Abstract
Goat domestication and human selection for valued traits have formed diverse breeds with characteristic phenotypes. This process led to the fixation of causative genetic variants controlling breed-specific traits within regions of reduced genetic diversity—so-called “selection signatures”. We previously reported an analysis of selection signatures based on pooled whole-genome sequencing data of 20 goat breeds and bezoar goats. In the present study, we reanalyzed the data and focused on a subset of eight Pakistani goat breeds (Angora, Barbari, Beetal, Dera Din Panah, Kamori, Nachi, Pahari, Teddy). We identified 749 selection signatures based on reduced heterozygosity in these breeds. A search for signatures that are shared across large-sized goat breeds revealed that five medium-to-large-sized Pakistani goat breeds had a common selection signature on chromosome 6 in a region harboring the LCORL gene, which has been shown to modulate height or body size in several mammalian species. Fine-mapping of the region confirmed that all five goat breeds with the selection signature were nearly fixed for the same haplotype in a ~191 kb region spanning positions 37,747,447–37,938,449. From the pool sequencing data, we identified a frame-shifting single base insertion into an isoform-specific exon of LCORL as a potential candidate causal variant mediating the size-increasing effect. If this preliminary result can be confirmed in independent replication studies, genotyping of this variant might be used to improve breeding programs and the selection for stature in goats.
Keywords: Capra hircus, whole-genome sequence, body size, height, stature, QTL, selection signature, animal breeding, meat production
1. Introduction
There are 25 goat breeds in Pakistan [1] with an estimated population of 76.1 million heads, which are mostly raised for milk, meat, hair, and hide purposes. Goats produce ~940 thousand tons of milk annually in Pakistan and ~732 thousand tons of mutton (goat and sheep) [2]. Around 6.8 million people are involved in small ruminant farming in Pakistan and rearing goats under nomadic, transhumant, household and sedentary production systems [3]. Genomic selection and proper targeted breeding policies are considered promising tools for genetic improvement of this valued species. Body size is an important trait to investigate for the improvement of meat-purpose domestic animals. It is also marginally associated with milk production [4,5,6].
Human adult stature and skeletal frame size is controlled by more than 700 genes [7,8,9,10,11,12]. The LCORL gene encoding ligand-dependent nuclear receptor corepressor-like has been repeatedly found to be associated with human height [7,8,9,10,11,12]. LCORL (previously also called MLR1) is a putative transcription factor that utilizes a conserved helix-turn-helix motif for DNA binding [13]. There are several experimentally confirmed human LCORL transcript isoforms that encode vastly different proteins. Not all of these proteins share the DNA-binding domain, and they also differ in their other protein domains [14].
Size variation in livestock species is typically controlled by fewer genes with larger effects as compared to humans [15,16]. The LCORL locus has also been found associated with height and body size in many domestic animal species, including dogs [17], horses [18,19,20], pigs [21], and sheep [22,23].
In cattle, the genomic locus is also strongly associated with height and growth-related traits. However, the functional effect in cattle has been claimed to be mediated by the NCAPG gene, which is adjacent to LCORL [16,24,25,26,27]. Determining which of these two genes is responsible for variability in height has not been possible because of the close proximity of these genes and the high levels of linkage disequilibrium among markers in this genome region. Recently, the identification of a missense variant in bovine LCOR—a gene with very high homology and potentially similar function to LCORL—as being associated with stature provides some supporting evidence for LCORL as the causative gene [28].
We previously conducted a comprehensive screen for selection signatures in 20 genetically diverse goat breeds [29]. In the current study, we used this dataset to analyze loci under selection in large-sized Pakistani goat breeds with the aim of identifying important genes for meat production.
2. Materials and Methods
2.1. Ethics Statement
All goats in this study were privately owned and samples were collected with the consent of their owners. Extracted DNA from Pakistani goats was sent to Switzerland for downstream sequencing and genotyping.
2.2. Animal Selection
Eight Pakistani domestic goat breeds selected for this study belong to diverse geographical regions across Pakistan. Breed averages of the height at withers served as proxies for the size phenotypes (Table 1). Peripheral blood samples were collected from goats of the selected breeds in EDTA vacutainers and stored at −20 °C.
Table 1.
Phenotypic characteristics of eight goat breeds of Pakistan.
| Breed Name | Abbr. | WH (cm) | BW (kg) | Main Purposes | Geographic Distribution | Population Size (2006) a |
|---|---|---|---|---|---|---|
| Angora | ANG | 75 | 47 | Meat, hair | Punjab | NA |
| Barbari (Bari) | BAR | 68 | 30 | Meat, milk | Punjab, Sindh | 2306 |
| Beetal | BEE | NA | NA | Meat, milk | Punjab | 4214 |
| Dera Din Panah | DDP | 81 | 61.7 | Meat, milk, hair | Punjab | 1424 |
| Kamori | KAM | 90 | 55 | Meat, milk | Sindh | 5294 |
| Nachi (Bikaneri) | NAC | 71 | 35 | Meat, milk, hair | Punjab | 1135 |
| Pahari (Kajli) | PAH | 77 | 67 | Meat, milk, hair | Punjab, Balochistan | NA |
| Teddy | TED | 64 | 33.9 | Meat, milk | Punjab, Azad Jammu and Kashmir | 1342 |
WH = wither height, BW = body weight; data taken from FAO [http://www.fao.org/dad-is/data]. a Thousand heads.
Based on the personal experience of the authors and the recorded average wither height in the FAO database, Beetal (BEE), Dera Din Panah (DDP), and Kamori (KAM) were arbitrarily classified as large-sized breeds for this study. Angora (ANG), Barbari (BAR), Nachi (NAC), and Pahari (PAH) goats were considered medium in size, while Teddy (TED) goats were classified as small. Representative animals of each breed are shown in Figure 1.
Figure 1.
Representative animals of eight Pakistani goat breeds.
2.3. DNA Extraction and Identification of Selection Signatures
DNA was extracted from whole blood using TIANamp Genomic DNA Kit (Tiangen Biotech, Beijing, China) according to the manufacturer’s instructions. Pooled sequencing (pool-seq) and identification of selection signatures of these samples has been described in detail before [29].
Briefly, pooled heterozygosity scoring statistics were used as described [21,30] to identify the regions under selection, which was calculated as , where and are the sums of major and minor alleles for all single-nucleotide polymorphisms (SNPs) in each window. Then, individual values were -transformed by , where is the overall average heterozygosity, and is the standard deviation of all windows within each pool. We calculated and converted it to − values in 150-kb sliding windows. In this study, we applied a threshold of ≥ 4 for putative selection signatures. The underlying sequence data are publicly available under the study accession PRJEB23815 at the European Nucleotide Archive.
2.4. Fine Mapping
A selection window of 150 kb with > 4 was found common in three large and two medium-sized breeds, but was not present in the small breed group. In order to define the precise boundaries of the selection signature, we considered all variants in a 375 kb region spanning positions 37,650,001–38,025,000 on chromosome 6. We calculated the frequency of the minor allele at each variant in each breed pool. The selection signature was then defined as the region in which the five breeds with the selection signature showed a long consecutive stretch of variants with minor allele frequency < 0.2.
2.5. Gene Analysis
We used the ARS1 goat reference genome assembly [31], accession number: GCF_001704415.1, as reference for all downstream analyses. Numbering within the goat LCORL gene corresponds to the NCBI RefSeq accession numbers XM_018049322.1 (mRNA) and XP_017904811.1 (protein).
2.6. Sanger Sequencing
Seven LCORL variants were genotyped by direct Sanger sequencing after PCR amplification with the primers listed in Table 2.
Table 2.
Primer sequences to amplify seven LCORL variants. The first amplicon contains two variable positions.
| Primer Name | Primer Sequence | Product (bp) | Amplified Variants |
|---|---|---|---|
| LCORL_1F | CTTTCACCCAAGTCAGTGTCA | 332 | c.3480C>T |
| LCORL_1R | CCCCAGGTTGTGAAACAGAT | c.3360G>A | |
| LCORL_2F | TTGGATGCTTTATACCCTTCTGA | 213 | c.2513A>G |
| LCORL_2R | AAAATCCCCTAAGGC CAAAA | ||
| LCORL_3F | CATGTTGACTCAGCAATTCCA | 226 | c.1685A>G |
| LCORL_3R | ACAAATCAT GAAAAGGGTGAAAC | ||
| LCORL_4F | TGCTGGTGTCAGAGATGGAG | 215 | c.1298A>G |
| LCORL_4R | CAGGCTTTCAGAGTCCTCGT | ||
| LCORL_5F | AACAGCAAAGAGAAGCAGCA | 495 | c.828_829insA |
| LCORL_5R | TCCTTCTGAAGCACTTTCCA | ||
| LCORL_6F | GGGTTCAGTATAGATCTGAGAGACC | 479 | c.777-4235T>C |
| LCORL_6R | TGGGCAGTGCATTTTAACTTT |
The resulting amplicons were sequenced on an ABI 3730 DNA Analyzer after treating with exonuclease I and alkaline phosphatase. Finally, the obtained Sanger sequences were analyzed using the Sequencher 5.1 software (GeneCodes, Ann Arbor, MI, USA).
2.7. Association Analysis
We performed an allelic association study in 74 goats (Table S1) using the genotypes at the seven genotyped variants in a case–control design and the PLINK 1.07 software [32]. We considered goats from BAR (n = 8), BEE (n = 12), DDP (n = 12), KAM (n = 13), and NAC (n = 12) as cases and goats from ANG (n = 6), PAH (n = 5), and TED (n = 6) as controls (Table S1).
3. Results
3.1. Selection Signatures in Large-Sized Goats
Our pool-seq dataset comprised eight Pakistani goat breeds [28]. Each breed pool was composed of 12 animals except Angora, for which only 10 animals were contained in the pool. Pools were sequenced to 30× coverage and sequence data mapped to the ARS1 goat reference genome. On average ~12.7 million single nucleotide variants (SNVs) were observed in each breed from Pakistan as compared to ~11.7 million SNVs in Swiss goat breeds [29].
In the Pakistani goat breeds we identified a total of 2064 windows with > 4. After merging overlapping windows, this resulted in 749 putative selection signatures in the eight studied breeds (Table 3, Table S2).
Table 3.
Selection signatures in the Pakistani goat breeds.
| Breed | Animals per Pool | Windows under Selection | Selection Signatures | Genes in Selection Signatures |
|---|---|---|---|---|
| ANG | 10 | 140 | 61 | 144 |
| BAR | 12 | 95 | 51 | 109 |
| BEE | 12 | 126 | 58 | 142 |
| DDP | 12 | 132 | 66 | 178 |
| KAM | 12 | 1244 | 341 | 889 |
| NAC | 12 | 130 | 79 | 137 |
| PAH | 12 | 61 | 44 | 92 |
| TED | 12 | 136 | 49 | 120 |
| Total | 2064 | 749 | 1811 |
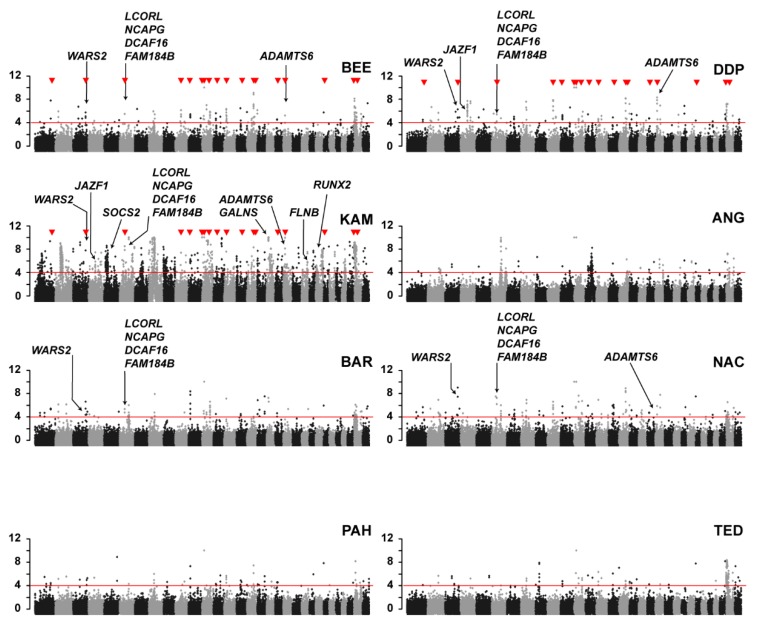
In order to identify selection signatures related to body size, we searched for signatures that were shared between all three large-sized goat breeds in the study (BEE, DDP, KAM). This identified 18 common selection signatures in these breeds (Figure 2; Table S3).
Figure 2.
Manhattan plots illustrating the observed selection signatures in eight Pakistani goat breeds. Red triangles indicate 18 selection signatures that are shared between three breeds of large-sized goats (BEE, DDP, KAM). Selection signatures that harbor genes known to be related to height or body size in other species are indicated.
One of these shared selection signatures was located on chromosome 6 at ~38 Mb and contained the LCORL gene, which has been shown to be associated with size in many species. Interestingly, two of the medium-sized breeds, BAR and NAC, also had this selection signature harboring the LCORL gene. Inspection of the pool-seq data revealed that all five breeds shared the same major haplotype in this region. This prompted us to hypothesize that a size-increasing allele caused by the same genetic variant and identical by descent (IBD) is under selection in the BAR, BEE, DDP, KAM, and NAC goat breeds.
3.2. Fine-Mapping of the LCORL Selection Signature
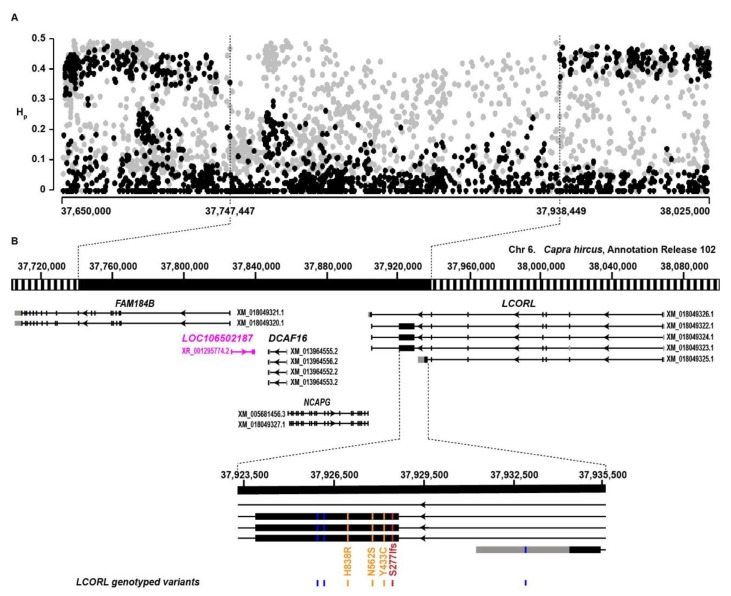
The window with the highest score spanned positions 37.80–37.95 Mb in all five breed-pools. In order to precisely define the boundaries of the selection signature, we also considered adjacent regions with slightly elevated scores and initially examined a 375-kb region spanning 37.600–38.025 Mb. Our dataset with all eight breed pools contained 1469 SNVs in this interval. We counted the minor and major alleles for each of these variants in each breed pool (Table S4). A consecutive stretch of 156 variants showed reduced variation with minor allele frequencies of less than 0.2 in the five breeds with the selection signature. The five breeds under selection were nearly fixed for a shared 191-kb haplotype ranging from 37,747,447 to 37,938,449 (Figure 3A; Table S4). We considered this as the critical interval harboring the hypothetical size-increasing genetic variant.
Figure 3.
(A) Pooled heterozygosity (Hp) distribution at the LCORL locus on chromosome 6 in five medium/large-sized Pakistani goat breeds (black dots) and three small/medium breeds (grey dots). A ~191 kb region from 37,747,447 to 37,938,449 showed greatly reduced heterozygosity in the breeds with the selection signature. (B) The gene annotation for the selection signature is illustrated (NCBI annotation release 102). The genomic locations of seven genotyped variants in the LCORL gene are indicated in red (putative frameshift variant), orange (putative missense variants), or blue (silent or non-coding variants).
3.3. Search for Candidate Causative Variants
The critical interval at this selection signature contained four protein-coding genes (FAM184B, DCAF16, NCAPG, LCORL) and one gene for a non-coding RNA (LOC106502187; Figure 3B). Visual inspection of the short read alignments did not reveal any structural variants that were private to the five breed pools with the selected haplotype. Based on literature data in other mammalian species [16,17,18,19,20,21,22,23,24], we considered LCORL the most likely functional candidate gene for size. We selected six SNVs and a single base insertion that were located in potential exons of the LCORL gene and genotyped them on 74 individual goats. We calculated the association to the phenotype and observed the most significant association for LCORL:c.828_829insA (Table 5). The insertion allele had a frequency of 0.95 in the five breeds with the selection signature vs. 0.35 in three goat breeds that are not specifically selected for size. This variant introduces a frameshift in three of the five annotated transcripts. On the longest protein isoforms, the single-base insertion, is predicted to result in the truncation of more than 85% of the open reading frame. The formal variant designation on the protein level is XP_017904811.1:p.(Ser277Ilefs*38).
Table 5.
Genotype phenotype association of selected SNVs in the LCORL gene.
| Chr. | Position | cDNA Variant XM_018049322.1 | Protein Variant XP_017904811.1 | Alternative Allele Frequency in Breeds with the Selection Signature a | Alternative Allele Frequency in Breeds without the Selection Signature b | p-Value |
|---|---|---|---|---|---|---|
| 6 | 37,925,990 | c.3480C>T | p.= | 0.05 | 0.26 | 3.0 × 10−04 |
| 6 | 37,926,110 | c.3360G>A | p.= | 0.93 | 0.35 | 2.1 × 10−13 |
| 6 | 37,926,957 | c.2513A>G | p.His838Arg | 1.00 | 1.00 | NA |
| 6 | 37,927,785 | c.1685A>G | p.Asn562Ser | 0.01 | 0.26 | 1.8 × 10−07 |
| 6 | 37,928,172 | c.1298A>G | p.Tyr433Cys | 0.96 | 0.53 | 3.9 × 10−10 |
| 6 | 37,928,645 | c.828_829insA | p.Ser277Ilefs*38 | 0.95 | 0.35 | 8.0 × 10−15 |
| 6 | 37,932,928 | c.777-4235T>C | intronic/3′-UTR of short isoform | 0.01 | 0.09 | 1.0 × 10−02 |
a BAR, BEE, DDP, KAM; NAC, b ANG, PAH, TED.
4. Discussion
We investigated selection signatures potentially related to body size due to the prime importance of this trait in the highly meat consuming society of Pakistan, which is influenced by religious and other personal choices as well as economic needs for marginal farmers in low-income countries [2,33,34,35]. Several Pakistani goat breeds have already been genotyped under the AdaptMap project for exploring goat diversity (e.g., local adaption and coat color genetics) [36,37,38], but to our knowledge the trait of skeletal frame size and adult height has not been addressed before.
Our comprehensive study identified 749 selection signatures in Pakistani goat breeds. We observed many selection signatures harboring genes that influence height or body size in other species, thereby validating our experimental approach. Examples include the WARS2 gene, which is under selection in BAR, BEE, DDP, KAM, and NAC and associated with body fat distribution in humans [39], or ADAMTS6, which is under selection in BEE, DDP, KAM, and NAC and associated with bone length in mice [40].
The LCORL locus has previously been found to be associated with height in at least six mammalian species [16,17,18,19,20,21,22,23,24,25,26,27]. It is therefore not surprising that this locus is apparently also under selection in several medium-to-large-sized Pakistani goat breeds. However, it should be noted that this region does not show any evidence of selection in any of the previously studied Swiss goat breeds [29]. While there is accumulating evidence that genetic variation at the LCORL locus is involved in the determination of height and body size in diverse mammalian species, the molecular mechanism remains elusive. The strongest associations are frequently seen within a region harboring DCAF16, NCAPG, and the 3′–end of the LCORL gene. The in vivo function of the LCORL gene is not fully understood. LCORL encodes several different transcript isoforms, which mostly differ by the alternative use of at least four different exons at the 3′-end of the gene. One of them is almost 5 kb in length and predicted to encode more than 1600 amino acids. A single-base insertion, XM_022416410.1:c.3661_3662_insA, in this giant exon has been postulated to increase size in dogs [17].
In our study, another frame-shifting single-base-insertion variant in this exon showed the strongest differentiation between large and small breeds. It is intriguing that two independent single-base insertions into homologous exons in dogs and goats are associated with large body size. Unfortunately, there is no available functional confirmation for the causality of the insertion in dogs [17], and we did not perform any attempts to analyze the functional mechanism by which this LCORL variant might lead to an increase in body size. Nonetheless, in light of the parallels between dogs and goats, it is tempting to speculate that LCORL:c.828_829insA might be the true causative variant for a size-increasing QTL in Pakistani goats. Unfortunately, our study relying on breed-average phenotypes cannot provide conclusive proof for this hypothesis. Further studies are required to confirm this preliminary finding. As some medium-sized goat breeds are apparently not fixed at the LCORL locus, the next logical step will be a within-breed association study correlating individually measured height phenotypes with the genotypes at c.828_829insA in a breed that segregates both alleles. We must also caution that our analysis exclusively focused on the LCORL gene and that we cannot exclude an additional functional effect by any of the other three genes in the selection signature.
To the best of our knowledge, this is one of the first studies to report body-size-related selection signatures in different Pakistani goat breeds. The provided pooled heterozygosity statistics may also be used to investigate other breed-specific selection signatures and traits. This should help to enable more efficient breeding strategies.
5. Conclusions
A total of 749 selection signatures were observed in Pakistani goat breeds using pooled heterozygosity statistics. One selection signature harbored the LCORL gene and was observed in three large-sized and two medium-sized breeds. Detailed analysis of the LCORL selection signature suggested that the effect may be mediated by a frame-shifting single base insertion into the giant exon encoding parts of the long LCORL protein isoforms. However, this preliminary finding requires additional confirmation before the causality of this particular variant should be considered proven.
Acknowledgments
Authors are thankful to Hamid Mustafa Pasha and other goat breeders for helping in the provision of goat photos. We would also pay our gratitude to Mushtaq Ahmad Gondal, Safdar Ali Fazlani, and Faisal Shahzad for helping in sample collection. This study was supported in part by a grant from the Swiss National Science Foundation (31003A_172964). Rashid Saif was supported by a Swiss Government Excellence Scholarship and a supplementary grant from the Hans Sigrist Foundation.
Supplementary Materials
The following are available online at https://www.mdpi.com/2073-4425/11/2/168/s1, Table S1. Genotypes of 74 goats for association study. Table S2. Selection signatures per breed pool. Table S3. Significant windows in three large-sized Pakistani goat breeds (BEE, DDP, and KAM). Table S4. Alternative allele frequency at 1469 SNVs in large and small breeds.
Author Contributions
Conceptualization, T.L.; Data curation, V.J.; Investigation, R.S., J.H.; Methodology, V.J., C.D., C.F.; Resources, R.S.; Supervision, T.L.; Visualization, R.S.; Writing—original draft, R.S., T.L.; Writing—review and editing, R.S., J.H., V.J., C.D., C.F., T.L. All authors have read and agreed to the published version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interest. The funders had no role in the design of the study; in the collection, analyses, or interpretation of data; in the writing of the manuscript, or in the decision to publish the results.
References
- 1.Khan M.S., Khan M., Mahmood S. Genetic resources and diversity in Pakistani goats. Int. J. Agric. Biol. 2008;10:227–231. [Google Scholar]
- 2.Government of Pakistan Pakistan Economic Survey 2018–2019. [(accessed on 29 August 2019)]; Available online: http://finance.gov.pk/survey/chapters_19/Economic_Survey_2018_19.pdf.
- 3.Khan M.F.U., Ashfaq F. Current status of dairy goat farming in Pakistan; Proceedings of the 4th Asian-Australasian Dairy Goat Conference; Tra Vinh, Vietnam. 17–19 October 2018; Tra Vinh, Vietnam: Tra Vinh University Press; 2018. pp. 102–107. [Google Scholar]
- 4.Mavrogenis A., Papachristoforou C. Genetic and phenotypic relationships between milk production and body weight in Chios sheep and Damascus goats. Livest. Prod. Sci. 2000;67:81–87. doi: 10.1016/S0301-6226(00)00187-1. [DOI] [Google Scholar]
- 5.Dickerson G. Animal size and efficiency: Basic concepts. Anim. Sci. 1978;27:367–379. doi: 10.1017/S0003356100036278. [DOI] [Google Scholar]
- 6.Blackmore D., McGilliard L., Lush J. Relationships between body measurements, meat conformation, and milk production. J. Dairy Sci. 1958;41:1050–1056. doi: 10.3168/jds.S0022-0302(58)91049-X. [DOI] [Google Scholar]
- 7.Soranzo N., Rivadeneira F., Chinappen-Horsley U., Malkina I., Richards J.B., Hammond N., Stolk L., Nica A., Inouye M., Hofman A. Meta-analysis of genome-wide scans for human adult stature identifies novel Loci and associations with measures of skeletal frame size. PLoS Genet. 2009;5:e1000445. doi: 10.1371/journal.pgen.1000445. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Liu J.Z., Medland S.E., Wright M.J., Henders A.K., Heath A.C., Madden P.A., Duncan A., Montgomery G.W., Martin N.G., McRae A.F. Genome-wide association study of height and body mass index in Australian twin families. Twin Res. Hum. Genet. 2010;13:179–193. doi: 10.1375/twin.13.2.179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Lango Allen H., Estrada K., Lettre G., Berndt S.I., Weedon M.N., Rivadeneira F., Willer C.J., Jackson A.U., Vedantam S., Raychaudhuri S., et al. Hundreds of variants clustered in genomic loci and biological pathways affect human height. Nature. 2010;467:832–838. doi: 10.1038/nature09410. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Horikoshi M., Yaghootkar H., Mook-Kanamori D.O., Sovio U., Taal H.R., Hennig B.J., Bradfield J.P., St Pourcain B., Evans D.M., Charoen P., et al. New loci associated with birth weight identify genetic links between intrauterine growth and adult height and metabolism. Nat. Genet. 2013;45:76–82. doi: 10.1038/ng.2477. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yengo L., Sidorenko J., Kemper K.E., Zheng Z., Wood A.R., Weedon M.N., Frayling T.M., Hirschhorn J., Yang J., Visscher P.M. Meta-analysis of genome-wide association studies for height and body mass index in ∼700000 individuals of European ancestry. Hum. Mol. Genet. 2018;27:3641–3649. doi: 10.1093/hmg/ddy271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 12.Carty C.L., Johnson N.A., Hutter C.M., Reiner A.P., Peters U., Tang H., Kooperberg C. Genome-wide association study of body height in African Americans: The women’s health initiative SNP health association resource (share) Hum. Mol. Genet. 2011;21:711–720. doi: 10.1093/hmg/ddr489. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Kunieda T., Park J.M., Takeuchi H., Kubo T. Identification and characterization of Mlr1,2: Two mouse homologues of Mblk-1, a transcription factor from the honeybee brain. FEBS Lett. 2003;535:61–65. doi: 10.1016/S0014-5793(02)03858-9. [DOI] [PubMed] [Google Scholar]
- 14.NCBI Genome Data Viewer Homo Sapiens (Human) Annotation Release 109. [(accessed on 22 August 2019)]; Available online: https://www.ncbi.nlm.nih.gov/genome/gdv/
- 15.Kemper K.E., Visscher P.M., Goddard M.E. Genetic architecture of body size in mammals. Genome Biol. 2012;13:244. doi: 10.1186/gb-2012-13-4-244. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Takasuga A. PLAG1 and NCAPG-LCORL in livestock. Anim. Sci. J. 2016;87:159–167. doi: 10.1111/asj.12417. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Plassais J., Kim J., Davis B.W., Karyadi D.M., Hogan A.N., Harris A.C., Decker B., Parker H.G., Ostrander E.A. Whole genome sequencing of canids reveals genomic regions under selection and variants influencing morphology. Nat. Comm. 2019;10:1489. doi: 10.1038/s41467-019-09373-w. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Signer-Hasler H., Flury C., Haase B., Burger D., Simianer H., Leeb T., Rieder S. A genome-wide association study reveals loci influencing height and other conformation traits in horses. PLoS ONE. 2012;7:e37282. doi: 10.1371/journal.pone.0037282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Makvandi-Nejad S., Hoffman G.E., Allen J.J., Chu E., Gu E., Chandler A.M., Loredo A.I., Bellone R.R., Mezey J.G., Brooks S.A., et al. Four loci explain 83% of size variation in the horse. PLoS ONE. 2012;7:e39929. doi: 10.1371/journal.pone.0039929. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Tetens J., Widmann P., Kühn C., Thaller G. A genome-wide association study indicates LCORL/NCAPG as a candidate locus for withers height in German Warmblood horses. Anim. Genet. 2013;44:467–471. doi: 10.1111/age.12031. [DOI] [PubMed] [Google Scholar]
- 21.Rubin C.-J., Megens H.-J., Barrio A.M., Maqbool K., Sayyab S., Schwochow D., Wang C., Carlborg Ö., Jern P., Jørgensen C.B. Strong signatures of selection in the domestic pig genome. Proc. Natl. Acad. Sci. USA. 2012;109:19529–19536. doi: 10.1073/pnas.1217149109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Ruiz-Larrañaga O., Langa J., Rendo F., Manzano C., Iriondo M., Estonba A. Genomic selection signatures in sheep from the Western Pyrenees. Genet. Sel. Evol. 2018;50:9. doi: 10.1186/s12711-018-0378-x. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23.Signer-Hasler H., Burren A., Ammann P., Drögemüller C., Flury C. Runs of homozygosity and signatures of selection: A comparison among eight local Swiss sheep breeds. Anim. Genet. 2019;50:512–525. doi: 10.1111/age.12828. [DOI] [PubMed] [Google Scholar]
- 24.Setoguchi K., Furuta M., Hirano T., Nagao T., Watanabe T., Sugimoto Y., Takasuga A. Cross-breed comparisons identified a critical 591-kb region for bovine carcass weight QTL (CW-2) on chromosome 6 and the Ile-442-Met substitution in NCAPG as a positional candidate. BMC Genet. 2009;10:43. doi: 10.1186/1471-2156-10-43. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Weikard R., Altmaier E., Suhre K., Weinberger K.M., Hammon H.M., Albrecht E., Setoguchi K., Takasuga A., Kühn C. Metabolomic profiles indicate distinct physiological pathways affected by two loci with major divergent effect on Bos taurus growth and lipid deposition. Physiol. Genom. 2010;42A:79–88. doi: 10.1152/physiolgenomics.00120.2010. [DOI] [PubMed] [Google Scholar]
- 26.Randhawa I.A., Khatkar M.S., Thomson P.C., Raadsma H.W. Composite selection signals for complex traits exemplified through bovine stature using multibreed cohorts of European and African Bos taurus. G3 (Bethesda) 2015;5:1391–1401. doi: 10.1534/g3.115.017772. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 27.Zhang W., Li J., Guo Y., Zhang L., Xu L., Gao X., Zhu B., Gao H., Ni H., Chen Y. Multi-strategy genome-wide association studies identify the DCAF16-NCAPG region as a susceptibility locus for average daily gain in cattle. Sci. Rep. 2016;6:38073. doi: 10.1038/srep38073. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Bouwman A.C., Daetwyler H.D., Chamberlain A.J., Ponce C.H., Sargolzaei M., Schenkel F.S., Sahana G., Govignon-Gion A., Boitard S., Dolezal M., et al. Meta-analysis of genome-wide association studies for cattle stature identifies common genes that regulate body size in mammals. Nat. Genet. 2018;50:362–367. doi: 10.1038/s41588-018-0056-5. [DOI] [PubMed] [Google Scholar]
- 29.Henkel J., Saif R., Jagannathan V., Schmocker C., Zeindler F., Bangerter E., Herren U., Posantzis D., Bulut Z., Ammann P., et al. Selection signatures in goats reveal copy number variants underlying breed-defining coat color phenotypes. PLoS Genet. 2019;15:e1008536. doi: 10.1371/journal.pgen.1008536. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Rubin C.-J., Zody M.C., Eriksson J., Meadows J.R., Sherwood E., Webster M.T., Jiang L., Ingman M., Sharpe T., Ka S., et al. Whole-genome resequencing reveals loci under selection during chicken domestication. Nature. 2010;464:587–591. doi: 10.1038/nature08832. [DOI] [PubMed] [Google Scholar]
- 31.Bickhart D.M., Rosen B.D., Koren S., Sayre B.L., Hastie A.R., Chan S., Lee J., Lam E.T., Liachko I., Sullivan S.T., et al. Single-molecule sequencing and chromatin conformation capture enable de novo reference assembly of the domestic goat genome. Nat. Genet. 2017;49:643–650. doi: 10.1038/ng.3802. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Purcell S., Neale B., Todd-Brown K., Thomas L., Ferreira M.A., Bender D., Maller J., Sklar P., De Bakker P.I., Daly M.J. PLINK: A tool set for whole-genome association and population-based linkage analyses. Am. J. Hum. Genet. 2007;81:559–575. doi: 10.1086/519795. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Sohaib M., Jamil F. An insight of meat industry in Pakistan with special reference to halal meat: A comprehensive review. Korean J. Food Sci. Anim. Resour. 2017;37:329–341. doi: 10.5851/kosfa.2017.37.3.329. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Skapetas B., Bampidis V. Goat production in the World: Present situation and trends. Livest. Res. Rural Dev. 2016;28:200. [Google Scholar]
- 35.Escareño L., Salinas-González H., Wurzinger M., Iñiguez L., Sölkner J., Meza-Herrera C. Dairy goat production systems. Trop. Anim. Health Prod. 2012;45:17–34. doi: 10.1007/s11250-012-0246-6. [DOI] [PubMed] [Google Scholar]
- 36.Bertolini F., Servin B., Talenti A., Rochat E., Kim E.S., Oget C., Palhière I., Crisà A., Catillo G., Steri R. Signatures of selection and environmental adaptation across the goat genome post-domestication. Genet. Sel. Evol. 2018;50:57. doi: 10.1186/s12711-018-0421-y. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Stella A., Nicolazzi E.L., Van Tassell C.P., Rothschild M.F., Colli L., Rosen B.D., Sonstegard T.S., Crepaldi P., Tosser-Klopp G., Joost S. AdaptMap: Exploring goat diversity and adaptation. Genet. Sel. Evol. 2018;50:61. doi: 10.1186/s12711-018-0427-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Talenti A., Bertolini F., Williams J., Moaeen-ud-Din M., Frattini S., Coizet B., Pagnacco G., Reecy J., Rothschild M.F., Crepaldi P. Genomic analysis suggests KITLG is responsible for a roan pattern in two Pakistani goat breeds. J. Hered. 2017;109:315–319. doi: 10.1093/jhered/esx093. [DOI] [PubMed] [Google Scholar]
- 39.Shungin D., Winkler T.W., Croteau-Chonka D.C., Ferreira T., Locke A.E., Mägi R., Strawbridge R.J., Pers T.H., Fischer K., Justice A.E., et al. New genetic loci link adipose and insulin biology to body fat distribution. Nature. 2015;518:187–196. doi: 10.1038/nature14132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Norgard E.A., Lawson H.A., Pletscher L.S., Wang B., Brooks V.R., Wolf J.B., Cheverud J.M. Genetic factors and diet affect long-bone length in the F 34 LG, SM advanced intercross. Mamm. Genome. 2011;22:178–196. doi: 10.1007/s00335-010-9311-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.