Abstract
Background
Chronic obstructive pulmonary disease (COPD) is a common disease characterized by persistent respiratory symptoms and airflow restriction. It is usually manifested as airway and/or alveolar abnormalities caused by significant exposure to harmful particulates or gases.
Objective
We aim to explore plasma metabolomic changes in the acute exacerbation stage of COPD (AECOPD) and stable stage of COPD (Stable COPD) to identify potential biomarkers for diagnosis or prognosis in clinical practice.
Methods
Untargeted metabolomics and lipidomics analyses were performed to investigate dysregulated molecules in blood plasma of AECOPD patients (n=48) and Stable COPD (n=48), and a cohort of healthy people were included as a control group (n=48). Statistical analysis and bioinformatics analysis were performed to reveal dysregulated metabolites and perturbed metabolic pathways. SVM-based multivariate ROC analysis was used for candidate biomarker screening.
Results
A total of 142 metabolites and 688 lipids were dysregulated in COPD patients. Pathway enrichment analysis showed that several metabolic pathways were perturbed after COPD onset. Several biomarker panels were proposed for diagnosis of COPD vs healthy control and AECOPD vs Stable COPD with AUC greater than 0.9.
Conclusion
Numerous plasma metabolites and several metabolic pathways were detected relevant to COPD disease onset or progression. These metabolites may be considered as candidate biomarkers for diagnosis or prognosis of COPD. The perturbed pathways involved in COPD provide clues for further pathological mechanism studies of COPD.
Keywords: metabolomics, lipidomics, COPD, biomarker
Introduction
Chronic obstructive pulmonary disease (COPD) is a common, preventable, and therapeutic disease characterized by persistent respiratory symptoms and airflow limitations, usually associated with exposure to toxic particles and gases. COPD is currently the fourth leading cause of death in the world and is expected to become the third leading cause of death by 2020.1 In 2012, more than 3 million people died of COPD worldwide, accounting for 6% of all deaths. Continuous exposure to risk factors and population aging will lead to an increase in the burden of COPD disease in the future.2 It is estimated that by 2020, the number of COPD patients will reach 384 million.3 In China, the latest research shows that the standardized prevalence of COPD is 13.6% for people over 40 years old, 8.1% for women, and 19.0% for men. As a chronic disease, early diagnosis and disease monitoring of COPD are urgent. In the clinic, COPD can be further divided into subtypes according to phenotypes.4 Acute exacerbation of COPD (AECOPD) is the aggravation of any symptoms (such as cough, sputum, wheezing) in COPD patients. It can be caused by bacterial or viral infection, environmental pollution, cold weather, or interrupted routine treatment, and is the main cause of admission and death. Every acute aggravation will aggravate the pulmonary function and complications of patients, and increase the risk of re-hospitalization.5 In 2011, the Society for Qualification of Biomarkers for Chronic Obstructive Pulmonary Disease was established to accelerate the research and development of biomarkers. COPD biomarkers have become an important and challenging area in COPD research.
The metabolome is defined as the total collection of small molecular metabolites present in a given type of cell or organism, and is the final downstream product of metabolism. The metabolome can provide a more exact reflection of the current metabolic status of the organic body compared to genomics, transcriptomics, or proteomics.6 NMR,7 GC-MS,8 and LC-MS9 are the main techniques used for metabolomics. Due to the advantages of the great metabolite coverage and reliable analytical performance,10–12 LC-MS-based lipidomics and metabolomics have been widely employed in biological studies or biomarker screening studies.13–15 The metabolomics technique has been widely applied in mechanism studies and biomarker screening using exhaled breath condensate16 or blood of COPD patients.17 A variety of metabolic pathways and biological molecules have been found to be involved in the progression of COPD, such as amino acid metabolism dysfunction,18–20 oxidative stress,21,22 energy metabolism dysfunction,23,24 and lipid metabolism dysfunction.25,26 But most of the previous studies focused on patients from African, American, and European populations, and to the best of our knowledge, no COPD related metabolomics study has been reported using large cohorts of the Chinese population. Considering the complexity of COPD pathology and different influences of environmental factors on COPD disease onset and progression, it is urgent and meaningful to systematically study the metabolic perturbance in Chinese patient cohorts.
In the present study, we aim to explore the metabolic changes of Chinese COPD patients in progressive and stable stages by applying comprehensive LC-MS-based metabolomic and lipidomic analysis to plasma samples. Our findings can not only provide clues for pathological studies and novel biomarker development but also provide important complementary data for previous studies using non-Chinese populations.
Materials and Methods
Chemicals and Reagents
Ammonium acetate was purchased from Sigma-Aldrich. Formic acid, HPLC grade isopropanol, acetonitrile, and methanol were purchased from Fisher Scientific. Deionized water was produced by a Milli-Q system. The chemical standards were of analytical grade with typical purity of >99%.
Study Population
Initially, a total of 234 subjects were retrospectively enrolled in this study, including 133 patients with COPD and 101 healthy volunteers, from January 2012 to June 2016 at Peking University Third Hospital. In all, 133 COPD cases were diagnosed according to the GOLD criteria, which are based on COPD risk factors, symptoms, and post-bronchodilator FEV1/FVC 70%, and patients with asthma, bronchiectasis, pleural effusion, history of exposure to noxious particles (such as founder’s pneumoconiosis, silicosis, and asbestosis) and diseases affecting activity were excluded. After the exclusion procedure, 48 patients in the AECOPD group and 72 patients in the Stable COPD group were left. Different GOLD airflow limitation stages were distinguished as COPD 1–4 according to post-bronchodilator FEV1% predicted. The included healthy controls were subjects with normal pulmonary function and without chronic heart and lung diseases or a recent history of respiratory tract infection. This study was conducted in accordance with the Declaration of Helsinki and with approval from the ethics committee of Peking University Third Hospital. All participants provided written informed consent. General information, past history, smoking history, complications, out-of-hospital basic medication, and home oxygen therapy were collected. The number of times of hospitalization was increased in the past year. The aggravating factors, such as fever, lower extremity edema, pulmonary function in stable period, serological indicators at admission, CAT score and hormone use were also collected. Information including usage of mechanical ventilation, length of stay, blood routine, blood gas analysis, CT score, and other clinical data were recorded at discharge. Three groups of plasma samples were included in the metabolomics analysis, including patients in acute exacerbation of COPD (AECOPD, n=48), stable stage of COPD (Stable COPD, n=48), and healthy controls (n=48). The patients or healthy subjects not included in the metabolomics analysis were no longer analyzed in the present study. Baseline characteristics and clinical parameters of the patients included in the metabolomics analysis are presented in Table 1A and B.
Table 1.
Baseline characteristics and clinical parameters of the patients
| (A) Baseline Characteristics of the Participants | ||||
|---|---|---|---|---|
| Control | Stable COPD | Acute Exacerbation of COPD | P value | |
| Number | 48 | 48 | 48 | |
| Clinical Features | ||||
| Male/female | 38/10 | 38/10 | 38/10 | 1 |
| Age | 63.72±6.12 | 67.34±8.45 | 66.19±7.34 | 0.893 |
| BMI | 23.76±5.12 | 24.85±4.65 | 23.09±6.34 | 0.854 |
| Course of COPD (year) | 0 | 4(1,7) | 6(2,9) | 0.712 |
| (B) Clinical Parameters of the Patients | |||
|---|---|---|---|
| Stable COPD | Acute Exacerbation of COPD | P value | |
| GOLD Classification | |||
| I | 1(2.08%) | 2(4.16%) | 0.868 |
| II | 9(18.95%) | 7(14.58%) | |
| III | 18(37.5%) | 20(41.67%) | |
| IV | 20(41.67%) | 19(39.58%) | |
| Complicated with cor pulmonale | 13(27.1%) | 15(31.25%) | 0.653 |
| Complication | |||
| Coronary heart disease | 24(50%) | 26(64.17) | 0.683 |
| Heart failure | 11(22.9%) | 14(29.2%) | 0.485 |
| Diabetes | 8(16.7%) | 6(12.5%) | 0.563 |
| Hypertension | 37(77.1%) | 39(81.25%) | 0.615 |
| OASAHS | 13(27.1%) | 12(25%) | 0.816 |
| Anemia | 4(8.3%) | 3(6.25%) | 0.695 |
| Smoking index | 800(200,1000) | 780(200,900) | 0.834 |
| Smoking | 12(25%) | 11(22.92%) | 0.811 |
| ICS | 46(95.83%) | 48(100%) | 0.153 |
| Family oxygen therapy | 35(72.9%) | 37(77.1%) | 0.637 |
| The number of aggravations in the past year | 1(0,1.75) | 1(0.1.25) | 0.712 |
| CAT score | 26.22±7.12 | 25.75±6.14 | 0.834 |
| Laboratory Examination | |||
| White blood cell (*109/L) | 7.45±2.13 | 13.54±3.15 | 0.033 |
| Neutrophil/lymphocyte count | 1.16(0.70–1.65) | 2.43(1.34,2.91) | 0.041 |
| Hemoglobin (g/L) | 131.13±22.14 | 134.23±12.07 | 0.791 |
| Platelet (*109/L) | 254.14±54.24 | 289.15±76.65 | 0.704 |
| Fibrinogen | 3.85(3.12,4.75) | 5.54(3.78, 4.76) | 0.056 |
| D-Dimer | 0.45(0.21,0.63) | 0.78(0.54,1.67) | 0.061 |
| Procalcitonin positive | 1(2.08%) | 43(89.58%) | <0.01 |
| C-reactive protein | 4.00(3.01,4.65) | 180.31(90.23,331.56) | <0.01 |
| FEV1%pred | 40.91±12.87% | 37.58±14.75 | 0.692 |
Sample Preparation
After an overnight fast, blood was collected in EDTA tubes and placed on ice. Plasma was separated by centrifugation at 3000 g for 20 min, and then stored at −80°C. All plasma samples included for metabolomic analyses were processed for metabolite extraction as one batch. Metabolites and lipids were extracted from the plasma samples using liquid–liquid extraction as follows: 100 μL plasma was extracted by fourfold volume of cold chloroform: methanol (v/v=2:1). The mixture was centrifuged at 13,000 g for 15 min and then the upper phase (hydrophilic metabolites) and lower organic phase (hydrophobic metabolites) were separately collected and evaporated at room temperature under vacuum. The dried samples were stored at −80°C for about 2 days until LC-MS analysis.
Liquid Chromatography
Metabolomics and lipidomics were performed on an Ultimate 3000 UHPLC system coupled with Q-Exactive MS (Thermo Scientific). For the aqueous phase (metabolomics), an Xbridge amide column (100 × 2.1 mm i.d., 2.5 μm; Waters) was employed for compound separation at 30°C. The mobile phase A consisted of 5 mM ammonium acetate in water with 5% acetonitrile, and mobile phase B was acetonitrile. The flow rate was 0.35 mL/min with the following linear gradient: 0 min, 95% B; 3 min, 90% B; 13 min, 50% B, 14 min, 50% B; 15 min, 95% B, and 17 min, 95% B. The samples were suspended with 100 μL of acetonitrile:water (1:1, v/v) solution and the injection volume was 10 μL.
For the lipid, chromatographic separation was performed on a reversed phase X-select CSH C18 column (2.1 mm × 100 mm, 2.5 μm, Waters, USA) at 40°C. Two solvents, containing 10 mM ammonium acetate and 0.1% formic acid, were used for gradient elution: (A) ACN/water (3:2, v/v), (B) IPA/ACN (9:1, v/v). The gradient program was: 0 min 40% B; 2 min 43% B; 12 min 60% B; 12.1 75% B; 18 min 99% B; 19 min 99% B; 20 min 40% B. The flow rate was set to 0.4 mL/min. The samples were suspended with 100 μL of chloroform:methanol (1:1, v/v) solution and then diluted threefold with isopropanol:acetonitrile:H2O (2:1:1, v/v/v) solution. The injection volume was 10 μL.
Mass Spectrometry
Data-dependent acquisition (DDA) was performed using the Q-Exactive MS (Thermo Scientific) using positive-negative ion switching mode. Each acquisition cycle consists of 1 survey scan (MS1 scan) at 70,000 resolution from 60 to 900 m/z for the hydrophilic metabolites and mass range m/z 300 to 1200 for the lipids, followed by 10 MS/MS scans in HCD mode at 13,500 resolution using stepped normalized collision energy (step-NCE) of 15, 30, and 45. The dynamic exclusion was set to 8 s. The automatic gain control (AGC) target was set to 5e6 (maximum injection time 30 ms) and 2e5 (maximum injection time 100 ms) for the MS1 and MS/MS scan. The parameters of ion source were: spray voltage 3.3 kV for positive ion mode and 3.0 kV for negative ion mode; ion source sheath gas 40; aux gas 10; capillary temperature 320°C; probe heater temperature 300°C; S-lens RF level 55. Samples (n=144 in total) were analyzed in random order. Quality control (QC) samples were prepared by pooling equally volumes of all study samples, and were analyzed between every 15 samples during the entire LC-MS analytical sequence.
Data Processing
Raw data collected from the DDA-MS were processed on MS-DIAL software v3.6 (http://prime.psc.riken.jp/Metabolomics_Software/MS-DIAL/) according to the user guide. Briefly, the raw MS data (.raw) were converted from into the common file format of Reifycs Inc. (.abf) using the Reifycs ABF converter (http://www.reifycs.com/AbfConverter/index.html). After conversion, the MS-DIAL software was used for feature detection, spectra deconvolution, metabolite identification and peak alignment between samples. The MS2 spectra based metabolite identification was performed in MS-DIAL by searching the acquired MS2 spectra against the MassBank database provided by MS-DIAL software, containing MS1 and MS/MS information of metabolites (8068 records in positive ion mode and 4782 records in negative ion mode). The MS2 spectra based lipid identification was performed in MS-DIAL by searching the acquired MS2 spectra against the software’s internal in silico MS/MS spectra database (version: LipidDBs-VS23-FiehnO), which includes MS1 and MS/MS information of common lipid species. The tolerances for MS1 and MS/MS search were set to 0.01 Da and 0.05 Da, separately. Peak alignment was performed using retention time tolerance of 0.2 min and MS1 tolerance of 0.01 Da. Other parameters used in MS-DIAL were set as default.
Statistical Analysis and Multivariate ROC Analysis
Statistical analysis (including PCA analysis, ANOVA, and hierarchical cluster analysis) and bioinformatics (pathway enrichment analysis) were carried out by the MetaboAnalyst web service (https://www.metaboanalyst.ca/). For each sample, the peak area of each metabolite was normalized to the total peak areas of all metabolites. Significance was analyzed using ANOVA and an FDR-adjusted p-value of less than 0.01 was considered as significant, and Tukey’s HSD was used as a post hoc test. For pathway enrichment analysis, dysregulated metabolites were used as input to search against the KEGG and SMP database, and a p-value of less than 0.01 was considered significant. Multivariate ROC curve analyses were performed using the MetaboAnalyst web service.27 The analysis was based on support vector machine (SVM) algorithms. ROC curves are generated by Monte-Carlo cross-validation (MCCV) using balanced sub-sampling, to visualize the outcome of multivariate modeling. In each MCCV, two-thirds of the samples are used to evaluate the feature importance. The top 5, 10, 15, 25, 50, and 100 important features are then used to build classification models that are validated on the one-third of the samples that were left out. The procedure was repeated multiple times to calculate the performance and confidence interval of each model. The feature ranking method was set to “SVM built-in.”
Results
Study Design and Overview of the Study
In this study, we systematically investigated the expressive profiles of metabolites and lipids in the plasma of COPD patients and healthy people in a Chinese population. Two subgroups were separately analyzed in COPD patient cohorts according to COPD disease stage (ie AECOPD and Stable COPD). After metabolite identification and quantification, statistical analyses were used to retrieve dysregulated molecules between groups. Pathway enrichment analysis was also used to find relevant metabolic pathways to COPD pathology. Finally, we performed multivariate ROC analysis to reveal potential biomarker panels that could be used for diagnosis of COPD disease onset or stages. Our findings provide, for the first time, a comprehensive view of the plasma metabolic profile of COPD patients in a Chinese population, and thus provide valuable information for further pathological research and biomarker development studies.
A total of 96 patients with COPD (including 48 AECOPD patients, 48 Stable COPD patients) and 48 age-sex matched healthy volunteers were retrospectively enrolled in this study. Their demographic data are summarized in Table 1. There were no significant differences in gender, age, BMI, smoking history, pulmonary heart disease, complications, basic ICS use, home oxygen therapy, or the number of aggravations in the past year between the two groups (Table 1A). For other clinical parameters, there were significantly higher level of white blood cell (13.54 ± 3.15 vs 7.45 ± 2.13, p=0.033), ratio of neutrophil/lymphocyte (5.54 (3.78, 4.76) vs 3.85 (3.12, 4.75), p=0.041), procalcitonin positive (89.58% vs 2.08%, p<0.01) and C-reactive protein level (180.31 (90.23, 331.56) vs 4.00 (3.01, 4.65), p<0.01) in AECOPD than those in Stable COPD (Table 1b).
Metabolomics Revealed Dysregulated Metabolites Between COPD Stages
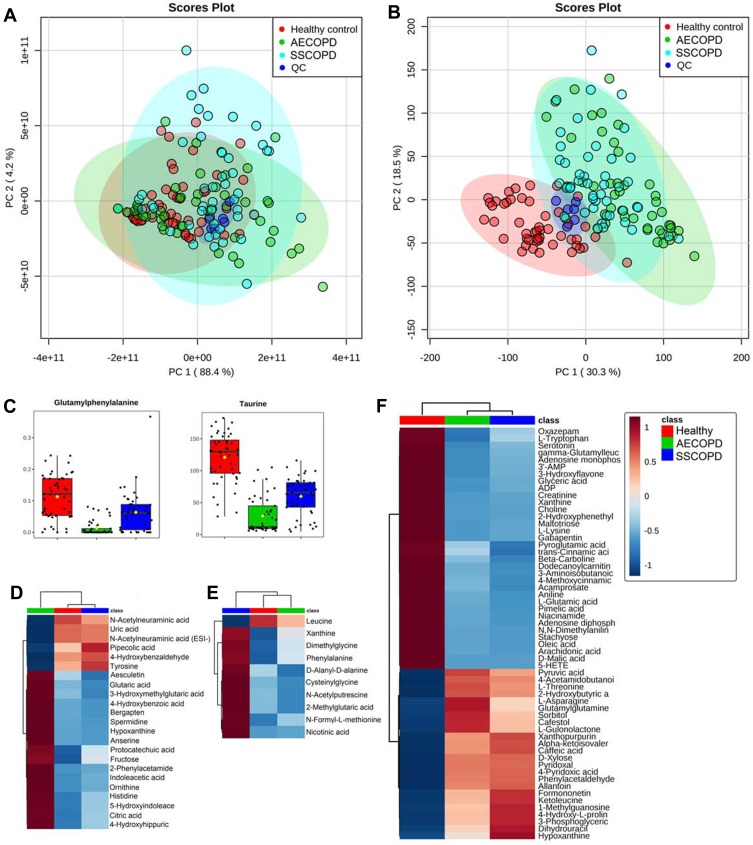
Principle component analysis (PCA) was performed to get a comprehensive view of the metabolomics data. PCA score plots of all sample groups (ie AECOPD, Stable COPD, Healthy control, and QC) in positive (Figure 1A) and negative ion mode (Figure 1B) are shown. QC samples clustered well in both ion modes, proving the reliability of the metabolomics data, thus no QC-based drift correction or data cleaning were performed. No obvious trends of separation were observed in positive ion modes, but the healthy control group showed clear trends of separation with COPD groups in negative ion mode. Further ANOVA analysis revealed a total of 142 dysregulated metabolites between three sample groups, as presented in Table S1. Dysregulated metabolites belonging to four different expression patterns are shown in Figure 1C–F. Glutamylphenylalanine and taurine were found to be dysregulated in three groups (Figure 1C). Metabolites dysregulated in only one group are shown by the heat map in Figure 1D (AECOPD), E (Stable COPD) and F (Healthy control).
Figure 1.
Dysregulated metabolites revealed by metabolomics. (A and B) Principle component analysis score plots of all sample groups in positive (A) and negative ion mode (B). Groups are presented in different colors (AECOPD, green; SSCOPD (Stable COPD), blue; healthy control, red; QC, dark blue). (C) Grouped scatter plots presenting levels of glutamylphenylalanine and taurine. (D) Heat map presenting metabolites that are only dysregulated in AECOPD. (E) Heat map presenting metabolites that are only dysregulated in SSCOPD (Stable COPD). (F) Heat map presenting metabolites that are only dysregulated in the Healthy control.
Lipidomics Revealed Dysregulated Metabolites Between COPD Stages
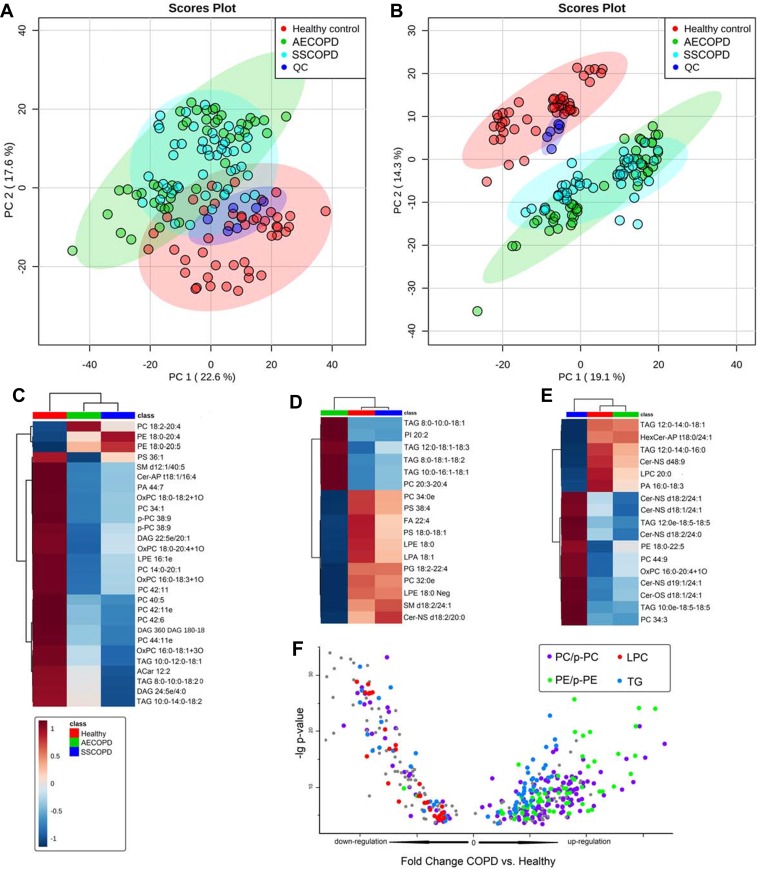
For the lipidomics data, PCA was performed to get a comprehensive view. PCA score plots of all sample groups (ie AECOPD, Stable COPD, Healthy control, and QC) in positive (Figure 2A) and negative ion mode (Figure 2B) were shown. QC samples clustered well in both ion modes, proving the reliability of the lipidomics data, thus no QC-based drift correction or data cleaning were performed. The healthy control group showed clear trends of separation with COPD groups in both positive and negative ion modes, but no obvious trends of separation were observed between two COPD subgroups. Further ANOVA analysis revealed a total of 688 dysregulated lipids, as presented in Table S2. Dysregulated lipids belonging to four different expression patterns are shown in Figure 1C–F. Lipids that dysregulated between three groups are shown in Figure 2C, while lipids that only dysregulated in AECOPD or Stable COPD are shown in Figure 2D and E. A scatter plot (Figure 2F) was used to present lipids only dysregulated in healthy control but not dysregulated between COPD subgroups.
Figure 2.
Dysregulated metabolites revealed by lipidomics. (A and B) Principle component analysis score plots of all sample groups in positive (A) and negative ion mode (B). Groups are presented in different colors (AECOPD, green; SSCOPD (Stable COPD), blue; healthy control, red; QC, dark blue). (C) Heat map presenting lipids dysregulated in three groups. (D) Heat map presenting lipids that are only dysregulated in AECOPD. (E) Heat map presenting lipids that are only dysregulated in SSCOPD (Stable COPD). (F) Heat map presenting lipids that are only dysregulated in the Healthy control.
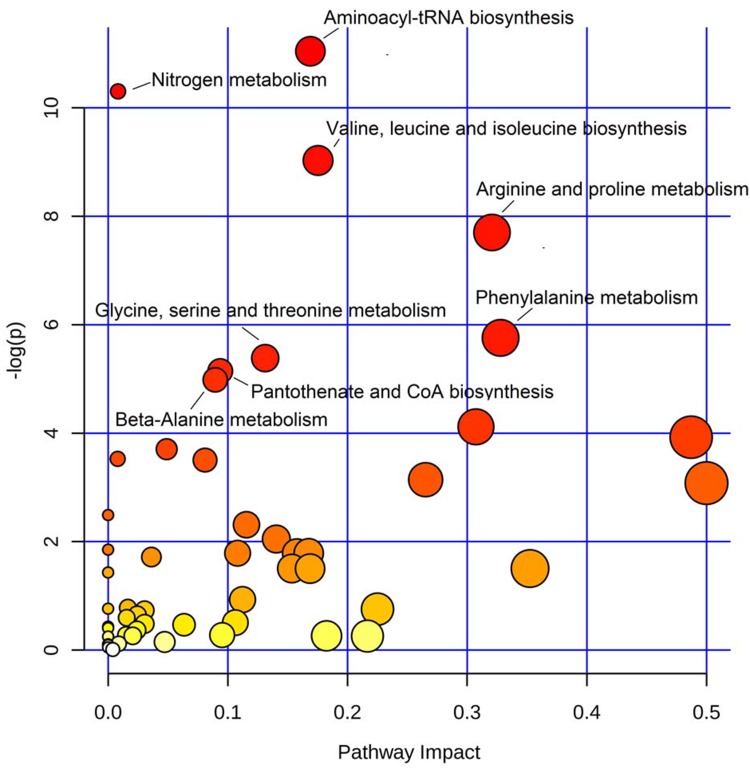
Pathway Enrichment Analysis Revealed Perturbed Metabolic Pathways
To find the metabolic pathways involved in COPD disease onset or progression, we performed pathway enrichment analysis using all the dysregulated metabolites as inputs. As shown in the scatter plot in Figure 3, dysregulated metabolites were enriched in eight pathways with a p-value of less than 0.01 (Table S3). Five of the dysregulated pathways belonged to amino acid metabolic pathways involving amino acids of valine, leucine, isoleucine, arginine, proline, phenylalanine, glycine, serine, threonine, and beta-alanine.
Figure 3.
Metabolic pathways involved in COPD disease onset and progression. Scatter plot presenting enriched metabolic pathways. The color gradient indicates the significance of the pathway ranked by p-value (y-axis; yellow: higher p-values and red: lower p-values), and circle size indicates the pathway impact score (x-axis; the larger circle the higher impact score). Significantly affected pathways with p-value less than 0.01 were marked by names.
ROC Analysis Revealed Candidate Biomarkers for COPD Onset
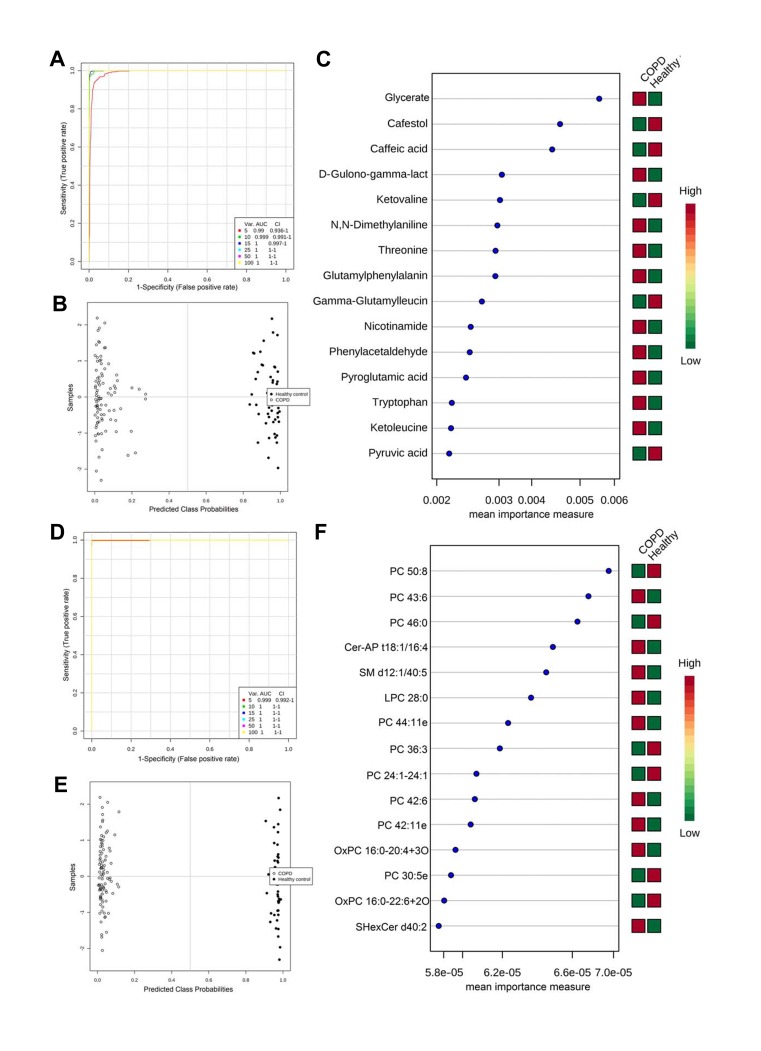
We employed SVM-based multivariate ROC curve analysis to retrieve metabolites or lipids that can distinguish COPD patients from healthy controls, which can be potential biomarkers for COPD onset. AECOPD and Stable COPD were combined as the disease group and compared against healthy control. Multivariate ROC curves constructed with 5–100 metabolites or lipids are shown in Figure 4A and D. The predicted class probabilities (average of the cross-validation) for each sample using the 15 feature model are shown in Figure 4B and E, in which the COPD disease group showed clear separation from the healthy control. The top 15 metabolites or lipids contributing to the prediction model are shown in Figure 4C and F, ranked by mean importance measure.
Figure 4.
ROC analysis revealed candidate biomarkers for COPD diagnosis. (A and D) Multivariate ROC curve constructed with 5–100 metabolites (A) or lipids (D) based on the cross-validation (CV) performance. (B and E) The predicted class probabilities (average of the cross-validation) for each sample using the 15 feature model of metabolites (B) or lipids (E). The top 15 metabolites (C) or lipids (F) contributed to the prediction model ranked by mean importance measure and the expressive levels are presented aside by color (red, high; green, low).
ROC Analysis Revealed Candidate Biomarkers for COPD Stages
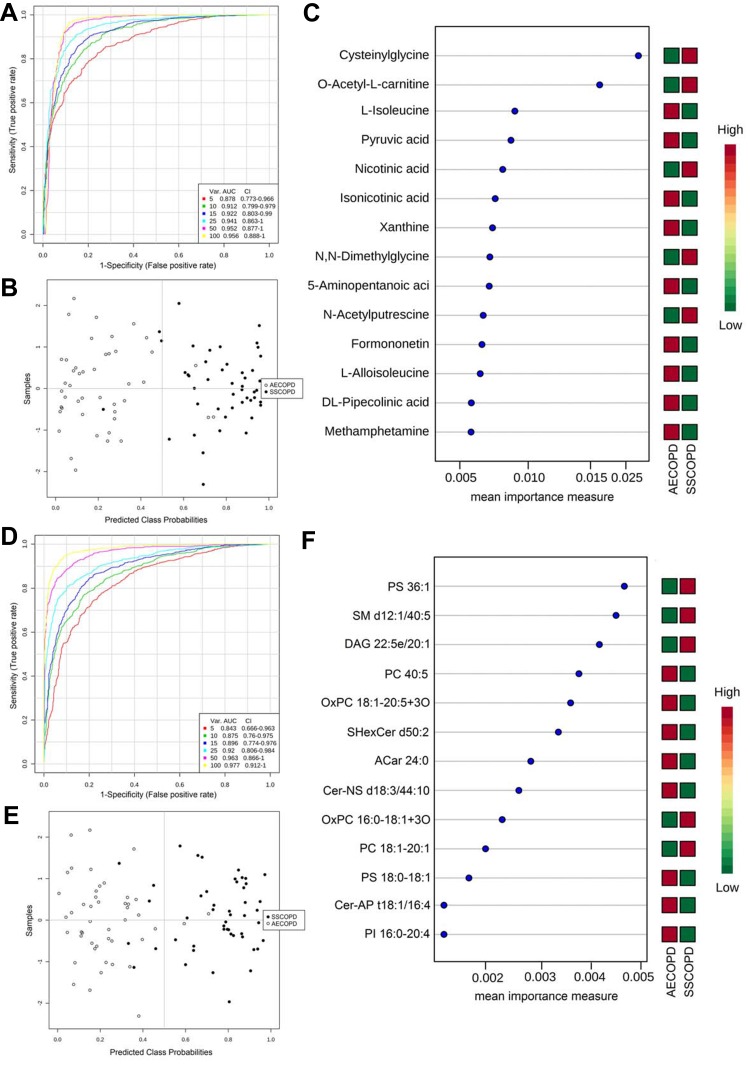
Multivariate ROC curve analysis was also used to retrieve metabolites or lipids that can distinguish AECOPD from Stable COPD, which can be potential biomarkers for the diagnosis of COPD stages. Multivariate ROC curves constructed with 5–100 metabolites or lipids are shown in Figure 5A and D. The predicted class probabilities for each sample using the 15-feature model are shown in Figure 5B and E, in which AECOPD and Stable COPD did not show absolute separation, but most of the samples could be distinguished correctly. The top 15 metabolites or lipids contributed to the prediction model are shown in Figure 5C and F ranked by mean importance measure.
Figure 5.
ROC analysis revealed candidate biomarkers for COPD disease stage diagnosis. (A and D) Multivariate ROC curve constructed with 5–100 metabolites (A) or lipids (D) based on the cross-validation (CV) performance. (B and E) The predicted class probabilities (average of the cross-validation) for each sample using the 15 feature model of metabolites (B) or lipids (E). The top 15 metabolites (C) or lipids (F) contributed to the prediction model ranked by mean importance measure and the expressive levels are presented aside by color (red, high; green, low). SSCOPD stands for Stable COPD group.
Discussion
COPD is a common disease characterized by persistent respiratory symptoms and airflow limitations, usually associated with exposure to toxic particles and gases, and airflow is not fully reversible.28 As a chronic disease, early diagnosis and disease monitoring of COPD are urgent. Several previous studies have explored the metabolic changes between COPD patients in different disease stages as well as healthy people, but few studies have used the Chinese population as participants. Our present study provides a comprehensive view of the metabolic and lipidomic profile of plasma in Chinese COPD patients, thus can provide not only clues for pathological mechanism studies and novel diagnostic biomarker development, but also complementary data to previous COPD related studies focusing on non-Chinese populations.
Among the dysregulated metabolites, four different expression patterns can be concluded as shown in Figure 1C–F. Glutamylphenylalanine and taurine were found significantly dysregulated in three groups (highly expressed in the healthy group and less expressed in AECOPD), thus may be the key metabolites involved in the disease onset and progress, and our study was the first to report the relationship between COPD and these two metabolites. Glutamylphenylalanine is a dipeptide composed of glutamate and phenylalanine, and is a proteolytic breakdown product of larger proteins; thus, the different plasma levels of glutamylphenylalanine in different COPD stages may be the result of different proteolytic activities. Taurine is a sulfur amino acid and is a lesser-known amino acid because it is not incorporated into the structural building blocks of protein.29 Taurine has many diverse biological functions, serving as a neurotransmitter in the brain, a stabilizer of cell membranes and a facilitator in the transport of ions such as sodium, potassium, calcium, and magnesium.30 In the pathway enrichment analysis (Figure 3 and Table S3), five of the perturbed pathways belonged to amino acid metabolism, showing the close link between amino acid metabolism and COPD, and this finding was in accordance with previous studies.17 Phenylalanine is one of the essential amino acids, and the activity level of the phenylalanine metabolism pathway may reflect the synthesis and breakdown state of the systematic protein. Previous studies indicated that its level may be related to the severity of COPD.31 The three BCAAs included in the valine, leucine, and isoleucine biosynthesis pathway can promote protein anabolism and maintain glucose homeostasis in skeletal muscle.20 Reduced BCAA levels in COPD progression may be a result of protein malnutrition, and in some cases hypermetabolism caused by COPD exacerbation is another possible reason for that.19 The pathways of arginine and proline metabolism and nitrogen metabolism may influence COPD progression via regulation of oxidative stress. It has been reported that NO, a common and highly reactive free radical in living systems, is converted by arginine oxidation and nitrite reduction, and the dysregulation of these pathways may contribute to airway obstruction and chronical airway remodeling in COPD.32
In lipidomics results, several lipids showed a significant difference between three groups (Figure 2C), which can be used to distinguish not only COPD patients from healthy people, but also COPD patients in different disease stages. For the lipids dysregulated only between healthy control and COPD groups (Figure 2F), different lipid species showed different changing trends, for example, all lipids belonged to LPC were downregulated in COPD groups, and all PE and p-PE were upregulated; PCs and TGs were partially upregulated and others were downregulated in COPD groups. These results hinted about the complexity of lipid metabolism involved in COPD, and lipid molecules belonging to the same lipid species may play different roles in COPD pathology. Among the total of 18 lipid species we found dysregulated between groups, some of them have been reported as relevant to COPD, such as LPC, LPE, and PE,25,33,34 and some lipid species, such as ceramide, PS, and PI, were newly found to be involved in COPD.
For the purpose of providing new biomarkers for diagnosis, we firstly explored the potential biomarkers to discriminate COPD patients from healthy controls. As shown in Figure 4, many dysregulated metabolites and lipids showed excellent discriminative power; the AUC of the ROC curve constructed with only 5 molecules is higher than 0.99. Using the SVM-based discriminative mode constructed with 15 metabolites or lipids, samples in the COPD group and healthy control could be entirely separated (Figure 4B and E). These candidate markers provide a new choice for further biomarker development for COPD diagnosis. COPD is a heterogeneous disease, and the diagnosis of COPD is mainly based on the pulmonary function test, but some patients cannot fully finish the pulmonary function test due to poor lung function or body condition.35 Thus, the candidate biomarkers we found in this study may be considered as a complementary way to perform a pulmonary function test for the diagnosis of COPD because the blood test was relatively noninvasive and more practicable. What is more, patients with COPD diagnosed by pulmonary function have an irreversible decline in pulmonary function,36 and the warning effect of pulmonary function on COPD is limited. The blood-based molecular biomarkers may be detected at an early stage of COPD, and thus have a better warning effect.
In the past decades, researchers have found that COPD has different phenotypes, such as eosinophilic phenotype, overlapping phenotype of COPD asthma, and acute exacerbation. Some important biomarkers can identify the characteristics and severity of the disease, and monitor the therapeutic effect.37 In the present study, we also tried to find potential biomarkers for discrimination between AECOPD and Stable COPD. The metabolic and lipidomic profiles between these two stages showed less difference than COPD vs healthy control, and the ROCs constructed also had less discriminative performance (Figure 5A and D). With the increased number of selected features, the AUC increases from 0.8 to 0.9. But considering the increased feature number may result in the complexity of analytical assay methods, so we chose 15 metabolites or lipids as targets of the biomarker panel. Even though total separation cannot be achieved using these 15 biomarker panels (Figure 5B and E), most samples can still be assigned correctly to AECOPD and Stable COPD groups. These two biomarker panels could be considered for the diagnosis of COPD disease stages. Further, the biomarkers we found for COPD vs Healthy and AECOPD vs Stable COPD merely overlapped except private acid, hinting that COPD onset and development may involve different metabolic and pathological procedures.
There were some limitations to our study that need to be addressed. First, the metabolite and lipid expressive levels in plasma are a comprehensive reflection of body metabolism, and considering the fact that cellular metabolism is highly flexible, and varies with tissue of origin, environment factors, and diets, we cannot directly assess the exact metabolic changes in lung tissue from results of plasma analysis. Our study aims to provide clues for changed metabolism relevant to COPD disease onset and progression, and the detailed molecular regulative mechanism in lung tissue needs to be further validated using more biological experiments before a final conclusion can be made. Second, the expressive levels of metabolites and lipids provided in our study are determined by relative quantification; for biomarkers used for clinical practice, absolute quantification is usually needed. Thus, further validation studies with absolute quantitative analysis of these candidate biomarkers (or biomarker panels) in larger COPD patient cohorts need to be done before these candidate biomarkers can finally be applied in clinical practice.
In summary, this study provides a comprehensive view of dysregulated plasma metabolites and lipids in AECOPD and Stable COPD in a Chinese population. We found that expression levels of a variety of metabolites and lipids are different between COPD disease stages or healthy people. Our study provides clues for further COPD pathological studies and several potential biomarker panels for diagnosis of COPD disease stages.
Acknowledgments
This study is supported by the National Key Research and Development Program of China (2016YFA0500302), the Interdisciplinary Medicine Seed Fund of Peking University (Grant No. BMU2017MX006), the National Natural Science Foundation of China (Grant No. 81900641), the Medjaden Academy and Research Foundation for Young Scientists (Grant No. MJR20180030), and the Lam Chung Nin Foundation for Systems Biomedicine.
Abbreviations
COPD, chronic obstructive pulmonary disease; AECOPD, acute exacerbation stage of COPD; stable COPD/SSCOPD, stable stage of COPD; LC-MS, liquid chromatography–mass spectrometry; MS/MS, tandem mass spectrometry; QC, quality control; CE, cholesteryl ester; Cer, ceramide; FFA, free fatty acid; LPC, lysophosphatidylcholine; LPE, lysophosphatidylethanolamine; PC, phosphatidylcholine; PE, phosphatidylethanolamine; p-PC, ether-linked phosphatidylcholine; p-PE, ether-linked phosphatidylethanolamine; PI, phosphatidylinositol; SM, sphingomyelin; DAG, diacylglycerol; TAG, triacylglycerol; ACar, acylcarnitine; OxPC, oxidized phosphatidylcholine; OxFA, oxidized fatty acid.
Author Contributions
J.Z., Q.L. and Y.Y. designed the research. Q.L. collected plasma samples. J.Z. and C.L. performed sample preparations and LC-MS analysis. R.P. analyzed the data. All authors made substantial contributions to conception and design, acquisition of data, or analysis and interpretation of data; took part in drafting the article or revising it critically for important intellectual content; gave final approval of the version to be published; and agree to be accountable for all aspects of the work.
Disclosure
The authors declare no competing financial interests in this work.
References
- 1.Buist AS, McBurnie MA, Vollmer WM, et al. International variation in the prevalence of COPD (the BOLD Study): a population-based prevalence study. Lancet. 2007;370:741–750. doi: 10.1016/s0140-6736(07)61377-4 [DOI] [PubMed] [Google Scholar]
- 2.Kessler R, Partridge MR, Miravitlles M, et al. Symptom variability in patients with severe COPD: a pan-European cross-sectional study. Eur Respir J. 2011;37:264–272. doi: 10.1183/09031936.00051110 [DOI] [PubMed] [Google Scholar]
- 3.Jackson H, Hubbard R. Detecting chronic obstructive pulmonary disease using peak flow rate: cross sectional survey. BMJ. 2003;327:653–654. doi: 10.1136/bmj.327.7416.653 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Kovacs G, Agusti A, Barberà JA, et al. Pulmonary vascular involvement in chronic obstructive pulmonary disease is there a pulmonary vascular phenotype? Am J Respir Crit Care Med. 2018;198:1000–1011. doi: 10.1164/rccm.201801-0095PP [DOI] [PubMed] [Google Scholar]
- 5.Vogelmeier CF, Criner GJ, Martínez FJ, et al. Global strategy for the diagnosis, management, and prevention of chronic obstructive lung disease 2017 report: GOLD executive summary. Arch Bronconeumol. 2017;53:128–149. doi: 10.1016/j.arbres.2017.02.001 [DOI] [PubMed] [Google Scholar]
- 6.Dunn WB, Bailey NJ, Johnson HE. Measuring the metabolome: current analytical technologies. Analyst. 2005;130:606–625. doi: 10.1039/b418288j [DOI] [PubMed] [Google Scholar]
- 7.de Laurentiis G, Paris D, Melck D, et al. Separating smoking-related diseases using NMR-based metabolomics of exhaled breath condensate. J Proteome Res. 2013;12:1502–1511. doi: 10.1021/pr301171p [DOI] [PubMed] [Google Scholar]
- 8.Rodriguez-Perez R, Cortés R, Guamán A, et al. Instrumental drift removal in GC-MS data for breath analysis: the short-term and long-term temporal validation of putative biomarkers for COPD. J Breath Res. 2018;12:036007. doi: 10.1088/1752-7163/aaa492 [DOI] [PubMed] [Google Scholar]
- 9.Fiehn O. Metabolomics–the link between genotypes and phenotypes. Plant Mol Biol. 2002;48:155–171. doi: 10.1023/A:1013713905833 [DOI] [PubMed] [Google Scholar]
- 10.Zhou J, Yin Y. Strategies for large-scale targeted metabolomics quantification by liquid chromatography-mass spectrometry. Analyst. 2016;141:6362–6373. doi: 10.1039/c6an01753c [DOI] [PubMed] [Google Scholar]
- 11.Kim YM, Heyman HM. Mass spectrometry-based metabolomics. Methods Mol Biol. 2018;1775:107–118. doi: 10.1007/978-1-4939-7804-5_10 [DOI] [PubMed] [Google Scholar]
- 12.Dunn WB, Ellis DI. Metabolomics: current analytical platforms and methodologies. TrAC Trend Anal Chem. 2005;24:285–294. doi: 10.1016/j.trac.2004.11.021 [DOI] [Google Scholar]
- 13.Ortmayr K, Dubuis S, Zampieri M. Metabolic profiling of cancer cells reveals genome-wide crosstalk between transcriptional regulators and metabolism. Nat Commun. 2019;10:1841. doi: 10.1038/s41467-019-09695-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Luo P, Yin P, Hua R, et al. A Large-scale, multicenter serum metabolite biomarker identification study for the early detection of hepatocellular carcinoma. Hepatology. 2018;67:662–675. doi: 10.1002/hep.29561 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Calzada E, Avery E, Sam PN, et al. Phosphatidylethanolamine made in the inner mitochondrial membrane is essential for yeast cytochrome bc1 complex function. Nat Commun. 2019;10:1432. doi: 10.1038/s41467-019-09425-1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Maniscalco M, Paris D, Melck DJ, et al. Differential diagnosis between newly diagnosed asthma and COPD using exhaled breath condensate metabolomics: a pilot study. Eur Respir J. 2018;51:1701825. doi: 10.1183/13993003.01825-2017 [DOI] [PubMed] [Google Scholar]
- 17.Ran N, Pang Z, Gu Y, et al. An updated overview of metabolomic profile changes in chronic obstructive pulmonary disease. Metabolites. 2019;9:111. doi: 10.3390/metabo9060111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Engelen MP, De Castro CLN, Rutten EPA, et al. Enhanced anabolic response to milk protein sip feeding in elderly subjects with COPD is associated with a reduced splanchnic extraction of multiple amino acids. Clin Nutr. 2012;31:616–624. doi: 10.1016/j.clnu.2012.04.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Yoneda T, Yoshikawa M, Fu A, et al. Plasma levels of amino acids and hypermetabolism in patients with chronic obstructive pulmonary disease. Nutrition. 2001;17:95–99. doi: 10.1016/s0899-9007(00)00509-8 [DOI] [PubMed] [Google Scholar]
- 20.Vahid I, Abdolali B, Fatemeh M, Alireza N, Mehdi S. The effects of branch-chain amino acids on fatigue in the athletes. Interv Med Appl Sci. 2018;10:233–235. doi: 10.1556/1646.10.2018.10 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Rahman I. The role of oxidative stress in the pathogenesis of COPD. Treat Respir Med. 2005;4:175–200. doi: 10.2165/00151829-200504030-00003 [DOI] [PubMed] [Google Scholar]
- 22.Ji Y, Wu Z, Dai Z, et al. Nutritional epigenetics with a focus on amino acids: implications for the development and treatment of metabolic syndrome. J Nutr Biochem. 2016;27:1–8. doi: 10.1016/j.jnutbio.2015.08.003 [DOI] [PubMed] [Google Scholar]
- 23.Sugawara K, Takahashi H, Kasai C, et al. Effects of nutritional supplementation combined with low-intensity exercise in malnourished patients with COPD. Respir Med. 2010;104:1883–1889. doi: 10.1016/j.rmed.2010.05.008 [DOI] [PubMed] [Google Scholar]
- 24.Ishikawa S, Matsumura K, Kitamura N, Takanami Y, Ito S. Multi-omics analysis: repeated exposure of a 3D bronchial tissue culture to whole-cigarette smoke. Toxicol in Vitro. 2019;54:251–262. doi: 10.1016/j.tiv.2018.10.001 [DOI] [PubMed] [Google Scholar]
- 25.Ren X, Zhang J, Fu X, et al. LC-MS based metabolomics identification of novel biomarkers of tobacco smoke-induced chronic bronchitis. Biomed Chromatogr. 2016;30:68–74. doi: 10.1002/bmc.3620 [DOI] [PubMed] [Google Scholar]
- 26.Navarrete A, Rupérez FJ, Mendes TO, et al. A metabolomic approach shows sphingosine 1-phosphate and lysophospholipids as mediators of the therapeutic effect of liver growth factor in emphysema. J Pharm Biomed Anal. 2017;139:238–246. doi: 10.1016/j.jpba.2017.02.045 [DOI] [PubMed] [Google Scholar]
- 27.Xia J, Sinelnikov IV, Han B, Wishart DS. MetaboAnalyst 3.0–making metabolomics more meaningful. Nucleic Acids Res. 2015;43:W251–W257. doi: 10.1093/nar/gkv380 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Wang C, Xu J, Yang L, et al. Prevalence and risk factors of chronic obstructive pulmonary disease in China (the China Pulmonary Health [CPH] study): a national cross-sectional study. Lancet. 2018;391:1706–1717. doi: 10.1016/s0140-6736(18)30841-9 [DOI] [PubMed] [Google Scholar]
- 29.Wright CE, Tallan HH, Lin YY, Gaull GE. Taurine: biological update. Annu Rev Biochem. 1986;55:427–453. doi: 10.1146/annurev.bi.55.070186.002235 [DOI] [PubMed] [Google Scholar]
- 30.Huxtable RJ. Physiological actions of taurine. Physiol Rev. 1992;72:101–163. doi: 10.1152/physrev.1992.72.1.101 [DOI] [PubMed] [Google Scholar]
- 31.Ubhi BK, Riley JH, Shaw PA, et al. Metabolic profiling detects biomarkers of protein degradation in COPD patients. Eur Respir J. 2012;40:345–355. doi: 10.1183/09031936.00112411 [DOI] [PubMed] [Google Scholar]
- 32.Xu W, KANEKO FT, ZHENG S, et al. Increased arginase II and decreased NO synthesis in endothelial cells of patients with pulmonary arterial hypertension. FASEB J. 2004;18:1746–1748. doi: 10.1096/fj.04-2317fje [DOI] [PubMed] [Google Scholar]
- 33.Kilk K, Aug A, Ottas A, et al. Phenotyping of chronic obstructive pulmonary disease based on the integration of metabolomes and clinical characteristics. Int J Mol Sci. 2018;19:666. doi: 10.3390/ijms19030666 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.De Benedetto F, Pastorelli R, Ferrario M, et al. Supplementation with Qter((R)) and creatine improves functional performance in COPD patients on long term oxygen therapy. Respir Med. 2018;142:86–93. doi: 10.1016/j.rmed.2018.08.002 [DOI] [PubMed] [Google Scholar]
- 35.Schols AMWJ, Soeters PB, Dingemans AMC, et al. Prevalence and characteristics of nutritional depletion in patients with stable COPD eligible for pulmonary rehabilitation. Am Rev Respir Dis. 1993;147:1151–1156. doi: 10.1164/ajrccm/147.5.1151 [DOI] [PubMed] [Google Scholar]
- 36.Peruzza S, Sergi G, Vianello A, et al. Chronic obstructive pulmonary disease (COPD) in elderly subjects: impact on functional status and quality of life. Respir Med. 2003;97:612–617. doi: 10.1053/rmed.2003.1488 [DOI] [PubMed] [Google Scholar]
- 37.Chalmers JD, Aliberti S, Filonenko A, et al. Characterization of the “frequent exacerbator phenotype” in bronchiectasis. Am J Respir Crit Care Med. 2018;197:1410–1420. doi: 10.1164/rccm.201711-2202OC [DOI] [PubMed] [Google Scholar]