Abstract
Objective
Leishmaniasis is caused by different Leishmania spp. Treatment failure (TF) of cutaneous leishmaniasis (CL) is a serious issue that may be due to various reasons, previous studies suggested Leishmania RNA virus (LRV) as a potential cause of TF. Two variant groups of LRV1 and LRV2 are reported. In this study, the presence of LRV1/LRV2 was compared in TF with treatment response (TR) isolates of L. major. Clinical isolates of 15 TF and 15 TR were collected from CL patients referred to the Health Centers of Isfahan. Genomic DNA was extracted to identify Leishmania spp. using ITS1-PCR–RFLP. Identification of LRV1/LRV2 was performed using SYBR Green Real-Time PCR. The statistical analysis to test relationship between the treatment response with Glucantime and the presence of LRV were performed using SPSS 16.0 with Fisher’s Exact test. P value of less than 0.05 was considered significant.
Results
ITS1-PCR–RFLP results showed that every isolate was identified as L. major. The results showed no LRV1 in any of the samples but 7 TR isolates and 2 TF isolates showed positive for LRV2. Statistical analysis showed no significant difference between the presence of LRV2 and response to Glucantime (p-value = 0.1086). Therefore, other mechanisms might be responsible for TF.
Keywords: Cutaneous Leishmaniasis, Leishmania RNA Virus, Treatment Failure, Leishmania major, SYBR Green Real-Time PCR
Introduction
Different species of Leishmania cause a wide range of clinical manifestation, including a simple self-healing skin lesion called cutaneous leishmaniasis (CL), a systemic fatal form called visceral leishmaniasis (VL), and mucocutaneous leishmaniasis (MCL). The diseases are transmitted by Phlebotomus spp. Most cases of CL occur in Afghanistan, Algeria, Brazil, Colombia, the Islamic Republic of Iran, Peru, Saudi Arabia, and the Syrian Arab Republic [1–3]. The causative agents of CL in the old world are L. major, L. tropica, and L. aethiopica [4].
Sodium stibogluconate and meglumine antimoniate are the first-line treatment [5]. The occurrence of low response, treatment failure (TF), and resistance are reported from some endemic regions of the world [6]. Different mechanisms have been proposed to explain the lack of response to treatment drugs [7] such as presence of Leishmania RNA virus (LRV) (Family: Totiviridae) which infects–Leishmania [8–10]. LRV is a double-stranded RNA (dsRNA), which exists in some Leishmania isolates [8–11] and suggested as a virulence factor in some reports [12]. Two species of LRV1 and LRV2 are known [12–14]. So far, no LRV1 has been found in Leishmania spp. from new world countries, with the exception of L. panamensis, evidence potentially related to the virulence of the parasite. The LRV2 has been reported from the old world human cases of leishmaniasis [8, 15]. LRV2 has also been found in L. aethiopica isolated from the biopsy of Ethiopian CL patients [16].
It is reported that the presence of LRV viruses in Leishmania spp. might lead to destructive hyper-inflammation resulted in disease severity and parasite metastasis [17]. In addition, previous studies on human cases suggested that LRV in Leishmania spp. may exacerbate clinical prognosis of CL, and induce MCL development [18, 19] and possibly TF [14–17].
Despite recent findings of LRV role on Leishmania pathogenicity, a few reports exist on TF in old and new world cases of leishmaniasis (16-18). The aim of the present study was to study the presence of Leishmania RNA virus in human cases of CL caused by L. major and compare the rate of TF to treatment response.
Main text
Ethical consideration
This study was approved by Ethics Committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran. The informed consent was signed by each patient recruited based on Helsinki declaration.
Study population
The analytical cross-sectional study included a population of patients, with zoonotic CL/caused by L. major, who were referred to the Health Centers of Isfahan, Iran, from September 2017 to December 2018. All cases agreed to participate and signed an informed consent form. Patients who discontinued treatment with Glucantime were excluded. Leishmania parasites have been isolated from two population of CL patients who responded to the treatment referred to as treatment response (TR) and patients not responded to treatment as treatment failure (TF); the first category includes 15 isolates from the patients with lesion(s) cured after one full course of treatment, the second group consists of 15 isolates collected from the patients who had active lesion(s), after three courses of treatment with Glucantime (20 mg/kg/day for 14 days in each course) and were considered as TF.
Sampling
Samples were collected by scrubbing the lesion edge after disinfecting by using 70% alcohol. About 10 mg of each sample was transferred on a slide for direct microscopic examination and subsequent DNA extraction in order to detect and identify Leishmania spp. About 10 mg of each sample was transferred into RNA later and stored at − 20 °C for search for the presence of Leishmania RNA virus (LRV1 and LRV2).
Microscopic examination
The smear was fixed using methyl alcohol (methanol), then, the slide was stained using Giemsa. Microscopic examination was carried out using 100 × magnifications to find the Leishman bodies. The slides with positive Leishman body were used for molecular identification.
DNA extraction
DNA was isolated from Leishmania positive smears using DNA extraction kit (GenAll, South Korea) based on manufacturer’s instructions. The quality and quantity of the isolated DNA were evaluated using agarose gel electrophoresis and spectrophotometer using nanodrop (ABI, USA).
Detection and identification
To detect and identify Leishmania spp., a diagnostic key based on restriction banding pattern of ITS1 was used based on specific primer pair of LITSr-F 5′-CTGGATCATTTTCCgATg-3′ and L5.8 s-R 5′-TGATACCACTTATCGCACTT-3′ [20]. Amplification reaction mixture included 1x PCR buffer, 0.2 mM dNTP, 1.5 mM MgCl2, 1.5 U Taq DNA polymerase, 0.5 µM each primer pair, and 100 ng DNA. The amplification protocol was: 35 cycles of 94 °C for 45 s, 50 °C for 45 s, and 72 °C for 45 s. The final extension was done by 72 °C for 5 min. Positive and negative control with DNA of L. major (MRHO/IR/75/ER) and ddH2O, respectively, were applied in each amplification reaction. The amplification analysis was done using agarose gel electrophoresis (1%) alongside with 50 bp DNA ladder and was visualized by gel documentation system (ATP, Iran, Tehran, G940401). The expected amplicon was about 300–50 bp for detection of Leishmania genus.
Molecular identification was performed on positive amplicons digested using Hae III (Bsu RI) for 3 h at 37 °C. The digestion banding pattern analysis was assessed using agarose gel electrophoresis 3% alongside with 50 bp DNA ladder and L major (MRHO/IR/75/ER) as positive control. A banding pattern was visualized using gel documentation system revealing two main bands of 220 and 140 bp pattern considered as L. major. All tests were done in triplicate.
The patients with L. major were followed for 3 months. The cases with no responses to Glucantime treatment were considered as TF and the cases with complete cure were considered as TR.
RNA extraction
Total RNA extraction was performed using the Gf-1 Total RNA extraction kit (Vivantis, South Korea) according to the manufacture’s instruction. The quality and quantity of the isolated RNA were checked using agarose gel electrophoresis (1%) and spectrophotometer by nanodrop (ABI, USA).
cDNA synthesis
Synthesis of cDNA was obtained using cDNA synthesis kit (Fermentas, USA) in accordance with the manufacture’s instruction. The cDNA was amplified using specific primer pair of kmp11-F 5′-GCC TGG ATG AGG AGT TCA ACA-3′ and kmp-11-R 5′-GTC CTC CTT CAT CTC GGG-3′ [21]. The reaction was performed by a mixture of 1x PCR buffer, 0.2 mM dNTP, 1.5 mM MgCl2, 1.5 U Taq DNA polymerase, and 0.5 µM each primer pair of kmp11. The reaction protocol was done by 40 cycles of 94 °C for 45 s, 60 °C for 45 s, and 72 °C for 45 s. The final extension was done by 72 °C for 5 min. Positive and negative control was applied in each amplification by L. major (MRHO/IR/75/ER) and ddH2O. The amplification analysis was realized with agarose gel electrophoresis (3%) alongside with positive control (L. major MRHO/IR/75/ER), negative control (ddH2O), and 50 bp DNA ladder. The experiments were done triplicate.
Detection of LRV1/LRV2
The presence of LRV1 performed using SYBR Green Real-Time PCR by two sets of specific primers of LRV1-setA-F 5′-CTG ACT GGA CGG GGG GTA AT-3′ and LRV1-setA-R 5′-CAA AAC ACT CCC TTA CGC-3′, and LRV1-setB-F 5′-GTC TGT TTC GTA CCC GCC G-3′ and LRV1-setB-R 5′-AAG CTC AGG ATG TGC ATG TTC CA-3′ [21]. For LRV2 detection the specific primer pair of LRV2-F 5′-GCC ATT ACC CAG CCA GCC AT-3′ and LRV2-R 5′-GCC GTC ACC AGC TCT GTT GT-3′ (this study). The Kmp11 was considered as endogenous control with the primer pair of 5′-GCC TGG ATG AGG AGT TCA ACA-3′ and 5′-GTG CTC.
CTT CAT CTC GGG-3′ [19]. The reaction mixture for all primer pairs was done in 20 µl volume, including 200 nM each primer pair. The reaction temperature was 95 °C for 5 min in one cycle, followed by 40 cycles of 95 °C for 10 s and 60 °C for 1 min in order to annealing and extension. The melting curve was designed after the mentioned cycle.
Statistical analysis
The data was statistically analyzed using SPSS 16.0 by Fisher’s Exact test for possible relationship between the response to Glucantime treatment and the presence of LRV (LRV1 and LRV2).
Results
Patients
Upon the results pf PCR, only the L. major isolates were included in this study. A total of 30 isolates, 15 TF (Fig. 1) and 15 TR isolates were included in the study. The number of lesions in the TF and TR isolates was approximately the same with an average of 2. The mean age of the patients was 26 years (M = 23, F = 31 years). There was no significant difference between the age of the patients with TR isolates (25 years) and TR isolates (26 years).
Fig. 1.
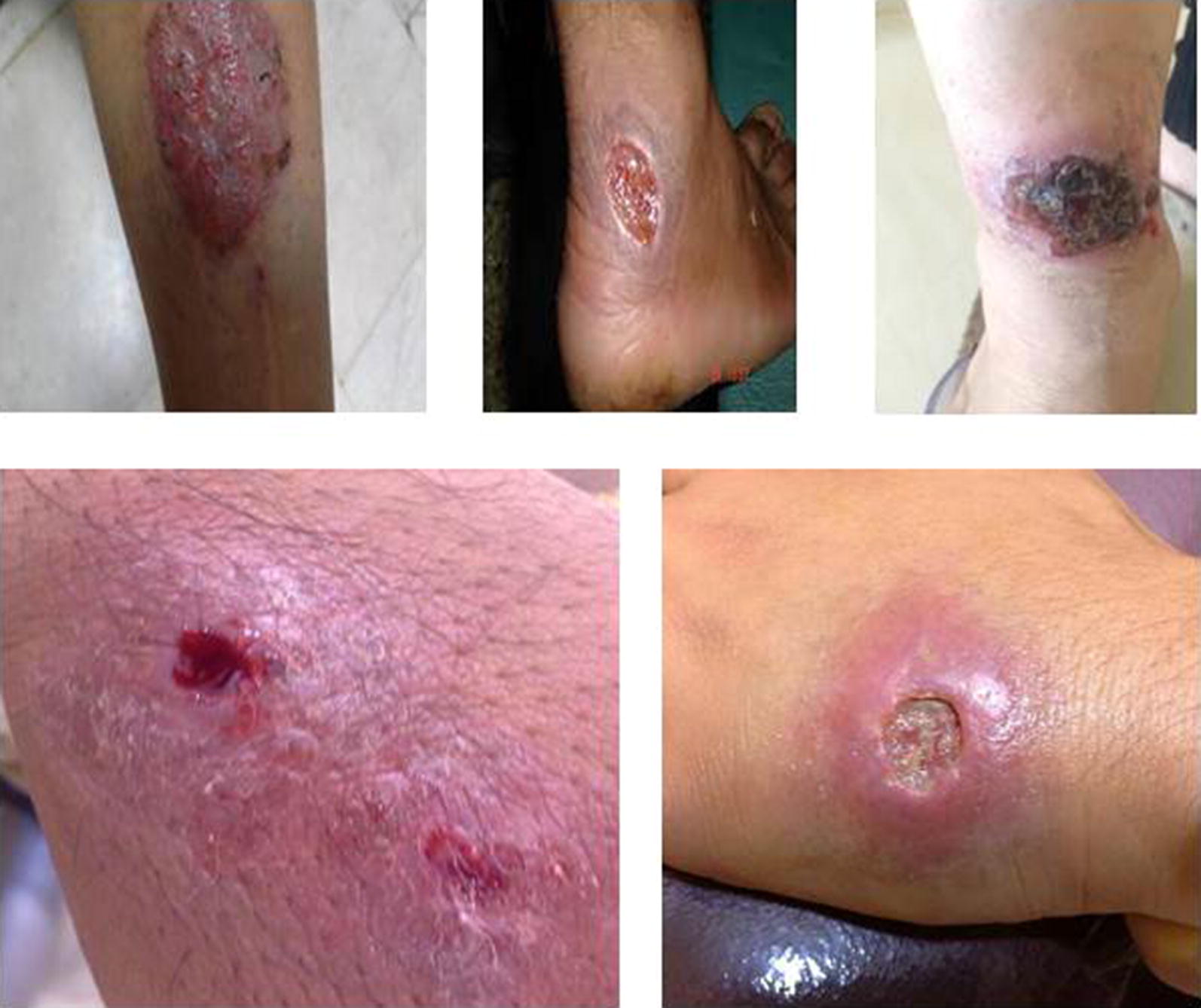
Lesions from patients harboring zoonotic cutaneous leishmaniasis with treatment failure to Glucantime
Molecular diagnosis and identification
The results of molecular detection confirmed that 30 isolates were positive for Leishmania and the species identification showed all 30 isolates belonging to L. major. The expected size of PCR amplicons was around350 bp (Additional file 1: Figure S1). The expected size of the bands after enzymatic digestion was of 127 and 220 bp (Additional file 2: Figure S2).
Assessing of the extracted RNA
The mean of 260/280 ratio at spectrophotometer analyses the extracted RNA was 1.98 ± 0.2 with the mean concentration of 136.2 ± 7.45 ng/μl. After extraction of RNA and synthesis of cDNA, in order to control the system for amplifiability, PCR was done using the specific primer pair of kmp11. All the synthesized cDNA were successfully amplified.
Present of LRV1/LRV2
The presence of LRV1 and LRV2 were checked using specific primer pairs using SYBR Green Real-Time PCR. The amplification results showed LRV2 in 7 TR isolates and 2 TR isolates collected from patients. Statistical analysis showed no relationship between response to Glucantime treatment and the presence of LRV2 (p value = 0.1086).
No LRV1 was obtained in any sample.
Melting curve analysis
For verification of the specific amplification for kmp11 and LRV2, the melting curve analysis was done. The results showed that the amplifications were done specific with the melting temperature of 82.6 °C for kmp11 and 79.8 °C for LVR2.
Discussion
In this study, 30 CL cases were included, 15 with TF to Glucantime and 15 with TR to Glucantime. All 30 isolates were checked for LRV1 and LRV2, 7 TR isolates and 2 TF isolates showed positive amplification for LRV2. Every clinical isolate was negative for LRV1. Various modalities are used to treat CL [22], World Health Organization standard recommended treatment is to use pentavalent antimoniate derivatives, but a portion of CL patients do not respond to this chemotherapy [23–25]. There are a few reports about the relation of Leishmania RNA virus infection and TF [26, 27]. It was previously demonstrated that presence of LRV1 may affect the Leishmania virulence [28]. According to Pereira et al. [21] study, there is no relationship between LRV and L. braziliensis response to treatment, which agreed with the results of the current study. The study by Hajjaran et al. [8] showed that not only LRV2 inside the Leishmania causative agents of CL, also LRV inside the causative agent of kala-azar have no relation with response to treatment similar to the current study. On the contrary, some studies showed a significant relationship between the presence of LRV1 and TF [6, 7, 27].
Hartley et al. [17], Parmentier et al. [28], and Ives et al. [19] showed resistance patterns to chemotherapy in Leishmania isolates infected with LRV1, that the presence of LRV1 affects the inflammatory responses and parasites metastatic behavior. In particular, LRV1 in L. guyanensis activates strong immunogenicity and triggers host immune system inflammatory factors [25]. The consequences of the last event are related to a more acute form of the disease and also resistance to chemotherapy. The effect of LRV1 on immune response is via toll-like receptor 3 (TLR3) and activation of pro-inflammatory factors and chemokines and therefore increases tissue damages [29]. In the present study, the TF isolates showed no infection with LRV1 but two TF isolates showed to be infected with LRV2, suggesting that other mechanisms might be related to TF. Cantanhêde et al. [18] found that LRV1 is one of the most important factors associated with exacerbation of the lesion and increases the risk of mucosal involvment.
Limitations
The results of the current study suggested no relationship between L. major infected with LRV2 and response to treatment although the sample size is small and further studies needed to clarify the role of LRV in response to treatment.
Supplementary information
Additional file 1: Figure S1. Agarose gel electrophoresis for molecular detection of Leishmania genus using L5.8 s and LITSR primer pairs. Line 1: 50 bp DNA ladder, line 2: negative control, line 3: positive control: L. major (MRHO/IR/75/ER), lines 4 and 5: clinical isolates with Leishmania genus. The expected fragment was around 300–350 bp for Leishmania spp. detection.
Additional file 2: Figure S2. Agarose gel electrophoresis for RFLP analysis. Line 1: 50 bp DNA ladder, line 2: positive control: L. major (MRHO/IR/75/ER), lines 3 and 4: clinical isolates of L. major. The fragments with the size of 220 and 127 bp was considered as L. major.
Acknowledgements
This research was done by Research Center of Food Hygiene and Safety, Shahid Sadoughi University of Medical Sciences, Yazd, Iran. We sincerely thank the technical supports of the staff of Research Center of Food Hygiene and Safety, Shahid Sadoughi University of Medical Sciences, Yazd, Iran.
Abbreviations
- CL
Cutaeous Leishmaniasis
- LRV
Leishmania RNA virus
- TF
Treatment failure
- TR
Treatment response
Authors’ contributions
MA collected the samples and did the experiments, GE designed and supervised the project, and wrote the draft of the manuscript, SC designed the protocol, MV analyzed the statistical analysis, SSH set up the protocol, SA helped in experiment and collecting the samples, MJB helped in writing the draft of the manuscript, AK designed the project and revised the manuscript All the authors read the manuscript and revised and accepted it. All authors read and approved the final manuscript.
Funding
This research was financially supported by Shahid Sadoughi University of Medical Sciences, Yazd, Iran. None of the roles of design of the study and collection, analysis, and interpretation of data and in writing the manuscript were for Shahid Sadoughi University of Medical Sciences, Yazd, Iran (Grant Number 4523).
Availability of data and materials
All data generated or analyzed during this study are included in this published article.
Ethics approval and consent to participate
All experiments and sampling procedures were ethically performed in accordance with standard protocols approved by Ethics Committee of Shahid Sadoughi University of Medical Sciences, Yazd, Iran. The informed consent was written and completed by each patient participating in this study before recording the information and sampling based on Helsinki declaration.
Consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing interests.
Footnotes
Publisher's Note
Springer Nature remains neutral with regard to jurisdictional claims in published maps and institutional affiliations.
Supplementary information
Supplementary information accompanies this paper at 10.1186/s13104-020-04973-y.
References
- 1.Alvar J, Vélez ID, Bern C, Herrero M, Desjeux P, Cano J, et al. Leishmaniasis worldwide and global estimates of its incidence. PLoS ONE. 2012;7:e35671. doi: 10.1371/journal.pone.0035671. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 2.Torres-Guerrero E, Quintanilla-Cedillo MR, Ruiz-Esmenjaud J, Arenas R. Leishmaniasis: a review. F1000Research. 2017;6:750. doi: 10.12688/f1000research.11120.1. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 3.https://www.who.int/news-room/fact-sheets/detail/leishmaniasis. Accessed 14 Mar 2019.
- 4.Steverding D. The history of Leishmaniasis. Parasit Vectors. 2017;10:82. doi: 10.1186/s13071-017-2028-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 5.Singh N, Kumar M, Singh RK. Leishmaniasis: current status of available drugs and new potential drug targets. Asian Pac J Trop Med. 2012;5:485–497. doi: 10.1016/S1995-7645(12)60084-4. [DOI] [PubMed] [Google Scholar]
- 6.Ponte-Sucre A, Gamarro F, Dujardin J-C, Barrett MP, Lo ´pez-Ve ´lez R, Garcı ´a-Herna ´ndez R, et al. Drug resistance and treatment failure in leishmaniasis: a 21st century challenge. PLoS Negl Trop Dis. 2017;11:e0006052. doi: 10.1371/journal.pntd.0006052. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 7.Vanaerschot M, Dumetz F, Roy S, Ponte-Sucre A, Arevalo J, Dujardin JC. Treatment failure in leishmaniasis: drug-resistance or another (epi-) phenotype? Expert Rev Anti Infect Ther. 2014;12:937–946. doi: 10.1586/14787210.2014.916614. [DOI] [PubMed] [Google Scholar]
- 8.Hajjaran H, Mahdi M, Mohebali M, Samimi-Rad K, Ataei-Pirkooh A, Kazemi-Rad E, et al. Detection and molecular identification of Leishmania RNA virus (LRV) in Iranian Leishmania species. Arch Virol. 2016;161:3385–3390. doi: 10.1007/s00705-016-3044-z. [DOI] [PubMed] [Google Scholar]
- 9.Zangger H, Ronet C, Desponds C, Kuhlmann FM, Robinson J, Hartley MA, et al. Detection of Leishmania RNA virus in Leishmania parasites. PLoS Negl Trop Dis. 2013;7:e2006. doi: 10.1371/journal.pntd.0002006. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 10.Scheffter SM, Ro YT, Chung IK, Patterson JL. The complete sequence of Leishmania RNA virus LRV2-1, a virus of an Old World parasite strain. Virology. 1995;212:84–90. doi: 10.1006/viro.1995.1456. [DOI] [PubMed] [Google Scholar]
- 11.Guilbride L, Myler PJ, Stuart K. Distribution and sequence divergence of LRV1 viruses among different Leishmania species. Mol Biochem Parasitol. 1992;54:101–104. doi: 10.1016/0166-6851(92)90099-6. [DOI] [PubMed] [Google Scholar]
- 12.Widmer G, Dooley S. Phylogenetic analysis of Leishmania RNA virus and Leishmania suggests ancient virus-parasite association. Nucleic Acids Res. 1995;23:2300–2304. doi: 10.1093/nar/23.12.2300. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 13.Cadd TL, Keenan MC, Patterson JL. Detection of Leishmania RNA virus 1 proteins. J Virol. 1993;67:5647–5650. doi: 10.1128/jvi.67.9.5647-5650.1993. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 14.Stuart KD, Weeks R, Guilbride L, Myler PJ. Molecular organization of Leishmania RNA virus 1. Proc Natl Acad Sci USA. 1992;89:8596–8600. doi: 10.1073/pnas.89.18.8596. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 15.Kleschenko Y, Grybchuk D, Matveeva NS, Macedo DH, Ponirovsky EN, Lukashev AN, et al. Molecular characterization of Leishmania RNA virus 2 in Leishmania major from Uzbekistan. Genes. 2019;10:E830. doi: 10.3390/genes10100830. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 16.Zangger H, Hailu A, Desponds C, Lye L-F, Akopyants NS, Dobson DE, et al. Leishmania aethiopica field isolates bearing an endosymbiontic dsRNA virus induce pro-inflammatory cytokine response. PLoS Negl Trop Dis. 2014;8:e2836. doi: 10.1371/journal.pntd.0002836. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 17.Hartley MA, Bourreau E, Rossi M, Castiglioni P, Eren RO, Prevel F, et al. Leishmania virus-dependent metastatic leishmaniasis is prevented by blocking IL-17A. PLoS Pathog. 2016;12:e1005852. doi: 10.1371/journal.ppat.1005852. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 18.Cantanhêde LM, da Silva Júnior CF, Ito MM, Felipin KP, Nicolete R, Salcedo JMV, et al. Further evidence of an association between the presence of Leishmania virus 1 and the mucosal manifestations in tegumentary leishmaniasis patients. PLoS Negl Trop Dis. 2015;15:e0004079. doi: 10.1371/journal.pntd.0004079. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 19.Ives A, Ronet C, Prevel F, Ruzzante G, Fuertes-Marraco S, Schutz F, et al. Leishmania RNA virus controls the severity of mucocutaneous leishmaniasis. Science. 2011;331:775–778. doi: 10.1126/science.1199326. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 20.Eslami G, Hajimohammadi B, Jafari AA, Mirzaei F, Gholamrezai M, Anvari H, et al. Molecular identification of Leishmania tropica infections in patients with cutaneous leishmaniasis from an endemic central of Iran. Trop Biomed. 2014;31:592–599. [PubMed] [Google Scholar]
- 21.Pereira Lde O, Maretti-Mira AC, Rodrigues KM, Lima RB, Oliveira-Neto MP, Cupolillo E, et al. Severity of tegumentary leishmaniasis is not exclusively associated with Leishmania RNA virus 1 infection in Brazil. Mem Inst Oswaldo Cruz. 2013;108:665–667. doi: 10.1590/0074-0276108052013021. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 22.Hejazi H, Eslami G, Dalimi A. The parasiticidal effect of electricity on Leishmania major, both in vitro and in vivo. Ann Trop Med Parasitol. 2004;98:37–42. doi: 10.1179/136485913X13789813917661. [DOI] [PubMed] [Google Scholar]
- 23.Ajamein V, Eslami G, Ahmadian S, Khamesipour A, Elloumi M. Gene expression of RNAP II, JBP1 and JBP2 in Leishmania major exposed to antimonials, amphotericin B and paromomycin. Ann Parasitol. 2018;64:181–187. doi: 10.17420/ap6403.149. [DOI] [PubMed] [Google Scholar]
- 24.Eslami G, Zarchi MV, Moradi A, Hejazi SH, Sohrevardi SM, Vakili M, et al. Aquaglyceroporin1 gene expression in antimony resistance and susceptible Leishmania major isolates. J Vector Borne Dis. 2016;53:370–374. [PubMed] [Google Scholar]
- 25.Ahmadian S, Eslami G, Fatahi A, Hosseini SS, Vakili M, Ajamein Fahadan V, et al. J-binding protein 1 and J-binding protein 2 expression in clinical Leishmania major no response-antimonial isolates. J Parasit Dis. 2019;43:39–45. doi: 10.1007/s12639-018-1052-5. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 26.Bourreau E, Ginouves M, Prévot G, Hartley MA, Gangneux JP, Robert-Gangneux F, et al. Presence of Leishmania RNA virus 1 in Leishmania guyanensis increases the risk of first-line treatment failure and symptomatic relapse. J Infect Dis. 2016;213:105–111. doi: 10.1093/infdis/jiv355. [DOI] [PubMed] [Google Scholar]
- 27.Adaui V, Lye LF, Akopyants NS, Zimic M, Llanos-Cuentas A, Garcia L, et al. Association of the endobiont double-stranded RNA virus LRV1 with treatment failure for human leishmaniasis caused by Leishmaniabraziliensis in Peru and Bolivia. J Infect Dis. 2016;213:112–121. doi: 10.1093/infdis/jiv354. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 28.Parmentier L, Cusini A, Müller N, Zangger H, Hartley MA, Desponds C, et al. Severe cutaneous leishmaniasis in a human immunodeficiency virus patient coinfected with Leishmania braziliensis and its endosymbiotic virus. Am J Trop Med Hyg. 2016;94:840–843. doi: 10.4269/ajtmh.15-0803. [DOI] [PMC free article] [PubMed] [Google Scholar]
- 29.de Carvalho RVH, Lima-Junior DS, da Silva MVG, Dilucca M, Rodrigues TS, Horta CV, et al. Publisher Correction: Leishmania RNA virus exacerbates leishmaniasis by subverting innate immunity via TLR3-mediated NLRP3 inflammasome inhibition. Nat Commun. 2020;11:203. doi: 10.1038/s41467-019-13900-0. [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.
Supplementary Materials
Additional file 1: Figure S1. Agarose gel electrophoresis for molecular detection of Leishmania genus using L5.8 s and LITSR primer pairs. Line 1: 50 bp DNA ladder, line 2: negative control, line 3: positive control: L. major (MRHO/IR/75/ER), lines 4 and 5: clinical isolates with Leishmania genus. The expected fragment was around 300–350 bp for Leishmania spp. detection.
Additional file 2: Figure S2. Agarose gel electrophoresis for RFLP analysis. Line 1: 50 bp DNA ladder, line 2: positive control: L. major (MRHO/IR/75/ER), lines 3 and 4: clinical isolates of L. major. The fragments with the size of 220 and 127 bp was considered as L. major.
Data Availability Statement
All data generated or analyzed during this study are included in this published article.


