Abstract
Background
Alterations in the epidermal growth factor receptor and PI3K pathways in head and neck squamous cell carcinomas (HNSCCs) are frequent events that promote tumor progression. Ectopic expression of the epidermal growth factor receptor–targeting microRNA (miR), miR-27a* (miR-27a-5p), inhibits tumor growth. We sought to identify mechanisms mediating repression of miR-27a* in HNSCC, which have not been previously identified.
Methods
We quantified miR-27a* in 47 oral cavity squamous cell carcinoma patient samples along with analysis of miR-27a* in 73 oropharyngeal and 66 human papillomavirus–positive (HPV+) samples from The Cancer Genome Atlas. In vivo and in vitro TP53 models engineered to express mutant TP53, along with promoter analysis using chromatin immunoprecipitation and luciferase assays, were used to identify the role of TP53 and TP63 in miR-27a* transcription. An HNSCC cell line engineered to conditionally express miR-27a* was used in vitro to determine effects of miR-27a* on target genes and tumor cells.
Results
miR-27a* expression was repressed in 47 oral cavity tumor samples vs matched normal tissue (mean log2 difference = −0.023, 95% confidence interval = −0.044 to −0.002; two-sided paired t test, P = .03), and low miR-27a* levels were associated with poor survival in HPV+ and oropharyngeal HNSCC samples. Binding of ΔNp63α to the promoter led to an upregulation of miR-27a*. In vitro and in vivo findings showed that mutant TP53 represses the miR-27a* promoter, downregulating miR-27a* levels. ΔNp63α and nucleoporin 62, a protein involved in ΔNP63α transport, were validated as novel targets of miR-27a*.
Conclusion
Our results characterize a negative feedback loop between TP63 and miR-27a*. Genetic alterations in TP53, a frequent event in HNSCC, disrupt this regulatory loop by repressing miR-27a* expression, promoting tumor survival.
Epidermal growth factor receptor (EGFR) is frequently overexpressed in head and neck squamous cell carcinomas (HNSCCs) and is thought to influence therapeutic response. As an important component of a multimodal therapeutic strategy, molecular inhibitors targeting EGFR showed initial promise (1,2). The minimal clinical benefits observed in patients, despite strong biological justification for the use of EGFR inhibitors, suggests that other cellular pathway alterations may lead to baseline or acquired resistance of HNSCC to EGFR-targeted agents (3).
Although HNSCC exhibits high levels of genomic instability, alterations in a relatively small number of pathways act as drivers of tumor progression (4–6). Alterations in the TP53 family of proteins (TP53, TP63, and TP73) are frequent events in HNSCC (2,4,5), and changes in one member of the TP53 family affects the function of other members (7). Mutations and/or deletions in TP53 are the leading genetic alteration in patients and directly affect response to therapy and overall survival (8,9). ΔNp63α, one of six transcripts of the TP63 gene, is overexpressed in HNSCC and is essential for its survival (10). ΔNp63α transcriptionally regulates a host of microRNAs (miRNAs) and genes affecting a wide spectrum of processes, including apoptosis, wound healing, and proliferation (7).
Dysregulation of miRNA expression in cancers including HNSCC has been linked to tumor development (11–13). We previously identified EGFR as a target of miR-27a (miR-27a-3p) and its complementary strand, miR-27a* (miR-27a-5p) (14). Both miRNAs are transcribed from the miR-23a-24∼2-27a locus on chromosome 19 and, depending on the context and cell type, these miRNAs act as oncomiRs or as tumor suppressors (12,13). AKT1 and mTOR, components of the PI3K/AKT1/mTOR pathway, which is the most activated mitogenic network both in human papillomavirus–positive (HPV+) and HPV-negative (HPV–) HNSCC, are direct targets of miR-27a* but not miR-27a (6,14–16). The high frequency of mutations in PI3K, AKT1, and mTOR promotes tumor growth (6,16). Targeting of multiple oncogenes, including EGFR, relevant to HNSCC progression underlies the molecular mechanism for miR-27a* but not miR-27a, affecting HNSCC survival (14).
Our earlier findings in a small patient cohort demonstrated that the expression of miR-27a and miR-27a* is repressed in HNSCC (14). However, the mechanisms mediating miR-27a* repression in HNSCC have not been defined. The goal of the current study was to define mechanisms regulating miR-27a* expression and their effects on HNSCC. We found that increased miR-27a* levels were associated with improved survival in patients with HPV+ HNSCC. We identified ΔNp63α as an activator of miR-27a* transcription. ΔNp63α and nucleoporin 62 (NUP62) were validated as novel miR-27a* targets. Thus, ΔNp63α-miR-27a* forms a feedback loop, whereas mutant TP53-mediated repression of miR-27a* disrupts this regulatory loop.
Methods
Patient Samples
Formalin-fixed, paraffin-embedded tissue samples were obtained using a protocol approved by the institutional review board. We obtained 47 matched normal and tumor samples from oral cavity squamous cell carcinoma (OCSCC) patients with primary disease (n = 40) and locoregional recurrence (n = 7) who were treated at The University of Texas MD Anderson Cancer Center. Patients were selected to provide a balance of clinical characteristics including age, stage, and sex. Informed consent from all patients was obtained for tissue used in the study. Tissue sections were examined, selected, and marked for microdissection by a board-certified pathologist (MW).
Quantitative Real-Time Polymerase Chain Reaction (qPCR)
qPCR analysis on RNA was carried out on a Biorad CFX 96 real-time PCR machine (Biorad, Carlsbad, CA) and data analyzed using Biorad CFX Manager software (Biorad). Each measurement was carried out in triplicate and each experiment was repeated independently for reproducibility. Additional details are provided in the Supplementary Methods (available online).
MTT Assay
Cells were plated in 96-well plates. The next morning cells were treated with the indicated concentration of doxycycline (Dox and incubated for 72 hours. MTT reagent was then added for 4 hours, after which the supernatant was replaced with dimethyl sulfoxide, and optical density values were read at 570 nm on a Spectrostar plate reader (BMG Labtech, Cary, NC). Data represents three experiments with error bars representing SD.
Luciferase Assay
Luciferase assays were carried out in 24-well plates in triplicate and repeated independently. Cells were cotransfected with indicated reporter constructs and 5 ng of Renilla using Lipofectamine 2000 (Thermo Fisher, Carlsbad, CA). Lysates were assayed with the Dual-Glo Luciferase Assay System (Promega, Madison, WI). Firefly activity was normalized to Renilla luciferase activity.
Clonogenic Assay
Clonogenic assays were carried out in triplicate and repeated three times. A representative experiment is shown with error bars representing SD. Details in the Supplementary Methods (available online).
Statistical Analysis
Experimental data was analyzed with GraphPad Prism (GraphPad Software, La Jolla, CA), Excel (Microsoft Corp, Seattle, WA) and R (version 3.4.1). Analysis of The Cancer Genome Atlas (TCGA) data was performed in R (version 3.4.1). All tests were two-sided and the statistical significance was defined as a P value less .05. Further details are presented in the Supplementary Methods (available online).
Results
Repression of miR-27a* and Poor Patient Survival in HNSCC
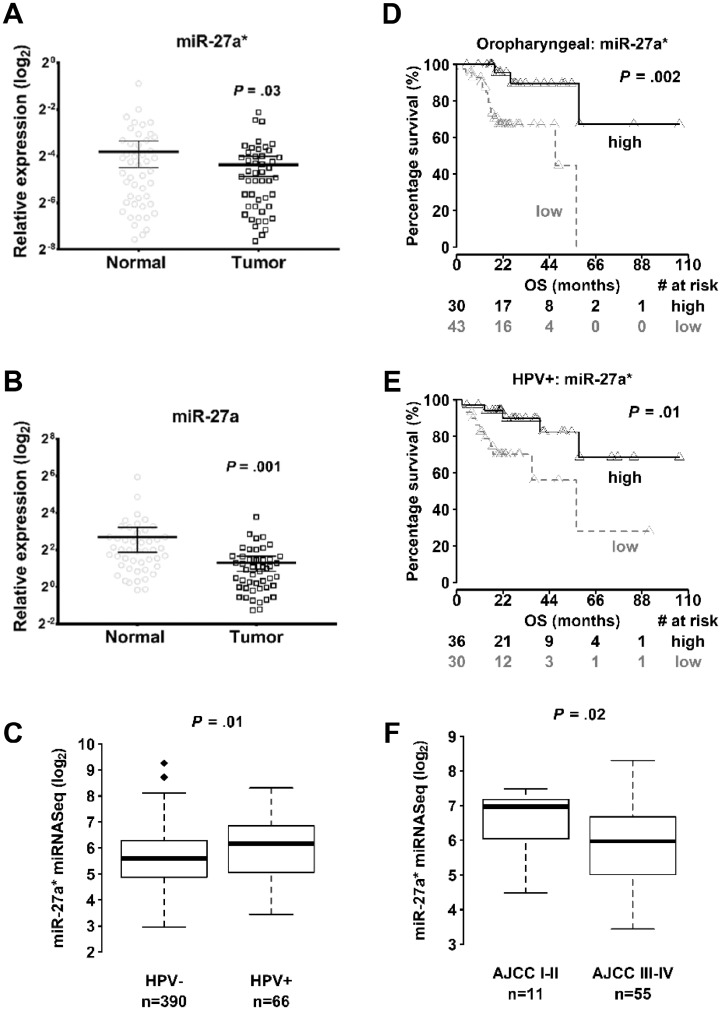
To expand on our previous findings of miR-27a and miR-27a* repression in HNSCC (14), we identified a well-annotated cohort of matched normal and tumor samples from 47 patients with OCSCC. qPCR analysis showed that expression of miR-27a* and miR-27a was lower in tumors than in matched normal samples (miR-27a* mean log2 difference = −0.023, 95% confidence interval [CI] = −0.044 to −0.002; two-sided paired t test, P = .03; Figure 1A; miR-27a mean log2 difference = −4.01, 95% CI = −6.37 to −1.66; two-sided paired t test, P = .001; Figure 1B).
Figure 1.
Expression of microRNA-27a* (miR-27a-5p) in head and neck squamous cell carcinoma (HNSCC). Quantitative real-time polymerase chain reaction (qPCR) analysis of miRNA levels from matched normal and tumor samples (n = 47) of (A) miR-27a* and (B) miR-27a (miR-27a-3p) expression in oral cavity squamous cell carcinoma (OCSCC) tumors. P values calculated by two-tailed Student t test. Error bars represent mean (95% confidence intervals [CI]). RNU 44 used as an endogenous control. C) Expression levels of miR-27a* in human papillomavirus–negative (HPV–) (n = 390) and HPV-positive (HPV+) (n = 66) HNSCC samples from The Cancer Genome Atlas (TCGA) database. Kaplan-Meier overall survival (OS) curves for (D) oropharyngeal squamous cell carcinoma (OPSCC) (n = 73) and (E) HPV+ HNSCC (n = 66) patients from TCGA expressing high miR-27a* or low miR-27a* levels; # = number. The P value was calculated using the log-rank test. (F) Expression of miR-27a* in American Joint Committee on Cancer (AJCC) stage I–II (n = 11) and stage III–IV (n = 55) HPV+ tumors in TCGA. The P value was calculated using the Mann-Whitney test. All statistical tests were two-sided.
The number of patients with HPV+ HNSCC has sharply increased (2,17,18). Analysis of HNSCC samples in TCGA showed higher levels of miR-27a* in HPV+ tumors (n = 66), which have a better prognosis than do HPV– tumors (n = 390) (P = .01) (Figure 1C). We also found that decreased levels of miR-27a* were associated with poor survival in patients with HPV+ (n = 66, P = .01) HNSCC, who mainly present with oropharyngeal squamous cell carcinoma (SCC; n = 73, P = .002) (OPSCC; Figure 1, D and E, and Supplementary Tables 1–7). In HPV+ tumors, higher levels of miR-27a* were observed in low-grade tumors (n = 11) (American Joint Committee on Cancer stage I/II) compared with high-grade (stage III/IV) (n = 55) tumors in the TCGA (P = .02) (Figure 1F).
ΔNp63α Regulation of miR-27a* Transcription
Transcriptional regulators of the miR-23a-24∼2-27a locus have not been defined in HNSCCs. We found that levels of miR-27a were substantially higher than miR-27a* in normal tissue (P < .001) and OCSCC samples (n = 47) (P < .001) (Supplementary Figures S1A and B, available online) and HNSCC cell lines (Supplementary Figure 2A, available online). Having established previously (14) and in the current study (Supplementary Figure 2B, and C, available online) that ectopic expression of miR-27a*, but not miR-23a, -24, or -27a, in HNSCC cells inhibited cellular survival, we focused on identifying mechanisms repressing miR-27a* expression in HNSCC.
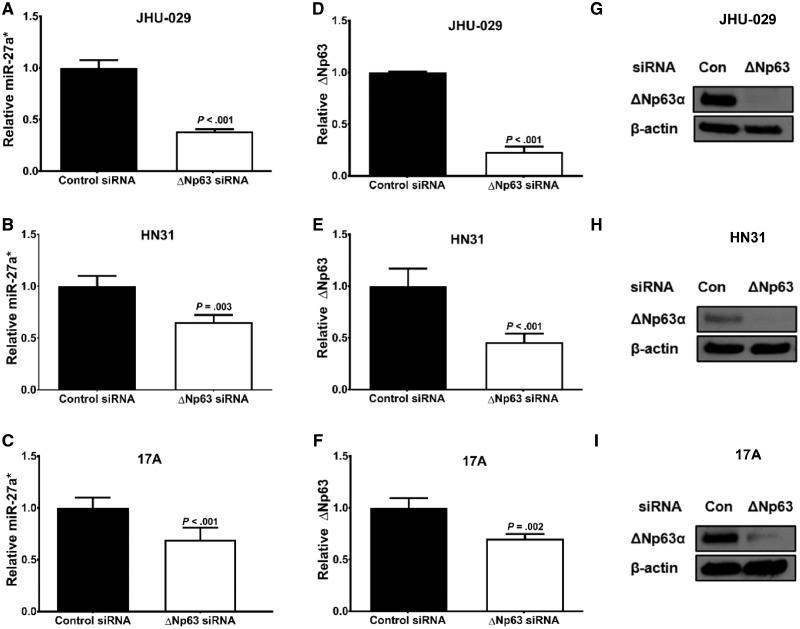
Robust expression of ΔNp63α in HNSCC cells is a primary driver of tumor progression by regulating a miRNA program essential to tumor survival (19). To determine if ΔNp63α regulates miR-27a*, we suppressed ΔNp63 expression using a ΔNp63-specific small interfering RNA (siRNA) in JHU-029 (TP53 Null) (20), HN31 (TP53 mutant), and UM-SCC-17A (17A; TP53 wild-type) cells resulting in a decrease in miR-27a* expression (Figure 2, A–C). Knockdown was specific for the ΔNp63 isoform in all cell lines as seen by a depletion of ΔNp63 mRNA (Figure 2, D–F) and ΔNp63α protein (Figure 2, G–I). An increase in TAp63 mRNA levels was seen in the JHU-029 and HN31 cells (Supplementary Figure 3A, and B available online), and a decrease in TAp63 was seen in 17A cells (Supplementary Figure 3C, available online). To confirm that the change in miR-27a* levels was not due to an increase in TAp63 levels, we used a TP63 siRNA targeting both TA and ΔN isoforms in JHU-029 cells. A decrease in TP63 and TAp63 (Supplementary Figure 3D, available online) was observed, along with a concomitant decrease in miR-27a* levels (Supplementary Figure 3E, available online). This suggests that ΔNp63α, the dominant isoform expressed in HNSCC (21), regulates miR-27a* expression.
Figure 2.
Effects of ΔNp63α levels on microRNA (miR)-27a* expression in head and neck squamous cell carcinoma (HNSCC) cells. ΔNp63α was depleted using a ΔNp63-targeting small interfering RNA (siRNA) in the indicated HNSCC cell lines. Expression of miR-27a* in (A) JHU-029, (B) HN31, and (C) UM-SCC-17A (17A) cells and ΔNp63 mRNA in (D) JHU-029, (E) HN31, and (F) 17A cells were analyzed by quantitative real-time polymerase chain reaction (qPCR). Error bars represent mean (SEM). Protein levels were analyzed by immunoblotting in (G) JHU-029, (H) HN31, and (I) 17A cells treated with either control (Con) or ΔNp63 siRNA with β-actin as an endogenous control. Endogenous controls used in qPCR analysis were RNU44/RNU6B for miRNA and glyceraldehyde-3-phosphate dehydrogenase for mRNA.
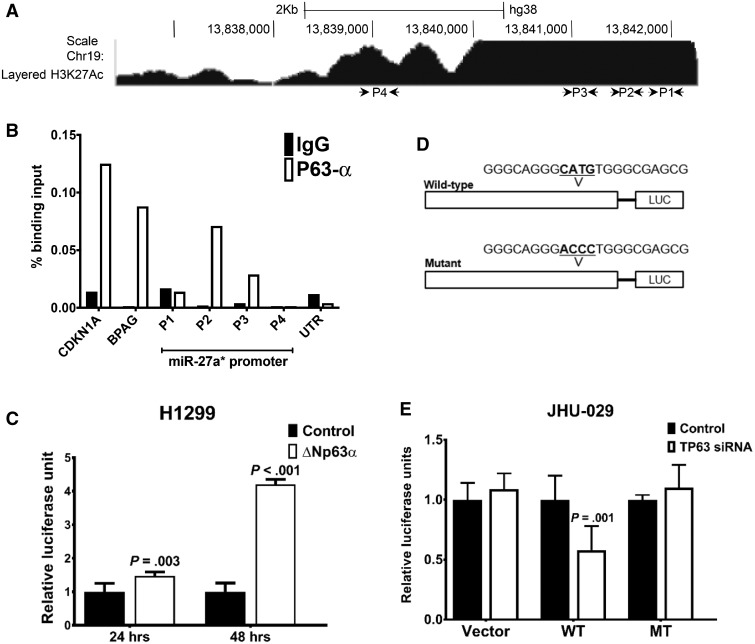
In silico analysis of the miR-23a-24∼2-27a promoter revealed a number of high-affinity putative TP53 and TP63 binding sites (Figure 3A). Quantitative chromatin immunoprecipitation analysis in UM-SCC-22A (22A) cells, which express high levels of ΔNp63α, showed that ΔNp63α was recruited to a region upstream of the transcription start site (Figure 3B). HNSCC cells express robust levels of ΔNp63α, therefore, H1299 cells, which do not have detectable levels of ΔNp63α protein (22), were cotransfected with luciferase constructs encompassing the ΔNp63α binding site on the miR-27a* promoter (miR-27a*-luc) and a vector control or ΔNp63α expression vector. Upregulation of luciferase activity was seen when the ΔNp63α vector was transfected compared with control vector (Figure 3C). Mutations that abrogated ΔNp63α binding were introduced in the putative binding site on the mutant miR-27a*-luc (miR-27a*-luc MT) (Figure 3D). JHU-029 cells were cotransfected with the wild-type (WT) or mutant (MT) miR-27a*-luc along with control or TP63 siRNA. Cotransfection of TP63 siRNA resulted in a decrease in WT miR-27a*-luc activity compared with control siRNA; a similar change was not observed with the miR-27a*-luc MT construct (Figure 3E), demonstrating that ΔNp63α binds to the miR-27a* promoter activating miR-27a* transcription.
Figure 3.
Transcriptional regulation of microRNA (miR)-27a* expression. (A) Schematic representation of the miR-27a* promoter with putative TP53 and TP63 binding sites (P1-P4) along with chromatin immunoprecipitation-RNA sequencing (ChIP-seq) enrichment of H3K27ac from ENCODE. B) Binding of endogenous TP63α in UM-SCC-22A (22A) cells to the miR-27a* promoter element was analyzed by quantitative ChIP. Representative experiment was performed in triplicate and carried out three times independently. C) Results of luciferase assays in H1299 cells with the miR-27a* promoter luciferase (miR-27a*-luc) in the presence of vector control (control) or ΔNp63α cDNA. D) Schematic of wild-type (WT) and mutant (MT) miR-27a*-luc constructs with indicated alterations in the TP63 binding site. E) Results of luciferase assays, carried out using the WT and MT miR-27a*-LUC constructs in JHU-029 cells with control or TP63 small interfering RNA (siRNA). Empty luciferase vector (vector) was transfected along with control or TP63 siRNA in JHU-029 cells as a control, and data were normalized to Renilla luciferase. Luciferase experiments represent the average of two independent experiments with error bars representing mean (SD). P value was calculated using a two-sided Student t test.
Mutant TP53 Repression of miR-27a* Expression
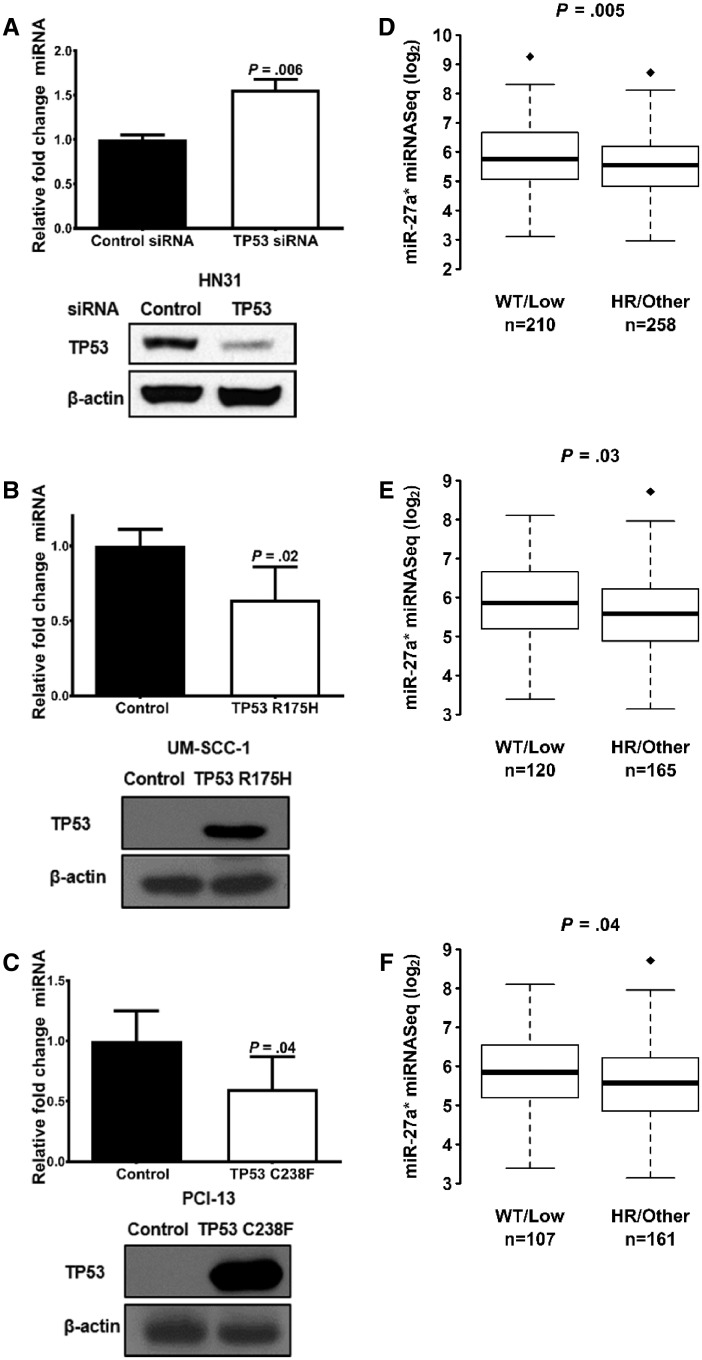
Mutations in TP53, a frequent event in HNSCC, can greatly affect TP63 activity (4,9,23). TP53-targeting siRNA was used to suppress mutant TP53 in HN31 cells to determine whether mutant TP53 affects ΔNp63α-directed miR-27a* transcription. A reduction in TP53 mutant protein resulted in an increase in miR-27a* levels (Figure 4A). UM-SCC-1 and PCI-13 cells devoid of TP53 protein were engineered to express mutant TP53, allowing us to explore the role of mutant TP53 in miR-27a* regulation. Expression of mutant TP53 in cells led to a decrease in miR-27a* levels (Figure 4, B and C). These findings strongly suggest that mutant TP53 represses miR-27a* expression.
Figure 4.
Effect of mutant TP53 on microRNA (miR)-27a* in head and neck squamous cell carcinoma (HNSCC). A) Analysis of miR-27a* and TP53 protein levels in HN31 cells transfected with control small interfering RNA (siRNA) or TP53 siRNA. miR-27a* and TP53 protein levels were examined in (B) UM-SCC-1 and (C) PCI-13 cells engineered to express TP53 R175H and TP53 C238F, respectively. Error bars represent mean (SEM). RNU6B was used as an endogenous control for miRNA analysis. Mutation status of HNSCC tumors in TCGA was classified according to the EAp53 scoring system into wild-type (WT), low-risk (LR), high-risk (HR), and other. miR-27a* expression analyzed in (D) HNSCC cohort, (E) oral cavity cohort, (F) and oral cavity HPV– cohort. Box-and-whisker plots are shown (box plot represents first [lower bound] and third [upper bound] quartiles; whiskers represent 1.5 times the interquartile range). P values are indicated. All statistical tests were two-sided.
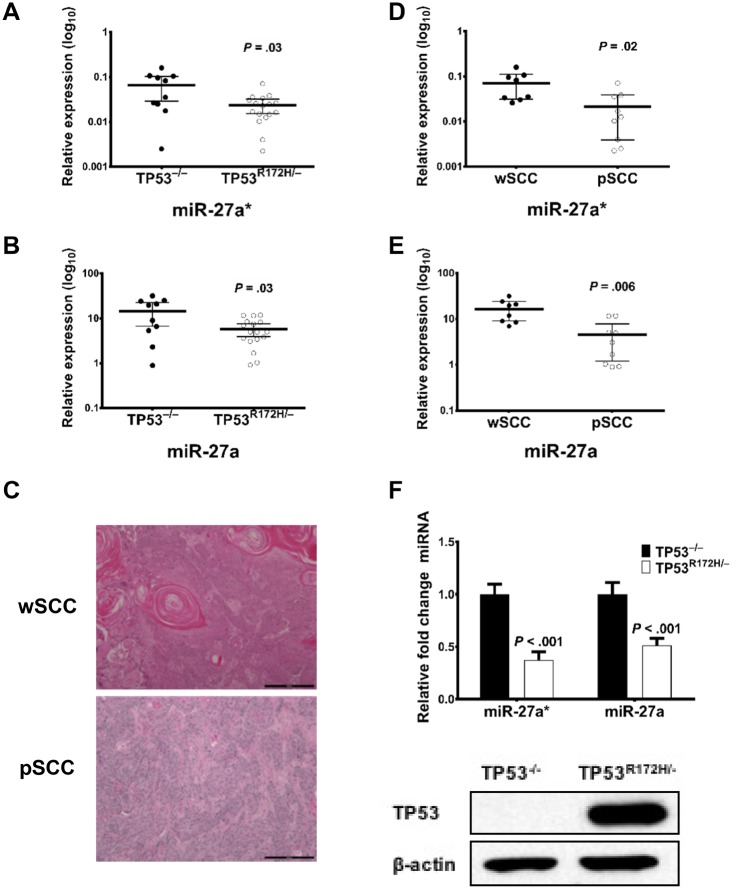
Mutant TP53 has been reported to promote tumor progression (24). Certain mutations are more disruptive in some cancers than others (25,26). We used the Evolutionary Action (EAp53) scoring system, which has been validated in HNSCC patient datasets, murine models, and in vitro studies (27,28). EAp53 stratifies tumors with TP53 mutations associated with poor outcomes—that is, high risk (HR)—from mutations with outcomes similar to patients with WT TP53—that is, low risk (LR). As has been previously published (29), we grouped the patient tumors in the HNSCC TCGA cohort into two clusters based on TP53 mutations: HR/other and LR/WT. miR-27a* expression levels are lower in the HR/other group compared with the LR/WT group in the HNSCC (WT/LR n = 210, HR/other n = 258; P = .005; Figure 4D), OCSCC (WT/LR n = 120, HR/other n = 165; P = .03; Figure 4E), and the HPV–OCSCC (WT/LR n = 107, HR/other n = 161; P = .04; Figure 4F) cohorts (Figure 4, D–F). To analyze the role of TP53 mutations on miR-27a* expression in SCCs in an in vivo setting, we analyzed miR-27a* expression in cutaneous SCCs that developed in mice generated by conditional activation of TP53 deletion (TP53–/–) or the TP53 R172H mutation (TP53R172H/–) in stratified epithelia, as previously described (30). qPCR analysis established lower levels of miR-27a* and miR-27a in tumors from TP53R172H/– mice than in tumors from TP53–/– mice (Figure 5, A and B) (miR-27a* log10 difference between means = −0.042, 95% CI = −0.07907 to −0.004923; two-sided Welch t test, P = .03; Figure 5A; miR-27a log10 difference between means = −8.708, 95% CI = −16.6 to −0.8168; two-sided Welch t test, P = .03; Figure 5B).
Figure 5.
TP53R172H/– activation in murine skin. Quantitative polymerase chain reaction (qPCR) analysis of (A) microRNA (miR)-27a* and (B) miR-27a levels in RNA extracted from tumors obtained from TP53–/– and TP53R172H/– mice. Error bars represent mean 95% confidence intervals. C) Hematoxylin and eosin stain of tumor sections showing a gross appearance of well-differentiated squamous cell carcinoma (wSCC) and poorly differentiated SCC (pSCC) (scale bars = 200 µm). Analysis of (D) miR-27a* and (E) miR-27a levels in wSCC tumors and pSCC tumors. Error bars represent 95% confidence intervals of the mean. P value determined using a two-sided Student t test. (F) Expression levels of TP53 protein and mRNA in cells generated from SCC obtained from TP53 –/– and TP53R172H/– mice. qPCR data were normalized to Sno234. Error bars represent mean (SEM).
Tumors generated in the TP53R172H/– and TP53–/– mice were either well to moderately differentiated (wSCC) or poorly differentiated SCCs (pSCC; Figure 5C), or spindle cell carcinomas in numbers as described previously (30). To examine whether levels of miR-27a* were associated with the differentiation status of the tumors, we grouped the tumors into two categories (wSCCs and pSCCs) irrespective of genotype and examined the miR-27a and miR-27a* levels in the two groups. We found that pSCCs had lower levels of miR-27a* and miR-27a than did wSCCs (Figure 5, D and E) (miR-27a* log10 difference between means = −0.05, 95% CI = −0.09139 to −0.007982; two-sided Welch t test, P = 0.02; Figure 5D; miR-27a log10 difference between means = −11.99, 95% CI = −19.66 to −4.32; two-sided Welch t test, P = .006; Figure 5E). Consistent with these findings, analysis of miR-27a* expression in cell lines generated from tumors showed that miR-27a* levels were lower in TP53R172H/– cells than in TP53–/– cells (Figure 5F).
Expression of miR-27a* and Survival
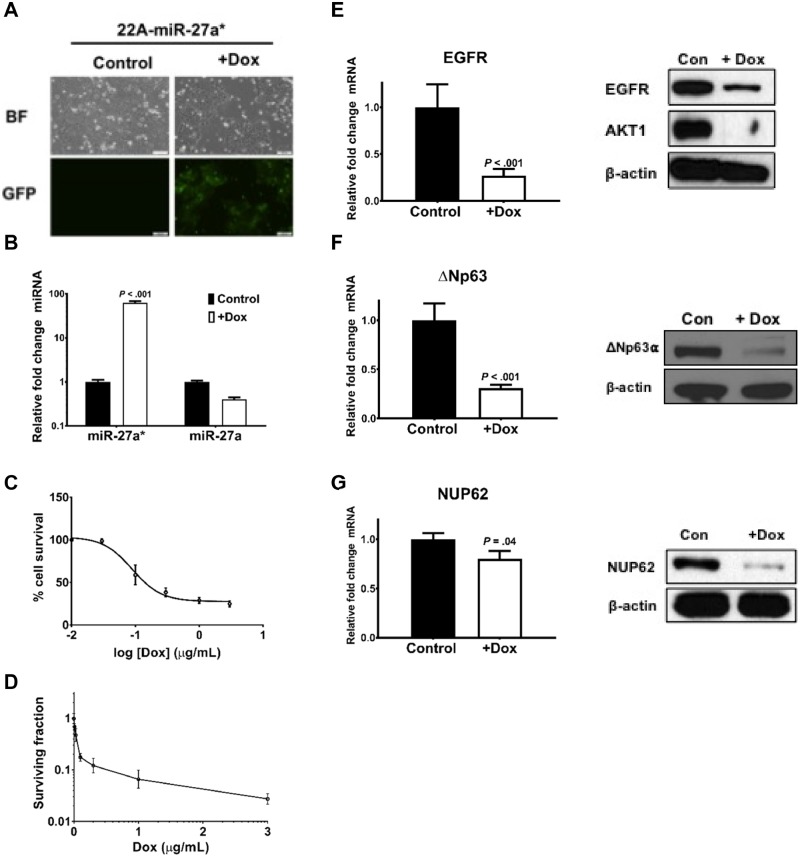
Viability of HNSCC cells is drastically affected by transient transfections of miR-27a* (14). To create a more controlled environment to study the biological activity of miR-27a*, we generated 22A cells with doxycycline-inducible miR-27a* (22A-miR-27a*) construct coexpressing green fluorescent protein. Addition of doxycycline to 22A-miR-27a* cells resulted in green fluorescent protein expression and an increase in levels of miR-27a* but not miR-27a (Figure 6, A and B). Increased miR-27a* expression resulted in a decrease in cell viability as measured by MTT and clonogenic assays (Figure 6, C and D; Supplementary Figure 4, available online).
Figure 6.
ΔNp63α expression levels following microRNA (miR)-27a* induction. (A) Green fluorescent protein (GFP) expression in the 22A-miR-27a* cells is induced only on the addition of doxycycline (Dox) (BF: brightfield) (scale bars = 100 µm). (B) Expression of miR-27a* and miR-27a following treatment with Dox as measured by quantitative polymerase chain reaction (qPCR). Cell viability assessed using the (C) MTT assay and (D) clonogenic assay following miR-27a* induction. Error bars represent mean (SD). Quantification of mRNA and protein levels of (E) epidermal growth factor receptor (EGFR), (F) ΔNp63, and (G) NUP62 as measured by qPCR and immunoblotting, respectively, on induction of miR-27a* in 22A-miR-27a* cells. Error bars represent mean (SEM). TATA-box binding protein (TBP) was used as endogenous control in qPCR. All statistical tests were two-sided.
Transcription factors are often targets of the miRNAs they transcriptionally regulate, forming regulatory feedback loops (31). Additionally, ΔNp63α has been shown to play an important role in promoting HNSCC cell survival, and lowered levels of ΔNp63α greatly affect cell viability (10). To assess the presence of a miR-27a*-ΔNp63α loop, we induced miR-27a* in 22A-miR-27a* cells, which resulted in a substantial decrease in not only EGFR, a previously identified miR-27a* target (14), but also ΔNp63 mRNA and ΔNp63α protein (Figure 6, E and F). Ectopic expression of miR-27a* mimic in HN31 cells resulted in similar findings (Supplementary Figure 5, available online).
We identified a novel miR-27a* target, NUP62, which regulates ΔNp63α transport to the nucleus. NUP62 promotes ΔNp63α activity and is overexpressed in SCC and HNSCC (Supplementary Figure 6, available online: normal n = 39, tumor n = 466; P < .001) (32). Loss of NUP62 does not affect total ΔNp63α protein levels, but has been shown to affect ΔNp63α levels in the nucleus of SCC cells, thereby affecting its ability to transcriptionally regulate target genes, inhibiting proliferation of SCC cells (32). Induction of miR-27a* expression in 22A-miR-27a* cells resulted in a decrease in NUP62 mRNA and protein (Figure 6G).
Multiple Components of the ΔNp63 Network as Direct miR-27a* Targets
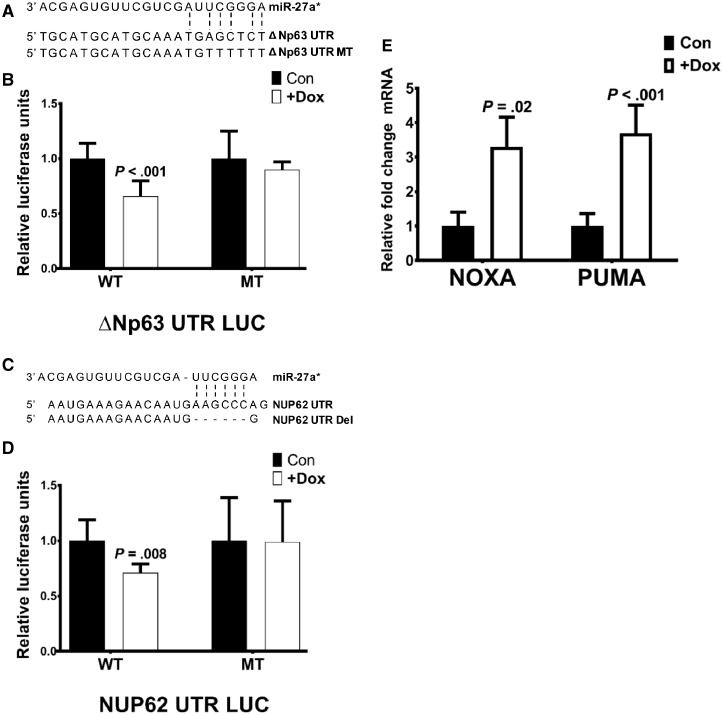
To validate ΔNp63α and NUP62 as direct targets of miR-27a*, we generated luciferase constructs to encompass the putative miR-27a* binding sites on target genes along with the respective companion mutant constructs with alterations and/or deletions of the miR-27a* binding site (Figure 7, A and C). Luciferase assays with the WT ΔNp63 (ΔNp63 UTR LUC) and NUP62 (NUP62 UTR LUC) constructs in 22A-miR-27a* cells showed a decrease in luciferase activity on induction of miR-27a*. This decrease in luciferase activity was not observed with the respective mutant constructs on miR-27a* induction because of abrogation of miR-27a* binding caused by mutation or deletion of the binding site (Figure 7, B and D).
Figure 7.
Interaction of microRNA (miR)-27a* with the ΔNp63α network. Schematic representation of the miR-27a* binding sites on the 3’UTR of (A) ΔNp63 and (C) NUP62 with the mutations (MT) or deletions (Del) generated in the respective miR-27a* binding site sequences. Luciferase assays with the (B) ΔNp63α and (D) NUP62 wild-type (WT) and MT UTR LUC constructs in 22A-miR-27a* cells. Renilla was used as a normalization control. Data represent the average of two independent experiments carried out in triplicate. Error bars represent mean (SD). E) Expression levels of ΔNp63α target genes NOXA and PUMA on induction of miR-27a* in 22A-miR-27a* cells were analyzed by quantitative polymerase chain reaction. Error bars represent mean (SEM). TATA-box binding protein (TBP) was used as endogenous control in qPCR. P value calculated using two-sided tests.
Maintenance of high levels of ΔNp63 is crucial to HNSCC survival because ΔNp63 represses the transcription of many genes involved in apoptosis and survival (10). Expression of miR-27a* not only affects ΔNp63α expression but also targets NUP62, essential to transport of ΔNp63α to the nucleus. To examine the effects of inhibition of ΔNp63α function on miR-27a* induction, we examined the expression of ΔNp63α-regulated genes involved in HNSCC survival in 22A-miR-27a* cells. An increase in NOXA and PUMA, proapoptotic genes repressed by ΔNp63, was observed on miR-27a* induction (Figure 7E). This suggests that miR-27a* inhibits ΔNp63α function, including its ability to bind to the promoters of genes and transcriptionally regulate them.
Discussion
In the current study, we confirmed repression of miR-27a* expression in tumors from patients with OCSCC. Furthermore, we identified two particular features of miR-27a* expression: 1) increased expression of miR-27a* was associated with survival in OPSCC and HPV+ HNSCC patients and 2) repression of miR-27a* was associated with poorly differentiated and advanced HPV+ tumors. Introduction of miR-27a* induces cell death in HNSCC (14) and prostate cancer (14,33) and sensitizes multiple myeloma cells to bortezomib (34), demonstrating the therapeutic potential for miR-27a*.
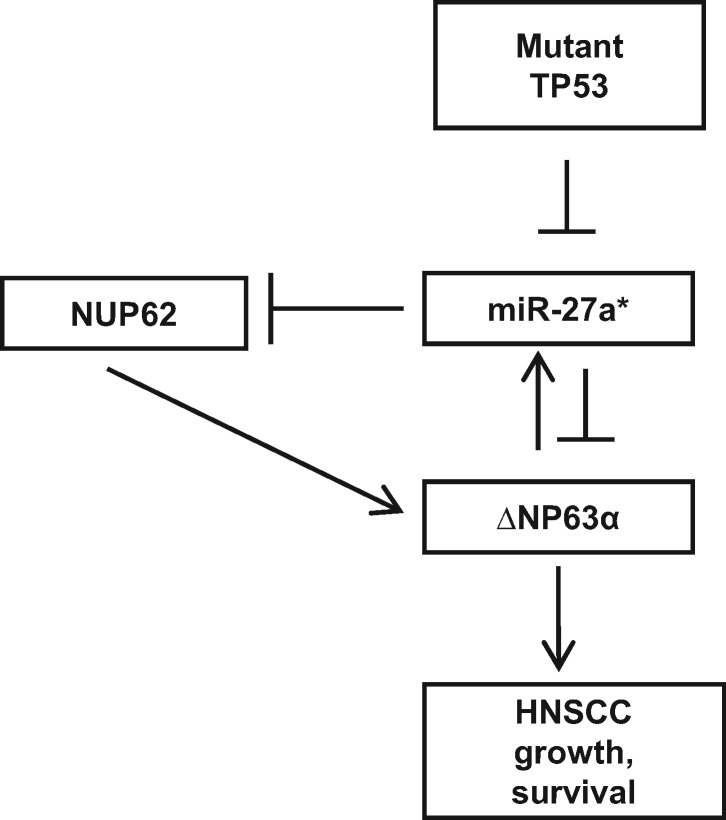
Our studies to elucidate the mechanism underlying miR-27a* expression identified ΔNp63α as a transcriptional activator of miR-27a*. Knockdown experiments using siRNA targeting either all TP63 isoforms or only ΔNp63 isoforms suggest that the TA isoforms of TP63, present at reduced levels in HNSCC, are not involved (35). We also found that miR-27a* directly targets ΔNp63α. This work supports a feedback loop that balances levels of the prosurvival ΔNp63α and proapoptotic miR-27a*, maintaining cellular homeostasis (Figure 8). Additionally, miR-27a* targets NUP62, a protein essential for nuclear translocation of ΔNp63α in SCCs and overexpressed in HNSCC (32). Thus, in addition to ΔNp63α, miR-27a* targets NUP62, which is essential for ΔNp63α function and might have an independent role as an oncogene in SCC progression (Figure 8) (32). The ΔNp63α-miR-27a* balance is disrupted in HNSCCs by mutations in TP53 (Figure 8). Knockdown of mutant TP53 in HNSCC cells resulted in an increase in miR-27a* expression; similarly, levels of miR-27a* were lower in tumor cells engineered to express mutant TP53, suggesting that mutant TP53 represses miR-27a* expression. This repressive effect is also seen in TCGA, in which disruptive TP53 mutations are shown to repress miR-27a* expression in OCSCC, including HPV-OCSCC, which has a very poor prognosis. This suggests that mutant TP53 probably disrupts ΔNp63 control over target genes as has been previously observed (36).
Figure 8.
Schematic of microRNA (miR)-27a* regulation in head and neck squamous cell carcinoma (HNSCC). ΔNp63α transcriptionally upregulates miR-27a*. ΔNp63α is a direct target of miR-27a*, forming a feedback loop. NUP62 regulates ΔNp63α protein transport into the nucleus and is a direct target of miR-27a*. Mutant TP53 disrupts the ΔNp63α-miR-27a* autoregulatory loop by repressing miR-27a*.
Furthermore, the ability of mutant TP53 to lower miR-27a* levels was also observed in the gain-of-function mutant TP53R172H/– mouse model, which developed cutaneous SCCs; lower levels of miR-27a* were found in tumors from TP53R172H/– mice than in tumors from TP53–/– mice. An interesting finding is that regardless of genomic background, pSCCs, which were the only tumors that developed metastases, had lower levels of miR-27a* than did wSCCs. This finding complements our findings in HPV+ patient tumors in which advanced-stage tumors had reduced levels of miR-27a*. Cutaneous SCC, like HNSCC, presents with high levels of ΔNp63α and a preponderance of TP53 mutations, suggesting that our findings might have a broader relevance to other SCCs beyond HNSCC.
A number of transcriptional and posttranscriptional programs tightly control ΔNp63α levels in tissues (37–39). Enforced expression of ΔNp63α in murine in vivo models promotes tumorigenesis, whereas deletion of ΔNp63α results in apoptosis and regression of tumors, suggesting that survival in SCCs is dependent on ΔNp63α (40). The current lack of ΔNp63α-targeting molecules precludes testing this hypothesis in patients. Disruption of the miR-27a*-ΔNp63α loop has clinical relevance because ΔNp63α mediates cisplatin resistance via AKT1 upregulation (41) and further regulates a miRNA program that mediates cisplatin response in HNSCCs (42). There is a high overlap between mutant TP53 and TP63-binding sites (43), with studies suggesting that mutant TP53 uses ΔNp63α as a chaperone to affect transcriptional programs and promote invasion (44). Mutant TP53 suppression of miR-27a* could represent a mechanism that allows ΔNp63α accumulation.
Advanced HNSCC tumors can have mutations in multiple components of the same pathway, as observed in the PI3K/AKT1/mTOR pathway (6). This has led to evaluation of simultaneous targeting of EGFR and mTOR in HNSCC preclinical models with promising results (45,46). A PanCancer Atlas study across multiple tumor types in the TCGA, which aimed at identifying features distinguishing SCC, found a number of important molecular pathways relevant to SCCs (35). A notable genomic feature, typical of SCC, is increased expression of ΔNp63α. Copy number alterations in several components of the PI3K/AKT1/mTOR axis across several chromosomes were also a characteristic feature of SCCs compared with other tumor types. Additional mutations were found in other genes that enhance oncogenic potential of the PI3K and TP63 networks. Alterations in TP63 in SCCs can affect therapeutic response and disease progression (35). Overexpression of ΔNp63α was found to be associated with alterations in PI3KCA, SOX2, and ACTLA6 pathways, which are involved in growth, stemness, and survival (35). All of this suggests that in addition to targeting multiple components of a single pathway, targeting of a focused subset of pathways may be required to overcome adaptive and innate resistance in HNSCCs.
Outcomes for HPV–HNSCC patients with advanced disease remain poor (17). Even with the advent of EGFR-targeted therapies, cisplatin remains a mainstay in the treatment of advanced HNSCC (47). TP53 mutations can often be indicative of cisplatin resistance (8,27). As we have comprehensively demonstrated in the current study, miR-27a* targets multiple components of the ΔNp63α pathway essential to its oncogenic role in SCC, in addition to the ΔNp63α protein itself. Our earlier work has clearly detailed that multiple components of the PI3K/AKT1/mTOR pathway (AKT1 and mTOR) and EGFR are direct targets of miR-27a*, causing tumor inhibition in murine orthotopic xenograft models (14). Although we have defined a set of miR-27a* target proteins essential to HNSCC viability, further work is needed to validate other targets that might be important in HNSCC biology.
In conclusion, although others have shown that mutant TP53 affects miRNA function in other tumor types (48,49), to our knowledge, this study for the first time provides a regulatory framework of miR-27a* expression in HNSCC. miR-27a* connects and regulates the TP63/TP53 pathways and mTOR/AKT1 networks, which are fundamental to SCC progression. The importance of miR-27a*-regulated targets necessitates repression of miR-27a* for tumor survival. Thus, our study presents not only a regulatory model for miR-27a* expression but also a new modality to disrupt multiple oncogenic pathways essential to tumor survival, potentially having a profound impact on HNSCC therapy.
Funding
This work was partially supported by grants from the Brandon C. Gromada Head and Neck Cancer Foundation, Elsa U. Pardee Foundation, Institutional Startup Funds, and The University of Texas MD Anderson Cancer Center-Oropharynx Cancer Program generously supported by Mr and Mrs Charles W. Stiefel awarded to SYL. This work was supported in part by National Institutes of Health/National Cancer Institute Cancer Center Support Grant grant P30CA016672, including CBP (Biostatistics Shared Resource).
Notes
The authors declare no conflicts of interest.
Conception and design: SYL and NSC. Acquisition of data (provided reagent, acquired and managed patients, provided facilities, etc): NSC, AAP, AAO, JNM, MDW, XL, JL, CC, GAC. Analysis and interpretation of data (eg, statistical analysis, biostatistics, computational analysis): NSC, CI, CBP, SYL. NSC and SYL wrote the manuscript, and all authors were involved in manuscript review.
We would like to acknowledge the Department of Scientific Publications at The University of Texas MD Anderson Cancer Center for review of the manuscript.
Supplementary Material
References
- 1. Cohen RB. Current challenges and clinical investigations of epidermal growth factor receptor (EGFR)- and ERBB family-targeted agents in the treatment of head and neck squamous cell carcinoma (HNSCC). Cancer Treat Rev. 2014;40(4):567–577. [DOI] [PubMed] [Google Scholar]
- 2. Leemans CR, Braakhuis BJ, Brakenhoff RH.. The molecular biology of head and neck cancer. Nat Rev Cancer. 2011;11(1):9–22. [DOI] [PubMed] [Google Scholar]
- 3. Fojo T, Parkinson DR.. Biologically targeted cancer therapy and marginal benefits: are we making too much of too little or are we achieving too little by giving too much? Clin Cancer Res. 2010;16(24):5972–5980. [DOI] [PubMed] [Google Scholar]
- 4. Pickering CR, Zhang J, Yoo SY, et al. Integrative genomic characterization of oral squamous cell carcinoma identifies frequent somatic drivers. Cancer Discov. 2013;3(7):770–781. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Agrawal N, Frederick MJ, Pickering CR, et al. Exome sequencing of head and neck squamous cell carcinoma reveals inactivating mutations in Notch1. Science. 2011;333(6046):1154–1157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Lui VW, Hedberg ML, Li H, et al. Frequent mutation of the PI3K pathway in head and neck cancer defines predictive biomarkers. Cancer Discov. 2013;3(7):761–769. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Ory B, Ellisen LW.. A microRNA-dependent circuit controlling p63/p73 homeostasis: P53 family cross-talk meets therapeutic opportunity. Oncotarget. 2011;2(3):259–264. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Poeta ML, Manola J, Goldwasser MA, et al. Tp53 mutations and survival in squamous-cell carcinoma of the head and neck. N Engl J Med. 2007;357(25):2552–2561. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Balz V, Scheckenbach K, Gotte K, et al. Is the p53 inactivation frequency in squamous cell carcinomas of the head and neck underestimated? Analysis of p53 exons 2-11 and human papillomavirus 16/18 e6 transcripts in 123 unselected tumor specimens. Cancer Res. 2003;63(6):1188–1191. [PubMed] [Google Scholar]
- 10. Rocco JW, Leong CO, Kuperwasser N, et al. P63 mediates survival in squamous cell carcinoma by suppression of p73-dependent apoptosis. Cancer Cell. 2006;9(1):45–56. [DOI] [PubMed] [Google Scholar]
- 11. Bartel DP. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116(2):281–297. [DOI] [PubMed] [Google Scholar]
- 12. Ma L. MicroRNA and metastasis. Adv Cancer Res. 2016;132:165–207. https://doi.org/10.1016/bs.acr.2016.07.004 [DOI] [PubMed] [Google Scholar]
- 13. Haier J, Strose A, Matuszcak C, et al. Mir clusters target cellular functional complexes by defining their degree of regulatory freedom. Cancer Metastasis Rev. 2016;35(2):289–322. [DOI] [PubMed] [Google Scholar]
- 14. Wu X, Bhayani MK, Dodge CT, et al. Coordinated targeting of the EGFR signaling axis by microRNA-27a*. Oncotarget. 2013;4(9):1388–1398. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15. Molinolo AA, Marsh C, El Dinali M, et al. mTOR as a molecular target in HPV-associated oral and cervical squamous carcinomas. Clin Cancer Res. 2012;18(9):2558–2568. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Molinolo AA, Hewitt SM, Amornphimoltham P, et al. Dissecting the AKT/mammalian target of rapamycin signaling network: emerging results from the head and neck cancer tissue array initiative. Clin Cancer Res. 2007;13(17):4964–4973. [DOI] [PubMed] [Google Scholar]
- 17. Vigneswaran N, Williams MD.. Epidemiologic trends in head and neck cancer and aids in diagnosis. Oral Maxillofac Surg Clin North Am. 2014;26(2):123–141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Gillison ML, Koch WM, Capone RB, et al. Evidence for a causal association between human papillomavirus and a subset of head and neck cancers. J Natl Cancer Inst. 2000;92(9):709–720. [DOI] [PubMed] [Google Scholar]
- 19. Ory B, Ramsey MR, Wilson C, et al. A microrna-dependent program controls p53-independent survival and chemosensitivity in human and murine squamous cell carcinoma. J Clin Invest. 2011;121(2):809–820. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20. Raimondi I, Ciribilli Y, Monti P, et al. P53 family members modulate the expression of PRODH, but not PRODH2, via intronic p53 response elements. PLoS One. 2013;8(7):e69152.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. Sniezek JC, Matheny KE, Westfall MD, et al. Dominant negative p63 isoform expression in head and neck squamous cell carcinoma. Laryngoscope. 2004;114(12):2063–2072. [DOI] [PubMed] [Google Scholar]
- 22. Rosenbluth JM, Johnson K, Tang L, et al. Evaluation of p63 and p73 antibodies for cross-reactivity. Cell Cycle. 2009;8(22):3702–3706. [DOI] [PubMed] [Google Scholar]
- 23. Ferraiuolo M, Di Agostino S, Blandino G, et al. Oncogenic intra-p53 family member interactions in human cancers. Front Oncol. 2016;6(77):doi.org/10.3389/fonc.2016.00077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Oren M, Rotter V.. Mutant p53 gain-of-function in cancer. Cold Spring Harb Perspect Biol. 2010;2(2):a001107.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Robles AI, Harris CC.. Clinical outcomes and correlates of TP53 mutations and cancer. Cold Spring Harb Perspect Biol. 2010;2(3):a001016. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26. Stengel A, Kern W, Haferlach T, et al. The impact of TP53 mutations and TP53 deletions on survival varies between AML, ALL, MDS and CLL: an analysis of 3307 cases. Leukemia. 2017;31(3):705–711. [DOI] [PubMed] [Google Scholar]
- 27. Osman AA, Neskey DM, Katsonis P, et al. Evolutionary action score of TP53 coding variants is predictive of platinum response in head and neck cancer patients. Cancer Res. 2015;75(7):1205–1215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Neskey DM, Osman AA, Ow TJ, et al. Evolutionary action score of TP53 identifies high-risk mutations associated with decreased survival and increased distant metastases in head and neck cancer. Cancer Res. 2015;75(7):1527–1536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29. Sandulache VC, Michikawa C, Kataria P, et al. High-risk TP53 mutations are associated with extranodal extension in oral cavity squamous cell carcinoma. Clin Cancer Res. 2018;24(7):1727–1733. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Li Z, Gonzalez CL, Wang B, et al. Cdkn2a suppresses metastasis in squamous cell carcinomas induced by the gain-of-function mutant p53(r172h). J Pathol. 2016;240(2):224–234. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Tsang J, Zhu J, van Oudenaarden A.. Microrna-mediated feedback and feedforward loops are recurrent network motifs in mammals. Mol Cell. 2007;26(5):753–767. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Hazawa M, Lin DC, Kobayashi A, et al. ROCK-dependent phosphorylation of NUP62 regulates p63 nuclear transport and squamous cell carcinoma proliferation. EMBO Rep. 2018;19(1):73–88. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33. Barros-Silva D, Costa-Pinheiro P, Duarte H, et al. MicroRNA-27a-5p regulation by promoter methylation and MYC signaling in prostate carcinogenesis. Cell Death Dis. 2018;9(2):167.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34. Ballabio E, Armesto M, Breeze CE, et al. Bortezomib action in multiple myeloma: microrna-mediated synergy (and miR-27a/CDK5 driven sensitivity)? Blood Cancer J. 2012;2(8):e83.. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Campbell JD, Yau C, Bowlby R, et al. Genomic, pathway network, and immunologic features distinguishing squamous carcinomas. Cell Rep. 2018;23(1):194–212.e196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Cordani N, Pozzi S, Martynova E, et al. Mutant p53 subverts p63 control over KLF4 expression in keratinocytes. Oncogene. 2011;30(8):922–932. [DOI] [PubMed] [Google Scholar]
- 37. Chari NS, Romano RA, Koster MI, et al. Interaction between the TP63 and SHH pathways is an important determinant of epidermal homeostasis. Cell Death Differ. 2013;20(8):1080–1088. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Yoh K, Prywes R.. Pathway regulation of p63, a director of epithelial cell fate. Front Endocrinol (Lausanne). 2015;6:51. doi.org/10.3389/fendo.2015.00051 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Lena AM, Shalom-Feuerstein R, Rivetti di Val Cervo P, et al. miR-203 represses ‘stemness’ by repressing DeltaNp63. Cell Death Differ. 2008;15(7):1187–1195. [DOI] [PubMed] [Google Scholar]
- 40. Ramsey MR, Wilson C, Ory B, et al. FGRF2 signaling underlies p63 oncogenic function in squamous cell carcinoma. J Clin Invest. 2013;123(8):3525–3538. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Sen T, Sen N, Brait M, et al. Deltanp63alpha confers tumor cell resistance to cisplatin through the AKT1 transcriptional regulation. Cancer Res. 2011;71(3):1167–1176. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Huang Y, Chuang A, Hao H, et al. Phospho-deltanp63alpha is a key regulator of the cisplatin-induced micrornaome in cancer cells. Cell Death Differ. 2011;18(7):1220–1230. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Martynova E, Pozzi S, Basile V, et al. Gain-of-function p53 mutants have widespread genomic locations partially overlapping with p63. Oncotarget. 2012;3(2):132–143. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Neilsen PM, Noll JE, Suetani RJ, et al. Mutant p53 uses p63 as a molecular chaperone to alter gene expression and induce a pro-invasive secretome. Oncotarget. 2011;2(12):1203–1217. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Wang Z, Martin D, Molinolo AA, et al. mTOR co-targeting in cetuximab resistance in head and neck cancers harboring PIK3CA and RAS mutations. J Natl Cancer Inst. 2014;106(9):dju215. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Janku F, Yap TA, Meric-Bernstam F.. Targeting the PI3K pathway in cancer: are we making headway? Nat Rev Clin Oncol. 2018;15(5):273–291. [DOI] [PubMed] [Google Scholar]
- 47. Pignon JP, Le Maître A, Maillard E, et al. Meta-analysis of chemotherapy in head and neck cancer (MACH-NC): an update on 93 randomised trials and 17, 346 patients. Radiother Oncol. 2009;92(1):4–14. [DOI] [PubMed] [Google Scholar]
- 48. Jones M, Lal A.. MicroRNAs, wild-type and mutant p53: more questions than answers. RNA Biol. 2012;9(6):781–791. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Wang W, Cheng B, Miao L, et al. Mutant p53-R273H gains new function in sustained activation of EGFR signaling via suppressing miR-27a expression. Cell Death Dis. 2013;4:e574 https://www.nature.com/articles/cddis201397 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.