Abstract
Cholangiocarcinoma (CCA) is a primary malignancy, which is often diagnosed as locally advanced or metastatic. Previous studies have revealed genomic characteristics of CCA in Western patients, however comprehensive genomic features of CCA in Chinese patients have not been well understood. To explore the specific genomic characteristics of Chinese patients with CCA, a total of 66 patients with CCA, including 44 intrahepatic CCA (iCCA) and 22 extrahepatic CCA (exCCA) cases, were studied. The most commonly altered genes in CCAs were TP53 (62.12%, 41/66), KRAS (36.36%, 24/66), SMAD4 (24.24%, 16/66), TERT (21.21%, 14/66), ARID1A (19.70%, 13/66), CDKN2A (19.70%, 13/66), KMT2C (9.09%, 6/66) and RBM10 (9.09%, 6/66), ERBB2 (7.58%, 5/66) and BRAF (7.58%, 5/66). Many gene mutations, including STK11, CCND1 and FGF19, were only found in iCCA. RBM10 mutations were found to be significantly higher in exCCA. The gene mutations of neurofibromin 1, STK11, CCND1 and FBXW7 specifically occurred in males, whereas gene mutations of ERBB2, AXIN2 and CREBBP specifically occurred in females. ERBB2 mutations were significantly associated with the sex of patients with CCA. Mutations in PIK3CA, FGFR2 and ZNF750 were significantly associated with the age of patients with CCA and TERT mutations were significantly associated with tumor differentiation. Alterations in KMT2C, PBRM1, AXIN2, MAGI2, BRCA2 and SPTA1 were associated with tumor mutational burden. The findings of the present study suggest that targeted sequencing, using next-generation sequencing technology, provides comprehensive and accurate information on genomic alterations, which will provide novel potential biomarkers for the diagnosis of CCA and may guide precise therapeutic strategies for Chinese patients with CCA.
Keywords: comprehensive genomic profile, cholangiocarcinoma, tumor mutational burden, biomarker
Introduction
Cholangiocarcinoma (CCA) is a primary malignancy originating from the intrahepatic (iCCA) or extrahepatic (exCCA) bile duct epithelium (1). Although the incidence of exCCA remains stable, the incidence of iCCA has increased over the last 15 years, worldwide (1). Due to atypical clinical presentation and imaging, CCA is often diagnosed at advanced stages (2). CCA is a multifactorial disease based on a combination of genetic and environmental factors (3). Some biomarkers are considered to be an effective strategy for the diagnosis and treatment of cancer, however new biomarkers for CCA still need to be developed and explored for earlier identification and diagnosis.
Next-generation sequencing (NGS) technology provides a more accurate and efficient genome sequencing method and has enabled large-scale cancer genomics projects, such as The Cancer Genome Atlas (4) and the International Cancer Genome Consortium (5), to characterize the cancer genome and genomic alterations. The identification and exploration of cancer-associated mutations has been greatly facilitated by NGS, due to the success of the aforementioned projects. Genomic sequencing studies in CCA have identified many driver gene alterations, such as in ELF3, ARID1 and ARID2 (6). Lin et al (7) previously screened survival-associated genes of CCA and found that genes were significantly enriched in the Wnt signaling pathway, the apoptotic process and a number of oncogenic pathways, which may be altered in patients with poorer survival. The target genes SGSH, EIF5A, BET1L, GCNT4 and PLCG2 were identified, which may be associated with the prognosis of CCA (8). Furthermore, mutation profiling of iCCA and exCCA was identified by NGS, and notable differences included IDH1 mutations, which exclusively occurred in iCCA, and ERBB2 mutations, which occurred in exCCA (9). Moreover, KRAS mutations and the MAP/ERK pathway were significantly associated with progression-free survival (PFS) in iCCA, whereas BAP1 mutations and aberrations in the fibroblast growth factor (FGF) pathway were significantly correlated with PFS in exCCA (9). Based on the comprehensive molecular profiling of 194 patients with CCA, including Caucasian, Asian and African American patients, Lowery et al (10) demonstrated that almost 50% of the patients were accompanied by therapeutic somatic alterations. These studies indicate that molecular profiling can facilitate biomarker-based clinical trials in patients with CCA. Although several studies have revealed the genomic characterization of CCA in Western patients, the comprehensive genomic features of CCA in Chinese patients have not been well understood.
The present study characterized the comprehensive genomic features of 66 cases of Chinese patients with CCA by using NGS, and aimed to identify the specific biomarkers for early diagnosis and prognosis, and for the development of potential therapeutic targets for CCA.
Materials and methods
Patient enrollment and sample collection
Between December 2017 and March 2019, a total of 66 Chinese patients with CCA, aged between 43 and 82 years, (mean age of 62.38 years), including 45 males and 21 females, were enrolled from two hospitals located in North China, Tianjin Medical University General Hospital (3 cases) and the Affiliated Hospital of Qingdao University (63 cases). Informed consent was obtained in writing from each patient. Formalin-fixed paraffin-embedded (FFPE) tumor tissues and matched blood samples were collected and transferred to OrigiMed, Shanghai for genetic variation detection. Genomic DNA was prepared by using the QIAamp DNA FFPE Tissue kit and QIAamp DNA Blood Midi kit (Qiagen GmbH), according to the manufacturer's instructions. The concentration of DNA was measured by Qubit and normalized to 20–50 ng/µl.
Identification of genomic alterations and tumor mutational burden (TMB)
The genomic information was produced using the NGS-based YuanSu™450 gene panel (OrigiMed), which covers all the coding exons of 450 cancer-associated genes and 64 selected introns in 39 genes that are frequently rearranged in solid tumors. The genes were captured and sequenced with a mean depth of 800X, using Illumina NextSeq 500 (Illumina, Inc.). Genomic alterations (GAs) were identified by the alignment of sequences from tumor tissues and matched blood samples, following previously reported methods (11). Single nucleotide variants (SNVs) were identified using MuTect (v1.7) (12). Insertion-deletion polymorphisms (Indels) were identified using PINDEL (v0.2.5) (13). The functional impact of each GA was annotated by SnpEff v3.0 (14). Copy number variation (CNV) regions were identified using Control-FREEC (v9.7) (15) with the following parameters: Window=50,000 and step=10,000. Gene fusions were detected through an in-house pipeline. In brief, aligned reads with a distance of over 2,000 bp or 0 bp (where 2 aligned reads are located on different chromosomes) were collected and used as discordant reads. Discordant reads with a distance <500 bp formed clusters that were further assembled to identify potential rearrangement breakpoints. The breakpoints were reconfirmed using the BLAST-like alignment tool (http://genome.cse.ucsc.edu/cgi-bin/hgBlat) and the resulting chimeric gene candidates were annotated. Gene rearrangements were assessed by Integrative Genomics Viewer (16). TMB was estimated by counting the somatic mutations, including SNVs and Indels, per megabase of the sequence examined in each patient. Driver mutations and known germline alterations were not counted.
Statistical analysis
Statistical analyses were performed using SPSS version 22.0 (SPSS Inc.). Fisher's exact test was used to analyze the significance of the differences. Mann-Whitney U test from the Wilcox.test of R basic package (v3.5.0; http://cran.r-project.org/src/base/R-3/) was used to analyze the association between gene variations and TMB. P<0.05 was considered indicate a statistically significant difference.
Results
Clinical characteristics of patients with CCA
The cohort of 66 Chinese patients with CCA included 45 males and 21 females, with a median age of 62 years (range, 43–82 years). The CCA samples included 44 (66.67%) iCCA and 22 (33.33%) exCCA cases, of which 64 (96.97%) cases were primary tumors from the bile duct epithelium and 2 (3.03%) cases were metastatic CCA samples from the liver and abdominal cavity. Clinical or pathological information was obtained and is presented in Table I. Histologically, 35 (53.03%) samples were well/moderately differentiated and 24 (36.36%) samples were poorly/undifferentiated. Histological information was not available for 7 (10.61%) samples (Table I).
Table I.
Clinicopathologic features of Chinese cholangiocarcinoma cohort.
| Characteristics | Number |
|---|---|
| Sex, (%) | |
| Male | 45 (68.18) |
| Female | 21 (31.82) |
| Median age, years (range) | 62 (43–82) |
| Median TMB, (range) | 5.40 (0–36.7) |
| Sample, (%) | |
| Primary | 64 (96.97) |
| Metastases | 2 (3.03) |
| Histology (%) | |
| Intrahepatic cholangiocarcinoma | 44 (66.67) |
| Extrahepatic cholangiocarcinoma | 22 (33.33) |
| Histological grade (%) | |
| Well/moderately differentiated | 35 (53.03) |
| Poorly/undifferentiated | 24 (36.36) |
| Not available | 7 (10.61) |
TMB, tumor mutational burden.
Genomic alterations in CCA
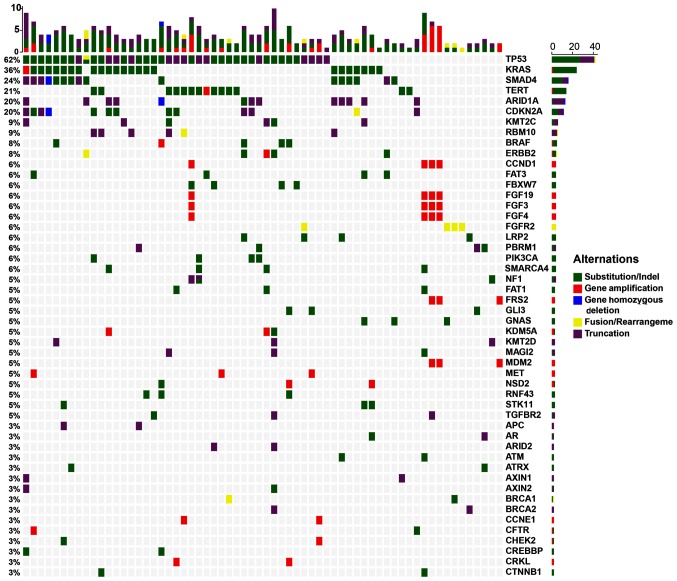
A total of 431 clinically relevant GAs were identified using NGS sequencing targeting 450 cancer genes, with a mean of 2.58 alterations per sample (range, 0–42) in 176 genes (Fig. 1). Of the 431 GAs, 316 (73.32%) were SNV/ShortIndel, 96 (22.27%) were CNV, 10 (2.32%) were fusion and 9 (2.09%) were LongIndel (Fig. 1; Table SI). The most frequent GAs were found in TP53 (62.12%, 41/66), KRAS (36.36%, 24/66), SMAD4 (24.24%, 16/66), TERT (21.21%, 14/66), ARID1A (19.70%, 13/66), CDKN2A (19.70%, 13/66), KMT2C (9.09%, 6/66), RBM10 (9.09%, 6/66, ERBB2 (7.58%, 5/66) and BRAF (7.58%, 5/66). All other gene mutations were detected <5 times (Table SI). Most genes were mutated once, whereas a few genes, including STK11, NF1, SMAD4, TP53, LRP1B, KEAP1 and ATM, had 2 or 3 mutations. Among them, NF1 and SMAD4 had multiple mutation sites and were detected in 2 cases of CCA. The most common GAs, TP53 and KRAS, were detected in 17 cases (25.76%). TP53 was detected in a total of 42 GAs in 41 CCA samples, including 41 SNV and 1 gene-fusion. Of these GAs, R248Q was detected in 4 samples, R273C (L), R342*, R213*, R175H, and 993 + 1G > A (splice site) were detected in 2 samples, and the rest were detected only in 1 sample (Table SII). KRAS GAs were detected in 24 samples, including 23 SNV and 1 amplification. Among them, substitution of the 12th amino acid was detected 15 times, including 6 G12D, 6 G12V, 1 G12C, 1 G12R, and 1 G12S. Substitution of the 13th amino acid was detected 5 times (2 G13C and 3 G13D), substitution of the 61st amino acid mutation was detected twice (Q61H), substitution of the 146th amino acid (A146P) was detected once and gene amplification was detected once (Table SII).
Figure 1.
Most common genomic alterations of 66 cholangiocarcinoma. The x-axis represents each case sample and the y-axis represents each mutated gene. The top bar graph shows the gene mutation frequency of each sample, the percentages on the left represents the mutation frequency of each mutated gene, and the bar graph on the right shows the number of each mutated gene. Green represents substitution/Indel mutations, red represents gene amplification mutations, blue represents gene homozygous deletion mutations, yellow represents fusion/rearrangement mutations, and purple represents truncation mutations.
Genomic alterations in iCCA and exCCA
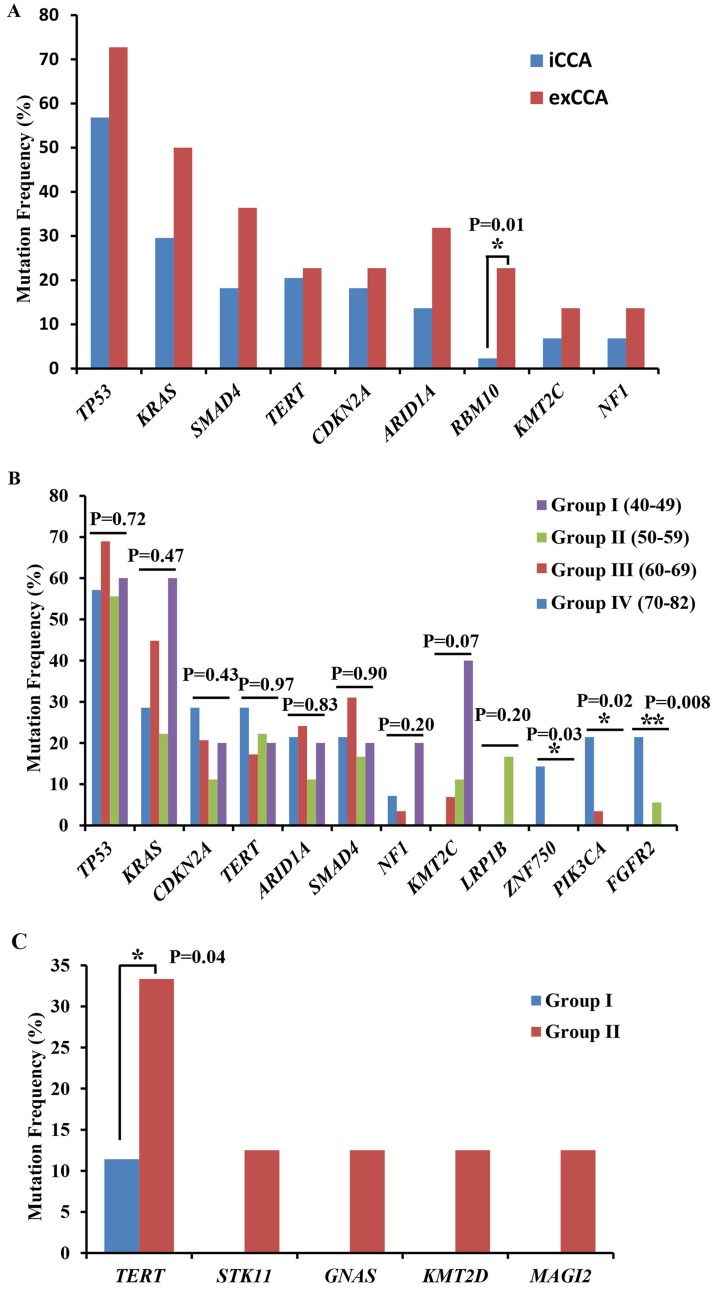
In 44 iCCA patients, 290 GAs from 145 genes were detected. The most common gene mutations in iCCA were TP53 (56.82%, 25/44), KRAS (29.55%, 13/44), SMAD4 (18.18%, 8/44), TERT (20.45%, 9/44), CDKN2A (18.18%, 8/44) and ARID1A (13.64%, 6/44) (Fig. 2A; Table SIII). By contrast, in 22 exCCA patients, a total of 141 GAs from 70 genes were detected. The most common mutations were TP53 (72.73%, 16/22), KRAS (50%, 11/22), SMAD4 (36.36%, 8/22), ARID1A (31.82%, 7/22), CDKN2A (22.73%, 5/22), RBM10 (22.73%, 5/22) and TERT (22.73%, 5/22) (Fig. 2A; Table SIII). The mutations of TP53, KRAS, SMAD, CDKN2A, TERT and ARID1A were common in both iCCA and exCCA. The mutation frequency of RBM10 was significantly higher in exCCA compared with iCCA (5/22 vs. 1/44, respectively; P=0.01). Other gene mutations, such as STK11 (6.82%, 3/44), CCND1, FGF19, FGF3, FGF4, FGFR2 and PBRM1 (all 9.09%, 4/44), were found only in iCCA (Table SIII).
Figure 2.
Most common mutated genes (>10%) in different patient groups. Most common mutated genes in (A) iCCA and exCCA, (B) different age groups, and (C) among the differentiated tumor groups (Group I, well/moderately differentiated CCA; Group II: Poorly/undifferentiated CCA). *P<0.05, **P<0.01. iCCA, intrahepatic cholangiocarcinoma; exCCA, extrahepatic cholangiocarcinoma.
Genomic alterations of CCA in male and female patients
In 45 male patients with CCA, 279 mutations were detected in 134 genes, and the most common mutated genes were TP53 (60%, 27/45), KRAS (33.33%, 15/45), TERT (24.44%, 11/45), ARID1A (20%, 9/45), CDKN2A (15.56%, 7/45) and SMAD4 (15.56%, 7/45) (Table SIV). In 21 female patients, 152 mutations were detected in 89 genes. The most common mutations were TP53 (66.67%, 14/21), SMAD4 (42.86%, 9/21) and KRAS (42.86%, 9/21), CDKN2A (28.57%, 6/21), ERBB2 (23.81%, 5/21), ARID1A (19.05%, 4/21) and KMT2C (19.05%, 4/21), RBM10 (14.29%, 3/21) and TERT (14.29%, 3/21) (Table SIV). Gene mutations in CCND1, FBXW7, FGF3/4/19, PIK3C1, NF1 and STK11 were specific to male patients (Table II), whereas gene mutations in ERBB2, AXIN2, CREBBP, ERBB3, MTOR and MYB were specific to female patients (Table II).
Table II.
Specific mutated genes in male and female patients with cholangiocarcinoma.
| Gene | Sex | Mutant number | Male/female ratio, % | P-value |
|---|---|---|---|---|
| ERBB2 | Female | 5 | 23.81 | 0.006331 |
| AXIN2 | Female | 2 | 9.52 | 0.140075 |
| CREBBP | Female | 2 | 9.52 | 0.140075 |
| ERBB3 | Female | 2 | 9.52 | 0.140075 |
| MTOR | Female | 2 | 9.52 | 0.140075 |
| MYB | Female | 2 | 9.52 | 0.140075 |
| CCND1 | Male | 4 | 8.89 | 0.293039 |
| FBXW7 | Male | 4 | 8.89 | 0.293039 |
| FGF19 | Male | 4 | 8.89 | 0.293039 |
| FGF3 | Male | 4 | 8.89 | 0.293039 |
| FGF4 | Male | 4 | 8.89 | 0.293039 |
| PIK3CA | Male | 4 | 8.89 | 0.293039 |
| NF1 | Male | 3 | 6.67 | 0.286891 |
| STK11 | Male | 3 | 6.67 | 0.286891 |
Genomic alterations of CCA in different age groups
In order to study GAs in different age ranges, patients were divided into 4 age groups: Group I, patients aged 40–49 (7.58%, 5/66); group II, patients aged 50–59 (27.27%, 18/66); group III, patients aged 60–69 (39.39%, 29/66); and group IV, patients aged 70–82 (21.21%, 14/66). In group I, 36 GAs from 29 genes were detected (7.2 GAs/patient and 5.8 genes/patient). In group II, 119 GAs genes from 84 genes were detected (6.6 GAs/patient and 4.7 genes/patient). In group III, 188 GAs from 95 genes were detected (6.5 GAs genes/patient and 3.3 genes/patient). In group IV, 88 GAs from 59 genes were detected (6.3 GAs/patient and 4.1 genes/patient) (Table SV).
The mutation frequencies of CDKN2A, TERT ZNF750, PIK3CA and FGFR2 were highest in group IV, while the mutation frequencies of TP53, ARID1A and SMAD4 were highest in group III. The mutations in LRP1B specifically occurred in group II, and the mutation frequencies of KRAS, NF1 and KMT2C were highest in group I (Fig. 2B). Statistical analysis revealed that the mutation frequencies of PIK3CA, FGFR2 and ZNF750 were significantly associated with age in patients with CCA (Fig. 2B).
Genomic alterations in different differentiated CCAs
To investigate the association between GAs and the differentiation of CCA, patients were divided into 2 groups, based on the pathological diagnosis: Group I, patients with well/moderately differentiated CCA; and group II, patients with poorly/undifferentiated CCA. Among the most frequently mutated genes, TERT mutations were significantly higher in group II, and STK11, GNAS2, KMT2D and MAGI2 mutations were only detected in group II patients (Fig. 2C).
Association between genetic variations and TMB
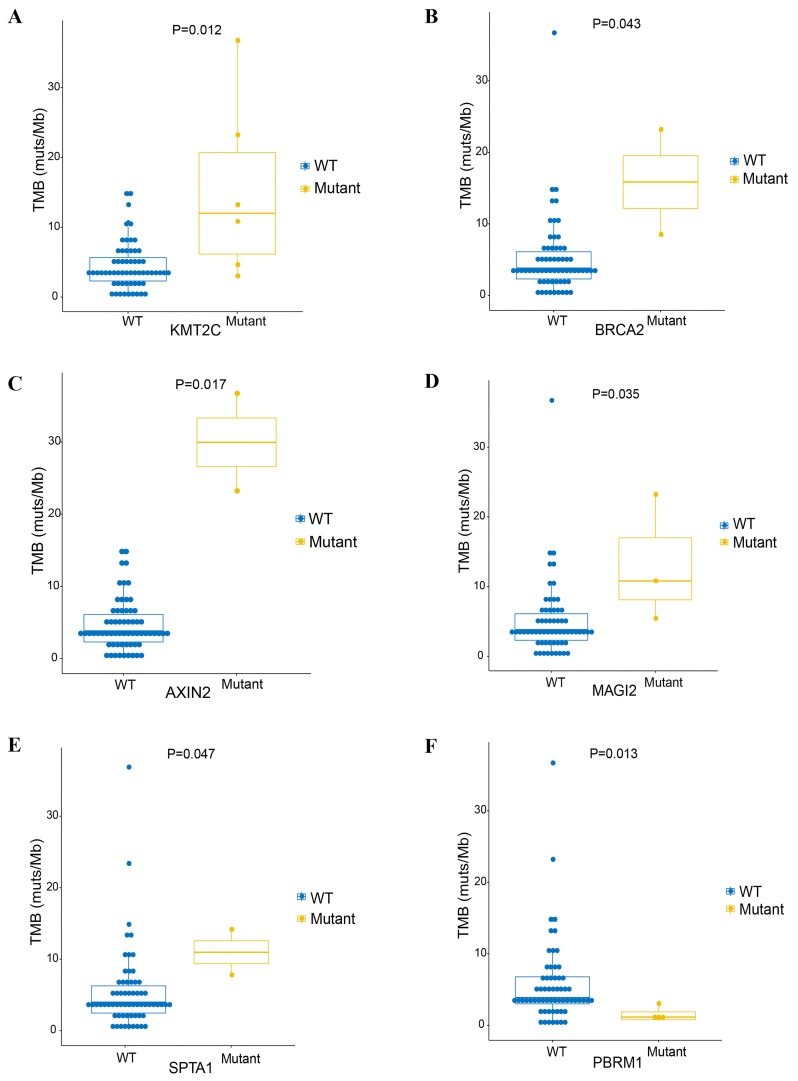
TMB is an emerging genomic biomarker that measures the number of gene mutations in the tumor genome (17). To explore the association between TMB and clinically relevant GAs, the TMB was determined in all samples. The median TMB of 66 samples was 5.4 mutations/Mb (range 0–36.7 muts/Mb; Table I). A high TMB (>10 mutations/Mb) was found in 9 patients (13.6%, 9/66). High TMB (TMB-H) was significantly associated with the GAs of KMT2C (P=0.012), AXIN2 (P=0.017), MAGI2 (P=0.035), SPTA1 (P=0.047) and BRCA2 (P=0.043), whereas low TMB was significantly associated with mutation of PBRM1 (P=0.013; Fig. 3). There were no significant associations between TMB and GAs of the other genes (data not shown).
Figure 3.
Median TMB value of 66 patients. Cholangiocarcinoma was significantly associated with (A) KMT2C, (B) BRCA2, (C) AXIN2, (D) MAGI2, (E) SPTA1 and (F) PBRM1. TMB, tumor mutational burden; WT, wild-type.
Difference in GAs between Chinese and Western patients with CCA
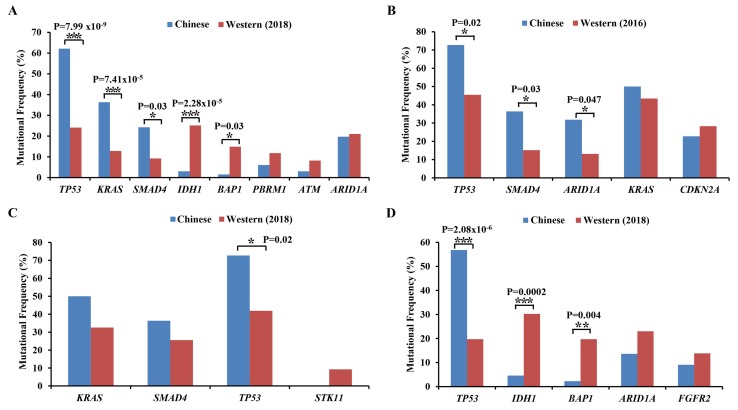
The most common mutations were IDH1, TP53, ARID1A, BAP1, KRAS, PBKM1, SMAD4 and ATM in Western patients with CCA (10). In 66 Chinese patients with CCA, the mutations of TP53, ARID1A, KRAS and SMAD4 also frequently occurred. Based on statistical analysis, the mutations of TP53, KRAS and SMAD4 were significantly higher in Chinese patients, whereas the mutations of IDH1 and BAP1 were significantly lower compared with Western patients (Fig. 4A).
Figure 4.
Comparative analysis of high frequency mutant genes in Chinese and Western patients with CCA. (A) Comparative analysis between Chinese patients and 194 reported (2018) Western patients with CCA (10). (B) Comparative analysis between Chinese patients with exCCA and 99 reported (2016) Western exCCA patients (18). (C) Comparative analysis between Chinese patients with exCCA and 43 reported (2018) Western patients with exCCA (10). (D) Comparative analysis between Chinese patients with iCCA and 152 reported (2018) Western patients with iCCA (10). *P<0.05, **P<0.01, ***P<0.001. iCCA, intrahepatic cholangiocarcinoma; exCCA, extrahepatic cholangiocarcinoma.
The differences in mutation frequencies of exCCA and iCCA between Chinese and Western patients were also determined. Compared with the results of Lee et al (18), based on 99 patients with exCCA, the mutation frequencies of TP53, SMAD4 and ARID1A in Chinese patients were higher compared with those in Western patients (Fig. 4B). Compared with a recent study by Lowery et al (10) on 43 patients with exCCA, the mutation frequencies of KRAS, SMAD4 and STK11 were similar in Chinese and Western patients, however, the mutation frequency of TP53 was significantly higher in Chinese patients compared with Western patients (Fig. 4C). In comparison with 152 Western patients with iCCA, a significantly higher frequency of TP53 mutations and a lower frequency of IDH1 and BAP1 mutations were detected in Chinese patients with iCCA (Fig. 4D).
Discussion
Since CCA is associated with poor prognosis, largely due to late diagnosis, the identification and exploration of biomarkers for early diagnosis has become important (19,20). The rapid development of NGS technologies allows for genomic profiling detection of multiple samples from different patients with multiplexed panels (21). The alignment of the sequences from tumor tissue samples and matched blood samples were considered to be an effective method for detecting somatic mutations (22–24). To date, many studies on potential biomarkers for CCA have been reported (25–27). Ong et al (28) identified and validated 206 somatic mutations in 187 genes, including somatic mutations in 10 newly implicated genes, using Sanger sequencing. Jiao et al (29) discovered frequent inactivating mutations of BAP1, ARID1A and PBRM1, with a high mutation rate in the sequenced tumor. Recently, Lowery et al (10) reported the genome profiling of patients with CCA, including 152 iCCA and 43 exCCA cases, from Caucasian (89.2%, 174/195), Asian (7.1%, 14/195), and African American (3.6%, 7/195) patients, and found that the most common mutations were IDH1, TP53, ARID1A, BAP1, KRAS, PBKM1, SMAD4 and ATM. In the present study, common high frequency mutations (>10%) of TP53, KRAS, SMAD4, TERT, ARID1A and CDKN2A were detected in Chinese patients with CCA. In comparison with Western patients, TP53, KRAS and SMAD4 mutations were significantly higher, however, IDH1 and BAP1 mutations were significantly lower in Chinese patients with CCA.
Changes in the p53 gene have been reported in >50% of patients with cancer, including those with CCA (30,31). Studies have reported TP53 as a potential diagnostic biomarker in esophageal cancer and head and neck cancer (32,33). In iCCA, Simbolo et al (34) also demonstrated that the mutation of TP53 is an independent predictor of poor prognosis. In the present study, TP53 mutations were detected at amino acid residues R273 and R248. Previous reports have shown that mutations at amino acid residues R273 and R248 of TP53 both resulted in oncogenic gain-of-function phenotypes (35). Based on these similar mutation types, it was suggested that the mutation of TP53 in this cohort might be a potential biomarker for predicting prognosis in Chinese patients with CCA. KRAS, the main subtype of the RAS gene family, has been reported to promote a variety of lethal human tumors, such as lung, colon and pancreatic cancer (36,37). Several studies have reported KRAS as a tumor biomarker (37–39) and that the co-mutation of TP53 and KRAS in many tumors (30,40). Consistent with previous studies (41–43), the most frequent mutations of KRAS occurred at the 12 and 13th amino acid sites in Chinese patients with CCA, and co-mutations of TP53 and KRAS were also detected with a high frequency, suggesting that KRAS mutations are conserved in most tumors.
CCA can be divided into iCCA and exCCA based on the origin. iCCA usually originates from intrahepatic bile duct wall cells, whereas exCCA originates from tumors in the left and right hepatic ducts, common hepatic ducts and common bile ducts. Several studies have identified the different GAs in iCCA and exCCA. The most common mutations in iCCA were found to be IDH1/2, BAP1, KRAS, TP53, SMAD4 and ARID1A, whereas common mutations in exCCA were identified in KRAS, TP53, SMAD4, SKT11, ERBB2, PTEN, ATM and NF1 (10,44). TP53 may play a key role in enabling hepatocyte-derived iCCA (30,18), and KRAS mutations in the absence of TP53 mutations can drive exCCA (41,45). The present study demonstrated that the most frequent alterations of TP53, KRAS, SMAD4, CDKN2A, TERT and ARID1A were found in both iCCA and exCCA. CDKN2A, which is associated with the cell cycle process, and TERT, which is involved in DNA stability, were reported to be associated with the tumor progression, invasion, and metastasis (46,47). To the best of our knowledge, this is the first study that detected CDKN2A and TERT alterations in Chinese patients with CCA. In contrast to previous studies on iCCA, SKT11 mutations occurred more frequently, whereas IDH1/2 and BAP1 mutations occurred less frequently; whereas in exCCA, both SKT11 and PTEN mutations were not detected. This implies that the genome profiling of Chinese CCA patients may differ from Western patients. RBM10 has been reported to regulate the Notch pathway by interacting with NUMB in cancer (48). The present study demonstrated a significantly higher mutation frequency of RBM10 in exCCA compared with iCCA, suggesting that RBM10 mutations may be specifically associated with exCCA in Chinese patients with CCA. In addition, mutations of STK11, CCND1, FGF19, FGF3, FGF4, FGFR2 and PBRM1 were found only in iCCA, suggesting that the mutations of these genes may be specific and are potential tumor biomarkers in iCCA.
Previously, Bhagat and Somasundar (49) assessed the risk of CCA incidence in patients of different genders. However, little is known about gender-associated specific genomic characteristics in patients with CCA. The present study identified CCND1, FBXW7 FGF3/4/19, PIK3C1, NF1 and STK11 mutations that were specific to male patients with CCA, and that the mutations in ERBB2, AXIN2, CREBBP, ERBB3, MTOR and MYB were specific to female patients. Notably, a significantly higher mutation rate of ERBB2 was found in female patients. Thus, it can be speculated that ERBB2 may be a potential biomarker for female patients with CCA.
Different age groups have different risks of cancer (50). The CCA patients were divided into 4 groups, based on age and analyzed gene mutations. TP53, KRAS, CDKN2A, TERT and ARID1A mutations were detected in all 4 age groups, however, PIK3CA, FGFR2, and ZNF750 mutations were mainly detected in group IV (>70 years), implying that these genes may be potential predictors and therapeutic diagnostic biomarkers in elderly patients (>70 years) with CCA. PIK3CA mutations have been reported to be associated with age, tobacco use and clinical stage in Chinese patients diagnosed with esophageal squamous cell carcinoma (ESCC) (51). Furthermore, it was reported that PIK3CA mutations frequently occur in Chinese patients with CCA, determined by PCR method (52). Although the number of cases was limited, a significant association was detected between PIK3CA mutations and age in the present study. This result also confirms the reliability of our large PANEL test for genetic variation. The FGFR2 GTGT haplotype was reported to be associated with breast cancer in younger women, but not older women (53), indicating that FGFR2 may be associated with the age of patients with cancer. FGFR2 mutations have been reported in iCCA (18,54). The present study also detected FGFR2 mutations in iCCA, and demonstrated that they mainly occurred in patients older than 70 years, which implies that the mutation of FGFR2 was associated with the age of Chinese patients with CCA. ZNF750 has been reported as a prognostic biomarker in ESCC (55). Although only two mutations of ZNF750 were detected in the present study, notably, both mutations occurred in group IV (>70 years).
STK11 is a tumor suppressor gene and the methylation of its promoter in renal cell carcinoma was reported as a risk factor for prognosis (56). The mutations of KMT2D and TERT have been reported to be associated with prognosis in different tumors (57–59). Histopathological grade is considered to be associated with the clinical characteristics of lymphovascular invasion, perineural invasion, tumor differentiation, lymph node and metastasis, as well as the prediction of prognosis (60,61). In the present study, the mutations of STK11, GNAS, KMT2D, MAGI2 and TERT were mainly detected in poorly differentiated CCA, indicating that these genes might be potential biomarkers to predict the differentiation of CCA tumors.
Similar to Western patients with exCCA (18), the most frequently mutated genes that were detected in 66 Chinese patients were TP53, KRAS, SMAD4 and ARID1A. However, the mutation rate of TP53, SMAD4 and ARID1A in the present study was significantly higher compared with Western patients. Multiple studies have investigated the mutational landscape in CCA by NGS. Borger et al (62) investigated the mutational profiling of 287 gastrointestinal tumors from Massachusetts General Hospital, including a total of 50 iCCA and 22 exCCA cases, and found that the IDH mutations represent the first hotspot variants in iCCA patients. Jiao et al (29) demonstrated a total of 1,128 mutated genes in 32 American patients with iCCA, and confirmed several inactivating alterations of BAP1, ARID1A, IDH1 and IDH2. Lowery et al (10) also reported high mutation frequency of IDH1, ARID1A, TP53, BAP1 and FGFR2 in 152 Caucasian patients with iCCA. Compared with previous studies, a significantly higher frequency of TP53 mutations was detected in Chinese patients with both iCCA and exCCA, however, a lower frequency of IDH1 and BAP1 mutations. Notably, Zou et al (63) detected high mutation frequencies of IDH1/IDH2 in 103 Chinese patients with iCCA. Since the patients enrolled in the present study were mainly from North China, the difference in IDH1 and BAP1 mutations appears to be a local feature of iCCA in China; however, further studies are required with a larger cohort to confirm this finding.
TMB is a measure of the number of somatic mutations occurring in a tumor. The TMB pattern may further guide the selection of checkpoint inhibitors (CPI) for patients (64). TMB is correlated with clinical outcome in multiple cancer types treated with CPI (65,66). Previous studies indicated that TMB levels correlated with gene mutations in tumors (25,40,67,68). TMB-H has been reported to correlate with the generation of neoantigens and potential clinical responses to immunotherapies (69). Mou et al (70) reported that a Chinese patient with iCCA, TMB-H and a high expression of PD-L1 responded to the combination of chemotherapy and pembrolizumab. In the present study, the association between TMB and gene mutations was analyzed in Chinese patients CCA, which identified that mutations in KMT2C, BRCA2, AXIN2, MAGI2, PBRM1 and SPTA1 were significantly associated with TMB. TMB-H was found to be associated with the response rate, PFS and overall survival of patients with cancer (71). KMT2C, AXIN2 and MAGI1 mutations were also reported to be associated with poor prognosis (72–74). These reports support the association between TMB and the mutations of KMT2C, AXIN2 and MAGI1. Furthermore, the significant association of these genes with TMB suggests that they may be potential predictive biomarkers in Chinese patients with CCA.
In conclusion, the comprehensive genomic features of 66 Chinese patients with CCA was investigated, which identified that TP53 mutations were significantly higher in Chinese patients compared with Western patients with CCA. This suggests that TP53 may be more suitable as a predictive and diagnostic biomarker in Chinese patients with CCA. Furthermore, gene mutations that were associated with age, disease staging and high TMB were detected. Overall, these data provide an insight into the diagnosis, prognosis and therapeutic strategies, both targeted and immune therapies, in Chinese patients with CCA.
Supplementary Material
Acknowledgements
Not applicable.
Glossary
Abbreviations
- CCA
cholangiocarcinoma
- iCCA
intrahepatic CCA
- exCCA
extrahepatic CCA
- NGS
next-generation sequencing
- GA
genomic alterations
- TMB
tumor mutational burden
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.
Authors' contributions
WT and WH recruited patients, collected samples, analyzed data, and wrote the manuscript. PL, XM, WZ, and LQ recruited patients, collected samples and analyzed data. XS, AL, SZ and WS processed samples, performed experiments, analyzed data and reviewed the manuscript. JC designed and supervised the study. All authors read and approved the final manuscript.
Ethics approval and consent to participate
The project was approved by the Ethics Committee of both Tianjin Medical University General Hospital and Affiliated Hospital of Qingdao University. Written informed consent for participation was obtained from all subjects.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
References
- 1.Hoyos S, Navas MC, Restrepo JC, Botero RC. Current controversies in cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1461–1467. doi: 10.1016/j.bbadis.2017.07.027. [DOI] [PubMed] [Google Scholar]
- 2.Chong YS, Kim YK, Lee MW, Kim SH, Lee WJ, Rhim HC, Lee SJ. Differentiating mass-forming intrahepatic cholangiocarcinoma from atypical hepatocellular carcinoma using gadoxetic acid-enhanced MRI. Clin Radiol. 2012;67:766–773. doi: 10.1016/j.crad.2012.01.004. [DOI] [PubMed] [Google Scholar]
- 3.Songserm N, Promthet S, Pientong C, Ekalaksananan T, Chopjitt P, Wiangnon S. Gene-environment interaction involved in cholangiocarcinoma in the Thai population: Polymorphisms of DNA repair genes, smoking and use of alcohol. BMJ Open. 2014;4:e005447. doi: 10.1136/bmjopen-2014-005447. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Cancer Genome Atlas Research Network. Weinstein JN, Collisson EA, Mills GB, Shaw KR, Ozenberger BA, Ellrott K, Shmulevich I, Sander C, Stuart JM. The cancer genome atlas pan-cancer analysis project. Nat Genet. 2013;45:1113–1120. doi: 10.1038/ng.2764. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cancer Genome Atlas Research Network. Kandoth C, Schultz N, Cherniack AD, Akbani R, Liu Y, Shen H, Robertson AG, Pashtan I, Shen R, et al. Integrated genomic characterization of endometrial carcinoma. Nature. 2013;497:67–73. doi: 10.1038/nature12113. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Nakamura H, Arai Y, Totoki Y, Shirota T, Elzawahry A, Kato M, Hama N, Hosoda F, Urushidate T, Ohashi S, et al. Genomic spectra of biliary tract cancer. Nat Genet. 2015;47:1003–1010. doi: 10.1038/ng.3375. [DOI] [PubMed] [Google Scholar]
- 7.Lin P, Zhong XZ, Wang XD, Li JJ, Zhao RQ, He Y, Jiang YQ, Huang XW, Chen G, He Y, Yang H. Survival analysis of genome-wide profiles coupled with connectivity map database mining to identify potential therapeutic targets for cholangiocarcinoma. Oncol Rep. 2018;40:3189–3198. doi: 10.3892/or.2018.6710. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Mertens JC, Rizvi S, Gores GJ. Targeting cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1454–1460. doi: 10.1016/j.bbadis.2017.08.027. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Churi CR, Shroff R, Wang Y, Rashid A, Kang HC, Weatherly J, Zuo M, Zinner R, Hong D, Meric-Bernstam F, et al. Mutation profiling in cholangiocarcinoma: Prognostic and therapeutic implications. PLoS One. 2014;9:e115383. doi: 10.1371/journal.pone.0115383. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Lowery MA, Ptashkin R, Jordan E, Berger MF, Zehir A, Capanu M, Kemeny NE, O'Reilly EM, El-Dika I, Jarnagin WR, et al. Comprehensive molecular profiling of intrahepatic and extrahepatic cholangiocarcinomas: potential targets for intervention. Clin Cancer Res. 2018;24:4154–4161. doi: 10.1158/1078-0432.CCR-18-0078. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Zhao P, Chen H, Wen D, Mou S, Zhang F, Zheng S. Personalized treatment based on mini patient-derived xenografts and WES/RNA sequencing in a patient with metastatic duodenal adenocarcinoma. Cancer Commun (Lond) 2018;38:54. doi: 10.1186/s40880-018-0323-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Cibulskis K, Lawrence MS, Carter SL, Sivachenko A, Jaffe D, Sougnez C, Gabriel S, Meyerson M, Lander ES, Getz G. Sensitive detection of somatic point mutations in impure and heterogeneous cancer samples. Nat Biotechnol. 2013;31:213–219. doi: 10.1038/nbt.2514. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Ye K, Schulz MH, Long Q, Apweiler R, Ning Z. Pindel: A pattern growth approach to detect break points of large deletions and medium sized insertions from paired-end short reads. Bioinformatics. 2009;25:2865–2871. doi: 10.1093/bioinformatics/btp394. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Cingolani P, Platts A, Wang le L, Coon M, Nguyen T, Wang L, Land SJ, Lu X, Ruden DM. A program for annotating and predicting the effects of single nucleotide polymorphisms, SnpEff: SNPs in the genome of Drosophila melanogaster strain w1118; iso-2; iso-3. Fly (Austin) 2012;6:80–92. doi: 10.4161/fly.19695. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Boeva V, Popova T, Bleakley K, Chiche P, Cappo J, Schleiermacher G, Janoueix-Lerosey I, Delattre O, Barillot E. Control-FREEC: A tool for assessing copy number and allelic content using next-generation sequencing data. Bioinformatics. 2012;28:423–425. doi: 10.1093/bioinformatics/btr670. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Thorvaldsdóttir H, Robinson JT, Mesirov JP. Integrative genomics viewer (IGV): High-performance genomics data visualization and exploration. Brief Bioinform. 2013;14:178–192. doi: 10.1093/bib/bbs017. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Steuer CE, Ramalingam SS. Tumor mutation burden: Leading immunotherapy to the era of precision medicine? J Clin Oncol. 2018;36:631–632. doi: 10.1200/JCO.2017.76.8770. [DOI] [PubMed] [Google Scholar]
- 18.Lee H, Wang K, Johnson A, Jones DM, Ali SM, Elvin JA, Yelensky R, Lipson D, Miller VA, Stephens PJ, et al. Comprehensive genomic profiling of extrahepatic cholangiocarcinoma reveals a long tail of therapeutic targets. J Clin Pathol. 2016;69:403–408. doi: 10.1136/jclinpath-2015-203394. [DOI] [PubMed] [Google Scholar]
- 19.Macias RIR, Banales JM, Sangro B, Muntané J, Avila MA, Lozano E, Perugorria MJ, Padillo FJ, Bujanda L, Marin JJG. The search for novel diagnostic and prognostic biomarkers in cholangiocarcinoma. Biochim Biophys Acta Mol Basis Dis. 2018;1864:1468–1477. doi: 10.1016/j.bbadis.2017.08.002. [DOI] [PubMed] [Google Scholar]
- 20.Chan TA, Yarchoan M, Jaffee E, Swanton C, Quezada SA, Stenzinger A, Peters S. Development of tumor mutation burden as an immunotherapy biomarker: Utility for the oncology clinic. Ann Oncol. 2019;30:44–56. doi: 10.1093/annonc/mdy495. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Loree JM, Kopetz S. Recent developments in the treatment of metastatic colorectal cancer. Ther Adv Med Oncol. 2017;9:551–564. doi: 10.1177/1758834017714997. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Zheng B, Qu Y, Wang J, Shi Y, Yan W. Pathogenic and targetable genetic alterations in resected recurrent undifferentiated pleomorphic sarcomas identified by targeted next-generation sequencing. Cancer Genomics Proteomics. 2019;16:221–228. doi: 10.21873/cgp.20127. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Hu J, Wang Y, Zhang Y, Yu Y, Chen H, Liu K, Yao M, Wang K, Gu W, Shou T. Comprehensive genomic profiling of small cell lung cancer in Chinese patients and the implications for therapeutic potential. Cancer Med. 2019;8:4338–4347. doi: 10.1002/cam4.2199. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Fang W, Huang Y, Hong S, Zhang Z, Wang M, Gan J, Wang W, Guo H, Wang K, Zhang L. EGFR exon 20 insertion mutations and response to osimertinib in non-small-cell lung cancer. BMC Cancer. 2019;19:595. doi: 10.1186/s12885-019-5820-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Zheng B, Jeong S, Zhu Y, Chen L, Xia Q. miRNA and lncRNA as biomarkers in cholangiocarcinoma (CCA) Oncotarget. 2017;8:100819–100830. doi: 10.18632/oncotarget.19044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Nakaoka T, Saito Y, Saito H. Aberrant DNA methylation as a biomarker and a therapeutic target of cholangiocarcinoma. Int J Mol Sci. 2017;18(pii):E1111. doi: 10.3390/ijms18061111. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Betesh L, Comunale MA, Wang M, Liang H, Hafner J, Karabudak A, Giama NH, Moser CD, Miyoshi E, Roberts LR, et al. Identification of fucosylated Fetuin-A as a potential biomarker for cholangiocarcinoma. Proteomics Clin Appl. 2017;11 doi: 10.1002/prca.201600141. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Ong CK, Subimerb C, Pairojkul C, Wongkham S, Cutcutache I, Yu W, McPherson JR, Allen GE, Ng CC, Wong BH, et al. Exome sequencing of liver fluke-associated cholangiocarcinoma. Nat Genet. 2012;44:690–693. doi: 10.1038/ng.2273. [DOI] [PubMed] [Google Scholar]
- 29.Jiao Y, Pawlik TM, Anders RA, Selaru FM, Streppel MM, Lucas DJ, Niknafs N, Guthrie VB, Maitra A, Argani P, et al. Exome sequencing identifies frequent inactivating mutations in BAP1, ARID1A and PBRM1 in intrahepatic cholangiocarcinomas. Nat Genet. 2013;45:1470–1473. doi: 10.1038/ng.2813. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Hill MA, Alexander WB, Guo B, Kato Y, Patra K, O'Dell MR, McCall MN, Whitney-Miller CL, Bardeesy N, Hezel AF. Kras and Tp53 mutations cause cholangiocyte- and hepatocyte-derived cholangiocarcinoma. Cancer Res. 2018;78:4445–4451. doi: 10.1158/0008-5472.CAN-17-1123. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Hedau S, Jain N, Husain SA, Mandal AK, Ray G, Shahid M, Kant R, Gupta V, Shukla NK, Deo SS, Das BC. Novel germline mutations in breast cancer susceptibility genes BRCA1, BRCA2 and p53 gene in breast cancer patients from India. Breast Cancer Res Treat. 2004;88:177–186. doi: 10.1007/s10549-004-0593-8. [DOI] [PubMed] [Google Scholar]
- 32.van Ginkel JH, de Leng WW, de Bree R, van Es RJ, Willems SM. Targeted sequencing reveals TP53 as a potential diagnostic biomarker in the post-treatment surveillance of head and neck cancer. Oncotarget. 2016;7:61575–61586. doi: 10.18632/oncotarget.11196. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Kandioler D, Schoppmann SF, Zwrtek R, Kappel S, Wolf B, Mittlböck M, Kührer I, Hejna M, Pluschnig U, Ba-Ssalamah A, et al. The biomarker TP53 divides patients with neoadjuvantly treated esophageal cancer into 2 subgroups with markedly different outcomes. A p53 Research Group study. J Thorac Cardiovasc Surg. 2014;148:2280–2286. doi: 10.1016/j.jtcvs.2014.06.079. [DOI] [PubMed] [Google Scholar]
- 34.Simbolo M, Vicentini C, Ruzzenente A, Brunelli M, Conci S, Fassan M, Mafficini A, Rusev B, Corbo V, Capelli P, et al. Genetic alterations analysis in prognostic stratified groups identified TP53 and ARID1A as poor clinical performance markers in intrahepatic cholangiocarcinoma. Sci Rep. 2018;8:7119. doi: 10.1038/s41598-018-25669-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Shtraizent N, Matsui H, Polotskaia A, Bargonetti J. Hot spot mutation in TP53 (R248Q) causes oncogenic gain-of-function phenotypes in a breast cancer cell line derived from an African American patient. Int J Environ Res Public Health. 2015;13:ijerph13010022. doi: 10.3390/ijerph13010022. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Yang K, Li Y, Lian G, Lin H, Shang C, Zeng L, Chen S, Li J, Huang C, Huang K, Chen Y. KRAS promotes tumor metastasis and chemoresistance by repressing RKIP via the MAPK-ERK pathway in pancreatic cancer. Int J Cancer. 2018;142:2323–2334. doi: 10.1002/ijc.31248. [DOI] [PubMed] [Google Scholar]
- 37.Nakano Y, Kitago M, Matsuda S, Nakamura Y, Fujita Y, Imai S, Shinoda M, Yagi H, Abe Y, Hibi T, et al. KRAS mutations in cell-free DNA from preoperative and postoperative sera as a pancreatic cancer marker: A retrospective study. Br J Cancer. 2018;118:662–669. doi: 10.1038/bjc.2017.479. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Addeo A, Torralvo J, Dietrich PY. 192P KRAS as predictive biomarker of response to checkpoint inhibitors in NSCLC. J Thorac Oncol. 2018;13(4 Suppl):S114–S115. doi: 10.1016/S1556-0864(18)30465-9. [DOI] [Google Scholar]
- 39.Petrelli F, Coinu A, Cabiddu M, Ghilardi M, Barni S. KRAS as prognostic biomarker in metastatic colorectal cancer patients treated with bevacizumab: A pooled analysis of 12 published trials. Med Oncol. 2013;30:650. doi: 10.1007/s12032-013-0650-4. [DOI] [PubMed] [Google Scholar]
- 40.Dong ZY, Zhong WZ, Zhang XC, Su J, Xie Z, Liu SY, Tu HY, Chen HJ, Sun YL, Zhou Q, et al. Potential predictive value of TP53 and KRAS mutation status for response to PD-1 blockade immunotherapy in lung adenocarcinoma. Clin Cancer Res. 2017;23:3012–3024. doi: 10.1158/1078-0432.CCR-16-2554. [DOI] [PubMed] [Google Scholar]
- 41.O'Dell MR, Huang JL, Whitney-Miller CL, Deshpande V, Rothberg P, Grose V, Rossi RM, Zhu AX, Land H, Bardeesy N, Hezel AF. Kras(G12D) and p53 mutation cause primary intrahepatic cholangiocarcinoma. Cancer Res. 2012;72:1557–1567. doi: 10.1158/0008-5472.CAN-11-3596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Ma ES, Wong CL, Law FB, Chan WK, Siu D. Detection of KRAS mutations in colorectal cancer by high-resolution melting analysis. J Clin Pathol. 2009;62:886–891. doi: 10.1136/jcp.2008.063677. [DOI] [PubMed] [Google Scholar]
- 43.Thobe MN, Yan SB, Credille KM, Nasir A, Baker JAR, Lajiness M, Brooks NA, Ballard DW, Farley DM, Peek VL, et al. Abstract 2339: Prevalence of MET expression, activating mutations of KRAS and IDH1/2, and ROS1 fusions in cholangiocarcinoma patient tumor samples. Cancer Res. 2013;73:2339. [Google Scholar]
- 44.Jusakul A, Cutcutache I, Yong CH, Lim JQ, Huang MN, Padmanabhan N, Nellore V, Kongpetch S, Ng AWT, Ng LM, et al. Whole-genome and epigenomic landscapes of etiologically distinct subtypes of cholangiocarcinoma. Cancer Discov. 2017;7:1116–1135. doi: 10.1158/2159-8290.CD-17-0368. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Ikenoue T, Terakado Y, Nakagawa H, Hikiba Y, Fujii T, Matsubara D, Noguchi R, Zhu C, Yamamoto K, Kudo Y, et al. A novel mouse model of intrahepatic cholangiocarcinoma induced by liver-specific Kras activation and Pten deletion. Sci Rep. 2016;6:23899. doi: 10.1038/srep23899. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Foulkes WD, Flanders TY, Pollock PM, Hayward NK. The CDKN2A (p16) gene and human cancer. Mol Med. 1997;3:5–20. doi: 10.1007/BF03401664. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Heidenreich B, Rachakonda PS, Hemminki K, Kumar R. TERT promoter mutations in cancer development. Curr Opin Genet Dev. 2014;24:30–37. doi: 10.1016/j.gde.2013.11.005. [DOI] [PubMed] [Google Scholar]
- 48.Hernández J, Bechara E, Schlesinger D, Delgado J, Serrano L, Valcárcel J. Tumor suppressor properties of the splicing regulatory factor RBM10. RNA Biol. 2016;13:466–472. doi: 10.1080/15476286.2016.1144004. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Bhagat V, Somasundar P. 16 Impact of gender and race/ethnicity on hepatocellular carcinoma, intrahepatic cholangiocarcinoma and extrahepatic cholangiocarcinoma. Gastroenterology. 2016;150:S5. doi: 10.1016/S0016-5085(16)30151-2. [DOI] [Google Scholar]
- 50.Taber JM, Klein WM, Suls JM, Ferrer RA. Lay awareness of the relationship between age and cancer risk. Ann Behav Med. 2016;51:214–225. doi: 10.1007/s12160-016-9845-1. [DOI] [PubMed] [Google Scholar]
- 51.Lin JW, Li X, Qiu ML, Luo RG, Lin JB, Liu B. PI3K overexpression and PIK3CA mutations are associated with age, tumor staging, and other clinical characteristics in Chinese patients with esophageal squamous cell carcinoma. Genet Test Mol Biomarkers. 2017;21:236–241. doi: 10.1089/gtmb.2016.0316. [DOI] [PubMed] [Google Scholar]
- 52.Xu RF, Sun JP, Zhang SR, Zhu GS, Li LB, Liao YL, Xie JM, Liao WJ. KRAS and PIK3CA but not BRAF genes are frequently mutated in Chinese cholangiocarcinoma patients. Biomed Pharmacother. 2011;65:22–26. doi: 10.1016/j.biopha.2010.06.009. [DOI] [PubMed] [Google Scholar]
- 53.Barnholtz-Sloan JS, Shetty PB, Guan X, Nyante SJ, Luo J, Brennan DJ, Millikan RC. FGFR2 and other loci identified in genome-wide association studies are associated with breast cancer in African-American and younger women. Carcinogenesis. 2010;31:1417–1423. doi: 10.1093/carcin/bgq128. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Katoh M. Cancer genomics and genetics of FGFR2 (Review) Int J Oncol. 2008;33:233–237. [PubMed] [Google Scholar]
- 55.Otsuka R, Akutsu Y, Sakata H, Hanari N, Murakami K, Kano M, Toyozumi T, Takahashi M, Matsumoto Y, Sekino N, et al. ZNF750 expression is a potential prognostic biomarker in esophageal squamous cell carcinoma. Oncology. 2018;94:142–148. doi: 10.1159/000484932. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Zheng F, Yuan X, Chen E, Ye Y, Li X, Dai Y. Methylation of STK11 promoter is a risk factor for tumor stage and survival in clear cell renal cell carcinoma. Oncol Lett. 2017;14:3065–3070. doi: 10.3892/ol.2017.6534. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Ardeshir-Larijani F, Bhateja P, Lipka MB, Sharma N, Fu P, Dowlati A. KMT2D mutation is associated with poor prognosis in non-small-cell lung cancer. Clin Lung Cancer. 2018;19:e489–e501. doi: 10.1016/j.cllc.2018.03.005. [DOI] [PubMed] [Google Scholar]
- 58.Bahrami A, Lee S, Schaefer IM, Boland JM, Patton KT, Pounds S, Fletcher CD. TERT promoter mutations and prognosis in solitary fibrous tumor. Mod Pathol. 2016;29:1511–1522. doi: 10.1038/modpathol.2016.126. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Shen N, Lu Y, Wang X, Peng J, Zhu Y, Cheng L. Association between rs2853669 in TERT gene and the risk and prognosis of human cancer: A systematic review and meta-analysis. Oncotarget. 2017;8:50864–50872. doi: 10.18632/oncotarget.15140. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Elston CW. The assessment of histological differentiation in breast cancer. Aust N Z J Surg. 1984;54:11–15. doi: 10.1111/j.1445-2197.1984.tb06677.x. [DOI] [PubMed] [Google Scholar]
- 61.Padma R, Kalaivani A, Sundaresan S, Sathish P. The relationship between histological differentiation and disease recurrence of primary oral squamous cell carcinoma. J Oral Maxillofac Pathol. 2017;21:461. doi: 10.4103/jomfp.JOMFP_241_16. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Borger DR, Tanabe KK, Fan KC, Lopez HU, Fantin VR, Straley KS, Schenkein DP, Hezel AF, Ancukiewicz M, Liebman HM, et al. Frequent mutation of isocitrate dehydrogenase (IDH)1 and IDH2 in cholangiocarcinoma identified through broad-based tumor genotyping. Oncologist. 2012;17:72–79. doi: 10.1634/theoncologist.2011-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 63.Zou S, Li J, Zhou H, Frech C, Jiang X, Chu JS, Zhao X, Li Y, Li Q, Wang H, et al. Mutational landscape of intrahepatic cholangiocarcinoma. Nat Commun. 2014;5:5696. doi: 10.1038/ncomms6696. [DOI] [PubMed] [Google Scholar]
- 64.Riaz N, Havel JJ, Makarov V, Desrichard A, Urba WJ, Sims JS, Hodi FS, Martín-Algarra S, Mandal R, Sharfman WH, et al. Tumor and microenvironment evolution during immunotherapy with nivolumab. Cell. 2017;171:934–949.e16. doi: 10.1016/j.cell.2017.09.028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Yarchoan M, Hopkins A, Jaffee EM. Tumor mutational burden and response rate to PD-1 inhibition. N Engl J Med. 2017;377:2500–2501. doi: 10.1056/NEJMc1713444. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Rizvi NA, Hellmann MD, Snyder A, Kvistborg P, Makarov V, Havel JJ, Lee W, Yuan J, Wong P, Ho TS, et al. Cancer immunology. Mutational landscape determines sensitivity to PD-1 blockade in non-small cell lung cancer. Science. 2015;348:124–128. doi: 10.1126/science.aaa1348. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Goswami B, Mittal P, Gupta N. Correlation of levels of IL-6 with tumor burden and receptor status in patients of locally advanced carcinoma breast. Indian J Clin Biochem. 2013;28:90–94. doi: 10.1007/s12291-012-0234-7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Frampton AE, Prado MM, Lopez-Jimenez E, Fajardo-Puerta AB, Jawad ZAR, Lawton P, Giovannetti E, Habib NA, Castellano L, Stebbing J, et al. Glypican-1 is enriched in circulating-exosomes in pancreatic cancer and correlates with tumor burden. Oncotarget. 2018;9:19006–19013. doi: 10.18632/oncotarget.24873. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Yuza K, Nagahashi M, Watanabe S, Takabe K, Wakai T. Hypermutation and microsatellite instability in gastrointestinal cancers. Oncotarget. 2017;8:112103–112115. doi: 10.18632/oncotarget.22783. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Mou H, Yu L, Liao Q, Hou X, Wu Y, Cui Q, Yan N, Ma R, Wang L, Yao M, Wang K. Successful response to the combination of immunotherapy and chemotherapy in cholangiocarcinoma with high tumour mutational burden and PD-L1 expression: A case report. BMC Cancer. 2018;18:1105. doi: 10.1186/s12885-018-5021-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Goodman AM, Kato S, Bazhenova L, Patel SP, Frampton GM, Miller V, Stephens PJ, Daniels GA, Kurzrock R. Tumor mutational burden as an independent predictor of response to immunotherapy in diverse cancers. Mol Cancer Ther. 2017;16:2598–2608. doi: 10.1158/1535-7163.MCT-17-0386. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Zhang G, Liu T, Wang Z. Downregulation of MAGI1 associates with poor prognosis of hepatocellular carcinoma. J Invest Surg. 2012;25:93–99. doi: 10.3109/08941939.2011.606875. [DOI] [PubMed] [Google Scholar]
- 73.Tseng RC, Lin RK, Wen CK, Tseng C, Hsu HS, Hsu WH, Wang YC. Epigenetic silencing of AXIN2/betaTrCP and deregulation of p53-mediated control lead to wild-type beta-catenin nuclear accumulation in lung tumorigenesis. Oncogene. 2008;27:4488–4496. doi: 10.1038/onc.2008.83. [DOI] [PubMed] [Google Scholar]
- 74.Cho SJ, Yoon C, Lee JH, Chang KK, Lin JX, Kim YH, Kook MC, Aksoy BA, Park DJ, Ashktorab H, et al. KMT2C mutations in diffuse-type gastric adenocarcinoma promote epithelial-to-mesenchymal transition. Clin Cancer Res. 2018;24:6556–6569. doi: 10.1158/1078-0432.CCR-17-1679. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Data Availability Statement
The datasets used and/or analyzed during the current study are available from the corresponding author on reasonable request.