ABSTRACT
Many filamentous pathogens invade plant cells through specialized hyphae called haustoria. These infection structures are enveloped by a newly synthesized plant-derived membrane called the extrahaustorial membrane (EHM). This specialized membrane is the ultimate interface between the plant and pathogen, and is key to the success or failure of infection. Strikingly, the EHM is reminiscent of host-derived membrane interfaces that engulf intracellular metazoan parasites. These perimicrobial interfaces are critical sites where pathogens facilitate nutrient uptake and deploy virulence factors to disarm cellular defenses mounted by their hosts. Although the mechanisms underlying the biogenesis and functions of these host–microbe interfaces are poorly understood, recent studies have provided new insights into the cellular and molecular mechanisms involved. In this Cell Science at a Glance and the accompanying poster, we summarize these recent advances with a specific focus on the haustorial interfaces associated with filamentous plant pathogens. We highlight the progress in the field that fundamentally underpin this research topic. Furthermore, we relate our knowledge of plant–filamentous pathogen interfaces to those generated by other plant-associated organisms. Finally, we compare the similarities between host–pathogen interfaces in plants and animals, and emphasize the key questions in this research area.
KEY WORDS: Defense-related autophagy, Effector translocation, Extrahaustorial membrane, Haustorium, Host–pathogen interface, Plant–pathogen interaction
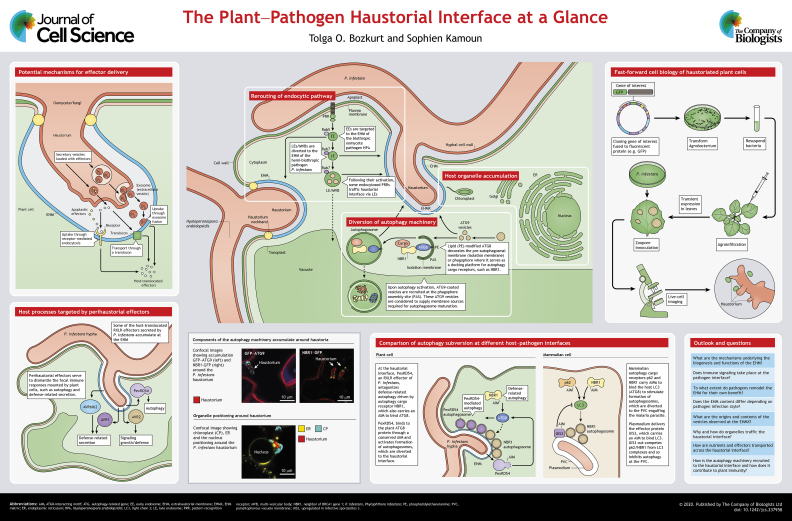
Summary: A discussion of the distinct host cell transport routes that underpin the biogenesis and function of plant–pathogen interfaces.
Introduction
Plant pathogens produce specialized cellular structures that invade host cells but remain enveloped by host-derived membranes. One such structure is the haustorium produced by many species of fungi and oomycetes (herein referred to as filamentous pathogens) (Panstruga and Dodds, 2009). Haustoria form tight membrane interfaces between these plant pathogens and their invaded host cells (haustoriated cells) (Bozkurt et al., 2015, 2014; Whisson et al., 2016; Bozkurt et al., 2011) and resemble to some degree host-derived membrane interfaces that engulf intracellular metazoan parasites (Haldar et al., 2006). These interfaces are a key cellular site of the tug-of-war between pathogens and their hosts, which ends in either host colonization or pathogen arrest.
Our understanding of the biogenesis and functions of plant–pathogen interfaces remains somewhat superficial, but recent advances have yielded new insights into cellular and molecular mechanisms. Here, we summarize this new knowledge with a focus on the haustorial interfaces associated with filamentous pathogens. We emphasize the two major questions that underpin this research topic, how are plant–pathogen membrane interfaces formed and what are the functions of haustoria? We also relate our understanding of plant-filamentous pathogen interfaces to other interfaces generated by other plant-associated organisms (see Box 1).
Box 1. Parasitic plants form haustoria too!
Parasitic plants produce specialized structures – also known as haustoria –to acquire water and nutrients from their hosts (Yoshida and Shirasu, 2012; Kokla and Melnyk, 2018). Although the haustoria of parasitic plants appear to be functionally analogous to those of filamentous pathogens, they result in very distinct interfaces with the host plants. In contrast to filamentous pathogens, haustoria of parasitic plants are multicellular organs that differentiate from stems and roots to penetrate host tissue and directly connect the parasite vasculature to that of its host. Haustoria thus enable the parasite to siphon nutrients and create an interface that facilitates bidirectional exchanges of macromolecules. Among the trafficking trans-species molecules are various types of RNA, including mRNAs and microRNAs (miRNAs). Although the precise functions of these RNAs are still being elucidated, trans-species RNA produced by the parasite mediate the cleavage of host mRNAs to modulate host gene expression presumably to the parasite's advantage (Shahid et al., 2018; Johnson and Axtell, 2019).
The haustorial interface
Haustoria are thought to facilitate exchange of macromolecules between the host and the pathogen. These specialized infection compartments are typically separated from the host cytoplasm through a newly synthesized plant-derived membrane called the extrahaustorial membrane (EHM) (Bozkurt et al., 2015, 2014; Whisson et al., 2016; Bozkurt et al., 2011). The haustorial interface is demarcated on one side by the EHM and on the other by the pathogen membrane and cell wall that surround the haustorium (see poster). These are separated by an extracellular matrix called the extrahaustorial matrix (EHMX) (Peresypkin et al., 1979; Baka, 2002). The number of haustoria per haustoriated host cell varies depending on the pathogen. Both oomycetes and fungi can form multiple haustoria in an individual plant cell (Bindschedler et al., 2009). However, unlike in oomycetes, the fungal haustorium is typically a separate cell that has its own nucleus with a haustorial neckband marking the cell border. Unlike intracellular hypha that can grow relentlessly and invade neighboring cells, haustoria remain restricted to infected host cells and are a terminal hyphal state.
Phylogenetically unrelated filamentous pathogens, such as the oomycetes, powdery mildew ascomycetes and rust basidiomycetes have evolved the haustorial lifestyle independently (Latijnhouwers et al., 2003). Despite their common physiological identity, the precise molecular features of the haustorial interfaces produced by these different classes of filamentous pathogens are unlikely to be the same even though some common features have been noted, as described herein.
Haustoria are not limited to filamentous pathogens. Strikingly, parasitic plants also form haustoria to tap into nutrient resources of their host plants (see Box 1). The convergent evolution of haustoria in divergent filamentous pathogens and parasitic plants further points to their importance for successful parasitism on plants.
Molecular traffic across the haustorial interface
The specific accumulation of a sugar transporter at fungal haustoria provided the evidence that haustoria can mediate nutrient uptake (Mendgen and Nass, 1988; Hahn et al., 1997; Voegele et al., 2001). However, direct evidence for the channeling of nutrients through the haustorial interface is still generally lacking. More recently, haustorial interfaces have emerged as delivery sites of pathogen-encoded virulence factors known as effectors (Kemen et al., 2005; Whisson et al., 2007; Wang et al., 2018); these not only include proteins, but also various species of RNAs with immunomodulatory functions produced by parasitic plants (Box 1; see poster). Fungal and filamentous pathogens also appear to deploy small RNAs inside their host cells to subvert host immunity (Sperschneider et al., 2018preprint; Dunker et al., 2019, preprint). However, it is not clear whether these nucleic acids are specifically transported through the haustorial interface. More importantly, the inter-organismal transport mechanisms across the haustorial interface remain uncharacterized. One possible transport mechanism could employ extracellular vesicles (EVs), which have established roles in cell-to-cell communication (see poster). Supporting this view, EVs with unknown identity have been observed at the EHMX during fungal invasion of plant cells (Micali et al., 2011). Furthermore, the finding that both pathogens and plant can discharge EVs with immunomodulatory functions (Bahar et al., 2016; Wang et al., 2017a,b; Cai et al., 2018; Baldrich et al., 2019) has sparked renewed interest in dissecting the contents and functions of the EVs deployed at the haustorial interface.
Effector delivery through the EHM
Specialized filamentous pathogens deliver effectors inside host cells to downregulate plant immunity and promote infection (Bozkurt et al., 2012; Thordal-Christensen et al., 2018). However, how these effectors enter plant cells remains a mystery. The majority of host-translocated oomycete effectors carry a conserved amino acid region defined by the RXLR motif that follows the N-terminal secretion signal (Whisson et al., 2007). The RXLR domain is dispensable for effector activities inside the host cells and mediates host translocation, similar to the PEXEL element found in plasmodium effectors (Hiller et al., 2004; Bozkurt et al., 2012). Like the PEXEL element, the RXLR motif undergoes proteolytic cleavage inside the parasite, with mature effectors lacking the motif (Boddey et al., 2016; Wawra et al., 2017). However, the precise mechanism by which the RXLR domain mediates effector translocation is still under debate, as the proposed models lack conclusive experimental evidence (Petre and Kamoun, 2014). For instance, the hypothesis that effector uptake takes place via binding of the RXLR motif to plant-derived phospholipids at the plant cell surface contradicts the finding that the RXLR motif is cleaved inside the pathogen prior to secretion (Wawra et al., 2017). In addition, more recent findings point to non-conventional secretory routes for host-translocation of RXLR effectors through the haustorial interface (Wang et al., 2017a,b; Wang et al., 2018).
The process of effector delivery is likely to have emerged multiple times throughout the evolution of filamentous pathogens. Unlike what is seen for oomycetes, conserved cell entry motifs and domains have not been identified in fungal effectors (Petre and Kamoun, 2014). The process is likely to be different in fungi as fungal haustoria are separate cells with nuclei and other organelles. (Petre and Kamoun, 2014). Because the fungal haustorium cell is accommodated inside the plant cell, it is assumed that the majority of the proteins secreted by fungal haustorium are either host-translocated or function at the EHMX. The translocation of effectors through the haustorial interface could possibly occur by (1) receptor-mediated endocytosis, (2) fusion of EVs loaded with effectors, or (3) through active transport facilitated by a pathogen-encoded translocon (see poster), as is the case in the apicomplexan parasite Plasmodium (Matthews et al., 2019).
EHM composition is different from the plasma membrane
One striking observation, originally made over a decade ago, is that the protein and lipid composition of the EHM contrasts sharply with that of the adjacent plasma membrane. Most of the proteins embedded in the plasma membrane, such as surface immune receptors, are excluded from the EHM (Koh et al., 2005; Micali et al., 2011; Lu et al., 2012). The few exceptions include the membrane-associated remorin protein REM1.3 and the vesicle fusion protein SYT1. Particularly, REM1.3 and SYT1 are exclusively localized to discrete micro-domains along the EHM, revealing that the EHM is not a uniform interface (Bozkurt et al., 2014) (see poster). Furthermore, the plasma membrane-localized pattern recognition receptor FLS2 is found to label the EHM of the oomycete pathogen Hyaloperonospora arabidopsidis (HPA) but not that of Phytophthora infestans. This indicates that the EHM composition varies depending on the pathosystem, although experimental differences between systems cannot be totally ruled out (Lu et al., 2012).
In some cases, the EHM remains isolated from the rest of the cytosol through encasements that are formed by defense-related focal deployment of the plant cell wall material callose (Micali et al., 2011; Caillaud et al., 2014). In contrast, haustoria of the oomycete pathogen P. infestans are generally not fully encased, with only 20% of the haustoria showing a ‘collar’ of callose around the haustorial neck (Bozkurt et al., 2014), indicating that pathogens can further modify the perihaustorial niche or that the host prevents encasement. It is conceivable that the callose encasements contribute to the overall defense mechanisms by preventing pathogen access to host resources and defense systems. However, the degree to which pathogens suppress haustoria-related defense processes, such as callose encasements, is not understood.
Rerouting of host-endocytic pathways to the haustorial interface
How infected plant cells selectively sort proteins into the EHM is poorly understood. The emerging paradigm is that diverse vesicular pathways may converge toward the EHM to generate a mosaic membrane interface (see poster). The EHM appears to accommodate proteins from diverse origins, including the plasma membrane, the vacuolar membrane, endocytic vesicles, plasmodesmata and the ER (Wang et al., 2009; Bozkurt et al., 2014, 2015; Caillaud et al., 2014; Inada et al., 2016; Kwaaitaal et al., 2017; Dagdas et al., 2018). It is plausible that redirection of multiple stress-related transport routes accounts for EHM biogenesis and maturation. Consistent with this notion, the vacuole-targeted late endocytic pathway marked by the small GTPase RabG3c (a Rab7 family member) is diverted toward the EHM during P. infestans infection of the solanaceous model plant Nicotiana benthamiana (Lu et al., 2012; Bozkurt et al., 2015). Upon activation, some PRRs are re-routed to the EHM through late endosomes (Bozkurt et al., 2015). However, it is unknown whether these PRRs are active in signaling or trapped at the haustorial interface by the pathogen in order to prevent their recycling back to cell surface, thus helping to suppress the host immune response.
Differential rerouting of the early endosomes (marked by Rab5), but not late endosomes (marked by RabG3f) towards the EHM has been observed in Arabidopsis leaves infected by two different oomycete pathogens (Lu et al., 2012). In contrast, the early endosomal marker Rab5 is excluded from the EHM engulfing the hemibiothrophic fungal pathogen Colletotrichum higginsianum (Inada et al., 2016). These findings further highlight that EHM composition varies in different pathosystems, which could be due to the divergent strategies employed by pathogens to manipulate the EHM to support virulence. In support of this notion, several host-translocated RXLR effectors of Phytophthora accumulate and probably target the EHM (Bozkurt et al., 2011; Wang et al., 2019) (see poster), but how they reconfigure the EHM for the benefit of the pathogen remains to be elucidated.
Interestingly, REM1.3 and RabG3C label only about half of the EHM enveloping the P. infestans haustoria, suggesting that the EHM is a dynamic interface that undergoes maturation (Bozkurt et al., 2014, 2015). In agreement with this notion, Arabidopsis PLASMODESMATA-LOCATED PROTEIN 1 (PDLP1) localizes only to the non-encased EHM of the oomycete pathogen H. arabidopsidis (Caillaud et al., 2014). Thus, it is possible that the EHM is modified gradually, starting from the initial haustoria formation to its subsequent maturation and ultimate encasement, and pathogens could actively manipulate this process through host-translocated effectors.
Diversion of autophagy machinery to the pathogen interface
Autophagy is a conserved eukaryotic trafficking process, in which cellular components and microbes are removed or relocated after engulfment in vesicular double-membrane-enclosed structures called autophagosomes (Lamb et al., 2013; Dagdas et al., 2018). Interestingly, selective forms of autophagy are induced at the perimicrobial interfaces in both plant and metazoan cells to counteract pathogen invasion (Thurston et al., 2012; Choi et al., 2014; Haldar et al., 2014; Schmuckli-Maurer et al., 2017; Wacker et al., 2017; Dagdas et al., 2018; Real et al., 2018). In plants, a defense-related autophagy machinery comprising the autophagy cargo receptor NBR1 (also known as Joka2) and the core autophagy adaptor ATG8 (ATG8CL isoform) target the EHM during P. infestans infection (see poster) (Dagdas et al., 2016). The pathogen counteracts this by deploying an RXLR effector called PexRD54. PexRD54 antagonizes NBR1 function by outcompeting it for ATG8CL binding, thereby neutralizing the defense-related autophagy at the haustorial interface (Dagdas et al., 2016, 2018). Thus, the autophagy machinery appears to participate in complex immune functions at perimicrobial membrane interfaces.
Unlike the many effectors of metazoan parasites that inhibit autophagy, PexRD54 stimulates formation of autophagosomes that accumulate at the haustorial interface (see poster). Why this is the case and what cargoes these autophagosomes carry remains uncharacterized. One hypothesis is that PexRD54 co-opts the host autophagy machinery as a molecular sink to absorb nutrients through the haustorial interface.
Organelle trafficking to the pathogen interface
Early work showed that some plant organelles accumulate around the haustorial interface (Heath et al., 1997; Koh et al., 2005) (see poster). However, the mechanisms by which organelles are recruited to pathogen interface and how they function at these sites are unknown. Positioning the plant endomembrane system (nucleus, ER, Golgi and secretory vesicles) around the haustorial interface is considered to aid localized deployment of defense-related compounds (Schmelzer, 2002; Underwood and Somerville, 2008). Interestingly, the ER surrounding fungal haustoria in Arabidopsis has a different morphology from the remainder of the ER network, for example by exhibiting swollen tubes (Micali et al., 2011). Altered ER morphology correlates with restricted intra-luminal ER transport (Tolley et al., 2008). These changes in ER morphology could be possibly triggered by pathogens to counteract the focal deployment of secretory components to the haustorial interface.
Intriguingly, host mitochondria have also been reported to accumulate around the EHM during fungal invasion of barley (Kunoh and Ishizaki, 1973; Micali et al., 2011; Fuchs et al., 2016). Although how and why mitochondria are targeted to the haustorial interface is unknown, electron microscopy images revealed intimate interactions between mitochondria and the EHM, such as membrane fusions (Kunoh and Ishizaki, 1973). Likewise, chloroplasts also accumulate at the haustorial interface and form tubular extensions embracing the EHM (Toufexi et al., 2019 preprint). Notably, the chloroplast photorelocation protein CHLOROPLAST UNUSUAL POSITIONING 1 (CHUP1) is required for the perihaustorial positioning of chloroplasts and immunity against P. infestans (Toufexi et al., 2019preprint). These findings implicate chloroplasts in plant immunity, but the exact defense-related functions of perihaustorial chloroplasts remain to be elucidated.
Similarities between plant–pathogen and animal–parasite interfaces
The differences in EHM composition compared to the plasma membrane are reminiscent of the perimicrobial membrane interfaces that engulf metazoan parasites (Haldar et al., 2006). Intracellular mammalian parasites typically deploy a variety of effector proteins to divert the trafficking of Rab GTPases to the pathogen interface (Asrat et al., 2014). Interestingly, these Rab GTPases include Rab5 and Rab7 family proteins, which are also found to localize to the EHM of filamentous plant pathogens as discussed above. The host-derived membranes that engulf Salmonella enterica are marked by Rab5, whereas Rab7 is recruited during later stages of infection (Drecktrah et al., 2007). Such a stepwise maturation of the perimicrobial membrane interfaces could also be the underlying reason for partial labelling of the EHM with Rab7 (∼50%) we observed during P. infestans infection (Bozkurt et al., 2015). Strikingly, a time-dependent accumulation of Rab7 also occurs during maturation of the Leishmania-containing parasitophorous vacuole membrane (PVM) in mammalian cells, where Rab7 labels 70% of the PVMs within 30 min after infection and reaching complete coverage within 48 h (Courret et al., 2002). Interestingly, the early endosomal marker Rab5 was found to be excluded from Leishmania PVM in mammalian cells (Courret et al., 2002), which is similar to what is found for the C. higginsianum (hemibiotrhopic fungus) EHM in Arabidopsis (Inada et al., 2016). However, there are also differences in Rab requirement, as Mycobacterium phagosomal compartments in mammalian cells are Rab5 positive but lack Rab7 in mammalian cells (Via et al., 1997), which is similar to the EHM of biotrophic filamentous plant pathogens infecting Arabidopsis (Inada et al., 2016).
Another similarity between plant–pathogen and mammalian–parasite interfaces is the induction of autophagy responses that are directed towards the pathogens, which are contained in modified phagosomal compartments, similar to recent observations with P. infestans (Dagdas et al., 2016, 2018). For instance, components of mammalian autophagy machinery such as ATG8 (the LC3/GABRAP family in mammalian cells) as well as the autophagy cargo receptors p62 (also known as SQSTM) and NBR1 target the peri-microbial membrane interface engulfing the Plasmodium parasite (Schmuckli-Maurer et al., 2017; Wacker et al., 2017; Real et al., 2018) (see poster). Interestingly, similar to the P. infestans RXLR effector PexRD54, one of the PVM-embedded plasmodium effector proteins, called UIS3, binds to the mammalian ATG8 isoform LC3 to avoid being degraded by autophagy (Real et al., 2018). Similar to antagonistic relationship between PexRD54 and the plant autophagy receptor NBR1 that occurs at the EHM, UIS3 outcompetes the mammalian autophagy cargo receptors for ATG8 (LC3) binding at the PVM (Real et al., 2018). Plant NBR1 has a similar domain architecture and shares functional features of the mammalian autophagy receptors NBR1 and p62 (Svenning et al., 2011). It is not clear whether these autophagy cargo receptors convergently evolved to counteract microbial penetration of host cells. Nevertheless, it appears that both plant and mammalian parasites have developed similar strategies to disarm host cargo receptors at the pathogen interface.
Conclusions and outlook
Despite the fact that phylogenetically diverse filamentous pathogens have convergently evolved the capacity to form haustoria and trigger the EHM interface with their plant hosts, there are some common principles. One common strategy employed by filamentous pathogens for successful invasion of the host cells is the reprogramming of host membrane trafficking pathways to avoid destruction by the host cellular defenses and facilitate efficient uptake of nutrients and possibly effector delivery. In addition, there are striking similarities in the processes that accommodate pathogens between plants and animals, some of which could possibly have originated from the ancestral eukaryotic cell. Future studies and emerging experimental systems, such as the fast-forward cell biology depicted in the poster, will help to further determine commonalities and differences across pathosystems and address pertinent questions about the haustorial interface. What are the mechanisms underlying the biogenesis and functions of the EHM? Does immune signaling take place at the pathogen interface? To what extent do pathogens manipulate the EHM for their own benefit? Does the EHM content differ depending on pathogen infection style? What are the origins and contents of the vesicles observed at the EHMX? Why and how do organelles traffic to the haustorial interface? How are nutrients and effectors transported across the haustorial interface? How is the autophagy machinery recruited to haustorial interface and how does it contribute to plant immunity? Answering these questions will further unveil the complex molecular and cellular processes that take place at the haustorial interface.
Acknowledgements
We thank Cian Duggan and Zachary Savage for comments on drafts.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
We thank the Gatsby Charitable Foundation, European Research Council (ERC), and Biotechnology and Biological Sciences Research Council (BBSRC) for funding. Open access funding provided by Imperial College London. Deposited in PMC for immediate release.
Cell science at a glance
A high-resolution version of the poster and individual poster panels are available for downloading at http://jcs.biologists.org/lookup/doi/10.1242/jcs.237958.supplemental
References
- Asrat S., de Jesús D. A., Hempstead A. D., Ramabhadran V. and Isberg R. R. (2014). Bacterial pathogen manipulation of host membrane trafficking. Annu. Rev. Cell Dev. Biol. 30, 79-109. 10.1146/annurev-cellbio-100913-013439 [DOI] [PubMed] [Google Scholar]
- Bahar O., Mordukhovich G., Luu D. D., Schwessinger B., Daudi A., Jehle A. K., Felix G. and Ronald P. C. (2016). Bacterial outer membrane vesicles induce plant immune responses. Mol. Plant-Microbe Interact. 29, 374-384. 10.1094/MPMI-12-15-0270-R [DOI] [PubMed] [Google Scholar]
- Baka Z. A. (2002). Ultrastructure of intercellular hypha and haustorium of the rust fungus, Uromyces euphorbiae. Mycopathologia 156, 215-221. [PubMed] [Google Scholar]
- Baldrich P., Rutter B. D., Karimi H. Z., Podicheti R., Meyers B. C. and Innes R. W. (2019). Plant extracellular vesicles contain diverse small RNA species and are enriched in 10- to 17-nucleotide “Tiny” RNAs. Plant Cell 31, 315-324. 10.1105/tpc.18.00872 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bindschedler L. V., Burgis T. A., Mills D. J. S., Ho J. T. C., Cramer R. and Spanu P. D. (2009). In planta proteomics and proteogenomics of the biotrophic barley fungal pathogen Blumeria graminis f. sp. hordei. Mol. Cell. Proteomics 8, 2368-2381. 10.1074/mcp.M900188-MCP200 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Boddey J. A., O'Neill M. T., Lopaticki S., Carvalho T. G., Hodder A. N., Nebl T., Wawra S., van West P., Ebrahimzadeh Z., Richard D. et al. (2016). Export of malaria proteins requires co-translational processing of the PEXEL motif independent of phosphatidylinositol-3-phosphate binding. Nat. Commun. 7, 10470 10.1038/ncomms10470 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Win J., Shindo T., Ilyas M., Oliva R., Cano L. M., Jones A. M. E., Huitema E., van der Hoorn R. A. L. et al. (2011). Phytophthora infestans effector AVRblb2 prevents secretion of a plant immune protease at the haustorial interface. Proc. Natl. Acad. Sci. USA 108, 20832-20837. 10.1073/pnas.1112708109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T. O., Schornack S., Banfield M. J. and Kamoun S. (2012). Oomycetes, effectors, and all that jazz. Curr. Opin. Plant Biol. 15, 483-492. 10.1016/j.pbi.2012.03.008 [DOI] [PubMed] [Google Scholar]
- Bozkurt T. O., Richardson A., Dagdas Y. F., Mongrand S., Kamoun S. and Raffaele S. (2014). The plant membrane-associated REMORIN1.3 accumulates in discrete perihaustorial domains and enhances susceptibility to Phytophthora infestans. Plant Physiol. 165, 1005-1018. 10.1104/pp.114.235804 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bozkurt T. O., Belhaj K., Dagdas Y. F., Chaparro-Garcia A., Wu C.-H., Cano L. M. and Kamoun S. (2015). Rerouting of plant late endocytic trafficking toward a pathogen interface. Traffic 16, 204-226. 10.1111/tra.12245 [DOI] [PubMed] [Google Scholar]
- Cai Q., Qiao L., Wang M., He B., Lin F.-M., Palmquist J., Huang S.-D. and Jin H. (2018). Plants send small RNAs in extracellular vesicles to fungal pathogen to silence virulence genes. Science 360, 1126-1129. 10.1126/science.aar4142 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Caillaud M.-C., Wirthmueller L., Sklenar J., Findlay K., Piquerez S. J. M., Jones A. M. E., Robatzek S., Jones J. D. G. and Faulkner C. (2014). The plasmodesmal protein PDLP1 localises to haustoria-associated membranes during downy mildew infection and regulates callose deposition. PLoS Pathog. 10, e1004496 10.1371/journal.ppat.1004496 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Carella P., Gogleva A., Tomaselli M., Alfs C. and Schornack S. (2018). Phytophthora palmivora establishes tissue-specific intracellular infection structures in the earliest divergent land plant lineage. Proc. Natl. Acad. Sci. USA 115, E3846-E3855. 10.1073/pnas.1717900115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Choi J., Park S., Biering S. B., Selleck E., Liu C. Y., Zhang X., Fujita N., Saitoh T., Akira S., Yoshimori T. et al. (2014). The parasitophorous vacuole membrane of toxoplasma gondii is targeted for disruption by ubiquitin-like conjugation systems of autophagy. Immunity 40, 924-935. 10.1016/j.immuni.2014.05.006 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Courret N., Frehel C., Gouhier N., Pouchelet M., Pina E., Roux P. and Antoine J. C. (2002). Biogenesis of Leishmania-harbouring parasitophorous vacuoles following phagocytosis of the metacyclic promastigote or amastigote stages of the parasites. J. Cell Sci. 115, 2303-2316. [DOI] [PubMed] [Google Scholar]
- Dagdas Y. F., Belhaj K., Maqbool A., Chaparro-Garcia A., Pandey P., Petre B., Tabassum N., Cruz-Mireles N., Hughes R. K., Sklenar J. et al. (2016). An effector of the Irish potato famine pathogen antagonizes a host autophagy cargo receptor. eLife 5, e10856 10.7554/eLife.10856 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dagdas Y. F., Pandey P., Tumtas Y., Sanguankiattichai N., Belhaj K., Duggan C., Leary A. Y., Segretin M. E., Contreras M. P., Savage Z. et al. (2018). Host autophagy machinery is diverted to the pathogen interface to mediate focal defense responses against the Irish potato famine pathogen. eLife 7, e37476 10.7554/eLife.37476 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Drecktrah D., Knodler L. A., Howe D. and Steele-Mortimer O. (2007). Salmonella trafficking is defined by continuous dynamic interactions with the endolysosomal system. Traffic 8, 212-225. 10.1111/j.1600-0854.2006.00529.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dunker F., Trutzenberg A., Rothenpieler J., Kuhn S., Pröls R., Schreiber T., Tissier A., Hückelhoven R. and Weiberg A. (2019). Oomycete small RNAs invade the plant RNA-induced silencing complex for virulence. bioRxiv, 689190. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fuchs R., Kopischke M., Klapprodt C., Hause G., Meyer A. J., Schwarzländer M., Fricker M. D. and Lipka V. (2016). Immobilized subpopulations of leaf epidermal mitochondria mediate PENETRATION2-dependent pathogen entry control in arabidopsis. Plant Cell 28, 130-145. 10.1105/tpc.15.00887 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hahn M., Neef U., Struck C., Göttfert M. and Mendgen K. (1997). A putative amino acid transporter is specifically expressed in haustoria of the rust fungus Uromyces fabae. Mol. Plant Microbe Interact. 10, 438-445. 10.1094/MPMI.1997.10.4.438 [DOI] [PubMed] [Google Scholar]
- Haldar K., Kamoun S., Hiller N. L., Bhattacharje S. and van Ooij C. (2006). Common infection strategies of pathogenic eukaryotes. Nat. Rev. Microbiol. 4, 922-931. 10.1038/nrmicro1549 [DOI] [PubMed] [Google Scholar]
- Haldar A. K., Piro A. S., Pilla D. M., Yamamoto M. and Coers J. (2014). The E2-like conjugation enzyme Atg3 promotes binding of IRG and Gbp proteins to chlamydia- and toxoplasma-containing vacuoles and host resistance. PLoS ONE 9, e86684 10.1371/journal.pone.0086684 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heath M. C., Nimchuk Z. L. and Xu H. X. (1997). Plant nuclear migrations as indicators of critical interactions between resistant or susceptible cowpea epidermal cells and invasion hyphae of the cowpea rust fungus. New Phytol. 135, 689-700. 10.1046/j.1469-8137.1997.00710.x [DOI] [Google Scholar]
- Hiller N. L., Bhattacharjee S., van Ooij C., Liolios K., Harrison T., Lopez-Estrano C. and Haldar K. (2004). A host-targeting signal in virulence proteins reveals a secretome in malarial infection. Science 306, 1934-1937. 10.1126/science.1102737 [DOI] [PubMed] [Google Scholar]
- Inada N., Betsuyaku S., Shimada T. L., Ebine K., Ito E., Kutsuna N., Hasezawa S., Takano Y., Fukuda H., Nakano A. et al. (2016). Modulation of plant RAB GTPase-mediated membrane trafficking pathway at the interface between plants and obligate biotrophic pathogens. Plant Cell Physiol. 57, 1854-1864. 10.1093/pcp/pcw107 [DOI] [PubMed] [Google Scholar]
- Johnson N. R. and Axtell M. J. (2019). Small RNA warfare: exploring origins and function of trans-species microRNAs from the parasitic plant Cuscuta. Curr. Opin. Plant Biol. 50, 76-81. 10.1016/j.pbi.2019.03.014 [DOI] [PubMed] [Google Scholar]
- Kemen E., Kemen A. C., Rafiqi M., Hempel U., Mendgen K., Hahn M. and Voegele R. T. (2005). Identification of a protein from rust fungi transferred from haustoria into infected plant cells. Mol. Plant Microbe Interact. 18, 1130-1139. 10.1094/MPMI-18-1130 [DOI] [PubMed] [Google Scholar]
- Koh S., André A., Edwards H., Ehrhardt D. and Somerville S. (2005). Arabidopsis thaliana subcellular responses to compatible Erysiphe cichoracearum infections. Plant J. 44, 516-529. 10.1111/j.1365-313X.2005.02545.x [DOI] [PubMed] [Google Scholar]
- Kokla A. and Melnyk C. W. (2018). Developing a thief: Haustoria formation in parasitic plants. Dev. Biol. 442, 53-59. 10.1016/j.ydbio.2018.06.013 [DOI] [PubMed] [Google Scholar]
- Kunoh H. and Ishizaki H. (1973). Incorporation of host mitochondria into the haustorial encapsulation of barley powdery mildew (II). Ann. Phytopath. Soc. Japan 39, 42-48. 10.3186/jjphytopath.39.42 [DOI] [Google Scholar]
- Kwaaitaal M., Nielsen M. E., Böhlenius H. and Thordal-Christensen H. (2017). The plant membrane surrounding powdery mildew haustoria shares properties with the endoplasmic reticulum membrane. J. Exp. Bot. 68, 5731-5743. 10.1093/jxb/erx403 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lamb C. A., Yoshimori T. and Tooze S. A. (2013). The autophagosome: origins unknown, biogenesis complex. Nat. Rev. Mol. Cell Biol. 14, 759-774. 10.1038/nrm3696 [DOI] [PubMed] [Google Scholar]
- Latijnhouwers M., de Wit P. J. G. M. and Govers F. (2003). Oomycetes and fungi: similar weaponry to attack plants. Trends Microbiol. 11, 462-469. 10.1016/j.tim.2003.08.002 [DOI] [PubMed] [Google Scholar]
- Lu Y.-J., Schornack S., Spallek T., Geldner N., Chory J., Schellmann S., Schumacher K., Kamoun S. and Robatzek S. (2012). Patterns of plant subcellular responses to successful oomycete infections reveal differences in host cell reprogramming and endocytic trafficking. Cell. Microbiol. 14, 682-697. 10.1111/j.1462-5822.2012.01751.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matthews K. M., Pitman E. L. and de Koning-Ward T. F. (2019). “Illuminating how malaria parasites export proteins into host erythrocytes. Cell. Microbiol. 21, e13009 10.1111/cmi.13009 [DOI] [PubMed] [Google Scholar]
- Mendgen K. and Nass P. (1988). The activity of powdery-mildew haustoria after feeding the host cells with different sugars, as measured with a potentiometric cyanine dye. Planta 174, 283-288. 10.1007/BF00394782 [DOI] [PubMed] [Google Scholar]
- Micali C. O., Neumann U., Grunewald D., Panstruga R. and O'Connell R. (2011). Biogenesis of a specialized plant-fungal interface during host cell internalization of Golovinomyces orontii haustoria. Cell. Microbiol. 13, 210-226. 10.1111/j.1462-5822.2010.01530.x [DOI] [PubMed] [Google Scholar]
- Panstruga R. and Dodds P. N. (2009). Terrific protein traffic: the mystery of effector protein delivery by filamentous plant pathogens. Science 324, 748-750. 10.1126/science.1171652 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peresypkin V. F., Loban V. L. and Voloshin N. V. (1979). [Ultrastructure of the haustoria and intercellular hyphae of Puccinia triticina Erikss mycelia]. Mikrobiol. Zh. 41, 245-247. [PubMed] [Google Scholar]
- Petre B. and Kamoun S. (2014). How do filamentous pathogens deliver effector proteins into plant cells? PLoS Biol. 12, e1001801 10.1371/journal.pbio.1001801 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Real E., Rodrigues L., Cabal G. G., Enguita F. J., Mancio-Silva L., Mello-Vieira J., Beatty W., Vera I. M., Zuzarte-Luís V., Figueira T. N. et al. (2018). Plasmodium UIS3 sequesters host LC3 to avoid elimination by autophagy in hepatocytes. Nat. Microbiol. 3, 17-25. 10.1038/s41564-017-0054-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Schmelzer E. (2002). Cell polarization, a crucial process in fungal defence. Trends Plant Sci. 7, 411-415. 10.1016/S1360-1385(02)02307-5 [DOI] [PubMed] [Google Scholar]
- Schmuckli-Maurer J., Reber V., Wacker R., Bindschedler A., Zakher A. and Heussler V. T. (2017). Inverted recruitment of autophagy proteins to the Plasmodium berghei parasitophorous vacuole membrane. PLoS ONE 12, e0183797 10.1371/journal.pone.0183797 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shahid S., Kim G., Johnson N. R., Wafula E., Wang F., Coruh C., Bernal-Galeano V., Phifer T., dePamphilis C. W., Westwood J. H. et al. (2018). MicroRNAs from the parasitic plant Cuscuta campestris target host messenger RNAs. Nature 553, 82 10.1038/nature25027 [DOI] [PubMed] [Google Scholar]
- Sperschneider J., Jacques S., Xu B., Upadhyaya N. M., Mago R., Singh K. B., Stone E. A., Wang M.-B., Dodds P. N. and Taylor J. M. (2018). The stem rust fungus Puccinia graminis f. sp. tritici induces waves of small RNAs with opposing profiles during wheat infection. bioRxiv, 469338. [Google Scholar]
- Svenning S., Lamark T., Krause K. and Johansen T. (2011). Plant NBR1 is a selective autophagy substrate and a functional hybrid of the mammalian autophagic adapters NBR1 and p62/SQSTM1. Autophagy 7, 993-1010. 10.4161/auto.7.9.16389 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thordal-Christensen H., Birch P. R. J., Spanu P. D. and Panstruga R. (2018). Why did filamentous plant pathogens evolve the potential to secrete hundreds of effectors to enable disease? Mol. Plant Pathol. 19, 781-785. 10.1111/mpp.12649 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Thurston T. L. M., Wandel M. P., von Muhlinen N., Foeglein A. and Randow F. (2012). Galectin 8 targets damaged vesicles for autophagy to defend cells against bacterial invasion. Nature 482, 414-418. 10.1038/nature10744 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tolley N., Sparkes I. A., Hunter P. R., Craddock C. P., Nuttall J., Roberts L. M., Hawes C., Pedrazzini E. and Frigerio L. (2008). Overexpression of a plant reticulon remodels the lumen of the cortical endoplasmic reticulum but does not perturb protein transport. Traffic 9, 94-102. 10.1111/j.1600-0854.2007.00670.x [DOI] [PubMed] [Google Scholar]
- Toufexi A., Duggan C., Pandey P., Savage Z., Segretin M. E., Yuen L. H., Gaboriau D. C. A., Leary A. Y., Khandare V., Ward A. D. et al. (2019). Chloroplasts navigate towards the pathogen interface to counteract infection by the Irish potato famine pathogen. bioRxiv, 516443. [Google Scholar]
- Underwood W. and Somerville S. C. (2008). Focal accumulation of defences at sites of fungal pathogen attack. J. Exp. Bot. 59, 3501-3508. 10.1093/jxb/ern205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Via L. E., Deretic D., Ulmer R. J., Hibler N. S., Huber L. A. and Deretic V. (1997). Arrest of mycobacterial phagosome maturation is caused by a block in vesicle fusion between stages controlled by rab5 and rab7. J. Biol. Chem. 272, 13326-13331. 10.1074/jbc.272.20.13326 [DOI] [PubMed] [Google Scholar]
- Voegele R. T., Struck C., Hahn M. and Mendgen K. (2001). The role of haustoria in sugar supply during infection of broad bean by the rust fungus Uromyces fabae. Proc. Natl. Acad. Sci. USA 98, 8133-8138. 10.1073/pnas.131186798 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wacker R., Eickel N., Schmuckli-Maurer J., Annoura T., Niklaus L., Khan S. M., Guan J.-L. and Heussler V. T. (2017). LC3-association with the parasitophorous vacuole membrane of Plasmodium berghei liver stages follows a noncanonical autophagy pathway. Cell. Microbiol. 19, e12754 10.1111/cmi.12754 [DOI] [PubMed] [Google Scholar]
- Wang W., Wen Y., Berkey R. and Xiao S. (2009). Specific targeting of the Arabidopsis resistance protein RPW8.2 to the interfacial membrane encasing the fungal Haustorium renders broad-spectrum resistance to powdery mildew. Plant Cell 21, 2898-2913. 10.1105/tpc.109.067587 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang M., Weiberg A., Dellota E., Yamane D. and Jin H. L. (2017a). Botrytis small RNA Bc-siR37 suppresses plant defense genes by cross-kingdom RNAi. RNA Biol. 14, 421-428. 10.1080/15476286.2017.1291112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. M., Boevink P. C., Welsh L., Zhang R. F., Whisson S. C. and Birch P. R. J. (2017b). Delivery of cytoplasmic and apoplastic effectors from Phytophthora infestans haustoria by distinct secretion pathways. New Phytol. 216, 205-215. 10.1111/nph.14696 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S. M., Welsh L., Thorpe P., Whisson S. C., Boevink P. C. and Birch P. R. J. (2018). The Phytophthora infestans haustorium is a site for secretion of diverse classes of infection-associated proteins. Mbio 9, e01216-18 10.1128/mBio.01216-18 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wang S., McLellan H., Bukharova T., He Q., Murphy F., Shi J., Sun S., van Weymers P., Ren Y., Thilliez G. et al. (2019). Phytophthora infestans RXLR effectors act in concert at diverse subcellular locations to enhance host colonization. J. Exp. Bot. 70, 343-356. 10.1093/jxb/ery360 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wawra S., Trusch F., Matena A., Apostolakis K., Linne U., Zhukov I., Stanek J., Koźmiński W., Davidson I., Secombes C. J. et al. (2017). The RxLR motif of the host targeting effector AVR3a of Phytophthora infestans is cleaved before secretion. Plant Cell 29, 1184-1195. 10.1105/tpc.16.00552 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whisson S. C., Boevink P. C., Moleleki L., Avrova A. O., Morales J. G., Gilroy E. M., Armstrong M. R., Grouffaud S., van West P., Chapman S. et al. (2007). A translocation signal for delivery of oomycete effector proteins into host plant cells. Nature 450, 115 10.1038/nature06203 [DOI] [PubMed] [Google Scholar]
- Yoshida S. and Shirasu K. (2012). Plants that attack plants: molecular elucidation of plant parasitism. Curr. Opin. Plant Biol. 15, 708-713. 10.1016/j.pbi.2012.07.004 [DOI] [PubMed] [Google Scholar]