Abstract
The incidence of pancreatic neuroendocrine tumor (PNET) is increasing, and it presents with various clinical manifestations and an unfavorable survival rate. A better understanding of the drivers of PNET tumorigenesis is urgently needed. Distinct miRNA signatures have been identified for different stages of tumorigenesis in both human and mouse PNETs. The functions of these miRNAs are poorly understood. miR-431 is the most up-regulated miRNA in the metastatic signature. However, it is unknown whether miR-431 contributes to metastasis of PNETs. Herein, we show that miR-431 overexpression activates Ras/extracellular signal-regulated kinase (Erk) signaling and promotes epithelial-mesenchymal transition, migration/invasion in vitro, and metastasis in both xenograft and spontaneous mouse models of PNET. Treatment of PNET cells with Erk inhibitor or locked nucleic acids sequestering miR-431 inhibits invasion. Four target prediction modules and dual-luciferase reporter assays were used to identify potential mRNA targets of miR-431. A Ras GTPase activating protein tumor suppressor (RasGAP), DAB2 interacting protein (DAB2IP), was discovered as an miR-431 target. Overexpression of DAB2IP's rat homolog, but not its mutant defective in Ras GTPase activating protein activity, reverses miR-431's effect on promoting invasion, Erk phosphorylation, and epithelial-mesenchymal transition of PNETs. Taken together, miR-431 silences DAB2IP to active Ras/Erk and promote metastasis of PNETs. miR-431 may be targeted to manage metastatic PNETs.
Pancreatic neuroendocrine tumors (PNETs) represent one-third of gastroenteropancreatic neuroendocrine tumors and are the second malignancy of the pancreas. The 5-year survival rate is approximately 55% when the tumors are localized and resected, but only approximately 15% when the tumors are not resectable.1 However, PNETs often display an indolent phenotype, resulting in late diagnosis at advanced stages when surgical resection with curative intent is no longer an alternative.2,3 Sunitinib (a multitargeted protein tyrosine kinase inhibitor), everolimus (a mammalian target of rapamycin inhibitor), and lutetium Lu 177 dotatate (a radioactive peptide targeted to somatostatin receptor) are approved for the treatment of unresectable and progressive or metastatic PNETs.2,4,5 All three drugs have their limitations. Both sunitinib and everolimus only extend median survival of PNET patients by approximately 6 months, and all patients developed resistance for both drugs.4,6 Lutetium Lu 177 dotatate can only be used for tumors expressing somatostatin receptors. As the incidences of PNETs are increasing,7 a better understanding of the mechanisms that underline PNET metastasis is vitally needed to improve therapeutic options.
miRNAs are small RNAs that regulate the expression of complementary messenger RNAs and play multiple roles in cellular functions, including cell growth, differentiation, and development.8 Metastasis promoter miRNAs9 and suppressor miRNAs10 were first discovered in breast cancer. Subsequent studies revealed more miRNAs with functions in tumorigenesis and metastasis of other cancer types. A cross-species study of miRNAs has identified stage-specific miRNA expression signatures in human PNETs and the RIP1-Tag2 (RIP-Tag) transgenic mouse model of PNETs.11 Another comprehensive cross-species analysis of mRNA and miRNA transcriptomes of PNETs from the mouse model and human patients supports for the RIP-Tag mouse model as representative of human PNETs.12 Among those miRNAs expressed in different stages of PNET tumorigenesis, miR-431 is one of the up-regulated miRNAs in the metastasis-specific miRNA signature.11 However, the role of miR-431 during PNET tumorigenesis is unknown. In this study, we aim to determine whether miR-431 contributes to PNET metastasis.
Materials and Methods
Cloning miRNA into Replication Competent Avian Leukosis Virus Long Terminal Repeat with a Splice, Bryan-Strain Pol Gene (RCASBP or RCABP) Vectors
RCASBP-Y-DEST has been described.13 After KpnI site was added before pCMV and the first BamHI site was destroyed in pcDNA6.2 GW-EmGFP-miR-lacZ vector,14 the pCMV-GFP-miR-lacZ part was subcloned from the modified pcDNA6.2 GW-EmGFP-miR-lacZ vector into pENTR3C using KpnI and XhoI sites. PCR-generated miR-431 was cloned into pENTR3C-GFP-lacZ using BamHI and BglII sites. pENTR3C-pCMV-GFP-miR-LacZ and pENTR3C-pCMV-GFP-miR-431were recombined with RCASBP-Y-DEST using LR Clonase (Invitrogen, Carlsbad, CA) to generate RCASBP-GFP-miR-LacZ and RCASBP-GFP-miR-431. Inserted regions in the new plasmids were confirmed by DNA sequencing.
Cell Lines and Reagents
Chicken fibroblast DF115,16 and N134 cell lines17 have been described. N134/GFP-miR-LacZ and N134/GFP-miR-431 were generated following previously described procedure.18 QGP1 cell line was kindly provided by Chris Harris (Rutgers University Cancer Institute of New Jersey, New Brunswick, NJ).19,20 QGP1 cells were infected with viruses carrying thymidine kinase/green fluorescent protein (GFP)/luciferase fusion reporter,21 which was kindly provided by Drs. Inna Serganova and Ronald Blasberg (Memorial Sloan Kettering Cancer Center, New York, NY). GFP+ N134/GFP-miR-LacZ, N134/GFP-miR-431, and QGP1/TGL cells were sorted using FACS-DiVa Cell Sorter (BD Biosciences, San Jose, CA) in the Weill Cornell Medicine (WCM) Flow Cytometry Core.
Rat DAB2IP expression construct, GTPase activating protein (GAP) mutant of rat DAB2IPexpression construct, and the control vector (pCI-neo) were gifts from Dr. Jer-Tsong Hsieh (University of Texas Southwestern Medical Center, Dallas, TX)22 and they were transiently transfected into N134/GFP-miR-431 cells by Lipofectamine 3000 (Invitrogen) following the instructions of the manufacturer. Forty-eight hours after transfection, cells were either used for the invasion assay described below or lysed in radioimmunoprecipitation assay buffer (50 mmol/L Tris, pH 7.4, 150 mmol/L NaCl, 1% NP-40, and 0.5% sodium deoxycholate) supplemented with a protease inhibitor mixture and PhosSTOP (Roche, Pleasanton, CA) for Western blot analysis.
DF1 and N134 cells were cultured in Dulbecco's modified Eagle's medium (DMEM) supplemented with 10% fetal bovine serum (FBS), 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. QGP1 cells were cultured in RPMI-1640 medium supplemented with 10% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin.
DNA synthesis was measured by Click-iT Plus EdU Alexa Fluor 647 Flow Cytometry Assay Kit (Invitrogen), according to the manufacturer's protocol.
Two miRCURY LNA Power miRNA inhibitors (Qiagen/Exiqon, Germantown, MD) were number 4103450102 for has-miR-431-5p (sequence: 5’-GCATGACGGCCTGCAAGAC-3’) and negative control A, number 199006102 (sequence: 5’-TAACACGTCTATACGCCCA-3’).
Analysis of miRNA and mRNA Expression
For miRNA analyses, total RNA was extracted from various cell lines using microRNeasy mini kit (Qiagen). miRNAs were reverse transcribed by TaqMan miRNA reverse transcription kit (number 4366596; Applied Biosystems, Foster City, CA) using 10 ng of total RNA, and the expression levels of mature miRNAs were quantified using the TaqMan miRNA expression assay (number 4427975; Applied Biosystems). Mouse snoRNA202 and human SNORD48 (previously known as RNU48) were used as endogenous controls for normalization.
For mRNA expression analyses, mRNA was isolated from cells grown on 6- or 10-cm plates using Trizol with DNase I treatment (Invitrogen) or RNeasy mini kit (Qiagen) containing genomic DNA Eliminator spin columns. Total RNA (5 μg) was reverse transcribed using cDNA First-Strand Synthesis Kit (Invitrogen), and 20 ng of the resulting cDNA was then mixed with SYBR Green PCR Master Mix (Applied Biosystems) and the appropriate primers. Each reaction was performed in quadruplicate, and mRNA expression was quantified by performing real-time PCR amplification using an ABI Prism 7900HT Real-Time PCR System (Applied Biosystems). The primer sequences are as follows: CDH1 (mouse, forward: 5′-CAGGTCTCCTCATGGCTTTGC-3′; reverse: 5′-CTTCCGAAAAGAAGGCTGTCC-3′), CDH2 (mouse, forward: 5′-CATCAACCGGCTTAATGGTG-3′; reverse: 5′-ACTTTCACACGCAGGATGGA-3′), ZEB2 (mouse, forward: 5′-ATGGCAACACATGGGTTTAGTGGC-3′; reverse: 5′-ATTGGACTCTGAGCAGATGGGTGT-3′), SNAI2 (mouse, forward: 5′-CACATTCGAACCCACACATTGCCT-3′; reverse: 5′-TGTGCCCTCAGGTTTGATCTGTCT-3′), ACTB (mouse, forward: 5′-ATAGGAGTCCTTCTGACCCATTCC-3′; reverse: 5′-ATGACGATATCGCTGCGCTGGT-3′), and B2M (mouse, forward: 5′-ATGCTGAAGAACGGGAAAAA-3′; reverse: 5′-CAGTCTCAGTGGGGGTGAAT-3′) with the comparative CT method (ΔΔCT). Both ACTB and B2M were used as endogenous controls for normalization.
Animal Experiments, Tissue Preparation, and Immunohistochemical Analysis
Generation of RIP-Tag; RIP-tva mice has been described.17 Nonobese diabetic/scid-IL2Rgc knockout mice were generated by the Jackson Laboratory (Bar Harbor, ME). All mice were housed in accordance with institutional guidelines. All procedures involving mice were approved by the institutional animal care and use committee. For the experimental metastasis assay, 1 × 106 N134 cells in 150 μL phosphate-buffered saline were injected into the tail veins of male nonobese diabetic/scid-IL2Rgc knockout mice at the age of 7 to 8 weeks. For in vivo infection, viral supernatant was passed through 0.45-μm filters and was concentrated by high-speed ultracentrifugation at 95,400 × g for 1.5 hours before intracardiac injection into RIP-Tag; RIP-tva mice at 7 weeks of age. Mice were sacrificed when they were 16 weeks of age or sick. Intracardiac injection of concentrated RCAS viruses to RIP-Tag; RIP-tva mice and viral titer determination by end-point dilution of DF1 producer cells were performed as described.17,18
Tissues were removed and fixed in 10% buffered formalin overnight at room temperature. Formalin-fixed, paraffin-embedded sections (5 μm thick) were deparaffinized and rehydrated by passage through a graded xylene/ethanol series before staining. Hematoxylin and eosin staining was performed. Immunochemistry was performed using VECTASTAIN Elite ABC kit, following the manufacturer's instructions. The primary antibody used was rabbit antisynaptophysin (1:500; VP-S284; Vector Labs, Burlingame, CA).
Active Ras Pull-Down Assay
Cells at 80% confluency from 4- to 10-cm plates were combined and lysed in 1 mL lysis buffer (25 mmol/L Tris-HCl, pH 7.2, 150 mmol/L NaCl, 5 mmol/L MgCl2, 1% NP-40, and 5% glycerol). Of total lysate of each cell line, 6% was saved before the pull-down assay. Protein of each cell line (500 μg) was processed with the Active Ras Pull-Down and Detection Kit (number 16117; Thermo Scientific, Waltham, MA). Activated Ras proteins were precipitated with a GST-fusion protein of the Ras-binding domain of Raf1 along with glutathione agarose resin, following the manufacturer's instruction. Precipitated proteins were eluted from beads using 2 × loading buffer (12 mmol/L Tris, pH 6.8, 5% glycerol, 0.4% SDS, 140 mmol/L 2-mercaptoethanol, and 0.02% bromophenol blue), separated by SDS-PAGE, and analyzed by Western blot analysis with anti-Ras antibody provided in the kit. The intensities of bands are quantified by Fiji software version 2.0.0-rc-41/1.40d (NIH, Bethesda, MD). The relative ratios were normalized to bands from the first lane.
Western Blot Analysis and Immunoprecipitation
Cells were lysed in NP-40 buffer (100 mmol/L NaCl, 100 mmol/L Tris, pH 8.2, and 0.5% NP-40) supplemented with a protease inhibitor mixture and PhosSTOP (Roche). Proteins were quantified by Bradford assay (Bio-Rad, Hercules, CA). Equal amounts of proteins were separated by SDS-PAGE and transferred to nitrocellulose membranes. To visualize equal protein loading, blots were stained with Ponceau S. Blots were incubated in 5% nonfat milk in Tris-buffered saline and Tween 20, probed with primary antibodies to phosphorylated extracellular signal-regulated kinase 1/2 (p-Erk1/2; 1:1000; 4376; Cell Signaling Technology, Danvers, MA), total Erk1/2 (1:1000; 9102; Cell Signaling Technology), cadherin 1 (1:1000; 610181; BD Biosciences), α-tubulin (1:1000; T5168; Sigma, St. Louis, MO), Slug (1:1000; ab27568; Abcam, Cambridge, MA), and DAB2 interacting protein (DAB2IP; 1:800; gift from Dr. Jer-Tsong Hsieh), and then incubated with horseradish peroxidase–conjugated secondary antibodies. Protein bands were visualized by enhanced chemical luminescence (Pierce, Rockford, IL).
In Vitro Transwell Migration and Invasion Assay
For migration assays, 0.5 × 106 or 1 × 106 of cells were seeded in the upper chambers of 8-μm porous polycarbonate membranes with DMEM containing 2% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. The lower chambers were filled with DMEM containing 10% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. After 72 hours, cells migrating to the bottom chambers were fixed, stained with 0.1% crystal violet for 30 minutes, and counted in eight fields under ×10 magnification.
For invasion assays, 2.5 to 5 × 104 cells were seeded in the Matrigel-coated transwell chambers (BD Biosciences) with DMEM containing 1% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. The lower chambers were filled with DMEM containing 10% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. After 48 hours, cells on the opposite side of the chambers were fixed with 4% paraformaldehyde for 20 minutes, stained with 0.1% crystal violet for 30 minutes, and counted in eight fields under ×20 magnification.
For SCH772984 (Selleck Chemicals LLC, Houston, TX) treatment, N134 cells were pretreated with dimethyl sulfoxide (vehicle control) or 1 μmol/L SCH772984 for 7 days. A total of 2.5 × 104 cells were seeded in Matrigel-coated transwell chambers with either dimethyl sulfoxide or 1 μmol/L SCH772984 in both the upper and lower chambers.
For invasion assays of transiently transfected cells, 106 cells were seeded in the Matrigel-coated transwell chambers (BD Biosciences) with DMEM containing 1% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. The lower chambers were filled with DMEM containing 10% FBS, 0.2 mmol/L l-glutamine, and 1% penicillin/streptomycin. After 72 hours of incubation, cells on the opposite side of the chambers were fixed by 4% paraformaldehyde for 20 minutes, stained with 0.1% crystal violet for 30 minutes, and counted in all the fields under ×20 magnification. Experiment was conducted in triplicates. To analyze cell proliferation, 0.5 × 106 cells were seeded directly onto 24-well plates in triplicate. After 72 hours, cells were fixed by 4% paraformaldehyde for 20 minutes, stained with 0.1% crystal violet for 30 minutes, and lysed by 500 μL methanol with gentle shaking at the speed of 200 rpm for 20 minutes. Cell lysate from each well (200 μL) was transferred to a 96-well plate, and OD560 was read.
For QGP1 cell invasion assay, a control inhibitor (LNA-CTRL) or miR-431 inhibitor (LNA-miR-431) was transfected to QGP1 cells. After 4 days, 7.5 × 104 cells were seeded in Matrigel-coated transwell chambers. Forty-eight hours later, cells were counted as described.23
Total RNA Extraction, RNA-Seq Analysis, and Computational Analysis
Total RNA was extracted using the RNeasy Plus mini kit (74134; Qiagen) and QIAshredder kit (79654; Qiagen), according to the manufacturer's protocol. After isolation, the total RNA integrity was checked using an Agilent Technologies (Santa Clara, CA) 2100 Bioanalyzer. cDNA libraries were generated using the Illumina TruSeq RNA Sample Preparation kit and sequenced with pair-end 51 bp on HiSeq4000 sequencer (Illumina, San Diego, CA). Tophat version 2.0.1124 was used to align sequencing reads to the mm9 mouse reference genome, and Cufflinks version 2.1.125,26 was used to measure transcript abundances in fragments per kilobase of exon model per million mapped reads and to identify differentially expressed genes. The RNA-sequencing (RNA-Seq) data sets of this study are available in Gene Expression Omnibus (https://www.ncbi.nlm.nih.gov/geo; accession number GSE139119). Gene set enrichment analysis was used to determine whether an a priori defined set of genes shows statistically significant, concordant differences between N134 cells expressing GFP-miR-431 and N134 cells expressing GFP-miR-LacZ (control).
RPPA Profiling and Analysis
Reverse phase protein array (RPPA) was performed as described previously with minor modifications.27, 28, 29, 30 Protein lysates were prepared from cultured cells with modified Tissue Protein Extraction Reagent (Life Technologies, Carlsbad, CA) and a cocktail of protease and phosphatase inhibitors (Roche). The lysates were diluted into 0.5 mg/mL in SDS sample buffer and denatured on the same day. The Aushon 2470 Arrayer (Aushon BioSystems, Billerica, MA) with a 40-pin (185-μm) configuration was used to spot samples and control lysates onto nitrocellulose-coated slides (Grace Bio-Labs, Bend, OR) using an array format of 960 lysates/slide (2880 spots/slide). The slides were processed as described and probed with a set of 224 antibodies against total proteins and phosphoproteins using an automated slide stainer Autolink 48 (Dako, Santa Clara, CA). Each slide was incubated with one specific primary antibody, and a negative control slide was incubated with antibody diluent without any primary antibody. Primary antibody binding was detected using a biotinylated secondary antibody followed by streptavidin-conjugated IRDye680 fluorophore (LI-COR Biosciences, Lincoln, NE). Total protein content of each spotted lysate was assessed by fluorescent staining with Sypro Ruby Protein Blot Stain, according to the manufacturer's instructions (Molecular Probes, Eugene, OR).
Fluorescence-labeled slides were scanned on a GenePix 4400 AL scanner, along with accompanying negative control slides, at an appropriate photomultiplier tubes voltage setting to obtain optimal signal for this specific set of samples. The images were analyzed with GenePix Pro 7.2 (Molecular Devices, San Jose, CA). Total fluorescence signal intensities of each spot were obtained after subtraction of the local background signal for each slide and were then normalized for variation in total protein, background, and non-specific labeling using a group-based normalization method, as described.27 For each spot on the array, the background-subtracted foreground signal intensity was subtracted by the corresponding signal intensity of the negative control slide (omission of primary antibody) and then normalized to the corresponding signal intensity of total protein for that spot. Each image, along with its normalized data, was evaluated for quality through manual inspection and control samples. Antibody slides that failed the quality inspection were either repeated at the end of the staining runs or removed before data reporting. Total 211 antibodies remained in the list.
Transfection and 3′ UTR Luciferase Reporter Assay
The 3′ untranslated region (UTR) of mouse DAB2IP was amplified by PCR and cloned into a pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega, Madison, WI). HEK 293T cells were cotransfected with miRNA mimic (Invitrogen) and pmirGLO Dual-Luciferase 3′ UTR vector using the Lipofectamine 2000 Transfection Agent following the manufacturer's instructions (Invitrogen). Scrambled miRNA mimic (number 4464066; Invitrogen) was a control. 3′ UTR mutant was a mutant form of the DAB2IP 3′ UTR within the miR-431 binding site. Forty-eight hours after transfection, cells were analyzed for luciferase activity using the Dual-Glo Luciferase Assay System (Promega). QGP1 cells were transfected with pmirGLO Dual-Luciferase 3′ UTR vector using Lipofectamine 3000 Transfection Agent, following the manufacturer's instructions (Invitrogen).
Statistical Analysis
To compare the survival outcome between groups, Kaplan-Meier method was used to estimate the survival probability, followed by a log-rank test. Two-sample t-test was used to compare numerical outcomes. Fisher exact test was used to compare categorical outcomes. All statistical tests were two sided, with an α level of 0.05 as the significance cutoff. Statistical analyses were performed in statistical software SAS version 9.4 (SAS Institute, Cary, NC).
The normalized values were used in RPPA data analysis. The three replicas of each sample were log2 transformed and then averaged before analysis as a biological replicate. R package limma version 3.28.21 (https://bioconductor.org/packages/3.2/bioc/html/limma.html)31 was used to identify the differentially expressed protein between treatment and control with a Benjamini-Hochberg false discovery rate of 0.05 as the significance cutoff.
Results
miR-431 Promotes Metastasis in a Spontaneous Mouse Model of PNETs in Vivo
We have developed a novel somatic gene transfer system using RIP-Tag; RIP-tva mouse model to study the function of genes in metastasis.17,18 To evaluate whether miR-431 can promote PNET metastasis, the RIP-Tag; RIP-tva mouse model was used. RCASBP viruses encoding the cDNA for GFP-miR-431 or GFP-miR-LacZ were generated. High titer virus stocks (0.1 mL; >108 infectious units per milliliter) were injected into 7-week–old RIP-Tag; RIP-tva mice intracardially. RCASBP-GFP-miR-LacZ was used as a negative control for adverse effects of viral infection per se. Nine weeks after infection, the pancreas and other organs were harvested for histologic staging and grading of the lesions. To aid the search for PNET metastases, one pancreatic tissue section per mouse was subjected to immunohistochemical staining for synaptophysin, a neuroendocrine marker. Approximately three pancreatic lymph nodes were found per pancreatic tissue section. Pancreatic lymph node metastases were readily observed by synaptophysin staining in 7 of 20 mice (10 males and 10 females) injected with RCASBP-GFP-miR-431, but not in any of the control mice (six males and four females) injected with RCASBP-GFP-miR-LacZ (Figure 1A and Table 1). Among the seven mice that developed pancreatic lymph node metastases by miR-431 overexpression, six of them were males. There was an increasing trend approaching significance that miR-431 promoted more pancreatic lymph node metastasis in male RIP-Tag; RIP-tva mice than in female mice (P = 0.0573; Fisher exact test).
Figure 1.
miR-431 promotes lymph node metastasis in the spontaneous pancreatic neuroendocrine tumor (PNET) mouse model. A:RIP-Tag; RIP-tva mice were infected with the indicated RCASBP retroviruses at 7 weeks of age and sacrificed at 16 weeks of age. Lymph node metastasis was only observed in mice infected with RCASBP-GFP-miR-431, not in mice infected with RCASBP-GFP-miR-LacZ. Images show representative synaptophysin staining of metastatic PNET in pancreatic lymph nodes. B: Kaplan-Meier survival curve for RIP-Tag; RIP-tva mice infected with RCASBP-GFP-miR-LacZ and RCASBP-GFP-miR-431. C: Blood glucose levels of RIP-Tag; RIP-tva mice infected with RCASBP-GFP-miR-LacZ and RCASBP-GFP-miR-431 at the end point. D: Tumor burden of RIP-Tag; RIP-tva mice infected with RCASBP-GFP-miR-LacZ and RCASBP-GFP-miR-431 at the end point. *P < 0.05, ***P < 0.001 versus miR-LacZ. Scale bars = 20 μm (A).
Table 1.
Impact of miR-431 Overexpression on Spontaneous Metastasis of RIP-Tag; RIP-tva in Vivo
| RCASBP- | Lymph node metastases |
|---|---|
| GFP-miR-LacZ | 0/10 mice (0%) |
| GFP-miR-431 | 7/20 mice (35%)∗ |
GFP, green fluorescent protein; RCASBP, replication competent ALV LTR with a splice, bryan-strain pol gene.
P < 0.05 (Fisher exact test).
miR-431 group had 5.53 times higher risk of death than miR-LacZ control group, and the survival difference was significantly different (P = 0.0003; log-rank test) (Figure 1B). Because PNETs in RIP-Tag; RIP-tva mice secreted insulin and lowered blood glucose levels, blood glucose levels of the mice were also measured as a severity of illness indicator. The nonfasting glucose levels of mice infected with RCASBP-GFP-miR-431 were significantly lower than those of the mice injected with RCASBP-GFP-miR-LacZ at the end point (Figure 1C). On the other hand, the primary tumor burden of mice infected with RCASBP-GFP-miR-431 was not significantly different from that of control mice at the end point (Figure 1D). These data suggested that miR-431 promotes spontaneous lymph node metastasis, but not the growth of primary PNETs.
miR-431 Increases Migration and Invasion of PNETs in Vitro and Promotes PNET Metastasis in a Tail Vein Assay of Metastasis in Vivo
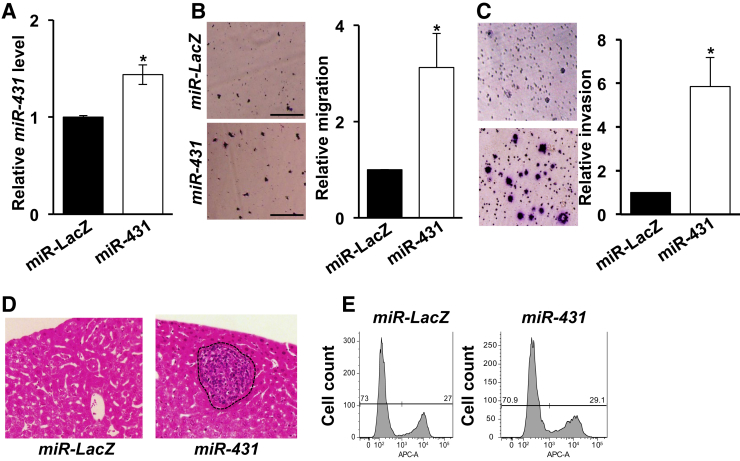
For in vitro studies, mouse PNET N134 cells expressing either GFP-miR-431 or control GFP-miR-LacZ were generated. The expression levels of miR-431 in these cell lines were determined by the TaqMan miRNA expression assay, and miR-431 was increased approximately 1.5-fold in GFP-miR-431–expressing cells than in control GFP-miR-LacZ–expressing cells (Figure 2A). To examine whether miR-431 has a role in PNET migration and invasion, in vitro transwell migration assays and invasion assays were used. Cell migration or invasion was measured through Matrigel along a serum gradient through the membrane for 48 or 72 hours. GFP-miR-431–expressing cells increased migration and invasion compared with GFP-miR-LacZ–expressing cells (Figure 2, B and C), suggesting that miR-431 promotes migration and invasion of mouse PNETs.
Figure 2.
miR-431 facilitates pancreatic neuroendocrine tumor migration, invasion, and metastasis in vitro and in vivo.A: Increased levels of miR-431 in N134/GFP-miR-431 compared with N134/GFP-miR-LacZ (control). B: Migration of N134/GFP-miR-LacZ and N134/GFP-miR-431 cells was determined using in vitro transwell migration chambers with a serum gradient (1% to 10%). Representative images of the migrated cells are shown. Data are expressed as the normalized number of migrated cells in the bottom chambers in eight fields under ×10 magnification after 72 hours relative to that of control cells from five independent experiments. C: Invasion of N134/GFP-miR-LacZ and N134/GFP-miR-431 cells was determined using in vitro Matrigel invasion chambers with a serum gradient (1% to 10%). Representative images of the invaded cells are shown. Data are expressed as the normalized number of invaded cells in the bottom surface of chambers in eight fields under ×10 magnification after 48 hours relative to that of control cells from four independent experiments. D:miR-431 increases the incidence of metastasis formation compared with miR-LacZ in tail vein metastasis assay in nonobese diabetic/scid-IL2Rgc knockout mice. Liver sections from mice injected with N134/GFP-miR-LacZ or N134/GFP-miR-431 through tail vein were stained with hematoxylin and eosin. The dashed line indicates macrometastasis. E: Flow cytometric analysis of N134/GFP-miR-LacZ and N134/GFP-miR-431 cells showing the number of proliferating [5-ethynyl-2′-deoxyuridine (EdU)-positive] cells after 1-hour treatment of 10 μmol/L EdU. Representative images are shown; two-tailed t-test was used. Data are expressed as means ± SEM (B and C). Error bars represent SDs (A). n = 5 (E). *P < 0.05 versus miR-LacZ (two-tailed t-test). Scale bars = 200 μm (B). Original magnification: ×10 (C); ×20 (D).
To further investigate the metastatic effect of miR-431 in vivo, an experimental metastasis assay was performed. A total of 1.5 million of GFP-miR-LacZ–expressing N134 cells or GFP-miR-431 cells were injected into the tail vein of male nonobese diabetic/scid-IL2Rgc knockout immunodeficient mice. Six weeks after the injection, livers of the recipient mice were harvested to survey for metastasis. The four major liver lobes of each mouse were processed into histologic section with hematoxylin and eosin staining. One section of each major liver lobe was examined for the presence of metastases. Although only two of seven mice receiving control GFP-miR-LacZ–expressing N134 cells developed metastases, and six of six mice receiving GFP-miR-431–expressing N134 cells developed liver metastases (Table 2 and Figure 2D). These results suggested that miR-431 promotes metastasis of PNET cells.
Table 2.
Impact of miR-431 Overexpression on Experimental Metastasis in Vivo
| N134 cells | Liver metastases |
|---|---|
| GFP-miR-LacZ | 2/7 mice (28.6%) |
| GFP-miR-431 | 6/6 mice (100%)∗ |
P < 0.05 (Fisher exact test).
To determine the effect of miR-431 on cell proliferation, 5-ethynyl-2´-deoxyuridine (EdU) assay was performed. The flow cytometric analysis of cells labeled with Alexa Fluor EDU demonstrated that miR-431 did not significantly increase cell proliferation (Figure 2E). This was consistent with no significant change of primary tumor burden in mice infected with RCASBP-GFP-miR-431 (Figure 1D).
miR-431 Increases Ras/Erk Signaling and Induces EMT of PNETs
To investigate potential pathways regulated by miR-431 at the mRNA expression levels and the functional protein levels, RNA-Seq was performed using N134 cells expressing GFP-miR-LacZ (control) or GFP-miR-431. Gene set enrichment analysis showed that the 16 gene sets were significantly enriched at nominal P = 0.01 in N134-expressing GFP-miR-431, five of them were oncogenic KRAS signatures, and one of them was a constitutively active MEK signature (Figure 3A and Supplemental Figure S1). To validate the effect of miR-431 on Ras signaling activation, an active Ras pull-down assay was performed. Cell lysates were prepared from N134 cells expressing GFP-miR-LacZ or GFP-miR-431 and a human pancreatic ductal adenocarcinoma (PDAC) cell line, AsPC1. AsPC1, which has an activating KRASG12D mutation32 and active p-Erk1/2 signaling, served as a positive control (Figure 3B). A GST-fusion protein of the Ras-binding domain of Raf1, along with glutathione agarose resin, was used to specifically pull down active Ras. Western blot analysis using an anti-Ras antibody showed higher levels of active Ras and p-Erk1/2 in GFP-miR-431–expressing N134 cells than in GFP-miR-LacZ–expressing N134 cells (Figure 3B).
Figure 3.
miR-431 activates Erk and EMT. A: RNA-sequencing (RNA-Seq) and gene set enrichment analysis shows a correlation between miR-431 expression and KRAS signature in N134 cells. B: Cell lysates were prepared from N134/GFP-miR-LacZ and N134/GFP-miR-431 and a human pancreatic ductal adenocarcinoma cell line, AsPC1. Western blot analysis shows precipitated active Ras protein, total Ras, phosphorylated extracellular signal-regulated kinase 1/2 (p-Erk1/2), and total Erk1/2 (t-Erk1/2). The intensity of the Ras protein bands was quantified and normalized (Norm.) to the Ras bands in the N134/GFP-miR-LacZ cells. C: Western blot analysis for p-Erk1/2, total Erk1/2, cadherin 1, Slug, and tubulin. D: RNA-Seq analysis in fragments per kilobase of transcript per million mapped reads (FPKM) reveals up-regulation of mesenchymal markers, CDH2, ZEB2, and SNAI2, in N134/GFP-miR-431 cells compared with those in the control N134/GFP-miR-LacZ cells. t-Test was used. E: Real-time PCR analysis of CDH1, CDH2, ZEB2, and SNAI2 mRNAs in N134/GFP-miR-LacZ and N134/GFP-miR-431 cells. Two-tailed t-test was used. Data are expressed as means ± SEM (D and E). n = 7 independent experiments (E). *P < 0.05 versus miR-LacZ. NES, normalized enrichment score.
Moreover, RPPA analysis, a high-throughput antibody-based technique for simultaneously measuring multiple protein expression levels, was performed.28 This led to an identification of 10 differentially up-regulated proteins by miR-431 that function in epithelial-mesenchymal transition (EMT), migration, invasion, and metastasis in N134 cells (Table 3). Among them, p-Erk was the most up-regulated protein in cells expressing GFP-miR-431 (fold change, 5.2). It was independently verified that p-Erk levels were higher in GFP-miR-431–expressing N134 cells than in the control cells by Western blot analysis (Figure 3C).
Table 3.
Protein Up-Regulated in N134/GFP-miR-431 Cells Compared with N134/GFP-miR-LacZ Cells in RPPA Analysis
| Protein name | Fold change (miR-431 vs miR-LacZ) | Adjusted P value |
|---|---|---|
| p-Erk1/2 (T202/Y204) | 5.20 | 0.031 |
| Slug | 1.72 | 0.000 |
| Cadherin 2 | 1.70 | 0.015 |
| PKM1/2 | 1.31 | 0.004 |
| p-FAK (Y576/577) | 1.30 | 0.004 |
| p-Stat3 (S727) | 1.29 | 0.004 |
| Sin3b | 1.24 | 0.018 |
| Stat6 | 1.23 | 0.007 |
| Ezh2 | 1.19 | 0.007 |
| Erk1/2 | 1.17 | 0.047 |
Erk1/2, extracellular signal-regulated kinase 1/2; p-Erk1/2, phosphorylated Erk1/2; RPPA, reverse phase protein array.
It has been reported that Ras/Erk2 signaling induced EMT.33 GFP-miR-431–expressing N134 cells were more mesenchymal-like, with up-regulation of CDH2, ZEB2, and SNAI2 in the RNA-Seq data (Figure 3D). Real-time PCR also demonstrated that miR-431 down-regulated an epithelial marker, CDH1, and up-regulated mesenchymal markers, including CDH2, SIP1, and SLUG (Figure 3E). In addition, Western blot analysis showed down-regulation of cadherin 1 and up-regulation of Slug in GFP-miR-431–expressing N134 cells compared with GFP-miR-LacZ–expressing N134 cells (Figure 3C). Taken together, the results of RNA-Seq, Western blot analysis, and RPPA revealed the function of GFP-miR-431 in increasing Ras/Erk signaling and inducing EMT of PNET cells.
To determine whether Erk signaling plays a role for PNET cell invasion, N134/GFP-miR-431 cells were treated with an Erk inhibitor, SCH772984, or dimethyl sulfoxide vehicle control; and the effect on cell invasion was measured using transwell invasion assay. SCH772984 greatly reduced p-Erk levels (Figure 4A) and reduced cell invasion (Figure 4B), suggesting that active Erk signaling is required for PNET cell invasion.
Figure 4.
Extracellular signal-regulated kinase (Erk) inhibition suppresses invasion induced by miR-431 in vitro. A: Western blot analysis for phosphorylated Erk1/2 (p-Erk1/2) and total Erk1/2 (t-Erk1/2) in N134/GFP-miR-LacZ and N134/GFP-miR-431 cells treated with dimethyl sulfoxide (DMSO; vehicle control) or SCH772984 (SCH). B: Invasion of cells pretreated with DMSO (vehicle control) or SCH772984 (1 μmol/L) using in vitro Matrigel invasion chambers with a serum gradient (1% to 10%). Data are expressed as the normalized number of invaded cells in the bottom surface of chambers in eight fields under ×10 magnification after 48 hours relative to that of vehicle treated control cells. Data are expressed as means ± SEM (B). *P < 0.05 (two-tailed t-test). Scale bars = 200 μm (B).
miR-431 Directly Targets DAB2IP, a RasGAP
The mature sequence of miR-431 (5’-UGUCUUGCAGGCCGUCAUGCA-3’) is identical among human, rat, and mouse (Figure 5A). To identify mRNA targets of miR-431, four target prediction modules, including miRWalk,34 miRnada,35 RNA22,36 and Targetscan, were searched.37 The putative miR-431 target genes that were predicted by at least three modules and showed an inverse correlation with miR-431 expression in the RNA-Seq described above were selected. This approach identified 202 potential miR-431 target genes (Figure 5B and Supplemental Table S1). They were subjected to DAVID gene-enrichment and functional annotation analysis. Functional annotation clustering showed enrichment of GTPase activator activity among the top three clusters (Supplemental Table S2). DAB2IP was identified as the top potential target of miR-431 in PNETs (Figure 5, B and C).
Figure 5.
miR-431 directly targets DAB2IP. A: Sequence alignment of miR-431 from human, rat, and mouse. Asterisks indicate identical nucleotides. B:miR-431 target sequence and the potential binding site at the 3′ untranslated region (UTR) of mouse DAB2IP. DAB2IP 3′ UTR mutant (Mut) indicates the mutated sites in the seeding sequence. C: Alignment of miR-431 targeting sequence (red: seed region) in DAB2IP 3′ UTR and the sequence of Mut DAB2IP 3′UTR. D:DAB2IP expression was measured by RNA sequencing in fragments per kilobase of transcript per million mapped reads (FPKM). **q = 0.004, q values represent false discovery rate–adjusted P values outputted by Cuffdiff. E: Western blot analysis for DAB2IP to demonstrate the repression of DAB2IP in N134/GFP-miR-431 cells. F: Dual-luciferase assays of pmirGLO or wild-type (WT) 3′ UTR or 3′ UTR Mut of DAB2IP after cotransfection with miR-431 mimic or scrambled miRNA. G: Dual-luciferase assays after transfection with pmirGLO or DAB2IP WT 3′ UTR or 3′ UTR Mut in QGP1 cells. F and G: Normalized firefly luciferase activity (firefly luciferase activity/Renilla luciferase activity) for each construct was compared with that of the pmirGLO vector no-insert control. Three independent measurements were performed. Data are expressed as means ± SEM (D, F, and G). **P < 0.01 versus miR-LacZ; ††P < 0.01; ‡‡P < 0.01 versus vector alone or the mutant 3’ UTR of DAB2IP.
DAB2IP is a RasGAP, and it acts as a tumor suppressor gene in prostate cancer.22 The mRNA expression of DAB2IP was down-regulated [log2 (fold change) = −0.427264] in GFP-miR-431–expressing N134 cells compared with GFP-miR-LacZ–expressing N134 cells in the above RNA-Seq analysis (Figure 5D). DAB2IP protein levels were also down-regulated in GFP-miR-431–expressing N134 cells compared with control GFP-miR-LacZ–expressing N134 cells (Figure 5E).
To validate whether DAB2IP is a direct target of miR-431, a luciferase reporter construct containing a partial sequence of the DAB2IP 3′ UTR encompassing the miR-431 site was used. Wild-type or mutant 3′ UTR of mouse DAB2IP (Figure 5C) was cloned into a pmirGLO dual-luciferase miRNA target expression vector. Unexpectedly, a putative miR-431 target sequence was found in pmirGLO vector, so cotransfection of miR-431 reduced the luciferase activity of the control pmirGLO vector compared with cotransfection of scrambled miRNA control (Figure 5F). Cotransfection of miR-431 and the vector carrying wild-type 3′ UTR of mouse DAB2IP further repressed the luciferase activity in 293T cells (Figure 5F). Consistent with its direct targeting, mutating the miR-431-5p complementary seed region (Figure 5C) on the 3′ UTR of DAB2IP abrogated miRNA-mediated regulation (Figure 5F).
Moreover, the luciferase reporter–based activity assay was performed in human PNET QGP1 cell line with its endogenous miR-431. The pmirGLO vector, the vector carrying wild-type 3′ UTR of DAB2IP, or the vector carrying mutant 3′ UTR was transfected to QGP1 cells, and their luciferase activities were measured. Consistent with the cotransfection results in 293T cells, luciferase reporter activity of wild-type 3′ UTR of DAB2IP was significantly lower than that of the vector alone or the mutant 3′ UTR of DAB2IP (Figure 5G). Taken together, these findings established DAB2IP as a direct target of miR-431.
GAP Activity of DAB2IP Interferes with the Metastatic Function of miR-431
DAB2IP protein identity is >80% among human, rat, and mouse. To determine whether the metastatic function of miR-431 is mediated by targeting DAB2IP, rat DAB2IP homolog, or a mutant defective in its GAP activity22,38 was expressed in N134/GFP-miR-431 cells. Both of cDNAs did not have the 3′ UTR binding site of miR-431 and therefore could not be down-regulated by miR-431 (miR-431 resistant). Overexpression of miR-431–resistant rat DAB2IP cDNA reversed the invasive effect of miR-431 without affecting cell proliferation (Figure 6, A and B). Moreover, miR-431–resistant rat DAB2IP reduced p-Erk and Slug levels and increased cadherin 1 (Figure 6C). These effects required the GAP activity, because overexpression of the GAP mutant in N134 cells expressing GFP-miR-431 did not sufficiently inhibit cell invasion, reduce p-Erk, or block EMT (Figure 6, A–C). The data suggested that the GAP activity of DAB2IP is important to inhibit PNET invasion.
Figure 6.
Expression of human DAB2IP and rat homolog blocks the invasiveness effect of miR-431, and miR-431 inhibition suppresses invasion of human pancreatic neuroendocrine tumor (PNET) cells. A: Invasion of N134/GFP-miR-431 expressing a control vector (pCI-neo), rat DAB2IP or GTPase activating protein (GAP) mutant of DAB2IP was determined using in vitro Matrigel invasion chamber with a serum gradient (1% to 10%). Forty-eight hours after transfection of DNA, cells were trypsinized and 106 cells were seeded onto Matrigel invasion chamber. After 72 hours of incubation, cells on the opposite side of the chambers were fixed, stained, and counted. Experiment was conducted in triplicate. Data are expressed as the normalized number of invaded cells in the bottom surface of chambers in all fields under ×20 magnification. B: Proliferation of N134/GFP-miR-431 expressing a control vector (pCI-neo), rat DAB2IP or GAP mutant was compared. C: Western blot analysis for rat DAB2IP using DAB2IP antibodies, phosphorylated extracellular signal-regulated kinase 1/2 (p-Erk1/2), total Erk1/2 (t-Erk1/2), E-cadherin, and Slug in N134/GFP-miR-431 cells expressing a control vector (pCI-neo), rat DAB2IP, or GAP mutant. D: Expression levels of miR-431, determined by TaqMan miRNA expression assay, in three human pancreatic ductal adenocarcinoma (PDAC) cell lines and two human PNET cell lines. E: Invasion of human PNET cells (QGP1) treated with a control inhibitor (LNA-CTRL) or a miR-431 inhibitor (LNA-miR-431) was determined using in vitro Matrigel invasion chambers with a serum gradient (1% to 10%). F: Western blot analysis for DAB2IP and tubulin in human PNET cells (QGP1) treated with control inhibitor (LNA-CTRL) or miR-431 inhibitor (LNA-miR-431). G: Model depicting the mechanism by which miR-431 promotes metastasis of PNETs. Data are expressed as means ± SEM (A, B, D, and E). n = 5 (E). *P < 0.05 versus Panc1, MIA Paca-2, and BxPC3 (two-tailed t-test); †P < 0.05 versus LNA-CTRL (two-tailed t-test). Scale bars = 200 μm (E). EMT, epithelial-mesenchymal transition; GEF, guanine nucleotide exchange factor.
miR-431 as a Therapeutic Target in PNET Metastasis
By real-time PCR, it was found that two human PNET cell lines, BON1 and QGP1, expressed higher endogenous miR-431 levels than three human PDAC cell lines, Panc1, MIA Paca-2, and BxPC3 (Figure 6D). Especially, QGP1 had the highest endogenous miR-431 levels among these five cell lines (Figure 6D). Consistent with our findings that DAB2IP is a direct target of miR-431 in pancreatic cancer cells, Western blot analysis showed an inverse correlation between the expression levels of miR-431 and the DAB2IP protein levels in these five human PDAC and PNET cell lines (Figure 6D).
To determine therapeutic value of targeting miR-431, QGP1 cells were treated with antisense locked nucleic acids (LNAs).39 LNAs sequester their target miRNAs in highly stable heteroduplexes, thereby effectively preventing the miRNA from hybridizing with its normal cellular targets. Transfection of an LNA inhibitor for human miR-431-5p significantly inhibited invasiveness of QGP1 compared with transfection of a control LNA (Figure 6E), restored the expression of DAB2IP, and down-regulated Erk signaling (Figure 6F).
Discussion
The incidence of PNETs is increasing in recent years. Because of the indolent nature of the disease, most PNET patients are diagnosed with liver metastasis. The drivers of metastatic PNETs are largely unknown, which makes the development of treatment difficult. Through a cross-species analysis in human PNETs and mouse PNETs in RIP-Tag mouse models, a metastatic-specific miRNA signature has been revealed.11 miR-431 is the most up-regulated miRNA in this signature. The function of miR-431 in tumorigenesis appears to be context dependent. miR-431 acts as tumor suppressor in colorectal cancer cells and hepatocellular carcinoma. It inhibits migration of colorectal cancer cells and hepatocellular carcinoma cells.40,41 In contrast, miR-431 demonstrated promoted migration and invasion of PNET in vitro, and metastasis in both spontaneous PNET and experimental metastasis mouse models, suggesting its oncogenic role in PNETs. The disparity of miR-431 functions could be because of the availability of its targets in different cancer types. Further studies are needed to address this possibility.
Although oncogenic KRAS is found in nearly every PDAC to drive tumorigenesis,42,43 KRAS mutations are virtually nonexistent in PNETs.44 On the other hand, miR-431 overexpression is not detected in PDAC,45 suggesting that the two mechanisms are redundant and likely mutually exclusive in pancreatic tumorigenesis. This study demonstrated that metastatic PNETs up-regulated miR-431 to activate the Ras/Erk pathway through targeting DAB2IP. Our study suggested that the Ras signaling pathway is also important in PNET progression. DAB2IP, a RasGAP, negatively controls Ras-dependent mitogenic signals.46 DAB2IP has recently been found significantly down-regulated in multiple types of cancer,47 including prostate cancer,48 breast cancer,49,50 colon cancer,51 and bladder cancer.52 Down-regulation of DAB2IP has been shown to promote prostate cancer progression53 and colorectal cancer invasion and metastasis.51 The aberrant alteration of DAB2IP in cancer is caused by a variety of mechanisms, including targeted by miRNAs, aberrant promoter methylation, histone deacetylation, and others. Herein, it was shown that DAB2IP suppression plays an oncogenic role in PNET metastasis. It requires further scrutiny to determine the frequency of human metastatic PNET with down-regulation of DAB2IP.
Given the ability of miRNAs to target multiple genes, they are appealing therapeutic agents.54 The therapeutic strategies either blocking oncogenic miRNAs or introducing tumor suppressor miRNAs have developed rapidly in recent years.55 It is proposed that targeting miR-431 may be a novel therapeutic strategy for PNETs. Besides RasGAPs, genes involved in zinc transport were among the top putative miR-431 target genes in PNETs from our analysis (Supplemental Table S2). Zinc is an essential micronutrient that plays a role in the functions of many cellular proteins, and zinc is emerging as an important signaling molecule in the progression of cancer. Further studies are required to investigate whether these candidates are miR-431 targets and whether zinc dyshomeostasis contributes to PNET metastasis.
In summary, we have identified that overexpression of miR-431 promotes PNET cell migration and invasion in vitro and metastasis in both spontaneous and xenograft mouse models of PNETs (Figure 6G). miR-431 directly silenced RasGAP tumor suppressor, DAB2IP. Down-regulation of DAB2IP led to the activation of Ras/Erk signaling, EMT, and cell invasion.
Acknowledgments
We thank Drs. Inna Serganova and Ronald Blasberg for viruses carrying thymidine kinase/green fluorescent protein/luciferase fusion reporter; Jason T. Huse for RCASBP constructs for miRNA expression; Jer-Tsong Hsieh for DAB2 interacting protein (DAB2IP) antibodies, DAB2IP plasmids, and discussion; Chris Harris for QGP1 cell line; Andrea Ventura, Yi Li, Pengbo Zhou, and Irene Min for advice; Danny Huang for mouse database design; George Zhang, Megan Wong, and Joseph Na for mouse colony management; Kimal Rajapakshe and Cristian Coarfa for reverse phase protein array (RPPA) data processing and normalization; and Fuli Jia and Danli Wu (Antibody-Based Proteomics Core/Shared Resource) for excellent technical assistance in performing RPPA experiments.
Footnotes
Supported by US Department of Defense grants W81XWH-13-1-0331 (S.C. and Y.-C.N.D.) and W81XWH-16-1-0619 (T.Z. and Y.-C.N.D.), NIH grant 1R01CA204916 (T.Z., Z.C., Y.C., and Y.-C.N.D.), Goldhirsh-Yellin Foundation (S.C. and Y.-C.N.D.), Cancer Prevention and Research Institute of Texas Proteomics and Metabolomics Core Facility Support Award RP170005 (S.H.), and National Cancer Institute Cancer Center support grant to Antibody-Based Proteomics Core/Shared Resource P30CA125123 (S.H.).
T.Z. and S.C. contributed equally to this work.
Disclosures: None declared.
Supplemental material for this article can be found at https://doi.org/10.1016/j.ajpath.2019.11.007.
Author Contributions
T.Z., S.C., and Y.C., designed and performed the experiments and analyzed the data; T.Z. contributed to bioinformatics analysis; Z.C. performed statistical analysis; S.H. directed reverse phase protein array; J.Z.X. directed RNA sequencing; Y.C.N.D. designed the experiments, analyzed the data, and wrote the manuscript.
Supplemental Data
miR-431 increases Ras/extracellular signal-regulated kinase signaling. RNA sequencing and gene set enrichment analysis shows a correlation between miR-431 expression and KRAS signature and constitutively active MEK signature in mouse pancreatic neuroendocrine tumor N134 cells. NES, normalized enrichment score; Norm., normalized.
References
- 1.Edge S., Byrd D.R., Compton C.C., Fritz A.G., Greene F.L., Trotti A. ed 7. Springer-Verlag; New York, NY: 2010. Exocrine and endocrine pancreas. AJCC Cancer Staging Manual; pp. 241–249. [Google Scholar]
- 2.Zhang J.F.R., Iyer R., Seshadri M., Zajac-Kaye M., Hochwald S.N. Current understanding of the molecular biology of pancreatic neuroendocrine tumors. J Natl Cancer Inst. 2013;105:1005–1017. doi: 10.1093/jnci/djt135. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Buicko J.L., Finnerty B.M., Zhang T., Kim B.J., Fahey T.J., 3rd, Du Y.C. Insights into the biology and treatment strategies of pancreatic neuroendocrine tumors. Ann Pancreat Cancer. 2019;2:12. doi: 10.21037/apc.2019.06.02. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Blumenthal G.M.C.P., Zhang J.J., Tang S., Sridhara R., Murgo A., Justice R., Pazdur R. FDA approval summary: sunitinib for the treatment of progressive well-differentiated locally advanced or metastatic pancreatic neuroendocrine tumors. Oncologist. 2012;17:1108–1113. doi: 10.1634/theoncologist.2012-0044. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Cives M., Strosberg J. Radionuclide therapy for neuroendocrine tumors. Curr Oncol Rep. 2017;19:9. doi: 10.1007/s11912-017-0567-8. [DOI] [PubMed] [Google Scholar]
- 6.Yao J.C.S.M., Ito T., Bohas C.L., Wolin E.M., Van Cutsem E., Hobday T.J., Okusaka T., Capdevila J., de Vries E.G., Tomassetti P., Pavel M.E., Hoosen S., Haas T., Lincy J., Lebwohl D., Öberg K. Everolimus for advanced pancreatic neuroendocrine tumors. N Engl J Med. 2011;364:514–523. doi: 10.1056/NEJMoa1009290. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Dasari A., Shen C., Halperin D., Zhao B., Zhou S., Xu Y., Shih T., Yao J.C. Trends in the incidence, prevalence, and survival outcomes in patients with neuroendocrine tumors in the United States. JAMA Oncol. 2017;3:1335–1342. doi: 10.1001/jamaoncol.2017.0589. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bartel D.P. MicroRNAs: genomics, biogenesis, mechanism, and function. Cell. 2004;116:281–297. doi: 10.1016/s0092-8674(04)00045-5. [DOI] [PubMed] [Google Scholar]
- 9.Ma L., Teruya-Feldstein J., Weinberg R.A. Tumour invasion and metastasis initiated by microRNA-10b in breast cancer. Nature. 2007;449:682–688. doi: 10.1038/nature06174. [DOI] [PubMed] [Google Scholar]
- 10.Tavazoie S.F., Alarcon C., Oskarsson T., Padua D., Wang Q., Bos P.D., Gerald W.L., Massague J. Endogenous human microRNAs that suppress breast cancer metastasis. Nature. 2008;451:147–152. doi: 10.1038/nature06487. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Olson P., Lu J., Zhang H., Shai A., Chun M.G., Wang Y., Libutti S.K., Nakakura E.K., Golub T.R., Hanahan D. MicroRNA dynamics in the stages of tumorigenesis correlate with hallmark capabilities of cancer. Genes Dev. 2009;23:2152–2165. doi: 10.1101/gad.1820109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Sadanandam A., Wullschleger S., Lyssiotis C.A., Grotzinger C., Barbi S., Bersani S., Korner J., Wafy I., Mafficini A., Lawlor R.T., Simbolo M., Asara J.M., Blaker H., Cantley L.C., Wiedenmann B., Scarpa A., Hanahan D. A cross-species analysis in pancreatic neuroendocrine tumors reveals molecular subtypes with distinctive clinical, metastatic, developmental, and metabolic characteristics. Cancer Discov. 2015;5:1296–1313. doi: 10.1158/2159-8290.CD-15-0068. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Loftus S.K., Larson D.M., Watkins-Chow D., Church D.M., Pavan W.J. Generation of RCAS vectors useful for functional genomic analyses. DNA Res. 2001;8:221–226. doi: 10.1093/dnares/8.5.221. [DOI] [PubMed] [Google Scholar]
- 14.Huse J.T., Brennan C., Hambardzumyan D., Wee B., Pena J., Rouhanifard S.H., Sohn-Lee C., le Sage C., Agami R., Tuschl T., Holland E.C. The PTEN-regulating microRNA miR-26a is amplified in high-grade glioma and facilitates gliomagenesis in vivo. Genes Dev. 2009;23:1327–1337. doi: 10.1101/gad.1777409. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Himly M., Foster D.N., Bottoli I., Iacovoni J.S., Vogt P.K. The DF-1 chicken fibroblast cell line: transformation induced by diverse oncogenes and cell death resulting from infection by avian leukosis viruses. Virology. 1998;248:295–304. doi: 10.1006/viro.1998.9290. [DOI] [PubMed] [Google Scholar]
- 16.Schaefer-Klein J., Givol I., Barsov E.V., Whitcomb J.M., VanBrocklin M., Foster D.N., Federspiel M.J., Hughes S.H. The EV-O-derived cell line DF-1 supports the efficient replication of avian leukosis-sarcoma viruses and vectors. Virology. 1998;248:305–311. doi: 10.1006/viro.1998.9291. [DOI] [PubMed] [Google Scholar]
- 17.Du Y.C., Lewis B.C., Hanahan D., Varmus H. Assessing tumor progression factors by somatic gene transfer into a mouse model: Bcl-xL promotes islet tumor cell invasion. PLoS Biol. 2007;5:e276. doi: 10.1371/journal.pbio.0050276. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Zhang G., Chi Y., Du Y.C. Identification and characterization of metastatic factors by gene transfer into the novel RIP-Tag; RIP-tva Murine model. J Vis Exp. 2017;128:55890. doi: 10.3791/55890. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Tang L.H., Contractor T., Clausen R., Klimstra D.S., Du Y.C., Allen P.J., Brennan M.F., Levine A.J., Harris C.R. Attenuation of the retinoblastoma pathway in pancreatic neuroendocrine tumors due to increased cdk4/cdk6. Clin Cancer Res. 2012;18:4612–4620. doi: 10.1158/1078-0432.CCR-11-3264. [DOI] [PubMed] [Google Scholar]
- 20.Evers B.M., Townsend C.M., Jr., Upp J.R., Allen E., Hurlbut S.C., Kim S.W., Rajaraman S., Singh P., Reubi J.C., Thompson J.C. Establishment and characterization of a human carcinoid in nude mice and effect of various agents on tumor growth. Gastroenterology. 1991;101:303–311. doi: 10.1016/0016-5085(91)90004-5. [DOI] [PubMed] [Google Scholar]
- 21.Ponomarev V., Doubrovin M., Serganova I., Vider J., Shavrin A., Beresten T., Ivanova A., Ageyeva L., Tourkova V., Balatoni J., Bornmann W., Blasberg R., Gelovani Tjuvajev J. A novel triple-modality reporter gene for whole-body fluorescent, bioluminescent, and nuclear noninvasive imaging. Eur J Nucl Med Mol Imaging. 2004;31:740–751. doi: 10.1007/s00259-003-1441-5. [DOI] [PubMed] [Google Scholar]
- 22.Wang Z., Tseng C.P., Pong R.C., Chen H., McConnell J.D., Navone N., Hsieh J.T. The mechanism of growth-inhibitory effect of DOC-2/DAB2 in prostate cancer: characterization of a novel GTPase-activating protein associated with N-terminal domain of DOC-2/DAB2. J Biol Chem. 2002;277:12622–12631. doi: 10.1074/jbc.M110568200. [DOI] [PubMed] [Google Scholar]
- 23.Choi S., Chen Z., Tang L.H., Fang Y., Shin S.J., Panarelli N.C., Chen Y.T., Li Y., Jiang X., Du Y.C. Bcl-xL promotes metastasis independent of its anti-apoptotic activity. Nat Commun. 2016;7:10384. doi: 10.1038/ncomms10384. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Kim D., Pertea G., Trapnell C., Pimentel H., Kelley R., Salzberg S.L. TopHat2: accurate alignment of transcriptomes in the presence of insertions, deletions and gene fusions. Genome Biol. 2013;14:R36. doi: 10.1186/gb-2013-14-4-r36. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Trapnell C., Williams B.A., Pertea G., Mortazavi A., Kwan G., van Baren M.J., Salzberg S.L., Wold B.J., Pachter L. Transcript assembly and quantification by RNA-Seq reveals unannotated transcripts and isoform switching during cell differentiation. Nat Biotechnol. 2010;28:511–515. doi: 10.1038/nbt.1621. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Trapnell C., Hendrickson D.G., Sauvageau M., Goff L., Rinn J.L., Pachter L. Differential analysis of gene regulation at transcript resolution with RNA-seq. Nat Biotechnol. 2013;31:46–53. doi: 10.1038/nbt.2450. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Chang C.H., Zhang M., Rajapakshe K., Coarfa C., Edwards D., Huang S., Rosen J.M. Mammary stem cells and tumor-initiating cells are more resistant to apoptosis and exhibit increased DNA repair activity in response to DNA damage. Stem Cell Reports. 2015;5:378–391. doi: 10.1016/j.stemcr.2015.07.009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Creighton C.J., Huang S. Reverse phase protein arrays in signaling pathways: a data integration perspective. Drug Des Devel Ther. 2015;9:3519–3527. doi: 10.2147/DDDT.S38375. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Holdman X.B., Welte T., Rajapakshe K., Pond A., Coarfa C., Mo Q., Huang S., Hilsenbeck S.G., Edwards D.P., Zhang X., Rosen J.M. Upregulation of EGFR signaling is correlated with tumor stroma remodeling and tumor recurrence in FGFR1-driven breast cancer. Breast Cancer Res. 2015;17:141. doi: 10.1186/s13058-015-0649-1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Welte T., Kim I.S., Tian L., Gao X., Wang H., Li J., Holdman X.B., Herschkowitz J.I., Pond A., Xie G., Kurley S., Nguyen T., Liao L., Dobrolecki L.E., Pang L., Mo Q., Edwards D.P., Huang S., Xin L., Xu J., Li Y., Lewis M.T., Wang T., Westbrook T.F., Rosen J.M., Zhang X.H. Oncogenic mTOR signalling recruits myeloid-derived suppressor cells to promote tumour initiation. Nat Cell Biol. 2016;18:632–644. doi: 10.1038/ncb3355. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Ritchie M.E., Phipson B., Wu D., Hu Y., Law C.W., Shi W., Smyth G.K. limma powers differential expression analyses for RNA-sequencing and microarray studies. Nucleic Acids Res. 2015;43:e47. doi: 10.1093/nar/gkv007. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Deer E.L., Gonzalez-Hernandez J., Coursen J.D., Shea J.E., Ngatia J., Scaife C.L., Firpo M.A., Mulvihill S.J. Phenotype and genotype of pancreatic cancer cell lines. Pancreas. 2010;39:425–435. doi: 10.1097/MPA.0b013e3181c15963. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Shin S., Dimitri C.A., Yoon S.O., Dowdle W., Blenis J. ERK2 but not ERK1 induces epithelial-to-mesenchymal transformation via DEF motif-dependent signaling events. Mol Cell. 2010;38:114–127. doi: 10.1016/j.molcel.2010.02.020. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Dweep H., Sticht C., Pandey P., Gretz N. miRWalk-database: prediction of possible miRNA binding sites by “walking” the genes of three genomes. J Biomed Inform. 2011;44:839–847. doi: 10.1016/j.jbi.2011.05.002. [DOI] [PubMed] [Google Scholar]
- 35.John B., Enright A.J., Aravin A., Tuschl T., Sander C., Marks D.S. Human microRNA targets. PLoS Biol. 2004;2:e363. doi: 10.1371/journal.pbio.0020363. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Miranda K.C., Huynh T., Tay Y., Ang Y.S., Tam W.L., Thomson A.M., Lim B., Rigoutsos I. A pattern-based method for the identification of microRNA binding sites and their corresponding heteroduplexes. Cell. 2006;126:1203–1217. doi: 10.1016/j.cell.2006.07.031. [DOI] [PubMed] [Google Scholar]
- 37.Agarwal V., Bell G.W., Nam J.W., Bartel D.P. Predicting effective microRNA target sites in mammalian mRNAs. eLife. 2015;4:e05005. doi: 10.7554/eLife.05005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Chen H., Karam J.A., Schultz R., Zhang Z., Duncan C., Hsieh J.T. Cloning of mouse Dab2ip gene, a novel member of the RasGTPase-activating protein family and characterization of its regulatory region in prostate. DNA Cell Biol. 2006;25:232–245. doi: 10.1089/dna.2006.25.232. [DOI] [PubMed] [Google Scholar]
- 39.Elmen J., Lindow M., Schutz S., Lawrence M., Petri A., Obad S., Lindholm M., Hedtjarn M., Hansen H.F., Berger U., Gullans S., Kearney P., Sarnow P., Straarup E.M., Kauppinen S. LNA-mediated microRNA silencing in non-human primates. Nature. 2008;452:896–899. doi: 10.1038/nature06783. [DOI] [PubMed] [Google Scholar]
- 40.Su W.B., Liu Z.Y. MiR-431 inhibits colorectal cancer cell invasion via repressing CUL4B. Eur Rev Med Pharmacol Sci. 2018;22:3047–3052. doi: 10.26355/eurrev_201805_15062. [DOI] [PubMed] [Google Scholar]
- 41.Sun K., Zeng T., Huang D., Liu Z., Huang S., Liu J., Qu Z. MicroRNA-431 inhibits migration and invasion of hepatocellular carcinoma cells by targeting the ZEB1-mediated epithelial-mensenchymal transition. FEBS Open Bio. 2015;5:900–907. doi: 10.1016/j.fob.2015.11.001. [DOI] [PMC free article] [PubMed] [Google Scholar] [Retracted]
- 42.Almoguera C., Shibata D., Forrester K., Martin J., Arnheim N., Perucho M. Most human carcinomas of the exocrine pancreas contain mutant c-K-ras genes. Cell. 1988;53:549–554. doi: 10.1016/0092-8674(88)90571-5. [DOI] [PubMed] [Google Scholar]
- 43.Hingorani S.R., Petricoin E.F., Maitra A., Rajapakse V., King C., Jacobetz M.A., Ross S., Conrads T.P., Veenstra T.D., Hitt B.A., Kawaguchi Y., Johann D., Liotta L.A., Crawford H.C., Putt M.E., Jacks T., Wright C.V., Hruban R.H., Lowy A.M., Tuveson D.A. Preinvasive and invasive ductal pancreatic cancer and its early detection in the mouse. Cancer Cell. 2003;4:437–450. doi: 10.1016/s1535-6108(03)00309-x. [DOI] [PubMed] [Google Scholar]
- 44.Jiao Y., Shi C., Edil B.H., de Wilde R.F., Klimstra D.S., Maitra A., Schulick R.D., Tang L.H., Wolfgang C.L., Choti M.A., Velculescu V.E., Diaz L.A., Jr., Vogelstein B., Kinzler K.W., Hruban R.H., Papadopoulos N. DAXX/ATRX, MEN1, and mTOR pathway genes are frequently altered in pancreatic neuroendocrine tumors. Science. 2011;331:1199–1203. doi: 10.1126/science.1200609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Tesfaye A.A., Azmi A.S., Philip P.A. miRNA and gene expression in pancreatic ductal adenocarcinoma. Am J Pathol. 2019;189:58–70. doi: 10.1016/j.ajpath.2018.10.005. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Bellazzo A., Di Minin G., Collavin L. Block one, unleash a hundred: mechanisms of DAB2IP inactivation in cancer. Cell Death Differ. 2017;24:15–25. doi: 10.1038/cdd.2016.134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Liu L., Xu C., Hsieh J.T., Gong J., Xie D. DAB2IP in cancer. Oncotarget. 2016;7:3766–3776. doi: 10.18632/oncotarget.6501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Chen H., Pong R.C., Wang Z., Hsieh J.T. Differential regulation of the human gene DAB2IP in normal and malignant prostatic epithelia: cloning and characterization. Genomics. 2002;79:573–581. doi: 10.1006/geno.2002.6739. [DOI] [PubMed] [Google Scholar]
- 49.Olsen S.N., Wronski A., Castano Z., Dake B., Malone C., De Raedt T., Enos M., DeRose Y.S., Zhou W., Guerra S., Loda M., Welm A., Partridge A.H., McAllister S.S., Kuperwasser C., Cichowski K. Loss of RasGAP tumor suppressors underlies the aggressive nature of luminal B breast cancers. Cancer Discov. 2017;7:202–217. doi: 10.1158/2159-8290.CD-16-0520. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Dote H., Toyooka S., Tsukuda K., Yano M., Ouchida M., Doihara H., Suzuki M., Chen H., Hsieh J.T., Gazdar A.F., Shimizu N. Aberrant promoter methylation in human DAB2 interactive protein (hDAB2IP) gene in breast cancer. Clin Cancer Res. 2004;10:2082–2089. doi: 10.1158/1078-0432.ccr-03-0236. [DOI] [PubMed] [Google Scholar]
- 51.Zhu X.H., Wang J.M., Yang S.S., Wang F.F., Hu J.L., Xin S.N., Men H., Lu G.F., Lan X.L., Zhang D., Wang X.Y., Liao W.T., Ding Y.Q., Liang L. Down-regulation of DAB2IP promotes colorectal cancer invasion and metastasis by translocating hnRNPK into nucleus to enhance the transcription of MMP2. Int J Cancer. 2017;141:172–183. doi: 10.1002/ijc.30701. [DOI] [PubMed] [Google Scholar]
- 52.Feng C., Sun P., Hu J., Feng H., Li M., Liu G., Pan Y., Feng Y., Xu Y., Feng K., Feng Y. miRNA-556-3p promotes human bladder cancer proliferation, migration and invasion by negatively regulating DAB2IP expression. Int J Oncol. 2017;50:2101–2112. doi: 10.3892/ijo.2017.3969. [DOI] [PubMed] [Google Scholar]
- 53.Wu K., Liu J., Tseng S.F., Gore C., Ning Z., Sharifi N., Fazli L., Gleave M., Kapur P., Xiao G., Sun X., Oz O.K., Min W., Alexandrakis G., Yang C.R., Hsieh C.L., Wu H.C., He D., Xie D., Hsieh J.T. The role of DAB2IP in androgen receptor activation during prostate cancer progression. Oncogene. 2014;33:1954–1963. doi: 10.1038/onc.2013.143. [DOI] [PubMed] [Google Scholar]
- 54.Garzon R., Marcucci G., Croce C.M. Targeting microRNAs in cancer: rationale, strategies and challenges. Nat Rev Drug Discov. 2010;9:775–789. doi: 10.1038/nrd3179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Ji W., Sun B., Su C. Targeting microRNAs in cancer gene therapy. Genes (Basel) 2017;8:21. doi: 10.3390/genes8010021. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
miR-431 increases Ras/extracellular signal-regulated kinase signaling. RNA sequencing and gene set enrichment analysis shows a correlation between miR-431 expression and KRAS signature and constitutively active MEK signature in mouse pancreatic neuroendocrine tumor N134 cells. NES, normalized enrichment score; Norm., normalized.