Abstract
The genomic, bipartite replication promoter of Ebola virus (EBOV) consists of elements 1 (PE1) and 2 (PE2). PE1 (55 nt at the 3′-terminus) is separated from PE2 (harboring eight 3′-UN5 hexamers) by the transcription start sequence (TSS) of the first nucleoprotein (NP) gene plus a spacer sequence. Insertions or deletions in the spacer were reported to support genome replication if comprising 6 or 12, but not 1/2/3/5/9 nt. This gave rise to the formulation of the “rule of 6” for the EBOV replication promoter. Here, we studied the impact of such hexamer phasing on viral transcription using a series of replication-competent and -deficient monocistronic minigenomes, in which the spacer of the NP gene was mutated or replaced with that of internal EBOV genes and mutated variants thereof. Beyond reporter gene assays, we conducted qRT-PCR to determine the levels of mRNA, genomic and antigenomic RNA. We demonstrate that hexamer phasing is also essential for viral transcription, that UN5 hexamer periodicity extends into PE1 and that the spacer region can be expanded by 48 nt without losses of transcriptional activity. Making the UN5 hexamer phasing continuous between PE1 and PE2 enhanced the efficiency of transcription and replication. We show that the 2 nt preceding the TSS are essential for transcription. We further propose a role for UN5 hexamer phasing in positioning NP during initiation of RNA synthesis, or in dissociation/reassociation of NP from the template RNA strand while threading the RNA through the active site of the elongating polymerase during replication and transcription.
Keywords: viral transcription, transcription promoter, minigenome systems, 3′-leader mutants, 3′-UNNNNN hexamer phasing
INTRODUCTION
The family Filoviridae in the order Mononegavirales (Bukreyev et al. 2014) includes five genera (Cuevavirus, Ebolavirus, Marburgvirus, Striavirus, and Thamnovirus) and a total of nine species. Members of this family form variously shaped, often filamentous, enveloped virions containing linear non-segmented, negative-sense (NNS) RNA genomes of 15–19 kb (Kuhn et al. 2019).
Filoviruses are causative agents of severe hemorrhagic fever in humans and non-human primates with reported case fatality rates between 34% and 81% in humans (Burk et al. 2016). The 2014/15 outbreak in West Africa and the current EBOV epidemic in the Democratic Republic of Congo suggest that filoviruses have become a constant threat of public health.
The negative-sense RNA genome of EBOV has a length of 19 kb with regulatory regions at the 5′- and 3′-end (termed 5′-trailer and 3′-leader) that harbor replication promoters and, specific for the 3′-leader, the first transcription start sequence (TSS) for the synthesis of the nucleoprotein (NP) mRNA (Sanchez et al. 1993; Mühlberger 2007). The helical viral nucleocapsid, consisting of the viral genome in complex with the viral proteins NP, VP35, and VP24, serves as the template for replication and transcription. Recent cryo-EM structures of NP:RNA complexes verified previous predictions that six RNA residues are bound per NP monomer (Wan et al. 2017; Sugita et al. 2018). For viral transcription, the polymerase L and its cofactor VP35 require association with the EBOV transcription factor VP30 (Mühlberger et al. 1999). Evidence obtained for the Mumps virus phosphoprotein (P) (Cox et al. 2014), the putative ortholog of VP35, suggests that VP35 may target L to the nucleocapsid and unwind the latter to allow the polymerase to gain access to the genomic RNA. Additionally, a novel NTPase and helicase function was revealed for VP35, with a preference for unwinding of dsRNA helices with 5′ overhangs (Shu et al. 2019). Inhibition of helicase activity resulted in a decrease of viral transcription and replication in cells, suggesting that RNA secondary structures need to be resolved during viral transcription/replication. Transcription is positively influenced by interaction of VP35 and VP30 (Biedenkopf et al. 2013) which was shown to be mediated by RNA and to depend on the dsRNA-binding capacity of VP35 (Biedenkopf et al. 2016b).
The phosphoprotein VP30 preferentially binds single-stranded RNA (ssRNA) of mixed base composition in conjunction with stem–loop structures (Schlereth et al. 2016). VP30 supports transcription in its non-phosphorylated state, whereas VP30 phosphorylation particularly at the key serine 29 inhibits viral transcription (Modrof et al. 2002; Biedenkopf et al. 2016a; Kruse et al. 2018). Phosphorylation of VP30 strengthens its interaction with NP and weakens the interaction with the polymerase cofactor VP35, which is thought to result in release of VP30 from the viral transcription complex (Biedenkopf et al. 2013). There is evidence that multiple phosphorylation/dephosphorylation events mediate the protein's dynamic association and dissociation with the nucleocapsid and transcription complexes throughout the viral life cycle (Biedenkopf et al. 2016a).
The EBOV replication promoters could be confined to the 3′-terminal 156 nt of the genomic leader and the 3′-terminal 177 nt of the antigenomic RNA (trailer promoter) (Calain et al. 1999). In this context, it is worth mentioning that antigenome synthesis was found to be initiated opposite to the penultimate C residue of the genome's 3′-end (Deflubé et al. 2019).
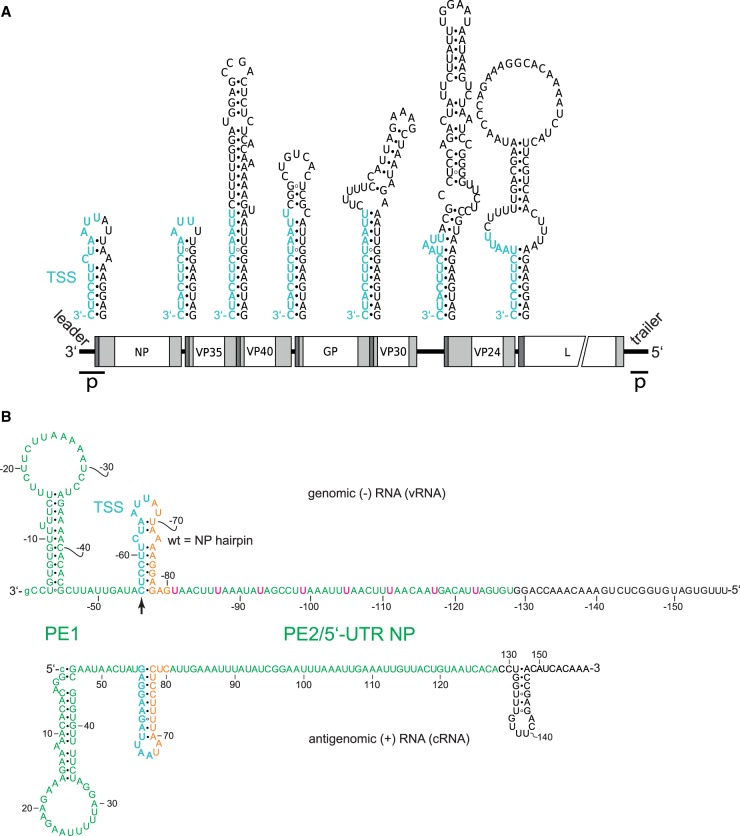
Mühlberger and coworkers (Weik et al. 2005) performed a mutational investigation of the genomic 3′-leader promoter in a replication-competent minigenome system. They concluded that the EBOV replication promoter is bipartite (Fig. 1B): Promoter element 1 (PE1) comprises the 3′-terminal 55 nt of the genomic leader up to the TSS directing transcription of the first NP mRNA; promoter element 2 (PE2) comprises eight consecutive 3′-UN5 hexamers in the region of nt −81 to −128. Mutational analysis of UN5 hexamers suggested that the five 5′-proximal UN5 hexamers in PE2 are sufficient for normal replication activity and the 5′-proximal three for residual replication activity (Weik et al. 2005). Finally, deletions and insertions of up to 12 nt in the spacer region between TSS and PE2 (Fig. 1B) revealed that only insertions or deletions of 6 or 12 nt, but not 1, 2, 3, 5, or 9 nt, are compatible with replication (Weik et al. 2005). This motivated Weik et al. (2005) to propose the “rule of 6” for the EBOV replication promoter, although the EBOV genome is not divisible by 6. The term “rule of 6” was originally coined in the paramyxovirus field to indicate the dependency of viral polymerase activity on hexamer phasing of the entire genome (Calain and Roux 1993; Kolakofsky et al. 1998). To avoid terminological confusion, we have preferentially used the term “hexamer phasing” instead of “rule of 6” for the EBOV system.
FIGURE 1.
Secondary structure formation potential at (A) transcription start regions of EBOV genes and (B) specifically in the genomic 3′-leader and the complementary antigenomic RNA. (A) Genomic sequence elements required for transcription (re)initiation are shown in cyan at the top; transcription is initiated opposite to the 3′-terminal C residue of the TSS. Schematic white boxes at the bottom mark the reading frames for EBOV proteins NP, VP35, VP40, GP, VP30, VP24 and L; light gray boxes indicate 5′- and 3′-UTRs, with dark gray areas depicting the position of the predicted secondary structures illustrated above the genome; p, location of leader and trailer promoters (Calain et al. 1999). (B) Validated secondary structures forming in the naked genomic leader RNA (top) and its complementary antigenomic RNA (bottom) (Weik et al. 2002, 2005; Schlereth et al. 2016); in the text and following figures, the genomic RNA is abbreviated as vRNA for viral RNA, and the antigenomic RNA as cRNA for complementary or copy RNA; the numbering of nucleotides in the genomic RNA strand is indicated by a minus sign preceding the nucleotide number. Proposed promoter elements 1 and 2 (PE1, PE2) are shown in green letters and the 3′-U residues of the eight UN5 hexamers in PE2 are highlighted in pink in the genomic RNA; orange nucleotides mark the spacer sequence between the transcription start sequence (TSS, in cyan) and PE2. The transcription initiation site on the genomic RNA is marked by the vertical arrow. Nucleotide numbering of the genomic RNA starts at the 3′-terminal nt (position −1) that is complementary to position 1 (5′-terminus) of the antigenome. The 3′-terminal G of the genome is shown as small letter to consider the recent finding that the presence of this nucleotide is not essential as the EBOV RNA polymerase initiates polymerization at the C residue preceding the 3′-terminal G residue (Deflubé et al. 2019).
However, the reference point for hexamer phasing in the promoter region remained unclear in the study by Weik et al. (2005), and the results left open whether hexamer phasing also pertains to viral transcription. In the present study, we sought to define such a reference point for hexamer phasing, how this pattern may functionally link PE1 and PE2 and if there are additional length, sequence and structural constraints for the region connecting PE1 and PE2. We addressed these questions by investigating a series of replication-competent and -deficient monocistronic minigenomes (MGs), in which the terminal part of the 5′-UTR coding region of the NP gene was mutated or replaced with that of internal EBOV genes and mutated variants thereof. In addition to reporter gene assays as a measure of mRNA synthesis by the EBOV polymerase complex, we conducted qRT-PCR to determine the levels of mRNA, genomic RNA (abbreviated as vRNA for viral RNA) and antigenomic RNA (abbreviated as cRNA for complementary or copy RNA) to assess effects on transcription and replication.
RESULTS
The EBOV genome and the potential RNA secondary structures harboring the seven EBOV TSS elements (highlighted in cyan) are depicted in Figure 1A. The current state of knowledge regarding PE1 and PE2 of the 3′-leader, including the eight 3′-UN5 hexamers in PE2, the TSS, the spacer sequence (in orange) and experimentally verified secondary structures (Weik et al. 2005; Schlereth et al. 2016) of the (naked) genomic RNA and its antigenomic copy are summarized in Figure 1B.
Replacing the NP hairpin structure with hairpin structures of internal EBOV genes
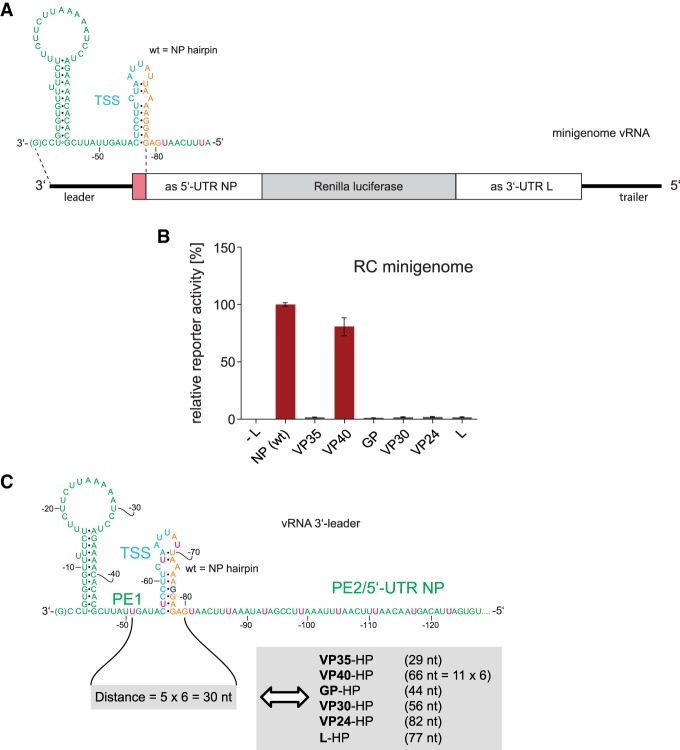
As a first approach to study the sequence, structure and spacing constraints for replication and transcription in the region between PE1 and PE2, we replaced the hairpin (HP) structure harboring the TSS of the first NP gene with the corresponding HP structures of internal EBOV genes in the replication-competent (RC) monocistronic MG (Fig. 2A). Remarkably, only the MG with the VP40 HP substitution gave rise to wild type-like reporter activity, whereas constructs with HP structures derived from the VP35, GP, VP30, VP24, and L genes failed to do so (Fig. 2B). When reinspecting the sequence of the 3′-leader, we noticed that the pattern of 3′-UN5 hexamers (termed UN5 hexamers in the following) could be extended from PE2 to position −51 in PE1 with a single interruption at G−75 (Fig. 2C). This perspective suggests that hexamer periodicity may already begin in PE1 and thus may functionally link PE1 and PE2. Indeed, the distance of U−51 to position −80 has a length of 30 nt in case of the wild type (wt) NP hairpin (Fig. 2C). In the construct with the VP40 hairpin, this distance is a multiple of 6 as well (66 nt), but in all other aforementioned constructs the distance deviates from this hexamer phase (Fig. 2C). This finding made it plausible to define U−51 as a reference point for UN5 hexamer phasing in the 3′-leader.
FIGURE 2.
(A) Schematic representation of the RC MG used in the presented study. The sequence complementary to the mRNA encoding the Renilla luciferase reporter mRNA is indicated by the gray box; the red segment corresponds to the hairpin structure shown at the top (TSS plus spacer sequence); white boxes mark sequences antisense (as) to the 5′-UTR of the NP mRNA and antisense to the 3′-UTR of the L mRNA. (B) Relative reporter activity of RC MGs in which the wild type (wt) NP hairpin was replaced with hairpin structures (see Figs. 1A, 3 for details) derived from the transcription start region of internal EBOV genes. Data (±standard error of the mean, SEM), normalized to the native NP (wt) construct, are based on at least three biological replicates with two or three technical replicates each. (C) Schematic representation of the 3′-leader promoter region depicting an extended UN5 hexamer pattern from PE2 to position −51 in PE1 with a single interruption at G−75, thus defining a reference point (nt −51) in PE1 and linking UN5 hexamer phasing between PE1 and PE2. Accordingly, the distance between nt −51 and −80 is 5 × 6 = 30 nt in the wt MG (light gray box on the left). Only the mutant construct with the VP40 HP maintained a hexamer phasing (66 nt) between PE1 and PE2, whereas mutant MG constructs harboring the VP35, GP, VP30, VP24, or L HP, respectively, resulted in spacings not divisible by 6 (light gray box on the right). For color coding of the sequence, see legend to Figure 1B.
Engineering of hairpins to comply with hexamer phasing
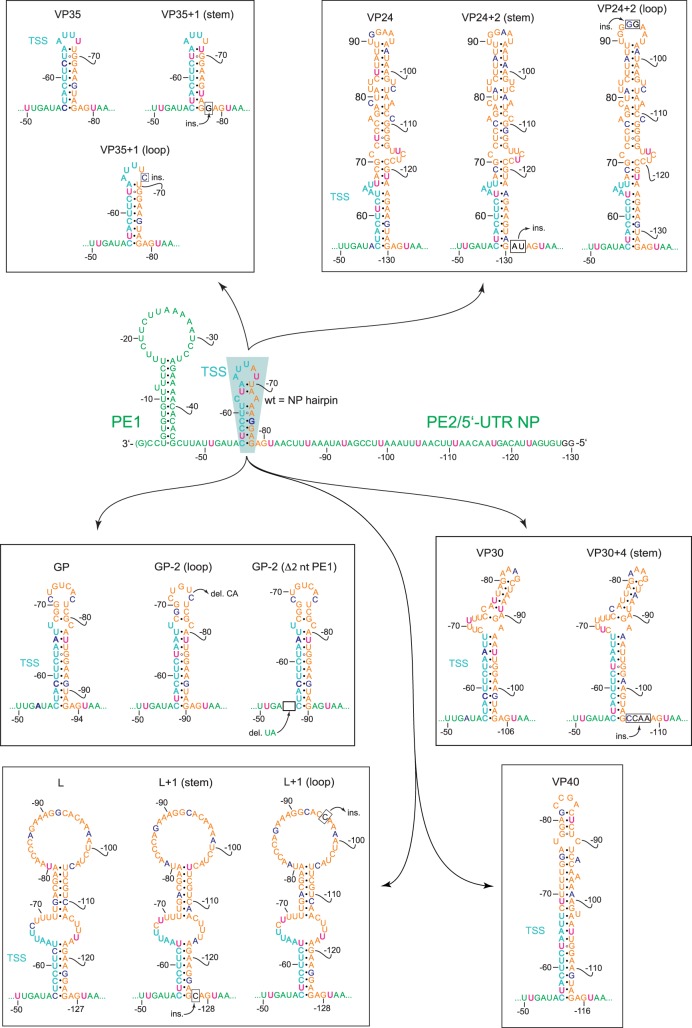
We next examined if the inactive constructs could be reactivated by adjusting the HP structures to a length that conforms to hexamer phasing (mutant constructs are illustrated in Fig. 3). Inserting 1 nt in the stem or loop of the VP35 HP to elongate the distance between U−51 and the first UN5 hexamer in PE2 from 29 to 30 nt indeed fully restored reporter gene activity (Fig. 4A). Basically the same pertains to the engineered VP30 HP (insertion of 4 nt in the stem), the L HP (1 nt insertion in the stem or loop) and the GP HP (deletion of 2 nt in the loop; restoration of reporter activity to ∼50% of wt). In contrast, deletion of nt −54 and −55 preceding the GP stem (Fig. 3) failed to restore activity (Fig. 4A, construct GP-2 [Δ2 nt PE1]), indicating inactivation of the promoter by deletion of nucleotides in PE1. This is in line with the finding that mutation of nucleotides −53 to −55 abolished the production of replicated RNA (Weik et al. 2005). For the VP24 HP construct, 2-nt insertions in the stem or loop to adjust hexamer phasing only weakly restored reporter activity. As the engineered VP24 HP expands the spacer region by 54 nt relative to the wt NP HP, this result may be attributable to length limitations for the distance between PE1 and PE2. The results shown in Figure 4A support the concept of using position −51 in PE1 as reference point for hexamer phasing in the 3′-leader promoter (Fig. 2C), which functionally links PE1 to PE2. In addition, the results demonstrate considerable tolerance for variation in the spacer sequence between PE1 and PE2 in terms of sequence (except for the conserved TSS), length and structure as long as hexamer phasing is maintained.
FIGURE 3.
Illustration of EBOV 3′-leader MG constructs in which the NP HP (turquoise-shaded area in the center) consisting of the TSS and spacer sequence was replaced with corresponding HP structures derived from the transcription reinitiation sites of internal EBOV genes. For the basic color coding and promoter structure, see legend to Figure 1B. Wild-type MG in the center: U residues that extend the PE2 hexamer phasing 3′ of nt −81 are shown in pink; G−75 interrupting UN5 hexamer continuity is shown in boldface blue; (ins.) insertion; (del.) deletion; insertions and deletions are indicated as boxes. The peripheral mutant constructs are depicted accordingly, with residues disrupting UN5 hexamer continuity highlighted in boldface blue as well.
FIGURE 4.
(A) Engineering of the spacer region to conform to hexamer phasing and (B) reverse engineering of the NP (wt) spacer region to violate hexamer phasing in RC MGs. (A) Luciferase reporter activities as a function of sequence/structure variation in the TSS/spacer region of MG 3′-leaders. Leaders containing the NP (wt) HP hairpin or native HP structures of internal EBOV genes instead are indicated by blue bars; HP structures violating hexamer phasing were engineered to restore the hexamer phase by deletion or insertion of nucleotides in the spacer region, either in the apical loop, stem, or between the stem and PE2 (gray bars). (B) Luciferase reporter activities of RC MGs carrying the NP (wt) HP or variants with 1-nt deletions or insertions, illustrated in the NP HP structure on the right (+: insertion; Δ: deletion) resulting in deviation from hexamer phasing. Values in (A) and (B) were normalized to the NP (wt) 3′-leader as control (100%). As a negative control (−L), the plasmid encoding the L gene was omitted in transfection of cells with the set of plasmids encoding EBOV proteins relevant to replication and transcription. Mean values (±SEM) are based on three independent experiments with at least three technical replicates each.
We further pursued a reverse engineering approach, where we inserted or deleted a single nucleotide in the native NP HP (Fig. 4B). This was done to exclude any influence of sequence and length variation that might have contributed to the functional readout of the constructs in which the wt NP HP was replaced with hairpins of internal EBOV genes. This 1-nt insertion or deletion, either in the stem or loop of the NP HP, abolished reporter activity, verifying that conformance with hexamer phasing is a basic prerequisite for reporter activity.
Hexamer phasing in the context of replication-deficient minigenomes
In reverse genetic systems using RC MGs, the amplitude of reporter gene expression is not only a measure of viral transcription, but also includes contributions from viral replication that leads to the synthesis of new vRNA nucleocapsids that are efficient templates for transcription (Hoenen et al. 2010). To provide further evidence that hexamer phasing also applies to transcription, we tested the constructs described above in the context of a replication-deficient (RD) MG (Hoenen et al. 2010). In the RD MG system, where only transcription takes place, viral mRNA levels are much lower than in the RC MG system, most likely due to low-efficiency assembly of functional nucleocapsids from minigenomic vRNAs produced by T7 RNA polymerase and coexpressed NP protein in the absence of viral replication (Blakqori et al. 2003). Only replicative synthesis of new vRNA by the viral polymerase in the RC MG system will direct the assembly of functionally more competent nucleocapsids that are much more efficient templates for viral transcription. We tried to quantify the viral mRNA levels for the RD NP (wt) MG construct by qRT-PCR, but could not determine viral mRNA-specific CT values sufficiently above background (−L control): This might be attributable to T7 RNA polymerase also synthesizing some antiminigenomic RNAs that are recognized by the RT primers specific for the viral RNAs (cRNA, mRNA). Comparable experiences were made in the previous study (Hoenen et al. 2010). Nevertheless, viral mRNA levels in the RD MG system were sufficient for reporter enzyme activity measurements reliably above background. As a result, we observed the same picture in the RD versus RC MG setup: The VP40 construct gave rise to NP (wt)-like reporter activity (and, by inference, to viral mRNA transcription), whereas other replacement constructs, such as those harboring the VP35, GP or VP30 HP, failed to do so (Fig. 5A). Likewise, engineered HP constructs designed to restore hexamer periodicity also restored reporter activity to similar relative extents as in the RC system, as exemplified for the VP35, GP and VP30 hairpins (cf. Figs. 4A, 5B). Conversely, 1-nt deletions or insertions in the wt NP HP abolished reporter activity (Fig. 5C). As reporter activities in the RD MG system depend on viral transcription in the absence of replicative vRNA synthesis, and assuming that translation efficiencies of the tested mRNA variants as well as their half-lifes were comparable, our findings indicate, for the first time, that hexamer phasing is mandatory for EBOV transcription.
FIGURE 5.
Luciferase reporter activities as a function of compliance with hexamer phasing in the 3′-leader of RD MGs. (A) Performance of MG 3′-leaders in which the NP (wt) HP was replaced with HP structures derived from the transcription start region of internal EBOV genes, normalized to the MG carrying the NP (wt) 3′-leader; bar color: conforming (blue) or not conforming (gray) to hexamer phasing. (B) MG 3′-leaders with HP structures of internal EBOV genes, which were mutated by deletion or insertion of nucleotides to conform to hexamer phasing (red bars). (C) RD MGs carrying the NP (wt) HP or variants with 1-nt deletions or insertions (for more details, see Fig. 4B). Mean values (±SEM) are based on three independent experiments with at least three technical replicates each. (***) P < 0.001 (unpaired t-test).
Residual polymerase activity upon disruption of hexamer phasing
Using qRT-PCR, we quantified the levels of mRNA, cRNA, and vRNA in the RC MG system for the wt, VP35, VP35 + 1 (loop), NP-1 (stem), and the GP-2 (Δ2 nt PE1) constructs relative to the L omission control. The reporter activities measured for these variants in the RC and RD MG systems, already shown in Figures 4, 5, were included in Figure 6A,B for immediate comparison with the qRT-PCR data. Individual constructs showed very similar relative differences in reporter activity in the RD and RC MG setups (Fig. 6A,B). However, the absolute amounts of viral mRNA were two to three orders of magnitude higher (this study; Hoenen et al. 2010) in the RC versus RD MG system as discussed above, indicating that viral replication yields new copies of genomic vRNA, each of which is efficiently transcribed multiple times. Thus, viral replication boosts transcription, although the majority of RNA synthesis products are viral mRNAs as illustrated by comparison of 2−ΔCT values for mRNA versus cRNA and vRNA in the RC MG system (Fig. 6C). The relative levels of mRNA, which could only be reliably quantified in the RC system (Fig. 6D), matched the pattern seen for reporter activity: mRNA levels in cells transfected with the VP35, NP-1 (stem), or GP-2 (Δ2 nt PE1) construct were as low (GP-2 [Δ2 nt PE1]) or almost as low as in the background control (L omission), whereas the VP35 + 1 (loop) construct, reengineered to conform to hexamer phasing, restored wt-like mRNA levels (Fig. 6D). The reduction of mRNA levels essentially to background levels in the case of the GP-2 (Δ2 nt PE1) construct reveals that deletion of nt −54 and −55 in PE1 essentially prevents any viral transcription activity in the 3′-leader. In turn, mRNA levels above background for the other two constructs with disrupted hexamer phasing (VP35, NP-1 [stem]) suggests residual activity of the viral RNA polymerase despite noncompliance with hexamer phasing. However, the extent of this hexamer phasing-independent transcription seems to be context-dependent; this is suggested by higher mRNA levels for the VP35 versus NP-1 (stem) variant (Fig. 6D).
FIGURE 6.
Impact of hexamer phasing in the 3′-leader on transcription and replication. (A) Relative reporter activity of RC MGs with replacements of the NP (wt) HP as indicated (for structural details of leader variants, see Figs. 3, 4B). (B) As in panel A, but the same 3′-leader variants integrated into the RD MG scaffold. Mean values (±SEM), normalized to the native NP (wt) leader construct, are based on three biological replicates with two or three technical replicates each. Above each column, we calculated the activity (in %) of the individual mutant MG relative to the NP (wt) MG (=100%) after subtraction of the −L background from both. The diagrams in panels A and B combine results already shown in Figures 4 and 5. (C) Mean 2−ΔCT values obtained with RNA isolated from cells transfected with the NP (wt) MG to illustrate the abundance of viral mRNA (pink), cRNA (magenta), and vRNA (green), measured by a two-step strand-specific qRT-PCR of RC MG samples using the same cells as in panel A; the presence (+) or absence (−) of L is indicated below the x-axis. 2−ΔCT values were calculated for each experiment individually; mean 2−ΔCT values (±SEM) were derived from six to 16 independent experiments with three technical replicates each. 2−ΔCT values for mRNA were calculated as follows, based on the mean CT (±SEM) values of 14.52 ± 0.15 for mRNA + cRNA, 20.97 ± 0.11 for cRNA, and 23.42 ± 0.09 for firefly luciferase mRNA: CT (mRNA + cRNA) minus CT (firefly luciferase mRNA), that is, 14.52 − 23.42 = −8.9, corresponding to a 2−ΔCT value of 477.7; CT (cRNA) minus CT (firefly luciferase mRNA), i.e. 20.97 − 23.42 = −2.45, corresponding to a 2−ΔCT value of 5.46; subtracting 5.46 from 477.7 then gives the 2−ΔCT value of 472.24 for mRNA. Above the cRNA and vRNA columns (+L), we calculated the mole percentage for cRNA and vRNA relative to mRNA (pink column) after subtraction of the corresponding −L background controls. For the binding sites of primer pairs, see Supplemental Figure S2. (D–F) Relative levels of viral mRNA, cRNA, and vRNA using the same cells as in panel A and normalized to the NP (wt) construct. Mean 2−ΔΔCT values (±SEM) were derived from at least three independent experiments with two or three technical replicates each. Dotted horizontal lines mark the level of the −L control. For experimental details, see Materials and Methods.
Among the two variants with disrupted hexamer phasing, cRNA and vRNA levels, after subtraction of background (−L control), were higher for the VP35 than for the NP-1 (stem) variant (Fig. 6E,F), in line with the corresponding hierarchy of mRNA levels (Fig. 6D). Surprisingly, particularly the vRNA levels for the GP-2 (Δ2 nt PE1) variant were substantial (Fig. 6F), in contrast to the essentially insignificant mRNA levels (Fig. 6D). This may suggest that the deletion of nt −54/55 impairs transcription more severely than replication.
In summary, the results presented in Figure 6C illustrate that the majority of RNA products generated by the viral RNA polymerase in the RC MG system are mRNAs and viral replication boosts transcription. The data in Figure 6C suggest that each vRNA nucleocapsid newly synthesized by the viral polymerase serves as template for ∼13 rounds of mRNA synthesis. Polymerization activity is highly sensitive to violation of hexamer phasing, but residual and context-dependent polymerase activity still occurs upon deviation from the hexamer phase; a dinucleotide deletion in PE1, immediately preceding the TSS (construct GP-2 [Δ2 nt PE1]), essentially abolished transcription.
Extension of the UN5 hexamer pattern
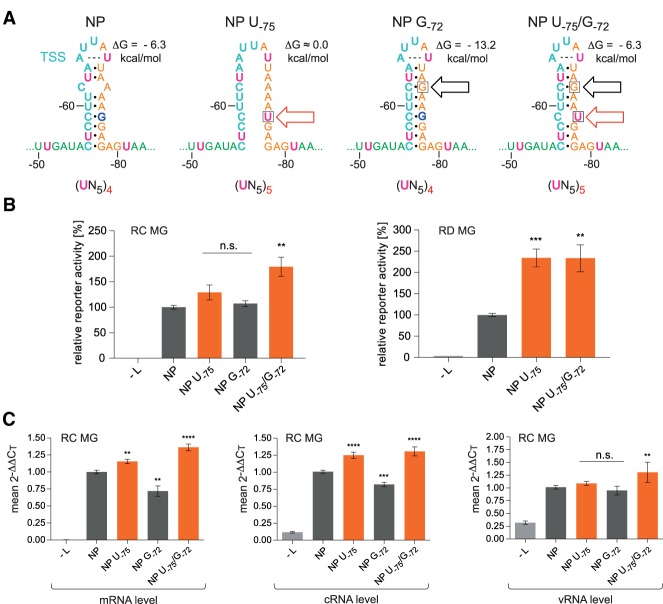
We next addressed the question whether extension of the UN5 hexamer pattern between PE1 and PE2 may influence reporter activity. UN5 hexamer phasing is continuous from nt −51 in PE1 to PE2 with a single interruption at G−75 (Fig. 2C). We thus mutated G−75 to U, resulting in variant NP U−75. As RNAfold and Mfold predicted almost complete disruption of secondary structure for this variant, we constructed variant NP U−75/G−72, in which a second mutation (G−72) introduced a G:C bp into the upper stem region to compensate for the destabilizing effect of the U−75 mutation (Fig. 7A). The hairpin stability of variant NP U−75/G−72 is predicted to be similar to that of the wt NP hairpin. We further constructed variant NP G−72 carrying only the G−72 mutation (Fig. 7A) to control for stability effects. Variants NP U−75 and NP U−75/G−72 with continuous UN5 hexamer phasing gave rise to increased reporter activity in RC and RD MG systems relative to the NP and NP G−72 controls (Fig. 7B). This correlated with increased mRNA and cRNA levels in the RC system; corresponding increases in the vRNA level were less pronounced (Fig. 7C). The latter result might be attributable to the narrower dynamic range of variations in the amount of vRNAs that are presynthesized by the T7 RNA polymerase in the MG system. Relative increases in reporter activity for the U−75 variants were particularly pronounced in the RD MG system, again consistent with the notion that continuous UN5 hexamer phasing strongly impacts viral transcription.
FIGURE 7.
UN5 hexamer continuity between PE1 and PE2 increases viral transcription and replication. (A) Sequence and predicted structure of the wt NP HP (left), variant NP U−75 and the stabilized variants NP G−72 and NP U−75/G−72; the red arrow indicates the mutation that extends UN5 hexamer phasing and the black one marks the mutation at position −72 that stabilizes the HP stem by converting the A,C mismatch to a G:C bp. The number of UN5 hexamers between nt −51 and −80 is indicated below each individual structure. The ΔG values of the stem–loops are the minimum free energies of the centroid secondary structures predicted by RNAfold for the sequence of nt −52 to −81 using the default parameters (http://rna.tbi.univie.ac.at/cgi-bin/RNAWebSuite/RNAfold.cgi). The dashed line connecting A−65 and U−69 indicates base-pairing as suggested by RNAfold. (B) Corresponding luciferase activities of RC (left panel) or RD (right panel) MGs carrying the NP variants illustrated in panel A. Activity values are given in % relative to the native 3′-leader (NP [wt] = 100%). As a negative control, the plasmid encoding the L gene was omitted (−L) during transfection. (C) Corresponding levels of viral mRNA, cRNA, and vRNA measured by a two-step strand-specific qRT-PCR of RC MG samples using the same cells as in panel B. For more details, see Materials and Methods. In panels B and C, mean 2−ΔΔCT values (±SEM) were derived from three independent experiments with at least three technical replicates each. (**) P < 0.01; (***) P < 0.001; (****) P < 0.0001; n.s., not significant (unpaired t-test).
We conclude that increasing the number of UN5 hexamers between nt −51 and −80 can enhance transcriptional but also replicative activity of the viral polymerase.
DISCUSSION
The 3′-leader regions of NNS RNA viruses harbor essential information for viral RNA synthesis: promoters that allow either transcription of viral mRNAs or enable replication into full-length genomes (vRNA) via antigenomes (cRNA) as intermediates. Genomic promoter architectures of NNS viruses were divided into two groups: monopartite replication promoters confined to the leader 3′-end (Rhabdoviridae and Pneumoviridae) and bipartite replication promoters (Paramyxoviridae, Filoviridae) harboring a second more internal element separated from the first by the TSS and a spacer sequence of variable length (Tapparel et al. 1998; Mühlberger 2007; Fearns and Plemper 2017). Here we studied the length, sequence and structural constraints in the region connecting PE1 and PE2 of the bipartite EBOV promoter, with the primary focus on viral transcription. We could demonstrate, for the first time, that UN5 hexamer phasing is also essential for EBOV transcription, that UN5 hexamer periodicity can be extended into PE1 and that the spacer region can be expanded by at least 48 nt without losses of transcriptional activity. Making the UN5 hexamer phasing continuous between PE1 and PE2 enhanced the efficiency of genome transcription and replication.
The EBOV 3′-leader has the potential to form stable secondary structures (Fig. 1B), raising the question whether not only sequence but also structure of PE1 contributes to promoter recognition by the EBOV polymerase. Point mutations in the 3′-terminal hairpin stem (nt −1 to −45; Fig. 1B, genomic RNA) which disrupt single base pairs had no or only a moderate effect on viral polymerase activity; the strongest effect was seen for a G-9 to C mutation. Likewise, double and triple mutations weakening this hairpin stem showed at best moderate effects on viral transcription/replication (Crary et al. 2003). Using a minigenome setup similar to the one used in the study presented here, Mühlberger and coworkers (Weik et al. 2005) found that changing four consecutive U residues (−10 to −13, Fig. 1B) to adenosines abolished replication; this also pertained to a construct with four additional compensatory mutations (A to U; nt −36 to −39 in Fig. 1B) restoring base-pairing in the stem. Likewise, changing the base identity of nucleotides 44–46, 50–52, or 53–55 abolished the production of replicated RNA (Weik et al. 2005). Together with the results of Crary et al. (2003), these findings led to the conclusion that the 3′-terminal 55 nt (=PE1) of the genomic leader up to the TSS of the first (NP) gene are essential for replication, but it appears that the PE1 sequence rather than its secondary structure is crucial for promoter function.
In the presented study, we observed that deletion of the two 5′-terminal nucleotides in PE1 (nt −54 and −55; construct GP-2 [Δ2 nt PE1], Fig. 5B) immediately preceding the TSS abolished viral transcription, although this construct adhered to hexamer phasing. One possible interpretation is that PE1 has not only replication but also transcription promoter function, similar to what is known for other NNS RNA viruses (for review, see Noton and Fearns 2015). In view of the finding by Weik et al. (2005) that changing the base identity of nucleotides 44–46, 50–52, or 53–55 in particular abolishes the production of replicated RNA, it is not unlikely that sequences in PE1 are recognized by the viral polymerase in its replication and transcription mode. To our knowledge, the structural requirements of a filoviral transcription promoter were not specifically addressed in any previous studies. Yet another interpretation of the Δ −54/−55 phenotype could be that the transcription start site must be in the correct hexamer phase relative to the genome 3′-end. This possibility will be tested in future experiments by single nucleotide substitutions at positions −54 and −55. Generally, the features of PE1 that are recognized by the EBOV polymerase in the replication versus transcription mode need to be unraveled. Remarkably, the EBOV transcription promoter appears more similar to promoters of rhabdoviruses, such as VSV, than that of paramyxoviruses with respect to the following aspect: Comparable to EBOV, the entire 3′-terminal 50 nt of the VSV genome, including nt −47 to −50 immediately preceding the TSS, harbor signals for transcription (Whelan and Wertz 1999). In paramyxoviruses, however, a core transcription promoter could be defined spanning only the 3′-terminal 12 nt of the viral genome (Fearns et al. 2002; Cowton and Fearns 2005; Tremaglio et al. 2013). Substitution of nt −13 to −36 was neutral and nucleotide identities at positions −37 to −44 immediately preceding the TSS were not essential for transcription, although contributing to transcription activity (McGivern et al. 2005).
Current models of bipartite 3′-leader promoters, developed for paramyxoviruses (Murphy et al. 1998; Tapparel et al. 1998; for reviews, see Noton and Fearns 2015; le Mercier and Kolakofsky 2019), predict that 3′-terminal and internal promoter elements (resembling PE1 and PE2, Fig 1B) need to be juxtaposed on the same vertical face of the nucleocapsid helix for concerted recognition by the viral polymerase. This model is based on the finding that the paramyxoviral nucleocapsid contains 13 NP molecules per helical turn and NP molecules 14–16 (counted from the 3′-end) bind to the three PE2 hexamers (3′-CN5 in the case of Sendai virus [SeV]). Displacement of the three PE2 hexamers by deletion of 6 nt in the spacer between PE1 and PE2 abolished SeV genome replication (Tapparel et al. 1998). Similar defects were observed for Simian virus 5 (SV5) upon deletion of 6 to 18 nt in the spacer between PE1 and PE2 (Murphy et al. 1998). On the other hand, 6-nt insertions in this region of the SeV promoter were tolerated, but insertion of 12 nt largely ablated promoter activity (Pelet et al. 1996).
For EBOV, relaxed nucleocapsids formed in the presence of NP alone were inferred to have 26 ± 2 NP subunits per helical turn, whereas those assembled in the presence of NP, VP24, VP35, and VP40 are condensed having 13 or 14 per turn (Wan et al. 2017; Sugita et al. 2018). The relaxed nucleocapsids are considered to be present during active transcription and replication (Hoenen et al. 2019), consistent with viral RNA synthesis taking place in the MG system lacking VP24 and VP40. Taking into account that the EBOV NP protein binds 6 nt per NP monomer (Wan et al. 2017; Sugita et al. 2018) as in other NNS systems, the model of concerted polymerase binding to PE1 and PE2 juxtaposed on the same vertical face of the nucleocapsid helix appears questionable for filoviruses: The spacer preceding PE2 can be expanded by up to 48 nt or 8 NP monomers (L + 1 variants, Fig. 4A) without apparent losses in viral transcription; in this construct, PE1 and PE2 are expected to be considerably displaced from each other in the nucleocapsid helix, making the concomitant recognition of PE1 and PE2 hard to imagine. Alternatively, the filoviral RNA polymerase might be more flexible than its paramyxoviral counterparts in being able to loop out longer spacer insertions during recognition of PE1 and PE2; or the extended HP structures can form and protrude from the nucleocapsid in the process of promoter recognition to bring PE1 and PE2 closer to each other.
It is interesting to mention that a Gln202 to Ala mutation in the NP protein of the human parainfluenza virus type 2 (hPIV2, paramyxovirus) allowed promoter utilization of the viral polymerase in the absence of the PE2 element which was then largely independent of a genome length divisible by 6. In contrast, promoter utilization was strictly dependent on PE2 and genome hexamer phasing in the presence of wild type NP (Matsumoto et al. 2017, 2018). This observation provides evidence that PE2 is not essential for promoter function but may rather exert a regulatory function. The authors proposed that Gln202 of hPIV2 NP can form a base-specific contact to the 3′-terminal base of a bound RNA hexamer (particularly to the uracil at the genome 3′-end) to control that viral replication/transcription can only take place when the correct hexamer register is ensured. The finding of 3′-UN5 hexamer periodicity in the EBOV system suggests that such base-specific contacts of NP might also control promoter utilization by the filoviral polymerase.
Making UN5 hexamer periodicity continuous between nt −51 and −80 by a single point mutation in the context of the native NP construct increased reporter activity as well as the levels of mRNA and cRNA (Fig. 7). This effect was very similar for a largely destabilized NP HP structure and for a variant predicted to be of similar stability as the native NP HP, suggesting that UN5 hexamer periodicity is functionally more important than the potential to form a stable hairpin structure. It will be interesting to explore if the enhancement of polymerase activity upon completing UN5 hexamer periodicity between PE1 and PE2 may occur on the level of polymerase initiation frequency, kinetics of elongation or reduced premature termination of polymerization.
It was reported that EBOV mRNAs contain upstream AUG (uAUG) codons that could potentially attenuate translation initiation at the main AUG. This kind of regulation was found to be most pronounced at the L gene with a relatively short 5′-UTR (80 nt), where the upstream ORF (uORF) coding for a 22-aa peptide overlaps with the L ORF (Shabman et al. 2013). The uORF mitigated L translation under normal conditions but elevated L expression under stress conditions. Such translational effects of uORFs could also potentially affect reporter activities measured in the MG system. In our MG, the Rluc gene was preceded by the ∼400 nt long 5′-UTR of the NP gene and structural alterations were confined to the very 5′-end of this 5′-UTR. In the case of our L MG constructs (Fig. 3), the uAUG introduced by the L hairpin encodes peptides of 22 aa (L and L + 1 [loop] constructs) and 16 aa (L + 1 [stem] construct) that are terminated 360–380 nt upstream of the Rluc AUG and thus do not overlap with the main ORF as it is the case in the native L gene (Shabman et al. 2013). We do not see evidence that the uORF in our L constructs might have had a substantial effect on the presented results, considering that reporter activities of the L + 1 (loop) and L + 1 (stem) constructs were in the same range as that of the NP (wt) construct (Fig. 4A). As the Rluc gene was preceded by the ∼400 nt long 5′-UTR of the NP gene in all of our MG constructs, any uORFs introduced by insertions or deletions in the transcription start region have their stop codons at considerable distance to the AUG of the Rluc gene, making their direct interference with Rluc translation unlikely. Another possibility is that the sequence and structural alterations introduced into the 3′-leader of our mutant minigenomes might have affected mRNA stability (or translation efficiency) rather than viral transcription per se. At present, we cannot exclude that such an effect might have contributed to the phenotypes of some variants such as VP24 + 2 (stem) and VP24 + 2 (loop) (Fig. 4A).
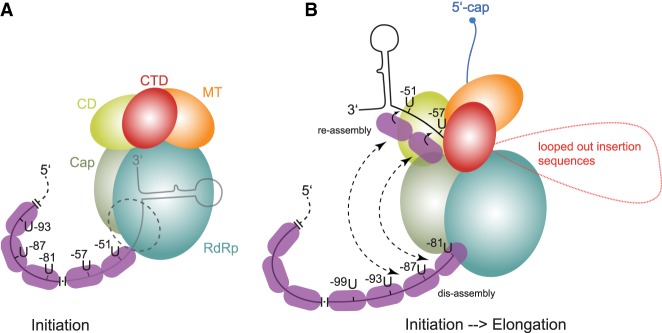
We found that hexamer phasing in the EBOV 3′-leader promoter not only applies to replication, but also transcription, and that UN5 hexamer periodicity can be extended, with interruption, into PE1. So far, we have shown that the distance between PE1 and PE2 can be expanded by up to 48 nt without substantially affecting viral polymerase activity. How can the results be explained? It appears obvious that the hexamer phase is linked to the capacity of each NP protein to bind exactly 6 nt (Wan et al. 2017; Sugita et al. 2018). In addition, the hexamer phasing pertains to transcription and replication, suggesting that a functional aspect common to both processes is affected. We like to put up two models for discussion. In both models we consider that the discrepancy between the hexamer phase in the leader promoter and EBOV genome length being unequal to n × 6 may imply that those NP molecules binding to the genome ends may either cover <6 nt (le Mercier and Kolakofsky 2019), or up to a few of the terminal nucleotides remain uncovered by NP. In the first model we assume that there is essentially no replication or transcription activity (not even abortive transcription) of the EBOV polymerase when hexamer phasing in the promoter is violated. Here, the polymerase binds the ∼55 3′-terminal nucleotides (PE1) of the genome to position the genome 3′-end in the active site to be able to initiate RNA synthesis (Fig. 8A). Proper positioning of the enzyme on the template strand is only achieved when NP is assembled in the correct register, directed by the UN5 hexamers, explaining why 6 or 12 nt of the spacer can be deleted or the spacer be expanded by multiples of 6 (Weik et al. 2005; this study). The mechanism how NP monomers are directed into this register during genomic RNA synthesis is yet unclear as NP is thought to bind RNA in a sequence-independent manner (Sugita et al. 2018). A possibility is that the UN5 hexamers are preferred NP binding sites and consecutive UN5 hexamers are cooperatively bound by several NP monomers, based on evidence that each NP monomer may form an H bond to the 3′-nucleotide of the hexamer bound by the neighboring NP molecule in the nucleocapsid (Sugita et al. 2018).
FIGURE 8.
Models to explain the hexamer phasing, suggesting a key role for NP which binds 6 nt per monomer. The sketches are drawn according to the cryo-EM structure of the VSV polymerase (Liang et al. 2015). (A) This model assumes that there is essentially no replication or transcription activity (not even abortive transcription) of the EBOV polymerase when hexamer phasing is abolished. Here, the polymerase binds the ∼55 3′-terminal nucleotides (PE1) of the genome. Proper positioning of the enzyme on the template strand is only achieved when NP is assembled in the correct register, directed by the UN5 hexamers, explaining why the spacer can be expanded by multiples of 6. (B) This model, which is mechanistically largely speculative, may reflect the transition from the initiation to the elongation mode of the polymerase. It assumes that the enzyme starts to polymerize upon positioning of the genome 3′-end in the active site; when the genome 3′-end leaves the template exit channel, the enzyme may hold on to the first UN5 hexamers (U-51, U-57); as soon as UN5 hexamers enter the template entry path, they will be bound as well to trigger communication between the entry and exit path; hexamer periodicity may synchronize reassembly at the exit site and dis-assembly at the entry site; expansion sequences in the spacer region that may loop out are shown in red. The nascent mRNA transcript is shown in blue; in the VSV system, the nascent mRNA transcript is capped when reaching a length of 31 nt (Tekes et al. 2011). For more details, see text.
The second model, which is more speculative, may reflect the transition from the initiation to the elongation mode of the polymerase. (Fig. 8B). It assumes that the enzyme starts to polymerize upon positioning of the genome 3′-end in the active site. When the genome 3′-end leaves the template exit channel, the enzyme may hold on to the first UN5 hexamers (U-51, U-57); as soon as UN5 hexamers enter the template entry path, they will be bound as well to trigger communication between the entry and exit path; the hexamer periodicity may synchronize reassembly at the exit site and dis-assembly at the entry site, possibly involving transfer of NP molecules, which are separated from the template RNA at the entry site, to the protruding UN5 hexamers at the template exit site. It is further conceivable that NP molecules, dis-assembled at the template entry channel, form a transiently RNA-free NP chain that interacts with the polymerase's surface to mediate coordinated translocation of the enzyme in a hexamer phase-dependent manner (le Mercier and Kolakofsky 2019). Expansion sequences in the spacer region may loop out as shown in red. It is unclear whether the 3′-terminal RNA hairpin shown in both models forms transiently or at all. Both models shown in Figure 8 are consistent with the fact that template strand passage through the active site is a commonality of viral replication and transcription, whereas other functional aspects largely differ between the two processes. In contrast to the replication mode, the transcribing polymerase recognizes gene start and end sequences, adds a 5′-cap and monomethylates the mRNA's 5′-end (formation of a cap 1 structure; Fearns and Plemper 2017), polyadenylates the transcript and releases the newly synthesized mRNA without NP packaging. In contrast to viral mRNAs, replicative cRNA is fully encapsidated by NP during RNA synthesis in NNS viral systems (Ogino and Green 2019).
MATERIALS AND METHODS
Cell culture and EBOV-specific minigenome systems
Human embryo kidney cells (HEK293; authenticated as cell line 293 [DMSZ ACC 305]) were cultivated at 37°C and 5% CO2 in a humidified atmosphere in Dulbecco's modified Eagle medium (DMEM) containing 50 U/mL penicillin, 50 µg/mL streptomycin, 2 mM l-glutamine and 10% FCS (all from Thermo Fisher Scientific [TFS]). For transfection, 8 × 105 HEK293 cells were seeded in each well of a six-well plate (Greiner) using the aforementioned medium (3 mL/well) and cultivated for 18–24 h until ∼60%–80% of cell confluency was reached. Then transfection of plasmid mixtures was conducted in the same growth medium using the TransIT-LT1 Reagent (Mirus) without media exchange post-transfection according to the manufacturer's recommendations. As described previously (Mühlberger et al. 1999), the transfection mixtures included plasmids encoding the EBOV nucleocapsid proteins NP (125 ng), VP35 (125 ng), VP30 (100 ng), L (1000 ng), the respective T7 promoter-driven EBOV-specific minigenome variant coding for Renilla luciferase (250 ng) as well as a plasmid encoding T7 RNA polymerase (250 ng). Additional transfection of 100 ng pGL4.13 (Promega) encoding a firefly luciferase was performed for normalization of transfection efficiencies. For TransIT-LT1:DNA complex formation, 6 µL TransIT-LT1 Reagent were added to a premix of 1950 ng plasmid DNA and 200 µL Opti-MEM I Reduced-Serum Medium (TFS), followed by incubation at room temperature for 15–30 min before drop-wise addition to the HEK293 cells. Replication-competent minigenomes (RC MGs) included the native 3′-leader and 5′-trailer sequences. In contrast, replication-deficient minigenomes (RD MGs) lacked the terminal 55 nt of the antigenomic trailer promoter, thus restricting the viral polymerase to transcription and synthesis of antigenomic copy RNA (cRNA), but excluding the production of new genomic vRNA. As a consequence, measured reporter activities are solely based on viral transcription in RD MG systems. Reporter activities were measured 48 h post-transfection (p.t.) in Renilla and Firefly luciferase assays (both purchased from PJK).
Plasmids
Derivatives of plasmid pCAGGS coding for the Zaire EBOV nucleocapsid proteins (VP30, NP, VP35, and L), the T7 RNA polymerase, the EBOV-specific MG (pANDY 3E5E), as well as an EBOV RD MG, were described earlier (Hoenen et al. 2006, 2010). The E. coli DH5α strain was used for cloning and propagating of plasmids applying standard microbiological procedures.
Construction of replication-competent (RC) and replication-deficient (RD) minigenome variants
All 3′-leader MG variants are based on the negative-sense EBOV-specific MG pANDY 3E5E (Hoenen et al. 2006). The native NP (wt) hairpin, whose 3′-end coincides with the transcription initiation site, was exchanged or mutated in pANDY 3E5E using Dpn I-based site-directed mutagenesis techniques (see Supplemental Fig. S1 for details). The used primers are specified in the Supplemental Table S1. Corresponding RD MGs were constructed by inserting the 3′-leader variants into a pANDY 3E5E derivative plasmid lacking the terminal 55 nt of the trailer replication promoter at the 3′ end of the antigenome (Hoenen et al. 2010) by use of standard restriction enzyme-based approaches or site-directed mutagenesis. Nucleotide numbers refer to the EBOV Mayinga/Zaire/1976 sequence, GenBank accession no. AF086833. All constructs were verified by DNA sequencing.
Luciferase assay
For MG activity measurements, cells were lysed in 200 µL 1× Reaction Lysis Buffer (2× Lysis-Juice; PJK). For measurement of Renilla Luciferase activity, the lysates were diluted 1:50 in ddH20 in case of RC MG samples. Diluted lysates (10 µL) were mixed with 50 µL of Renilla Luciferase Reagent (Renilla-Juice Fluid, mixed with coelenterazine substrate in reconstruction buffer according to the manufacturer's protocol; PJK). For cells transfected with RD MGs, 10 µL of undiluted lysate were mixed with 50 µL of Firefly Luciferase Reagent (Beetle-Juice; PJK). Luciferase activities were measured using a Centro LB 960 luminometer (Berthold Technologies). To account for potential differences in transfection efficiency, Renilla luciferase values were normalized to Firefly luciferase values. Results obtained for MGs carrying the native NP (wt) leader were set to 100%.
RNA extraction and purification for qRT-PCR experiments
HEK293 cells were transfected with the components of an EBOV-specific MG system (Biedenkopf et al. 2013). Cells were harvested at 48 h p.t. and RNA was isolated using the RNeasy mini kit (QIAGEN) according to the manufacturer's protocol, including a first on-column digestion for 1 h using the RNase-Free DNase Set (QIAGEN). RNA was eluted in 40 µL RNase-free ddH2O. A second DNase treatment in solution was performed using Ambion DNase I in the presence of 20 U RiboLock RNase Inhibitor (both from TFS) at 37°C for 1 h in a total reaction volume of 60 µL, followed by purification with Roti-Phenol/Chloroform/Isoamyl alcohol (Carl Roth). RNA was precipitated by addition of three volumes EtOH containing 0.1 M NaOAc (pH 5). The RNA pellet was washed with 70% EtOH, air-dried and finally redissolved in RNase-free water.
Reverse transcription
The strategies for the detection and quantification of the different virus-derived RNAs are schematically illustrated in Supplemental Figure S2. Five hundred nanograms of RNA were used for reverse transcription (RT) of either negative strand RNA (vRNA, primer luc [+] 5′-GGCCTCTTCTTATTTATGGCGA-3′) or positive strand RNAs (cRNA/mRNA, primer luc [−] 5′-AGAACCATTACCAGATTTGCCTGA-3′). Primer sequences for reverse transcription of vRNA and cRNA/mRNA were adapted from Kruse et al. (2018). In addition, the primer RT_cRNA 5′-CAGTCCTGCCTTTTCTTTTAATTTTATC-3′, specific for the cRNA trailer region, was used, and Firefly luciferase mRNA (encoded by the cotransfected pGL4.13 plasmid used for normalization of transfection efficiency) was reverse-transcribed with the Random Hexamer Primer set (TFS). Reactions were conducted with the RevertAid H Minus Reverse Transcriptase (TFS) according to the manufacturer's protocol, including a denaturation step at 65°C for 5 min as recommended for structured RNAs; cDNAs were diluted 1:10 for quantitative real-time PCR and 2 µL of the diluted cDNA (∼5 ng) were used in the respective PCR reaction.
Quantitative real-time PCR
Quantitative real-time PCR was performed in a total volume of 10 µL on a QuantStudio3 Real-Time PCR System (TFS) using the PowerUp SYBR Green Master Mix 2× (TFS). For amplification of vRNA and (+)RNA (cRNA and mRNA), primers luc (+) and luc (−) were used (see above), yielding a PCR product of 112 bp. For specific amplification of cRNA, primers RT_cRNA (see above) and qPCR_cRNA (5′-CGGTGATAGCCTTAATCTTTGTG-3′) were used, yielding a PCR product of 118 bp (see Supplemental Fig. S2 for primer annealing sites). For amplification of Firefly luciferase mRNA, primers qPCR_FF_fwd (5′-CGTGCAAAAGAAGCTACCG-3′) and qPCR_FF_rev (5′-GGTGGCAAATGGGAAGTCAC-3′) were combined, yielding a PCR product of 108 bp. PCR conditions were chosen according to the manufacturer's fast cycling mode protocol: Uracil-DNA glycosylase (UDG) activation at 50°C for 2 min, initial denaturation of cDNA at 95°C for 2 min, followed by 40 cycles with denaturation at 95°C for 1 sec, annealing and extension at 60°C for 30 sec. RNA levels were quantified using the 2−ΔΔCT values method. Firefly luciferase mRNA was used as reference RNA (internal standard) to calculate ΔCT values. 2−ΔΔCT values were determined for each independent experiment (mostly conducted as technical triplicates) and mean values were derived from three or more independent experiments. The mean 2−ΔCT value determined for the NP (wt) MG was then subtracted from the mean 2−ΔCT value measured for the mutant MG construct to derive the specific mean 2−ΔΔCT value. To determine mRNA levels, 2−ΔCT values obtained with the specific cRNA primer set were first subtracted from 2−ΔCT values obtained with the (+)RNA (cRNA + mRNA) primer set. Note that a 2−ΔΔCT value of 1 would correspond to equal RNA levels for NP (wt) and the mutant MG, whereas values below 1 correspond to higher RNA levels for the NP (wt) MG. Ten independent experiments with three NP (wt) replicates each were analyzed for primer efficiency determination based on fluorescence in exponential phase (Ramakers et al. 2003). The real-time PCR efficiencies were 1.96 for Firefly luciferase mRNA, 1.98 for vRNA and cRNA and 2.00 for (+)RNA (cRNA and mRNA).
Quantification and statistical analysis
All statistical analyses were performed using GraphPad Prism version 8.1.1 for Windows. Statistical details and definition of parameters can be found in the figure legends. The statistical significance level was chosen as 0.05; calculated P-values are indicated in the respective figure legends. Measured values that are normally distributed were analyzed by applying the unpaired t-test using two-tailed P-values. Measured values not normally distributed were analyzed by the nonparametric Mann–Whitney test.
SUPPLEMENTAL MATERIAL
Supplemental material is available for this article.
Supplementary Material
ACKNOWLEDGMENTS
We acknowledge experimental and technical support by Hannes Huber and Sören Seidler (initial cloning experiments) and Astrid Herwig (cell culture). This work was supported by the German Research Foundation (DFG), grant CRC 1021, to S. Becker and R.K.H.
Footnotes
Article is online at http://www.rnajournal.org/cgi/doi/10.1261/rna.073718.119.
REFERENCES
- Biedenkopf N, Hartlieb B, Hoenen T, Becker S. 2013. Phosphorylation of Ebola virus VP30 influences the composition of the viral nucleocapsid complex: impact on viral transcription and replication. J Biol Chem 288: 11165–11174. 10.1074/jbc.M113.461285 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkopf N, Lier C, Becker S. 2016a. Dynamic phosphorylation of VP30 is essential for Ebola virus life cycle. J Virol 90: 4914–4925. 10.1128/JVI.03257-15 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Biedenkopf N, Schlereth J, Grünweller A, Becker S, Hartmann RK. 2016b. RNA binding of Ebola virus VP30 is essential for activating viral transcription. J Virol 90: 7481–7496. 10.1128/JVI.00271-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Blakqori G, Kochs G, Haller O, Weber F. 2003. Functional L polymerase of La Crosse virus allows in vivo reconstitution of recombinant nucleocapsids. J Gen Virol 84: 1207–1214. 10.1099/vir.0.18876-0 [DOI] [PubMed] [Google Scholar]
- Bukreyev AA, Chandran K, Dolnik O, Dye JM, Ebihara H, Leroy EM, Mühlberger E, Netesov SV, Patterson JL, Paweska JT, et al. 2014. Discussions and decisions of the 2012–2014 International Committee on Taxonomy of Viruses (ICTV) Filoviridae Study Group, January 2012–June 2013. Arch Virol 159: 821–830. 10.1007/s00705-013-1846-9 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Burk R, Bollinger L, Johnson JC, Wada J, Radoshitzky SR, Palacios G, Bavari S, Jahrling PB, Kuhn JH. 2016. Neglected filoviruses. FEMS Microbiol Rev 40: 494–519. 10.1093/femsre/fuw010 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P, Roux L. 1993. The rule of six, a basic feature for efficient replication of Sendai virus defective interfering RNA. J Virol 67: 4822–4830. 10.1128/JVI.67.8.4822-4830.1993 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Calain P, Monroe MC, Nichol ST. 1999. Ebola virus defective interfering particles and persistent infection. Virology 262: 114–128. 10.1006/viro.1999.9915 [DOI] [PubMed] [Google Scholar]
- Cowton VM, Fearns R. 2005. Evidence that the respiratory syncytial virus polymerase is recruited to nucleotides 1 to 11 at the 3′ end of the nucleocapsid and can scan to access internal signals. J Virol 79: 11311–11322. 10.1128/JVI.79.17.11311-11322.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Cox R, Pickar A, Qiu S, Tsao J, Rodenburg C, Dokland T, Elson A, He B, Luo M. 2014. Structural studies on the authentic mumps virus nucleocapsid showing uncoiling by the phosphoprotein. Proc Natl Acad Sci 111: 15208–15213. 10.1073/pnas.1413268111 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Crary SM, Towner JS, Honig JE, Shoemaker TR, Nichol ST. 2003. Analysis of the role of predicted RNA secondary structures in Ebola virus replication. Virology 306: 210–218. 10.1016/S0042-6822(02)00014-4 [DOI] [PubMed] [Google Scholar]
- Deflubé LR, Cressey TN, Hume AJ, Olejnik J, Haddock E, Feldmann F, Ebihara H, Fearns R, Mühlberger E. 2019. Ebolavirus polymerase uses an unconventional genome replication mechanism. Proc Natl Acad Sci 116: 8535–8543. 10.1073/pnas.1815745116 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearns R, Plemper RK. 2017. Polymerases of paramyxoviruses and pneumoviruses. Virus Res 234: 87–102. 10.1016/j.virusres.2017.01.008 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Fearns R, Peeples ME, Collins PL. 2002. Mapping the transcription and replication promoters of respiratory syncytial virus. J Virol 76: 1663–1672. 10.1128/JVI.76.4.1663-1672.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Kolesnikova L, Theriault S, Ebihara H, Hartlieb B, Bamberg S, Feldmann H, Ströher U, Becker S. 2006. Infection of naive target cells with virus-like particles: implications for the function of Ebola virus VP24. J Virol 80: 7260–7264. 10.1128/JVI.00051-06 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Hoenen T, Jung S, Herwig A, Groseth A, Becker S. 2010. Both matrix proteins of Ebola virus contribute to the regulation of viral genome replication and transcription. Virology 403: 56–66. 10.1016/j.virol.2010.04.002 [DOI] [PubMed] [Google Scholar]
- Hoenen T, Groseth A, Feldmann H. 2019. Therapeutic strategies to target the Ebola virus life cycle. Nat Rev Microbiol 17: 593–606. 10.1038/s41579-019-0233-2 [DOI] [PubMed] [Google Scholar]
- Kolakofsky D, Pelet T, Garcin D, Hausmann S, Curran J, Roux L. 1998. Paramyxovirus RNA synthesis and the requirement for hexamer genome length: the rule of six revisited. J Virol 72: 891–899. 10.1128/JVI.72.2.891-899.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Kruse T, Biedenkopf N, Hertz EPT, Dietzel E, Stalmann G, López-Méndez B, Davey NE, Nilsson J, Becker S. 2018. The Ebola virus nucleoprotein recruits the host PP2A-B56 phosphatase to activate transcriptional support activity of VP30. Mol Cell 69: 136–145.e6. 10.1016/j.molcel.2017.11.034 [DOI] [PubMed] [Google Scholar]
- Kuhn JH, Amarasinghe GK, Basler CF, Bavari S, Bukreyev A, Chandran K, Crozier I, Dolnik O, Dye JM, Formenty PBH, et al. 2019. ICTV virus taxonomy profile: Filoviridae. J Gen Virol 100: 911–912. 10.1099/jgv.0.001252 [DOI] [PMC free article] [PubMed] [Google Scholar]
- le Mercier P, Kolakofsky D. 2019. Bipartite promoters and RNA editing of paramyxoviruses and filoviruses. RNA 25: 279–285. 10.1261/rna.068825.118 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Liang B, Li Z, Jenni S, Rahmeh AA, Morin BM, Grant T, Grigorieff N, Harrison SC, Whelan SPJ. 2015. Structure of the L protein of vesicular stomatitis virus from electron cryomicroscopy. Cell 162: 314–327. 10.1016/j.cell.2015.06.018 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Ohta K, Kolakofsky D, Nishio M. 2017. A point mutation in the RNA-binding domain of human parainfluenza virus type 2 nucleoprotein elicits abnormally enhanced polymerase activity. J Virol 91: e02203-16 10.1128/JVI.02203-16 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Matsumoto Y, Ohta K, Kolakofsky D, Nishio M. 2018. The control of paramyxovirus genome hexamer length and mRNA editing. RNA 24: 461–467. 10.1261/rna.065243.117 [DOI] [PMC free article] [PubMed] [Google Scholar]
- McGivern DR, Collins PL, Fearns R. 2005. Identification of internal sequences in the 3′ leader region of human respiratory syncytial virus that enhance transcription and confer replication processivity. J Virol 79: 2449–2460. 10.1128/JVI.79.4.2449-2460.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Modrof J, Mühlberger E, Klenk HD, Becker S. 2002. Phosphorylation of VP30 impairs Ebola virus transcription. J Biol Chem 227: 33099–33104. 10.1074/jbc.M203775200 [DOI] [PubMed] [Google Scholar]
- Mühlberger E. 2007. Filovirus replication and transcription. Future Virol 2: 205–215. 10.2217/17460794.2.2.205 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Mühlberger E, Weik M, Volchkov VE, Klenk HD, Becker S. 1999. Comparison of the transcription and replication strategies of Marburg virus and Ebola virus by using artificial replication systems. J Virol 73: 2333–2342. 10.1128/JVI.73.3.2333-2342.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Murphy SK, Ito Y, Parks GD. 1998. A functional antigenomic promoter for the paramyxovirus simian virus 5 requires proper spacing between an essential internal segment and the 3′ terminus. J Virol 72: 10–19. 10.1128/JVI.72.1.10-19.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Noton SL, Fearns R. 2015. Initiation and regulation of paramyxovirus transcription and replication. Virology 479–480: 545–554. 10.1016/j.virol.2015.01.014 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Ogino T, Green TJ. 2019. RNA synthesis and capping by non-segmented negative strand RNA viral polymerases: lessons from a prototypic virus. Front Microbiol 10: 1490 10.3389/fmicb.2019.01490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Pelet T, Delenda C, Gubbay O, Garcin D, Kolakofsky D. 1996. Partial characterization of a Sendai virus replication promoter and the rule of six. Virology 224: 405–414. 10.1006/viro.1996.0547 [DOI] [PubMed] [Google Scholar]
- Ramakers C, Ruijter JM, Deprez RH, Moorman AF. 2003. Assumption-free analysis of quantitative real-time polymerase chain reaction (PCR) data. Neurosci Lett 339: 62–66. 10.1016/S0304-3940(02)01423-4 [DOI] [PubMed] [Google Scholar]
- Sanchez A, Kiley MP, Holloway BP, Auperin DD. 1993. Sequence analysis of the Ebola virus genome: organization, genetic elements, and comparison with the genome of Marburg virus. Virus Res 29: 215–240. 10.1016/0168-1702(93)90063-S [DOI] [PubMed] [Google Scholar]
- Schlereth J, Grünweller A, Biedenkopf N, Becker S, Hartmann RK. 2016. RNA binding specificity of Ebola virus transcription factor VP30. RNA Biol 13: 783–798. 10.1080/15476286.2016.1194160 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shabman RS, Hoenen T, Groseth A, Jabado O, Binning JM, Amarasinghe GK, Feldmann H, Basler CF. 2013. An upstream open reading frame modulates Ebola virus polymerase translation and virus replication. PLoS Pathog 9: e1003147 10.1371/journal.ppat.1003147 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Shu T, Gan T, Bai P, Wang X, Qian Q, Zhou H, Cheng Q, Qiu Y, Yin L, Zhong J, et al. 2019. Ebola virus VP35 has novel NTPase and helicase-like activities. Nucleic Acids Res 47: 5837–5851. 10.1093/nar/gkz340 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Sugita Y, Matsunami H, Kawaoka Y, Noda T, Wolf M. 2018. Cryo-EM structure of the Ebola virus nucleoprotein–RNA complex at 3.6 Å resolution. Nature 563: 137–140. 10.1038/s41586-018-0630-0 [DOI] [PubMed] [Google Scholar]
- Tapparel C, Maurice D, Roux L. 1998. The activity of Sendai virus genomic and antigenomic promoters requires a second element past the leader template regions: a motif (GNNNNN)3 is essential for replication. J Virol 72: 3117–3128. 10.1128/JVI.72.4.3117-3128.1998 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tekes G, Rahmeh AA, Whelan SP. 2011. A freeze frame view of vesicular stomatitis virus transcription defines a minimal length of RNA for 5′ processing. PLoS Pathog 7: e1002073 10.1371/journal.ppat.1002073 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Tremaglio CZ, Noton SL, Deflube LR, Fearns R. 2013. Respiratory syncytial virus polymerase can initiate transcription from position 3 of the leader promoter. J Virol 87: 3196–3207. 10.1128/JVI.02862-12 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Wan W, Kolesnikova L, Clarke M, Koehler A, Noda T, Becker S, Briggs JAG. 2017. Structure and assembly of the Ebola virus nucleocapsid. Nature 551: 394–397. 10.1038/nature24490 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weik M, Modrof J, Klenk HD, Becker S, Mühlberger E. 2002. Ebola virus VP30-mediated transcription is regulated by RNA secondary structure formation. J Virol 76: 8532–8539. 10.1128/JVI.76.17.8532-8539.2002 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Weik M, Enterlein S, Schlenz K, Mühlberger E. 2005. The Ebola virus genomic replication promoter is bipartite and follows the rule of six. J Virol 79: 10660–10671. 10.1128/JVI.79.16.10660-10671.2005 [DOI] [PMC free article] [PubMed] [Google Scholar]
- Whelan SP, Wertz GW. 1999. Regulation of RNA synthesis by the genomic termini of vesicular stomatitis virus: identification of distinct sequences essential for transcription but not replication. J Virol 73: 297–306. 10.1128/JVI.73.1.297-306.1999 [DOI] [PMC free article] [PubMed] [Google Scholar]
Associated Data
This section collects any data citations, data availability statements, or supplementary materials included in this article.