Abstract
Significance: Elucidation of the central importance of mitophagy in homeostasis of cells and organisms emphasizes that mitochondrial functions extend far beyond short-term needs for energy production. In mitochondria systems biology, the mitochondrial genome, proteome, and metabolome operate as a functional network in coordination of cell activities. Organization occurs through subnetworks that are interconnected by membrane potential, transport activities, allosteric and cooperative interactions, redox signaling mechanisms, rheostatic control by post-translational modifications, and metal ion homeostasis. These subnetworks enable use of varied energy precursors, defense against environmental stressors, and macromolecular rewiring to titrate energy production, biosynthesis, and detoxification according to cell-specific needs. Rewiring mechanisms, termed mitochondrial reprogramming, enhance fitness to respond to metabolic resources and challenges from the environment. Maladaptive responses can cause cell death. Maladaptive rewiring can cause disease. In cancer, adaptive rewiring can interfere with effective treatment.
Recent Advances: Many recent advances have been facilitated by the development of new omics tools, which create opportunities to use data-driven analysis of omics data to address these complex adaptive and maladaptive mechanisms of mitochondrial reprogramming in human disease.
Critical Issues: Application of omics integration to model systems reveals a critical role for metal ion homeostasis broadly impacting mitochondrial reprogramming. Importantly, data show that trans-omics associations are more robust and biologically relevant than single omics associations.
Future Directions: Application of omics integration to mitophagy research creates new opportunities to link the complex, interactive functions of mitochondrial form and function in mitochondria systems biology.
Keywords: mitochondria, omics, systems biology, biological network, integration
What is cause, what is effect,
If the effect causes the cause to change?
Where effect and cause, and cause and effect,
Are entangled in a complex web,
The mind is lost
And ponders in vain
The principal cause of all.
—Eberhard Voit
Introduction
Mitochondria are directly or indirectly critical for all cellular functions in aerobic eukaryotes. As the so-called powerhouse of the cell, mitochondrial function as the central organizational hub of metabolism is often overshadowed by their primary role in oxidative phosphorylation (OXPHOS) and ATP production (156). Their roles in metabolism go far beyond, however, and include the tricarboxylic acid (TCA) cycle and fatty acid oxidation systems (94) to support NAD supply for the electron transport chain (ETC), as well as amino acid catabolism and anabolism (31, 126), nitrogen balance and elimination (179), biosynthesis of heme (139, 147), steroid hormones (114), pyrimidines and purines (46), and other pathways, and control of Ca2+ (54) and other critical metal ion homeostasis (132).
Mitochondria also contain components of enzyme system and metabolic intermediates to support mitochondrial replication (174), transcription (45), translation (124), and post-translational modifications (16, 27), which are essential to adaptation to environmental resources and challenges. Knowledge of mitochondria is largely built by the vast efforts and discoveries of the many great scientists outlined in the text of Nelson and Cox (121a). The research to isolate and study each component to understand the whole, termed Cartesian reductionism, has provided extensive knowledge regarding mitochondrial bioenergetics, metabolism, and regulation, and has effectively defined mitochondrial components and functions.
This complex and dynamic nature of mitochondria is being increasingly recognized through studies demonstrating dynamic and branched structures that continuously undergo fission and fusion (115), communicating with the nucleus (144) and other compartments (176) to adjust their number, form, and function to cellular needs. Omics methods have enabled new approaches to study the functional network of mitochondria and their adaptive (as well as maladaptive) response to nutrient availability, physiologic needs, environmental challenges, and aging (55, 187). Whole genome, exome sequencing in large cohorts of patients has expedited the discovery of mitochondrial disease genes (135).
This detailed information is complemented by high-resolution mass spectrometry (MS) and metabolic pathway analysis to provide global metabolic phenotyping through high-resolution metabolomics (60, 61). Despite the challenges of varied coverage and scalability in the available omics profiles, integration of high-resolution metabolomics with other omics technologies [epigenomics (25), transcriptomics (93), proteomics (23, 160, 171), interactomics (103)] has started to reveal the central organizational structures of mitochondria functional networks derived from the complex data with multiple layers of interaction and regulation.
In this article, we review applications of integrative omics to advance understanding of mitochondrial systems in metabolic organization and cell functions. The complex metabolic and signaling networks reiterate the concept outlined by Loscalzo et al. (106) that human pathobiology involves complex systems biology that cannot be classified by simple cause–effect mechanisms. Metabolic reprogramming of mitochondria in many cell states shows major reorganization of activities to support cell demands (26, 40, 65), and new bioinformatics tools have been advancing understanding in this area.
In our review, we use pathways to refer to linear and branched paths for transfer of molecules or information. These pathways have largely been defined by using reductionist approaches and are described in textbooks and knowledge bases. Proof often relies on isolation of the components and reconstitution of the pathway. In biologic systems, however, components of pathways are interconnected with other biological elements, sometimes in extensive and complex networks; such interconnections may be very important in control of function and be lost or not represented by isolation and reconstitution of a pathway.
For data-driven omics integration analyses discussed later, we operationally define networks in terms of associations present among measured parameters of omics data sets. In this, biologic network information can be confounded by associations introduced by sampling, analytic, and bioinformatic methods (166). Thus, optimal use of data-driven approaches requires use of biology knowledge bases and experimental validation (41). Biological pathway analysis software (30, 85, 102) allows understanding omics data in the form of pathway diagrams structured on prior knowledge, and network analysis software (110, 165, 197) utilizes graphical methods to represent complex connections among diverse types of cellular components such as genes, proteins, and metabolites (10).
Data-driven analysis of multiple types of omics data and use of network analysis tools to integrate multiple omics layers with independent variables and phenotypic measures allows visualization of the hierarchy of network structures (41, 48, 55). Further mining of the network structure with systematic experimental challenges in model systems provides insight into adaptive and maladaptive responses. Such effects are illustrated by transcriptome and metallome integration with the metabolome, showing connectivity and organization of nuclear transcription with mitochondrial transport systems in cellular responses to mitochondria-mediated metal toxicity. Combined redox proteomics and metabolomics further clarifies underlying mechanisms using an integrative approach. Thus, these recently available data-driven integrated omics approaches provide powerful methods and create important new opportunities for development of mitochondria systems biology.
Mitochondrial Functional Network in Metabolism
Mitochondria have a central role in metabolism. Although containing only about 10% of the proteins encoded by the nuclear genome, the mitochondrial metabolome includes representative metabolites of up to 90% of the whole cellular metabolome (61). This is shown by high-resolution metabolomics of isolated mitochondria that captures intermediates from 136 out of 154 mammalian metabolic pathways (141). Early mitochondrial metabolomics research has mainly focused on the TCA cycle and energy metabolism, but mitochondria contain metabolic precursors and intermediates for mitochondrial replication, transcription, translation, and post-translational modifications (154), as well as ancillary systems that assist in amino acid, peptide, and RNA modification (70, 122).
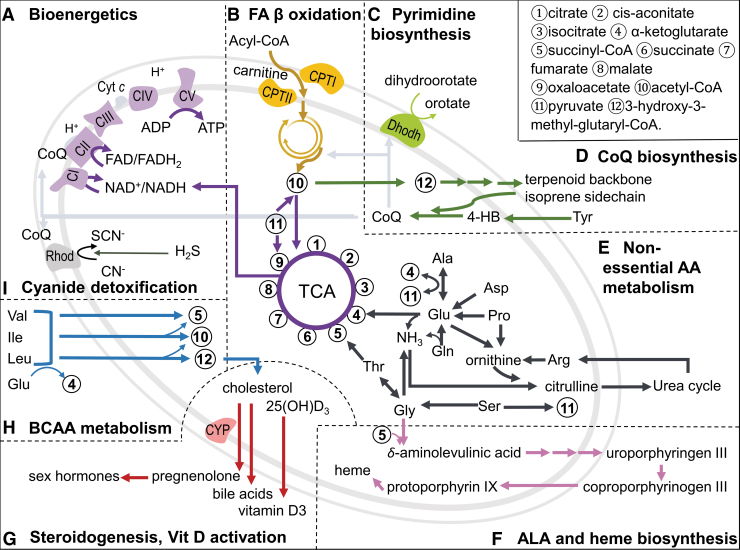
The OXPHOS system is the center of bioenergetics (Fig. 1A), consisting of five multimeric enzymes (complexes I–V) in the inner mitochondrial membrane (IMM), and two mobile electron carriers, coenzyme Q10 (CoQ, ubiquinone) and cytochrome c (cyt c). Complexes I–IV constitute the ETC, and during the respiration, electron flow within these transmembrane complexes creates a proton gradient. This proton-motive force is critical for F1Fo-ATP synthase (complex V) to phosphorylate ADP to ATP, the major energy currency of the cell (68). Each of these complexes is composed of multiple subunit proteins, some at variable stoichiometries and many with regulatory activities (151). Some of the subunits produce the superoxide anion radical, which also dismutates to hydrogen peroxide and functions in redox signaling. A spectrum of metabolic pathways also supports the key bioenergetic function, and mitochondria also includes a broad array of signaling systems to integrate mitochondrial function with other cell functions (Fig. 1).
FIG. 1.
Diversity of mitochondrial metabolism. Mitochondria contain a remarkable array of metabolic functions that converge to key intermediates that are coupled to the TCA cycle and support bioenergetics, anabolic pathways, and detoxification (A–I). Metabolomics methods support measurement of metabolites in most pathways, enabling network analysis of mitochondrial function by coupling with other omics and phenotypic measures. 4-HB, 4-hydroxybenzoaldehyde; ALA, δ-aminolevulinic acid; CI through CV, complex I through V; CoQ, ubiquinone, coenzyme Q10; CPT, carnitine palmitoyltransferase; CYP, cytochrome P450; cyt c, cytochrome c; Dhodh, dihydroorotate dehydrogenase; FA, fatty acid; Rhod, rhodanese; TCA, tricarboxylic acid.
All of the major nutritional fuels (fat, carbohydrate, protein) provide oxidizable precursors for ATP production through interaction with the TCA cycle to maintain NADH supply (Fig. 1, center). Fatty acid oxidation (Fig. 1B) is especially important because of the high ATP production per gram of fat, and some cell types rely extensively on fatty acid oxidation (15, 32, 154). Fatty acid oxidation must be optimized in the context of carbohydrate and amino acid oxidation and integrated with peroxisomal fatty acid metabolism. In addition, mitochondria supply precursors for both lipid biosynthesis in the cytoplasm and a less characterized mitochondrial fatty acid pathway (71). The import of fatty acids via the carnitine shuttle is tightly regulated and deranged in many disease processes (11, 92). Long-term incorporation and turnover of lipids in cardiolipin is important for mitochondrial structure and function but their role in mitochondrial reprogramming has not been fully elucidated.
A complex array of changes in free fatty acids and other intermediates such as acyl-coenzyme A (CoAs) and acyl-carnitines have been detected in mitochondria, cells, and plasma, and new integrative methods are available to mechanistically link these modulations to changes in gene expression and protein abundance and activities (2, 73, 74). For instance, monounsaturated fatty acid and saturated fatty acid cause differential effects in steatosis versus apoptosis through distinct activation of transcription factors (138). Along similar lines, by complementing the layers of sequence variant, targeted proteomics and metabolomics, and phenotype data, Wu et al. characterized metabolome, transcriptome, and proteome from 40 strains of the BXD mouse genetic reference population, and they identified molecules associated with diabetes status in both mice and humans (187). These studies demonstrate the value of an integrated multilayered omics method that advances our understanding of complex biological systems and molecular mechanisms.
Anabolic systems in mitochondria are essential for biosynthesis of key molecules to maintain nucleotides for nucleic acid synthesis and turnover (46). These include steps for de novo pyrimidine and purine biosynthesis (Fig. 1C). Biosynthesis of isoprene and related terpenoid products, such as cholesterol, CoQ, and squalene precursors for vitamin D, depend on acetyl-CoA produced by mitochondria (Fig. 1D). For CoQ biosynthesis, key steps for tail polymerization, tail attachment to 4-hydroxybenzaldehyde and head group modifications, occur within mitochondria (162). These reactions are critical because CoQ is a cofactor for OXPHOS, fatty acid oxidation, mitochondrial uncoupling proteins, cyanide detoxification, and pyrimidine biosynthesis.
Central pathways balancing the use of lipids with other fuel sources for NADH supply, as well as integrating these needs with biosynthesis of nucleotides, NADPH and anabolic and detoxification needs, occur through intermediates linked to the TCA cycle (Fig. 1E). For example, α-ketoglutarate (α-KG) is oxidized in the TCA cycle to support NADH while also being a precursor to maintain the intracellular pool of nonessential amino acids used in the synthesis of proteins and nucleotides (40). The transamination metabolite of α-KG, glutamate, is interconverted to glutamine (the most abundant amino acids in mammals), which connects to many metabolic processes, including nitrogen balance (69, 190). These intermediates are measured by metabolomics, and extensive information is available from both basic and applied research (53, 66).
Other pathways in mitochondria include porphyrin and heme biosynthesis (Fig. 1F) (139, 147), and pathways in coenzyme metabolism, such as lipoic acid biosynthesis (152) and assembly of iron–sulfur clusters (139). Mitochondria also contain critical cytochrome P450 (CYP) enzymes (Fig. 1G), functioning in catabolism of cholesterol for the synthesis of pregnenolone, the precursor for all sex hormones, mineralocorticoids and glucocorticoids (114), as well as synthesis of bile acid precursors and activation of vitamin D (22). Considerable information is available from targeted studies of each of these pathways, especially for related disease mechanisms, but functional integration into holistic models for personalized health is lacking.
Other mitochondrial pathways include branched-chain amino acid (BCAA) metabolism (Fig. 1H), which occurs through branched-chain fatty acid intermediates (36) and is essential to prevent neurotoxicity from BCAA (35). Accumulation of these intermediates is important in metabolic disorders, including type 2 diabetes (112), and mounting evidence suggests that aberrations in related acyl-carnitines are sensitive and mechanistic markers for mitochondrial dysfunction (92). Mitochondria contain detoxification systems, including rhodanese (thiosulfate sulfurtransferase) for cyanide detoxification (6) (Fig. 1I), aldehyde dehydrogenases, glutathione (GSH) peroxidases, and GSH S-transferases (120, 148). Mitochondria also have a critical function in maintaining ionic homeostasis by protecting against cytoplasmic Ca2+ overload (140).
Mitochondrial Communication with Cytoplasmic Components
Research in the past two decades has shown that mitochondria communicate with the rest of cellular components and serve as a signaling organelle beyond their metabolic functions. Due to the impermeability of the IMM gated by specific ion channels, mitochondrial signaling is often mediated by changes of metabolite levels and ion flow (e.g., Ca2+) or structural changes of the organelle itself such as fission and fusion. Signaling systems between mitochondria and the nucleus are distinguished as anterograde signaling (nucleus to mitochondria) and retrograde signaling (mitochondria to nucleus), together comprising a bi-directional communication between the two compartments (104). Because bidirectional communication can occur simultaneously in response to exogenous stressors, simple cause–effect relationships may not occur. Despite this caveat, targeted studies of anterograde and retrograde signaling have defined key systems controlling nuclear regulation of mitochondrial biogenesis as well as integration of mitochondrial metabolism with extramitochondrial functions. For simplicity in the following consideration of mitochondria-cell signaling, unless otherwise designated, we use cytoplasm globally to denote non-mitochondrial components of the cell.
The anterograde regulation of mitochondrial biogenesis by the nuclear genome has been extensively studied and covered in recent reviews (43, 62, 149). A well-studied example is the peroxisome-proliferator-activated receptor coactivator-1 (PGC-1) as a universal regulatory system for mitochondrial DNA replication and transcription as well as nuclear gene expression for synthesis of mitochondrial proteins (144). PGC-1α is a transcriptional co-activator of many transcription factors that target mitochondrial biology, including nuclear respiratory factor 1 (NRF1) activating mitochondrial DNA replication and transcription, nuclear respiratory factor 2 (NRF2; GA binding protein) regulating cyt c oxidase assembly, peroxisome proliferator-activated receptors (PPARs) regulating fatty acid oxidation, and many more (86). Cellular PGC-1α level is closely correlated with the number of mitochondria, and both are highly inducible in specific tissues (e.g., skeletal muscle, brown adipose tissue) in response to environmental cues (189).
One must bear in mind that mitochondria are also highly variable in characteristics that are not described by change in mitochondrial number. For example, the mitochondrial forkhead box protein O1 (FoxO1) can respond to starvation mediated by mitochondrial phosphatase PTPMT1 (protein tyrosine phosphatase mitochondrial 1) (100); translocation of cyclin B1 to mitochondria is similarly redox regulated in radiation-induced oxidative stress (83). Such fine-tuning of mitochondrial characteristics requires a dynamic mitochondria-driven retrograde signaling to adjust and integrate the mitochondrial function with cytoplasmic components at different regulation levels.
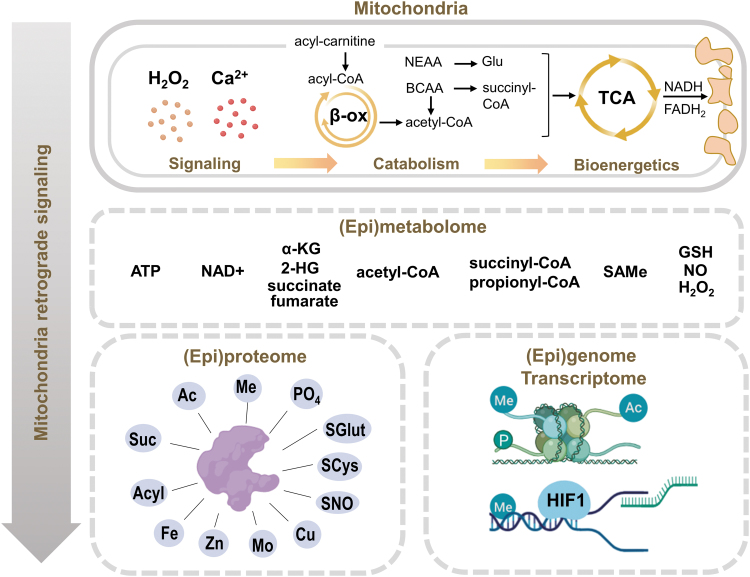
Mitochondria have a major role in controlling two universal signaling molecules for cellular homeostasis, H2O2 and Ca2+, both of which are tightly maintained in the cytoplasm at low nanomolar concentrations (Fig. 2, top). As a catabolic powerhouse, mitochondria also export a panel of metabolic components with relatively high potential energies and flux to couple bioenergetics and mitochondrial metabolism to the cellular macromolecular organization and function, that is, genome, transcriptome, and proteome (Fig. 2, bottom). These include ATP, acetyl-CoA, NAD(H), S-adenosyl-l-methionine (SAMe), TCA intermediates (α-KG, citrate, fumarate, succinate), and acyl-CoAs (Fig. 2, middle) (25, 79).
FIG. 2.
Mitochondrial retrograde signaling regulates upstream controllers of biologic activities. Mitochondrial catabolism and bioenergetics are controlled by universal signaling molecules, which, in turn, generate a panel of key metabolic intermediates (epimetabolome). These intermediates support control of activity and function through covalent modification of the proteome (epiproteome) and chromatin changes that control gene expression other than alteration of gene code (epigenome). Through the metabolome, cellular bioenergetics is thus coupled with structural and functional organization of macromolecules dictating the cell function and fate. In this sense, mitochondria are a mediator translating the external signal into intracellular functional changes to adjust the cells based on the environment. 2-HG, 2-hydroxyglutarate; α-KG, α-ketoglutarate; β-ox, β-oxidation; Ac, acetylation; BCAA, branched-chain amino acids; GSH, glutathione; HIF-1, hypoxia-inducible factor-1; Me, methylation; NEAA, non-essential amino acids; PO4, phosphorylation; SAMe, S-adenosyl-l-methionine; Suc, succinylation.
Physiologic fluctuation of these molecules provides efficient signaling by supporting covalent modifications of the epiproteome [defined as covalent modifications to provide function and regulation beyond that of the translated sequence of amino acids (56)] and epigenome (defined as chromatin changes causing modification of gene expression without altering the genetic code itself) to control proteome and genome structure and organization and regulate kinetics of enzymes, transporters, and receptors, ultimately controlling transcription, replication, and repair (76, 89, 107). A conserved series of molecule motifs from mitochondrial activity is suggested as the signature of mitochondrial damage in dysregulated processes, termed as damage-associated molecular patterns (DAMPs) (196).
An important principle of mitochondria being a redox regulation hub is provided in the “redox code,” where cellular bioenergetics and metabolism are organized through high-flux, NAD, and NADP systems (80). The NAD system supports catabolic reactions using nutrient fuels and oxygen for ATP generation, whereas the NADP system functions in biosynthetic processes (anabolism), defense, cell signaling, and energy storage. The NAD system uses powerful dehydrogenases and catalyzes reactions transferring two electrons to biomolecules with covalent bonds of C, O, S, N, and H (80). Many NAD+-coupled enzyme-catalyzed reactions are included in the Enzyme Commission Class 1 enzymes, including pyruvate dehydrogenase, isocitrate dehydrogenase, α-ketoglutarate dehydrogenase (αKGDH), and malate dehydrogenase from the citric acid cycle; 2-hydroxyfatty acyl-CoA dehydrogenase from β-oxidation of fatty acids; and glyceraldehyde 3-phosphate dehydrogenase and lactate dehydrogenase from the glycolytic pathway. Thus, the NADH/NAD+ (reduced/oxidized) couple is at a stable steady state redox potential maintained by a large number of oxidizable substrates that reduce NAD+ to NADH (80).
Mitochondrial-localized NAD is also used for protein post-translational modification, such as protein mono- or poly(ADP ribosyl)ation, adenylylation, or deacetylation (42). Other compartments rely on mitochondria to re-oxidize NADH to NAD+ to sustain function such as cytosolic glycolysis and peroxisomal fatty acid oxidation. Thus, the steady-state potential of the NAD system serves as an integrative hub for diverse biologic processes (80, 158), ultimately maintaining the bioenergetic conditions that are essential to supply the precursors for the integrated array of epiproteomic modifications needed to coordinate complex cell functions (Fig. 2).
Epimetabolome
In mitochondria systems biology, the group of high-energy intermediates that serve to maintain the epiproteome and epigenome can be considered as components of an “epi-metabolome,” or epimetabolome, defined here as central hub metabolites of critical organizational importance because they have broad, integrative roles in downstream protein, genetic, and hormonal control of metabolism. ATP, acetyl-CoA, NAD+/NADH, SAMe, TCA intermediates (α-KG, citrate, fumarate, succinate), their derivatives (2-hydroxyglutarate [2-HG]), and acyl-CoAs have such roles in post-translational modifications of proteins (Fig. 2, middle). Although considerable cross-talk between systems is known, integrated analyses are only beginning to become available.
Systems involved in mitochondrial redox regulation provide examples of such integrated functions linked to the proteome. The second principle of the redox code is that kinetically controlled sulfur switches of cysteine residues are linked to bioenergetics and metabolism through H2O2 production by mitochondria and by NADPH oxidases (28, 98). Mitochondrial generation of H2O2 has been well documented and linked to physiologic signaling (56, 57) and also to toxicologic responses. Importantly, the redox proteome network includes oxidation, glutathionylation, nitrosylation, and a range of other protein modifications (56), which are linked to the broader spectrum of epiproteome modifications, including phosphorylation, methylation. acetylation, succinylation, and other acylations from acyl-CoA precursors (Fig. 2). Integrated models of the epimetabolome and epiproteome are needed to enhance understanding of mitochondria systems biology.
This integrated view of retrograde signaling extends throughout the cell. For example, mitochondria control the levels of key signaling molecules (epimetabolome) that regulate and feed to chromatin-modifying enzymes. An emerging field studies the role of mitochondrial genome and metabolism in shaping epigenetic modulation of nuclear gene expression. ATP participates in chromatin modification through phosphorylation of histone tails. ATP depletion increases AMP:ATP ratio, leading to activation of AMP-activated kinase (AMPK), a principal cellular bioenergetics and nutrient sensor (198). The consequent alteration in the NAD+/NADH ratio activates sirtuins, a major class of deacetylases.
N-Acetylation neutralizes the lysine residues on the histone tails and releases DNA for transcription factor binding (25). This process relies on acetyl-CoA pool from mitochondria-derived citrate. Similarly, mitochondria-derived serine is important in the methionine cycle that maintains SAMe as a substrate for histone and DNA methyl transferases (108). In opposition to methylation, the dioxygenases responsible for demethylation of DNA and histones are regulated by TCA cycle intermediates α-KG, succinate, and fumarate (191). A related metabolite, 2-HG, is a competitive inhibitor of α-KG-dependent dioxygenases (192). Integrative maps of other histone modifications, such as propionylation, butyrylation, crotonylation, hydroxy-isobutyrylation, and succinylation (144), will improve understanding of these modifications in disease pathobiology.
The TCA cycle intermediates also regulate nuclear transcription factor activity. An example is hypoxia-inducible factor-1 (HIF-1) mediating the physiological response to hypoxia (Fig. 2, bottom). Two key regulators of HIF-1, Egl-Nine (EGLN, also called PHD) prolyl hydroxylase and HIF-1AN (hypoxia-inducible factor 1, alpha subunit inhibitor; also called FIH) use the TCA cycle intermediates α-KG as a co-substrate to catalyze Fe(II)-dependent hydroxylation, which can be inhibited by succinate, fumarate, and 2-HG (90, 193). Metabolic reprogramming, including adaptive changes in the physiology and metabolism of an organism in response to a nutritional stress/stimulus (127, 131), plays an important role in development, cell proliferation, and cancer.
Mitochondrial interaction with cytoplasmic components is important in maintaining Ca2+ regulation that ultimately determines cell activity and fate. Close interactions between the endoplasmic reticulum (ER) and mitochondria are essential for rapid and sustained Ca2+ signaling at nanomolar concentrations despite extracellular Ca2+ at millimolar concentrations. The structural interface between the ER and mitochondria, the so-called mitochondria-associated membrane (MAM), is enriched with mitofusin (Mfn), a component of the mitochondrial fusion and fission machinery, which helps tether ER and mitochondria to a close distance of about 50 nm (140). Maintenance of this interface is critical for compartmentalization of cellular metabolism and signaling processes, as discussed next.
Mitochondrial Metallome
Mitochondrial function in metal ion homeostasis extends far beyond maintenance of Ca2+ balance. Mitochondria contain a dynamic network of transporters (Fig. 3) with complex and incompletely understood selectivity and interactions. Considerable information is available for individual metal ions, such as iron, but the dynamic metallomic interactions under different pathophysiologic conditions have not yet been addressed by using an integrated systems biology approach.
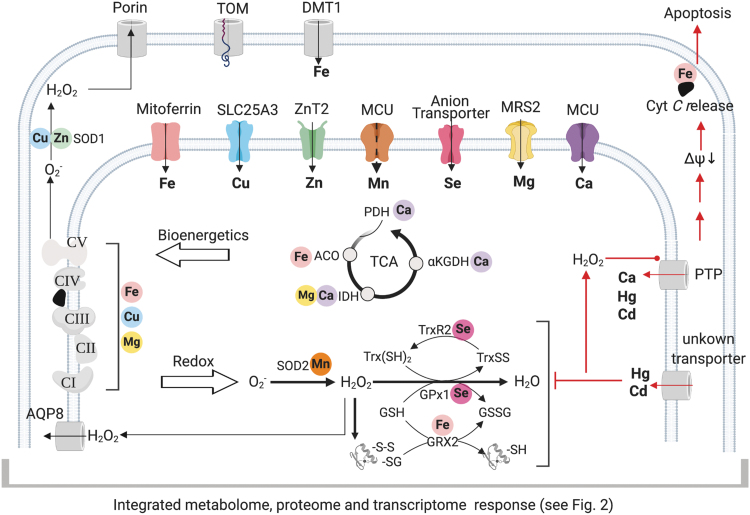
FIG. 3.
Functional network of transporters regulating mitochondrial metallome. Metals, as essential cofactors and structural elements of mitochondrial metalloproteins, play essential roles in the bioenergetics and redox regulation. Although the outer mitochondrial membrane is permeable to free ions, the availabilities of essential metals are controlled by selective transporters in the inner membrane (color coded to each metal). The availabilities in the matrix determine the activities of the TCA cycle, respiratory chain, redox regulation, and cell fate. Importantly, the membrane potential also drives environmental toxic metals into the mitochondrial matrix and causes oxidative stress and inhibition of antioxidant defense systems. Accumulation of H2O2 (and prolonged Ca2+ overload) activates PTP, which can cause non-selective influx of metal ions. PTP activation disrupts membrane integrity and, ultimately, causes apoptosis or necrosis. αKGDH, α–ketoglutarate dehydrogenase; ACO, aconitase; AQP, aquaporin; DMT, divalent metal transporter; GPx, glutathione peroxidase; GRX, glutaredoxin; IDH, isocitrate dehydrogenase; MCU, mitochondrial calcium uniporter; MRS2, mitochondrial RNA splicing 2; PDH, pyruvate dehydrogenase; PTP, permeability transition pore; SLC, solute carrier; SOD, superoxide dismutase; TOM, translocase of outer membrane; Trx, thioredoxin; TrxR, thioredoxin reductase; ZnT, zinc transporter.
Regulation in physiologic and pathologic conditions is controlled by highly conserved channels and transporters that are located in the outer and IMM (117, 128, 163). The outer mitochondrial membrane (OMM) is permeable to solutes and ions by porins, whereas the transport across the IMM is highly regulated (Fig. 3). Metal transporters are included within an array of 53 transporters in the SLC25A (solute carrier) family, among which 31 transporters have identified functions (128) for transport of TCA cycle intermediates, amino acids, nucleotides, vitamins, and metals. A lesser number of ABC (ATP-binding cassette) transporters have been identified localizing in the mitochondria, most without known substrates or functions (60). Recent studies show functions in porphyrin transport, Fe/S cluster export to cytoplasm (ABCB7) (133), and GSH polysulfide export (150) for cytosolic cofactor biosynthesis.
Figure 3 (top) includes a few of the best characterized metal transporters located on the IMM. Heme and copper serve as prosthetic groups for several proteins that constitute the complexes of ETC, whereas iron–sulfur clusters constantly undergo oxidation–reduction reactions to shuttle electrons (Fig. 3, left). The metabolic support of mitochondria to these critical components (e.g., synthesis of heme and ubiquinone) is tightly coupled to transport of required metals, and a large amount of knowledge has come from elegant studies in yeast (39). Recent work examining mitochondrial diseases (e.g., Friedreich's ataxia) in the engineered yeast model has established that communication exists between iron metabolism in the mitochondrion and the cytosol (19, 51, 139). Mitoferrins are well-studied essential Fe(II) importers residing on the IMM (139).
Other transporters, including a symporter, divalent metal transporter 1 (DMT1), have been indicated to safely transport redox active metal across the OMM (186). Metallation of cytochrome c oxidase (COX), a mitochondrial copper/heme metalloenzyme as the terminal enzyme of respiratory chain, occurs within the mitochondrial intermembrane space yet the source of the copper comes from the mitochondrial matrix pool (34). Import of copper into the mitochondrial matrix is mediated by Pic2 in yeast (170), and its metazoan ortholog, SLC25A3 (a previously known phosphate carrier) (18). Depletion of SLC25A3 causes deficiency in COX and mitochondrial copper, and mutations in humans cause muscle hypotonia, cardiomyopathy, and lactic acidosis (183). Interestingly, yeast studies showed that mitochondria contain a pool of copper distinct from the two known mitochondrial cuproenzymes, superoxide dismutase (SOD) and COX, suggesting mitochondria as a component of cellular copper buffering (33).
As indicated earlier, mitochondrial Ca2+ uptake from the ER is tightly controlled by MCU (mitochondrial calcium uniporter) complex, a highly selective ion channel (88). The selectivity to transport Ca2+ instead of a smaller ion Mn2+ is discriminated by the Ca2+-sensing inhibitory subunit (MICU), and the absence of MICU may indicate a new mechanism of metal toxicity (84). Sustained overload of Ca2+, as well as excessive loading of other divalent cations, leads to the opening of high-conductance PTP (permeability transition pore), rapid collapse of the membrane potential, and release of cyt c for apoptosis (Fig. 3, right) (54, 140).
Mitochondrial Ca2+ efflux is directed by the Na+-Ca2+-Li+ exchanger (NCLX) and relies on the tightly controlled matrix Na+ level, which is near equilibrium with the proton gradient via the sodium-proton exchanger (49). Ca2+ also regulates at least three mitochondrial matrix dehydrogenases (Fig. 3, center). On cytoplasmic Ca2+ signaling, increased mitochondrial Ca2+ activates these dehydrogenases to increase NADH availability through the TCA cycle and adjust ATP synthesis to the needs of the stimulated cell. MRS2 (mitochondrial RNA splicing 2) is a primary channel that is used to import magnesium (Mg2+) to mitochondria (8). As a calcium antagonist, Mg2+ also contributes to regulation of energy metabolism through effects on TCA cycle, maintenance of ETC, and bio-activation of ATP (101). Such effects illustrate the important metallome–metabolome interactions required for metabolic and bioenergetic homeostasis.
Other mitochondrial metalloenzymes are critical as redox regulators. The anionic free-radical, superoxide, is constantly generated by the respiratory chain and converted to H2O2 by Mn-containing SOD (SOD2) in the matrix and Cu,Zn-containing SOD1 in IMS. The H2O2 availability is kinetically controlled by selenium (Se)-dependent selenoenzymes in the mitochondrial matrix (129) (Fig. 3, bottom center). For reduction, the thioredoxin reductases have a central role in reducing oxidized proteins. For oxidation, the GSH peroxidases, as well as the thioredoxin-dependent peroxiredoxins, function to regulate the H2O2 supply for protein oxidation (48). However, uptake mechanisms controlling matrix contents of Se, Zn, and Mn remain unclear. Selenite/selenate appears to traverse the IMM by using anion transporters such as phosphate carriers (96). Two Zn transporters (ZIP8 and ZIP9) play a role in mitochondrial uptake (7), yet their physical localization is unknown. ZnT2 (zinc transporter) is the first identified as directly localized within mitochondria in mammary cells (155). Insight into the key regulatory mechanisms for these metals will benefit from systematic studies of the metallome with simultaneous analysis of the metabolome, transcriptome, and proteome.
Such studies are needed to understand homeostatic control with limited selectivity of proteins toward various metals (175). Interaction with cytoplasmic metal ion concentrations is vitally important, because almost half of all enzymes must associate with a particular metal to function, estimated by a systemic bioinformatics survey of 1371 different enzymes (4). Metals such as iron, zinc, copper, or manganese are essential as cofactors for mitochondrial metalloenzymes and metalloproteins. The Irving-Williams series indicates that copper and zinc form the tightest complexes with proteins, followed by nickel and copper, and then iron and manganese (77). Thus, tight control by metal importers, metal exporters, and metal stores is necessary to overcome this trend (175). For example, under impaired iron availability, increased zinc protoporphyrin (95) is formed as one of the first biochemical responses. In SOD2 overexpressed cells or mice fed with Mn-deficient diet, FeSOD forms with Fe instead of Mn. This converts an antioxidant defense into a prooxidant enzyme (52).
Similar to other mitochondria carrier systems, the metal transporters are functionally redundant in that they often show overlapping substrate selectivity (163). This is particularly important for accumulation of metals with molecular mimicry to calcium. Cellular uptake occurs by DMT1 (9), multidrug resistance associated protein-1 (MRP1) (16), and calcium channels (169) across cell membranes. Toxic cationic metals, such as Pb, Cd, and Hg, are preferentially attracted to the negative charge and slightly alkaline environment of mitochondrial matrix, potentially interfering with homeostasis of essential metals (113). Accumulation of heavy metals by mitochondria can not only protect against cytoplasmic toxicity but also cause mitochondrial toxicity through oxidative effects on the major thiol antioxidant systems, including GSH, cytoplasmic thioredoxin-1 and mitochondrial thioredoxin-2, and activated apoptosis (Fig. 3) (67).
Even nutritionally required metals can be toxic, however, as shown by Mn; in excess, brain mitochondria accumulate Mn2+ with Parkinson disease-like neurotoxicity (5). Specificities of many transporters remain unknown, and 16 putative transporters (60) do not have known functions. The wealth of information on individual metals coupled with the incomplete knowledge of transporters and network interactions create an important opportunity for the study of metallomics in mitochondria systems biology.
It is worth underscoring the importance of compartmentalization in cellular metabolism and signaling processes. One of the most remarkable features of Ca2+ signaling is the ability to regulate entirely different processes in the same cell and this almost completely relies on compartmentalization and efficient control of the opening of Ca2+-permeable channels. The very low resting Ca2+ concentration in the cytosol is ideal for signaling purposes; the substantial storage in the ER, 1000–5000 times higher than in the surrounding cytosol, can easily produce intracellular Ca2+ signals without reliance on extracellular Ca2+; and the channel gates creating sharp and short-lasting Ca2+ signals are activated by various metabolic signals [e.g., inositol 1,4,5-trisphosphate, cyclic ADP-ribose, and nicotinic acid adenine dinucleotide phosphate (24)]. Similarly, compartmentalization of membranes to retard H2O2 diffusion, often with transport facilitated across membranes by aquaporin (184), is critical to its signaling role. Although mitochondria-specific metabolites were studied in mitochondria (142, 159), only a few studies (29, 111, 141) using modern MS to study compartmentalized metabolomics and proteomics are available.
Mitochondrial Reprogramming
In development and throughout life, genetically encoded systems undergo phenotypic adaptation and responses to environmental exposures, diet, and infectious agents. Such exposures and biologic responses are collectively termed the exposome. The mitochondrial exposome has only begun to be systematically studied. Importantly, the adaptive response structure of anterograde and retrograde signaling changes mitochondrial properties over time. In antagonistic pleiotropy, adaptive responses can improve adaptability to one stress while compromising tolerance to others (97). Mitochondrial DNA damage can accumulate and cause long-term changes in mitochondrial function and signaling (134, 173).
Changes in the epiproteome can be reversible, but reactive chemicals cause irreversible modifications, termed the protein adductome, which can disrupt mitochondrial function and proteostasis (80, 104). Long-term accumulation of toxic metals, such as Cd, can permanently alter mitochondrial function because effective elimination mechanisms are not available (58, 59, 74). Considerable evidence has accumulated to show that mitochondrial reprogramming allows mitochondria to adapt to environmental exposures. As discussed later, integrated omics offers considerable opportunity to elucidate common processes of mitochondrial reprogramming and their roles in health and disease.
Mitochondrial reprogramming is part of a broader spectrum of long-term responses to the exposome. Sustained impact of the exposome occurs through changes in cell populations, often with cell loss, senescence, trans-differentiation, or malignant transformation. At sub-cellular levels, metabolic reprogramming occurs through changes in cellular organelles, including mitochondria, and also through impacts on macromolecular super-structures and regulation at the level of DNA, RNA, proteins, and metabolites (57). Mitochondria adjust mitochondrial DNA, organelle structure, metabolic functions, and signaling mechanisms in response to diet, exercise, disease, and aging. Indeed, live cell imaging and electron microscopy studies show mitochondrial dynamics executed by mitochondrial fission and fusion (13). These dynamics are usually initiated by the cellular demands of bioenergetics and anabolic processes, yet it ultimately results in a coordinated reprogramming of mitochondria through multiple regulatory levels and mechanisms (181). Concomitant changes in the metabolome and transcriptome are available for some conditions; integrated systems level analyses are not yet available but technically feasible. Such analyses will enable network understanding by connecting morphology and epiproteomic changes to metabolism and transcriptional responses.
Mitochondrial morphology differs among tissue and cell types, yet normal healthy mitochondria share a characteristic rapid turnover by fission and fusion (Fig. 4). This is maintained by several large guanosine triphosphatases (GTPases) in the dynamin family, fission by Drp1 (dynamin-1-like protein [major regulator of mitochondria fission]), and fusion by Mfn1/2 (outer membrane) and Opa1 (inner membrane). In growing and dividing cells, mitochondrial fission is essential to achieve adequate numbers of mitochondria for daughter cells; whereas in quiescent cells, this turnover [usually within 20 min shown by live cell imaging (167)] is important to eliminate disrupted membrane potential through mitophagy.
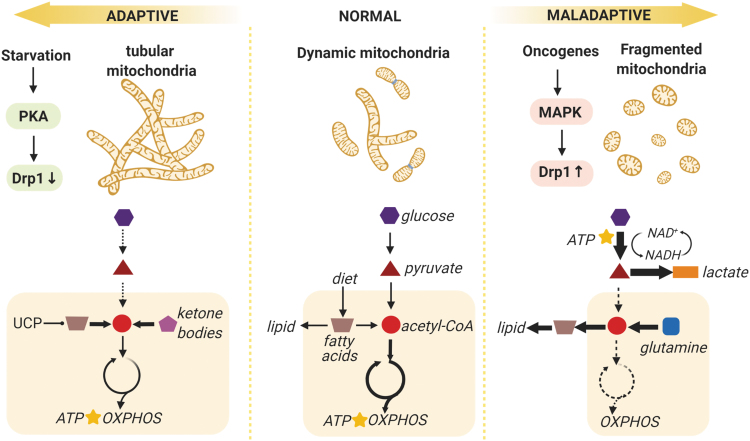
FIG. 4.
Mitochondrial reprogramming involves a reorganization of signaling systems, structure, and metabolic activities into a network with altered energy production, biosynthesis, and detoxification capabilities. This reprogramming can represent adaptive responses to preserve energy and maximize respiration efficiency under starvation stress, or maladaptive response to prioritize the anabolic demand for unlimited proliferation and tumor growth. Mitochondrial morphology is coupled to its metabolic activity (enclosed in light yellow boxes): When the bioenergetic state becomes critical, highly fused mitochondria are formed to increase OXPHOS and lipid catabolism is upregulated by UCP; when the TCA cycle is used as a hub for biosynthesis in malignant cells, respiratory activity is diminished and mitochondria fission is required for removal of damaged organelles. The major (thick arrows) and minor (dashed arrows) metabolic pathways for ATP production (star) are delineated in a simplified scheme. Drp1, dynamin-1-like protein (major regulator of mitochondria fission); MAPK, mitogen-activated protein kinase; OXPHOS, oxidative phosphorylation; PKA, protein kinase A; UCP, uncoupling proteins.
Most cells in resting condition show a balanced energy metabolism phenotype and produce most ATP by mitochondrial respiration. For example, for the resting brain, the rates of cytosolic glycolysis match well with mitochondrial consumption of oxygen. As described later, the rates of mitochondrial fission and fusion respond to changes in energy metabolism that is directed by availability of glucose and need for ATP generation in adaptation to the environment. A disturbance represents a maladaptive response and often leads to pathological reprogramming of mitochondria.
Mitochondria become more fused when cells are forced to rely on OXPHOS to produce ATP (Fig. 4.). This occurs with deprivation of glucose (starvation or exercise) or high energy need (3). Cyclic AMP (cAMP) level rises rapidly in starving cells, activates protein kinase A (PKA), and, in turn, sequesters Drp1 in cytosol (181, 182). The consequent unopposed mitochondrial fusion protects the mitochondria from autophagic elimination (63, 136) and generates denser cristae (91). This maximizes OXPHOS, boosts ATP production, and promotes cell survival (182). When glucose is limited, alternative fuels become dominant, such as fatty acids from lipid degradation and ketone bodies synthesized in liver (14, 125). In mitochondria, uncoupling proteins also respond to the stress by regulating lipid metabolism and oxidant signals from the respiratory chain (121, 143). Resulting hyperfused mitochondrial syncytia (Fig. 4) are proposed to deliver appropriate membrane potential to oxygen-poor regions of cells to maintain energy production (182). Under chronic energy crisis of low ATP level, serine/threonine kinase AMPK can promote fission and, ultimately, mitophagy (181, 182).
Although considerable progress has been made in understanding these central mechanisms governing energy production, less is known about adaptive changes in other fundamental metabolic systems required for mitochondria and cellular homeostasis. For instance, mitochondrial fatty acid biosynthesis of lipoate is essential for entry of pyruvate into the TCA cycle (195); heme biosynthesis, CoQ biosynthesis, and iron–sulfur cluster formation are essential for a functional ETC (139), and a functional ETC is essential for nucleotide biosynthesis (17). Microbiome products such as butyrate impact mitochondrial energy metabolism (44), and pantothenic acid is essential for CoA biosynthesis, carbohydrate, fatty acid, and amino acid metabolism, but these are rarely examined as determinants in mitochondrial reprogramming. Under short-term studies, especially in vitro, dysfunction of such systems may not be evident. In long-lived organisms, however, dysfunction and adaptations in these other diverse and critical molecular components (e.g., see Fig. 1) could be key to mitochondrial reprogramming and health outcomes.
Among the most complex and intriguing mitochondrial reprogramming described to date is the fragmented mitochondria phenotype, which is frequently found with maladaptive (e.g., malignant transformation, senescence) (Fig. 4) and adaptive (e.g., helper T cells, pluripotent stem cells) status. This morphology usually associates with a glycolysis-dominant energy metabolism. For instance, oncogenic K-Ras mutations can activate mitogen-activated protein kinase (MAPK) signaling, resulting in a coordinated reprogramming of mitochondrial structure and metabolic functions including upregulation of glycolysis, inhibition of complex I in respiratory chain, and upregulation of Drp1 to promote fission and mitophagy (173). The consequent metabolic shift is striking: Given the high energy demand, rapidly proliferating cells generate ATP via glycolysis (partial metabolism of glucose to pyruvate) rather than by mitochondrial respiration, as described by Otto Warburg a century ago (178).
The precise reason for cells to use a low-efficient energy strategy (2 ATP vs. 36 ATP from 1 glucose molecule) remains controversial, and evidence is available for other reprogramming mechanisms such as upregulated lactate dehydrogenase to regenerate NAD+ in the face of a relentless glycolytic flux (40). In contrast to use of dietary fatty acids by normal cells, transformed cells rely more on de novo fatty acid synthesis (180). Use of citrate for lipid synthesis results in loss of oxaloacetate from the TCA cycle; the replenishment of the TCA cycle (anaplerosis) is fulfilled by glutamine (40, 134, 173). Thus, the major metabolic function of fragmented mitochondria is shifted from a bioenergetic hub to an anabolic hub. This reprogrammed metabolic function connects mitochondria to other facets of the cell phenotype, that is, the PI3K/Akt/mTOR pathway for protein synthesis and enhanced glucose uptake, HIF-1α activity targeting upregulation of glucose metabolism and inhibition of pyruvate metabolism to acetyl-CoA, and increased MYC activity with upregulated glycolysis and nucleotide biosynthesis (37, 50, 81).
Mitochondrial reprogramming can also involve abnormal accumulation of metabolites, including fumarate, succinate, and 2-HG, which may be sufficient to initiate malignant transformation under some conditions. These have been termed oncometabolites because all three inhibit α-KG-dependent epigenetic switches controlling an oncogenic transcriptional program (123). Degradation of HIF-1α, a growth-factor inducible transcription factor, is impaired by the mitochondrial oncometabolites. Succinylation of kelch-like ECH-associated protein-1 (KEAP1) can also activate the oncogenic transcription factor nuclear factor (nuclear factor erythroid 2-related factor 2 [NRF2]) (87). Indeed, several oncogenic mechanisms involve mitochondrial reprogramming as a component initiating malignant transformation (109, 146, 153).
The extensive literature shows that even though mitochondria display a highly plastic metabolic rewiring capacity, the mechanisms that integrate signal transduction and cell metabolism are largely conserved among cells. For instance, inhibition of mitochondrial fusion by Mfn1/2 ablation facilitates the induction of pluripotency through the restructuring of mitochondrial dynamics and reprogramming to a tumor-like glycolytic metabolic phenotype (161). This mitochondrial flexibility provides adaptive power for somatic cell reprogramming and, in principle, also provides a central logic for regenerative medicine. The connection between the signaling of lung development and those responding to lung injury has been noted (57). Systematic studies will be needed to address the multiple molecular pathways, anatomic diversities, and spatiotemporal organization. This will require combining molecular biology approaches with metabolic flux analysis and ultra-resolution omics to address the complexity of the mitochondrial systems.
Integrative Mitochondrial Network Analysis
Mitochondria research includes many aspects of biologic regulation and is inherently holistic. Mitochondrial research has, however, largely relied on the logic of Descartes (Cartesian reductionism), with hypotheses focused on component parts and often limited to single pathways (Fig. 5A). This reductionist approach has led to considerable success in mapping out functions, interactions, and fates of mitochondrial components, and it has effectively provided a foundation of knowledge for construction of biochemical regulation pathways and networks. The challenge for mitochondria systems biology is that there are about 1018 elements that can vary in abundance and strengths of interactions, making bottom-up assembly intractable (55).
FIG. 5.
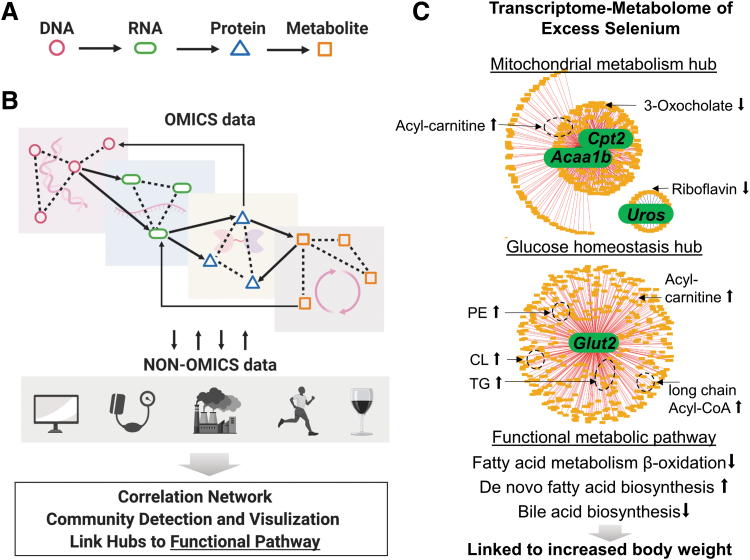
Holistic approach informs the complex mitochondrial network operating within a dynamic cellular environment. (A, B) Cartesian reductionism has provided a critical foundation of knowledge of metabolites, pathways, and regulatory mechanisms yet it often focuses on limited data-driven integration tools; investigation of network-level interactions enables detection of the connections across multiple omics layers (solid arrows), which can be biologically more relevant to biologic function than the connections within a single omics layer (dotted lines). The data-driven integration strategy also allows incorporation of non-omics phenotypic and clinical data such as imaging and blood test results, as well as exposure data such as air pollution, exercise, and diet. (C) In vivo study of Se supplementation in mice shows the utility of data-driven transcriptome–metabolome integration in mouse liver to identify central response hubs in liver [modified with permission from Hu et al. (73)]. A total of 12,421 metabolic features and 80 genes that were identified as significantly changed transcriptomic pathways by Gene Set Enrichment Analysis were integrated by xMWAS and the network was visualized at a correlation threshold of 0.9. The three network hubs represent highly correlated metabolites and genes (|ρ| ≥ 0.9). The two major hubs centered on FA β oxidation enzymes and glucose transporter and the minor hub centered on heme biosynthesis enzyme highlighted the crucial role of mitochondrial energy metabolism in Se effects. Annotated metabolites correlating with the genes were enriched in FA metabolism and were linked to increased body weight gain. Acaa1b, acetyl-CoA acyltransferase 1b; CL, cardiolipin; Cpt2, carnitine palmitoyltransferase 2; Glut2, glucose transporter 2; PE, phosphatidyl ethanolamine; Se, selenium; TG, triglyceride.
The failure of antioxidants to prevent morbidity and mortality in large-scale double-blinded interventional trials in humans amply illustrates the weaknesses inherent in extrapolation of findings from Cartesian reductionism to complex systems. Evidence for oxidative stress in disease was evident from many observational studies, and large numbers of targeted studies showed that antioxidants protected against oxidative stress. The rigorously conducted, double-blind interventional trials with antioxidants showed, however, no health benefit (20, 64, 105). These studies failed to address the complex adaptive network of redox systems that could differ among individuals and obscure and/or alter critical sites for interventions, leaving unresolved questions regarding individual benefits and risks, which could potentially be addressed with integrative omics methods. In particular, without detailed investigation of the response network, the flexibly to rewire mitochondrial metabolism could subvert targeted intervention. Thus, mitochondrial reprogramming may underlie multiple instances of pathobiology and therapeutic failure (130, 145, 168).
As outlined by Loscalzo et al. (106), simple additive and synergistic cause-effect mechanisms cannot explain most diseases. Biologic systems are genetically designed to utilize a range of energy precursors and have multiple defense mechanisms to adapt to environmental conditions. Omics technologies, as now widely available, provide an opportunity to gain systems-level understanding (82). Available methods include polynucleotide sequencing by next-generation sequencers for genome, transcriptome, and chromatin sequencing (1, 12, 119); MS-based proteomics with post-translational modification quantification (21, 78); and high-resolution metabolomics based on ultra-high-resolution MS and liquid and gas chromatography (166, 185). Strengths and challenges of individual omics have been nicely reviewed (116). Sequencing-based technologies are still the most advanced of the omics technologies in availability of laboratory reagents, standardized protocols, consensus analytical tools, and public databases for data sharing. In contrast, although proteomics has moved toward more system-wide screening approaches with quantitative steps that generate favorable results, debates remain on data formatting and normalization. Similar challenges exist in both metabolomics and proteomics on the coverage of different chemical properties and variability of data annotation and analysis. Nevertheless, omics approaches share common challenges of handling large datasets, annotation of biomolecules, and sensitivity of signal versus noise.
Bioinformatic workflows have been refined to support efficient analysis of omics data, typically through data preprocessing, statistical testing, clustering analysis, pathway enrichment analysis, and network mapping (118). Single-omics approaches are well developed and very powerful but have limited ability to capture interaction across multiple interacting layers (194). Although direct interactions within single omics layers are well known, such as transcription factors and target gene expression, and the relationship between substrates and products, interactions between omics layers are often overlooked at global levels. This is critically important because metabolites impact activities of transcription factors; transcription factors impact expression of messenger RNA (mRNA), which, in turn, impacts synthesis and abundance of proteins that control metabolite levels. Further, elements of one omics layer can impact other omics layers in multiple ways. For example, individual metabolites regulate enzymatic activity through allosteric sites and also compete in binding to multiple transcription factors and epigenetic enzymes. In this sense, the connections across omics layers are indeed more robust and stronger than the connections within a single omics layer (Fig. 5B, dotted vs. solid arrows).
Several tools and approaches are now available for the integrative omics, based on existing biological knowledge, or empirical correlation analysis [reviewed in Wanichthanarak et al. (177)]. The advantage of the knowledge-driven approach relying on predefined biochemical pathways and networks is the ease of interpreting in a biological meaningful context (BIND, HPRD, Kyoto Encyclopedia of Genes and Genomes: KEGG, and the Biochemical Genetic and Genomics knowledgebase: BIGG) (10). This approach has provided a framework to identify differentially active biological processes between synthetic yeast strains and wild type by using yeast KEGG pathways and Gene Ontology (GO) annotations to integrate transcriptomics, proteomics, and metabolomics data (157).
However, a knowledge-driven approach may yield limited insight due to insufficient domain knowledge of the biological interactions. Integration of omics data with clinical and epidemiological exposure data is also crucial to understand disease etiology, yet it is challenging due to high heterogeneity of the data types (Fig. 5B). These challenges have led to development of data-driven approaches to derive network structures and utilize knowledgebases as downstream tools to enhance understanding of network structures. The R package mixOmics supports empirical correlation analysis between two high-dimensional datasets; weighted gene correlation network analysis (WGCNA) extends the concept of correlations to graph topology models and, thus, is widely used to analyze gene coexpression networks. In a recent clinical trial testing calorie restriction to improve insulin sensitivity, this data-driven instead of knowledge-driven approach reconstructed a biological network linking insulin sensitivity, serum branched chain amino acids, subcutaneous adipose fat genes, and gut microbiota species, demonstrating the value of the data-driven approach in discovery of unknown interactions (38).
xMWAS is a data-driven approach that statistically infers associations and correlations between molecules based on multi-omics data (164). Compared with other data-driven integration software [mixOmics, WGCNA, 3Omics, MetaboAnalyst and many more (164)], xMWAS allows integration of more than two datasets and identification of community structure to reveal topological modules comprising functionally related biomolecules (10). In a study of 16-week Se supplementation to C57BL/6J male mice, mice supplemented with Se had greater body mass gain from baseline in the absence of oxidative stress (81).
An integrated transcriptome–metabolome wide association study (TMWAS) of mouse liver using xMWAS showed two major gene–metabolite interaction clusters after integrating 12,421 metabolic features and 80 genes from significant transcriptomic pathways identified by gene set enrichment analysis. The outcome of integrative analysis with high correlation cutoff (r ≥ 0.9) yielded that one was centered on the transcript for the bidirectional glucose transporter 2 (Glut2) and the other on the transcripts for carnitine palmitoyltransferase 2 (Cpt2) and acetyl-CoA acyltransferase 1 (Acaa1), suggesting perturbations in both glucose homeostasis and fatty acid oxidation (73). Importantly, expression of Cpt2, as an essential metabolic component of mitochondria (Fig. 1), was highly correlated with lipid and fatty acid intermediates, visualized in Figure 5C. Indeed, Se supplementation caused accumulation of acyl-carnitines in mouse liver and conversely decreased acetyl-CoA and other TCA cycle intermediates, suggesting that supplemental Se disrupts mitochondrial energy metabolism by decreasing capability for fatty acid β-oxidation. Thus, this data-driven integrated omics approach provides novel insight into previously reported disease risk of type 2 diabetes, obesity, and hyperlipidemia associated with high Se status in humans (48, 137).
Increasing evidence shows that important functional changes occur with minor variation in a dietary nutrient. Exposure of human neuroblastoma (SH-SY5Y) cells to increasing Mn concentration showed that Mn dose–response characteristics differed for H2O2 production and mitochondrial respiration (47). TMWAS visualized distinct metabolite-transcript clusters associated with basal and proton-leak dependent oxygen consumption rate, and clusters associated with redox measures including levels of H2O2, thiols, and mitochondrial oxidants (55). The results demonstrate the fundamental importance of subtle variation in Mn concentration in mitochondrial biology, and they establish an analytical platform to incorporate bioenergetics and metabolism for integrated analysis of mitochondrial and cell functions (55).
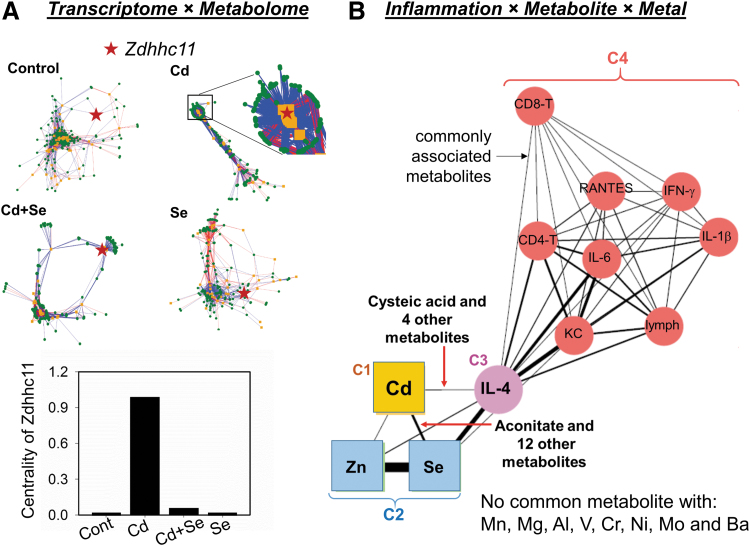
Such interactions are also illustrated by studies of toxic interactions of environmental levels of Cd exposure in mice. As observed with Se and Mn shown earlier, low-level Cd has systems-level effects on mitochondria (58, 59, 74). Visualization of key interactions by topological analysis identifies nodes that undergo network changes under different conditions, and contributions can be quantified by eigenvector centrality (164) (Fig. 6A). Results for lungs of Cd-treated mice showed that the gene-metabolite network was more condensed, indicating a more rigid and less adaptable structure. This was especially exemplified by Zdhhc11 (a zinc-finger enzyme responsible for S-acylation of proteins), which showed a switch from a peripheral to central position and much greater centrality in the network structure after Cd treatment. This remarkable increase in association between Zdhhc11 and the metabolome suggested a novel mechanism in Cd-induced lung toxicity. Importantly, co-treatment with Se reversed the network structure caused by Cd and reduced the centrality of Zdhhc11 to baseline (Fig. 6A), showing an antagonistic effect of low-level Se on low-level Cd-dependent lung toxicity (72).
FIG. 6.
Network analysis strategies advance understanding of complex biologic responses in metal–metal interaction studies. (A) Eigenvector centrality measures allow identification of key hubs in network change in a study of Se antagonizing Cd lung toxicity. Integration of transcriptome (orange) and metabolome (green) shows a collapsed network structure of Cd-treated mouse lung compared with control. Zdhhc11 (red star) switched from a peripheral position in the control network to a central position associating with a large number of metabolites after Cd treatment (inset showing zoomed-in structure), reflected by centrality measures (bottom). Se co-treatment restored the network structure to an adaptive and flexible structure similar to control. (B) Community detection allows visualization of relations between metals and inflammation markers via commonly associated metabolites in a study of Cd potentiating RSV-induced lung inflammation. The thickness of the edges connecting nodes (circles, inflammation markers; squares, metals) are proportional to the number of associated metabolites shared by the two nodes. Communities C1 through C4 are from unsupervised topology analysis and show a strong interaction of Cd, Se, and Zn with IL-4 but not with other inflammation markers (C4, red circles). This interaction among Cd, Se, Zn, and IL-4 was specific as no common metabolite association was found with IL-4 and other metals (not shown in the network). (A, B) Modified with permission from Hu et al. (72) and Hu et al. (75), respectively. IL-4, interleukin 4; RSV, respiratory syncytial virus.
Use of community detection has also been useful to dissect complex interactions of environmental Cd and viral infection (75). Associations among inflammation markers, lung metabolomics, and the lung metallome, including 11 nutritional and toxic metals, provided insight into mechanisms by which Cd exposure worsens lung inflammation caused by respiratory syncytial virus (RSV) infection. The network structure showed a strong interaction between the community of Cd, Se, and Zn and a community containing interleukin 4 (IL-4) occurred through metabolite associations (Fig. 6B). This community relationship confirmed IL-4 linkage to unique mitochondrial metabolic perturbations induced by Cd. The results further demonstrate that metal dysregulation has a broad effect on mitochondrial functional networks. Collectively, the examples show that data-driven omics integration provides a powerful approach to identify and visualize the hierarchy of mitochondrial network structures and most central modules in complex interactions involving metals, metabolites, and other macromolecules.
Concluding Remarks
Mitochondrial dysfunction has been implicated in human diseases impacting most major organ systems. Participation of mitochondria in various pathobiology (e.g., senescence, malignant proliferation, dysregulated differentiation) involves maladaptive responses at a systems level and encompasses modifications of epigenome, epiproteome, epimetabolome, and metallome. In particular, the function of mitochondria extends far beyond energy production, and key metabolites, termed the epimetabolome, represent central hub metabolites of critical organizational importance. These metabolites support switching mechanisms to control structural and functional organization of proteins, dictating control of gene expression, cell activities, and cell fate.
Importantly, high-resolution metabolomics methods now provide capability to measure most metabolic pathways linked to mitochondrial function, and integration of metabolomics, transcriptomics, and proteomics shows that network organization involves trans-omics connections. Data-driven integration of mitochondrial phenotypic measures and cell fate occurring as a consequence of systematic variation in metal ions show that the powerful tools enable identification of central hubs of the dynamic mitochondrial functional networks. Although therapeutic strategies to target these hubs and protect against mitochondrial network dysfunctions are not yet available, the results provide a foundation for such development.
Applications to address mitochondrial dysfunction in human disease will require improved delineation of the mitochondrial phenotypes in different cell and organ systems. Mitochondria are highly flexible and efficient in titrating activities and morphology to cellular needs. This rewiring of mitochondria, termed mitochondrial reprogramming, enables an organism to adapt to environmental resources and challenges. The pleotropic roles and reprogramming of mitochondria show that, rather than a bystander, mitochondria can be an important contributor to disease. Application of omics tools to measure the integrated functions of the metabolome, proteome, and transcriptome now enables detailed assessment of the mitochondrial phenotype. This greatly extends capabilities beyond the small number of commonly used indicators, such as high lactate or high lactate/pyruvate ratio, and it improves the opportunity to understand maladaptive reorganization of network structures and associated signaling mechanisms needed to support the broad spectrum of cell functions supported by mitochondria.
Tools and approaches are now available to advance understanding of complex interactive molecular structures and functions through development of mitochondria systems biology. Broad, systems-based approaches will be needed to integrate information from multiomics with mitochondrial phenotypic and functional measures and link these to health outcomes. High-resolution metabolomics provides an important tool to support this development, because the mitochondrial metabolome communicates with cytoplasmic and plasma metabolome, allowing plasma measures to provide insight into mitochondrial function, reprogramming, and dysfunction. Expansion of knowledge bases, such as MitoCarta (23), and tools such as xMWAS (164), will be needed to support efficient integration of omics data for study of mitochondrial network responses. Systematic studies of mitochondrial network responses in model systems and human disease will be critical to tease apart the major functional relationships and guide development of therapeutic interventions. The recent progress has been dramatic, and it sets the stage for substantial progress in development of mitochondria systems biology to improve human health and decrease the costly burden of disease.
Acknowledgment
The poem by Eberhard Voit was from a first course in systems biology (172).
Abbreviations Used
- 2-HG
2-hydroxyglutarate
- α-KG
α-ketoglutarate
- ABC
ATP-Binding Cassette
- AMPK
AMP-activated kinase
- BCAA
branched-chain amino acids
- CoA
coenzyme A
- CoQ
coenzyme Q10
- COX
cytochrome c oxidase
- Cpt2
carnitine palmitoyltransferase 2
- cyt c
cytochrome c
- DMT
divalent metal transporter
- Drp1
dynamin-1-like protein (major regulator of mitochondria fission)
- ER
endoplasmic reticulum
- ETC
electron transport chain
- Glut2
glucose transporter 2
- GSH
glutathione
- HIF-1
hypoxia-inducible factor-1
- IL-4
interleukin 4
- IMM
inner mitochondrial membrane
- KEGG
Kyoto Encyclopedia of Genes and Genomes
- Mfn
mitofusin
- Mg2+
magnesium
- MS
mass spectrometry
- OMM
outer mitochondrial membrane
- OXPHOS
oxidative phosphorylation
- PGC-1
peroxisome-proliferator-activated receptor coactivator-1
- SAMe
S-adenosyl-l-methionine
- Se
selenium
- SLC
solute carrier
- SOD
superoxide dismutase
- TCA
tricarboxylic acid
- TMWAS
transcriptome-metabolome wide association study
- WGCNA
weighted gene correlation network analysis
Funding Information
This study was supported by NIH grants R01 ES023485, U2C ES030163, P30 ES019776, and S10 OD018006.
References
- 1. Adli M and Bernstein BE. Whole-genome chromatin profiling from limited numbers of cells using nano-ChIP-seq. Nat Protoc 6: 1656, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Aguer C, McCoin CS, Knotts TA, Thrush AB, Ono-Moore K, McPherson R, Dent R, Hwang DH, Adams SH, and Harper M-E. Acylcarnitines: potential implications for skeletal muscle insulin resistance. FASEB J 29: 336–345, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3. Amchenkova AA, Bakeeva LE, Chentsov YS, Skulachev VP, and Zorov DB. Coupling membranes as energy-transmitting cables. I. Filamentous mitochondria in fibroblasts and mitochondrial clusters in cardiomyocytes. J Cell Biol 107: 481–495, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 4. Andreini C, Bertini I, Cavallaro G, Holliday GL, and Thornton JM. Metal ions in biological catalysis: from enzyme databases to general principles. J Biol Inorg Chem 13: 1205–1218, 2008 [DOI] [PubMed] [Google Scholar]
- 5. Aschner M, Erikson KM, Herrero Hernández E, and Tjalkens R. Manganese and its role in Parkinson's disease: from transport to neuropathology. Neuromolecular Med 11: 252–266, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 6. Aussignargues C, Giuliani M-C, Infossi P, Lojou E, Guiral M, Giudici-Orticoni M-T, and Ilbert M. Rhodanese functions as sulfur supplier for key enzymes in sulfur energy metabolism. J Biol Chem 287: 19936–19948, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7. Avila DS, Puntel RL, and Aschner M. Manganese in health and disease. In: Interrelations Between Essential Metal Ions and Human Diseases, edited by Sigel A, Sigel H, and Sigel RKO. Dordrecht: Springer Netherlands, 2013, pp. 199–227 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. de Baaij JH, Hoenderop JGJ, and Bindels RJM. Magnesium in Man: implications for Health and Disease. Physiol Rev 95: 1–46, 2015 [DOI] [PubMed] [Google Scholar]
- 9. Bannon DI, Abounader R, Lees PS, and Bressler JP. Effect of DMT1 knockdown on iron, cadmium, and lead uptake in Caco-2 cells. Am J Physiol Cell Physiol 284: C44–C50, 2003 [DOI] [PubMed] [Google Scholar]
- 10. Barabási A-L, Gulbahce N, and Loscalzo J. Network medicine: a network-based approach to human disease. Nat Rev Genet 12: 56–68, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Bartlett K and Eaton S. Mitochondrial β-oxidation. Eur J Biochem 271: 462–469, 2004 [DOI] [PubMed] [Google Scholar]
- 12. Bentley DR, Balasubramanian S, Swerdlow HP, Smith GP, Milton J, Brown CG, Hall KP, Evers DJ, Barnes CL, and Bignell HR. Accurate whole human genome sequencing using reversible terminator chemistry. Nature 456: 53, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13. Bereiter-Hahn J. Behavior of mitochondria in the living cell. In: International Review of Cytology, edited by Jean KW and Friedlander M. Amsterdam, Netherlands: Elsevier, 1990, pp. 1–63 [DOI] [PubMed] [Google Scholar]
- 14. Berg J, Tymoczko J, and Stryer L. Food intake and starvation induce metabolic changes. In: Biochemistry, 5th ed., edited by Berg JM, Tymoczko JL, and Stryer L. New York, NY: W.H. Freeman, 2002 [Google Scholar]
- 15. Bergseth S, Lund H, Poisson J-P, Bremer J, Davis-Van Thienen W, and Davis EJ. Carnitine palmitoyltransferase: activation and inactivation in liver mitochondria from fed, fasted, hypo-and hyperthyroid rats. Biochim Biophys Acta 876: 551–558, 1986 [DOI] [PubMed] [Google Scholar]
- 16. Bhattacharjee A, Majumdar U, Maity D, Sarkar TS, Goswami AM, Sahoo R, and Ghosh S. In vivo protein tyrosine nitration in S. cerevisiae: identification of tyrosine-nitrated proteins in mitochondria. Biochem Biophys Res Commun 388: 612–617, 2009 [DOI] [PubMed] [Google Scholar]
- 17. Birsoy K, Wang T, Chen WW, Freinkman E, Abu-Remaileh M, and Sabatini DM. An essential role of the mitochondrial electron transport chain in cell proliferation is to enable aspartate synthesis. Cell 162: 540–551, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18. Boulet A, Vest KE, Maynard MK, Gammon MG, Russell AC, Mathews AT, Cole SE, Zhu X, Phillips CB, Kwong JQ, Dodani SC, Leary SC, and Cobine PA. The mammalian phosphate carrier SLC25A3 is a mitochondrial copper transporter required for cytochrome c oxidase biogenesis. J Biol Chem 293: 1887–1896, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Branda SS, Yang Z-y, Chew A, and Isaya G. Mitochondrial intermediate peptidase and the yeast frataxin homolog together maintain mitochondrial iron homeostasis in Saccharomyces cerevisiae. Hum Mol Genet 8: 1099–1110, 1999 [DOI] [PubMed] [Google Scholar]
- 20. Brion LP, Bell EF, and Raghuveer TS. Vitamin E supplementation for prevention of morbidity and mortality in preterm infants. Cochrane Database Syst Rev 4: CD003665, 2003 [DOI] [PubMed] [Google Scholar]
- 21. Butterfield DA and Perluigi M. Redox proteomics: a key tool for new insights into protein modification with relevance to disease. Antioxid Redox Signal 26: 277–279, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22. Cali JJ and Russell DW. Characterization of human sterol 27-hydroxylase. A mitochondrial cytochrome P-450 that catalyzes multiple oxidation reaction in bile acid biosynthesis. J Biol Chem 266: 7774–7778, 1991 [PubMed] [Google Scholar]
- 23. Calvo SE, Clauser KR, and Mootha VK. MitoCarta2. 0: an updated inventory of mammalian mitochondrial proteins. Nucleic Acids Res 44: D1251–D1257, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Cancela JM, Gerasimenko OV, Gerasimenko JV, Tepikin AV, and Petersen OH. Two different but converging messenger pathways to intracellular Ca(2+) release: the roles of nicotinic acid adenine dinucleotide phosphate, cyclic ADP-ribose and inositol trisphosphate. EMBO J 19: 2549–2557, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 25. Castegna A, Iacobazzi V, and Infantino V. The mitochondrial side of epigenetics. Physiol Genomics 47: 299–307, 2015 [DOI] [PubMed] [Google Scholar]
- 26. Chan DC. Mitochondria: dynamic organelles in disease, aging, and development. Cell 125: 1241–1252, 2006 [DOI] [PubMed] [Google Scholar]
- 27. Chavez JD, Wu J, Bisson W, and Maier CS. Site-specific proteomic analysis of lipoxidation adducts in cardiac mitochondria reveals chemical diversity of 2-alkenal adduction. J Proteomics 74: 2417–2429, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Chen Q, Vazquez EJ, Moghaddas S, Hoppel CL, and Lesnefsky EJ. Production of reactive oxygen species by mitochondria central role of complex III. J Biol Chem 278: 36027–36031, 2003 [DOI] [PubMed] [Google Scholar]
- 29. Chen WW, Freinkman E, Wang T, Birsoy K, and Sabatini DM. Absolute quantification of matrix metabolites reveals the dynamics of mitochondrial metabolism. Cell 166: 1324. e11–1337. e11, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 30. Chong J, Soufan O, Li C, Caraus I, Li S, Bourque G, Wishart DS, and Xia J. MetaboAnalyst 4.0: towards more transparent and integrative metabolomics analysis. Nucleic Acids Res 46: W486–W494, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Christian BE and Spremulli LL. Mechanism of protein biosynthesis in mammalian mitochondria. Biochim Biophys Acta 1819: 1035–1054, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 32. Christiansen RZ. Regulation of palmitate metabolism by carnitine and glucagon in hepatocytes isolated from fasted and carbohydrate refed rats. Biochim Biophys Acta 488: 249–262, 1977 [DOI] [PubMed] [Google Scholar]
- 33. Cobine PA, Ojeda LD, Rigby KM, and Winge DR. Yeast contain a non-proteinaceous pool of copper in the mitochondrial matrix. J Biol Chem 279: 14447–14455, 2004 [DOI] [PubMed] [Google Scholar]
- 34. Cobine PA, Pierrel F, Bestwick ML, and Winge DR. Mitochondrial matrix copper complex used in metallation of cytochrome oxidase and superoxide dismutase. J Biol Chem 281: 36552–36559, 2006 [DOI] [PubMed] [Google Scholar]
- 35. Contrusciere V, Paradisi S, Matteucci A, and Malchiodi-Albedi F. Branched-chain amino acids induce neurotoxicity in rat cortical cultures. Neurotox Res 17: 392–398, 2010 [DOI] [PubMed] [Google Scholar]
- 36. Crown SB, Marze N, and Antoniewicz MR. Catabolism of branched chain amino acids contributes significantly to synthesis of odd-chain and even-chain fatty acids in 3T3–L1 adipocytes. PLoS One 10: e0145850, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Dang CV, Kim J-W, Gao P, and Yustein J. The interplay between MYC and HIF in cancer. Nat Rev Cancer 8: 51, 2008 [DOI] [PubMed] [Google Scholar]
- 38. Dao MC, Sokolovska N, Brazeilles R, Affeldt S, Pelloux V, Prifti E, Chilloux J, Verger EO, Kayser BD, Aron-Wisnewsky J, Ichou F, Pujos-Guillot E, Hoyles L, Juste C, Doré J, Dumas M-E, Rizkalla SW, Holmes BA, Zucker J-D, Clément K TM-OC, Cotillard A, Kennedy SP, Pons N, Chatelier EL, Almeida M, Quinquis B, Galleron N, Batto J-M, Renault P, Ehrlich SD, Blottière H, Leclerc M, de Wouters T, and Lepage P. A data integration multi-omics approach to study calorie restriction-induced changes in insulin sensitivity. Front Physiol 9: 1958, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. De Freitas J, Wintz H, Hyoun Kim J, Poynton H, Fox T, and Vulpe C. Yeast, a model organism for iron and copper metabolism studies. Biometals 16: 185–197, 2003 [DOI] [PubMed] [Google Scholar]
- 40. DeBerardinis RJ, Lum JJ, Hatzivassiliou G, and Thompson CB. The biology of cancer: metabolic reprogramming fuels cell growth and proliferation. Cell Metab 7: 11–20, 2008 [DOI] [PubMed] [Google Scholar]
- 41. Dennis KK, Go Y-M, and Jones DP. Redox systems biology of nutrition and oxidative stress. J Nutr 149: 553–565, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 42. Di Stefano M and Conforti L.. Diversification of NAD biological role: the importance of location. FEBS J 280: 4711–4728, 2013 [DOI] [PubMed] [Google Scholar]
- 43. Dominy JE and Puigserver P. Mitochondrial biogenesis through activation of nuclear signaling proteins. Cold Spring Harb Perspect Biol 5: a015008, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Donohoe DR, Garge N, Zhang X, Sun W, O'Connell TM, Bunger MK, and Bultman SJ. The microbiome and butyrate regulate energy metabolism and autophagy in the mammalian colon. Cell Metab 13: 517–526, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Edwards JC, Levens D, and Rabinowitz M. Analysis of transcriptional initiation of yeast mitochondrial DNA in a homologous in vitro transcription system. Cell 31: 337–346, 1982 [DOI] [PubMed] [Google Scholar]
- 46. Eriksson S and Wang L. Molecular mechanisms of mitochondrial DNA depletion diseases caused by deficiencies in enzymes in purine and pyrimidine metabolism. Nucleosides Nucleotides Nucleic Acids 27: 800–808, 2008 [DOI] [PubMed] [Google Scholar]
- 47. Fernandes J, Hao L, Bijli KM, Chandler JD, Orr M, Hu X, Jones DP, and Go YM. From the cover: manganese stimulates mitochondrial H2O2 production in SH-SY5Y human neuroblastoma cells over physiologic as well as toxicologic range. Toxicol Sci 155: 213–223, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 48. Fernandes J, Hu X, Ryan Smith M, Go Y-M, and Jones DP. Selenium at the redox interface of the genome, metabolome and exposome. Free Radic Biol Med 127: 215–227, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 49. Finkel T, Menazza S, Holmström KM, Parks RJ, Liu J, Sun J, Liu J, Pan X, and Murphy E. The ins and outs of mitochondrial calcium. Circ Res 116: 1810–1819, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Finley LW, Carracedo A, Lee J, Souza A, Egia A, Zhang J, Teruya-Feldstein J, Moreira PI, Cardoso SM, and Clish CB. SIRT3 opposes reprogramming of cancer cell metabolism through HIF1α destabilization. Cancer Cell 19: 416–428, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Foury F and Talibi D. Mitochondrial control of iron homeostasis: a genome wide analysis of gene expression in a yeast frataxin-deficient strain. J Biol Chem 276: 7762–7768, 2001 [DOI] [PubMed] [Google Scholar]
- 52. Ganini D, Santos JH, Bonini MG, and Mason RP. Switch of mitochondrial superoxide dismutase into a prooxidant peroxidase in manganese-deficient cells and mice. Cell Chem Biol 25: 413.e6–425.e6, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 53. Gheni G, Ogura M, Iwasaki M, Yokoi N, Minami K, Nakayama Y, Harada K, Hastoy B, Wu X, Takahashi H, Kimura K, Matsubara T, Hoshikawa R, Hatano N, Sugawara K, Shibasaki T, Inagaki N, Bamba T, Mizoguchi A, Fukusaki E, Rorsman P, and Seino S. Glutamate acts as a key signal linking glucose metabolism to incretin/cAMP action to amplify insulin secretion. Cell Rep 9: 661–673, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Giorgi C, Marchi S, and Pinton P. The machineries, regulation and cellular functions of mitochondrial calcium. Nat Rev Mol Cell Biol 19: 713–730, 2018 [DOI] [PubMed] [Google Scholar]
- 55. Go Y-M, Fernandes J, Hu X, Uppal K, and Jones DP. Mitochondrial network responses in oxidative physiology and disease. Free Radic Biol Med 116: 31–40, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Go Y-M and Jones DP. The redox proteome. J Biol Chem 288: 26512–26520, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 57. Go Y-M and Jones DP. Exposure memory and lung regeneration. Ann Am Thorac Soc 13: S452–S461, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 58. Go Y-M, Orr M, and Jones DP. Actin cytoskeleton redox proteome oxidation by cadmium. Am J Physiol Lung Cell Mol Physiol 305: L831–L843, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Go Y-M, Sutliff RL, Chandler JD, Khalidur R, Kang B-Y, Anania FA, Orr M, Hao L, Fowler BA, and Jones DP. Low-dose cadmium causes metabolic and genetic dysregulation associated with fatty liver disease in mice. Toxicol Sci 147: 524–534, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 60. Go Y-M, Uppal K, Walker DI, Tran V, Dury L, Strobel FH, Baubichon-Cortay H, Pennell KD, Roede JR, and Jones DP. Mitochondrial metabolomics using high-resolution Fourier-transform mass spectrometry. In: Mass Spectrometry in Metabolomics. Methods in Molecular Biology (Methods and Protocols), vol. 1198, edited by Raftery D. New York, NY: Humana Press, 2014, pp. 43–73 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 61. Go YM, Uppal K, and Jones DP. Central mitochondrial signaling mechanisms in response to environmental agents. In: Mitochondrial Dysfunction Caused by Drugs and Environmental Toxicants, edited by Will Y and Dykens JA. 2018, pp. 639–654 [Google Scholar]
- 62. Goffart S and Wiesner R. Regulation and co-ordination of nuclear gene expression during mitochondrial biogenesis. Exp Physiol 88: 33–40, 2003 [DOI] [PubMed] [Google Scholar]
- 63. Gomes LC, Di Benedetto G, and Scorrano L. During autophagy mitochondria elongate, are spared from degradation and sustain cell viability. Nat Cell Biol 13: 589–598, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Goodman GE, Thornquist MD, Balmes J, Cullen MR, Meyskens FL Jr., Omenn GS, Valanis B, and Williams JH Jr.. The beta-carotene and retinol efficacy trial: incidence of lung cancer and cardiovascular disease mortality during 6-year follow-up after stopping β-carotene and retinol supplements. J Natl Cancer Inst 96: 1743–1750, 2004 [DOI] [PubMed] [Google Scholar]
- 65. Guarente L. Mitochondria—a nexus for aging, calorie restriction, and sirtuins? Cell 132: 171–176, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Hallan S, Afkarian M, Zelnick LR, Kestenbaum B, Sharma S, Saito R, Darshi M, Barding G, Raftery D, Ju W, Kretzler M, Sharma K, and de Boer IH. Metabolomics and gene expression analysis reveal down-regulation of the citric acid (TCA) cycle in non-diabetic CKD patients. EBioMedicine 26: 68–77, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Hansen JM, Zhang H, and Jones DP. Differential oxidation of thioredoxin-1, thioredoxin-2, and glutathione by metal ions. Free Radic Biol Med 40: 138–145, 2006 [DOI] [PubMed] [Google Scholar]
- 68. Hatefi Y. The mitochondrial electron transport and oxidative phosphorylation system. Annu Rev Biochem 54: 1015–1069, 1985 [DOI] [PubMed] [Google Scholar]
- 69. Haüssinger D. Nitrogen metabolism in liver: structural and functional organization and physiological relevance. Biochem J 267: 281, 1990 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 70. He W, Newman JC, Wang MZ, Ho L, and Verdin E. Mitochondrial sirtuins: regulators of protein acylation and metabolism. Trends Endocrinol Metab 23: 467–476, 2012 [DOI] [PubMed] [Google Scholar]
- 71. Hiltunen JK, Schonauer MS, Autio KJ, Mittelmeier TM, Kastaniotis AJ, and Dieckmann CL. Mitochondrial fatty acid synthesis type II: more than just fatty acids. J Biol Chem 284: 9011–9015, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 72. Hu X, Chandler JD, Fernandes J, Orr ML, Hao L, Uppal K, Neujahr DC, Jones DP, and Go Y-M. Selenium supplementation prevents metabolic and transcriptomic responses to cadmium in mouse lung. Biochim Biophys Acta 1862: 2417–2426, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Hu X, Chandler JD, Orr ML, Hao L, Liu K, Uppal K, Go Y-M, and Jones DP. Selenium supplementation alters hepatic energy and fatty acid metabolism in mice. J Nutr 148: 675–684, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 74. Hu X, Chandler JD, Park S, Liu K, Fernandes J, Orr M, Smith MR, Ma C, Kang S-M, Uppal K, Jones DP, and Go Y-M. Low-dose cadmium disrupts mitochondrial citric acid cycle and lipid metabolism in mouse lung. Free Radic Biol Med 131: 209–217, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Hu X, Kim KH, Lee Y, Fernandes J, Smith MR, Jung Y, Orr M, Kang SM, Jones DP, and Go YM. Environmental cadmium enhances lung injury by respiratory syncytial virus infection. Am J Pathol 189: 1513–1525, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Hung YP, Albeck JG, Tantama M, and Yellen G. Imaging cytosolic NADH-NAD+ redox state with a genetically encoded fluorescent biosensor. Cell Metab 14: 545–554, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Irving H and Williams R. Order of stability of metal complexes. Nature 162: 746, 1948 [Google Scholar]
- 78. Ishihama Y, Sato T, Tabata T, Miyamoto N, Sagane K, Nagasu T, and Oda Y. Quantitative mouse brain proteomics using culture-derived isotope tags as internal standards. Nat Biotechnol 23: 617, 2005 [DOI] [PubMed] [Google Scholar]
- 79. Janssen JJE, Grefte S, Keijer J, and de Boer VCJ. Mito-nuclear communication by mitochondrial metabolites and its regulation by B-vitamins. Front Physiol 10, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Jones DP and Sies H. The redox code. Antioxid Redox Signal 23: 734–746, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Jones RG and Thompson CB. Tumor suppressors and cell metabolism: a recipe for cancer growth. Genes Dev 23: 537–548, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Joyce AR and Palsson BØ. The model organism as a system: integrating “omics” data sets. Nat Rev Mol Cell Biol 7: 198–210, 2006 [DOI] [PubMed] [Google Scholar]
- 83. Kalen AL, Ahmad IM, Abdalla MY, O'Malley YQ, Goswami PC, and Sarsour EH. MnSOD and cyclin B1 coordinate a mito-checkpoint during cell cycle response to oxidative stress. Antioxidants (Basel) 6: 92, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 84. Kamer KJ, Sancak Y, Fomina Y, Meisel JD, Chaudhuri D, Grabarek Z, and Mootha VK. MICU1 imparts the mitochondrial uniporter with the ability to discriminate between Ca2+ and Mn2+. Proc Natl Acad Sci U S A 115: E7960–E7969, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 85. Kanehisa M and Goto S. KEGG: Kyoto encyclopedia of genes and genomes. Nucleic Acids Res 28: 27–30, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 86. Kelly DP and Scarpulla RC. Transcriptional regulatory circuits controlling mitochondrial biogenesis and function. Genes Dev 18: 357–368, 2004 [DOI] [PubMed] [Google Scholar]
- 87. Kinch L, Grishin NV, and Brugarolas J. Succination of Keap1 and activation of Nrf2-dependent antioxidant pathways in FH-deficient papillary renal cell carcinoma type 2. Cancer Cell 20: 418–420, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 88. Kirichok Y, Krapivinsky G, and Clapham DE. The mitochondrial calcium uniporter is a highly selective ion channel. Nature 427: 360–364, 2004 [DOI] [PubMed] [Google Scholar]
- 89. Knoefler D, Thamsen M, Koniczek M, Niemuth NJ, Diederich A-K, and Jakob U. Quantitative in vivo redox sensors uncover oxidative stress as an early event in life. Mol Cell 47: 767–776, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Koivunen P, Lee S, Duncan CG, Lopez G, Lu G, Ramkissoon S, Losman JA, Joensuu P, Bergmann U, and Gross S. Transformation by the (R)-enantiomer of 2-hydroxyglutarate linked to EGLN activation. Nature 483: 484, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Kosta A, Luciani M-F, Geerts WJC, and Golstein P. Marked mitochondrial alterations upon starvation without cell death, caspases or Bcl-2 family members. Biochim Biophys Acta 1783: 2013–2019, 2008 [DOI] [PubMed] [Google Scholar]
- 92. Koves TR, Ussher JR, Noland RC, Slentz D, Mosedale M, Ilkayeva O, Bain J, Stevens R, Dyck JR, and Newgard CB. Mitochondrial overload and incomplete fatty acid oxidation contribute to skeletal muscle insulin resistance. Cell Metab 7: 45–56, 2008 [DOI] [PubMed] [Google Scholar]
- 93. Kremer LS, Bader DM, Mertes C, Kopajtich R, Pichler G, Iuso A, Haack TB, Graf E, Schwarzmayr T, and Terrile C. Genetic diagnosis of Mendelian disorders via RNA sequencing. Nat Commun 8: 15824, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Kunau W-H, Dommes V, and Schulz H. β-Oxidation of fatty acids in mitochondria, peroxisomes, and bacteria: a century of continued progress. Prog Lipid Res 34: 267–342, 1995 [DOI] [PubMed] [Google Scholar]
- 95. Labbé RF, Vreman HJ, and Stevenson DK. Zinc protoporphyrin: a metabolite with a mission. Clin Chem 45: 2060–2072, 1999 [PubMed] [Google Scholar]
- 96. Lalitha K and Rani P. Mitochondrial selenium-75 uptake and regulation revealed by kinetic analysis. Biol Trace Elem Res 49: 21–42, 1995 [DOI] [PubMed] [Google Scholar]
- 97. Lambeth JD. Nox enzymes, ROS, and chronic disease: an example of antagonistic pleiotropy. Free Radic Biol Med 43: 332–347, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 98. Lassègue B, Sorescu D, Szöcs K, Yin Q, Akers M, Zhang Y, Grant SL, Lambeth JD, and Griendling KK. Novel gp91 phox homologues in vascular smooth muscle cells: nox1 mediates angiotensin II–induced superoxide formation and redox-sensitive signaling pathways. Circ Res 88: 888–894, 2001 [DOI] [PubMed] [Google Scholar]
- 99. This reference has been deleted.
- 100. Lettieri-Barbato D, Ioannilli L, Aquilano K, Ciccarone F, Rosina M, and Ciriolo MR. FoxO1 localizes to mitochondria of adipose tissue and is affected by nutrient stress. Metabolism 95: 84–92, 2019 [DOI] [PubMed] [Google Scholar]
- 101. Levine BS and Coburn JW. Magnesium, the mimic/antagonist of calcium. N Engl J Med 310: 1253–1255, 1984 [DOI] [PubMed] [Google Scholar]
- 102. Li S, Park Y, Duraisingham S, Strobel FH, Khan N, Soltow QA, Jones DP, and Pulendran B. Predicting network activity from high throughput metabolomics. PLoS Comput Biol 9: e1003123, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 103. Liu F, Lössl P, Rabbitts BM, Balaban RS, and Heck AJ. The interactome of intact mitochondria by cross-linking mass spectrometry provides evidence for coexisting respiratory supercomplexes. Mol Cell Proteomics 17: 216–232, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Liu Z and Butow RA. Mitochondrial retrograde signaling. Annu Rev Genet 40: 159–185, 2006 [DOI] [PubMed] [Google Scholar]
- 105. Lonn E, Bosch J, Yusuf S, Sheridan P, Pogue J, Arnold J, Ross C, Arnold A, Sleight P, and Probstfield J. Effects of long-term vitamin E supplementation on cardiovascular events and cancer: a randomized controlled trial. JAMA 293: 1338–1347, 2005 [DOI] [PubMed] [Google Scholar]
- 106. Loscalzo J, Kohane I, and Barabasi AL.. Human disease classification in the postgenomic era: a complex systems approach to human pathobiology. Mol Syst Biol 3, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 107. Love NR, Chen Y, Ishibashi S, Kritsiligkou P, Lea R, Koh Y, Gallop JL, Dorey K, and Amaya E. Amputation-induced reactive oxygen species are required for successful Xenopus tadpole tail regeneration. Nat Cell Biol 15: 222–228, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 108. Lucas S, Chen G, Aras S, and Wang J. Serine catabolism is essential to maintain mitochondrial respiration in mammalian cells. Life Sci Alliance 1: e201800036, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Masgras I, Ciscato F, Brunati AM, Tibaldi E, Indraccolo S, Curtarello M, Chiara F, Cannino G, Papaleo E, and Lambrughi M. Absence of neurofibromin induces an oncogenic metabolic switch via mitochondrial ERK-mediated phosphorylation of the chaperone TRAP1. Cell Rep 18: 659–672, 2017 [DOI] [PubMed] [Google Scholar]
- 110. Matthews L, Gopinath G, Gillespie M, Caudy M, Croft D, de Bono B, Garapati P, Hemish J, Hermjakob H, and Jassal B. Reactome knowledgebase of human biological pathways and processes. Nucleic Acids Res 37: D619–D622, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Matuszczyk J-C, Teleki A, Pfizenmaier J, and Takors R. Compartment-specific metabolomics for CHO reveals that ATP pools in mitochondria are much lower than in cytosol. Biotechnol J 10: 1639–1650, 2015 [DOI] [PubMed] [Google Scholar]
- 112. Menni C, Fauman E, Erte I, Perry JR, Kastenmüller G, Shin S-Y, Petersen A-K, Hyde C, Psatha M, and Ward KJ. Biomarkers for type 2 diabetes and impaired fasting glucose using a nontargeted metabolomics approach. Diabetes 62: 4270–4276, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 113. Meyer JN, Leung MC, Rooney JP, Sendoel A, Hengartner MO, Kisby GE, and Bess AS. Mitochondria as a target of environmental toxicants. Toxicol Sci 134: 1–17, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Miller WL. Steroid hormone synthesis in mitochondria. Mol Cell Endocrinol 379: 62–73, 2013 [DOI] [PubMed] [Google Scholar]
- 115. Mishra P and Chan DC. Mitochondrial dynamics and inheritance during cell division, development and disease. Nat Rev Mol Cell Biol 15: 634, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Misra BB, Langefeld C, Olivier M, and Cox LA. Integrated omics: tools, advances and future approaches. J Mol Endocrinol 62: R21–R45, 2019 [DOI] [PubMed] [Google Scholar]
- 117. Monné M and Palmieri F. Antiporters of the mitochondrial carrier family. In: Current Topics in Membranes, edited by Bevensee MO. Amsterdam, Netherlands: Elsevier, 2014, pp. 289–320 [DOI] [PubMed] [Google Scholar]
- 118. Mühlberger I, Wilflingseder J, Bernthaler A, Fechete R, Lukas A, and Perco P. Computational analysis workflows for omics data interpretation. In: Bioinformatics for Omics Data: Methods and Protocols, edited by Mayer B. Totowa, NJ: Humana Press, 2011, pp. 379–397 [DOI] [PubMed] [Google Scholar]
- 119. Nagalakshmi U, Wang Z, Waern K, Shou C, Raha D, Gerstein M, and Snyder M. The transcriptional landscape of the yeast genome defined by RNA sequencing. Science 320: 1344–1349, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 120. Nebert DW and Russell DW. Clinical importance of the cytochromes P450. Lancet 360: 1155–1162, 2002 [DOI] [PubMed] [Google Scholar]
- 121. Nègre-Salvayre A, Hirtz C, Carrera G, Cazenave R, Troly M, Salvayre R, Pénicaud L, and Casteilla L. A role for uncoupling protein-2 as a regulator of mitochondrial hydrogen peroxide generation. FASEB J 11: 809–815, 1997 [PubMed] [Google Scholar]
- 121a. Nelson DL and Cox MM. Lehninger Principles of Biochemistry, 6th ed. New York, NY: Macmillan Learning, 2012 [Google Scholar]
- 122. Ngo JK and Davies KJ. Importance of the lon protease in mitochondrial maintenance and the significance of declining lon in aging. Ann N Y Acad Sci 1119: 78–87, 2007 [DOI] [PubMed] [Google Scholar]
- 123. Nowicki S and Gottlieb E. Oncometabolites: tailoring our genes. FEBS J 282: 2796–2805, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 124. Ostrander DB, Zhang M, Mileykovskaya E, Rho M, and Dowhan W. Lack of mitochondrial anionic phospholipids causes an inhibition of translation of protein components of the electron transport chain. A yeast genetic model system for the study of anionic phospholipid function in mitochondria. J Biol Chem 276: 25262–25272, 2001 [DOI] [PubMed] [Google Scholar]
- 125. Owen OE. Ketone bodies as a fuel for the brain during starvation. Biochem Mol Biol Educ 33: 246–251, 2005 [Google Scholar]
- 126. Owen OE, Kalhan SC, and Hanson RW. The key role of anaplerosis and cataplerosis for citric acid cycle function. J Biol Chem 277: 30409–30412, 2002 [DOI] [PubMed] [Google Scholar]
- 127. Ozanne SE. Metabolic programming—knowns, unknowns and possibilities. Nat Rev Endocrinol 11: 67–68, 2015 [DOI] [PubMed] [Google Scholar]
- 128. Palmieri F. The mitochondrial transporter family SLC25: identification, properties and physiopathology. Mol Aspects Med 34: 465–484, 2013 [DOI] [PubMed] [Google Scholar]
- 129. Papp LV, Lu J, Holmgren A, and Khanna KK. From selenium to selenoproteins: synthesis, identity, and their role in human health. Antioxid Redox Signal 9: 775–806, 2007 [DOI] [PubMed] [Google Scholar]
- 130. Park S, Chang C-Y, Safi R, Liu X, Baldi R, Jasper JS, Anderson GR, Liu T, Rathmell JC, and Dewhirst MW. ERRα-regulated lactate metabolism contributes to resistance to targeted therapies in breast cancer. Cell Rep 15: 323–335, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 131. Patel MS and Srinivasan M. Metabolic programming: causes and consequences. J Biol Chem 277: 1629–1632, 2002 [DOI] [PubMed] [Google Scholar]
- 132. Pierrel F, Cobine PA, and Winge DR. Metal Ion availability in mitochondria. Biometals 20: 675–682, 2007 [DOI] [PubMed] [Google Scholar]
- 133. Pondarré C, Antiochos BB, Campagna DR, Clarke SL, Greer EL, Deck KM, McDonald A, Han A-P, Medlock A, and Kutok JL. The mitochondrial ATP-binding cassette transporter Abcb7 is essential in mice and participates in cytosolic iron–sulfur cluster biogenesis. Hum Mol Genet 15: 953–964, 2006 [DOI] [PubMed] [Google Scholar]
- 134. Porporato PE, Filigheddu N, Pedro JMB-S, Kroemer G, and Galluzzi L. Mitochondrial metabolism and cancer. Cell Res 28: 265–280, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 135. Rahman J and Rahman S. Mitochondrial medicine in the omics era. Lancet 391: 2560–2574, 2018 [DOI] [PubMed] [Google Scholar]
- 136. Rambold AS, Kostelecky B, Elia N, and Lippincott-Schwartz J. Tubular network formation protects mitochondria from autophagosomal degradation during nutrient starvation. Proc Natl Acad Sci U S A 108: 10190–10195, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 137. Rayman MP. Selenium and human health. Lancet 379: 1256–1268, 2012 [DOI] [PubMed] [Google Scholar]
- 138. Ricchi M, Odoardi MR, Carulli L, Anzivino C, Ballestri S, Pinetti A, Fantoni LI, Marra F, Bertolotti M, Banni S, Lonardo A, Carulli N, and Loria P. Differential effect of oleic and palmitic acid on lipid accumulation and apoptosis in cultured hepatocytes. J Gastroenterol Hepatol 24: 830–840, 2009 [DOI] [PubMed] [Google Scholar]
- 139. Richardson DR, Lane DJ, Becker EM, Huang ML-H, Whitnall M, Rahmanto YS, Sheftel AD, and Ponka P. Mitochondrial iron trafficking and the integration of iron metabolism between the mitochondrion and cytosol. Proc Natl Acad Sci U S A 107: 10775–10782, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 140. Rizzuto R, De Stefani D, Raffaello A, and Mammucari C. Mitochondria as sensors and regulators of calcium signalling. Nat Rev Mol Cell Biol 13: 566, 2012 [DOI] [PubMed] [Google Scholar]
- 141. Roede JR, Park Y, Li S, Strobel FH, and Jones DP. Detailed mitochondrial phenotyping by high resolution metabolomics. PLoS One 7: e33020, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 142. Ross-Inta C, Tsai C-Y, and Giulivi C. The mitochondrial pool of free amino acids reflects the composition of mitochondrial DNA-encoded proteins: indication of a post-translational quality control for protein synthesis. Biosci Rep 28: 239–249, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 143. Rousset S, Alves-Guerra M-C, Mozo J, Miroux B, Cassard-Doulcier A-M, Bouillaud F, and Ricquier D. The biology of mitochondrial uncoupling proteins. Diabetes 53: S130–S135, 2004 [DOI] [PubMed] [Google Scholar]
- 144. Ryan MT and Hoogenraad NJ. Mitochondrial-nuclear communications. Annu Rev Biochem 76: 701–722, 2007 [DOI] [PubMed] [Google Scholar]
- 145. Sancho P, Burgos-Ramos E, Tavera A, Kheir TB, Jagust P, Schoenhals M, Barneda D, Sellers K, Campos-Olivas R, and Graña O. MYC/PGC-1α balance determines the metabolic phenotype and plasticity of pancreatic cancer stem cells. Cell Metab 22: 590–605, 2015 [DOI] [PubMed] [Google Scholar]
- 146. Sandoval IT, Delacruz RGC, Miller BN, Hill S, Olson KA, Gabriel AE, Boyd K, Satterfield C, Van Remmen H, and Rutter J. A metabolic switch controls intestinal differentiation downstream of Adenomatous polyposis coli (APC). Elife 6: e22706, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 147. Sano S, Inoue S, Tanabe Y, Sumiya C, and Koike S. Significance of mitochondria for porphyrin and heme biosynthesis. Science 129: 275–276, 1959 [DOI] [PubMed] [Google Scholar]
- 148. Sato R, Atsuta Y, Imai Y, Taniguchi S, and Okuda K. Hepatic mitochondrial cytochrome P-450: isolation and functional characterization. Proc Natl Acad Sci U S A 74: 5477–5481, 1977 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 149. Scarpulla RC. Nuclear activators and coactivators in mammalian mitochondrial biogenesis. Biochim Biophys Acta 1576: 1–14, 2002 [DOI] [PubMed] [Google Scholar]
- 150. Schaedler TA, Thornton JD, Kruse I, Schwarzländer M, Meyer AJ, van Veen HW, and Balk J. A conserved mitochondrial ATP-binding cassette transporter exports glutathione polysulfide for cytosolic metal cofactor assembly. J Biol Chem 289: 23264–23274, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 151. Scherz-Shouval R and Elazar Z. ROS, mitochondria and the regulation of autophagy. Trends Cell Biol 17: 422–427, 2007 [DOI] [PubMed] [Google Scholar]
- 152. Schonauer MS, Kastaniotis AJ, Kursu VA, Hiltunen JK, and Dieckmann CL. Lipoic acid synthesis and attachment in yeast mitochondria. J Biol Chem 284: 23234–23242, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 153. Sciacovelli M, Guzzo G, Morello V, Frezza C, Zheng L, Nannini N, Calabrese F, Laudiero G, Esposito F, and Landriscina M. The mitochondrial chaperone TRAP1 promotes neoplastic growth by inhibiting succinate dehydrogenase. Cell Metab 17: 988–999, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 154. Seitz H, Müller M, Krone W, and Tarnowski W. Coordinate control of intermediary metabolism in rat liver by the insulin/glucagon ratio during starvation and after glucose refeeding: regulatory significance of long-chain acyl-CoA and cyclic AMP. Arch Biochem Biophys 183: 647–663, 1977 [DOI] [PubMed] [Google Scholar]
- 155. Seo YA, Lopez V, and Kelleher SL. A histidine-rich motif mediates mitochondrial localization of ZnT2 to modulate mitochondrial function. Am J Physiol Cell Physiol 300: C1479–C1489, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 156. Shadel Gerald S and Horvath Tamas L. Mitochondrial ROS signaling in organismal homeostasis. Cell 163: 560–569, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 157. Shen Y, Wang Y, Chen T, Gao F, Gong J, Abramczyk D, Walker R, Zhao H, Chen S, Liu W, Luo Y, Müller CA, Paul-Dubois-Taine A, Alver B, Stracquadanio G, Mitchell LA, Luo Z, Fan Y, Zhou B, Wen B, Tan F, Wang Y, Zi J, Xie Z, Li B, Yang K, Richardson SM, Jiang H, French CE, Nieduszynski CA, Koszul R, Marston AL, Yuan Y, Wang J, Bader JS, Dai J, Boeke JD, Xu X, Cai Y, and Yang H. Deep functional analysis of synII, a 770-kilobase synthetic yeast chromosome. Science 355: eaaf4791, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 158. Sies H. Nicotinamide nucleotide compartmentation. In: Metabolic Compartmentation, edited by Sies H. New York, NY: Academic Press, 1982, pp. 205–231 [Google Scholar]
- 159. Siess EA, Brocks DG, and Wieland OH. Distribution of metabolites between the cytosolic and mitochondrial compartments of hepatocytes isolated from fed rats. Hoppe Seylers Z Physiol Chem 359: 785–798, 1978 [DOI] [PubMed] [Google Scholar]
- 160. Smith AC and Robinson AJ. MitoMiner v3. 1, an update on the mitochondrial proteomics database. Nucleic Acids Res 44: D1258–D1261, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 161. Son M, Kwon Y, Son M, Seol B, Choi H, Ryu S, Choi C, and Cho Y. Mitofusins deficiency elicits mitochondrial metabolic reprogramming to pluripotency. Cell Death Differ 22: 1957, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 162. Stefely JA and Pagliarini DJ. Biochemistry of mitochondrial coenzyme Q biosynthesis. Trends Biochem Sci 42: 824–843, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 163. Taylor EB. Functional properties of the mitochondrial carrier system. Trends Cell Biol 27: 633–644, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 164. Uppal K, Ma C, Go YM, Jones DP, and Wren J. xMWAS: a data-driven integration and differential network analysis tool. Bioinformatics 34: 701–702, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 165. Uppal K, Soltow QA, Promislow DE, Wachtman LM, Quyyumi AA, and Jones DP. MetabNet: an R package for metabolic association analysis of high-resolution metabolomics data. Front Bioeng Biotechnol 3: 87, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 166. Uppal K, Walker DI, Liu K, Li S, Go Y-M, and Jones DP. Computational metabolomics: a framework for the million metabolome. Chem Res Toxicol 29: 1956–1975, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 167. Van der Bliek AM, Shen Q, and Kawajiri S. Mechanisms of mitochondrial fission and fusion. Cold Spring Harb Perspect Biol 5: a011072, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 168. Vellinga TT, Borovski T, de Boer VC, Fatrai S, van Schelven S, Trumpi K, Verheem A, Snoeren N, Emmink BL, and Koster J. SIRT1/PGC1α-dependent increase in oxidative phosphorylation supports chemotherapy resistance of colon cancer. Clin Cancer Res 21: 2870–2879, 2015 [DOI] [PubMed] [Google Scholar]
- 169. Verbost P, Flik G, Lock R, and Bonga SW. Cadmium inhibits plasma membrane calcium transport. J Membr Biol 102: 97–104, 1988 [DOI] [PubMed] [Google Scholar]
- 170. Vest KE, Leary SC, Winge DR, and Cobine PA. Copper import into the mitochondrial matrix in Saccharomyces cerevisiae is mediated by Pic2, a mitochondrial carrier family protein. J Biol Chem 288: 23884–23892, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 171. Vögtle F-N, Burkhart JM, Gonczarowska-Jorge H, Kücükköse C, Taskin AA, Kopczynski D, Ahrends R, Mossmann D, Sickmann A, and Zahedi RP. Landscape of submitochondrial protein distribution. Nat Commun 8: 290, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 172. Voit E. A First Course in Systems Biology. New York:Garland Science, 2018 [Google Scholar]
- 173. Vyas S, Zaganjor E, and Haigis MC. Mitochondria and cancer. Cell 166: 555–566, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 174. Walberg M and Clayton DA. In vitro transcription of human mitochondrial DNA. Identification of specific light strand transcripts from the displacement loop region. J Biol Chem 258: 1268–1275, 1983 [PubMed] [Google Scholar]
- 175. Waldron KJ, Rutherford JC, Ford D, and Robinson NJ. Metalloproteins and metal sensing. Nature 460: 823, 2009 [DOI] [PubMed] [Google Scholar]
- 176. Wanders RJA, Waterham HR, and Ferdinandusse S.. Metabolic interplay between peroxisomes and other subcellular organelles including mitochondria and the endoplasmic reticulum. Front Cell Dev Biol 3, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 177. Wanichthanarak K, Fahrmann JF, and Grapov D. Genomic, proteomic, and metabolomic data integration strategies. Biomark Insights 10: 1–6, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 178. Warburg O. The metabolism of carcinoma cells. Cancer Res 9: 148–163, 1925 [Google Scholar]
- 179. Waterlow J. The mysteries of nitrogen balance. Nutr Res Rev 12: 25–54, 1999 [DOI] [PubMed] [Google Scholar]
- 180. Weinberg SE and Chandel NS. Targeting mitochondria metabolism for cancer therapy. Nat Chem Biol 11: 9, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 181. Westermann B. Mitochondrial fusion and fission in cell life and death. Nat Rev Mol Cell Biol 11: 872, 2010 [DOI] [PubMed] [Google Scholar]
- 182. Westermann B. Bioenergetic role of mitochondrial fusion and fission. Biochim Biophys Acta 1817: 1833–1838, 2012 [DOI] [PubMed] [Google Scholar]
- 183. Winge DR. Filling the mitochondrial copper pool. J Biol Chem 293: 1897–1898, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 184. Winterbourn CC. The biological chemistry of hydrogen peroxide. In: Methods in Enzymology, edited by Cadenas E and Packer L. Amsterdam, Netherlands: Elsevier, 2013, pp. 3–25 [DOI] [PubMed] [Google Scholar]
- 185. Wishart DS, Tzur D, Knox C, Eisner R, Guo AC, Young N, Cheng D, Jewell K, Arndt D, Sawhney S, Fung C, Nikolai L, Lewis M, Coutouly MA, Forsythe I, Tang P, Shrivastava S, Jeroncic K, Stothard P, Amegbey G, Block D, Hau DD, Wagner J, Miniaci J, Clements M, Gebremedhin M, Guo N, Zhang Y, Duggan GE, Macinnis GD, Weljie AM, Dowlatabadi R, Bamforth F, Clive D, Greiner R, Li L, Marrie T, Sykes BD, Vogel HJ, and Querengesser L. HMDB: the human metabolome database. Nucleic Acids Res 35: D521–D526, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 186. Wolff NA, Ghio AJ, Garrick LM, Garrick MD, Zhao L, Fenton RA, and Thévenod F. Evidence for mitochondrial localization of divalent metal transporter 1 (DMT1). FASEB J 28: 2134–2145, 2014 [DOI] [PubMed] [Google Scholar]
- 187. Wu Y, Williams EG, Dubuis S, Mottis A, Jovaisaite V, Houten SM, Argmann CA, Faridi P, Wolski W, Kutalik Z, Zamboni N, Auwerx J, and Aebersold R. Multilayered genetic and omics dissection of mitochondrial activity in a mouse reference population. Cell 158: 1415–1430, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 188. This reference has been deleted
- 189. Wu Z, Puigserver P, Andersson U, Zhang C, Adelmant G, Mootha V, Troy A, Cinti S, Lowell B, Scarpulla RC, and Spiegelman BM. Mechanisms controlling mitochondrial biogenesis and respiration through the thermogenic coactivator PGC-1. Cell 98: 115–124, 1999 [DOI] [PubMed] [Google Scholar]
- 190. Xiao D, Zeng L, Yao K, Kong X, Wu G, and Yin Y. The glutamine-alpha-ketoglutarate (AKG) metabolism and its nutritional implications. Amino Acids 48: 2067–2080, 2016 [DOI] [PubMed] [Google Scholar]
- 191. Xiao M, Yang H, Xu W, Ma S, Lin H, Zhu H, Liu L, Liu Y, Yang C, Xu Y, Zhao S, Ye D, Xiong Y, and Guan K-L. Inhibition of α-KG-dependent histone and DNA demethylases by fumarate and succinate that are accumulated in mutations of FH and SDH tumor suppressors. Genes Dev 26: 1326–1338, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 192. Xu W, Yang H, Liu Y, Yang Y, Wang P, Kim S-H, Ito S, Yang C, Wang P, and Xiao M-T. Oncometabolite 2-hydroxyglutarate is a competitive inhibitor of α-ketoglutarate-dependent dioxygenases. Cancer Cell 19: 17–30, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 193. Yang M, Soga T, and Pollard PJ. Oncometabolites: linking altered metabolism with cancer. J Clin Invest 123: 3652–3658, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 194. Yugi K, Kubota H, Hatano A, and Kuroda S. Trans-omics: how to reconstruct biochemical networks across multiple “omic” layers. Trends Biotechnol 34: 276–290, 2016 [DOI] [PubMed] [Google Scholar]
- 195. Zachar Z, Marecek J, Maturo C, Gupta S, Stuart SD, Howell K, Schasuble A, Lem J, Piramzadian A, Karnik S, Lee K, Rodriguez R, Shorr R, and Bingham PM. Non-redox-active lipoate derivates disrupt cancer cell mitochondrial metabolism and are potent anticancer agents in vivo. J Mol Med 89: 1137, 2011 [DOI] [PubMed] [Google Scholar]
- 196. Zhang Q, Raoof M, Chen Y, Sumi Y, Sursal T, Junger W, Brohi K, Itagaki K, and Hauser CJ. Circulating mitochondrial DAMPs cause inflammatory responses to injury. Nature 464: 104, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 197. Zhou G and Xia J. OmicsNet: a web-based tool for creation and visual analysis of biological networks in 3D space. Nucleic Acids Res 46: W514–W522, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 198. Zong H, Ren JM, Young LH, Pypaert M, Mu J, Birnbaum MJ, and Shulman GI. AMP kinase is required for mitochondrial biogenesis in skeletal muscle in response to chronic energy deprivation. Proc Natl Acad Sci U S A 99: 15983–15987, 2002 [DOI] [PMC free article] [PubMed] [Google Scholar]