Short abstract
Recently, multiple studies regarding the human microbiota and its role on the development of disease have emerged. Current research suggests that the nasal cavity is a major reservoir for opportunistic pathogens, which can then spread to other sections of the respiratory tract and be involved in the development of conditions such as allergic rhinitis, chronic rhinosinusitis, asthma, pneumonia, and otitis media. However, our knowledge of how nasal microbiota changes originate nasopharyngeal and respiratory conditions is still incipient. Herein, we describe how the nasal microbiome in healthy individuals varies with age and explore the effect of nasal microbiota changes in a range of infectious and immunological conditions. We also describe the potential health benefits of human microbiota modulation through probiotic use, both in disease prevention and as adjuvant therapy. Current research suggests that patients with different chronic rhinosinusitis phenotypes possess distinct nasal microbiota profiles, which influence immune response and may be used in the future as biomarkers of disease progression. Probiotic intervention may also have a promising role in the prevention and adjunctive treatment of acute respiratory tract infections and allergic rhinitis, respectively. However, further studies are needed to define the role of probiotics in the chronic rhinosinusitis.
Keywords: allergic rhinitis, chronic rhinosinusitis, dysbiosis, microbiome, microbiota, nose, probiotics
Introduction
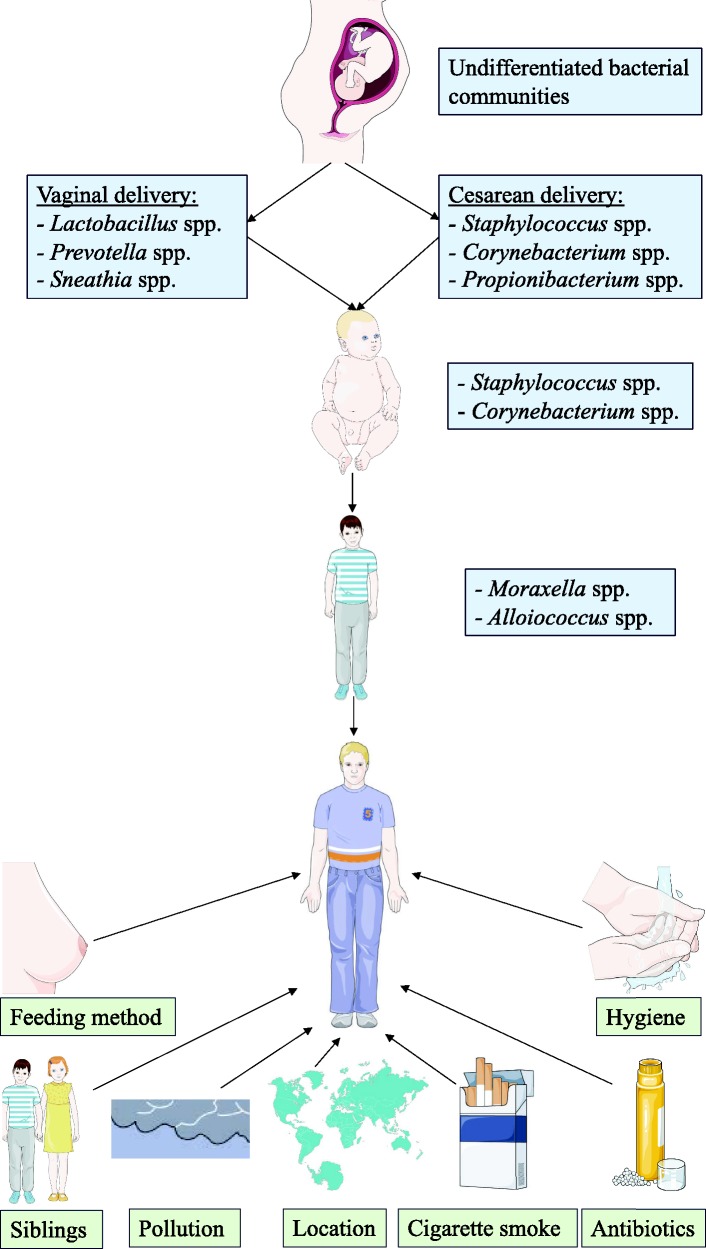
The human microbiome consists of the collective genomes of commensal, symbiotic, and pathogenic microorganisms living on the human body and plays a key role in health and immunity.1 Each body habitat harbors a characteristic bacterial community,2,3 which is not constant throughout life, but rather changes with age.4 The human nasal cavity harbors commensal bacteria that suppress opportunistic pathogen colonization by competing for limited space and nutrients and can even produce toxic compounds that directly inhibit or kill competing microorganisms.5 As depicted in Figure 1, many environmental factors that have been shown to modulate the composition of the nose microbiome.6–8 However, the distinction between commensal and pathogenic bacteria is often ambiguous, as some microorganisms, such as Staphylococcus aureus, can be both commensal and a versatile opportunistic pathogen, causative of significant morbidity and even mortality.9–11
Figure 1.
Factors that modulate nasal microbiota throughout life.
Dysbiosis, a dysfunction or imbalance in the microbial communities, can therefore greatly impact human health.12 Research into the bacterial communities of the nasal cavity has revealed that the nose harbors opportunistic pathogens, which can then spread to other sections of the respiratory tract and be involved in the development of conditions such as allergic rhinitis (AR), chronic rhinosinusitis (CRS), acute respiratory tract infections (ARTI), otitis media (OM), and asthma.13,14 This review aims to describe how changes in the nasal microbiota and microbiome are linked to several immune system disorders and infectious diseases. We also aim to define how probiotics can increase health through the modulation of the nasal microbiota and microbiome, both as adjuvant therapy and in prevention.
The Nasal Microbiome
Nasal Microbiome and CRS
Even though the causal role of bacterial infection in the development of CRS is yet to be proven, it is hypothesized that bacterial infection triggers an inflammatory response within the sinus cavity, with resulting chronic changes and symptoms.6,15 Previous descriptive studies have shown that the nasal microbiome of CRS patients most frequently includes coagulase-negative Staphylococcus, Pseudomonas aeruginosa, and S. aureus.16–21
More recently, Lal et al. compared the microbiota of the middle meatus and inferior meatus CRS patients with and without nasal polyps and in healthy controls. They found that samples collected from the middle meatus of CRS patients without nasal polyps were enriched in Streptococcus, Haemophilus, and Fusobacterium spp. but exhibited loss of bacterial diversity in comparison to healthy controls. Samples collected from the middle meatus of CRS patients with nasal polyps were enriched in Staphylococcus, Alloiococcus, and Corynebacterium spp.22
Cope et al. used 16S rRNA gene sequencing to analyze samples collected during endoscopic sinus surgery in CRS patients and healthy controls and determined that CRS patients can be divided into distinct subgroups, with specific patterns of bacterial colonization, uniquely enriched gene pathways and distinct host immune responses.23 The subgroup predominantly containing Streptococcaceae evoked pro-inflammatory, TH1 responses and mostly encoded an ansamycin biosynthesis gene pathway.23 The subgroup mainly containing Pseudomonadaceae also evoked pro-inflammatory, TH1 responses, but encoded tryptophan metabolism gene pathways.23 The subgroup predominantly containing Corynebacteriaceae encoded peroxisome proliferator-activated receptor-γ signaling pathways, with enhanced IL-5 expression and significantly increased incidence of nasal polyps.23 These findings support the idea that the nose microbiome in CRS patients can not only influence CRS phenotype but also modulate immune response. As summarized in Table 1, nasal dysbiosis plays a role in the development of CRS and the formation of nasal polyps, but further studies are required to clarify the pathophysiologic mechanisms involved.
Table 1.
Nasal Microbiota Alterations in CRS Patients.
| Disorder | Nasal Microbiota Alterations in Comparison to Healthy Individuals |
|---|---|
| Chronic rhinosinusitis | |
| In general | ↑ Coagulase-negative Staphylococcus16,17,19 |
| ↑ Pseudomonas aeruginosa16,17,19 | |
| ↑ Staphylococcus aureus16,17,19 | |
| ↓ Bacterial diversity6 | |
| With nasal polyps | ↑↑ Staphylococcus aureus20 |
| ↑ Alloiococcus spp.22 | |
| ↑ Corynebacterium spp.22 | |
| ↓ Bacterial diversity20 | |
| Without nasal polyps | ↑ Staphylococcus spp.20 |
| ↑ Streptococcus spp.22 | |
| ↑ Haemophilus spp.22 | |
| ↑ Fusobacterium spp.22 | |
| ↓ Bacterial diversity22 | |
| ↓ Prevotella spp.20 | |
| With asthma | ↑ Coagulase-negative Staphylococcus21 |
| ↑ Staphylococcus aureus21 | |
| ↓ Bacterial diversity21 | |
Nasal Microbiome and Asthma
Although the relationship between nasal microbiota and asthma phenotypes and severity is still poorly defined,24–26 recent studies have demonstrated that the nasal microbiota plays a significant role in the onset, development, and severity of asthma.27–30 Culture-independent sequencing methods have shown that the composition of the nasal microbiome is different in adult patients with exacerbated asthma, nonexacerbated asthma, and healthy controls.31–33 In comparison with healthy individuals, the nasal microbiota of asthma patients was enriched with taxa from Bacteroidetes and Proteobacteria, with Prevotella buccalis and Gardnerella vaginalis in particular being more abundant in asthma patients.33 P. buccalis, G. vaginalis, Dialister invisus, and Alkanindiges hongkongensis species were differentially abundant depending on asthma activity, and metagenomic inference revealed differences in bacterial glycerolipid metabolism.33
Teo et al. investigated the nasopharynx microbiome during the first year of life and found that the nasopharynx microbiome was a determinant of risk of future asthma development and severity of accompanying inflammatory symptoms, with early asymptomatic colonization with Streptococcus in particular being a strong predictor for the future development of asthma.30 Pérez-Losada et al. also found that the composition and structure of the nasal microbiota of children and adolescents with asthma was significantly different than that of healthy controls and varied between different asthma phenotypic clusters.34
In a separate study, Pérez-Losada et al. used targeted 16S rRNA sequencing methods to characterize the nasopharyngeal microbiota in asthmatic children and found that the nasal microbiome was dominated by Moraxella, Staphylococcus, Corynebacterium, Haemophilus, Fusobacterium, Prevotella, and Dolosigranulum, which were different from those described in the nasopharynx of adults.29 Further investigation is needed to determine how the nose and nasopharynx microbiome ultimately influences asthma pathophysiology and to develop custom-fit treatment options, which target nasal microbiome dysbiosis in asthma patients.
Nasal Microbiome and AR
Although there is still limited research into the relationship between nasal microbiome dysbiosis and the development of AR, the nasal microbiome potentially holds an important role in the modulation of localized immune responses, pathophysiology, and development of AR. Choi et al. studied the nasal microbiota before and during corresponding pollen seasons in seasonal AR (SAR) patients and healthy controls.35 Samples were collected from the middle meatus and vestibule using endoscopy. During pollen season, there was a correlation between increase in symptoms and nasal eosinophils in SAR patients and a significant increase bacterial biodiversity in the middle meatus compared with healthy controls.35 This pattern is distinct from that found in CRS patients, where disease severity seems to correlate with a decrease in bacterial biodiversity. No significant changes were found in the nasal vestibule.35 Lal et al. compared the microbiota of the middle meatus and inferior meatus AR patients and in healthy controls.22 However, Lal et al. were unable to reproduce the findings of increased biodiversity in AR patients compared to healthy controls, likely due to smaller sample size and not collecting the nasal samples during a specific allergy season.22 Further research is needed to fully comprehend the role of nasal microbiome dysbiosis in AR.
Nasal Microbiome and ARTI
Multiple studies have progressively defined the impact of the nose microbiome in host immunity and protection against opportunistic pathogens, such as S. aureus, rhinovirus, and influenza virus.1,36–38 The interaction between the influenza virus and epithelial cells is mediated by complex bacterial communities, among other factors, which regulate innate and adaptive host immune response and influence risk of infection.1,36 In turn, this host immune response against respiratory viruses can provoke protective changes in the respiratory and nasopharyngeal microbiome.39,40
The influenza A virus infection has also been shown to modify the community structure of the microbiome, with an increase in pathogenic bacteria.36,41 Salk et al. conducted a human experimental trial, in which healthy adults were administered intranasal live attenuated influenza vaccine.42 A significant increase in taxa richness was observed as well as a variation in influenza-specific immunoglobulin A (IgA) antibody production.42 De Lastours et al. showed that adults with influenza virus infection expressed increased nasal carriage of Streptococcus pneumoniae and S. aureus.43 Specifically, S. pneumoniae has been shown to establish a mutually beneficial relationship with the influenza virus, with studies suggesting that influenza A virus infection can enhance the transmission of S. pneumoniae.44,45 Wen et al. found that the nasopharyngeal microbiota of patients with influenza A virus infection differed from healthy controls, with influenza A virus patients showing a predominance of Streptococcus, Phyllobacterium, Moraxella, Staphylococcus, Corynebacterium, and Dolosigranulum.46 In turn, S. pneumoniae can secrete proteases that activate the viral hemagglutinin and even modulate the host innate immune response of the host in order to facilitate influenza A virus infection.44,47
Other viruses are also associated with changes in the nose microbiome. Fan et al. observed an increase in pneumococcal density following rhinovirus infection.48 Wolter et al. found that following a respiratory virus infection by influenza virus, adenovirus, or rhinovirus, there was an elevation in colonization density by S. pneumoniae and therefore an increased risk of invasive pneumococcal pneumonia.49
The nasal microbiome can also influence and modulate infection by other viruses. Rosas-Salazar et al. found significant differences in the taxonomic composition and abundance between infants infected with human rhinovirus and respiratory syncytial virus.50 Toivonen et al. found that the nasopharyngeal microbiota influenced infection by different rhinovirus species.51 Infants with a predominant Haemophilus microbiota profile were more likely to have infection by rhinovirus-A species, whereas infants with a predominant Moraxella microbiota profile were more likely to have infection by rhinovirus-C species.51 Mansbach et al. found an association between a predominant Haemophilus nasopharyngeal microbiota and delayed clearance of respiratory syncytial virus in infants hospitalized for bronchiolitis.52 As shown in Table 2, nasal bacterial communities influence host susceptibility to ARTI, emphasizing the need to develop novel prophylactic and therapeutic probiotic interventions.
Table 2.
Nasal Microbiota Alterations in Patients With ARTI.
| Disorder | Nasal Microbiota Alterations in Comparison to Healthy Individuals |
|---|---|
| Acute respiratory tract infections | |
| Influenza A virus | ↑ Pathogenic bacteria36,41,42 |
| ↑ Streptococcus pneumoniae43 | |
| ↑ Staphylococcus aureus43,46 | |
| ↑ Phyllobacterium spp.46 | |
| ↑ Moraxella spp.46 | |
| ↑ Corynebacterium spp.46 | |
| ↑ Dolosigranulum spp.46 | |
| ↓ Pseudomonas spp.45 | |
| Rhinovirus-A | ↑ Streptococcus pneumoniae48,49 |
| ↑ Haemophilus spp.51 | |
| Rhinovirus-C | ↑ Streptococcus pneumoniae48,49 |
| ↑ Moraxella spp.51 | |
| Adenovirus | ↑ Streptococcus pneumoniae49 |
| Respiratory syncytial virus | ↑ Haemophilus spp.51 |
Nasal Microbiome and OM
As displayed in Table 3, the nose microbiome influences the development of OM, with an increase in carriage of pathogenic bacteria elevating the risk of invasive disease, probably due to bacterial migration to the middle ear.10,53,54 Hilty et al. studied the nasopharyngeal microbiota of 163 infants with or without acute OM using nasopharyngeal swabs and multiplexed pyrosequencing of 16S rRNA and found that commensal bacteria were less prevalent in infants with acute OM in comparison to healthy controls.55 Moraxella, Streptococcus, and Pasteurella spp. were most frequent in acute OM patients in comparison to healthy controls.55
Table 3.
Nasal Microbiota Alterations in Patients With OM.
| Disorder | Nasal Microbiota Alterations in Comparison to Healthy Individuals |
|---|---|
| Otitis media | |
| In general | ↑ Moraxella spp.56,59 |
| ↑ Streptococcus pneumoniae56,57,59 | |
| ↑ Pasteurella spp.56 | |
| ↑ Haemophilus spp.57,59 | |
| ↑ Actinomyces spp.57 | |
| ↑ Rothia spp.57 | |
| ↑ Neisseria spp.57 | |
| ↑ Veillonella spp.57 | |
| ↓ Commensal bacteria56 | |
| ↓ Bacterial diversity57,59 | |
| ↓ Lactococcus spp.57 | |
| ↓ Propionibacterium spp.57 | |
| ↓ Corynebacterium spp.57 | |
| ↓ Dolosigranulum spp.57 | |
| ↓ Staphylococcus spp.57 | |
| ↓ Pseudomonas spp.59 | |
| ↓Myroides spp.59 | |
| ↓ Yersinia spp.59 | |
| ↓ Sphingomonas spp.59 | |
| Recurrent | ↑ Gemella spp.58 |
| ↑ Neisseria spp.58 | |
| ↓ Corynebacterium spp.58 | |
| ↓ Dolosigranulum spp.58 | |
Laufer et al. found that S. pneumoniae colonization was significantly more frequent in children with OM, and nasal colonization with Haemophilus, Actinomyces, Rothia, Neisseria, and Veillonella was also associated with an increased risk of OM.56 On the other hand, colonization with Corynebacterium and Dolosigranulum and Propionibacterium, Lactococcus, and Staphylococcus was associated with a decreased risk of pneumococcal colonization and OM.56 Recently, Lappan et al. found that the nasopharyngeal microbiomes of patients with recurrent acute OM and healthy controls were distinct, with Gemella and Neisseria being more prevalent in the nasopharynx of patients with recurrent acute OM in comparison with healthy controls and Corynebacterium and Dolosigranulum being significantly more abundant in the nasopharynx of controls.57
Chonmaitree et al. analyzed 971 nasopharyngeal samples collected during episodes of acute OM and found an increased abundance in 3 otopathogen genera: Moraxella, Haemophilus, and Streptococcus, with lower bacterial diversity.58 During acute OM episodes, there were increases in abundance of otopathogen genera and decreases in Pseudomonas, Myroides, Yersinia, and Sphingomonas.58 Further studies are needed to define how probiotics could be used to modulate the nasal and nasopharyngeal microbiome and protect against OM.
The Role of Probiotic Intervention in Nasal Microbiome Dysbiosis
Probiotic Intervention in AR
Even though the role of dysbiosis in the pathophysiology of AR is poorly understood, there have been promising developments in the use of probiotics as adjuvant treatment. Ishida et al. found that the administration of Lactobacillus acidophilus 92 in fermented milk significantly improved nasal symptom scores in participants with perennial AR (PAR), in comparison with the administration of milk without lactic acid bacteria.59 In a double-blind, placebo-controlled randomized study with 60 children with PAR, half were treated with the anti-histamine agent levocetirizine, together with Lactobacillus paracasei, and half were treated with levocetirizine with placebo. The group treated with L. paracasei reported an improvement in pediatric rhinoconjunctivitis quality-of-life scores and a significant improvement in nasal itching and sneezing scores in comparison with the placebo.60
Jerzynska et al. studied the effect of Lactobacillus rhamnosus GG and vitamin D supplementation on the immunologic effectiveness of grass-specific sublingual immunotherapy (SLIT) in children with AR with sensitization to grass pollen. They found that there was a decrease in the symptom-medication score in all groups treated with 5-grass SLIT as well as a significant increase in CD4+CD25+Fox3+ cells in the children receiving SLIT with L. rhamnosus, compared with children treated with SLIT and vitamin D, resulting in a better immunologic response.61 These results suggest that probiotics do have a potential important role in the adjunctive treatment of PAR and SAR.
Probiotic Intervention in CRS
In contrast with AR, current research into the use of probiotics in the adjunctive treatment of CRS is limited and does not support a prominent role of probiotics in the treatment of CRS. In a double-blind, randomized, placebo-controlled trial by Mukerji et al., there were no significant clinical improvements in sinonasal quality-of-life scores with oral administration of a preparation containing L. rhamnosus in 77 patients with CRS.62 Furthermore, the topical administration of honeybee lactic acid bacteria (HLAB), a mixture of 9 Lactobacillus spp. and 4 Bifidobacterium spp., directly into the nasal passage was safe and well tolerated63 but did not produce any changes in symptoms, inflammatory biomarkers in nasal lavage fluids, or commensal bacteria of the nasal cavity in patients with CRS without nasal polyps.64 Although further research is needed in this field, probiotics are currently not recommended in the adjuvant treatment of CRS.
Probiotic Intervention in Respiratory Tract Infections
Given the significant role of the nose and throat microbiome in the susceptibility to influenza virus infection,1,36,38 there has been some investigation into the potential role of probiotics in the modulation of the nasal microbiome and innate and adaptive host immune responses. Salk et al. measured changes to the nasal microbiome in young adults who received intranasal administration of live attenuated influenza vaccine (LAIV) and found that LAIV altered the nasal microbiome, with increased taxa richness and variations in influenza-specific IgA antibody production.42
Furthermore, molecular profiling studies have recently revealed that species-specific interactions play an important role in avoiding S. aureus nasal colonization and persistence.5 Specifically, the Corynebacterium genus interacts with S. aureus in the nasal cavity65 and artificial inoculation of Corynebacterium pseudodiphtheriticum into the nasal cavity appears to eradicate S. aureus nasal colonization.66–68 Hardy et al. determined that this is because C. pseudodiphtheriticum selectively targets S. aureus for killing, mainly through Agr QS, its primary virulence factor. This may confer selective advantage to S. aureus strains deficient in Agr QS, inducing a switch from a pathogenic state to a commensal state.5 Further studies are needed to characterize the therapeutic use of C. pseudodiphtheriticum-derived factors as bactericidal agents against S. aureus.
The effects of a 1-week administration of a probiotic product composed of a combination of Streptococcus salivarius 24SMBc and Streptococcus oralis 89a, on the nostril microbiota were evaluated using a next-generation sequencing approach. A significant decrease in S. aureus abundance was detected immediately after the probiotic administration, and there was an increase in the total number of beneficial microorganisms which could limit the overgrowth of potential pathogens.69
In 2 separate randomized, double-blind, placebo-controlled trials, newborns who were administered prebiotics and probiotics had a significantly lower incidence of respiratory tract infections compared to the placebo.70,71 Specifically, the incidence of rhinovirus-induced episodes, which comprised 80% of all respiratory tract infections, was found to be significantly lower in the prebiotic and probiotic (groups compared with the placebo group.70 In another randomized symbiotic trial, 4556 infants were administered an oral preparation containing Lactobacillus plantarum. Significant reductions were observed in the incidence of respiratory tract infections.71
There are also encouraging results with regard to immunity enhancement through intranasal vaccines in experiments conducted on animals. The intranasal administration of Bacillus subtilis vaccines in the nasal mucosa and tonsils of piglets resulted in an increase in the number of dendritic cells, immunoglobulin A+ B cells and T cells in the nasal mucosa and tonsils, as well as an increase in toll-like receptor (TLR)-2 and TLR-9 mRNA expression. This lays the foundation for further study into the intranasal administration of B. subtilis in humans to enhance the immunity of human nasal mucosa to respiratory diseases.72
Conclusion
In healthy individuals, the nasal microbiome varies with age and is shaped by various factors. Interestingly, pathological conditions of the respiratory tract seem to be associated with a reduction in nasal microbiota biodiversity, a feature also observed in the gut. Nasal microbiome dysbiosis has been implicated in the pathophysiology of many diseases, including CRS, asthma, AR, bronchiolitis, the flu, and OM, although further studies are needed to further characterize the role of the nasal dysbiosis in AR. Current research suggests that the nasal microbiota profile influences immune response and may modulate CRS phenotype. Further studies are needed to explore the use potential use of the nasal microbiota profile as a prognostic tool in clinical practice.
However, when considering the potential clinical use of the characterization of the nasal microbiome profile, it is important to take into consideration the wide array of methodologies available, with distinct operating costs and potentially conflicting results. For instance, polymerase chain reaction methods, which are relatively cheap, can only identify a relatively small array of genes, in comparison with more expensive next-generation sequencing, a high throughput methodology with broad range applications which could allow for a more complete characterization of the microbiome profile in different individuals. Although these methods could be useful in clinical practice, allowing for personalized and optimized patient treatment and follow-up, the operational costs need to be taken into account, and presently compromise the use of these techniques as disease biomarkers and prognostic factors in patient care. Furthermore, the possibility of changes in nasal microbiome richness and diversity being secondary to inflammation in diseases such as CRS, AR, and ARTI must also be taken into consideration.
Currently, studies suggest that probiotic intervention may have a promising role in the adjunctive treatment of PAR and SAR, with improvements in immune response, symptom scores and quality of life, and in the prevention of ARTI, through species-specific interactions and immunological modulation. Further studies are needed to fully characterize how these interventions can be applied in clinical practice. However, it remains unclear whether probiotics could have a role in the treatment of CRS.
Author Contributions
P. B. contributed to concept and design. S. D.-P. was involved in search, analysis, and inclusion/exclusion of papers as well as drafting of the manuscript. All authors contributed to critical revision of the manuscript for important intellectual content. P. B. and R. S. contributed to supervision.
Authors’ Note
Images are created using Servier Medical ART®.
Declaration of Conflicting Interests
The author(s) declared no potential conflicts of interest with respect to the research, authorship, and/or publication of this article.
Ethical Approval
This study was approved by our institutional review board.
Funding
The author(s) disclosed receipt of the following financial support for the research, authorship, and/or publication of this article: This work was supported by the Norte Portugal Regional Programme (NORTE 2020), under the PORTUGAL 2020 Partnership Agreement, through the European Regional Development Fund (ERDF)—Bioengineered therapies for infectious disease and tissue regeneration (grant number NORTE-01-0145-FEDER-0000012).
Statement of Human and Animal Rights
This article does not contain any studies with human or animal subjects.
Statement of Informed Consent
There are no human subjects in this article and informed consent is not applicable.
ORCID iD
Sofia Dimitri-Pinheiro https://orcid.org/0000-0002-0583-2792
References
- 1.Abt MC, Osborne LC, Monticelli LA, et al. Commensal bacteria calibrate the activation threshold of innate antiviral immunity. Immunity. 2012; 37(1):158–170. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Costello EK, Lauber CL, Hamady M, Fierer N, Gordon JI, Knight R. Bacterial community variation in human body habitats across space and time. Science (New York, NY ) 2009; 326(5960):1694–1697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Huttenhower C, Gevers D, Knight R, et al. Structure, function and diversity of the healthy human microbiome. Nature. 2012; 486(7402):207–214. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Thomas S, Izard J, Walsh E, et al. The host microbiome regulates and maintains human health: a primer and perspective for non-microbiologists. Cancer Res. 2017; 77(8):1783–1812. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Hardy BL, Dickey SW, Plaut RD, et al. Corynebacterium pseudodiphtheriticum exploits Staphylococcus aureus virulence components in a novel polymicrobial defense strategy . mBio. 2019; 10(1):e02491–18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Rawls M, Ellis AK. The microbiome of the nose. Ann Allergy Asthma Immunol. 2019; 122(1):17–24. [DOI] [PubMed] [Google Scholar]
- 7.Man WH, de Steenhuijsen Piters WA, Bogaert D. The microbiota of the respiratory tract: gatekeeper to respiratory health. Nat Rev Microbiol. 2017; 15(5):259–270. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Arrieta MC, Stiemsma LT, Amenyogbe N, Brown EM, Finlay B. The intestinal microbiome in early life: health and disease. Front Immunol. 2014; 5:427. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Otto M. Basis of virulence in community-associated methicillin-resistant Staphylococcus aureus. Ann Rev Microbiol. 2010; 64:143–162. [DOI] [PubMed] [Google Scholar]
- 10.Wertheim HF, Melles DC, Vos MC, et al. The role of nasal carriage in Staphylococcus aureus infections. Lancet Infect Dis. 2005; 5(12):751–762. [DOI] [PubMed] [Google Scholar]
- 11.Kluytmans JA, Wertheim HF. Nasal carriage of Staphylococcus aureus and prevention of nosocomial infections. Infection. 2005; 33(1):3–8. [DOI] [PubMed] [Google Scholar]
- 12.DeGruttola AK, Low D, Mizoguchi A, Mizoguchi E. Current understanding of dysbiosis in disease in human and animal models. Inflamm Bowel Dis. 2016; 22(5):1137–1150. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Esposito S, Principi N. Impact of nasopharyngeal microbiota on the development of respiratory tract diseases. Eur J Clin Microbiol Infect Dis. 2018; 37(1):1–7. [DOI] [PubMed] [Google Scholar]
- 14.Huang YJ. Nasopharyngeal microbiota: gatekeepers or fortune tellers of susceptibility to respiratory tract infections? Am J Respir Crit Care Med. 2017; 196(12):1504–1505. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Desrosiers M. Refractory chronic rhinosinusitis: pathophysiology and management of chronic rhinosinusitis persisting after endoscopic sinus surgery. Curr Allergy Asthma Rep. 2004; 4(3):200–207. [DOI] [PubMed] [Google Scholar]
- 16.Bhattacharyya N, Kepnes LJ. Assessment of trends in antimicrobial resistance in chronic rhinosinusitis. Ann Otol Rhinol Laryngol. 2008; 117(6):448–452. [DOI] [PubMed] [Google Scholar]
- 17.Kingdom TT, Swain RE., Jr. The microbiology and antimicrobial resistance patterns in chronic rhinosinusitis. Am J Otolaryngol. 2004; 25(5):323–328. [DOI] [PubMed] [Google Scholar]
- 18.Nadel DM, Lanza DC, Kennedy DW. Endoscopically guided cultures in chronic sinusitis. Am J Rhinol. 1998; 12(4):233–241. [DOI] [PubMed] [Google Scholar]
- 19.Coffey CS, Sonnenburg RE, Melroy CT, Dubin MG, Senior BA. Endoscopically guided aerobic cultures in postsurgical patients with chronic rhinosinusitis. Am J Rhinol. 2006; 20(1):72–76. [PubMed] [Google Scholar]
- 20.Choi EB, Hong SW, Kim DK, et al. Decreased diversity of nasal microbiota and their secreted extracellular vesicles in patients with chronic rhinosinusitis based on a metagenomic analysis. Allergy. 2014; 69(4):517–526. [DOI] [PubMed] [Google Scholar]
- 21.Feazel LM, Robertson CE, Ramakrishnan VR, Frank DN. Microbiome complexity and Staphylococcus aureus in chronic rhinosinusitis. Laryngoscope. 2012; 122(2):467–472. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Lal D, Keim P, Delisle J, et al. Mapping and comparing bacterial microbiota in the sinonasal cavity of healthy, allergic rhinitis, and chronic rhinosinusitis subjects. Int Forum Allergy Rhinol. 2017; 7(6):561–569. [DOI] [PubMed] [Google Scholar]
- 23.Cope EK, Goldberg AN, Pletcher SD, Lynch SV. Compositionally and functionally distinct sinus microbiota in chronic rhinosinusitis patients have immunological and clinically divergent consequences. Microbiome. 2017; 5(1):53. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Chung KF. Potential role of the lung microbiome in shaping asthma phenotypes. Ann Am Thorac Soc. 2017; 14(Supplement_5):S326–S331. [DOI] [PubMed] [Google Scholar]
- 25.Zhang Q, Cox M, Liang Z, et al. Airway microbiota in severe asthma and relationship to asthma severity and phenotypes. PLoS One. 2016; 11(4):e0152724. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Taylor SL, Leong LEX, Choo JM, et al. Inflammatory phenotypes in patients with severe asthma are associated with distinct airway microbiology. J Allergy Clin Immunol. 2018; 141(1):94–103.e115. [DOI] [PubMed] [Google Scholar]
- 27.Huang YJ, Boushey HA. The microbiome in asthma . J Allergy Clin Immunol. 2015; 135(1):25–30. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Pérez-Losada M, Alamri L, Crandall KA, Freishtat RJ. Nasopharyngeal microbiome diversity changes over time in children with asthma. PLoS One. 2017; 12(1):e0170543. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Perez-Losada M, Crandall KA, Freishtat RJ. Two sampling methods yield distinct microbial signatures in the nasopharynges of asthmatic children. Microbiome. 2016; 4(1):25. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Teo SM, Mok D, Pham K, et al. The infant nasopharyngeal microbiome impacts severity of lower respiratory infection and risk of asthma development. Cell Host Microbe. 2015; 17(5):704–715. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Dickson RP, Erb-Downward JR, Huffnagle GB. The role of the bacterial microbiome in lung disease. Exp Rev Respir Med. 2013; 7(3):245–257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Perez-Losada M, Castro-Nallar E, Bendall ML, Freishtat RJ, Crandall KA. Dual transcriptomic profiling of host and microbiota during health and disease in pediatric asthma. PLoS One. 2015; 10(6):e0131819. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 33.Fazlollahi M, Lee TD, Andrade J, et al. The nasal microbiome in asthma. J Allergy Clin Immunol. 2018; 142(3):834–843.e832. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 34.Pérez-Losada M, Authelet KJ, Hoptay CE, Kwak C, Crandall KA, Freishtat RJ. Pediatric asthma comprises different phenotypic clusters with unique nasal microbiotas. Microbiome. 2018; 6(1):179. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35.Choi CH, Poroyko V, Watanabe S, et al. Seasonal allergic rhinitis affects sinonasal microbiota. Am J Rhinol Allergy. 2014; 28(4):281–286. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36.Ichinohe T, Pang IK, Kumamoto Y, et al. Microbiota regulates immune defense against respiratory tract influenza A virus infection. Proc Natl Acad Sci U S A. 2011; 108(13):5354–5359. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Levy M, Blacher E, Elinav E. Microbiome, metabolites and host immunity. Curr Opin Microbiol. 2017; 35:8–15. [DOI] [PubMed] [Google Scholar]
- 38.Lee KH, Gordon A, Shedden K, et al. The respiratory microbiome and susceptibility to influenza virus infection. PLoS One. 2019; 14(1):e0207898. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Lynch SV. Viruses and microbiome alterations. Ann Am Thorac Soc. 2014; 11(Suppl 1):S57–S60. [DOI] [PubMed] [Google Scholar]
- 40.Lee KH, Gordon A, Foxman B. The role of respiratory viruses in the etiology of bacterial pneumonia: An ecological perspective. Evol Med Public Health. 2016; 2016(1):95–109. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Cauley LS, Vella AT. Why is coinfection with influenza virus and bacteria so difficult to control? Discov Med. 2015; 19(102):33–40. [PMC free article] [PubMed] [Google Scholar]
- 42.Salk HM, Simon WL, Lambert ND, et al. Taxa of the nasal microbiome are associated with influenza-specific IgA response to live attenuated influenza vaccine. PLoS One. 2016; 11(9):e0162803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.De Lastours V, Malosh R, Ramadugu K, et al. Co-colonization by Streptococcus pneumoniae and Staphylococcus aureus in the throat during acute respiratory illnesses. Epidemiol Infect. 2016; 144(16): 3507–3519. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Short KR, Habets MN, Hermans PW, Diavatopoulos DA. Interactions between Streptococcus pneumoniae and influenza virus: a mutually beneficial relationship? Future Microbiol. 2012; 7(5):609–624. [DOI] [PubMed] [Google Scholar]
- 45.Borges L, Giongo A, Pereira LM, et al. Comparison of the nasopharynx microbiome between influenza and non-influenza cases of severe acute respiratory infections: A pilot study. Health Sci Rep. 2018; 1(6):e47. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Wen Z, Xie G, Zhou Q, et al. Distinct nasopharyngeal and oropharyngeal microbiota of children with influenza A virus compared with healthy children. BioMed Res Int. 2018; 2018:6362716. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.McCullers JA, McAuley JL, Browall S, Iverson AR, Boyd KL, Henriques Normark B. Influenza enhances susceptibility to natural acquisition of and disease due to Streptococcus pneumoniae in ferrets. J Infect Dis. 2010; 202(8):1287–1295. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Fan RR, Howard LM, Griffin MR, et al. Nasopharyngeal pneumococcal density and evolution of acute respiratory illnesses in young children, Peru, 2009-2011. Emerg Infect Dis. 2016; 22(11):1996–1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Wolter N, Tempia S, Cohen C, et al. High nasopharyngeal pneumococcal density, increased by viral coinfection, is associated with invasive pneumococcal pneumonia. J Infect Dis. 2014; 210(10):1649–1657. [DOI] [PubMed] [Google Scholar]
- 50.Rosas-Salazar C, Shilts MH, Tovchigrechko A, et al. Differences in the nasopharyngeal microbiome during acute respiratory tract infection with human rhinovirus and respiratory syncytial virus in infancy. J Infect Dis. 2016; 214(12):1924–1928. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Toivonen L, Camargo CA, Jr, Gern JE, et al. Association between rhinovirus species and nasopharyngeal microbiota in infants with severe bronchiolitis. J Allergy Clin Immunol. 2019; 143(5):1925–1928.e7. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Mansbach JM, Hasegawa K, Piedra PA, et al. Haemophilus-dominant nasopharyngeal microbiota is associated with delayed clearance of respiratory syncytial virus in infants hospitalized for bronchiolitis. J Infect Dis. 2018; 219(11):1804–1808. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Krone CL, van de Groep K, Trzcinski K, Sanders EA, Bogaert D. Immunosenescence and pneumococcal disease: an imbalance in host-pathogen interactions. Lancet Respir Med. 2014; 2(2):141–153. [DOI] [PubMed] [Google Scholar]
- 54.Bogaert D, De Groot R, Hermans PW. Streptococcus pneumoniae colonisation: the key to pneumococcal disease. Lancet Infect Dis. 2004; 4(3):144–154. [DOI] [PubMed] [Google Scholar]
- 55.Hilty M, Qi W, Brugger SD, et al. Nasopharyngeal microbiota in infants with acute otitis media. J Infect Dis. 2012; 205(7):1048–1055. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Laufer AS, Metlay JP, Gent JF, Fennie KP, Kong Y, Pettigrew MM. Microbial communities of the upper respiratory tract and otitis media in children. mBio. 2011; 2(1):e00245–00210. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Lappan R, Imbrogno K, Sikazwe C, et al. A microbiome case-control study of recurrent acute otitis media identified potentially protective bacterial genera. BMC Microbiol. 2018; 18(1):13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58.Chonmaitree T, Jennings K, Golovko G, et al. Nasopharyngeal microbiota in infants and changes during viral upper respiratory tract infection and acute otitis media. PLoS One. 2017; 12(7):e0180630. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Ishida Y, Nakamura F, Kanzato H, et al. Clinical effects of Lactobacillus acidophilus strain L-92 on perennial allergic rhinitis: a double-blind, placebo-controlled study. J Dairy Sci. 2005; 88(2):527–533. [DOI] [PubMed] [Google Scholar]
- 60.Lin WY, Fu LS, Lin HK, Shen CY, Chen YJ. Evaluation of the effect of Lactobacillus paracasei (HF.A00232) in children (6-13 years old) with perennial allergic rhinitis: a 12-week, double-blind, randomized, placebo-controlled study. Pediatr Neonatol. 2014; 55(3):181–188. [DOI] [PubMed] [Google Scholar]
- 61.Jerzynska J, Stelmach W, Balcerak J, et al. Effect of Lactobacillus rhamnosus GG and vitamin D supplementation on the immunologic effectiveness of grass-specific sublingual immunotherapy in children with allergy. Allergy Asthma Proc. 2016; 37(4):324–334. [DOI] [PubMed] [Google Scholar]
- 62.Mukerji SS, Pynnonen MA, Kim HM, Singer A, Tabor M, Terrell JE. Probiotics as adjunctive treatment for chronic rhinosinusitis: a randomized controlled trial. Otolaryngol Head Neck Surg. 2009; 140(2):202–208. [DOI] [PubMed] [Google Scholar]
- 63.Martensson A, Greiff L, Lamei SS, et al. Effects of a honeybee lactic acid bacterial microbiome on human nasal symptoms, commensals, and biomarkers. Int Forum Allergy Rhinol. 2016; 6(9):956–963. [DOI] [PubMed] [Google Scholar]
- 64.Martensson A, Abolhalaj M, Lindstedt M, et al. Clinical efficacy of a topical lactic acid bacterial microbiome in chronic rhinosinusitis: a randomized controlled trial. Laryngosc Investigat Otolaryngol. 2017; 2(6):410–416. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65.Johnson RC, Ellis MW, Lanier JB, Schlett CD, Cui T, Merrell DS. Correlation between nasal microbiome composition and remote purulent skin and soft tissue infections. Infect Immun. 2015; 83(2):802–811. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66.Kiryukhina NV, Melnikov VG, Suvorov AV, Morozova YA, Ilyin VK. Use of Corynebacterium pseudodiphtheriticum for elimination of Staphylococcus aureus from the nasal cavity in volunteers exposed to abnormal microclimate and altered gaseous environment. Probiot Antimicrob Proteins. 2013; 5(4):233–238. [DOI] [PubMed] [Google Scholar]
- 67.Yan M, Pamp SJ, Fukuyama J, et al. Nasal microenvironments and interspecific interactions influence nasal microbiota complexity and S. aureus carriage. Cell Host Microbe. 2013; 14(6):631–640. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68.Uehara Y, Nakama H, Agematsu K, et al. Bacterial interference among nasal inhabitants: eradication of Staphylococcus aureus from nasal cavities by artificial implantation of Corynebacterium sp. J Hosp Infect. 2000; 44(2):127–133. [DOI] [PubMed] [Google Scholar]
- 69.De Grandi R, Drago L, Bidossi A, Bottagisio M, Gelardi M, De Vecchi E. Putative microbial population shifts attributable to nasal administration of Streptococcus salivarius 24SMBc and Streptococcus oralis 89a. Probiot Antimicrob Proteins. 2019; 11(4):1219–1226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70.Luoto R, Ruuskanen O, Waris M, Kalliomaki M, Salminen S, Isolauri E. Prebiotic and probiotic supplementation prevents rhinovirus infections in preterm infants: a randomized, placebo-controlled trial. J Allergy Clin Immunol. 2014; 133(2):405–413. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Panigrahi P, Parida S, Nanda NC, et al. A randomized synbiotic trial to prevent sepsis among infants in rural India. Nature. 2017; 548(7668):407–412. [DOI] [PubMed] [Google Scholar]
- 72.Yang Y, Jing Y, Yang J, Yang Q. Effects of intranasal administration with Bacillus subtilis on immune cells in the nasal mucosa and tonsils of piglets. Exp Therapeut Med. 2018; 15(6):5189–5198. [DOI] [PMC free article] [PubMed] [Google Scholar]