Abstract
Purpose:
Sessile serrated lesions (SSL) are precursors to colon carcinoma (CRC), and their distinction from other polyps, in particular hyperplastic polyps (HPs), presents significant diagnostic challenges. We evaluated expression patterns in colonic polyps of previously identified CRC-associated extracellular matrix (ECM) proteins to identify markers distinguishing SSLs from other polyps.
Experimental Design:
Gene-expression analyses of ECM proteins were performed using publicly available data on pre-neoplastic colonic polyps. In parallel, we evaluated by immunohistochemistry the expression of agrin (AGRN) in over 400 colonic polyps, including HP, SSL with and without dysplasia, traditional serrated adenomas (TSA), tubular adenomas (TA) and compared the consistency of standard histological diagnosis of SSLs by experienced gastrointestinal pathologists with that of AGRN immunohistochemistry.
Results:
Differential gene expression analysis and immunohistochemistry identified AGRN, serine peptidase inhibitor (SERPINE2) and TIMP metallopeptidase inhibitor 1 (TIMP1) elevated in SSLs and HPs but decreased in TAs and absent in normal colon. AGRN-positive basal laminae (BL) were noted in all TA, TSA, HP and SSL in distinguishable patterns, whereas other polyps and normal mucosa were negative. SSL with or without dysplasia consistently showed immunohistochemical staining for AGRN in the muscularis mucosae (MM), which was absent in HP, TSA, TA, and other polyps. In contrast, histological evaluation showed only weak inter-observer agreement (kappa value = 0.493) in distinguishing SSLs.
Conclusions:
Muscularis-mucosae-based AGRN immunostaining is a novel biomarker to distinguish SSL from HP, TSA and TA with a specificity of 97.1% and sensitivity of 98.9% and can assist in diagnosis of morphologically challenging colonic polyps.
Keywords: Agrin, Biomarker, Hyperplastic polyps, Immunohistochemistry, Sessile serrated lesions
Introduction
Colonic polyps differ in their risk for progression to cancer; consequently, recommendations for removal and further management are dependent on polyp classification (1). While tubular adenomas (TA) were traditionally considered to be the sole precursor lesions of colorectal carcinoma (CRC), the recognition of the serrated polyp route to carcinoma has been an important milestone (2,3).
The World Health Organization (WHO) recognizes three categories of serrated polyps: traditional serrated adenomas (TSA), hyperplastic polyps (HP) and sessile serrated lesions (SSL) (4–8). HPs, predominantly arising in rectum and sigmoid colon, lack malignant potential and are by far the most common colonic polyps, accounting for approximately 80% of all serrated polyps (6). In contrast, SSLs, which do have malignant potential, are estimated to represent up to 20% of serrated polyps with a predilection for the right colon (9,10). TSAs also have malignant potential but represent only 1% of all serrated lesions (11).
While SSLs show abnormal basal crypt architecture, HPs lack these specific features (9). However, uncertainty exists regarding the minimum criteria for diagnosis of SSL and their distinction from HP. The number of abnormal crypts required has varied among guidelines from 1 to 3 SSL-like crypts (5–7). Furthermore, there is significant inter-observer variability as to what constitutes an abnormal crypt and poor specimen orientation and other biopsy artifacts accentuate the challenge (12–14). Indeed, substantial inter-observer variation among gastrointestinal (GI) pathologists remains and agreement among experts is moderate at best (13,15–20). These uncertainties often result in the inability to definitively distinguish SSL from HP. The diagnosis of SSL prompts more aggressive surveillance, generally 1-3 years, while a diagnosis of HP and small TAs (<10 mm) indicates less aggressive surveillance, every 5-10 years (5,18). Collectively, the evidence suggests a need to identify biomarkers for SSLs to guide appropriate management. Suggested biomarkers evaluated, MUC6, ANXA10 and HES1 (21–25) have not been rigorously tested nor are they used in routine practice.
To identify novel biomarkers for SSLs, we evaluated the expression of extracellular matrix (ECM) proteins upregulated in CRC (26). We investigated human colonic polyps and found differential AGRN positivity in the basal lamina (BL) of TA, TSA, HP and SSL whereas hamartomatous and mucosal prolapse polyps were negative. Importantly, SSL showed AGRN reactivity in the muscularis mucosae (MM) whereas other colonic polyps and controls were negative. Our study suggests that MM-based AGRN immunostaining represents a novel biomarker to differentiate SSL from HP, TSA and TA with a high specificity of 97.1% and sensitivity of 98.8%.
Materials and Methods
Patients and Samples
Patients were not involved in the study. Formalin-fixed, paraffin-embedded (FFPE) biopsies of normal colon and colonic lesions/polyps were identified from patients (between 2 and 89 years of age) undergoing colonoscopy between 2006 and 2019. Hematoxylin-eosin (H&E) slides were examined by two pathologists (VD, OHY) before inclusion in this study. We analyzed 408 colonic polyps, including hyperplastic polyps (HP; n=71), sessile serrated lesions (SSL) with dysplasia (n=23) and without dysplasia (n=166), traditional serrated adenomas (TSA; n=25), tubular adenomas (TA; n=64), dysplasia related to inflammatory bowel disease (IBD, n=29), sporadic adenomas arising in IBD (TA in IBD; n=9), mucosal prolapse polyps (n=3). Colonic hamartomatous polyps (n=18) including juvenile polyps (n=10) and Peutz-Jeghers polyps (n=8) served as controls (Supplementary Fig. S9). The splenic flexure was used to differentiate left from right sided polyps; rectal polyps were categorized separately (see Supplementary Fig. S2 and Table S2). The SSL samples (n=189) included 132 (69.9%) polyps from the right colon, 49 (25.9%) from the left colon, and 8 (4.2%) rectal polyps. Among the 71 HP cases, 9 (12.7%) polyps were from the right colon, 32 (45.1%) from the left colon, and 30 (42.2%) from the rectum. HPs were further characterized as microvesicular HPs (MVHP; n= 38) and goblet cell HPs (GCHP; n= 33) by two pathologists (AN, VD, Supplementary Table S2) (27).
To define the serrated polyposis syndrome (SPS) we used criteria proposed by Syngal et al. (28). Briefly, diagnosis was based on the following criteria: (i) at least 5 serrated polyps proximal to the sigmoid colon with ≥2 of these being >10 mm; (ii) any number of serrated polyps proximal to the sigmoid colon in an individual who has a first-degree relative with serrated polyposis; or (iii) >20 serrated polyps of any size, distributed throughout the large intestine. Additional SPS patient samples were provided by the Department of Pathology, University of Utah (Salt Lake City, UT) (29).
Ethics
The study was approved by the Partners Institutional Review Board protocol (IRB #2017P000061) and the MIT IRB (Protocol #1408006568).
Majority-based pathology classification approach
Approach for majority agreement on fifty diagnostically challenging serrated polyps
We used a majority approach for the diagnosis of SSL. From our cohort of 408 colonic polyps and within the 100 cases validated by the 4 non-pathologists (see Supplementary Materials and Methods), two gastrointestinal (GI) pathologists identified fifty diagnostically challenging polyps with a low level of inter-observer agreement. Digital whole-slide H&E stained slides were evaluated by nine GI pathologists (DP, IB, ARM, LZ, QZ, RC, GL, OHY, VD) using the prior WHO guidelines (6). The pathologists were blinded to endoscopic size, endoscopic appearance, and location of the individual samples and classified the polyps into one of the following four categories: HP, SSL with or without dysplasia, TSA, or unclassified.
Approach for near-universal agreement on fifty SSLs
Given the existing lack of consensus among pathologists, we strove to identify fifty SSLs with near-universal agreement from within the cohort of 408 polyps. Among 65 additional polyps evaluated by nine GI pathologists (DP, IB, ARM, LZ, QZ, RC, GL, OHY, VD), all blinded to endoscopic size, endoscopic appearance, and location of the samples, we identified fifty SSLs with near-universal agreement.
Inter-observer variability among community pathologists
To assess the inter-observer variability on selected morphologically challenging polyps (see above), we further assessed fifty-five community-based pathologists. Based on representative images of H&E-stained slides from eight polyps the pathologists recorded their diagnosis using an audience response system. Each pathologist was required to characterize the polyp into one of 5 categories: HP, SSL with or without dysplasia, TSA, TA or Rather not say (Supplementary Fig. S10).
Antibodies, Immunohistochemistry, Immunofluorescence, In situ hybridization and RNA-seq data analysis
Antibodies, standard techniques, RNA-seq data analysis and detailed information are provided in the Supplementary Materials and Methods and in Rickelt and Hynes (2018, ref. 30).
Results
Identification of extracellular matrix proteins in colonic polyps
To identify novel biomarkers to distinguish pre-invasive colorectal neoplasms we investigated the presence of 67 ECM proteins recently identified as increased in tumor samples from CRC patients (26). We used two parallel approaches to elucidate their value as potential biomarkers for detection of precancerous colonic lesions. First, we analyzed their expression levels in a publicly available RNA-seq dataset (29) on colonic polyp and control samples (Fig. 1). Second, we performed extensive immunohistochemical (IHC) screening on FFPE samples of diverse polyp types.
Figure 1. Summary of selection of biomarker candidates.
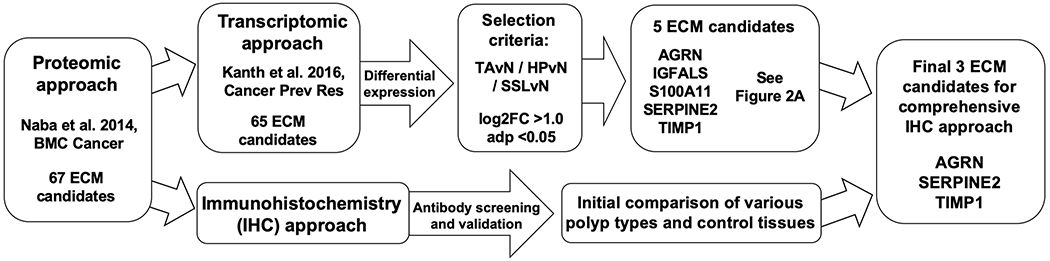
Strategies to identify candidate extracellular matrix (ECM) genes and proteins involved in colonic polyp development. ECM biomarker proteins upregulated in colorectal cancer (CRC) and CRC-derived liver metastasis (Naba et al. 2014, ref. 26) were investigated for their presence in colonic polyps using two independent approaches; [1] Screening by immunohistochemistry (IHC) using reliable antibodies applicable to FFPE patient samples and [2] differential expression analyses of publicly available patient RNA-seq datasets (Kanth et al. 2016, ref. 29). These approaches identified three relevant ECM proteins: AGRN, TIMP1 and SERPINE2.
Gene expression analyses of sixty-five ECM molecules identified by proteomics (26) were performed using RNA-seq data (29) from forty-one colonic polyps including SSL (n=21), HP (n=10), TA (n=10) and colonic control samples (n=20). Most individual SSL samples clustered separately from the other polyp and control samples (Supplementary Fig. S1) clearly indicating that this class of polyps is distinct from others. To determine the most abundant ECM candidates upregulated in common colonic polyps, we performed differential-expression analysis comparing individual polyps to normal control tissues using cut-off selection criteria of log2-fold change (FC) >1.0 and adjusted p value (adp) <0.05 (Fig. 2; Supplementary Table S1). This approach identified five ECM genes; agrin (AGRN), insulin-like growth factor binding protein, acid-labile subunit (IGFALS), S100 calcium-binding protein A11 (S100A11), serine peptidase inhibitor (SERPINE2) and TIMP metallopeptidase inhibitor 1 (TIMP1), all of which were upregulated in both SSLs and HPs (Figs. 1,2 and Supplementary Table S1) while their expression was lower in TAs.
Figure 2. Differential expression analysis to identify candidate extracellular matrix genes in human colon polyps.
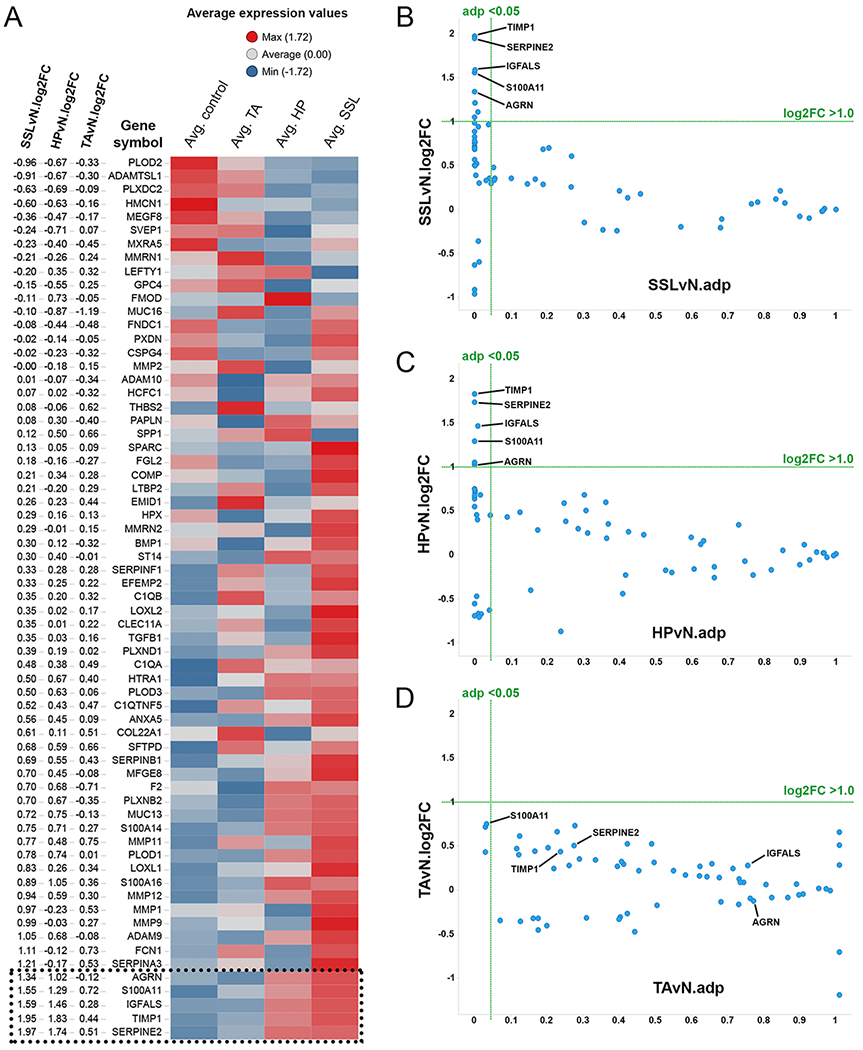
Differential expression analysis of sixty-five genes encoding ECM proteins previously identified to be upregulated in patients with colorectal cancer (26) using RNA-seq data (29) from a variety of colonic polyps including sessile serrated lesions (SSL; n=21), hyperplastic polyp (HP; n=10), tubular adenoma (TA; n=10) and normal colon controls (N; n=20).
A) Row-centered average expression values for all three polyp types and normal controls are plotted in the heatmap. The rows are rank-ordered according to SSLvN log2 fold changes (log2FC) as shown in the first three columns. Five ECM genes which meet the threshold [log2FC >1.0 and adjusted p value (adp) <0.05] in SSL and HP samples are indicated at the bottom of the heatmap. See Supplementary Fig. S1 for gene expression data of the individual samples.
B-D) Scatterplots showing the distribution of expression levels of the sixty-five ECM-protein genes and selection criteria; log2FC >1.0 and adp <0.05 (green lines) among SSL (B), HP (C) and TA (D) samples analyzed. Highlighted are the five genes (AGRN, IGFALS, S100A11, SERPINE2, TIMP1) which meet the selection threshold and are overexpressed in SSLs and HPs but not TAs.
In parallel, we screened and validated (on FFPE material) commercially available antibodies specific for CRC-associated ECM proteins (30). The combined IHC and transcriptomics approaches highlighted three ECM-associated proteins (AGRN, SERPINE2 and TIMP1) with reliable antibodies, which were investigated further (Fig. 1).
AGRN and TIMP1 immunostaining in colorectal polyps
To investigate the potential diagnostic utility of AGRN, SERPINE2 and TIMP1we performed IHC on a cohort of TAs, HPs, SSLs and TSAs (Supplementary Fig. S2) with antibodies to AGRN, SERPINE2 and TIMP1 (Supplementary Fig. S3). We detected strong staining for AGRN in the basal laminae (BL) of blood vessels and crypts of the polyps, and TIMP1 in the cytoplasm of all polyp types investigated (Supplementary Figs. S2A and S3). In the normal colon, AGRN was only present in the BL of blood vessels and negative in the normal colonic crypts. TIMP1 was also negative in the normal colon, however, neuroendocrine cells were positive for TIMP1, as previously described (31). In contrast, variable SERPINE2 cytoplasmic and ECM staining was noted in normal and lesional crypts (Supplementary Fig. S3). Given the lack of a consistent and clear differential reactivity with TIMP1 and SERPINE2 in normal versus polyp tissue, we elected to evaluate AGRN further.
Differential localization of AGRN as a biomarker of SSLs
In total, we evaluated four-hundred-eight colonic polyps representing the following pathologic diagnoses: HP (n=71), SSL with (n=23) and without (n=166) dysplasia, TSA (n=25), TA (n=64), colonic hamartomatous (n=18) and mucosal prolapse polyps (n=3) (Supplementary Figs. S2B–E, S9, Table S2). We found positive AGRN staining of the BL in all colonic lesions, whereas hamartomatous, mucosal prolapse polyps and normal colon mucosa were negative. However, differential patterns of BL localization were noted (Fig. 3); a top-heavy pattern of AGRN positivity was consistently observed in TAs, whereas in TSAs the BL reactivity was uniformly distributed along the length of the colonic crypt. In contrast, in SSL and HP samples, AGRN BL positivity was more prominent in the basal crypts, with weaker staining consistently seen in HPs.
Figure 3. Differential localization of AGRN as a biomarker to distinguish colorectal polyps.
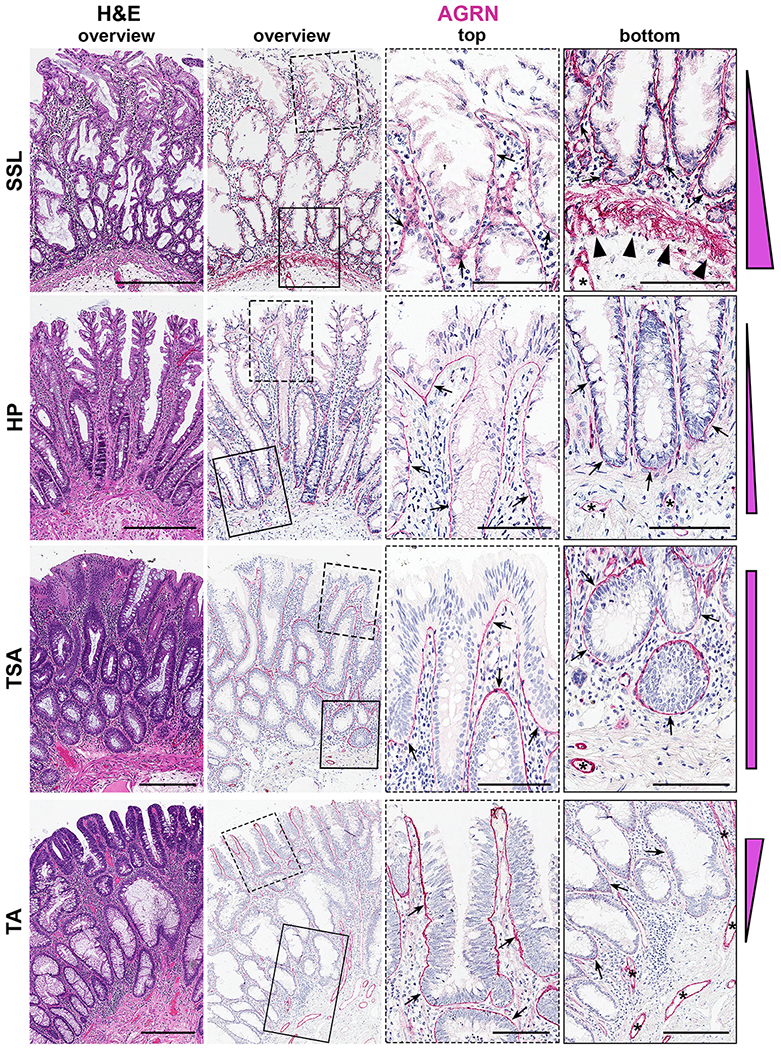
Representative H&E and agrin (AGRN) IHC images of colonic polyps. Presented are overview images (left two columns) and enlarged images for AGRN IHC (boxed areas) from the top and the bottom of the crypt (right two columns) for the individual polyp types.
Note the positive stain in the basal lamina (BL) of all blood vessels (*) and the differential localization patterns of AGRN in the BL (indicated by arrows) of the different types of polyps (also presented schematically at the right edge of each polyp panel). AGRN reactivity is consistently observed as follows: SSLs and HPs (basal crypt predominance); TSAs (top- and bottom-high); TAs (top-high-to-bottom-low). Also note the presence of AGRN in the muscularis mucosa (MM) exclusively in SSL (arrowheads). Scale bars: 300 μm (overviews) and 100 μm (magnifications)
To examine further the biological relevance of these findings, we assessed the BL AGRN positivity in a range of mouse models with conventional adenoma (Supplementary Fig. S4A–D) and those with serrated-like (Supplementary Fig. S4E) morphology. In concordance with the human samples, AGRN immunostaining with two anti-AGRN antibodies confirmed the consistent presence of this protein in the BL of murine colonic polyps while the BL of adjacent normal colonic mucosa was negative (Supplementary Fig. S4). More importantly, the BL distribution patterns of AGRN in the mouse models paralleled the human data (Supplementary Fig. S4). Collectively, the genetically defined mouse models of colonic polyps recapitulate the distribution patterns of AGRN seen in human TAs and TSAs.
AGRN is expressed in epithelial cells and deposited into the muscularis mucosae in SSL
Strikingly, we also noted the selective presence of muscularis mucosa (MM)-based AGRN in human SSLs with and without dysplasia but not in other polyps (Fig. 3 and Supplementary Fig. S3). IHC and confocal immunofluorescence microscopy on consecutive sections of SSLs demonstrated AGRN co-localization with SMA and DES (both muscle-specific markers) in the MM (Fig. 4A–D); AGRN was localized to the upper half of the MM (Fig. 4D). AGRN mRNA expression was predominantly localized to crypt-base cells with stronger expression in SSL as compared with HP, TA and TSA (Supplementary Fig. S5A–H), while the adjacent MM of SSLs was negative (Supplementary Fig. S5E,F, arrowheads). Collectively, these results suggest that AGRN, expressed by colonic epithelial cells, is deposited in the MM in SSLs.
Figure 4. AGRN reactivity in the muscularis mucosae of sessile serrated lesions.
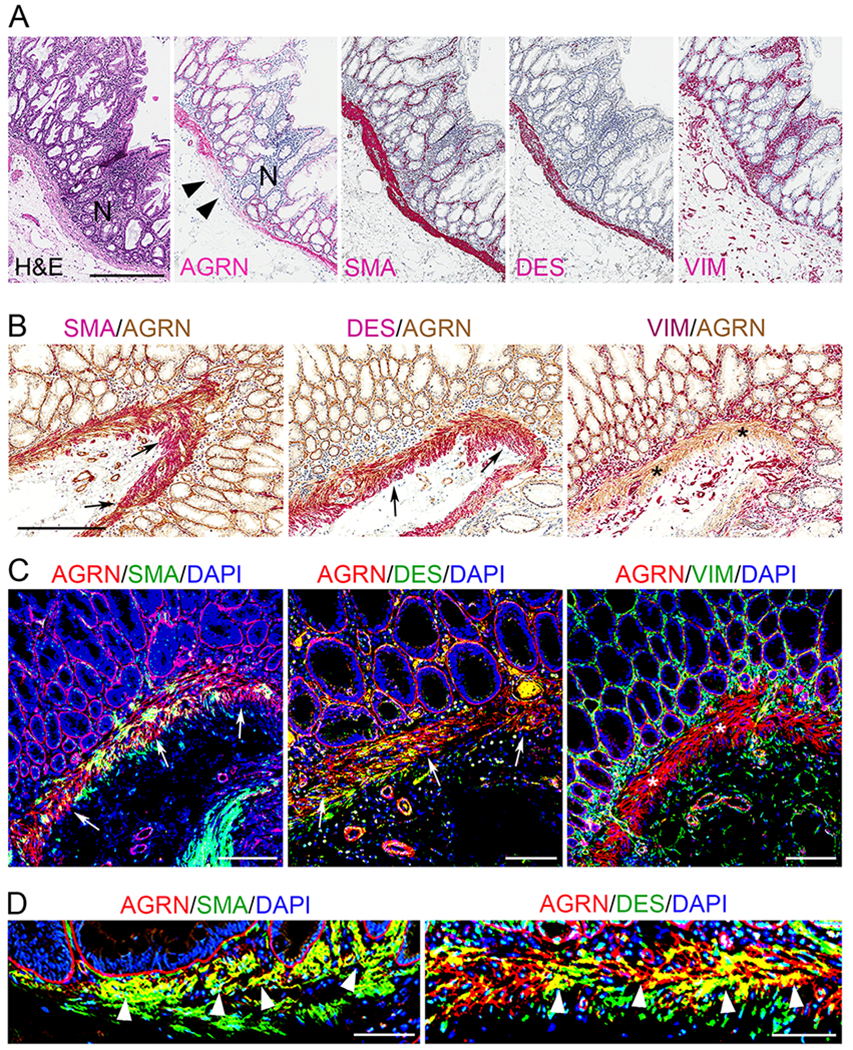
A) Representative H&E and IHC images of parallel sections of SSL comparing AGRN reactivity to smooth-muscle actin (SMA), desmin (DES) and vimentin (VIM). Scale bar: 300 μm
B) IHC images of immunostains for AGRN (brown), SMA, DES and VIM (red) performed on consecutive sections. AGRN colocalizes with SMA and DES but not with VIM. Scale bar: 300 μm
C and D) Confocal immunofluorescence microscopy of SSL tissue sections, comparing the localization of AGRN (red) with that of SMA, DES and VIM (all in green); yellow signal shows colocalization. Cell nuclei are shown in blue. Scale bars: 100 μm (C), 50 μm (D)
AGRN is localized to MM, which is also positive for SMA and DES (A-D) but negative for VIM (*; B and C). Arrows in B and C mark the co-localization of AGRN with SMA and DES (color overlay in B, and yellow merged color in C and D). Also note, AGRN is exclusively present in the MM adjacent to the abnormal crypts of SSL and ends, often abruptly, in the adjacent normal (N) crypts (black arrowheads in A). Magnified images reveal that AGRN mainly localizes to the upper half of the MM (white arrowheads in D).
AGRN positivity of the muscularis mucosae as a biomarker for SSLs
To investigate further the utility of MM-based AGRN reactivity in diagnosing SSLs we investigated twelve anti-AGRN antibodies on human FFPE samples (30), however, we identified only one additional reliable anti-AGRN antibody. Both anti-AGRN antibodies showed similar staining patterns (Supplementary Fig. S5I–L); and selectively stained the MM of SSL.
We next identified fifty SSLs with near-universal diagnostic agreement among nine expert GI pathologists. MM-based AGRN reactivity was noted in all samples (data not shown). In addition, we tested 18 SSL polyps from ten serrated polyposis syndrome (SPS) patients (a single polyp in two patients and 2 polyps in eight patients) and all showed a strong MM-based positivity for AGRN (Supplementary Fig. S6).
To control for bias, since experienced pathologists can recognize a SSL without an AGRN stain, four non-pathologists were asked to blindly validate the MM-positivity for AGRN on one-hundred samples (Supplementary Fig. S7A, Supplementary Table S2), including all cases used in the validation cohort (see below). Notably, there was complete agreement with regard to MM-based reactivity among the four observers (Supplementary Fig. S7B), validating the robustness of this criterion.
Finally, we selected fifty diagnostically challenging HPs, SSLs and TSAs (Supplementary Table S2) and compared the results of AGRN staining with the evaluation performed by nine GI pathologists on H&E sections (see Materials and Methods). In total, samples #1-46 were scored (Fig. 5A); four samples had to be excluded (samples #47-50, Supplementary Fig. S7C). Complete concordance among the pathologists was achieved in only 13 cases (26%), kappa value = 0.493, indicating only weak overall agreement (Fig. 5A, green). The principal source of disagreement lay in the distinction of HP (Fig. 5A, grey) from SSL (Fig. 5A, orange). Based on majority opinion (at least 5 of 9 pathologists) the polyps were classified as follows: 25 SSLs with or without dysplasia, 14 HPs and 4 TSAs (Fig. 5A, purple). Three samples did not yield a majority and the opinions were widely divergent. The MM staining for AGRN was identified (Fig. 5A, red) in 24 of the 25 cases diagnosed as SSL. In contrast, 19 cases that lacked a majority opinion diagnosis of SSL were negative for AGRN in the MM (Fig. 5A, blue). In the three cases lacking a majority opinion (arrows in Fig. 5A), the results differed from those noted above and are illustrated in Fig. 5B–G. Although SSL represented the majority (5 of 9) opinion in case #25, no AGRN reactivity was noted (Fig. 5B,C). Two cases #26 and #27 both showed MM-based AGRN reactivity, however, for #26 no majority opinion was reached and #27 majority (5/9) opinion was HP, although both cases showed contiguous crypts with basal crypt dilatation and basal serrations (Fig. 5D–G). Finally, we also tested fifty-five community pathologists by showing them representative images of H&E slides from eight cases and observed a virtually similar level of inter-observer agreement (Supplementary Fig. S10).
Figure 5. Majority-based polyp validation among expert gastrointestinal pathologists.
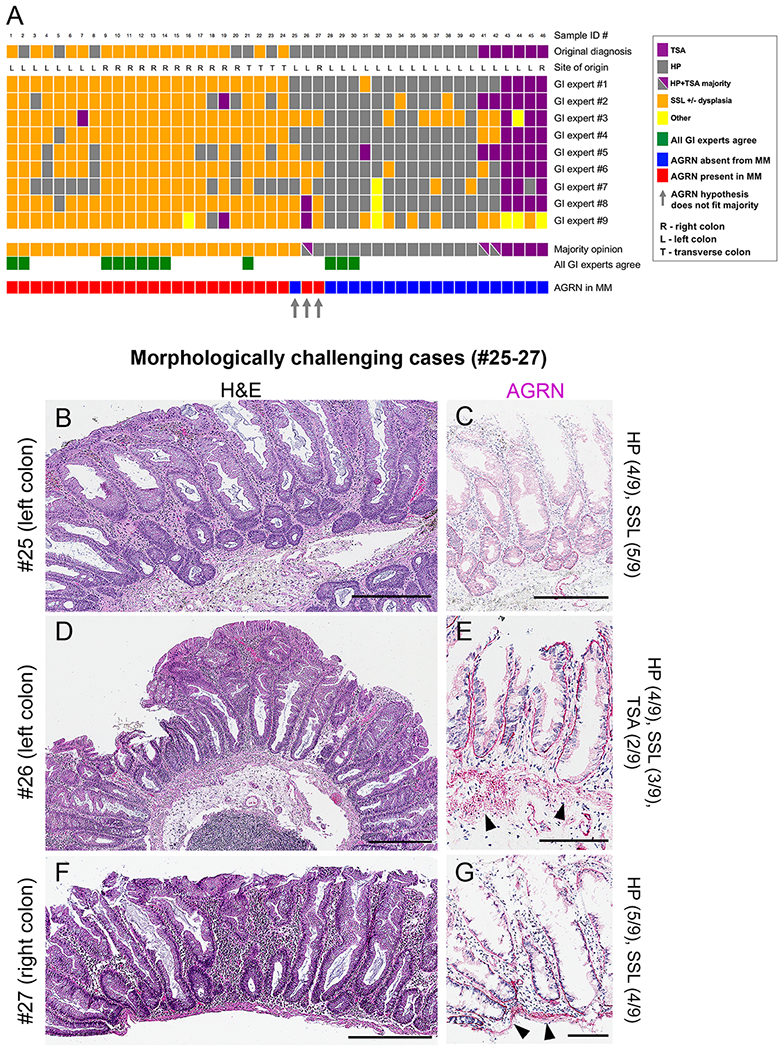
Nine experts in gastrointestinal (GI) pathology classified fifty diagnostically challenging polyps by H&E into: 1) TSA, 2) HP, 3) SSL) +/− dysplasia and 4) other, according to previous WHO criteria (see text for details).
A) Presented are the results of forty-six of these cases (for the four samples not shown see Supplementary Fig. S7). Indicated are the original diagnoses, the site of polyp origin and the individual GI expert opinions. For each case, the majority pathologist’s opinion is compared to the corresponding MM-positivity for AGRN as scored by 4 non-pathologists (Supplementary Fig. S7A). Note the diagnostic variability among the individual GI experts as compared with the high concurrence of AGRN-positive MM staining with SSL (majority opinion) whereas most HPs and TSAs are negative.
B-G) H&E (B,D,F) and corresponding AGRN IHC (C,E,G) images of three morphologically challenging cases (#25-27) as indicated by the three grey arrows in (A) in which the AGRN-positive stain of MM differs from the majority GI expert opinion, although those opinions were widely divergent. (C) Note the absence of AGRN from the MM of sample #25 (~1.8x4.8mm, left colon; 4/9 GI experts classified this as HP and 5/9 as SSL). (E and G) Note AGRN-positive MM (arrowheads) in samples #26 (~2.1x1.6mm, left colon); 4/9 GI experts classified this as HP, 3 as SSL and 2 as TSA and #27 (~0.55x1.8mm, right colon); 5/9 GI experts classified this as HP and 4 as SSL). Notably, these polyps show 2 contiguous crypts with basal dilatation, meeting the WHO definition of SSLs. See also Supplementary Table S2 for individual samples. Scale bars: 300 μm (B,D,F), 100 μm (C,E,G)
Collectively, for all samples investigated in this study, the majority reads of the AGRN staining used for final calculations were as follows: MM-based AGRN positive staining in 186/188 SSLs with (23/23) and without (163/165) dysplasia, 5/68 HPs, 0/64 TA, 0/25 TSA and 0/21 hamartomatous and mucosal prolapse polyps (Supplementary Figs. S7D, S9, Supplementary Table S2). AGRN reactivity also assisted in differentiating TA from SSL with dysplasia when the continuity between the SSL and dysplastic component was lost due to fragmentation of the specimen (Supplementary Fig. S8). Additionally, loss of BL-based AGRN reactivity was noted in the dysplastic portion of SSL (Supplementary Fig. S8D,I,J). In summary, these observations suggest that the MM-based AGRN stain could help to assist in identifying dysplasia arising within SSL in cases in which the SSL component is poorly represented. Given that SSL with dysplasia are more likely to progress to cancer it is important to accurately diagnose a dysplastic component of serrated polyps.
Finally, samples of both subtypes of HPs (MVHP, n=37 and GCHP, n=32) and 9 large HPs (>4.5 mm) were negative for MM-based AGRN (see also Supplementary Table S2).
Power analyses ensured sufficient precision of our estimation. Based on sensitivity and specificity, a 95% confidence interval as narrow as 7% was calculated. Collectively, this provides a reliable estimation of MM-based AGRN as a diagnostic marker for SSLs with sensitivity of 98.9% and specificity of 97.1% (Supplementary Fig. S7D).
RNA-seq-profiled colonic polyps corroborate the biomarker capability of AGRN immunoreactivity
We finally sought to validate our AGRN immunohistochemical findings on the cohort of polyps from the RNA-seq dataset (29) (Figs. 1, 2; Supplementary Fig. S1). Many SSL (n=14) and a few HP samples (n=4) showed higher AGRN expression, most other polyps and controls showed lower AGRN expression (Fig. 6A). We evaluated 10 SSL and 5 HP for MM-based AGRN reactivity (Fig. 6). Blinded evaluation identified 9 out of 10 SSL to be AGRN-positive (data not shown). The majority of these SSLs also showed elevated AGRN expression and formed a distinct cluster (Supplementary Fig. S1), three samples (#14, 18, 20) did not conform with our hypothesis (Fig. 6, Supplementary Fig. S1, see figure legend for details).
Figure 6. AGRN IHC validation on colonic polyp samples previously analyzed by RNA-seq.
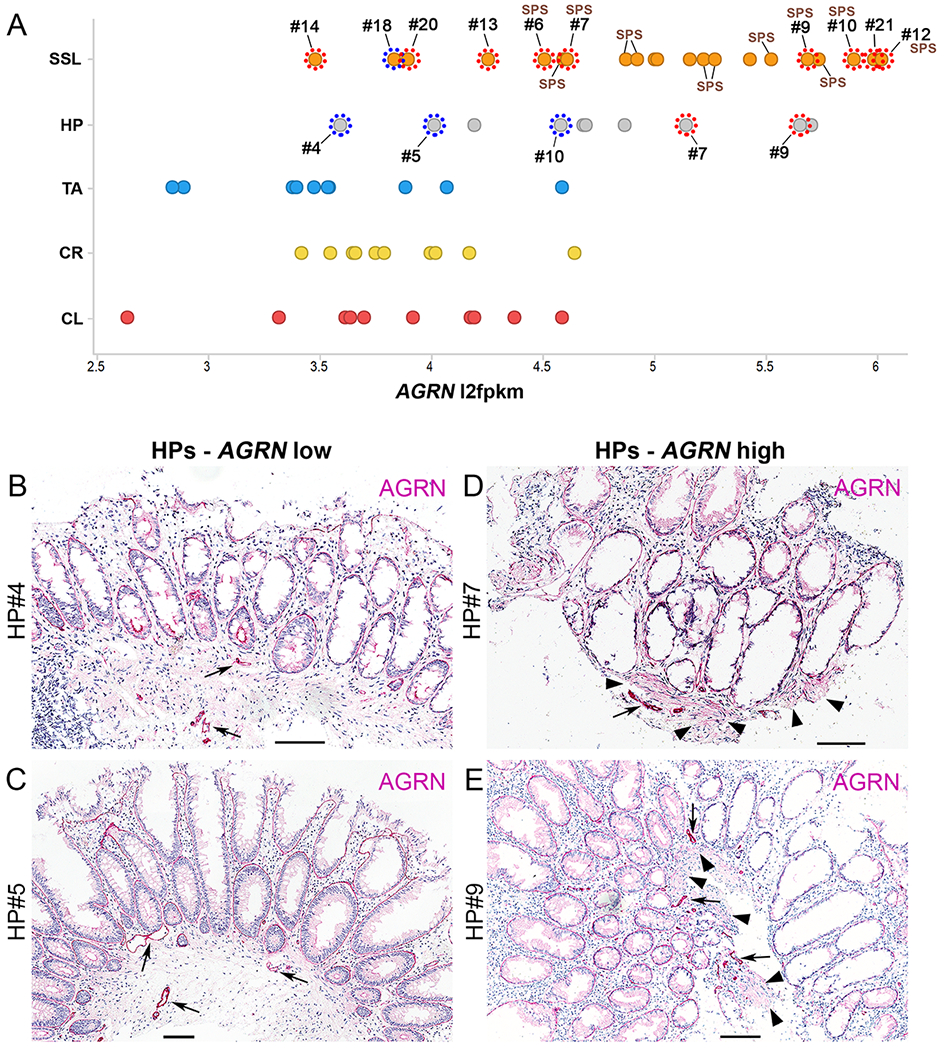
A) Scatter plot of AGRN mRNA expression values for SSL (n=21), HP (n=10), TA (n=10) and colon controls (CL, control left, n=10; CR, control right, n=10) using publicly available RNA-seq data (29) (see Supplementary Fig. S1). Expression values of AGRN (l2fpkm, log2 fragments per kilobase million) for the individual polyps and controls are indicated and diagnosis highlighted by a colored dot: orange (SSL), grey (HP), blue (TA), yellow (CR), red (CL). Note that many SSLs and a few HPs show high AGRN values (above 5) whereas all other samples show lower AGRN expression. Examples of SSL (n=10) and HP (n=5) for which sections were stained for AGRN are indicated by dotted circles; positive (red dotted circles) or negative (blue dotted circles) for MM-based AGRN reactivity. Note that the two designated HP samples showing high AGRN expression levels also scored positive for MM-based AGRN, whereas those with lower overall AGRN expression were also negative for AGRN MM staining. Most SSL samples scored positive for MM-based AGRN staining. SSL samples from serrated polyposis syndrome (SPS) patients are indicated.
B-E) AGRN IHC images of sections from four HPs samples from the RNA-seq dataset (29) presented in A. Shown are sections of HP samples in which the AGRN expression values are low [HP#4 (B), HP#5 (C)] or high [HP#7 (D), HP#9 (E)]. Note also in two of five HP samples the additional positivity for AGRN in the MM (HP#7, HP#9, arrowheads in D and E). Notably, these two cases cluster with the majority of SSLs whereas HPs#4 and HP#5 (negative for MM-based AGRN) cluster separately (Supplementary Fig. S1). Scale bars: 100 μm
Finally, in agreement with our prior observation (see above) all 12 samples from SPS patients in this cohort (29) clustered together and showed high AGRN expression (Fig. 6A, Supplementary Fig. S1). In addition, of these samples all six polyps that we stained were positive for MM-based AGRN reactivity (Fig. 6A).
Discussion
SSLs have a risk of malignant progression and require accelerated screening for CRC. However, in routine clinical practice, histopathological characterization of SSLs has met significant challenges; overlapping histological features with HPs, poor biopsy orientation, resulting in moderate-to-poor inter-observer agreement among pathologists (15–17,19,20). A biomarker specific for SSLs would clearly help pathologists and allow evidence-based surveillance of serrated polyps, and the results presented here suggest that MM-based AGRN reactivity could serve as such an objective marker for SSL.
There have been several prior attempts to identify SSL-specific biomarkers. Caruso et al (32) performed a gene-array study and compared SSLs to TAs and controls. SSLs showed upregulation of CTSE and TFF1. ANXA10 was identified in SSLs as compared to HPs and validated by IHC as a potential diagnostic marker with a sensitivity of 73% and specificity of 95% in the diagnosis of SSL (24). MUC6 was found to be expressed in SSL but not in HP (21), however, subsequent studies revealed a relatively low sensitivity (22) and lack of specificity for SSL (23). In addition, Delker et al (33) examined gene expression to discriminate between SSLs and HPs and found unique staining patterns for the cell junction protein VSIG1 and MUC17 in SSLs. Finally, the loss of the transcription factor HES1, was observed in the majority of SSLs compared to normal expression in HPs (25), however, TA and also TSA showed variable staining for HES1, diminishing its value as a marker of SSL. Notably, none of these potential biomarkers is currently applied in routine clinical practice.
ECM proteins play an important role in the initiation and progression of colon carcinomas, often related to poor prognosis (34,35). The diagnostic relevance of ECM proteins in CRC and their applicability to distinguish individual colonic polyps so far remains untested. Our results demonstrate the differential expression of several ECM genes among colonic polyps and show that AGRN, SERPINE2 and TIMP1 are overexpressed in SSL and HP samples (Figs. 1, 2; Supplementary Figs. S1, S3). Colonic polyps show distinct patterns of AGRN deposition in BL. Importantly, MM-based AGRN reactivity was restricted to SSLs (Figs. 3, 4, Supplementary Figs. S3, S5). We confirmed this finding by evaluating a cohort of fifty diagnostically challenging colonic polyps with only weak agreement among GI pathologists. Virtually all cases with a majority read of SSL showed MM-based AGRN reactivity, while non-SSL polyps lacked AGRN reactivity in the MM. Another strong validation of the diagnostic value of MM-based AGRN reactivity is the near-universal presence of MM-based AGRN in SPS patients.
The ECM molecule AGRN, a large multidomain heparan sulfate proteoglycan, is abundantly expressed in developing brain and in virtually all BL of developing organs. Functionally, AGRN aids in the formation of neuromuscular junctions and acetylcholine receptor clustering in the central nervous system (36). The non-neuronal functions of AGRN are only poorly understood. Although little is known about AGRN in carcinogenesis, in recent years, a tumor-promoting role has been reported for several cancer types, including hepatocellular and cholangiocellular carcinoma (37,38), prostate cancer (39) and oral squamous cell carcinoma (40). Functional studies have shown that AGRN is involved in proliferation, migration and invasion of liver cancer cells by regulating focal adhesion integrity and to relay mechanosensitive signals into cells to regulate YAP activity to promote tumorigenesis (41,42). In contrast, the role of AGRN in normal colorectal mucosa, colonic polyps and CRC has not been investigated extensively. By tissue secretome profiling, AGRN was one of seventy-six potential CRC protein biomarkers that may facilitate blood- or stool-based assay development to support clinical management of CRC (43). Another analysis of the extracellular proteome of CRC cells identified elevated levels of the C-terminal fragment of AGRN (44), however, its suitability as a biomarker has not yet been assessed.
Although expressed by colonic epithelial cells, AGRN is specifically localized in the MM of SSL. AGRN interacts with several other ECM proteins and a variety of growth factors. In addition, earlier studies have shown AGRN to bind cell-surface adhesion receptors, such as NCAM, integrins, α-dystroglycan and muscle-specific kinase (36). The determination of whether any of these proteins co-localize at the MM of SSLs and interact with AGRN will therefore be of interest.
In conclusion, our study supports the use of MM-based reactivity for AGRN as a novel biomarker to differentiate SSL from HP. Our study uses a consensus opinion as the ‘comparison standard’, and MM-based AGRN expression compared very favorably with this standard, outperforming H&E-based diagnosis by expert pathologists in a set of challenging SSL; whereas in a set of straightforward SSL the results of AGRN staining were concordant with virtually all pathologists. Finally, MM-based AGRN was also universally detected in SPS syndrome. Thus, MM-based immunostaining for AGRN may enable more accurate diagnosis of patients with SSLs and assist with morphologically challenging cases.
Supplementary Material
Translational Relevance.
Sessile serrated lesions (SSL) are precursors of colon carcinoma. Their distinction from other polyps with better prognoses, most notably hyperplastic polyps (HPs), often presents a significant diagnostic challenge and histological evaluation shows only weak inter-observer agreement among experts. We identified the extracellular matrix protein agrin in the muscularis-mucosae (MM) of SSL biopsies but not in other polyps or normal colonic tissue. Immunohistochemical staining of the MM for agrin presents a novel biomarker with high specificity and sensitivity and markedly improves the discrimination among polyp subtypes. Immunostaining for agrin in the muscularis mucosae enables pathologists towards more accurate diagnoses of patients with SSLs, can assist with morphologically challenging cases and may allow evidence-based surveillance of serrated polyps.
Acknowledgements
The authors wish to thank Chenxi Tian for assistance in using the confocal laser-scanning microscope, and all members of the Hynes laboratory for advice and discussions. We thank the Swanson Biotechnology Center at the Koch Institute/MIT, especially Mike S. Brown and Kathleen S. Cormier from the Hope Babette Tang (1983) Histology Facility and Jeff Wykoff in the Microscopy Facility for exceptional technical support, Duanduan Ma from the Barbara K. Ostrom Bioinformatics & Computing Facility for assistance with bioinformatic analyses and power calculations, and Sven Holder for sample sectioning. We also thank Lucia Suarez-Lopez (Koch Institute), Roderick T. Bronson (Harvard Medical School) and Mari Mino-Kenudson, Jeck Williams and Martin S. Taylor (Massachusetts General Hospital) for discussion, data collection and advice. The authors also thank Eric R. Fearon (Department of Internal Medicine, University of Michigan, Ann Arbor, MI) for kindly providing slides of colon sections from control and CDX2P-CreERT2 CDX2fl/fl;BrafLSL-V600E/+ mice and Mary Bronner (Department of Pathology, University of Utah, Salt Lake City, UT) for critically reading the manuscript and providing samples previously described by Kanth et al. (2016, ref. 29).
Funding:
This work was supported by NIH grants U54-CA163109 (Tumor Microenvironment Network to ROH) and R01 CA211184, R01 CA034992 (to OHY), the MIT Ludwig Center for Molecular Oncology and the Howard Hughes Medical Institute, of which ROH is an investigator. OHY was supported by the Pew Foundation, Sidney Kimmel Foundation and MIT Center for Stem Cell Research. Facility support was provided by the Koch Institute Swanson Biotechnology Center (Cancer Center Support Grant NIH-P30CA014051). SR was supported by postdoctoral fellowships from the Deutsche Forschungsgemeinschaft (DFG) RI2408/1-1 and the MIT Ludwig Center for Molecular Oncology. CP was supported by the MGH ECOR Tosteson and Fund for Medical Discovery Fellowship; and JR by NIH/NCI (1K08CA198002-01).
Abbreviations:
- AGRN
agrin
- BL
basal lamina
- CRC
colon carcinoma
- CL
control left
- CR
control right
- CRC
colorectal cancer
- DES
desmin
- ECM
extracellular matrix
- FFPE
formaldehyde-fixed and paraffin-embedded
- GCHP
goblet cell hyperplastic polyp
- GI
gastrointestinal
- H&E
hematoxylin-eosin
- HIER
heat-induced epitope-retrieval
- HP
hyperplastic polyp
- IBD
inflammatory bowel disease
- IGFALS
insulin-like growth factor binding protein acid labile subunit
- IHC
immunohistochemistry
- ISH
in situ hybridization
- JPS
juvenile polyps
- mAbs
monoclonal antibodies
- MM
muscularis mucosae
- MVHP
microvesicular hyperplastic polyp
- PJP
Peutz-Jeghers polyps
- RNA-seq
RNA sequencing
- S100A11
S100 calcium-binding protein A11
- SERPINE2
serine peptidase inhibitor, clade E, member 2
- SMA
smooth-muscle actin
- SPS
serrated polyposis syndrome
- SSL
sessile serrated lesions
- TA
tubular adenoma
- TIMP1
TIMP metallopeptidase inhibitor 1
- TSA
traditional serrated adenoma
- VIM
vimentin
Footnotes
Competing interests: The authors declare that they have no competing financial interests.
References
- 1.Limketkai BN, Lam-Himlin D, Arnold MA, Arnold CA. The cutting edge of serrated polyps: a practical guide to approaching and managing serrated colon polyps. Gastrointest Endosc 2013;77(3):360–75. [DOI] [PubMed] [Google Scholar]
- 2.Noffsinger AE. Serrated polyps and colorectal cancer: new pathway to malignancy. Annu Rev Pathol 2009;4:343–64. [DOI] [PubMed] [Google Scholar]
- 3.JE IJ, Medema JP, Dekker E. Colorectal neoplasia pathways: state of the art. Gastrointest Endosc Clin N Am 2015;25(2):169–82. [DOI] [PubMed] [Google Scholar]
- 4.Bosman FT, Carneiro F, Hruban R. World Health Organization classification of tumours of the digestive system. Lyon (France): IARC Press; 2010. [Google Scholar]
- 5.Rex DK, Ahnen DJ, Baron JA, Batts KP, Burke CA, Burt RW, et al. Serrated lesions of the colorectum: review and recommendations from an expert panel. Am J Gastroenterol 2012;107(9):1315–29; quiz 4, 30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Snover DC, Ahnen DJ, Burt RW, Odze RD. Serrated polyps of the colon and rectum and serrated polyposis In: Bosman FT, Carneiro F, Hruban RH, Theise ND, editors. WHO Classification of Tumours of the Digestive System. 4th ed. Volume 3, World Health Organization Classification of Tumours. Lyon, France: IARC Press; 2010. p 160–5. [Google Scholar]
- 7.WHO Classification of Tumours. Editorial Board Digestive System Tumours: WHO Classification of Tumours of the Digestive System, 5th Edition IARC; 2019. [Google Scholar]
- 8.East JE, Atkin WS, Bateman AC, Clark SK, Dolwani S, Ket SN, et al. British Society of Gastroenterology position statement on serrated polyps in the colon and rectum. Gut 2017;66(7):1181–96. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Torlakovic E, Skovlund E, Snover DC, Torlakovic G, Nesland JM. Morphologic reappraisal of serrated colorectal polyps. Am J Surg Pathol 2003;27(1):65–81. [DOI] [PubMed] [Google Scholar]
- 10.Snover DC. Update on the serrated pathway to colorectal carcinoma. Hum Pathol 2011;42(1):1–10. [DOI] [PubMed] [Google Scholar]
- 11.Bettington ML, Chetty R. Traditional serrated adenoma: an update. Hum Pathol 2015;46(7):933–8. [DOI] [PubMed] [Google Scholar]
- 12.Khalid O, Radaideh S, Cummings OW, O’Brien MJ, Goldblum JR, Rex DK. Reinterpretation of histology of proximal colon polyps called hyperplastic in 2001. World J Gastroenterol 2009;15(30):3767–70. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Wong NA, Hunt LP, Novelli MR, Shepherd NA, Warren BF. Observer agreement in the diagnosis of serrated polyps of the large bowel. Histopathology 2009;55(1):63–6. [DOI] [PubMed] [Google Scholar]
- 14.Mohammadi M, Garbyal RS, Kristensen MH, Madsen PM, Nielsen HJ, Holck S. Sessile serrated lesion and its borderline variant - Variables with impact on recorded data. Pathol Res Pract 2011;207(7):410–6. [DOI] [PubMed] [Google Scholar]
- 15.Glatz K, Pritt B, Glatz D, Hartmann A, O’Brien MJ, Blaszyk H. A multinational, internet-based assessment of observer variability in the diagnosis of serrated colorectal polyps. Am J Clin Pathol 2007;127(6):938–45. [DOI] [PubMed] [Google Scholar]
- 16.Sandmeier D, Seelentag W, Bouzourene H. Serrated polyps of the colorectum: is sessile serrated adenoma distinguishable from hyperplastic polyp in a daily practice? Virchows Arch 2007;450(6):613–8. [DOI] [PubMed] [Google Scholar]
- 17.Farris AB, Misdraji J, Srivastava A, Muzikansky A, Deshpande V, Lauwers GY, et al. Sessile serrated adenoma: challenging discrimination from other serrated colonic polyps. Am J Surg Pathol 2008;32(1):30–5. [DOI] [PubMed] [Google Scholar]
- 18.Lieberman DA, Rex DK, Winawer SJ, Giardiello FM, Johnson DA, Levin TR. Guidelines for colonoscopy surveillance after screening and polypectomy: a consensus update by the US Multi-Society Task Force on Colorectal Cancer. Gastroenterology 2012;143(3):844–57. [DOI] [PubMed] [Google Scholar]
- 19.Payne SR, Church TR, Wandell M, Rosch T, Osborn N, Snover D, et al. Endoscopic detection of proximal serrated lesions and pathologic identification of sessile serrated adenomas/polyps vary on the basis of center. Clin Gastroenterol Hepatol 2014;12(7):1119–26. [DOI] [PubMed] [Google Scholar]
- 20.Tinmouth J, Henry P, Hsieh E, Baxter NN, Hilsden RJ, Elizabeth McGregor S, et al. Sessile serrated polyps at screening colonoscopy: have they been under diagnosed? Am J Gastroenterol 2014;109(11):1698–704. [DOI] [PubMed] [Google Scholar]
- 21.Owens SR, Chiosea SI, Kuan SF. Selective expression of gastric mucin MUC6 in colonic sessile serrated adenoma but not in hyperplastic polyp aids in morphological diagnosis of serrated polyps. Mod Pathol 2008;21(6):660–9. [DOI] [PubMed] [Google Scholar]
- 22.Bartley AN, Thompson PA, Buckmeier JA, Kepler CY, Hsu CH, Snyder MS, et al. Expression of gastric pyloric mucin, MUC6, in colorectal serrated polyps. Mod Pathol 2010;23(2):169–76. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Gibson JA, Hahn HP, Shahsafaei A, Odze RD. MUC expression in hyperplastic and serrated colonic polyps: lack of specificity of MUC6. Am J Surg Pathol 2011;35(5):742–9. [DOI] [PubMed] [Google Scholar]
- 24.Gonzalo DH, Lai KK, Shadrach B, Goldblum JR, Bennett AE, Downs-Kelly E, et al. Gene expression profiling of serrated polyps identifies annexin A10 as a marker of a sessile serrated adenoma/polyp. J Pathol 2013;230(4):420–9. [DOI] [PubMed] [Google Scholar]
- 25.Cui M, Awadallah A, Liu W, Zhou L, Xin W. Loss of Hes1 Differentiates Sessile Serrated Adenoma/Polyp From Hyperplastic Polyp. Am J Surg Pathol 2016;40(1):113–9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Naba A, Clauser KR, Whittaker CA, Carr SA, Tanabe KK, Hynes RO. Extracellular matrix signatures of human primary metastatic colon cancers and their metastases to liver. BMC Cancer 2014;14:518. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.O’Brien MJ. Hyperplastic and serrated polyps of the colorectum. Gastroenterol Clin North Am 2007;36(4):947–68, viii. [DOI] [PubMed] [Google Scholar]
- 28.Syngal S, Brand RE, Church JM, Giardiello FM, Hampel HL, Burt RW, et al. ACG clinical guideline: Genetic testing and management of hereditary gastrointestinal cancer syndromes. Am J Gastroenterol 2015;110(2):223–62; quiz 63. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Kanth P, Bronner MP, Boucher KM, Burt RW, Neklason DW, Hagedorn CH, et al. Gene Signature in Sessile Serrated Polyps Identifies Colon Cancer Subtype. Cancer Prev Res (Phila) 2016;9(6):456–65. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rickelt S, Hynes RO. Antibodies and methods for immunohistochemistry of extracellular matrix proteins. Matrix Biol 2018;71-72:10–27. [DOI] [PubMed] [Google Scholar]
- 31.Sorensen IV, Fenger C, Winther H, Foged NT, Lademann U, Brunner N, et al. Characterization of anti-TIMP-1 monoclonal antibodies for immunohistochemical localization in formalin-fixed, paraffin-embedded tissue. J Histochem Cytochem 2006;54(10):1075–86. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Caruso M, Moore J, Goodall GJ, Thomas M, Phillis S, Tyskin A, et al. Over-expression of cathepsin E and trefoil factor 1 in sessile serrated adenomas of the colorectum identified by gene expression analysis. Virchows Arch 2009;454(3):291–302. [DOI] [PubMed] [Google Scholar]
- 33.Delker DA, McGettigan BM, Kanth P, Pop S, Neklason DW, Bronner MP, et al. RNA sequencing of sessile serrated colon polyps identifies differentially expressed genes and immunohistochemical markers. PLoS One 2014;9(2):e88367. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Calon A, Lonardo E, Berenguer-Llergo A, Espinet E, Hernando-Momblona X, Iglesias M, et al. Stromal gene expression defines poor-prognosis subtypes in colorectal cancer. Nat Genet 2015;47(4):320–9. [DOI] [PubMed] [Google Scholar]
- 35.Crotti S, Piccoli M, Rizzolio F, Giordano A, Nitti D, Agostini M. Extracellular Matrix and Colorectal Cancer: How Surrounding Microenvironment Affects Cancer Cell Behavior? J Cell Physiol 2017;232(5):967–75. [DOI] [PubMed] [Google Scholar]
- 36.Bezakova G, Ruegg MA. New insights into the roles of agrin. Nat Rev Mol Cell Biol 2003;4(4):295–308. [DOI] [PubMed] [Google Scholar]
- 37.Tatrai P, Dudas J, Batmunkh E, Mathe M, Zalatnai A, Schaff Z, et al. Agrin, a novel basement membrane component in human and rat liver, accumulates in cirrhosis and hepatocellular carcinoma. Lab Invest 2006;86(11):1149–60. [DOI] [PubMed] [Google Scholar]
- 38.Batmunkh E, Tatrai P, Szabo E, Lodi C, Holczbauer A, Paska C, et al. Comparison of the expression of agrin, a basement membrane heparan sulfate proteoglycan, in cholangiocarcinoma and hepatocellular carcinoma. Hum Pathol 2007;38(10):1508–15. [DOI] [PubMed] [Google Scholar]
- 39.Li X, Wang X, Song W, Xu H, Huang R, Wang Y, et al. Oncogenic Properties of NEAT1 in Prostate Cancer Cells Depend on the CDC5L-AGRN Transcriptional Regulation Circuit. Cancer Res 2018;78(15):4138–49. [DOI] [PubMed] [Google Scholar]
- 40.Rivera C, Zandonadi FS, Sanchez-Romero C, Soares CD, Granato DC, Gonzalez-Arriagada WA, et al. Agrin has a pathological role in the progression of oral cancer. Br J Cancer 2018;118(12):1628–38. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Chakraborty S, Lakshmanan M, Swa HL, Chen J, Zhang X, Ong YS, et al. An oncogenic role of Agrin in regulating focal adhesion integrity in hepatocellular carcinoma. Nat Commun 2015;6:6184. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Chakraborty S, Njah K, Pobbati AV, Lim YB, Raju A, Lakshmanan M, et al. Agrin as a Mechanotransduction Signal Regulating YAP through the Hippo Pathway. Cell Rep 2017;18(10):2464–79. [DOI] [PubMed] [Google Scholar]
- 43.de Wit M, Kant H, Piersma SR, Pham TV, Mongera S, van Berkel MP, et al. Colorectal cancer candidate biomarkers identified by tissue secretome proteome profiling. J Proteomics 2014;99:26–39. [DOI] [PubMed] [Google Scholar]
- 44.Klein-Scory S, Kubler S, Diehl H, Eilert-Micus C, Reinacher-Schick A, Stuhler K, et al. Immunoscreening of the extracellular proteome of colorectal cancer cells. BMC Cancer 2010;10:70. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.


