Abstract
Introduction
Exhaled-breath analysis of volatile organic compounds could detect lung cancer earlier, possibly leading to improved outcomes. Combining exhaled-breath data with clinical parameters may improve lung cancer diagnosis.
Methods
Based on data from a previous multi-centre study, this article reports additional analyses. 138 subjects with non-small cell lung cancer (NSCLC) and 143 controls without NSCLC breathed into the Aeonose. The diagnostic accuracy, presented as area under the receiver operating characteristic curve (AUC-ROC), of the Aeonose itself was compared with 1) performing a multivariate logistic regression analysis of the distinct clinical parameters obtained, and 2) using this clinical information beforehand in the training process of the artificial neural network (ANN) for the breath analysis.
Results
NSCLC patients (mean±sd age 67.1±9.1 years, 58% male) were compared with controls (62.1±7.0 years, 40.6% male). The AUC-ROC of the classification value of the Aeonose itself was 0.75 (95% CI 0.69–0.81). Adding age, number of pack-years and presence of COPD to this value in a multivariate regression analysis resulted in an improved performance with an AUC-ROC of 0.86 (95% CI 0.81–0.90). Adding these clinical variables beforehand to the ANN for classifying the breath print also led to an improved performance with an AUC-ROC of 0.84 (95% CI 0.79–0.89).
Conclusions
Adding readily available clinical information to the classification value of exhaled-breath analysis with the Aeonose, either post hoc in a multivariate regression analysis or a priori to the ANN, significantly improves the diagnostic accuracy to detect the presence or absence of lung cancer.
Short abstract
Adding readily available clinical information to the classification value of exhaled-breath analysis with the Aeonose significantly improves the diagnostic accuracy to detect the presence or absence of lung cancer http://bit.ly/38ps6fH
Introduction
Lung cancer remains the leading cause of cancer-related death worldwide, accounting for ∼5% of total mortality in many countries [1]. Unfortunately, most patients present with late-stage disease (stages III and IV) and there are limited effective treatment options with consequently high mortality rates (5-year survival rate <10%) [2, 3]. Currently, the only screening method leading to reduced lung cancer mortality in high-risk groups is annual low-dose computed tomography (LDCT) [4, 5]. However, LDCT screening for lung cancer has also resulted in a notable rate of false-positive cases, leading to unnecessary invasive procedures, risks due to radiation exposure and unnecessary anxiety [4–8]. In Europe, results of the Dutch–Belgian lung cancer screening trial (NELSON) are awaited before a decision on implementation of screening programmes in Europe will be made [9]. Hence, there is an increasing demand for innovative, noninvasive, point-of-care diagnostic tools to detect lung cancer at an early stage.
Exhaled-breath analysis with electronic nose technology is a technique based on detecting combinations of volatile organic compounds (VOCs) that are exhaled in very low concentrations. These VOCs reflect pathophysiological processes, such as infection, inflammation and neoplasms [10–12]. VOCs are of interest since they might be directly related to the presence of diseases, they can be tested noninvasively and pattern recognition techniques can serve as classifiers for diseases. Several studies on exhaled-breath analysis have supported the hypothesis that VOC patterns alter when lung cancer is present [13–20].
Recently, we reported results of a study (including 290 subjects) differentiating subjects with lung cancer, including classification into subtypes of lung cancer, from healthy individuals by means of exhaled-breath analysis with the Aeonose (The eNose Company, Zutphen, the Netherlands) [13]. In this study, an artificial neural network (ANN) was trained using exhaled-breath data only.
The Aeonose was able to diagnose patients with non-small cell lung cancer (NSCLC) with a sensitivity of 94%, a negative predictive value (NPV) of 85% and an area under the receiver operating characteristic curve (AUC-ROC) of 0.76. Subtyping NSCLC into adenocarcinoma and squamous cell carcinoma also showed promising results.
These diagnostic parameters were based on the analysis of exhaled VOCs only and did not take into account any of the subjects' risk factors, such as age, sex, smoking status (number of pack-years) and presence of COPD. This paper describes the potential of adding specific clinical information to the classification value obtained from the Aeonose on the diagnostic accuracy to diagnose lung cancer. The hypothesis was that adding clinical information would improve the diagnostic performance. This was assessed in two ways: first, the clinical information was added afterwards to the classification value of the Aeonose as obtained from the ANN by applying multivariate logistic regression analysis, and second, by using this clinical information a priori in the training process of the ANN.
Methods
Data were obtained from a previous prospective, multi-centre study where subjects suspected for lung cancer, as well as healthy volunteers, were asked to participate [13]. The originally collected breath samples were currently used for additional analyses. The four secondary teaching hospitals participating were Medisch Spectrum Twente (Enschede, the Netherlands), Bernhoven (Uden, the Netherlands), Medisch Centrum Leeuwarden (Leeuwarden, the Netherlands) and Deventer Ziekenhuis (Deventer, the Netherlands). For patients with confirmed lung cancer based on histopathology, staging was established according to the seventh edition of the American Joint Committee on Cancer TNM (tumour, node, metastasis) staging system [21]. The control group consisted of suspected subjects with a rejected diagnosis based on imaging and/or derived histopathology and healthy volunteers. Healthy volunteers with a minimum age of 50 years were recruited through an advertisement on the hospitals' websites. There were two exclusion criteria for all subjects: another active malignancy in the past 5 years or the inability to perform a complete Aeonose measurement. Demographic data were collected including age, sex, body mass index, smoking status, number of pack-years, and presence of COPD, hypertension and diabetes mellitus.
The Aeonose is a handheld electronic nose device containing three metal-oxide sensors [13, 22, 23]. This device is a noninvasive, easy-to-use, low-cost tool that is, once trained and validated, able to perform real-time analysis to detect lung cancer. Temporary storage of the breath sample is not required. Subjects were instructed to breathe through the Aeonose for 5 minutes with their nose clipped to prevent nose breathing.
The study protocol was approved by the medical ethics committee of Medisch Spectrum Twente and by the board of directors at each participating centre. All subjects gave informed consent.
Statistical analysis
Continuous variables are reported as mean with corresponding standard deviation or as median with interquartile range. Nominal variables are reported as numbers with corresponding percentages. To assess differences between the groups, either the t-test for normally distributed continuous variables, Mann–Whitney U-test for skewed distributed continuous or ordinal variables, or Chi-squared test for nominal and categorical variables was applied. Number of pack-years was categorised as 0, 1–20, 21–40 and >40 pack-years. Based on clinical reasoning, we assumed a strong relationship between smoking status and number of pack-years, which was confirmed (p<0.001). Number of pack-years contained most relevant information. Therefore, we excluded smoking status as clinical variable from the multivariate analysis.
Data from exhaled breath were analysed using Aethena, a proprietary, dedicated software package from The eNose Company. The software package comprises techniques for data pre-treatment, data compression methods, ANN training and classification to assess the probability of lung cancer, ranging a single value between −1 and 1 [23]. ANNs have been developed as an alternative statistical technique to perform multifactorial analyses by interconnecting nodes with weighted connection lines to predict outcomes or classifying values on an individualised basis [24]. Sensitivity, specificity, positive predictive value (PPV) and NPV were calculated for the diagnosis of lung cancer based on the classification by the Aeonose, and receiver operating characteristics (ROC) curves were composed with a corresponding AUC-ROC with 95% confidence interval.
Clinical variables that were univariately associated with the presence of lung cancer (p<0.15) were entered in a multivariate logistic regression analysis where variables with the highest p-values were eliminated step-by-step (backward method) until the fit of the model decreased significantly, based on the −2 log likelihood. This analysis was based on clinical variables only.
Subsequently, two types of multivariate analysis were performed where breath data were included. First, another multivariate logistic regression analysis, consistent with the aforementioned method, was performed together with the classification value of the Aeonose as obtained from the ANN. Second, clinical variables that were univariately associated with the presence of lung cancer (p<0.15) were added to the vector containing breath profile information once data compression had been realised. These extended vectors (one per subject) were used for training the ANN.
Sensitivity, specificity, positive predictive value (PPV), NPV and AUC-ROC were then calculated for the diagnosis of lung cancer according to the selected multivariate logistic regression model and the extended ANN. These outcomes were compared with the diagnostic accuracy obtained by the classification result of the exhaled-breath analysis only. The multivariate regression model was internally validated by 1000 iterations of bootstrap.
All statistical tests were two-sided with a significance level at 0.05. SPSS version 24.0 (IBM, Armonk, NY, USA) was used to perform statistical mathematics.
Results
A total of 281 subjects were included, of whom 138 had confirmed NSCLC. The control group consisted of 143 subjects without lung cancer of whom 59 were suspected for lung cancer but were considered negative after investigation, and 84 subjects were healthy volunteers (figure 1). Table 1 provides a description of the study participants including clinical characteristics for both groups.
FIGURE 1.
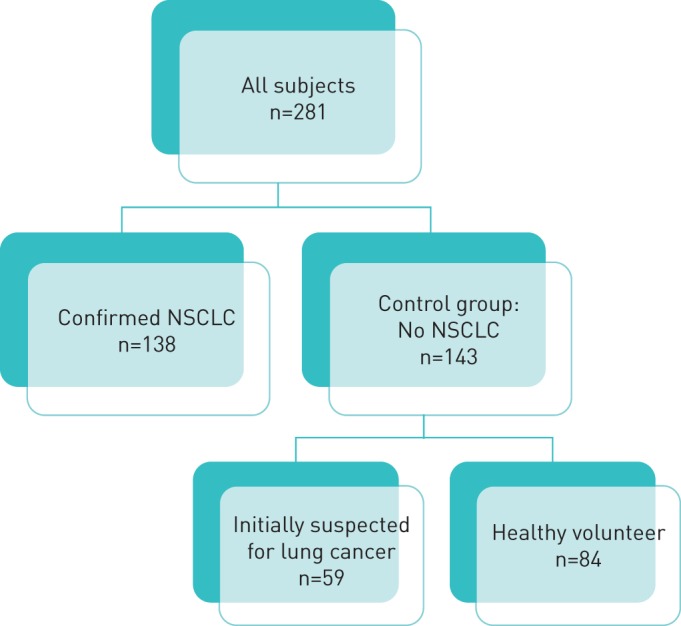
Flow chart showing the different groups. NSCLC: non-small cell lung cancer.
TABLE 1.
Clinical characteristics of subjects
| Confirmed NSCLC | Total control group | Suspected, proven negative | Healthy volunteer | p-value | |
| Subjects n | 138 | 143 | 59 | 84 | |
| Age years | 67.1±9.1 | 62.1±7.0 | 65.2±8.8 | 59.8±4.3 | <0.001¶ |
| Males | 80 (58.0%) | 58 (40.6%) | 31 (52.5%) | 27 (32.1%) | <0.001+ |
| Smoking status | |||||
| Current smokers | 49 (35.5%) | 19 (13.3%) | 13 (22.0%) | 6 (7.1%) | <0.001+ |
| Ex-smokers | 82 (59.4%) | 76 (53.1%) | 32 (54.2%) | 44 (52.4%) | |
| Never-smokers | 7 (5.1%) | 48 (33.6%) | 14 (23.7%) | 34 (40.5%) | |
| Smoking exposure pack-years | |||||
| 0 | 7 (5.1%) | 48 (33.6%) | 14 (23.7%) | 34 (40.5%) | |
| 1–20 | 30 (21.7%) | 53 (37.1%) | 18 (30.5%) | 35 (41.7%) | <0.001+ |
| 21–40 | 53 (38.4%) | 25 (17.5%) | 17 (28.8%) | 8 (9.5%) | |
| >40 | 48 (34.8%) | 17 (11.9%) | 10 (16.9%) | 7 (8.3%) | |
| COPD | 66 (47.8%) | 22 (15.4%) | 21 (35.6%) | 1 (1.2%) | <0.001+ |
| BMI kg·m−2 | 25.6±4.6 | 25.9±4.8 | 26.9±5.9 | 25.2±3.8 | 0.104 |
| Type of NSCLC | |||||
| Adenocarcinoma | 88 (63.8%) | ||||
| Squamous cell carcinoma | 41 (29.7%) | ||||
| Large cell carcinoma | 4 (2.9%) | ||||
| NOS | 5 (3.6%) | ||||
| NSCLC stage# | |||||
| I | 25 (14.5%) | ||||
| II | 15 (10.8%) | ||||
| III | 39 (28.3%) | ||||
| IV | 64 (46.4%) |
Data are presented as mean±sd or n (%), unless otherwise stated. NSCLC: non-small cell lung cancer; BMI: body mass index; NOS: not otherwise specified.#: according to the seventh edition of the American Joint Committee on Cancer TNM staging system; ¶: after Games–Howell correction, there was a significant difference between healthy volunteers and confirmed NSCLC and healthy volunteers and suspected, proven negative subjects; +: after Holm–Bonferroni correction, there was a significant difference between healthy volunteers and confirmed NSCLC and suspected proven negative subjects.
Lung cancer patients were significantly older (mean age 67.1±9.1 years), more likely to be male and a current or ex-smoker, had smoked more pack-years, and were more often diagnosed with COPD than subjects in the control group. Almost 75% of the NSCLC patients were classified as stage III or IV disease.
Table 2 shows that sex, age, smoking status, number of pack-years, presence of COPD and the classification value obtained by the Aeonose were univariately associated with the presence of lung cancer. Subsequently, we added these candidate variables to a multivariate regression analysis, which showed that age, number of pack-years, presence of COPD and the value of the Aeonose remained significantly predictive for the presence of lung cancer.
TABLE 2.
Results of the univariate and multivariate logistic regression analyses for diagnosing lung cancer
| Variable | Univariate analysis | Multivariate analysis | β# |
| Sex | 2.01 (1.26–3.20) | 1.42 (0.76–2.58) | 0.34 |
| Age | 1.08 (1.05–1.11) | 1.05 (1.02–1.09) | 0.05 |
| BMI | 0.99 (0.94–1.04) | − | |
| Smoking status | − | ||
| Current smoker | 17.49 (6.79–45.06) | ||
| Ex-smoker | 7.56 (3.23–17.69) | ||
| Never smoked | Ref. | ||
| Smoking exposure pack-years | |||
| 0 | Ref. | Ref. | |
| 1–20 | 3.88 (1.56–9.65) | 3.48 (1.25–9.66) | 1.25 |
| 21–40 | 14.77 (5.89–37.04) | 10.20 (3.66–28.46) | 2.32 |
| >40 | 19.36 (7.36–50.91) | 11.69 (4.04–33.87) | 2.46 |
| COPD | 4.90 (2.80–8.58) | 2.29 (1.18–4.43) | 0.83 |
| Diabetes mellitus | 0.70 (0.30–1.64) | − | |
| Aeonose classification value [13] | 24.20 (9.71–60.33) | 12.67 (4.48–35.83) | 2.54 |
Data are presented as odds ratio (95% confidence interval) unless otherwise stated. β: regression coefficient. BMI: body mass index; −: not added to the multivariate model. #: constant −5.54.
Each additional year of age was associated with a 5% higher chance of having lung cancer (OR 1.05, 95% CI 1.02–1.09). Subjects who had smoked 1–20 pack-years have a 3.5-fold higher chance of developing lung cancer (OR 3.5, 95% CI 1.25–9.66), whereas those who had smoked >40 pack-years have a 11.7-fold higher chance (OR 11.7, 95% CI 4.04–33.87). Patients with COPD had a 2.3-fold increased risk of having lung cancer (OR 2.27, 95% CI 1.18–4.43). The classification value of the Aeonose was also strongly associated with the presence of lung cancer (OR 12.7, 95% CI 4.48–35.83).
The multivariate logistic regression analysis based on clinical variables only showed a sensitivity of 93.5%, a specificity of 50%, a PPV of 64.5% and an NPV of 88.8%. This corresponded with an AUC-ROC of 0.80 (95% CI 0.75–0.85).
When the ANN was trained with exhaled-breath data from the Aeonose only, we found a sensitivity of 94.2%, a specificity of 44.1%, and a PPV and NPV of 61.9% and 88.7%, respectively, with an AUC-ROC of 0.75 (95% CI 0.69–0.81) (table 3). When applying the multivariate logistic regression model including the resulting value (−1 to 1) of the exhaled-breath data from the ANN in the exact same study population, we found an improved performance to distinguish NSCLC patients from controls with an AUC-ROC of 0.86 (95% CI 0.81–0.90). By choosing a relevant threshold value in the ROC curve, focusing on high sensitivity and high NPV, the analysis showed a sensitivity of 95.7%, a specificity of 59.7%, and a PPV and NPV of 69.5% and 92.5%, respectively. The bootstrap analysis for internal validation showed similar regression coefficients compared to our original model showing robustness of the model.
TABLE 3.
Diagnostic performance of the three investigated prediction models
| Positive/negative | Optimal cut-off | Sensitivity | Specificity | PPV | NPV | AUC-ROC (95% CI) | |
| Clinical variables only | 138/143 | 0.32 | 93.5% | 50.0% | 64.5% | 88.8% | 0.80 (0.75–0.85) |
| Aeonose result only | 138/143 | −0.38 | 94.2% | 44.1% | 61.9% | 88.7% | 0.75 (0.69–0.81) |
| Multivariate logistic regression model | 138/143 | 0.27 | 95.7% | 59.7% | 69.5% | 92.5% | 0.86 (0.81–0.90) |
| Extended ANN | 138/143 | −0.65 | 94.2% | 49.0% | 64.0% | 89.7% | 0.84 (0.79–0.89) |
PPV: positive predictive value; NPV: negative predictive value; AUC-ROC: area under the receiver operating curve; ANN: artificial neural network.
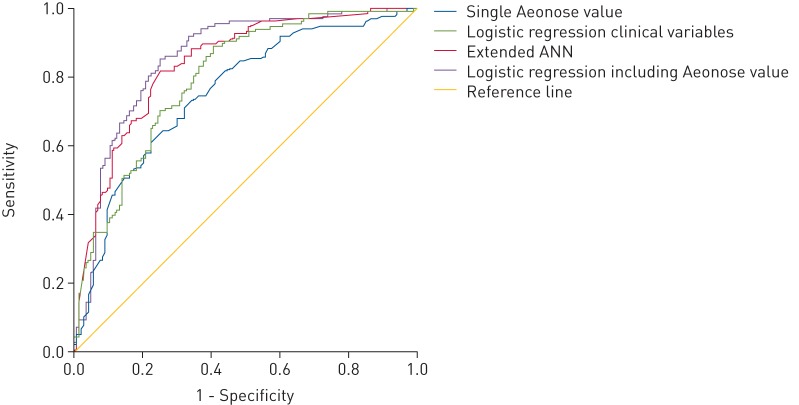
When training the ANN with exhaled-breath data together with the clinical variables that were univariately associated with the presence of lung cancer, we found an improved diagnostic performance as well to distinguish NSCLC patients from controls, showing an AUC-ROC of 0.84 (95% CI 0.79–0.89). By choosing an appropriate threshold value in the ROC diagram, we observed a sensitivity of 94.2%, a specificity of 49.0%, and a PPV and NPV of 64.0% and 89.7% respectively. Figure 2 shows the combined ROC curve showing the improved performance of both multivariate models.
FIGURE 2.
Combined receiver operating characteristic curve showing four predictive algorithms: logistic regression clinical variables, single Aeonose value, extended artificial neural network (ANN) and logistic regression including Aeonose value.
Discussion
This study assessed the impact of combining exhaled-breath analysis and clinical parameters in diagnosing lung cancer. We showed that adding readily available clinical information to the classification value of exhaled-breath analysis by the Aeonose in a relatively easy-to-perform multivariate regression model improved the diagnostic accuracy, expressed as AUC-ROC, from 0.75 to 0.86 to diagnose lung cancer in a noninvasive way. When extending the vector containing compressed breath data with clinical variables, and in this way training the ANN to distinguish between sick and healthy individuals, the diagnostic accuracy, expressed as AUC-ROC, increased from 0.75 to 0.84. Minor differences were observed compared to our previous performed analysis due to the somewhat smaller sample size because of missing information on pack-years (previous study: n=290, AUC-ROC 0.76) and the fact that outcomes of frequently training an ANN can slightly fluctuate.
It turns out that logistic regression analysis and ANN are equally capable of increasing classification quality of lung cancer diagnosis. We expected to see rather improved accuracy when entering a combination of clinical and exhaled-breath data directly to the untrained ANN, because it considers possible interactions. Independency of parameters, like breath profile, COPD and pack-years, cannot be fully assumed so far. As extending exhaled-breath data with clinical parameters followed by classification by an ANN is more complex than building a multivariate regression model out of single clinical parameters, the latter is to be recommended for practical use.
Besides ANN and logistic regression analysis, other statistical learning methods for classifying exhaled-breath data are available, such as random forest and support vector machines [25]. In a previous study, results from neural network analysis were compared to results obtained from random forest and support vector machine showing comparable diagnostic performance [26]. In this study, we only focused on the two statistical learning methods described: logistic regression analysis and neural network analysis.
Previous studies have shown that electronic nose technology based on pattern recognition of VOCs or identifying VOCs with gas chromatography/mass spectrometry can differentiate between subjects with and without lung cancer [13, 14, 17, 20, 27–29]. Several studies using techniques for VOC identification have used logistic regression analysis to identify lung cancer-specific VOCs [30, 31]. However, logistic regression analysis including clinical parameters in studies using pattern recognition techniques has not been shown often yet. Tirzïte et al. [19] used logistic regression analysis to predict the presence of lung cancer with the Cyranose 320 electronic nose mainly using segments of exhaled breath as input variables for the logistic regression analysis, but also including a few clinical parameters, such as age, smoking status, smoking history and ambient temperature. They were able to distinguish subjects with lung cancer from controls with a sensitivity of 96% in both smokers and nonsmokers, and a specificity >90% in both groups. To our knowledge, no studies have been performed using clinical variables and exhaled-breath data based on pattern recognition combined in an ANN to diagnose lung cancer. However, several studies have used ANNs to detect lung cancer without performing exhaled-breath analysis. These studies mainly focused on clinical parameters and biomarkers based on blood and genetic abnormalities [32, 33]. As described in our training study, the optimal cut-off point chosen determines the number of false-positive cases versus the number of missed cases concerning lung cancer [13]. We focused again on a high sensitivity and a high NPV, since lung cancer has an extremely high mortality rate if not detected early. By adding the clinical variables to the exhaled-breath data, we saw in both models that all diagnostic parameters improved, thereby reaching higher sensitivity and NPV compared to the training study, but we also observed fewer false-positive cases by achieving higher specificity.
In the near future, the results obtained could be proposed to add value in several ways. First, in the case of implementation of LDCT in Europe, the Aeonose may be deployed after suspicion of lung cancer has been raised with LDCT. Due to the high NPV with the Aeonose, subjects could be prevented from undergoing unnecessary invasive interventions and be monitored with prolonged intervals [8]. In addition, there is current debate about identifying at-risk groups relevant for LDCT screening [34–36]. Combining clinical parameters and exhaled-breath data in an ANN could indicate the degree of suspicion of lung cancer, and therefore serve as an adjunct for risk stratification in lung cancer screening, supporting clinical decision making.
Limitations of our study should also be mentioned. We did not analyse subjects with small cell lung cancer, nor did we analyse differences between the histological subtypes of NSCLC and lung cancer stages, since these subgroups were too small to include all relevant variables in the multivariate model. Moreover, we do not know the influence of food intake by subjects. Eating and drinking was not restricted before the exhaled-breath measurement. However, the neural network is being trained comparing breath profiles of positive and negative subjects regardless their food intake. When numbers of subjects are sufficiently large, it can be assumed that food intake is not relevant as it averages out. It should also be noted that most subjects had stage III and IV lung cancer (75%). This population differs from the high-risk asymptomatic subjects suitable for screening, where focus lies in early-stage lung cancer. However, the risk factors included in the multivariate analysis are applicable for both early- and late-stage lung cancer, so when exhaled-breath analysis is able to detect early-stage lung cancer, readily available clinical information should be incorporated in the analysis. Future analysis, including sufficient stage I and II NSCLC, should indicate whether breath patterns already change early in the course of the disease.
We should also note that in high-dimensional data sets as obtained with the Aeonose, the problem of overfitting can occur where a prediction model that looks appropriate on training data used to develop it, will perform poorly on future observations. Combining analytical techniques, such as data compression and cross-validation, partly overcomes this issue. Currently, an external validation study is performed where a complete new cohort of subjects is included to totally overcome the issue of overfitting.
Conclusion
Due to the aggressive nature of lung cancer, diagnostic accuracy should be as high as possible. This diagnostic accuracy to detect the presence or absence of lung cancer by exhaled-breath analysis with the Aeonose can be improved by adding readily available clinical information, either post hoc in a multivariate logistic regression model or a priori in the training process to the ANN, compared to the single classification value based on exhaled-breath data only. As both approaches yielded similar results, the multivariate logistic regression model should be preferred as its application is more convenient.
Footnotes
Conflict of interest: S. Kort reports an unrestricted research grant paid to her institution by The eNose Company, Zutphen, during the conduct of the study.
Conflict of interest: M. Brusse-Keizer has nothing to disclose.
Conflict of interest: J.W. Gerritsen is an employee of The eNose Company, Zutphen.
Conflict of interest: J.H. Schouwink has nothing to disclose.
Conflict of interest: E. Citgez has nothing to disclose.
Conflict of interest: F.H.C. de Jongh has nothing to disclose.
Conflict of interest: J. van der Maten has nothing to disclose.
Conflict of interest: S. Samii has nothing to disclose.
Conflict of interest: M. van den Bogart has nothing to disclose.
Conflict of interest: J. van der Palen has nothing to disclose.
References
- 1.Bray F, Ferlay J, Soerjomataram I, et al. Global cancer statistics 2018: GLOBOCAN estimates of incidence and mortality worldwide for 36 cancers in 185 countries. CA Cancer J Clin 2018; 68: 394–424. doi: 10.3322/caac.21492 [DOI] [PubMed] [Google Scholar]
- 2.Goldstraw P, Chansky K, Crowley J, et al. The IASLC Lung Cancer Staging Project: proposals for revision of the TNM stage groupings in the forthcoming (eighth) edition of the TNM classification for lung cancer. J Thorac Oncol 2016; 11: 39–51. doi: 10.1016/j.jtho.2015.09.009 [DOI] [PubMed] [Google Scholar]
- 3.Siegel RL, Miller KD, Jemal A. Cancer statistics, 2018. CA Cancer J Clin 2018; 68: 7–30. doi: 10.3322/caac.21442 [DOI] [PubMed] [Google Scholar]
- 4.Aberle DR, Adams AM, Berg CD, et al. Reduced lung-cancer mortality with low-dose computed tomographic screening. N Engl J Med 2011; 365: 395–409. doi: 10.1056/NEJMoa1102873 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Patz EF Jr, Greco E, Gatsonis C, et al. Lung cancer incidence and mortality in National Lung Screening Trial participants who underwent low-dose CT prevalence screening: a retrospective cohort analysis of a randomised, multicentre, diagnostic screening trial. Lancet Oncol 2016; 17: 590–599. doi: 10.1016/S1470-2045(15)00621-X [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Black WC, Keeler EB, Soneji SS. Cost-effectiveness of CT screening in the National Lung Screening Trial. N Engl J Med 2015; 372: 388. [DOI] [PubMed] [Google Scholar]
- 7.de Koning HJ, Meza R, Plevritis SK, et al. Benefits and harms of computed tomography lung cancer screening strategies: a comparative modeling study for the U.S. Preventive Services Task Force. Ann Intern Med 2014; 160: 311–320. doi: 10.7326/M13-2316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Huo J, Xu Y, Sheu T, et al. Complication Rates and downstream medical costs associated with invasive diagnostic procedures for lung abnormalities in the community setting. JAMA Intern Med 2019; 179: 324–332. doi: 10.1001/jamainternmed.2018.6277 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Oudkerk M, Devaraj A, Vliegenthart R, et al. European position statement on lung cancer screening. Lancet Oncol 2017; 18: e754–e766. doi: 10.1016/S1470-2045(17)30861-6 [DOI] [PubMed] [Google Scholar]
- 10.Boots AW, van Berkel JJ, Dallinga JW, et al. The versatile use of exhaled volatile organic compounds in human health and disease. J Breath Res 2012; 6: 027108. doi: 10.1088/1752-7155/6/2/027108 [DOI] [PubMed] [Google Scholar]
- 11.Buszewski B, Kesy M, Ligor T, et al. Human exhaled air analytics: biomarkers of diseases. Biomed Chromatogr 2007; 21: 553–566. doi: 10.1002/bmc.835 [DOI] [PubMed] [Google Scholar]
- 12.Nakhleh MK, Amal H, Jeries R, et al. Diagnosis and classification of 17 diseases from 1404 subjects via pattern analysis of exhaled molecules. ACS Nano 2017; 11: 112–125. doi: 10.1021/acsnano.6b04930 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kort S, Tiggeloven MM, Brusse-Keizer M, et al. Multi-centre prospective study on diagnosing subtypes of lung cancer by exhaled-breath analysis. Lung Cancer 2018; 125: 223–229. doi: 10.1016/j.lungcan.2018.09.022 [DOI] [PubMed] [Google Scholar]
- 14.van de Goor R, van Hooren M, Dingemans AM, et al. Training and validating a portable electronic nose for lung cancer screening. J Thorac Oncol 2018; 13: 676–681. 10.1016/j.jtho.2018.01.024. [DOI] [PubMed] [Google Scholar]
- 15.Phillips M, Altorki N, Austin JH, et al. Prediction of lung cancer using volatile biomarkers in breath. Cancer Biomark 2007; 3: 95–109. doi: 10.3233/CBM-2007-3204 [DOI] [PubMed] [Google Scholar]
- 16.Machado RF, Laskowski D, Deffenderfer O, et al. Detection of lung cancer by sensor array analyses of exhaled breath. Am J Respir Crit Care Med 2005; 171: 1286–1291. doi: 10.1164/rccm.200409-1184OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Bajtarevic A, Ager C, Pienz M, et al. Noninvasive detection of lung cancer by analysis of exhaled breath. BMC Cancer 2009; 9: 348. doi: 10.1186/1471-2407-9-348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Mazzone PJ, Wang XF, Lim S, et al. Progress in the development of volatile exhaled breath signatures of lung cancer. Ann Am Thorac Soc 2015; 12: 752–757. doi: 10.1513/AnnalsATS.201411-540OC [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tirzïte M, Bukovskis M, Strazda G, et al. Detection of lung cancer with electronic nose and logistic regression analysis. J Breath Res 2018; 13: 016006. doi: 10.1088/1752-7163/aae1b8 [DOI] [PubMed] [Google Scholar]
- 20.Gasparri R, Santonico M, Valentini C, et al. Volatile signature for the early diagnosis of lung cancer. J Breath Res 2016; 10: 016007. doi: 10.1088/1752-7155/10/1/016007 [DOI] [PubMed] [Google Scholar]
- 21.Goldstraw P, Crowley J, Chansky K, et al. The IASLC Lung Cancer Staging Project: proposals for the revision of the TNM stage groupings in the forthcoming (seventh) edition of the TNM classification of malignant tumours. J Thorac Oncol 2007; 2: 706–714. doi: 10.1097/JTO.0b013e31812f3c1a [DOI] [PubMed] [Google Scholar]
- 22.Bruins MGJ, van de Sande W, van Belkum A. Enabling a transferable calibration model for metal-oxide type electronic noses. Sens Actuators B Chem 2013; 188: 1187–1195. doi: 10.1016/j.snb.2013.08.006 [DOI] [Google Scholar]
- 23.Kort S, Brusse-Keizer M, Gerritsen JW, et al. Data analysis of electronic nose technology in lung cancer: generating prediction models by means of Aethena. J Breath Res 2017; 11: 026006. doi: 10.1088/1752-7163/aa6b08 [DOI] [PubMed] [Google Scholar]
- 24.Hanai T, Yatabe Y, Nakayama Y, et al. Prognostic models in patients with non-small-cell lung cancer using artificial neural networks in comparison with logistic regression. Cancer Sci 2003; 94: 473–477. doi: 10.1111/j.1349-7006.2003.tb01467.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leopold JH, Bos LD, Sterk PJ, et al. Comparison of classification methods in breath analysis by electronic nose. J Breath Res 2015; 9: 046002. doi: 10.1088/1752-7155/9/4/046002 [DOI] [PubMed] [Google Scholar]
- 26.Hauschild AC, Baumbach JI, Baumbach J. Integrated statistical learning of metabolic ion mobility spectrometry profiles for pulmonary disease identification. Genet Mol Res 2012; 11: 2733–2744. doi: 10.4238/2012.July.10.17 [DOI] [PubMed] [Google Scholar]
- 27.Antoniou SX, Gaude E, Ruparel M, et al. The potential of breath analysis to improve outcome for patients with lung cancer. J Breath Res 2019; 13: 034002. doi: 10.1088/1752-7163/ab0bee [DOI] [PubMed] [Google Scholar]
- 28.Gasparri R, Romano R, Sedda G, et al. Diagnostic biomarkers for lung cancer prevention. J Breath Res 2018; 12: 027111. doi: 10.1088/1752-7163/aa9386 [DOI] [PubMed] [Google Scholar]
- 29.Peng G, Tisch U, Adams O, et al. Diagnosing lung cancer in exhaled breath using gold nanoparticles. Nat Nanotechnol 2009; 4: 669–673. doi: 10.1038/nnano.2009.235 [DOI] [PubMed] [Google Scholar]
- 30.Corradi M, Poli D, Banda I, et al. Exhaled breath analysis in suspected cases of non-small-cell lung cancer: a cross-sectional study. J Breath Res 2015; 9: 027101. doi: 10.1088/1752-7155/9/2/027101 [DOI] [PubMed] [Google Scholar]
- 31.Mazzone PJ, Hammel J, Dweik R, et al. Diagnosis of lung cancer by the analysis of exhaled breath with a colorimetric sensor array. Thorax 2007; 62: 565–568. doi: 10.1136/thx.2006.072892 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Duan X, Yang Y, Tan S, et al. Application of artificial neural network model combined with four biomarkers in auxiliary diagnosis of lung cancer. Med Biol Eng Comput 2017; 55: 1239–1248. doi: 10.1007/s11517-016-1585-7 [DOI] [PubMed] [Google Scholar]
- 33.Hart GR, Roffman DA, Decker R, et al. A multi-parameterized artificial neural network for lung cancer risk prediction. PLoS One 2018; 13: e0205264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Katki HA, Kovalchik SA, Berg CD, et al. Development and validation of risk models to select ever-smokers for CT lung cancer screening. JAMA 2016; 315: 2300–2311. doi: 10.1001/jama.2016.6255 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Kovalchik SA, Tammemagi M, Berg CD, et al. Targeting of low-dose CT screening according to the risk of lung-cancer death. N Engl J Med 2013; 369: 245–254. doi: 10.1056/NEJMoa1301851 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Tammemagi MC, Katki HA, Hocking WG, et al. Selection criteria for lung-cancer screening. N Engl J Med 2013; 368: 728–736. doi: 10.1056/NEJMoa1211776 [DOI] [PMC free article] [PubMed] [Google Scholar]