ABSTRACT
Immune responses involve many types of leukocytes that traffic to the site of injury, recognize the insult and respond appropriately. Imaging of the immune system involves a set of methods and analytical tools that are used to visualize immune responses at the cellular and molecular level as they occur in real time. We will review recent and emerging technological advances in optical imaging, and their application to understanding the molecular and cellular responses of neutrophils, macrophages and lymphocytes. Optical live-cell imaging provides deep mechanistic insights at the molecular, cellular, tissue and organism levels. Live-cell imaging can capture quantitative information in real time at subcellular resolution with minimal phototoxicity and repeatedly in the same living cells or in accessible tissues of the living organism. Advanced FRET probes allow tracking signaling events in live cells. Light-sheet microscopy allows for deeper tissue penetration in optically clear samples, enriching our understanding of the higher-level organization of the immune response. Super-resolution microscopy offers insights into compartmentalized signaling at a resolution beyond the diffraction limit, approaching single-molecule resolution. This Review provides a current perspective on live-cell imaging in vitro and in vivo with a focus on the assessment of the immune system.
KEY WORDS: Imaging, Immune cells, Microscopy, FRET, Intravital imaging, Subcellular imaging, Molecular imaging
Summary: This Review provides a current perspective on live-cell imaging in vitro and in vivo with a focus on the understanding of the immune system at the subcellular and molecular level.
Introduction
The complexity of the immune system makes the imaging of various immune cells as a whole demanding but also worthwhile. Although non-optical methods, such as positron emission tomography (PET) and magnetic resonance imaging (MRI) exist (Willmann et al., 2008), their resolution is not at the single-cell level. Hence, this Review will be limited to optical approaches with higher resolution that have been developed in the last few years. We will place the focus on the understanding of immune cells and their signaling at the subcellular and molecular level using new and emerging microscopic techniques. The starting point for this Review are three recent reviews that cover multiphoton and confocal live-cell imaging (Germain et al., 2012; McArdle et al., 2016; Pittet et al., 2018). We will cover advances in imaging of the immune system towards a subcellular and molecular understanding through the application of total internal reflection fluorescence (TIRF) and quantitative dynamic footprinting (qDF) microscopy, Förster resonance energy transfer (FRET) imaging, intravital imaging, light-sheet microscopy (LSM) and super-resolution microscopy.
Imaging methods
TIRF and qDF microscopy
The contact region between a cell and its substrate is of great biological importance. A large number of molecular events occur at the cellular surface, where cells exchange materials and information with the surrounding environment. Examples include leukocyte rolling, adhesion (Ley et al., 2007), ligand–receptor interaction, T cell receptor (TCR) engagement (Dustin and Choudhuri, 2016), molecule transport, and exocytotic and endocytotic processes (Wu et al., 2014). TIRF microscopy is a technique that is best suited to study these membrane-associated processes at the cell–surface interface. Thus, TIRF is a high-resolution live-cell imaging method. However, TIRF is not suitable for in vivo studies, because it requires a glass–cell interface that does not exist in vivo.
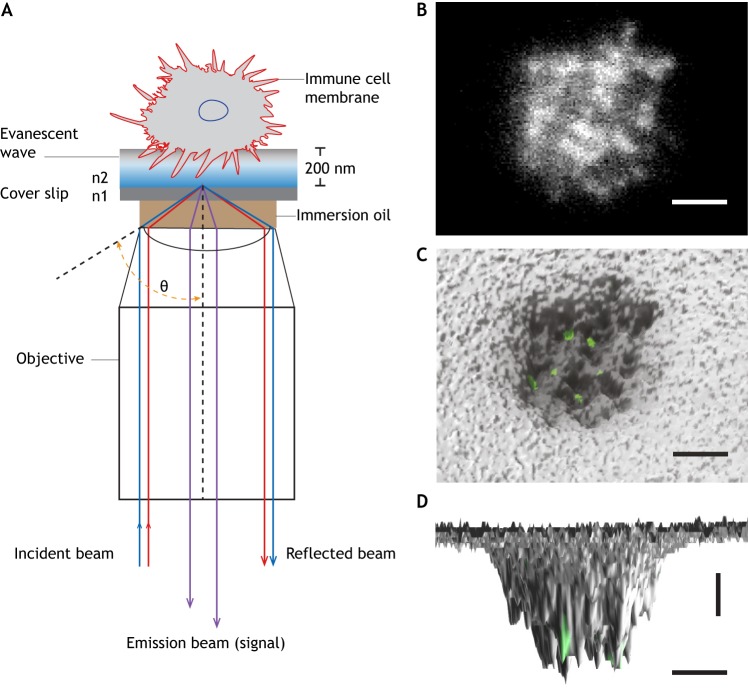
TIRF microscopy was first introduced by Daniel Axelrod (Axelrod, 1981). It allows visualization of cell and substrate contacts within a thin region of a specimen (∼200 nm from a coverslip). This is achieved by selectively illuminating and exciting fluorophores in a thin layer of the specimen in the immediate vicinity of the glass–water interface using an evanescent wave. The evanescent wave is generated when the incident light is totally reflected at the glass–water interface, because the electromagnetic fields cannot be discontinuous. The critical angle is the smallest angle of incidence that yields total reflection. The main purpose of TIRF in biology is to look at only the cell surface at the glass–water interface. TIRF completely eliminates the background fluorescence further into the cell.
One of the evolved versions of TIRF is based on variable angle TIRF (Stock et al., 2003), also called quantitative dynamic footprinting (qDF) (Fig. 1A) (Sundd et al., 2012, 2010). qDF was developed to image the footprints of live leukocytes rolling, adhering, spreading or migrating on a glass coverslip. qDF is diffraction-limited in the x- and y-axes, but affords single-digit nanometer resolution in the z-axis. Reasonable temporal resolution of better than one frame per second (fps) has been achieved (Sundd et al., 2012, 2010). Up to three fluorochromes have been imaged simultaneously (Fan et al., 2016; Sundd et al., 2011), thus allowing for simultaneous analysis of three molecular events. qDF is suitable for imaging small 3D membrane structures, such as microvilli, tethers and slings in living immune cells (Sundd et al., 2012, 2010). qDF has also been combined with super-resolution microscopy to achieve high resolution in x, y and z planes, showing that T cell receptors (TCRs) are concentrated in the microvilli of T cells (Jung et al., 2016). However, unlike qDF, super-resolution microscopy usually entails fixed specimens and long data collection times, which are unfavorable for live-cell imaging.
Fig. 1.
TIRF-based qDF imaging. (A) Schematic illustration of qDF imaging. An incident laser beam travels through the glass–cell interface at an angle (θ) with total internal reflection, (θ>θC=sin−1(n2/n1)). n1 and n2 are the refractive indices of glass and cell, respectively (n1>n2). The cell surface can be reconstructed by converting the cell membrane-labeled fluorescence into a 3D topographical map. (B) Footprints of a human neutrophil at the time of arrest on P-selectin and ICAM-1 in response to interleukin 8 (IL-8) at 6 dyn cm−2. Membrane was labeled with CellMask™ Deep Red. A top-view (C) and side-view (D) of the 3D topography of membrane (gray) was overlaid with high affinity integrin clusters (green) labeled with an antibody (mAb24). Images were taken in the Ley laboratory. Horizontal scale bars: 2 µm, vertical scale bar: 10 nm.
qDF combined with a homogeneous binding assay (Chigaev and Sklar, 2011) allows for reporting of dynamic antibody binding to molecules in the membrane of live cells. For example, antibodies reporting the conformation of integrin β2 have been introduced into flow chambers to show β2 integrin activation in live cells that move under shear stress (Fan et al., 2016). Here, exposure of human neutrophils to the chemokine IL-8 induces a β2 integrin conformation shift to a high-affinity form as reported by an activation-specific antibody (Fig. 1). qDF imaging revealed that β2 integrins can achieve this high-affinity conformation by first extending and then rearranging the β2 I-like domain (the canonical pathway of integrin activation) or by attaining high affinity first and extending subsequently (Fan et al., 2016). qDF is also suitable to track fluorescent protein (FP) reporter constructs to map proteins near the plasma membrane. Indeed, we (L.W. and K.L.) are currently using FP-fused kindlin-3 to analyze its recruitment to the plasma membrane during neutrophil rolling and arrest.
FRET imaging
FRET is a distance-dependent quantum mechanical phenomenon characterized by a non-radiative transfer of energy from a donor fluorophore to an acceptor fluorophore through dipole–dipole coupling (Zhang et al., 2002). FRET microscopy has become a powerful tool for measurements of molecular conformations or intermolecular interactions at the single-molecule level. The FRET efficiency (E) of the FRET process depends on the inverse sixth power of the distance between donor and acceptor,
where R0 (Förster distance) is the characteristic distance at which 50% of the energy is transferred (typically 2–6 nm), and r is the actual distance between donor and acceptor. R0 is dependent on the extent of spectral overlap, the quantum yield of the donor, and the relative orientation of donor and acceptor (Förster, 1948; Zhang et al., 2002). Owing to the 1/r6 dependence, the FRET efficiency is highly sensitive to r and practically limited to 10 nm.
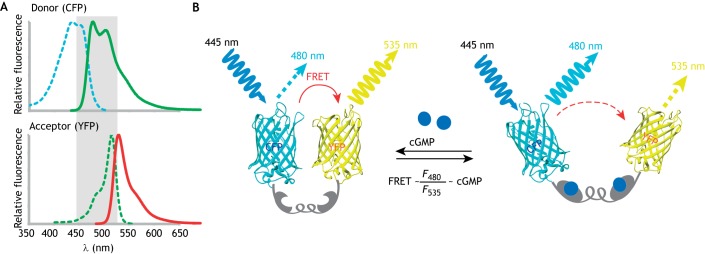
FRET requires (1) a substantial overlap (more than 30%) between the emission spectrum of the donor and the excitation spectrum of the acceptor; (2) that both excitation spectra must be separated enough to allow independent excitation and photobleaching (Fig. 2A); and (3) that the donor and acceptor fluorophores must be in a favorable spatial orientation and close proximity. Cyan fluorescent protein (CFP) and yellow fluorescent protein (YFP) are an established pair of donor and acceptor fluorescent proteins (Bajar et al., 2016; Lam et al., 2012). They can be engineered into the same protein, and so can also serve as force probes (Grashoff et al., 2010) or probes for biochemical events (O'Shaughnessy et al., 2019).
Fig. 2.
Working principles of FRET. (A) Overlap of the emission and excitation spectra of donor (CFP) and acceptor (YFP). (B) The intramolecular FRET biosensor; a cGMP biosensor is shown as an example. cGi500 (Russwurm et al., 2007) is a cGMP indicator consisting of the engineered cGMP-binding domain of the cGMP-dependent protein kinase type I (gray) sandwiched between CFP and YFP (Wen et al., 2018). The energy transfer occurs from excited CFP to YFP. Upon cGMP binding to the sensor, the FRET efficiency is reduced. Thus, light emission from YFP at 535 nm is decreased, while emission from CFP at 480 nm is increased. The ratio of emission at 480 nm and 535 nm (F480/F535) is related to the cGMP level.
FRET is outstanding in probing subcellular signaling because it can measure signaling with high spatial and temporal resolution in live cells. FRET can be performed with almost any type of fluorescent microscope. It works deep in the tissue, at low numerical aperture, and even in flow cytometry, where immune cells ‘fly’ by the excitation laser and detection photomultiplier tubes (PMTs) in a small fraction of a second (Fan et al., 2016; Lefort et al., 2009; Stadtmann et al., 2013). FRET multiphoton microscopy permits intravital imaging with deep tissue penetration and represents an emerging area of in vivo analysis of signaling events under physiological conditions (Pittet and Weissleder, 2011; Radbruch et al., 2015).
The methods to quantify the FRET efficiency include primarily (1) fluorescence intensity-based analysis, (2) fluorescence intensity-independent lifetime analysis, and (3) anisotropy-based analysis (Shrestha et al., 2015). Thus, FRET efficiency can be detected as a ratio (for example, FRET:CFP) or shortening of the donor fluorescence lifetime in FLIM analysis (Shrestha et al., 2015). Based on practical considerations, some general approaches have demonstrated their potential in studying biological samples, including two-color emission ratio imaging, acceptor photobleaching and FLIM-FRET (Box 1).
Box 1. FRET measurement methods.
Two-color emission ratio imaging, also known as sensitized emission
This is a popular method applied to probing protein–protein interactions or intramolecular biosensing in both microscopy and flow cytometry. Samples are illuminated at the donor excitation wavelength, and fluorescence is collected in both donor and acceptor (FRET) channels. The fluorescence intensity of the acceptor channel divided by the donor channel is defined as the FRET index. Ratiometric imaging can be practiced on a regular wide-field or confocal fluorescence microscope. Since the ratiometric imaging is intensity dependent, separating FRET from non-FRET signal can often be a major challenge. Controls and corrections for fluorescence cross-talk, bleed-through and photobleaching are required. A constant ratio of the donor to acceptor concentrations is important for reliable ratiometric FRET imaging. In an intramolecular FRET biosensor, the sensing domain is sandwiched by the donor and acceptor fluorophores. Thus, the expression level of donor to acceptor fluorophores is perfectly 1:1. One good example is the cGMP biosensor (see Fig. 2B) (Russwurm et al., 2007).
Acceptor photobleaching
This method measures increased donor emission when the acceptor is photobleached. Acceptor photobleaching is based on destroying acceptor fluorophores, which naturally abolishes FRET. After acceptor photobleaching, the donor intensity increases, since no FRET energy is transferred to the acceptor. This method is therefore straightforward, but destructive (single-time measurement) and thus not suitable for capturing molecular dynamics.
FLIM-FRET
The fluorescence lifetime (τ) is defined as the average time that a molecule remains in an excited state prior to emitting a photon and returning to the ground state. The presence of acceptor molecules within the local environment of the donor that permits energy transfer will influence the fluorescence lifetime of the donor. By measuring the donor lifetime in the presence and the absence of acceptor one can estimate the distance between the donor- and acceptor-labeled proteins.
FRET has also been used to investigate integrin activation. A spatial separation of the integrin αL and β2-subunit cytoplasmic tail has been detected during activation of K562 myeloid cells transfected with integrin αL–CFP and integrin β2–YFP (Kim et al., 2003). Here, acceptor photobleaching FRET was performed to assess the conformational changes that are reflected by the distance between the cytoplasmic tails of the two integrin subunits. In the resting state, these domains were found close to each other. A loss of FRET upon integrin activation was interpreted as spatial separation between integrin αL and β2 (Kim et al., 2003).
The formation of the immune synapse during antigen presentation is another area that has been intensely investigated using FRET methods. The adaptive immune response is initiated when the TCR is engaged by antigenic peptides bound by major histocompatibility complex (pMHC) proteins that are expressed on the surface of antigen-presenting cells (APCs). The interface between an APC and a lymphocyte is called immunological synapse (or immune synapse). The TCR–pMHC complex is surrounded by co-stimulatory and adhesion molecules on the membranes of both cells (Dustin and Choudhuri, 2016). Using flow cytometry-based FRET assays with fluorescein isothiocyanate (FITC) and tetramethylrhodamine (TRITC) as FRET pairs and conjugated monoclonal antibodies against the surface molecules [i.e. cluster of differentiation 3 (CD3), CD4 and CD45], FRET was detected between the CD3–TCR co-receptor complex and CD4, but not between CD3–TCR and CD45 when CD4+ T cells were activated using anti-CD3 monoclonal antibodies (mAbs) (Mittler et al., 1989). These experiments demonstrated that CD4 molecules on the surface of a murine T-cell hybridoma rapidly associate with the murine TCR–CD3 complex after crosslinking of the TCR complex with anti-CD3 mAbs. By using flow cytometric analysis to measure FRET between fluorochrome-labeled CD4 and CD3 molecules, it was determined that the association of the two receptors requires CD3 crosslinking and is dependent on the expression of the cytoplasmic domain of the CD4 molecule (Collins et al., 1992). It was later shown that the movement of CD3 to CD4 further depends on the tyrosine kinase Lck and the cytoplasmic domain of CD4 (Collins et al., 1992; Philipsen et al., 2017; Stirnweiss et al., 2013). These data are consistent with the idea that CD4 plays an active role in CD3-mediated signal transduction in T cells, whereas CD45 has an inhibitory role in TCR signaling (Dustin and Choudhuri, 2016).
Both acceptor photobleaching and ratiometric FRET were applied to investigate the association between CD3ζ and CD4 in the context of agonist or antagonist binding (Zal et al., 2002). Unlike photobleaching FRET microscopy, the ratiometric approach is compatible with 3D and time-lapse experiments. In this study, CFP (donor) was fused with CD3ζ, and YFP (acceptor) was fused to CD4. The authors detected an increase of FRET efficiency between CD3ζ–CFP and CD4–YFP in response to the agonist peptide presentation by MHC II. Thus, recognition of agonist pMHC complexes triggers an intermolecular interaction between CD4 and TCR, which is detectable across the T-hybridoma–APC contact area (Zal et al., 2002).
FLIM-FRET has been used to measure TCR clustering (Ma et al., 2017); to that end, the authors first developed membrane-anchored, single-molecule FRET pairs that underwent intermolecular FRET at high molecular density. This cluster sensor (named CliF) utilized the sensitivity of FRET to detect membrane protein clusters irrespective of the presence of non-clustered molecules. FLIM-FRET measured the increased clustering of TCR–CD3ζ upon T cell activation, which was correlated with cluster density (Ma et al., 2017).
One primary application of FRET is to use intramolecular biosensors to monitor activities of signaling molecules in cells in vivo. An emerging trend is to do this using intravital imaging; this has been facilitated by recent advances in the development of improved FRET biosensors with high FRET efficiency, good specificity and excellent response kinetics (Bajar et al., 2016; Greenwald et al., 2018; Lam et al., 2012). Mice have been engineered to express FRET-based biosensors, and intravital FRET imaging has been applied to study signaling molecules, such as cGMP (Thunemann et al., 2013; Wen et al., 2018), cAMP (Sprenger et al., 2015), Ca2+ (Yang et al., 2018), protein kinase A (PKA) and extracellular signal-regulated kinase 1/2 (ERK1/2) activity (Mizuno et al., 2014).
Confocal and multiphoton microscopy for intravital imaging
Studying immune cells in their native environment is the key benefit of intravital imaging. Intravital imaging is a very old technique, going back to the invention of the microscope (van Leeuwenhoek, 1719), but major advances included the introduction of fluorescence microscopy around 1980 (Sanderson et al., 2014), confocal microscopy (Minsky, 1988) and multiphoton microscopy (Larson, 2011). These methods are now well established and are commercially available from many manufacturers, and will not be covered here. Limitations include finite tissue transparency, light absorption and scattering, and the trauma associated with surgical access (McArdle et al., 2016). The use of intravital imaging to monitor cellular behavior has already been reviewed extensively (Germain et al., 2012; McArdle et al., 2016; Pittet et al., 2018). We will focus in this section on the advances towards understanding the molecular signaling by using biosensors.
One type of biosensing system based on single-color fluorescent proteins has been developed to detect molecular signaling events, such as Ca2+ mobilization (Zariwala et al., 2012). The GFP-based Ca2+ indicator GCaMP6 has been used to monitor T cell activation in vivo (Moseman et al., 2016). The single fluorescence-based method is easy to use, can generate very good signals and has good time resolution. However, it is sensitive to tissue movement and focal plane changes; thus, imaging must be performed on a relatively static object. In comparison, two-color ratiometric FRET imaging has many advantages, because the artifacts caused by tissue movement can be removed. For example, FRET-based Ca2+ biosensors have allowed high resolution functional tracking of T lymphocytes (Thestrup et al., 2014).
Two-photon FRET microscopy has been used to visualize activity changes of protein kinases in neutrophils during neutrophil recruitment to the inflamed intestine of transgenic mice expressing FRET biosensors that contained CFP and YFP (Mizuno et al., 2014). Ratiometric FRET imaging showed that ERK1/2 activity increased during neutrophil recruitment to the inflamed intestine in vivo, while high PKA activity suppressed neutrophil migration (Mizuno et al., 2014).
However, even in mechanically stable preparations, the tissue through which both excitation and emission signals must travel can cause problems because of absorption, scattering and autofluorescence. Because transmission of light with longer wavelengths (like YFP) in biological tissues is higher than that of light with shorter wavelength (like CFP), the apparent FRET efficiency calculated by the conventional ratiometric method becomes higher when the emitted light originates from deeper tissues. In addition, tissue autofluorescence undermines the net FRET efficiency (Sebestyén et al., 2002). Both problems can be solved by assessing FRET with FLIM. Because the lifetime of a fluorescence protein does not rely on the fluorescence intensity, the measurement is not affected by the transmission or light scattering in non-uniform environments. The detection of lifetime is accurate as long as the emitted light remains detectable at the PMT.
The FLIM system is compatible with multiphoton microscopy (Becker, 2012). In biosensor mice for the small GTPase RhoA (RhoA-FRET), FLIM-FRET imaging has been performed in highly motile cells, such as neutrophils. RhoA activity was monitored in swarming neutrophils in vivo in a laser ablation-induced tissue damage model in the mouse ear (Nobis et al., 2017). For each neutrophil, the RhoA activity was mapped from the front to the rear of the cell, relative to the direction of motion, indicating that RhoA is activated during neutrophil migration. Similar FRET-based biosensor mice have also been used for the evaluation of drug targeting efficacy in vivo (Nobis et al., 2017, 2018, 2013). The downside of FLIM-FRET is the lack of speed (more than 20 s per section) (Becker, 2012).
Light-sheet microscopy
LSM (Huisken et al., 2004; Mickoleit et al., 2014; Scherz et al., 2008; Siedentopf and Zsigmondy, 1902) offers relief from the light toxicity, low tissue penetration and close working distance of conventional confocal microscopy. LSM became practical with the advent of fast charge-coupled device (CCD) cameras (Huisken et al., 2004). While LSM is widely used for cleared tissues (Dodt et al., 2007), it is also suitable for live-cell imaging in transparent organisms and tissues, such as zebrafish, embryos or the mouse eye. Unlike other types of microscopy, the excitation is usually generated by a continuous wave laser and a cylindrical lens (Buytaert et al., 2012; Buytaert and Dirckx, 2009; Santi et al., 2009), and the excitation and emission pathways in LSM are orthogonal to each other. The resolution of LSM is diffraction limited, but acquisition is extremely fast (tens of milliseconds), thus limiting light toxicity and allowing high frame rates. LSM evolved to digitally scanned LSM with the use of a Gaussian laser beam (Keller et al., 2008), which enabled using two-photon excitation that is suitable for imaging deeper tissues (Truong et al., 2011). Four-lens systems have been developed to improve registration (Krzic et al., 2012). In addition, structured illumination reduced out-of-focus light (Huisken and Stainier, 2007).
A typical application of LSM in live-cell imaging is tracking genetically fluorescent cells in the zebrafish heart (Ding et al., 2017; Lee et al., 2016; Packard et al., 2017). Cell tracking is useful for studying development, inflammation and immunity. LSM has demonstrated the dynamics of cardiac valves during the cardiac cycle (Lee et al., 2013). Using a rainbow fluorescent protein system, the source of new cardiomyocytes was identified in mice at birth (Sereti et al., 2018). LSM is inherently 3D, allowing the imaging of an entire mouse heart in 60 s. Another study has demonstrated vascular calcification in the Apoe−/− mouse model of atherosclerosis (Hsu et al., 2018). Lungs and eyes have also been imaged by LSM. For example, LSM was employed to characterize fungal growth and the local immune response at cellular resolution in the context of whole lungs (Amich et al., 2020). LSM has not been used in live mice except in the eye, which is naturally clear (Prahst et al., 2019 preprint). More recently, LSM has been utilized for live-cell imaging of signaling events in cells transfected with a FRET biosensor for activity of the small GTPase Rap1; this allowed for a visualization of the rapid 3D dynamics of Rap1 in vesicles and cell protrusions (O'Shaughnessy et al., 2019).
Recent developments in LSM include adaptive optics to compensate for distortions (Liu et al., 2018a; Masson et al., 2015; Wilding et al., 2016), and the use of two-photon Bessel beams for single-cell imaging by LSM (Planchon et al., 2011). A very recent development is the advent of single-objective LSM (Millett-Sikking et al.; https://github.com/AndrewGYork/high_na_single_objective_lightsheet/tree/0.2.1). Unlike the conventional LSM, the single-objective LSM provides compatibility with standard confocal microscopy, better sample accessibility, reduced phototoxicity and improved volumetric imaging performance.
Super-resolution microscopy
Fluorescence microscopy is a powerful tool for imaging immune cells. However, its spatial resolution is limited by the physics of diffraction. This limitation is caused by the optical spot size in the lateral dimensions (in xy, d=λ/2NA), which is determined by the wavelength of the light being observed (λ) and the numerical aperture (NA) of the objective. In conventional fluorescence microscopy, the best lateral resolution is ∼200–300 nm. The extent of the focal spot is even larger (≥500 nm, and often more) in the axial direction (in z).
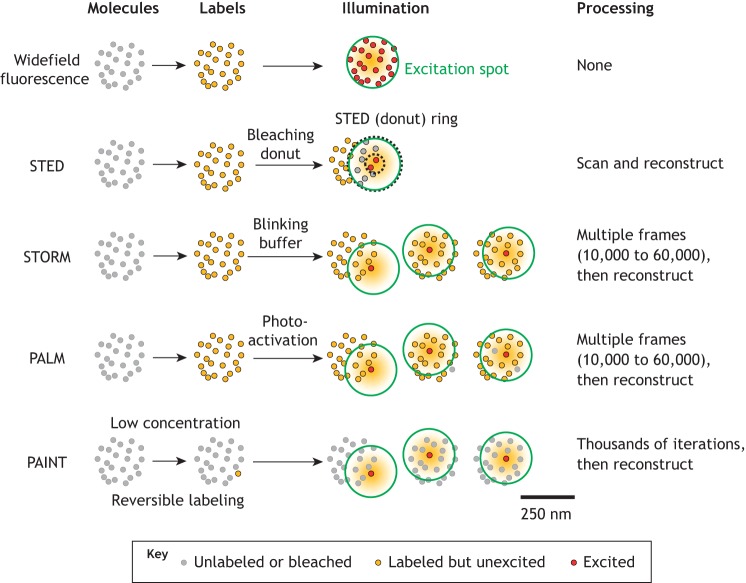
The diffraction limit was overcome by the invention of multiple microscopic techniques (Fig. 3), such as stimulated emission depletion (STED) (Hell and Wichmann, 1994; Klar and Hell, 1999; Klar et al., 2000), structured illumination microscopy (SIM) (Gustafsson, 2000; Heintzmann and Cremer, 1999), single-molecule localization microscopy (SMLM), which includes stochastic optical reconstruction microscopy (STORM) (Rust et al., 2006), photo-activated localization microscopy (PALM) (Betzig, 1995; Betzig et al., 2006; Dickson et al., 1997; Hess et al., 2006), points accumulation for imaging in nanoscale topography (PAINT) (Sharonov and Hochstrasser, 2006), and expansion microscopy (ExM) (Chen et al., 2015). In the past few years, STED, SIM, STORM and PALM super-resolution microscopes have all become commercially available and thus have been extensively used to investigate the distribution and function of different molecules found on a variety of leukocytes.
Fig. 3.
Super-resolution methods. Widefield fluorescence (top) excites all labeled molecules (yellow) within the diffraction-limited illuminated area (excitation spot, green circle). Thus, signal emanates from all labeled molecules (red) in the excited area (250 nm diameter). STED, although the excitation spot (green) is still diffraction-limited, most fluorochromes are reversibly bleached by the donut laser (ring between dotted black circles) so that only unbleached fluorochromes in the donut hole contribute to the image. This procedure is repeated for each location. STORM, the fluorochrome molecule is induced to blink using a special blinking buffer. Each field is recorded 10,000 to 60,000 times, enough that only one fluorochrome molecule within the illuminated area is ‘on’ in each frame. PALM, each fluorochrome molecule is activated by a UV laser (usually 405 nm). Activation is stochastic and sparse so that, on average, only one fluorochrome molecule within the illuminated area is ‘on’ in each frame. PAINT, a very small fraction of molecules is labeled reversibly (yellow) so that on average only one molecule per frame is ‘on’. This is repeated thousands of times.
Most studies published to-date apply super-resolution microscopy to fixed samples, but here, we focus on super-resolution imaging of live-cell samples (Cox, 2015; Godin et al., 2014). The major challenge in super-resolution live-cell imaging are speed, buffer and light toxicity. In super-resolution imaging (∼50 nm resolution), cells, cell processes or tracked molecules that move faster than 50 nm per acquisition frame will not be imaged accurately. Thus, faster super-resolution imaging techniques, such as SIM (at least 9.3 Hz) (Turcotte et al., 2019), Airyscan (30 Hz) (Huff, 2016) and STED (12.5 Hz for 2.9×2.9 μm) (Yang et al., 2014), are more suitable for live-cell imaging. SIM is the most commonly used super-resolution imaging technique for live-cell imaging (Chen et al., 2018; Fiolka et al., 2012; Turcotte et al., 2019). SIM has a good imaging speed, can use any labels used in conventional live-cell imaging, and has tolerable light toxicity, similar to conventional live-cell imaging. Airyscan is also widely used for live-cell imaging (Kolossov et al., 2018; Mishra et al., 2019; Scipioni et al., 2018) and here has similar properties compared to SIM. The limitation of both SIM and Airyscan is that the improvement in resolution compared to confocal imaging is small (less than a factor of two). Thus, SIM and Airyscan are at the lower end of super-resolution imaging. STED is attractive for live-cell imaging, because it has much better resolution (70 to 90 nm) compared to SIM and Airyscan. Accordingly, STED has been used in multiple live-cell imaging studies (Bottanelli and Schroeder, 2018; Heine et al., 2018; Kilian et al., 2018; Mishina and Belousov, 2017). However, the major limitation of STED live-cell imaging is phototoxicity. To some extent, this can be improved by adjusting the imaging conditions (Kilian et al., 2018). SMLM has the highest resolution and has been used in studies of live-cell imaging, for instance STORM (Jones et al., 2011; Shim et al., 2012), PALM (Deschout et al., 2016; Shroff et al., 2008), PAINT (Giannone et al., 2013). However, at least 1000 frames of raw data are required for the reconstruction of a single frame of the image. Thus, the temporal resolution is limited as one fluorescent molecule may generate multiple different localizations within one image. Live-cell STORM imaging is also limited by probes that can ‘blink’ in physiological media, because STORM buffer is toxic for live cells. A few dyes such as DiI, Mito-Tracker, ER-Tracker and Lyso-Tracker have been used in live-cell STORM (Shim et al., 2012). PALM is commonly used for single-molecule tracking in live cells (Katz et al., 2017; Katz et al., 2019), which can show the tracks of the sparse photo-activated molecules, but not all labeled molecules. Often, the surrounding structures are acquired using diffraction-limited microscopy (Katz et al., 2017; Katz et al., 2019). Phototoxicity is a problem for live-cell STORM and PALM imaging, as STORM needs a high-power laser, while PALM uses a 405 nm laser to trigger the photoactivable fluorescent proteins.
Overall, super-resolution microscopy techniques are able to break the diffraction limit to improve the resolution by a factor of two to ten. By using super-resolution imaging, investigators can thus visualize small structures of the cell organelles and cytoskeleton in more detail. Combining STORM with molecular modeling has achieved single-molecule resolution for integrins, including the distribution of molecule nanoclusters, co-localizations and spatial distribution patterns (Fan et al., 2019).
Immune cells imaged by super-resolution microscopy
Super-resolution microscopy has been used in studying the functions and molecular distributions of different immune cells, and has provided molecular details of many biological processes, including neutrophil recruitment (Fan et al., 2016; Kolaczkowska and Kubes, 2013; Ley et al., 2018; Margraf et al., 2019; Mócsai et al., 2015), exocytosis (Cassatella et al., 2019; Johnson et al., 2016b; Ley et al., 2018; Mollinedo, 2019), phagocytosis (Colucci-Guyon et al., 2011; Davies et al., 2019; Kambara et al., 2018; Manfredi et al., 2018; Matlung et al., 2018; Nordenfelt and Tapper, 2011) and NETosis, which involves the release of neutrophil extracellular traps (NETs) (Ali et al., 2019; Clark et al., 2007; Hsu et al., 2019; Manfredi et al., 2018; Neubert et al., 2018; Perdomo et al., 2019; Sollberger et al., 2018).
A recent study used STORM to investigate the spatial distribution and reorganization of CD44 (a selectin ligand) expressed on KG1a cells, an acute myelogenous leukemia cell line similar to neutrophil/monocyte progenitors, during their rolling under shear stresses of 1 dyn cm−2 (AbuZineh et al., 2018). Their results suggest that the spatial reorganization of CD44 and the actin cytoskeleton is the result of the concerted effect of E-selectin–ligand interactions, external shear stress and spatial clustering of the selectin ligands (AbuZineh et al., 2018).
The transition of rolling to arrest is an important step for immune cell recruitment. In neutrophils, β2 integrins undergo two major conformational changes during activation (Fan and Ley, 2015): the resting state has a bent extracellular domain and low-affinity headpiece (denoted E−H−). In the canonical switchblade model (Lefort and Ley, 2012; Luo et al., 2007; Moore et al., 2018; Nordenfelt et al., 2016, 2017) of integrin activation, an E−H− integrin will extend first (denoted E+H−), and then acquire the high-affinity conformation (denoted E+H+). Alternative models, such as the deadbolt model have been suggested. In this model, the β-tail domain CD loop regulates the H+ regardless of E+ (Arnaout et al., 2007; Gupta et al., 2007). By using qDF microscopy with conformation-reporting antibodies, we observed an integrin conformation with a bent extracellular domain and a high-affinity headpiece (E−H+) and its transition to the E+H+ conformation on primary live neutrophils (Fan et al., 2016). Furthermore, by combining molecular modeling-based spatial simulation and STORM, we recently developed a technique called SuperSTORM, which allowed us to visualize integrin activation at the single-molecule level and to identify the number, distribution and arrangement (clustering) of three active conformations (E+H−, E−H+ and E+H+) of β2 integrins (Fan et al., 2019).
Another study addressed the role of the tetraspanin CD37 in neutrophil arrest and showed that CD37 regulates β2 integrin-mediated adhesion and migration in neutrophils (Wee et al., 2015). STORM imaging revealed that CD37 and the β2 integrin do not co-cluster significantly in human neutrophils. Combined with their observation of actin polymerization and Rac-1 activation deficiencies in CD37-knockout cells, the authors conclude that CD37 promotes neutrophil adhesion and recruitment by facilitating cytoskeletal functions downstream of integrin-mediated adhesion (Wee et al., 2015).
Real-time SIM imaging revealed the release of exosomes (CD63–GFP) as vesicular trails from cells during the initial stages of migration (Szatmary et al., 2017), which may release leukotriene B4 to boost the recruitment of other neutrophils. SIM has also been used to image the actin cytoskeleton of HL-60 cells (Gustafsson et al., 2008; Millius and Weiner, 2009) and neutrophils (Tanaka et al., 2017), as well as to determine the distribution of ADP-ribosylation factor 1 (ARF1) and Ras-related C3 botulinum toxin substrate 1 (Rac1) during HL-60 migration (Mazaki et al., 2017), the distribution of Rac1 and RhoA during neutrophil migration (Tanaka et al., 2017), and interactions between neutrophils and platelets (Bennewitz et al., 2017).
Super-resolution STORM imaging has been used to study neutrophil endosome maturation (He et al., 2016), exocytosis (Johnson et al., 2016a) and migration (Ramadass et al., 2019). Munc13-4 associated with syntaxin 7, indicating a role of Munc13-4 in regulating late endosomal maturation, endosomal signaling and Toll-like receptor 9 (TLR9)-initiated cellular responses (He et al., 2016). STORM imaging also showed that Munc13-4 is associated with the small GTPase Rab11; thus, Munc13-4 can regulate the trafficking and docking of Rab11-positive vesicles at the cell membrane, which is critical for the killing of bacteria by neutrophils (Johnson et al., 2016a). The trafficking protein synaptotagmin-like 1 (SYTL1, also known as JCF1) appears to regulate the localization of Rac1-GFP during neutrophil chemotaxis, which is important for neutrophil recruitment during inflammation or infections (Ramadass et al., 2019).
During NETosis, neutrophils can release three-dimensional meshworks (NETs) of chromatin (Brinkmann et al., 2004). Super-resolution STED microscopy has recently been used to image chromatin during the late stage of neutrophil NETosis (P2, before rupture) (Neubert et al., 2018). However, the authors observed no differences in NET structures between STED imaging and confocal imaging, from which they concluded that the chromatin mesh size is below the resolution of STED (i.e. 120 nm as claimed by the authors) (Neubert et al., 2018). In another study, SIM imaging was used to show that NETs can trap human immunodeficiency virus-1 (HIV-1) (Saitoh et al., 2012). This prevents HIV-1 from spreading and ensures high local concentrations of antiviral factors to eliminate HIV-1 (Saitoh et al., 2012). SIM imaging was also used to show the presence of myeloperoxidase (MPO), citrullinated histone H3 (H3Cit), protein arginine deiminases 4 (PAD4) and histone H2A–H2B in NET-forming neutrophils in samples from patients with anti-neutrophil cytoplasmic antibody (ANCA)-associated glomerulonephritis, which is an unusual autoimmune disease caused by the release of MPO by neutrophils as an autoantigen in glomeruli (O'Sullivan et al., 2015). Populations of glycolipids change markedly during leukocyte differentiation, suggesting that these molecules are involved in biological functions. STED analysis showed that the lipid-raft marker phosphatidylglucoside and the glycosphingolipid lactosylceramide form different domains on plasma membranes of neutrophils (Ekyalongo et al., 2015), with phosphatidylglucoside preferentially expressed along the neutrophil differentiation pathway.
Different super-resolution techniques (SIM, STED and SMLM), have been used to image and compare microtubules in Drosophila macrophages (Wegel et al., 2016). All three techniques resolved well-separated microtubules, but not microtubule bundles (Wegel et al., 2016). Using super-resolution microscopy, the formation of podosome-like structures during Fc receptor-mediated phagocytosis could be observed (Ostrowski et al., 2019). Unlike conventional podosomes, these structures are short-lived and vectorial, expanding radially from the sites where phagocytic targets are initially engaged. Time-lapse SIM of phagocytosis of cultured microglia cells revealed different phases of macrophage phagocytic behavior, including initial active internalization, final digestion and autophagy (Fumagalli et al., 2019). Phagocytosis of Porphyromonas gingivalis by RAW 264.7 macrophages has also been assessed by SIM (Lam et al., 2016; Lenzo et al., 2016), showing that M1 and M2 macrophages have higher phagocytic capacity than naïve macrophages. Imaging of F-actin in wild-type (WT) and in myosin IXb (Myo9b)-knockout macrophages by SIM showed the loss of actin polarization in Myo9b-knockout cells and demonstrated the importance of Myo9b in regulating actin distribution and cell migration (Hanley et al., 2010). Another study also used SIM to image F-actin in WT and Cdc42-knockout macrophages, and showed that the defect in cell spreading is due to Cdc42 deficiency (Horsthemke et al., 2017). SIM was also used to evaluate the expression and distribution of ionized Ca2+-binding adaptor molecule 1 in macrophages in the cochlea, suggesting that the human auditory nerve is under the surveillance and possible neurotrophic stimulation of a well-developed resident macrophage system (Liu et al., 2018b). SIM also revealed that tunneling nanotubes (TNTs), membranous channels that connect cells, allow for the transfer of signals, vesicles and organelles (Hanna et al., 2017). Extracellular vesicles derived from oligodeoxynucleotide-stimulated macrophages can transfer and activate Cdc42 in the recipient cells and enhance immune responses (Zhang et al., 2019). SMLM imaging showed the distribution and clustering of Toll-like receptor 4 (TLR4) molecules on macrophages (Aaron et al., 2012; Kruger et al., 2017; Neumann et al., 2019). Lipopolysaccharide (LPS) stimulation increased the size of TLR4 clusters (Aaron et al., 2012). STORM imaging revealed nanoclusters of signal regulatory protein α (SIRPα), Fc gamma receptor I (FcγRI) and Fc gamma receptor II (FcγRII) on human macrophages (Lopes et al., 2017), and β2 integrins on human neutrophils (Fan et al., 2019). Nanoclusters of FcγRI, but not FcγRII, are constitutively associated with nanoclusters of SIRPα. Upon Fc receptor activation, Src family kinase signaling leads to segregation of FcγRI and SIRPα nanoclusters. The formation of cholesterol crystals in J774A.1 macrophage-like cells has also been followed by STORM imaging (Varsano et al., 2018). With regard to viral infection, PALM and STORM imaging have been used to visualize the process from cytoplasmic entry to HIV-1 to the integration of the reverse transcribed genome in macrophages (Peng et al., 2014). STED microscopy was used to identify cholesterol microdomains on the macrophage membrane, which can shed excess cholesterol (Jin et al., 2018). STED also showed the transfer of HIV-1 from macrophages to astrocytes (Russell et al., 2017). A STORM study showed that the number of histone H2A clusters in the nucleus is decreased in macrophages upon LPS stimulation (Nair et al., 2018), suggesting that histones may be released into the circulation by macrophages and promote distal inflammatory responses.
Of all immune cells, the imaging of lymphocytes is the most challenging, because (resting) lymphocytes are much smaller than other immune cells. Super-resolution microscopy has provided a superior means to investigate the function and regulation of lymphocytes, especially T lymphocytes. Using PALM imaging, the spatio-temporal dynamics of TCR complexes and linker for activation of T cells (Lat), a key adaptor molecule in the TCR signaling pathway, was determined in T cell membranes (Lillemeier et al., 2010). In quiescent T cells, both molecules existed in separate membrane domains (so-called protein islands) and these domains concatenated after T cell activation (Lillemeier et al., 2010). Their follow-up study provided evidence that TCR and CD4 molecules are clustered separately in resting T cells (Roh et al., 2015). Upon activation, TCR and CD4 begin to cluster together and develop into microclusters that then undergo a larger-scale redistribution to form supramolecular activation clusters (Roh et al., 2015). Another study also directly observed the organization of TCRs and their rearrangement on lymph node-resident T cells during an immune response by using STORM and SIM (Hu et al., 2016). They found that TCRs were preclustered into nanometer-scale protein islands, which were organized into larger membrane territories, and that T-cell activation induced the formation of microclusters (Hu et al., 2016). Using PALM-based single-molecule tracking, a cycle of recruitment, activation and release for Zeta-chain-associated protein kinase 70 (ZAP70) kinases was discovered at phosphorylated TCRs (Katz et al., 2017). Another study used STORM imaging to address the recruitment and phosphorylation of TCRζ, ZAP70, Lat and SLP76 (SH2-domain-containing leukocyte protein of 76 kDa), a key orchestrator of TCR signal transduction, in Jurkat T cells that were spread on CD3 antibody-coated substrate (Soares et al., 2013). A recent study challenged the view that TCRs are preclustered by using label-density-variation SMLM and STED (Rossboth et al., 2018); their findings suggest that TCRs are instead randomly distributed on the plasma membrane of resting antigen-experienced T cells (Rossboth et al., 2018). However, this interpretation is not fully supported by the images, which showed clustered TCRs in resting T cells and clustering disappeared after cells had spread. In non-spreading T cells, STORM combined with variable-angle total internal reflection microscopy showed that their TCRs are highly concentrated on microvilli (Jung et al., 2016). The authors interpreted this to mean that the immunological synapse may start to form at microvilli.
Similar to what was found in neutrophils as described above (Fan et al., 2019), integrin molecules are also important for the migration of T lymphocytes. Interferometric PALM and a constrained photoactivatable fluorescent protein fused to αLβ2 integrin was used to measure the displacement of the head of integrin LFA-1, as a result of its conformational change that is induced on the surface of migrating Jurkat T cells (Moore et al., 2018).
Recording cytoskeletal actin dynamics during the activation of resting T cells by STED microscopy (Fritzsche et al., 2017) showed that, following contact formation with activating surfaces, these cells sequentially rearrange their cortical actin across the entire cell, creating a previously unreported ramifying actin network above the immunological synapse. This network shows all the characteristics of an inward-growing transportation network and its dynamics correlate with T cell receptor rearrangements. SIM imaging has revealed the cortical actin and tubulin networks in cytotoxic T lymphocytes (CTLs) (Ritter et al., 2015). A super-resolution optical fluctuation imaging technique has been used to assess the CD4 clusters on the plasma membrane of Jurkat T cells (Lukeš et al., 2017). STORM imaging was also used to follow the infection of HIV to T cells (Roy et al., 2013). Finally, some super-resolution microscopy studies have focused on B lymphocytes. STORM identified B cell antigen receptor (BCR) microclusters on B cells (Maity et al., 2015). SIM and STORM revealed the distribution of the cytoskeletal protein vimentin in B cells (Tsui et al., 2018).
Concluding remarks
Imaging the immune system at the cell level has provided abundant information of how immune cells traffic and interact. Here, we highlight three key developments. With the advent of advanced FRET probes, it has become possible to image signaling events in live cells in real time. Deeper tissue penetration, 3D reconstruction and lower light toxicity has been achieved by light-sheet microscopy. Finally, imaging immune responses at the molecular level in subcellular domains and in real time is now on the horizon with the advent of various super-resolution approaches that are compatible with live-cell imaging. Further improvements in live-cell imaging by super-resolution microscopy will require more sensitive and faster detectors to reduce the light toxicity and increase acquisition speed.
Acknowledgements
We apologize to the authors whose work could not be cited because of space constraints.
Footnotes
Competing interests
The authors declare no competing or financial interests.
Funding
Our work in this area was supported by grants from the National Institutes of Health, USA (HL078784 to K.L., R01HL145454 to Z.F.), a Postdoctoral Fellowship (19POST34450228 to L.W.) and a Career Development Award (18CDA34110426 to Z.F.) from the American Heart Association, USA. Deposited in PMC for release after 12 months.
References
- Aaron J. S., Carson B. D. and Timlin J. A. (2012). Characterization of differential Toll-like receptor responses below the optical diffraction limit. Small 8, 3041-3049. 10.1002/smll.201200106 [DOI] [PMC free article] [PubMed] [Google Scholar]
- AbuZineh K., Joudeh L. I., Al Alwan B., Hamdan S. M., Merzaban J. S. and Habuchi S. (2018). Microfluidics-based super-resolution microscopy enables nanoscopic characterization of blood stem cell rolling. Sci. Adv. 4, eaat5304 10.1126/sciadv.aat5304 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ali R. A., Gandhi A. A., Meng H., Yalavarthi S., Vreede A. P., Estes S. K., Palmer O. R., Bockenstedt P. L., Pinsky D. J., Greve J. M. et al. (2019). Adenosine receptor agonism protects against NETosis and thrombosis in antiphospholipid syndrome. Nat. Commun. 10, 1916 10.1038/s41467-019-09801-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Amich J., Mokhtari Z., Strobel M., Vialetto E., Sheta D., Yu Y., Hartweg J., Kalleda N., Jarick K. J., Brede C., et al. (2020). Three-dimensional light sheet fluorescence microscopy of lungs to dissect local host immune-Aspergillus fumigatus interactions. mBio, 11 10.1128/mBio.02752-19 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Arnaout M. A., Goodman S. L. and Xiong J. P. (2007). Structure and mechanics of integrin-based cell adhesion. Curr. Opin. Cell Biol. 19, 495-507. 10.1016/j.ceb.2007.08.002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Axelrod D. (1981). Cell-substrate contacts illuminated by total internal reflection fluorescence. J. Cell Biol. 89, 141-145. 10.1083/jcb.89.1.141 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Bajar B. T., Wang E. S., Zhang S., Lin M. Z. and Chu J. (2016). A guide to fluorescent protein FRET pairs. Sensors (Basel) 16, 1488 10.3390/s16091488 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Becker W. (2012). Fluorescence lifetime imaging--techniques and applications. J. Microsc. 247, 119-136. 10.1111/j.1365-2818.2012.03618.x [DOI] [PubMed] [Google Scholar]
- Bennewitz M. F., Jimenez M. A., Vats R., Tutuncuoglu E., Jonassaint J., Kato G. J., Gladwin M. T. and Sundd P. (2017). Lung vaso-occlusion in sickle cell disease mediated by arteriolar neutrophil-platelet microemboli. JCI Insight 2, e89761 10.1172/jci.insight.89761 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Betzig E. (1995). Proposed method for molecular optical imaging. Opt. Lett. 20, 237-239. 10.1364/OL.20.000237 [DOI] [PubMed] [Google Scholar]
- Betzig E., Patterson G. H., Sougrat R., Lindwasser O. W., Olenych S., Bonifacino J. S., Davidson M. W., Lippincott-Schwartz J. and Hess H. F. (2006). Imaging intracellular fluorescent proteins at nanometer resolution. Science 313, 1642-1645. 10.1126/science.1127344 [DOI] [PubMed] [Google Scholar]
- Bottanelli F. and Schroeder L. (2018). Stimulated emission depletion (STED) imaging of clathrin-mediated endocytosis in living cells. Methods Mol. Biol. 1847, 189-195. 10.1007/978-1-4939-8719-1_14 [DOI] [PubMed] [Google Scholar]
- Brinkmann V., Reichard U., Goosmann C., Fauler B., Uhlemann Y., Weiss D. S., Weinrauch Y. and Zychlinsky A. (2004). Neutrophil extracellular traps kill bacteria. Science 303, 1532-1535. 10.1126/science.1092385 [DOI] [PubMed] [Google Scholar]
- Buytaert J. A. N. and Dirckx J. J. J. (2009). Tomographic imaging of macroscopic biomedical objects in high resolution and three dimensions using orthogonal-plane fluorescence optical sectioning. Appl. Opt. 48, 941-948. 10.1364/AO.48.000941 [DOI] [PubMed] [Google Scholar]
- Buytaert J. A., Descamps E., Adriaens D. and Dirckx J. J. (2012). The OPFOS microscopy family: high-resolution optical sectioning of biomedical specimens. Anat. Res. Int. 2012, 206238 10.1155/2012/206238 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cassatella M. A., Östberg N. K., Tamassia N. and Soehnlein O. (2019). Biological roles of neutrophil-derived granule proteins and cytokines. Trends Immunol. 40, 648-664. 10.1016/j.it.2019.05.003 [DOI] [PubMed] [Google Scholar]
- Chen F., Tillberg P. W. and Boyden E. S. (2015). Optical imaging. Expansion microscopy. Science 347, 543-548. 10.1126/science.1260088 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chen Y., Liu W., Zhang Z., Zheng C., Huang Y., Cao R., Zhu D., Xu L., Zhang M., Zhang Y.-H. et al. (2018). Multi-color live-cell super-resolution volume imaging with multi-angle interference microscopy. Nat. Commun. 9, 4818 10.1038/s41467-018-07244-4 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Chigaev A. and Sklar L. A. (2011). Overview: assays for studying integrin-dependent cell adhesion. Methods Mol. Biol. 757, 3-14. 10.1007/978-1-61779-166-6_1 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Clark S. R., Ma A. C., Tavener S. A., McDonald B., Goodarzi Z., Kelly M. M., Patel K. D., Chakrabarti S., McAvoy E., Sinclair G. D. et al. (2007). Platelet TLR4 activates neutrophil extracellular traps to ensnare bacteria in septic blood. Nat. Med. 13, 463-469. 10.1038/nm1565 [DOI] [PubMed] [Google Scholar]
- Collins T. L., Uniyal S., Shin J., Strominger J. L., Mittler R. S. and Burakoff S. J. (1992). p56lck association with CD4 is required for the interaction between CD4 and the TCR/CD3 complex and for optimal antigen stimulation. J. Immunol. 148, 2159-2162. [PubMed] [Google Scholar]
- Colucci-Guyon E., Tinevez J. Y., Renshaw S. A. and Herbomel P. (2011). Strategies of professional phagocytes in vivo: unlike macrophages, neutrophils engulf only surface-associated microbes. J. Cell Sci. 124, 3053-3059. 10.1242/jcs.082792 [DOI] [PubMed] [Google Scholar]
- Cox S. (2015). Super-resolution imaging in live cells. Dev. Biol. 401, 175-181. 10.1016/j.ydbio.2014.11.025 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Davies L. C., Rice C. M., McVicar D. W. and Weiss J. M. (2019). Diversity and environmental adaptation of phagocytic cell metabolism. J. Leukoc. Biol. 105, 37-48. 10.1002/JLB.4RI0518-195R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Deschout H., Lukeš T., Sharipov A., Szlag D., Feletti L., Vandenberg W., Dedecker P., Hofkens J., Leutenegger M., Lasser T. et al. (2016). Complementarity of PALM and SOFI for super-resolution live-cell imaging of focal adhesions. Nat. Commun. 7, 13693 10.1038/ncomms13693 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dickson R. M., Cubitt A. B., Tsien R. Y. and Moerner W. E. (1997). On/off blinking and switching behaviour of single molecules of green fluorescent protein. Nature 388, 355-358. 10.1038/41048 [DOI] [PubMed] [Google Scholar]
- Ding Y., Lee J., Ma J., Sung K., Yokota T., Singh N., Dooraghi M., Abiri P., Wang Y., Kulkarni R. P. et al. (2017). Light-sheet fluorescence imaging to localize cardiac lineage and protein distribution. Sci. Rep. 7, 42209 10.1038/srep42209 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Dodt H. U., Leischner U., Schierloh A., Jährling N., Mauch C. P., Deininger K., Deussing J. M., Eder M., Zieglgansberger W. and Becker K. (2007). Ultramicroscopy: three-dimensional visualization of neuronal networks in the whole mouse brain. Nat. Methods 4, 331-336. 10.1038/nmeth1036 [DOI] [PubMed] [Google Scholar]
- Dustin M. L. and Choudhuri K. (2016). Signaling and polarized communication across the T cell immunological synapse. Annu. Rev. Cell Dev. Biol. 32, 303-325. 10.1146/annurev-cellbio-100814-125330 [DOI] [PubMed] [Google Scholar]
- Ekyalongo R. C., Nakayama H., Kina K., Kaga N. and Iwabuchi K. (2015). Organization and functions of glycolipid-enriched microdomains in phagocytes. Biochim. Biophys. Acta 1851, 90-97. 10.1016/j.bbalip.2014.06.009 [DOI] [PubMed] [Google Scholar]
- Fan Z. and Ley K. (2015). Leukocyte arrest: Biomechanics and molecular mechanisms of beta2 integrin activation. Biorheology 52, 353-377. 10.3233/BIR-15085 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., McArdle S., Marki A., Mikulski Z., Gutierrez E., Engelhardt B., Deutsch U., Ginsberg M., Groisman A. and Ley K. (2016). Neutrophil recruitment limited by high-affinity bent beta2 integrin binding ligand in cis. Nat. Commun. 7, 12658 10.1038/ncomms12658 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fan Z., Kiosses W. B., Sun H., Orecchioni M., Ghosheh Y., Zajonc D. M., Arnaout M. A., Gutierrez E., Groisman A., Ginsberg M. H. et al. (2019). High-affinity bent beta2-integrin molecules in arresting neutrophils face each other through binding to ICAMs in cis. Cell Rep 26, 119-130.e115. 10.1016/j.celrep.2018.12.038 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fiolka R., Shao L., Rego E. H., Davidson M. W. and Gustafsson M. G. L. (2012). Time-lapse two-color 3D imaging of live cells with doubled resolution using structured illumination. Proc. Natl. Acad. Sci. USA 109, 5311-5315. 10.1073/pnas.1119262109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Förster T. (1948). Intermolecular energy migration and fluorescence. Ann. Physiol. 2, 55-75. 10.1002/andp.19484370105 [DOI] [Google Scholar]
- Fritzsche M., Fernandes R. A., Chang V. T., Colin-York H., Clausen M. P., Felce J. H., Galiani S., Erlenkamper C., Santos A. M., Heddleston J. M. et al. (2017). Cytoskeletal actin dynamics shape a ramifying actin network underpinning immunological synapse formation. Sci. Adv. 3, e1603032 10.1126/sciadv.1603032 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fumagalli S., Fiordaliso F., Perego C., Corbelli A., Mariani A., De Paola M. and De Simoni M. G. (2019). The phagocytic state of brain myeloid cells after ischemia revealed by superresolution structured illumination microscopy. J. Neuroinflammation 16, 9 10.1186/s12974-019-1401-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Germain R. N., Robey E. A. and Cahalan M. D. (2012). A decade of imaging cellular motility and interaction dynamics in the immune system. Science 336, 1676-1681. 10.1126/science.1221063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Giannone G., Hosy E., Sibarita J. B., Choquet D. and Cognet L. (2013). High-content super-resolution imaging of live cell by uPAINT. Methods Mol. Biol. 950, 95-110. 10.1007/978-1-62703-137-0_7 [DOI] [PubMed] [Google Scholar]
- Godin A. G., Lounis B. and Cognet L. (2014). Super-resolution microscopy approaches for live cell imaging. Biophys. J. 107, 1777-1784. 10.1016/j.bpj.2014.08.028 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Grashoff C., Hoffman B. D., Brenner M. D., Zhou R., Parsons M., Yang M. T., McLean M. A., Sligar S. G., Chen C. S., Ha T. et al. (2010). Measuring mechanical tension across vinculin reveals regulation of focal adhesion dynamics. Nature 466, 263-266. 10.1038/nature09198 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Greenwald E. C., Mehta S. and Zhang J. (2018). Genetically encoded fluorescent biosensors illuminate the spatiotemporal regulation of signaling networks. Chem. Rev. 118, 11707-11794. 10.1021/acs.chemrev.8b00333 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gupta V., Gylling A., Alonso J. L., Sugimori T., Ianakiev P., Xiong J.-P. and Arnaout M. A. (2007). The beta-tail domain (betaTD) regulates physiologic ligand binding to integrin CD11b/CD18. Blood 109, 3513-3520. 10.1182/blood-2005-11-056689 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Gustafsson M. G. L. (2000). Surpassing the lateral resolution limit by a factor of two using structured illumination microscopy. J. Microsc. 198, 82-87. 10.1046/j.1365-2818.2000.00710.x [DOI] [PubMed] [Google Scholar]
- Gustafsson M. G., Shao L., Carlton P. M., Wang C. J., Golubovskaya I. N., Cande W. Z., Agard D. A. and Sedat J. W. (2008). Three-dimensional resolution doubling in wide-field fluorescence microscopy by structured illumination. Biophys. J. 94, 4957-4970. 10.1529/biophysj.107.120345 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanley P. J., Xu Y., Kronlage M., Grobe K., Schon P., Song J., Sorokin L., Schwab A. and Bahler M. (2010). Motorized RhoGAP myosin IXb (Myo9b) controls cell shape and motility. Proc. Natl. Acad. Sci. USA 107, 12145-12150. 10.1073/pnas.0911986107 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hanna S. J., McCoy-Simandle K., Miskolci V., Guo P., Cammer M., Hodgson L. and Cox D. (2017). The Role of Rho-GTPases and actin polymerization during Macrophage Tunneling Nanotube Biogenesis. Sci. Rep. 7, 8547 10.1038/s41598-017-08950-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- He J., Johnson J. L., Monfregola J., Ramadass M., Pestonjamasp K., Napolitano G., Zhang J. and Catz S. D. (2016). Munc13-4 interacts with syntaxin 7 and regulates late endosomal maturation, endosomal signaling, and TLR9-initiated cellular responses. Mol. Biol. Cell 27, 572-587. 10.1091/mbc.e15-05-0283 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Heine J., Wurm C. A., Keller-Findeisen J., Schönle A., Harke B., Reuss M., Winter F. R. and Donnert G. (2018). Three dimensional live-cell STED microscopy at increased depth using a water immersion objective. Rev. Sci. Instrum. 89, 053701 10.1063/1.5020249 [DOI] [PubMed] [Google Scholar]
- Heintzmann R. and Cremer C. G. (1999). Laterally modulated excitation microscopy: improvement of resolution by using a diffraction grating. SPIE Proc. 3568, 185-196. 10.1117/12.336833 [DOI] [Google Scholar]
- Hell S. W. and Wichmann J. (1994). Breaking the diffraction resolution limit by stimulated emission: stimulated-emission-depletion fluorescence microscopy. Opt. Lett. 19, 780-782. 10.1364/OL.19.000780 [DOI] [PubMed] [Google Scholar]
- Hess S. T., Girirajan T. P. and Mason M. D. (2006). Ultra-high resolution imaging by fluorescence photoactivation localization microscopy. Biophys. J. 91, 4258-4272. 10.1529/biophysj.106.091116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Horsthemke M., Bachg A. C., Groll K., Moyzio S., Müther B., Hemkemeyer S. A., Wedlich-Söldner R., Sixt M., Tacke S., Bähler M. et al. (2017). Multiple roles of filopodial dynamics in particle capture and phagocytosis and phenotypes of Cdc42 and Myo10 deletion. J. Biol. Chem. 292, 7258-7273. 10.1074/jbc.M116.766923 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu J. J., Lu J., Umar S., Lee J. T., Kulkarni R. P., Ding Y., Chang C.-C., Hsiai T. K., Hokugo A., Gkouveris I. et al. (2018). Effects of teriparatide on morphology of aortic calcification in aged hyperlipidemic mice. Am. J. Physiol. Heart Circ. Physiol. 314, H1203-H1213. 10.1152/ajpheart.00718.2017 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hsu B. E., Tabariès S., Johnson R. M., Andrzejewski S., Senecal J., Lehuédé C., Annis M. G., Ma E. H., Völs S., Ramsay L. et al. (2019). Immature low-density neutrophils exhibit metabolic flexibility that facilitates breast cancer liver metastasis. Cell Rep 27, 3902-3915.e3906. 10.1016/j.celrep.2019.05.091 [DOI] [PubMed] [Google Scholar]
- Hu Y. S., Cang H. and Lillemeier B. F. (2016). Superresolution imaging reveals nanometer- and micrometer-scale spatial distributions of T-cell receptors in lymph nodes. Proc. Natl. Acad. Sci. USA 113, 7201-7206. 10.1073/pnas.1512331113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Huff J. (2016). The Fast mode for ZEISS LSM 880 with Airyscan: high-speed confocal imaging with super-resolution and improved signal-to-noise ratio. Nat. Methods 13, i-ii. 10.1038/nmeth.f.398 [DOI] [Google Scholar]
- Huisken J. and Stainier D. Y. (2007). Even fluorescence excitation by multidirectional selective plane illumination microscopy (mSPIM). Opt. Lett. 32, 2608-2610. 10.1364/OL.32.002608 [DOI] [PubMed] [Google Scholar]
- Huisken J., Swoger J., Del Bene F., Wittbrodt J. and Stelzer E. H. (2004). Optical sectioning deep inside live embryos by selective plane illumination microscopy. Science 305, 1007-1009. 10.1126/science.1100035 [DOI] [PubMed] [Google Scholar]
- Jin X., Dimitriadis E. K., Liu Y., Combs C. A., Chang J., Varsano N., Stempinski E., Flores R., Jackson S. N., Muller L. et al. (2018). Macrophages shed excess cholesterol in unique extracellular structures containing cholesterol microdomains. Arterioscler. Thromb. Vasc. Biol. 38, 1504-1518. 10.1161/ATVBAHA.118.311269 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., He J., Ramadass M., Pestonjamasp K., Kiosses W. B., Zhang J. and Catz S. D. (2016a). Munc13-4 Is a Rab11-binding Protein That Regulates Rab11-positive Vesicle Trafficking and Docking at the Plasma Membrane. J. Biol. Chem. 291, 3423-3438. 10.1074/jbc.M115.705871 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Johnson J. L., Ramadass M., He J., Brown S. J., Zhang J., Abgaryan L., Biris N., Gavathiotis E., Rosen H. and Catz S. D. (2016b). Identification of neutrophil exocytosis inhibitors (Nexinhibs), small molecule inhibitors of neutrophil exocytosis and inflammation: DRUGGABILITY OF THE SMALL GTPase Rab27a. J. Biol. Chem. 291, 25965-25982. 10.1074/jbc.M116.741884 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jones S. A., Shim S. H., He J. and Zhuang X. (2011). Fast, three-dimensional super-resolution imaging of live cells. Nat. Methods 8, 499-508. 10.1038/nmeth.1605 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Jung Y., Riven I., Feigelson S. W., Kartvelishvily E., Tohya K., Miyasaka M., Alon R. and Haran G. (2016). Three-dimensional localization of T-cell receptors in relation to microvilli using a combination of superresolution microscopies. Proc. Natl. Acad. Sci. USA 113, E5916-E5924. 10.1073/pnas.1605399113 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kambara H., Liu F., Zhang X., Liu P., Bajrami B., Teng Y., Zhao L., Zhou S., Yu H., Zhou W. et al. (2018). Gasdermin D exerts anti-inflammatory effects by promoting neutrophil death. Cell Rep 22, 2924-2936. 10.1016/j.celrep.2018.02.067 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Z. B., Novotná L., Blount A. and Lillemeier B. F. (2017). A cycle of Zap70 kinase activation and release from the TCR amplifies and disperses antigenic stimuli. Nat. Immunol. 18, 86-95. 10.1038/ni.3631 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Katz Z. B., Zhang C., Quintana A., Lillemeier B. F. and Hogan P. G. (2019). Septins organize endoplasmic reticulum-plasma membrane junctions for STIM1-ORAI1 calcium signalling. Sci. Rep. 9, 10839 10.1038/s41598-019-46862-w [DOI] [PMC free article] [PubMed] [Google Scholar]
- Keller P. J., Schmidt A. D., Wittbrodt J. and Stelzer E. H. (2008). Reconstruction of zebrafish early embryonic development by scanned light sheet microscopy. Science 322, 1065-1069. 10.1126/science.1162493 [DOI] [PubMed] [Google Scholar]
- Kilian N., Goryaynov A., Lessard M. D., Hooker G., Toomre D., Rothman J. E. and Bewersdorf J. (2018). Assessing photodamage in live-cell STED microscopy. Nat. Methods 15, 755-756. 10.1038/s41592-018-0145-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kim M., Carman C. V. and Springer T. A. (2003). Bidirectional transmembrane signaling by cytoplasmic domain separation in integrins. Science 301, 1720-1725. 10.1126/science.1084174 [DOI] [PubMed] [Google Scholar]
- Klar T. A. and Hell S. W. (1999). Subdiffraction resolution in far-field fluorescence microscopy. Opt. Lett. 24, 954-956. 10.1364/OL.24.000954 [DOI] [PubMed] [Google Scholar]
- Klar T. A., Jakobs S., Dyba M., Egner A. and Hell S. W. (2000). Fluorescence microscopy with diffraction resolution barrier broken by stimulated emission. Proc. Natl. Acad. Sci. USA 97, 8206-8210. 10.1073/pnas.97.15.8206 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kolaczkowska E. and Kubes P. (2013). Neutrophil recruitment and function in health and inflammation. Nat. Rev. Immunol. 13, 159-175. 10.1038/nri3399 [DOI] [PubMed] [Google Scholar]
- Kolossov V. L., Sivaguru M., Huff J., Luby K., Kanakaraju K. and Gaskins H. R. (2018). Airyscan super-resolution microscopy of mitochondrial morphology and dynamics in living tumor cells. Microsc. Res. Tech. 81, 115-128. 10.1002/jemt.22968 [DOI] [PubMed] [Google Scholar]
- Kruger C. L., Zeuner M. T., Cottrell G. S., Widera D. and Heilemann M. (2017). Quantitative single-molecule imaging of TLR4 reveals ligand-specific receptor dimerization. Sci. Signal. 10, eaan1308 10.1126/scisignal.aan1308 [DOI] [PubMed] [Google Scholar]
- Krzic U., Gunther S., Saunders T. E., Streichan S. J. and Hufnagel L. (2012). Multiview light-sheet microscope for rapid in toto imaging. Nat. Methods 9, 730-733. 10.1038/nmeth.2064 [DOI] [PubMed] [Google Scholar]
- Lam A. J., St-Pierre F., Gong Y., Marshall J. D., Cranfill P. J., Baird M. A., McKeown M. R., Wiedenmann J., Davidson M. W., Schnitzer M. J. et al. (2012). Improving FRET dynamic range with bright green and red fluorescent proteins. Nat. Methods 9, 1005-1012. 10.1038/nmeth.2171 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lam R. S., O'Brien-Simpson N. M., Holden J. A., Lenzo J. C., Fong S. B. and Reynolds E. C. (2016). Unprimed, M1 and M2 macrophages differentially interact with porphyromonas gingivalis. PLoS ONE 11, e0158629 10.1371/journal.pone.0158629 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Larson A. M. (2011). Multiphoton microscopy. Nat. Photonics 5, 1 10.1038/nphoton.an.2010.2 [DOI] [Google Scholar]
- Lee J., Moghadam M. E., Kung E., Cao H., Beebe T., Miller Y., Roman B. L., Lien C. L., Chi N. C., Marsden A. L. et al. (2013). Moving domain computational fluid dynamics to interface with an embryonic model of cardiac morphogenesis. PLoS ONE 8, e72924 10.1371/journal.pone.0072924 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lee J., Fei P., Sevag Packard R. R., Kang H., Xu H., Baek K. I., Jen N., Chen J., Yen H., Kuo C. C. et al. (2016). 4-Dimensional light-sheet microscopy to elucidate shear stress modulation of cardiac trabeculation. J. Clin. Invest. 126, 3158 10.1172/JCI89549 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort C. T. and Ley K. (2012). Neutrophil arrest by LFA-1 activation. Front. Immunol. 3, 157 10.3389/fimmu.2012.00157 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lefort C. T., Hyun Y.-M., Schultz J. B., Law F.-Y., Waugh R. E., Knauf P. A. and Kim M. (2009). Outside-in signal transmission by conformational changes in integrin Mac-1. J. Immunol. 183, 6460-6468. 10.4049/jimmunol.0900983 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lenzo J. C., O'Brien-Simpson N. M., Cecil J., Holden J. A. and Reynolds E. C. (2016). Determination of active phagocytosis of unopsonized porphyromonas gingivalis by macrophages and neutrophils using the pH-sensitive fluorescent dye pHrodo. Infect. Immun. 84, 1753-1760. 10.1128/IAI.01482-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ley K., Laudanna C., Cybulsky M. I. and Nourshargh S. (2007). Getting to the site of inflammation: the leukocyte adhesion cascade updated. Nat. Rev. Immunol. 7, 678-689. 10.1038/nri2156 [DOI] [PubMed] [Google Scholar]
- Ley K., Hoffman H. M., Kubes P., Cassatella M. A., Zychlinsky A., Hedrick C. C. and Catz S. D. (2018). Neutrophils: new insights and open questions. Sci. Immunol. 3, eaat4579 10.1126/sciimmunol.aat4579 [DOI] [PubMed] [Google Scholar]
- Lillemeier B. F., Mortelmaier M. A., Forstner M. B., Huppa J. B., Groves J. T. and Davis M. M. (2010). TCR and Lat are expressed on separate protein islands on T cell membranes and concatenate during activation. Nat. Immunol. 11, 90-96. 10.1038/ni.1832 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu T. L., Upadhyayula S., Milkie D. E., Singh V., Wang K., Swinburne I. A., Mosaliganti K. R., Collins Z. M., Hiscock T. W., Shea J. et al. (2018a). Observing the cell in its native state: Imaging subcellular dynamics in multicellular organisms. Science 360, eaaq1392 10.1126/science.aaq1392 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liu W., Molnar M., Garnham C., Benav H. and Rask-Andersen H. (2018b). Macrophages in the human cochlea: saviors or predators-a study using super-resolution immunohistochemistry. Front. Immunol. 9, 223 10.3389/fimmu.2018.00223 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lopes F. B., Bálint S., Valvo S., Felce J. H., Hessel E. M., Dustin M. L. and Davis D. M. (2017). Membrane nanoclusters of FcgammaRI segregate from inhibitory SIRPalpha upon activation of human macrophages. J. Cell Biol. 216, 1123-1141. 10.1083/jcb.201608094 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Lukeš T., Glatzová D., Kvíčalová Z., Levet F., Benda A., Letschert S., Sauer M., Brdička T., Lasser T. and Cebecauer M. (2017). Quantifying protein densities on cell membranes using super-resolution optical fluctuation imaging. Nat. Commun. 8, 1731 10.1038/s41467-017-01857-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Luo B.-H., Carman C. V. and Springer T. A. (2007). Structural basis of integrin regulation and signaling. Annu. Rev. Immunol. 25, 619-647. 10.1146/annurev.immunol.25.022106.141618 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ma Y., Pandzic E., Nicovich P. R., Yamamoto Y., Kwiatek J., Pageon S. V., Benda A., Rossy J. and Gaus K. (2017). An intermolecular FRET sensor detects the dynamics of T cell receptor clustering. Nat. Commun. 8, 15100 10.1038/ncomms15100 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Maity P. C., Blount A., Jumaa H., Ronneberger O., Lillemeier B. F. and Reth M. (2015). B cell antigen receptors of the IgM and IgD classes are clustered in different protein islands that are altered during B cell activation. Sci. Signal. 8, ra93 10.1126/scisignal.2005887 [DOI] [PubMed] [Google Scholar]
- Manfredi A. A., Ramirez G. A., Rovere-Querini P. and Maugeri N. (2018). The Neutrophil's choice: phagocytose vs make neutrophil extracellular traps. Front. Immunol. 9, 288 10.3389/fimmu.2018.00288 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Margraf A., Ley K. and Zarbock A. (2019). Neutrophil recruitment: from model systems to tissue-specific patterns. Trends Immunol. 40, 613-634. 10.1016/j.it.2019.04.010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Masson A., Escande P., Frongia C., Clouvel G., Ducommun B. and Lorenzo C. (2015). High-resolution in-depth imaging of optically cleared thick samples using an adaptive SPIM. Sci. Rep. 5, 16898 10.1038/srep16898 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matlung H. L., Babes L., Zhao X. W., van Houdt M., Treffers L. W., van Rees D. J., Franke K., Schornagel K., Verkuijlen P., Janssen H. et al. (2018). Neutrophils kill antibody-opsonized cancer cells by trogoptosis. Cell Rep 23, 3946-3959.e3946. 10.1016/j.celrep.2018.05.082 [DOI] [PubMed] [Google Scholar]
- Mazaki Y., Onodera Y., Higashi T., Horinouchi T., Oikawa T. and Sabe H. (2017). ARF1 recruits RAC1 to leading edge in neutrophil chemotaxis. Cell Commun. Signal 15, 36 10.1186/s12964-017-0193-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- McArdle S., Mikulski Z. and Ley K. (2016). Live cell imaging to understand monocyte, macrophage, and dendritic cell function in atherosclerosis. J. Exp. Med. 213, 1117-1131. 10.1084/jem.20151885 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mickoleit M., Schmid B., Weber M., Fahrbach F. O., Hombach S., Reischauer S. and Huisken J. (2014). High-resolution reconstruction of the beating zebrafish heart. Nat. Methods 11, 919-922. 10.1038/nmeth.3037 [DOI] [PubMed] [Google Scholar]
- Millius A. and Weiner O. D. (2009). Chemotaxis in neutrophil-like HL-60 cells. Methods Mol. Biol. 571, 167-177. 10.1007/978-1-60761-198-1_11 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Minsky M. (1988). Memoir on inventing the confocal scanning microscope. Scanning 10, 129-138. 10.1002/sca.4950100403 [DOI] [Google Scholar]
- Mishina N. M. and Belousov V. V. (2017). Live-Cell STED imaging with the HyPer2 biosensor. Methods Mol. Biol. 1663, 21-28. 10.1007/978-1-4939-7265-4_3 [DOI] [PubMed] [Google Scholar]
- Mishra A. K., Mondo J. A., Campanale J. P. and Montell D. J. (2019). Coordination of protrusion dynamics within and between collectively migrating border cells by myosin II. Mol. Biol. Cell 30, 2490-2502. 10.1091/mbc.E19-02-0124 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mittler R. S., Goldman S. J., Spitalny G. L. and Burakoff S. J. (1989). T-cell receptor-CD4 physical association in a murine T-cell hybridoma: induction by antigen receptor ligation. Proc. Natl. Acad. Sci. USA 86, 8531-8535. 10.1073/pnas.86.21.8531 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mizuno R., Kamioka Y., Kabashima K., Imajo M., Sumiyama K., Nakasho E., Ito T., Hamazaki Y., Okuchi Y., Sakai Y. et al. (2014). In vivo imaging reveals PKA regulation of ERK activity during neutrophil recruitment to inflamed intestines. J. Exp. Med. 211, 1123-1136. 10.1084/jem.20132112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mócsai A., Walzog B. and Lowell C. A. (2015). Intracellular signalling during neutrophil recruitment. Cardiovasc. Res. 107, 373-385. 10.1093/cvr/cvv159 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mollinedo F. (2019). Neutrophil degranulation, plasticity, and cancer metastasis. Trends Immunol. 40, 228-242. 10.1016/j.it.2019.01.006 [DOI] [PubMed] [Google Scholar]
- Moore T. I., Aaron J., Chew T. L. and Springer T. A. (2018). Measuring integrin conformational change on the cell surface with super-resolution microscopy. Cell Rep 22, 1903-1912. 10.1016/j.celrep.2018.01.062 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Moseman E. A., Wu T., de la Torre J. C., Schwartzberg P. L. and McGavern D. B. (2016). Type I interferon suppresses virus-specific B cell responses by modulating CD8+ T cell differentiation. Sci. Immunol. 1, eaah3565. [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nair R. R., Mazza D., Brambilla F., Gorzanelli A., Agresti A. and Bianchi M. E. (2018). LPS-challenged macrophages release microvesicles coated with histones. Front. Immunol. 9, 1463 10.3389/fimmu.2018.01463 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neubert E., Meyer D., Rocca F., Gunay G., Kwaczala-Tessmann A., Grandke J., Senger-Sander S., Geisler C., Egner A., Schon M. P. et al. (2018). Chromatin swelling drives neutrophil extracellular trap release. Nat. Commun. 9, 3767 10.1038/s41467-018-06263-5 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Neumann J., Ziegler K., Gelléri M., Fröhlich-Nowoisky J., Liu F., Bellinghausen I., Schuppan D., Birk U., Pöschl U., Cremer C. et al. (2019). Nanoscale distribution of TLR4 on primary human macrophages stimulated with LPS and ATI. Nanoscale 11, 9769-9779. 10.1039/C9NR00943D [DOI] [PubMed] [Google Scholar]
- Nobis M., McGhee E. J., Morton J. P., Schwarz J. P., Karim S. A., Quinn J., Edward M., Campbell A. D., McGarry L. C., Evans T. R. et al. (2013). Intravital FLIM-FRET imaging reveals dasatinib-induced spatial control of src in pancreatic cancer. Cancer Res. 73, 4674-4686. 10.1158/0008-5472.CAN-12-4545 [DOI] [PubMed] [Google Scholar]
- Nobis M., Herrmann D., Warren S. C., Kadir S., Leung W., Killen M., Magenau A., Stevenson D., Lucas M. C., Reischmann N. et al. (2017). A RhoA-FRET biosensor mouse for intravital imaging in normal tissue homeostasis and disease contexts. Cell Rep 21, 274-288. 10.1016/j.celrep.2017.09.022 [DOI] [PubMed] [Google Scholar]
- Nobis M., Herrmann D., Warren S. C., Strathdee D., Cox T. R., Anderson K. I., and Timpson P. (2018). Shedding new light on RhoA signalling as a drug target in vivo using a novel RhoA-FRET biosensor mouse. Small GTPases, doi: 10.1080/21541248.2018.1438024 10.1080/21541248.2018.1438024 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenfelt P. and Tapper H. (2011). Phagosome dynamics during phagocytosis by neutrophils. J. Leukoc. Biol. 90, 271-284. 10.1189/jlb.0810457 [DOI] [PubMed] [Google Scholar]
- Nordenfelt P., Elliott H. L. and Springer T. A. (2016). Coordinated integrin activation by actin-dependent force during T-cell migration. Nat. Commun. 7, 13119 10.1038/ncomms13119 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Nordenfelt P., Moore T. I., Mehta S. B., Kalappurakkal J. M., Swaminathan V., Koga N., Lambert T. J., Baker D., Waters J. C., Oldenbourg R. et al. (2017). Direction of actin flow dictates integrin LFA-1 orientation during leukocyte migration. Nat. Commun. 8, 2047 10.1038/s41467-017-01848-y [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Shaughnessy E. C., Stone O. J., LaFosse P. K., Azoitei M. L., Tsygankov D., Heddleston J. M., Legant W. R., Wittchen E. S., Burridge K., Elston T. C. et al. (2019). Software for lattice light-sheet imaging of FRET biosensors, illustrated with a new Rap1 biosensor. J. Cell Biol. 218, 3153-3160. 10.1083/jcb.201903019 [DOI] [PMC free article] [PubMed] [Google Scholar]
- O'Sullivan K. M., Lo C. Y., Summers S. A., Elgass K. D., McMillan P. J., Longano A., Ford S. L., Gan P. Y., Kerr P. G., Kitching A. R. et al. (2015). Renal participation of myeloperoxidase in antineutrophil cytoplasmic antibody (ANCA)-associated glomerulonephritis. Kidney Int. 88, 1030-1046. 10.1038/ki.2015.202 [DOI] [PubMed] [Google Scholar]
- Ostrowski P. P., Freeman S. A., Fairn G. and Grinstein S. (2019). Dynamic podosome-like structures in nascent phagosomes are coordinated by phosphoinositides. Dev. Cell 50, 397-410.e3. 10.1016/j.devcel.2019.05.028 [DOI] [PubMed] [Google Scholar]
- Packard R. R. S., Baek K. I., Beebe T., Jen N., Ding Y., Shi F., Fei P., Kang B. J., Chen P. H., Gau J. et al. (2017). Automated segmentation of light-sheet fluorescent imaging to characterize experimental doxorubicin-induced cardiac injury and repair. Sci. Rep. 7, 8603 10.1038/s41598-017-09152-x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Peng K., Muranyi W., Glass B., Laketa V., Yant S. R., Tsai L., Cihlar T., Müller B. and Kräusslich H. G. (2014). Quantitative microscopy of functional HIV post-entry complexes reveals association of replication with the viral capsid. Elife 3, e04114 10.7554/eLife.04114 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Perdomo J., Leung H. H. L., Ahmadi Z., Yan F., Chong J. J. H., Passam F. H. and Chong B. H. (2019). Neutrophil activation and NETosis are the major drivers of thrombosis in heparin-induced thrombocytopenia. Nat. Commun. 10, 1322 10.1038/s41467-019-09160-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Philipsen L., Reddycherla A. V., Hartig R., Gumz J., Kästle M., Kritikos A., Poltorak M. P., Prokazov Y., Turbin E., Weber A. et al. (2017). De novo phosphorylation and conformational opening of the tyrosine kinase Lck act in concert to initiate T cell receptor signaling. Sci. Signal. 10 10.1126/scisignal.aaf4736 [DOI] [PubMed] [Google Scholar]
- Pittet M. J. and Weissleder R. (2011). Intravital imaging. Cell 147, 983-991. 10.1016/j.cell.2011.11.004 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pittet M. J., Garris C. S., Arlauckas S. P. and Weissleder R. (2018). Recording the wild lives of immune cells. Sci. Immunol. 3 10.1126/sciimmunol.aaq0491 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Planchon T. A., Gao L., Milkie D. E., Davidson M. W., Galbraith J. A., Galbraith C. G. and Betzig E. (2011). Rapid three-dimensional isotropic imaging of living cells using Bessel beam plane illumination. Nat. Methods 8, 417-423. 10.1038/nmeth.1586 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Prahst C., Ashrafzadeh P., Harrington K., Venkatraman L., Richards M., Russo A. M., Cho K.-S., Chang K., Mead T., Chen D. F. et al. (2019). Mouse retinal cell behaviour in space and time using light sheet fluorescence microscopy. bioRxiv 686626 10.1101/686626 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Radbruch H., Bremer D., Mothes R., Gunther R., Rinnenthal J. L., Pohlan J., Ulbricht C., Hauser A. E. and Niesner R. (2015). Intravital FRET: probing cellular and tissue function in Vivo. Int. J. Mol. Sci. 16, 11713-11727. 10.3390/ijms160511713 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ramadass M., Johnson J. L., Marki A., Zhang J., Wolf D., Kiosses W. B., Pestonjamasp K., Ley K. and Catz S. D. (2019). The trafficking protein JFC1 regulates Rac1-GTP localization at the uropod controlling neutrophil chemotaxis and in vivo migration. J. Leukoc. Biol. 105, 1209-1224. 10.1002/JLB.1VMA0818-320R [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ritter A. T., Asano Y., Stinchcombe J. C., Dieckmann N. M. C., Chen B.-C., Gawden-Bone C., van Engelenburg S., Legant W., Gao L., Davidson M. W. et al. (2015). Actin depletion initiates events leading to granule secretion at the immunological synapse. Immunity 42, 864-876. 10.1016/j.immuni.2015.04.013 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roh K.-H., Lillemeier B. F., Wang F. and Davis M. M. (2015). The coreceptor CD4 is expressed in distinct nanoclusters and does not colocalize with T-cell receptor and active protein tyrosine kinase p56lck. Proc. Natl. Acad. Sci. USA 112, E1604-E1613. 10.1073/pnas.1503532112 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rossboth B., Arnold A. M., Ta H., Platzer R., Kellner F., Huppa J. B., Brameshuber M., Baumgart F. and Schütz G. J. (2018). TCRs are randomly distributed on the plasma membrane of resting antigen-experienced T cells. Nat. Immunol. 19, 821-827. 10.1038/s41590-018-0162-7 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Roy N. H., Chan J., Lambele M. and Thali M. (2013). Clustering and mobility of HIV-1 Env at viral assembly sites predict its propensity to induce cell-cell fusion. J. Virol. 87, 7516-7525. 10.1128/JVI.00790-13 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russell R. A., Chojnacki J., Jones D. M., Johnson E., Do T., Eggeling C., Padilla-Parra S. and Sattentau Q. J. (2017). Astrocytes resist HIV-1 fusion but engulf infected macrophage material. Cell Rep 18, 1473-1483. 10.1016/j.celrep.2017.01.027 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Russwurm M., Mullershausen F., Friebe A., Jäger R., Russwurm C. and Koesling D. (2007). Design of fluorescence resonance energy transfer (FRET)-based cGMP indicators: a systematic approach. Biochem. J. 407, 69-77. 10.1042/BJ20070348 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Rust M. J., Bates M. and Zhuang X. (2006). Sub-diffraction-limit imaging by stochastic optical reconstruction microscopy (STORM). Nat. Methods 3, 793-796. 10.1038/nmeth929 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Saitoh T., Komano J., Saitoh Y., Misawa T., Takahama M., Kozaki T., Uehata T., Iwasaki H., Omori H., Yamaoka S. et al. (2012). Neutrophil extracellular traps mediate a host defense response to human immunodeficiency virus-1. Cell Host Microbe 12, 109-116. 10.1016/j.chom.2012.05.015 [DOI] [PubMed] [Google Scholar]
- Sanderson M. J., Smith I., Parker I. and Bootman M. D. (2014). Fluorescence microscopy. Cold Spring Harb. Protoc. 2014, pdb top071795 10.1101/pdb.top071795 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Santi P. A., Johnson S. B., Hillenbrand M., GrandPre P. Z., Glass T. J. and Leger J. R. (2009). Thin-sheet laser imaging microscopy for optical sectioning of thick tissues. BioTechniques 46, 287-294. 10.2144/000113087 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Scherz P. J., Huisken J., Sahai-Hernandez P. and Stainier D. Y. R. (2008). High-speed imaging of developing heart valves reveals interplay of morphogenesis and function. Development 135, 1179-1187. 10.1242/dev.010694 [DOI] [PubMed] [Google Scholar]
- Scipioni L., Lanzanó L., Diaspro A. and Gratton E. (2018). Comprehensive correlation analysis for super-resolution dynamic fingerprinting of cellular compartments using the Zeiss Airyscan detector. Nat. Commun. 9, 5120 10.1038/s41467-018-07513-2 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sebestyén Z., Nagy P., Horváth G., Vámosi G., Debets R., Gratama J. W., Alexander D. R. and Szöllősi J. (2002). Long wavelength fluorophores and cell-by-cell correction for autofluorescence significantly improves the accuracy of flow cytometric energy transfer measurements on a dual-laser benchtop flow cytometer. Cytometry 48, 124-135. 10.1002/cyto.10121 [DOI] [PubMed] [Google Scholar]
- Sereti K. I., Nguyen N. B., Kamran P., Zhao P., Ranjbarvaziri S., Park S., Sabri S., Engel J. L., Sung K., Kulkarni R. P. et al. (2018). Analysis of cardiomyocyte clonal expansion during mouse heart development and injury. Nat. Commun. 9, 754 10.1038/s41467-018-02891-z [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sharonov A. and Hochstrasser R. M. (2006). Wide-field subdiffraction imaging by accumulated binding of diffusing probes. Proc. Natl. Acad. Sci. USA 103, 18911-18916. 10.1073/pnas.0609643104 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shim S.-H., Xia C., Zhong G., Babcock H. P., Vaughan J. C., Huang B., Wang X., Xu C., Bi G.-Q. and Zhuang X. (2012). Super-resolution fluorescence imaging of organelles in live cells with photoswitchable membrane probes. Proc. Natl. Acad. Sci. USA 109, 13978-13983. 10.1073/pnas.1201882109 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shrestha D., Jenei A., Nagy P., Vereb G. and Szöllősi J. (2015). Understanding FRET as a research tool for cellular studies. Int. J. Mol. Sci. 16, 6718-6756. 10.3390/ijms16046718 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shroff H., Galbraith C. G., Galbraith J. A. and Betzig E. (2008). Live-cell photoactivated localization microscopy of nanoscale adhesion dynamics. Nat. Methods 5, 417-423. 10.1038/nmeth.1202 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Siedentopf H. and Zsigmondy R. (1902). Uber Sichtbarmachung und Größenbestimmung ultramikoskopischer Teilchen, mit besonderer Anwendung auf Goldrubingläser. Ann. Phys. 315, 1-39. 10.1002/andp.19023150102 [DOI] [Google Scholar]
- Soares H., Henriques R., Sachse M., Ventimiglia L., Alonso M. A., Zimmer C., Thoulouze M. I. and Alcover A. (2013). Regulated vesicle fusion generates signaling nanoterritories that control T cell activation at the immunological synapse. J. Exp. Med. 210, 2415-2433. 10.1084/jem.20130150 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sollberger G., Tilley D. O. and Zychlinsky A. (2018). Neutrophil extracellular traps: the biology of chromatin externalization. Dev. Cell 44, 542-553. 10.1016/j.devcel.2018.01.019 [DOI] [PubMed] [Google Scholar]
- Sprenger J. U., Perera R. K., Steinbrecher J. H., Lehnart S. E., Maier L. S., Hasenfuss G. and Nikolaev V. O. (2015). In vivo model with targeted cAMP biosensor reveals changes in receptor-microdomain communication in cardiac disease. Nat. Commun. 6, 6965 10.1038/ncomms7965 [DOI] [PubMed] [Google Scholar]
- Stadtmann A., Germena G., Block H., Boras M., Rossaint J., Sundd P., Lefort C., Fisher C. I., Buscher K., Gelschefarth B. et al. (2013). The PSGL-1-L-selectin signaling complex regulates neutrophil adhesion under flow. J. Exp. Med. 210, 2171-2180. 10.1084/jem.20130664 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Stirnweiss A., Hartig R., Gieseler S., Lindquist J. A., Reichardt P., Philipsen L., Simeoni L., Poltorak M., Merten C., Zuschratter W. et al. (2013). T cell activation results in conformational changes in the Src family kinase Lck to induce its activation. Sci. Signal. 6, ra13 10.1126/scisignal.2003607 [DOI] [PubMed] [Google Scholar]
- Stock K., Sailer R., Strauss W. S., Lyttek M., Steiner R. and Schneckenburger H. (2003). Variable-angle total internal reflection fluorescence microscopy (VA-TIRFM): realization and application of a compact illumination device. J. Microsc. 211, 19-29. 10.1046/j.1365-2818.2003.01200.x [DOI] [PubMed] [Google Scholar]
- Sundd P., Gutierrez E., Pospieszalska M. K., Zhang H., Groisman A. and Ley K. (2010). Quantitative dynamic footprinting microscopy reveals mechanisms of neutrophil rolling. Nat. Methods 7, 821-824. 10.1038/nmeth.1508 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundd P., Gutierrez E., Petrich B. G., Ginsberg M. H., Groisman A. and Ley K. (2011). Live cell imaging of paxillin in rolling neutrophils by dual-color quantitative dynamic footprinting. Microcirculation 18, 361-372. 10.1111/j.1549-8719.2011.00090.x [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sundd P., Gutierrez E., Koltsova E. K., Kuwano Y., Fukuda S., Pospieszalska M. K., Groisman A. and Ley K. (2012). ‘Slings’ enable neutrophil rolling at high shear. Nature 488, 399-403. 10.1038/nature11248 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Szatmary A. C., Nossal R., Parent C. A., and Majumdar R. (2017). Modeling neutrophil migration in dynamic chemoattractant gradients: assessing the role of exosomes during signal relay. Mol. Biol. Cell 28, 3457-3470. 10.1091/mbc.e17-05-0298 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tanaka K., Kim S. E., Yano H., Matsumoto G., Ohuchida R., Ishikura Y., Araki M., Araki K., Park S., Komatsu T. et al. (2017). MiR-142 is required for staphylococcus aureus clearance at skin wound sites via small GTPase-mediated regulation of the neutrophil actin cytoskeleton. J. Invest. Dermatol. 137, 931-940. 10.1016/j.jid.2016.11.018 [DOI] [PubMed] [Google Scholar]
- Thestrup T., Litzlbauer J., Bartholomaus I., Mues M., Russo L., Dana H., Kovalchuk Y., Liang Y., Kalamakis G., Laukat Y. et al. (2014). Optimized ratiometric calcium sensors for functional in vivo imaging of neurons and T lymphocytes. Nat. Methods 11, 175-182. 10.1038/nmeth.2773 [DOI] [PubMed] [Google Scholar]
- Thunemann M., Wen L., Hillenbrand M., Vachaviolos A., Feil S., Ott T., Han X., Fukumura D., Jain R. K., Russwurm M. et al. (2013). Transgenic mice for cGMP imaging. Circ. Res. 113, 365-371. 10.1161/CIRCRESAHA.113.301063 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Truong T. V., Supatto W., Koos D. S., Choi J. M. and Fraser S. E. (2011). Deep and fast live imaging with two-photon scanned light-sheet microscopy. Nat. Methods 8, 757-760. 10.1038/nmeth.1652 [DOI] [PubMed] [Google Scholar]
- Tsui C., Maldonado P., Montaner B., Borroto A., Alarcon B., Bruckbauer A., Martinez-Martin N. and Batista F. D. (2018). Dynamic reorganisation of intermediate filaments coordinates early B-cell activation. Life Sci Alliance 1, e201800060 10.26508/lsa.201800060 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Turcotte R., Liang Y., Tanimoto M., Zhang Q., Li Z., Koyama M., Betzig E. and Ji N. (2019). Dynamic super-resolution structured illumination imaging in the living brain. Proc. Natl. Acad. Sci. USA 116, 9586-9591. 10.1073/pnas.1819965116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- van Leeuwenhoek A. (1719). Epistolae ad societatem regiam anglicam: et alio illustres viros seu continuatio mirandorum Arcanorum Naturae detectorum (Leiden: Joh. Arnold.).
- Varsano N., Beghi F., Elad N., Pereiro E., Dadosh T., Pinkas I., Perez-Berna A. J., Jin X., Kruth H. S., Leiserowitz L. et al. (2018). Two polymorphic cholesterol monohydrate crystal structures form in macrophage culture models of atherosclerosis. Proc. Natl. Acad. Sci. USA 115, 7662-7669. 10.1073/pnas.1803119115 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wee J. L., Schulze K. E., Jones E. L., Yeung L., Cheng Q., Pereira C. F., Costin A., Ramm G., van Spriel A. B., Hickey M. J. et al. (2015). Tetraspanin CD37 regulates beta2 integrin-mediated adhesion and migration in neutrophils. J. Immunol. 195, 5770-5779. 10.4049/jimmunol.1402414 [DOI] [PubMed] [Google Scholar]
- Wegel E., Gohler A., Lagerholm B. C., Wainman A., Uphoff S., Kaufmann R. and Dobbie I. M. (2016). Imaging cellular structures in super-resolution with SIM, STED and localisation microscopy: a practical comparison. Sci. Rep. 6, 27290 10.1038/srep27290 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wen L., Feil S., Wolters M., Thunemann M., Regler F., Schmidt K., Friebe A., Olbrich M., Langer H., Gawaz M. et al. (2018). A shear-dependent NO-cGMP-cGKI cascade in platelets acts as an auto-regulatory brake of thrombosis. Nat. Commun. 9, 4301 10.1038/s41467-018-06638-8 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wilding D., Pozzi P., Soloviev O., Vdovin G. and Verhaegen M. (2016). Adaptive illumination based on direct wavefront sensing in a light-sheet fluorescence microscope. Opt. Express 24, 24896-24906. 10.1364/OE.24.024896 [DOI] [PubMed] [Google Scholar]
- Willmann J. K., van Bruggen N., Dinkelborg L. M. and Gambhir S. S. (2008). Molecular imaging in drug development. Nat. Rev. Drug Discov. 7, 591-607. 10.1038/nrd2290 [DOI] [PubMed] [Google Scholar]
- Wu L. G., Hamid E., Shin W. and Chiang H. C. (2014). Exocytosis and endocytosis: modes, functions, and coupling mechanisms. Annu. Rev. Physiol. 76, 301-331. 10.1146/annurev-physiol-021113-170305 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Yang B., Przybilla F., Mestre M., Trebbia J.-B. and Lounis B. (2014). Large parallelization of STED nanoscopy using optical lattices. Opt. Express 22, 5581-5589. 10.1364/OE.22.005581 [DOI] [PubMed] [Google Scholar]
- Yang Y., Liu N., He Y., Liu Y., Ge L., Zou L., Song S., Xiong W. and Liu X. (2018). Improved calcium sensor GCaMP-X overcomes the calcium channel perturbations induced by the calmodulin in GCaMP. Nat. Commun. 9, 1504 10.1038/s41467-018-03719-6 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zal T., Zal M. A. and Gascoigne N. R. (2002). Inhibition of T cell receptor-coreceptor interactions by antagonist ligands visualized by live FRET imaging of the T-hybridoma immunological synapse. Immunity 16, 521-534. 10.1016/S1074-7613(02)00301-1 [DOI] [PubMed] [Google Scholar]
- Zariwala H. A., Borghuis B. G., Hoogland T. M., Madisen L., Tian L., De Zeeuw C. I., Zeng H., Looger L. L., Svoboda K. and Chen T. W. (2012). A Cre-dependent GCaMP3 reporter mouse for neuronal imaging in vivo. J. Neurosci. 32, 3131-3141. 10.1523/JNEUROSCI.4469-11.2012 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Zhang J., Campbell R. E., Ting A. Y. and Tsien R. Y. (2002). Creating new fluorescent probes for cell biology. Nat. Rev. Mol. Cell Biol. 3, 906-918. 10.1038/nrm976 [DOI] [PubMed] [Google Scholar]
- Zhang Y., Jin X., Liang J., Guo Y., Sun G., Zeng X. and Yin H. (2019). Extracellular vesicles derived from ODN-stimulated macrophages transfer and activate Cdc42 in recipient cells and thereby increase cellular permissiveness to EV uptake. Sci. Adv. 5, eaav1564 10.1126/sciadv.aav1564 [DOI] [PMC free article] [PubMed] [Google Scholar]