Abstract
This study was performed to investigate the changes in gene expression in periodontal ligament (PDL) cells following mechanical stimulus through RNA sequencing. In this study, premolars extracted for orthodontic treatment were used. To stimulate the PDL cells, an orthodontic force of 100× g was applied to the premolar (experimental group; n = 11), whereas the tooth on the other side was left untreated (control group; n = 11). After the PDL cells were isolated from the extracted teeth, gene set enrichment analysis (GSEA), differentially expressed gene (DEG) analysis, and real-time PCR were performed to compare the two groups. GSEA demonstrated that gene sets related to the cell cycle pathway were upregulated in PDL. Thirteen upregulated and twenty downregulated genes were found through DEG analysis. Real-time PCR results confirmed that five upregulated genes (CC2D1B, CPNE3, OPHN1, TANGO2, and UAP-1) and six downregulated genes (MYOM2, PPM1F, PCDP1, ATP2A1, GPR171, and RP1-34H18.1-1) were consistent with RNA sequencing results. We suggest that, from among these eleven genes, two upregulated genes, CPNE3 and OPHN1, and one downregulated gene, PPM1F, play an important role in PDL regeneration in humans when orthodontic force is applied.
Keywords: RNA-sequencing, orthodontic force, periodontal ligament, mechanical stimulus, prospective study
1. Introduction
Periodontal ligament (PDL), which is a group of connective tissue fibers, connects the tooth root to the adjacent alveolar bone. It protects blood vessels and nerves by absorbing mechanical force such as mastication force and provides proprioception. Moreover, it mediates orthodontic tooth movement under compressional or tensional force and plays a critical role in recovery from periodontal disease [1]. Mesenchymal stem cells were isolated from PDL [2] and have shown potential for periodontal regeneration [3]. PDL regeneration can also be enhanced under mechanical stimulus [4,5,6]. Low-magnitude, high frequency mechanical vibration promotes human PDL stem cells differentiation [7], and mechanical shear stress promotes the osteogenic differentiation of dental stem cells, including those derived from the pulp and the PDL [8]. An in vivo study using rats showed that occlusal stimulus promotes regeneration of PDL and prevents dentoalveolar ankylosis after autotransplantation [9].
Changes in PDL cells under mechanical stress have been reported to be critical in maintaining homeostasis of the periodontal tissue and enabling its remodeling [10,11,12,13]. Cyclic stretch applied to the PDL influences membrane protein expression [10] and upregulates and downregulates genes related to the extracellular matrix in cultured human PDL cells [11]. Orthodontic force activates the CC chemokine receptor 5 (CCR5)-CCR5 ligands axis in rats, which was suggested to be a key factor in PDL remodeling. Moreover, PDL induces favorable circumstances for remodeling by releasing cytokines and growth factors under orthodontic force in animal models [12,13]. However, there have been few in vivo studies investigating the relation between mechanical stimulus and PDL regeneration in humans. Although PDL exhibits different length, volume, mechanoreceptors, and remodeling capacity according to occlusal contact [14], it would be meaningful to investigate the transcriptional changes in PDL under mechanical stimulus.
Early application of the mechanical force can increase the success rate of autotransplantation by preventing ankylosis and external root resorption, which is based on PDL regeneration [15]. Under mechanical stress, expression of cell cycle regulatory proteins, such as proliferating cell nuclear antigen, cyclin-dependent kinases, and cyclin D1, are increased in human PDL cells [16], although transcriptional changes are barely unknown in humans. RNA sequencing (RNA-seq) is a useful technique for analyzing dynamic transcriptomes rather than static genomes [17] and discovering novel transcripts from a wide range of transcriptomes as well as providing quantitative analysis of gene expression [18].
In the present study, we simulated mechanical stimulus to the PDL by applying orthodontic force. If mechanical stimulus induces changes in PDL gene expression in humans, RNA-seq can show these changes in transcriptional expression patterns, which would prove valuable in understanding the changes occurring in PDL following mechanical stimulus such as mastication force, orthodontic force, or trauma to the teeth, and determining potential target genes. Therefore, the aims of this study were to investigate changes in gene expression in the PDL after mechanical stimulus and to search for novel genes through RNA sequencing.
2. Materials and Methods
2.1. Patient Selection
Subjects free from underlying disease were selected from orthodontic patients who had visited Yonsei University Dental Hospital between June 2017 and May 2018 for orthodontic extraction of at least two premolars. The inclusion criteria were as follows: a sound premolar in one quadrant without caries or restorations, and no history of taking any steroidal or non-steroidal anti-inflammatory drugs during force application. The exclusion criteria were root malformation such as dilaceration, which might result in difficult extraction. The minimum sample size of 10 subjects was estimated to overcome the individual variation in gene expression in PDL cells on the basis of a previous study using RNA-seq to analyze differences in gene expression between periodontitis-affected and healthy sides [19]. The study protocol was approved by the institutional review board of Yonsei University Dental Hospital (IRB No. 2-2017-0028) and performed after obtaining written informed consent from every participant.
Based on the inclusion/exclusion criteria, 14 patients were initially enrolled. Samples from three patients were excluded at initial quality check, and the samples from 11 subjects (5 men and 6 women; mean age, 22.4 years; age range, 17.5–31.0 years) were used in this study (Table A1). Two premolars were extracted in seven subjects; three premolars (two premolars in the experimental group and one premolar in the control group) were extracted in one subject (subject #4); and four premolars (two premolars in the experimental group and two premolars in the control group) were extracted in three subjects (subjects #7, #8, and #9). In cases of 3- or 4-premolar extraction, the two teeth on the same side were assigned to the same group.
2.2. Mechanical Stimulus (Applying Orthodontic Force)
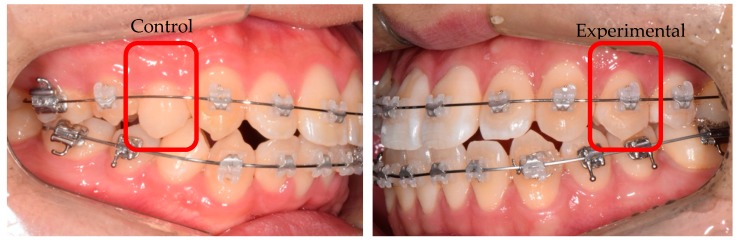
To stimulate the PDL, orthodontic force was applied to the first or second premolar on one side (the experimental group; n = 11), whereas the tooth on the other side was left untreated (the control group; n = 11). The study was a split-mouth, randomized, controlled trial with a 1:1 allocation ratio. A 0.016-inch nickel–titanium wire (Tomy International, Yokohama, Japan), which is known to deliver light continuous force (100× g) with a deflection of 0.5–1.8 mm, was used to apply mechanical stimulus in the experimental group (Figure 1). The orthodontic force was applied for three weeks [20], and thereafter, the teeth in both groups were carefully extracted. PDL cells were washed with cold PBS after gentle scrapping from the middle third of the root and prepared for RNA-seq.
Figure 1.
Split-mouth design: control group (left, no orthodontic force) and experimental group (right, orthodontic force applied).
2.3. RNA Sequencing
To evaluate changes in gene expression after mechanical stimulus, total RNA was extracted from PDL cells using TRIzol Reagent (Invitrogen, Waltham, MA, USA). The RNA was stored at −70 °C and measured at an optical density of 260 nm. The mixtures of total RNA were incubated with Oligo dT (Gibco BRL, Rockville, NY, USA).
The library was constructed, sequenced using an Illumina HiSeq2500 sequencer (Illumina, CA, USA), and the data obtained through RNA-seq. A gene set underlying mechanical stimulus was analyzed using gene set enrichment analysis (GSEA). Differentially expressed gene (DEG) analysis was also performed to determine differences in gene expression between the experimental and control groups. For GSEA, libraries from 11 subjects in each experimental or control group were merged into one library for each group, and then, the experimental and control groups in total, were compared. Based on the GSEA results, an enrichment map was obtained. DEG analyses were performed to compare the libraries between the experimental and control groups in each sample as well as in the merged samples. Based on the DEG results, we selected target genes based on gene expression fold-changes: more than 1.5 fold-change in gene expression in the experimental group compared to the control group was defined as upregulated, and less than −1.5 fold-change was defined as downregulated [21,22,23].
2.4. Real-Time PCR
Real-time PCR was performed for the selected upregulated and downregulated genes to verify RNA-seq results. A cDNA synthesis reaction was performed using a mixture of AccuPower PCR PreMix (Bioneer, Daejeon, Korea) and water. The cDNA was synthesized from total RNA obtained from both groups using the SuperScript First-Strand Synthesis System (Invitrogen, Carlsbad, CA, USA) and was amplified with an ABI-7300 (Applied Biosystems, Mortlake, Waltham, Massachusetts, USA). The amplified cDNA was detected with SYBR Green PCR Master Mix Reagent Kit (Takara, Seoul, Korea). PCR conditions were as follows: incubation for 10 min at 95 °C, followed by 40 cycles of 10 s denaturation at 95 °C, and annealing for 60 s at 60 °C. The reaction mixture lacking cDNA was used as a negative control in each run. Primer sequences are summarized in Table 1. Ratios of the intensities of the target genes and GAPDH signals were used as a relative measure of the expression level of the target genes. To ensure accuracy of the experiments, primer specificity was confirmed by the dissociation curve after PCR, and real-time PCR assays were performed in triplicate for each sample. The mean fold-change in expression in the experimental group compared with the control group was calculated from the △△Ct values, and the range of the fold-changes was represented by standard deviations of the values [21].
Table 1.
Primer sequences used for real-time quantitative PCR.
| Genes | Transcript ID | Forward | Reverse |
|---|---|---|---|
| CC2D1B | NM_032449.2 | GAGTCGCAGCTAGCCTCTGT | TCTGTCTCAGGGCTCCTGTT |
| TMEM253 | NM_001146683.1 | CTTGCTGAGCCAGAGGAAAC | CAAACCAGGAACCTCTTCCA |
| TENM4 | NM_001098816.2 | CCGTCTTCCTTTCTGACAGC | ATCAGCCCAAACTTGTCCAC |
| CPNE3 | NM_003909.5 | TCGACCACTGGTGATGAAAA | CCGATGAACCATTAGCCAGT |
| MYOM2 | NM_003970.4 | CGGTGAATACAAGGCAACCT | TCACATATCTGCAGCCAAGC |
| PPM1F | NM_014634.4 | GTACAGCAGGGACAGGTGGT | ACAGGCAAGCAGCAGGTAGT |
| PCDP1 | NM_001271049.2 | TCAACAAGTAGCACGCAAGG | ATCCGCCTCAGGAAGAATTT |
| ATP2A1 | NM_173201.3 | TGGCTCTTCTTCCGCTACAT | GCCTCGAAGACCTCACAGTC |
| GPR171 | NM_013308.3 | CAACCGTTGTGTGGCTAATG | TATGATGTAGCCCGTGGTCA |
| OPHN1 | NM_002547.3 | GTCCCCAAGCAGGCCTAT | GTCCATTGGTGGCCTTTG |
| TANGO2 | NM_152906.6 | TCCCTGGAGGAAGCTGTG | GCTGCGCCTCTTCATTGT |
| UAP1 | NM_001324116.1 | TCCAAAGCTGGGCAAGAG | GGTTCCATTCGTGCATCC |
| RP1-34H18.1 | ENST00000550042.1 | GCGGAGGAGGGAAGAAAG | AAAACCAACCGAGGCACA |
| GAPDH | TCCGCGGCTATATGAAAACAG | TCGTAGTGGGCTTGCTG AA |
2.5. Statistical Analysis
For differential expression analysis, gene level count data were generated using HTSeq-count v0.5.4p3 tool [22] with the option “-m intersection-nonempty” and -r option considering paired-end sequence. Based on the calculated read count data, DEGs were identified using the R package called TCC [23]. TCC package applies robust normalization strategies to compare tag count data. Normalization factors were calculated using the iterative DEGES/edgeR method. Q-value was calculated based on the p-value using the p.adjust function of R package with default parameter settings. Differentially expressed genes were identified based on the q value threshold less than 0.05.
3. Results
3.1. Extracted Premolar Samples
Two premolars were extracted in seven subjects; three premolars (two premolars in the experimental group and one premolar in the control group) were extracted in one subject (subject #4); and four premolars (two premolars in the experimental group and two premolars in the control group) were extracted in three subjects (subjects #7, #8, and #9). In cases of 3- or 4-premolar extraction, the two teeth on the same side were assigned to the same group.
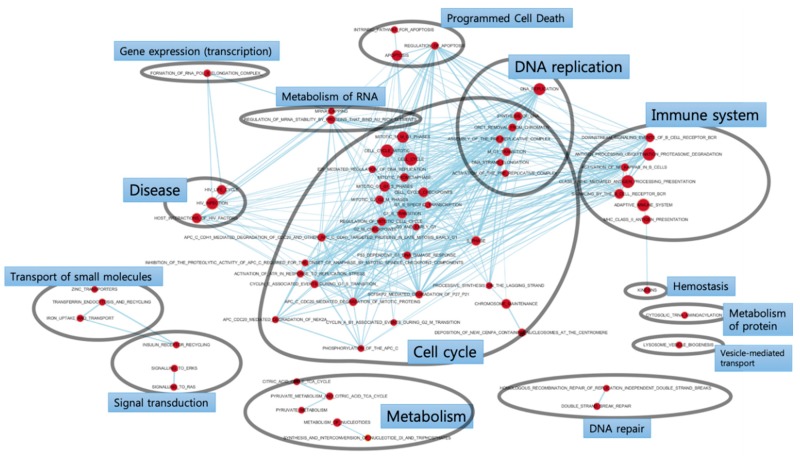
3.2. Gene Set Enrichment Analysis of Mechanically Stimulated PDL Cells
GSEA enrichment maps show enriched gene sets, most of which are related to the cell cycle pathway (Figure 2). Additionally, pathways for DNA replication, immune system, and metabolism were represented among upregulated genes. The node size, which indicates the number of enriched genes, was large in the following gene sets: cell cycle, cell cycle mitotic, DNA replication, antigen processing ubiquitination proteasome degradation, and class I MHC-mediated antigen processing presentation.
Figure 2.
Gene set enrichment analysis (GSEA) enrichment map. The enriched gene sets in the periodontal ligament underlying mechanical stimulus are mostly related to the cell cycle pathway.
3.3. DEG Analysis of Mechanically Stimulated PDL Cells
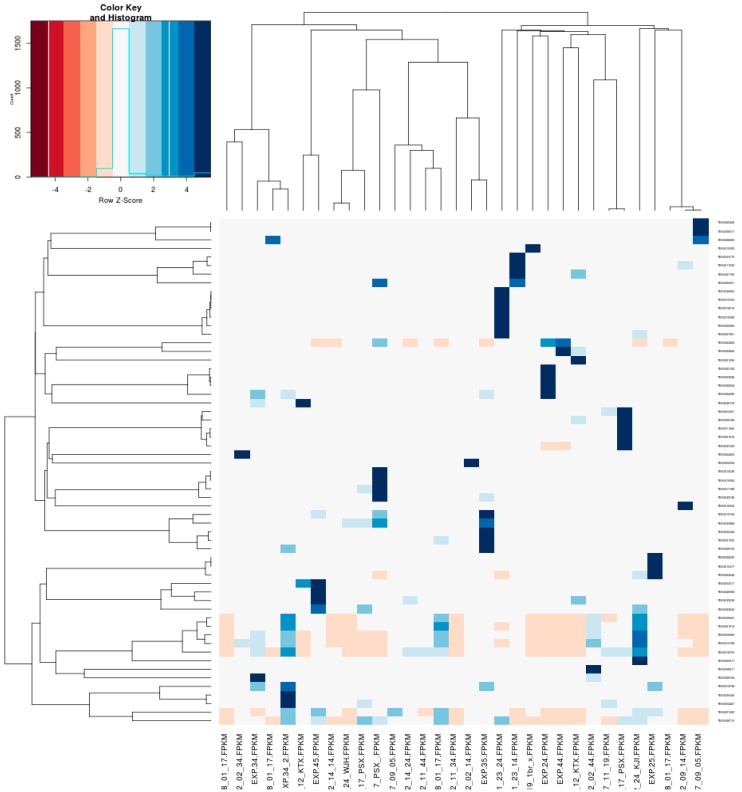
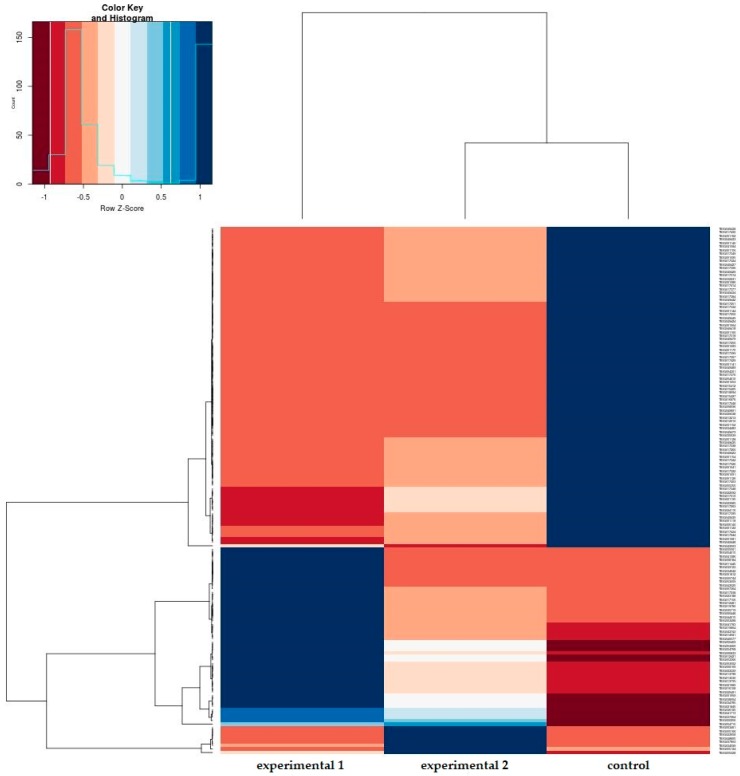
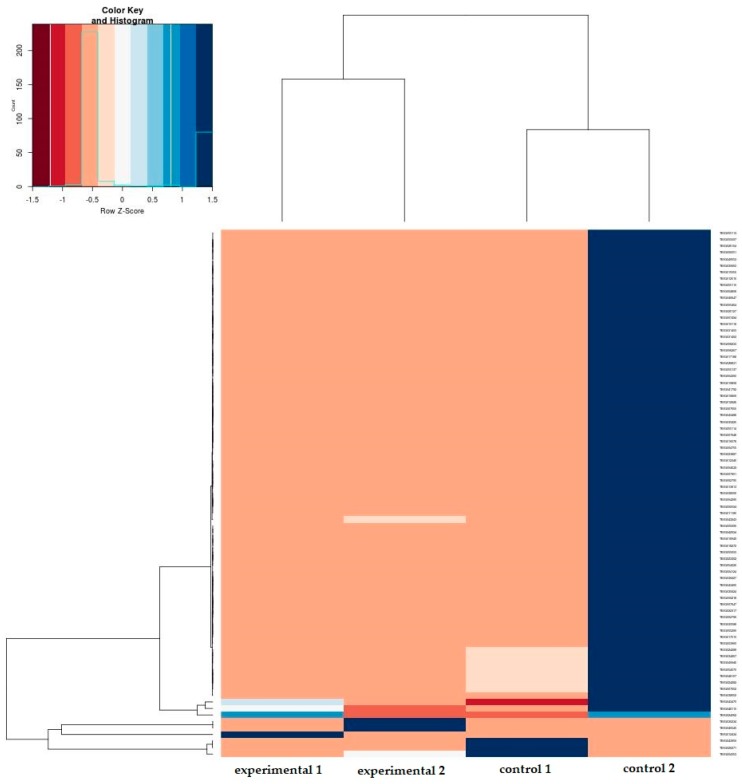
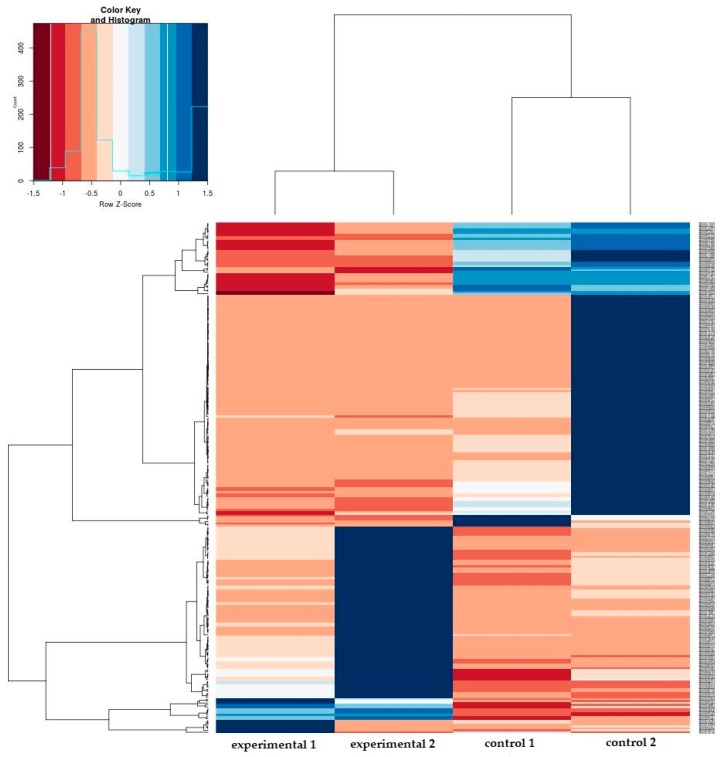
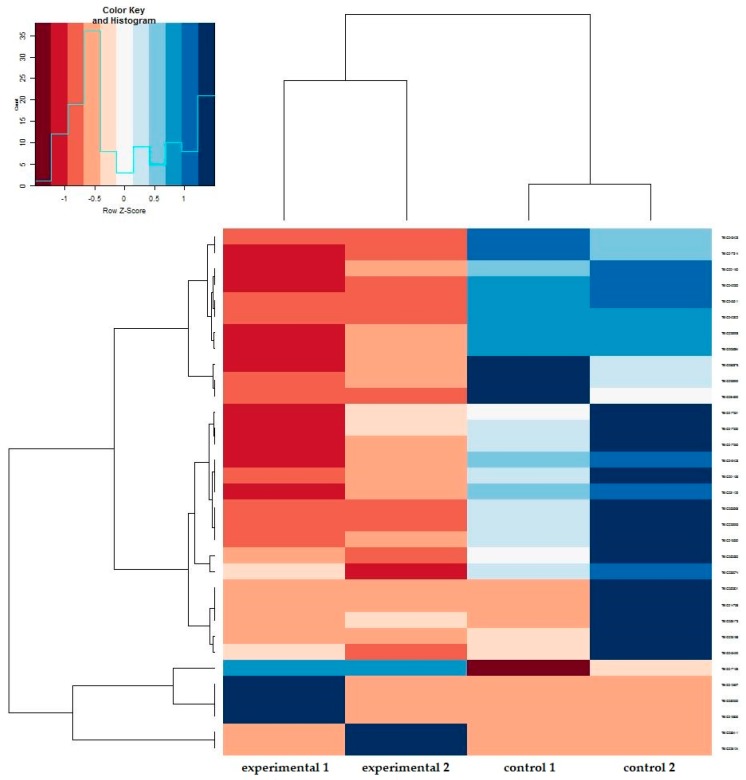
A heatmap was obtained by comparing the merged libraries of 11 subjects between the experimental and control groups (Figure 3). The 11 heatmaps obtained from each subject are presented in the Appendix Materials (Figure A1, Figure A2, Figure A3, Figure A4, Figure A5, Figure A6, Figure A7, Figure A8, Figure A9, Figure A10 and Figure A11). Fifty-nine genes were selected by q value (q < 0.05), then 13 up-regulated and 20 down-regulated genes were finally selected based on a fold-change of 1.5 (Table 2 and Table 3).
Figure 3.
Heatmap representing differential expression of the periodontal ligament underlying mechanical stimulus. This heatmap demonstrates the Fragments Per Kilobase Million (FPKM) value of the most significant differentially expressed genes. FPKM value indicates the relative expression of a transcript which is proportional to the number of cDNA fragments in RNA sequencing. A detailed heatmap for each subject is shown in the appendix.
Table 2.
RNA sequencing results: 13 upregulated genes after mechanical stimulus in the periodontal ligament (fold-change ≥ 1.5).
| Gene Name | Description | Control | Experimental | log2fc | p Value | q Value |
|---|---|---|---|---|---|---|
| OPHN1 | oligophrenin1 | 5.31 | 781 | 7.2 | 5.00E-05 | 0.0149 |
| TANGO2 | transport and golgi organization 2 homolog (Drosophila) | 6.66 | 86.1 | 3.69 | 5.00E-05 | 0.0149 |
| CC2D1B | coiled-coil and C2 domaincontaining 1B | 12 | 150 | 3.58 | 5.00E-05 | 0.0149 |
| UAP1 | UDP-N-acteylglucosaminepyrophosphorylase1 | 43 | 468 | 3.45 | 5.00E-05 | 0.0149 |
| TMEM253 | transmembrane protein 253 | 0.836 | 7.66 | 3.2 | 5.00E-05 | 0.0149 |
| TENM4 | teneurin transmembrane protein 4 | 13.5 | 93.6 | 2.8 | 5.00E-05 | 0.014911 |
| ABHD4 | abhydrolase domain containing 4 | 20.9 | 137 | 2.71 | 5.00E-05 | 0.0149 |
| CPNE3 | copine III | 23.7 | 134 | 2.5 | 5.00E-05 | 0.0149 |
| RP11-820L6.1 | - | 1.76 | 9.19 | 2.38 | 5.00E-05 | 0.0149 |
| SPATA22 | spermatogenesis associated 22 | 0.379 | 1.89 | 2.31 | 5.00E-05 | 0.0149 |
| SCUBE1 | signalpeptide, CUB domain, EGF-like1 | 0.217 | 0.88 | 2.02 | 0.0001 | 0.0277 |
| ARMC8 | armadillo repeat containing 8 | 13.5 | 54 | 2 | 5.00E-05 | 0.0149 |
| ERI2 | ERI1 exoribonuclease family member 2 | 6.69 | 26.5 | 1.99 | 5.00E-05 | 0.0149 |
Table 3.
RNA sequencing results: 20 downregulated genes after mechanical stimulus in the periodontal ligament (fold-change ≤ −1.5).
| Gene Name | Description | Control | Experimental | log2fc | p Value | q Value |
|---|---|---|---|---|---|---|
| ITCH | itchy E3 ubiquitin protein ligase | 41.6 | 14.4 | −1.53 | 5.00E-05 | 0.0149 |
| ARL14EP | ADP-ribosylation factor-like 14 effector protein | 71.8 | 22.8 | −1.65 | 5.00E-05 | 0.0149 |
| RFX1 | regulatory factor X, 1 (influences HLA class II expression) | 17.1 | 5.47 | −1.65 | 5.00E-05 | 0.0149 |
| PLEKHH1 | pleckstrin homology domain containing, family H (with MyTH4 domain) member1 | 6.91 | 2.18 | −1.66 | 5.00E-05 | 0.0149 |
| EHBP1L1 | EH domain binding protein1-like 1 | 162 | 46 | −1.81 | 5.00E-05 | 0.0149 |
| PIGQ | phosphatidylinositol glycan anchor biosynthesis, class Q | 286 | 81.7 | −1.81 | 5.00E-05 | 0.0149 |
| ASPG | asparaginase homolog (S.cerevisiae) | 56.8 | 15.2 | −1.9 | 5.00E-05 | 0.0149 |
| BOD1L1 | biorientation of chromosomes in cell division 1-like 1 | 92.5 | 22.5 | −2.04 | 5.00E-05 | 0.0149 |
| PTPN12 | protein tyrosine phosphatase, non-receptortype12 | 268 | 55.9 | −2.26 | 5.00E-05 | 0.0149 |
| USH1C | Usher syndrome 1C(autosomal recessive,severe) | 4.9 | 0.927 | −2.4 | 5.00E-05 | 0.0149 |
| MYOM2 | myomesin 2 | 18.2 | 3.03 | −2.58 | 5.00E-05 | 0.0149 |
| PRR11 | proline rich11 | 16.7 | 2.72 | −2.62 | 5.00E-05 | 0.0149 |
| CCDC91 | coiled-coil domain containing 91 | 166 | 26.8 | −2.63 | 5.00E-05 | 0.0149 |
| PPM1F | protein phosphatase,Mg2+/Mn2+ dependent, 1F | 165 | 24.5 | −2.75 | 5.00E-05 | 0.0149 |
| PCDP1 | Homo sapiens primary ciliary dyskinesia protein1 (PCDP1),transcript variant1, mRNA. | 2.35 | 0.254 | −3.21 | 5.00E-05 | 0.0149 |
| TSPAN8 | tetraspanin 8 | 34.8 | 3.1 | −3.49 | 5.00E-05 | 0.0149 |
| RCCD1 | RCC1 domain containing1 | 114 | 7.76 | −3.87 | 5.00E-05 | 0.0149 |
| ATP2A1 | ATPase, Ca++ transporting,cardiac muscle,fast twitch 1 | 13.7 | 0.916 | −3.91 | 5.00E-05 | 0.0149 |
| GPR171 | G protein-coupled receptor 171 | 49 | 0.782 | −5.97 | 5.00E-05 | 0.0149 |
| RP1-34H18.1 | - | 4.14E+03 | 0.774 | −12.4 | 5.00E-05 | 0.0149 |
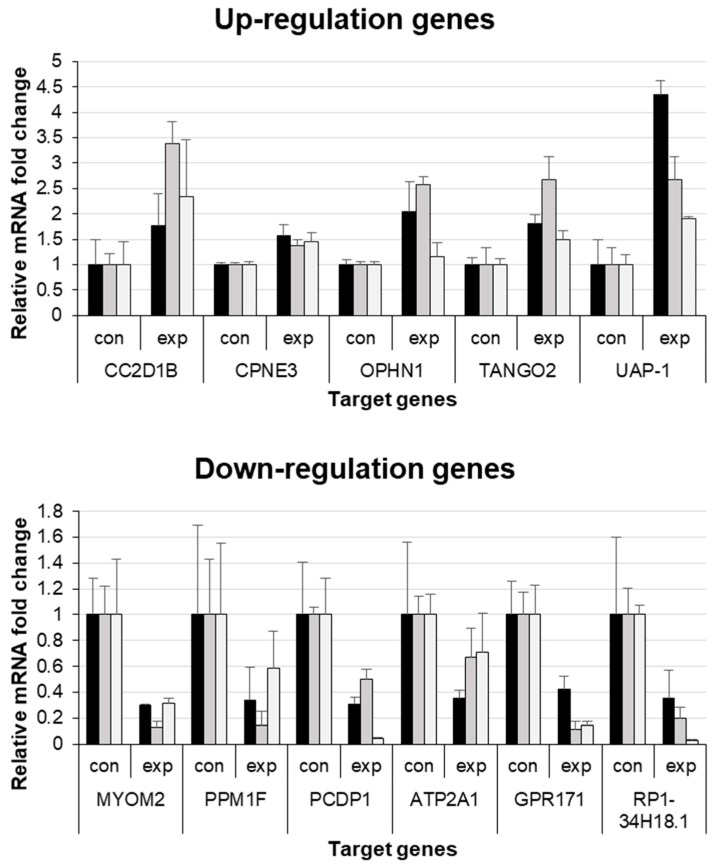
3.4. Validation of DEGs in Mechanically Stimulated PDL Cells
Among the 13 upregulated and 20 downregulated genes, we performed gene searches in the human gene database (www.genecard.org) and selected 5 upregulated (CC2D1B, CPNE3, OPHN1, TANGO2, and UAP-1) and 6 downregulated (MYOM2, PPM1F, PCDP1, ATP2A1, GPR171, and RP1-34H18.1-1) genes related to the cell cycle. Real-time PCR results for the 5 upregulated and 6 downregulated genes confirmed the corresponding RNA-seq results (Figure 4).
Figure 4.
Real-time quantitative PCR results of the 5 upregulated and 6 downregulated genes from RNA sequencing. Con, control group; exp, experimental group.
4. Discussion
This prospective study investigated the transcriptional expression in human PDL cells stimulated with an orthodontic force with RNA-seq. Because we intended to identify gene transcripts potentially related to the remodeling of PDL, this study focused on the differential expression of PDL genes using GSEA, DEG analysis, and real-time PCR. From these results, we found 11 significant DEGs when orthodontic force was applied on the PDL.
In the early phase of orthodontic tooth movement, mechanically stimulated PDL cells recruit local factors related to homeostasis and remodeling [16,24] of the surrounding periodontal tissue, which may result from the upregulated cell cycle pathways. Under tension force, PDL cells participate in DNA synthesis of osteoblasts; are differentiated to osteoblasts; or are released from G2 block, which contributes to the alveolar bone formation. Moreover, morphological deformation of the PDL induced by mechanical force plays a key role in recovery of its original shape via the remodeling process [25]. Therefore, the upregulated cell cycle of the PDLs would make orthodontic tooth movement possible by facilitating the remodeling process.
Among the 11 genes, CPNE3, OPHN1, and PPM1F are specially interesting, which were likely related to PDL remodeling based on the function and the signaling pathway of each gene. CPNE3 is a calcium-dependent membrane-binding protein that plays a major role in the synthesis of phospholipids and the immune system. CPNE3 belongs to the copine family, which can bind both calcium ions and phospholipids simultaneously. In particular, CPNE7 is involved in PDL regeneration [26]. The gene has a functional role in attachment of PDL cells to cementum and promotes the physiological arrangement of PDL fibers. Therefore, CPNE3, in the same gene family as CPNE7, may be involved in the regeneration of PDL. Further studies are needed to investigate whether and how CPNE3 affects PDL regeneration.
OPHN1 encodes the protein oligophrenin-1 that has a Rho-GAP domain shown to negatively regulate RhoA, Rac, and Cdc42 in vitro and in nonneuronal cells [27]. Rho GTPases participate in important cell biological processes, including cell growth control, cell motility, and development [28]. As expression of OPHN1 increases, the rhoA, Rac, and Cdc42 proteins, which are known to activate caspase 3 and finally activate apoptosis, decrease, which leads to reduced apoptosis of PDL cells. Therefore, upregulation of OPHN1 may indirectly contribute to the maintenance of PDL cells.
PPM1F is a member of the serine/threonine protein phosphatase family. This gene is known to be involved in caspase-dependent apoptosis [29]. When orthodontic force is applied, apoptosis in PDL cells may be inhibited by downregulation of PPM1F. It can be assumed that downregulation of PPM1F has a positive effect on PDL regeneration.
The upregulated and downregulated genes identified in the DEG analysis were confirmed by qPCR, although the qPCR ratio of each sample was different. Furthermore, there was a great variety of gene expression between subjects. It was assumed that limitations of the clinical study using human samples might cause the variety. As seen in the demographic features of the subjects (Table A1), different age, sex, tooth allocation, and occlusion of each subject would result in a wide variety of gene expressions unlikely in the animal models. In addition, the orthodontic force in the experimental group and the occlusal force in the control group would not be exactly the same in each tooth, which may result in differences in gene expression [4]. Moreover, the nonlinear properties of the PDL under different loading directions [30] would affect the outcome. The unstable nature of RNA and different elapsed times from tooth extraction to RNA extraction from the PDL may be another reason for the difference because proliferation of PDL fibers changes depending on post-extraction time [31].
The small numbers of DEG, 13 upregulated and 20 downregulated genes, were selected by comparing the merged libraries of 11 subjects between the experimental and control groups in the present study. Although the GSEA analysis demonstrated not only the cell cycle pathways but also the pathways related to metabolism, immune system, disease, and programmed cell death, the present study focused only on the genes related to the cell cycle. There are other factors that limited the number of DEG: high cutoff value, short duration of mechanical stimulation, and non-differentiation between compression and tension sides of the root surface, which might prevent the genes involved in PDL regeneration from expressing differentially. Moreover, the 33 genes could not be fully verified by qPCR in this study. Although 22 genes were discarded because they were not related to the cell cycle, some of the genes might play a role in PDL regeneration indirectly.
Mechanical stimulus contributes to PDL regeneration in vitro [4,7] and prevention of ankylosis after transplantation or replantation in in vivo animal models [9]. To the best of our knowledge, this is the first report of gene expression changes in the PDL following mechanical stimulus, in humans. We confirmed the upregulation of the cell cycle pathways after mechanical stimulus and found novel genes related to the process. Future studies are needed to identify the biological role of these genes and to obtain clinical relevancies such as saving the tooth using mechanical stimulus in trauma or transplantation cases.
5. Conclusions
When orthodontic force is applied, gene sets related to the cell cycle pathway were upregulated in PDL. The 5 upregulated (CC2D1B, CPNE3, OPHN1, TANGO2, and UAP-1) and 6 downregulated genes (MYOM2, PPM1F, PCDP1, ATP2A1, GPR171, and RP1-34H18.1-1) genes may play an important role in PDL regeneration after mechanical stimulus in humans.
Acknowledgments
The authors appreciate Dr. Sue Bean Cho for support in data preparation and illustration.
Appendix A
Figure A1.
Heatmap of subject #1. Experimental group vs. control group.
Figure A2.
Heatmap of subject #2. Experimental group vs. control group.
Figure A3.
Heatmap of subject #3. Experimental group vs. control group.
Figure A4.
Heatmap of subject #4. Experimental group vs. control group.
Figure A5.
Heatmap of subject #5. Experimental group vs. control group.
Figure A6.
Heatmap of subject #6. Experimental group vs. control group.
Figure A7.
Heatmap of subject #7. Experimental group vs. control group.
Figure A8.
Heatmap of subject #8. Experimental group vs. control group.
Figure A9.
Heatmap of subject #9. Experimental group vs. control group.
Figure A10.
Heatmap of subject #10. Experimental group vs. control group.
Figure A11.
Heatmap of subject #11. Experimental group vs. control group.
Table A1.
Demographic features of subjects.
| Subject No. | Sex | Age | Tooth allocation | |
|---|---|---|---|---|
| Experimental Group | Control Group | |||
| 1 | Female | 28 years 2 months | Mx Lt 2nd premolar | Mn Lt 2nd premolar |
| 2 | Male | 22 years 0 months | Mx Lt 1st premolar | Mn Lt 1st premolar |
| 3 | Male | 17 years 6 months | Mx Rt 1st premolar | Mx Lt 1st premolar |
| 4 | Male | 20 years 3 months | Mx Lt 2nd premolar, Mn Lt 2nd premolar | Mn Rt 1st premolar |
| 5 | Female | 20 years 9 months | Mn Lt 1st premolar | Mx Lt 1st premolar |
| 6 | Female | 25 years 7 months | Mn Lt 1st premolar | Mx Lt 1st premolar |
| 7 | Female | 31 years 0 months | Mx Lt 1st premolar, Mn Lt 1st premolar | Mx Rt 1st premolar, Mn Rt 1st premolar |
| 8 | Female | 20 years 8 months | Mx Rt 1st premolar, Mn Rt 1st premolar | Mx Lt 1st premolar, Mn Lt 1st premolar |
| 9 | Male | 18 years 4 months | Mx Lt 1st premolar, Mn Lt 1st premolar | Mx Rt 1st premolar, Mn Rt 1st premolar |
| 10 | Female | 18 years 6 months | Mx Lt 1st premolar | Mx Rt 1st premolar |
| 11 | Male | 25 years 1 months | Mx Rt 1st premolar | Mn Rt 1st premolar |
Mx, maxilla; Mn, mandible; Rt, right side; Lt, left side.
Author Contributions
Conceptualization, E.K. and Y.J.C.; Data curation, K.K., H.E.K. and Y.J.C.; Formal analysis, K.K. and H.E.K.; Funding acquisition, Y.J.C.; Investigation, K.K. and H.E.K.; Methodology, K.K., H.E.K., J.I.Y. and Y.J.C.; Project administration, E.K. and Y.J.C.; Resources, H.-S.Y., J.-Y.C. and Y.J.C.; Software, K.K. and H.E.K.; Supervision, J.I.Y., H.-S.Y., E.K., J.-Y.C. and Y.J.C.; Validation, J.-Y.C. and Y.J.C.; Visualization, K.K. and Y.J.C.; Writing—original draft, K.K., H.E.K., H.-S.Y. and Y.J.C.; Writing—review & editing, J.I.Y., E.K. and J.-Y.C. All authors have read and agreed to the published version of the manuscript.
Funding
This research was supported by a faculty research grant from Yonsei University College of Dentistry for 2018-0019.
Conflicts of Interest
The authors declare no conflict of interest.
References
- 1.Kirschneck C., Batschkus S., Proff P., Kostler J., Spanier G., Schroder A. Valid gene expression normalization by RT-qPCR in studies on hPDL fibroblasts with focus on orthodontic tooth movement and periodontitis. Sci. Rep. 2017;7:14751. doi: 10.1038/s41598-017-15281-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Seo B.M., Miura M., Gronthos S., Bartold P.M., Batouli S., Brahim J., Young M., Robey P.G., Wang C.Y., Shi S. Investigation of multipotent postnatal stem cells from human periodontal ligament. Lancet. 2004;364:149–155. doi: 10.1016/S0140-6736(04)16627-0. [DOI] [PubMed] [Google Scholar]
- 3.Bright R., Hynes K., Gronthos S., Bartold P.M. Periodontal ligament-derived cells for periodontal regeneration in animal models: A systematic review. J. Periodontal. Res. 2015;50:160–172. doi: 10.1111/jre.12205. [DOI] [PubMed] [Google Scholar]
- 4.Kraft D.C., Bindslev D.A., Melsen B., Abdallah B.M., Kassem M., Klein-Nulend J. Mechanosensitivity of dental pulp stem cells is related to their osteogenic maturity. Eur. J. Oral Sci. 2010;118:29–38. doi: 10.1111/j.1600-0722.2009.00709.x. [DOI] [PubMed] [Google Scholar]
- 5.Lekic P., McCulloch C.A. Periodontal ligament cell population: The central role of fibroblasts in creating a unique tissue. Anat. Rec. 1996;245:327–341. doi: 10.1002/(SICI)1097-0185(199606)245:2<327::AID-AR15>3.0.CO;2-R. [DOI] [PubMed] [Google Scholar]
- 6.Lee S.Y., Yoo H.I., Kim S.H. CCR5-CCL Axis in PDL during Orthodontic Biophysical Force Application. J. Dent. Res. 2015;94:1715–1723. doi: 10.1177/0022034515603926. [DOI] [PubMed] [Google Scholar]
- 7.Zhang C., Li J., Zhang L., Zhou Y., Hou W., Quan H., Li X., Chen Y., Yu H. Effects of mechanical vibration on proliferation and osteogenic differentiation of human periodontal ligament stem cells. Arch. Oral Biol. 2012;57:1395–1407. doi: 10.1016/j.archoralbio.2012.04.010. [DOI] [PubMed] [Google Scholar]
- 8.Pavlin D., Gluhak-Heinrich J. Effect of mechanical loading on periodontal cells. Crit. Rev. Oral Biol. Med. 2001;12:414–424. doi: 10.1177/10454411010120050401. [DOI] [PubMed] [Google Scholar]
- 9.Mine K., Kanno Z., Muramoto T., Soma K. Occlusal forces promote periodontal healing of transplanted teeth and prevent dentoalveolar ankylosis: An experimental study in rats. Angle Orthod. 2005;75:637–644. doi: 10.1043/0003-3219(2005)75[637:OFPPHO]2.0.CO;2. [DOI] [PubMed] [Google Scholar]
- 10.Xu C., Fan Z., Shan W., Hao Y., Ma J., Huang Q., Zhang F. Cyclic stretch influenced expression of membrane connexin 43 in human periodontal ligament cell. Arch. Oral Biol. 2012;57:1602–1608. doi: 10.1016/j.archoralbio.2012.07.002. [DOI] [PubMed] [Google Scholar]
- 11.Ma J., Zhao D., Wu Y., Xu C., Zhang F. Cyclic stretch induced gene expression of extracellular matrix and adhesion molecules in human periodontal ligament cells. Arch. Oral Biol. 2015;60:447–455. doi: 10.1016/j.archoralbio.2014.11.019. [DOI] [PubMed] [Google Scholar]
- 12.Sprogar S., Vaupotic T., Cor A., Drevensek M., Drevensek G. The endothelin system mediates bone modeling in the late stage of orthodontic tooth movement in rats. Bone. 2008;43:740–747. doi: 10.1016/j.bone.2008.06.012. [DOI] [PubMed] [Google Scholar]
- 13.Di Domenico M., D’Apuzzo F., Feola A., Cito L., Monsurro A., Pierantoni G.M., Berrino L., De Rosa A., Polimeni A., Perillo L. Cytokines and VEGF induction in orthodontic movement in animal models. J. Biomed. Biotechnol. 2012;2012:201689. doi: 10.1155/2012/201689. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Howard P.S., Kucich U., Taliwal R., Korostoff J.M. Mechanical forces alter extracellular matrix synthesis by human periodontal ligament fibroblasts. J. Periodontal. Res. 1998;33:500–508. doi: 10.1111/j.1600-0765.1998.tb02350.x. [DOI] [PubMed] [Google Scholar]
- 15.Kokai S., Kanno Z., Koike S., Uesugi S., Takahashi Y., Ono T., Soma K. Retrospective study of 100 autotransplanted teeth with complete root formation and subsequent orthodontic treatment. Am. J. Orthod. Dentofacial. Orthop. 2015;148:982–989. doi: 10.1016/j.ajodo.2015.06.018. [DOI] [PubMed] [Google Scholar]
- 16.Min H.-K.Y.H.-S.S.G.L.B.-M. Effect of Mechanical Stress on the Proliferation and Expression of Cell Cycle Regulators in Human Periodontal Ligament Cells. J. Korean Acad. Periodontol. 1999;29:593–605. [Google Scholar]
- 17.Kukurba K.R., Montgomery S.B. RNA Sequencing and Analysis. Cold Spring Harb. Protoc. 2015;2015:951–969. doi: 10.1101/pdb.top084970. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Qian X., Ba Y., Zhuang Q., Zhong G. RNA-Seq technology and its application in fish transcriptomics. OMICS. 2014;18:98–110. doi: 10.1089/omi.2013.0110. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Davanian H., Stranneheim H., Bage T., Lagervall M., Jansson L., Lundeberg J., Yucel-Lindberg T. Gene expression profiles in paired gingival biopsies from periodontitis-affected and healthy tissues revealed by massively parallel sequencing. PLoS ONE. 2012;7:e46440. doi: 10.1371/journal.pone.0046440. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Kurt G., Iseri H., Kisnisci R. Rapid tooth movement and orthodontic treatment using dentoalveolar distraction (DAD). Long-term (5 years) follow-up of a Class II case. Angle Orthod. 2010;80:597–606. doi: 10.2319/041209-209.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Fu W.J., Hu J., Spencer T., Carroll R., Wu G. Statistical models in assessing fold change of gene expression in real-time RT-PCR experiments. Comput. Biol. Chem. 2006;30:21–26. doi: 10.1016/j.compbiolchem.2005.10.005. [DOI] [PubMed] [Google Scholar]
- 22.Anders S., Pyl P.T., Huber W. HTSeq--a Python framework to work with high-throughput sequencing data. Bioinformatics. 2015;31:166–169. doi: 10.1093/bioinformatics/btu638. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Sun J., Nishiyama T., Shimizu K., Kadota K. TCC: An R package for comparing tag count data with robust normalization strategies. BMC Bioinformatics. 2013;14:219. doi: 10.1186/1471-2105-14-219. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Yee J.A., Kimmel D.B., Jee W.S. Periodontal ligament cell kinetics following orthodontic tooth movement. Cell Tissue Kinet. 1976;9:293–302. doi: 10.1111/j.1365-2184.1976.tb01276.x. [DOI] [PubMed] [Google Scholar]
- 25.Ullrich N., Schroder A., Jantsch J., Spanier G., Proff P., Kirschneck C. The role of mechanotransduction versus hypoxia during simulated orthodontic compressive strain-an in vitro study of human periodontal ligament fibroblasts. Int. J. Oral Sci. 2019;11:33. doi: 10.1038/s41368-019-0066-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Choung H.W., Lee D.S., Park Y.H., Lee Y.S., Bai S., Yoo S.H., Lee J.H., You H.K., Park J.C. The effect of CPNE7 on periodontal regeneration. Connect. Tissue Res. 2019:1–12. doi: 10.1080/03008207.2019.1574776. [DOI] [PubMed] [Google Scholar]
- 27.Billuart P., Bienvenu T., Ronce N., des Portes V., Vinet M.C., Zemni R., Roest Crollius H., Carrie A., Fauchereau F., Cherry M., et al. Oligophrenin-1 encodes a rhoGAP protein involved in X-linked mental retardation. Nature. 1998;392:923–926. doi: 10.1038/31940. [DOI] [PubMed] [Google Scholar]
- 28.Govek E.E., Newey S.E., Van Aelst L. The role of the Rho GTPases in neuronal development. Genes Dev. 2005;19:1–49. doi: 10.1101/gad.1256405. [DOI] [PubMed] [Google Scholar]
- 29.Ishida A., Okuno S., Kitani T., Kameshita I., Fujisawa H. Regulation of multifunctional Ca2+/calmodulin-dependent protein kinases by Ca2+/calmodulin-dependent protein kinase phosphatase. Biochem. Biophys. Res. Commun. 1998;253:159–163. doi: 10.1006/bbrc.1998.9771. [DOI] [PubMed] [Google Scholar]
- 30.Toms S.R., Lemons J.E., Bartolucci A.A., Eberhardt A.W. Nonlinear stress-strain behavior of periodontal ligament under orthodontic loading. Am. J. Orthod. Dentofacial. Orthop. 2002;122:174–179. doi: 10.1067/mod.2002.124997. [DOI] [PubMed] [Google Scholar]
- 31.Lin W.L., McCulloch C.A., Cho M.I. Differentiation of periodontal ligament fibroblasts into osteoblasts during socket healing after tooth extraction in the rat. Anat. Rec. 1994;240:492–506. doi: 10.1002/ar.1092400407. [DOI] [PubMed] [Google Scholar]