Abstract
The unfolded protein response (UPR) is a cellular stress response mechanism that is critical for cell survival. Pharmacological modulation of the ATPase activity of the chaperone Hsp70 can trigger UPR-mediated cell death, thus removing pathogenic cells in human malignancies, or, alternatively, stimulate survival, thereby preventing apoptosis in neuronal cells and slowing the progress of inflammation, neurodegeneration, and aging. This Viewpoint highlights the complexity of the protein homeostasis network and discusses different approaches for modulating Hsp70 activity, including the use of a chemical reaction development-inspired library of Hsp70 agonists and antagonists.
Keywords: Heat shock protein, Hsps, UPR, ERAD, protein folding, chaperones
While the 1,000 most highly expressed proteins in a cell can account for 80% of the proteome mass, mammalian cells typically contain 1010 protein molecules of >10,000 different types,1 establishing a highly complex, dynamic network.2 The endoplasmic reticulum (ER) serves as the site of maturation of millions of protein molecules per minute, and authentic protein folding and maintenance of native structure are vital for proper cell function.3 Protein quality control systems include proteases and molecular chaperones—specialized guardian proteins that prevent client protein aggregation and general malfunction in times of cellular stress.
Properly folded proteins are transferred to the Golgi complex, while misfolded proteins remain in the ER to complete folding or are disposed of in the endoplasmic reticulum-associated degradation (ERAD) pathway.4 Common disposal mechanisms include ubiquitylation (Ub) by E3 ubiquitin ligases and translocation to the proteasome by the AAA+ ATPase p97.5 ER stress may be triggered by nutrient restriction, changes in calcium, oxygen or redox homeostasis, errors in post-translational modifications, and spikes in protein synthesis.6
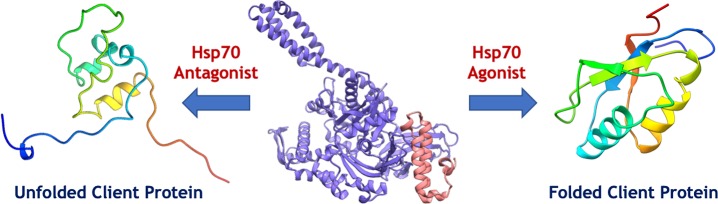
The accumulation of misfolded or unfolded proteins tagged with the Hsp70 chaperone, binding immunoglobulin protein (BiP, also known as heat shock 70 kDa protein 5, HSPA5), in the ER also induces the unfolded protein response (UPR),7 which is regulated through three stress sensors embedded in the ER membrane: PKR-like ER kinase (PERK), inositol-requiring enzyme one alpha (IRE1α), and activated transcription factor 6 (ATF6) (Figure 1).8 The UPR stress sensors may either promote cell survival or induce apoptosis.9 In the former case, the UPR response upregulates transcription of chaperones and enhances ERAD and autophagy machineries. Heat shock factor 1 (HSF1), Hsp70, Hsp90, and HDAC6 can form a complex in the cytosol, and as part of the heat shock response, HSF1 translocates to the nucleus and upregulates protein and chaperone transcription.8 Heat shock proteins (Hsps) are categorized by molecular mass, i.e. Hsps of 110 kDa (Hsp110s), Hsp100s, Hsp90s, Hsp70s, Hsp60s, and Hsp40s, consuming ATP to prevent protein misfolding and recognize misfolded polypeptide chains.10
Figure 1.
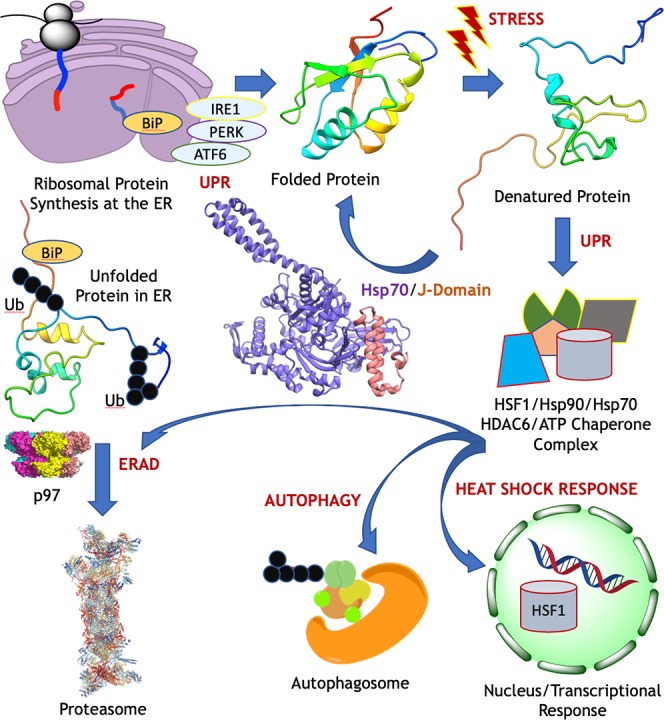
Schematic of protein quality control pathways. After ribosomal synthesis at the endoplasmic reticulum (ER), an Hsp70/Hsp40 (J-domain) complex12 assists in the folding of the protein chain as well as in the refolding of denatured13 proteins.
Both Hsp90 and Hsp70 are collaboratively engaged in protein refolding,11 and residual unfolded proteins are ubiquitylated and degraded in the proteasome. The J-domain of the cochaperone DnaJ, also known as Hsp40, binds tightly to a highly conserved interdomain linker on Hsp70 and significantly increases its catalytic turnover and substrate trapping efficiency (Figure 1).12
While the functions of Hsp90 and Hsp70 are correlated, leading to compensatory increases in expression if one of them is inhibited, a key feature of selective chaperone probe molecules as well as potential drugs is likely to prevent HSF1 from broadly activating the transcriptional heat shock response and thus upregulating all generic protein quality control pathways. Similarly, activation of Hsp90 or Hsp70 should not result in a downregulation of other proteotoxic defenses. Depletion or chemical inhibition of Hsp70 appears to be more readily decoupled from a general induction of stress response factors, and Hsp70 inhibitors have also been shown to act synergistically, suggesting that Hsp70 might be the more “druggable” chaperone.14 Another challenge for drug discovery is the nature of the ATP site in Hsps, which shares many structural features with other ATPases, thus potentially discouraging efforts to develop selective ATP-site inhibitors of Hsps.15 Therefore, protein–protein interaction (PPI) modulators are likely to represent a more fruitful medicinal chemistry strategy for targeting Hsps.
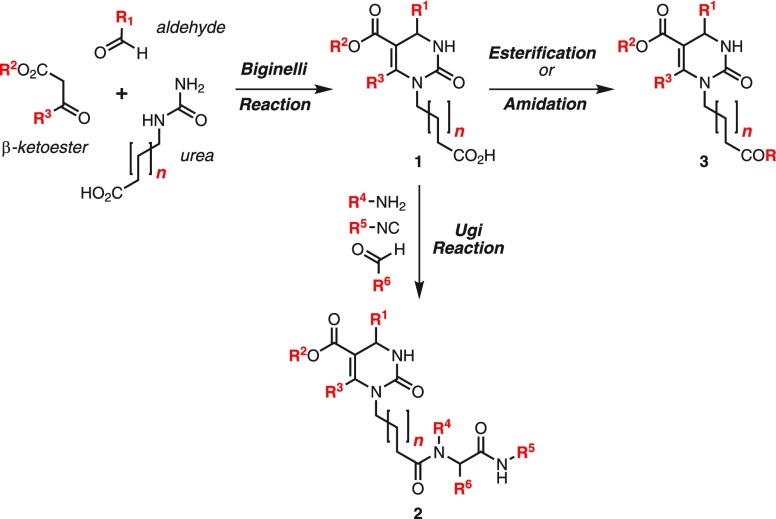
The historical context of the development of Hsps inhibitors,16 including the Hsp70 chaperone,17 has recently been reviewed. As an example for how new synthetic methods can enable chemical biology tool discovery,18 a consecutive Biginelli–Ugi multicomponent reaction (MCR) was used for the rapid synthesis of a library of screening samples for Hsp70 assays (Scheme 1).19 The acid-catalyzed cyclocondensation of readily available aldehydes, β-ketoesters, and urea carboxylates generates dihydropyrimidinones 1 with four sites of modification,20 which can immediately be subjected to an Ugi reaction with an amine, an isonitrile, and another aldehyde to yield the chain extended heterocycle/amino acid hybrid molecules 2 containing three additional sites for combinatorial structure variation. Alternatively, direct coupling with alcohols or amines converts carboxylates 1 into esters or amides 3. As a consequence of the presence of the densely functionalized dihydropyrimidinone core, the flexible linker region, and the peptoid terminus, scaffolds 2 and 3 are ideally suited to mimic small protein strands at the interface of PPIs. Several selective agonists and antagonists for Hsp70 have been discovered through this approach.17
Scheme 1. Biginelli–Ugi MCR for the Preparation of Hsp70 Modulators and Probes for PPI Interfaces.
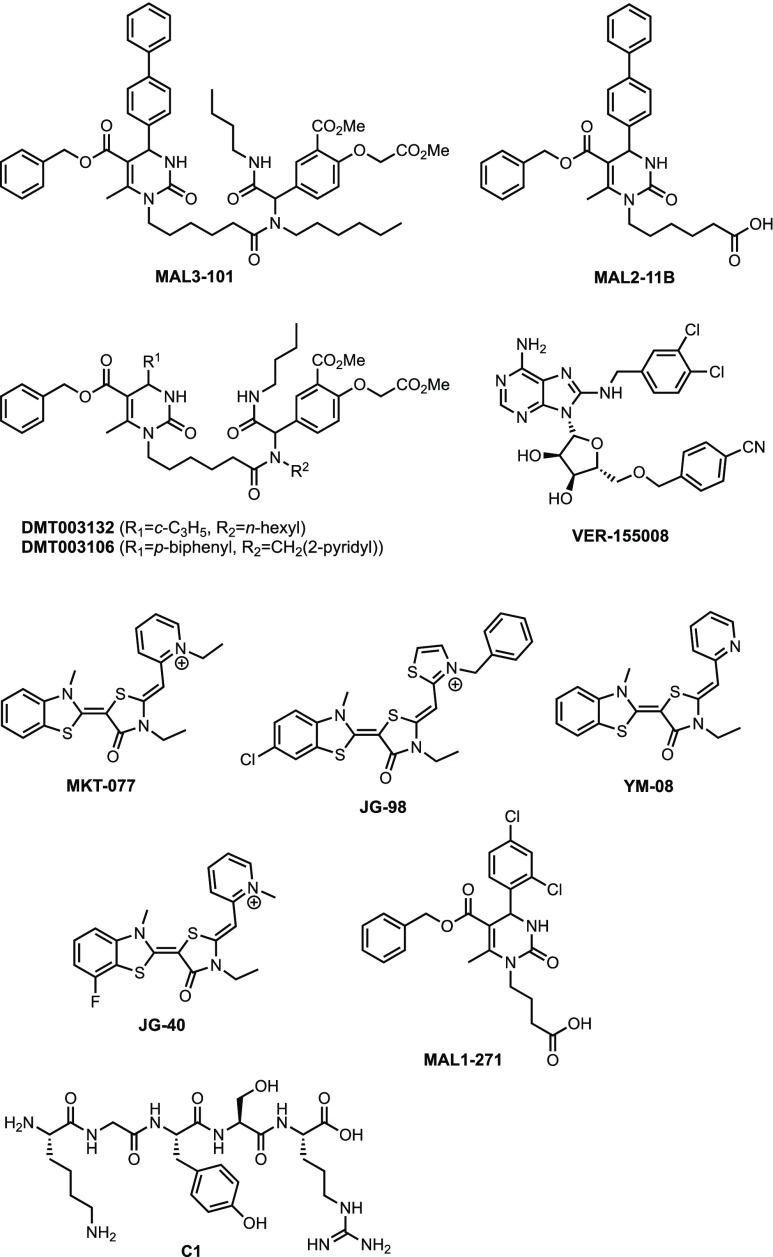
The selective Hsp70 inhibitor, MAL3-101 (Figure 2),21 has been shown to induce the UPR through PERK signaling, triggering apoptosis via activation of the C/EBP homologous protein (CHOP) transcription factor.22,23 In contrast to the ATP-competitive inhibitor VER-155008, MAL3-101 is an allosteric inhibitor of Hsp70 at high micromolar concentrations.24
Figure 2.
Structures of selected Hsp70 modulators, exemplifying four major chemotypes for allosteric and ATP-site ligands.
In order to inhibit Hsp70 ATPase activity, MAL3-101 requires the presence of a J-domain, suggesting a mechanism of action that involves binding to an allosteric region on Hsp70, quite likely in close vicinity to the J-domain binding site, as found by NMR analysis for the structurally related MAL2-11B.25 MAL3-101 has proven efficacious in multiple myeloma,26 rhabdomyosarcoma, and Merkel cell carcinoma27 models. Structure–activity relationship studies on the anticancer activity of MAL3-101 showed a dependence on aldehyde and amine components used in the Biginelli and Ugi reactions, respectively. In an SK-BR-3 breast cancer cell growth assay, for example, the GI50 for MAL3-101 was 27 μM, while cyclopropyl analog DMT003132 and 2-pyridyl analog DMT003106 had GI50’s of 6.2 and 7.1 μM, respectively (Figure 2).21
Another allosteric inhibitor chemotype, MKT-077, with affinity to the nucleotide-binding domain (NBD) of Hsp70, displayed anticancer activity in multiple cancer lines, including melanomas and carcinomas of the colon, breast, and pancreas.28 However, anticancer development of MKT-077 in a Phase 1 clinical study was stopped due to nephrotoxicity. The neutral pyridine analog YM-08, earmarked for neurodegenerative diseases, resulted in higher blood brain barrier (BBB) penetration, quicker kidney clearance, and retention of affinity for Hsp70. In contrast, it was less effective in antitau and anticancer assays. A detailed analysis of a second-generation YM-08 derivative, JG-98, included a gene expression profile; however, in this case, the UPR was not invoked.29 JG-98, in combination with a chalcone-based Hsp40 inhibitor, is effective in blocking androgen receptor (AR) signaling and inhibiting 22Rv1 castration-resistant prostate cancer growth (CRPC),30 suggesting opportunities to drug Hsp70 as an alternative strategy to standard antiandrogen therapy in CRPC.31 Finally, recent studies also demonstrate that close analogs of YM-08, such as the Hsp70 inhibitor JG-40, block zika virus replication in both human and mosquito cells without causing drug resistance and protect mice from viral infection.32
An alternative approach to generate a PPI-based Hsp70 modulator, specifically an agent that acts as a molecular glue on Hsp70 and its co-chaperone, heat shock organizing protein (HOP), recently yielded the pentapeptide C1.33 C1 stabilizes the HOP–Hsp70 complex and prevents protein folding at micromolar concentrations.
While our group’s initial interest was mainly in the development of Hsp70 inhibitors, in order to control cancer growth, viral infections, and parasitic infections such as malaria, screening of the readily synthesized library of dihydropyrimidinones 2 and 3 also identified agonists of the chaperone, which could be of significant therapeutic relevance in neurodegenerative diseases. In particular, MAL1-271 has proven to be an effective activator of ATP turnover in Hsp70/J-protein assays.34 In combination with the Hsp-70 antagonist, MAL3-101, MAL1-271 has been an important tool compound to test hypotheses on the effects of Hsp70 activity on synuclein aggregation35 and heat shock defenses in the cortex.36 MAL1-271 binds to the same allosteric, J-protein (Hsp40) pocket as MAL2-11B25 and, by inference, MAL3-101, making it a mechanistically relevant and particularly valuable positive control compound, as well as a potential lead structure for the development of antineurodegenerative, Hsp70 based therapeutics.
It is intriguing to speculate that the relatively low in vitro potency of both Hsp70 agonists and antagonists in ATP turnover assays, in contrast to their enhanced potency in cell-based assays and remarkable activity in the available in vivo models, is a result of the redundancies built into the mammalian protein homeostasis network, which lacks a single master control protein (MCP). In such a system of checks and balances, mildly potent but highly selective regulators that take advantage of natural PPI regulatory sites might well be biologically more relevant for disease control than highly potent, but less selective agents that elicit a strong compensatory counter reaction in the network. In particular in synergy with other proteostasis modulators, allosteric agents that modify the rate of Hsp70 ATP hydrolysis by a relatively modest −2 to +2-fold range at <100 μM concentration, such as MAL3-101 and MAL1-271, might still be highly relevant for clinical applications. However, since none of the multiple active compounds in this series, or in any other class of allosteric Hsps modulators, have successfully completed human trials yet, it remains to be established if “soft” allosteric control is sufficient to prevent or cure human diseases induced by a pathogenic breakdown of protein homeostasis.
Even though cellular folding and quality control pathways are highly redundant and exquisitely orchestrated, protein homeostasis is continuously challenged by cell growth, genomic mutations, environmental factors, viral and bacterial infections, the release of reaction oxygen species (ROS) species in mitochondria,37 and degenerative aging processes.7 Interestingly, proteotoxic defense systems have diverged between bacteria, fungi, protozoa, and plants versus metazoans.38 The former deploy a disaggregase complex composed of Hsp70, J-protein cochaperone, nucleotide exchange factor (NEF), and the AAA+ ATPase Hsp100. In contrast, metazoans lack cytosolic and nuclear Hsp100s but have vastly expanded cooperativity between multiple Hsp70 subtypes (12 in humans), J-protein family members (54 in humans), and NEFs (19 in humans), thus enabling >12,000 permutations of Hsp70-based disaggregase complexes, with the potential evolutionary advantage of providing far greater scope and specificity in protein refolding required in complex organisms.37 Allosteric control is central to the chaperone function of Hsp70s,39 but how PPIs can be harnessed in drug discovery for pathological conditions such as cancer, infection, and neurodegeneration is still poorly understood.40 Accordingly, small molecular weight probe molecules can serve as a valuable tool set to pharmacologically interrogate the vast chaperone network and suggest novel opportunities for drug development.39
Acknowledgments
The authors thank their biological collaborators in the Hsp70 program, in particular J. Brodsky (University of Pittsburgh) and R. Leak (Duquesne University).
Glossary
Abbreviations
- AAA
ATPases Associated with diverse cellular Activities
- AR
androgen receptor
- ATF6
activated transcription factor 6
- BBB
blood brain barrier
- BiP
binding immunoglobulin protein
- CHOP
C/EBP homologous protein
- CRPC
castration-resistant prostate cancer
- ER
endoplasmic reticulum
- ERAD
endoplasmic reticulum-associated degradation
- HOP
heat shock organizing protein
- HSF1
heat shock factor 1
- Hsp70
heat shock protein 70 kDa
- Hsps
heat shock proteins
- IRE1α
inositol-requiring enzyme one alpha
- MCR
multicomponent reaction
- NEF
nucleotide exchange factor
- PERK
PKR-like ER kinase
- Ub
ubiquitylation
- UPR
unfolded protein response
- MCP
master control protein
Author Contributions
The manuscript was written through contributions of all authors. All authors have given approval to the final version of the manuscript.
This work was supported in part by grant P30 DK79307 from the National Institutes of Health and a generous donation from the Moravitz family.
The authors declare no competing financial interest.
References
- Milo R. What Is the Total Number of Protein Molecules Per Cell Volume? A Call to Rethink Some Published Values. BioEssays 2013, 35, 1050–1055. 10.1002/bies.201300066. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boettcher T. From Molecules to Life: Quantifying the Complexity of Chemical and Biological Systems in the Universe. J. Mol. Evol. 2018, 86, 1–10. 10.1007/s00239-017-9824-6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dissmeyer N.; Coux O.; Rodriguez M. S.; Barrio R. Proteostasis: A European Network to Break Barriers and Integrate Science on Protein Homeostasis. Trends Biochem. Sci. 2019, 44, 383–387. 10.1016/j.tibs.2019.01.007. [DOI] [PubMed] [Google Scholar]
- Sun Z.; Brodsky J. L. Guardians of the ERAD Galaxy. Cell 2017, 171, 267–268. 10.1016/j.cell.2017.09.023. [DOI] [PubMed] [Google Scholar]
- Huryn D. M.; Kornfilt D. J. P.; Wipf P. p97: An Emerging Target for Cancer, Neurodegenerative Diseases, and Viral Infections. J. Med. Chem. 2019, 10.1021/acs.jmedchem.9b01318. [DOI] [PubMed] [Google Scholar]
- Kitamura M. The Unfolded Protein Response Triggered by Environmental Factors. Semin. Immunopathol. 2013, 35, 259–275. 10.1007/s00281-013-0371-y. [DOI] [PubMed] [Google Scholar]
- Frakes A. E.; Dillin A. The UPRER: Sensor and Coordinator of Organismal Homeostasis. Mol. Cell 2017, 66, 761–771. 10.1016/j.molcel.2017.05.031. [DOI] [PubMed] [Google Scholar]
- Sannino S.; Brodsky J. L. Targeting Protein Quality Control Pathways in Breast Cancer.. BMC Biol. 2017, 15, 109. 10.1186/s12915-017-0449-4. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ojha R.; Amaravadi R. K. Targeting the Unfolded Protein Response in Cancer. Pharmacol. Res. 2017, 120, 258–266. 10.1016/j.phrs.2017.04.003. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Alderson T. R.; Kim J. H.; Markley J. L. Dynamical Structures of Hsp70 and Hsp70-Hsp40 Complexes. Structure 2016, 24, 1014–1030. 10.1016/j.str.2016.05.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Genest O.; Wickner S.; Doyle S. M. Hsp90 and Hsp70 Chaperones: Collaborators in Protein Remodeling. J. Biol. Chem. 2019, 294, 2109–2120. 10.1074/jbc.REV118.002806. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kityk R.; Kopp J.; Mayer M. P. Molecular Mechanism of J-Domain-Triggered ATP Hydrolysis by Hsp70 Chaperones.. Mol. Cell 2018, 69, 227–237. 10.1016/j.molcel.2017.12.003. [DOI] [PubMed] [Google Scholar]
- Kukic P.; Pustovalova Y.; Camilloni C.; Gianni S.; Korzhnev D. M.; Vendruscolo M. Structural Characterization of the Early Events in the Nucleation-Condensation Mechanism in a Protein Folding Process. J. Am. Chem. Soc. 2017, 139, 6899–6910. 10.1021/jacs.7b01540. [DOI] [PubMed] [Google Scholar]
- Prince T.; Schreiter B.; Danella J.; Williams H.; Ackerman A.; Cavanaugh A.; Juengst B.; Andolino C.; Chernin M. Dual Targeting of Hsp70 Does Not Induce the Heat Shock Response and Synergistically Reduces Cell Viability in Muscle Invasive Bladder Cancer. Oncotarget 2018, 9, 32702–32717. 10.18632/oncotarget.26021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Massey A. J. ATPases as Drug Targets: Insights from Heat Shock Proteins 70 and 90. J. Med. Chem. 2010, 53, 7280–7286. 10.1021/jm100342z. [DOI] [PubMed] [Google Scholar]
- McAlpine S. R.; Edkins A. L. Heat Shock Protein Inhibitors. Top. Med. Chem. 2016, 19, 1–238. [Google Scholar]
- Manos-Turvey A.; Brodsky J. L.; Wipf P. The Effect of Structure and Mechanism of the Hsp70 Chaperone on the Ability to Identify Chemical Modulators and Therapeutics. Top. Med. Chem. 2016, 19, 81–129. 10.1007/7355_2015_90. [DOI] [Google Scholar]
- Huryn D. M.; Brodsky J. L.; Brummond K. M.; Chambers P. G.; Eyer B.; Ireland A. W.; Kawasumi M.; LaPorte M. G.; Lloyd K.; Manteau B.; Nghiem P.; Quade B.; Seguin S. P.; Wipf P. Chemical Methodology as a Source of Small-Molecule Checkpoint Inhibitors and Heat Shock Protein 70 (Hsp70) Modulators. Proc. Natl. Acad. Sci. U. S. A. 2011, 108, 6757–6762. 10.1073/pnas.1015251108. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Werner S.; Turner D. M.; Lyon M. A.; Huryn D. M.; Wipf P. A Focused Library of Tetrahydropyrimidinone Amides Via a Tandem Biginelli-Ugi Multi-Component Process. Synlett 2006, 2334–2338. 10.1055/s-2006-949648. [DOI] [Google Scholar]
- Wipf P.; Cunningham A. A Solid Phase Protocol of the Biginelli Dihydropyrimidine Synthesis Suitable for Combinatorial Chemistry. Tetrahedron Lett. 1995, 36, 7819–7822. 10.1016/0040-4039(95)01660-A. [DOI] [Google Scholar]
- Wright C. M.; Chovatiya R. J.; Jameson N. E.; Turner D. M.; Zhu G.; Werner S.; Huryn D. M.; Pipas J. M.; Day B. W.; Wipf P.; Brodsky J. L. Pyrimidinone-Peptoid Hybrid Molecules with Distinct Effects on Molecular Chaperone Function and Cell Proliferation. Bioorg. Med. Chem. 2008, 16, 3291–3301. 10.1016/j.bmc.2007.12.014. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sannino S.; Guerriero C. J.; Sabnis A. J.; Stolz D. B.; Wallace C. T.; Wipf P.; Watkins S. C.; Bivona T. G.; Brodsky J. L. Compensatory Increases of Select Proteostasis Networks after Hsp70 Inhibition in Cancer Cells. J. Cell Sci. 2018, 131, jcs217760 10.1242/jcs.217760. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sabnis A. J.; Guerriero C. J.; Olivas V.; Sayana A.; Shue J.; Flanagan J.; Asthana S.; Paton A. W.; Paton J. C.; Gestwicki J. E.; Walter P.; Weissman J. S.; Wipf P.; Brodsky J. L.; Bivona T. G. Combined Chemical-Genetic Approach Identifies Cytosolic Hsp70 Dependence in Rhabdomyosarcoma. Proc. Natl. Acad. Sci. U. S. A. 2016, 113, 9015–9020. 10.1073/pnas.1603883113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fewell S. W.; Smith C. M.; Lyon M. A.; Dumitrescu T. P.; Wipf P.; Day B. W.; Brodsky J. L. Small Molecule Modulators of Endogenous and Co-Chaperone-Stimulated Hsp70 ATPase Activity. J. Biol. Chem. 2004, 279, 51131–51140. 10.1074/jbc.M404857200. [DOI] [PubMed] [Google Scholar]
- Wisen S.; Bertelsen E. B.; Thompson A. D.; Patury S.; Ung P.; Chang L.; Evans C. G.; Walter G. M.; Wipf P.; Carlson H. A.; Brodsky J. L.; Zuiderweg E. R. P.; Gestwicki J. E. Binding of a Small Molecule at a Protein-Protein Interface Regulates the Chaperone Activity of Hsp70-Hsp40. ACS Chem. Biol. 2010, 5, 611–622. 10.1021/cb1000422. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Braunstein M. J.; Scott S. S.; Scott C. M.; Behrman S.; Walter P.; Wipf P.; Coplan J. D.; Chrico W.; Joseph D.; Brodsky J. L.; Batuman O. Antimyeloma Effects of the Heat Shock Protein 70 Molecular Chaperone Inhibitor MAL3-101. J. Oncol. 2011, 2011, 232037. 10.1155/2011/232037. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Adam C.; Baeurle A.; Houben R.; Brodsky J. L.; Wipf P.; Schrama D.; Becker J. C. The Hsp70 Modulator MAL3–101 Inhibits Merkel Cell Carcinoma. PLoS One 2014, 9, e92041 10.1371/journal.pone.0092041. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Miyata Y.; Li X.; Lee H.-F.; Jinwal U. K.; Srinivasan S. R.; Seguin S. P.; Young Z. T.; Brodsky J. L.; Dickey C. A.; Sun D.; Gestwicki J. E. Synthesis and Initial Evaluation of YM-08, a Blood-Brain Barrier Permeable Derivative of the Heat Shock Protein 70 (Hsp70) Inhibitor MKT-077, Which Reduces Tau Levels.. ACS Chem. Neurosci. 2013, 4, 930–939. 10.1021/cn300210g. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yaglom J. A.; Wang Y.; Li A.; Li Z.; Monti S.; Alexandrov I.; Lu X.; Sherman M. Y. Cancer Cell Responses to Hsp70 Inhibitor JG-98: Comparison with Hsp90 Inhibitors and Finding Synergistic Drug Combinations. Sci. Rep. 2018, 8, 3010. 10.1038/s41598-017-14900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moses M. A.; Kim Y. S.; Rivera-Marquez G. M.; Oshima N.; Watson M. J.; Beebe K. E.; Wells C.; Lee S.; Zuehlke A. D.; Shao H.; Bingman W. E. III; Kumar V.; Malhotra S. V.; Weigel N. L.; Gestwicki J. E.; Trepel J. B.; Neckers L. M. Targeting the Hsp40/Hsp70 Chaperone Axis as a Novel Strategy to Treat Castration-Resistant Prostate Cancer. Cancer Res. 2018, 78, 4022–4035. 10.1158/0008-5472.CAN-17-3728. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dong J.; Wu Z.; Wang D.; Pascal L. E.; Nelson J. B.; Wipf P.; Wang Z. Hsp70 Binds to the Androgen Receptor N-Terminal Domain and Modulates the Receptor Function in Prostate Cancer Cells. Mol. Cancer Ther. 2019, 18, 39–50. 10.1158/1535-7163.MCT-18-0432. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Taguwa S.; Yeh M.-T.; Rainbolt T. K.; Nayak A.; Shao H.; Gestwicki J. E.; Andino R.; Frydman J. Zika Virus Dependence on Host Hsp70 Provides a Protective Strategy against Infection and Disease. Cell Rep. 2019, 26, 906–920. 10.1016/j.celrep.2018.12.095. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zaiter S. S.; Huo Y.; Tiew F. Y.; Gestwicki J. E.; McAlpine S. R. Designing De Novo Small Molecules That Control Heat Shock Protein 70 (Hsp70) and Heat Shock Organizing Protein (HOP) within the Chaperone Protein-Folding Machinery. J. Med. Chem. 2019, 62, 742–761. 10.1021/acs.jmedchem.8b01436. [DOI] [PubMed] [Google Scholar]
- Chiang A. N.; Liang M.; Dominguez-Meijide A.; Masaracchia C.; Goeckeler-Fried J. L.; Mazzone C. S.; Newhouse D. W.; Kendsersky N. M.; Yates M. E.; Manos-Turvey A.; Needham P. G.; Outeiro T. F.; Wipf P.; Brodsky J. L. Synthesis and Evaluation of Esterified Hsp70 Agonists in Cellular Models of Protein Aggregation and Folding. Bioorg. Med. Chem. 2019, 27, 79–91. 10.1016/j.bmc.2018.11.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kilpatrick K.; Novoa J. A.; Hancock T.; Guerriero C. J.; Wipf P.; Brodsky J. L.; Segatori L. Chemical Induction of Hsp70 Reduces A-Synuclein Aggregation in Neuroglioma Cells. ACS Chem. Biol. 2013, 8, 1460–1468. 10.1021/cb400017h. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Posimo J. M.; Weilnau J. N.; Gleixner A. M.; Broeren M. T.; Weiland N. L.; Brodsky J. L.; Wipf P.; Leak R. K. Heat Shock Protein Defenses in the Neocortex and Allocortex of the Telencephalon. Neurobiol. Aging 2015, 36, 1924–1937. 10.1016/j.neurobiolaging.2015.02.011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Frantz M.-C.; Wipf P. Mitochondria as a Target in Treatment. Environ. Mol. Mutagen. 2010, 51, 462–475. 10.1002/em.20554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nillegoda N. B.; Bukau B. Metazoan Hsp70-Based Protein Disaggregases: Emergence and Mechanisms. Front. Mol. Biosci. 2015, 2, 57. 10.3389/fmolb.2015.00057. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rosenzweig R.; Nillegoda N. B.; Mayer M. P.; Bukau B. The Hsp70 Chaperone Network. Nat. Rev. Mol. Cell Biol. 2019, 20, 665–680. 10.1038/s41580-019-0133-3. [DOI] [PubMed] [Google Scholar]
- Gestwicki J. E.; Shao H. Inhibitors and Chemical Probes for Molecular Chaperone Networks. J. Biol. Chem. 2019, 294, 2151–2161. 10.1074/jbc.TM118.002813. [DOI] [PMC free article] [PubMed] [Google Scholar]