Abstract
Export of mRNA from the nucleus to the cytoplasm is a critical process for all eukaryotic gene expression. As mRNA is synthesized, it is packaged with a myriad of RNA-binding proteins to form ribonucleoprotein particles (mRNPs). For each step in the processes of maturation and export, mRNPs must have the correct complement of proteins. Much of the mRNA export pathway revolves around the heterodimeric export receptor yeast Mex67•Mtr2/ human NXF1•NXT1, which is recruited to signal the completion of nuclear mRNP assembly, mediates mRNP targeting/translocation through the nuclear pore complex (NPC), and is displaced at the cytoplasmic side of the NPC to release the mRNP into the cytoplasm. Directionality of the transport is governed by at least two DEAD-box ATPases, yeast Sub2/human UAP56 in the nucleus and yeast Dbp5/human DDX19 at the cytoplasmic side of the NPC, which respectively mediate the association and dissociation of Mex67•Mtr2/NXF1•NXT1 onto the mRNP. Here we review recent progress from structural studies of key constituents in different steps of nuclear mRNA export. These findings have laid the foundation for further studies to obtain a comprehensive mechanistic view of the mRNA export pathway.
Keywords: mRNA export, structural biology, mRNP remodeling, DEAD-box ATPase, export receptor
With the arrival of the nucleus in cellular evolution, intricate mechanisms to transport macromolecules between the nucleus and the cytoplasm became essential. Nucleocytoplasmic transport events occur through the nuclear pore complex (NPC), the cell’s largest and most versatile transport channel, which allows transport of proteins and various RNA families such as mRNA, rRNA, tRNA, miRNA, snRNA1,2. With respect to mRNAs, transport through the NPC is unidirectional: mRNAs are synthesized in the nucleus, and are exported to the cytoplasm for translation. The central channel of the NPC is filled with thousands of phenylalanine-glycine (FG) peptide repeats3. As a general principle for all transport across the NPC, cargo needs to acquire at least one transport receptor to overcome the permeability barrier of the FG milieu4. Most nucleocytoplasmic transport events employ the karyopherin family of transport receptors. To establish the transport directionality, the small GTPase Ran, via a concentration gradient of RanGTP across the nuclear envelope maintained by the NTF2 protein, drives the assembly/disassembly of the cargo-receptor complex4,5. Overall, Ran-dependent transport of protein and several RNA families is well understood (readers are referred to an excellent review by Güttler and Görlich6). Bulk mRNA export, however, employs a unique mechanism that does not depend on karyopherins or Ran, and the underlying molecular basis is less well understood. This review discusses ongoing research, mainly from a structural perspective, to provide an overview of the key steps in nuclear mRNA export.
1. The principal export receptor Mex67•Mtr2/NXF1•NXT1 mediates bulk mRNA export
As mRNA is synthesized and processed in the nucleus, it is packaged with RNA-binding proteins (RBPs) to form ribonucleoprotein particles (mRNPs)7. mRNPs are formidably diverse. Human cells carry tens of thousands of different mRNAs, and the protein composition of each individual mRNP is unique and highly dynamic throughout its life cycle. Despite the complexity in mRNP protein composition, export of the vast majority of mRNAs utilizes a non-karyopherin export receptor, the heterodimeric Mex67•Mtr2 in budding yeast/NXF1•NXT1 (TAP•p15) in humans8–11. NXF1•NXT1 was found to be an mRNA export receptor in light of the observation that NXF1•NXT1 mediates nuclear export of CTE (constitutive transport element) containing RNA, which resides in some retroviral genomes and promotes export of unspliced retroviral RNA9. Expression of NXF1•NXT1 in yeast rescues growth of the otherwise lethal mex67 mtr2 double null strain, revealing a conserved function of the export receptor from yeast to humans8.
Two key aspects of transport receptor function are cargo recognition and nucleoporin FG-repeat recognition. Both yeast Mex67 and human NXF1 have a modular architecture that includes an RRM (RNA recognition motif), LRR (leucine-rich region), NTF2L (NTF2-like), and UBA (ubiquitin-associated) domains. Mtr2/NXT1, which also exhibits an NTF2-like fold, binds to the NTF2L domain of Mex67/NXF1 (Figure 1A). With respect to cargo recognition, RRM, LRR, and NTF2L domains are all capable of binding RNA12,13. Mex67•Mtr2/NXF1•NXT1 binds to RNA without sequence specificity14,15, which is consistent with the role as a general export receptor. Transcriptome-wide analysis of the RNA-binding landscape of Mex67 suggests a uniform coverage on mRNAs14,15, but the exact binding mode and copy number of export receptor(s) on an individual mRNP remain unclear. In regard to FG-repeat recognition, Mex67/NXF1 features two FG-repeat binding sites on the NTF2L and UBA domains16,17 (Figures 1B and 1C). Binding between Mex67/NXF1 and FG-repeats is weak in nature17, enabling rapid exchange of FG-repeats from the export receptor as the mRNP translocates through the central channel of NPC.
Figure 1.
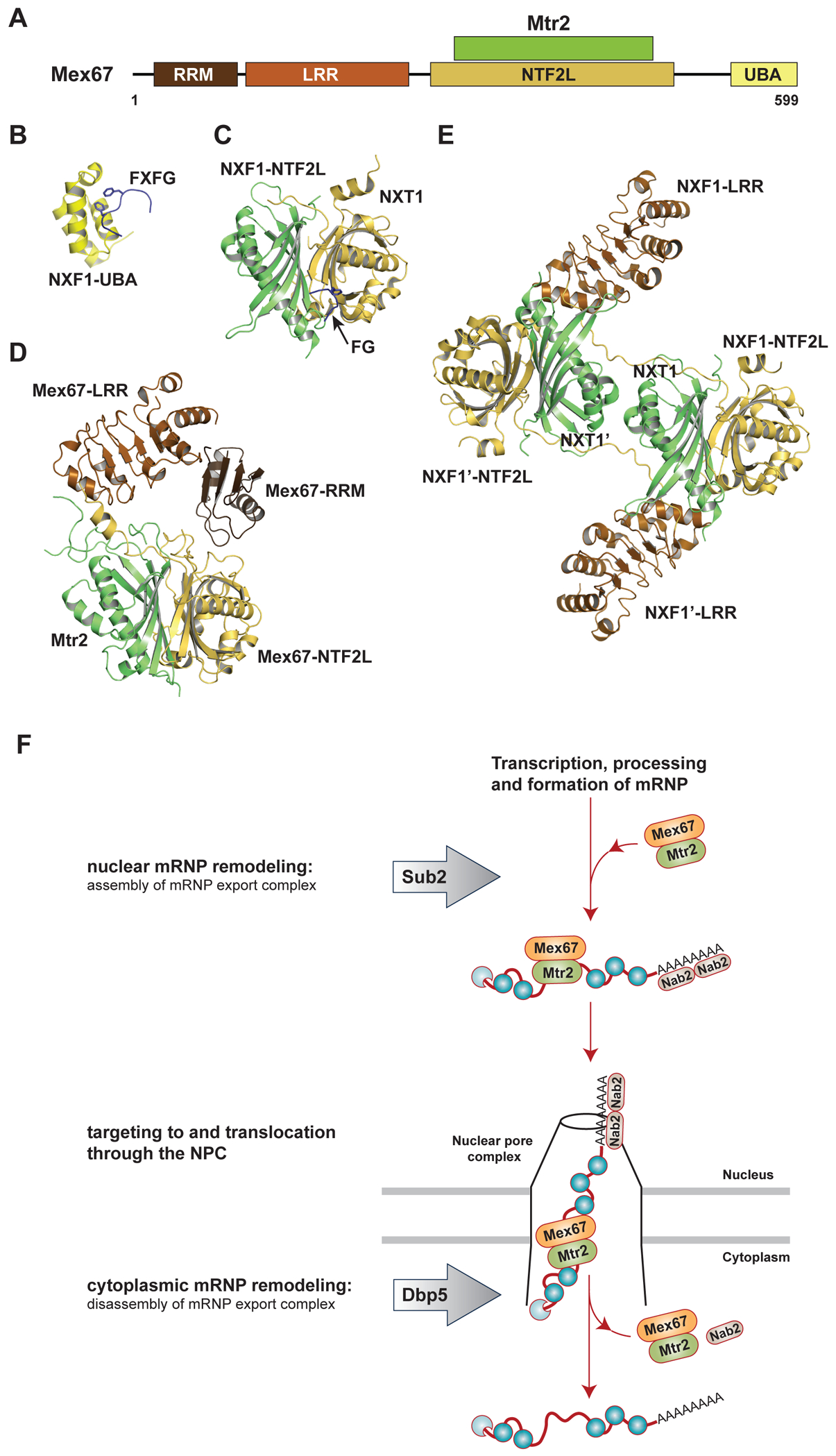
Nuclear mRNA export is mediated by the principal export receptor yeast Mex67•Mtr2/ human NXF1•NXT1. (A) Domain schematic of yeast Mex67•Mtr2. (B) Structure of the NXF1-UBA domain bound to a FXFG peptide (PDB ID 1OAI). (C) Structure of the NXF1-NTF2L domain bound to a FG peptide (PDB ID 1JN5). (D) Structure of yeast Mex67 (RRM, LRR, and NTF2L domains) associated with Mtr2 (PDB ID 4WWU). (E) Structure of NXF1 (LRR and NTF2L domains) associated with NXT1 (PDB ID 4WYK). (F) A working model of Mex67•Mtr2 mediated mRNA export. Export is driven by specific alterations of the mRNP protein composition. In the nucleus, Sub2 facilitates the assembly of export-competent mRNPs by recruiting the principal mRNA export receptor Mex67•Mtr2. At the cytoplasmic side of the NPC, Dbp5 mediates disassembly of the export complex by displacing proteins including Mex67•Mtr2 and the poly(A) RNA binding protein Nab2 from the mRNP.
For a long time, the four domains of NXF1 were thought to be arranged like beads on a string, and there was no clear understanding of how the export receptor functions as a whole. However, a recent structure of yeast Mex67•Mtr2 containing the RRM, LRR, and NTF2L domains of Mex67 has established that the LRR domain has a defined orientation relative to the NTF2L domain, while RRM and UBA domains are mobile12 (Figure 1D). Intriguingly, two copies of human NXF1•NXT1 can also form a higher-order assembly via domain swapping, where two NXT1 molecules are juxtaposed and the LRR and NTF2L of each NXF1 are connected by a linker that traverses along the surface of both NXT1s18 (Figure 1E). This configuration generates a 2-fold symmetric platform, featuring a continuous RNA binding surface on one side of the protein complex with the FG-repeat binding sites on the opposite side. Intriguingly, the higher-order NXF1•NXT1 assembly is targeted by the influenza A virus NS1 protein to block nuclear export of host mRNAs19. Mutations engineered to disrupt the formation of the higher-order NXF1•NXT1 assembly reduces the nuclear export of CTE-RNA in vivo, which also exhibits a 2-fold symmetry, but does not seem to affect bulk poly(A) RNA export. One hypothesis is that the higher-order NXF1•NXT1 assembly may facilitate nuclear export of a subset of structured RNAs, but the precise function and the prevalence of the higher-order form of NXF1•NXT1 in human cells remain unclear.
Although NXF1 can directly bind to RNA, its association and dissociation on the transcripts are regulated by DEAD-box ATPases. These energy-spending processes govern the directionality of the mRNA export process. Overall, nuclear mRNA export can be broken down into three steps: 1) Assembly of an export competent mRNP marked with Mex67•Mtr2/NXF1•NXT1, mediated by the Sub2/UAP56 ATPase in the nucleus; 2) mRNP targeting to and translocation through the NPC, and 3) Disassembly of the mRNP export complex by displacing factors including Mex67•Mtr2/NXF1•NXT1, mediated by the Dbp5/DDX19 ATPase at the cytoplasmic face of the NPC20–22 (Figure 1F and Table 1). Below we highlight recent work on each of these steps in mRNA export.
Table 1.
Key constituents of the mRNA export machinery
| Yeast | Human | Function |
|---|---|---|
| Mex67•Mtr2 | NXF1•NXT1 (TAP•p15) | Principal export receptor for bulk mRNA export |
| In the nucleus | ||
| Sub2 | UAP56 | DEAD-box ATPase that remodels nuclear mRNP, component of the TREX complex |
| Yra1 | ALY (REF, THOC4) | Adaptor for the export receptor, component of the TREX complex |
| THO | THO | A multi-subunit complex, component of the TREX complex |
| Nab2 | ZC3H14 | Poly(A) RNA binding protein |
| TREX-2 | TREX-2 | A multi-subunit complex that targets actively transcribed genes to the NPC |
| At the NPC cytoplasmic face | ||
| Dbp5 | DDX19 | DEAD-box ATPase that remodels mRNP at the cytoplasmic side of the NPC |
| Gle1 | GLE1 | Activator of Dbp5/DDX19, requires IP6 for mRNA export |
| Nup42 | NUP42 (hCG1) | FG-Nucleoporin to which Gle1/GLE1 binds |
| Nup159 | NUP214 (CAN) | FG-Nucleoporin to which Dbp5/DDX19 binds |
| Nup116 | NUP98 | FG-Nucleoporin, binds to the export receptor |
| Gle2 | RAE1 | mRNA export factor, binds to Nup116/NUP98 |
2. Assembly of export competent mRNP
2.1. Sub2 mediated nuclear mRNP remodeling
In the nucleus, transcription, basic pre-mRNA processing (capping, splicing, and 3’ end processing), and nuclear transport are integral steps in the nuclear phase of gene expression. A wealth of studies indicate that proper mRNA export is first set in motion at the steps of cotranscriptional RBP loading and pre-mRNA processing23–26. Association of Mex67/NXF1 onto a nuclear mRNP is driven by the RNA-dependent ATPase Sub2/UAP5627 with assistance of a multisubunit complex THO28–30 and an RBP Yra1/ALY31,32; Sub2/UAP56, THO, and Yra1/ALY together form the TRanscription-EXport (TREX) complex33 (Table 1). The Sub2/UAP56 ATPase belongs to the family of DEAD-box RNA helicases, named based on a characteristic motif Asp-Glu-Ala-Asp (DEAD in single-letter code)34. DEAD-box proteins participate in all steps of RNA metabolism, in a manner that resembles the activities of protein chaperones. In particular, DEAD-box proteins promote rearrangement of RNA structures or assembly/disassembly of RNA-protein complexes at the expense of ATP hydrolysis. In addition to mRNA export, Sub2/UAP56 has additional roles in splicing and piRNA biogenesis35–37. Sub2/UAP56 contains two RecA like domains (NTD and CTD) and a short N-terminal extension. In vitro, Sub2/UAP56 like many DEAD-box proteins has ATP-dependent RNA helicase activity in a non-processive manner, and RNA-dependent ATPase activity34,38,39.
Sub2-mediated nuclear mRNP remodeling occurs in a step wise fashion27,40 (Figure 2A and 2B). Within the yeast TREX complex, THO exists as a robust structural and functional unit comprised of the Tho2, Hpr1, Mft1, Thp2, and Tex1 proteins28,29. THO travels with transcribing RNA pol II by binding to phosphorylated Pol II CTD and recruits Sub2 to the transcription machinery23,24. Recombinant hetero-pentameric THO complex has been shown to stimulate Sub2 ATPase activity41. Crystal structure of a 360 kDa THO•Sub2 complex has been determined at 6 Å resolution, revealing the overall architecture of the hetero-pentameric THO complex and how THO activates the Sub2 ATPase41 (Figure 2C). In particular, the THO complex forms an elongated scaffold, approximately 25 nm in length. It makes contact with both Sub2-NTD and Sub2-CTD, and induces a “half-open” configuration, in which the conserved motifs for ATP/RNA binding are pre-aligned in Sub2. The configuration of Sub2 bound to THO is structurally similar to Dbp5 bound to its activator Gle142–44. Thus, the DEAD-box ATPases mediated nuclear and cytoplasmic mRNP remodeling share a similar activation mechanism. The THO•Sub2 crystal structure likely captures how THO recruits Sub2 to the transcription machinery, with the Sub2 primed in a “half-open” state for subsequent mRNP engagement.
Figure 2.
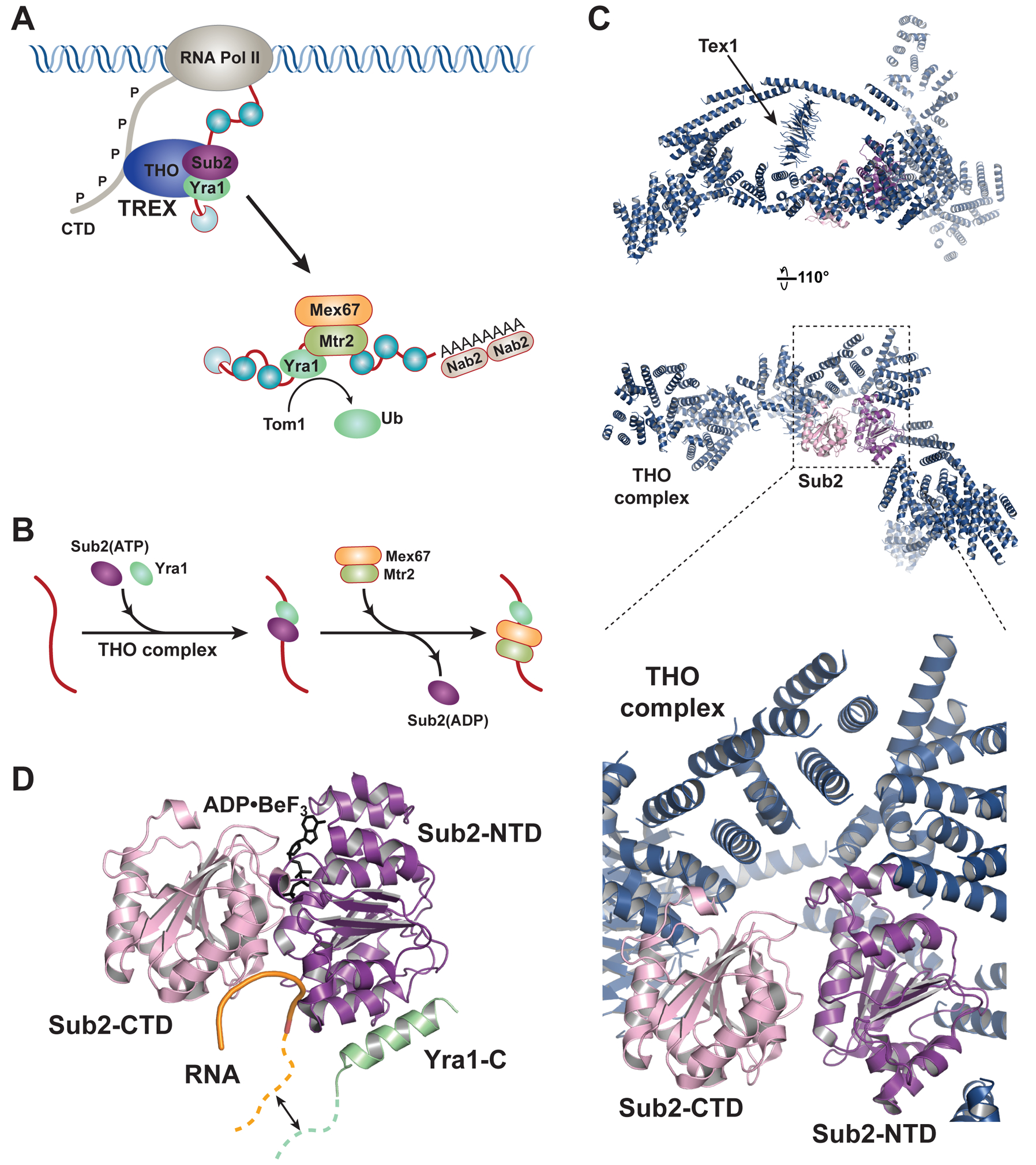
ATPase mediated nuclear mRNP assembly. (A) Yeast TREX complex THO•Sub2•Yra1 travels with RNA Pol II and facilitates loading of the export receptor onto mRNA. (B) A detailed view of the stepwise remodeling reactions driven by the Sub2 ATPase. THO is omitted from the RNA because the dynamics of THO association with mRNA is not known. (C) A 6.0 Å resolution structure of Sub2 bound to a THO core complex which contains S. cerevisiae Tho21–1207, Hpr11–603, Mft11–256, and Thp21–26, as well as S. bayanus Tex11–380 (PDB ID 5SUQ). Top two panels show the overall architecture of the complex. THO is represented by a polyalanine model and only the Tex1 subunit is assigned. The bottom panel highlights the THO-Sub2 binding interface. (D) Structure of Sub2 in association with a truncated Yra1 (Yra1-C, a.a. 208–226) and poly (U) RNA in the presence of ADP•BeF3 (PDB ID 5SUP). The bound RNA is sharply bent, which is characteristic of DEAD-box proteins. The Yra1 region preceding the crystallized fragment is capable of binding RNA (depicted by a green dashed line), and has been proposed to extend the RNA binding site in the Sub2•Yra1 complex.
Sub2 has been implicated in loading of Yra1 onto the mRNP, which in turn acts as an adaptor protein to recruit the export receptor Mex67•Mtr227. Sub2 and Yra1 cooperatively bind to RNA in vitro41. A crystal structure of Sub2 in complex with the C-terminal region of Yra1 (Yra1-C) and poly (U) RNA reveals that Yra1-C folds into a helix and binds to the Sub2-NTD (Figure 2D)41. Sub2, in a “closed” configuration, recognizes the sugar-phosphate backbone of RNA, indicating a sequence non-specific binding. Interestingly, the RNA binding region of Yra1 that precedes Yra1-C would be located close to the bound RNA. The juxtaposition of RNA binding regions of Sub2 and Yra1 could generate an extended RNA binding site, in line with their cooperative RNA binding in vitro. In addition, Yra1-C is able to stimulate Sub2 ATPase activity in vitro. Together, the Sub2•Yra1-C•RNA complex likely represents a key assembly for recruitment of Yra1 onto the mRNP, which is coupled to the Sub2 engagement with RNA. Sub2 is thought to be later displaced, presumably after ATP hydrolysis, by the export receptor Mex67•Mtr2, as binding of Sub2 and Mex67•Mtr2 to Yra1 is mutually exclusive (Figure 2B)27,31. Yra1 does not accompany the mRNP to the cytoplasm; it is ubiquitinated by the E3 ligase Tom1, resulting in Yra1 dissociation form the mRNP, by an yet to define mechanism, prior to nuclear export (Figure 2A)45.
To date, structural evidence for how the TREX complex facilitates the recruitment of the export receptor is still lacking. Recent studies of human TREX indicate that THO directly interacts with ALY and together they coordinately facilitate RNA binding of the export receptor46,47. This implies a more sophisticated mechanism than originally envisioned for this important step in mRNP maturation. When does THO associate with the mRNP and when is THO displaced? Although the THO•Sub2 structure suggests that THO does not favor binding to a “closed” form of Sub2, THO could remain anchored to one of the Sub2 domains when Sub2 is bound to RNA. In fact, evidence suggests that, as discussed in section 4.2, Gle1 can bind to the CTD of Dbp5 alone or to both the NTD and CTD together, indicating a dynamic nature of the Gle1-Dbp5 interaction. In addition, Yra1, as suggested by the human THO-ALY interaction, could potentially serve as another anchor for THO to associate with the mRNP. Furthermore, THO itself has been shown to bind RNA directly in vitro29. To date, the dynamics of the interaction between THO and the mRNP remain to be elucidated. Of note, THO makes up the most mass of the TREX complex, containing five subunits in yeast and six subunits in humans28–30,48. Yet surprisingly, no atomic resolution structure has been reported for any THO subunit from yeast to humans, underscoring the need for further structural studies.
Sub2/UAP56-powered nuclear mRNP remodeling is a conserved mechanism from yeast to humans. Perhaps due to the more complex gene expression in humans, multiple adaptors including UIF and CHTOP share similar features with ALY, and are considered dynamic components of the human TREX complex49,50. Intriguingly, TREX mediates the export of both intronless and intron-containing genes30,46,51–53. One major difference in gene expression between yeast and humans is the prevalence of splicing. While only a small population (~5%) of yeast genes contain introns, the opposite is true in humans54,55. The recruitment mechanism of TREX likely reflects this difference, as yeast TREX is recruited by the transcriptional machinery, while human TREX is recruited to mRNA in a splicing dependent manner23,24,30. Much remains to be studied to elucidate how the same core machineries are employed in different ways in yeast and humans.
2.2. Integration of pre-mRNA processing and nuclear mRNP remodeling
How does the cell ensure that only properly processed transcripts will be exported into the cytoplasm for translation? There is compelling evidence that TREX is physically and functionally linked to mRNA biogenesis factors involved in every step of pre-mRNA processing. For example, Human TREX is recruited to the 5’ end of the mRNP through the interaction between ALY and the mRNA Cap binding complex (CBC)25. CBC is one of the earliest factors deposited on a growing mRNA chain. It associates with various RNAs transcribed by RNA Pol II, including mRNA, snRNA, and miRNA. CBC recruits discrete factors to promote processing and nuclear export for different RNA families56. With respect to mRNA, ALY association may be the first step to direct mRNA to the UAP56 mediated export pathway. In addition, the connection of TREX function to splicing has been found in both yeast and humans. Yeast THO associates with the two SR (serine/arginine-rich) proteins Hrb1 and Gbp2, which have been proposed to function as surveillance factors for the selective export of spliced mRNAs57,58. In humans, loading of TREX to spliced mRNA occurs by a splicing-coupled mechanism30. Furthermore, TREX function is connected to pre-mRNA 3’-end processing. Yra1 interacts with Pcf11, the Pol II CTD binding subunit of the cleavage-polyadenylation factor CF1A49. The Yra1-Pcf11 interaction is conserved from yeast to humans, and has been suggested to modulate the assembly of the 3’-end processing machinery59,60. Recent work also shows that human THO interacts with the poly(A) RNA binding protein ZC3H14, and their interaction is required for proper control of bulk poly(A) tail length61. Together, this multitude of connections between TREX and pre-mRNA processing steps suggests that decision-making during mRNA export is concurrent with pre-mRNA processing. These connections may direct, step by step, the ATPase-powered remodeling reactions to ultimately mark mature transcripts with the export receptor, thereby ensuring the fidelity of gene expression specifically at the stage of mRNP nuclear maturation.
3. mRNP targeting to the NPC
Export competent mRNPs first encounter the nuclear basket of the NPC preceding translocation through the central channel. The nuclear basket is composed of Mlp1, Mlp2, Nup1, Nup2, and Nup60 in yeast62. One mechanism that targets export-competent mRNPs to the NPC nuclear basket is transcription-coupled mRNA export, mediated by the yeast TREX-2 complex63–66 (Figure 3A). TREX-2 is linked to transcription machinery via the SAGA complex, which is a chromatin-modifying transcriptional coactivator, and the Mediator complex, which is an essential regulator of RNA Pol II67,68. TREX-2 associates with the nuclear basket and promotes the targeting of actively transcribed genes to the NPC67,69. TREX-2 may thereby mediate a fast track from transcription to mRNA export for these transcripts. It remains unclear whether TREX-2 has a global function to tether mRNPs to the NPC, including those that are not transcribed at the NPC peripheral.
Figure 3.
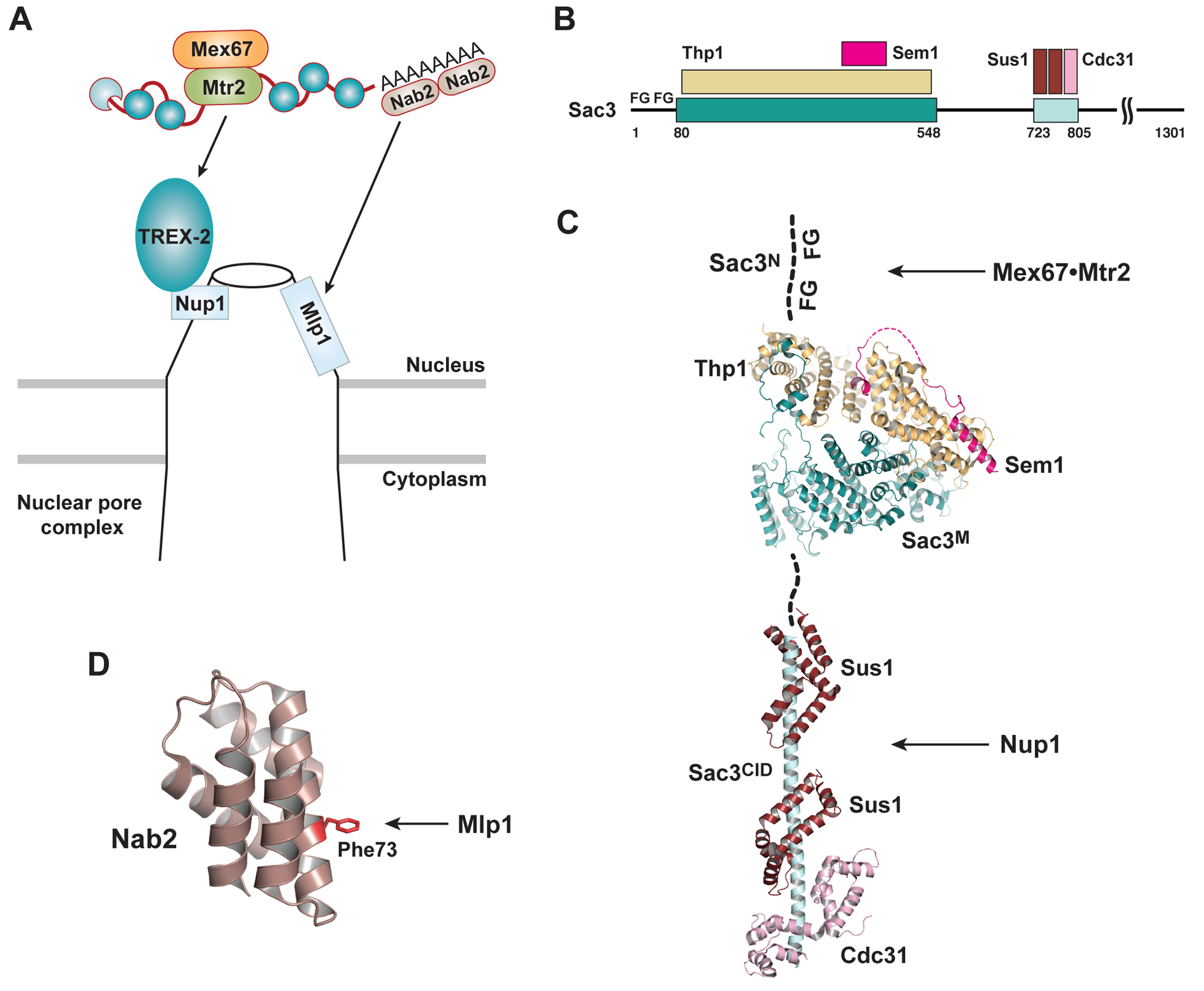
mRNP targeting to the nuclear basket of the NPC in yeast. (A) Schematic of mRNP targeting mediated by the TREX-2 complex and Mlp1 at the nuclear basket of the NPC. (B) Schematic of the TREX-2 complex. (C) Structural basis for the TREX-2 mediated mRNP targeting. TREX-2 complex is built on a scaffold of the Sac3 subunit. The N-terminal region of Sac3 (Sac3N) features FG-repeats that are recognized by the export receptor on mRNP. The middle region of Sac3 (Sac3M) binds to the Thp1 and Sem1 subunits of TREX-2 (PDB ID 5UBP). The C-terminal CID region of Sac3 (Sac3CID) binds to the Sus1 (two copies) and Cdc31 subunits of TREX-2 (PDB ID 3FWC), together mediating NPC association through interaction with the nuclear basket protein Nup1. (D) Structure of the N-terminal Mlp1-binding domain of Nab2 (PDB ID 2V75). The Phe73 residue is critical for the Nab2-Mlp1 interaction.
TREX-2 is composed of Sac3, Thp1, Sem1, Sus1, and Cdc31 in yeast63–66. The entire complex is arranged based on a Sac3 scaffold (Figure 3B and 3C). The Sac3 N-terminal region features FG-repeats that recognize Mex67•Mtr2 associated mRNPs70. The Sac3 middle region binds to Thp1 and Sem1, forming an architectural platform that can bind RNA in vitro71,72. In addition, the Sac3 C-terminal CID (cdc31 interacting domain) region folds into an extended helix where two Sus1 molecules and one Cdc31 molecule bind. This Sac3C•Sus1•Cdc31 subcomplex mediates the interaction with the basket nucleoporin Nup173,74. Together, structural characterization of TREX-2 reveals an architecture that is ideally suited to tether an export-competent mRNP to the nuclear basket of the NPC.
The nuclear basket of the NPC has been suggested to implement a quality control step for mRNA export. In particular, deletion of the coiled-coil protein Mlp1 causes leakage of intron-containing mRNAs into the cytoplasm75. Mlp1 interacts with the poly(A) RNA binding protein Nab2, which is required for proper poly(A) tail length control and mRNA export76. Nab2 contains an N-terminal PWI-like domain and C-terminal tandem zinc finger domains. The N-terminal domain of Nab2 interacts with Mlp1 and a key Phe73 residue is shown to be critical for their interaction77 (Figure 3A and 3D). The Nab2-Mlp1 interaction could serve as a means of mRNP targeting to the nuclear basket, and also contribute to the Mlp1-mediated quality control.
The molecular basis for mRNP targeting to the NPC and quality control in human cells is poorly understood. Human TREX-2 contains all the orthologous proteins corresponding to yeast including GANP, PCID, DSS1, ENY2, and CENT2/CENT3 (homologues of yeast Sac3, Thp1, Sem1, Sus1, and Cdc31, respectively)78. However, in contrast to yeast, most transcription in human cells takes place in the nucleoplasm79. Therefore, mRNP must travel from the nuclear interior to the nuclear periphery to find a NPC. Nevertheless, TREX-2 seems to have a conserved role in human cells. The Sac3 homologue GANP partitions between the nuclear interior and the NPC80. Of note, NPC association of GANP requires both the CID region and the C-terminal MCM3AP domain (not present in yeast Sac3)78. GANP depletion inhibits bulk mRNA export, with retention of mRNPs and NXF1 in punctate foci within the nucleus80. This observation is consistent with a model in which GANP contributes to the movement of NXF1-containing mRNPs from the nuclear interior to the NPC. In regard to the quality control mediated by the NPC nuclear basket, human TPR (Mlp1 homologue) appears to be the main player in retaining aberrant mRNAs like yeast Mlp181. However, it is not known how TPR distinguishes normal and aberrant mRNAs. Overall, the mechanism underlying mRNP concentration and quality control at the NPC nuclear face in humans will require further study.
4. Disassembly of the mRNP export complex
4.1. The mRNA export platform at the NPC cytoplasmic face
The actual translocation of mRNP through the NPC channel is not inherently directional. At the terminal step in nuclear mRNA export, the DEAD-box ATPase Dbp5/DDX19-mediated remodeling releases the export receptor and other RBPs from the exporting mRNP, prohibiting the mRNP from sliding back to the nucleus and thereby ensuring directional movement20,21. Dbp5/DDX19 localizes to the cytoplasmic side of the NPC, and is part of the mRNA export platform that also contains Gle1/GLE1, Nup42/NUP42, Nup159/NUP214, Nup116/NUP98, and Gle2/RAE182 (Figure 4A and Table 1). Among them, Nup42/NUP42, Nup159/NUP214, and Nup116/NUP98 all contain FG repeat domains, which facilitate docking of the exporting mRNP to the vicinity of Dbp5/DDX1983,84. Recent work demonstrates that the mRNA export platform is positioned right over the NPC’s central channel, in contrast to the traditional view of the export factors being localized at the distal end of the NPC cytoplasmic filament85. This spatial configuration allows efficient cargo capture and remodeling once the exporting mRNP emerges from the central channel of the NPC.
Figure 4.
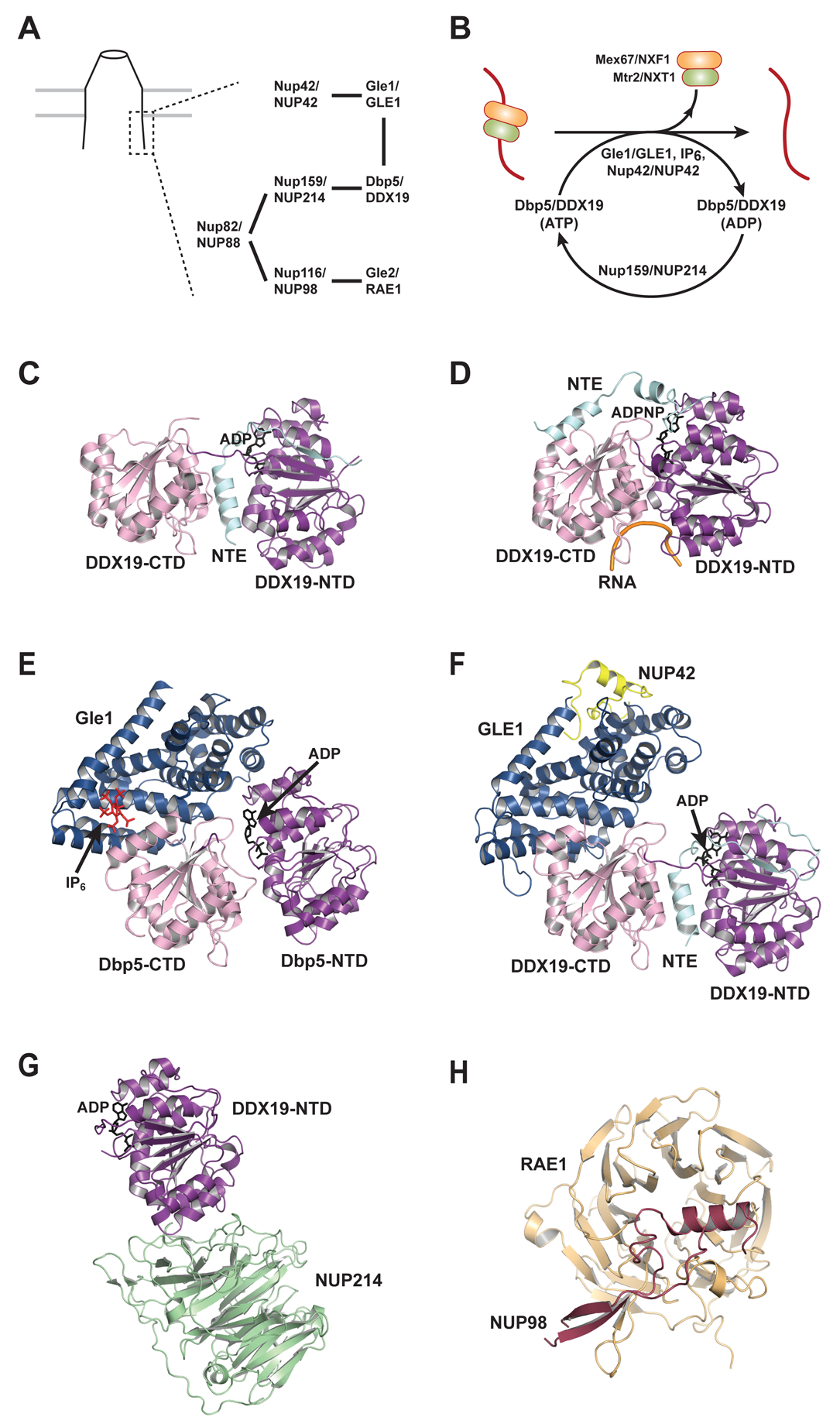
ATPase mediated mRNP remodeling at the cytoplasmic side of the NPC. (A) Schematic diagram of the interaction network of the mRNA export platform at the cytoplasmic side of the NPC. (B) Schematic of Dbp5/DDX19 mediated dissociation of the export receptor from the mRNP, and regulation of the Dbp5/DDX19 catalytic cycle. (C) Structure of DDX19 bound to ADP (PDB ID 3EWS). (D) Structure of DDX19 bound to an ATP-analogue and poly(U) RNA (PDB ID 3G0H). (E) Structure of the Dbp5•Gle1•IP6 complex in the presence of ADP (PDB ID 3RRN). (F) Structure of the DDX19•GLE1•NUP42 complex in the presence of ADP (PDB ID 6B4I). (G) Structure of the DDX19-NTD•NUP214 complex in the presence of ADP (PDB ID 3FMO). (H) Structure of RAE1 in complex with a NUP98 fragment (PDB ID 3MMY).
4.2. Dbp5/DDX19 activation by Gle1/GLE1, IP6, Nup42/NUP42
In the mRNA export platform, Gle1, IP6 (inositol hexakisphosphate), and Nup42 provide spatial and temporal regulation of Dbp5 by stimulating its ATPase activity42,43,86,87 (Figure 4B). Dbp5 contains two RecA like domains (NTD and CTD) and a short N-terminal extension (NTE). The NTE seems to be auto-inhibitory, as deletion of the NTE in human DDX19 yields a more active ATPase88. In line with this observation, structures of DDX19 reveal that an alpha-helix from the NTE occupies the cleft between NTD and CTD when DDX19 is bound to ADP, whereas this helix is displaced when DDX19 is bound to ATP and RNA88 (Figure 4C and 4D). Early evidence about the mechanism of Dbp5 activation came from a genetic screen with a gle1 mutant, which identified Ipk1 as the enzyme that phosphorylates IP5 to generate IP6, and revealed that Ipk1 is required for mRNA export89. Gle1 and IP6 were later shown to activate the ATPase activity of Dbp5 in vitro42,43. Of note, both Dbp5 and Gle1 are also required for protein translation, probably in a manner distinct from their roles in mRNA export90–92. Furthermore, Nup42 is recently found to be an integral component, along with Gle1 and IP6, to Dbp5 activation, and this mechanism is conserved from yeast to humans86.
Structural studies on the Dbp5•Gle1•IP6 complex reveal that Dbp5 is activated through a conserved mechanism that is shared with another DEAD-box ATPase eIF4A, which is activated by eIF4G during translation initiation93. This notion is reinforced by the subsequent structural studies on THO-mediated Sub2 activation in nuclear mRNP remodeling41, and CNOTI-mediated DDX6 stimulation in miRNA-mediated translational repression94. In particular, the C-terminal domain of Gle1 resembles the middle domain of eIF4G, and contacts both the NTD and CTD of Dbp544 (Figure 4E). A unique feature of the Gle1-Dbp5 interaction is that IP6 bridges the protein interaction at the interface of Gle1 and Dbp5-CTD. Gle1-mediated Dbp5 activation is conserved from yeast to humans86,87. However, IP6 activation of human DDX19 was only observed using recombinant DDX19 expressed in the baculovirus-insect cell expression system which carries most of the post-translational modification pathways present in mammalian systems, but not observed using DDX19 expressed in E. coli86. Future studies are needed to determine potential DDX19 modifications that enable the IP6 activation of human DDX19.
The mechanism by which Nup42 coordinates Gle1 stimulation of Dbp5 is not entirely clear. Nup42 interacts with Gle1, and they both are required for the export of heat shock mRNAs following stress86,95–97. Structure of the human DDX19•GLE1•NUP42 complex reveals that NUP42 does not contact DDX19 directly, and causes no significant conformational change in GLE1 (Figure 4F)87. The effect of NUP42 is suggested to be attributed to increasing GLE1 thermostability; the melting temperature of GLE1 increased from 37 to 50 °C in the presence of NUP4287. Of note, in the human DDX19•GLE1•NUP42 complex, only DDX19-CTD is engaged in GLE1 binding, whereas Gle1 contacts both Dbp5 NTD and CTD in the yeast Dbp5•Gle1•IP6 complex. This difference likely reflects the dynamic nature of the Dbp5/DDX19-Gle1/GLE1 interaction. The CTD of Dbp5/DDX19 serves as the primary anchor for Gle1/GLE1, and the NTD provides a secondary binding site that may sample between Gle1/GLE1 binding and non-binding modes. Interestingly, eIF4A also features a primary anchor on CTD and a secondary weaker binding site on NTD for eIF4G98. This configuration may be a conserved feature among the interactions between DEAD-box ATPases and their activators.
4.3. Dbp5/DDX19 catalytic cycle
Activation of Dbp5/DDX19 drives the mRNP remodeling and yields a ADP-bound Dbp5/DDX19 which needs to be recycled for the next round of remodeling events. ADP is not efficiently released from full length Dbp5 in vitro, and Nup159 has been shown to enhance the release of ADP through direct interaction with Dbp599. Consistently, a dbp5-R256D/R259D mutant with reduced ADP binding bypasses the need for Nup159 interaction in yeast. A structure of the NUP214•DDX19 complex reveals that NUP214, the human homologue of Nup159, binds to the NTD of DDX19, and NUP214 and RNA occupy overlapping binding sites on DDX19100,101 (Figure 4G). Together, these results place the role of Nup159/NUP214 in the post-ATP hydrolysis state(s), facilitating nucleotide exchange from ADP to ATP (Figure 4B) and allowing Dbp5/DDX19 to perform multiple cycles of mRNP remodeling.
While it is clear that Gle1, IP6, and Nup42 act in the pre-ATP hydrolysis state(s), and Nup159 functions in the post-ATP hydrolysis state(s) (Figure 4B), a consensus of how the catalytic cycle of Dbp5 is orchestrated has not been reached. In particular, the aforementioned dynamic nature of the interaction between Gle1 and Dbp5 introduces more variables to the system. Further studies will be needed to pinpoint the precise molecular steps during the Dbp5/DDX19 enzymatic cycle. To date, there is still a substantial gap between our biochemical understanding of the protein machinery and what happens in cells. For example, what proteins are displaced in vivo during Dbp5/DDX19-mediated remodeling? In addition to the export receptor, poly(A) RNA binding protein Nab2 is thought to be another physiological target of Dbp5. Recombinant Dbp5 is able to displace Nab2 from a Nab2-RNA complex in vitro102. In vivo, Nab2 accompanies mRNPs through the NPC, but is not found associated with mRNA in polysomes103–105. A dbp5 mutant shows an accumulation of Nab2 on poly(A) RNA102. Together, these studies are consistent with a model in which Dbp5 mediates release of Nab2 from mRNPs at the cytoplasmic side of the NPC. It remains to be determined how many more proteins are physiological targets for the Dbp5/DDX19-mediated remodeling.
4.4. NUP98 and RAE1
Nup116/NUP98 and Gle2/RAE1 are constituents of the mRNA export platform that have not been shown to directly regulate the activity of Dbp5/DDX19. Nup116/NUP98 contains an FG-repeat domain that can be recognized by nuclear transport receptors84,106–109. Nup116/NUP98 binds to Gel1/RAE1 and their interaction is required for Gle2/RAE1 localization at the NPC110–113 (Figure 4H). Yeast Gle2 was identified along with Gle1 in the same genetic screen for genes that are synthetically lethal with a nup100 null mutant114. Nup100 and Nup116 are highly homologous, with the exception that only Nup116 contains the Gle2 binding region. A further advance in our understanding of RAE1 function came from the observation that RAE1 is targeted by the Matrix (M) protein of the vesicular stomatitis virus (VSV) to block host mRNA export115. A structure of the VSV M protein in complex with RAE1 and NUP98 shows the M protein occupying the nucleic acid binding site on RAE1116, indicating that RAE1 function is RNA-binding dependent. The precise role of RAE1 in mRNA export remains to be determined. Given its spatial proximity to DDX19 and dependence on NUP98 for localization, together with the putative role NUP98 plays in docking mRNPs through interaction with the export receptor, it is plausible that RAE1 and NUP98 also contribute to DDX19-mediated terminal steps of mRNA export.
5. Future directions
To date, studies have elucidated the cast of proteins involved in bulk mRNA export and have laid out the core principles. In particular, the spatiotemporal regulation of Mex67•Mtr2/NXF1•NXT1 receptor association and dissociation on an mRNP by DEAD-box ATPases has been shown to be the key to mRNA export. Although considerable progress has been made to obtain structural snapshots of the DEAD-box ATPases in action, a comprehensive mechanistic understanding is lacking. Deciphering the precise nature of mRNP remodeling in the biological context is still a challenge. This will also require further structural knowledge of the mRNA export machinery and novel functional approaches to capture protein exchanges on mRNPs in cells.
A largely unexplored area in the field is how mRNA export is tuned to accommodate the specific needs of a cell with respect to developmental stages, tissue specificity, extracellular stimuli, etc. Accumulating evidence in recent years has revealed that post-transcriptional gene regulation plays a critical role in shaping gene expression profiles117. Indeed, mRNA export can selectively modulate critical biological processes such as DNA repair, stress response, maintenance of pluripotency, etc118–122. Our molecular understanding of the general mRNA export pathway has provided a valuable toolset to investigate the control of gene expression by selective mRNA export.
Not surprisingly, given the essential role of mRNA export in gene expression, the integrity of the pathway is critical for human health. For example, mutations in the gene encoding GLE1 are causally linked to human motor neuron diseases, including lethal congenital contracture syndrome 1 (LCCS-1), lethal arthrogryposis with anterior horn cell disease (LAAHD), and amyotrophic lateral sclerosis (ALS)123–126. In addition, aberrant expression of mRNA export factors has been found in many different forms of cancer127. A plausible hypothesis is that dysregulation of these factors may alter the export of specific transcripts that are critical for cell proliferation and oncogenesis. Furthermore, many viruses interfere with host mRNA export to block host gene expression and/or facilitate essential viral processes. Key constituents of the mRNA export machinery (NXF1•NXT1, UAP56, REF, RAE1•NUP98, etc) are exploited by a wide range of viruses including herpesvirus, adenovirus, VSV, and influenza virus128. Overall, further advances in elucidating the mRNA export pathway will shed light on the pathogenesis of relevant diseases.
Synopsis:
Nuclear export of mRNA, in the form of ribonucleoprotein particles (mRNPs), is governed by the highly regulated alterations in mRNP protein composition known as mRNP remodeling. Central to mRNP remodeling are two DEAD-box ATPases, Sub2 and Dbp5, which respectively mediate the assembly of export competent mRNPs in the nucleus and the release of mRNPs into the cytoplasm. Here we provide an overview on structural studies of key steps in mRNA export.
Acknowledgements
We would like to acknowledge the Ren laboratory, the Wente laboratory and Charles Cole for discussions and critical reading of the manuscript. We apologize to colleagues whose work could not be cited here due to space limitation. This manuscript is supported by funds from Vanderbilt University School of Medicine and NIH R35 GM133743.
Footnotes
Conflicts of interest
The authors have no conflicts of interest to report.
References
- 1.Kohler A, Hurt E. Exporting RNA from the nucleus to the cytoplasm. Nat Rev Mol Cell Biol. 2007;8(10):761–773. [DOI] [PubMed] [Google Scholar]
- 2.Wente SR, Rout MP. The nuclear pore complex and nuclear transport. Cold Spring Harb Perspect Biol. 2010;2(10):a000562. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.Terry LJ, Wente SR. Flexible gates: dynamic topologies and functions for FG nucleoporins in nucleocytoplasmic transport. Eukaryot Cell. 2009;8(12):1814–1827. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4.Smith A, Brownawell A, Macara IG. Nuclear import of Ran is mediated by the transport factor NTF2. Current Biology. 1998;8(25):1403–1406. [DOI] [PubMed] [Google Scholar]
- 5.Stewart M Ran in Nucleocytoplasmic Transport Ras Superfamily Small G Proteins: Biology and Mechanisms 2: Springer, Cham; 2014:109–124. [Google Scholar]
- 6.Guttler T, Gorlich D. Ran-dependent nuclear export mediators: a structural perspective. Embo Journal. 2011;30(17):3457–3474. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Singh G, Pratt G, Yeo GW, Moore MJ. The Clothes Make the mRNA: Past and Present Trends in mRNP Fashion. Annu Rev Biochem. 2015;84:325–354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8.Katahira J, Strasser K, Podtelejnikov A, Mann M, Jung JU, Hurt E. The Mex67p-mediated nuclear mRNA export pathway is conserved from yeast to human. EMBO J. 1999;18(9):2593–2609. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9.Gruter P, Tabernero C, von Kobbe C, et al. TAP, the human homolog of Mex67p, mediates CTE-dependent RNA export from the nucleus. Mol Cell. 1998;1(5):649–659. [DOI] [PubMed] [Google Scholar]
- 10.Segref A, Sharma K, Doye V, et al. Mex67p, a novel factor for nuclear mRNA export, binds to both poly(A)+ RNA and nuclear pores. EMBO J. 1997;16(11):3256–3271. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11.Yoon DW, Lee H, Seol W, DeMaria M, Rosenzweig M, Jung JU. Tap: A novel cellular protein that interacts with Tip of herpesvirus saimiri and induces lymphocyte aggregation. Immunity. 1997;6(5):571–582. [DOI] [PubMed] [Google Scholar]
- 12.Aibara S, Valkov E, Lamers M, Stewart M. Domain organization within the nuclear export factor Mex67:Mtr2 generates an extended mRNA binding surface. Nucleic Acids Res. 2015;43(3):1927–1936. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Katahira J, Dimitrova L, Imai Y, Hurt E. NTF2-like domain of Tap plays a critical role in cargo mRNA recognition and export. Nucleic Acids Res. 2015;43(3):1894–1904. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Baejen C, Torkler P, Gressel S, Essig K, Soding J, Cramer P. Transcriptome maps of mRNP biogenesis factors define pre-mRNA recognition. Mol Cell. 2014;55(5):745–757. [DOI] [PubMed] [Google Scholar]
- 15.Tuck AC, Tollervey D. A transcriptome-wide atlas of RNP composition reveals diverse classes of mRNAs and lncRNAs. Cell. 2013;154(5):996–1009. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Fribourg S, Braun IC, Izaurralde E, Conti E. Structural basis for the recognition of a nucleoporin FG repeat by the NTF2-like domain of the TAP/p15 mRNA nuclear export factor. Mol Cell. 2001;8(3):645–656. [DOI] [PubMed] [Google Scholar]
- 17.Grant RP, Neuhaus D, Stewart M. Structural basis for the interaction between the Tap/NXF1 UBA domain and FG nucleoporins at 1A resolution. J Mol Biol. 2003;326(3):849–858. [DOI] [PubMed] [Google Scholar]
- 18.Aibara S, Katahira J, Valkov E, Stewart M. The principal mRNA nuclear export factor NXF1:NXT1 forms a symmetric binding platform that facilitates export of retroviral CTE-RNA. Nucleic Acids Res. 2015;43(3):1883–1893. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Zhang K, Xie Y, Munoz-Moreno R, et al. Structural basis for influenza virus NS1 protein block of mRNA nuclear export. Nat Microbiol. 2019. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Carmody SR, Wente SR. mRNA nuclear export at a glance. J Cell Sci. 2009;122(Pt 12):1933–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21.Stewart M Nuclear export of mRNA. Trends Biochem Sci. 2010;35(11):609–617. [DOI] [PubMed] [Google Scholar]
- 22.Nino CA, Herissant L, Babour A, Dargemont C. mRNA nuclear export in yeast. Chem Rev. 2013;113(11):8523–8545. [DOI] [PubMed] [Google Scholar]
- 23.Zenklusen D, Vinciguerra P, Wyss JC, Stutz F. Stable mRNP formation and export require cotranscriptional recruitment of the mRNA export factors Yra1p and Sub2p by Hpr1p. Mol Cell Biol. 2002;22(23):8241–8253. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24.Meinel DM, Burkert-Kautzsch C, Kieser A, et al. Recruitment of TREX to the transcription machinery by its direct binding to the phospho-CTD of RNA polymerase II. PLoS Genet. 2013;9(11):e1003914. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25.Cheng H, Dufu K, Lee CS, Hsu JL, Dias A, Reed R. Human mRNA export machinery recruited to the 5’ end of mRNA. Cell. 2006;127(7):1389–1400. [DOI] [PubMed] [Google Scholar]
- 26.Rougemaille M, Dieppois G, Kisseleva-Romanova E, et al. THO/Sub2p functions to coordinate 3’-end processing with gene-nuclear pore association. Cell. 2008;135(2):308–321. [DOI] [PubMed] [Google Scholar]
- 27.Strasser K, Hurt E. Splicing factor Sub2p is required for nuclear mRNA export through its interaction with Yra1p. Nature. 2001;413(6856):648–652. [DOI] [PubMed] [Google Scholar]
- 28.Chavez S, Beilharz T, Rondon AG, et al. A protein complex containing Tho2, Hpr1, Mft1 and a novel protein, Thp2, connects transcription elongation with mitotic recombination in Saccharomyces cerevisiae. EMBO J. 2000;19(21):5824–5834. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.Pena A, Gewartowski K, Mroczek S, et al. Architecture and nucleic acids recognition mechanism of the THO complex, an mRNP assembly factor. EMBO J. 2012;31(6):1605–1616. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30.Masuda S, Das R, Cheng H, Hurt E, Dorman N, Reed R. Recruitment of the human TREX complex to mRNA during splicing. Genes Dev. 2005;19(13):1512–1517. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31.Strasser K, Hurt E. Yra1p, a conserved nuclear RNA-binding protein, interacts directly with Mex67p and is required for mRNA export. EMBO J. 2000;19(3):410–420. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32.Luo ML, Zhou Z, Magni K, et al. Pre-mRNA splicing and mRNA export linked by direct interactions between UAP56 and Aly. Nature. 2001;413(6856):644–647. [DOI] [PubMed] [Google Scholar]
- 33.Strasser K, Masuda S, Mason P, et al. TREX is a conserved complex coupling transcription with messenger RNA export. Nature. 2002;417(6886):304–308. [DOI] [PubMed] [Google Scholar]
- 34.Linder P, Jankowsky E. From unwinding to clamping - the DEAD box RNA helicase family. Nat Rev Mol Cell Biol. 2011;12(8):505–516. [DOI] [PubMed] [Google Scholar]
- 35.Fleckner J, Zhang M, Valcarcel J, Green MR. U2AF65 recruits a novel human DEAD box protein required for the U2 snRNP-branchpoint interaction. Genes Dev. 1997;11(14):1864–1872. [DOI] [PubMed] [Google Scholar]
- 36.Kistler AL, Guthrie C. Deletion of MUD2, the yeast homolog of U2AF65, can bypass the requirement for sub2, an essential spliceosomal ATPase. Genes Dev. 2001;15(1):42–49. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37.Zhang F, Wang J, Xu J, et al. UAP56 couples piRNA clusters to the perinuclear transposon silencing machinery. Cell. 2012;151(4):871–884. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38.Saguez C, Gonzales FA, Schmid M, et al. Mutational analysis of the yeast RNA helicase Sub2p reveals conserved domains required for growth, mRNA export, and genomic stability. Rna-a Publication of the Rna Society. 2013;19(10):1363–1371. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39.Shi H, Cordin O, Minder CM, Linder P, Xu RM. Crystal structure of the human ATP-dependent splicing and export factor UAP56. Proc Natl Acad Sci U S A. 2004;101(51):17628–17633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 40.Hautbergue GM, Hung ML, Golovanov AP, Lian LY, Wilson SA. Mutually exclusive interactions drive handover of mRNA from export adaptors to TAP. Proc Natl Acad Sci U S A. 2008;105(13):5154–5159. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41.Ren Y, Schmiege P, Blobel G. Structural and biochemical analyses of the DEAD-box ATPase Sub2 in association with THO or Yra1. Elife. 2017;6. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42.Alcazar-Roman AR, Tran EJ, Guo SL, Wente SR. Inositol hexakisphosphate and Gle1 activate the DEAD-box protein Dbp5 for nuclear mRNA export. Nature Cell Biology. 2006;8(7):711–U131. [DOI] [PubMed] [Google Scholar]
- 43.Weirich CS, Erzberger JP, Flick JS, Berger JM, Thorner J, Weis K. Activation of the DExD/H-box protein Dbp5 by the nuclear-pore protein Gle1 and its coactivator InsP(6) is required for mRNA export. Nature Cell Biology. 2006;8(7):668–U654. [DOI] [PubMed] [Google Scholar]
- 44.Montpetit B, Thomsen ND, Helmke KJ, Seeliger MA, Berger JM, Weis K. A conserved mechanism of DEAD-box ATPase activation by nucleoporins and InsP6 in mRNA export. Nature. 2011;472(7342):238–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45.Iglesias N, Tutucci E, Gwizdek C, et al. Ubiquitin-mediated mRNP dynamics and surveillance prior to budding yeast mRNA export. Genes Dev. 2010;24(17):1927–1938. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46.Chi B, Wang Q, Wu G, et al. Aly and THO are required for assembly of the human TREX complex and association of TREX components with the spliced mRNA. Nucleic Acids Res. 2013;41(2):1294–1306. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47.Viphakone N, Hautbergue GM, Walsh M, et al. TREX exposes the RNA-binding domain of Nxf1 to enable mRNA export. Nat Commun. 2012;3:1006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48.Rehwinkel J, Herold A, Gari K, et al. Genome-wide analysis of mRNAs regulated by the THO complex in Drosophila melanogaster. Nat Struct Mol Biol. 2004;11(6):558–566. [DOI] [PubMed] [Google Scholar]
- 49.Hautbergue GM, Hung ML, Walsh MJ, et al. UIF, a New mRNA export adaptor that works together with REF/ALY, requires FACT for recruitment to mRNA. Curr Biol. 2009;19(22):1918–1924. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50.Chang CT, Hautbergue GM, Walsh MJ, et al. Chtop is a component of the dynamic TREX mRNA export complex. EMBO J. 2013;32(3):473–486. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51.Abruzzi KC, Lacadie S, Rosbash M. Biochemical analysis of TREX complex recruitment to intronless and intron-containing yeast genes. EMBO J. 2004;23(13):2620–2631. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52.Lei HX, Zhai B, Yin SY, Gygi S, Reed R. Evidence that a consensus element found in naturally intronless mRNAs promotes mRNA export. Nucleic Acids Research. 2013;41(4):2517–2525. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53.Chi BK, Wang K, Du YH, et al. A Sub-Element in PRE enhances nuclear export of intronless mRNAs by recruiting the TREX complex via ZC3H18. Nucleic Acids Research. 2014;42(11):7305–7318. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54.Lander ES, Consortium IHGS, Linton LM, et al. Initial sequencing and analysis of the human genome. Nature. 2001;409(6822):860–921. [DOI] [PubMed] [Google Scholar]
- 55.Ares M, Grate L, Pauling MH. A handful of intron-containing genes produces the lion’s share of yeast mRNA. Rna-a Publication of the Rna Society. 1999;5(9):1138–1139. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56.Gonatopoulos-Pournatzis T, Cowling VH. Cap-binding complex (CBC). Biochem J. 2014;457(2):231–242. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57.Hackmann A, Wu H, Schneider UM, Meyer K, Jung K, Krebber H. Quality control of spliced mRNAs requires the shuttling SR proteins Gbp2 and Hrb1. Nat Commun. 2014;5:3123. [DOI] [PubMed] [Google Scholar]
- 58.Hurt E, Luo MJ, Rother S, Reed R, Strasser K. Cotranscriptional recruitment of the serine-arginine-rich (SR)-like proteins Gbp2 and Hrb1 to nascent mRNA via the TREX complex. Proc Natl Acad Sci U S A. 2004;101(7):1858–1862. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59.Johnson SA, Cubberley G, Bentley DL. Cotranscriptional recruitment of the mRNA export factor Yra1 by direct interaction with the 3’ end processing factor Pcf11. Mol Cell. 2009;33(2):215–226. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60.Johnson SA, Kim H, Erickson B, Bentley DL. The export factor Yra1 modulates mRNA 3 ‘ end processing. Nature Structural & Molecular Biology. 2011;18(10):1164–U1104. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61.Morris KJ, Corbett AH. The polyadenosine RNA-binding protein ZC3H14 interacts with the THO complex and coordinately regulates the processing of neuronal transcripts. Nucleic Acids Res. 2018;46(13):6561–6575. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 62.Strambio-De-Castillia C, Niepel M, Rout MP. The nuclear pore complex: bridging nuclear transport and gene regulation. Nat Rev Mol Cell Bio. 2010;11(7):490–501. [DOI] [PubMed] [Google Scholar]
- 63.Fischer T, Strasser K, Racz A, et al. The mRNA export machinery requires the novel Sac3p-Thp1p complex to dock at the nucleoplasmic entrance of the nuclear pores. Embo Journal. 2002;21(21):5843–5852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64.Fischer T, Rodriguez-Navarro S, Pereira G, Racz A, Schiebel E, Hurt E. Yeast centrin Cdc31 is linked to the nuclear mRNA export machinery. Nature Cell Biology. 2004;6(9):840–U844. [DOI] [PubMed] [Google Scholar]
- 65.Rodriguez-Navarro S, Fischer T, Luo MJ, et al. Sus1, a functional component of the SAGA histone acetylase complex and the nuclear pore-associated mRNA export machinery. Cell. 2004;116(1):75–86. [DOI] [PubMed] [Google Scholar]
- 66.Faza MB, Kemmler S, Jimeno S, et al. Sem1 is a functional component of the nuclear pore complex-associated messenger RNA export machinery. Journal of Cell Biology. 2009;184(6):833–846. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67.Cabal GG, Genovesio A, Rodriguez-Navarro S, et al. SAGA interacting factors confine sub-diffusion of transcribed genes to the nuclear envelope. Nature. 2006;441(7094):770–773. [DOI] [PubMed] [Google Scholar]
- 68.Schneider M, Hellerschmied D, Schubert T, et al. The Nuclear Pore-Associated TREX-2 Complex Employs Mediator to Regulate Gene Expression. Cell. 2015;162(5):1016–1028. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69.Akhtar A, Gasser SM. The nuclear envelope and transcriptional control. Nature Reviews Genetics. 2007;8(7):507–517. [DOI] [PubMed] [Google Scholar]
- 70.Dimitrova L, Valkov E, Aibara S, et al. Structural Characterization of the Chaetomium thermophilum TREX-2 Complex and its Interaction with the mRNA Nuclear Export Factor Mex67:Mtr2. Structure. 2015;23(7):1246–1257. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71.Ellisdon AM, Dimitrova L, Hurt E, Stewart M. Structural basis for the assembly and nucleic acid binding of the TREX-2 transcription-export complex. Nature Structural & Molecular Biology. 2012;19(3):328–U390. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72.Gordon JMB, Aibara S, Stewart M. Structure of the Sac3 RNA-binding M-region in the Saccharomyces cerevisiae TREX-2 complex. Nucleic Acids Research. 2017;45(9):5577–5585. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73.Jani DY, Lutz S, Marshall NJ, et al. Sus1, Cdc31, and the Sac3 CID Region Form a Conserved Interaction Platform that Promotes Nuclear Pore Association and mRNA Export. Molecular Cell. 2009;33(6):727–737. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74.Jani D, Valkov E, Stewart M. Structural basis for binding the TREX2 complex to nuclear pores, GAL1 localisation and mRNA export. Nucleic Acids Research. 2014;42(10):6686–6697. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75.Galy V, Gadal O, Fromont-Racine M, Romano A, Jacquier A, Nehrbass U. Nuclear retention of unspliced mRNAs in yeast is mediated by perinuclear Mlp1. Cell. 2004;116(1):63–73. [DOI] [PubMed] [Google Scholar]
- 76.Soucek S, Corbett AH, Fasken MB. The long and the short of it: The role of the zinc finger polyadenosine RNA binding protein, Nab2, in control of poly(A) tail length. Bba-Gene Regul Mech. 2012;1819(6):546–554. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77.Grant RP, Marshall NJ, Yang JC, et al. Structure of the N-terminal Mlp1-binding domain of the Saccharomyces cerevisiae mRNA-binding protein, Nab2. Journal of Molecular Biology. 2008;376(4):1048–1059. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78.Jani D, Lutz S, Hurt E, Laskey RA, Stewart M, Wickramasinghe VO. Functional and structural characterization of the mammalian TREX-2 complex that links transcription with nuclear messenger RNA export. Nucleic Acids Research. 2012;40(10):4562–4573. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79.Fraser P, Bickmore W. Nuclear organization of the genome and the potential for gene regulation. Nature. 2007;447(7143):413–417. [DOI] [PubMed] [Google Scholar]
- 80.Wickramasinghe VO, McMurtrie PIA, Mills AD, et al. mRNA Export from Mammalian Cell Nuclei Is Dependent on GANP. Current Biology. 2010;20(1):25–31. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81.Coyle JH, Bor YC, Rekosh D, Hammarskjold ML. The Tpr protein regulates export of mRNAs with retained introns that traffic through the Nxf1 pathway. Rna-a Publication of the Rna Society. 2011;17(7):1344–1356. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82.Kim SJ, Fernandez-Martinez J, Nudelman I, et al. Integrative structure and functional anatomy of a nuclear pore complex. Nature. 2018;555(7697):475–482. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 83.Adams RL, Terry LJ, Wente SR. Nucleoporin FG Domains Facilitate mRNP Remodeling at the Cytoplasmic Face of the Nuclear Pore Complex. Genetics. 2014;197(4):1213–U1277. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84.Radu A, Moore MS, Blobel G. The peptide repeat domain of nucleoporin Nup98 functions as a docking site in transport across the nuclear pore complex. Cell. 1995;81(2):215–222. [DOI] [PubMed] [Google Scholar]
- 85.Fernandez-Martinez J, Kim SJ, Shi Y, et al. Structure and Function of the Nuclear Pore Complex Cytoplasmic mRNA Export Platform. Cell. 2016;167(5):1215–1228 e1225. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86.Adams RL, Mason AC, Glass L, Aditi, Wente SR. Nup42 and IP6 coordinate Gle1 stimulation of Dbp5/DDX19B for mRNA export in yeast and human cells Traffic. 2018;18(12):776–790. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87.Lin DH, Correia AR, Cai SW, Huber FM, Jette CA, Hoelz A. Structural and functional analysis of mRNA export regulation by the nuclear pore complex. Nature Communications. 2018;9. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88.Collins R, Karlberg T, Lehtio L, et al. The DEXD/H-box RNA helicase DDX19 is regulated by an {alpha}-helical switch. J Biol Chem. 2009;284(16):10296–10300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 89.York JD, Odom AR, Murphy R, Ives EB, Wente SR. A phospholipase C-dependent inositol polyphosphate kinase pathway required for efficient messenger RNA export. Science. 1999;285(5424):96–100. [DOI] [PubMed] [Google Scholar]
- 90.Gross T, Siepmann A, Sturm D, et al. The DEAD-box RNA helicase Dbp5 functions in translation termination. Science. 2007;315(5812):646–649. [DOI] [PubMed] [Google Scholar]
- 91.Bolger TA, Folkmann AW, Tran EJ, Wente SR. The mRNA export factor Gle1 and inositol hexakisphosphate regulate distinct stages of translation. Cell. 2008;134(4):624–633. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92.Bolger TA, Wente SR. Gle1 Is a Multifunctional DEAD-box Protein Regulator That Modulates Ded1 in Translation Initiation. Journal of Biological Chemistry. 2011;286(46):39750–39759. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93.Schutz P, Bumann M, Oberholzer AE, et al. Crystal structure of the yeast eIF4A-eIF4G complex: An RNA-helicase controlled by protein-protein interactions. P Natl Acad Sci USA. 2008;105(28):9564–9569. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94.Mathys H, Basquin J, Ozgur S, et al. Structural and Biochemical Insights to the Role of the CCR4-NOT Complex and DDX6 ATPase in MicroRNA Repression. Molecular Cell. 2014;54(5):751–765. [DOI] [PubMed] [Google Scholar]
- 95.Saavedra CA, Hammell CM, Heath CV, Cole CN. Yeast heat shock mRNAs are exported through a distinct pathway defined by Rip1p. Gene Dev. 1997;11(21):2845–2856. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 96.Kendirgi F, Rexer DJ, Alcazar-Roman AR, Onishko HM, Wente SR. Interaction between the shuttling mRNA export factor Gle1 and the nucleoporin hCG1: A conserved mechanism in the export of Hsp70 mRNA. Molecular Biology of the Cell. 2005;16(9):4304–4315. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 97.Stutz F, Kantor J, Zhang D, McCarthy T, Neville M, Rosbash M. The yeast nucleoporin Rip1p contributes to multiple export pathways with no essential role for its FG-repeat region. Gene Dev. 1997;11(21):2857–2868. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98.Andreou AZ, Klostermeier D. The DEAD-box helicase eIF4A Paradigm or the odd one out? Rna Biology. 2013;10(1):19–32. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99.Noble KN, Tran EJ, Alcazar-Roman AR, Hodge CA, Cole CN, Wente SR. The Dbp5 cycle at the nuclear pore complex during mRNA export II: nucleotide cycling and mRNP remodeling by Dbp5 are controlled by Nup159 and Gle1. Gene Dev. 2011;25(10):1065–1077. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 100.von Moeller H, Basquin C, Conti E. The mRNA export protein DBP5 binds RNA and the cytoplasmic nucleoporin NUP214 in a mutually exclusive manner. Nature Structural & Molecular Biology. 2009;16(3):247–254. [DOI] [PubMed] [Google Scholar]
- 101.Napetschnig J, Kassube SA, Debler EW, Wong RW, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup214 and the DEAD-box helicase Ddx19. P Natl Acad Sci USA. 2009;106(9):3089–3094. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102.Tran EJ, Zhou Y, Corbett AH, Wente SR. The DEAD-box protein dbp5 controls mRNA export by triggering specific RNA: Protein remodeling events. Molecular Cell. 2007;28(5):850–859. [DOI] [PubMed] [Google Scholar]
- 103.Anderson JT, Wilson SM, Datar KV, Swanson MS. Nab2 - a Yeast Nuclear Polyadenylated Rna-Binding Protein Essential for Cell Viability. Molecular and Cellular Biology. 1993;13(5):2730–2741. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104.Green DM, Marfatia KA, Crafton EB, Zhang X, Cheng XD, Corbett AH. Nab2p is required for poly(A) RNA export in Saccharomyces cerevisiae and is regulated by arginine methylation via Hmt1p. Journal of Biological Chemistry. 2002;277(10):7752–7760. [DOI] [PubMed] [Google Scholar]
- 105.Windgassen M, Sturm D, Cajigas IJ, et al. Yeast shuttling SR proteins Npl3p, Gbp2p, and Hrb1p are part of the translating mRNPs, and Npl3p can function as a translational repressor. Molecular and Cellular Biology. 2004;24(23):10479–10491. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 106.Powers MA, Forbes DJ, Dahlberg JE, Lund E. The vertebrate GLFG nucleoporin, Nup98, is an essential component of multiple RNA export pathways. Journal of Cell Biology. 1997;136(2):241–250. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107.Blevins MB, Smith AM, Phillips EM, Powers MA. Complex formation among the RNA export proteins Nup98, Rae1/Gle2, and TAP. Journal of Biological Chemistry. 2003;278(23):20979–20988. [DOI] [PubMed] [Google Scholar]
- 108.Iovine MK, Watkins JL, Wente SR. The GLFG repetitive region of the nucleoporin Nup116p interacts with Kap95p, an essential yeast nuclear import factor. J Cell Biol. 1995;131(6 Pt 2):1699–1713. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109.Strawn LA, Shen TX, Wente SR. The GLFG regions of Nup116p and Nup100p serve as binding sites for both Kap95p and Mex67p at the nuclear pore complex. Journal of Biological Chemistry. 2001;276(9):6445–6452. [DOI] [PubMed] [Google Scholar]
- 110.Kraemer D, Blobel G. mRNA binding protein mrnp 41 localizes to both nucleus and cytoplasm. P Natl Acad Sci USA. 1997;94(17):9119–9124. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111.Ren Y, Seo HS, Blobel G, Hoelz A. Structural and functional analysis of the interaction between the nucleoporin Nup98 and the mRNA export factor Rae1. Proc Natl Acad Sci U S A. 2010;107(23):10406–10411. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 112.Bailer SM, Siniossoglou S, Podtelejnikov A, Hellwig A, Mann M, Hurt E. Nup116p and Nup100p are interchangeable through a conserved motif which constitutes a docking site for the mRNA transport factor Gle2p. Embo Journal. 1998;17(4):1107–1119. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113.Ho AK, Raczniak GA, Ives EB, Wente SR. The integral membrane protein Snl1p is genetically linked to yeast nuclear pore complex function. Molecular Biology of the Cell. 1998;9(2):355–373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114.Murphy R, Watkins JL, Wente SR. GLE2, a Saccharomyces cerevisiae homologue of the Schizosaccharomyces pombe export factor RAE1, is required for nuclear pore complex structure and function. Molecular Biology of the Cell. 1996;7(12):1921–1937. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115.Faria PA, Chakraborty P, Levay A, et al. VSV disrupts the Rae1/mrnp41 mRNA nuclear export pathway. Mol Cell. 2005;17(1):93–102. [DOI] [PubMed] [Google Scholar]
- 116.Quan B, Seo HS, Blobel G, Ren Y. Vesiculoviral matrix (M) protein occupies nucleic acid binding site at nucleoporin pair (Rae1 * Nup98). Proc Natl Acad Sci U S A. 2014;111(25):9127–9132. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 117.Glisovic T, Bachorik JL, Yong J, Dreyfuss G. RNA-binding proteins and post-transcriptional gene regulation. FEBS Lett. 2008;582(14):1977–1986. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118.Bretes H, Rouviere JO, Leger T, et al. Sumoylation of the THO complex regulates the biogenesis of a subset of mRNPs. Nucleic Acids Research. 2014;42(8):5043–5058. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119.Wickramasinghe VO, Laskey RA. Control of mammalian gene expression by selective mRNA export. Nat Rev Mol Cell Biol. 2015;16(7):431–442. [DOI] [PubMed] [Google Scholar]
- 120.Wickramasinghe VO, Savill JM, Chavali S, et al. Human inositol polyphosphate multikinase regulates transcript-selective nuclear mRNA export to preserve genome integrity. Mol Cell. 2013;51(6):737–750. [DOI] [PubMed] [Google Scholar]
- 121.Culjkovic B, Topisirovic I, Skrabanek L, Ruiz-Gutierrez M, Borden KLB. eIF4E is a central node of an RNA regulon that governs cellular proliferation. Journal of Cell Biology. 2006;175(3):415–426. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 122.Wang L, Miao YL, Zheng XF, et al. The THO Complex Regulates Pluripotency Gene mRNA Export and Controls Embryonic Stem Cell Self-Renewal and Somatic Cell Reprogramming. Cell Stem Cell. 2013;13(6):676–690. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 123.Nousiainen HO, Kestila M, Pakkasjarvi N, et al. Mutations in mRNA export mediator GLE1 result in a fetal motoneuron disease. Nat Genet. 2008;40(2):155–157. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124.Folkmann AW, Collier SE, Zhan XY, Ohi MD Aditi, Wente SR. Gle1 Functions during mRNA Export in an Oligomeric Complex that Is Altered in Human Disease. Cell. 2013;155(3):582–593. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 125.Folkmann AW, Dawson TR, Wente SR. Insights into mRNA export-linked molecular mechanisms of human disease through a Gle1 structure-function analysis. Adv Biol Regul. 2014;54:74–91. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 126.Kaneb HM, Folkmann AW, Belzil VV, et al. Deleterious mutations in the essential mRNA metabolism factor, hGle1, in amyotrophic lateral sclerosis. Hum Mol Genet. 2015;24(5):1363–1373. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 127.Culjkovic-Kraljacic B, Borden KL. Aiding and abetting cancer: mRNA export and the nuclear pore. Trends Cell Biol. 2013;23(7):328–335. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 128.Kuss SK, Mata MA, Zhang L, Fontoura BM. Nuclear imprisonment: viral strategies to arrest host mRNA nuclear export. Viruses. 2013;5(7):1824–1849. [DOI] [PMC free article] [PubMed] [Google Scholar]


