatory mechanisms and biological functions of NCAPG are still unknown. In particular, NCAPG’s role in the development of myoblasts in the fetal stage needs exploration. In this study, the role of NCAPG in the myogenic differentiation of myoblasts derived from fetal bovine tissue was determined using RNA interference to knock down NCAPG gene expression. Moreover, due to NCAPG’s conserved roles in other tissues, our study and its meanings may also be applied elsewhere in the body.
2. Results
2.1. Myogenic Differentiation of Myoblasts Derived from Fetal Bovine Tissue
After 24 h of differentiation, some myotubes could be observed. After 48 h, a majority of the myoblasts had fused into myotubes. After an additional two days of differentiation, both the size and length of the myotubes increased (Figure 1A). RNA and proteins were extracted from the cells on days 0, 2, and 4 to analyze the muscle-specific gene expression profile during proliferation and differentiation. Myoblast determination protein 1 (MYOD) and myogenic Factor 5 (MYF5) mRNA expression decreased during proliferation and differentiation, while MYOG expression increased until day 2 of differentiation, after which it decreased (Figure 1B).
Figure 1.
Myogenic differentiation of myoblasts derived from fetal bovine tissue. (A) Microscopic images of bovine myoblasts on days 0, 2, and 4 of differentiation. Scale bars = 100 µm. (B) Transcript levels of MYF5, MYOD, MYOG, MYH1, MYH2, MYH3 and MYH4 during proliferation (P) and myogenic differentiation (D0, D2, and D4). The results are represented as the mean ± the SEM from at least three independent experiments. (C) Western blot evaluating the protein levels of MYOD and MHCs in the cultured cells, as described in (B).
As for myosin heavy chain (MYH) isoforms in skeletal muscle, the expression of MYH1, MYH2, MYH3, and MYH4 was relatively low during proliferation and early differentiation, but it increased dramatically during late differentiation (Figure 1B). In particular, MYH2 expression increased nearly 200-fold compared to basal level on day 4. Consistently, western bolt analyses showed that MYH expression increased gradually during the differentiation of myoblasts, while MYOD expression decreased during differentiation (Figure 1C). From these results, it is clear that the use of myoblasts derived from fetal tissue is a reliable model for studying early myogenic differentiation in cattle.
2.2. NCAPG Inhibition Prolonged the Prometaphase and Metaphase of the Proliferating Myoblasts
To investigate the role of NCAPG, we first analyzed its expression (as well as that of other subunit genes of condensin I and II) during proliferation and differentiation. NCAPG was robustly expressed during proliferation and the beginning of differentiation. The mRNA condensin core subunits SMC2 and SMC4 had similar expression patterns, as did the non-SMC regulatory subunits NCAPG, NCAPH, NCAPD2, NCAPG2, NCAPH2, and NCAPD3. These genes had robust expression during proliferation, but then the mRNA level attenuated until day 2. After day 2 of differentiation, their expression increased slightly (Figure S1).
The regulatory mechanism of NCAPG in mitosis has previously been studied in rodent cells. To explore its function in bovine myoblasts, we designed three siRNAs targeting common exons of NCAPG transcript isoforms to knock down total NCAPG mRNA. Here, siRNA003 was selected for the knockdown experiments due to its high efficiency (Figure 2A). We first identified a morphological change in the myoblasts in the knowndown (KD) group, which appeared as a flattened stellate shape (Figure 2B).
Figure 2.
NCAPG KD elongated the prometaphase and metaphase. (A) The silencing efficiency of NCAPG siRNAs. We designed three siRNAs that were transfected into myoblasts, and the mRNA level was measured to determine the siRNA efficiency. The results are represented as the mean ± the SEM (three biological replicates), ** p < 0.01. (B) Cell morphology of NCAPG silencing myoblasts. NCAPG was depleted in the proliferating myoblasts. Scale bar = 100 µm. (C) An analysis of chromosome condensation during the prophase in live myoblasts depleted of NCAPG. To visualize chromatin, we stably expressed H2B–EGFP in the myoblasts. Prophase image sequences were extracted from long-term imaging experiments and aligned along a time axis according to nuclear envelope breakdown (NEBD). Imaging was done 48–72 h after siRNA transfection. Bar = 5 µm. (D) The time duration of the prometaphase and metaphase in the myoblasts. The time from NEBD to the onset of the anaphase was measured using at least six replicates, ** p < 0.01.
To confirm NCAPG’s role in bovine myoblast mitosis, we employed a lentivirus to express the fused bH2B–EGFP gene while knocking down NCAPG mRNA expression. About one day after the lentivirus infection, green fluorescent protein (GFP) expression was clustered in the nucleus. Then, the myoblasts were subcultured at a low density and transfected with siRNA. In order to observe the mitotic process, pictures were taken every 3 min during proliferation for two days after siRNA transfection (Figure 2C). The time between NEBD and the onset of the anaphase was determined. The siRNA control (NC) group required about 19.5 ± 4.1 min from NEBD to initiate of anaphase, whereas the siRNA group took about 25.8 ± 5.0 min (Figure 2D). Therefore, we found that the knockdown of NCAPG prolonged the procession of myoblast mitosis.
2.3. NCAPG Inhibition Impaired Myogenic Differentiation in Myoblasts
Until this study, the role of NCAPG in myogenic differentiation had not been explored. Intriguingly, during differentiation, both the mRNA and protein expression of NCAPG were relatively low compared to during proliferation (Figure 1C and Figure S1). First, we detected NCAPG localization in the myoblasts. The immunofluorescence (IF) results showed that NCAPG could be detected in both the nucleus and cytoplasm of the myoblasts on day 2 of differentiation (Figure S2A). Subsequently, in order to explore the distribution of NCAPG in the myoblasts, we analyzed its presence in the cytoplasm and nucleus (separately). Here, β-tubulin and H2B were used as a reference for cytoplasm proteins and nuclear proteins, respectively. The immunostaining results showed that NCAPG was localized in both the cytoplasm and nucleus (Figure S2B). This nuclear localization made it possible for NCAPG to access chromatin. This suggests that NCAPG took part in regulating myogenic differentiation.
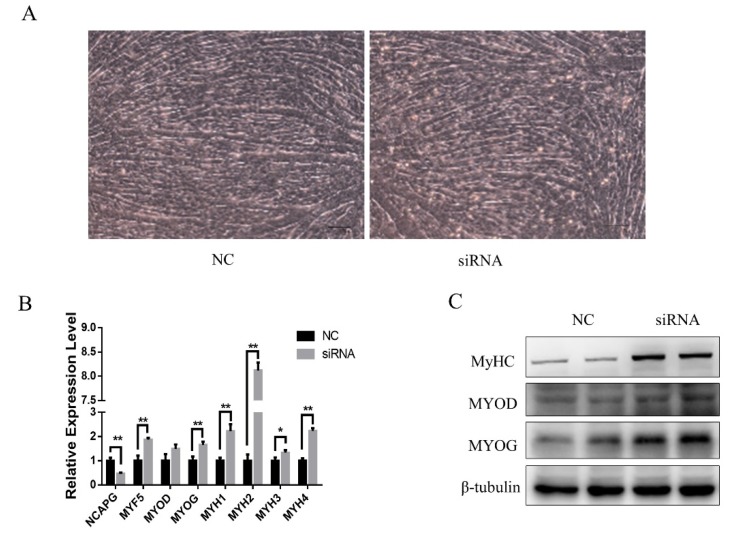
To investigate the role of NCAPG in myogenic differentiation, we interfered with its expression by transfecting siRNA003, while scrambled siRNA used as a control. Myotubes appeared around 48 h after differentiation. About four days after the induction of myogenesis, we could observe obvious myotubes in the NC group, whereas KD caused dysfunctional fusion (Figure 3A). Then, an IF test was done using an MYH antibody to identify the myotubes, and a similar difference was observed (Figure 3B). To further explore the alteration in gene expression, we extracted RNA and proteins from the myoblasts four days after NCAPG knockdown. NCAPG expression had decreased by about 70% by day 4. We detected the expression of some muscle-specific genes. For instance, MYH1, MYH2, MYH3, MYH4, MYOD, and MYOG expression was significantly decreased in the KD group (p < 0.01, Figure 3C). However, a difference in MYF5 expression between the two groups was not detected (Figure 3C). Immunostaining showed similar results: the protein levels of MYHs and NCAPG decreased after NCAPG knockdown (Figure 3D).
Figure 3.
Silencing NCAPG impaired the differentiation of myoblasts. (A) Cell morphology of NCAPG-silenced myoblasts on day 4 of differentiation. Scale bar = 100 µm. (B) NCAPG-silenced myoblasts on day 4 of differentiation. Myotubes were visualized using IF staining with MYH antibodies. Scale bar = 100 µm. (C) Transcript levels of muscle-specific genes on day 4 of differentiation. The results are presented as the mean ± the SEM from at least three independent experiments: ** p < 0.01. (D) Western blot evaluating the protein levels of NCAPG and MHCs in myoblasts on day 4 of differentiation.
2.4. NCAPG KD Facilitated Apoptosis
To explore the mechanism of NCAPG in impairing myogenesis, we detected apoptosis on day 4 using a TdT-mediated dUTP nick-end labeling (TUNEL) assay and a caspase 3 activity assay. As shown in Figure 4A, NCAPG knockdown increased the number of TUNEL-positive cells on day 4. About 23% of cells in the siRNA group were TUNEL-positive, while the apoptosis rate of the NC group was less than 1% (Figure 4B). The caspase 3 activity assay showed that NCAPG knockdown led to a three-fold increase in caspase-3 activity compared to the NC group (Figure 4C).
Figure 4.
NCAPG KD induced apoptosis. (A) Transcript levels of muscle-specific gene expression in AP-1 subunits (FOSL2) in depleted myoblasts. (B,C) Transcript levels of muscle-specific gene expression in AP-1 subunits (JUNB) in depleted myoblasts. The results are presented as the mean ± the SEM from at least three independent experiments, * p < 0.05, ** p < 0.01.
2.5. Myogenic Gene Expression was Boosted on Day 1.5 of Differentiation after NCAPG Knockdown
Interestingly, despite the reduced myogenesis in the NCAPG knockdown myoblasts after four days of myogenesis, we found that the myotube formation of the siRNA group was slightly higher than that of the NC group at 1.5–2 days of myogenesis (Figure 5A). At 36 h, we analyzed the muscle-specific gene expression as well as the condensin subunit genes. The results showed that in the siRNA group, the mRNA levels of MYH1, MYH2, MYH3, MYH4, MYF5, and MYOG were significantly higher than in the NC group. There was no difference in the expression of MYOD between groups (Figure 5B). Consistently, the protein expression analysis showed that the expression of MYHs and MYOG in the siRNA group was upregulated, while MYOD was not affected (Figure 5C).
Figure 5.
Myogenic gene expression was upregulated after 1.5 d of differentiation in NCAPG-silenced myoblast. (A) The cell morphology of NCAPG-silenced myoblasts on day 1.5 of differentiation. Scale bar = 100 µm. (B) Transcript levels of muscle-specific genes on day 1.5 of differentiation. The results are presented as the mean ± the SEM from at least three independent experiments: * p < 0.05, ** p < 0.01. (C) Western blot evaluating the protein levels of MYOD, MYOG, and MHCs in myoblasts on day 1.5 of differentiation.
2.6. NCAPG Knockdown Altered Chromatin Compaction and Accessibility
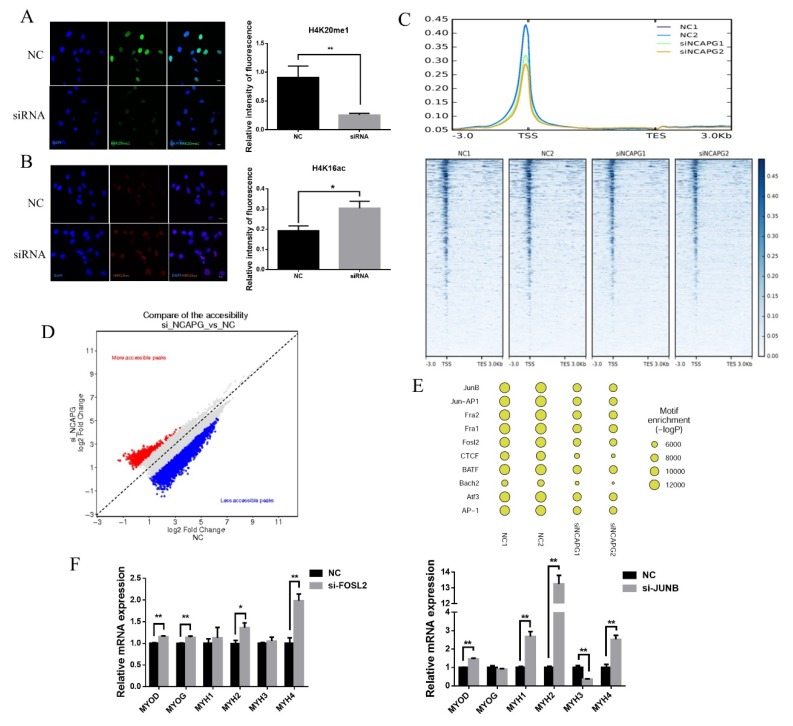
To investigate the relationship between altered gene expression and NCAPG knockdown, we conducted an IF test using H4K20me1 and H4K16ac antibodies. H4K20me1 modification is positively correlated with chromatin compaction, while H4K16ac is negatively correlated with chromatin compaction [20,21]. The results showed that the fluorescence intensity of H4K20me1 in the siRNA group attenuated significantly, while that of H4K16ac increased significantly after knockdown on day 1.5 (Figure 6A,B). These results demonstrated that NCAPG knockdown reduced chromatin compaction. To further study chromatin compaction after NCAPG knockdown, we used an ATAC-seq to isolate the accessible chromatin on day 1.5. The sequencing reads were mapped to the bovine genome, and summarized statistics of the read mapping are listed in Table S1. The reads from both the NC and si-NCAPG groups were enriched around transcription start sites (TSS) (Figure 6C), and the peaks were called using MACS2. The peak number from the NC group was around 110,000, while the peak number from the si-NCAPG group varied between 71,000 and 75,000 (Table S2). The annotation of peaks revealed that silencing NCAPG resulted in the proportion of the peaks located in −1000 bp (Figure S3). However, compared to NC, silencing NCAPG led to more of a decrease in the accessibility of chromatin sites (Figure 6D). Pearson correlation coefficients between the NC and si-NCAPG groups were calculated based on the Log10 RPM matrix (Figure S4). A Gene Ontology analysis revealed that silencing NCAPG affected multiple processes, including development, cell differentiation, and the positive regulation of biological processes (Figure S5). The Kyoto Encyclopedia of Genes and Genomes (KEGG) analysis also revealed the pathways regulated by NCAPG, including the metabolic pathway, the MAPK pathway, the regulation of actin cytoskeleton pathway, and the calcium signaling pathway (Figure S5). The metabolic pathway had the highest proportion of genes with altered accessibility, including PRKAA1 and SIRT1 (Figure S6). The 10 most enriched motifs are listed in Figure 6E. A motif analysis revealed that silencing NCAPG induced the decreased accessibility of chromatin fragments containing transcription factors (TF) binding sites such as AP-1 subunits (JUNB, FOSL2, and FOSL1), CCCTC-binding factor (CTCF), and basic leucine zipper ATF-Like transcription factor (BATF) (Figure 6E).
Figure 6.
NCAPG knockdown altered chromatin compaction and accessibility. (A,B) (Left) Representative immunofluorescence images of myoblasts transfected with a negative control or si-NCAPG and stained with antibodies against H4K20me1 (green) or H4K16ac (red). DNA (DAPI) is shown in blue. Scale bars = 10 μm. (Right) Fluorescence intensity quantification. The results are presented as the mean ± the SEM, * p < 0.05, ** p < 0.01. (C) (top) A histogram of the intensity of enriched reads (from TSS −3 kb to TES +3 kb) for each replicate. (bottom) A heatmap of ATAC-seq signal mapping with annotated TSS. (D) Scatterplots showing changes in chromatin accessibility between the NC and si-NCAPG groups. (E) TF motifs identified from ATAC-seq peaks. (F) (Left) Transcript levels of muscle-specific gene expression in AP-1 subunits (FOSL2) in depleted myoblasts. (Right) Transcript levels of muscle-specific gene expression in AP-1 subunits (JUNB) in depleted myoblasts. The results are presented as the mean ± the SEM from at least three independent experiments, * p < 0.05, ** p < 0.01.
To analyze the relationship between AP-1 binding and muscle-specific gene expression, we knocked down the AP-1 subunit genes FOSL2 and JUNB using siRNA. FOSL2 silencing increased the expression of MYOD, MYOG, MYH2, and MYH4, while JUNB silencing improved the expression of MYOD, MYH1, MYH2, and MYH4 but repressed MYH3 expression (Figure 6F).
3. Discussion
The location of condensins in the cytoplasm and their role in mitosis have been intensively investigated [14,15,16]. Condensin II primarily contributes to the axial shortening of chromatids, while condensin I has a role in lateral compaction [22]. In one study, depletion of the condensin I subunit prolonged prometaphase-to-anaphase progression by more than 150% [15]. In our study, we found a similar delay after silencing NCAPG. However, prometaphase-to-anaphase progress was delayed by about 32% compared to the control group. The difference may have been due to the cell type, the siRNA concentration and efficiency, and the condensin subunit selected.
The common view of condensin localization is that condensin I is cytoplasmic during the interphase and, after the prophase and NEBD, has access to chromatin [15,23]. However, recent reports have indicated that a small amount of condensin I can be found in the nucleus during the G1 phase, which is gradually lost during the S and G2 phases [24]. In those reports, silencing the subunits of condensin I in the G0/G1-phase cells resulted in gene misregulation [24,25]. In our study, all condensin subunits, including NCAPG, decreased dramatically after the initiation of myogenic differentiation and remained at a relatively low level late in the differentiation process. We also found NCAPG expressed in the nuclei of differentiating myogenic myoblast cells. This implies that condensin I may function not only during mitosis, but also during other stages of the cell cycle.
We found that knocking down NCAPG during the myogenic differentiation of fetal bovine muscle myoblasts promoted apoptosis. The association between apoptosis and NCAPG or condensin I depletion has been reported in several studies. In zebrafish, a mutation in NCAPG resulted in increased genomic imbalances and in an increased rate of apoptosis in the retina [26]. In another work, silencing NCAPG in hepatocellular carcinoma cells inhibited proliferation and induced apoptosis [27,28]. Condensin I disruption has also led to apoptosis in the germline [29]. Further, knocking down NCAPD2, a subunit of condensin I, has led to apoptosis in triple-negative breast cancer cells [30].
In contrast to its established role in chromatin condensation in mitosis, our knowledge about condensin I in regulating gene expression is still limited. Condensin I is largely absent from heterochromatic regions and binds predominantly to promoter sequences of active genes in mitotic chicken DT40 cells [31]. In human cell mitosis, TATA-binding protein (TBP) transmits active gene memory to daughter cells by directly interacting with NCAPG in the vicinity of these promoters via its associated protein phosphatase 2A (PP2A), thereby inhibiting the compaction of these regions [32]. In Drosophila melanogaster, condensin I has been implicated in the repression of homeotic genes [33]. In yeast, Hocquet et al. have reported that condensins play no direct role in the maintenance of the transcriptome either during interphase or during mitosis: in that study, the gene expression changes in postmitotic fission yeast cells were largely a consequence of chromosome mis-segregation during the anaphase [34]. Condensin depletion causes genome decompaction without altering the level of global gene expression in saccharomyces cerevisiae [35]. In one study, condensin II physically interacted with TFIIIC, and they both colocalized at active gene promoters (in mouse and human genomes): this was facilitated by the interaction between NCAPD3 and the epigenetic mark H3K4me3 [36]. Thus far, previous studies have not provided conclusive evidence showing how condensin I regulates gene expression. In our study, chromatin condensation decreased after NCAPG silencing. We also found that muscle-specific genes were first upregulated after the silencing of NCAPG, but then they decreased. Because accessibility is related to chromatin condensation [37] and histone acetylation [38], we suspect that NCAPG might affect chromatin accessibility.
Using an ATAC-seq and motif analysis, we found that AP-1 enrichment (and that of its subunits) at accessible chromatin regions decreased after NCAPG silencing. AP-1 transcription is a dimeric complex involved in diverse cellular processes such as proliferation, differentiation, and apoptosis [39]. This dimeric complex is composed of Jun (c-Jun, JunB, and JunD) and Fos (c-Fos, FosB, Fra-1, and Fra-2) proto-oncogene proteins that bind to a cis-element called the TRE (12-O-tetradecanoylphorbol-13-acetate response element) [39]. Different AP-1 dimer combinations are crucial for AP-1 binding activities and their biological function [40,41,42]. In one study of skeletal muscle cells, AP-1 binding sites were also enriched in a large subset of MyoD-regulated genes, many of which were downregulated during differentiation [43]. During myogenic differentiation, Fra-2 is a major component of the AP-1 complex in differentiating cells [44]. For instance, silencing Fra-2 increases the protein expression of terminal differentiation markers such as muscle creatine kinase (MCK) and MYHC [45]. Previous research has reported that c-Jun can inhibit differentiation by directly binding to MYOD [46]. MYOD can act as a negative regulator in c-fos transcription by binding with serum-responsive elements in the c-fos promoter [47]. JunB is also involved in the early steps of the inhibition of myogenic differentiation. An increase in JunB mRNA expression is highly correlated with AP-1 binding activities and inhibits the expression of myoblast differentiation markers in C2C12 cells [48]. Here, we used siRNA to silence the AP-1 subunits JunB and Fra-2 and found increased expression in most muscle-specific genes.
In conclusion, NCAPG is indispensable in myogenic differentiation, and the lack of NCAPG induces apoptosis. NCAPG also regulates chromatin accessibility to AP-1, a ubiquitously expressed TF complex, and subsequently affects gene expression. These results provide new evidence for NCAPG’s function in myogenesis, as well as its potential role in gene expression.
4. Materials and Methods
4.1. Ethics Statement
The animal experiments were performed according to the guidelines established by the Regulations for the Administration of Affairs Concerning Experimental Animals (Ministry of Science and Technology, China, 2004). All animal experimental protocols in this study were carried out in strict conformance with the rules of the Animal Ethics Committee of the Institute of Animal Sciences, Chinese Academy of Agricultural Sciences (No. IAS2019-48; 12/9/2019). Pregnant cows were raised by the Inner Mongolia Aokesi Agriculture Co., Ltd. (Wulagai, China). All efforts were made to minimize the cows’ suffering.
4.2. Primary Cell Isolation and Cell Culture
The myoblast cells were enzymatically isolated from longissimus dorsi tissues obtained from bovine fetuses 90 to 120 days old, as described previously [49]. At 70%–80% confluence, the cells were passaged with 0.25% trypsin-EDTA (Gibco, Grand Island, NY, USA). After reaching 100% confluence, cells were induced in differentiation medium (DM) consisting of DMEM containing 5% horse serum (Gibco). The medium was exchanged every 2 days.
4.3. siRNA Transfection and Gene Knockdown
The transfection of siRNA into myoblast cells was performed using Lipofectamine™ RNAiMAX transfection reagent (Invitrogen Life Technologies, Carlsbad, CA, USA) when the cells reached 100% confluence.
Small interfering RNA (siRNA) was obtained from RiboBio (Guangzhou, China), and the sequences are shown in Table S3: siRNA transfection was performed by following the manufacturer′s recommended procedure. Cells were transfected with siRNA against NCAPG (siNCAPG) or siRNA control (NC) at a final concentration of 50 nM.
4.4. RNA Extraction, cDNA Synthesis, and Quantitative Real-Time PCR
Total RNA was extracted from cells using TRIzol reagent (Invitrogen Life Technologies). RNA concentration and quality were assessed by a NanoPhotometer N50 (Implen, Munich, Germany) and 1.5% agarose gel electrophoresis. A cDNA synthesis for mRNA was performed using PrimeScript RT Master Mix (Perfect Real Time) (TaKaRa, Kusatsu, Japan). In addition, qRT-PCR was performed on a QuantStudio 7 Flex Real-Time PCR System (Life, Carlsbad, CA, USA) with a KAPA SYBR® FAST qPCR Kit (KAPABiosystems, Wilmington, MA, USA). Samples from at least three independent experiments were assayed following the manufacturer′s instructions. The sequence of qRT-PCR detection primers can be found in CS4.
4.5. Immunofluorescence
The immunofluorescence tests were performed in myoblast cells cultured in 12-well plates. Cells were fixed in 4% paraformaldehyde for 15 min and then washed three times with phosphate-buffered saline (PBS). Subsequently, cells were incubated in 0.1% Triton X-100 diluted by PBS for 10 min at room temperature, and the cells were blocked with 1% albumin bovine serum (Beyotime, Shanghai, China) for 30 min in order to reduce the nonspecific binding of primary antibodies. After incubation with the primary antibodies overnight at 4 °C, secondary antibodies were added, and the cells were incubated at room temperature for 1 h. The cell nuclei were stained with 4′,6-diamidino-2-phenylindole (DAPI) (Sigma-Aldrich, St. Louis, MO, USA) for 5 min, and images were obtained with a confocal microscope (TCS SP8, Leica, Wetzlar, Germany). The following antibodies were used: MHC antibody (MF20, Developmental Studies Hybridoma Bank, Iowa, USA, 1:100); NCAPG (sc-101014, Santa Cruz, Delaware Ave Santa Cruz, CA, USA, 1:1000); NCAPD3 (16828-1-AP, Proteintech, Chicago, IL, USA, 1:500); H4K20me1 (ab9051, Abcam, Cambridge, UK, 1:1000); and H4K16ac (13534, Cell Signaling Technology, Danvers, MA, USA, 1:1600).
4.6. Western Blot
Cells were digested with low-concentration trypsin and collected into tubes. We used a Nuclear and Cytoplasm Protein Extraction Kit (Beyotime) to separate nuclear and cytoplasm proteins. Cells were lysed in an ice cold radio immunoprecipitation assay (RIPA) lysis buffer with 1 mM phenylmethyl sulfonyl fluoride (Sigma-Aldrich), which was used for cell protein extraction. Protein concentration was determined using a BCA KIT (Beyotime). Proteins were separated by 4%–12% SurePAGE gels (GenScript, Nanjing, China), transferred to a nitrocellulose membrane (Pall, Mexico), and then detected using antibodies according to standard procedures. Primary antibodies were applied overnight at 4 °C for western blot tests, and their dilutions were as follows: NCAPG (sc-101014, Santa Cruz, 1:1000); MYHC (MF20, Developmental Studies Hybridoma Bank, 1:50); MYOD (sc-377460, Santa Cruz, 1:1000); MYOG (sc-12732, Santa Cruz, 1:1000) and β-tubulin (10094-1-AP, Proteintech, 1:2000). Finally, secondary antibodies were visualized with HRP-conjugated secondary antibodies that were applied for 1 h at room temperature. ECL western blotting detection reagent (Beyotime) was used to visualize the protein bands.
4.7. Lentivirus Infection
Cells were seeded in 3.5-cm culture dishes (Corning, Corning, NY, USA) with a 40%~50% confluence. A lentivirus expressing GFP and H2B was purchased from HanBio (Shanghai, China; www.hanbio.net). The lentivirus-coated GFP–H2B was transfected into cells with polybrene (HanBio). After 48 h of infection at 37 °C, the medium was replaced by fresh DMEM. The mitosis process was observed with a confocal microscope (TCS SP8, Leica, Wetzlar, Germany) at day 1.5.
4.8. TUNEL Staining
The apoptosis of myoblasts was examined using a TUNEL assay (Beyotime). The TUNEL assay was performed in accordance with the manufacturer’s protocols. TUNEL-positive cells (indicated by red fluorescent staining) were defined as having undergone apoptotic cell death. In terms of cell counts, TUNEL-positive cells were counted in three random fields of each section. The apoptosis index was calculated according to the following formula: the number of apoptotic cells/the total number of nucleated cells × 100%.
4.9. Caspase-3 Activity Detection
Caspase-3 activity was measured spectrophotometrically via the detection of pNA cleavage by caspase-3-specific substrates. These experiments were completed using a Caspase-3 Assay Kit (Beyotime). After the cell lysates were incubated with Ac-DEVD-pNA for 2 h at 37 °C, the samples were read at 405 nm.
4.10. ATAC-Seq and Data Analysis
The ATAC-seq libraries were constructed with a TruePrep DNA Library Prep Kit V2 for Illumina (Vazyme, Nanjing, China). Briefly, 50,000 collected cells (counted using trypan blue exclusion) were lysed in cold lysis buffer (10 mM Tris-HCl, pH 7.4, 10 mM NaCl, 3 mM MgCl2, 0.1% NP40, 0.1% Tween-20, and 0.01% Digitalis saponin) for 10 min on ice. After centrifugation at 500 g for 5 min, the nuclei were pelleted and resuspended in Transposase buffer. The transposition reaction was carried out for 30 min at 37 °C. Following purification, the libraries were amplified for 16 cycles and purified using VAHTS RNA Clean Beads (Vazyme). Libraries were quantitated using a Qubit 4 Fluorometer (Invitrogen, Singapore). Quality control of the libraries was performed with a Bioanalyzer 2100 (Agilent Technologies, Santa Clara, CA, USA, D1000 screentapes and reagents, 5067-5582). ATAC libraries were sequenced and multiplexed on an Illumina HiSeq X Ten with 150-bp paired ends. Raw sequence reads were initially processed for quality control by a FastQC (0.11.5), and then a Skewer (0.2.2) was used to remove the adapter sequences and poor-quality reads. Subsequently, the remaining reads were mapped onto the bovine reference genome of ARS-UCD1.2 using Burrows–Wheeler Alignment (BWA) (0.7.12). SAM files were converted into a BAM format using Samtools and were used for peak calling. A consensus map was created for each group by merging all samples using the BEDTools merge command. MACS2 (2.1.2) was used to call peaks. Correlations between libraries were calculated using the deepTools (3.0.2) bamCorrelate bins command. ATAC-seq peaks were annotated using a PeakAnnotator. A TF-binding motif analysis of ATAC-seq data was performed using HOMER (v4.9.1). Only known motifs from HOMER’s motif database were considered. We studied the motifs enriched by ATAC-seq peaks using findMotifsGenome.pl. A GO enrichment analysis of differential peaks was performed using the Goseq R package. GOseq was applied to assess enrichment, and topGO was used for plot directed acyclic graph bases of significantly enriched genes. We used KOBAS software (3.0) to determine whether differential peaks were significantly enriched in KEGG pathways. A motif enrichment analysis was performed with the findMotifsGenome.pl command in the HOMER package (4.9.1). The raw sequence data reported in this paper have been deposited in the Genome Sequence Archive (Genomics, Proteomics & Bioinformatics 2017) at the BIG Data Center (Nucleic Acids Res 2019), Beijing Institute of Genomics (BIG), Chinese Academy of Sciences, under accession numbers CRA002306 and CRA002306. They are publicly accessible at https://bigd.big.ac.cn/gsa.
4.11. Statistical Analyses
All data are presented as the mean ± the SEM from at least three independent experiments for each treatment. Data were analyzed using Student’s t-test and incorporated into GraphPad Prism version 6.0 software (GraphPad Inc., San Diego, CA, USA). p < 0.05 was considered to be statistically significant.
Acknowledgments
The authors would like to thank all of the staff at the cattle experimental unit in Beijing for sharing their insights.
Abbreviations
| NCAPG | Non-SMC condensin I complex subunit G |
| SMC | Structural maintenance of chromosomes |
| CAP-G | Chromosome-associated protein G |
| CAP-H | Non-SMC condensin I complex subunit H |
| CAP-D2 | Non-SMC condensin I complex subunit D2 |
| MYH | Myosin heavy chain |
| MYOG | Myogenin |
| MYOD | Myoblast determination protein 1 |
| MYF5 | Myogenic Factor 5 |
| NEBD | Nuclear envelope breakdown |
| H2B | Histone H2B-like |
| IF | Immunofluorescence |
| KD | Knockdown |
| GFP | Green fluorescent protein |
| TUNEL | TdT-mediated dUTP nick-end labeling |
| H4K20me1 | Histone 4 lysine 20 monomethylation |
| H4K16ac | Acetylation of histone H4 on lysine 16 |
| H3K4me3 | Histone H3 trimethylated at lysine 4 |
| TSS | Transcription start site |
| KEGG | Kyoto Encyclopedia of Genes and Genomes |
| MAPK | Mitogen-activated protein kinase |
| NC | Negative control |
| MCK | Muscle creatine kinase |
| siRNA | Small interfering RNAs |
| ATAC- | Assay for Transposase-Accessible Chromatin |
| seq | High-throughput sequencing |
| TSS | Transcription start sites |
| AP-1 | Activating protein 1 |
| c-Jun | Transcription factor AP-1-like |
| JUNB | JunB proto-oncogene |
| JUND | JunD proto-oncogene |
| FOSL2 | Fos-related antigen 2 |
| FOSL1 | Fos-related antigen 1 |
| PRKAA1 | Protein kinase AMP-activated catalytic subunit alpha 1 |
| SIRT1 | Sirtuin 1 |
| CTCF | CCCTC-Binding Factor |
| BATF | Basic Leucine Zipper ATF-Like Transcription Factor |
| TBP | TATA-binding protein |
| PP2A | Protein phosphatase 2A |
| DAPI | 4′,6-diamidino-2-phenylindole |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/4/1248/s1.
Author Contributions
Conceptualization, L.Z.; methodology X.F.; software, Q.L.; validation, L.Z. and X.H.; formal analysis, X.H. and Y.X.; investigation, X.H. and Y.X.; data curation, X.H., Y.X., and Q.L.; writing—original draft preparation, X.H., L.Z., and Y.X.; writing—review and editing, Q.Y. and X.F.; visualization, Y.W. and L.R.; supervision, L.Z. and J.L.; project administration, L.Z.; funding acquisition, J.L. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by the National Natural Science Foundation of China (No. 31672384), the Cattle Breeding Innovative Research Team (ASTIPIAS03), and the Program of National Beef Cattle and Yak Industrial Technology Systems (CARS-37).
Conflicts of Interest
The authors declare no conflicts of interest.
References
- 1.Hoshiba H., Setoguchi K., Watanabe T., Kinoshita A., Mizoshita K., Sugimoto Y., Takasuga A. Comparison of the effects explained by variations in the bovine PLAG1 and NCAPG genes on daily body weight gain, linear skeletal measurements and carcass traits in Japanese Black steers from a progeny testing program. Anim. Sci. J. 2013;84:529–534. doi: 10.1111/asj.12033. [DOI] [PubMed] [Google Scholar]
- 2.Lindholm-Perry A.K., Sexten A.K., Kuehn L.A., Smith T.P., King D.A., Shackelford S.D., Wheeler T.L., Ferrell C.L., Jenkins T.G., Snelling W.M., et al. Association, effects and validation of polymorphisms within the NCAPG - LCORL locus located on BTA6 with feed intake, gain, meat and carcass traits in beef cattle. BMC Genet. 2011;12:103. doi: 10.1186/1471-2156-12-103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Zhang W., Li J., Guo Y., Zhang L., Xu L., Gao X., Zhu B., Gao H., Ni H., Chen Y. Multi-strategy genome-wide association studies identify the DCAF16-NCAPG region as a susceptibility locus for average daily gain in cattle. Sci. Rep. 2016;6:38073. doi: 10.1038/srep38073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lindholm-Perry A.K., Kuehn L.A., Oliver W.T., Sexten A.K., Miles J.R., Rempel L.A., Cushman R.A., Freetly H.C. Adipose and muscle tissue gene expression of two genes (NCAPG and LCORL) located in a chromosomal region associated with cattle feed intake and gain. PLoS ONE. 2013;8:e80882. doi: 10.1371/journal.pone.0080882. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Eberlein A., Takasuga A., Setoguchi K., Pfuhl R., Flisikowski K., Fries R., Klopp N., Furbass R., Weikard R., Kuhn C. Dissection of genetic factors modulating fetal growth in cattle indicates a substantial role of the non-SMC condensin I complex, subunit G (NCAPG) gene. Genetics. 2009;183:951–964. doi: 10.1534/genetics.109.106476. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Setoguchi K., Watanabe T., Weikard R., Albrecht E., Kuhn C., Kinoshita A., Sugimoto Y., Takasuga A. The SNP c.1326T>G in the non-SMC condensin I complex, subunit G (NCAPG) gene encoding a p.Ile442Met variant is associated with an increase in body frame size at puberty in cattle. Anim. Genet. 2011;42:650–655. doi: 10.1111/j.1365-2052.2011.02196.x. [DOI] [PubMed] [Google Scholar]
- 7.Liu Y., Duan X., Chen S., He H., Liu X. NCAPG is differentially expressed during longissimus muscle development and is associated with growth traits in Chinese Qinchuan beef cattle. Genet. Mol. Biol. 2015;38:450–456. doi: 10.1590/S1415-475738420140287. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Al-Mamun H.A., Kwan P., Clark S.A., Ferdosi M.H., Tellam R., Gondro C. Genome-wide association study of body weight in Australian Merino sheep reveals an orthologous region on OAR6 to human and bovine genomic regions affecting height and weight. Genet. Sel. Evol. GSE. 2015;47:66. doi: 10.1186/s12711-015-0142-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Tetens J., Widmann P., Kuhn C., Thaller G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 2013;44:467–471. doi: 10.1111/age.12031. [DOI] [PubMed] [Google Scholar]
- 10.Signer-Hasler H., Flury C., Haase B., Burger D., Simianer H., Leeb T., Rieder S. A Genome-Wide Association Study Reveals Loci Influencing Height and Other Conformation Traits in Horses. PLoS ONE. 2012;7:e37282. doi: 10.1371/journal.pone.0037282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Carty C.L., Johnson N.A., Hutter C.M., Reiner A.P., Peters U., Tang H., Kooperberg C. Genome-wide association study of body height in African Americans: The Women’s Health Initiative SNP Health Association Resource (SHARe) Hum. Mol. Genet. 2012;21:711–720. doi: 10.1093/hmg/ddr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Kimura K., Cuvier O., Hirano T. Chromosome condensation by a human condensin complex in Xenopus egg extracts. J. Biol. Chem. 2001;276:5417–5420. doi: 10.1074/jbc.C000873200. [DOI] [PubMed] [Google Scholar]
- 13.Hirano M., Hirano T. Opening closed arms: Long-distance activation of SMC ATPase by hinge-DNA interactions. Mol. Cell. 2006;21:175–186. doi: 10.1016/j.molcel.2005.11.026. [DOI] [PubMed] [Google Scholar]
- 14.Ono T., Fang Y., Spector D.L., Hirano T. Spatial and temporal regulation of Condensins I and II in mitotic chromosome assembly in human cells. Mol. Biol. Cell. 2004;15:3296–3308. doi: 10.1091/mbc.e04-03-0242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Hirota T., Gerlich D., Koch B., Ellenberg J., Peters J.M. Distinct functions of condensin I and II in mitotic chromosome assembly. J. Cell. Sci. 2004;117 Pt 26:6435–6445. doi: 10.1242/jcs.01604. [DOI] [PubMed] [Google Scholar]
- 16.Ono T., Losada A., Hirano M., Myers M.P., Neuwald A.F., Hirano T. Differential contributions of condensin I and condensin II to mitotic chromosome architecture in vertebrate cells. Cell. 2003;115:109–121. doi: 10.1016/S0092-8674(03)00724-4. [DOI] [PubMed] [Google Scholar]
- 17.Stickland N.C. A quantitative study of muscle development in the bovine foetus (Bos indicus) Anat. Histol. Embryol. 1978;7:193–205. doi: 10.1111/j.1439-0264.1978.tb00795.x. [DOI] [PubMed] [Google Scholar]
- 18.Du M., Tong J., Zhao J., Underwood K.R., Zhu M., Ford S.P., Nathanielsz P.W. Fetal programming of skeletal muscle development in ruminant animals. J. Anim. Sci. 2010;88(Suppl. S13):E51–E60. doi: 10.2527/jas.2009-2311. [DOI] [PubMed] [Google Scholar]
- 19.Du M., Yan X., Tong J.F., Zhao J.X., Zhu M.J. Maternal Obesity, Inflammation, and Fetal Skeletal Muscle Development. Biol. Reprod. 2010;82:4–12. doi: 10.1095/biolreprod.109.077099. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Shogren-Knaak M., Ishii H., Sun J.-M., Pazin M.J., Davie J.R., Peterson C.L. Histone H4-K16 acetylation controls chromatin structure and protein interactions. Science. 2006;311:844–847. doi: 10.1126/science.1124000. [DOI] [PubMed] [Google Scholar]
- 21.Oda H., Okamoto I., Murphy N., Chu J., Price S.M., Shen M.M., Torres-Padilla M.E., Heard E., Reinberg D. Monomethylation of histone H4-lysine 20 is involved in chromosome structure and stability and is essential for mouse development. Mol. Cell. Biol. 2009;29:2278–2295. doi: 10.1128/MCB.01768-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Shintomi K., Hirano T. The relative ratio of condensin I to II determines chromosome shapes. Genes Dev. 2011;25:1464–1469. doi: 10.1101/gad.2060311. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gerlich D., Hirota T., Koch B., Peters J.M., Ellenberg J. Condensin I stabilizes chromosomes mechanically through a dynamic interaction in live cells. Curr. Biol. 2006;16:333–344. doi: 10.1016/j.cub.2005.12.040. [DOI] [PubMed] [Google Scholar]
- 24.Zhang T., Paulson J.R., Bakhrebah M., Kim J.H., Nowell C., Kalitsis P., Hudson D.F. Condensin I and II behaviour in interphase nuclei and cells undergoing premature chromosome condensation. Chromosome Res. 2016;24:243–269. doi: 10.1007/s10577-016-9519-7. [DOI] [PubMed] [Google Scholar]
- 25.Li W., Hu Y., Oh S., Ma Q., Merkurjev D., Song X., Zhou X., Liu Z., Tanasa B., He X., et al. Condensin I and II Complexes License Full Estrogen Receptor alpha-Dependent Enhancer Activation. Mol. Cell. 2015;59:188–202. doi: 10.1016/j.molcel.2015.06.002. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Seipold S., Priller F.C., Goldsmith P., Harris W.A., Baier H., Abdelilah-Seyfried S. Non-SMC condensin I complex proteins control chromosome segregation and survival of proliferating cells in the zebrafish neural retina. BMC Dev. Biol. 2009;9:40. doi: 10.1186/1471-213X-9-40. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Liu K., Li Y., Yu B., Wang F., Mi T., Zhao Y. Silencing non-SMC chromosome-associated polypeptide G inhibits proliferation and induces apoptosis in hepatocellular carcinoma cells. Can. J. Physiol. Pharmacol. 2018;96:1246–1254. doi: 10.1139/cjpp-2018-0195. [DOI] [PubMed] [Google Scholar]
- 28.Liu W., Liang B., Liu H., Huang Y., Yin X., Zhou F., Yu X., Feng Q., Li E., Zou Z., et al. Overexpression of nonSMC condensin I complex subunit G serves as a promising prognostic marker and therapeutic target for hepatocellular carcinoma. Int. J. Mol. Med. 2017;40:731–738. doi: 10.3892/ijmm.2017.3079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Hernandez M.R., Davis M.B., Jiang J., Brouhard E.A., Severson A.F., Csankovszki G. Condensin I protects meiotic cohesin from WAPL-1 mediated removal. PLoS Genet. 2018;14:e1007382. doi: 10.1371/journal.pgen.1007382. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Zhang Y., Liu F., Zhang C., Ren M., Kuang M., Xiao T., Di X., Feng L., Fu L., Cheng S. Non-SMC condensin I complex subunit D2 is a prognostic factor in triple-negative breast cancer for the ability to promote cell cycle and enhance invasion. Am. J. Pathol. 2019;190:37–47. doi: 10.1016/j.ajpath.2019.09.014. [DOI] [PubMed] [Google Scholar]
- 31.Kim J.H., Zhang T., Wong N.C., Davidson N., Maksimovic J., Oshlack A., Earnshaw W.C., Kalitsis P., Hudson D.F. Condensin I associates with structural and gene regulatory regions in vertebrate chromosomes. Nat. Commun. 2013;4:2537. doi: 10.1038/ncomms3537. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Xing H., Vanderford N.L., Sarge K.D. The TBP-PP2A mitotic complex bookmarks genes by preventing condensin action. Nat. Cell Biol. 2008;10:1318–1323. doi: 10.1038/ncb1790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Lupo R., Breiling A., Bianchi M.E., Orlando V. Drosophila chromosome condensation proteins Topoisomerase II and Barren colocalize with Polycomb and maintain Fab-7 PRE silencing. Mol. Cell. 2001;7:127–136. doi: 10.1016/S1097-2765(01)00161-7. [DOI] [PubMed] [Google Scholar]
- 34.Hocquet C., Robellet X., Modolo L., Sun X.M., Burny C., Cuylen-Haering S., Toselli E., Clauder-Munster S., Steinmetz L., Haering C.H., et al. Condensin controls cellular RNA levels through the accurate segregation of chromosomes instead of directly regulating transcription. Elife. 2018;7:e38517. doi: 10.7554/eLife.38517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Paul M.R., Markowitz T.E., Hochwagen A., Ercan S. Condensin Depletion Causes Genome Decompaction Without Altering the Level of Global Gene Expression in Saccharomyces cerevisiae. Genetics. 2018;210:331–344. doi: 10.1534/genetics.118.301217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yuen K.C., Slaughter B.D., Gerton J.L. Condensin II is anchored by TFIIIC and H3K4me3 in the mammalian genome and supports the expression of active dense gene clusters. Sci. Adv. 2017;3:e1700191. doi: 10.1126/sciadv.1700191. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Martin R.M., Cardoso M.C. Chromatin condensation modulates access and binding of nuclear proteins. FASEB J. Off. Publ. Fed. Am. Soc. Exp. Biol. 2010;24:1066–1072. doi: 10.1096/fj.08-128959. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Gorisch S.M., Wachsmuth M., Toth K.F., Lichter P., Rippe K. Histone acetylation increases chromatin accessibility. J. Cell. Sci. 2005;118 Pt 24:5825–5834. doi: 10.1242/jcs.02689. [DOI] [PubMed] [Google Scholar]
- 39.Angel P., Karin M. The role of Jun, Fos and the AP-1 complex in cell-proliferation and transformation. Biochim. Biophys. Acta. 1991;1072:129–157. doi: 10.1016/0304-419X(91)90011-9. [DOI] [PubMed] [Google Scholar]
- 40.Ryseck R.P., Bravo R. c-JUN, JUN B, and JUN D differ in their binding affinities to AP-1 and CRE consensus sequences: Effect of FOS proteins. Oncogene. 1991;6:533–542. [PubMed] [Google Scholar]
- 41.Hai T., Curran T. Cross-family dimerization of transcription factors Fos/Jun and ATF/CREB alters DNA binding specificity. Proc. Natl. Acad. Sci. USA. 1991;88:3720–3724. doi: 10.1073/pnas.88.9.3720. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Boise L.H., Petryniak B., Mao X., June C.H., Wang C.Y., Lindsten T., Bravo R., Kovary K., Leiden J.M., Thompson C.B. The NFAT-1 DNA binding complex in activated T cells contains Fra-1 and JunB. Mol. Cell. Biol. 1993;13:1911–1919. doi: 10.1128/MCB.13.3.1911. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Cao Y., Yao Z., Sarkar D., Lawrence M., Sanchez G.J., Parker M.H., MacQuarrie K.L., Davison J., Morgan M.T., Ruzzo W.L., et al. Genome-wide MyoD binding in skeletal muscle cells: A potential for broad cellular reprogramming. Dev. Cell. 2010;18:662–674. doi: 10.1016/j.devcel.2010.02.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Andreucci J.J., Grant D., Cox D.M., Tomc L.K., Prywes R., Goldhamer D.J., Rodrigues N., Bedard P.A., McDermott J.C. Composition and function of AP-1 transcription complexes during muscle cell differentiation. J. Biol. Chem. 2002;277:16426–16432. doi: 10.1074/jbc.M110891200. [DOI] [PubMed] [Google Scholar]
- 45.Alli N.S., Yang E.C., Miyake T., Aziz A., Collins-Hooper H., Patel K., McDermott J.C. Signal-dependent fra-2 regulation in skeletal muscle reserve and satellite cells. Cell Death Dis. 2013;4:e692. doi: 10.1038/cddis.2013.221. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bengal E., Ransone L., Scharfmann R., Dwarki V.J., Tapscott S.J., Weintraub H., Verma I.M. Functional antagonism between c-Jun and MyoD proteins: A direct physical association. Cell. 1992;68:507–519. doi: 10.1016/0092-8674(92)90187-H. [DOI] [PubMed] [Google Scholar]
- 47.Trouche D., Grigoriev M., Lenormand J.L., Robin P., Leibovitch S.A., Sassone-Corsi P., Harel-Bellan A. Repression of c-fos promoter by MyoD on muscle cell differentiation. Nature. 1993;363:79–82. doi: 10.1038/363079a0. [DOI] [PubMed] [Google Scholar]
- 48.Chalaux E., Lopez-Rovira T., Rosa J.L., Bartrons R., Ventura F. JunB is involved in the inhibition of myogenic differentiation by bone morphogenetic protein-2. J. Biol. Chem. 1998;273:537–543. doi: 10.1074/jbc.273.1.537. [DOI] [PubMed] [Google Scholar]
- 49.Guan L., Hu X., Liu L., Xing Y.S., Zhou Z.K., Liang X.W., Yang Q.Y., Jin S.Y., Bao J.S., Gao H.J., et al. bta-miR-23a involves in adipogenesis of progenitor cells derived from fetal bovine skeletal muscle. Sci. Rep. 2017;7:43716. doi: 10.1038/srep43716. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.