Abstract
Research on non‐coding RNA (ncRNA) is a rapidly expanding field. Providing an official gene symbol and name to ncRNA genes brings order to otherwise potential chaos as it allows unambiguous communication about each gene. The HUGO Gene Nomenclature Committee (HGNC, http://www.genenames.org) is the only group with the authority to approve symbols for human genes. The HGNC works with specialist advisors for different classes of ncRNA to ensure that ncRNA nomenclature is accurate and informative, where possible. Here, we review each major class of ncRNA that is currently annotated in the human genome and describe how each class is assigned a standardised nomenclature.
Keywords: gene nomenclature, gene symbols, non‐coding RNA
Subject Categories: RNA Biology
Working with experts in the field, the HUGO Gene Nomenclature Committee describes the standardized nomenclature assigned to all major classes of ncRNA currently annotated in the human genome.
Introduction
The HUGO Gene Nomenclature Committee (HGNC) works under the auspices of Human Genome Organisation (HUGO) and is the only worldwide authority that assigns standardised symbols and names to human genes (Braschi et al, 2019). A unique symbol for every gene is essential to enable unambiguous scientific communication, and approved symbols should be used ubiquitously in research papers, conference talks and posters, and biomedical databases. The HGNC endeavours to approve symbols for all classes of genes that are supported by gene annotation projects and began working on non‐coding RNA (ncRNA) nomenclature in the mid‐1980s with the approval of initial gene symbols for mitochondrial transfer RNA (tRNA) genes. Since then, we have worked closely with experts in the ncRNA field to develop symbols for many different kinds of ncRNA genes.
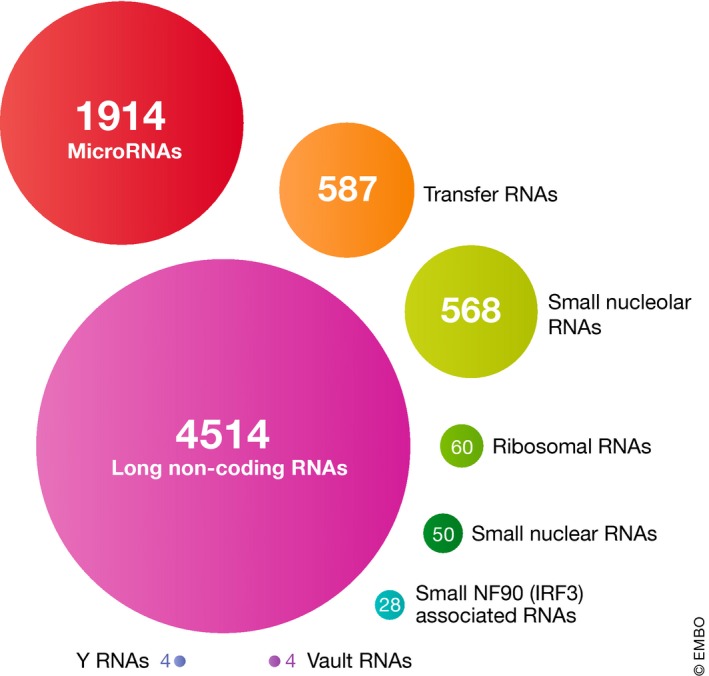
The number of genes that the HGNC has named per ncRNA class is shown in Fig 1, and ranges in number from over 4,500 long ncRNA (lncRNA) genes and over 1,900 microRNA genes, to just four genes in the vault and Y RNA classes. Every gene symbol has a Symbol Report on our website, http://www.genenames.org, which displays the gene symbol, gene name, chromosomal location and also includes links to key resources such as Ensembl (Zerbino et al, 2018), NCBI Gene (O'Leary et al, 2016) and GeneCards (Stelzer et al, 2016). We collaborate directly with these biomedical databases and, importantly, these databases always use our gene symbols as the primary symbol for the gene. Due to the relative completeness of the HGNC ncRNA gene set, our data have been chosen as the canonical human dataset in the RNAcentral database (The RNAcentral Consortium, 2019), an RNA sequence database resource. For microRNAs, we work with the specialist resource miRBase (Kozomara et al, 2019), and for tRNAs, we work with the specialist resource GtRNAdb (Chan & Lowe, 2016). We display links to these resources from the relevant Symbol Report. Where available, for lncRNAs we provide specialist links to LNCipedia (Volders et al, 2019), a key lncRNA resource that displays HGNC gene symbols (Box 1).
Figure 1. The number of HGNC gene symbols by type of ncRNA.
A full list of locus types, along with numbers of genes per category, can be found at our Statistics & Downloads webpage (https://www.genenames.org/download/statistics-and-files/).
Box 1. Useful resources for non‐coding RNA genes used by the HGNC.
| RNA resource | Resource URL | Description |
|---|---|---|
| RNAcentral | https://rnacentral.org/ | Centralised database of non‐coding RNA sequences collated from expert non‐coding RNA member databases, model organism databases and sequence accession databases |
| miRBase | http://www.mirbase.org/ | Searchable database of microRNA sequences and annotations. Also hosts the miRBase registry where researchers can submit prospective new microRNAs |
| GtRNAdb | http://gtrnadb.ucsc.edu/ | The genomic tRNA database, which contains predicted tRNA genes by the tRNAscan‐SE program for many different species |
| snoRNABase | https://www-snorna.biotoul.fr/ | Database of human snoRNA genes; useful resource but no longer being updated |
| LNCipedia | https://lncipedia.org/ | Database of human long non‐coding RNA sequences and manually curated lncRNA articles |
| Ensembl | http://www.ensembl.org/ | Genome browser for vertebrate genomes that hosts the GENCODE annotation models for non‐coding RNA genes for mouse and human genes |
| NCBI Gene | https://www.ncbi.nlm.nih.gov/gene/ | Integrated annotation and related information for many different genomes. Incudes RefSeq manual annotation of human and mouse non‐coding RNA genes |
For each class of ncRNA, we host curated gene group pages on http://www.genenames.org—a list of URLs for these is shown in Table 1.
Table 1.
The HGNC hosts gene group pages for different types of non‐coding RNA genes. These pages follow a hierarchical structure and all pages can be browsed starting at the highest‐level gene group page labelled “Non‐coding RNAs”
| Gene group name | Gene group URL | Description |
|---|---|---|
| Non‐coding RNAs | https://www.genenames.org/data/genegroup/#!/group/475 | Overview page of all non‐coding RNAs in the HGNC project. Can be used as a starting point to browse through all types of named ncRNAs |
| MicroRNAs | https://www.genenames.org/data/genegroup/#!/group/476 | Starting page for all microRNAs, which are split into curated human families where possible. MicroRNAs not in a defined family are listed on the first page |
| MicroRNA host genes | https://www.genenames.org/data/genegroup/#!/group/1690 | A curated list of microRNA host genes, which is split into protein coding and non‐coding subgroups |
| Transfer RNAs | https://www.genenames.org/data/genegroup/#!/group/478 | Starting page for all transfer RNA genes, with subgroups “Mitochondrially encoded transfer RNAs” and “Cytoplasmic transfer RNAs” (this page also has the subsets “Cytoplasmic transfer RNA pseudogenes” and “Low confidence cytoplasmic transfer RNAs”) |
| Small nuclear RNAs | https://www.genenames.org/data/genegroup/#!/group/1819 | Lists all canonical small nuclear RNA genes; variant snRNA genes are shown as a subgroup |
| Small nucleolar RNAs | https://www.genenames.org/data/genegroup/#!/group/844 | Starting page for snoRNAs with the subgroups “Small Cajal body‐specific RNAs”, “Small nucleolar RNAs, C/D box” and “Small nucleolar RNAs, H/ACA box” |
| Small nucleolar RNA host genes | https://www.genenames.org/data/genegroup/#!/group/1838 | A curated list of snoRNA host genes, which is split into protein coding and non‐coding subgroups |
| Ribosomal RNAs | https://www.genenames.org/data/genegroup/#!/group/848 | Starting page for all ribosomal RNAs, split into the major subgroups “Mitochondrially encoded ribosomal RNAs” and “Cytoplasmic ribosomal RNAs”, which is further split into subtypes of rRNAs |
| Vault RNAs | https://www.genenames.org/data/genegroup/#!/group/852 | Full list of vault RNA genes |
| Y RNAs | https://www.genenames.org/data/genegroup/#!/group/853 | Full list of Y RNA genes |
| Small NF90 (ILF3) associated RNAs | https://www.genenames.org/data/genegroup/#!/group/1624 | Full list of SNAR genes |
| Long non‐coding RNAs | https://www.genenames.org/data/genegroup/#!/group/788 | Starting page for all long non‐coding RNA gene. Divided into subgroups: Long intergenic non‐protein coding RNAs, MicroRNA non‐coding host genes, Overlapping transcripts, Intronic transcripts, Antisense RNAs, Divergent transcripts, Small nucleolar RNA non‐coding host genes, Long non‐coding RNAs with non‐systematic symbols, Long non‐coding RNAs with FAM root symbol |
The aim of this paper was to provide an overview for each of the main types of ncRNA that we have named, as well as a guide to how we name them. Each section has been written in collaboration with our specialist advisors for each ncRNA class: Sam Griffiths‐Jones of the University of Manchester for microRNAs, Todd Lowe of the University of California, Santa Cruz for tRNAs, Dawn O'Reilly of the University of Oxford for small nuclear RNAs (snRNAs), Peter Stadler of the University of Leipzig for small nucleolar and vault RNAs, Andrew Pierce currently at AstraZeneca, Cambridge for ribosomal RNAs (rRNAs), Sandra Wolin of the NIH for Y RNAs, Michael Mathews of Rutgers New Jersey Medical School for small NF90 (ILF3) associated RNAs, and Igor Ulitsky of the Weizmann Institute of Science and Ling‐Ling Chen of the Shaghai Institute for Biochemistry and Cell Biology for long non‐coding RNAs. We finish by outlining recommendations for the nomenclature of circular and circular intronic RNAs, which are currently lacking official nomenclature.
MicroRNAs
MicroRNAs are transcripts of ~ 22 nucleotides that mediate the post‐transcriptional regulation of genes via direct binding to messenger RNA (mRNA) molecules. In animal cells, microRNA (miRNA) genes are usually transcribed as long primary transcripts (pri‐miRNAs), which are processed by the Drosha microprocessor complex into precursor hairpin stem‐loop sequences (pre‐miRNAs). These hairpins are exported from the nucleus to the cytoplasm, where the stem‐loop is cleaved by the Dicer enzyme to produce a ~ 22 nt duplex. One strand of the duplex associates with an Argonaute (AGO) protein and this microRNA ribonucleoprotein complex (miRNP) binds to sites in mRNAs that are complementary to the miRNA sequence, usually in the 3′ untranslated region (UTR). The Ago‐miRNP complex then recruits other proteins, which typically mediate either the degradation or translational repression of the mRNA [for a review, see (Bartel, 2018)]. Approximately 60% of all human genes produce mRNAs that can be bound by miRNAs (Friedman et al, 2009), so these small RNAs provide regulation for diverse biological processes across all tissue types and stages of life. As such, miRNA genes have been implicated in many human diseases including rheumatoid arthritis (Guggino et al, 2018), deafness (Mencía et al, 2009), stroke (Panagal et al, 2019), psoriasis (Yan et al, 2019), cirrhosis (Fernández‐Ramos et al, 2018) and several forms of cancer (Kwok et al, 2017).
The name “microRNA” to reflect the small size of the active RNA molecule was agreed upon and first used by three Caenorhabditis elegans research groups that published in the same 2001 issue of Science (Lagos‐Quintana et al, 2001; Lau et al, 2001; Lee & Ambros, 2001). Once the field of miRNA research started to expand, experts came together to publish guidelines on how to name these transcripts across species (Ambros et al, 2003), and the miRNA Registry was founded to ensure that the same symbols were not mistakenly used by different research groups for different miRNAs (Griffiths‐Jones, 2004). The miRNA Registry evolved into the dedicated online miRNA resource miRBase, which has continued to be responsible for providing unique identifiers for miRNAs as well as acting as a database of sequences and curated publications (Kozomara et al, 2019). Researchers submit hairpin and mature microRNA sequences to miRBase, which are then publicly assigned new symbols after manuscript acceptance. miRBase assigns each microRNA stem‐loop sequence a symbol in the format “mir‐#” and each mature miRNA a symbol in the format “miR‐#” followed by a unique sequential number that reflects order of submission to the database. The HGNC then approves a gene symbol for human miRNA genes in the format MIR#; for example, as shown in Fig 2 and Box 2, MIR17 represents the miRNA gene, mir‐17 represents the stem‐loop, and miR‐17 represents the mature miRNA. However, the complete extent of the miRNA gene and primary transcript is not often known, so the entity associated with an HGNC name and entry is frequently the length of the hairpin precursor miRNA, rather than the primary transcript. For genes that encode identical mature miRNAs, the same unique identifier is used followed by a hyphenated numerical suffix; e.g., MIR1‐1 and MIR1‐2 are distinct genomic loci that encode identical mature miRNAs. For paralogous genes that encode mature miRNAs, which differ by only one or two nucleotides, the same unique identifier is used followed by a letter suffix, e.g. MIR10A and MIR10B. The HGNC does not accept any direct requests for miRNA gene symbols, and all requests must go to miRBase first (please see http://www.mirbase.org/registry.shtml).
Figure 2. The microRNA gene MIR17 is part of a cluster of microRNA genes that are hosted within an intron of the long non‐coding RNA gene MIR17HG (miR‐17‐92a‐1 cluster host gene).
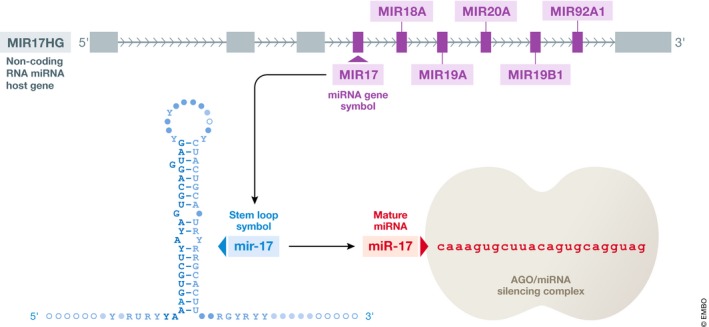
The symbol MIR17 represents the gene; the symbol mir‐17 represents the miRNA precursor stem‐loop structure; and the symbol miR‐17 represents the active mature microRNA, which interacts with an AGO protein to form the AGO/miRNA silencing complex.
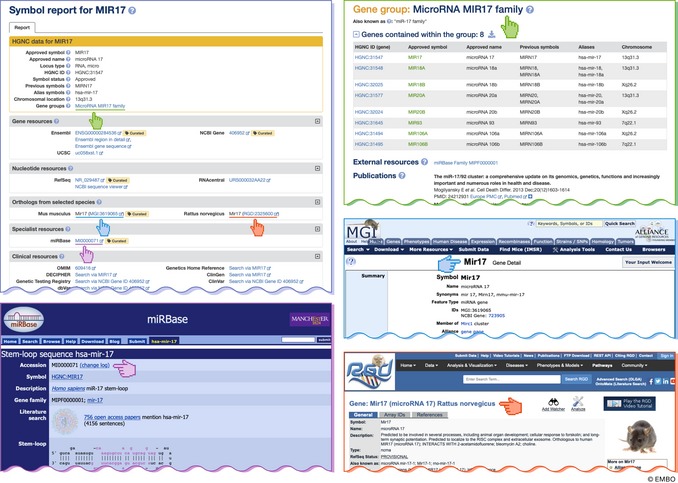
Box 2. The HGNC Symbol Report for MIR17 provides more than gene nomenclature: as highlighted here there is a link to the HGNC “MIR17 microRNA family group page”; a link out to the relevant microRNA report on miRBase; and where possible a link to the mouse ortholog at MGI and the rat ortholog at RGD.
In accordance with miRBase, the HGNC provides one gene symbol per miRNA gene, even though miRNAs are sometimes processed from the same transcripts as proteins or other miRNAs, and therefore might not be considered separate genes in the canonical sense. For example, many miRNAs are hosted in the introns, or less frequently the exons, of protein coding genes or long non‐coding RNA genes (Fig 2 and Box 2). The HGNC has curated gene group pages listing these host genes (Table 1), and the naming conventions for non‐coding miRNA host genes are discussed in the long non‐coding RNA section below.
Recently, there have been a few ideas published on how to “improve” miRNA nomenclature, including correcting the identifiers of particular miRNA genes to show evolutionary relationships (e.g. Desvignes et al, 2015; Fromm et al, 2015; Budak et al, 2016). As nomenclature advisors, we understand the desire to perfect nomenclature systems once more information becomes available. At the same time, experience has taught us that such revised systems are often not fully adopted and may cause considerable confusion in the community. It can therefore be more appropriate to find other ways to represent relationships between genes, in order to maintain stable gene symbols. The HGNC has recently curated gene groups to show paralogous relationships between human miRNA genes, based on the family groups at miRBase and information in publications. For example, the “MicroRNA MIR1/206 family” contains the family members MIR1‐1, MIR1‐2 and MIR206. The miRNA symbol miR‐206 has already been used in over 600 papers so it would be unhelpful to try to alter this symbol. However, the MIR206 Symbol Report now provides a link to the curated MicroRNA MIR1/206 family gene group page, where there are also associated publications and a link through to the corresponding miRBase Family MIPF0000038 page, which lists orthologous and paralogous miRNAs in different species. Where possible, the miRNA Symbol Reports on genenames.org also display the mouse and rat miRNA orthologs, with links to the relevant gene report on the Mouse Genomic Database (http://www.informatics.jax.org/) and Rat Genome Database (https://rgd.mcw.edu/), see Box 2.
Transfer RNAs
Transfer RNA was the first type of non‐coding RNA to be characterised over 60 years ago (Hoagland et al, 1958). The term “transfer” (Smith et al, 1959) represents the function of this RNA in transferring amino acids from the cytosol of the cell to the ribosome where the amino acids are bonded together to form a peptide according to the sequence of the mRNA being translated. Typical tRNAs vary in size from 73 to 93 nucleotides (Rich & RajBhandary, 1976) and have a distinctive cloverleaf secondary structure that folds into an L‐shaped tertiary structure (Kim et al, 1973). At one end of the L is the CCA acceptor site where the tRNA binds to the relevant amino acid (Hou, 2010) and at the other end is a loop that contains the three‐nucleotide anticodon which precisely pairs to the codons of mRNA (Kim et al, 1973). The first two nucleotides of the anticodon form Watson‐Crick base pairs with the corresponding mRNA codon, while the third nucleotide can form “wobble” pairing which allows one tRNA to recognise more than one mRNA codon. Post‐transcriptional modifications at the “wobble” position can influence binding to a particular mRNA codon (Agris et al, 2018).
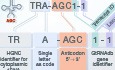
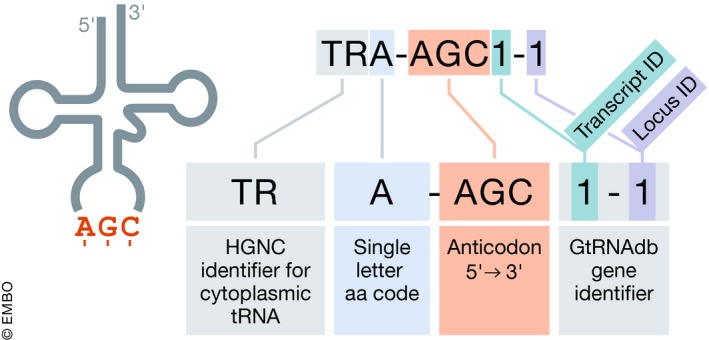
Transfer RNA genes share characteristics that make it possible to predict them from genomic sequence. The Genomic tRNA Database (GtRNAdb) (Chan & Lowe, 2016) contains predicted tRNA gene sets for thousands of species across Eukaryota, Archaea and Bacteria, including a set of 429 high confidence tRNA genes for the most current human reference genome, GRCh38. tRNA gene predictions are made using the tRNAscan‐SE analysis pipeline (Lowe & Chan, 2016), which uses probabilistic tRNA primary sequence and secondary structure “covariance models” to determine the gene loci and the functional identity (i.e. tRNA isotype and anticodon) for each putative tRNA gene. The predicted tRNA genes then undergo further analysis by comparison with isotype‐specific covariance models to give confirmation of isotype classification. The GtRNAdb assigns a unique ID to each tRNA gene in the format tRNA‐[three letter amino acid code]‐[anticodon]‐[GtRNAdb gene identifier], e.g. tRNA‐Ala‐AGC‐1‐1. (Note the “GtRNAdb gene identifier” is actually made up of two numbers, the first is a “transcript ID”, the second a “locus ID”, such that multiple gene loci producing identical tRNA transcripts share the same transcript ID, but each have a different locus numbers; e.g., Ala‐AGC‐1‐1 and Ala‐AGC‐1‐2 are two different gene loci producing identical mature tRNAs, whereas Ala‐AGC‐2‐1 and Ala‐AGC‐3‐1 are genes that each produce different tRNA transcripts.) The HGNC assigns a slightly condensed but equivalent tRNA gene symbol in the format TR[one letter amino acid code]‐[anticodon][GtRNAdb gene identifier], e.g. TRA‐AGC1‐1 (Fig 3). tRNAscan‐SE analysis also predicts tRNA pseudogenes and candidate genes that include atypical tRNA features and may not be transcribed and/or may not be capable of ribosomal translation. To reflect these different sets, the HGNC displays the gene groups “Cytosolic transfer RNAs”, “Low confidence cytosolic transfer” RNAs and “Transfer RNA pseudogenes on genenames.org” (Table 1).
Figure 3. An annotated tRNA gene symbol explaining what each part of the approved gene symbol represents.
The human mitochondrial genome contains 22 tRNA genes (Anderson et al, 1981) that encode tRNAs with both canonical and non‐canonical cloverleaf structures which enable translation within mitochondrial ribosomes in the mitochondria. While pathological mutations in cytosolic tRNA genes have not yet been discovered, mutations in mitochondrial tRNA genes cause a variety of well‐studied mitochondrial diseases such as MELAS (mitochondrial encephalomyopathy, lactic acidosis and stroke‐like episodes) and MERRF (myoclonic epilepsy with ragged red fibres) (Suzuki & Nagao, 2011; Abbott et al, 2014). Mitochondrial tRNA genes were named in collaboration with the MitoMap resource (Lott et al, 2013); gene symbols are of the format “MT‐T + one letter amino acid code”; e.g., MT‐TA represents the mitochondrial tRNA gene that recruits alanine. Most amino acids are decoded by just one human mitochondrial tRNA, but there are two mitochondrial leucine and serine tRNA genes—these gene symbols therefore include numbers to distinguish the individual loci: MT‐TL1, MT‐TL2, MT‐TS1 and MT‐TS2.
Small nuclear RNAs
Small nuclear RNAs are abundant transcripts of around 150 nucleotides that end in a 3′ stem loop (Matera et al, 2007). While the name of this RNA class is based on cellular location, each individual snRNA has a “U” identifier that stems from the historical name “U‐RNA” which was derived from early observations of their high uridine content (Hodnett & Busch, 1968). The U‐RNAs were numbered according to their apparent abundance when discovered (Chen & Moore, 2015). Some of these were subsequently found to be small nucleolar RNAs (snoRNAs) resulting in the following numbering for the snRNAs: U1, U2, U4, U5, U6, U7, U11 and U12.
Most snRNAs are involved in the splicing of introns from pre‐mRNA as part of either the major or minor spliceosome. The major spliceosome features U1, U2, U4, U5 and U6 snRNPs, plus many other non‐snRNP proteins, and performs splicing of U2‐type introns. Here, the U1 and U2 snRNPs assemble on introns and are joined by the preassembled U4/U6.U5 tri‐snRNP. This is followed by a series of rearrangements resulting in the formation of the U2/U6 catalytic core and the splicing reaction (Anokhina et al, 2013), and finally release of the spliced RNA and disassembly of the spliceosome. The minor spliceosome splices U12‐type introns, which make up < 0.5% of introns in the genome (Turunen et al, 2013). It contains the same U5 snRNA as the major spliceosome, but in contrast consists of the snRNAs U11, U12, U4atac and U6atac, which are functional analogs of the major spliceosome U1, U2, U4 and U6 snRNAs. Minor spliceosome snRNAs can fold into similar structures to their equivalent major spliceosome snRNAs, but display limited sequence similarity to them (Will & Lührmann, 2005). The term “atac” in U4atac and U6atac refers to the AT/AC splice sites found in the first U12‐type introns to be discovered (Tarn & Steitz, 1996). Instead of splicing, U7 snRNA is involved in processing the distinctive 3′ end stem loop of histone mRNA by binding to the histone downstream element and recruiting proteins, some of which shared with the spliceosome (Strub et al, 1984; Marz et al, 2007). Most snRNAs are transcribed by RNA polymerase II, with the exception of U6 and U6atac, which are transcribed by RNA polymerase III (Singh & Reddy, 1989; Younis et al, 2013).
All snRNA genes are named with the root symbol “RNU” for “RNA, U# small nuclear”. The GRCh38 human reference genome contains four annotated U1‐encoding loci: RNU1‐1, RNU1‐2, RNU1‐3 and RNU1‐4, although individuals may have around 30 copies of tandemly repeated U1 genes (Lund & Dahlberg, 1984). The GRCh38 reference also contains a single U2 gene (RNU2‐1), which resides in a 6 kb region that is organised as a tandem array of 10–20 copies in many individuals (Van Arsdell & Weiner, 1984). The U7 (RNU7‐1), U11 (RNU11), U12 (RNU12), U4atac (RNU4ATAC) and U6atac (RNU6ATAC) snRNAs are each encoded by a single gene. There are two U4 and five U6 genes, which have numerical identifiers in the same format as the U1 genes, e.g. RNU4‐1, RNU6‐2, while the five U5 genes have letter identifiers based on the scientific literature (Sontheimer & Steitz, 1992): RNU5A‐1, RNU5B‐1, RNU5D‐1, RNU5E‐1 and RNU5F‐1.
The human genome contains over 1,000 divergent gene copies of snRNA genes (Vazquez‐Arango & O'Reilly, 2018), most of which are presumed to be unexpressed pseudogenes. In the case of the U1 family, some of the genes present on the 1q21.1 cluster have been shown to be expressed, undergo 3′ end processing and bind U1‐specific proteins to form snRNPs in vivo (O'Reilly et al, 2013). These genes have been named with the root symbol RNVU1 for “RNA, variant U1 small nuclear”. The snRNA vU1.8, encoded by RNVU1‐8, has been shown to be capable of processing the 3′ end of pre‐mRNAs expressed from a subset of target genes (O'Reilly et al, 2013). Moreover, snRNAs encoded by RNVU1‐3, RNVU1‐8 and RNVU1‐20 are implicated in stem cell maintenance and neuromuscular disease (Vazquez‐Arango et al, 2016).
SnoRNAs
Small nucleolar RNAs are transcripts of around 60–170 nucleotides that can be divided into three major classes: C/D box snoRNAs (SNORDs), H/ACA box snoRNAs (SNORAs) and small Cajal body‐specific RNAs (scaRNAs). Although some are transcribed from independent promoters, most snoRNAs are encoded within the introns of either protein coding or long non‐coding “host” genes (see Table 1 for details on accessing gene groups listing these). C/D box snoRNAs are named after their two conserved box motifs: C (sequence: RUGAUGA) and D (sequence: CUGA) (Tyc & Steitz, 1989); these snoRNAs primarily function in the nucleolus within small nucleolar ribonucleoprotein (snoRNP) complexes to direct target site‐specific 2′‐O‐methylation of rRNAs (Kiss‐László et al, 1996). H/ACA box snoRNAs share a common secondary structure and contain the AnAnnA sequence known as the “hinge” or “H” box and the trinucleotide “ACA” box (Ganot et al, 1997a, 1997b). H/ACA snoRNAs also function with snoRNP complexes in the nucleolus to guide modification of rRNAs, but in this case the modification is pseudouridylation of target uridines (Ganot et al, 1997a, 1997b). Small Cajal body‐specific RNAs function in the Cajal body, a nuclear organelle named after its discoverer Santiago Ramón y Cajal (Gall et al, 1999). ScaRNAs contain either H/ACA boxes, C/D boxes or a mixture of both types, and function as guides for the same type of RNA modifications as the nucleolar snoRNAs—guiding RNP complexes to catalyse pseudouridylation or 2′‐O‐methylation—but for modification of snRNAs instead of rRNAs. The major difference in sequence between scaRNAs and snoRNAs is thought to be the presence of Cajal body targeting sequences, the CAB box in H/ACA scaRNAs (Richard et al, 2003) or the G.U/U.G wobble stems in C/D scaRNAs (Marnef et al, 2014). Some snoRNAs show no sequence complementarity to either rRNAs or snRNAs, suggesting they have an alternative function to the canonical snoRNAs described above. For example, there have been recent reports of snoRNAs involved in diverse functions such as activation of enzymes, or regulation of alternative splicing and mRNA levels (Falaleeva et al, 2017).
When snoRNAs were first discovered, they were initially not distinguished from other snRNAs and were therefore assigned “U” numbers, e.g. U3, U8 and U13 (Tyc & Steitz, 1989), which are the identifiers still in use for snRNAs (see small nuclear RNAs section above). Once the H/ACA and C/D boxes were identified, a convention of using the root ACA# (Kiss et al, 2004) or HB‐I# for human H/ACA box snoRNAs and HB‐II# for C/D box snoRNAs (Cavaillé et al, 2000) was established, which then formed a “rival” nomenclature to the U# system that was still in use. Originally, scaRNAs were not discernible from other snoRNAs by symbol; e.g., the first identified scaRNA was referred to as U85 (Jády & Kiss, 2001) and another as ACA26 (Tycowski et al, 2009). In 2007, the HGNC worked with snoRNABase (Lestrade & Weber, 2006) to devise a standardised, easily recognisable nomenclature for all three types of snoRNA: SNORD# for “small nucleolar RNA, C/D box” genes; SNORA# for “small nucleolar RNA, H/ACA box” genes; and SCARNA# for “small Cajal body‐specific RNA” genes. Unfortunately, the snoRNABase resource, although still valuable, is no longer being updated. The HGNC now works with the Stadler Bioinformatics Leipzig group to assign symbols to newly identified snoRNA genes (Jorjani et al, 2016), and as such, the HGNC snoRNA gene group pages (Table 1) provide an up‐to‐date list of canonical human snoRNA and scaRNA genes.
A potential issue for nomenclature is that snoRNAs and scaRNAs cannot always be distinguished unambiguously without evidence of localisation. Thus, ncRNAs of these classes are by default named as SNORA# or SNORD# unless evidence for Cajal body specificity is available. Some snoRNAs are a source of miRNA‐like small RNAs; in a few cases, these small RNAs function in post‐transcriptional gene silencing like miRNAs (Scott & Ono, 2011). Interestingly, H/ACA snoRNAs are processed by Dicer, while small RNAs derived from box C/D snoRNA appear to use a different processing pathway (Langenberger et al, 2013). At present, HGNC does not provide a nomenclature for the small RNAs derived from snoRNA and scaRNAs.
Ribosomal RNAs
The ribosome is responsible for the synthesis of peptides using mRNA as a template. The term “ribosome” was coined by Richard B. Roberts to provide a more user‐friendly version of “ribonucleoprotein particles of the microsome fraction” (Roberts, 1958). The ribosome, its subunits and rRNAs have all been assigned unique identifiers in Svedberg units based on their sedimentation rate in a centrifuge—the eukaryotic ribosome is referred to as the 80S ribosome and comprises a large (60S) subunit that contains 28S, 5S and 5.8S rRNA and a small (40S) subunit that contains 18S rRNA. Both subunits also contain a large number of ribosomal proteins (Khatter et al, 2015). The 28S rRNA forms the core of the large subunit and contains the catalytic peptidyl transferase centre (Polacek & Mankin, 2005) that forms bonds between amino acids to create peptides, meaning that the ribosome is also a ribozyme. 5S rRNA is necessary for translation (Ciganda & Williams, 2011) although its exact role is unclear, while 5.8S rRNA appears to have a role in ribosome translocation (Abou Elela & Nazar, 1997). 18S rRNA is at the core of the small subunit and binds directly to mRNA during translation initiation (Martin et al, 2016) and translation elongation (Tranque et al, 1998; Demeshkina et al, 2000).
Cytoplasmic rRNAs are transcribed from multicopy gene clusters (Fig. 4)—the 5S rRNA cluster on chromosome 1q42.13 (Sørensen & Frederiksen, 1991) transcribed by RNA polymerase III and the 45S rRNA clusters that encode 18S, 5.8S and 28S rRNA on the p arms of the five human acrocentric chromosomes in the cytogenetically visible nucleolar organising regions (NORs) (Gonzalez & Sylvester, 2001) transcribed by RNA polymerase I. There is great variation in the number of rRNA repeats within all of these clusters both within and between different individuals. The 5S cluster is the only one in which individual genes have been annotated on the current GRCh38 human reference genome, although currently there are only 17 annotated 5S rRNA genes, while the average individual has around 98 5S genes (Stults et al, 2008). The HGNC has named the 17 annotated genes RNA5S1‐RNA5S17. The 45S rRNA genes have a highly repetitive nature, which has made accurate sequence assembly difficult, and as a result, no individual 45S rRNA gene is present within the NORs on the GRCh38 reference genome. The number of 45S rRNA genes per cluster differs between individuals and varies from a single gene to more than 140 repeated genes, which are usually arranged in a head‐to‐tail orientation (Stults et al, 2008). The HGNC has approved a gene symbol for each of the acrocentric 45S rRNA clusters: RNR1 (13p12), RNR2 (14p12), RNR3 (15p12), RNR4 (21p12) and RNR5 (22p12; Fig 4). The 45S rRNA repeats are post‐transcriptionally processed into the rRNAs 18S, 5.8S and 28S by a series of cleavage events. The HGNC has reserved the stem symbols RNA45S for pre‐45S transcription units, and RNA18S, RNA5‐8S and RNA28S for each processed rRNA. Each acrocentric 45S rRNA cluster in turn has a set of stem symbols reserved using the same numerical identifier as the RNR cluster symbol; e.g., the symbols RNA45S1, RNA18S1, RNA5‐8S1 and RNA28S1 are stem symbols for rRNA copies from the RNR1 acrocentric cluster. In the future, when the 45S rRNA clusters are added to the reference genome we will assign numbers to each individual gene annotated in each cluster; e.g., RNA45S1‐1, RNA28S1‐1, RNA18S1‐1 and RNA5‐8S1‐1 will represent the pre‐rRNA and the processed rRNAs from the first sequenced gene on RNR1; RNA45S2‐3, RNA28S2‐3, RNA18S2‐3 and RNA5‐8S2‐3 will represent the pre‐rRNA and the processed rRNAs from the third sequenced gene on RNR2.
Figure 4. Schematic showing the two types of ribosomal RNA (rRNA) gene cluster found within the human genome.

The 5S cluster has a variable copy number between individuals, with 98 being the average copy number, while the current human reference genome, GRCh38, has just 17 copies. The HGNC has approved symbols for the 17 annotated copies as shown above. There are five separate 45S rRNA clusters, which are named RNR1‐RNR5. These clusters are not currently represented on GRCh38. The HGNC has approved root symbols for each 45S rRNA genes and their post‐transcriptionally processed transcripts (root symbols shown in dark blue text). The light blue symbols show the format that will be approved in the future for individual 45S rRNA genes and transcripts once the clusters are included and annotated on the human reference genome.
While there are many 45S rRNA pseudogenes located throughout the reference genome, interestingly there are just five 45S rRNA genes that are located outside of the acrocentric 45S clusters, which appear to be transcribed and have no obvious mutations. Because these genes are outside of the 45S rRNA clusters around which the nucleolus forms and rRNA transcription takes place (reviewed in (Lam et al, 2005), it is unclear as to whether these genes could be transcribed into functional rRNA molecules. We have approved gene symbols for these genes, which include the letter “N” before the numerical identifier for rRNA cluster “number unspecified”; e.g., RNA45SN1 is located at 21p11.2 and could potentially produce the rRNAs represented by the symbols RNA18SN1, RNA28SN1 and RNA5‐8SN1.
Mitochondria contain their own ribosomes, known as mitoribosomes, that comprise a large subunit containing 16S rRNA and over 50 mitochondrial ribosomal proteins (MRPs) (Koc et al, 2001) and a small subunit containing 12S rRNA and over 35 MRPs (Cavdar Koc et al, 2001). While the MRPs are encoded by the nucleus, the 16S and 12S rRNAs are encoded by the mitochondrial genome (Anderson et al, 1981). As for the mitochondrial tRNA genes, the mitochondrial rRNA genes were named in collaboration with Mitomap (Lott et al, 2013)—the gene encoding 12S rRNA has the symbol MT‐RNR1 for “mitochondrially encoded 12S rRNA” and that encoding 16S rRNA has the symbol MT‐RNR2 for “mitochondrially encoded 16S rRNA”.
Vault RNAs
Vault RNAs are small transcripts of roughly 100 nucleotides with a conserved panhandle‐like secondary structure that are transcribed by RNA polymerase III (Stadler et al, 2009). This class of ncRNA was originally discovered as part of a large ribonucleoprotein complex in rat liver that was named the vault complex due to its characteristic arches, which reminded the researchers of the arches found in the vaults of cathedrals (Kedersha & Rome, 1986). The current nomenclature for human vault RNA genes—using the root symbol “VTRNA” for “vault RNA”—was approved by the HGNC in coordination with the publication of two papers (Nandy et al, 2009; Stadler et al, 2009). The human genome contains a cluster of 3 vault genes on 5q31.3: VTRNA1‐1, VTRNA1‐2, and VTRNA1‐3; one VTRNA2‐1 gene on chromosome 5q31.1; and a pseudogene, VTRNA3‐1P on Xp11.22. Association of vault RNAs with the vault complex depends upon binding and stabilisation by the TEP1 protein (Kickhoefer et al, 2001). The molecular function of the vault complex has remained elusive, while VTRNA1‐1 has been found to function separately from the complex as a regulator of autophagy (Horos et al, 2019) and an inhibitor of apoptosis (Amort et al, 2015).
VTRNA2‐1 has manifold functions unrelated to the vault complex, in particular in inflammation as binding partner of EIF2AK2 (also known as PKR) (Jeon et al, 2012; Kunkeaw et al, 2013). It is also a source of derived functional small RNAs (Kong et al, 2015). In earlier literature, it was mistakenly identified as “mirRNA‐886” and sometimes appears as “nc866”; it is, however, clearly a mammalian‐specific paralog of VTRNA1.
Y RNAs
Y RNAs are small transcripts of ~ 100 nucleotides with distinctive secondary structures that are largely bound by the Ro60 protein, which is similar in structure to the TEP1 protein that binds vault RNAs (Bateman & Kickhoefer, 2003), hinting at an evolutionary relationship between these two classes of ncRNPs. These RNAs were first identified in RNP complexes that were immunoprecipitated with anti‐Ro60 antibodies from patients with systemic lupus erythematosus (Hendrick et al, 1981; Lerner et al, 1981) and were designated “Y” RNAs because they are mostly cytoplasmic, in contrast to the U class of small nuclear RNAs (Lerner et al, 1981). The human genome encodes 4 active Y RNA genes, which are all located on 7q36.1 and are transcribed by RNA polymerase III (Wolin & Steitz, 1983; Maraia et al, 1994, 1996). While the transcripts are referred to as Y1, Y3, Y4, and Y5, the equivalent approved gene symbols are RNY1, RNY3, RNY4 and RNY5 for “RNA, Ro60‐associated Y#”. Note there is no Y2 as this symbol was used for a short transcript that was subsequently found to be a truncated form of Y1 (Hendrick et al, 1981; Wolin & Steitz, 1983).
All Y RNAs contain a stem, formed by base pairing of the 5′ and 3′ ends, that includes the Ro60 binding site (Wolin & Steitz, 1984; Pruijn et al, 1991; Green et al, 1998). At the other end of this stem are one or more internal loops and stem loops that interact with other proteins to generate specialised RNPs (Sim et al, 2012; Chen et al, 2013). Y RNAs can influence the subcellular location of Ro60 (Sim et al, 2009, 2012) and may regulate the ability of Ro60 to bind misfolded RNAs (Stein et al, 2005; Fuchs et al, 2006; Wolin et al, 2013), a function supported by work in bacteria (Chen et al, 2007, 2013). There have also been reports of a Ro60‐independent function for mammalian Y RNAs in DNA replication (Christov et al, 2006; Krude et al, 2009), although mouse cell lines depleted of Y RNAs show no growth defects (Sim et al, 2009, 2012; Reed et al, 2013).
SNARs
Small NF90 (ILF3)‐associated RNAs (snaRs) were first identified following immunoprecipitation of ribonucleoproteins with antibodies against NF90, an abundant protein isoform expressed from the ILF3 gene (Parrott & Mathews, 2007). The snaR transcripts are around 117 nucleotides, show highest expression in immortalised cell lines and testis and are transcribed by RNA polymerase III (Parrott & Mathews, 2007). snaR genes are specific to great apes and evolved from an Alu repeat element followed by genomic duplication (Parrott & Mathews, 2009). Bioinformatic analysis identified nine subsets of snaRs based on sequence similarity and the HGNC agreed on the root symbol SNAR, for “small NF90 (ILF3)‐associated RNA”, followed by a unique letter for each subset and a unique number for each gene in a subset, e.g. SNAR‐A1, SNAR‐B2 and SNAR‐C3. SnaRs are the least well‐characterised category of small RNAs named by the HGNC and their function remains to be determined but snaR‐A transcripts bind to ribosomes, suggesting these RNAs could have a role in translational control (Parrott & Mathews, 2009).
Long non‐coding RNAs
Before the human genome was sequenced, a small number of functional non‐coding transcripts had been identified that could not be placed into any of the categories described so far in this paper: 7SK (encoded by RN7SK) (Zieve & Penman, 1976; Diribarne & Bensaude, 2009) and 7SL (encoded by three human loci: RNA7SL1, RN7SL2 and RN7SL3) (Walker et al, 1974; Walter & Blobel, 1982) in the 1970s, and H19 (Brannan et al, 1990), BCYRN1 (BC200) (Tiedge et al, 1993), and XIST (Brown et al, 1991, 1992) in the early 1990s. 7SK, 7SL and BCYRN1 are all transcribed by RNA polymerase III and function via forming complexes with proteins: 7SK is an RNA scaffold in a complex that regulates the P‐TEFb transcription factor (Diribarne & Bensaude, 2009); 7SL is the RNA component of the signal recognition particle that targets proteins with a signal peptide to the endoplasmic reticulum (Walter & Blobel, 1982); BCYRN1 inhibits translation via binding to eIF4A and PABP (Muddashetty et al, 2002; Lin et al, 2008). In contrast, H19 and XIST, like protein coding transcripts, are transcribed by RNA polymerase II. While XIST has a defined molecular function in binding to and silencing the inactive X chromosome (Chow et al, 2005), the exact molecular function of H19 is still not clear—it has been associated with many types of cancer and regulates several target genes by post‐transcriptional mechanisms (Gabory et al, 2010).
Large‐scale studies made possible following the release of the sequenced human genome in 2001 (Lander et al, 2001) revealed the existence of large numbers of transcripts that appear to be untranslated and, like those above, do not belong to previously defined classes of non‐coding RNAs (Kapranov et al, 2002; Bertone et al, 2004; Cheng et al, 2005). These were initially referred to as mRNA‐like ncRNAs because they are generally transcribed by RNA polymerase II, and are capped, spliced and polyadenylated like protein‐coding mRNAs (Erdmann et al, 1999; Lottin et al, 2002; Bompfünewerer et al, 2005; Széll et al, 2008). A 2007 study on non‐coding transcripts in human and mouse first used the term “long” to refer to transcripts of over 200 nucleotides (Kapranov et al, 2007), and this classification became widespread with the term “long non‐coding RNA” (or “long non‐coding RNA”) appearing in the title of 18 papers in a 2010 PubMed search, increasing to 123 papers by the year 2013 and 1,517 papers in 2018. Although the term “long non‐coding RNA” (abbreviated to lncRNA) does not truly represent a class of non‐coding RNA, it has become a useful shorthand for such transcripts of varied/unknown function and is entrenched in the scientific literature.
Functional studies have been performed for a relatively small subset of lncRNAs. The modes of action that have been described can be grouped into several different categories (see (Chen, 2016) for a comprehensive review of lncRNA by category):
Cis regulation of a neighbouring protein coding locus, which can be either positive or negative regulation; e.g., TARID binds to the promoter of, and activates, the TCF21 gene (Arab et al, 2014); PLUT upregulates transcription of PDX1 by affecting local 3D chromatin structure (Akerman et al, 2017); FLICR represses FOXP3 transcription by modifying chromatin accessibility (Zemmour et al, 2017).
Trans regulation, i.e. regulating loci away from the site of transcription of the lncRNA, e.g. NRON, represses NFAT trafficking as part of an RNA–protein complex (Willingham et al, 2005); RMST is a transcriptional coregulator of SOX2 that influences the transcription of genes involved in neurogenesis (Ng et al, 2013); THRIL regulates TNF gene expression by binding to the hnRNPL protein (Li et al, 2014).
Acting as structural components, e.g. NEAT1, is a core RNA component of nuclear paraspeckles (Clemson et al, 2009); MALAT1 has been associated with nuclear speckles (Tripathi et al, 2010); FIRRE influences nuclear architecture by binding to several different chromosomes (Hacisuleyman et al, 2014).
Acting as molecular “decoys” to titrate proteins or small RNAs away from other binding partners, e.g. the abundant lncRNA NORAD sequesters PUM1 and PUM2 proteins (Lee et al, 2016; Tichon et al, 2016); GAS5 binds to the glucocorticoid receptor NR3C1 thus preventing its binding to glucocorticoid response elements in promoters (Kino et al, 2010). There are many papers on the binding and sequestering of microRNAs by lncRNAs (Grüll & Massé, 2019) although there is some debate over whether lncRNAs would usually be at high enough levels within cells to effectively compete for microRNAs (Ulitsky, 2018).
The HGNC provides unique gene symbols so that lncRNA genes can be discussed unambiguously. Akin to newly characterised protein coding genes, a symbol may be chosen by research groups working on a lncRNA gene if it is unique and follows the guidelines of the HGNC. The HGNC requests that all authors contact us prior to publication so that we can check any proposed new nomenclature conforms to our guidelines and, once the symbol is accepted by us, reserve it. This ensures that the approved symbol on the HGNC website (http://www.genenames.org), and on the Ensembl, NCBI Gene and LNCipedia websites, will be exactly the same as the lncRNA symbol that appears in the literature. Failure to contact the HGNC prior to publication may result in the approval of a symbol that does not match the first published symbol, e.g. PANDAR instead of PANDA (Hung et al, 2011), DANCR instead of ANCR (Kretz et al, 2012), THORLNC instead of THOR (Ye et al, 2018). In cases like these where we are unable to approve a symbol that appears in a publication, we contact the corresponding author of that paper to discuss an appropriate alternative. As shown by the symbols listed here, we try to approve a symbol similar to the original published symbol.
The primary rule for naming human genes is that gene symbols must be unique; i.e., the symbol does not overlap a symbol used for another human gene and ideally does not generate a high number of false‐positive hits on literature search engines. Symbols should be a short form representation of a meaningful gene name, should not be the same as a common word in the English language, should not be named after a person or place and should not include “H” for human. Although the HGNC discourages the use of punctuation in gene symbols, hyphens may sometimes be used in lncRNA gene symbols. Where known, the gene name should represent the normal function of the lncRNA gene; e.g., the full name of NEAT1 is “nuclear paraspeckle assembly transcript 1”, and the full name of NRON is “non‐coding repressor of NFAT”. We appreciate that for lncRNA genes this is not always possible, and we do permit reference to expression, e.g. BMNCR for “bone marrow associated non‐coding RNA” (Li et al, 2018a), and, in some cases, disease where the association of the lncRNA with the disease is based on more than a change in expression, e.g. PRINS for “psoriasis associated non‐protein coding RNA induced by stress” (Sonkoly et al, 2005), NBAT1 for “neuroblastoma associated transcript 1” (Pandey et al, 2014). Again, such cases should be discussed individually with the HGNC prior to publication.
In addition to providing a unique symbol for each named lncRNA gene, the HGNC records other alternative symbols used by different research groups, which we refer to as alias symbols. For example, the lncRNA gene LINC00261 was first approved by the HGNC in 2012; this unique symbol first appeared in a publication in 2013 (Cao et al, 2013) and has since appeared in more than 25 publications. A different symbol, ALIEN, was used in a 2015 publication (Kurian et al, 2015) with no reference to the approved symbol and the symbol DEANR1 appeared the same year (Jiang et al, 2015) with reference to the approved symbol in the paper but not in the title or abstract. Using or referencing the approved symbols in the title or abstract allows all papers on a particular gene to be found easily and ensures that key information will not be missed. The HGNC symbol report for LINC00261 shows all alias symbols so that searching our database with any of the published symbols will retrieve the correct gene in our database and in other major biomedical databases such as NCBI Gene and Ensembl. Although the HGNC endeavours to record alias symbols, there is always the possibility that these may be missed and valuable data on genes lost to future interested parties if the approved symbol is not referenced anywhere else in the publication.
Where possible, we coordinate with the Mouse Genomic Nomenclature Committee to assign the equivalent symbol for orthologous human and mouse lncRNA genes. For example, the mouse ortholog of the human lncRNA gene NEAT1 has the symbol Neat1, while the mouse ortholog of human XIST has the symbol Xist (see Table 2 for further selected examples). However, it is not always straight forward to determine orthology between human and mouse lncRNA genes (Ulitsky, 2016). We require the two genes to be at a conserved syntenic location and to have detectable sequence similarity.
Table 2.
Selected examples of lncRNA genes with equivalent approved symbols in human and mouse. For human and mouse lncRNA genes to be considered orthologous and named as such, the HGNC requires that the genes are at a conserved syntenic location and have detectable sequence similarity. Note that human gene symbols are uppercase while mouse symbols are title case, and mouse gene symbols do not contain hyphens
| Human symbol | Human gene name | Mouse symbol | References |
|---|---|---|---|
| AIRN | antisense of IGF2R non‐protein coding RNA | Airn | Yotova et al (2008) |
| DANCR | Differentiation antagonising non‐protein coding RNA | Dancr | Chalei et al (2014) |
| EGOT | Eosinophil granule ontogeny transcript | Egot | Rose and Stadler (2011) |
| FENDRR | FOXF1 adjacent non‐coding developmental regulatory RNA | Fendrr | Grote et al (2013) |
| FIRRE | Firre intergenic repeating RNA element | Firre | Hacisuleyman et al (2014) |
| GAS5 | Growth arrest specific 5 | Gas5 | Smith and Steitz (1998) |
| HOTAIR | HOX transcript antisense RNA | Hotair | Rinn et al (2007) |
| HOTTIP | HOXA distal transcript antisense RNA | Hottip | Wang et al (2011) |
| KCNQ1OT1 | KCNQ1 opposite strand/antisense transcript 1 | Kcnq1ot1 | Gicquel et al (2003) |
| MALAT1 | Metastasis associated lung adenocarcinoma transcript 1 | Malat1 | Ji et al (2003) |
| MEG3 | Maternally expressed 3 | Meg3 | Miyoshi et al (2000) |
| MIR17HG | miR‐17‐92a‐1 cluster host gene | Mir17hg | Dews et al (2010) |
| NEAT1 | Nuclear paraspeckle assembly transcript 1 | Neat1 | Clemson et al (2009) |
| NRON | Non‐coding repressor of NFAT | Nron | Willingham et al (2005) |
| PANTR1 | POU3F3 adjacent non‐coding transcript 1 | Pantr1 | Clark and Blackshaw (2017) |
| PAUPAR | PAX6 upstream antisense RNA | Paupar | Vance et al (2014) |
| PEG13 | Paternally expressed 13 | Peg13 | Court et al (2014) |
| PVT1 | Pvt1 oncogene | Pvt1 | Carramusa et al (2007) |
| RN7SK | RNA component of 7SK nuclear ribonucleoprotein | Rn7sk | Driscoll et al (1994) |
| SNHG3 | Small nucleolar RNA host gene 3 | Snhg3 | Pelczar and Filipowicz (1998) |
| SOX1‐OT | SOX1 overlapping transcript | Sox1ot | Ahmad et al (2017) |
| TSIX | TSIX transcript, XIST antisense RNA | Tsix | Lee et al (1999) |
| TUG1 | Taurine up‐regulated 1 | Tug1 | Young et al (2005) |
| TUNAR | TCL1 upstream neural differentiation‐associated RNA | Tunar | Ulitsky et al (2011) |
| XIST | X inactive specific transcript | Xist | Brown et al (1991) |
| ZFAS1 | ZNFX1 antisense RNA 1 | Zfas1 | Askarian‐Amiri et al (2011) |
As mentioned above, a relatively small fraction of the predicted total number of lncRNA genes have been cited in publications. In addition to naming published lncRNA genes, the HGNC names genes that have been annotated by the RefSeq (O'Leary et al, 2016) and GENCODE (Frankish et al, 2019) projects. These projects initially annotated lncRNA genes based on EST, cDNA and mRNA data, which provided a set of relatively high stringency, but not necessarily full‐length, transcripts. Both projects have since started to incorporate long read RNA‐Seq data, e.g. (Lagarde et al, 2017). Genes are annotated as lncRNAs where there is sufficient transcriptional support for a locus, but there is not sufficient evidence of protein coding potential. Assessment of protein coding potential includes assessing cross‐species conservation of a putative open reading frame (ORF), length of a putative ORF, presence/absence of encoded features such as protein domains, ribosome profiling data and evidence of peptides via mass spectrometry. Due to the constant emergence of new data, there is a certain amount of flux between the protein coding and lncRNA gene sets. Some protein coding genes have subsequently been reannotated as lncRNA genes; e.g., the gene formerly known as C6orf48 was reannotated as a lncRNA gene and therefore renamed by the HGNC as SNHG32. Equally, some lncRNA genes have been reannotated as protein coding genes either due to a reassessment based on new metrics such as phyloCSF (Lin et al, 2011) or based on emerging evidence from new publications. For example, LINC00083 was reannotated as protein coding gene CLEC20A because the ORF is conserved and exhibits a C‐type lectin domain, while LINC01420 was reannotated as protein coding and renamed NBDY based on published data (D'Lima et al, 2017).
Feedback from conferences and research groups informed us that the lncRNA community finds genomic context with respect to protein coding genes a useful metric when considering lncRNA genes on a genomic scale. Therefore, working with the lncRNA annotation classification used by the GENCODE group, we devised a nomenclature system using the following categories (see Fig 5):
Figure 5. LncRNA naming schema for lncRNA genes with no published information at the time of naming.
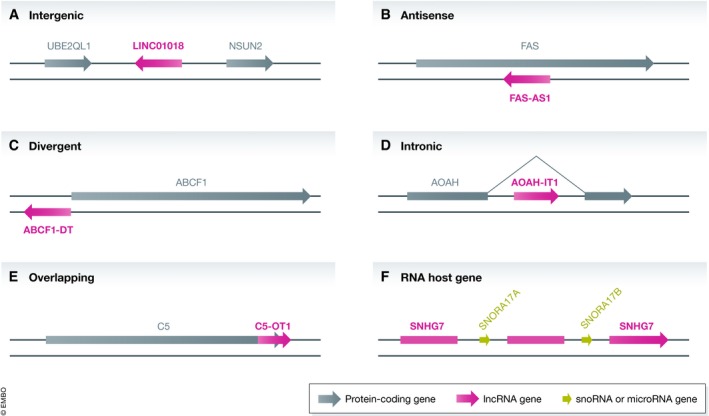
- LncRNAs that are intergenic with respect to protein coding genes are assigned the root symbol LINC# followed by a 5‐digit number.
- LncRNAs that are antisense to the genomic span of a protein coding gene are assigned the symbol format [protein coding gene symbol]‐AS#.
- LncRNAs that are divergent to (share a bidirectional promoter with) a protein coding gene are assigned the symbol format [protein coding gene symbol]‐DT.
- LncRNAs that are contained within an intron of a protein coding gene on the same strand are assigned the symbol format [protein coding gene symbol]‐IT#.
- LncRNAs that overlap a protein coding gene on the same strand are assigned the symbol format [protein gene coding symbol]‐OT#.
- LncRNAs that contain microRNA or snoRNA genes within introns or exons are named as host genes. See the main text for details on how these microRNA host genes and snoRNA host genes are named.
Box 3.
Intergenic lncRNA genes are assigned the root symbol “LINC” for “long intergenic non‐protein coding RNA” followed by a unique 5‐digit number, e.g. LINC01018 (Fig 5A). A lncRNA gene is considered intergenic (meaning between protein coding genes in this context) if it does not overlap a protein coding gene on either strand, does not share a bidirectional promoter with a protein coding gene and is not a host gene for a microRNA or snoRNA.
Antisense lncRNA genes are named using the format [protein coding gene symbol] with the suffix ‐AS and a sequential number, e.g. FAS‐AS1 for “FAS antisense RNA 1” (Fig 5B). A lncRNA gene is considered antisense if it overlaps the genomic coordinates of a protein coding gene on the opposite strand. There does not need to be exon–exon overlap. These symbols are not intended to imply that there is a regulatory role between the protein coding and lncRNA gene. If the lncRNA is antisense to more than one protein coding gene, the symbol of the most 5′ protein coding gene will be chosen as the basis of the lncRNA gene symbol, unless there is exon–exon overlap between a more 3′ protein coding gene, which would be chosen in preference.
Divergent transcripts that are transcribed from a bidirectional promoter in the opposite direction to a protein coding gene are named using the format [protein coding gene symbol] with the suffix ‐DT, e.g. ABCF1‐DT for “ABCF1 divergent transcript” (Fig 5C). A lncRNA is considered divergent if it is within 300–500 nucleotides of the 5′ end of a protein coding gene on the other strand. Usually evidence of bidirectional transcription can be seen with cap analysis gene expression tags, although this is not a requirement. If a protein coding gene has multiple transcription start sites, the lncRNA will be named as a divergent transcript only if it shares the 5′ most promoter; otherwise, it will overlap the genomic span of the protein coding gene and be considered antisense.
Intronic transcripts that are transcribed entirely from within an intron of a protein coding gene on the same strand are named using the format [protein coding gene symbol] with the suffix ‐IT and a sequential number, e.g. AOAH‐IT1 for “AOAH intronic transcript” (Fig 5D). This category accounts for a small number of our named lncRNA genes and is applied sparingly because we have found that future evidence may reveal that these loci are alternative exons or rare intron degradation intermediates of the protein coding locus.
Overlapping transcripts that overlap a protein coding gene on the same strand are named using the format [protein coding gene symbol] with the suffix ‐OT and a sequential number, e.g. C5‐OT1 for “C5 3′ UTR overlapping transcript 1” (Fig 5E). As for the intronic transcripts above, this category is applied with caution because experience has shown us that such lncRNA genes may eventually be merged into the protein coding locus when further transcriptional evidence becomes available.
Host genes for microRNAs or snoRNAs. The small RNA may be in an exon or intron but must be on the same strand as the lncRNA (Fig 5F). LncRNA genes that host a microRNA gene are named using the format [microRNA gene symbol]HG, e.g. MIR122HG for “MIR122 host gene”. Where there are several microRNA genes hosted by the same lncRNA gene, the lncRNA is named after the 5′ most microRNA. If the lncRNA gene hosts a cluster, this is shown in the gene name; e.g., MIR17HG has the full gene name “miR‐17‐92a‐1 cluster host gene”. MIR200CHG hosts the microRNA genes MIR200C and MIR141; this is shown in the full gene name “MIR200C and MIR141 host gene”.
LncRNA genes that host a snoRNA gene are named using the root symbol SNHG for snoRNAs host gene followed by a unique number, e.g. SNHG1. This lncRNA hosts seven different snoRNA genes, so an early decision was taken to not include reference to individual snoRNA genes at the gene symbol level.
Please see the previous sections on snoRNA and microRNA genes above for more information on these small RNAs and their host genes.
In future, the HGNC will explore the possible annotation and naming of sno‐lncRNAs, a new class of transcript with a snoRNA at each end (Yin et al, 2012; Xing et al, 2017). These are processed from the introns of snoRNA host genes that host more than one snoRNA within an intron. We will also explore transcripts derived from snoRNA host genes that have a 5′ snoRNA and a poly(A) tail, which have been referred to as 5′ snoRNA capped and 3′ polyadenylated (SPAs) (Wu et al, 2016; Lykke‐Andersen et al, 2018).
The HGNC names genes and not alternative transcripts, so we assign only one name per lncRNA gene and do not provide separate symbols for non‐coding transcripts that are part of protein coding loci. Please note that the symbols in the above scheme do not mean that the lncRNA genes they represent have no function—the symbols are systematically applied where no other informative data are available at the time of naming. The HGNC will only change such symbols once future information is available where there is a consensus from groups working on these genes to do so. In some cases, our systematic symbols are already becoming well used in the literature, e.g. LINC00473, MIR17HG, LOXL1‐AS1. We have already named over 4,300 lncRNA genes, but we are still a long way from naming all annotated lncRNA genes; we are currently working on naming a dataset of intergenic lncRNA genes that are consistently annotated by both the GENCODE and RefSeq projects.
Circular RNAs
Circular RNAs (circRNAs) and circular intronic RNAs (ciRNAs) are both produced during the splicing of pre‐mRNA—the major difference being that circRNAs are derived from exonic sequence, while ciRNAs are derived from intronic sequence. Currently, there are no approved symbols for circRNAs or ciRNAs; this may be a future task for the HGNC if a consensus is found in the community. CircRNAs are the result of back‐splicing of exons from pre‐mRNA, which creates a circRNA joined to itself by a 3′,5′‐phosphodiester bond (Wu et al, 2017; Li et al, 2018b). Although most of these RNAs are expressed at low levels, there are examples where the circRNA is more highly expressed than the spliced linear mRNA (Salzman et al, 2013). Recent studies have suggested roles for circRNAs in competitive regulation of pre‐mRNA splicing (Ashwal‐Fluss et al, 2014; Zhang et al, 2014), competitive binding to microRNAs (Hansen et al, 2013; Memczak et al, 2013), regulation of RNA polymerase II (Li et al, 2015) and involvement in innate immunity (Liu et al, 2019a). CiRNAs are derived from spliced‐out intron lariats that have escaped cleavage by the debranching enzyme. These RNAs have 2′,5′‐phosphodiester bonds between their 5′ ends and the intronic branching site creating a circular structure. Sequence analysis shows that the generation of ciRNAs is not random but depends on the presence of a consensus RNA motif containing a seven nucleotide GU‐rich element near the 5′ splice site and an 11 nucleotide C‐rich element near the intron branch point of the parent mRNA (Zhang et al, 2013). Knockdown of ciRNAs has been shown to reduce expression of the genes from which they are derived (Zhang et al, 2013).
While there is no current standardised system for naming circRNAs or ciRNAs, we suggest the following nomenclature schemes:
For circRNAs:
circ[gene symbol]‐n where the gene symbol represents the unspliced “host” gene and n is an iterative five digit number; e.g., the first circRNA named for the host gene PARN would be circPARN‐00001
For ciRNAs:
ci[gene symbol]‐n where the gene symbol represents the unspliced “host” gene and n is an iterative five digit number; e.g., the first ciRNA named for the host gene PARN would be ciPARN‐00001
There are currently huge numbers of circRNAs listed in public databases such as CIRCpedia (Dong et al, 2018), circBank (Liu et al, 2019b) and circBase (Glažar et al, 2014) all using different identifiers. We call on the community to come together to discuss standards creating a consensus set of circRNAs and ciRNAs that could be given standardised nomenclature in the future.
Conclusion
In summary, the HGNC works directly with specialist advisors in the ncRNA field to ensure that appropriate and informative gene symbols are approved for ncRNA genes. We urge all ncRNA researchers to use, or at least mention, HGNC‐approved gene symbols in publications. This will ensure that ncRNA genes are correctly cited and will prevent confusion in the field. To discuss any aspect of ncRNA nomenclature, please contact the HGNC via our email address hgnc@genenames.org.
Conflict of interest
The authors declare that they have no conflict of interest.
Acknowledgements
We would like to thank Dr. Patricia Chan for her input on transfer RNA nomenclature and the HGNC team, both past and present, for their helpful discussions. This work was supported by the National Human Genome Research Institute (NHGRI) grant U24HG003345, Wellcome Trust grant 208349/Z/17/Z and Germany Ministry Education and Research grant no 031A538A, de.NBI‐RBC.
The EMBO Journal (2020) 39: e103777
References
- Abbott JA, Francklyn CS, Robey‐Bond SM (2014) Transfer RNA and human disease. Front Genet 5: 158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Abou Elela S, Nazar RN (1997) Role of the 5.8S rRNA in ribosome translocation. Nucleic Acids Res 25: 1788–1794 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Agris PF, Eruysal ER, Narendran A, Väre VYP, Vangaveti S, Ranganathan SV (2018) Celebrating wobble decoding: half a century and still much is new. RNA Biol 15: 537–553 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ahmad A, Strohbuecker S, Tufarelli C, Sottile V (2017) Expression of a SOX1 overlapping transcript in neural differentiation and cancer models. Cell Mol Life Sci 74: 4245–4258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Akerman I, Tu Z, Beucher A, Rolando DMY, Sauty‐Colace C, Benazra M, Nakic N, Yang J, Wang H, Pasquali L et al (2017) Human pancreatic β cell lncRNAs control cell‐specific regulatory networks. Cell Metab 25: 400–411 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ambros V, Bartel B, Bartel DP, Burge CB, Carrington JC, Chen X, Dreyfuss G, Eddy SR, Griffiths‐Jones S, Marshall M et al (2003) A uniform system for microRNA annotation. RNA 9: 277–279 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amort M, Nachbauer B, Tuzlak S, Kieser A, Schepers A, Villunger A, Polacek N (2015) Expression of the vault RNA protects cells from undergoing apoptosis. Nat Commun 6: 7030 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Anderson S, Bankier AT, Barrell BG, de Bruijn MH, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F et al (1981) Sequence and organization of the human mitochondrial genome. Nature 290: 457–465 [DOI] [PubMed] [Google Scholar]
- Anokhina M, Bessonov S, Miao Z, Westhof E, Hartmuth K, Lührmann R (2013) RNA structure analysis of human spliceosomes reveals a compact 3D arrangement of snRNAs at the catalytic core. EMBO J 32: 2804–2818 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arab K, Park YJ, Lindroth AM, Schäfer A, Oakes C, Weichenhan D, Lukanova A, Lundin E, Risch A, Meister M et al (2014) Long noncoding RNA TARID directs demethylation and activation of the tumor suppressor TCF21 via GADD45A. Mol Cell 55: 604–614 [DOI] [PubMed] [Google Scholar]
- Ashwal‐Fluss R, Meyer M, Pamudurti NR, Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N, Kadener S (2014) circRNA biogenesis competes with pre‐mRNA splicing. Mol Cell 56: 55–66 [DOI] [PubMed] [Google Scholar]
- Askarian‐Amiri ME, Crawford J, French JD, Smart CE, Smith MA, Clark MB, Ru K, Mercer TR, Thompson ER, Lakhani SR et al (2011) SNORD‐host RNA Zfas1 is a regulator of mammary development and a potential marker for breast cancer. RNA 17: 878–891 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bartel DP (2018) Metazoan MicroRNAs. Cell 173: 20–51 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bateman A, Kickhoefer V (2003) The TROVE module: a common element in Telomerase, Ro and Vault ribonucleoproteins. BMC Bioinformatics 4: 49 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bertone P, Stolc V, Royce TE, Rozowsky JS, Urban AE, Zhu X, Rinn JL, Tongprasit W, Samanta M, Weissman S et al (2004) Global identification of human transcribed sequences with genome tiling arrays. Science 306: 2242–2246 [DOI] [PubMed] [Google Scholar]
- Bompfünewerer AF, Flamm C, Fried C, Fritzsch G, Hofacker IL, Lehmann J, Missal K, Mosig A, Müller B, Prohaska SJ et al (2005) Evolutionary patterns of non‐coding RNAs. Theory Biosci 123: 301–369 [DOI] [PubMed] [Google Scholar]
- Brannan CI, Dees EC, Ingram RS, Tilghman SM (1990) The product of the H19 gene may function as an RNA. Mol Cell Biol 10: 28–36 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braschi B, Denny P, Gray K, Jones T, Seal R, Tweedie S, Yates B, Bruford E (2019) Genenames.org: the HGNC and VGNC resources in 2019. Nucleic Acids Res 47: D786–D792 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Brown CJ, Ballabio A, Rupert JL, Lafreniere RG, Grompe M, Tonlorenzi R, Willard HF (1991) A gene from the region of the human X inactivation centre is expressed exclusively from the inactive X chromosome. Nature 349: 38–44 [DOI] [PubMed] [Google Scholar]
- Brown CJ, Hendrich BD, Rupert JL, Lafrenière RG, Xing Y, Lawrence J, Willard HF (1992) The human XIST gene: analysis of a 17 kb inactive X‐specific RNA that contains conserved repeats and is highly localized within the nucleus. Cell 71: 527–542 [DOI] [PubMed] [Google Scholar]
- Budak H, Bulut R, Kantar M, Alptekin B (2016) MicroRNA nomenclature and the need for a revised naming prescription. Brief Funct Genomics 15: 65–71 [DOI] [PubMed] [Google Scholar]
- Cao WJ, Wu HL, He BS, Zhang YS, Zhang ZY (2013) Analysis of long non‐coding RNA expression profiles in gastric cancer. World J Gastroenterol 19: 3658–3664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carramusa L, Contino F, Ferro A, Minafra L, Perconti G, Giallongo A, Feo S (2007) The PVT‐1 oncogene is a Myc protein target that is overexpressed in transformed cells. J Cell Physiol 213: 511–518 [DOI] [PubMed] [Google Scholar]
- Cavaillé J, Buiting K, Kiefmann M, Lalande M, Brannan CI, Horsthemke B, Bachellerie JP, Brosius J, Hüttenhofer A (2000) Identification of brain‐specific and imprinted small nucleolar RNA genes exhibiting an unusual genomic organization. Proc Natl Acad Sci USA 97: 14311–14316 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cavdar Koc E, Burkhart W, Blackburn K, Moseley A, Spremulli LL (2001) The small subunit of the mammalian mitochondrial ribosome. Identification of the full complement of ribosomal proteins present. J Biol Chem 276: 19363–19374 [DOI] [PubMed] [Google Scholar]
- Chalei V, Sansom S, Kong L, Lee S, Montiel J, Vance K (2014) The long non‐coding RNA Dali is an epigenetic regulator of neural differentiation. Elife 3: e04530 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chan PP, Lowe TM (2016) GtRNAdb 2.0: an expanded database of transfer RNA genes identified in complete and draft genomes. Nucleic Acids Res 44: D184–D189 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Wurtmann EJ, Van Batavia J, Zybailov B, Washburn MP, Wolin SL (2007) An ortholog of the Ro autoantigen functions in 23S rRNA maturation in D. radiodurans . Genes Dev 21: 1328–1339 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen X, Taylor DW, Fowler CC, Galan JE, Wang HW, Wolin SL (2013) An RNA degradation machine sculpted by Ro autoantigen and noncoding RNA. Cell 153: 166–177 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen W, Moore MJ (2015) Spliceosomes. Curr Biol 25: R181–R183 [DOI] [PubMed] [Google Scholar]
- Chen LL (2016) Linking long noncoding RNA localization and function. Trends Biochem Sci 41: 761–772 [DOI] [PubMed] [Google Scholar]
- Cheng J, Kapranov P, Drenkow J, Dike S, Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G et al (2005) Transcriptional maps of 10 human chromosomes at 5‐nucleotide resolution. Science 308: 1149–1154 [DOI] [PubMed] [Google Scholar]
- Chow JC, Yen Z, Ziesche SM, Brown CJ (2005) Silencing of the mammalian X chromosome. Annu Rev Genomics Hum Genet 6: 69–92 [DOI] [PubMed] [Google Scholar]
- Christov CP, Gardiner TJ, Szüts D, Krude T (2006) Functional requirement of noncoding Y RNAs for human chromosomal DNA replication. Mol Cell Biol 26: 6993–7004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ciganda M, Williams N (2011) Eukaryotic 5S rRNA biogenesis. Wiley Interdiscip Rev RNA 2: 523–533 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark BS, Blackshaw S (2017) Understanding the role of lncRNAs in nervous system development. Adv Exp Med Biol 1008: 253–282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clemson CM, Hutchinson JN, Sara SA, Ensminger AW, Fox AH, Chess A, Lawrence JB (2009) An architectural role for a nuclear noncoding RNA: NEAT1 RNA is essential for the structure of paraspeckles. Mol Cell 33: 717–726 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Court F, Camprubi C, Garcia CV, Guillaumet‐Adkins A, Sparago A, Seruggia D, Sandoval J, Esteller M, Martin‐Trujillo A, Riccio A et al (2014) The PEG13‐DMR and brain‐specific enhancers dictate imprinted expression within the 8q24 intellectual disability risk locus. Epigenetics Chromatin 7: 5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Demeshkina N, Repkova M, Ven'yaminova A, Graifer D, Karpova G (2000) Nucleotides of 18S rRNA surrounding mRNA codons at the human ribosomal A, P, and E sites: a crosslinking study with mRNA analogs carrying an aryl azide group at either the uracil or the guanine residue. RNA 6: 1727–1736 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Desvignes T, Batzel P, Berezikov E, Eilbeck K, Eppig JT, McAndrews MS, Singer A, Postlethwait JH (2015) miRNA nomenclature: a view incorporating genetic origins, biosynthetic pathways, and sequence variants. Trends Genet 31: 613–626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dews M, Fox JL, Hultine S, Sundaram P, Wang W, Liu YY, Furth E, Enders GH, El‐Deiry W, Schelter JM et al (2010) The myc‐miR‐17~92 axis blunts TGF{beta} signaling and production of multiple TGF{beta}‐dependent antiangiogenic factors. Cancer Res 70: 8233–8246 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Diribarne G, Bensaude O (2009) 7SK RNA, a non‐coding RNA regulating P‐TEFb, a general transcription factor. RNA Biol 6: 122–128 [DOI] [PubMed] [Google Scholar]
- D'Lima NG, Ma J, Winkler L, Chu Q, Loh KH, Corpuz EO, Budnik BA, Lykke‐Andersen J, Saghatelian A, Slavoff SA (2017) A human microprotein that interacts with the mRNA decapping complex. Nat Chem Biol 13: 174–180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong R, Ma XK, Li GW, Yang L (2018) CIRCpedia v2: an updated database for comprehensive circular RNA annotation and expression comparison. Genomics Proteomics Bioinformatics 16: 226–233 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Driscoll CT, Darlington GJ, Maraia RJ (1994) The conserved 7SK snRNA gene localizes to human chromosome 6 by homolog exclusion probing of somatic cell hybrid RNA. Nucleic Acids Res 22: 722–725 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Erdmann VA, Szymanski M, Hochberg A, de Groot N, Barciszewski J (1999) Collection of mRNA‐like non‐coding RNAs. Nucleic Acids Res 27: 192–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Falaleeva M, Welden JR, Duncan MJ, Stamm S (2017) C/D‐box snoRNAs form methylating and non‐methylating ribonucleoprotein complexes: old dogs show new tricks. BioEssays 39: 6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fernández‐Ramos D, Fernández‐Tussy P, Lopitz‐Otsoa F, Gutiérrez‐de‐Juan V, Navasa N, Barbier‐Torres L, Zubiete‐Franco I, Simón J, Fernández AF, Arbelaiz A et al (2018) MiR‐873‐5p acts as an epigenetic regulator in early stages of liver fibrosis and cirrhosis. Cell Death Dis 9: 958 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frankish A, Diekhans M, Ferreira AM, Johnson R, Jungreis I, Loveland J, Mudge JM, Sisu C, Wright J, Armstrong J et al (2019) GENCODE reference annotation for the human and mouse genomes. Nucleic Acids Res 47: D766–D773 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Friedman RC, Farh KK, Burge CB, Bartel DP (2009) Most mammalian mRNAs are conserved targets of microRNAs. Genome Res 19: 92–105 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fromm B, Billipp T, Peck LE, Johansen M, Tarver JE, King BL, Newcomb JM, Sempere LF, Flatmark K, Hovig E et al (2015) A uniform system for the annotation of vertebrate microRNA genes and the evolution of the human microRNAome. Annu Rev Genet 49: 213–242 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs G, Stein AJ, Fu C, Reinisch KM, Wolin SL (2006) Structural and biochemical basis for misfolded RNA recognition by the Ro autoantigen. Nat Struct Mol Biol 13: 1002–1009 [DOI] [PubMed] [Google Scholar]
- Gabory A, Jammes H, Dandolo L (2010) The H19 locus: role of an imprinted non‐coding RNA in growth and development. BioEssays 32: 473–480 [DOI] [PubMed] [Google Scholar]
- Gall JG, Bellini M, Wu Z, Murphy C (1999) Assembly of the nuclear transcription and processing machinery: Cajal bodies (coiled bodies) and transcriptosomes. Mol Biol Cell 10: 4385–4402 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ganot P, Bortolin ML, Kiss T (1997a) Site‐specific pseudouridine formation in preribosomal RNA is guided by small nucleolar RNAs. Cell 89: 799–809 [DOI] [PubMed] [Google Scholar]
- Ganot P, Caizergues‐Ferrer M, Kiss T (1997b) The family of box ACA small nucleolar RNAs is defined by an evolutionarily conserved secondary structure and ubiquitous sequence elements essential for RNA accumulation. Genes Dev 11: 941–956 [DOI] [PubMed] [Google Scholar]
- Gicquel C, Gaston V, Mandelbaum J, Siffroi JP, Flahault A, Le Bouc Y (2003) In vitro fertilization may increase the risk of Beckwith‐Wiedemann syndrome related to the abnormal imprinting of the KCN1OT gene. Am J Hum Genet 72: 1338–1341 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Glažar P, Papavasileiou P, Rajewsky N (2014) circBase: a database for circular RNAs. RNA 20: 1666–1670 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gonzalez IL, Sylvester JE (2001) Human rDNA: evolutionary patterns within the genes and tandem arrays derived from multiple chromosomes. Genomics 73: 255–263 [DOI] [PubMed] [Google Scholar]
- Green CD, Long KS, Shi H, Wolin SL (1998) Binding of the 60‐kDa Ro autoantigen to Y RNAs: evidence for recognition in the major groove of a conserved helix. RNA 4: 750–765 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Griffiths‐Jones S (2004) The microRNA registry. Nucleic Acids Res 32: D109–D111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grote P, Wittler L, Hendrix D, Koch F, Währisch S, Beisaw A, Macura K, Bläss G, Kellis M, Werber M et al (2013) The tissue‐specific lncRNA Fendrr is an essential regulator of heart and body wall development in the mouse. Dev Cell 24: 206–214 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grüll MP, Massé E (2019) Mimicry, deception and competition: the life of competing endogenous RNAs. Wiley Interdiscip Rev RNA 10: e1525 [DOI] [PubMed] [Google Scholar]
- Guggino G, Orlando V, Saieva L, Ruscitti P, Cipriani P, La Manna MP, Giacomelli R, Alessandro R, Triolo G, Ciccia F et al (2018) Downregulation of miRNA17‐92 cluster marks Vγ9Vδ2 T cells from patients with rheumatoid arthritis. Arthritis Res Ther 20: 236 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hacisuleyman E, Goff LA, Trapnell C, Williams A, Henao‐Mejia J, Sun L, McClanahan P, Hendrickson DG, Sauvageau M, Kelley DR et al (2014) Topological organization of multichromosomal regions by the long intergenic noncoding RNA Firre. Nat Struct Mol Biol 21: 198–206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hansen TB, Jensen TI, Clausen BH, Bramsen JB, Finsen B, Damgaard CK, Kjems J (2013) Natural RNA circles function as efficient microRNA sponges. Nature 495: 384–388 [DOI] [PubMed] [Google Scholar]
- Hendrick JP, Wolin SL, Rinke J, Lerner MR, Steitz JA (1981) Ro small cytoplasmic ribonucleoproteins are a subclass of La ribonucleoproteins: further characterization of the Ro and La small ribonucleoproteins from uninfected mammalian cells. Mol Cell Biol 1: 1138–1149 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoagland MB, Stephenson ML, Scott JF, Hecht LI, Zamenik PC (1958) A soluble ribonucleic acid intermediate in protein synthesis. J Biol Chem 231: 241–257 [PubMed] [Google Scholar]
- Hodnett JL, Busch H (1968) Isolation and characterization of uridylic acid‐rich 7 S ribonucleic acid of rat liver nuclei. J Biol Chem 243: 6334–6342 [PubMed] [Google Scholar]
- Horos R, Büscher M, Kleinendorst R, Alleaume AM, Tarafder AK, Schwarzl T, Dziuba D, Tischer C, Zielonka EM, Adak A et al (2019) The small non‐coding vault RNA1‐1 acts as a riboregulator of autophagy. Cell 176: 1054–1067.e12 [DOI] [PubMed] [Google Scholar]
- Hou YM (2010) CCA addition to tRNA: implications for tRNA quality control. IUBMB Life 62: 251–260 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hung T, Wang Y, Lin MF, Koegel AK, Kotake Y, Grant GD, Horlings HM, Shah N, Umbricht C, Wang P et al (2011) Extensive and coordinated transcription of noncoding RNAs within cell‐cycle promoters. Nat Genet 43: 621–629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jády BE, Kiss T (2001) A small nucleolar guide RNA functions both in 2′‐O‐ribose methylation and pseudouridylation of the U5 spliceosomal RNA. EMBO J 20: 541–551 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jeon SH, Lee K, Lee KS, Kunkeaw N, Johnson BH, Holthauzen LM, Gong B, Leelayuwat C, Lee YS (2012) Characterization of the direct physical interaction of nc886, a cellular non‐coding RNA, and PKR. FEBS Lett 586: 3477–3484 [DOI] [PubMed] [Google Scholar]
- Ji P, Diederichs S, Wang W, Böing S, Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E et al (2003) MALAT‐1, a novel noncoding RNA, and thymosin beta4 predict metastasis and survival in early‐stage non‐small cell lung cancer. Oncogene 22: 8031–8041 [DOI] [PubMed] [Google Scholar]
- Jiang W, Liu Y, Liu R, Zhang K, Zhang Y (2015) The lncRNA DEANR1 facilitates human endoderm differentiation by activating FOXA2 expression. Cell Rep 11: 137–148 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jorjani H, Kehr S, Jedlinski DJ, Gumienny R, Hertel J, Stadler PF, Zavolan M, Gruber AR (2016) An updated human snoRNAome. Nucleic Acids Res 44: 5068–5082 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kapranov P, Cawley SE, Drenkow J, Bekiranov S, Strausberg RL, Fodor SP, Gingeras TR (2002) Large‐scale transcriptional activity in chromosomes 21 and 22. Science 296: 916–919 [DOI] [PubMed] [Google Scholar]
- Kapranov P, Cheng J, Dike S, Nix DA, Duttagupta R, Willingham AT, Stadler PF, Hertel J, Hackermüller J, Hofacker IL et al (2007) RNA maps reveal new RNA classes and a possible function for pervasive transcription. Science 316: 1484–1488 [DOI] [PubMed] [Google Scholar]
- Kedersha NL, Rome LH (1986) Isolation and characterization of a novel ribonucleoprotein particle: large structures contain a single species of small RNA. J Cell Biol 103: 699–709 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Khatter H, Myasnikov AG, Natchiar SK, Klaholz BP (2015) Structure of the human 80S ribosome. Nature 520: 640–645 [DOI] [PubMed] [Google Scholar]
- Kickhoefer VA, Liu Y, Kong LB, Snow BE, Stewart PL, Harrington L, Rome LH (2001) The Telomerase/vault‐associated protein TEP1 is required for vault RNA stability and its association with the vault particle. J Cell Biol 152: 157–164 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim SH, Quigley GJ, Suddath FL, McPherson A, Sneden D, Kim JJ, Weinzierl J, Rich A (1973) Three‐dimensional structure of yeast phenylalanine transfer RNA: folding of the polynucleotide chain. Science 179: 285–288 [DOI] [PubMed] [Google Scholar]
- Kino T, Hurt DE, Ichijo T, Nader N, Chrousos GP (2010) Noncoding RNA gas5 is a growth arrest‐ and starvation‐associated repressor of the glucocorticoid receptor. Sci Signal 3: ra8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss AM, Jády BE, Bertrand E, Kiss T (2004) Human box H/ACA pseudouridylation guide RNA machinery. Mol Cell Biol 24: 5797–5807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kiss‐László Z, Henry Y, Bachellerie JP, Caizergues‐Ferrer M, Kiss T (1996) Site‐specific ribose methylation of preribosomal RNA: a novel function for small nucleolar RNAs. Cell 85: 1077–1088 [DOI] [PubMed] [Google Scholar]
- Koc EC, Burkhart W, Blackburn K, Moyer MB, Schlatzer DM, Moseley A, Spremulli LL (2001) The large subunit of the mammalian mitochondrial ribosome. Analysis of the complement of ribosomal proteins present. J Biol Chem 276: 43958–43969 [DOI] [PubMed] [Google Scholar]
- Kong L, Hao Q, Wang Y, Zhou P, Zou B, Zhang YX (2015) Regulation of p53 expression and apoptosis by vault RNA2‐1‐5p in cervical cancer cells. Oncotarget 6: 28371–28388 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kozomara A, Birgaoanu M, Griffiths‐Jones S (2019) miRBase: from microRNA sequences to function. Nucleic Acids Res 47: D155–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kretz M, Webster DE, Flockhart RJ, Lee CS, Zehnder A, Lopez‐Pajares V, Qu K, Zheng GX, Chow J, Kim GE et al (2012) Suppression of progenitor differentiation requires the long noncoding RNA ANCR. Genes Dev 26: 338–343 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Krude T, Christov CP, Hyrien O, Marheineke K (2009) Y RNA functions at the initiation step of mammalian chromosomal DNA replication. J Cell Sci 122: 2836–2845 [DOI] [PubMed] [Google Scholar]
- Kunkeaw N, Jeon SH, Lee K, Johnson BH, Tanasanvimon S, Javle M, Pairojkul C, Chamgramol Y, Wongfieng W, Gong B et al (2013) Cell death/proliferation roles for nc886, a non‐coding RNA, in the protein kinase R pathway in cholangiocarcinoma. Oncogene 32: 3722–3731 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kurian L, Aguirre A, Sancho‐Martinez I, Benner C, Hishida T, Nguyen TB, Reddy P, Nivet E, Krause MN, Nelles DA et al (2015) Identification of novel long noncoding RNAs underlying vertebrate cardiovascular development. Circulation 131: 1278–1290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kwok GT, Zhao JT, Weiss J, Mugridge N, Brahmbhatt H, MacDiarmid JA, Robinson BG, Sidhu SB (2017) Translational applications of microRNAs in cancer, and therapeutic implications. Noncoding RNA Res 2: 143–150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagarde J, Uszczynska‐Ratajczak B, Carbonell S, Pérez‐Lluch S, Abad A, Davis C, Gingeras TR, Frankish A, Harrow J, Guigo R et al (2017) High‐throughput annotation of full‐length long noncoding RNAs with capture long‐read sequencing. Nat Genet 49: 1731–1740 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lagos‐Quintana M, Rauhut R, Lendeckel W, Tuschl T (2001) Identification of novel genes coding for small expressed RNAs. Science 294: 853–858 [DOI] [PubMed] [Google Scholar]
- Lam YW, Trinkle‐Mulcahy L, Lamond AI (2005) The nucleolus. J Cell Sci 118: 1335–1337 [DOI] [PubMed] [Google Scholar]
- Lander ES, Linton LM, Birren B, Nusbaum C, Zody MC, Baldwin J, Devon K, Dewar K, Doyle M, FitzHugh W et al (2001) Initial sequencing and analysis of the human genome. Nature 409: 860–921 [DOI] [PubMed] [Google Scholar]
- Langenberger D, Çakir MV, Hoffmann S, Stadler PF (2013) Dicer‐processed small RNAs: rules and exceptions. J Exp Zool B Mol Dev Evol 320: 35–46 [DOI] [PubMed] [Google Scholar]
- Lau NC, Lim LP, Weinstein EG, Bartel DP (2001) An abundant class of tiny RNAs with probable regulatory roles in Caenorhabditis elegans . Science 294: 858–862 [DOI] [PubMed] [Google Scholar]
- Lee JT, Davidow LS, Warshawsky D (1999) Tsix, a gene antisense to Xist at the X‐inactivation centre. Nat Genet 21: 400–404 [DOI] [PubMed] [Google Scholar]
- Lee RC, Ambros V (2001) An extensive class of small RNAs in Caenorhabditis elegans . Science 294: 862–864 [DOI] [PubMed] [Google Scholar]
- Lee S, Kopp F, Chang TC, Sataluri A, Chen B, Sivakumar S, Yu H, Xie Y, Mendell JT (2016) Noncoding RNA NORAD regulates genomic stability by sequestering PUMILIO proteins. Cell 164: 69–80 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lerner MR, Boyle JA, Hardin JA, Steitz JA (1981) Two novel classes of small ribonucleoproteins detected by antibodies associated with lupus erythematosus. Science 211: 400–402 [DOI] [PubMed] [Google Scholar]
- Lestrade L, Weber MJ (2006) snoRNA‐LBME‐db, a comprehensive database of human H/ACA and C/D box snoRNAs. Nucleic Acids Res 34: D158–D162 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Chao TC, Chang KY, Lin N, Patil VS, Shimizu C, Head SR, Burns JC, Rana TM (2014) The long noncoding RNA THRIL regulates TNFα expression through its interaction with hnRNPL. Proc Natl Acad Sci USA 111: 1002–1007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li Z, Huang C, Bao C, Chen L, Lin M, Wang X, Zhong G, Yu B, Hu W, Dai L et al (2015) Exon‐intron circular RNAs regulate transcription in the nucleus. Nat Struct Mol Biol 22: 256–264 [DOI] [PubMed] [Google Scholar]
- Li CJ, Xiao Y, Yang M, Su T, Sun X, Guo Q, Huang Y, Luo XH (2018a) Long noncoding RNA Bmncr regulates mesenchymal stem cell fate during skeletal aging. J Clin Invest 128: 5251–5266 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Li X, Yang L, Chen LL (2018b) The biogenesis, functions, and challenges of circular RNAs. Mol Cell 71: 428–442 [DOI] [PubMed] [Google Scholar]
- Lin D, Pestova TV, Hellen CU, Tiedge H (2008) Translational control by a small RNA: dendritic BC1 RNA targets the eukaryotic initiation factor 4A helicase mechanism. Mol Cell Biol 28: 3008–3019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lin MF, Jungreis I, Kellis M (2011) PhyloCSF: a comparative genomics method to distinguish protein coding and non‐coding regions. Bioinformatics 27: i275–i282 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu CX, Li X, Nan F, Jiang S, Gao X, Guo SK, Xue W, Cui Y, Dong K, Ding H et al (2019a) Structure and degradation of circular RNAs regulate PKR activation in innate immunity. Cell 177: 865–880.e21 [DOI] [PubMed] [Google Scholar]
- Liu M, Wang Q, Shen J, Yang BB, Ding X (2019b) Circbank: a comprehensive database for circRNA with standard nomenclature. RNA Biol 16: 899–905 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lott MT, Leipzig JN, Derbeneva O, Xie HM, Chalkia D, Sarmady M, Procaccio V, Wallace DC (2013) mtDNA variation and analysis using mitomap and mitomaster. Curr Protoc Bioinformatics 44: 1.23.1‐26 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lottin S, Vercoutter‐Edouart AS, Adriaenssens E, Czeszak X, Lemoine J, Roudbaraki M, Coll J, Hondermarck H, Dugimont T, Curgy JJ (2002) Thioredoxin post‐transcriptional regulation by H19 provides a new function to mRNA‐like non‐coding RNA. Oncogene 21: 1625–1631 [DOI] [PubMed] [Google Scholar]
- Lowe TM, Chan PP (2016) tRNAscan‐SE On‐line: integrating search and context for analysis of transfer RNA genes. Nucleic Acids Res 44: W54–W57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lund E, Dahlberg JE (1984) True genes for human U1 small nuclear RNA. Copy number, polymorphism, and methylation. J Biol Chem 259: 2013–2021 [PubMed] [Google Scholar]
- Lykke‐Andersen S, Ardal BK, Hollensen AK, Damgaard CK, Jensen TH (2018) Box C/D snoRNP autoregulation by a cis‐acting snoRNA in the NOP56 pre‐mRNA. Mol Cell 72: 99–111.e5 [DOI] [PubMed] [Google Scholar]
- Maraia RJ, Sasaki‐Tozawa N, Driscoll CT, Green ED, Darlington GJ (1994) The human Y4 small cytoplasmic RNA gene is controlled by upstream elements and resides on chromosome 7 with all other hY scRNA genes. Nucleic Acids Res 22: 3045–3052 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maraia R, Sakulich AL, Brinkmann E, Green ED (1996) Gene encoding human Ro‐associated autoantigen Y5 RNA. Nucleic Acids Res 24: 3552–3559 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marnef A, Richard P, Pinzón N, Kiss T (2014) Targeting vertebrate intron‐encoded box C/D 2′‐O‐methylation guide RNAs into the Cajal body. Nucleic Acids Res 42: 6616–6629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Martin F, Ménétret JF, Simonetti A, Myasnikov AG, Vicens Q, Prongidi‐Fix L, Natchiar SK, Klaholz BP, Eriani G (2016) Ribosomal 18S rRNA base pairs with mRNA during eukaryotic translation initiation. Nat Commun 7: 12622 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Marz M, Mosig A, Stadler BM, Stadler PF (2007) U7 snRNAs: a computational survey. Genomics Proteomics Bioinformatics 5: 187–195 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matera AG, Terns RM, Terns MP (2007) Non‐coding RNAs: lessons from the small nuclear and small nucleolar RNAs. Nat Rev Mol Cell Biol 8: 209–220 [DOI] [PubMed] [Google Scholar]
- Memczak S, Jens M, Elefsinioti A, Torti F, Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer M et al (2013) Circular RNAs are a large class of animal RNAs with regulatory potency. Nature 495: 333–338 [DOI] [PubMed] [Google Scholar]
- Mencía A, Modamio‐Høybjør S, Redshaw N, Morín M, Mayo‐Merino F, Olavarrieta L, Aguirre LA, del Castillo I, Steel KP, Dalmay T et al (2009) Mutations in the seed region of human miR‐96 are responsible for nonsyndromic progressive hearing loss. Nat Genet 41: 609–613 [DOI] [PubMed] [Google Scholar]
- Miyoshi N, Wagatsuma H, Wakana S, Shiroishi T, Nomura M, Aisaka K, Kohda T, Surani MA, Kaneko‐Ishino T, Ishino F (2000) Identification of an imprinted gene, Meg3/Gtl2 and its human homologue MEG3, first mapped on mouse distal chromosome 12 and human chromosome 14q. Genes Cells 5: 211–220 [DOI] [PubMed] [Google Scholar]
- Muddashetty R, Khanam T, Kondrashov A, Bundman M, Iacoangeli A, Kremerskothen J, Duning K, Barnekow A, Hüttenhofer A, Tiedge H et al (2002) Poly(A)‐binding protein is associated with neuronal BC1 and BC200 ribonucleoprotein particles. J Mol Biol 321: 433–445 [DOI] [PubMed] [Google Scholar]
- Nandy C, Mrázek J, Stoiber H, Grässer FA, Hüttenhofer A, Polacek N (2009) Epstein‐barr virus‐induced expression of a novel human vault RNA. J Mol Biol 388: 776–784 [DOI] [PubMed] [Google Scholar]
- Ng SY, Bogu GK, Soh BS, Stanton LW (2013) The long noncoding RNA RMST interacts with SOX2 to regulate neurogenesis. Mol Cell 51: 349–359 [DOI] [PubMed] [Google Scholar]
- O'Leary NA, Wright MW, Brister JR, Ciufo S, Haddad D, McVeigh R, Rajput B, Robbertse B, Smith‐White B, Ako‐Adjei D et al (2016) Reference sequence (RefSeq) database at NCBI: current status, taxonomic expansion, and functional annotation. Nucleic Acids Res 44: D733–D745 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Reilly D, Dienstbier M, Cowley SA, Vazquez P, Drozdz M, Taylor S, James WS, Murphy S (2013) Differentially expressed, variant U1 snRNAs regulate gene expression in human cells. Genome Res 23: 281–291 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Panagal M, Biruntha M, Vidhyavathi RM, Sivagurunathan P, Senthilkumar SR, Sekar D (2019) Dissecting the role of miR‐21 in different types of stroke. Gene 681: 69–72 [DOI] [PubMed] [Google Scholar]
- Pandey GK, Mitra S, Subhash S, Hertwig F, Kanduri M, Mishra K, Fransson S, Ganeshram A, Mondal T, Bandaru S et al (2014) The risk‐associated long noncoding RNA NBAT‐1 controls neuroblastoma progression by regulating cell proliferation and neuronal differentiation. Cancer Cell 26: 722–737 [DOI] [PubMed] [Google Scholar]
- Parrott AM, Mathews MB (2007) Novel rapidly evolving hominid RNAs bind nuclear factor 90 and display tissue‐restricted distribution. Nucleic Acids Res 35: 6249–6258 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Parrott AM, Mathews MB (2009) snaR genes: recent descendants of Alu involved in the evolution of chorionic gonadotropins. Cold Spring Harb Symp Quant Biol 74: 363–373 [DOI] [PubMed] [Google Scholar]
- Pelczar P, Filipowicz W (1998) The host gene for intronic U17 small nucleolar RNAs in mammals has no protein‐coding potential and is a member of the 5′‐terminal oligopyrimidine gene family. Mol Cell Biol 18: 4509–4518 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Polacek N, Mankin AS (2005) The ribosomal peptidyl transferase center: structure, function, evolution, inhibition. Crit Rev Biochem Mol Biol 40: 285–311 [DOI] [PubMed] [Google Scholar]
- Pruijn GJ, Slobbe RL, van Venrooij WJ (1991) Analysis of protein–RNA interactions within Ro ribonucleoprotein complexes. Nucleic Acids Res 19: 5173–5180 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Reed JH, Sim S, Wolin SL, Clancy RM, Buyon JP (2013) Ro60 requires Y3 RNA for cell surface exposure and inflammation associated with cardiac manifestations of neonatal lupus. J Immunol 191: 110–116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rich A, RajBhandary UL (1976) Transfer RNA: molecular structure, sequence, and properties. Annu Rev Biochem 45: 805–860 [DOI] [PubMed] [Google Scholar]
- Richard P, Darzacq X, Bertrand E, Jády BE, Verheggen C, Kiss T (2003) A common sequence motif determines the Cajal body‐specific localization of box H/ACA scaRNAs. EMBO J 22: 4283–4293 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rinn J, Kertesz M, Wang J, Squazzo S, Xu X, Brugmann S, Goodnough L, Helms J, Farnham P, Segal E et al (2007) Functional demarcation of active and silent chromatin domains in human HOX loci by Noncoding RNAs. Cell 129: 1311–1323 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roberts RB (1958) Introduction In Microsomal particles and protein synthesis, Roberts RB. (ed.), pp vii–viii. New York, NY: Pergamon Press Inc; [Google Scholar]
- Rose D, Stadler PF (2011) Molecular evolution of the non‐coding eosinophil granule ontogeny transcript. Front Genet 2: 69 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Salzman J, Chen RE, Olsen MN, Wang PL, Brown PO (2013) Cell‐type specific features of circular RNA expression. PLoS Genet 9: e1003777 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scott MS, Ono M (2011) From snoRNA to miRNA: dual function regulatory non‐coding RNAs. Biochimie 93: 1987–1992 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Weinberg DE, Fuchs G, Choi K, Chung J, Wolin SL (2009) The subcellular distribution of an RNA quality control protein, the Ro autoantigen, is regulated by noncoding Y RNA binding. Mol Biol Cell 20: 1555–1564 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sim S, Yao J, Weinberg DE, Niessen S, Yates JR, Wolin SL (2012) The zipcode‐binding protein ZBP1 influences the subcellular location of the Ro 60‐kDa autoantigen and the noncoding Y3 RNA. RNA 18: 100–110 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Singh R, Reddy R (1989) Gamma‐monomethyl phosphate: a cap structure in spliceosomal U6 small nuclear RNA. Proc Natl Acad Sci USA 86: 8280–8283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Smith KC, Cordes E, Schweet RS (1959) Fractionation of transfer ribonucleic acid. Biochim Biophys Acta 33: 286–287 [DOI] [PubMed] [Google Scholar]
- Smith C, Steitz J (1998) Classification of gas5 as a multi‐small‐nucleolar‐RNA (snoRNA) host gene and a member of the 5 ‘‐terminal oligopyrimidine gene family reveals common features of snoRNA host genes. Mol Cell Biol 18: 6897–6909 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sonkoly E, Bata‐Csorgo Z, Pivarcsi A, Polyanka H, Kenderessy‐Szabo A, Molnar G, Szentpali K, Bari L, Megyeri K, Mandi Y et al (2005) Identification and characterization of a novel, psoriasis susceptibility‐related noncoding RNA gene, PRINS. J Biol Chem 280: 24159–24167 [DOI] [PubMed] [Google Scholar]
- Sontheimer EJ, Steitz JA (1992) Three novel functional variants of human U5 small nuclear RNA. Mol Cell Biol 12: 734–746 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sørensen PD, Frederiksen S (1991) Characterization of human 5S rRNA genes. Nucleic Acids Res 19: 4147–4151 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stadler PF, Chen JJ, Hackermüller J, Hoffmann S, Horn F, Khaitovich P, Kretzschmar AK, Mosig A, Prohaska SJ, Qi X et al (2009) Evolution of vault RNAs. Mol Biol Evol 26: 1975–1991 [DOI] [PubMed] [Google Scholar]
- Stein AJ, Fuchs G, Fu C, Wolin SL, Reinisch KM (2005) Structural insights into RNA quality control: the Ro autoantigen binds misfolded RNAs via its central cavity. Cell 121: 529–539 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stelzer G, Rosen N, Plaschkes I, Zimmerman S, Twik M, Fishilevich S, Stein TI, Nudel R, Lieder I, Mazor Y et al (2016) The GeneCards suite: from gene data mining to disease genome sequence analyses. Curr Protoc Bioinformatics 54: 1.30.1–1.30.33 [DOI] [PubMed] [Google Scholar]
- Strub K, Galli G, Busslinger M, Birnstiel ML (1984) The cDNA sequences of the sea urchin U7 small nuclear RNA suggest specific contacts between histone mRNA precursor and U7 RNA during RNA processing. EMBO J 3: 2801–2807 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stults DM, Killen MW, Pierce HH, Pierce AJ (2008) Genomic architecture and inheritance of human ribosomal RNA gene clusters. Genome Res 18: 13–18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Suzuki T, Nagao A (2011) Human mitochondrial tRNAs: biogenesis, function, structural aspects, and diseases. Annu Rev Genet 45: 299–329 [DOI] [PubMed] [Google Scholar]
- Széll M, Bata‐Csörgo Z, Kemény L (2008) The enigmatic world of mRNA‐like ncRNAs: their role in human evolution and in human diseases. Semin Cancer Biol 18: 141–148 [DOI] [PubMed] [Google Scholar]
- Tarn WY, Steitz JA (1996) Highly diverged U4 and U6 small nuclear RNAs required for splicing rare AT‐AC introns. Science 273: 1824–1832 [DOI] [PubMed] [Google Scholar]
- The RNAcentral Consortium (2019) RNAcentral: a hub of information for non‐coding RNA sequences. Nucleic Acids Res 47: D221–D229 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tichon A, Gil N, Lubelsky Y, Havkin Solomon T, Lemze D, Itzkovitz S, Stern‐Ginossar N, Ulitsky I (2016) A conserved abundant cytoplasmic long noncoding RNA modulates repression by Pumilio proteins in human cells. Nat Commun 7: 12209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tiedge H, Chen W, Brosius J (1993) Primary structure, neural‐specific expression, and dendritic location of human BC200 RNA. J Neurosci 13: 2382–2390 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tranque P, Hu MC, Edelman GM, Mauro VP (1998) rRNA complementarity within mRNAs: a possible basis for mRNA‐ribosome interactions and translational control. Proc Natl Acad Sci USA 95: 12238–12243 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tripathi V, Ellis JD, Shen Z, Song DY, Pan Q, Watt AT, Freier SM, Bennett CF, Sharma A, Bubulya PA et al (2010) The nuclear‐retained noncoding RNA MALAT1 regulates alternative splicing by modulating SR splicing factor phosphorylation. Mol Cell 39: 925–938 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turunen JJ, Niemelä EH, Verma B, Frilander MJ (2013) The significant other: splicing by the minor spliceosome. Wiley Interdiscip Rev RNA 4: 61–76 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tyc K, Steitz JA (1989) U3, U8 and U13 comprise a new class of mammalian snRNPs localized in the cell nucleolus. EMBO J 8: 3113–3119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tycowski KT, Shu MD, Kukoyi A, Steitz JA (2009) A conserved WD40 protein binds the Cajal body localization signal of scaRNP particles. Mol Cell 34: 47–57 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I, Shkumatava A, Jan CH, Sive H, Bartel DP (2011) Conserved function of lincRNAs in vertebrate embryonic development despite rapid sequence evolution. Cell 147: 1537–1550 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ulitsky I (2016) Evolution to the rescue: using comparative genomics to understand long non‐coding RNAs. Nat Rev Genet 17: 601–614 [DOI] [PubMed] [Google Scholar]
- Ulitsky I (2018) Interactions between short and long noncoding RNAs. FEBS Lett 592: 2874–2883 [DOI] [PubMed] [Google Scholar]
- Van Arsdell SW, Weiner AM (1984) Human genes for U2 small nuclear RNA are tandemly repeated. Mol Cell Biol 4: 492–499 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vance KW, Sansom SN, Lee S, Chalei V, Kong L, Cooper SE, Oliver PL, Ponting CP (2014) The long non‐coding RNA Paupar regulates the expression of both local and distal genes. EMBO J 33: 296–311 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Arango P, Vowles J, Browne C, Hartfield E, Fernandes HJ, Mandefro B, Sareen D, James W, Wade‐Martins R, Cowley SA et al (2016) Variant U1 snRNAs are implicated in human pluripotent stem cell maintenance and neuromuscular disease. Nucleic Acids Res 44: 10960–10973 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Vazquez‐Arango P, O'Reilly D (2018) Variant snRNPs: new players within the spliceosome system. RNA Biol 15: 17–25 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Volders PJ, Anckaert J, Verheggen K, Nuytens J, Martens L, Mestdagh P, Vandesompele J (2019) LNCipedia 5: towards a reference set of human long non‐coding RNAs. Nucleic Acids Res 47: D135–D139 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walker TA, Pace NR, Erikson RL, Erikson E, Behr F (1974) The 7S RNA common to oncornaviruses and normal cells is associated with polyribosomes. Proc Natl Acad Sci USA 71: 3390–3394 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Walter P, Blobel G (1982) Signal recognition particle contains a 7S RNA essential for protein translocation across the endoplasmic reticulum. Nature 299: 691–698 [DOI] [PubMed] [Google Scholar]
- Wang K, Yang Y, Liu B, Sanyal A, Corces‐Zimmerman R, Chen Y, Lajoie B, Protacio A, Flynn R, Gupta R et al (2011) A long noncoding RNA maintains active chromatin to coordinate homeotic gene expression. Nature 472: 120‐U158 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Will CL, Lührmann R (2005) Splicing of a rare class of introns by the U12‐dependent spliceosome. Biol Chem 386: 713–724 [DOI] [PubMed] [Google Scholar]
- Willingham AT, Orth AP, Batalov S, Peters EC, Wen BG, Aza‐Blanc P, Hogenesch JB, Schultz PG (2005) A strategy for probing the function of noncoding RNAs finds a repressor of NFAT. Science 309: 1570–1573 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA (1983) Genes for two small cytoplasmic Ro RNAs are adjacent and appear to be single‐copy in the human genome. Cell 32: 735–744 [DOI] [PubMed] [Google Scholar]
- Wolin SL, Steitz JA (1984) The Ro small cytoplasmic ribonucleoproteins: identification of the antigenic protein and its binding site on the Ro RNAs. Proc Natl Acad Sci USA 81: 1996–2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wolin SL, Belair C, Boccitto M, Chen X, Sim S, Taylor DW, Wang HW (2013) Non‐coding Y RNAs as tethers and gates: insights from bacteria. RNA Biol 10: 1602–1608 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wu H, Yin QF, Luo Z, Yao RW, Zheng CC, Zhang J, Xiang JF, Yang L, Chen LL (2016) Unusual processing generates SPA LncRNAs that sequester multiple RNA binding proteins. Mol Cell 64: 534–548 [DOI] [PubMed] [Google Scholar]
- Wu H, Yang L, Chen LL (2017) The diversity of long noncoding RNAs and their generation. Trends Genet 33: 540–552 [DOI] [PubMed] [Google Scholar]
- Xing YH, Yao RW, Zhang Y, Guo CJ, Jiang S, Xu G, Dong R, Yang L, Chen LL (2017) SLERT regulates DDX21 rings associated with Pol I transcription. Cell 169: 664–678.e16 [DOI] [PubMed] [Google Scholar]
- Yan JJ, Qiao M, Li RH, Zhao XT, Wang XY, Sun Q (2019) Downregulation of miR‐145‐5p contributes to hyperproliferation of keratinocytes and skin inflammation in psoriasis. Br J Dermatol 180: 365–372 [DOI] [PubMed] [Google Scholar]
- Ye XT, Huang H, Huang WP, Hu WL (2018) LncRNA THOR promotes human renal cell carcinoma cell growth. Biochem Biophys Res Commun 501: 661–667 [DOI] [PubMed] [Google Scholar]
- Yin QF, Yang L, Zhang Y, Xiang JF, Wu YW, Carmichael GG, Chen LL (2012) Long noncoding RNAs with snoRNA ends. Mol Cell 48: 219–230 [DOI] [PubMed] [Google Scholar]
- Yotova I, Vlatkovic I, Pauler F, Warczok K, Ambros P, Oshimura M, Theussl H, Gessler M, Wagner E, Barlow D (2008) Identification of the human homolog of the imprinted mouse Air non‐coding RNA. Genomics 92: 464–473 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Young TL, Matsuda T, Cepko CL (2005) The noncoding RNA taurine upregulated gene 1 is required for differentiation of the murine retina. Curr Biol 15: 501–512 [DOI] [PubMed] [Google Scholar]
- Younis I, Dittmar K, Wang W, Foley SW, Berg MG, Hu KY, Wei Z, Wan L, Dreyfuss G (2013) Minor introns are embedded molecular switches regulated by highly unstable U6atac snRNA. Elife 2: e00780 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zemmour D, Pratama A, Loughhead SM, Mathis D, Benoist C (2017) A long noncoding RNA, modulates Foxp3 expression and autoimmunity. Proc Natl Acad Sci USA 114: E3472–E3480 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zerbino DR, Achuthan P, Akanni W, Amode MR, Barrell D, Bhai J, Billis K, Cummins C, Gall A, Girón CG et al (2018) Ensembl 2018. Nucleic Acids Res 46: D754–D761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang Y, Zhang XO, Chen T, Xiang JF, Yin QF, Xing YH, Zhu S, Yang L, Chen LL (2013) Circular intronic long noncoding RNAs. Mol Cell 51: 792–806 [DOI] [PubMed] [Google Scholar]
- Zhang XO, Wang HB, Zhang Y, Lu X, Chen LL, Yang L (2014) Complementary sequence‐mediated exon circularization. Cell 159: 134–147 [DOI] [PubMed] [Google Scholar]
- Zieve G, Penman S (1976) Small RNA species of the HeLa cell: metabolism and subcellular localization. Cell 8: 19–31 [DOI] [PubMed] [Google Scholar]


