Abstract
The genus Listeria now comprises up to now 21 recognized species and six subspecies, with L. monocytogenes and L. innocua as the most prevalent sensu stricto associated species. Reports focusing on the challenges in Listeria detection and confirmation are available, especially from food-associated environmental samples. L. innocua is more prevalent in the food processing environment (FPE) than L. monocytogenes and has been shown to have a growth advantage in selective enrichment and agar media. Until now, the adaptive nature of L. innocua in FPEs has not been fully elucidated and potential persistence in the FPE has not been observed. Therefore, the aim of this study is to characterize L. innocua (n = 139) and L. monocytogenes (n = 81) isolated from FPEs and cheese products collected at five dairy processing facilities (A–E) at geno- and phenotypic levels. Biochemical profiling was conducted for all L. monocytogenes and the majority of L. innocua (n = 124) isolates and included a rhamnose positive reaction. L. monocytogenes isolates were most frequently confirmed as PCR-serogroups 1/2a, 3a (95%). Pulsed-field gel electrophoresis (PFGE)-typing, applying the restriction enzymes AscI, revealed 33 distinct Listeria PFGE profiles with a Simpson’s Index of Diversity of 0.75. Multi-locus sequence typing (MLST) resulted in 27 STs with seven new L. innocua local STs (ST1595 to ST1601). L. innocua ST1597 and ST603 and L. monocytogenes ST121 and ST14 were the most abundant genotypes in dairy processing facilities A–E over time. Either SSI-1 (ST14) or SSI-2 (ST121, all L. innocua) were present in successfully FPE-adapted strains. We identified housekeeping genes common in Listeria isolates and L. monocytogenes genetic lineage III. Wherever there are long-term contamination events of L. monocytogenes and other Listeria species, subtyping methods are helpful tools to identify niches of high risk.
Keywords: Listeria monocytogenes, Listeria innocua, dairy processing, subtyping, persistence
1. Introduction
The genus Listeria, assigned to the phylum Firmicutes, comprises Gram-positive facultative anaerobe bacteria that are found ubiquitously in environments such as soil, water, or plant material [1,2]. The genus comprises up to 21 recognized species and six subspecies, with L. monocytogenes and L. innocua as the most prevalent sensu stricto associated species [3] (http://www.bacterio.net/listeria.html). L. monocytogenes has been implicated in human listeriosis outbreaks, most often associated with ready-to-eat (RTE) food products (https://www.cdc.gov/listeria/outbreaks/index.html) [4,5]. Food is most commonly contaminated by L. monocytogenes from niches in the food processing environment (FPE) [6]. As an environmental saprophyte, L. monocytogenes is highly adapted to harsh conditions, such as those associated with osmotic and cold stress, low pH, desiccation, and competitive microflora [7]. The adaptive strategies of L. monocytogenes to ecological niches are clearly divergent. Genetic lineage I (serovar 1/2b and 4b) has a tropism to human and animal host tissues and cell types, whereas genetic lineage II (serovar 1/2a and 1/2c) is more adapted to environmental conditions [8,9].
The ecological co-habitation, genomic synteny, and phenotypic similarities of L. monocytogenes and L. innocua qualify the latter as surrogate for L. monocytogenes behavior prediction in FPEs [10]. Nevertheless, the prfA-virulence gene cluster that was present in the common ancestor of Listeria species was lost in two separate recombination events with L. innocua and L. welshimeri [11]. In fact, L. innocua comprises two major subgroups A and B, with one atypical subgroup D (e.g., hemolytic, LIPI regions and inlJ positive) serving as a link between L. monocytogenes and L. innocua in the evolutionary chain [12]. Internalin genes (lin0354, lin0661, lin1204, and lin2539) have been suggested as potential genetic markers for L. innocua subgroups A-D [13]. Hemolytic L. innocua strains have now been characterized by whole genome sequencing (WGS) and virulence testing. The strains harbor a pathogenic island, LIPI-1, and internalin inlA, required for mammalian cell invasion. These findings will challenge risk management in the food chain, as L. innocua was often assessed in the past as non-hazardious [12]. Currently, L. innocua genome sequences are still limited [14,15]. Eleven L. innocua genome assemblies are currently available from the National Center for Biotechnology Information (NCBI) (https://www.ncbi.nlm.nih.gov/genome/genomes/1024?).
The loss of full virulence in L. monocytogenes lineage II strains (e.g., sequence type (ST) 9, ST121), due to point mutations in prfA, inlA, inlB, and plcA, indicates the formation of monophyletic groups present in FPEs [16]. The latter strains were mentioned, amongst others (e.g., ST5, ST7, ST8), in past long-term FPE contamination scenarios, which were then referred to as in-house clones or persisters [17,18,19,20]. Until now, the adaptive nature of L. innocua in FPEs has not been fully characterized and potential persistence has not been observed. Accordingly, the aim of this study is to characterize potential L. innocua persistent isolates originating from FPEs and cheese products on geno- and phenotypic levels and to compare them to L. monocytogenes strains from the same habitat.
2. Materials and Methods
2.1. Isolate Selection and Listeria Species Confirmation
In total, 220 Listeria isolates (139 L. innocua and 81 L. monocytogenes) originating from different cheese types, product-associated liquids (PAL; smear, brine), product-associated samples (culture, enrichment), raw material (RM), food contact surfaces (FCS), non-food contact surfaces (NFCS), and environmental liquid samples (EL; floor and drain water) were included in this study. The isolates were collected during a Listeria monitoring program between 1987 and 2010 from five dairy producers (A–E) [21,22]. The selection criteria for this study were as following: (1) producer with a history of frequent L. innocua or L. monocytogenes isolation, and (2) L. monocytogenes previously identified as potentially persistent in the FPE. Producer A was mainly involved with ripening cheese and packaging grated and sliced semi-hard cheeses. Producers B and C manufactured a broad range of white dairy and fresh products and a product line of semi-hard and hard cheeses. Producer D manufactured a range of semi-hard and red smear cheeses and producer E manufactured acid curd cheeses (Table 1). The L. innocua and L. monocytogenes isolates were stored at −80 °C at the Listeria collection located at the Unit of Food Microbiology, University of Veterinary Medicine Vienna, Austria. L. innocua and L. monocytogenes isolates were re-cultivated on tryptic soy agar (TSA; Merck KgA, Darmstadt, Germany) at 37 °C overnight. DNA isolation followed a rapid protocol using Chelex 100-Resin (Bio-Rad Laboratories Inc., Hercules, CA, USA), as published by Walsh et al. [23]. For Listeria species differentiation, a multiplex PCR targeting the invasion-associated protein (iap) gene was performed [24]. L. monocytogenes isolates were subtyped by serogroup PCR, targeting the Listeria spp. specific prs, and lmo0737, lmo1118, ORF2819, and ORF2110 [25]. Biochemical profiling was performed for each Listeria isolate by applying the API-Listeria identification system (Biomérieux, Marcy l’Etoile, France).
Table 1.
L. innocua (n = 139) and L. monocytogenes (n = 81) isolate characteristics included in this study.
| PRODUCER | SOURCE | OCCURRENCE | TIME- FRAME |
ISOLATES (n) | PFGE PROFILE (ASCI/APAI) | CC f | ST (Serogroup) |
abcZ | bglA | cat | dapE | dat | ldh | lhkA | SSI-1 + g | SSI-1- h | SSI-2 i |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| L. monocytogenes (API profile 6-5-1-0) | |||||||||||||||||
| C | milk filter (PA) | sporc | once | 1 | M9[C] | 1 (I) | 1 (4b, 4d, 4e) | 3 | 1 | 1 | 1 | 3 | 1 | 3 | 0 | 1 | 0 |
| D | smear (PAL) | spor | once | 1 | M6[D] | 3 (I) | 3 (1/2b, 3b) | 4 | 4 | 4 | 3 | 2 | 1 | 5 | 1 | 0 | 0 |
| A | soft cheese (P) | spor | once | 1 | M8[A] | 7 (II) | 7 (1/2a, 3a) | 5 | 8 | 5 | 7 | 6 | 404 | 1 | 1 | 0 | 0 |
| E | smear (PAL) | spor | once | 1 | M4[E] | 398 (II) | 398 (1/2a, 3a) | 7 | 13 | 19 | 6 | 1 | 7 | 1 | 0 | 1 | 0 |
| A | smear (PAL) | spor | once | 1 | M12[A] | ST529 (III) | 529 (4b, 4d, 4e) | 25 | 73 | 82 | 96 | 45 | 211 | 67 | 0 | 1 | 0 |
| E | acid curd cheese (P)/smear (PAL) | rec b | 2 mo d | 2 | M1[E] | 59 (I) | 59 (1/2b, 3b) | 11 | 1 | 12 | 16 | 3 | 1 | 7 | 0 | 1 | 0 |
| C | swab (FCS) | recc | 1 yr d | 2 | M10[C] | 155 (II) | 155 (1/2a, 3a) | 7 | 10 | 16 | 7 | 5 | 2 | 1 | 1 | 0 | 0 |
| E | smear (PAL) | recc | 7 mo | 2 | M3[E] | 403 (II) | 403 (1/2a, 3a) | 7 | 7 | 10 | 4 | 5 | 24 | 1 | 1 | 0 | 0 |
| A/D | drain water (EL)/smear (PAL) | recc/spor | 11 yr/once | 11/1 | M11[A]=M7[D] | 14 (II) | 14 (1/2a, 3a) | 8 | 6 | 13 | 6 | 5 | 2 | 1 | 1 | 0 | 0 |
| E | acid curd cheese (P)/culture (PA)/environment (NFCS) | rec b | 1 mo | 3 | M2[E] | 121 (II) | 121 (1/2a, 3a) | 7 | 6 | 8 | 8 | 6 | 37 | 1 | 0 | 0 | 1 |
| B/D/E | smear (PAL) | recc/recb/spor | 7 yr/4mo/once | 47/7/1 | M5[B]=M5[D]=M5[E] | 121 (II) | 121 (1/2a, 3a) | 7 | 6 | 8 | 8 | 6 | 37 | 1 | 0 | 0 | 1 |
| L. innocua (API profile 7-5-1-0) | |||||||||||||||||
| C | smear (PAL) | spor | once | 1 | IN10[C] | ST1596 | 1596 | 26 | 21 | 33 | 33 | 48 | 213 | 216 | 0 | 0 | 1 |
| C | cheese (P) | spor | once | 1 | IN18[C] | ST530 | 530 | 28 | 62 | 40 | 97 | 45 | 214 | 53 | 0 | 0 | 1 |
| A | smear (PAL) | spor | once | 1 | IN12[A] | 140 | 637 | 28 | 23 | 33 | 35 | 23 | 192 | 16 | 0 | 0 | 1 |
| C | floor water (NFCS) | spor | once | 1 | IN15[C] | 140 | 637 | 28 | 23 | 33 | 35 | 23 | 192 | 16 | 0 | 0 | 1 |
| E | smear (PAL) | rec b | 4 mo | 2 | IN2[E] | 140 | 637 | 28 | 23 | 33 | 35 | 23 | 192 | 16 | 0 | 0 | 1 |
| E | smear (PAL) | spor | once | 1 | IN4ST[E] | 600 | 603 | 36 | 21 | 40 | 108 | 65 | 243 | 81 | 0 | 0 | 1 |
| C/E | hard cheese (P)/acid curd cheese (P)/smear (PAL) environment (NFCS | spor/recc | once/5.6 yr | 1/12 | IN4[C]=IN4[E] /n. t.e | 600 | 603a | 36 | 21 | 40 | 108 | 65 | 243 | 81 | 0 | 0 | 1 |
| A/C/D/E | grating cheese (P)/smear, brine(PAL)/floor water (EL) | rec c/rec c/rec c/rec c | 1 yr/6.8yr/6.2yr/1yr | 2/22/68/2 | IN5[A]=IN5[C]=IN5[D]=IN5[E]/n. t. | ST1597 | 1597 a | 36 | 23 | 30 | 96 | 195 | 19 | 16 | 0 | 0 | 1 |
| A | smear (PAL) | spor | once | 1 | IN9[A] | 448 | 448 | 65 | 21 | 40 | 33 | 45 | 170 | 53 | 0 | 0 | 1 |
| C | raw milk (RM) | spor | once | 1 | IN14[C] | ST1598 | 1598 | 79 | 21 | 33 | 97 | 20 | 356 | 58 | 0 | 0 | 1 |
| A | smear (PAL) | spor | once | 1 | IN16[A] | ST43 | 43 | 143 | 21 | 40 | 167 | 55 | 307 | 16 | 0 | 0 | 1 |
| C | hard cheese (P) | spor | once | 1 | IN21[C] | ST43 | 43 | 143 | 21 | 40 | 167 | 55 | 307 | 16 | 0 | 0 | 1 |
| A/C/D | soft cheese (P)/smear (PAL) | rec b/spor/rec b | 5 mo/once/4mo | 02.01.2003 | IN6[A]=IN6[C]=IN6[D] | ST1599 | 1599a | 143 | 95 | 30 | 96 | 55 | 180 | 16 | 0 | 0 | 1 |
| L. innocua (API profile 7-1-1-0) | |||||||||||||||||
| E | acid curd cheese (P)/enrichment (PAL)/drain water (EL) | rec c | 6 mo | 3 | IN1[E] | ST1595 | 1595 a | 25 | 73 | 237 | 130 | 55 | 19 | 16 | 0 | 0 | 1 |
| D | semi-hard cheese (P) | spor | once | 1 | IN8[D] | ST1482 | 1482 | 26 | 21 | 40 | 33 | 45 | 19 | 53 | 0 | 0 | 1 |
| C | cheese (P) | spor | once | 1 | IN17[C] | ST605 | 605 | 36 | 21 | 30 | 35 | 45 | 69 | 17 | 0 | 0 | 1 |
| C | cheese (P) | spor | once | 1 | IN20[C] | ST605 | 605 | 36 | 21 | 30 | 35 | 45 | 69 | 17 | 0 | 0 | 1 |
| C | cheese (P) | spor | once | 1 | IN19[C] | ST1087 | 1087 | 191 | 21 | 184 | 110 | 45 | 356 | 16 | 0 | 0 | 1 |
| C | smear (PAL) | rec c | 1.3 yr | 2 | IN7[C] | ST1085 | 1085 a | 188 | 157 | 182 | 223 | 136 | 353 | 148 | 0 | 0 | 1 |
| E | acid curd cheese (P) smear (PAL) | rec c | 6 mo | 4 | IN3[E] | ST1601 | 1601 a | 250 | 140 | 73 | 223 | 136 | 341 | 214 | 0 | 0 | 1 |
| L. innocua (API profile 7-5-3-0) | |||||||||||||||||
| A | smear (PAL) | spor | once | 1 | IN13[A] | ST1600 | 1600 | 40 | 62 | 30 | 33 | 55 | 356 | 17 | 0 | 0 | 1 |
| A | smear (PAL) | spor | once | 1 | IN11[A] | ST1008 | 1008 | 173 | 140 | 173 | 208 | 136 | 341 | 138 | 0 | 0 | 1 |
a sporadically isolated Listeria spp. genotypes; b recurrently isolated Listeria spp. genotypes; c recurrently isolated Listeria spp. genotypes over a period of ≥6 months defined as persistent; d mo, month; yr, year; e non-typable; f CC, clonal complexes; genetic lineages are provided in brackets; g SSI-1+, stress survival islet positive strains; h SSI-1-, stress survival islet negative strains, F2365_0481 homologous gene; i SSI-2, stress survival islet 2 positive genotypes. Abbreviations: Product associated liquids (PAL; smear, brine), product associated samples (PA, culture, and enrichment), raw material (RM), food contact surfaces (FCS), non-food contact (NFCS) surfaces, and environmental (E) liquid samples (floor and drain water). Red marked housekeeping genes are present in L. monocytogenes genetic lineage III and L. innocua.
To evaluate a possible association between the categorical variables, i.e., Listeria species, rhamnose (yes/no), and sporadic/persistence, the association coefficient (Cramer’s V) was calculated (Microsoft Excel 2010, Microsoft Corp., Redmond, WA, USA).
2.2. Molecular Epidemiological Analysis
DNA macrorestriction digest, applying AscI and ApaI of 220 Listeria isolates (139 L. innocua and 81 L. monocytogenes), was performed according to the latest CDC PulseNet International PFGE protocol (https://www.cdc.gov/pulsenet/pdf/listeria-pfge-protocol-508c.pdf). In brief, the cell suspension was standardized to an optical density of 1.0 in sterile TE buffer (10 mM Tris, 1 mM EDTA, pH 8; Sigma-Aldrich Corp, St. Louis, MO, USA) and lysed by adding 20 mg/mL lysozyme (Sigma-Aldrich Corp) to an incubation step at 55 °C for 30 min. After incubation, 20 mg/mL proteinase K (Roche Diagnostics GmbH, Mannheim, Germany) was added to the cell suspension followed by 1% SeaKem Gold agarose (1:1; each 400 µL; Lonza Group, Basel, Switzerland). The suspension was poured into plug molds and the solidified plugs were lysed overnight in 5 mL of cell lysis buffer (50 mM Tris, 50 mM EDTA, pH 8, 1% Sarcosyl + 0.1 mg/mL proteinase K; Sigma-Aldrich Corp). Subsequently, the plugs were washed with sterile water (twice; Mayrhofer Pharmazeutika, Leonding, Austria) and TE buffer (three times). The Listeria DNA macrorestriction digest was performed by applying 50 U each of AscI and ApaI at 37 °C and 30 °C for 4 h (Thermo Fisher Scientific Inc., Waltham, MA, USA). The universal standard Salmonella ser. Braenderup H9812 was digested with 50 U XbaI (Thermo Fisher Scientific Inc.) at 37 °C for 4 h (https://www.cdc.gov/pulsenet/pdf/ecoli-shigella-salmonella-pfge-protocol-508c.pdf). The restriction digested plugs were loaded into 1% SeaKem Gold Agarose gel in 0.5 × Tris borate EDTA buffer (45 mM Tris, 45 mM borate, 1 mM EDTA; Sigma-Aldrich Corp) and electrophorezed for 22.5 h at 6 V/cm with a linear ramping factor and pulse times from 4.0 to 40.0 s at 14 °C and an included angle of 120° (CHEF DR III system; Bio-Rad Laboratories Inc., Hercules, CA, USA).
The gel was stained with ethidium bromide (Sigma-Aldrich Corp) and digitally photographed with Gel Doc 2000 (Bio-Rad Laboratories, Inc.). The TIFF images were normalized with BioNumerics 6.6 software package (Applied Math NV, Sint-Martens-Latem, Belgium) to the universal standard Salmonella ser. Braenderup H9812. Pattern clustering utilized the unweighted pair group method with arithmetic mean (UPGMA) and the dice correlation coefficient with a position tolerance of 1.5%. PFGE types were considered identical when the patterns were indistinguishable. The Simpson’s Index of diversity was calculated with the online tool of Comparing Partitions (http://www.comparingpartitions.info/).
The L. monocytogenes multi-locus sequence typing (MLST) scheme included the following seven housekeeping genes: ABC transporter (acbZ, lmo2752), beta glucosidase (bglA, lmo0319), catalase (cat, lmo2785), succinyl diaminopimelate desuccinylase (dapE, lmo0265), D-amino acid aminotransferase (dat, lmo1617), L-lactate dehydrogenase (ldh, lmo0210), and histidine kinase (lhkA, lmo1508). Protocols for target-specific primers and PCR conditions are provided at https://bigsdb.web.pasteur.fr/listeria/primers_used.html. Target-specific PCR products were sequenced with universal sequencing primers (oF: GTT TTC CCA GTC ACG ACG TTG TA; oR: TTG TGA GCG GAT AAC AAT TTC; LGC Genomics, Berlin, Germany) and allele-specific sequences were submitted to the Institute Pasteur sequence and profile database (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_listeria_seqdef). The sequence types (ST) were determined by the combination of the seven housekeeping loci. The STs were compared to the Institute Pasteur isolate database (https://bigsdb.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_listeria_isolates_public) to estimate their global presence and potential niche attribution.
2.3. Screening for Stress Survival Islets (SSI-1 and SSI-2)
L. innocua and L. monocytogenes isolates were screened for the presence of SSI-1+ (9.7 kbp fragment), SSI-1-(F2365_0481 homologous gene; 1.1 kbp fragment) and SSI-2 (2.2 kbp fragment). PCR primers targeting the L. monocytogenes flanking genes lmo0443 and lmo0449 were used according to Ryan et al. [26]. The homologous genes related to L. innocua lin0464 and lin0465 (2.2 kbp fragment) were investigated according to Hein et al. [27]. PCR reactions contained 0.2 μM each primer, 2 mM MgCl2, 1 mM deoxynucleoside triphosphates (dNTPs; Thermo Fisher Scientific), 1 U Platinum Taq DNA polymerase (Thermo Fisher Scientific), 10× PCR buffer, diethyl pyrocarbonate (DEPC)-treated water (Thermo Fisher Scientific), and 1 μL DNA template in a final volume of 25 μL.
The PCR reaction for the detection of SSI-1+ (9.7 kbp) and SSI-1+ (1.1 kbp) differed from the latter mix by the following components: 2.5 U long range DNA polymerase and 2 μL DNA template in a final volume of 25 μL. The gel electrophoresis of PCR-reactions was determined in a 1.5% agarose gel containing 0.5 × Tris–borate–EDTA (TBE) buffer and 3.5 μL peqGREEN DNA gel stain (VWR International, Radnor, PA, USA). The DNA standard Thermo Scientific™ GeneRuler™ 100 bp and 1 kb plus (Thermo Fisher Scientific Inc.) were applied for fragment length comparison.
2.4. L. Monocytogenes and L. Innocua Minimum Inhibitory Concentration (MIC) towards Biocides
The minimum inhibitory concentrations (MIC) of five disinfectant compounds (peracetic acid, benzalkonium chloride, sodium hypochlorite, hydrogen peroxide, and isopropanol; all supplied by Sigma AldrichCorp) were determined for 10 recurrent L. monocytogenes and L. innocua genotypes: M1[E] (ST59, 1/2b); M5[B]=M5[D]=M5[E] (ST121, 1/2a); M10[C] (ST155, 1/2a); M11[A]=M7[D] (ST14, 1/2a); IN1[E] (ST1595); IN2[E] (ST637); IN3[E] (ST1601); IN4[C]=IN4 [E] (ST603); IN5[A]=IN5[C]=IN5[D]=IN5[E] (ST1597); IN7[C] (ST1085).
The disinfectant components were tested at concentration ranges of 31.3–1000 mg/L for peracetic acid and hydrogen peroxide, 0.5–1000 mg/L for benzalkonium chloride and 125–10,000 mg/L for sodium hypochlorite. An agar dilution method was performed in duplicates to determine the minimal inhibitory concentration (MIC) of the disinfectants against L. innocua and L. monocytogenes strains. As previously described, 5 µL of bacterial culture was spotted onto Mueller–Hinton agar (Oxoid, Basingstoke, UK) containing the disinfectants to be tested [28,29]. Plates were incubated at 37 °C for 24 to 48 h. Following incubation, the lowest disinfectant concentration that showed no bacterial growth was recorded as MIC. Mean MIC values were calculated using Excel (Microsoft Corporation, Redmond, WA, USA).
3. Results
3.1. Isolate Characteristics
The isolate set comprised 139 L. innocua and 81 L. monocytogenes PCR-confirmed isolates targeting the iap (invasion associated protein p60) gene [24]. Listeria spp. originated from the following Austrian cheese processing facilities: A (L. innocua/L. monocytogenes: (n = 9/13), B (n = 0/47), C (n = 34/3), D (n = 72/9), and E (n = 24/9). The Listeria isolate collection was established over a monitoring period of 23 years (1987–2010). Listeria spp. were isolated from cheese samples (24 L. innocua and 3 L. monocytogenes), product associated samples (PA, PAL; 100 L. innocua and 64 L. monocytogenes), production environment (FCS, NFCS, environmental liquids (EL); 14 L. innocua and 14 L. monocytogenes), and one L. innocua isolate from raw milk (RM) (Table 1).
The biochemical profiling resulted for all L. monocytogenes isolates in the typical naphthylamidase (DIM; Differentiation/Innocua/Monocytogenes) negative and rhamnose positive profile (API profile 6510). L. innocua isolates exhibited three different biochemical profiles: API profile 7510 with a DIM and rhamnose positive reaction (n = 124), API profile 7110 with a DIM positive and rhamnose negative reaction (n = 13), and API profile 7531 with a DIM, D-ribose, and D-tagatose positive reaction (n = 2) (Table 1).
L. monocytogenes isolates (n = 81) were most frequently confirmed as PCR-serogroups 1/2a, 3a (n = 76; 95%), followed by 1/2b, 3b (n = 3; 3.75%), and 4b, 4d, 4e (n = 2; 1.25%).
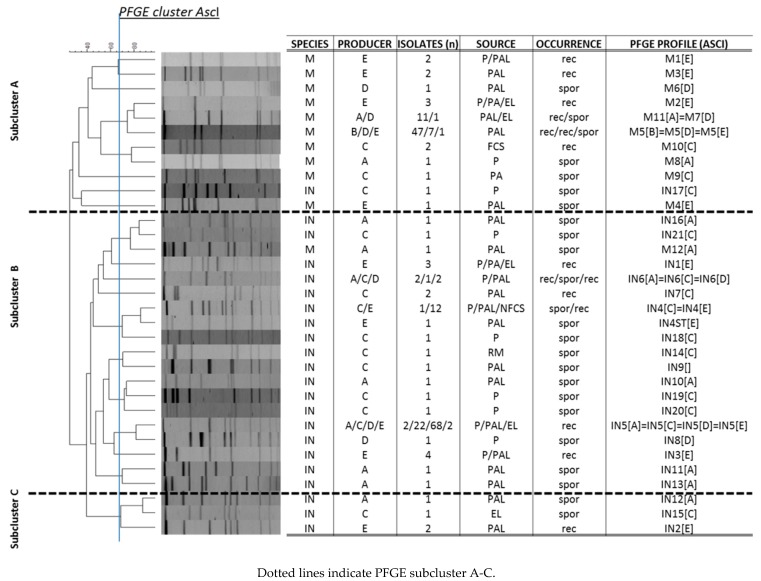
The PFGE-typing, applying the restriction enzyme AscI, revealed 33 distinct Listeria PFGE profiles with a Simpson’s Index of Diversity of 0.75. Thereof, 11 and 22 AscI were specific for L. monocytogenes and L. innocua, respectively (Simpson´s Index 0.519 and 0.533). The ApaI macrorestriction digest resulted in fewer PFGE profiles for L. innocua (n = 20). L. innocua PFGE profiles IN5[A]=IN5[C]=IN5[D]=IN5[E] and IN4[C]=IN4[E] were non-typable by ApaI.
PFGE-types with identical AscI profiles assigned to dairy producers A-E and isolated during two or more sampling events were classified as recurrent, whereas Listeria isolates with unique AscI profiles and isolated once were defined as sporadic genotypes. Listeria spp. genotypes recurrently isolated over a period of six or more months were defined as persistent. Generally, L. monocytogenes clustered together in a distinct subcluster A and could be clearly distinguished at a similarity level of 25% from L. innocua subcluster B and C (similarity level 40%) (Table 1, Figure 1).
Figure 1.
PFGE cluster analysis (AscI) of L. innocua (n = 139) and L. monocytogenes (n = 81) isolates included in this study.
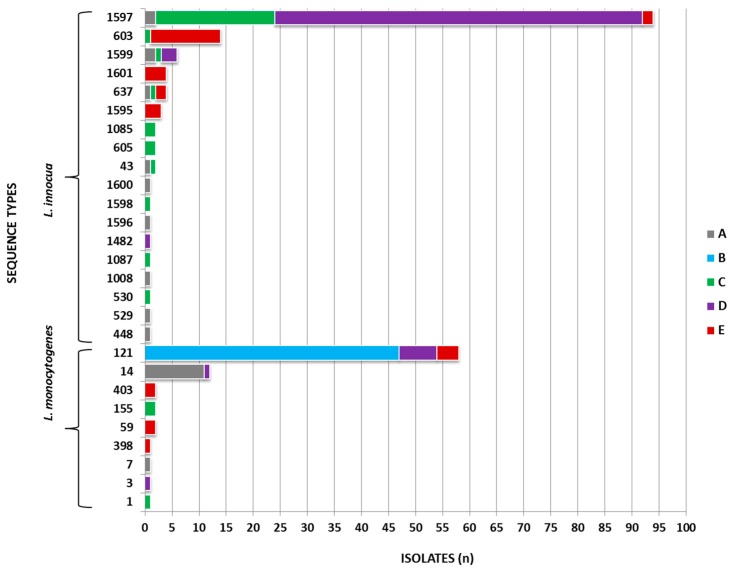
The MLST typing resulted in 27 STs (Simpson´s Index of Diversity of 0.742). In total, 9 and 3 clonal complexes (CCs) and 1 and 14 singletons were identified among L. monocytogenes and L. inncoua isolates, respectively. The discriminatory power of MLST analysis was comparable to PFGE-typing for L. monocytogenes (10 STs; Simpson´s Index 0.469). ST121 could be differentiated by applying PFGE typing into two L. monocytogenes distinct fingerprints. MLST analysis for L. innocua isolates was less discriminative in comparison to PFGE (AscI), resulting in 17 STs (Simpson´s Index 0.531).
Seven new L. innocua STs (ST1595 to ST1601) were defined by submitting the sequences to the Institute Pasteur MLST database (https://bigsdb.web.pasteur.fr/listeria/listeria.html; Table 1). The ST attribution to dairy processing facilities A–E is depicted in Figure 2.
Figure 2.
Comparison of the L. innocua and L. monocytogenes STs identified in this study to the Institute Pasteur MLST isolate database (https://bigsdb.pasteur.fr/listeria/listeria.html) was performed to identify highly abundant housekeeping genes, CC and STs (Table S1). Interestingly, some housekeeping genes were not specific for L. monocytogenes genetic lineage III or L. innocua.
The stress survival islet (SSI-1) inserted into intergenic region lmo0443 to lmo0449 in L. monocytogenes was present in ST3 (genetic lineage I; PCR serogroups 1/2b, 3b), ST7, ST14, ST155, and ST403 (genetic lineage II; PCR serogroup 1/2a, 3a). The L. monocytogenes homologous gene to F2365_0481 (1.1 kbp) was present in genetic lineage I isolates assigned to ST1 (serogroup 4b, 4d, 4e) and ST59 (1/2b, 3b), ST398 (genetic lineage II (PCR-serogroup 1/2a, 3a), and ST529 (genetic lineage III, PCR-serogroup 4b, 4d, 4e). L. monocytogenes ST121 and all L. innocua isolates harbored the SSI-2 (2.2 kbp fragment; Table 1).
3.2. Molecular Epidemiological Interpretation
The majority of L. innocua and L. monocytogenes PFGE types were isolated once (n = 20/33; 60.61%), but certain genotypes were recurrently isolated from process associated samples and cheese for a short period of time (n = 6/33; 18.18%). These genotypes were present in PAL (smear) and environmental samples and after contamination events in cheese for between one and six months and were successfully eliminated. Other Listeria spp. PFGE types were persistent in the dairy processing environment for a long period and somehow adapted to niches (smear, brine, drain water). The latter L. monocytogenes and L. innocua PFGE types cross-contaminated the surface of cheeses (e.g., hard cheese). Almost all of the persistent L. innocua and L. monocytogenes isolates were rhamnose positive (API profiles 7510 and 6510), except for one genotype with a rhamnose negative profile (IN7[C], ST1085). Almost all of the persistent L. innocua and L. monocytogenes isolates were rhamnose positive (API profiles 7510 and 6510) except for one genotype with a rhamnose negative profile (IN7[C], ST1085). The association coefficient Cramer’s V showed a weak association between L. innocua, L. monocytogenes, rhamnose positive, rhamnose negative sporadic and persistent occurrences, although the result “for persistence and rhamnose positive” was highly significant (p = 0.0065; Cramer’s V rV = WERT, p < 0.01).
The most abundant L. monocytogenes genotypes related to persistence in cheese processing associated samples and environments were PFGE profiles M5[B]=M5[D]=M5[E] (ST121, 1/2a, 3a) and M11[A]=M7[D] (ST14, 1/2a, 3a), which were present for 7 and 11 years at dairy processing facility B and A in smear and drain water, respectively. The latter profiles were also isolated once and after four months at producers D and E. PFGE profiles M10[C] (ST155, 1/2a, 3a), M3[E] (ST403, 1/2a, 3a) were present at producers C and E for one year and seven months. M1 (ST59, 1/2b, 3b) and M2 (ST121, 1/2a, 3a) were recurrently isolated during a shorter timeframe (one and two months), both at producer E. Other sporadically isolated L. monocytogenes genotypes (ST1, ST3, ST7, and ST398) were isolated once during the monitoring period.
The most common L. innocua genotypes were IN5[A]=IN5[C]=IN5[D]=IN5[E] (newly identified ST1597), IN4[C]=IN4[E] (ST603), and IN7[C] (ST1085), which were repeatedly isolated for a year up to 6.8 years in the same dairy processing environment (floor water) and product-associated liquids (smear, brine). IN1[E] (newly identified ST1595) and IN 3[E] (newly identified ST1601) were recurrently isolated during a six-month period at processing facility E.
IN6[A]=IN6[C]=IN6[D] was present during a short contamination event (four and five months) at processing facilities A and D. The L. monocytogenes and L. innocua PFGE clusters were heterogeneous (25% similarity). Interestingly, the persistence-related genotypes L. monocytogenes ST14 (M11[A]=M7[D]), ST155 (M10[C]), and ST121 (M5[B]=M5[D]=M5[E]) clustered together in subcluster A at a 75% and 80% similarity level. L. innocua ST43 (IN16[A], IN21[C]), ST603 (IN4ST[E], IN4[C]=IN4[E]), and ST637 (IN12[A], IN15[C]) related PFGE profiles clustered in subcluster B and C at a similarity level ≥80%.
L. innocua ST1597, ST603 and L. monocytogenes ST121 and ST14 were the most abundant genotypes in dairy producing facilities A–E over time (n = 178/220 isolates). The highest genotype diversity was identified in dairy producing facilities A, C, and E (n = 11, 12, and 9 different genotypes; Figure 2).
The following housekeeping genes were common in Listeria isolates included in this study and L. monocytogenes genetic lineage III: ST529, ST1595 (abcZ 25, bglA 73), ST529, ST1597 and ST1599 (dapE 96), ST529, ST530, ST448, ST1482, ST605, and ST1087 (dat 45) (Table 1). Furthermore, the housekeeping gene abcz 40 in L. innocua ST1600 was identified in the Institute Pasteur MLST database to be more related to L. monocytogenes genetic lineage III (ST267). Further details concerning the prevalence of housekeeping genes present in the MLST database are provided in Table S1.
3.3. Susceptibility to Biocides
The MIC towards biocides was determined for four recurrent L. monocytogenes and six L. innocua genotypes. All test strains except L. innocua genotype IN5[A]=IN5[C]=IN5[D]=IN5[E] (ST1597) and IN3[E] (ST1601), both 141 mg/L, were adapted to higher concentrations of peracetic acid (250 mg/L; 1.7 fold higher). M5[B]=M5[D]=M5[E] (ST121, 1/2a), IN5[A]=IN5[C]=IN5[D]=IN5[E] (ST1597), and IN2[E] (ST637) were better adapted to benzalkonium chloride (1.3- and 2-fold higher; mean MIC 15.6 mg/L in contrast to 11.7 and 7.8 mg/L). M5[B]=M5[D]=M5[E] (ST121, 1/2a) and M1[E] (ST59, 1/2b) were slightly better adapted to hydrogen peroxide (1.5-fold higher; 188 mg/L in contrast to 125 mg/L). IN1[E] (ST1595) and IN2[E] (ST637) were better adapted to sodium hypochlorite (2.7–5.7-fold higher; 10,000 mg in contrast to 1750–3750 mg/L).
4. Discussion
Dairy and cheese processing environments are frequently colonized by Listeria spp., including pathogenic L. monocytogenes. Even newly established dairy processing facilities become colonized after a short period of time [29,30].
Generally, prevalence and concentrations of L. monocytogenes in cheeses and cheese processing environments are low. Its growth is supported by the presence of fresh, ripened, veined, and smear cheeses (0.8%–5.1% prevalence in cheese lots). Brined cheeses are most often contaminated by L. monocytogenes (11.8%), according to a meta-analysis-based literature review [31]. This suggests that product-associated liquids (smear, brine) contribute to L. monocytogenes contamination of cheese lots [21,32]. In our study, smear and brine samples were indeed most often associated with L. monocytogenes and L. innocua surface contamination and supported the persistence of certain genotypes (ST121, ST14, ST603, and ST1597) in the dairy environment.
Floor drains are further niches for efficient Listeria spp. colonization of the FPE and hot-spots for cross and recontamination events [33,34]. These niches were also identified in our study. To a certain extent, drain waters harbored recurrent and persistent Listeria spp. (ST14, ST637, ST1595, ST1597; Table 1). Despite having a cleaning potential, the introduction of high-pressure water from hoses into a contaminated drain can cause the airborne spread of Listeria and further contribute to the successful establishment of persistent Listeria spp. strains in a facility [35].
As reports about dairy processing environment contamination scenarios in the literature are sparse, our goal was to identify any potential long-term contamination with certain L. innocua and L. monocytogenes genotypes.
Some studies indeed allude to wider contamination of dairy facilities. Parisi et al. [36] isolated Listeria spp. at 19/34 cheese factories (55.8%). Occasionally, L. innocua and L. monocytogenes were detected at the same sampling site (2/19 plants) and persisted in floor drains, which were identified as ideal sampling sites to be included in a monitoring system.
Relevantly, we clearly identified a higher fluctuation of L. innocua and L. monocytogenes genotypes in parallel in 4/5 dairy processing facilities (Figure 1, Table 1). One dairy plant (B) harbored a persistent L. monocytogenes genotype (ST121) for seven years without further introduction of other Listeria spp. genotypes.
Lomonaco et al. [37] reported two persistent L. monocytogenes genotypes in the Gorgonzola processing chain. About 88% of the L. monocytogenes strains were serotype 1/2a, which is consistent with our findings (95% of the isolates were serotype 1/2a). However, genotypes were not comparable due to the lack of common nomenclature in Listeria spp. subtyping, which is urgently in need of rectification [38]. In this respect, Jagadeesan et al. [39] highlighted the need to include metadata for genotypic approaches, which should be sufficiently cleaned with the removal of replicates and unintended information. Actual studies indicate that whole-genome sequencing (WGS) and core genome (cg) MLST approaches already contribute to the real-time exchange of information on the emergence and geographic dispersal of clones [40,41,42]. Maury et al. [43] reported that L. monocytogenes CC1 are strongly associated with dairy products, whereas hypovirulent clones, CC9 and CC121, are related to meat products.
In the presented study, several disease-related genotypes that are globally distributed were introduced into the dairy processing environment (ST1, ST3, ST7, ST59, ST155, ST398, and ST403). However, ST14 and ST121 established themselves for a longer time (7 and 11 years, respectively) in the FPE and tended to be persistent. Almost all of the persistent L. innocua and L. monocytogenes isolates that we identified were rhamnose-positive (API profiles 7510 and 6510; Table 1). Rhamnose is a naturally occurring monosaccharide present in plant material and important for saprophytes such as Listeria spp.
Atypical L. innocua and L. monocytogenes lacking the ability to ferment rhamnose (where the pdu operon for propanediol utilization is missing) are potentially less capable of exploiting nutritional sources important for adaptation to the FPE [44].
Further, atypical strains with deficient rhamnose fermentation have been reported to be attenuated in virulence and have reduced resistance to temperature changes [45]. In contrast, L. monocytogenes serotype 1/2a mutants confer phage-resistance due to a loss of rhamnose. This is important when there is a field application of lytic phage cocktails as biocontrol measures [46]. Therefore, detailed characterizations of L. monocytogenes and L. innocua rhamnose-positive and -negative field strains in respect of persistence in the FPE and resistance to biocontrol measures are important.
Pasquali et al. [47] also identified ST14 and ST121 as persisters in a rabbit meat processing plant. Several strain-specific features, such as a stronger biofilm-forming potential (ST14) and the presence of the qacH gene associated with adaptation to BAC in ST121, contributed to successful establishment in the FPE.
The stress survival islet (SSI-1) inserted into the intergenic region lmo0443 to lmo0449 in L. monocytogenes is present in long-term persister ST14 and is related to acid tolerance [26]. L. monocytogenes ST121 and all L. innocua isolates that we identified harbored the SSI-2 (2.2 kbp fragment), which is related to elevated tolerance to oxidative and alkaline stress (Table 1) [48].
The entrance and establishment of certain successful genotypes in the FPE are influenced by several factors. These include strain properties (e.g., prophage diversification, transposons, plasmids), the possibility of reintroduced genotypes by raw material, over-diluted biocides on wet surfaces, and a permanent change in cleaning regimes by external cleaning companies and consultants [49]. Consequentially, Muhterem-Uyar et al. (2018) reported that in a heavily contaminated cheese processing environment, the strain variability (ST1, ST7, ST21, and ST37) was reduced to a persistent genotype (ST5) harboring a plasmid type (plM80 related), which is present in successful clones worldwide [20].
In corroboration, we observed higher MICs towards biocides in M5[B]=M5[D]=M5[E] (ST121, 1/2a), IN5[A]=IN5[C]=IN5[D]=IN5[E] (ST1597), and IN2[E] (ST637), which were better adapted to benzalkonium chloride (up to two-fold higher). IN1[E] (ST1595) and IN2[E] (ST637) were better adapted to sodium hypochlorite (2.7–5.7-fold higher).
In FPEs, resident Listeria are frequently exposed to sublethal concentrations of biocides due to the dilution effect of wet surfaces and the presence of food soil [50]. Therefore, testing the sensitivity of Listeria spp. to sublethal concentrations of biocides should be performed routinely to identify potential strain adaptations. Particularly, L. monocytogenes and L. innocua isolated from the pork processing chain have been shown to harbor efflux pumps and resistance genes (cadA1-cadA4, arsA1, arsA2) that confer resistance to benzalkonium chloride and heavy metals [51].
The exchange of genetic material between L. innocua and L. monocytogenes has been observed in a few studies. The possibility of horizontal gene transfer (HGT) of plasmids, including heavy metal resistance, enhanced tolerance to QACs and DNA intercalating dyes between L. welshimeri, L. innocua and L. monocytogenes, has been described by sequence analysis in experimental settings and by comparison of FPE isolates [52,53,54].
The presence of atypical hemolytic L. innocua strains in the food chain might also have been introduced by HGT, and this constitutes a reservoir of virulence genes transferable to other species [12,55]. This could be the same for genetic exchange between L. innocua and L. monocytogenes related to environmental adaptation. More research focusing on the uptake of genetic material by Listeria spp. in the FPE is warranted.
We identified housekeeping genes common in novel identified L. innocua ST1595, ST1597, ST1599, and ST1601 in STs 605 and ST1085 that are recorded in the MLST database, and L. monocytogenes genetic lineage III (abcZ 25, bglA 73, dapE 96, dat 45) (Table 1, Table S1).
Comparing L. innocua STs from this study to the MLST Institute Pasteur database, we identified ST603 (CC600) and ST637 (CC140) (Figure 2), commonly isolated from different niches (e.g., silage, food, human blood samples). Comparing the NCBI available strains (n = 11) to L. innocua from this study, no complete match was found based on STs. The nearest match was L. innocua reference strain CLIP 11,262 assigned to CC140, which differed by the ldh housekeeping gene (ldh 74) in comparison to ST637 (ldh 192) (https://www.ncbi.nlm.nih.gov/genome/genomes/1024?; https://bigsdb.web.pasteur.fr/cgi-bin/bigsdb/bigsdb.pl?db=pubmlst_listeria_isolates).
In fact, the draft genome of Listeria innocua UAM003-1A, available from NCBI, is also related to highly abundant CC140 [14].
We did not identify any atypical hemolytic ST188 or ST437 L. innocua strains (LIP-1 positive, hly positive), which have recently been isolated from wild bird feces in Finland [12] and previously described by Volokhov [56]. The L. innocua strains included in our study showed no relationship to the newly announced MEZLIS26 genome, due to their different housekeeping genes. The latter L. innocua was assigned to the highly abundant CC537 [15].
L. innocua has been reported to be more commonly found in the FPE than L. monocytogenes [57], which is supported by our findings. We fully agree with Jemmi and Stephan [58] who suggest that L. innocua is a good hygiene indicator and also a marker for unrecognized L. monocytogenes contamination events in the FPE. What should now be considered in retrospect is the demanding nature of the microbiological reference method, for example, ISO 11290-1, concerning the differentiation of L. innocua and L. monocytogenes [59]. Reports focusing on the challenges associated with Listeria detection and confirmation are available, including atypical strains or L. monocytogenes present in lower concentrations due to competitive L. innocua strains during enrichment or that mask detection of L. monocytogenes on selective agar plates such as ALOA medium [60,61] (https://www.fda.gov/food/laboratory-methods-food/guidelines-bam-users-identification-atypical-hemolytic-listeria-isolates). This might also have contributed to a higher isolation rate of L. innocua compared to L. monocytogenes in our study.
In respect of long-term contamination events with L. monocytogenes and other Listeria species, subtyping methods are helpful tools to identify the true nature of persister candidates.
5. Conclusions
L. monocytogenes is a foodborne pathogen of significance to human health, and it is able to co-survive in the dairy FPE in microbial communities with other Listeria species and with other bacteria (e.g., Proteobacteria, lactic acid bacteria) [33,62]. Our study identified for the first time the recurrent isolation and persistence of L. innocua in L. monocytogenes-colonized habitats. Novel local L. innocua sequence types (ST1595 to ST1601) were identified, which shared, to a certain extent, the housekeeping genes that are also common in L. monocytogenes genetic lineage III. Either SSI-1 (ST14) or SSI-2 (ST121, all L. innocua) were present in strains successfully adapted to the FPE. There is a great need for further insight into the processes of FPE adaptation and exchange of genetic information between Listeria species so that appropriate food safety control measures can be designed.
Acknowledgments
We thank the team of curators of the Institute Pasteur MLST and genome databases for curating the data and making them publicly available at https://bigsdb.pasteur.fr. We thank Cameron McCulloch for assistance with the manuscript. Open Access Funding by the University of Veterinary Medicine Vienna.
Supplementary Materials
The following are available online at https://www.mdpi.com/2076-2607/8/2/234/s1.
Author Contributions
B.S., M.W. conceived and designed the experiments. I.K.-R., A.M. and S.M.-K. performed the experiments. I.K.-R., K.B. and B.S. analyzed the data. B.S. and I.K.-R. Drafted the manuscript. K.-H.W. reviewed the manuscript. All authors read and approved the final version of the manuscript.
Conflicts of Interest
The authors declare no conflict of interests.
References
- 1.Linke K., Rückerl I., Brugger K., Karpiskova R., Walland J., Muri-Klinger S., Tichy A., Wagner M., Stessl B. Reservoirs of Listeria species in three environmental ecosystems. Appl. Environ. Microbiol. 2014 doi: 10.1128/AEM.01018-14. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Vivant A.L., Garmyn D., Piveteau P. Listeria monocytogenes, a down-to-earth pathogen. Front. Cell. Infect. Microbiol. 2013 doi: 10.3389/fcimb.2013.00087. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Orsi R.H., Wiedmann M. Characteristics and distribution of Listeria spp., including Listeria species newly described since 2009. Appl. Microbiol. Biotechnol. 2016 doi: 10.1007/s00253-016-7552-2. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Lüth S., Boone I., Kleta S., Al Dahouk S. Analysis of RASFF notifications on food products contaminated with Listeria monocytogenes reveals options for improvement in the rapid alert system for food and feed. Food Control. 2019 doi: 10.1016/j.foodcont.2018.09.033. [DOI] [Google Scholar]
- 5.Ricci A., Allende A., Bolton D., Chemaly M., Davies R., Fernández Escámez P.S., Girones R., Herman L., Koutsoumanis K., Nørrung B., et al. Listeria monocytogenes contamination of ready-to-eat foods and the risk for human health in the EU. EFSA J. 2018 doi: 10.2903/j.efsa.2018.5134. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6.Buchanan R.L., Gorris L.G.M., Hayman M.M., Jackson T.C., Whiting R.C. A review of Listeria monocytogenes: An update on outbreaks, virulence, dose-response, ecology, and risk assessments. Food Control. 2017 doi: 10.1016/j.foodcont.2016.12.016. [DOI] [Google Scholar]
- 7.NicAogáin K., O’Byrne C.P. The role of stress and stress adaptations in determining the fate of the bacterial pathogen Listeria monocytogenes in the food chain. Front. Microbiol. 2016 doi: 10.3389/fmicb.2016.01865. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Bierne H., Milohanic E., Kortebi M. To be cytosolic or vacuolar: The double life of Listeria monocytogenes. Front. Cell. Infect. Microbiol. 2018 doi: 10.3389/fcimb.2018.00136. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Maury M.M., Tsai Y.H., Charlier C., Touchon M., Chenal-Francisque V., Leclercq A., Criscuolo A., Gaultier C., Roussel S., Brisabois A., et al. Uncovering Listeria monocytogenes hypervirulence by harnessing its biodiversity. Nat. Genet. 2016 doi: 10.1038/ng.3501. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Milillo S.R., Friedly E.C., Saldivar J.C., Muthaiyan A., O’Bryan C., Crandall P.G., Johnson M.G., Ricke S.C. A Review of the Ecology, Genomics, and Stress Response of Listeria innocua and Listeria monocytogenes. Crit. Rev. Food Sci. Nutr. 2012 doi: 10.1080/10408398.2010.507909. [DOI] [PubMed] [Google Scholar]
- 11.Schmid M.W., Ng E.Y.W., Lampidis R., Emmerth M., Walcher M., Kreft J., Goebel W., Wagner M., Schleifer K.H. Evolutionary history of the genus Listeria and its virulence genes. Syst. Appl. Microbiol. 2005 doi: 10.1016/j.syapm.2004.09.005. [DOI] [PubMed] [Google Scholar]
- 12.Moura A., Disson O., Lavina M., Thouvenot P., Huang L., Leclercq A., Fredriksson-Ahomaa M., Eshwar A.K., Stephan R., Lecuit M. Atypical hemolytic Listeria innocua isolates are virulent, albeit less than Listeria monocytogenes. Infect. Immun. 2019 doi: 10.1128/IAI.00758-18. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Chen J., Chen Q., Jiang L., Cheng C., Bai F., Wang J., Mo F., Fang W. Internalin profiling and multilocus sequence typing suggest four Listeria innocua subgroups with different evolutionary distances from Listeria monocytogenes. BMC Microbiol. 2010 doi: 10.1186/1471-2180-10-97. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Parsons C., Chen Y., Niedermeyer J., Hernandez K., Kathariou S. Draft Genome Sequence of Multidrug-Resistant Listeria innocua Strain UAM003-1A, Isolated from a Wild Black Bear (Ursus americanus) Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.01281-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.El Zowalaty M.E., Hickman R.A., Moura A., Lecuit M., Zishiri O.T., Noyes N., Järhult J.D. Genome Sequence of Listeria innocua Strain MEZLIS26, Isolated from a Goat in South Africa. Microbiol. Resour. Announc. 2019;8 doi: 10.1128/MRA.00991-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Roche S.M., Grépinet O., Kerouanton A., Ragon M., Leclercq A., Témoin S., Schaeffer B., Skorski G., Mereghetti L., Le Monnier A., et al. Polyphasic characterization and genetic relatedness of low-virulence and virulent Listeria monocytogenes isolates. BMC Microbiol. 2012 doi: 10.1186/1471-2180-12-304. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Martín B., Perich A., Gómez D., Yangüela J., Rodríguez A., Garriga M., Aymerich T. Diversity and distribution of Listeria monocytogenes in meat processing plants. Food Microbiol. 2014 doi: 10.1016/j.fm.2014.05.014. [DOI] [PubMed] [Google Scholar]
- 18.Schmitz-Esser S., Müller A., Stessl B., Wagner M. Genomes of sequence type 121 Listeria monocytogenes strains harbor highly conserved plasmids and prophages. Front. Microbiol. 2015 doi: 10.3389/fmicb.2015.00380. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Knudsen G.M., Nielsen J.B., Marvig R.L., Ng Y., Worning P., Westh H., Gram L. Genome-wide-analyses of Listeria monocytogenes from food-processing plants reveal clonal diversity and date the emergence of persisting sequence types. Environ. Microbiol. Rep. 2017 doi: 10.1111/1758-2229.12552. [DOI] [PubMed] [Google Scholar]
- 20.Muhterem-Uyar M., Ciolacu L., Wagner K.H., Wagner M., Schmitz-Esser S., Stessl B. New aspects on Listeria monocytogenes ST5-ECVI predominance in a heavily contaminated cheese processing environment. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.00064. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Asperger H., Wagner M., Brandl E. An approach towards public health and foodborne human listeriosis —The Austrian Listeria monitoring. Berl. Munch. Tierarztl. Wochenschr. 2001 doi: 10.1016/J.FOODCONT.2007.04.009. [DOI] [PubMed] [Google Scholar]
- 22.Wagner M., Stessl B. Sampling the Food Processing Environment: Taking Up the Cudgel for Preventive Quality Management in Food Processing Environments. Methods Mol. Biol. 2014;1157:275–283. doi: 10.1007/978-1-4939-0703-8_23. [DOI] [PubMed] [Google Scholar]
- 23.Walsh P.S., Metzger D.A., Higushi R. Chelex 100 as a medium for simple extraction of DNA for PCR-based typing from forensic material. Biotechniques. 2013 doi: 10.2144/000114018. [DOI] [PubMed] [Google Scholar]
- 24.Bubert A., Hein I., Rauch M., Lehner A., Yoon B., Goebel W., Wagner M. Detection and differentiation of Listeria spp. by a single reaction based on multiplex PCR. Appl. Environ. Microbiol. 1999;65:4688–4692. doi: 10.1128/AEM.65.10.4688-4692.1999. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Leclercq A., Chenal-Francisque V., Dieye H., Cantinelli T., Drali R., Brisse S., Lecuit M. Characterization of the novel Listeria monocytogenes PCR serogrouping profile IVb-v1. Int. J. Food Microbiol. 2011 doi: 10.1016/j.ijfoodmicro.2011.03.010. [DOI] [PubMed] [Google Scholar]
- 26.Ryan S., Begley M., Hill C., Gahan C.G.M. A five-gene stress survival islet (SSI-1) that contributes to the growth of Listeria monocytogenes in suboptimal conditions. J. Appl. Microbiol. 2010 doi: 10.1111/j.1365-2672.2010.04726.x. [DOI] [PubMed] [Google Scholar]
- 27.Hein I., Klinger S., Dooms M., Flekna G., Stessl B., Leclercq A., Hill C., Allerberger F., Wagner M. Stress survival islet 1 (SSI-1) survey in Listeria monocytogenes reveals an insert common to Listeria innocua in sequence type 121 L. monocytogenes strains. Appl. Environ. Microbiol. 2011 doi: 10.1128/AEM.02159-10. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Mereghetti L., Quentin R., Marquet-Van Der Mee N., Audurier A. Low sensitivity of Listeria monocytogenes to quaternary ammonium compounds. Appl. Environ. Microbiol. 2000 doi: 10.1128/AEM.66.11.5083-5086.2000. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Rückerl I., Muhterem-Uyar M., Muri-Klinger S., Wagner K.H., Wagner M., Stessl B.L. Monocytogenes in a cheese processing facility: Learning from contamination scenarios over three years of sampling. Int. J. Food Microbiol. 2014 doi: 10.1016/j.ijfoodmicro.2014.08.001. [DOI] [PubMed] [Google Scholar]
- 30.Melero B., Stessl B., Manso B., Wagner M., Esteban-Carbonero Ó.J., Hernández M., Rovira J., Rodriguez-Lázaro D. Listeria monocytogenes colonization in a newly established dairy processing facility. Int. J. Food Microbiol. 2019 doi: 10.1016/j.ijfoodmicro.2018.09.003. [DOI] [PubMed] [Google Scholar]
- 31.Martinez-Rios V., Dalgaard P. Prevalence of Listeria monocytogenes in European cheeses: A systematic review and meta-analysis. Food Control. 2018 doi: 10.1016/j.foodcont.2017.07.020. [DOI] [Google Scholar]
- 32.Wagner M., Skandamis P., Allerberger F., Schoder D., Lassnig C., Müller M., Rychli K. The impact of shelf life on exposure as revealed from quality control data associated with the quargel outbreak. Int. J. Food Microbiol. 2018 doi: 10.1016/j.ijfoodmicro.2018.04.031. [DOI] [PubMed] [Google Scholar]
- 33.Dzieciol M., Schornsteiner E., Muhterem-Uyar M., Stessl B., Wagner M., Schmitz-Esser S. Bacterial diversity of floor drain biofilms and drain waters in a Listeria monocytogenes contaminated food processing environment. Int. J. Food Microbiol. 2016 doi: 10.1016/j.ijfoodmicro.2016.02.004. [DOI] [PubMed] [Google Scholar]
- 34.Stessl B., Szakmary-Brändle K., Vorberg U., Schoder D., Wagner M. Temporal analysis of the Listeria monocytogenes population structure in floor drains during reconstruction and expansion of a meat processing plant. Int. J. Food Microbiol. 2019 doi: 10.1016/j.ijfoodmicro.2019.108360. [DOI] [PubMed] [Google Scholar]
- 35.Berrang M.E., Frank J.F. Generation of airborne Listeria innocua from model floor drains. J. Food Prot. 2012 doi: 10.4315/0362-028X.JFP-12-021. [DOI] [PubMed] [Google Scholar]
- 36.Parisi A., Latorre L., Fraccalvieri R., Miccolupo A., Normanno G., Caruso M., Santagada G. Occurrence of Listeria spp. in dairy plants in Southern Italy and molecular subtyping of isolates using AFLP. Food Control. 2013 doi: 10.1016/j.foodcont.2012.05.036. [DOI] [Google Scholar]
- 37.Lomonaco S., Decastelli L., Nucera D., Gallina S., Manila Bianchi D., Civera T. Listeria monocytogenes in Gorgonzola: Subtypes, diversity and persistence over time. Int. J. Food Microbiol. 2009 doi: 10.1016/j.ijfoodmicro.2008.10.009. [DOI] [PubMed] [Google Scholar]
- 38.Moura A., Criscuolo A., Pouseele H., Maury M.M., Leclercq A., Tarr C., Björkman J.T., Dallman T., Reimer A., Enouf V., et al. Whole genome-based population biology and epidemiological surveillance of Listeria monocytogenes. Nat. Microbiol. 2016 doi: 10.1038/nmicrobiol.2016.185. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Jagadeesan B., Baert L., Wiedmann M., Orsi R.H. Comparative analysis of tools and approaches for source tracking Listeria monocytogenes in a food facility using whole-genome sequence data. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.00947. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Cabal A., Pietzka A., Huhulescu S., Allerberger F., Ruppitsch W., Schmid D. Isolate-Based Surveillance of Listeria monocytogenes by Whole Genome Sequencing in Austria. Front. Microbiol. 2019 doi: 10.3389/fmicb.2019.02282. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Hurley D., Luque-Sastre L., Parker C.T., Huynh S., Eshwar A.K., Nguyen S.V., Andrews N., Moura A., Fox E.M., Jordan K., et al. Whole-Genome Sequencing-Based Characterization of 100 Listeria monocytogenes Isolates Collected from Food Processing Environments over a Four-Year Period. mSphere. 2019 doi: 10.1128/mSphere.00252-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Painset A., Björkman J.T., Kiil K., Guillier L., Mariet J.F., Felix B., Amar C., Rotariu O., Roussel S., Perez-Reche F., et al. Liseq—Whole-genome sequencing of a cross-sectional survey of Listeria monocytogenes in ready-to-eat foods and human clinical cases in Europe. Microb. Genomics. 2019 doi: 10.1099/mgen.0.000257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43.Maury M.M., Bracq-Dieye H., Huang L., Vales G., Lavina M., Thouvenot P., Disson O., Leclercq A., Brisse S., Lecuit M. Hypervirulent Listeria monocytogenes clones’ adaption to mammalian gut accounts for their association with dairy products. Nat. Commun. 2019 doi: 10.1038/s41467-019-10380-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44.Xue J., Murrieta C.M., Rule D.C., Miller K.W. Exogenous or L-rhamnose-derived 1,2-propanediol is metabolized via a pduD-dependent pathway in Listeria innocua. Appl. Environ. Microbiol. 2008 doi: 10.1128/AEM.01074-08. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Salazar J.K., Wu Z., David McMullen P., Luo Q., Freitag N.E., Tortorello M.L., Hu S., Zhanga W. prfa-like transcription factor gene lmo0753 contributes to l-rhamnose utilization in Listeria monocytogenes strains associated with human food-borne infections. Appl. Environ. Microbiol. 2013 doi: 10.1128/AEM.01812-13. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Trudelle D.M., Bryan D.W., Hudson L.K., Denes T.G. Cross-resistance to phage infection in Listeria monocytogenes serotype 1/2a mutants. Food Microbiol. 2019 doi: 10.1016/j.fm.2019.06.003. [DOI] [PubMed] [Google Scholar]
- 47.Pasquali F., Palma F., Guillier L., Lucchi A., Cesare A.D., Manfreda G. Listeria monocytogenes sequence types 121 and 14 repeatedly isolated within one year of sampling in a rabbit meat processing plant: Persistence and ecophysiology. Front. Microbiol. 2018 doi: 10.3389/fmicb.2018.00596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Harter E., Wagner E.M., Zaiser A., Halecker S., Wagner M., Rychli K. Stress survival islet 2, predominantly present in Listeria monocytogenes strains of sequence type 121, is involved in the alkaline and oxidative stress responses. Appl. Environ. Microbiol. 2017 doi: 10.1128/AEM.00827-17. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49.Harrand A.S., Jagadeesan B., Baert L., Wiedmann M., Orsi R.H. Evolution of Listeria monocytogenes in a food-processing plant involves limited single nucleotide substitutions, but considerable diversification by gain and loss of prophages. Appl. Environ. Microbiol. 2020 doi: 10.1128/AEM.02493-19. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Bansal M., Nannapaneni R., Sharma C.S., Kiess A. Listeria monocytogenes response to sublethal chlorine induced oxidative stress on homologous and heterologous stress adaptation. Front. Microbiol. 2018:2050. doi: 10.3389/fmicb.2018.02050. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Xu D., Deng Y., Fan R., Shi L., Bai J., Yan H. Coresistance to Benzalkonium Chloride Disinfectant and Heavy Metal Ions in Listeria monocytogenes and Listeria innocua Swine Isolates from China. Foodborne Pathog. Dis. 2019 doi: 10.1089/fpd.2018.2608. [DOI] [PubMed] [Google Scholar]
- 52.Katharios-Lanwermeyer S., Rakic-Martinez M., Elhanafi D., Ratani S., Tiedje J.M., Kathariou S. Coselection of cadmium and benzalkonium chloride resistance in conjugative transfers from nonpathogenic Listeria spp. to other Listeriae. Appl. Environ. Microbiol. 2012 doi: 10.1128/AEM.02245-12. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Korsak D., Chmielowska C., Szuplewska M., Bartosik D. Prevalence of plasmid-borne benzalkonium chloride resistance cassette bcrABC and cadmium resistance cadA genes in nonpathogenic Listeria spp. isolated from food and food-processing environments. Int. J. Food Microbiol. 2019 doi: 10.1016/j.ijfoodmicro.2018.10.019. [DOI] [PubMed] [Google Scholar]
- 54.Parsons C., Lee S., Kathariou S. Heavy metal resistance determinants of the foodborne pathogen Listeria monocytogenes. Genes. 2019;10:11. doi: 10.3390/genes10010011. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 55.Moreno L.Z., Paixão R., Gobbi D.D., Raimundo D.C., Ferreira T.P., Hofer E., Matte M.H., Moreno A.M. Characterization of atypical Listeria innocua isolated from swine slaughterhouses and meat markets. Res. Microbiol. 2012 doi: 10.1016/j.resmic.2012.02.004. [DOI] [PubMed] [Google Scholar]
- 56.Volokhov D.V., Duperrier S., Neverov A.A., George J., Buchrieser C., Hitchins A.D. The presence of the internalin gene in natural atypically hemolytic Listeria innocua strains suggests descent from L. monocytogenes. Appl. Environ. Microbiol. 2007;73:1928–1939. doi: 10.1128/AEM.01796-06. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Petran R.L., Swanson K.M.J. Simultaneous Growth of Listeria monocytogenes and Listeria innocua. J. Food Prot. 1993 doi: 10.4315/0362-028X-56.7.616. [DOI] [PubMed] [Google Scholar]
- 58.Jemmi T., Stephan R. Listeria monocytogenes: Food-borne pathogen and hygiene indicator. OIE Rev. Sci. Tech. 2006 [PubMed] [Google Scholar]
- 59.ISO . Microbiology of the Food Chain-Horizontal Method for the Detection and Enumeration of Listeria monocytogenes and of Listeria spp-Part 1: Detection Method. ISO; Geneva, Switzerland: 2017. [Google Scholar]
- 60.Keys A.L., Dailey R.C., Hitchins A.D., Smiley R.D. Postenrichment population differentials using buffered Listeria enrichment broth: Implications of the presence of Listeria innocua on Listeria monocytogenes in food test samples. J. Food Prot. 2013 doi: 10.4315/0362-028X.JFP-13-089. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Pusztahelyi T., Szabó J., Dombrádi Z., Kovács S., Pócsi I. Foodborne Listeria monocytogenes: A Real Challenge in Quality Control. Scientifica. 2016 doi: 10.1155/2016/5768526. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Rodríguez-López P., Bernárdez M., Rodríguez-Herrera J.J., Comesaña Á.S., Cabo M.L. Identification and metagenetic characterisation of Listeria monocytogenes-harbouring communities present in food-related industrial environments. Food Control. 2019 doi: 10.1016/j.foodcont.2018.07.023. [DOI] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.