Abstract
Significance: S-nitrosylation, the post-translational modification by nitric oxide (NO) to form S-nitrosothiols (SNOs), regulates diverse aspects of cellular function, and aberrant S-nitrosylation (nitrosative stress) is implicated in disease, from neurodegeneration to cancer. Essential roles for S-nitrosylation have been demonstrated in microbes, plants, and animals; notably, bacteria have often served as model systems for elucidation of general principles.
Recent Advances: Recent conceptual advances include the idea of a molecular code through which proteins sense and differentiate S-nitrosothiol (SNO) from alternative oxidative modifications, providing the basis for specificity in SNO signaling. In Escherichia coli, S-nitrosylation relies on an enzymatic cascade that regulates, and is regulated by, the transcription factor OxyR under anaerobic conditions. S-nitrosylated OxyR activates an anaerobic regulon of >100 genes that encode for enzymes that both mediate S-nitrosylation and protect against nitrosative stress.
Critical Issues: Mitochondria originated from endosymbiotic bacteria and generate NO under hypoxic conditions, analogous to conditions in E. coli. Nitrosative stress in mitochondria has been implicated in Alzheimer's and Parkinson's disease, among others. Many proteins that are S-nitrosylated in mitochondria are also S-nitrosylated in E. coli. Insights into enzymatic regulation of S-nitrosylation in E. coli may inform the identification of disease-relevant regulatory machinery in mammalian systems.
Future Directions: Using E. coli as a model system, in-depth analysis of the anaerobic response controlled by OxyR may lead to the identification of enzymatic mechanisms regulating S-nitrosylation in particular, and hypoxic signaling more generally, providing novel insights into analogous mechanisms in mammalian cells and within dysfunctional mitochondria that characterize neurodegenerative diseases.
Keywords: OxyR, S-nitrosylation, nitrosative stress, Hcp, hypoxia, mitochondrial disorders
Introduction
Bacteria perpetually encounter and respond to redox stresses in their environment, particularly oxidative and nitrosative stresses encountered during infection. Facultative bacteria such as Escherichia coli also experience endogenous redox stress: oxidative stress when growing aerobically in an oxygen-rich environment and nitrosative stress when growing anaerobically in the presence of nitrate. The bacterial transcription factor OxyR acts as a regulator of both stresses, undergoing oxidative or nitrosative modifications of a single regulatory Cys (S-sulfenylation/disulfide or S-nitrosylation, respectively) to direct distinct transcriptional responses. The OxyR oxidative stress regulon of ∼30 genes is well characterized, and includes genes with anti-oxidative functions. Similarly, nitrosative stress promotes the OxyR-dependent activation of a much larger, but less well-characterized regulon of ∼100 genes, including the gene for a master regulator of S-nitrosylation, the S-nitrosylase hybrid cluster protein (Hcp). More specifically, unlike oxidation of OxyR, which is a consequence of high levels of oxidants acting relatively nonspecifically, S-nitrosylation is mediated enzymatically. Targeted S-nitrosylation of proteins (including of OxyR itself) by Hcp confers a survival advantage. Thus, remarkably, targeted S-nitrosylation of specific proteins protects cells from the effects of aberrant and nontargeted S-nitrosylation due to external nitrosative stress. Here, we review this central function for OxyR—the classic aerobic regulator of oxidative stress—in protecting against nitrosative stress under anaerobic conditions, and reveal parallels between regulation of S-nitrosylation in E. coli and hypoxic signaling by nitric oxide (NO) in mammalian cells, and in mitochondria in particular.
Bacteria serve as model organisms that allow for identification and characterization of redox stress responses due to the ease of genetic manipulation coupled with the flexibility in growth conditions. Facultative bacteria such as E. coli can be grown aerobically or anaerobically; in the absence of oxygen, E. coli can extract energy through respiration by using any of a number of electron acceptors, with different metabolic outcomes, or they can even utilize fermentation in the absence of electron acceptors (105). Bacteria can encounter both oxidative and nitrosative stresses resulting from their environment or their own metabolism, and studies in bacteria have led to identification and/or better understanding of key enzymes that counter these stresses (9, 61).
During evolution, mitochondria in eukaryotic cells originated from endosymbiotic bacteria (84). Bacteria have been used as model systems to study mitochondrial functions, especially for studies involving respiratory complexes (80), and it is highly likely that at least some mechanisms of generation of and protection from redox stress are conserved. Mitochondria have long been known to be the source of damaging reactive oxygen species (ROS), especially superoxide (O2−) produced by the one-electron reduction of oxygen (67). Moreover, dismutation of superoxide leads to the formation of hydrogen peroxide (H2O2) that can reversibly oxidize proteins and can lead to redox signaling (67). More recently, it has been appreciated that mitochondria are also a source of NO, primarily under hypoxic conditions (54), and that S-nitrosothiol (SNO)-based signaling is prevalent within mitochondria. Accumulating evidence suggests that aberrant S-nitrosylation of mitochondrial proteins may play an important role in neurodegenerative diseases. In this review, we focus on the consequences of NO generation by bacteria under hypoxia, and by analogy in mitochondria (54).
Respiration-Mediated Generation of NO and ROS
In all organisms, aerobic growth is associated with the obligate production of reactive oxygen intermediates from the incomplete reduction of oxygen during metabolism (32, 44). The production of oxygen radicals and reactive species, including O2−, H2O2, and hydroxyl radicals (HO•), leads to endogenous oxidative stress and to consequent induction of enzymatic defense mechanisms (32, 43). Early studies in bacteria demonstrated the essential importance of these protective enzymes for cellular function. Complete lack of superoxide dismutase activity in E. coli inhibits growth in air; exogenous amino acids rescue growth by overcoming the oxidative inhibition of enzymes involved in amino acid biosynthesis (9). Similarly, bacteria in which H2O2-consuming activities, catalase (KatG) and alkyl hydroperoxide reductase (AhpC), are absent have a growth defect during aerobic conditions (89). In this case, growth inhibition is a result of oxidative damage at iron–sulfur clusters (29), DNA damage (8), and direct oxidation of reactive thiols in proteins (111). Thus, bacteria contain enzymatic activities to counter these inevitable stressors.
In the absence of oxygen, facultative anaerobes such as E. coli preferentially utilize nitrate as the electron acceptor for respiration (105). During anaerobic respiration on nitrate (ARN), the nitrate reductase NarGHI is induced (92), which serves as a source of NO by reducing nitrite (a reaction secondary to its function in nitrate reduction) (45, 79). NO produced during ARN exerts an endogenous nitrosative stress (90, 112) and, consequently, leads to the induction of genes previously shown to be activated by toxic amounts of NO, including hmp, hcp-hcr, yeaR-yoaG, and ytfE (27, 42, 71, 75). Notably, nitrosative stress by NO also disrupts iron–sulfur clusters in proteins by formation of iron nitrosyl species (19), damages DNA (109), and modifies Cys residues, mainly via S-nitrosylation (to form SNOs) (39). Thus, the endogenous nitrosative stress during ARN appears analogous to the endogenous oxidative stress incurred during aerobic growth.
Redox-Based Sensors and the Transcription Factor OxyR
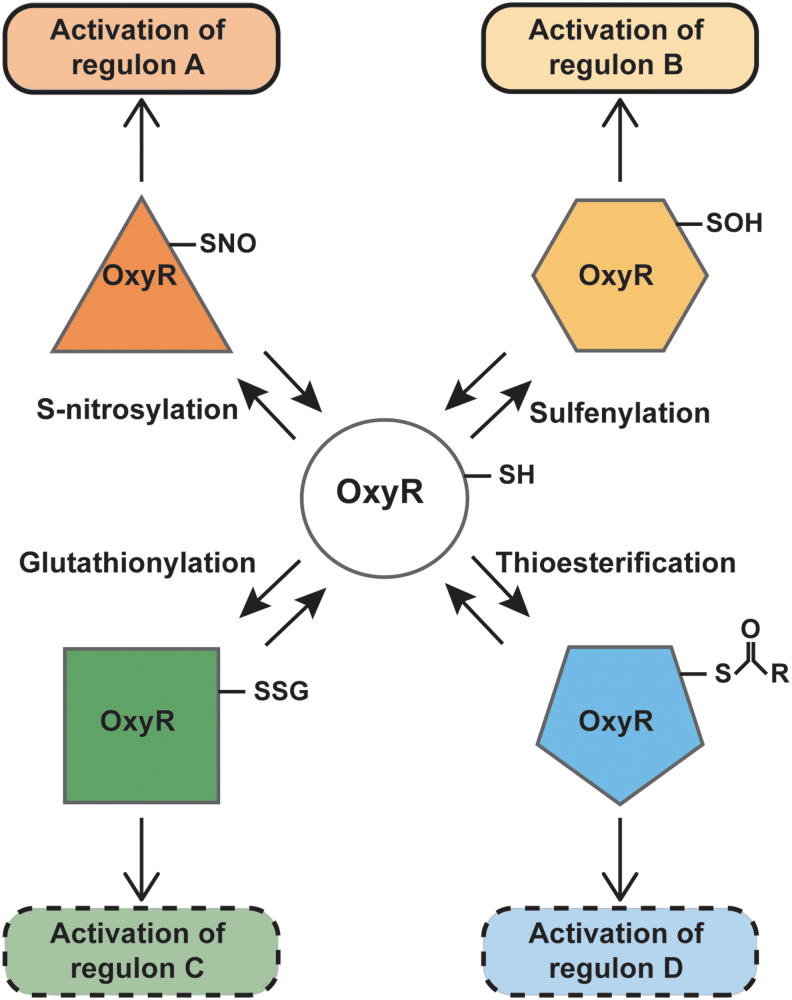
The analogy to oxidative stress centers on thiol groups, which may undergo several redox-based modifications in the context of redox-based stress—some specific to the nature of the stress and some shared across stressors. Reactive cysteines generally have a low pKa and are ionized at physiological pH; the thiolate is often stabilized by hydrogen bonding, by proximity to positively charged residues or by location at the N-terminus of an α-helix (85). The bacterial transcription factor OxyR, a member of the LysR family of transcription factors (88), possesses one such reactive cysteine, Cys199. Based on the crystal structure of OxyR, it has been suggested that interaction with a positively charged arginine residue contributes to the lowering of the pKa of Cys199 (12) to make it particularly susceptible to redox modifications. It has been further proposed that alternative redox modifications of OxyR (52) can lead to distinct transcriptional responses, thereby forming the basis of a molecular redox code (Fig. 1). We review here new evidence in favor of the concept that OxyR senses oxidative and nitrosative stresses through alternative modifications of Cys199, leading to distinct transcriptional consequences. We further discuss the protective function of an OxyR-regulated gene, hcp, in endogenous S-nitrosylation, and hypothesize that similar enzymatic mechanisms may function in mitochondria.
FIG. 1.
Conceptual basis for redox-based molecular code. OxyR can sense and differentiate among Cys-based modifications, providing the basis for a molecular code for redox signaling. Different redox modifications of OxyR can lead to transcription of unique regulons. The SOH and SNO-dependent regulons have been identified, whereas regulons activated by glutathionylation and thioesterification are hypothetical. SNO, S-nitrosylated; SOH, S-sulfenylation.
E. coli as a Model for Nitrosative Stress
Nitrosative stress is characterized by excessive S-nitrosylation of proteins. Intracellular protein–SNO levels are in equilibrium with low-molecular-weight SNOs such as S-nitrosoglutathione (GSNO). One mechanism by which cells can regulate SNO–protein levels is by regulating intracellular GSNO. The bacterial enzyme that metabolizes GSNO (thereby reducing SNO–protein levels) was identified as a glutathione-dependent formaldehyde dehydrogenase (ADH5) (61). This enzyme is conserved from E. coli to yeast to humans, and it was renamed S-nitrosoglutathione reductase (GSNOR). Since its discovery in E. coli, numerous studies have implicated GSNOR in human diseases, including Alzheimer's disease (118), asthma (76), cardiovascular disease (60), and several cancers (108) [reviewed in Rizza and Filomeni (82)]. Further, GSNOR has emerged as a therapeutic target, and GSNOR inhibitors have shown potential where increased S-nitrosylation is beneficial (18, 40, 77). This is one example of how E. coli has been used as a model to identify SNO-regulating enzymatic activity that is conserved across phylogeny with implications in human health and disease. One limitation in using E. coli to identify potential SNO-regulating enzymes in humans is the lack of homology between many E. coli and mammalian proteins; however, general principles of enzymatic regulation of S-nitrosylation can still be studied in E. coli and extrapolated to mammalian systems, as described in the Endogenous Oxidative and Nitrosative Stress section.
Endogenous Oxidative and Nitrosative Stress
Bacteria are exposed to exogenous redox stresses in their environment, and even more so during infection where the host innate immune system targets invaders by producing ROS and reactive nitrogen species (RNS) via NADPH oxidases and inducible nitric oxide synthase (iNOS), respectively [reviewed in Nathan and Shiloh (68)]. However, as discussed earlier, growth in an aerobic environment also leads to endogenous ROS-mediated oxidative stress, and thus bacteria have evolved shared mechanisms to protect against both endogenous and exogenous ROS stresses. Similarly, anaerobic growth in the presence of nitrate (the preferred electron acceptor for respiration in the absence of oxygen) leads to the production of NO/RNS, and hence endogenous nitrosative stress. Although the endogenous levels of RNS are far lower than those generated by the host immune system, defenses against constitutive RNS also help protect bacteria during infection (3). For example, bacterial flavohemoglobin (Hmp), an enzyme that consumes NO under both aerobic and anaerobic conditions (37), is essential for growth on nitrate (17) and is protective against host NO in infection (100). Indeed, it seems likely that bacteria lacking these endogenous responses would be more sensitive to exogenous nitrosative stress-induced killing. Thus, understanding how bacteria cope with endogenous oxidative and nitrosative stresses may allow insights into their ability to survive in a host during infection.
There are several similarities between the endogenous oxidative stress generated in bacteria during growth in oxygen and the endogenous nitrosative stress derived from anaerobic growth on nitrate, which we will draw attention to in this review. First, just as oxidative stress is an inevitable consequence of aerobic growth, endogenously generated NO can be considered an inevitable consequence of ARN. Second, nitrosative stress manifests as high levels of protein S-nitrosylation (39, 90), just as oxidative stress manifests as high levels of S-oxidation (26). Third, E. coli respond to these stresses by inducing genes that encode for proteins that scavenge ROS or NO or otherwise counter their effects. Fourth, the transcription factor OxyR is induced by both oxidative and nitrosative stresses, under either aerobic or anaerobic conditions, and confers protection against each, although through distinct responses.
Notably, OxyR is essential for bacterial survival in air (under certain conditions) (24, 36), and also for anaerobic growth on nitrate (90), indicative of the central role played by this sensor/transcription factor in protecting against both stresses. It is perhaps not surprising, therefore, that OxyR controls many of the genes previously identified with protection from hydrogen peroxide and from NO.
OxyR as a Sensor of Oxidative and Nitrosative Stress
First identified in Salmonella typhimurium and E. coli, the transcriptional factor OxyR is the primary sensor for hydrogen peroxide in bacteria (14). Reduced OxyR is inactive as a transcription factor, whereas oxidation of OxyR at the active site cysteine residue, Cys199, leads to transcription of the OxyR regulon of ∼30 genes, which includes the anti-oxidative genes catalase (katG), alkyl hydroperoxide reductase (ahpCF), and glutathione reductase (96) (Figs. 1 and 2). E. coli lacking OxyR are highly sensitive to hydrogen peroxide and other oxidative stress-inducing agents. Importantly, not only does the OxyR regulon ensure direct scavenging and removal of hydrogen peroxide (via katG) and organic hydroperoxides (via ahpC), but it also counters consequences of oxidative injury, including through dps, which encodes an iron (Fe2+)-sequestering protein that protects DNA from Fenton chemistry, and glutaredoxin and trxA (thioredoxin), which remove oxidative modifications from proteins (96). Surprisingly, OxyR functions in anaerobes as well, including Porphyromonas gingivalis, Bacteroides fragilis, and Bacteroides thetaiotaomicron (22, 65, 83). At first glance, the role and function of OxyR in oxidative stress would appear to preclude a direct role under anaerobic conditions, so it has been assumed that OxyR must protect against adventitious oxygen or H2O2 in these anaerobes.
FIG. 2.
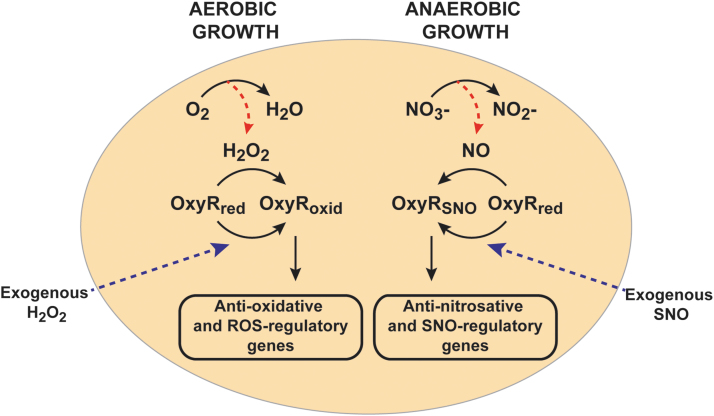
Role of OxyR in the induction of endogenous stress-specific responses. OxyR is either oxidized or SNO at reactive site Cys199 in response to oxidative or nitrosative stress, respectively. Alternative modifications of OxyR lead to the transcriptional induction of different regulons. H2O2, hydrogen peroxide; NO, nitric oxide; oxid, oxidized; red, reduced; ROS, reactive oxygen species.
Biology is nothing if not innovative in reusing mechanisms that are efficient, especially in bacteria, and OxyR is a prime example. This transcription factor is regulated by a sensor thiol Cys199 that is subject to sulfenylation (S-OH) and consequent disulfide formation (52, 96, 119). But thiols may undergo multiple modifications, including S-nitrosylation, S-glutathionylation, and thioesterification, which may form the basis of a molecular code through which alternative Cys modifications produce different functional responses (15, 113). It has, thus, been proposed that OxyR may be capable of transducing distinct transcriptional responses (35, 52, 90). Although distinct S-sulfenylation OxyR and SNO-OxyR regulons have been identified (90, 117, 120), glutathionylated- and thioesterified-OxyR regulons are still hypothetical (Fig. 1).
S-nitrosylation of OxyR, first demonstrated in anaerobically growing E. coli treated with the SNO donor S-nitrosocysteine (39), has served as proof-of-concept. S-nitrosylated OxyR (SNO-OxyR) is in an activated state that is able to promote transcription at target genes (39, 52, 90) (Fig. 2). Initial studies presumed that SNO-OxyR activated transcription of the same well-characterized oxidative stress-induced regulon of OxyR, including, for example, katG and oxyS, although to a lesser degree than hydrogen peroxide. However, although NO donors induced transcription of katG and oxyS (52, 119), S-nitrosylated OxyR was quite different from S-oxidized OxyR in its affinity for previously identified DNA target sites (52). Moreover, under truly physiological conditions where endogenously generated NO leads to S-nitrosylation, SNO-OxyR actually promotes transcription of a unique set of genes, almost entirely nonoverlapping with the previously known regulon induced by oxidized OxyR (90). This nitrosative stress-specific OxyR regulon of >100 genes includes numerous enzymes that are protective against nitrosative stress and/or well known to be induced by NO, including hcp-hcr (27, 42, 71, 75, 90). Figure 2 shows these distinct functions of OxyR under aerobic (or oxidative stress) conditions versus anaerobic (or nitrosative stress) conditions, which result in oxidized and S-nitrosylated forms of OxyR, respectively. This largely nonoverlapping nature of the oxidized and S-nitrosylated OxyR regulons explains why an earlier study that looked at NO-dependent gene expression of oxidized-OxyR genes concluded that OxyR is not a primary regulator of the bacterial response to nitrosative stress (66).
Thus, OxyR acts as a redox sensor and switch: It senses both ROS and RNS, and it transduces distinct transcriptional responses that are designed to remove the specific stresses that modified and activated OxyR in the first place. For example, oxidation of OxyR by H2O2 induces the OxyR-regulated gene katG, which encodes for catalase that effectively removes H2O2, whereas S-nitrosylation of OxyR induces an unknown denitrosylase enzyme that removes SNOs (Fig. 3A). More specifically, OxyR binds DNA as a dimer of dimers, using a helix-turn-helix motif for binding, with each monomer binding on an adjacent part of the DNA duplex (12, 102). On oxidative activation, OxyR recruits RNA polymerase to specific promoters (55, 90). Alternative post-translational modifications of OxyR at Cys199 (oxidation vs. S-nitrosylation) lead to different conformations (52) that bind to different DNA sequences and activate transcription at distinct promoters (Fig. 1). Oxidation of OxyR Cys199 to sulfenic acid is, in fact, sufficient for transcriptional activation (52), but structural studies have shown further transition of this Cys199 sulfenic acid to a disulfide bond between Cys199 and Cys208 (12). S-nitrosylation and S-sulfenylation of Cys199 lead to different structural changes in OxyR, as revealed by circular dichroism and by differences in DNA-binding affinity (52). Thus, these different modifications may alter the interactions between OxyR monomers, or the OxyR interaction with specific DNA sequences or with RNA polymerase. A careful structural analysis of each of these modified forms will be required to reveal exactly how the distinct redox-based modifications of a single cysteine in OxyR lead to differential transcriptional activity.
FIG. 3.
Distinct OxyR-dependent transcriptional regulons ameliorate different stresses, and metabolism of SNO is aberrant in the absence of OxyR. (A) OxyR can be oxidized by H2O2, and genes in the OxyR-oxidative regulon, exemplified in katG, degrade H2O2. Similarly, SNOs S-nitrosylate OxyR, and member(s) of the OxyR nitrosative stress regulon are expected to metabolize SNO, evidenced by increased SNO levels in ΔoxyR Escherichia coli. (B) Capillary electrophoretic traces of spent media from SNO-treated E. coli show accumulation of nitrite in ΔoxyR but not wild type E. coli, demonstrating a defect in OxyR-dependent SNO metabolism. (C) SNO-proteins pulled down (using the SNO-RAC method) from lysates of wild type and ΔoxyR E. coli grown anaerobically on nitrate show an aberrant increase in protein SNO levels in the absence of OxyR. SNO-RAC, S-nitrosothiol resin-assisted capture; WT, wildtype.
Notably, the OxyR gene itself is not transcriptionally induced by either oxidative or nitrosative stress (90, 97), although its transcription has been shown to be regulated by the growth phase (34). The induction of OxyR-regulated genes is solely a result of OxyR activation by post-translational modification, which is designed for rapid responses.
Role of OxyR in SNO Metabolism Under Aerobic Conditions
OxyR regulates the bacterial response to nitrosative stress not only under anaerobic conditions but also under aerobic conditions. E. coli rapidly metabolizes S-nitrosothiols to nitrate and nitrite aerobically, and it is notable that in the absence of OxyR there is an aberrant accumulation of nitrite when compared with the wild type strain (39) (Fig. 3B). Nitrite is known to be toxic to bacteria (10) and hence may be a factor causing the growth inhibitory effects of SNOs on ΔoxyR strains (39, 90). The exact mechanism by which OxyR influences metabolism of SNOs remains to be elucidated.
Role of OxyR in Regulating S-Nitrosylation During Anaerobic Growth
OxyR-deficient bacteria are sensitized to the growth inhibitory effects of NO/RNS. An E. coli oxyR knockout strain demonstrates growth inhibition during ARN (as well as after exposure to NO/SNO under aerobic conditions) (39, 90). When grown under ARN in the absence of OxyR, E. coli show overall increases in protein S-nitrosylation (SNO–protein levels) (Fig. 3C) (90). Nonetheless, deletion of individual genes within the OxyR regulon can lead to decreases (rather than increases) in S-nitrosylation (91), indicating that some genes may actually function to S-nitrosylate target proteins. That is, the OxyR regulon encodes for proteins that appear to catalyze the S-nitrosylation of other proteins. Hence, the protective effects of OxyR via induction of its nitrosative stress regulon is evidently achieved not only by decreasing overall S-nitrosylation but also by promoting S-nitrosylation of certain proteins, potentially reflecting enzymatic mechanisms of SNO targeting.
Enzymatic S-Nitrosylation in E. coli
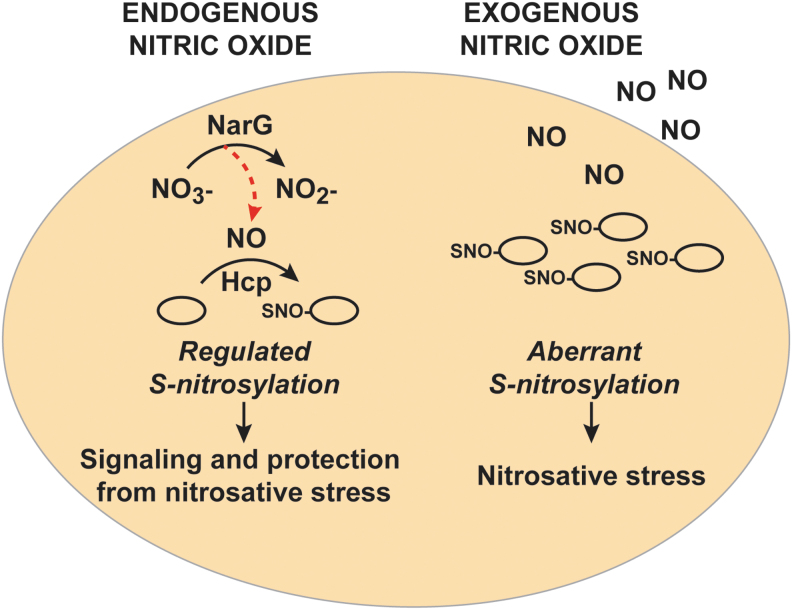
One member of the OxyR regulon, the hybrid cluster protein Hcp, is a critical element of a three-component enzymatic machinery that is responsible for specific and regulated S-nitrosylation of proteins during ARN (91). Hcp is one of the maximally induced genes when E. coli are grown on nitrate, and this induction is abrogated in the absence of OxyR. Hcp functions as a protein S-nitrosylase that mediates de novo synthesis of SNO from NO produced by nitrate reductase (NarG) (91). Hcp in E. coli contains a [2Fe-2S] cluster as well as a [4Fe-2S-2O] “hybrid” cluster (106), which are essential for its SNO-synthase activity (91). In fact, prior treatment of cells with the divalent metal cation chelator 1,10-phenanthroline decreases the formation of overall SNOs during ARN (90). The iron clusters can serve as electron acceptors for the conversion of NO to NO+, and dinitrosyl iron complexes (DNICs) have been shown to support the oxidative requirements for S-nitrosylation by NO (7, 57, 95, 107). Mechanistically, Hcp S-nitrosylates itself and then dimerizes in the presence of NO. The Hcp dimer serves as a nucleus for a large interactome; SNO-Hcp is then able to S-nitrosylate multiple partner proteins, including OxyR. The Hcp interactome also includes a class of enzymes known as transnitrosylases: proteins such as glyceraldehyde-3-phosphate dehydrogenase (GAPDH) and TrxA that can transfer their own SNO to additional substrates through direct interactions. Thus, under anaerobic growth conditions that generate endogenous NO, Hcp promotes the S-nitrosylation of OxyR, which is, in turn, activated to protect against nitrosative stress, as well as S-nitrosylation of many other proteins required for growth (Fig. 4). For example, Hcp-dependent S-nitrosylation regulates the activity of key metabolic enzymes, including dihydrolipoamide dehydrogenase (LpdA). Lack of Hcp also profoundly inhibits swimming motility during ARN (91).
FIG. 4.
A comparison of enzymatically regulated versus stochastic S-nitrosylation. Under conditions where NO is generated intracellularly by nitrate reductase, S-nitrosylation of proteins is enzymatically regulated by the S-nitrosylase, Hcp. S-nitrosylation by Hcp plays an essential role in bacterial motility and metabolism. By contrast, unregulated and aberrant S-nitrosylation by exogenous NO/RNS, which may occur via multiple chemical routes such as during host defense against infection, leads to nitrosative stress. RNS, reactive nitrogen species.
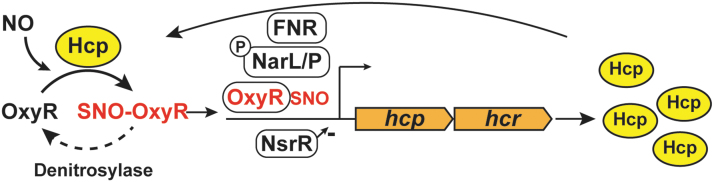
Even though the induction of Hcp in ARN is eightfold lower in the oxyR knockout strain, Hcp remains basally transcribed in the presence of nitrate independent of OxyR, ensuring that OxyR can become nitrosylated rapidly when NO is first present (90). The major transcription factors regulating gene expression during ARN include fumarate-nitrate regulation (FNR) and nitrate/nitrite response regulator proteins (NarL/NarP). FNR activates transcription of genes required during anaerobiosis and inhibits transcription of genes required for aerobic growth (49, 87), whereas NarL/NarP respond to levels of available nitrate to induce genes required for nitrate metabolism such as nitrate reductase, NarG (78). It is interesting to note that besides OxyR, transcription of hcp is regulated by FNR and NarL/NarP, in response to anaerobiosis and nitrite/nitrate, respectively (28). In addition, in the presence of NO, repression of hcp by the transcriptional repressor HTH-type transcriptional repressor (NsrR) is relieved (27). Besides hcp, the NsrR regulon also includes several other genes that protect against NO and RNS, such as hmp (27). In sum, hcp is part of the protective machinery against NO: basal levels of Hcp are controlled during ARN by FNR, NarL, NarP, and NsrR; whereas OxyR regulates further induction of Hcp to create a positive feedback loop (Fig. 5).
FIG. 5.
Regulation of OxyR S-nitrosylation. In vivo S-nitrosylation of OxyR by Hcp activates OxyR-mediated transcription, which, in turn, upregulates Hcp. Additional transcriptional regulators of hcp (FNR, NarL/P, and NsrR) are shown. SNO-OxyR is denitrosylated by enzymes (denitrosylases) that have not yet been identified. - symbol denotes inhibition. FNR, fumarate-nitrate regulation.
Until recently, S-nitrosylation in bacteria had been studied in the context of nitrosative stress, with no consideration for any role in signaling. Bacterial responses to NO/SNO were viewed as aimed at removing NO or SNO and mitigating cellular injury. These responses involved enzymes that directly consume NO, namely flavohemoglobin (Hmp) (38) and flavorubredoxin (NorV) (33), or that metabolize SNOs (GSNOR) (61). Our new data suggest a different perspective: The growth advantage conferred by Hcp is mediated not only by the removal of NO/SNO but also by promoting S-nitrosylation of specific proteins that confer benefit on metabolism and growth (Fig. 4), reflective of a primary signaling function for NO that is analogous to its role in yeast, plants, and mammals (1, 41, 116). Further, it should be noted that mitochondria are, of course, major players in cellular metabolism. Thus, endogenous signaling in bacteria draws analogy to the metabolic function of NO in mammalian cells. Inasmuch as signaling by S-nitrosylation is enzymatic in E. coli, one might anticipate the same in mammalian cells. Indeed, although Hcp does not have homologues in higher organisms, Fe-S cluster center-containing enzymes are commonplace in mitochondria, as discussed in the Regulation of OxyR S-Nitrosylation section.
Regulation of OxyR S-Nitrosylation
As noted earlier, S-nitrosylation of OxyR is enzymatically regulated by Hcp. However, SNO-OxyR levels are also regulated by an unidentified mechanism of denitrosylation, consistent with an enzymatic process. That is, after E. coli treatment with an exogenous NO donor, S-nitrosylated OxyR is rapidly denitrosylated (90). Additional studies will be required to determine whether this denitrosylation is due to known denitrosylases, including the thioredoxin system [reviewed in Benhar et al. (6)] or SNO reductases (1, 61), or to novel enzymes.
Role of the OxyR-Dependent Anaerobic Regulon
A gene ontology pathway analysis of the anaerobic OxyR regulon using STRING (101) revealed that these genes belong to pathways ranging from cellular respiration and amino acid biosynthesis to stress responses (Table 1). Many of these genes are metabolic enzymes involved in respiration, and they may be upregulated to maintain normal metabolism and growth. For example, NO inhibits branched-chain amino acid synthesis by inhibiting the enzyme IlvD, leading to auxotrophy (42). Upregulation of multiple genes in the branched-chain amino acid synthetic pathway (ilvA, ilvD, ilvE, ilvM, ilvN, leuB, and leuD) (Table 1) may, therefore, compensate for this inhibition.
Table 1.
Gene Ontology Pathway Enrichment Analysis of OxyR Regulon Genes
| Pathway | False discovery rate | Genes |
|---|---|---|
| Oxidation–reduction process | 2.02E-09 | aceA, aceB, aceE, acnA, acnB, cyoB, cyoC, cyoD, cysD, cysI, dadA, fdnG, fdnH, fdnI, fdoG, fdoI, fumA, gcd, gcvP, glpD, gltA, hcp, hcr, katG, leuB, lipA, lpdA, maeB, msrB, napB, narI, nrdD, sdhB, ssuE, sthA, sucD, tpx, ybiC |
| Generation of precursor metabolites and energy | 2.12E-09 | aceA, aceB, aceE, acnA, acnB, cyoB, cyoC, cyoD, fdnG, fdnH, fdnI, fdoG, fdoI, fumA, gltA, gpmA, lpdA, napB, narI, sdhB, sucD |
| Cellular respiration | 2.12E-09 | aceA, aceB, acnA, acnB, cyoB, cyoC, cyoD, fdnG, fdnH, fdnI, fdoG, fdoI, fumA, gltA, nap, narI, sdhB, sucD |
| Aerobic respiration | 2.01E-06 | aceA, aceB, acnA, acnB, cyoB, cyoC, cyoD, fumA, gltA, sdhB, sucD |
| Tricarboxylic acid cycle | 1.51E-05 | aceA, aceB, acnA, acnB, fumA, gltA, sdhB, sucD |
| Branched-chain amino acid biosynthetic process | 0.000131 | ilvA, ilvD, ilvE, ilvM, ilvN, leuB, leuD |
| Single-organism metabolic process | 0.000247 | aceA, aceB, aceE, acnA, acnB, allE, cyoB, cyoC, cyoD, cyoE, cysD, cysI, dadA, dadB, fdnG, fdnH, fdnI, fdoG, fdoI, fumA, gcd, gcvP, glpD, glpE, gltA, gntT, gpmA, hchA, hcp, hcr, ilvA, ilvD, ilvE, ilvM, ilvN, katG, leuB, leuD, lipA, lpdA, maeB, metH, msrB, napB, narI, nrdD, sdhB, speD, ssuE, sthA, sucD, thiH, tpx, ybiC, ynhG |
| Oxoacid metabolic process | 0.000523 | aceA, aceB, aceE, acnA, acnB, cysI, dadA, dadB, fumA, gcvP, gltA, gntT, gpmA, hchA, ilvA, ilvD, ilvE, ilvM, ilvN, leuB, leuD, lipA, lpdA, maeB, metH, narI, sdhB, sucD |
| Carboxylic acid metabolic process | 0.000592 | aceA, aceB, aceE, acnA, acnB, cysI, dadA, dadB, fumA, gcvP, gltA, gntT, gpmA, hchA, ilvA, ilvD, ilvE, ilvM, ilvN, leuB, leuD, lipA, lpdA, maeB, metH, sdhB, sucD |
| Isoleucine biosynthetic process | 0.00063 | ilvA, ilvD, ilvE, ilvM, ilvN |
| Anaerobic respiration | 0.00102 | acnA, fdnG, fdnH, fdnI, fdoG, fdoI, napB, narI |
| Glyoxylate cycle | 0.00244 | aceA, aceB, acnA, acnB |
| Small-molecule metabolic process | 0.00453 | aceA, aceB, aceE, acnA, acnB, allE, cysI, dadA, dadB, fumA, gcvP, glpD, glpE, gltA, gntT, gpmA, hchA, ilvA, ilvD, ilvE, ilvM, ilvN, leuB, leuD, lipA, lpdA, maeB, metH, narI, nrdD, sdhB, speD, sthA, sucD, thiH |
| Valine biosynthesis | 0.00635 | ilvD, ilvE, ilvM, ilvN |
| Cellular response to starvation | 0.0284 | leuB, psiE, ssuE, tauA, tauB |
| Response to oxidative stress | 0.0361 | acnA, clpA, cysD, hcp, katG, lpdA, msrB, tpx |
| Electron transport coupled proton transport | 0.0405 | cyoB, cyoC, cyoD |
| Respiratory electron transport chain | 0.0419 | cyoB, cyoC, cyoD, fdnI, fdoI |
| Alpha-amino acid metabolic process | 0.0419 | cysI, dadA, dadB, gcvP, ilvA, ilvD, ilvE, ilvM, ilvN, leuB, leuD, lpdA, metH |
| Taurine transport | 0.0485 | tauA, tauB |
| Leucine biosynthesis | 0.0498 | ilvE, leuB, leuD |
The OxyR-dependent genes induced under anaerobic growth on nitrate were grouped by biological processes by using STRING.
Interestingly, the SNO-OxyR regulon also includes genes with anti-oxidative functions (katG, catalase; msrB, methionine sulfoxide reductase; lpdA, lipoamide dehydrogenase; tpx, thiol peroxidase) (Table 1). One possibility is that these genes have yet-to-be discovered anti-nitrosative functions. This idea is supported by the fact that strains deficient in tpx or lpd have significant decreases in SNO levels during ARN (91). Another possibility is that the regulon is generally aimed at counteracting host defenses during infection, and it therefore provides coverage against both nitrosative and oxidative stresses.
OxyR-Dependent Regulons Under Different Growth Conditions
Facultative bacteria such as E. coli precisely and specifically regulate gene expression depending on growth conditions (aerobic vs. anaerobic) and availability of electron acceptors (nitrate, fumarate, dimethyl sulfoxide, or trimethylamine oxide) (105). The transcription factor FNR is required for the expression of many genes that are necessary for anaerobic metabolism, whereas aerobic respiration control protein regulates genes for aerobic metabolism. Other transcription factor systems such as NarL/P and nitrate/nitrite sensor proteins activate gene transcription in the presence of nitrate. Many bacterial genes have binding sites for multiple transcription factors, allowing complex regulation by growth conditions.
It is highly likely that the complete OxyR regulons, both oxidative and nitrosative stress specific, are much larger than those currently identified because the bacterial transcriptome can be very different under different growth conditions. One such example has been reported for the cytochrome c peroxidase (ccp) gene, which is only expressed under anaerobic conditions (because it requires FNR for expression), but it is further induced by H2O2 in an OxyR-dependent manner (50). Transcriptome analysis of H2O2-treated wild type and ΔoxyR E. coli growing anaerobically will undoubtedly identify additional genes that are regulated such as ccp. By the same token, the nitrosative stress-specific OxyR regulon was identified under anaerobic conditions in E. coli growing on nitrate (91). We expect exogenous NO to lead to a distinct transcriptional response, and additional SNO-OxyR targets likely will be identified in cells growing anaerobically with fumarate or DMSO as electron acceptors. Also, different sources of nitrosative stress, endogenous versus exogenous (NO donors or SNO donors), will likely generate varied transcriptional responses (48, 66, 90). Further, host innate immune responses can generate both oxidative and nitrosative stress at the same time, which should activate both oxidized-OxyR and SNO-OxyR regulons. Since OxyR operates as a multimer, combinatorial effects of SNO and SOH modifications of OxyR might activate additional sets of genes.
Role of OxyR Under Anaerobic/Microaerobic Conditions Within the Host
During infection of multicellular hosts with low oxygen/anaerobic microenvironments, facultative bacteria (e.g., E. coli) can transition from aerobic to anaerobic respiration. For example, during urinary tract infections (UTI), the concentration of oxygen within the bladder lumen is insufficient for aerobic respiration (62, 86), requiring invading bacteria traveling up the urethra to switch from aerobic to anaerobic metabolism. Various studies have shown that hmp (flavohemoglobin), norV (flavorubredoxin), and hcp (hybrid cluster protein), genes that are known to protect against nitrosative stress (33, 38, 90), are induced in uropathogenic strains during UTI (64, 86, 100), implying that bacteria are exposed to sufficient NO during UTI to induce SNO-OxyR-dependent transcription. Despite the pathophysiological relevance of low tissue oxygen concentrations in the presence of nitrate, this feature of infection is frequently neglected.
In the large bowel, which is generally believed to be anaerobic, it has been shown that the ability to respire on nitrate provides facultative anaerobes with an advantage compared with strictly aerobic or strictly anaerobic species, or to facultative anaerobes in which nitrate reductase is not induced (110). These conditions of anaerobic growth in the presence of nitrate are analogous to the conditions under which the induction of a large regulon by SNO-OxyR has been documented (90). Further, OxyR has been shown to be essential for growth in various mouse models of infection that are characterized by anaerobic/microaerobic conditions that are likely rich in nitrate (derived from host RNS), including intra-abdominal abscess, UTI, and sepsis (47, 99). Although OxyR function has been ascribed to protection from host-derived ROS, deletion of the neutrophil phagocyte oxidase, a major source of host-derived ROS, did not rescue the ΔoxyR phenotype in UTI models (46). Thus, it is likely that OxyR is also serving to protect against nitrosative stress under those conditions.
NO Production in Mitochondria Parallels Anaerobic NO Generation in E. coli
Recent studies suggest many parallels between anaerobic NO generation in bacteria and NO production by mitochondria, including the potential for enzymatic generation of SNOs. By way of reminder, nitric oxide synthases (NOSs) are the major source of aerobic NO generation in mammalian cells (30). A mitochondrial nitric oxide synthase (NOS) has also been described in the inner mitochondrial membrane (5), but studies regarding its activity have led to conflicting results and its existence has been questioned (56). In 1999, Kozlov et al. reported on an NOS-independent source of mitochondria-generated NO (54) under hypoxic conditions, whereby nitrite undergoes a one-electron reduction by the respiratory chain (11, 54). Different proteins in the respiratory chain have been proposed to be the site of NO production, including ubiquinone/cytochrome bc1 (69) and cytochrome c oxidase (11). In such cases, NO generation was observed under hypoxic conditions when the respective enzymes utilize nitrite instead of oxygen as an electron acceptor (11, 69). Other mitochondrial proteins with nitrite reductase activity include cytochrome c (4) and mitochondrial amidoxime reducing component proteins (94). Generation of NO in mitochondria is, thus, remarkably similar to anaerobic respiration in E. coli, where NOS-independent production of NO occurs due to a secondary activity of nitrate reductase under anaerobic conditions, as described earlier.
S-Nitrosylation in Mitochondria and Its Role in Regulating Mitophagy
In the earlier days of NO biology, S-nitrosylation was believed to require oxygen. Studies of the mitochondrion were the first to establish otherwise: Proteins were found to become S-nitrosylated in anoxic mitochondria after addition of NO (31). Many mitochondrial proteins are now known to be regulated by S-nitrosylation (13, 23, 74). These include enzymes involved in the tricarboxylic acid cycle, fatty acid synthesis, and the electron transport chain (13, 23). How proteins are S-nitrosylated in the mitochondria, however, is not well understood. One model is that protein-bound DNICs, formed from nitrite-derived NO, could act as SNO synthases in a manner similar to Hcp in E. coli.
Mitochondrial DNA (mtDNA) encodes for 13 subunits of the respiratory chain, 2 rRNAs and 22 tRNAs, independently of nuclear DNA (2), and therefore mtDNA transcription will regulate mitochondrial oxidative phosphorylation. Two mitochondrial transcription factors encoded by nuclear DNA, transcription factor A, mitochondrial (TFAM) and dimethyladenosine transferase 2, mitochondrial, as well as an RNA polymerase, DNA-directed RNA polymerase, mitochondrial are known to subserve mitochondrial gene expression [reviewed in D'Souza and Minczuk (20)]. Although S-nitrosylation of these proteins has not been studied, Hsp60, a chaperone required for the transport of TFAM to the mitochondria, is S-nitrosylated by iNOS, thereby facilitating its binding to TFAM and its transport to mitochondria (98). mtDNA transcription is also regulated by additional SNO-proteins, including cAMP response element-binding protein (21, 93) and signal transducer and activator of transcription 3 (51, 63), although the exact effect of S-nitrosylation on mitochondrial transcription is unclear. It is quite possible that akin to OxyR, a mitochondrial protein(s) could act as a sensor of both SNO and S-oxidation to regulate transcriptional responses. This is an especially tantalizing idea given that mitochondria generate superoxide/H2O2 versus NO under high and low oxygen tension, respectively.
Although there is no mitochondrial protein homologous to Hcp, the features and components of endogenous enzymatic S-nitrosylation observed in E. coli are present in mitochondria. A hypothetical enzymatic cascade in mitochondria akin to the one described in E. coli is shown in Figure 6. In this scenario, NO is generated in mitochondria by respiratory chain enzymes under hypoxic conditions (11, 54). Further, experiments done in mice demonstrated that the NO produced in mitochondria from dietary nitrite is associated with the formation of DNICs (25, 103) and SNOs (103). Also, GAPDH has been demonstrated within mitochondria of mouse hearts and exhibits transnitrosylase activity there (53). SNO-GAPDH may be transported into mitochondria (53), or possibly S-nitrosylated in situ by a DNIC-containing protein acting as an “SNO-synthase.” Thus, all three components required for de novo S-nitrosylation, namely an enzymatic NO source, a potential SNO-synthase, and a transnitrosylase, are present within mitochondria, as they are in E. coli.
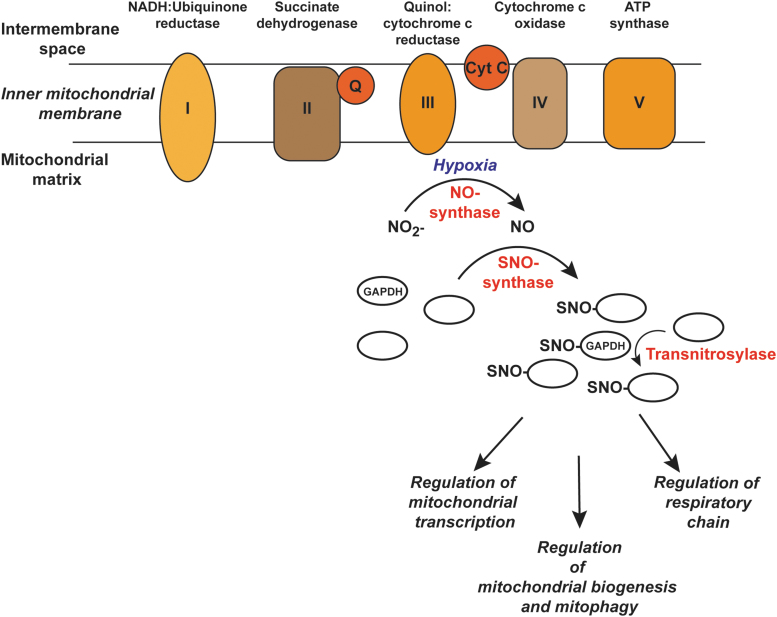
FIG. 6.
S-nitrosylation in mitochondria. Based on the similarities between NO generation in E. coli and mitochondria, a hypothetical SNO-cascade is shown where under hypoxic conditions NO is produced from nitrite by the respiratory chain. Metal-bound proteins can serve as potential SNO-synthases to S-nitrosylate specific mitochondrial proteins, leading to regulation of downstream metabolic processes. Transnitrosylases further propagate the SNO signal. GAPDH, glyceraldehyde-3-phospate dehydrogenase.
Mitochondria are dynamic, and they can undergo regulated proliferation, fission, fusion, and degradation (mitophagy) (104). Mitophagy refers to the selective autophagic removal of mitochondria (59), and regulated mitophagy is essential for normal cellular functioning whereas its dysregulation can lead to pathophysiology (73). One important pathway that mediates mitophagy is the PTEN-inducible putative kinase 1 (PINK1)-Parkin pathway whereby accumulation of PINK1 in damaged mitochondria leads to the phosphorylation and recruitment of the ubiquitin E3 ligase Parkin to the mitochondrial outer membrane to promote the recruitment of autophagy receptors (58, 73, 115). S-nitrosylation of Parkin was first reported in neurons and was associated with Parkinson's disease (16, 114). One study reported that Parkin S-nitrosylation inhibits ubiquitin ligase activity (16), whereas another showed that S-nitrosylation of Parkin at an alternate site (Cys323) activates the E3 ligase leading to enhanced mitophagy (72). PINK1 is also S-nitrosylated and this inhibits its protein kinase activity, leading to impaired mitophagy (70). Studies with mice lacking the GSNOR enzyme and in aging humans in whom GSNOR levels decline show higher SNO-levels and increased S-nitrosylation of Parkin (81) that is associated with mitochondrial dysfunction. Overall, these data suggest that excessive S-nitrosylation inhibits mitophagy, which may have implications not only for aging and Parkinson's but also for Alzheimer's disease where aberrant S-nitrosylation (e.g., of dynamin-related protein-1) has also been implicated in mitochondrial dysfunction. These interesting data notwithstanding, the key SNO-players in mitophagy and mitochondrial dysfunction remain to be identified. Such an understanding should lead to a more targeted approach to regulating mitochondrial S-nitrosylation therapeutically.
Conclusion
Although OxyR has served as a classic model for understanding the cellular response to oxidative stress, the physiological role of OxyR in situ also includes protection against nitrosative stress. Thus, E. coli has evolved a common transcription element in OxyR to protect itself against both aerobic and anaerobic stressors. In effect, facultative anaerobes such as E. coli can sense different stresses through alternative modifications in OxyR and tailor their genetic responses accordingly. This newly discovered role for OxyR in regulating anaerobic stress has also revealed new physiological functions mediated by S-nitrosylation, as well as enzymatic machinery subserving endogenous S-nitrosylation. Most notably, OxyR-regulated, Hcp-mediated S-nitrosylation is required for bacterial motility and metabolism under anaerobic conditions. Further examination of genes in the nitrate-dependent OxyR regulon may reveal targetable vulnerabilities in these strategies employed by bacteria to survive infections on the one hand and loci of dysfunction that underlie human disease on the other hand. Enzymes first characterized in bacteria that regulate S-nitrosylation may serve as models for identifying and understanding homologous or comparable mechanisms present in mammalian systems. Given that dysregulated S-nitrosylation of specific SNO-proteins is now recognized as a factor in many diseases, including neurological disorders mediated by mitochondrial dysfunction, elucidation of enzymatic mechanisms involved in S-nitrosylation/denitrosylation may suggest new therapeutic approaches to human disease.
Abbreviations Used
- ahpC
Escherichia coli gene encoding alkyl hydroperoxide reductase C
- ARN
anaerobic respiration on nitrate
- ccp
E. coli gene encoding cytochrome c oxidase YhjA
- DNIC
dinitrosyl iron complex
- dps
E. coli gene encoding DNA protection during starvation protein, Dps
- FNR
fumarate-nitrate regulation
- GAPDH
glyceraldehyde-3-phospate dehydrogenase
- GSNO
S-nitrosoglutathione
- GSNOR
S-nitrosoglutathione reductase
- H2O2
hydrogen peroxide
- hcp
E. coli gene encoding hybrid cluster protein
- hcr
E. coli gene encoding NADH oxidoreductase, Hcr
- hmp
E. coli gene encoding flavohemoglobin
- ilvA
E. coli gene encoding l-threonine dehydratase
- ilvD
E. coli gene encoding dihydroxy-acid dehydratase
- ilvE
E. coli gene encoding branched chain amino acid aminotransferase
- ilvM
E. coli gene encoding acetolactate synthase isozyme
- ilvN
E. coli gene encoding acetolactate synthase isozyme
- iNOS
inducible nitric oxide synthase
- katG
E. coli gene encoding catalase-peroxidase
- leuB
E. coli gene encoding 3-isopropylmalate dehydrogenase
- leuD
E. coli gene encoding 3-isopropylmalate dehydratase small subunit
- lpdA
E. coli gene encoding dihydrolipoyl dehydrogenase
- msrB
E. coli gene encoding peptide methionine sulfoxide reductase
- mtDNA
mitochondrial DNA
- narGHI
E. coli operon encoding nitrate reductase A
- NarL/P
nitrate/nitrite response regulator proteins
- NO
nitric oxide
- NorV
anaerobic nitric oxide reductase flavorubredoxin
- NOS
nitric oxide synthase
- NsrR
HTH-type transcriptional repressor
- PINK1
PTEN-inducible putative kinase 1
- RNS
reactive nitrogen species
- ROS
reactive oxygen species
- SNO
S-nitrosothiol
- TFAM
transcription factor A, mitochondrial
- tpx
E. coli gene encoding thiol peroxidase
- trxA
E. coli gene encoding thioredoxin 1
- UTI
urinary tract infections
- yeaR-yoaG
E. coli operon encoding YeaR-YoaG
- ytfE
E. coli gene encoding iron-sulfur cluster repair protein YtfE
Funding Information
This work was supported by the National Institutes of Health under grants GM099921, HL075443, HL128192, HL126900, and DK119506.
References
- 1. Anand P, Hausladen A, Wang YJ, Zhang GF, Stomberski C, Brunengraber H, Hess DT, and Stamler JS. Identification of S-nitroso-CoA reductases that regulate protein S-nitrosylation. Proc Natl Acad Sci U S A 111: 18572–18577, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2. Anderson S, Bankier AT, Barrell BG, Debruijn MHL, Coulson AR, Drouin J, Eperon IC, Nierlich DP, Roe BA, Sanger F, Schreier PH, Smith AJH, Staden R, and Young IG. Sequence and organization of the human mitochondrial genome. Nature 290: 457–465, 1981 [DOI] [PubMed] [Google Scholar]
- 3. Bang IS, Liu LM, Vazquez-Torres A, Crouch ML, Stamler JS, and Fang FC. Maintenance of nitric oxide and redox homeostasis by the Salmonella flavohemoglobin Hmp. J Biol Chem 281: 28039–28047, 2006 [DOI] [PubMed] [Google Scholar]
- 4. Basu S, Azarova NA, Font MD, King SB, Hogg N, Gladwin MT, Shiva S, and Kim-Shapiro DB. Nitrite reductase activity of Cytochrome c. J Biol Chem 283: 32590–32597, 2008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5. Bates TE, Loesch A, Burnstock G, and Clark JB. Immunocytochemical evidence for a mitochondrially located nitric-oxide synthase in brain and liver. Biochem Biophys Res Comm 213: 896–900, 1995 [DOI] [PubMed] [Google Scholar]
- 6. Benhar M, Forrester MT, and Stamler JS. Protein denitrosylation: enzymatic mechanisms and cellular functions. Nat Rev Mol Cell Biol 10: 721–732, 2009 [DOI] [PubMed] [Google Scholar]
- 7. Bosworth CA, Toledo JC, Zmijewski JW, Li Q, and Lancaster JR. Dinitrosyliron complexes and the mechanism(s) of cellular protein nitrosothiol formation from nitric oxide. Proc Natl Acad Sci U S A 106: 4671–4676, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 8. Cadet J and Wagner JR.. DNA base damage by reactive oxygen species, oxidizing agents, and UV radiation. Cold Spring Harb Perspect Biol 5, a012559, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 9. Carlioz A and Touati D. Isolation of superoxide dismutase mutants in Escherichia coli: is superoxide dismutase necessary for aerobic life? EMBO J 5: 623–630, 1986 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10. Castellani AG and Niven CF. Factors affecting the bacteriostatic action of sodium nitrite. Appl Microbiol 3: 154–159, 1955 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 11. Castello PR, David PS, McClure T, Crook Z, and Poyton RO. Mitochondrial cytochrome oxidase produces nitric oxide under hypoxic conditions: implications for oxygen sensing and hypoxic signaling in eukaryotes. Cell Metab 3: 277–287, 2006 [DOI] [PubMed] [Google Scholar]
- 12. Choi HJ, Kim SJ, Mukhopadhyay P, Cho S, Woo JR, Storz G, and Ryu SE. Structural basis of the redox switch in the OxyR transcription factor. Cell 105: 103–113, 2001 [DOI] [PubMed] [Google Scholar]
- 13. Chouchani ET, Hurd TR, Nadtochiy SM, Brookes PS, Fearnley IM, Lilley KS, Smith RAJ, and Murphy MP. Identification of S-nitrosated mitochondrial proteins by S-nitrosothiol difference in gel electrophoresis (SNO-DIGE): implications for the regulation of mitochondrial function by reversible S-nitrosation. Biochem J 430: 49–59, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14. Christman MF, Morgan RW, Jacobson FS, and Ames BN. Positive control of a regulon for defenses against oxidative stress and some heat-shock proteins in Salmonella typhimurium. Cell 41: 753–762, 1985 [DOI] [PubMed] [Google Scholar]
- 15. Chung HS, Wang SB, Venkatraman V, Murray CI, and Van Eyk JE. Cysteine oxidative posttranslational modifications: emerging regulation in the cardiovascular system. Circ Res 112: 382–392, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16. Chung KKK, Thomas B, Li XJ, Pletnikova O, Troncoso JC, Marsh L, Dawson VL, and Dawson TM. S-nitrosylation of Parkin regulates ubiquitination and compromises Parkin's protective function. Science 304: 1328–1331, 2004 [DOI] [PubMed] [Google Scholar]
- 17. Corker H and Poole RK. Nitric oxide formation by Escherichia coli—dependence on nitrite reductase, the NO-sensing regulator FNR, and flavohemoglobin Hmp. J Biol Chem 278: 31584–31592, 2003 [DOI] [PubMed] [Google Scholar]
- 18. Cox AG, Saunders DC, Kelsey PB, Conway AA, Tesmenitsky Y, Marchini JF, Brown KK, Stamler JS, Colagiovanni DB, Rosenthal GJ, Croce KJ, North TE, and Goessling W. S-Nitrosothiol signaling regulates liver development and improves outcome following toxic liver injury. Cell Rep 6: 56–69, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19. Crack JC, Green J, Thomson AJ, and Le Brun NE. Iron-sulfur clusters as biological sensors: the chemistry of reactions with molecular oxygen and nitric oxide. Accounts Chem Res 47: 3196–3205, 2014 [DOI] [PubMed] [Google Scholar]
- 20. D'Souza AR and Minczuk M. Mitochondrial transcription and translation: overview. Essays Biochem 62: 309–320, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 21. De Rasmo D, Signorile A, Roca E, and Papa S. cAMP response element-binding protein (CREB) is imported into mitochondria and promotes protein synthesis. FEBS J 276: 4325–4333, 2009 [DOI] [PubMed] [Google Scholar]
- 22. Diaz PI, Slakeski N, Reynolds EC, Morona R, Rogers AH, and Kolenbrander PE. Role of OxyR in the oral anaerobe Porphyromonas gingivalis. J Bacteriol 188: 2454–2462, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 23. Doulias PT, Tenopoulou M, Greene JL, Raju K, and Ischiropoulos H. Nitric oxide regulates mitochondrial fatty acid metabolism through reversible protein S-nitrosylation. Sci Signal 6: rs1, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 24. Dukan S and Nystrom T. Oxidative stress defense and deterioration of growth-arrested Escherichia coli cells. J Biol Chem 274: 26027–26032, 1999 [DOI] [PubMed] [Google Scholar]
- 25. Dungel P, Perlinger M, Weidinger A, Redl H, and Kozlov AV. The cytoprotective effect of nitrite is based on the formation of dinitrosyl iron complexes. Free Radic Biol Med 89: 300–310, 2015 [DOI] [PubMed] [Google Scholar]
- 26. Ezraty B, Gennaris A, Barras F, and Collet JF. Oxidative stress, protein damage and repair in bacteria. Nat Rev Microbiol 15: 385–396, 2017 [DOI] [PubMed] [Google Scholar]
- 27. Filenko N, Spiro S, Browning DF, Squire D, Overton TW, Cole J, and Constantinidou C. The NsrR regulon of Escherichia coli K-12 includes genes encoding the hybrid cluster protein and the periplasmic, respiratory nitrite reductase. J Bacteriol 189: 4410–4417, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28. Filenko NA, Browning DF, and Cole JA. Transcriptional regulation (prismane) protein of a hybrid cluster. Biochem Soc Trans 33: 195–197, 2005 [DOI] [PubMed] [Google Scholar]
- 29. Flint DH, Tuminello JF, and Emptage MH. The inactivation of Fe-S cluster containing hydro-lyases by superoxide. J Biol Chem 268: 22369–22376, 1993 [PubMed] [Google Scholar]
- 30. Forstermann U and Sessa WC. Nitric oxide synthases: regulation and function. Eur Heart J 33: 829–837, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 31. Foster MW and Stamler JS. New insights into protein S-nitrosylation—mitochondria as a model system. J Biol Chem 279: 25891–25897, 2004 [DOI] [PubMed] [Google Scholar]
- 32. Fridovich I. Biology of oxygen radicals. Science 201: 875–880, 1978 [DOI] [PubMed] [Google Scholar]
- 33. Gomes CM, Giuffrè A, Forte E, Vicente JB, Saraiva LgM, Brunori M, and Teixeira M. A novel type of nitric-oxide reductase Escherichia coli flavorubredoxin. J Biol Chem 277: 25273–25276, 2002 [DOI] [PubMed] [Google Scholar]
- 34. GonzalezFlecha B and Demple B. Transcriptional regulation of the Escherichia coli oxyR gene as a function of cell growth. J Bacteriol 179: 6181–6186, 1997 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 35. Haridas V, Kim SO, Nishimura G, Hausladen A, Stamler JS, and Gutterman JU. Avicinylation (thioesterification): a protein modification that can regulate the response to oxidative and nitrosative stress. Proc Natl Acad Sci U S A 102: 10088–10093, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 36. Hassett DJ, Alsabbagh E, Parvatiyar K, Howell ML, Wilmott RW, and Ochsner UA. A protease-resistant catalase, KatA, released upon cell lysis during stationary phase is essential for aerobic survival of a Pseudomonas aeruginosa oxyR mutant at low cell densities. J Bacteriol 182: 4557–4563, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 37. Hausladen A, Gow A, and Stamler JS. Flavohemoglobin denitrosylase catalyzes the reaction of a nitroxyl equivalent with molecular oxygen. Proc Natl Acad Sci U S A 98: 10108–10112, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 38. Hausladen A, Gow AJ, and Stamler JS. Nitrosative stress: metabolic pathway involving the flavohemoglobin. Proc Natl Acad Sci U S A 95: 14100–14105, 1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 39. Hausladen A, Privalle CT, Keng T, DeAngelo J, and Stamler JS. Nitrosative stress: activation of the transcription factor OxyR. Cell 86: 719–729, 1996 [DOI] [PubMed] [Google Scholar]
- 40. Hayashida K, Bagchi A, Miyazaki Y, Hirai S, Seth D, Silverman MG, Rezoagli E, Marutani E, Mori N, Magliocca A, Liu XW, Berra L, Hindle AG, Donnino MW, Malhotra R, Bradley MO, Stamler JS, and Ichinose F. Improvement in outcomes after cardiac arrest and resuscitation by inhibition of S-nitrosoglutathione reductase. Circulation 139: 815–827, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 41. Hess DT, Matsumoto A, Kim SO, Marshall HE, and Stamler JS. Protein S-nitrosylation: purview and parameters. Nat Rev Mol Cell Biol 6: 150–166, 2005 [DOI] [PubMed] [Google Scholar]
- 42. Hyduke DR, Jarboe LR, Tran LM, Chou KJY, and Liao JC. Integrated network analysis identifies nitric oxide response networks and dihydroxyacid dehydratase as a crucial target in Escherichia coli. Proc Natl Acad Sci U S A 104: 8484–8489, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 43. Imlay JA. The molecular mechanisms and physiological consequences of oxidative stress: lessons from a model bacterium. Nat Rev Microbiol 11: 443–454, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 44. Imlay JA, Sethu R, and Rohaun SK. Evolutionary adaptations that enable enzymes to tolerate oxidative stress. Free Radic Biol Med 140: 4–13, 2019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 45. Ji XB and Hollocher TC. Mechanism for nitrosation of 2,3-diaminonaphthalene by Escherichia coli: enzymatic production of NO followed by O2-dependent chemical nitrosation. Appl Environ Microbiol 54: 1791–1794, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 46. Johnson JR, Clabots C, and Rosen H. Effect of inactivation of the global oxidative stress regulator oxyR on the colonization ability of Escherichia coli O1: K1: H7 in a mouse model of ascending urinary tract infection. Infect Immun 74: 461–468, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 47. Johnson JR, Russo TA, Drawz SM, Clabots C, Olson R, Kuskowski MA, and Rosen H. OxyR contributes to the virulence of a clonal group A Escherichia coli strain (O17:K+:H18) in animal models of urinary tract infection, subcutaneous infection, and systemic sepsis. Microb Pathog 64: 1–5, 2013 [DOI] [PubMed] [Google Scholar]
- 48. Justino MC, Vicente JB, Teixeira M, and Saraiva LM. New genes implicated in the protection of anaerobically grown Escherichia coli against nitric oxide. J Biol Chem 280: 2636–2643, 2005 [DOI] [PubMed] [Google Scholar]
- 49. Kang YS, Weber KD, Yu Q, Kiley PJ, and Blattner FR. Genome-wide expression analysis indicates that FNR of Escherichia coli K-12 regulates a large number of genes of unknown function. J Bacteriol 187: 1135–1160, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 50. Khademian M and Imlay JA. Escherichia coli cytochrome c peroxidase is a respiratory oxidase that enables the use of hydrogen peroxide as a terminal electron acceptor. Proc Natl Acad Sci U S A 114: E6922–E6931, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 51. Kim J, Won JS, Singh AK, Sharma AK, and Singh I. STAT3 regulation by S-nitrosylation: implication for inflammatory disease. Antioxid Redox Signal 20: 2514–2527, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 52. Kim SO, Merchant K, Nudelman R, Beyer WF, Keng T, DeAngelo J, Hausladen A, and Stamler JS. OxyR: a molecular code for redox-related signaling. Cell 109: 383–396, 2002 [DOI] [PubMed] [Google Scholar]
- 53. Kohr MJ, Murphy E, and Steenbergen C. Glyceraldehyde-3-phosphate dehydrogenase acts as a mitochondrial trans-S-nitrosylase in the heart. PLoS One 9: e111448, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 54. Kozlov AV, Staniek K, and Nohl H. Nitrite reductase activity is a novel function of mammalian mitochondria. FEBS Lett 454: 127–130, 1999 [DOI] [PubMed] [Google Scholar]
- 55. Kullik I, Toledano MB, Tartaglia LA, and Storz G. Mutational analysis of the redox-sensitive transcriptional regulator OxyR—regions important for oxidation and transcriptional activation. J Bacteriol 177: 1275–1284, 1995 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 56. Lacza Z, Pankotai E, Csordas A, Gero D, Kiss L, Horvath EM, Kollai M, Busija DW, and Szabo C. Mitochondrial NO and reactive nitrogen species production: does mtNOS exist? Nitric Oxide 14: 162–168, 2006 [DOI] [PubMed] [Google Scholar]
- 57. Lancaster JR. How are nitrosothiols formed de novo in vivo? Arch Biochem Biophys 617: 137–144, 2017 [DOI] [PubMed] [Google Scholar]
- 58. Lazarou M, Sliter DA, Kane LA, Sarraf SA, Wang CX, Burman JL, Sideris DP, Fogel AI, and Youle RJ. The ubiquitin kinase PINK1 recruits autophagy receptors to induce mitophagy. Nature 524: 309–314, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 59. Lemasters JJ. Perspective—selective mitochondrial autophagy, or mitophagy, as a targeted defense against oxidative stress, mitochondrial dysfunction, and aging. Rejuvenation Res 8: 3–5, 2005 [DOI] [PubMed] [Google Scholar]
- 60. Liu L, Yan Y, Zeng M, Zhang J, Hanes MA, Ahearn G, McMahon TJ, Dickfeld T, Marshall HE, Que LG, and Stamler JS. Essential roles of S-nitrosothiols in vascular horneostasis and endotoxic shock. Cell 116: 617–628, 2004 [DOI] [PubMed] [Google Scholar]
- 61. Liu LM, Hausladen A, Zeng M, Que L, Heitman J, and Stamler JS. A metabolic enzyme for S-nitrosothiol conserved from bacteria to humans. Nature 410: 490–494, 2001 [DOI] [PubMed] [Google Scholar]
- 62. Lundberg JON, Carlsson S, Engstrand L, Morcos E, Wiklund NP, and Weitzberg E. Urinary nitrite: more than a marker of infection. Urology 50: 189–191, 1997 [DOI] [PubMed] [Google Scholar]
- 63. Macias E, Rao D, Carbajal S, Kiguchi K, and DiGiovanni J. STAT3 Binds to mtDNA and regulates mitochondrial gene expression in keratinocytes. J Invest Dermatol 134: 1971–1980, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 64. Mehta HH, Liu Y, Zhang MQ, and Spiro S. Genome-wide analysis of the response to nitric oxide in uropathogenic Escherichia coli CFT073. Microb Genom 1: e000031, 2015 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 65. Mishra S and Imlay JA. An anaerobic bacterium, Bacteroides thetaiotaomicron, uses a consortium of enzymes to scavenge hydrogen peroxide. Mol Microbiol 90: 1356–1371, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 66. Mukhopadhyay P, Zheng M, Bedzyk LA, LaRossa RA, and Storz G. Prominent roles of the NorR and Fur regulators in the Escherichia coli transcriptional response to reactive nitrogen species. Proc Natl Acad Sci U S A 101: 745–750, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 67. Murphy MP. How mitochondria produce reactive oxygen species. Biochem J 417: 1–13, 2009 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 68. Nathan C, and Shiloh MU. Reactive oxygen and nitrogen intermediates in the relationship between mammalian hosts and microbial pathogens. Proc Natl Acad Sci U S A 97: 8841–8848, 2000 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 69. Nohl H, Staniek K, Sobhian B, Bahrami S, Redl H, and Kozlov AV. Mitochondria recycle nitrite back to the bioregulator nitric monoxide. Acta Biochim Pol 47: 913–921, 2000 [PubMed] [Google Scholar]
- 70. Oh CK, Sultan A, Platzer J, Dolatabadi N, Soldner F, McClatchy DB, Diedrich JK, Yates JR, Ambasudhan R, Nakamura T, Jaenisch R, and Lipton SA. S-nitrosylation of PINK1 attenuates PINK1/Parkin-dependent mitophagy in hiPSC-based Parkinson's disease models. Cell Rep 21: 2171–2182, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 71. Overton TW, Griffiths L, Patel MD, Hobman JL, Penn CW, Cole JA, and Constantinidou C. Microarray analysis of gene regulation by oxygen, nitrate, nitrite, FNR, NarL and NarP during anaerobic growth of Escherichia coli: new insights into microbial physiology. Biochem Soc Trans 34: 104–107, 2006 [DOI] [PubMed] [Google Scholar]
- 72. Ozawa K, Komatsubara AT, Nishimura Y, Sawada T, Kawafune H, Tsumoto H, Tsuji Y, Zhao J, Kyotani Y, Tanaka T, Takahashi R, and Yoshizumi M. S-nitrosylation regulates mitochondrial quality control via activation of parkin. Sci Rep 3: 2202, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 73. Palikaras K, Lionaki E, and Tavernarakis N. Mechanisms of mitophagy in cellular homeostasis, physiology and pathology. Nat Cell Biol 20: 1013–1022, 2018 [DOI] [PubMed] [Google Scholar]
- 74. Piantadosi CA. Regulation of mitochondrial processes by protein S-nitrosylation. Biochim Biophys Acta 1820: 712–721, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 75. Pullan ST, Gidley MD, Jones RA, Barrett J, Stevanin TA, Read RC, Green J, and Poole RK. Nitric oxide in chemostat-cultured Escherichia coli is sensed by FNR and other global regulators: unaltered methionine biosynthesis indicates lack of S nitrosation. J Bacteriol 189: 1845–1855, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 76. Que LG, Liu LM, Yan Y, Whitehead GS, Gavett SH, Schwartz DA, and Stamler JS. Protection from experimental asthma by an endogenous bronchodilator. Science 308: 1618–1621, 2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 77. Que LG, Yang ZH, Lugogo NL, Katial RK, Shoemaker SA, Troha JM, Rodman DM, Tighe RM, and Kraft M. Effect of the S-nitrosoglutathione reductase inhibitor N6022 on bronchial hyperreactivity in asthma. Immun Inflamm Dis 6: 322–331, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 78. Rabin RS and Stewart V. Dual response regulators (NarL and NarP) interact with dual sensors (NarX and NarQ) to control nitrate-regulated and nitrite-regulated gene-expression in Escherichia coli K-12. J Bacteriol 175: 3259–3268, 1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 79. Ralt D, Wishnok JS, Fitts R, and Tannenbaum SR. Bacterial catalysis of nitrosation: involvement of the nar operon of Escherichia coli. J Bacteriol 170: 359–364, 1988 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 80. Rea SL, Graham BH, Nakamaru-Ogiso E, Kar A, and Falk MJ. Bacteria, yeast, worms, and flies: exploiting simple model organisms to investigate human mitochondrial diseases. Dev Disabil Res Rev 16:200–218, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 81. Rizza S, Cardaci S, Montagna C, Di Giacomo G, De Zio D, Bordi M, Maiani E, Campello S, Borreca A, Puca AA, Stamler JS, Cecconi F, and Filomeni G. S-nitrosylation drives cell senescence and aging in mammals by controlling mitochondrial dynamics and mitophagy. Proc Natl Acad Sci U S A 115: E3388–E3397, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 82. Rizza S and Filomeni G. Chronicles of a reductase: biochemistry, genetics and physio-pathological role of GSNOR. Free Radic Biol Med 110:19–30, 2017 [DOI] [PubMed] [Google Scholar]
- 83. Rocha ER, Herren CD, Smalley DJ, and Smith CJ. The complex oxidative stress response of Bacteroides fragilis: the role of OxyR in control of gene expression. Anaerobe 9: 165–173, 2003 [DOI] [PubMed] [Google Scholar]
- 84. Roger AJ, Munoz-Gomez SA, and Kamikawa R. The origin and diversification of mitochondria. Curr Biol 27: R1177–R1192, 2017 [DOI] [PubMed] [Google Scholar]
- 85. Roos G, Foloppe N, and Messens J. Understanding the pK(a) of redox cysteines: the key role of hydrogen bonding. Antioxid Redox Signal 18: 94–127, 2013 [DOI] [PubMed] [Google Scholar]
- 86. Roos V and Klemm P. Global gene expression profiling of the asymptomatic bacteriuria Escherichia coli strain 83972 in the human urinary tract. Infect Immun 74: 3565–3575, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 87. Salmon K, Hung SP, Mekjian K, Baldi P, Hatfield GW, and Gunsalus RP. Global gene expression profiling in Escherichia coli K12—the effects of oxygen availability and FNR. J Biol Chem 278: 29837–29855, 2003 [DOI] [PubMed] [Google Scholar]
- 88. Schell MA. Molecular biology of the LysR Family of transcriptional regulators. Annu Rev Microbiol 47: 597–626, 1993 [DOI] [PubMed] [Google Scholar]
- 89. Seaver L, Imlay JA. Alkyl hydroperoxide reductase is the primary scavenger of endogenous hydrogen peroxide in Escherichia coli. J Bacteriol 183: 7173–7181, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 90. Seth D, Hausladen A, Wang YJ, and Stamler JS. Endogenous protein S-nitrosylation in E. coli: regulation by OxyR. Science 336: 470–473, 2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 91. Seth D, Hess DT, Hausladen A, Wang LW, Wang YJ, and Stamler JS. A multiplex enzymatic machinery for cellular protein S-nitrosylation. Mol Cell 69: 451–464, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 92. Showe MK and DeMoss JA. Localization and regulation of synthesis of nitrate reductase in Escherichia coli. J Bacteriol 95: 1305–1313, 1968 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 93. Smith JG, Aldous SG, Andreassi C, Cuda G, Gaspari M, and Riccio A. Proteomic analysis of S-nitrosylated nuclear proteins in rat cortical neurons. Sci Signal 11: eaar3396, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 94. Sparacino-Watkins CE, Tejero J, Sun B, Gauthier MC, Thomas J, Ragireddy V, Merchant BA, Wang J, Azarov I, Basu P, and Gladwin MT. Nitrite reductase and Nitric-oxide synthase activity of the mitochondrial molybdopterin enzymes mARC1 and mARC2. J Biol Chem 289: 10345–10358, 2014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 95. Stojanovic S, Stanic D, Nikolic M, Spasic M, and Niketic V. Iron catalyzed conversion of NO into nitrosonium (NO+) and nitroxyl (HNO/NO-) species. Nitric Oxide 11: 256–262, 2004 [DOI] [PubMed] [Google Scholar]
- 96. Storz G, Tartaglia LA, and Ames BN. The OxyR regulon. Antonie Van Leeuwenhoek 58: 157–161, 1990 [DOI] [PubMed] [Google Scholar]
- 97. Storz G, Tartaglia LA, and Ames BN. Transcriptional regulator of oxidative stress-inducible genes—direct activation by oxidation. Science 248: 189–194, 1990 [DOI] [PubMed] [Google Scholar]
- 98. Suliman HB, Babiker A, Withers CM, Sweeney TE, Carraway MS, Tatro LG, Bartz RR, Welty-Wolf KE, and Piantadosi CA. Nitric oxide synthase-2 regulates mitochondrial Hsp60 chaperone function during bacterial peritonitis in mice. Free Radic Biol Med 48: 736–746, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 99. Sund CJ, Rocha ER, Tzinabos AO, Wells WG, Gee JM, Reott MA, O'Rourke DP, and Smith CJ. The Bacteroides fragilis transcriptome response to oxygen and H2O2: the role of OxyR and its effect on survival and virulence. Mol Microbiol 67: 129–142, 2008 [DOI] [PubMed] [Google Scholar]
- 100. Svensson L, Poljakovic M, Save S, Gilberthorpec N, Schon T, Strid S, Corker H, Poole RK, and Persson K. Role of flavohemoglobin in combating nitrosative stress in uropathogenic Escherichia coli—implications for urinary tract infection. Microb Pathog 49: 59–66, 2010 [DOI] [PubMed] [Google Scholar]
- 101. Szklarczyk D, Morris JH, Cook H, Kuhn M, Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, Jensen LJ, and von Mering C. The STRING database in 2017: quality-controlled protein-protein association networks, made broadly accessible. Nucleic Acids Res 45: D362–D368, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 102. Tartaglia LA, Gimeno CJ, Storz G, and Ames BN. Multidegenerate DNA recognition by the OxyR transcriptional regulator. J Biol Chem 267: 2038–2045, 1992 [PubMed] [Google Scholar]
- 103. Thomas DD, Corey C, Hickok J, Wang YN, and Shiva S. Differential mitochondrial dinitrosyliron complex formation by nitrite and nitric oxide. Redox Biol 15: 277–283, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 104. Tilokani L, Nagashima S, Paupe V, and Prudent J. Mitochondrial dynamics: overview of molecular mechanisms. Essays Biochem 62: 341–360, 2018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 105. Unden G, Bongaerts J. Alternative respiratory pathways of Escherichia coli: energetics and transcriptional regulation in response to electron acceptors. Biochim Biophys Acta 1320: 217–234, 1997 [DOI] [PubMed] [Google Scholar]
- 106. van den Berg WA, Hagen WR, and van Dongen WM. The hybrid-cluster protein (‘prismane protein’) from Escherichia coli. Characterization of the hybrid-cluster protein, redox properties of the [2Fe-2S] and [4Fe-2S-2O] clusters and identification of an associated NADH oxidoreductase containing FAD and [2Fe-2S]. Eur J Biochem 267: 666–676, 2000 [DOI] [PubMed] [Google Scholar]
- 107. Vanin AF, Malenkova IV, and Serezhenkov VA. Iron catalyzes both decomposition and synthesis of S-nitrosothiols: optical and electron paramagnetic resonance studies. Nitric Oxide 1: 191–203, 1997 [DOI] [PubMed] [Google Scholar]
- 108. Wei W, Li B, Hanes MA, Kakar S, Chen X, and Liu LM.. S-nitrosylation from GSNOR deficiency impairs DNA repair and promotes hepatocarcinogenesis. Sci Transl Med 2, 19ra13, 2010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 109. Weiss B. Evidence for mutagenesis by nitric oxide during nitrate metabolism in Escherichia coli. J Bacteriol 188: 829–833, 2006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 110. Winter SE, Winter MG, Xavier MN, Thiennimitr P, Poon V, Keestra AM, Laughlin RC, Gomez G, Wu J, Lawhon SD, Popova IE, Parikh SJ, Adams LG, Tsolis RM, Stewart VJ, and Baumler AJ. Host-derived nitrate boosts growth of E. coli in the inflamed gut. Science 339: 708–711, 2013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 111. Winterbourn CC and Metodiewa D. Reactivity of biologically important thiol compounds with superoxide and hydrogen peroxide. Free Radic Biol Med 27: 322–328, 1999 [DOI] [PubMed] [Google Scholar]
- 112. Wojdyla K, Williamson J, Roepstorff P, and Rogowska-Wrzesinska A. The SNO/SOH TMT strategy for combinatorial analysis of reversible cysteine oxidations. J Proteomics 113: 415–434, 2015 [DOI] [PubMed] [Google Scholar]
- 113. Yang J, Carroll KS, and Liebler DC. The expanding landscape of the thiol redox proteome. Mol Cell Proteomics 15: 1–11, 2016 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 114. Yao DD, Gu ZZ, Nakamura T, Shi ZQ, Ma YL, Gaston B, Palmer LA, Rockenstein EM, Zhang ZH, Masliah E, Uehara T, and Lipton SA. Nitrosative stress linked to sporadic Parkinson's disease: S-nitrosylation of parkin regulates its E3 ubiquitin ligase activity. Proc Natl Acad Sci U S A 101: 10810–10814, 2004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 115. Youle RJ and Narendra DP. Mechanisms of mitophagy. Nat Rev Mol Cell Biol 12: 9–14, 2011 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 116. Zaffagnini M, De Mia M, Morisse S, Di Giacinto N, Marchand CH, Maes A, Lemaire SD, and Trost P. Protein S-nitrosylation in photosynthetic organisms: a comprehensive overview with future perspectives. Biochim Biophys Acta 1864: 952–966, 2016 [DOI] [PubMed] [Google Scholar]
- 117. Zeller T, Mraheil MA, Moskvin OV, Li KY, Gomelsky M, and Klug G. Regulation of hydrogen peroxide-dependent gene expression in Rhodobacter sphaeroides: regulatory functions of OxyR. J Bacteriol 189: 3784–3792, 2007 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 118. Zhang YY, Wu KY, Su WT, Zhang DF, Wang P, Qiao XH, Yao Q, Yuan ZQ, Yao YG, Liu GH, Zhang C, Liu LM, and Chen C. Increased GSNOR expression during aging impairs cognitive function and decreases S-nitrosation of CaMKII alpha. J Neurosci 37: 9741–9758, 2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- 119. Zheng M, Aslund F, and Storz G. Activation of the OxyR transcription factor by reversible disulfide bond formation. Science 279: 1718–1721, 1998 [DOI] [PubMed] [Google Scholar]
- 120. Zheng M, Wang X, Templeton LJ, Smulski DR, LaRossa RA, and Storz G. DNA microarray-mediated transcriptional profiling of the Escherichia coli response to hydrogen peroxide. J Bacteriol 183: 4562–4570, 2001 [DOI] [PMC free article] [PubMed] [Google Scholar]