Abstract
Introduction
Non-small cell lung cancer (NSCLC) is a worldwide malignance threatening human life. TGF-β/Smad signaling is known to regulate cell proliferation, differentiation, migration and growth. As the only co-Smad playing crucial roles in TGF-β signaling, Smad4 is reported to be frequently mutated or to occur as alternatively spliced in tumor cells. Smad4 was reported to be involved in the TGF-β-induced EMT process. However, whether the alternative splicing occurs in the TGF-β-induced EMT process in NSCLC was not clear.
Methods
In our current study, we explored the alternative splicing of Smad4 during the process of TGF-β-induced EMT in A549 cells. 10 ng/mL TGF-β was used to induce EMT. Then, nest-PCR and agarose electrophoresis were performed to detect the expression of Smad4 variants and sequencing to get the variant DNA sequences. For recombinant expression of variants of Smad4 in A549 cells, we used lentiviral variants to infect cells. In order to explore the effects of variants on the proliferation and migration of A549 cells, the MTT assay, colony formation assay and wound-healing assay were done. The effects of variants on E-cad and VIM protein expression were explored through Western blot.
Results
There were several novel gene fragments expressed in TGF-β-induced A549 cells, and the sequencing results showed that they were indeed the Smad4 variants that were not reported. For recombinant expression of Smad4 variants in A549 cells, we found that they have significant effects on the proliferation and migration of cells, and also regulated the E-cad and VIM protein expression.
Conclusion
Our results indicated that novel Smad4 variants were expressed in TGF-β-induced EMT process. The functional study showed that these novel variants regulate cell proliferation and migration and affect E-cad and VIM protein expression, showing the potential as targets for cancer therapy.
Keywords: Smad4, EMT, TGF-β, alternatively spliced variants, non-small cell lung cancer
Introduction
Lung cancer, especially non-small cell lung cancer (NSCLC), is a common malignancy, with about 15% 5-year survival rate.1,2 Smoking was thought to be the main factor inducing lung cancer, and almost two-thirds of Chinese adult men were smokers.3,4 However, nonsmoking females also suffered from lung cancer.5 Treatments currently used for lung cancer ranged from surgery, radiotherapy and chemotherapy to target therapy and immunotherapy depending on tumor stage.1,5 The well-studied molecular targets were EGFR mutations and ALK rearrangement in patients with lung cancer, and the EGFR and ALK inhibitors used to treat patients with EGFR mutation or ALK rearrangement showed successful outcomes.5,6 The treatments with checkpoint inhibitors, such as PD-1 or CTLA-4 inhibitor, have been proven to be more effective than conventional drugs.7–9
The classical TGF-β/Smad signaling regulates proliferation, differentiation, migration and growth of tumors, in which Smad4 plays important roles.10 However, Smad4 is frequently mutated or lost in types of cancers, which brings big challenges for Smad4-related cancer therapies.11–14 Transforming growth factor beta 1 (TGF-β1)-induced epithelial–mesenchymal transition (EMT) process was reported to be involved in tumor metastasis and invasion.15,16 Smad4 was reported to be participating in regulating TGF-β-induced EMT process in lung cancer.17,18
The alternative splicing is the splicing machinery recognizes and splice-in different sets of exons forming a mature mRNA through removing the introns and ligating the exons, thus increasing mRNA and protein diversity.19 Alternative splicing frequently occurs in human gene encoding.20 It is worthy to mention that alternative splicing is another form of Smad4 diversification. Reports have shown that Smad4 occurred alternatively splicing in normal and tumor tissues. More than 10 years ago, scientists studied the alternative splicing of Smad4, and indeed some alternatively spliced variants were found in the mRNA level.21 Benefiting from the development and application of sequencing technology, more and more variants of Smad4 were reported.22–24 However, there was little knowledge about the functions of the alternatively spliced variants of Smad4. Recently, some researchers have put efforts on the functional study of Smad4 variants and found that some novel variants of Smad4 involved in tumor development. The variants that termed as rs12455792 CT or TT were reported promoting the process of thoracic aortic aneurysm and dissection.25 Irfan Ullah et al26 reported that novel alternatively spliced variants of Smad4 expressed in HaCat cells under the UVB irradiation and the variants showed effects on the E-cadherin (E-cad) and N-cadherin expression, which was considered as markers of EMT process. However, the functions of Smad4 variants in regulating tumor cell development were still unclear, and more work was needed to uncover the mystery.
In our current study, we explored the alternative splicing of Smad4 during the process of TGF-β-induced EMT in A549 cells. There were several variants of Smad4 expressed after induction. The functional study showed that these variants have effects on the proliferation and migration of A549 cells and also regulate the protein expression of E-cad and vimentin (VIM). These findings indicated that the alternatively spliced variants could be considered as the targets for cancer therapy.
Methods and Materials
Cell Culture
A549 cells and HEK293T cells were purchased from the cell bank of Chinese Academy of Sciences. A549 cells were cultured in DMEM/F-12K (Gibco, Cat#C11330500BT) medium containing 10% FBS, 100 Unit penicillin and 100 Unit streptomycin and cultured in 25 cm2 flasks (Corning, Cat#3815). HEK293T cells were cultured in DMEM medium containing 10% FBS, 100 unit penicillin and 100 unit streptomycin, and cultured in 10 cm dishes. Cells were incubated in a humidified cell culture incubator with 5% CO2 at 37°C.
RT-PCR and NEST-PCR
PrimeSTAR DNA polymerase (Takara, Code No.R050Q) was used to do RT-PCR and NEST-PCR. A549 cells were treated with or without 10 ng/mL TGF-β for 72-h culturing with DMEM/F12-K medium containing 0.5% FBS. Then,- total RNA extraction (RNAiso Plus, Takara, Code No.9108) and RT-PCR (PrimeScriptTM RT Reagent Kit with gDNA Eraser, Takara, Code No.RR047A) were done to get the cDNA. The cDNA was used as a template for the first-step NEST-PCR or other related PCR. Then, the first-step NEST-PCR products were used as a template for the second-step NEST-PCR. To confirm the EMT induction, the RT-PCR was performed to measure the mRNA expression level of E-cad and VIM. The primers used to amplify human E-cad were E-cad-F: 5ʹ-GTGGTCAAAGAGCCCTTACTGC-3ʹ and E-cad-R: 5ʹ-ACCGCTTCCTTCATAGTCAAACA-3ʹ. The primers used to amplify human VIM were VIM-F: 5ʹ-TGCCGTTGAAGCTGCTAACTA-3ʹ and VIM-R: 5ʹ-CCAGAGGGAGTGAATCCAGATTA-3ʹ. To detect the splicing variants of Smad4, the NEST-PCR was done. The primers used for the first step of NEST-PCR were Smad4-1-F: 5ʹ-TGCCATAGACAAGGTGGAGAGAGTG-3ʹ and Smad4-1-R: 5ʹ-CTAAAGGTTGTGGGTCTGCAATCGG-3ʹ. The primers used for the second step of NEST-PCR were Smad4-2-F: 5ʹ-GGTCGGAAAGGATTTCC-3ʹ and Smad4-2-R: 5ʹ-CCAACTGCACACCTTTGCCTA-3ʹ. The primers used to amplify GAPDH were GAPDH-F: 5ʹ-GGAGCGAGATCCCTCCAAAAT-3ʹ and GAPDH-R: 5ʹ-GGCTGTTGTCATACTTCTCATGG-3ʹ.
DNA Constructs
In order to induce lentivirus, Smad4 and its variants, gene fragments were inserted into pLenti-CMV vectors. The primers used for amplifying the gene fragments were Smad4-F: CGCGGATCCGCCACCATGGACAATATGTCTATTACGAAT (contains BamHI cutting site) and Smad4-R: ACGCGTCGACTCAGTCTAAAGGTTGTGGGTCTG (contains SalI cutting site). PrimeSTAR DNA polymerase was used to amplify target gene fragments following manufacturer's instructions. BamHI (Takara, Code No.1605) and SalI (Takara, Code No.1636) restrict enzyme were used to digest backbone plasmids and insertions. T4 DNA ligase (Thermo Fisher, Cat#EL0016) was used to ligate the backbone plasmids and insertions. Ligation products were transfected into Stbl3 competent cells. Plasmid sequencing was performed to confirm the sequence of constructed plasmids.
Lenti Virus Induction and Infection
To induce the lentiviruses, HEK293T cells were co-transfected with pLenti-CMV plasmids contained the target gene fragments (10 μg), pCMVΔR (5 μg) and pCMV–VSVG (5 μg) using effectene transfection reagent (QIAGEN, Cat#301425) in 10 cm dish. Before transfection, the HEK293T cells were changed with fresh DMEM culture medium without antibiotics. The virus was collected and filtered with a 0.22-μm filter after 72 h.
For infection, A549 cells were cultured in a 6-well plate. In the following day, cells were changed with 1 mL fresh culture medium and added 1 mL virus supernatant. 72 h later, F-12K medium containing 1 μg/mL puromycin was used to select the infected cells. The selection lasted for 1 week. Then, the RT-PCR and Western blot were performed to confirm the recombinant expression. The A549 cells stably expressed wildtype Smad4 or its variants, which were used for further research.
Western Blot
The A549 cells stably expressed wildtype Smad4 and its variants, which were used for the Western blot experiments. Collect and lyse cells with RIPA buffer for 30 min on ice. Total protein lysis was used to measure target protein expression. The anti-E-cad (Santa Cruz, Cat#sc-8426) and anti-VIM (Santa Cruz, Cat#sc-373717) antibodies were purchased from Santa Cruz. The anti-GAPDH (Cat#10494-1-AP) and Smad4 (Cat#10231-1-AP) antibodies were purchased from Proteintech. Load 30 μg total protein into 10% SDS-PAGE gel and run for 90 min at 120 V. Transfer the gel on the PVDF membrane and then incubate with 5% nonfat milk for 30 min by slowly shaking at room temperature. We then incubated the primary antibodies at 4°C in a freezer overnight and washed with TBST for 10 min, repeating 3 times. The secondary antibodies were then incubated at room temperature for 1 h by slowly shaking. The images of membrane were taken with a standard chemiluminescence.
Wound-Healing Assay
To explore the effects of Smad4 and its variants on the migration of A549 cells, we performed the wound-healing assay. A549 cells that recombinantly expressed wildtype Smad4 or its variants were cultured in a 12-well plate at a cell density of about 90% the next day in RPMI1640 medium without FBS. We then used 200μL tips to make healing. PBS was used to wash 3 times to remove the detached cells, and images were taken at time point 0 h. 72 h later, images were taken to record the migration. The migration distance was analyzed with Image J software.
MTT Assay
To measure the proliferation of cells, MTT assay was performed. Count and seed 3000 cells in each well of 96-well plate with 200 ul culture medium. Each sample repeated 6 wells, three wells as 0 h time point and the other three wells as 12 h time point. Put the seeded plate into incubator immediately and cultured for 6 h and then added 20 μL 5mg/mL MTT, incubated in an incubator for another 4 h. Abandoned the medium and then added 150 μL DMSO, incubating for 10 min at room temperature and then measure the OD495 as starting point (0 h). 12 h later, add the MTT, incubate for 4 h and then measure the OD495. OD495 increment was compared to indicate the proliferation of cells.
Colony Formation Assay
To further explore the effects of Smad4 and its variants on the proliferation of A549 cells, we performed the colony formation assay. We first counted 500 cells for each group and seeded them in dishes. The culture medium was changed every 2 days. 14 days later, we used a crystal violet dye to label colonies and took photos. The number of colonies was counted using Image J software.
Statistical Analysis
Two-tailed Student’s t-tests with unequal variants were used for statistical analysis. All the results shown in figures had at least three independent repeats.
Results
Novel Variants of Smad4 Expressed in A549 Cells During TGF-β-Induced EMT
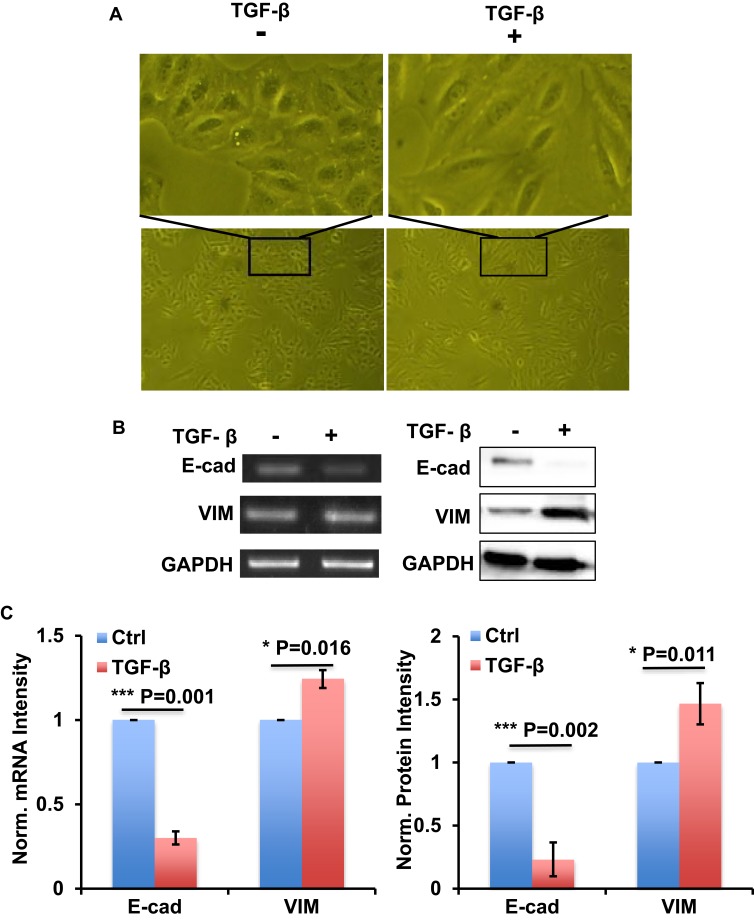
In order to establish the EMT model, we used 10 ng/mL TGF-β to treat A549 cells for 72 h with RPMI1640 medium containing 0.5% FBS. Cell images showed the morphology change of A549 cells transforming from epithelioid cells to fusiform mesenchymal cells (Figure 1A). RT-PCR and Western blot results showed that EMT-related gene E-cad was downregulated, and VIM was upregulated in mRNA and protein levels, respectively (Figure 1B and C). These data indicated that TGF-β could successfully establish the EMT process in A549 cells.
Figure 1.
TGF-β successfully induced EMT process in A549 cells. (A) The images show the morphology change of A549 cells with or without TGF-β treatment as indicated. (B) RT-PCR and Western blot show the expression of E-cad and VIM at mRNA level (left) and protein level (right) in A549 cells with or without TGF-β treatment as indicated. (C) Bar graph shows the intensity quantification of the expression of E-cad and VIM at mRNA level (left) and protein level (right) as indicated in panel B. n=4, Error bars: Mean±SD. *P<0.05, ***P<0.005.
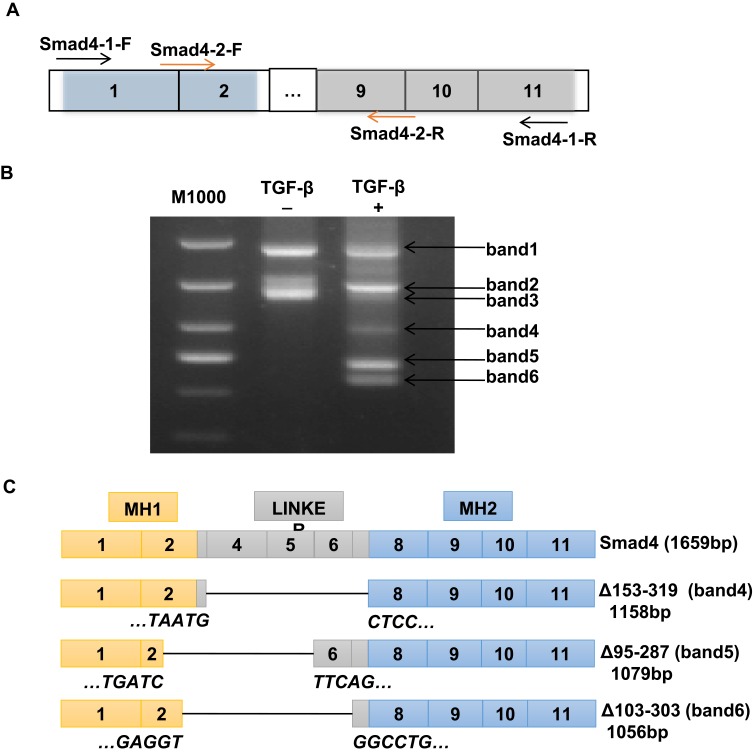
TGF-β/Smad signaling was involved in regulating the EMT process, in which Smad4 plays crucial roles.27 In order to detect whether there were variants of Smad4 expressed in the TGF-β-induced EMT process in A549 cells, NEST-PCR was performed. As shown in Figure 2A, the primer pairs used for NEST-PCR were indicated as arrows. The Smad4-1-F/Smad4-1-R primer pair was used for the first step of NEST-PCR and the Smad4-2-F/Smad4-2-R primer pair was used for the second step of NEST-PCR. A549 cells were treated with or without TGF-β for 72-h culturing with F-12K medium containing 0.5% FBS. And then the total RNA was extracted and did reverse transcription PCR to get cDNA used as a template in NEST-PCR. The agarose electrophoresis showed that there were several gene fragments expressed after TGF-β treatment (Figure 2B). We named them as band1, band2, band3, band4, band5 and band6 for convenience based on their size. Surprisingly, compared with the nontreated group, there were three novel gene fragments, band4, band5 and band6, expressed in TGF-β treated group that were totally not expressed in the nontreated group (Figure 2B).
Figure 2.
Novel splicing variants of Smad4 expressed during TGF-β-induced EMT in A549 cells. (A) Schematic drawing shows the primer pairs used for NEST-PCR. (B) Representative image of RT-PCR to show the variant expression of Smad4 in A549 cells with or without TGF-β treatment as indicated. More than 3 times, independent experiment repeats were done. (C) Schematic drawing to show the structure of Smad4 and its variants based on sequencing results.
To explore whether these novel expressed gene fragments were Smad4 variants, we did gel extraction to recover these three novel gene fragments and sent them to the company to do sequencing. After analyzing the sequencing results, we found that these three novel gene fragments were indeed variants of Smad4. Compared with wildtype Smad4, these three variants were mainly deleted the linker domain (Figure 2C). Δ153-319 (band4, 1158 bp) almost deleted the whole linker region, exon3 to exon7 (Figure 2C). Δ95-287 (band5, 1079 bp) deleted the linker region except the exon6 and exon7, and also deleted most part of exon2, which belongs to the MH1 domain (Figure 2C). Δ103-303 (band6, 1056 bp) deleted most parts of the linker region and part of exon2, but retains exon7 (Figure 2C). Δ95-287 and Δ103-303 splicing variants of Smad4 were not reported before. And Δ153-319 was reported in HaCat cells at a low amount.28
These data indicated that TGF-β could be used to establish the EMT model in A549 cells. During the TGF-β-induced EMT process, Smad4 occurred alternatively splicing and novel splicing variants of Smad4 were expressed.
Novel Splicing Variants of Smad4 Affect the E-Cadherin and VIM Expression in A549 Cells
To explore whether these novel splicing variants have effects on the EMT process in A549 cells, we used lenti-viral infection to recombinantly express wildtype Smad4 and its variants in A549 cells, respectively. Three days after infection, we used 1 μg/mL puromycin to select infected cells. RT-PCR results showed that all the target genes were expressed successfully in A549 cells (Figure S1A and B). Western blot results showed the successfully expressed wildtype Smad4 in A549 cells (Figure S1C and D). Because the sequence that anti-Smad4 antibody recognized was 13–227 amino acids, which was covered MH1 domain (14-138aa) and linker region (139-319aa) of wildtype Smad4. However, the variants mainly deleted these regions, which might be the reason why we could not detect the protein expression of Smad4 variants.
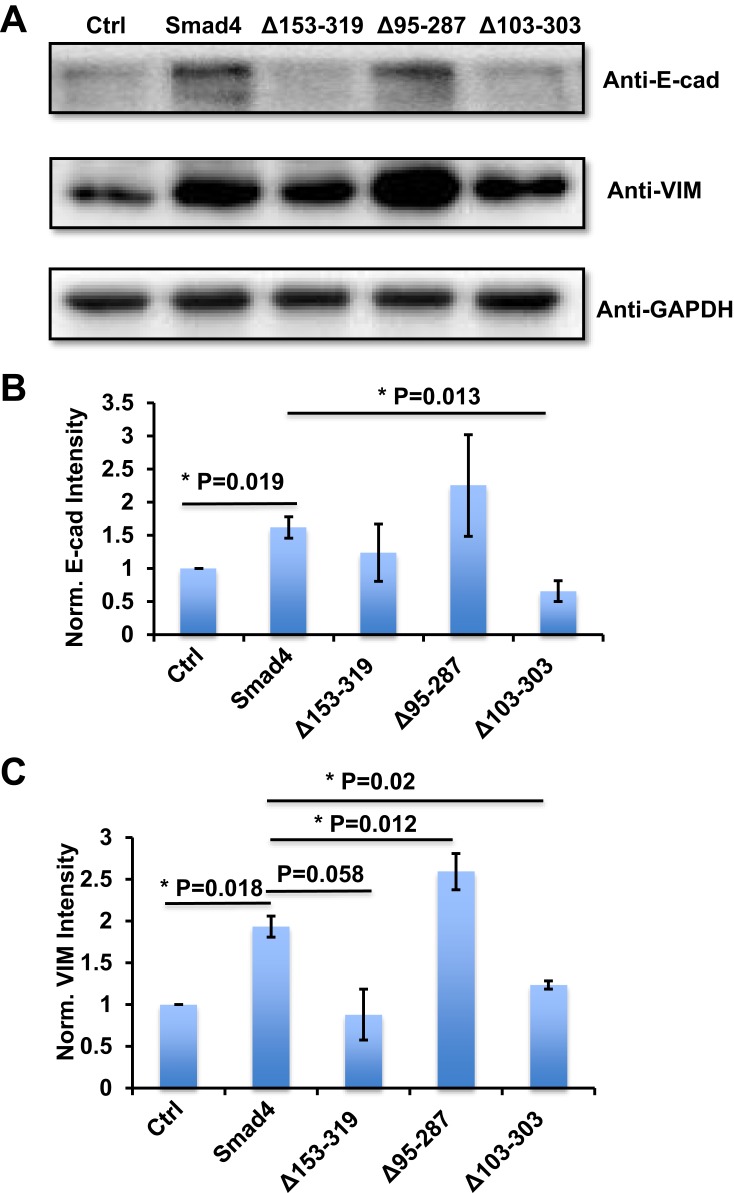
We then used these infected cells to measure the protein expression of E-cad and VIM. Compared with the control group, the A549 cells that expressed wildtype Smad4 showed significantly higher E-cad expression, and Δ153-319 and Δ95-287 group expressed a comparable level of E-cad with wildtype Smad4 group (Figure 3A and B). The A549 cells that recombinantly express Δ103-303 showed significantly lower E-cad expression than wildtype Smad4 (Figure 3A and B). Similarly, wildtype Smad4 and Δ95-287 group have higher expression of VIM compared with the control group (Figure 3A and C). The Δ153-319 and Δ103-303 groups have lower VIM expression compared with wildtype Smad4 and Δ95-287 group; however, they were comparably expressed as the control group (Figure 3A and C). These data indicated that Δ95-287 has similar effects on the expression of E-cad and VIM compared with wild type Smad4, while the Δ153-319 and Δ103-303 showed opposite effects on the expression of E-cad and VIM. After analyzing the sequences, we found that the main difference among Δ153-319, Δ95-287 and Δ103-303 was that Δ95-287 retained the exon6, while Δ153-319 and Δ103-303 were not (Figure 2C). Hence, we speculated that the exon6 plays vital roles in the functions of Smad4 in regulating the E-cad and VIM expression.
Figure 3.
Variants of Smad4 affect E-cad and VIM protein expression in A549 cells. (A) Representative images show E-cad and VIM protein expression in A549 cells that recombinantly express wildtype Smad4 or its variants as indicated. (B) Bar graphs to show the intensity-based quantification of E-cad expression in panel A. (C) Bar graphs show the intensity-based quantification of VIM expression in panel A. n=4, Error bars: Mean±SD. *P<0.05.
Δ153-319 Inhibits the Migration of A549 Cells
E-cad regulates the cell–cell adhesion, and the high expression of E-cad is accompanied with strong cell–cell adhesion. The downregulation of E-cad resulting in reduced cell–cell adhesion was important during tumor metastasis.29
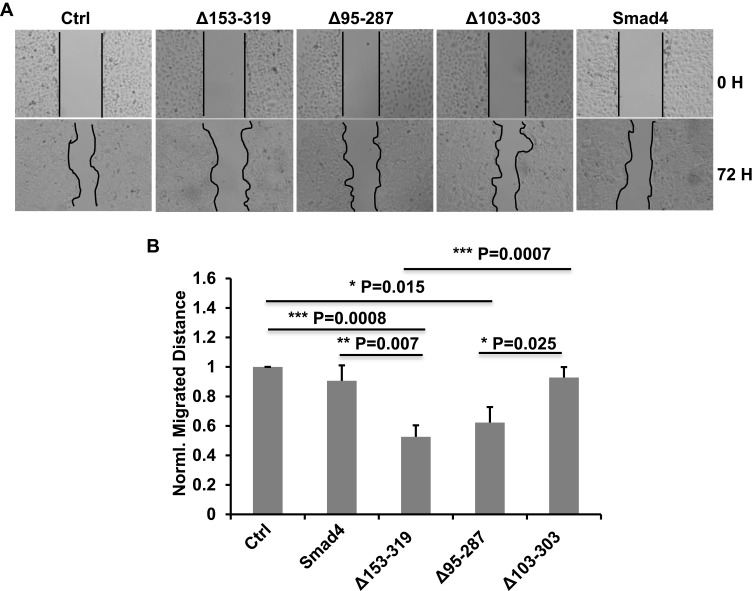
Our previous results showed that the novel splicing variants of Smad4 have effects on the expression of E-cad. We inferred that these variants possibly have effects on the migration of A549 cells. To confirm it, we did the wound-healing assay in A549 cells that recombinantly expressed wildtype Smad4 or variants to measure the migration ability of different groups of cells. The A549 cells were infected with lentiviral wild type Smad4, Δ153-319, Δ95-287 or Δ103-303 and selected with puromycin. Successfully infected cells were cultured in F-12K medium without FBS. As shown in Figure 4, the migration of A549 cells expressed Δ153-319 was significantly inhibited compared with both control and wild type Smad4 groups (Figure 4). While the migration ability of A549 cells that expressed Δ103-303 variant was not different compared with both wild type Smad4 and control groups (Figure 4). Moreover, the expression of Δ103-303 significantly facilitated the migration of A549 cells compared with Δ153-319 and Δ95-287 groups (Figure 4).
Figure 4.
Smad4 variants affect the migration of A549 cells. (A) Representative images show the wound-healing detected on time point 0 h and 72 h in A549 cells that recombinantly express wildtype Smad4 or its variants as indicated. (B) Bar graphs show the normalized migrated distance of A549 cells as indicated in panel A. n=4, Error bars: Mean±SD. *P<0.05, **P<0.01, ***P<0.001.
These data demonstrated that the novel splicing variants of Smad4 have different effects on the migration of A549 cells. Specifically, Δ153-319 significantly inhibited the A549 cell migration, while Δ103-303 has little effects on A549 cell migration.
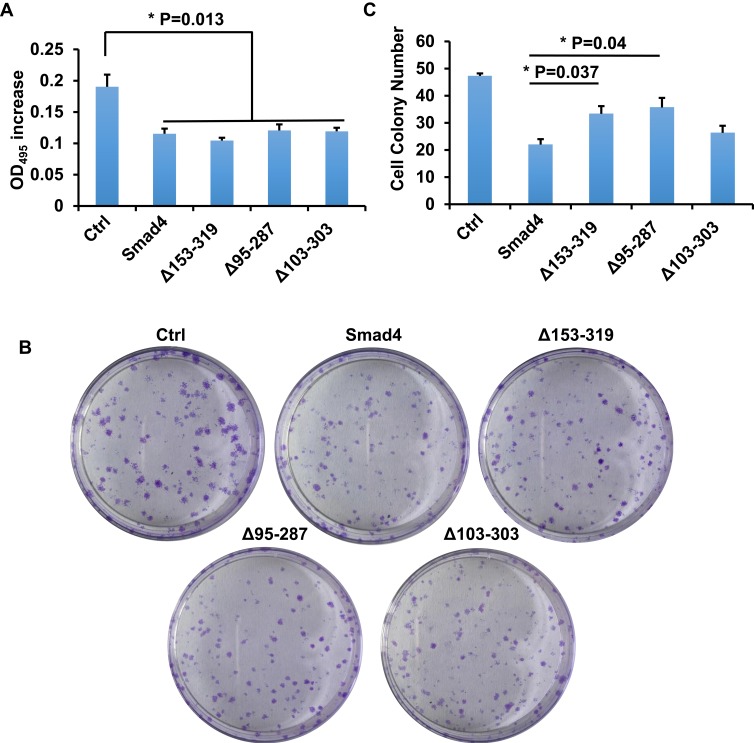
Δ95-287 Promotes the Proliferation of A549 Cells
TGF-β/Smad signaling mainly regulate the proliferation and invasion of cells.8 The splicing variants of Smad4 were expressed after the TGF-β treatment in A549 cells, and we asked whether these variants affect the proliferation of A549 cells. To answer this question, MTT assay and colony formation assay were done. We first performed the MTT assay to measure the proliferation of cells. Cells were cultured for 12 h and OD495 was measured. As shown in Figure 5A, all the variants and wildtype Smad4 inhibited the proliferation of A549 cells compared with the control group. However, there was no significant difference among all variants and wildtype Smad4. In order to zoom in the effects, we performed a colony formation assay. 500 cells were seeded in culture dishes, and 14 days later the colony number was measured. Compared with wildtype Smad4 group, the Δ153-319 and Δ95-287 groups formed more colonies (Figure 5B and C), indicating that these two variants facilitate tumor growth in long terms.
Figure 5.
Smad4 variants affect the proliferation of A549 cells. (A) Bar graphs show the OD495 increment of A549 cells that recombinantly express wild type Smad4 or its variants as indicated. (B) Representative images show colony formation of A549 cells that recombinantly express wildtype Smad4 or its variants as indicated. (C) Bar graphs show cell colony numbers of A549 cells as indicated in panel B. n=3, Error bars: Mean±SD. *P<0.05.
Discussion
Smad4 is the only co-Smad in Smads protein family playing crucial roles in TGF-β/Smad signaling pathway. It was reported that Smad4 has variants expressed in both normal and tumor tissues. All the six variants that Pierreux et al28 detected in HaCat cells were expressed in embryonic stem cells and embryo fibroblast cells as well. Two variants that Kageyama et al30 found were also expressed in normal tissue at a low level. Another two novel genetic variants in the promoter of Smad4 had been found in 85% of pancreatic cancer tissues. One was −462T (14)/-4T (10) affecting the activity of Smad4 and the other was −462T (15)/-4T (12), which might regulate gene expression of Smad4.31 We also detected three novel expressed variants of Smad4 in lung cancer cells (Figure 2). No matter in tumors or in normal tissues, most of Smad4 variants are detected at the mRNA level.
The exon deletion is the main pattern forming Smad4 variants, and most of the reported variants of Smad4 have different lengths of the linker region. Wang et al20 have found eight cDNAs of Smad4 encoding four proteins in Cyprinus carpio, and these isoforms mainly differ in the linker domain, which may not result from allelic variations. In human neuroblastoma, Kageyama et al30 had found at least two spliced variants of Smad4 mRNAs, one was deleted exon4-6 and the other was deleted exon5-6.30 In our works, we detected three variants of Smad4, and Δ153-319 mainly deleted exon4-7, Δ95-287 mainly deleted exon3-5 and Δ103-303 mainly deleted exon3-6 (Figure 2). Furthermore, the advanced-stage neuroblastomas expressed more Smad4 spliced variants than the low-stage tumors. In thyroid tumors, four abnormal-spliced variants losing part of the linker region were reported.21 Two of them were similar to what Kageyama et al found, and others were novel. Detecting the mRNA expression of Smad4 in HaCat cells, Pierreux et al28 found six widely expressed alternatively spliced variants of Smad4 resulting from different exon deletion in the linker region. They also proved that all these spliced variants could interact with activated R-Smads and form complexes.28
In our studies, we detected three novel expressed variants of Smad4 in A549 cells after TGF-β induction. These variants were deleted exons mainly in the linker region. Recombinant expression of these variants in A549 cells and detecting their effects on cell migration, proliferation and protein expression of E-cad and VIM, we found that Δ153-319 significantly inhibits the migration of A549 cells and downregulates the VIM protein expression (Figures 3 and 4). These results indicated that Δ153-319 probably inhibited the EMT process. However, Δ95-287 slightly promoted the cell proliferation in MTT assay and significantly facilitates the colony formation (Figure 5). Compared to Smad4, Δ95-287 inhibited cell migration without a statistical difference (Figure 4). Western blot results showed increased protein expression of both E-cad and VIM (Figure 3). These results indicated that Δ95-287 probably promoted the EMT process, and more studies are needed to prove this point. Hence, we predicted that Δ95-287 facilitated tumor growth mainly through enhancing the proliferation ability of tumor cells, and might promote tumor metastasis. Recombinant expression of Δ103-303 showed no significant effects on cell migration and proliferation compared with wildtype Smad4 group, no matter in MTT assay or colony formation assay.
MH1 and MH2 domain were always against each other because MH1 interferes the MH2 interaction with phosphorylated R-Smads, and MH2 prevents MH1 from binding to DNAs.32 The linker region between them is often deleted during alternatively splicing, which may change the interaction between MH1 and MH2. Recently, Demagny et al have found three GSK3 phosphorylation sites, which were related to Smad4 degradation, in the linker region, and the mutation or deletions of these sites decreased the degradation of Smad4.33,34 Those alternatively spliced variants deleting the GSK3 phosphorylation sites that are in the linker region may have the potential ability against the degradation. The three variants we detected in lung cells were also mainly deleted in the linker region. Depending on their different effects on cell proliferation and migration, we postulate that exon3 and exon6 are probably related to the regulation of cell migration and proliferation, respectively. It is possible that these three variants have the potential against degradation since their deletions in the linker region.
In summary, we successfully induced EMT process in A549 cells with TGF-β and detected three novel expressed alternatively spliced variants of Smad4, namely Δ153-319, Δ95-287 and Δ103-303. To explore their functions, we measured their effects on cell proliferation, migration and protein expression of E-cad and VIM. According to the results, we predicted that Δ153-319 inhibits the EMT process through inhibiting VIM expression, and further against the migration of cells. Δ95-287 facilitated the proliferation of cells to promote tumor growth. However, Δ103-303 showed no significant effects on the proliferation and migration of cells. These findings give new sights into the alternatively spliced variants. Considering them, such as Δ153-319 and Δ95-287, as targets may give new sights into the treatment strategy for NSCLCs.
Funding Statement
This research was funded by the National Natural Science Foundation of China (No. 31670952, No.81902796), and the Natural Science Foundation of Guangdong Province (No. 2019A1515011854).
Disclosure
The authors declare that they have no potential conflict of interests in this work.
References
- 1.Zappa C, Mousa SA. Non-small cell lung cancer: current treatment and future advances. Transl Lung Cancer Res. 2016;5(3):288–300. doi: 10.21037/tlcr [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Singhal S, Miller D, Ramalingam S, et al. Gene expression profiling of non-small cell lung cancer. Lung Cancer. 2008;60(3):313–324. doi: 10.1016/j.lungcan.2008.03.007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Molina JR, Yang P, Cassivi SD, et al. Non-small cell lung cancer: epidemiology, risk factors, treatment, and survivorship. Mayo Clin Proc. 2008;83(5):584–594. doi: 10.1016/S0025-6196(11)60735-0 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Jemal A, Bray F, Center MM, et al. Global cancer statistics. CA Cancer J Clin. 2011;61(2):69–90. doi: 10.3322/caac.v61:2 [DOI] [PubMed] [Google Scholar]
- 5.Saito S, Espinoza-Mercado F, Liu H, et al. Current status of research and treatment for non-small cell lung cancer in never-smoking females. Cancer Biol Ther. 2017;18(6):359–368. doi: [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Brigatinib outperforms crizotinib as first-line therapy. Cancer Discov. 2019;10(2):OF5. [DOI] [PubMed] [Google Scholar]
- 7.Suzuki S, Ishida T, Yoshikawa K, et al. Current status of immunotherapy. Jpn J Clin Oncol. 2016;46(3):191–203. doi: 10.1093/jjco/hyv201 [DOI] [PubMed] [Google Scholar]
- 8.Borghaei H, Paz-ares L, Horn L, et al. Nivolumab versus docetaxel in advanced nonsquamous non-small-cell lung cancer. N Engl J Med. 2015;373(17):1627–1639. doi: 10.1056/NEJMoa1507643 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Herbst RS, Baas P, Kim D-W, et al. Pembrolizumab versus docetaxel for previously treated, PD-L1-positive, advanced non-small-cell lung cancer (KEYNOTE-010): a randomised controlled trial. Lancet. 2016;387(10027):1540–1550. doi: 10.1016/S0140-6736(15)01281-7 [DOI] [PubMed] [Google Scholar]
- 10.Zhao M, Mishra L, Deng CX. The role of TGF-β/SMAD4 signaling in cancer. Int J Biol Sci. 2018;14(2):111–123. doi: 10.7150/ijbs.23230 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Wang F, Xia X, Yang C, et al. SMAD4 gene mutation renders pancreatic cancer resistance to radiotherapy through promotion of autophagy. Clin Cancer Res. 2018;24(13):3176–3185. doi: 10.1158/1078-0432.CCR-17-3435 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Mizuno T, Cloyd JM, Vicente D, et al. SMAD4 gene mutation predicts poor prognosis in patients undergoing resection for colorectal liver metastases. Eur J Surg Oncol. 2018;44(5):684–692. doi: 10.1016/j.ejso.2018.02.247 [DOI] [PubMed] [Google Scholar]
- 13.Vorselaars VMM, Diederik A, Prabhudesai V, et al. SMAD4 gene mutation increases the risk of aortic dilation in patients with hereditary haemorrhagic telangiectasia. Int J Cardiol. 2017;245:114–118. doi: 10.1016/j.ijcard.2017.06.059 [DOI] [PubMed] [Google Scholar]
- 14.Wu L. Functional characteristics of a novel SMAD4 mutation from thoracic aortic aneurysms (TAA). Gene. 2017;628:129–133. doi: 10.1016/j.gene.2017.07.042 [DOI] [PubMed] [Google Scholar]
- 15.Karlsson MC, Gonzalez SF, Welin J, et al. Epithelial-mesenchymal transition in cancer metastasis through the lymphatic system. Mol Oncol. 2017;11(7):781–791. doi: 10.1002/mol2.2017.11.issue-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Iwano M. EMT and TGF-beta in renal fibrosis. Front Biosci (Schol Ed). 2010;2:229–238. doi: 10.2741/s60 [DOI] [PubMed] [Google Scholar]
- 17.Zhang C, Hao Y, Wang Y, et al. TGF-β/SMAD4-regulated LncRNA-LINP1 inhibits epithelial-mesenchymal transition in lung cancer. Int J Biol Sci. 2018;14(12):1715–1723. doi: 10.7150/ijbs.27197 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zeng Y, Zhu J, Shen D, et al. Repression of Smad4 by miR-205 moderates TGF-β-induced epithelial-mesenchymal transition in A549 cell lines. Int J Oncol. 2016;49(2):700–708. doi: 10.3892/ijo.2016.3547 [DOI] [PubMed] [Google Scholar]
- 19.Ast G. How did alternative splicing evolve? Nat Rev Genet. 2004;5(10):773–782. doi: 10.1038/nrg1451 [DOI] [PubMed] [Google Scholar]
- 20.Wang ET, Sandberg R, Luo S, et al. Alternative isoform regulation in human tissue transcriptomes. Nature. 2008;456(7221):470–476. doi: 10.1038/nature07509 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Lazzereschi D, Nardi F, Turco A, et al. A complex pattern of mutations and abnormal splicing of Smad4 is present in thyroid tumours. Oncogene. 2005;24(34):5344–5354. doi: 10.1038/sj.onc.1208603 [DOI] [PubMed] [Google Scholar]
- 22.Watson CM, Camm N, Crinnion LA, et al. Characterization and genomic localization of a SMAD4 processed pseudogene. J Mol Diagn. 2017;19(6):933–940. doi: 10.1016/j.jmoldx.2017.08.002 [DOI] [PubMed] [Google Scholar]
- 23.Ashktorab H, Mokarram P, Azimi H, et al. Targeted exome sequencing reveals distinct pathogenic variants in Iranians with colorectal cancer. Oncotarget. 2017;8(5):7852–7866. doi: 10.18632/oncotarget.v8i5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Norris AL, Workman RE, Fan Y, et al. Nanopore sequencing detects structural variants in cancer. Cancer Biol Ther. 2016;17(3):246–253. doi: 10.1080/15384047.2016.1139236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Wang Y, Huang H-Y, Bian G-L, et al. A functional variant of SMAD4 enhances thoracic aortic aneurysm and dissection risk through promoting smooth muscle cell apoptosis and proteoglycan degradation. EBioMedicine. 2017;21:197–205. doi: 10.1016/j.ebiom.2017.06.022 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Ullah I, Liao Y, Wan R, et al. Alternative splicing of SMAD4 and its function in HaCaT cells in response to UVB irradiation. J Cancer. 2018;9(17):3177–3186. doi: 10.7150/jca.24756 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Wang X-L, Huang C. Difference of TGF-β/Smads signaling pathway in epithelial-mesenchymal transition of normal colonic epithelial cells induced by tumor-associated fibroblasts and colon cancer cells. Mol Biol Rep. 2019;46(3):2749–2759. doi: 10.1007/s11033-019-04719-5 [DOI] [PubMed] [Google Scholar]
- 28.Pierreux CE, Nicolás FJ, Hill CS. Transforming growth factor beta-independent shuttling of Smad4 between the cytoplasm and nucleus. Mol Cell Biol. 2000;20(23):9041–9054. doi: 10.1128/MCB.20.23.9041-9054.2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Beavon IR. The E-cadherin-catenin complex in tumour metastasis: structure, function and regulation. Eur J Cancer. 2000;36(13):1607–1620. doi: 10.1016/S0959-8049(00)00158-1 [DOI] [PubMed] [Google Scholar]
- 30.Kageyama H, Seki N, Yamada S, et al. DPC4 splice variants in neuroblastoma. Cancer Lett. 1998;122(1–2):187–193. doi: 10.1016/S0304-3835(97)00389-3 [DOI] [PubMed] [Google Scholar]
- 31.Nikolic A, Kojic S, Knezevic S, et al. Structural and functional analysis of SMAD4 gene promoter in malignant pancreatic and colorectal tissues: detection of two novel polymorphic nucleotide repeats. Cancer Epidemiol. 2011;35(3):265–271. doi: 10.1016/j.canep.2010.10.002 [DOI] [PubMed] [Google Scholar]
- 32.Hata A, Lo RS, Wotton D, et al. Mutations increasing autoinhibition inactivate tumour suppressors Smad2 and Smad4. Nature. 1997;388(6637):82–87. doi: 10.1038/40424 [DOI] [PubMed] [Google Scholar]
- 33.Demagny H, Araki T, De Robertis EM. The tumor suppressor Smad4/DPC4 is regulated by phosphorylations that integrate FGF, Wnt, and TGF-β signaling. Cell Rep. 2014;9(2):688–700. doi: 10.1016/j.celrep.2014.09.020 [DOI] [PubMed] [Google Scholar]
- 34.Demagny H, De Robertis EM. Point mutations in the tumor suppressor Smad4/DPC4 enhance its phosphorylation by GSK3 and reversibly inactivate TGF-β signaling. Mol Cell Oncol. 2016;3(1):e1025181. doi: 10.1080/23723556.2015.1025181 [DOI] [PMC free article] [PubMed] [Google Scholar]