Abstract
Following photosynthesis, sucrose is translocated to sink organs, where it provides the primary source of carbon and energy to sustain plant growth and development. Sugar transporters from the SWEET (sugar will eventually be exported transporter) family are rate-limiting factors that mediate sucrose transport across concentration gradients, sustain yields, and participate in reproductive development, plant senescence, stress responses, as well as support plant–pathogen interaction, the focus of this study. We identified 25 SWEET genes in the walnut genome and distinguished each by its individual gene structure and pattern of expression in different walnut tissues. Their chromosomal locations, cis-acting motifs within their 5′ regulatory elements, and phylogenetic relationship patterns provided the first comprehensive analysis of the SWEET gene family of sugar transporters in walnut. This family is divided into four clades, the analysis of which suggests duplication and expansion of the SWEET gene family in Juglans regia. In addition, tissue-specific gene expression signatures suggest diverse possible functions for JrSWEET genes. Although these are commonly used by pathogens to harness sugar products from their plant hosts, little was known about their role during Xanthomonas arboricola pv. juglandis (Xaj) infection. We monitored the expression profiles of the JrSWEET genes in different tissues of “Chandler” walnuts when challenged with pathogen Xaj417 and concluded that SWEET-mediated sugar translocation from the host is not a trigger for walnut blight disease development. This may be directly related to the absence of type III secretion system-dependent transcription activator-like effectors (TALEs) in Xaj417, which suggests different strategies are employed by this pathogen to promote susceptibility to this major aboveground disease of walnuts.
Keywords: SWEET sugar transporters, gene family, phylogeny, TAL effector, gene expression, walnut blight, Xanthomonas
1. Introduction
Following photosynthesis, sugars like sucrose, transported from their photosynthetic “sources” to heterotrophic “sinks”, enable plant growth and development [1,2,3]. Sucrose is the predominant form of sugar translocated in many plant species and is driven by hydrostatic pressure generated in the phloem to mediate long-distance distribution throughout the plant [4,5,6]. However, sucrose transport across the plant cell membrane requires the assistance of appropriate sugar transporters. A proton-coupled sucrose transporter (SUT) that acts as a sugar/H+ symporter is essential for translocation [7], while a sieve element–companion cell complex (SE/CC) is required for translocation into the phloem. Another type of sugar transporter known as SWEETs (sugar will eventually be exported transporters) effuse sucrose from inside the cell into the cell wall as a prerequisite for SUT1-mediated phloem loading [1]. Since these sugar transporters regulate the sucrose pools in plants, several microorganisms vie for carbohydrates from plants through this pathway. Increasing evidence suggests that both bacterial and fungal pathogens hijack sugar transporters to gain access to pools of sucrose for growth [8,9,10]. Unlike SUTs, which are expressed at a low level, require energy, and are therefore a rate-limiting factor for sucrose transport, SWEET proteins function as key transporters for sucrose, hexose, and fructose along a concentration gradient and become a priority target in plant–pathogen interactions [11,12,13].
The SWEET protein family members typically contain seven transmembrane domains (TMDs) harboring two MtN3/saliva domains, also referred to as the PQ-loop-repeat (PFAM code PF0383). This family was first identified with 17 members in Arabidopsis and 21 in rice [1,14,15]. Phylogenetic analysis revealed that SWEET proteins can be divided into four clades according to the type of substrate transported. Clades I, II, and IV members are monosaccharide efflux transporters, of which clades I and II are located on the plasma membrane and preferentially transport hexoses, while characterized SWEETs from clade IV are fructose transporters located on the vacuolar membrane [1,16,17]. Clade III SWEETs are sucrose efflux transporters located on the plasma membrane. The genes that encode these transporters in rice are targeted by transcription activator-like effectors (TALEs) from Xanthomonas oryzae pv. oryzae (Xoo), such as seen in the interaction of PthXo1 TALE with cis-acting elements within the 5′ regulatory region of OsSWEET11 and of PthXo2 with OsSWEET13 [18,19]. The pathogen Xoo can upregulate expression of the corresponding SWEET genes effectively, recruiting a specific SWEET protein to deliver sucrose to the apoplast and provide the bacteria with a carbon source [20,21,22,23]. Furthermore, clade III SWEET proteins GhSWEET10 in cotton and MeSWEET10a in cassava are also activated by TALEs from X. citri subsp. malvacearum (Xcm) and X. axonopodis pv. manihotis (Xam), respectively [24,25]. Notably, CsSWEET1 is the only SWEET protein from a different clade (clade I) that can be recruited by X. citri subsp. citri (Xcc), but in a TALE-dependent manner when connected directly to the target gene LOB1 [26]. Other SWEET proteins, including SWEETs in other clades, can be induced during plant interaction with fungi, symbiotic bacteria, and oomycetes [9,23,27]. Host SWEET proteins are induced in many other systems such as the infection of A. thaliana by Pseudomonas syringae pv. tomato DC3000, and by the obligate biotrophic powdery mildew pathogen Golovinomyces cichoracearum [28], the infection of grapes with Botrytis cinerea [29], Medicago truncatula with Sinorhizobium meliloti [30], oilseed rape by Sclerotinia sclerotiorum [31], wheat by Puccinia [32], banana by Fusarium oxysporum f. sp. cubense TR4 [33], and cabbage by Plasmodiophora brassicae [34]. It is thus reasonable to speculate that SWEETs play a pivotal role in plant–pathogen interactions and may serve as host susceptibility factors for many pathogens.
Walnut blight is the most serious aboveground disease in Juglans regia and is caused by the bacterial pathogen X. arboricola pv. juglandis (Xaj) (Figure 1). Since little is known about this plant–pathogen interaction, the current study examined the significance of SWEET genes as possible susceptibility genes, as occurs in rice bacterial leaf blight and other diseases also caused by Xanthomonas. Pathogens from Xanthomonas species are well known for modulating plant immune responses by secretion of effector proteins directly inside host cells through a highly conserved protein secretion system (T3SS) [35]. Comparative sequence analysis of T3SS effectors from Xanthomonas spp. demonstrates that they are divided into conserved protein families such as members of YopJ/AvrRxv, a family of predicted cysteine proteases, and members of the AvrBs3/PthA transcription factors, also designated transcription activator-like effectors (TALEs). TALEs have a nuclear localization signal and modulate host gene expression by direct interaction with the promoter region of specific genes in the host [36]. While vascular pathogens like X. campestris and oryzae typically use TALEs to modulate host gene expression, less is known about mesophillic pathogens such as X. arboricola and X. citri, although exceptions like Xcm are known to also employ TALEs [24]. Based on previous examples of pathogen TALEs or other effector types modulating host SWEET expression, we hypothesized that this interaction may also occur during walnut blight. To determine whether there is a causal relationship between JrSWEET genes and walnut blight, we first identified the SWEET family in the Juglans regia genome and analyzed gene structures, conserved domains, and established phylogenetic relationships. Next, we performed a comprehensive analysis of JrSWEET gene expression patterns in different walnut tissues in response to infection with Xaj and their correlation with other biomarkers of infection, such as polyphenol oxidase (PPO), which is induced during the interaction between walnut and Xaj [37,38]. In addition, we searched for TALEs in the Xaj genome and attempted to predict the target JrSWEET genes that would be induced during controlled in vitro inoculations.
Figure 1.
Bacterial walnut blight symptoms. (a) A healthy nut still with hull tissue is shown on top, contrasted with a walnut blight symptomatic nut on the bottom. (b) Blight symptoms can also be observed in microshoots maintained in tissue culture under controlled laboratory conditions. Top panel shows mock-inoculated shoots and bottom panel shows Xaj417-infected shoots four days post inoculation.
2. Results
2.1. Genome-Wide Identification of SWEET Family Genes in Walnut
Based on HMMER, 37 putative SWEET genes were matched with an E-value cutoff of 0.0001 using typical SWEET domains (PFAM: PF03083) as queries. Meanwhile, based on BLAST searches, 29 putative SWEET genes were identified using AtSWEET sequences as queries. Finally, 25 non-redundant JrSWEET genes were confirmed with conserved core domains and complete gene structure by KEGG, CDD, and SMART (Table 1). The homologous genes of AtSWEETs were used to name the corresponding SWEET genes in J. regia. The JrSWEET genes were named using numerals and small letters, followed by capital letters to distinguish paralogous genes on the basis of their position in chromosomal pseudomolecules.
Table 1.
Description of walnut SWEET genes.
| Clade | Gene Name | Locus | Size (aa) a | Mw (kDa) | pI | Genomic Location | MtN3/Saliva (PQ-Loop Repeat) Domain Position | Loc b |
|---|---|---|---|---|---|---|---|---|
| I | JrSWEET1 | 108999697 | 254 | 28.22 | 9.43 | NW_017444989.1:303-3465 | 7–94, 131–213 | PM |
| I | JrSWEET1A | 109004166 | 258 | 28.22 | 6.3 | NW_017388835.1:280022-280798 | 6–95, 129–215 | PM |
| I | JrSWEET1B | 109004173 | 261 | 29.01 | 9.35 | NW_017388835.1:318614-319399 | 7–96, 130–216 | PM |
| I | JrSWEET2 | 108985341 | 235 | 26.11 | 9.27 | NW_017442681.1:95892-99748 | 18–102, 139–222 | PM |
| I | JrSWEET2aA | 108987476 | 235 | 25.72 | 8.84 | NW_017442906.1:10127-13031 | 18–102, 138–219 | PM |
| I | JrSWEET2aB | 108998082 | 235 | 26.01 | 9.34 | NW_017443613.1:377035-380456 | 18–102, 138–220 | VM |
| I | JrSWEET3 | 108985064 | 246 | 27.72 | 9.15 | NW_017442611.1:20354-22484 | 9–95, 133–216 | PM |
| I | JrSWEET3A | 108982726 | 292 | 33.24 | 9.32 | NW_017441679.1:10593-14236 | 9–95, 121–201 | PM |
| II | JrSWEET4 | 108997137 | 241 | 26.75 | 9.01 | NW_017388976.1:192220-194338 | 10–97, 136–217 | PM |
| II | JrSWEET4A | 109002076 | 265 | 29.06 | 9.13 | NW_017443554.1:481695-483478 | 10–97, 133–217 | PM |
| II | JrSWEET4B | 108993759 | 301 | 33.19 | 9.23 | NW_017443600.1:567007-569556 | 49–136, 172–256 | PM |
| II | JrSWEET5A | 108999161 | 231 | 26.37 | 8.9 | NW_017443629.1:533788-536495 | 10–95, 133–216 | PM |
| II | JrSWEET6bA | 108992796 | 225 | 24.89 | 9.3 | NW_017443540.1:669544-671368 | 9–98, 134–220 | PM |
| III | JrN3 | 108984368 | 295 | 33.08 | 7.63 | NW_017442395.1:48585-51197 | 12–96, 132–217 | PM |
| III | JrN3A | 109004781 | 294 | 32.77 | 8.29 | NW_017389221.1:55377-57616 | 13–97, 133–219 | PM |
| III | JrNEC1A | 108992134 | 294 | 32.72 | 6.99 | NW_017443525.1:147356-149392 | 14–97, 136–219 | PM |
| III | JrSWEET10A | 108990746 | 281 | 31.88 | 9.06 | NW_017443364.1:192481-194495 | 10–95, 131–215 | PM |
| III | JrSWEET12A | 108990744 | 298 | 32.84 | 8.27 | NW_017443364.1:211540-213673 | 13–98, 134–218 | PM |
| III | JrSWEET14A | 108998489 | 272 | 30.98 | 9 | NW_017388807.1:776356-778642 | 10–96, 132–216 | PM |
| IV | JrSWEET16A | 109006766 | 154 | 16.63 | 9.72 | NW_017389361.1:1228957-1230806 | 6–93 | PM |
| IV | JrSWEET16B | 108979882 | 277 | 30.37 | 9.52 | NW_017439469.1:3236-6113 | 7–90, 128–211 | PM |
| IV | JrSWEET17 | 108994709 | 247 | 27.37 | 5.87 | NW_017443571.1:1587292-1596052 | 6–95, 133–214 | VM |
| IV | JrSWEET17A | 108987463 | 259 | 28.83 | 6.59 | NW_017442898.1:55816-57539 | 16–97, 136–218 | CL |
| IV | JrSWEET17B | 108987468 | 247 | 27.03 | 7.73 | NW_017442898.1:62119-64578 | 7–90, 128–212 | PM |
| IV | JrSWEET17C | 109012696 | 253 | 28.11 | 5.85 | NW_017389857.1:1048384-1050148 | 9–90, 129–211 | PM |
a aa: length of the protein’s amino acid chain; pI: isoelectric point; and Mw: molecular weight in kDa. b The Loc is the most probable subcellular location predicted by WoLFSORT and TargetP. PM: plasma membrane; VM: vacuolar membrane; and CL: chloroplast.
The 25 predicted JrSWEET proteins range from 154 (JrSWEET16A) to 301 (JrSWEET4B) amino acid residues long, with molecular masses between 16.63 (JrSWEET16A) and 33.24 kDa (JrSWEET3A) and isoelectric points from 5.85 (JrSWEET17C) to 9.72 (JrSWEET16A). The predicted subcellular localizations of the JrSWEET genes are primarily the plasma membrane, but also include vacuolar membrane (JrSWEET2aB and JrSWEET17) and chloroplast membrane (JrSWEET17A) (Table 1). The highly variable characteristics of JrSWEETs suggest different functional roles in different biological processes.
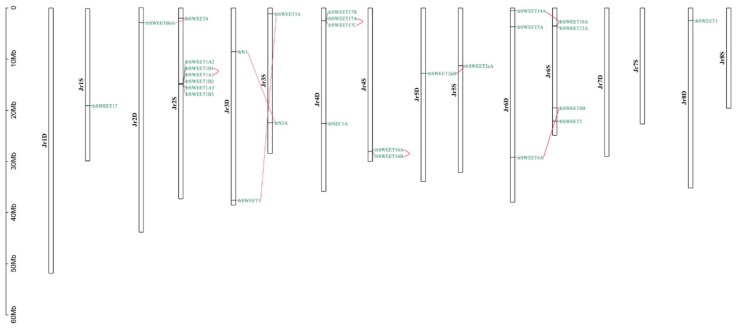
The JrSWEET genes are distributed on 12 of 16 chromosomes; no SWEET genes were found on chromosomes Jr1D, Jr7D, Jr7S, or Jr8S (Figure 2). Five chromosomes (Jr1S, 2D, 5D, 5S, and 8D) harbored only a single gene, chromosomes Jr3D, Jr3S, and Jr4S had two genes each, and chromosome Jr6D had three genes. Chromosomes Jr4D and Jr6S had four genes each. Four groups of genes were linked closely on four chromosomes. Two pairs occurred on chromosomes Jr4S (JrSWEET16A and 16B) and Jr6S (JrSWEET10A and 12A). Chromosome Jr4D had three genes (JrSWEET17A, 17B, and 17C) and Jr2S had six genes (JrSWEET1A1, 1B1, 1A2, 1B2, 1A3, and 1B3).
Figure 2.
Positions of SWEET gene family members on the Juglans regia chromosomes. Duplicated genes are indicated with red lines.
2.2. Gene Structure Analysis, Transmembrane Helix, and Motif Recognition of JrSWEETs
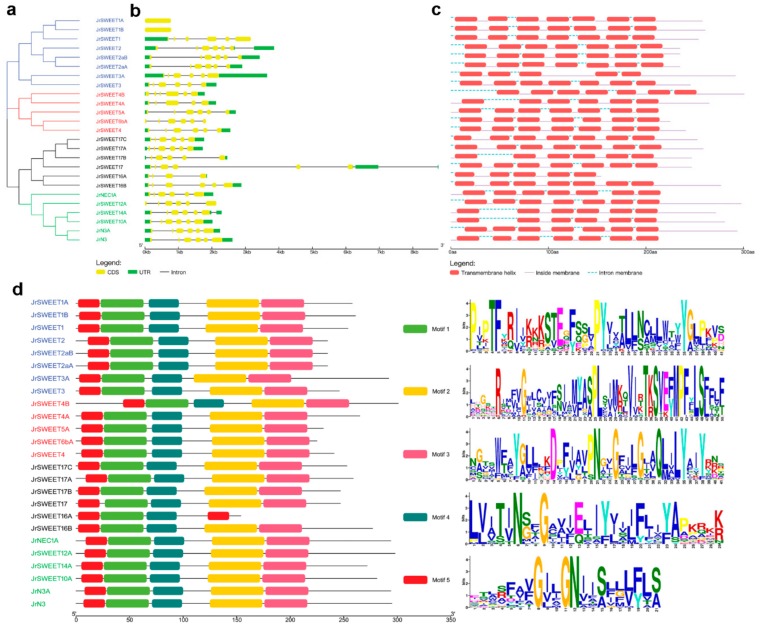
The phylogenetic tree generated by cluster analysis classified the 25 JrSWEETs into four groups (Figure 3a). To better understand the genesis of the JrSWEET gene family, we analyzed the gene structure, including exon and intron distribution, examined the conserved regions and motifs, and predicted the transmembrane helices for each gene. We mapped gene structures using the GSDS (gene structure display server) and found that the coding regions of JrSWEET genes were interrupted with introns of varying number and size (Figure 3b). Most JrSWEET genes had five introns, with the exceptions that no introns were present in JrSWEET1A and JrSWEET1B, four in JrSWEET4A, six in JrSWEET2 and JrSWEET14A, and seven in JrSWEET17 (Figure 3b).
Figure 3.
Phylogenetic relationship, gene structure, and distribution of conserved motifs of SWEET family proteins in Juglans regia. (a) The unrooted neighbor-joining phylogenetic tree of SWEET family members. Different clades are indicated by different colors: blue, clade I; red, clade II; green, clade III; and black, clade IV. (b) The exon/intron structures of 25 SWEET genes identified in walnut. Exons are represented by yellow boxes; introns by black lines; and upstream or downstream untranslated regions by green boxes. (c) The transmembrane helixes in JrSWEET proteins predicted by TMHMM. Red boxes: transmembrane helixes; purple lines: cytosol localization; and blue dashed lines: apoplast localization. (d) Conserved motifs in JrSWEET proteins. Five putative motifs are indicated in different colored boxes and detailed information is shown with logos obtained from the MEME Suite website. The bit score is proportional to the frequency of the corresponding amino acid at each position.
In addition to the nucleotide sequence analysis, the amino acid sequences of JrSWEETs revealed transmembrane helices and conserved motifs in the protein sequence (Figure 3c), as predicted by the MEME program. JrSWEET3A and JrSWEET16A encoded five transmembrane helices; JrSWEET4A, 17B, 14A, and 10A, six transmembrane helices; and the remaining JrSWEET family members encoded seven transmembrane helices. Five amino acid motifs were identified in all 25 JrSWEETs, all in the same order (motif 5, motif 1, motif 4, motif 2, and motif 3), except in JrSWEET16A, which lacked motifs 2 and 3, but contained an additional motif 5 (Figure 3d).
2.3. Phylogenetic Analysis of JrSWEET Proteins
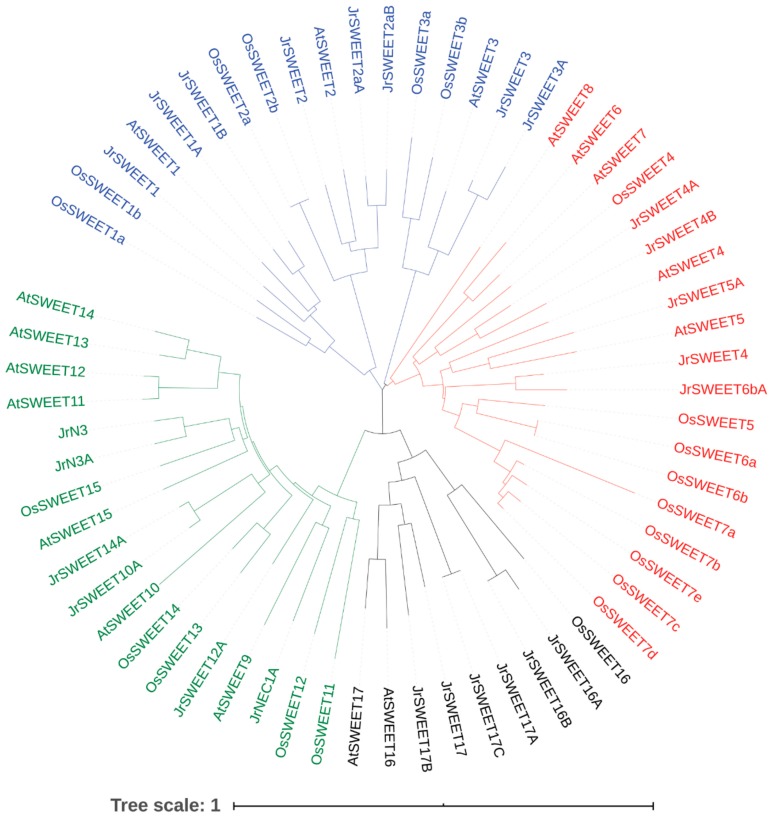
To better understand the evolutionary relationships among SWEET genes in walnut, Arabidopsis, and rice, the 25 JrSWEET proteins, 17 AtSWEET proteins, and 21 OsSWEET proteins were aligned by ClustalX and used to construct an unrooted phylogenetic tree using neighbor-joining implemented in MEGA 7.0. All 63 SWEET proteins fell into four clades. Clade I, clade II, and clade III contained 17, 19, and 18 proteins, respectively, while clade IV had only nine. Among the JrSWEET proteins, clade I contained eight members, followed by clades III and IV with six each, and clade II with only five. Walnut had more proteins in clade IV and fewer in clade II than Arabidopsis and rice (Figure 4).
Figure 4.
Unrooted neighbor-joining phylogenetic tree of SWEET proteins in Arabidopsis, rice, and walnut. Different colors represent the four different clades: blue, clade I; red, clade II; green, clade III; and black, clade IV. The scale bar represents 0.1 substitutions per amino acid position.
2.4. Expression Profiles of JrSWEETs in Different Tissues and Organs and at Different Developmental Stages
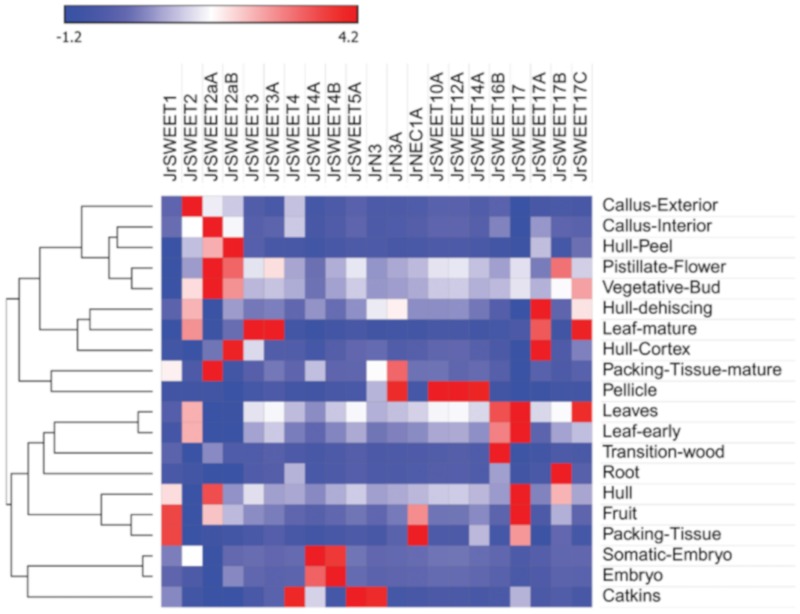
To understand the roles of JrSWEETs during plant growth and development, expression patterns of the 25 JrSWEETs in different tissues and at different developmental stages were extracted from the 20 library walnut RNA-seq dataset [39,40]. The expression of four genes, namely JrSWEET1A, JrSWEET1B, JrSWEET6bA, and JrSWEET16A, was either very low or undetectable, which suggests that they may be expressed in highly specific tissues and/or under specific conditions. Transcriptomic analysis revealed that the other 21 genes, including orthologs, had varied expression patterns in different tissues and at different developmental stages (Figure 5). In clade I, JrSWEET1 was highly expressed in fruit and packing tissues. JrSWEET2 and orthologs JrSWEET2aA and JrSWEET2aB were expressed in most tissues and highly expressed in the callus, but JrSWEET2 was present in the callus-exterior, JrSWEET2aA was present in the callus-interior, pistillate flower, vegetative bud, and mature packing tissue, while JrSWEET2aB was mainly present in hull peel and hull cortex. JrSWEET3 and its paralog, JrSWEET3A, both had high expression in mature leaf. Two genes from clade II, namely JrSWEET4 and JrSWEET5aA, were highly expressed in catkins, while the other genes of clade II, namely orthologs JrSWEET4A and JrSWEET4B, were both expressed most in somatic embryo and zygotic embryo. In clade III, JrSWEET10A, JrSWEET12A, and JrSWEET14A all displayed uniquely strong expression in the pellicle, JrN3A (an ortholog of AtSWEET15) showed strong expression both in mature packing tissue and pellicle, but JrN3, the paralog of JrN3A, was strongly expressed in catkins. JrNEC1A (an ortholog of AtSWEET9) was expressed most strongly in packing tissue and fruit. In clade IV, the five expressed genes were differentially expressed in all tissues. Among them, JrSWEET16B was highly expressed in transition wood, leaves, and early leaves. JrSWEET17B had the greatest expression in root. JrSWEET17 was highly expressed in many tissues, including leaves, early leaves, hull, and fruit. Orthologs JrSWEET17A and JrSWEET17B both were highly expressed in dehiscing hull and mature leaf, but JrSWEET17A also had strong expression in hull cortex, while JrSWEET17B was highly expressed in vegetative buds and leaves. These results suggest that all JrSWEETs have differential expression in different tissues, which may contribute to the functional diversity of JrSWEETs.
Figure 5.
Hierarchical clustering and heat map of JrSWEET genes expressed in different tissues. The FPKM values obtained from RNA-sequencing were used to represent relative expression. Data shown in the heat map was standardized by Z-score.
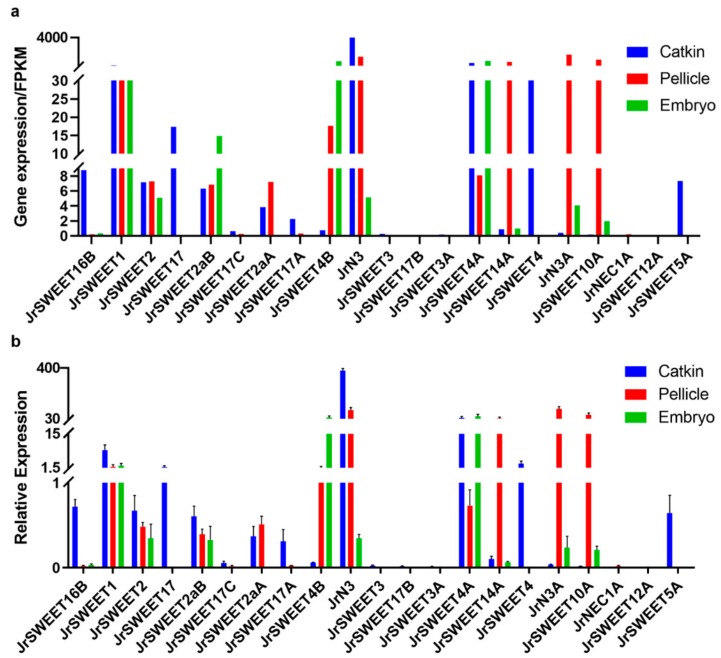
Expression of JrSWEET genes was further validated by qRT-PCR in catkins, pellicle, and embryo tissues (Figure 6). Expression of 21 JrSWEET genes was detected by qRT-PCR, with almost the same expression patterns and profiles obtained from RNA-seq data except for JrSWEET2aB, which was highly expressed in catkins instead of pellicle. In addition, JrSWEET16B, JrSWEET17, JrSWEET4, and JrSWEET5A exhibited a unique high expression in catkins, while JrSWEET14A, JrN3A, and JrSWEET10A were expressed in pellicle. The expression pattern was clearly different for individual clade members and further functional analysis of JrSWEET genes is necessary to define more clearly the roles of individual members of the family.
Figure 6.
Expression of JrSWEET genes in catkin, pellicle, and embryo tissues. (a) Gene expression in three tissues, with FPKM values obtained from RNA-seq libraries containing five replicas of each tissue type. (b) Relative gene expression compared to Jr18S-11 in three tissues, with expression data obtained by qRT-PCR. Mean values and error bars were calculated from triplicate measurements.
2.5. Expression Analysis of JrSWEET Genes after Xaj417 Infection
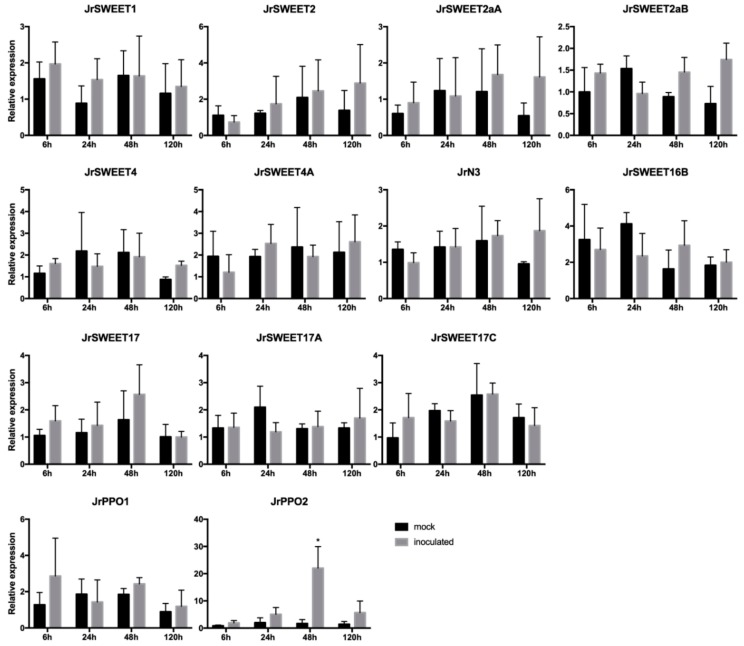
To better understand the response of JrSWEET genes to Xaj, qRT-PCR was used to examine the differences in expression of individual JrSWEET genes at different times after infection with Xaj417. As a base line for these studies, we initially examined the expression pattern in the RNA-seq data of JrSWEET genes in leaf tissues that showed expression for 14 of them (Figure 7). As the original RNA-seq libraries did not contain tissues infected with Xaj, in a following set of experiments walnut microshoots were inoculated with Xaj417 and gene expression of JrSWEET genes analyzed by qRT-PCR. In this set, expression of only 11 JrSWEETs was detected (Figure 8). Most JrSWEET genes detected in one dataset was also detected in the other. The exceptions were JrSWEET3 and JrSWEET3A, which were highly expressed in mature leaves in the RNA-seq data, but undetectable by qRT-PCR on leaves of Xaj-infected microshoots. Different types of leaf tissues were examined in the two datasets, which could explain the difference in expression patterns. The RNA-seq data was obtained from mature leaves harvested from trees in the field, while the inoculated tissues were leaves obtained from in vitro-grown shoots. JrSWEET4A was detected in in vitro shoots, but not in leaves from field grown trees, although the RNA-seq dataset showed extremely high expression in catkins and embryo.
Figure 7.
Expression of JrSWEET genes in leaves, leaf-mature, and leaf-early, with FPKM values obtained from RNA-sequencing. Expression of 21 of the 25 JrSWEET genes was detected in the complete 20 library RNA-seq data, but only 14 were expressed in at least one of the 3 leaf libraries analyzed.
Figure 8.
Relative expression of 11 JrSWEET genes detected at different times after inoculation with Xaj417. The other 14 JrSWEET genes were not detected (not shown). Mock treatments received water, and inoculated treatments were infected with Xaj417 bacteria. Relative gene expression in mock and inoculated tissues was measured at 0, 24, 48, and 120 h post inoculation. Asterisks indicate a significant difference (p < 0.05 in multiple T tests) in expression between mock and inoculated treatments, as seen for JrPPO2 used as a reference for responsive genes.
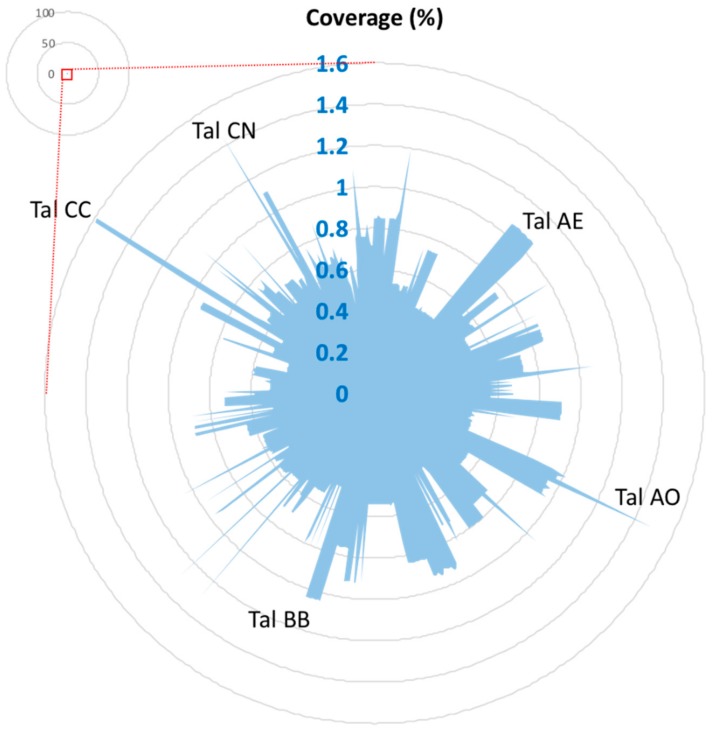
Unexpectedly, no JrSWEET genes were induced by Xaj when assayed by qRT-PCR, although JrPPO2 showed a 20.2-fold upregulation at 48 h post inoculation (Figure 8). To further examine this lack of JrSWEET response to Xaj417 infection, we analyzed the genome of the pathogen to identify candidate TALEs in Xaj that could possibly bind to the 5′ regulatory region of one or more JrSWEET genes. Interestingly, the results from the AnnoTALE search [41] showed that there were no TALEs in Xaj. Further confirmation was obtained by comparing all 516 known TALEs to the Xaj417 genome via BLAST. None displayed sequence coverage greater than 2% except for TalAE/AO/BB/CC/CN, confirming that there is no sequence in the Xaj417 genome with enough coverage and high identity that could be a genuine TALE homolog (Figure 9, Supplementary Materials Table S1). The lack of TALEs in Xaj may be connected to the reason why no JrSWEET genes were induced by Xaj and confirm there is no TALE–SWEET interactions during walnut blight development.
Figure 9.
Blast results of known transcription activator-like effectors (TALEs) to the Xaj417 genome. Note the low percentage coverage of each 516 putative reported TALE in the Xaj417 genome (shown in blue).
2.6. Prediction and Functional Analysis of Cis-Regulatory Elements in Putative Promoter Regions
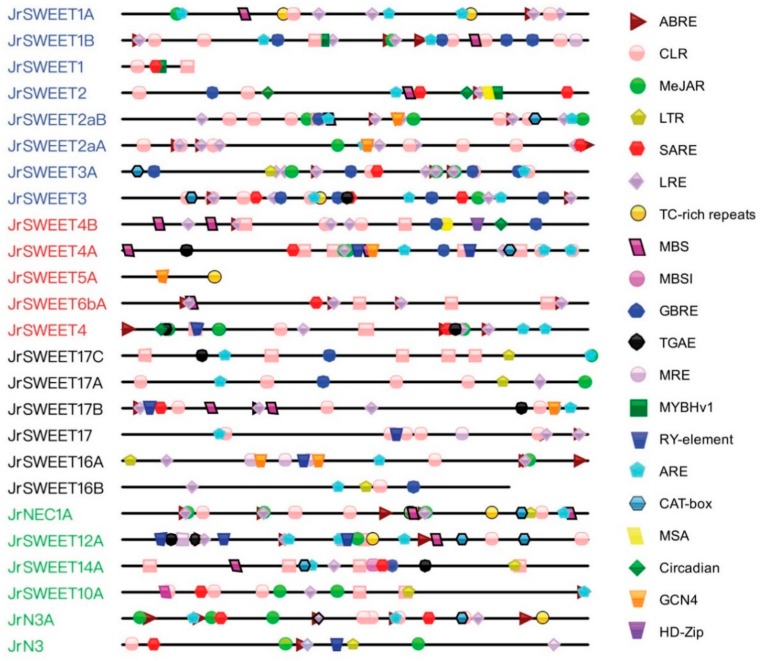
To further understand the transcriptional regulation and potential functions of JrSWEETs, 5′ regulatory elements in the 2000 bp sequences upstream of the translation start sites were determined and analyzed on the PlantCARE server [42]. For JrSWEET1, JrSWEET5A, and JrSWEET16B, only smaller fragments could be obtained from the databases due to the short intergenic space between genes. Twenty different types of cis-regulatory elements were detected in 5′ regulatory regions of JrSWEET genes. These included light response (elements involved in light responsiveness, light response element, MYB binding sites), stress response (low temperature, drought, wound, anaerobic induction, defense, and stress), hormone responsive (auxin, abscisic acid, gibberellic acid, jasmonic acid, and salicylic acid), and growth and development regulation elements (seed-specific regulation, meristem expression, cell cycle regulation, circadian control, endosperm expression, and palisade mesophyll cells elements). Each gene contained from two to twenty-four elements (Figure 10). The most frequent elements were light response elements. All 25 JrSWEET 5′ regulatory regions contained at least one light responsive element except for JrSWEET5A. Secondly, 76%, 72%, and 64% of JrSWEET 5′ regulatory regions contained elements related to anaerobic induction, abscisic acid, and jasmonic acid responsiveness, respectively. MYB binding sites mostly concentrated in clade I, but appeared twice separately in clades IV and III, with none in clade II. Similarly, low-temperature responsiveness was absent from clade II; elements related to endosperm expression, from clade III; elements related to meristem expression, defense and stress, from clade IV; and auxin responsive elements, from clade I. Salicylic acid responsiveness, drought inducibility elements, gibberellic acid responsiveness, and seed-specific regulation elements occurred randomly in all clades. Only the 5′ regulatory region of JrSWEET4B contained an element related to palisade mesophyll cells; JrSWEET2 and JrSWEET4B contained an element related to circadian control; and JrSWEET2, JrSWEET4B, and JrSWEET4 contained elements related to cell cycle regulation. These data indicate that JrSWEET clades may have specialization, and further analysis is necessary to understand the function of each SWEET gene.
Figure 10.
Predicted cis-elements responding to plant growth regulation, hormone response, and stresses response present in JrSWEET 5′ regulatory regions. The cis-elements are identified by function: ABRE, cis-acting element involved in abscisic acid responsiveness; CLR, part of a conserved DNA module involved in light responsiveness; MeJAR, cis-acting regulatory element involved in MeJA (methyl jasmonic acid)-responsiveness; LTR, cis-acting element involved in low-temperature responsiveness; SARE, cis-acting element involved in salicylic acid responsiveness; LRE, cis-acting regulatory element involved in light responsiveness; TC-rich repeats, cis-acting element involved in defense and stress responsiveness; MBS, MYB binding site involved in drought-inducibility; MBSI, MYB binding site involved in flavonoid biosynthetic gene regulation; GBRE, gibberellin-responsive element; TGAE, auxin-responsive element; MRE, MYB binding site involved in light responsiveness; MYBHv1, MYBHv1 binding site; RY-element, cis-acting regulatory element involved in seed-specific regulation; ARE, cis-acting regulatory element essential for anaerobic induction; CAT-box, cis-acting regulatory element related to meristem expression; MSA, cis-acting element involved in cell cycle regulation; Circadian, cis-acting regulatory element involved in circadian control; GCN4, cis-regulatory element involved in endosperm expression; and HD-Zip, element involved in differentiation of the palisade mesophyll cells.
3. Discussion
Sugar transporters play a key role in growth and development of many plant species [34,43,44]. SWEETs are critically important for sugar efflux, phloem loading, and nectar secretion [11,45]. These transporters are also important susceptibility genes facilitating interaction between host plants and Xanthomonas pathogens [21,22]. Once secreted into host cells through T3SS, TALEs bind to the promoter region of specific sugar transporter genes favoring bacterial growth [20]. Different Xanthomonas strains have different repertoires of effector protein including TALEs that are important for virulence in several plant diseases [36]. Distribution of TALEs varies in Xanthomonas species depending on host specificity and geographic origin [46]. X. oryzae pathogens typically harbor large TALE repertoires [47], followed by X. campestris [48]. Interestingly, among X. arboricola, only pathovar corylina presents a TALE homolog [49]. To better understand the susceptibility of walnut to Xaj, the causative agent of walnut blight disease, we characterized the SWEET gene family in walnut as a potential source of susceptibility and their response to Xaj417 infection. This study represents the first detailed characterization of the SWEET gene family in a woody plant.
Twenty-five SWEET genes were detected in the J. regia genome, all highly conserved to their respective orthologs in other plants. All contained two MtN3 units and five motifs except JrSWEET16A, which had only four motifs. In general, the number of SWEET genes is highly variable within the plant kingdom, ranging from one to three copies in green algae to 18 to 68 in vascular plants [34]. Comparing the set of 25 SWEET genes in walnut to 17 in Arabidopsis suggests the possibility of functional redundancy or diversification among SWEET gene family members. It also implies that SWEET genes expanded during the course of evolution. In this study, five pairs of JrSWEET genes were detected as tandem duplications. This finding agrees with other reports of SWEET gene duplications, including segmental, tandem, or whole genome duplications [32,33,50]. However, gene loss during expansion was also found in the J. regia genome, compared to the AtSWEET and OsSWEET genes. There was no homolog of AtSWEET7, AtSWEET8, AtSWEET11, AtSWEET13, or AtSWEET18 in the J. regia genome. All AtSWEET genes in clade III except AtSWEET15 (ortholog of JrN3 and JrN3A), AtSWEET5, and AtSWEET6 have only one respective orthologous gene in the J. regia genome [28]. Expansion and/or loss of SWEET genes have also occurred in other species, indicating possible functional variation in the evolution of this gene family. For example, the absence of SWEET10 in rice and wheat may suggest different roles of SWEET genes in dicot and monocot plants [15,32]. The expansion of SWEET genes in cucumber suggests possible functional differentiations in response to environmental conditions [51]. Additionally, five transmembrane helixes (TMHs) were observed in JrSWEET3A, JrSWEET16A, JrSWEET10A, and JrSWEET14A or six in JrSWEET4A and JrSWEET17B, which is less than the seven TMHs in eukaryotic SWEETs reported in previous studies. Gene loss or expansion and occurrence of some SWEETs with only five or six TMHs imply that duplication and fusion of SWEETs may be ongoing in the walnut genome. Our results support the proposal that the SWEET protein family was generated through duplication and fusion of SemiSWEETs, which contain three TMHs [12], rather than fusion of an archaeal with a bacterial SemiSWEET [52].
Interestingly, there was significant variation among SWEET proteins in length, predicted isoelectric points, and classification into four clades. This indicates that different SWEET proteins may function in different microenvironments, consistent with previous studies of SWEET functions [43]. Due to their functional diversity, we examined expression patterns of JrSWEET genes in different tissues. Expression of four JrSWEET genes was undetectable in any tissues/organs. This suggests that these genes may have degenerated or lost function after gene duplication during evolution. The expression patterns of the other SWEET genes were diverse among subfamilies, members of the same clade, and even between orthologous genes. The expression pattern of the SWEET gene family in sweet orange, banana, and other species suggests that SWEETs may undergo functionalization in many higher plant species [33,53,54]. Consistent with expectations, JrSWEET genes were expressed widely in all tested tissues, implying functional redundancy and an important role in both vegetative and reproductive growth. JrSWEET genes from clade I varied greatly among members. JrSWEET1 was strongly expressed in fruits and packing-tissues, indicating it may regulate sugar allocation during fruit ripening by functioning as a bidirectional and low-affinity glucose transporter. However, expression of JrSWEET2 and its orthologous genes in a variety of tissues, especially callus, indicates an important role in differentiation and development. SWEET genes from clade II generally function as bidirectional, low-affinity glucose transporters and are expressed mainly in floral organs [50,51,52]. JrSWEET4, JrSWEET5, and their orthologs are strongly expressed in catkins and embryos, providing evidence that they may play a role in reproductive development. AtSWEET5 is expressed at different stages of pollen development [55], AtSWEET8 is strongly expressed in microspores and tapetum during male meiosis, and AtSWEET13 can partially complement the defective pollen phenotype at later reproductive stages, indicating a broader role for SWEETs in pollen maturation [56,57]. This also explains the strong expression of JrN3 in catkins. Clade IV members are strongly expressed in root cortex and encode proteins that function as fructose-specific uniporters in the root tonoplast [58]. This explains the specific and strong expression of JrSWEET17B in root. Most of the 25 JrSWEET genes were predicted on the basis of structure to be localized in the plasma membrane, except for JrSWEET2aB and JrSWEET17, which are present in the vacuolar membrane, and JrSWEET17A, present in the chloroplast. These were validated by differential expression in different tissues, further suggesting that JrSWEET genes have developed a diversity of roles for sugar transport.
Members of clade III SWEET genes transport sucrose, which likely serves as a carbon source for plant pathogens, and show highly induced expression as sugar transporters during disease development. Several studies report involvement of SWEET genes in various plant pathogenic systems [20,21,22,23,25,26]. OsSWEET11, OsSWEET12, OsSWEET13, OsSWEET14, and their orthologs are susceptibility genes in rice infected by Xoo. MeSWEET10a in the dicot cassava is induced by infection with X. axonopodis, and CsSWEET1 in citrus is induced by infection with Xcc. SWEET genes are also induced by other bacterial and fungal pathogens, suggesting that SWEET induction may be a frequent strategy adopted by diverse pathogens to steal sugar from plants. In walnut, most JrSWEET genes in clade III are highly expressed in pellicle, a tissue typically attacked by microorganisms. We detected expression of 11 SWEET genes in microshoots’ leaves during infection by Xaj417, but interestingly, none showed increased expression in response to infection. Considering that this particular pathogen strain has a functional type III secretion system that in other Xanthomonas pathogens is able to secrete TALEs into the plant cell and influence SWEET genes expression [59], a logical hypothesis was that Xaj417 would adopt this strategy. If this was the case, a disease control strategy could encompass the inactivation of a particular responsive SWEET member in walnut to deprive the pathogen from this sugar source, as recently shown for the rice blight pathosystem [60,61]. Since we observed no induction of specific SWEET genes in response to Xaj417 infection, we investigated the pathogen’s genome in search of any known TALEs. Surprisingly, none were found, which may be connected with the observed lack of SWEET gene induction in response to Xaj417 infection. On the other hand, it cannot be excluded that a significant response would be detected at other time points or conditions (e.g., pot-experiment), although at up to 168 hpi no induction was observed (data not shown). Our results suggest that SWEET genes do not play a critical role in walnut blight susceptibility, and the absence of TALEs in Xaj417 indicates some other infection strategies are the source of susceptibility to walnut blight disease. The type II secretion system of Xaj417, for example, may play a greater role in increasing susceptibility to walnut blight disease [62].
We identified and characterized 25 SWEET genes in walnut, using their unique DNA sequences to determine a variety of parameters such as genomic location, predicted number of amino acids (length) and molecular weight, isoelectric point, sub-cellular localization, exon–intron structures and conserved motifs, phylogeny, expression analysis, and presence of cis-elements. The results from this study provide a basic understanding of the JrSWEET genes, facilitate further detailed research on SWEET genes in walnut, and provide a platform for identification and comprehensive functional characterization of SWEET gene families from other woody plants. The non-participation of SWEET genes in increasing susceptibility to walnut blight, along with the observation that no TALEs are present in the Xaj417 genome, suggest that other avenues of research into plant–pathogen interactions other than those mediated by TALE–SWEET genes may be more fruitful in developing strategies to improve resistance to walnut blight.
4. Materials and Methods
4.1. Genome-Wide Identification of SWEET Family Genes in Walnut
To identify walnut SWEET genes, two different approaches were used to compile a complete list. We downloaded the latest version of the whole Juglans regia genome annotation from the National Center for Biotechnology Information (NCBI). In one approach, we obtained putative SWEET genes by searching for the Hidden Markov Models (HMMER. http://hmmer.org) profile of the core domain MtN3/saliva protein (PF03083) from the Pfam database (Pfam. http://pfam.xfam.org) [63,64]. The other approach was to retrieve all 17 known SWEET proteins in A. thaliana from the Arabidopsis Information Resource (TAIR. https://www.arabidopsis.org/). Putative walnut SWEET proteins were identified by blast searches against the walnut genome using A. thaliana SWEET protein sequences as queries. In addition, we used “bidirectional sugar transporter” as a query keyword to search all walnut paralog genes listed in the Kyoto Encyclopedia of Genes and Genomes database (KEGG. https://www.genome.jp/kegg/) [65]. To confirm the core domain and completeness, all candidate proteins were verified manually with the Conserved Domain Database (CDD, https://www.ncbi.nlm.nih.gov/cdd) and SMART (SMART. http://smart.embl-heidelberg.de/) [66,67]. Some candidates were abandoned as too short or incomplete. The remaining proteins were reconfirmed by length of sequence, isoelectric point (pI), and molecular weight (Mw) using the ExPASy tool (ExPASy. https://web.expasy.org/compute_pi/) [68]. The subcellular localization of JrSWEET proteins was predicted through the TargetP 1.1 server (TargetP. http://www.cbs.dtu.dk/services/TargetP/) and WoLF PSORT (WoLF PSORT. https://wolfpsort.hgc.jp/) [69,70]. Moreover, we recovered the locational information of every single putative gene from the walnut genome databases (http://aegilops.wheat.ucdavis.edu/Walnut/data.php) and sketched all SWEET genes onto their respective chromosomes using Microsoft PowerPoint 2017 [71].
4.2. Gene Structure Analysis, Transmembrane Helix, and Motif Recognition of JrSWEETs
Information on untranslated regions (UTR), exons, and introns were obtained from NCBI by aligning the cDNA sequences with their corresponding mRNA sequence or genomic DNA sequences. Identification of predicted transmembrane helices in proteins was by TMHMM Server 2.0 (TMHMM. http://www.cbs.dtu.dk/services/TMHMM/) [72]. The gene and protein structures were constructed using the Gene Structure Display Server (GSDS. http://gsds.cbi.pku.edu.cn) [73]. The phylogenetic tree of walnut SWEET family members was constructed using unrooted neighbor-joining analysis in MEGA 7.0 (MEGA. https://www.megasoftware.net/), with bootstrap tests carried out 1000 times. The conserved motifs in SWEET proteins and the logos of motifs were generated by the MEME suite (MEME. http://meme-suite.org) and redrawn by TBtools [74].
4.3. Phylogenetic Analysis of JrSWEET Proteins
The full-length amino acid sequences of 17 AtSWEET genes derived from TAIR and 21 OsSWEET genes from the rice genome database in the rice genome annotation project (TIGR. http://rice.tigr.org) were combined with newly identified JrSWEET genes for phylogenetic analysis. All acquired protein sequences were first aligned by ClustalW with default parameters, then an unrooted neighbor-joining analysis was used to construct the phylogenetic tree using MEGA 7.0 with bootstrap test carried out 1000 times. The tree was further edited by iTOL (iTOL. https://itol.embl.de) [75].
4.4. Expression Profiles of JrSWEETs in Different Tissues, Organs, and Developmental Stages from RNA-Seq Data
Expression data of JrSWEET transcripts was obtained from RNA-seq data for twenty different tissues and/or developmental stages of ‘Chandler’ walnut [76]. Transcript abundance of 25 JrSWEET genes in thirteen tissues (callus, catkins, zygotic embryo, pistillate flower, hull, fruit, leaves, packing tissue, pellicle, root, somatic embryo, wood, and bud) and developmental stages of five tissues (callus, hull, leaf, packing tissue, and zygotic embryo) was analyzed. The expression data was used to generate a heat map using the Morpheus heat map program (MORPHEUS. https://software.broadinstitute.org/morpheus/) package with hierarchical clustering for twenty different tissues.
4.5. Expression of JrSWEET Genes after Inoculation with Xaj417
Tissue-cultured ‘Chandler’ walnut microshoots were transferred to fresh medium in individual culture tubes two days prior to inoculation with Xaj417, a copper-resistant strain isolated from diseased vegetative buds in California and sequenced at the University of California, Davis [77]. A frozen stock culture (stored in 60% glycerol at −80 °C) was grown on yeast extract peptone (YEP) agar for 48 h and then transferred to liquid culture for 16 h. Bacterial suspensions were diluted in 0.01% (v/v) Tween 80 solution to ~2 × 108 CFU/mL using a spectrophotometer at 600 nm, then diluted to 2 × 107 CFU/mL for inoculation. To inoculate the microshoots, 30 mL of bacterial suspension was poured into each tube, and whole microshoots were immersed for 20 min in the inoculum and then drained for an additional 10 min. The mock inoculation used 0.01% (v/v) Tween 80 solution instead of bacterial suspension. Forty-eight inoculated shoots were grown in individual tubes at 26 °C with a 16 h/8 h (light/dark) photoperiod. A leaf was harvested from each shoot at the time of inoculation, then another leaf was harvested from each of three shoot replicates at 6, 24, 48, and 120 h after inoculation per treatment (time points selected according to previous experiments). Harvested leaves were frozen immediately in liquid nitrogen and stored at −80 °C until use. All inoculation experiments were repeated at least three times.
4.6. Total RNA Isolation and Gene Expression by Quantitative Real-Time PCR (qRT-PCR)
Pellicles, catkins, and mature zygotic embryos were obtained from a field-grown ‘Chandler’ tree located at Davis, CA. Roots were obtained from embryos grown in tissue culture. Embryos were allowed to germinate, and leaves were harvested for gene expression analysis after inoculation with Xaj417. After collection, all tissues were frozen immediately in liquid nitrogen and stored at −80 °C until use. Total RNA was extracted using Plant RNA Purification Reagent (Thermo Fisher Scientific) and purified using the RNeasy MinElute Clean-up Kit (Qiagen, Hilden, Germany) following the manufacturer’s instructions. The quality of RNA was determined using a nanodrop and also by agarose gel electrophoresis. Complementary DNA (cDNA) was synthesized using a QuantiTect Reverse Transcription Kit (Qiagen) according to the manufacturer’s instructions. Quantitative real-time PCR was performed with SYBR Green PCR Master Mix (Applied Biosystems, Foster City, CA, USA) using an ABI StepOne Real-Time PCR system and software (Applied Biosystems). The primers used for each gene analyzed are listed in Supplementary Table S2. The protocol was initiated with a step of 10 s at 95 °C, followed by 40 cycles of 95 °C for 10 s and 62 °C for 30 s, ending with a melting curve that was used to verify the specificity of each reaction. All reactions for each gene, including negative controls (water instead of template), were performed in triplicate. All gene expression bar graphs were draw by Prism 7.0 software package with multiple T tests for significance analysis (p < 0.05) [78].
4.7. Searching for TALEs in Xaj417
To seek potential target JrSWEET genes through predicted TALE binding sites, we used three ways to identify TALEs in the whole Xaj417 genome downloaded from NCBI (https://www.ncbi.nlm.nih.gov/genome/11823?genome_assembly_id=244764). First, we used AnnoTALE to search for TALEs in the whole genome [41]. Next, all known TALEs and their sequences in Xanthomonas that were listed at The Xanthomonas Resource (http://www.xanthomonas.org/tools.html) were downloaded. Finally, a blast search using all 516 TALEs in The Xanthomonas Resource was performed against the Xanthomonas arboricola pv. juglandis strain 417 genome.
4.8. Prediction and Functional Analysis of Cis-Regulatory Elements in Putative Promoter Regions
The 2000 bp genomic DNA sequences upstream of the translation start sites of the JrSWEET genes were searched for 5′ regulatory sequences using PlantCARE (PlantCARE. http://bioinformatics.psb.ugent.be/webtools/plantcare/html/) to identify cis-acting elements [42]. Twenty regulatory elements involved in plant growth regulation, hormone response, and stress responses were identified in the 5′ regulatory regions and diagrammed in Figure 10 using Illustrator for Biological Sequences (IBS. http://ibs.biocuckoo.org/) [79].
Acknowledgments
We would like to extend our special thanks to Steven Lee for his help in propagating walnut shoots used in our experiments. Thanks to Lauro Ângelo de Moraes and Izinara Rosse from the Multi-user Bioinformatics Laboratory at Federal University of Ouro Preto, Brazil for support with TALEs analysis.
Abbreviations
| SWEET | Sugar will eventually be exported transporters |
| Xaj | Xanthomonas arboricola pv. juglandis |
| TALEs | Transcription activator-like effectors |
| SUTs | Sucrose transporters |
| TMDs | Transmembrane domains |
| TMHs | Transmembrane helixes |
| PPO2 | Polyphenol oxidase |
| qRT-PCR | Quantitative real-time PCR |
| pI | Isoelectric point |
Supplementary Materials
Supplementary materials can be found at https://www.mdpi.com/1422-0067/21/4/1251/s1.
Author Contributions
S.J. and A.M.D. conceived and designed the experiments; S.J. performed the experiments and analysis and wrote a first draft, which was edited by A.M.D.; B.B. conducted RNA-seq analysis; R.d.A.B.A. performed the TALEs blast with L.M.M. and helped with the experiments together with C.H.D.S.; X.W. and S.H. offered suggestions on the experiments and writing; L.W. drew the figures of chromosomal location; L.Z. and P.A.Z. helped collect the materials; S.L.W., A.J., S.H.L., and C.A.L. helped with the tissue culture and other materials; A.M.D. and all others revised the final manuscript. All authors have read and agreed to the published version of the manuscript.
Funding
This work was supported by funds obtained from the California Walnut Board. S.J. was supported by the China Scholarship Council.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Chen L.Q., Qu X.Q., Hou B.H., Sosso D., Osorio S., Fernie A.R., Frommer W.B. Sucrose Efflux Mediated by SWEET Proteins as a Key Step for Phloem Transport. Science. 2012;335:207–211. doi: 10.1126/science.1213351. [DOI] [PubMed] [Google Scholar]
- 2.Comtet J., Turgeon R., Stroock A.D. Phloem Loading through Plasmodesmata: A Biophysical Analysis. Plant Physiol. 2017;175:904–915. doi: 10.1104/pp.16.01041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Lalonde S., Wipf D., Frommer W.B. Transport Mechanisms for Organic Forms of Carbon and Nitrogen between Source and Sink. Annu. Rev. Plant Biol. 2004;55:341–372. doi: 10.1146/annurev.arplant.55.031903.141758. [DOI] [PubMed] [Google Scholar]
- 4.Comtet J., Jensen K.H., Turgeon R., Stroock A.D., Hosoi A.E. Passive Phloem Loading and Long Distance Transport in a Synthetic Tree on a chip. Nat. Plants. 2017;3:1–8. doi: 10.1038/nplants.2017.32. [DOI] [PubMed] [Google Scholar]
- 5.Ayre B.G. Membrane-Transport Systems for Sucrose in Relation to Whole-Plant Carbon Partitioning. Mol. Plant. 2011;4:377–394. doi: 10.1093/mp/ssr014. [DOI] [PubMed] [Google Scholar]
- 6.Zhang C., Turgeon R. Mechanisms of Phloem Loading. Curr. Opin. Plant Biol. 2018;43:71–75. doi: 10.1016/j.pbi.2018.01.009. [DOI] [PubMed] [Google Scholar]
- 7.Lalonde S., Frommer W.B. SUT Sucrose and MST Monosaccharide Transporter Inventory of the Selaginella Genome. Front. Plant Sci. 2012;3:24. doi: 10.3389/fpls.2012.00024. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Frank Baker R., Leach K.A., Braun D.M. SWEET as Sugar: New Sucrose Effluxers in Plants. Mol. Plant. 2012;5:766–768. doi: 10.1093/mp/SSS054. [DOI] [PubMed] [Google Scholar]
- 9.Doidy J., Grace E., Kühn C., Simon-Plas F., Casieri L., Wipf D. Sugar transporters in plants and in their interactions with fungi. Trends Plant Sci. 2012;17:413–422. doi: 10.1016/j.tplants.2012.03.009. [DOI] [PubMed] [Google Scholar]
- 10.Chandran D. Co-option of developmentally regulated plant SWEET transporters for pathogen nutrition and abiotic stress tolerance: Role of SWEET in Pathogen Nutrition and Srtress Tolerence. IUBMB Life. 2015;67:461–471. doi: 10.1002/iub.1394. [DOI] [PubMed] [Google Scholar]
- 11.Chen L.Q. SWEET Sugar Transporters for Phloem Transport and Pathogen Nutrition. New Phytol. 2014;201:1150–1155. doi: 10.1111/nph.12445. [DOI] [PubMed] [Google Scholar]
- 12.Xuan Y.H., Hu Y.B., Chen L.Q., Sosso D., Ducat D.C., Hou B.-H., Frommer W.B. Functional Role of Oligomerization for Bacterial and Plant SWEET Sugar Transporter Family. PNAS. 2013;110:E3685–E3694. doi: 10.1073/pnas.1311244110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Xu Y., Tao Y., Cheung L.S., Fan C., Chen L.Q., Xu S., Perry K., Frommer W.B., Feng L. Structures of Bacterial Homologues of SWEET Transporters in Two Distinct Conformations. Nature. 2014;515:448–452. doi: 10.1038/nature13670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Braun D.M. SWEET! The Pathway Is Complete. Science. 2012;335:173–174. doi: 10.1126/science.1216828. [DOI] [PubMed] [Google Scholar]
- 15.Yuan M., Zhao J., Huang R., Li X., Xiao J., Wang S. Rice MtN3/saliva/SWEET gene family: Evolution, Expression Profiling, and Sugar Transport. J. Integr. Plant Biol. 2014;56:559–570. doi: 10.1111/jipb.12173. [DOI] [PubMed] [Google Scholar]
- 16.Chardon F., Bedu M., Calenge F., Klemens P.A.W., Spinner L., Clement G., Chietera G., Léran S., Ferrand M., Lacombe B., et al. Leaf Fructose Content Is Controlled by the Vacuolar Transporter SWEET17 in Arabidopsis. Curr. Biol. 2013;23:697–702. doi: 10.1016/j.cub.2013.03.021. [DOI] [PubMed] [Google Scholar]
- 17.Liu X., Zhang Y., Yang C., Tian Z., Li J. AtSWEET4, a Hexose Facilitator, Mediates Sugar Transport to Axial Sinks and Affects Plant Development. Sci. Rep. 2016;6:1–12. doi: 10.1038/srep24563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Yang B., Sugio A., White F.F. Os8N3 is a Host Disease-susceptibility Gene for Bacterial Blight of Rice. Proc. Natl. Acad. Sci. 2006;103:10503–10508. doi: 10.1073/pnas.0604088103. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhou J., Peng Z., Long J., Sosso D., Liu B., Eom J.S., Huang S., Liu S., Vera Cruz C., Frommer W.B., et al. Gene targeting by the TAL effector PthXo2 reveals cryptic resistance gene for bacterial blight of rice. Plant J. 2015;82:632–643. doi: 10.1111/tpj.12838. [DOI] [PubMed] [Google Scholar]
- 20.Blanvillain-Baufumé S., Reschke M., Solé M., Auguy F., Doucoure H., Szurek B., Meynard D., Portefaix M., Cunnac S., Guiderdoni E., et al. Targeted Promoter Editing for Rice Resistance to Xanthomonas oryzae pv. oryzae reveals differential activities for SWEET14 Inducing TAL effectors. Plant Biotechnol. J. 2017;15:306–317. doi: 10.1111/pbi.12613. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Yuan M., Wang S. Rice MtN3/Saliva/SWEET Family Genes and Their Homologs in Cellular Organisms. Mol. Plant. 2013;6:665–674. doi: 10.1093/mp/sst035. [DOI] [PubMed] [Google Scholar]
- 22.Streubel J., Pesce C., Hutin M., Koebnik R., Boch J., Szurek B. Five Phylogenetically Close Rice SWEET Genes Confer TAL effector-mediated Susceptibility to Xanthomonas oryzae pv. oryzae. New Phytol. 2013;200:808–819. doi: 10.1111/nph.12411. [DOI] [PubMed] [Google Scholar]
- 23.Liu Q., Yuan M., Zhou Y., Li X., Xiao J., Wang S. A Paralog of the MtN3/saliva Family Recessively Confers Race-specific Resistance to Xanthomonas oryzae in Rice: MtN3/saliva-type Gene in Rice Disease Resistance. Plantcell Environ. 2011;34:1958–1969. doi: 10.1111/j.1365-3040.2011.02391.x. [DOI] [PubMed] [Google Scholar]
- 24.Cox K.L., Meng F., Wilkins K.E., Li F., Wang P., Booher N.J., Carpenter S.C.D., Chen L.Q., Zheng H., Gao X., et al. TAL effector Driven Induction of a SWEET gene Confers Susceptibility to Bacterial Blight of Cotton. Nat. Commun. 2017;8:15588. doi: 10.1038/ncomms15588. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cohn M., Bart R.S., Shybut M., Dahlbeck D., Gomez M., Morbitzer R., Hou B.H., Frommer W.B., Lahaye T., Staskawicz B.J. Xanthomonas axonopodis Virulence Is Promoted by a Transcription Activator-Like Effector–Mediated Induction of a SWEET Sugar Transporter in Cassava. MPMI. 2014;27:1186–1198. doi: 10.1094/MPMI-06-14-0161-R. [DOI] [PubMed] [Google Scholar]
- 26.Hu Y., Zhang J., Jia H., Sosso D., Li T., Frommer W.B., Yang B., White F.F., Wang N., Jones J.B. Lateral Organ Boundaries 1 is a Disease Susceptibility Gene for Citrus Bacterial Canker Disease. Proc. Natl. Acad. Sci. USA. 2014;111:E521–E529. doi: 10.1073/pnas.1313271111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Boevink P.C., McLellan H., Gilroy E.M., Naqvi S., He Q., Yang L., Wang X., Turnbull D., Armstrong M.R., Tian Z., et al. Oomycetes Seek Help from the Plant: Phytophthora infestans Effectors Target Host Susceptibility Factors. Mol. Plant. 2016;9:636–638. doi: 10.1016/j.molp.2016.04.005. [DOI] [PubMed] [Google Scholar]
- 28.Chen L.-Q., Hou B.H., Lalonde S., Takanaga H., Hartung M.L., Qu X.Q., Guo W.J., Kim J.-G., Underwood W., Chaudhuri B., et al. Sugar transporters for intercellular exchange and nutrition of pathogens. Nature. 2010;468:527–532. doi: 10.1038/nature09606. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Chong J., Piron M.C., Meyer S., Merdinoglu D., Bertsch C., Mestre P. The SWEET Family of Sugar Transporters in Grapevine: VvSWEET4 is Involved in the Interaction with Botrytis cinerea. J. Exp. Bot. 2014;65:6589–6601. doi: 10.1093/jxb/eru375. [DOI] [PubMed] [Google Scholar]
- 30.Gamas P., de Carvalho Niebel F., Lescure N., Cullimore J. Use of a Subtractive Hybridization Approach to Identify New Medicago truncatula Genes Induced During Root Nodule Development. Mol. Plant Microbe Interact. 1996;9:233–242. doi: 10.1094/MPMI-9-0233. [DOI] [PubMed] [Google Scholar]
- 31.Jian H., Lu K., Yang B., Wang T., Zhang L., Zhang A., Wang J., Liu L., Qu C., Li J. Genome-Wide Analysis and Expression Profiling of the SUC and SWEET Gene Families of Sucrose Transporters in Oilseed Rape (Brassica napus L.) Front. Plant Sci. 2016;7:1464. doi: 10.3389/fpls.2016.01464. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Gao Y., Wang Z.Y., Kumar V., Xu X.F., Yuan D.P., Zhu X.F., Li T.Y., Jia B., Xuan Y.H. Genome-wide Identification of the SWEET Gene Family in Wheat. Gene. 2018;642:284–292. doi: 10.1016/j.gene.2017.11.044. [DOI] [PubMed] [Google Scholar]
- 33.Miao H., Sun P., Liu Q., Miao Y., Liu J., Zhang K., Hu W., Zhang J., Wang J., Wang Z., et al. Genome-wide Analyses of SWEET Family Proteins Reveal Involvement in Fruit Development and Abiotic/biotic Stress Responses in Banana. Sci. Rep. 2017;7:3536. doi: 10.1038/s41598-017-03872-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Li H., Li X., Xuan Y., Jiang J., Wei Y., Piao Z. Genome Wide Identification and Expression Profiling of SWEET Genes Family Reveals Its Role During Plasmodiophora brassicae Induced Formation of Clubroot in Brassica rapa. Front. Plant Sci. 2018;9:207. doi: 10.3389/fpls.2018.00207. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Schornack S., Meyer A., Römer P., Jordan T., Lahaye T. Gene-for-gene-mediated recognition of nuclear-targeted AvrBs3-like bacterial effector proteins. J. Plant Physiol. 2006;163:256–272. doi: 10.1016/j.jplph.2005.12.001. [DOI] [PubMed] [Google Scholar]
- 36.Kay S., Hahn S., Marois E., Hause G., Bonas U. A Bacterial Effector Acts as a Plant Transcription Factor and Induces a Cell Size Regulator. Science. 2007;318:648–651. doi: 10.1126/science.1144956. [DOI] [PubMed] [Google Scholar]
- 37.Khodadadi F., Tohidfar M., Mohayeji M., Dandekar A.M., Leslie C.A., Kluepfel D.A., Butterfield T., Vahdati K. Induction of Polyphenol Oxidase in Walnut and Its Relationship to the Pathogenic Response to Bacterial Blight. J. Am. Soc. Hortic. Sci. 2016;141:119–124. doi: 10.21273/JASHS.141.2.119. [DOI] [Google Scholar]
- 38.Araji S., Grammer T.A., Gertzen R., Anderson S.D., Mikulic-Petkovsek M., Veberic R., Phu M.L., Solar A., Leslie C.A., Dandekar A.M., et al. Novel Roles for the Polyphenol Oxidase Enzyme in Secondary Metabolism and the Regulation of Cell Death in Walnut. Plant Physiol. 2014;164:1191–1203. doi: 10.1104/pp.113.228593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Chakraborty S., Britton M., Wegrzyn J., Butterfield T., Rao B.J., Leslie C.A., Aradhaya M., Neale D., Woeste K., Dandekar A.M. YeATS—A Tool Suite for Analyzing RNA-seq Derived Transcriptome Identifies a Highly Transcribed Putative Extensin in Heartwood/sapwood Transition Zone in Black Walnut. F1000Research. 2015;4:155. doi: 10.12688/f1000research.6617.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Martínez-García P.J., Crepeau M.W., Puiu D., Gonzalez-Ibeas D., Whalen J., Stevens K.A., Paul R., Butterfield T.S., Britton M.T., Reagan R.L., et al. The walnut (Juglans regia) Genome Sequence Reveals Diversity in Genes Coding for the Biosynthesis of Non-structural Polyphenols. Plant J. 2016;87:507–532. doi: 10.1111/tpj.13207. [DOI] [PubMed] [Google Scholar]
- 41.Grau J., Reschke M., Erkes A., Streubel J., Morgan R.D., Wilson G.G., Koebnik R., Boch J. AnnoTALE: Bioinformatics Tools for Identification, Annotation and Momenclature of TALEs from Xanthomonas Genomic Sequences. Sci. Rep. 2016;6 doi: 10.1038/srep21077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Lescot M. PlantCARE, a Database of Plant Cis-acting Regulatory Elements and a Portal to Tools for in Silico Analysis of Promoter Sequences. Nucleic Acids Res. 2002;30:325–327. doi: 10.1093/nar/30.1.325. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Zhao L., Yao J., Chen W., Li Y., LÜ Y., Guo Y., Wang J., Yuan L., Liu Z., Zhang Y. A Genome-wide Analysis of SWEET Gene Family in Cotton and Their Expressions Under Different Stresses. J. Cotton Res. 2018;1:7. doi: 10.1186/s42397-018-0007-9. [DOI] [Google Scholar]
- 44.Li J., Qin M., Qiao X., Cheng Y., Li X., Zhang H., Wu J. A New Insight into the Evolution and Functional Divergence of SWEET Transporters in Chinese White Pear (Pyrus bretschneideri) Plant Cell Physiol. 2017;58:839–850. doi: 10.1093/pcp/pcx025. [DOI] [PubMed] [Google Scholar]
- 45.Eom J.S., Chen L.Q., Sosso D., Julius B.T., Lin I., Qu X.Q., Braun D.M., Frommer W.B. SWEETs, transporters for intracellular and intercellular sugar translocation. Curr. Opin. Plant Biol. 2015;25:53–62. doi: 10.1016/j.pbi.2015.04.005. [DOI] [PubMed] [Google Scholar]
- 46.Triplett L.R., Hamilton J.P., Buell C.R., Tisserat N.A., Verdier V., Zink F., Leach J.E. Genomic Analysis of Xanthomonas oryzae Isolates from Rice Grown in the United States Reveals Substantial Divergence from Known X. oryzae Pathovars. Appl. Environ. Microbiol. 2011;77:3930–3937. doi: 10.1128/AEM.00028-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Erkes A., Reschke M., Boch J., Grau J. Evolution of Transcription Activator-Like Effectors in Xanthomonas oryzae. Genome Biol. Evol. 2017;9:1599–1615. doi: 10.1093/gbe/evx108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Denancé N., Szurek B., Doyle E.L., Lauber E., Fontaine-Bodin L., Carrère S., Guy E., Hajri A., Cerutti A., Boureau T., et al. Two ancestral genes shaped the Xanthomonas campestris TAL effector gene repertoire. New Phytol. 2018;219:391–407. doi: 10.1111/nph.15148. [DOI] [PubMed] [Google Scholar]
- 49.Hajri A., Pothier J.F., Fischer-Le Saux M., Bonneau S., Poussier S., Boureau T., Duffy B., Manceau C. Type Three Effector Gene Distribution and Sequence Analysis Provide New Insights into the Pathogenicity of Plant-Pathogenic Xanthomonas arboricola. Appl. Environ. Microbiol. 2012;78:371–384. doi: 10.1128/AEM.06119-11. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Feng C.Y., Han J.X., Han X.X., Jiang J. Genome-wide Identification, Phylogeny, and Expression Analysis of the SWEET Gene Family in Tomato. Gene. 2015;573:261–272. doi: 10.1016/j.gene.2015.07.055. [DOI] [PubMed] [Google Scholar]
- 51.Li Y., Feng S., Ma S., Sui X., Zhang Z. Spatiotemporal Expression and Substrate Specificity Analysis of the Cucumber SWEET Gene Family. Front. Plant Sci. 2017;8:1855. doi: 10.3389/fpls.2017.01855. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Jia B., Zhu X.F., Pu Z.J., Duan Y.X., Hao L.J., Zhang J., Chen L.Q., Jeon C.O., Xuan Y.H. Integrative View of the Diversity and Evolution of SWEET and SemiSWEET Sugar Transporters. Front. Plant Sci. 2017;8:2178. doi: 10.3389/fpls.2017.02178. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Wang W., Liu J.H. Genome-wide Identification and Expression Analysis of the Polyamine Oxidase Gene Family in sweet orange (Citrus sinensis) Gene. 2015;555:421–429. doi: 10.1016/j.gene.2014.11.042. [DOI] [PubMed] [Google Scholar]
- 54.Filyushin M.A., Kochieva E.Z., Shchennikova A.V., Beletsky A.V., Mardanov A.V., Ravin N.V., Skryabin K.G. SWEET Uniporter Gene Family Expression Profile in the Pitcher Development in the Carnivorous Plant Nepenthes sp. Russ. J. Genet. 2019;55:692–700. doi: 10.1134/S1022795419050089. [DOI] [Google Scholar]
- 55.Engel M.L., Holmes-Davis R., McCormick S. Green Sperm. Identification of Male Gamete Promoters in Arabidopsis. Plant Physiol. 2005;138:2124–2133. doi: 10.1104/pp.104.054213. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Guan Y.F., Huang X.Y., Zhu J., Gao J.F., Zhang H.X., Yang Z.N. RUPTURED POLLEN GRAIN1, a Member of the MtN3/saliva Gene Family, Is Crucial for Exine Pattern Formation and Cell Integrity of Microspores in Arabidopsis. Plant Physiol. 2008;147:852–863. doi: 10.1104/pp.108.118026. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Sun M.-X., Huang X.Y., Yang J., Guan Y.F., Yang Z.N. Arabidopsis RPG1 is Important for Primexine Deposition and Functions Redundantly with RPG2 for Plant Fertility at the Late Reproductive Stage. Plant Reprod. 2013;26:83–91. doi: 10.1007/s00497-012-0208-1. [DOI] [PubMed] [Google Scholar]
- 58.Guo W.J., Nagy R., Chen H.Y., Pfrunder S., Yu Y.C., Santelia D., Frommer W.B., Martinoia E. SWEET17, a Facilitative Transporter, Mediates Fructose Transport Across the Tonoplast of Arabidopsis Roots and Leaves. Plant Physiol. 2014;164:777–789. doi: 10.1104/pp.113.232751. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Boch J., Bonas U., Lahaye T. TAL effectors pathogen strategies and plant resistance engineering. New Phytol. 2014;204:823–832. doi: 10.1111/nph.13015. [DOI] [PubMed] [Google Scholar]
- 60.Oliva R., Ji C., Atienza-Grande G., Huguet-Tapia J.C., Perez-Quintero A., Li T., Eom J.S., Li C., Nguyen H., Liu B., et al. Broad-spectrum resistance to bacterial blight in rice using genome editing. Nat. Biotechnol. 2019;37:1344–1350. doi: 10.1038/s41587-019-0267-z. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Eom J.S., Luo D., Atienza-Grande G., Yang J., Ji C., Thi Luu V., Huguet-Tapia J.C., Char S.N., Liu B., Nguyen H., et al. Diagnostic kit for rice blight resistance. Nat. Biotechnol. 2019;37:1372–1379. doi: 10.1038/s41587-019-0268-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.De AB Assis R., Polloni L.C., Patané J.S.L., Thakur S., Felestrino É.B., Diaz-Caballero J., Digiampietri L.A., Goulart L.R., Almeida N.F., Nascimento R., et al. Identification and analysis of seven effector protein families with different adaptive and evolutionary histories in plant-associated members of the Xanthomonadaceae. Sci. Rep. 2017;7:1–17. doi: 10.1038/s41598-017-16325-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Finn R.D., Bateman A., Clements J., Coggill P., Eberhardt R.Y., Eddy S.R., Heger A., Hetherington K., Holm L., Mistry J., et al. Pfam: the protein families database. Nucleic Acids Res. 2014;42:D222–D230. doi: 10.1093/nar/gkt1223. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Finn R.D., Clements J., Eddy S.R. HMMER Web Server: Interactive Sequence Similarity Searching. Nucleic Acids Res. 2011;39:W29–W37. doi: 10.1093/nar/gkr367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Kanehisa M., Goto S. KEGG: Kyoto Encyclopedia of Genes and Genomes. Nucleic Acids Res. 2000;28:27–40. doi: 10.1093/nar/28.1.27. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Marchler-Bauer A. CDD: A Database of Conserved Domain Alignments with Links to Domain Three-dimensional Structure. Nucleic Acids Res. 2002;30:281–283. doi: 10.1093/nar/30.1.281. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Letunic I., Bork P. 20 Years of the SMART Protein Domain Annotation Resource. Nucleic Acids Res. 2018;46:D493–D496. doi: 10.1093/nar/gkx922. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Gasteiger E. ExPASy: The Proteomics Server for in-depth Protein Knowledge and Analysis. Nucleic Acids Res. 2003;31:3784–3788. doi: 10.1093/nar/gkg563. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Horton P., Park K.J., Obayashi T., Fujita N., Harada H., Adams-Collier C.J., Nakai K. WoLF PSORT: Protein Localization Predictor. Nucleic Acids Res. 2007;35:W585–W587. doi: 10.1093/nar/gkm259. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Emanuelsson O., Brunak S., von Heijne G., Nielsen H. Locating Proteins in the Cell using TargetP, SignalP and Related Tools. Nat. Protoc. 2007;2:953–971. doi: 10.1038/nprot.2007.131. [DOI] [PubMed] [Google Scholar]
- 71.Zhu T., Wang L., You F.M., Rodriguez J.C., Deal K.R., Chen L., Li J., Chakraborty S., Balan B., Jiang C.Z., et al. Sequencing a Juglans regia × J. microcarpa hybrid yields high-quality genome assemblies of parental species. Hortic. Res. 2019;6:55. doi: 10.1038/s41438-019-0139-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Krogh A., Larsson B., von Heijne G., Sonnhammer E.L.L. Predicting Transmembrane Protein Topology with a Hidden Markov Model: Application to Complete Genomes11Edited by F. Cohen. J. Mol. Biol. 2001;305:567–580. doi: 10.1006/jmbi.2000.4315. [DOI] [PubMed] [Google Scholar]
- 73.Hu B., Jin J., Guo A.Y., Zhang H., Luo J., Gao G. GSDS 2.0: An Upgraded Gene Feature Visualization Server. Bioinformatics. 2015;31:1296–1297. doi: 10.1093/bioinformatics/btu817. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Chen C., Xia R., Chen H., He Y. TBtools, a Toolkit for Biologists Integrating Various HTS-Data Handling Tools with a User-friendly Interface. BioRxiv. 2018:289660. [Google Scholar]
- 75.Letunic I., Bork P. Interactive Tree of Life (iTOL) v3: an Online Tool for the Display and Annotation of Phylogenetic and Other Trees. Nucleic Acids Res. 2016;44:W242–W245. doi: 10.1093/nar/gkw290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76.Chakraborty S., Britton M., Martínez-García P.J., Dandekar A.M. Deep RNA-Seq Profile Reveals Biodiversity, Plant–microbe Interactions and a Large Family of NBS-LRR Resistance genes in walnut (Juglans regia) Tissues. Amb Express. 2016;6:12. doi: 10.1186/s13568-016-0182-3. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Pereira U.P., Gouran H., Nascimento R., Adaskaveg J.E., Goulart L.R., Dandekar A.M. Complete Genome Sequence of Xanthomonas arboricola pv. juglandis 417, a Copper-Resistant Strain Isolated from Juglans regia L. Genome Announc. 2015;3:e01126–e01215. doi: 10.1128/genomeA.01126-15. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Swift M.L. GraphPad Prism, Data Analysis, and Scientific Graphing. J. Chem. Inf. Comput. Sci. 1997;37:411–412. doi: 10.1021/ci960402j. [DOI] [Google Scholar]
- 79.Liu W., Xie Y., Ma J., Luo X., Nie P., Zuo Z., Lahrmann U., Zhao Q., Zheng Y., Zhao Y., et al. IBS: An Illustrator for the Presentation and Visualization of Biological Sequences: Fig. 1. Bioinformatics. 2015;31:3359–3361. doi: 10.1093/bioinformatics/btv362. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.